TO THE EDITOR:
Driver mutations in the tyrosine kinase receptor FLT3, including internal tandem duplications (FLT3-ITD) and tyrosine kinase domain (FLT3-TKD) mutations, occur in approximately 30% of newly diagnosed cases of acute myeloid leukemia (AML) and enable constitutive receptor activation, leading to poor prognosis. Multiple tyrosine kinase inhibitors have shown clinical activity in patients and have improved overall treatment outcomes1, 2. These inhibitors are generally classified as type I or type II inhibitors, depending on whether they bind the active or inactive conformation of FLT33, respectively. Type I inhibitors include midostaurin, crenolanib, and gilteritinib, while type II inhibitors include quizartinib, sorafenib, and ponatinib. Despite their initial efficacy, the long-term durability of these inhibitors is curtailed by the development of drug resistance. To date, several modes of FLT3 inhibitor resistance have been identified3. Secondary mutations in the TKD, particularly at the D835 residue, are commonly reported as a mechanism of resistance to type II FLT3 inhibitors4-6. These mutations destabilize the inactive conformation of FLT3 required for the binding of type II inhibitors quizartinib4 and sorafenib5. Newer type I inhibitors, including crenolanib and gilteritinib, retain efficacy against most TKD mutations, and resistance commonly occurs through mutations in the RAS or MAPK signaling pathway2, 7. Resistance to both type I and II inhibitors can also occur through the FLT3F691L gatekeeper mutation2, 4, 6-8, although clinically this is less common than FLT3-TKD and RAS mutations.
We recently characterized in vitro gilteritinib resistance and found that resistance emerges in a dynamic, stepwise manner where microenvironmental support led to an early phase of resistance characterized by changes in cell cycle and metabolism, that later evolved to a late resistance phase concomitant with the notable expansion of NRAS mutations9, consistent with clinical data2. In addition to NRAS mutations, we also identified a novel FLT3N701K mutation in three resistant cultures that promoted gilteritinib resistance10. We demonstrated that FLT3N701K effectively acts as a gatekeeper mutation, blocking gilteritinib from binding to FLT3. Notably, FLT3N701K did not block quizartinib binding, indicating that FLT3N701K mutations are more specific for type I FLT3 inhibitors and, thereby serve as noncanonical gatekeepers unlike canonical gatekeepers, such as FLT3F691L, which block binding of type I and II inhibitors4, 6. Herein, we report two AML patients who developed a FLT3ITD+N701K mutation while on the combination of gilteritinib and venetoclax. We show that primary cells from one patient (patient #1, Figure 1A) with the FLT3ITD+N701K mutation were resistant to gilteritinib ex vivo, but remained exquisitely sensitive to quizartinib, highlighting the potential utility of switching type I to type II FLT3 inhibitors to restore clinical response.
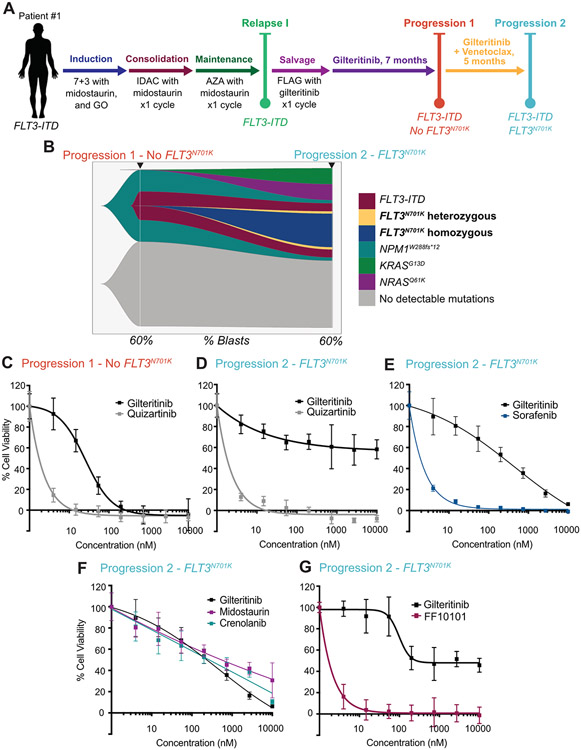
Figure 1: FLT3N701K confers resistance to type I inhibitor, gilteritinib in primary cells but retains sensitivity to the type II inhibitor, quizartinib.
A) Treatment time course is outlined with pertinent details about FLT3 mutation status. The FLT3N701K mutation was detected at Progression 2 via GeneTrails (clinical mutational panel at OHSU). Complete mutational profiling data are available in Supplemental Table 1. B) Fishplot illustrating clonal architecture and evolution of 1,330 single cells from cryopreserved bone marrow aspirate as captured by single cell DNA sequencing prior to gilteritinib and venetoclax (Progression 1) and at time of clinical resistance (Progression 2). Top x-axis indicates clinical timepoint and bottom x-axis indicates percent blasts on corresponding clinical bone marrow biopsy report. This patient demonstrated complex on- and off-target clonal evolution with therapeutic pressure, including de novo development of a FLT3N701K hetero- and homozygous mutation within the FLT3-ITD mutated subclone, de novo development of a NRASQ61K mutated subclone, and selection for a pre-existing KRASG13D mutated subclone. The gray area indicates cells that did not have detectable mutations, although the single cell mutation panel via the Tapestri platform is more limited (Supplemental Table 2) than the sequencing panel used in Supplemental Table 1, so these cells may still have mutations. Primary AML cells at C) Progression 1 and D-G) Progression 2 were enriched with CD33+/CD34+ selection and treated with gilteritinib, quizartinib, sorafenib, midostaurin, crenolanib, or F10101 as single agents (0 – 10000 nM). Cell viability was measured after 72 hours using a tetrazolamine-based viability assay and normalized to untreated cells. Mean of triplicates ± standard deviation are shown for all viability results.
The first patient initially presented with AML with NPM1, DNMT3A and FLT3-ITD mutations and received induction with 7+3 with midostaurin and gemtuzumab ozogomacin (GO) on study. He achieved a remission, but had difficulty tolerating consolidation and maintenance therapies and relapsed 6 months later (Figure 1A). He then received salvage with FLAG + gilteritinib 120 mg daily starting on day 8, but gilteritinib was held after chemotherapy due to prolonged neutropenia and infectious complications. A later marrow demonstrated persistence of FLT3-ITD, so gilteritinib monotherapy at 120 mg daily was initiated, with periodic interruptions for infections and neutropenia. Gilteritinib effectively suppressed his disease, but did not clear the FLT3-ITD clone and blasts slowly increased in his marrow (Figure 1A, Progression 1). Venetoclax 400 mg daily was added to improve response, but there were frequent interruptions and dose reductions of both gilteritinib and venetoclax due to neutropenia. The combination of gilteritinib + venetoclax was continued for 5 months until peripheral blasts increased rapidly. The FLT3N701K mutation was detected at high variant allele frequency (VAF) of 35% at this point (Figure 1A, Progression 2), suggesting that it was likely propelled by the selective pressure of gilteritinib and venetoclax treatment. We confirmed the presence of FLT3N701K in genomic DNA via Sanger sequencing at Progression 2 (Supplemental Figure 1). KRASG13D and NRASQ61K mutations were also detected at lower VAFs at Progression 2 (Supplemental Table 1). To reconstruct the clonal architecture and evolution of this patient’s AML, we performed single-cell DNA sequencing on bone marrow mononuclear cells from Progression 1 and Progression 2 timepoints. Single-cell DNA analysis revealed de novo development of a FLT3N701K population which was co-mutated with FLT3-ITD in the same cells. Surpisingly, the majority of cells with FLT3N701K mutations were homozygous, while only a minority were heterozygous (Figure 1B). Single cell and clonal analysis also revealed that mutations in KRAS and NRAS sublones were not co-mutated with FLT3-ITD or FLT3N701K (Figure 1B).
To evaluate the relative sensitivity of the FLT3N701K mutation to FLT3 inhibitors, Ba/F3 cells harboring FLT3-ITD and a concomitant N701K mutation (i.e., FLT3ITD+N701K) were exposed to titrations of gilteritinib or quizartinib, with IC50 values of 82.86 and 2.906 nM, respectively (Supplemental Figure 2A-B), indicating that the FLT3N701K mutation remains sensitive to quizartinib in this model system. To evaluate the sensitivity in primary cells, frozen viable cells from patient 1 at Progression 1 and Progression 2 were thawed and enriched for blasts via CD33+/CD34+ bead selection and then treated with gilteritinib and quizartinib ex vivo. Prior to the FLT3N701K mutation, the patient’s blasts responded to gilteritinib with IC50 of 24.38 nM and quizartinib with IC50 of 1.733 nM, reflecting the higher potency of quizartinib (Figure 1C). Although the patient had already been treated with gilteritinib at this timepoint, the ex vivo response to FLT3 inhibition does not account for the contribution of the marrow microenvironment, which we and others have shown can impact clinical response9, 11, 12. However, after expansion of the FLT3N701K mutation, the blasts developed increased resistance to gilteritinib ex vivo, with an IC50 of >10000 nM (>400 fold increase). Despite this resistance to gilteritinib, they remained exquisitely sensitive to quizartinib with a nearly unchanged IC50 of 1.771 nM (Figure 1D). Primary AML blasts harboring FLT3N701K also retained sensitivity to the type II inhibitor sorafenib, demonstrating the efficiacy of type II inhibitors more broadly in the setting of FLT3N701K (Figure 1E). In contrast, primary cells with FLT3N701K were resistant to both type I inhibitors, crenolanib and midostaurin (Figure 1F). In parallel, we also tested the activity of the allosteric FLT3 inhibitor FF10101, which overcomes FLT3 gatekeeper mutations by binding at a site outside of the gatekeeper region in FLT313. Similar to quizartinib, FF10101 remained very potent in primary AML cells harboring the FLT3ITD+N701K mutation with an approximate 150-fold greater potency by IC50 compared to gilteritinib (Figure 1G). Ba/F3 cells with FLT3ITD+N701K exhibited a similar response with a 19-fold increase in potency relative to gilteritinib (Supplemental Figure 2C).
While revising this manuscript, we became aware of a second patient (patient #2) with a FLT3N701K mutation. This patient initially presented with AML with a normal karyotype and mutations in RUNX1, SETBP1, WT1, and FLT3-ITD. He achieved complete remission after induction with 7+3 and midostaurin and subsequently received two cycles of high dose cytarabine and midostaurin followed by single-agent gilteritinib at 120 mg daily until admission for a myeloablative allogeneic transplantation from his brother. After engraftment, he restarted gilteritinib as post-transplant maintenance. However, the FLT3-ITD mutation reappeared on his day 100 bone marrow biopsy by PCR, and morphologic relapse followed five weeks later. NGS panel at first relapse showed the same mutations from diagnosis. Salvage therapy with venetoclax and gilteritinib was attempted but not tolerated due to prolonged cytopenias after cycle 2 and the marrow showed persistent low level marrow blasts (5-10%). After three cycles of azacitidine plus gilteritinib, circulating blasts and leukocytosis reappeared. NGS of the peripheral blood showed all prior mutations from diagnosis, but also a new FLT3N701K mutation at high VAF (41%).
Resistance to FLT3 inhibitors via acquisition or clonal expansion of secondary mutations is a well-appreciated phenomenon in AML2, 4, 7. Multiple studies have shown that mutations in the activation loop of FLT3 (most commonly D835) perturb the kinase to adopt an active, type I conformation, rendering them insensitive to type II FLT3 inhibitors, such as quizartinib or sorafenib, but these mutations remain sensitive to type I inhibitors, such as gilteritinib2, 14. However, secondary mutations in FLT3 that trigger a sensitivity switch in the reverse direction, from type I to type II inhibitors, is largely an underappreciated mechanism of resistance, although retained sensitivity to quizartinib has been described in midostaurin resistant cell lines harboring RAS mutations15. To our knowledge we are the first to report this observation in a patient sample with FLT3-AML. Our data has important clinical implications as the use of FLT3 inhibitors continues to expand in the clinic, and undoubtedly, the number of resistance mutations will increase as well. Functional validation of mutations is imperative to predict the efficacy of TKI-switching, similar to what has been described for other oncogenic kinases such as BCR-ABL116, TRK17, ROS118, EGFR19, MET20, and KIT21.
FLT3 gatekeeper mutations, most commonly the F691L mutation, have also been reported in patients treated with both type I and type II FLT3 inhibitors2, 4, 7. In the ADMIRAL trial, F691L mutations were the most common FLT3 mutation associated with resistance, although less frequent than activating mutations in the RAS pathway22. Although FLT3N701K mutations act as a gatekeeper for gilteritinib, the mutation retains a surprising sensitivity to quizartinib and other type II inhibitors10. Although we initially found this mutation in gilteritinib-resistant cell lines, we now report a patient with the FLT3ITD+N701K mutation with ex vivo sensitivity that mirrors our cell lines. In silico modeling suggests that the N701K mutation is predicted to sterically disrupt the binding of gilteritinib, and trigger a conformational switch that favors adoption of the inactive kinase conformation10. In vitro mutagenesis screening has also revealed a few other FLT3 mutations near F691 that confer moderate gilteritinib resistance23. An alternative approach is to use an allosteric inhibitor such as FF10101, that retains activity against F691L24 and other gatekeeper mutations. Indeed, we found that FLT3ITD+N701K mutation remained sensitive to this inhibitor, however further clinical testing is required to determine the efficacy and tolerability of FF10101 in patients25.
In summary, we describe two AML patients with FLT3ITD+N701K mutations that arose during development of clinical resistance to gilteritinib. Single-cell analysis confirmed this mutation was co-mutated with FLT3-ITD and was predominantly homozygous, at least in the one patient we tested. Ex vivo assays reveal that primary cells with FLT3N701K were resistant to gilteritinib, but retained ex vivo sensitivity to quizartinib, consistent with our work in cell line models. As quizartinib26 was recently approved by the FDA for frontline AML treatment, our data provides compelling evidence that switching to quizartinib with FLT3N701K mutations may restore sensitivity to FLT3 inhibitors in the setting of relapsed/refractory disease, extending the duration of response to therapy.
Supplementary Material
Supplemental Table 1: Genetrails Sequencing of patient at different clinical time-points.
Supplemental Figure 2: FLT3N701K is relatively sensitive to type II inhibitor, quizartinib and allosteric inhibitor, FF10101 in the BA/F3 cell line model. A-C. Six replicates of FLT3WT, FLT3ITD, FLT3N701K, FLT3ITD+F701K, and FLT3ITD+F691L Ba/F3 cells were plated with a dose gradient (0 – 1000 nM) of gilteritinib (type I inhibitor; A), quizartinib (type II inhibitor; B), and FF10101 (allosteric inhibitor; C). FLT3WT cells were plated in media supplemented with IL-3. Cell viability was determined using a tetrazolamine-based viability assay. Viability is represented as a percentage of the untreated control. The average mean ± SEM are shown.
Supplemental Figure 1: Sanger sequencing confirmation of FLT3N701K mutation. Electropherograms from sanger sequencing of patient genomic DNA at A) Progression 1 and B) Progression 2 confirm the presence of FLT3N701K mutation. Peaks correspond to the following nucleotides: A (green), T (red), C (blue), and G (black). Arrows indicate direction of sequencing.
Supplemental Table 2: Custom Mission Bio Amplicon Panel
ACKNOWLEDGEMENTS:
We are extremely thankful to our patients for their precious tissue samples. We also thank Sammantha Avalon for assistance in acquiring patient material from the Beat AML sample repository. This work was supported by the Acquired Resistance to Therapy Network (ARTNet), National Institutes of Health (NIH), National Cancer Institute (NCI) grant U54CA224019 awarded to B.J.D. and J.W.T (E.T. co-investigator). E.T is supported by the American Cancer Society Mission Boost Grant (MBGI-21-106-01). S.K.J. is supported by the ARCS Scholar Foundation, The Paul & Daisy Soros Fellowship, and the National Cancer Institute (F30CA239335). This research was supported in part by the West Charitable Trust. C.C.S. is a Leukemia & Lymphoma Society Scholar in Clinical Research and a Damon Runyon-Richard Lumsden Foundation Clinical Investigator supported (in part) by the Damon Runyon Cancer Research Foundation (CI-99–18).
Footnotes
DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST:
B.J.D. potential competing interests – SAB: Adela Bio, Aileron Therapeutics (inactive), Therapy Architects/ALLCRON (inactive), Cepheid, DNA SEQ, Nemucore Medical Innovations, Novartis, RUNX1 Research Program; SAB & Stock: Aptose Biosciences, Blueprint Medicines, Enliven Therapeutics, Iterion Therapeutics, GRAIL, Recludix Pharma; Board of Directors & Stock: Amgen, Vincerx Pharma; Board of Directors: Burroughs Wellcome Fund, CureOne; Joint Steering Committee: Beat AML LLS; Advisory Committee: Multicancer Early Detection Consortium; Founder: VB Therapeutics; Sponsored Research Agreement: Enliven Therapeutics, Recludix Pharma; Clinical Trial Funding: Novartis, Astra-Zeneca; Royalties from Patent 6958335 (Novartis exclusive license) and OHSU and Dana-Farber Cancer Institute (one Merck exclusive license, one CytoImage, Inc. exclusive license, and one Sun Pharma Advanced Research Company non-exclusive license); US Patents 4326534, 6958335, 7416873, 7592142, 10473667, 10664967, 11049247.
E.T. potential competing interests – Advisory Board/Consulting: Abbvie, Astellas, Daiichi-Sankyo, Servier, Rigel. Research Funding: Prelude Therapeutics, Schrodinger, Incyte, Astra-Zeneca.
C.C.S. potential competing interests – Advisory Board/Consulting: Abbvie, Genentech. Research Funding: Revolution Medicines, Erasca, Abbvie, Celgene.
All other authors report no COIs.
REFERENCES:
- 1.Stone RM, Mandrekar SJ, Sanford BL, Laumann K, Geyer S, Bloomfield CD, et al. , Midostaurin plus Chemotherapy for Acute Myeloid Leukemia with a FLT3 Mutation. N Engl J Med, 2017. 377(5): p. 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McMahon CM, Ferng T, Canaani J, Wang ES, Morrissette JJD, Eastburn DJ, et al. , Clonal Selection with RAS Pathway Activation Mediates Secondary Clinical Resistance to Selective FLT3 Inhibition in Acute Myeloid Leukemia. Cancer Discov, 2019. 9(8): p. 1050–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alotaibi AS, Yilmaz M, Kanagal-Shamanna R, Loghavi S, Kadia TM, DiNardo CD, et al. , Patterns of Resistance Differ in Patients with Acute Myeloid Leukemia Treated with Type I versus Type II FLT3 inhibitors. Blood Cancer Discov, 2021. 2(2): p. 125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith CC, Wang Q, Chin CS, Salerno S, Damon LE, Levis MJ, et al. , Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature, 2012. 485(7397): p. 260–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Man CH, Fung TK, Ho C, Han HH, Chow HC, Ma AC, et al. , Sorafenib treatment of FLT3-ITD(+) acute myeloid leukemia: favorable initial outcome and mechanisms of subsequent nonresponsiveness associated with the emergence of a D835 mutation. Blood, 2012. 119(22): p. 5133–43. [DOI] [PubMed] [Google Scholar]
- 6.Smith CC, Zhang C, Lin KC, Lasater EA, Zhang Y, Massi E, et al. , Characterizing and Overriding the Structural Mechanism of the Quizartinib-Resistant FLT3 "Gatekeeper" F691L Mutation with PLX3397. Cancer Discov, 2015. 5(6): p. 668–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H, Savage S, Schultz AR, Bottomly D, White L, Segerdell E, et al. , Clinical resistance to crenolanib in acute myeloid leukemia due to diverse molecular mechanisms. Nat Commun, 2019. 10(1): p. 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peretz CAC, McGary LHF, Kumar T, Jackson H, Jacob J, Durruthy-Durruthy R, et al. , Single-cell DNA sequencing reveals complex mechanisms of resistance to quizartinib. Blood Adv, 2021. 5(5): p. 1437–1441. Mission Bio, Inc. M.J.L. reports consultancy and research funding FujiFilm, consultancy and honoraria from Daiichi Sankyo Inc, consultancy and honoraria from Agios and Menarini, consultancy and research funding from Novartis and Astellas, and consultancy and honoraria from Amgen. A.P. reports honoraria, advisory board, research funding from Novartis; consultancy, honoraria, and research funding from Astellas and Daiichi Sankyo; consultancy and honoraria from NewLink Genetics; consultancy from Arog; research funding from FujiFilm; consultancy and honoraria from AbbVie; consultancy from Forma; and advisory board and research funding from Loxo and Syndax. C.C.S. reports research funding from Astellas Pharma, Abbvie, Revolution Medicines, and FujiFilm. The remaining authors declare no competing financial interests. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joshi SK, Nechiporuk T, Bottomly D, Piehowski PD, Reisz JA, Pittsenbarger J, et al. , The AML microenvironment catalyzes a stepwise evolution to gilteritinib resistance. Cancer Cell, 2021. 39(7): p. 999–1014 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joshi SK, Sharzehi S, Pittsenbarger J, Bottomly D, Tognon CE, McWeeney SK, et al. , A noncanonical FLT3 gatekeeper mutation disrupts gilteritinib binding and confers resistance. Am J Hematol, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Traer E, Martinez J, Javidi-Sharifi N, Agarwal A, Dunlap J, English I, et al. , FGF2 from Marrow Microenvironment Promotes Resistance to FLT3 Inhibitors in Acute Myeloid Leukemia. Cancer Res, 2016. 76(22): p. 6471–6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato T, Yang X, Knapper S, White P, Smith BD, Galkin S, et al. , FLT3 ligand impedes the efficacy of FLT3 inhibitors in vitro and in vivo. Blood, 2011. 117(12): p. 3286–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaura T, Nakatani T, Uda K, Ogura H, Shin W, Kurokawa N, et al. , A novel irreversible FLT3 inhibitor, FF-10101, shows excellent efficacy against AML cells with FLT3 mutations. Blood, 2018. 131(4): p. 426–438. [DOI] [PubMed] [Google Scholar]
- 14.Smith CC, Lasater EA, Lin KC, Wang Q, McCreery MQ, Stewart WK, et al. , Crenolanib is a selective type I pan-FLT3 inhibitor. Proc Natl Acad Sci U S A, 2014. 111(14): p. 5319–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aikawa T, Togashi N, Iwanaga K, Okada H, Nishiya Y, Inoue S, et al. , Quizartinib, a selective FLT3 inhibitor, maintains antileukemic activity in preclinical models of RAS-mediated midostaurin-resistant acute myeloid leukemia cells. Oncotarget, 2020. 11(11): p. 943–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lovera S, Morando M, Pucheta-Martinez E, Martinez-Torrecuadrada JL, Saladino G, and Gervasio FL, Towards a Molecular Understanding of the Link between Imatinib Resistance and Kinase Conformational Dynamics. PLoS Comput Biol, 2015. 11(11): p. e1004578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cocco E, Lee JE, Kannan S, Schram AM, Won HH, Shifman S, et al. , TRK xDFG mutations trigger a sensitivity switch from type I to II kinase inhibitors. Cancer Discov, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drilon A, Somwar R, Wagner JP, Vellore NA, Eide CA, Zabriskie MS, et al. , A Novel Crizotinib-Resistant Solvent-Front Mutation Responsive to Cabozantinib Therapy in a Patient with ROS1-Rearranged Lung Cancer. Clin Cancer Res, 2016. 22(10): p. 2351–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown BP, Zhang YK, Westover D, Yan Y, Qiao H, Huang V, et al. , On-target Resistance to the Mutant-Selective EGFR Inhibitor Osimertinib Can Develop in an Allele-Specific Manner Dependent on the Original EGFR-Activating Mutation. Clin Cancer Res, 2019. 25(11): p. 3341–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bahcall M, Sim T, Paweletz CP, Patel JD, Alden RS, Kuang Y, et al. , Acquired METD1228V Mutation and Resistance to MET Inhibition in Lung Cancer. Cancer Discov, 2016. 6(12): p. 1334–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laine E, Auclair C, and Tchertanov L, Allosteric communication across the native and mutated KIT receptor tyrosine kinase. PLoS Comput Biol, 2012. 8(8): p. e1002661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith CC, Levis MJ, Perl AE, Hill JE, Rosales M, and Bahceci E, Molecular profile of FLT3-mutated relapsed/refractory patients with AML in the phase 3 ADMIRAL study of gilteritinib. Blood Adv, 2022. 6(7): p. 2144–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tarver TC, Hill JE, Rahmat L, Perl AE, Bahceci E, Mori K, et al. , Gilteritinib is a clinically active FLT3 inhibitor with broad activity against FLT3 kinase domain mutations. Blood Adv, 2020. 4(3): p. 514–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferng TT, Terada D, Ando M, Tarver TC, Chaudhary F, Lin KC, et al. , The Irreversible FLT3 Inhibitor FF-10101 Is Active Against a Diversity of FLT3 Inhibitor Resistance Mechanisms. Mol Cancer Ther, 2022. 21(5): p. 844–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levis MJ, Smith CC, Perl AE, Schiller GJ, Fathi AT, Roboz GJ, et al. , Phase 1 first-in-human study of irreversible FLT3 inhibitor FF-10101-01 in relapsed or refractory acute myeloid leukemia. Journal of Clinical Oncology, 2021. 39(15_suppl): p. 7008–7008. [Google Scholar]
- 26.Erba HP, Montesinos P, Kim HJ, Patkowska E, Vrhovac R, Zak P, et al. , Quizartinib plus chemotherapy in newly diagnosed patients with FLT3-internal-tandem-duplication-positive acute myeloid leukaemia (QuANTUM-First): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet, 2023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1: Genetrails Sequencing of patient at different clinical time-points.
Supplemental Figure 2: FLT3N701K is relatively sensitive to type II inhibitor, quizartinib and allosteric inhibitor, FF10101 in the BA/F3 cell line model. A-C. Six replicates of FLT3WT, FLT3ITD, FLT3N701K, FLT3ITD+F701K, and FLT3ITD+F691L Ba/F3 cells were plated with a dose gradient (0 – 1000 nM) of gilteritinib (type I inhibitor; A), quizartinib (type II inhibitor; B), and FF10101 (allosteric inhibitor; C). FLT3WT cells were plated in media supplemented with IL-3. Cell viability was determined using a tetrazolamine-based viability assay. Viability is represented as a percentage of the untreated control. The average mean ± SEM are shown.
Supplemental Figure 1: Sanger sequencing confirmation of FLT3N701K mutation. Electropherograms from sanger sequencing of patient genomic DNA at A) Progression 1 and B) Progression 2 confirm the presence of FLT3N701K mutation. Peaks correspond to the following nucleotides: A (green), T (red), C (blue), and G (black). Arrows indicate direction of sequencing.
Supplemental Table 2: Custom Mission Bio Amplicon Panel