Abstract
Purpose:
The integrated stress response (ISR) kinase PERK serves as a survival factor for both proliferative and dormant cancer cells. We aim to validate PERK inhibition as a new strategy to specifically eliminate solitary disseminated cancer cells (DCCs) in secondary sites that eventually reawake and originate metastasis.
Experimental design:
A novel clinical-grade PERK inhibitor (HC4) was tested in mouse syngeneic and PDX models which present quiescent/dormant DCCs or growth-arrested cancer cells in micro-metastatic lesions that upregulate ISR.
Results:
HC4 significantly blocks metastasis, by killing quiescent/slow-cycling ISRhigh, but not proliferative ISRlow DCCs. HC4 blocked expansion of established micro-metastasis that contained ISRhigh slow-cycling cells. Single-cell gene expression profiling and imaging revealed that a significant proportion of solitary DCCs in lungs were indeed dormant and displayed an unresolved ER stress as revealed by high expression of a PERK-regulated signature. In human breast cancer metastasis biopsies, GADD34 expression (PERK-regulated gene) and quiescence were positively correlated. HC4 effectively eradicated dormant bone marrow DCCs, which usually persist after rounds of therapies. Importantly, treatment with CDK4/6 inhibitors (to force a quiescent state) followed by HC4 further reduced metastatic burden. In HNSCC and HER2+ cancers HC4 caused cell death in dormant DCCs. In HER2+ tumors, PERK inhibition caused killing by reducing HER2 activity because of sub-optimal HER2 trafficking and phosphorylation in response to EGF.
Conclusions:
Our data identify PERK as a unique vulnerability in quiescent or slow-cycling ISRhigh DCCs. The use of PERK inhibitors may allow targeting pre-existing or therapy-induced growth arrested “persister” cells that escape anti-proliferative therapies.
INTRODUCTION
Under stress conditions, the accumulation of unfolded proteins in the endoplasmic reticulum (ER) lumen activates three main pathways: PERK, IRE1α and ATF6. These pathways are part of a survival and adaptive mechanism known as the unfolded protein response (UPR) (1). However, other kinases that for example sense aminoacidic deficiency or double stranded RNAs, GCN2 and PKR, respectively, can phosphorylate eIF2α (1). Thus, because the UPR is only a subset of signals that can control eIF2α and other adaptation mechanisms during stress, the definition has been expanded to account for all mechanisms that might integrate via eIF2α various stress signals and it is termed integrated stress response (ISR) (2). Recent evidence suggests that in various cancer types the ISR allows tumor cells to respond to increased demands on the ER and greater oxidative conditions imposed by an enhanced translational load caused by oncogenes, hypoxia, and other stress conditions, including anti-cancer agents (3-5). Oncogene-activated pathways increase ER client protein load by activating mTOR signaling and translation initiation (5-7). Other studies have shown that PERK and the IRE1α-XBP-1 pathways contribute to the ability of cancer cells to adapt to hypoxia and microenvironmental stress (4,8-13), suggesting that the ISR enables critical adaptation mechanisms necessary to survive within a changing cellular milieu.
PERK activation initiates an antioxidant and autophagic response that coordinates a protection mechanism for mammary epithelial cells during loss of adhesion to the basement membrane (14). This survival response involves an ATF4 and CHOP transcriptional program (14) coupled to a rapid activation of the LKB1-AMPK-TSC2 pathway that inhibits mTOR (15). Human ductal carcinoma in situ (DCIS) lesions that displayed enhanced PERK phosphorylation and autophagy (14,16) showed, upon conditional ablation of PERK in the mammary epithelium, delayed mammary carcinogenesis promoted by the HER2 oncogene (17,18). Furthermore, HER2 increases the levels of proteotoxicity in tumor cells thereby activating JNK and ISR signaling and allowing HER2+ cancer cells to cope with this stress (19). Interestingly, 14 % of HER2-amplified human breast tumors display an upregulation of PERK mRNA (breast invasive carcinoma TCGA, Nature 2012) (20)), and enrichment for a PERK activity signature correlates with shorter metastasis-free periods in all breast cancer subtypes combined (21). These data support the notion that certain cancer cells in HER2+ and other breast tumor subtypes are dependent on PERK for survival.
We also reported that dormant (quiescent) cancer cells were dependent on PERK and ATF6 signaling for survival (13,22,23). Furthermore, solitary quiescent pancreatic disseminated cancer cells (DCCs) to the liver of mice also display a PERK-dependent ISR that was linked to loss of E-cadherin expression and downregulation of MHC-I, which favors immune evasion during dormancy (24). In the MMTV-HER2 model, quiescent DCCs in bone marrow and lungs were also found in a mesenchymal-like state and to express an ISR signature (25,26) but the link to the ISR was not tested. Together, these data suggest that the ISR may serve as a stress and immune microenvironmental adaptive survival mechanism for DCCs. However, until now no clinical-grade inhibitor of the PERK pathway has been available to test how targeting of this kinase may affect DCC fate and metastatic progression.
Here we report that a selective and potent inhibitor of PERK, HC-5404 (henceforth HC4), (27), currently in a human phase 1a clinical trial in solid tumors (NCT04834778), can block HER2-driven breast cancer metastasis or HER2-independent HNSCC initiated metastasis through the eradication of dormant DCCs or by targeting slow cycling ISRhigh cells in established metastasis. Imaging and single cell gene expression profiling revealed the existence of an ISRhigh/CDK inhibitorhigh quiescent population of cancer cells existing as solitary DCCs or as part of established metastasis. In addition, CDK4/6 inhibition, which heightened the ISR pathway, followed by HC4 treatment further decreased metastatic burden. Our work reveals a new application of a PERK inhibitor with anti-metastatic activity that has been vetted for specificity and selectivity for clinical trials use. This inhibitor, alone or in combination with anti-proliferative therapies, may represent a new strategy to target dormant ISRhigh cells existing as minimal residual disease or as part of established metastasis and help prevent and treat lethal metastases against which we lack effective therapies to date.
METHODS
Reagents.
EGF was obtained from PeproTech and used at 100 ng/ml. Thapsigargin was from Sigma and used at 0.1 μM, 0.2 μM or 2nM as indicated in the legends. HC4 and Abemaciclib were provided by HiberCell and Eli Lilly, respectively. AP20187 was from ARIAD Pharmaceuticals, Cambridge, MA.
Cell culture.
SKBR3 (RRID:CVCL_0033), ZR-75-1 (RRID:CVCL_0588), MCF10A (RRID:CVCL_0598) and EMT6 (RRID:CVCL_1923) cells were obtained from ATCC and cultured for a maximum of 10 passages. T-HEp3 and D-Hep3 cells are an Aguirre-Ghiso Lab proprietary PDX maintained in vivo and not available from ATCC. The PDX lines are used for a maximum of 5 passages and quarterly tested for phenotype consistency following historical records. All cell lines were monthly tested for mycoplasma using PCR Myco-plasma test kit. For 3D cultures, MCF10A-HER2, SKBR3 and ZR-75-1 cells were plated in growth factor-reduced Matrigel (Corning) and grown as described previously (15). Treatments with vehicle (DMSO) or HC4 (2 μM) were replaced every 24 h for 2D and every 48 h for 3D cultures.
Animal work and tissue processing.
Institutional Animal Care and Use Committees (IACUC) at Mount Sinai School of Medicine (MSSM) and Albert Einstein College of Medicine (AECOM) approved all animal studies. Protocol number: 08-0417. The FVB/N-Tg (MMTV-HER2) mouse strain (IMSR_JAX:005038) was obtained from Jackson Laboratories. These mice express the un-activated neu (HER2) form under the transcriptional control of the mouse mammary tumor virus promoter/enhancer. Before being used in any experiment, females underwent one round of pregnancy and at least two weeks of no lactation after weaning. Females between 24-32 weeks of age were injected intraperitoneally (IP) with vehicle (90% corn oil, 10% ethanol) or HC4 (50 mpk) daily, for two weeks. For the combination treatment, females 24-32 weeks of age were treated daily by oral gavage with Abemaciclib (50 mpk) for 4 weeks before starting the treatment described earlier with HC4. FvB wild type mice were injected with MMTV-HER2 cells (5.0x104/animal) from primary tumor lesions via the tail vein, allowed them to expand into established metastatic lesions for 3 weeks and treated with HC4 as described earlier for 2 weeks. 2.5x105 human HNSCC PDX T-HEp3 cells as single cell suspensions were injected i.v in Balb/c nu/nu mice and 24 h after injection we treated the mice with vehicle or HC4 (50mg/kg) for 10 days. 106 EMT6 cells were injected in fad pad of Balb/c mice and 9 days after implantation mice were treated twice per day with HC4 at 10 mg/kg for 17 days. Tumor volumes were measured using the formula (Dxd2)/2, where D is the longest and d is the shortest diameter. For circulating cancer cell (CCC) count, animals were anesthetized and whole blood was extracted by cardiac puncture. Mammary glands, tumors and lungs were collected and fixed in 10% buffered formalin overnight before paraffin embedding. The bone marrow from the two lower limbs was flushed with a 26G needle and further processed by Ficoll density gradient centrifugation. For CCC as well as for DCCs detection in bone marrow, tissues were depleted of mature hematopoietic cells by anti-mouse antibody-labeled magnetic bead separation (Miltenyi Biotec) before fixation in formalin for 20 min at 4 °C.
Ovo Chick Chorioallantoic Membrane (CAM) assay.
Tumor growth on chicken embryo CAMs was performed as described previously (28). 5x105 Dormant D-HEp3 cells were inoculated on the chorioallantoic membrane of 10-day old chick embryos. 24 h later CAM tumors were treated daily with vehicle or HC4 (20 μM) for 6 days. Nodules were collected after treatment, minced, and digested with collagenase and fixed with 4% paraformaldehyde (PFA) prior counting and staining.
Mammary gland whole mount staining.
Mammary glands fixed in 10% buffered formalin were incubated in Carmine Alum stain (Carmine 0.2%, Aluminum potassium sulfate 0.5%) (Sigma) for 2 days. Then, they were dehydrated and transfer to methyl salicylate solution before imaging using a stereomicroscope.
Immunohistochemistry and immunofluorescence.
Immunohistochemistry (IHC) and immunofluorescence (IF) from paraffin-embedded sections was performed as previously described (15). Briefly, slides were dewaxed and serially rehydrated. Heat-induced antigen retrieval was performed in either citrate buffer (10 mM, pH6), EDTA buffer (1 mM, pH 8) or Tris/EDTA (pH 9). Slides were further permeabilized in 0.1% Triton-X100, blocked and incubated with primary antibody overnight at 4 °C at 1:50-1:200 dilution. For IHC, an additional step of endogenous peroxidase and avidin/biotin quenching was performed before primary antibody incubation. Primary antibodies used were anti-cytokeratin 8/18 (Progen, RRID:AB_1541064), smooth muscle actin-Cy3 (Sigma, RRID:AB_476856), P-PERK (T980, RRID:AB_2095853) (29), P-EIF2A (Ser51, RRID:AB_2096481), Cleaved Caspase 3 (RRID:AB_2341188), P-H3 (S10, RRID:AB_331535 ) and P-HER2 (Y1221/1222, RRID:AB_490899) (Cell signaling), HER2 (Abcam, RRID:AB_2893179) and HER2 (Millipore, RRID:AB_310172), Ki67 (eBioscience, RRID:AB_10854564), cytokeratin cocktail (C11 and ck7, Abcam, RRID:AB_306047 and RRID:AB_2783822; AE1 and AE3, Millipore, RRID:AB_94853), Vimentin (R&D, RRID:AB_2241653), P-Rb (S249/T252) and GADD34 (Santa Cruz). Next, slides were incubated in secondary antibodies (Life Technologies) and mounted. The cells positive for each marker were counted and percentage of total cancer cells was calculated. For IHC, sections were processed using VectaStain ABC Elite kit (Vector Laboratories) and DAB Substrate kit for peroxidase labelling (Vector Laboratories) and mounted in VectaMount medium (Vector laboratories). For IF, sections were mounted in ProLong Gold Antifade aqueous medium (Thermo Fisher). In the case of immunocytofluorescence, cytospins of fixed cells (100,00-200,000 cells/cytospin) were prepared by cyto-centrifugation at 500 rpm for 3 min on poly-prep slides, and the staining protocol was performed as explained below from the permeabilization onward. For the staining of 3D cultures, acini were fixed in 4% PFA for 20 min at 4 °C, permeabilized with 0.5% Triton-X100 in PBS for 20 min at room temperature, washed in PBS-glycine and then blocked with 10% normal goat serum for 1 h at 37 °C, before performing immunofluorescence staining.
The scoring for P-HER2 levels is explained in Supplementary Fig. S5a. For the scoring of CK8/18 and SMA in mammary gland ducts, 20 low magnification fields/animal were evaluated for the expression of CK8/18 as negative (0), low (1) or high (2) and the same for SMA and the sum of the two scores was considered as the final score (from 0 to 4).
Microscopy.
Images were captured by using a Nikon Eclipse TS100 microscope, a Leica DM5500 or Leica SP5 confocal microscope. For whole mount mammary gland quantification, slides were scanned using a Leica BioSystems Versa Z1.0 slide scanner. Scanned images were analyzed in ImageJ / FIJI (NIH) with custom-written scripts. Briefly, the images were background corrected and processed with median filter, then skeletonized. The “skeleton 2D / 3D” plugin in ImageJ was used to count the number of branches, normalized to the total lengths of the ducts.
TUNEL in situ cell death detection.
Apoptosis levels were evaluated using the In situ Cell Death Detection kit, AP (Roche). Paraffin sections from tumors were dewaxed, rehydrated and permeabilized in phosphate buffered saline (PBS) 0.2% Triton-X100 for 8 min. Then, slides were washed and blocked in 20% normal goat serum for 1 h at 37 °C. The TUNEL reaction mixture was then added and let go for 1 h at 37 °C. The reaction was stopped by incubating with Buffer I (0.3 M Sodium chloride, 30 mM Sodium citrate). Next, the slides were incubated with anti-fluorescein-AP antibody for 30 min at 37 °C. After three washes in Tris buffered saline (TBS), slides were incubated in alkaline phosphatase substrate in 0.1% Tween-20 for 20 min at room temperature. Finally, the slides were mounted using aqueous mounting medium. The percentage of TUNEL positive cells was calculated using Image J software (NIH).
Immunoblot analysis.
Cells were lysed in RIPA buffer and protein analyzed by immunoblotting as described previously (23). Membranes were blotted using the additional following antibodies: P-PERK (T980) (29), PERK (Santa Cruz, RRID:AB_2762850), P-EGFR (Y1148, RRID:AB_331127 ), EGFR (RRID:AB_331707), P-AKT (S473, RRID:AB_329825), P-S6 (S235/236, RRID:AB_331679), P-ERK (Y204, RRID:AB_331646), P-HER2 (Y1221/1222 RRID:AB_490899, Y1112, Y877, RRID:AB_2099407), HSP90 (RRID:AB_2121214), eIF2α, (RRID:AB_2230924), P-eIF2α (S51, RRID:AB_330951) (Cell signaling), GAPDH (Millipore, RRID:AB_10615768) and β-Tubulin (Abcam, RRID:AB_2210370). For induction of ER stress, MCF10A-HER2 cells were plated in low adhesion plates for 24 h before collection.
Cell surface biotinylation and endocytosis assay.
For cell surface biotinylation, we used Pierce cell surface protein isolation kit following manufacturer’s instructions with minor changes. Briefly, MCF10A-HER2 cells were serum- and EGF-starved and treated +/− HC4 for 24 h before being stimulated with +/− EGF (100 ng/ml) for 20 min. Then, cells were washed with ice-cold PBS and surface proteins biotinylated for 30 min at 4 °C. After quenching, cells were harvested and lysed using RIPA buffer. Protein lysates were incubated with NeutrAvidin agarose beads and the bound proteins were released by incubation with SDS-PAGE sample buffer containing DTT (50 mM). For endocytosis assays (30), cells were treated similarly but before treatment with EGF cell surface proteins were biotinylated. After 20 min incubation +/− EGF (100 ng/ml) at 37 °C (to induce endocytosis), cells were washed with ice-cold PBS and incubated with stripping buffer (to remove cell surface biotinylation: 75 mM NaCl, 1mM MgCl2, 0.1mM CaCl2, 50 mM glutathione and 80 mM NaOH, pH 8.6) for 30 min. To control for stripping efficiency, cells were stripped without 37 °C incubation (t=0). Cell lysates were prepared and processed for biotinylated protein isolation as described before.
Single cell targeted gene expression analysis.
Primary tumors from MMTV-HER2 28-30-week old females were digested with collagenase into a single cell suspension. Lungs from MMTV-/HER2 24-30-week old females were digested into a single cell suspension with collagenase and resuspended in FACS buffer. Cells were then stained with anti-HER2-PE, anti-CD45-APC and DAPI and the HER2+/CD45− population of cells sorted using a BDFACSAria sorter. Sorted cells were resuspended at a 312,500cells/ml concentration in media and 80 μl were mixed with 20 μl suspension reagent (C1 Fluidigm). A C1 Single-cell Preamp IFC 10-17 μm was used for the single cell separation. Pre-amplification was run using Ambion Single Cell-to-CT qRT-PCR kit and 20x TaqMan Gene expression FAM-MGB assays. Resulting cDNA was further diluted in C1 DNA dilution reagent 1/3 and used for gene expression analysis using 96.96 IFCs (Fluidigm), Juno System controller and Biomark HD for high-throughput qPCR. TaqMan Fast Advanced Master Mix was used for the qPCR reactions. Analysis was performed using Fluidigm Real-Time PCR Analysis Software and Clustergrammer web-based tool (31) for hierarchical clustering heatmaps.
Quantitative Real Time PCR.
RNA was extracted using RNeasy kit (Qiagen) following manufacturer’s protocol. Quantitative PCR was performed using SYBR Green Master Mix, GAPDH was used as housekeeping control. GAPDH primers: CTTTGTCAAGCTCATTTCCTGG (FW), TCTTGCTCAGTGTCCTTGC (R); GADD34 primers: TCGGAAGGTACACTTCGCTGAG (FW) TCGATCTCGTGCAAACTGCT (R).
Kinetic Solubility Assay.
Test compound in 1% DMSO was incubated in PBS pH7.4 for 1 h, followed by determination via UV/Vis absorbance of dissolved concentration against a calibration curve.
Caco-2 Permeability Assay.
Caco-2 cells (clone cells (clone C2BBe1) were obtained from American Type Culture Collection (Manassas, VA). Cell monolayers were grown to confluence on collagen-coated, microporous membranes in 12-well assay plates. The permeability assay buffer was Hanks’ balanced salt solution (HBSS) containing 10 mM HEPES and 15 mM glucose at a pH of 7.4. The buffer in the receiver chamber also contained 1% bovine serum albumin. The dosing solution concentration was 5 μM of test article in the assay buffer +/− 1 μM valspodar. Cells were first pre-incubated for 30 min with HBSS +/− 1 μM valspodar. Cell monolayers were dosed on the apical side (A-to-B) or basolateral side (B-to-A) and incubated at 37 °C with 5% CO2 in a humidified incubator. Samples were taken from the donor and receiver chambers at 120 min. Each determination was performed in duplicate. The flux of lucifer yellow was also measured post-experimentally for each monolayer to ensure no damage was inflicted to the cell monolayers during the flux period. All samples were assayed by LC-MS/MS using electrospray ionization. The apparent permeability (Papp) was calculated as follows: Papp= (dCr / dt) x Vr / (A x CA), where dCr / dt is the slope of the cumulative concentration in the receiver compartment vs time in μM s−1; Vr is the volume of the receiver compartment in cm3; A is the area of the insert (1.13 cm2 for 12-well); CA is the average of the nominal dosing concentration and the measured 120-minute donor concentration in μM.
Protein Plasma Binding Assay.
Test compound was prepared at 1 μM concentration in incubation buffer containing 0.5% acetonitrile and incubated with plasma for 4 h prior to LC-MS/MS compound determination. Protein binding of the test compound was measured using Rapid Equilibrium Dialysis (RED).
Hepatocyte Intrinsic Clearance Assay.
Human hepatocytes pooled from 10 donors were incubated (106 viable cells/ml) with test compound at 1 μM concentration (0.1 % DMSO) and metabolic clearance determined by measuring percent remaining of test compound at multiple time points during incubation (15, 30, 60, 90 and 120 minutes). Compound concentration was determined by LC-MS/MS.
KINOMEscan™ Kinase Selectivity Assay.
The kinase selectivity assay was outsourced to DiscoverRX and the methodological details were provided by the company as described below. Kinase-tagged T7 phage strains were grown in parallel in 24-well blocks in an E. coli host derived from the BL21 strain. E. coli were grown to log-phase and infected with T7 phage from a frozen stock (multiplicity of infection = 0.4) and incubated with shaking at 32 °C until lysis (90-150 min). The lysates were centrifuged (6,000 x g) and filtered (0.2 μm) to remove cell debris. The remaining kinases were produced in HEK-293 cells and subsequently tagged with DNA for qPCR detection. Streptavidin-coated magnetic beads were treated with biotinylated small molecule ligands for 30 mins room temperature to generate affinity resins for kinase assays. The liganded beads were blocked with excess biotin and washed with blocking buffer (SeaBlock (Pierce), 1 % BSA, 0.05 % Tween 20, 1 mM DTT) to remove unbound ligand and to reduce non-specific phage binding. Binding reactions were kinases, liganded affinity beads, and test compounds in 1x binding buffer (20 % SeaBlock, 0.17x PBS, 0.05 % Tween 20, 6 mM DTT). Test compounds were prepared as 40x stocks in 100% DMSO and directly diluted into the assay. All reactions were performed in polypropylene 384-well plates in a final volume of 0.02 ml. The assay plates were incubated at room temperature with shaking for 1 h and the affinity beads were washed with wash buffer (1x PBS, 0.05 % Tween 20). The beads were then re-suspended in elution buffer (1x PBS, 0.05 % Tween 20, 0.5 μM non-biotinylated affinity ligand) and incubated at room temperature with shaking for 30 min. The kinase concentration in the eluates was measured by qPCR. Compounds that bind the kinase active site and directly (sterically) or indirectly (allosterically) prevent kinase binding to the immobilized ligand, will reduce the amo of kinase captured on the solid support. Screening hits are identified by measuring the amount of kinase captured in test vs control samples and reported as “% Ctrl”, where lower numbers indicate stronger hits.
Selectivity Score or S-score is a quantitative measure of compound selectivity. It is calculated by dividing the number of kinases that compounds bind to by the total number of distinct kinases tested, excluding mutant variants.
This value can be calculated using %Ctrl as a potency threshold (below) and provides a quantitative method of describing compound selectivity to facilitate comparison of different compounds.
In vivo Pharmacokinetic Study.
Female CD1 mice (6-8 weeks of age) were dosed with vehicle (10 % ethanol, 90 % corn oil) or HC4 at 50 mg/kg once and blood samples col into tubes containing K2-EDTA at different timepoints (15 min, 30 min, 1 h, 2 h, 4 h, 8 h, 12 h and 24 h). Plasma samples were prepared by centrifugation. The plasma concentration of test compound was determined by protein precipitation and LC-MS/MS.
Database:
TCGA data on mRNA expression levels of EIF2AK3 was accessed and analyzed through cBioPortal (RRID:SCR_014555) (https://bit.ly/3yBhw2b).
KM plotter software (RRID:SCR_018753) was used to analyze the prognostic value of the CSPS expression in 958 all-subtype and 119 HER2+ breast cpatients with available data (32). The best cut-off value was used to identify the high and low expression groups. The hazard ratio with 95 % CI and P log-rank value were calculated.
Statistical analysis.
All points represent independent biological samples with error bars representing standard deviations and statistical significance was determined using one-sided Mann–Whitney test or t test using the Graph Pad Prism Software (RRID:SCR_002798).
Data availability.
Previously published RNAseq of MMTV-Neu EL and PT spheres is available under the accession code GSE165431. All data needed to evaluate the conclusions are included in the paper and Supplementary Materials. Other data supporting the findings are available from the corresponding authors on reasonable request.
RESULTS
Quiescent HER2+ DCCs display activation of the ISR
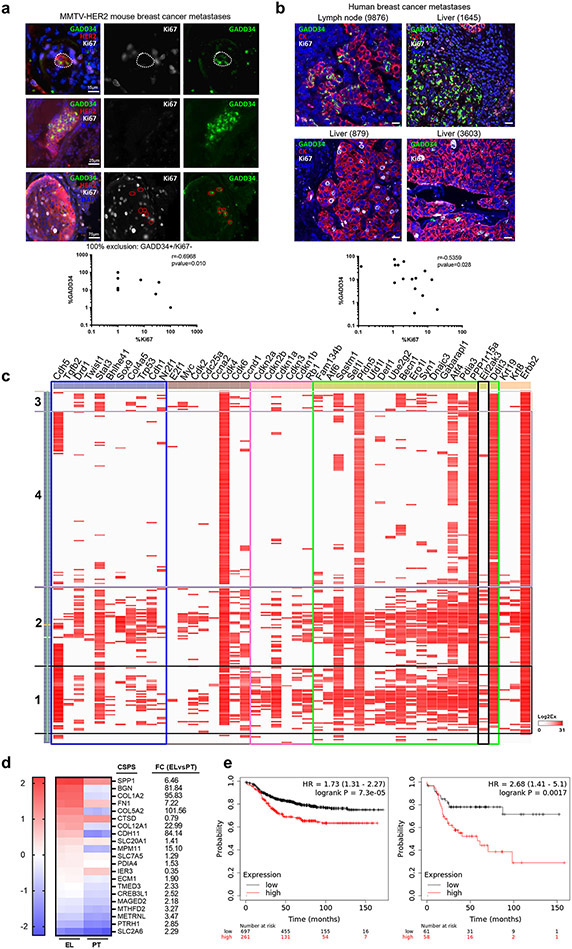
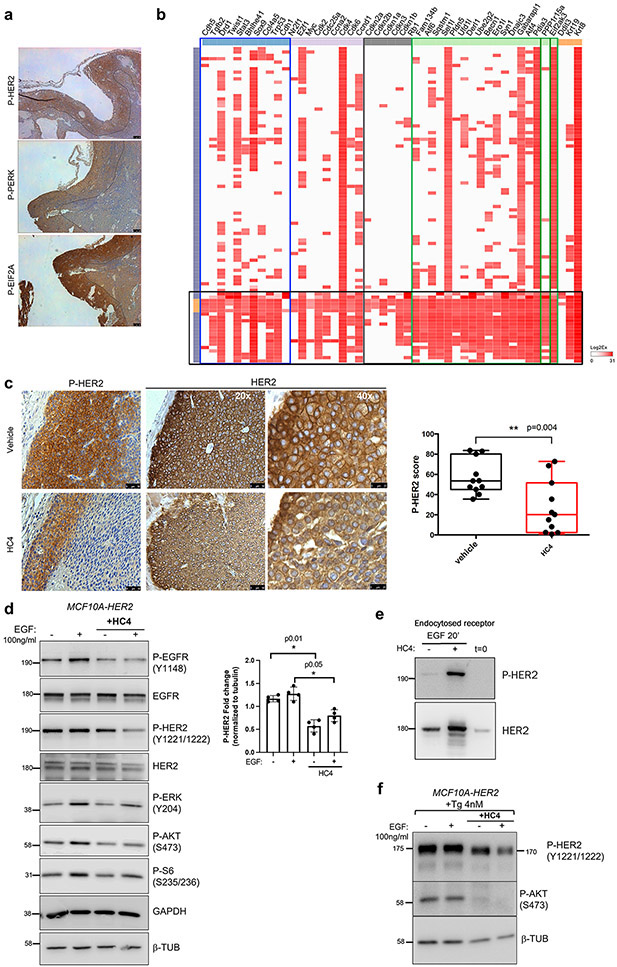
In the MMTV-HER2 (henceforth HER2+) mouse model, a highly utilized model of HER2+ human breast cancer, a high percentage of mice develop metastases to the lungs, which can be initiated by early or late DCCs (25,26,33,34). Dormant HER2+ DCCs display loss of an epithelial-like phenotype with E-cadherin as a main marker and gain expression of a mesenchymal-like program characterized by increased expression and function of the transcription factors (TF) ZFP281 and Twist1 (26). E-cadherin-negative DCCs in pancreatic cancer models were also shown to be quiescent and displayed upregulation of CHOP, a PERK-induced gene (24). We set out to determine whether in the mouse HER2+ spontaneous metastasis model this same correlation exists between ISRhigh levels and cell cycle arrest. The correlation was evaluated by two different approaches, high resolution imaging using IF and single cell resolution gene expression analysis of DCCs. We performed IF of MMTV-HER2 lung tissue sections of animals bearing large tumors and thus bearing dormant and proliferative DCCs (26). Tissues were co-stained to detect DCCs positive for HER2, Ki67 (as a marker of proliferation) and GADD34 (or PPP1R15A) as a surrogate marker of PERK activation. GADD34 is a PERK-inducible stress gene responsible for the programmed shift from translational repression (due to eIF2α phosphorylation) to stress-induced gene expression (35). However, GADD34 can be induced by other eIF2α kinases (e.g., GCN2, HRI, PKR) in addition to PERK, suggesting it may not only report on PERK activity and this is why we termed these DCCs and micro-metastatic lesions ISRhigh. Image analysis showed that HER2+ metastatic lesions or solitary DCCs with a low proliferative index (Ki67low) presented high levels of ER stress as shown by high levels of GADD34 expression (Fig. 1a upper panels and graph). On the other hand, highly proliferative DCCs or lesions showed very low levels of GADD34 staining (Fig. 1a lower panels and graph). The two markers, Ki67 and GADD34, did not co-occur in 100 % of the cells and proportion of Ki67+ and GADD34+ cells were anti-correlated (Spearman test, p=0.01), supporting that in our HER2+ model ISRhigh and quiescent DCCs and metastatic lesions can be identified via GADD34 detection. Inhibition of PERK with three potent and specific inhibitors of PERK (HC4, HC19, HC28) in MCF10A breast pre-malignant cells and E0771 luminal B breast cancer cells completely eliminated thapsigargin induced GADD34 expression, suggesting that while not a unique PERK-regulated gene GADD34 can be highly dependent on this kinase in ISRhigh breast premalignant and malignant cells (Supplementary Fig. S1a).
Figure 1. Quiescent disseminated HER2+ cells display high levels of ISR PERK pathway activation.
(a) Lung sections of MMTV-HER2 animals were stained for HER2, Ki67 (proliferation marker) and GADD34 (ER stress marker). The cells/met positive for either marker was quantified and shown as percentage of total cells and correlation was plotted (N=13). (b) Human breast cancer metastases from different locations (lymph node, liver, lung) were stained for cytokeratins, Ki67 (proliferation) and GADD34 (ER stress). The cells/met positive for either marker was quantified and shown as percentage of total cells and correlation was plotted (N=10), scale bar 25 μm. (c) Hierarchical clustering of the high-throughput targeted-gene expression (columns) profile of single cells (lung DCCs) (rows). Blue box, dormancy genes; brown box, cell cycle up genes; pink box, cell cycle down genes; green box, ER stress genes; black box, EIF2AK3 (PERK) gene. (d) Heat map showing the mRNA expression and fold-change (FC) of the previously published cancer-specific PERK signature (CSPS) genes (20) from bulk RNAseq analysis in MMTV-HER2 mammary gland early lesions (EL) and mammary primary tumors (PT) (26). (e) Distant metastasis-free survival Kaplan-Meier curve of all subtypes (left, N=958) and HER2+ (right, N=119) breast cancer patients stratified by CSPS expression using KM Plotter software.
We next tested if these correlations would also hold true in human breast metastatic lesions. HER2+ human breast cancer metastases (n=10) to lymph node and additional 7 metastatic samples from different subtypes and tissues (lymph node, lung, liver) (Supplementary Table S1) were stained with a pan-cytokeratin cocktail to identify the metastatic lesions, and for Ki67 and GADD34. We observed that advanced human metastatic lesions displayed a more heterogeneous pattern of staining for both markers between different patients and in-between different areas of the same lesion than in the mouse model. However, a consistent negative correlation between levels of proliferation (Ki67+) and GADD34 expression was found in HER2+ lymph node metastasis (Fig. 1b) or other target organs as well (Spearman test, p<0.028) (Supplementary Table S1). This analysis validates the findings in the mouse model and that GADD34 may help identify ISRhigh/quiescent tumor cells in human metastatic sites.
Next, the analysis was expanded to markers of proliferation, quiescence, dormancy and ISR in metastatic cells in the HER2+ mouse model. To this end, we performed single cell targeted-gene expression analysis of solitary DCCs, micro- and macro-metastases lodged in lungs of HER2+ mice. Lungs from MMTV-HER2 females were processed into single cell suspensions and HER2+/CD45− cells were sorted (Supplementary Fig. S1b). The sorted cells were then processed for single cell separation, lysis, RT and pre-amplification using the C1 (Fluidigm) technology as shown in Supplementary Fig. S1b. This technique allowed us to isolate and process with a high degree of confidence (IF and molecular confirmation of HER2+ single cell) and quality 255 single DCCs and 90 primary tumor cells and their corresponding pools. Subsequently, high-throughput qPCR was used to analyze the expression of ISR genes, cell cycle genes (both activators and inhibitors) and dormancy genes based on the literature (25,36,37) (Supplementary Fig. S1c). Single cell resolution gene expression of DCCs revealed the existence of two populations of cells that are enriched for ISR genes (groups 1 and 2, 41 % of DCCs) (Fig. 1c). Group 1 (approximately 19 % of the DCCs) showed concomitant and strong upregulation of all the ISR genes tested (including PERK itself) (green box) along with negative regulators of cell proliferation such as Rb1 and TP53 and CDK inhibitors p21, p27, p16 and p15 (pink box) (Fig. 1c). We also observed in these cells enrichment in the expression of dormancy genes such as NR2F1, DEC2 (Bhlhe41), TWIST1, CDH5, STAT3 and COL4A5 (25,37) (blue box). DCCs group 2 (22 %) also showed high levels of ISR genes expression along with the quiescence marker p21. In a third group (group 3 (6 %)) ISR, cell cycle inhibitors and dormancy genes were less prevalent, suggesting these might represent cells transiting out of dormancy or in cycling mode. In total, around 40 % of the DCCs showed high to intermediate level of ISR genes expression, concurrent with cell cycle inhibitors or dormancy genes. Importantly, independent published validation in the same HER2+ mouse model showed that ~40 % of solitary DCCs express the dormancy regulators ZFP281 and NR2F1 (26) . Thus, our findings in this study are in range with the percentage of dormant DCCs detected in advanced progression MMTV-HER2 animals previously reported by our lab using phospho-Histone H3, phospho-Rb, Ki67 and ZFP281 detection (25,26). These data illustrate that animals with detectable metastasis are comprised of ~40 % DCCs that display high expression of cell cycle inhibitory genes. Importantly, we show that dormant DCC subpopulations display an activated ISR with prominent expression of PERK pathway genes.
To strengthen our claim that dormant HER2+ DCCs we profiled in Figure 1c are upregulating a PERK signaling program, we used a published PERK activity signature of 23 genes that predict for shorter metastasis-free periods of breast cancer patients (21) and cross-referenced it with independently published data derived from bulk and single cell DCC RNAseq profiling (26). In agreement with our findings, this PERK activation signature was highly enriched in primary MMTV-HER2 early lesions (those that seed dormant DCCs more frequently (26)) compared to primary overt tumors in our bulk RNAseq analysis (Fig. 1d). The PERK activity signature was also enriched in early DCCs in lungs that adopt a mesenchymal-like and dormant phenotype (Fn1, Bgn, Spp1, Col5a2, Ecm1, Maged2, Col1a2) (26). Thus, the human-derived PERK activity signature (21) was specifically enriched in cancer cells prone to enter dormancy or that are dormant as DCCs (26). We further tested whether this signature informed on metastasis-free survival in the different subgroups of breast cancer patients. We found that the patients that displayed an enrichment of the PERK signature progressed faster to metastasis in all analyzed subtypes. Importantly, this was also true for the HER2+ subtype modeled in this study and the hazard ratio was higher than in the other subtypes (Fig. 1e). Our results, collectively provide a strong basis to support that quiescent DCCs and slow-cycling cells within metastasis are enriched for a ISRhigh signature (21).
HC4 specifically inhibits PERK kinase activity without toxicity
The above findings opened the possibility of using selective PERK inhibitors to test whether inhibition of PERK could affect dormant DCC fate and metastasis formation. We used a PERK inhibitor derived from a 2-amino-3-amido-5-aryl-pyridine scaffold that is currently in clinical development (27). Briefly, HC4 is a potent and selective PERK inhibitor with appropriate drug-like properties to support in vivo animal and human clinical studies (Supplementary Table S2, S3, S4 and S5). KINOMEscan™ kinase profiling of HC4 (along other variants from the amino-pyridine-mandelic acid-derived series, HC19 and HC28) (Supplementary Tables S3, S4, S6, S7) displayed exceedingly high selectivity compared to other PERK inhibitors described in the literature and specifically GSK2656157 (38), even at a very high concentration (10 μM). The high specificity of HC4 for EIF2AK3 (PERK) over the eIF2α kinase family, EIF2AK1 (also known as HRI), EIF2AK2 (also known as PKR) and EIF2AK4 (also known as GCN2) or HER2 (Supplementary Tables S3) indicates that the measured activity on eIF2α was highly specific to PERK inhibition.
In cellular assays, HC4 effectively decreased P-PERK (P-T980) levels in MCF10A cells overexpressing HER2 (MCF10A-HER2) and in HEK293 cells stimulated with Tunicamycin (Supplementary Fig. S1d) as well as GADD34 induction upon thapsigargin treatment (Supplementary Fig. S1a). Importantly, treatment with HC4 rendered MCF10A-HER2 cells sensitive to low dose thapsigargin treatment (Supplementary Fig. S1e), further supporting the idea that PERK activation is necessary to support survival of cancer cells in a stressed environment. Furthermore, specificity of HC4 was tested using an Fv2E-ΔNPERK fusion construct-expressing cell line described previously (14,22,39). This system allows to induce PERK activation via dimerization with the divalent small molecule AP20187. We used a dose of AP20187 that in this case we knew would induce rather than prevent apoptosis via strong pro-apoptotic PERK signaling (14,22,39). We next determined whether HC4 would rescue the pro-apoptotic effect. HC4 fully rescued the pro-apoptotic function of strong PERK activation supporting it is a specific inhibitor (Supplementary Fig. S1f).
In vivo studies showed that single dose administration of HC4 in CD1 female mice demonstrated bioavailability following IP administration and that the levels of HC4 in the plasma achieve a threshold that is well above that needed for PERK inhibition based on biochemical and cellular P-PERK IC50 values over a 24 h time window (Supplementary Fig. S1g). Repeated dosing of HC4 from 30 to 100 mg/kg was well tolerated as shown by no significant changes in body weight (Supplementary Fig. S1h).
Having confirmed the HC4 inhibitor specificity and in vivo drug-like properties, we treated 24–32-week-old uniparous MMTV-HER2 female mice (which present an incidence of lung metastases of around 80 %) with vehicle (see methods) or HC4 (50 mpk) IP daily, for two weeks, and collected mammary glands, lungs, pancreas, bone marrow and overt primary tumors for further analyses. The inhibitor did not have a significant effect on bone marrow cell homeostasis or on peripheral blood white cells as shown by no effect on total cell counts in MMTV-HER2 females (Supplementary Fig. S1i) as an additional indication of safety.
HC4 treatment caused a significant decrease in P-PERK and P-eIF2α levels in tissues such as the mammary gland ducts and in pancreatic tissue (although only partial inhibition was observed at this dose, especially in pancreatic islets), as shown by IHC (Fig. 2a). Similarly, around 50 % decrease in P-PERK levels was detected by western blot of tumor lysates (Fig. 2b; see also Fig. 4a for the effect of HC4 on primary tumor growth). We conclude that systemic HC4 delivery effectively inhibits PERK activation and eIF2α phosphorylation in tissues. The use of the inhibitor did not fully deplete PERK activity, which allows mice to control their pancreatic function and glucose levels (40). Moreover, in a separate study in mice dosed orally with HC4 for 28 days, where similar exposures were achieved to the 50 mpk IP dose noted above, no deleterious effects on the pancreas or with clinical chemistry (i.e. insulin and glucose levels) were observed (data not shown).
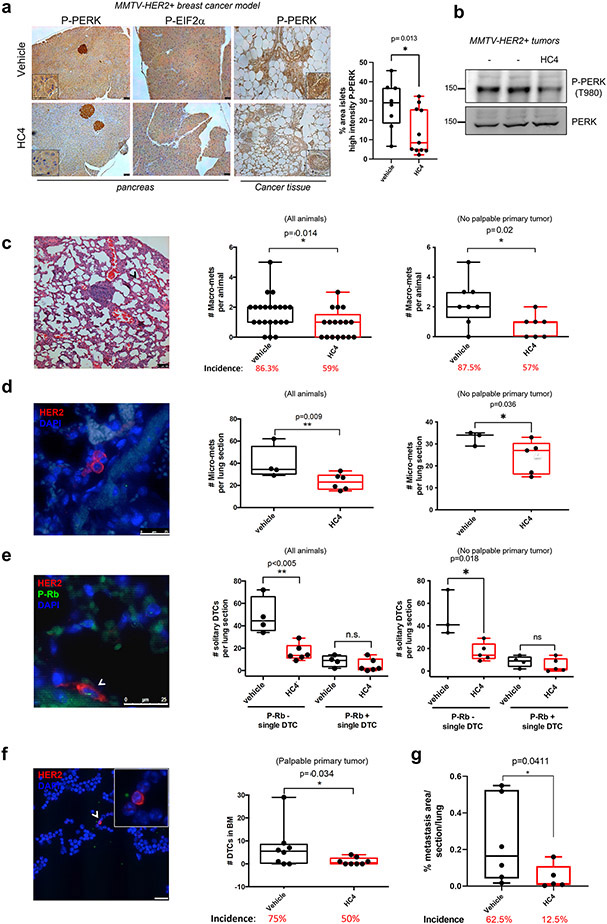
Figure 2. HC4 PERK inhibition decreases metastatic disease in lungs and bone marrow through elimination of quiescent DCCs.
(a) MMTV-HER2 females (24-week-old) were injected daily with vehicle or HC4 (50 mpk) for 2 weeks. IHC of pancreas and mammary gland sections with antibodies to P-PERK and P-EIF2α. Inserts show higher magnifications. Scale bars, 100 μm (left panel). Quantitation of the percentage area in the pancreas islets that show high intensity P-PERK staining (threshold method) (vehicle N=8, HC4 N=11) (right panel). (b) MMTV-HER2 tumors from control and HC4 treated mice were analyzed for P-PERK as well as total PERK levels by Western blot. Representative blot of three is shown. (c) Macro-metastases (>100 cells) were detected by H&E staining and quantified in 5 lung sections/animal ± s.d (vehicle N=22, HC4 N=17). Scale bar, 100 μm. P by Mann-Whitney test. (d) Micro-metastases (2-100 cells) were detected by IF staining using an anti-HER2 antibody and quantified per lung section/animal ± s.d. (vehicle N=6, HC4 N=6). Scale bar, 25 μm. P by Mann-Whitney test. (e) Solitary DCCs were detected by IF staining for HER2, classified as P-Rb+ or P-Rb− and quantified per lung section ± s.d. (vehicle N=5, HC4 N=6). Scale bar, 25 μm. P by Mann-Whitney test. (f) DCCs in bone marrow were detected by IF staining for HER2 in cytospins from mature hematopoietic cell-depleted bone marrow tissue (N=8 per condition). Scale bar, 25 μm. P by Mann-Whitney test. Mice were injected with MMTV-HER2 cells from primary tumor lesions via the tail vein, allowed them to expand into established metastatic lesions for 3 weeks and treated with vehicle or HC4 (50 mg/kg) for two additional weeks. (g) Metastasis were detected by H&E or IF (for HER2 detection) staining in 5 lung section/animal ± s.d (vehicle N=6, HC4 N=5) and total metastatic burden was calculated. P by Mann-Whitney test.
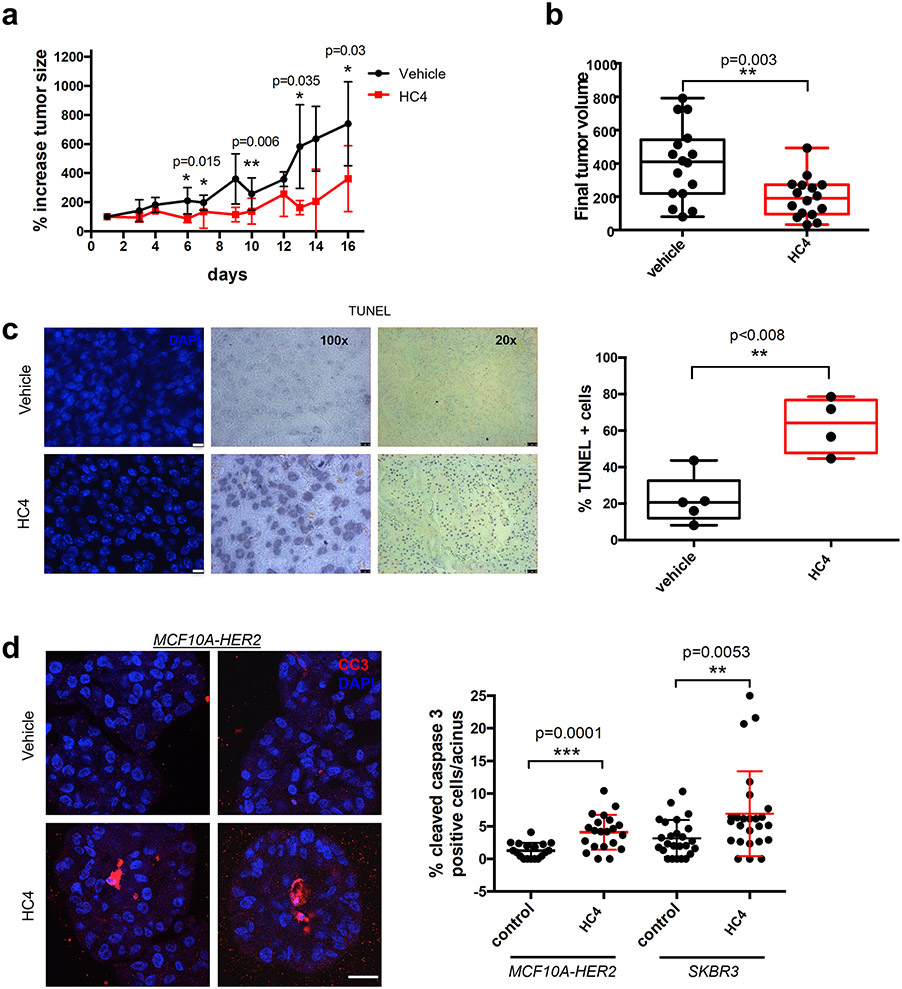
Figure 4. PERK inhibition impairs tumor growth in MMTV-HER2 females.
(a) MMTV- HER2 females (24- to 32-week-old) presenting overt tumors were injected daily with vehicle or HC4 (50 mpk) for 2 weeks. Percentage variation of tumor size in vehicle- and HC4-treated animals ± s.d. (N=16 per condition). P by Mann-Whitney test. (b) Final tumor volume (mm3). The whiskers represent the min and max of the data (N=16 per condition). P by Mann-Whitney test. (c) Representative IHC of TUNEL staining to measure apoptosis levels in tumor sections. Scale bars, 10 and 50 μm. Graph, percentage TUNEL positive cells in vehicle- and HC4-treated tumor sections (vehicle N=5, HC4 N=4). P by Mann-Whitney test. (d) HER2+ MCF10A-HER2 or SKBR3 cells were seeded on Matrigel and after acinus establishment (day 4) wells were treated with vehicle (control) or HC4 (2 μM) for 10 days. Representative confocal images of MCF10A-HER2 acini IF-stained for cleaved caspase-3. Scale bar 20 μm. Graph, percentage of cleaved caspase-3 positive cells per acini (MCF10A-HER2 N=20 per condition, SKBR3 control N=24, HC4 N=22) ± s.d. P by Student’s t test.
HC4 treatment eradicates quiescent DCCs suppressing lung metastasis and bone marrow DCC burden
MMTV-HER2 animals develop metastases to the lungs, which can be initiated early in progression, even during stages of non-palpable tumor (25,33,34). Thus, we next monitored the effect of HC4 on metastatic disease in these animals. More than 80 % of the vehicle-treated animals presented metastases detectable in sections stained with H&E (Fig. 2c). Lesions that displayed >100 cells were categorized as macro-metastases as they are also commonly positive for proliferation markers (Fig. 1a & (25,41)). The quantification of macro-metastases per animal (5 non-consecutive lung sections) revealed that, after just a two-week treatment period, HC4 reduced the number and the incidence of macro-metastases in the whole population of animals (Fig. 2c, left graph). This effect was independent on the primary tumor size since the quantification of macro-metastases in animals showing non-palpable tumors at this stage (as reported for this model (25,42)) also revealed a similar decrease in number and incidence in metastases (Fig. 2c, right graph). The effect on the number of macro-metastases was not accompanied by a significant effect on the area of these metastases (Supplementary Fig. S2a). This suggested that PERK inhibition might be acting on the initial steps of metastasis (solitary DCCs or micro-metastases) rather than eliminating established macro-metastases.
To expand these findings to another model of cancer not dependent on HER2 we chose a human HNSCC PDX (T-HEp3) previously shown to enter dormancy in the lung after tail vein injection (43-47) (Supplementary Fig. S2b). To this end, we injected i.v. 2.5x105 T-HEp3 cells as single cell suspensions in Balb/c nu/nu mice and 24 h after injection we treated the mice with vehicle or 50 mg/kg of HC4 for 10 days to primarily capture solitary dormant DCCs. At the end of the experiment mice were euthanized and lungs sectioned to score the frequency of T-HEp3 DCCs in the lung detected via a human specific anti-vimentin antibody, as reported previously (43,44,46). These results revealed that HC4 caused a statistically significant ~3-fold reduction in the number of solitary HEp3 DCCs in the lung (Supplementary Fig. S2b). At this stage of lung colonization HEp3 cells are in a quiescent phase (43), supporting that HC4 can target the survival of these dormant DCCs as in the HER2+ mouse model. In addition, to further expand the repertoire of models we employed the Balb/c syngeneic EMT6 model that spontaneously metastasizes to lung (48). In this model treatment with HC4 at 10 mg/kg 9 days after implanting 106 cells in the fat pad also strongly inhibited metastatic incidence and multiplicity of lesions per lung/animal after 17 days of treatment without affecting primary tumor growth (not shown) (Supplementary Fig. S2c).
It is possible that the effect of HC4 could be on the intravasation process and thus less metastasis is simply due to less DCCs reaching the lung from circulation. To test this hypothesis, we checked whether HC4 treatment might be affecting the intravasation of tumor cells from the primary site to circulation. Detection of HER2+ circulating tumor cells (CTCs) directly in blood samples showed no significant differences between vehicle and HC4-treated animals (Supplementary Fig. S2d), indicating that HC4 is not grossly affecting the intravasation of tumor cells. We next tested whether HC4 treatment affected the transition from solitary DCC to micro-metastasis (containing 2-100 cells) in the lung tissue. Detection of micro-metastasis and single DCCs in lung sections using HER2 detection via IF revealed a significant decrease in the number of micro-metastases in HC4-treated females (Fig. 2d). Again, this effect was not due to a potential direct effect on the primary tumor since in animals not showing any palpable tumors the effect on micro-metastases was similar (Fig. 2d, right graph).
More than 80 % of single DCCs found in lungs were negative for P-Rb, indicating that they are mostly out of cycle and dormant as reported in independent studies (25,26,41). HC4 significantly reduced the number of non-proliferating (P-Rb negative) single DCCs that are commonly associated with blood vessels in lung sections, while not affecting the number of P-Rb positive solitary DCCs (Fig. 2e) or in micro-metastases (Supplementary Fig. S2e). As with macro- and micro-metastases, this effect was independent on the presence or size of a primary tumor (Fig. 2e, right graph).
In this mouse model, the majority (>99 %) of single DCCs in the bone marrow are negative for P-Rb or Ki67 (25) and they never form spontaneous bone metastasis (33), indicating that they are mostly out of cycle and dormant (25), making it an ideal model to assess the effect of the PERK inhibitor on disseminated dormant disease. Importantly, HC4 significantly decreased the number of DCCs found in bone marrow (Fig. 2f). To incorporate another model of tumor cell dormancy and test the effect of HC4, we turned to the dormant variant of HNSCC PDX T-HEp3 cells, D-HEp3. D-HEp3 cells have been shown to depend on PERK but also ATF6α signaling for survival (13,23). These cells are prone to enter dormancy in vivo, adopting a G0/G1 quiescent state 48 h after inoculation on the chicken embryo CAM system or nude mice (28,49). These experiments revealed that HC4 caused an eradication of dormant D-HEp3 cells in vivo after a 6-day treatment with HC4 (20 μM, used at 10x dose due to dilution in CAM experiments), and this reduction correlated with a 5-fold increase in the accumulation of cleaved caspase-3 positive D-HEp3 cells in the nodules, also detected via a human specific vimentin antibody (Supplementary Fig. S2f). All together, these results argue that PERK inhibition can selectively target and eradicate dormant DCCs.
HC4 treatment suppresses HER2+ breast cancer lung metastasis also by targeting established micro-metastasis
The data in Figure 1a and b showed that in established micro-metastasis and to some extent macro-metastatic lesions we could find ISRhigh cancer cells. This led us to hypothesize that some cancer cells in established metastatic lesions may show a persistent unresolved ISR and/or that other cells may transit through that state and resolve the ER stress. Regardless of the time spent as ISRhigh cells, if this state enables fitness and survival for established lesions, targeting this ISRhigh population of cells could also affect progression of these metastatic lesions. To this end, we injected MMTV-HER2 cells from primary tumor lesions and via the tail vein and allowed them to expand into established metastatic lesions for 3 weeks as reported (25), which allows for conversion of all late DCCs to form micro-metastasis. We treated one group of animals with vehicle and another with HC4 (50 mg/kg) for two additional weeks. As shown in Figure 2g, HC4 treatment for 2 weeks was sufficient to strongly reduce macro-metastatic lesion incidence and total metastatic burden, while not affecting the frequency of already established micro-metastases (Supplementary Fig. S2g). These data support that the effect of HC4 on metastasis is due to a combined effect of targeting ISRhigh solitary DCCs (Fig. 2c-f) and micro-metastasis progression to larger lesions (Fig. 2g).
PERK inhibition blocks HER2-driven early and late mammary primary tumor progression
We next tested the effect of HC4 on primary tumor lesions during the spectrum of HER2+ mammary cancer in mice. HER2-driven progression was found to be genetically dependent on the PERK kinase in the MMTV-HER2 model previously (17) and another study showed that HER2+ tumors are sensitive to proteotoxicity and dependent on ERAD for survival (19). Further, TCGA database analysis (20) showed that ~14 % of HER2-amplified human breast tumors (Breast Invasive Carcinoma, TCGA, Nature 2012 dataset) display upregulation of the mRNA for PERK. This population may be a subset of a larger population of patients that we found in Figure 1 have a strong enrichment in a PERK high signature (48.74 % of tumors for the HER2+ subtype) and have also poor prognosis. Together these data led us to investigate whether HC4 affected HER2-induced breast tumor progression in primary lesions where the different stages of progression from hyperplastic mammary glands through DCIS and invasive cancer can be dissected (50,51).
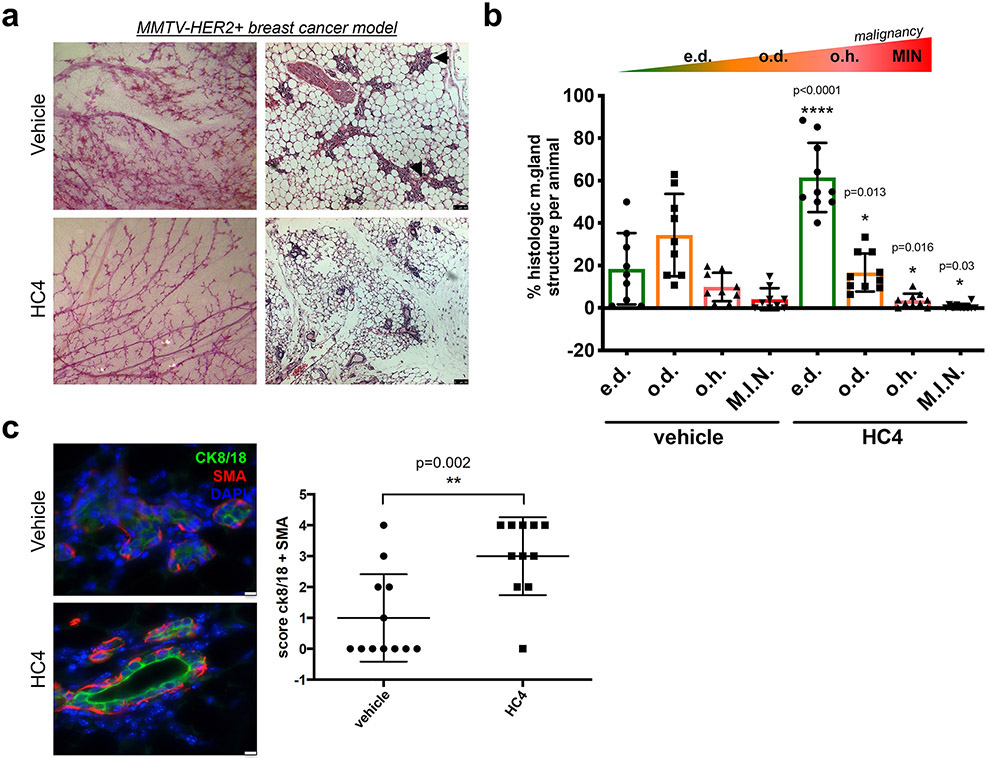
Analysis of 24-week-old uniparous female mammary glands showed that vehicle-treated MMTV-HER2 animals exhibited ducts with secondary and tertiary dense branching (Fig. 3a, left panels), and histological analysis showed frequent mammary hyperplastic lesions (Fig. 3a, right panels, black arrows). In contrast, HC4-treated animals showed a “normalized” glandular architecture with less dense branching, resembling the mammary tree of non-transgenic normal FVB mice (Supplementary Fig. S3a). HC4-treated animals also showed a dramatic increase in the number of hollow lumen mammary gland ducts, constituting more than 60 % of the structures compared with around 20 % in control females (Fig. 3b and Supplementary Fig. S3c). The number of occluded hyperplasias and DCIS-like lesions was also reduced to less than half of that observed in vehicle-treated animals. Hyperplastic lesions in control HER2+ animals showed varying degrees of luminal differentiation as assessed by the uneven levels of cytokeratin 8/18 expression (Fig. 3c, upper panel). The myoepithelial cells (detected as smooth muscle actin, SMA, positive) were unevenly distributed in the vehicle-treated hyperplasias in the MMTV-HER2 mice. In contrast, HC4-treated MMTV-HER2 animals presented increased expression of cytokeratin 8/18 in the luminal layer, frequently surrounding an empty lumen, and an external continuous layer of myoepithelial cells (Fig. 3c, lower panel and graph). Treatment of non-tumor bearing healthy mice with HC4 for 2 weeks did not affect the normal branching and morphology of the mammary gland ductal tree (Supplementary Fig. S3b). This data indicates that HC4 treatment leads to a delayed progression of HER2-driven early lesions in the mammary gland.
Figure 3. The PERK inhibitor HC4 causes mammary gland “normalization” in the MMTV-HER2+ breast cancer model.
(a) Representative images of carmine staining of whole mount mammary glands and H&E-stained mammary gland sections from vehicle- and HC4-treated animals. Scale bar, 100 μm (b) Quantification of histological structures (empty duct e.d., occluded duct o.d., occluded hyperplasia o.h. and DCIS-like mammary intraepithelial neoplasia M.I.N) present in H&E-stained mammary gland sections (N=50/animal, animals vehicle N=9, HC4 N=10) found in vehicle- and HC4-treated animals ± s.e.m. P by Mann-Whitney test. (c) IF for epithelial luminal marker cytokeratin 8/18 (CK8/18) and myoepithelial marker Smooth Muscle actin (SMA) in mammary gland sections. Score for CK8/18+ and SMA+ structures per animal, vehicle N=12, HC4 N=11. P by Mann-Whitney test. Scale bar, 75 μm.
We next treated animals once they displayed tumors, ranging from 30 to 200 mm3 volume (two tumors were >200 mm3) for two weeks with HC4. In the vehicle treatment group, tumors grew steadily (Fig. 4a), reaching up to 10 times its original volume in two weeks (Supplementary Fig. S4a, left graph). In contrast, HC4-treated tumors showed a reduced growth rate (Fig. 4a), with some tumors remaining in complete cytostasis (defined as doubling tumor volume only once in the 2-week period, 43 % in HC4-treated vs 7 % in controls) (Supplementary Fig. S4a, right graph). Four tumors showed regression in the 2-week window treatment (Supplementary Fig. S4b). This led to a significant decrease in median final tumor volume (Fig. 4b). While the levels of proliferation (phospho-histone H3 IHC) were not different between vehicle- and HC4-treated tumors (Supplementary Fig. S4c), TUNEL staining of tumor sections showed a significant increase in the levels of DNA fragmentation present in HC4-treated animals (Fig. 4c). Thus, in overt primary lesions, HC4 treatment induced apoptosis of established HER2+ tumors, arguing for context-dependent fitness-promoting functions of PERK during tumor progression.
Treatment of HER2-overexpressing (MCF10A-HER2 or ZR-75-1) or HER2-amplified (SKBR3) breast cancer cells (Fig. 4d and Supplementary Fig. 4d) in 3D acini cultures in Matrigel showed that a 10-day treatment with HC4 (2 μM) significantly increased levels of apoptosis (cleaved caspase-3) in these spheroids, especially in the inner cell mass that is deprived from contact with the ECM (Fig. 4d). As in the in vivo conditions, we did not detect a significant change in the levels of proliferation as detected by phospho-histone H3 levels (Supplementary Fig. S4e). We conclude that early MMTV-HER2 lesions require PERK for HER2-driven alterations in ductal epithelial organization and, eventually, tumor growth.
PERK signaling is required for optimal HER2 phosphorylation, localization and AKT and ERK activation
While we showed effects of HC4 in HER2-dependent and -independent tumors (HEp3), we wondered about the functional relationship between PERK and HER2-driven biology. This was because the K-M curves in Figure 1 showed a strong association between poor prognosis and a PERKhigh signature in HER2+ breast cancers, and also because HER2+ tumors are sensitive to proteotoxicity (19). Thus, HER2 signaling might depend on PERK and inhibition of PERK might affect optimal HER2 activity due to increased ER client protein load. Detection of HER2 phosphorylation at residues Y1221/1222 in tumors showed that the area positive for P-HER2 reported by others (52) overlapped with the staining for P-PERK and P-eIF2α (Fig. 5a), showing a distinctive rim staining pattern. This finding indicated that the activation of PERK and HER2 pathways is not generalized and co-localize in the same area of the tumor tissue. Single cell targeted-gene expression profiling of primary tumor cells showed a population of primary tumor cells (around 25 %) with high levels of ER stress genes expression (Fig. 5b), which could correspond to the ones localized in the rim of the tumor. Importantly, when we scored the P-HER2 levels in the tumors (taking into account both the area and the intensity of the staining (Supplementary Fig. S5a)) we found that HC4-treated tumors showed significantly lower levels of P-HER2 than control animals (Fig. 5c). HC4 does not target HER2 (or ErbB family) kinase activity directly as shown in Supplementary Table S6 and (27,53). In vitro treatment of MCF10A-HER2 cells with HC4 revealed that PERK inhibition decreased both the basal and EGF-induced levels of P-EGFR and P-HER2, along with downregulation of the survival pathway P-AKT, P-S6 and P-ERK1/2 levels (Fig. 5d and Supplementary Fig. S5b). No obvious effect was observed under these conditions on total HER2 levels or heterodimerization with EGFR as determined by surface biotinylation and co-immunoprecipitation studies (data not shown). As mentioned above, since HC4 does not have a direct inhibitory effect on the active site of any of the ErbB family members, AKT or S6 kinases (Supplementary Table S6), we hypothesized that this effect must be due to an indirect effect of PERK inhibition on HER2 signaling. In contrast to other ErbB family members, HER2 is known to remain at the plasma membrane after ligand binding and dimerization (54,55). We thus tested if HC4 might be disturbing the mechanism of activation of HER2 receptors. To this end, we performed surface biotinylation assays to measure the presence of the receptor on the cell surface, and reversible surface biotinylation to measure receptor endocytosis (30). Our data showed that while HC4 treatment did not significantly alter the abundance of P-HER2 and total HER2 in the cell surface (not shown), it clearly increased the endocytosed phospho- and total HER2 (Fig. 5e). That the surface levels did not change may be due to the dynamic nature of the process and high expression of HER2 that may preclude detecting a decrease in surface levels but not an endocytic retention. Previous reports (56) have shown the importance of PERK for the proper maturation of several tyrosine kinase receptors, by partially blocking their trafficking from the ER to the Golgi in conditions of stress. We, as well, observed a lower molecular weight of phosphorylated HER2 protein when treating cells with HC4 in the presence of low levels of stress (Thapsigargin, 4 nM), along with a decrease in the levels of P-HER2 and P-AKT (Fig. 5f). The lower molecular weight phospho-HER2 protein could correspond to the previously reported 170 kDa precursor, containing high mannose-type oligosaccharide chains located in the ER compartment (57). Our data suggest that PERK signaling and proper ISR function is required to maintain proper HER2 downstream signaling by affecting optimal receptor localization and activation.
Figure 5. HC4 treatment decreases the levels of phospho-HER2 and downstream signaling pathways.
(a) Representative images of IHC for P-HER2, P-PERK and P-EIF2α in a MMTV-HER2 breast tumor section. Note that the rim positive for P-HER2 overlaps with P-PERK and P-EIF2α staining. Scale bar, 100 μm. (b) Hierarchical clustering of the high-throughput targeted-gene expression (columns) profile of single cells (primary breast tumor) (rows) from MMTV-HER2 females. Blue box, dormancy genes; grey box, cell cycle down genes; green box, ER stress genes and EIF2AK3 (PERK) gene; black box, population of primary tumor cells (around 25%) with high levels of ER stress genes expression (c) Representative P-HER2 and total HER2 IHC staining in vehicle- and HC4-treated breast tumors. Quantification of P-HER2 levels in tumor sections, by IHC intensity and area scoring (N=11 per condition) (See Supplementary Fig. S5a). Scale bar, 50 μm in 20X and 25 μm in 40X. P by Mann-Whitney test. (d) MCF10A-HER2 cells were starved o/n and treated +/−HC4 (2 μM), after which +/−EGF (100 ng/ml) was added for 15 min before collection. The levels of P-HER2, P-EGFR, P-AKT, P-S6 and P-ERK, as well as total HER2 and EGFR were assessed by Western blot. GAPDH and ß-tubulin were used as loading controls. Representative blot of three is shown. Densitometry analysis for P-HER2 (N=3) ± s.d. P by Student’s t test. (e) MCF10A-HER2 cells were treated as in (d) and reversible surface receptor biotinylation assay was performed. Endocytosed levels of total HER2 and P-HER2 were assessed. One of two experiments shown. (f) MCF10A-HER2 cells were treated as in (d) in presence of thapsigargin (4 nM). The levels of P-HER2 and P-AKT. ß-tubulin was used as loading control. Representative blot of three is shown.
Sequential treatment with CDK4/6 inhibitors and HC4 further reduces metastatic disease
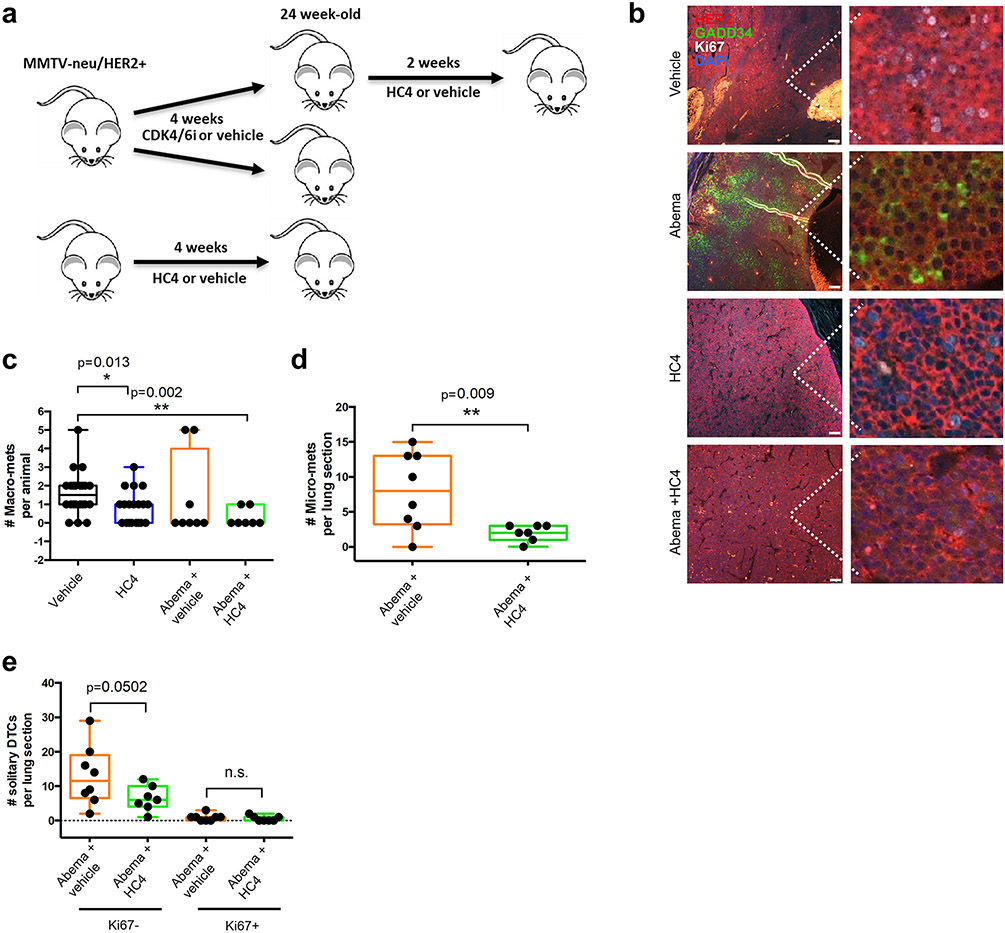
We next tested the hypothesis that treatments that may mimic dormancy could render the growth-arrested cells sensitive to HC4 treatment. In Fig. 6a, 20-week-old MMTV-HER2 mice with tumors were treated for 4 weeks with the CDK4/6 inhibitor Abemaciclib (Abema) at 50 mg/kg or control treatment. In parallel, mice were treated with control or HC4 for 4 weeks. Then, some of the mice treated with Abema were switched to treatment with HC4 for 2 weeks. We processed the tissues for analysis of ISR markers, metastasis and DCC burden.
Figure 6. Sequential CDK4/6 inhibitor followed by PERK inhibition enhances the anti-metastatic effect of Abemaciclib.
(a) MMTV-HER2 females (24-week-old) were distributed in groups and treated daily with the CDK4/6 inhibitor Abemaciclib (50 mpk), HC4 (50 mpk) or vehicle for 4 weeks. Abemaciclib treatment was followed by +/−HC4 (50 mpk) for 2 weeks. (b) IF of tumor sections for HER2, Ki67 (proliferation) and GADD34 (ER stress). Scale bars, 100 μm. (c) Macro-metastases (>100 cells) were detected by H&E staining and quantified in 5 lung sections/animal. P by Mann-Whitney test. (d) Micro-metastases (2-100 cells) were detected by IF staining using an anti-HER2 antibody and quantified per lung section/animal ±s.d. P by Mann-Whitney test. (e) Solitary DCCs were detected by IF staining for HER2, classified as Ki67+ or Ki67-and quantified per lung section±s.d. Vehicle N=18, HC4 N=17, Abema+ vehicle N+8, Abema +HC4 N=7. P by Mann-Whitney test.
The results in Figure 6b show that Abema alone reduces in primary tumors Ki67 staining and increases GADD34 expression, supporting that as we found in spontaneously dormant DCC clusters and micro-metastasis, acquisition of a forced growth arrest also triggers a ISR signal. Figure 6b also shows that HC4 treatment alone does not dramatically affect Ki67 signal as expected from our previous results but completely erased the GADD34 induction, supporting GADD34 is a good readout for PERK signaling as supported by Figure 1-2 data. Importantly, Abema + HC4 significantly decreased the number of macro-metastasis per animal compared to vehicle and a larger N would be required to draw significance from the combination vs Abema alone (Fig. 6c). Nevertheless, the results support that growth-arrested cells that enter that state in response to Abema and possibly other growth arrest inducers can be further targeted by HC4 via PERK inhibition. We further quantified micro-metastasis and solitary DCCs in different proliferative states. When scoring micro-metastasis, it was clear that HC4 further reduced micro-metastatic burden following Abema treatment (Fig. 6d). In Figure 6e we also show that HC4 can target Ki67− DCCs after Abema treatment, reproducing our prior results but now under forced induction of growth arrest. As before, we did not find an effect on Ki67+ DCCs but these were almost absent due to the Abema treatment. Overall, these data support that inducing a growth arrest program, spontaneously as in Figure 2 or in response to a CDK4/6 inhibitor, can render DCCs and likely established metastatic lesions susceptible to PERK inhibition with HC4.
DISCUSSION
Breast cancer models have suggested that HER2+ breast cancer tumorigenesis is dependent on PERK signaling for survival and adaptation (17,19). We had found that quiescent tumor cells that exist within surgical margins or as dormant DCCs in target organs (46,47,58,59) enhance their survival via PERK signaling as well as other ER stress pathways (13,22,23,60,61). Pommier et al. validated this finding in their studies demonstrating that pancreatic DCCs lodged in the liver also activate a ISR during quiescence (24). This level of concordance across a variety of tumor types and models, including the HER2 mammary models and human HNSCC HEp3 PDX tested here, supports the more general requisite of this stress adaptation biology across the cancer landscape.
We now show that pharmacological PERK inhibition with a very selective PERK inhibitor HC4 can effectively target HER2+ (and HEp3) solitary DCCs in the lung and bone marrow, established micro-metastasis and primary lesions. HC4 could also target spontaneously dormant D-HEp3 HNSCC cells, which also display dependency on PERK for survival (23). A salient finding to discuss is the inhibitory effect of PERK inhibition on metastasis. In the MMTV-HER2 and HEp3 HNSCC models PERK inhibition reduced metastasis independent of the primary tumor development timeline including those initiated early (before overt tumors were palpable). This is important because it argues that the effect on metastasis was not simply due to reduced primary tumor burden caused by HC4. These effects are further supported by the effect of HC4 on T-HEp3 HNSCC cells in the lung, HER2+ DCCs in the bone marrow and D-HEp3 cells in the CAM assay. Importantly, metastatic burden was reduced by HC4 treatment via eliminating non-proliferative solitary or small clusters of P-Rb-negative cells, but also ISRhigh established micro-metastasis. Imaging and single cell multiplex qPCR robustly revealed that HER2+ DCCs showed an upregulation of GADD34 (protein) and a larger set of ER stress and PERK-regulated genes, while also expressing genes representative of the quiescent phenotype as revealed by upregulation of several negative regulators of cell proliferation and dormancy markers. It should be taken into account that part of the PERK-induced ER stress response is transcriptional in nature while also having a key component of preferential translation of upstream ORF-containing genes, such as ATF4 and GADD34. Similarly, ISR-induced G1 arrest has been shown to be caused by inhibiting the translation of cyclin D1 (62). Our data strengthen the argument that, like overt tumors that contain ISRhigh cells, also quiescent DCCs are likely to rely on PERK signaling for survival. This supports the idea that HC4 PERK inhibition may benefit the efficacy of anti-proliferative therapies, as it would allow targeting those that divide (e.g., with anti-HER2, chemo or radiation) and the quiescent cancer cells that escape conventional treatment with HC4. A sub-population of human metastatic cells from breast cancer patients also showed a negative correlation between GADD34 and Ki67, supportive of the association seen in this study in mouse models. Our data suggest that along with NR2F1 (63), GADD34 alone or in combination with NR2F1 may serve as a robust biomarker set for dormant/ISRhigh DCCs and thus guide patient selection for treatment. A limitation of this study is related to the fact that we used GADD34 as a marker for PERK activation in human and mouse samples as we were not able to quantify PERK phosphorylation in DCCs. GADD34 can also be induced by other EIF2AK-dependent pathways limiting the claim that it is a solely readout for PERK. However, given our results with the HC4 compound and collectively with all results we are confident that GADD34 is in this system reading out PERK reliably as GADD34 expression was fully eliminated by treatment with PERK inhibitors. However, we cannot support that GADD34 is a linear reporter of PERK in vivo, without a specific control inhibiting PERK. An open question is related to the identification of the source of PERK activation in quiescent DCCs, which remains unknown. However, a recent publication revealed that dormant cells are avid producers of their own collagen-rich ECM niche (64). Translation folding and secretion of collagen impose a high demand on the ER, and thus, a heightened ISR may allow dormant cells to survive the ISR and oxidative stress imposed by such niche remodeling.
We also demonstrate that cytostatic therapies such as CDK4/6 inhibitors (Abemaciclib) not only decrease proliferation substantially, but at the same time result in concomitant activation of the ISR as shown by high GADD34 levels upon CDK4/6 inhibition. This observation would support the possibility of combining such CDK4/6 targeting therapies with PERK inhibitors to have an even more profound control of both proliferating and non-proliferating cancer cells. Encouragingly, the doses of HC4 we used did not significantly affect glucose levels, bone marrow or peripheral blood cell counts, drinking and feeding behavior of non-tumor or cancer bearing mice. A limitation of the experiments combing HC4 and Abemaciclib was that the inhibitory effect of the combination on the number of macro-metastasis per animal was not significant, likely due to reduced n. However, that HC4 + Abemaciclib reduced significantly the frequency of micro-metastasis and a similar trend was seen for Ki67− DCCs, further supports that perhaps larger studies or longer treatments would further reduce metastatic growth over the effect of each drug alone. Nevertheless, collectively, these data support the dose range evaluated in which we illustrate that a significant blockade of tumor growth and metastasis is possible through the elimination of dormant and micro-metastatic lesions and that HC4 does not adversely affect the host’s normal organ function at the doses tested.
In early lesions, our work shows that HC4 treatment precludes the development of hyperplasias and mammary intraepithelial neoplasias, indicating that HC4 treatment could have a potential preventive effect on the progression of early lesions such as DCIS to more aggressive invasive breast cancer. In established tumors, HC4 used as a single agent pushed tumors into stasis or regression via apoptosis. The full details of the mechanisms by which HC4 blocks tumor cell survival continue to be elucidated. It is possible that reduced adaptation to stress imposed by proteotoxicity in cancer cells (19) is a mechanism. Our data also revealed that HC4 reduced phospho-HER2 levels in vivo and decreased the abundance of active receptor in the membrane through enhanced endocytosis, but we did not see changes in HER2 protein degradation. However, it is still unclear how exactly PERK controls HER2 membrane localization or endocytosis. It is possible that internalization allows for better or faster de-phosphorylation of the receptor or decreases the chances of it being activated; hence resulting in decreased downstream signaling. This possibility is supported by the finding that shows that receptor endocytosis can reduce the signaling output of many plasma membrane receptors by physically reducing the concentration of the receptors at the cell surface (65). Nevertheless, HC4 also induced cell death in HNSCC cells that do not depend on HER2 suggesting PERK may enable adaptation to stress under different oncogenic pathways.
Discovering a target and drug that can eradicate dormant DCCs in addition to cells that undergo short- or long-term quiescence in metastatic lesions is highly significant because dormant DCCs are known to evade anti-proliferative therapies via active and passive mechanisms (66-72). Our work opens the door to the use of anti-dormant DCC survival therapies as a new way to target metastatic disease. This would allow targeting the full phenotypic heterogeneity of disseminated disease that may include proliferative, slow-cycling, and dormant DCCs (70). The eradication of DCCs in the bone marrow, where these cells also commonly reside in a dormant state (33,47,59,73,74), further strengthens the notion of PERK inhibition as an anti-dormant DCC therapy that may be used in the adjuvant setting to eliminate dormant minimal residual disease (70).
Supplementary Material
Translational Relevance.
Our study focuses on eradicating dormant disseminated cancer cells (DCCs) that persist in M0 or M1 patients and fuel relapse. We identified a new clinical grade inhibitor (HC-5404 or HC4 in short) of the endoplasmic reticulum kinase PERK, currently tested in a clinical trial (NCT04834778). We show that quiescent/dormant solitary DCCs or growth arrested cancer cells in micro-metastatic lesions upregulate theintegrated stress response . Treatment with HC4 eradicates only the dormant cancer cells, which in turn results in an impressive blockade of metastatic disease. We also report an almost complete eradication of metastatic disease by combining the standard of care CDK4/6 inhibitor Abemaciclib (to induce quiescence), followed by HC4 treatment that selectively kills quiescent DCCs. The future practice of cancer medicine may combine standard of care anti-proliferative therapies with HC4, to target any dormant DCCs that pre-date the treatment or “persister” cancer cells that emerge in response to conventional therapies.
ACKNOWLEDGEMENTS:
We thank the Aguirre-Ghiso and HiberCell teams for useful discussions. We thank Brian Lee for help with Clustergrammer use. Grant Support: Eli Lilly to J.A.A-G, LIFA Fellowship (Eli Lilly) to V.C., Ramon Areces Postdoctoral fellowship to A.A-A, NIH/NCI (CA109182, CA196521, CA216248), to J.A.A-G, HiberCell and DoD-BCRP Breakthrough Award (BC132674) to J.A.A-G. JA.A-G is a Samuel Waxman Cancer Research Foundation Investigator.
Footnotes
Conflict of Interests
JAAG is a scientific co-founder of, scientific advisory board member and equity owner in HiberCell and receives financial compensation as a consultant for HiberCell, a Mount Sinai spin-off company focused on therapeutics that prevent or delay cancer recurrence. VC, EFF, SF, DL, AN, MES and MM are HiberCell employees. The remaining authors declare no conflicts of interest.
REFERENCES
- 1.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science 2011;334(6059):1081–6 doi 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 2.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 2003;11(3):619–33 doi 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 3.Chevet E, Hetz C, Samali A. Endoplasmic reticulum stress-activated cell reprogramming in oncogenesis. Cancer Discov 2015;5(6):586–97 doi 10.1158/2159-8290.CD-14-1490. [DOI] [PubMed] [Google Scholar]
- 4.Blais JD, Filipenko V, Bi M, Harding HP, Ron D, Koumenis C, et al. Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol Cell Biol 2004;24(17):7469–82 doi 10.1128/MCB.24.17.7469-7482.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tameire F, Verginadis II, Koumenis C. Cell intrinsic and extrinsic activators of the unfolded protein response in cancer: Mechanisms and targets for therapy. Semin Cancer Biol 2015;33:3–15 doi 10.1016/j.semcancer.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hart LS, Cunningham JT, Datta T, Dey S, Tameire F, Lehman SL, et al. ER stress-mediated autophagy promotes Myc-dependent transformation and tumor growth. J Clin Invest 2012;122(12):4621–34 doi 10.1172/JCI62973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ozcan U, Ozcan L, Yilmaz E, Duvel K, Sahin M, Manning BD, et al. Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Mol Cell 2008;29(5):541–51 doi 10.1016/j.molcel.2007.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest 2010;120(1):127–41 doi 10.1172/JCI40027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. Embo J 2005;24(19):3470–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romero-Ramirez L, Cao H, Regalado MP, Kambham N, Siemann D, Kim JJ, et al. X box-binding protein 1 regulates angiogenesis in human pancreatic adenocarcinomas. Transl Oncol 2009;2(1):31–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X, Iliopoulos D, Zhang Q, Tang Q, Greenblatt MB, Hatziapostolou M, et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1alpha pathway. Nature 2014;508(7494):103–7 doi 10.1038/nature13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, et al. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. Embo J;29(12):2082–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schewe DM, Aguirre-Ghiso JA. ATF6alpha-Rheb-mTOR signaling promotes survival of dormant tumor cells in vivo. Proc Natl Acad Sci U S A 2008;105(30):10519–24 doi 10.1073/pnas.0800939105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Avivar-Valderas A, Salas E, Bobrovnikova-Marjon E, Diehl JA, Nagi C, Debnath J, et al. PERK integrates autophagy and oxidative stress responses to promote survival during extracellular matrix detachment. Mol Cell Biol 2011;31(17):3616–29 doi 10.1128/MCB.05164-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Avivar-Valderas A, Bobrovnikova-Marjon E, Alan Diehl J, Bardeesy N, Debnath J, Aguirre-Ghiso JA. Regulation of autophagy during ECM detachment is linked to a selective inhibition of mTORC1 by PERK. Oncogene 2013; 32, 4932–4940 doi.org/ 10.1038/onc.2012.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espina V, Mariani BD, Gallagher RI, Tran K, Banks S, Wiedemann J, et al. Malignant precursor cells pre-exist in human breast DCIS and require autophagy for survival. PloS one 2010;5(4):e10240 doi 10.1371/journal.pone.0010240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bobrovnikova-Marjon E, Grigoriadou C, Pytel D, Zhang F, Ye J, Koumenis C, et al. PERK promotes cancer cell proliferation and tumor growth by limiting oxidative DNA damage. Oncogene 2010;29(27):3881–95 doi 10.1038/onc.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bobrovnikova-Marjon E, Hatzivassiliou G, Grigoriadou C, Romero M, Cavener DR, Thompson CB, et al. PERK-dependent regulation of lipogenesis during mouse mammary gland development and adipocyte differentiation. Proc Natl Acad Sci U S A 2008;105(42):16314–9 doi 10.1073/pnas.0808517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh N, Joshi R, Komurov K. HER2-mTOR signaling-driven breast cancer cells require ER-associated degradation to survive. Sci Signal 2015;8(378):ra52 doi 10.1126/scisignal.aaa6922. [DOI] [PubMed] [Google Scholar]
- 20.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery 2012;2(5):401–4 doi 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng YX, Jin DX, Sokol ES, Reinhardt F, Miller DH, Gupta PB. Cancer-specific PERK signaling drives invasion and metastasis through CREB3L1. Nat Commun 2017;8(1):1079 doi 10.1038/s41467-017-01052-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ranganathan AC, Ojha S, Kourtidis A, Conklin DS, Aguirre-Ghiso JA. Dual function of pancreatic endoplasmic reticulum kinase in tumor cell growth arrest and survival. Cancer Res 2008;68(9):3260–8 doi 10.1158/0008-5472.CAN-07-6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ranganathan AC, Zhang L, Adam AP, Aguirre-Ghiso JA. Functional coupling of p38-induced up-regulation of BiP and activation of RNA-dependent protein kinase-like endoplasmic reticulum kinase to drug resistance of dormant carcinoma cells. Cancer Res 2006;66(3):1702–11 doi 10.1158/0008-5472.CAN-05-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pommier A, Anaparthy N, Memos N, Kelley ZL, Gouronnec A, Yan R, et al. Unresolved endoplasmic reticulum stress engenders immune-resistant, latent pancreatic cancer metastases. Science 2018;360(6394) doi 10.1126/science.aao4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harper KL, Sosa MS, Entenberg D, Hosseini H, Cheung JF, Nobre R, et al. Mechanism of early dissemination and metastasis in Her2(+) mammary cancer. Nature 2016;540(7634):588–92 doi 10.1038/nature20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nobre AR, Dalla E, Yang J, Huang X, Wullkopf L, Risson E, et al. ZFP281 drives a mesenchymal-like dormancy program in early disseminated breast cancer cells that prevents metastatic outgrowth in the lung. Nat Cancer 2022:3(10):1165–1180. doi: 10.1038/s43018-022-00424-8 [DOI] [PubMed] [Google Scholar]
- 27.Calvo V, Surguladze D, Li AH, Surman MD, Malibhatla S, Bandaru M, et al. Discovery of 2-amino-3-amido-5-aryl-pyridines as highly potent, orally bioavailable, and efficacious PERK kinase inhibitors. Bioorg Med Chem Lett 2021;43:128058 doi 10.1016/j.bmcl.2021.128058. [DOI] [PubMed] [Google Scholar]
- 28.Aguirre Ghiso JA, Kovalski K, Ossowski L. Tumor dormancy induced by downregulation of urokinase receptor in human carcinoma involves integrin and MAPK signaling. J Cell Biol 1999;147(1):89–104 doi 10.1083/jcb.147.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tenkerian C, Krishnamoorthy J, Mounir Z, Kazimierczak U, Khoutorsky A, Staschke KA, et al. mTORC2 Balances AKT Activation and eIF2alpha Serine 51 Phosphorylation to Promote Survival under Stress. Mol Cancer Res 2015;13(10):1377–88 doi 10.1158/1541-7786.MCR-15-0184-T. [DOI] [PubMed] [Google Scholar]
- 30.Cihil KM, Swiatecka-Urban A. The cell-based L-glutathione protection assays to study endocytosis and recycling of plasma membrane proteins. J Vis Exp 2013(82):e50867 doi 10.3791/50867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandez NF, Gundersen GW, Rahman A, Grimes ML, Rikova K, Hornbeck P, et al. Clustergrammer, a web-based heatmap visualization and analysis tool for high-dimensional biological data. Sci Data 2017;4:170151 doi 10.1038/sdata.2017.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gyorffy B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput Struct Biotechnol J 2021;19:4101–9 doi 10.1016/j.csbj.2021.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Husemann Y, Geigl JB, Schubert F, Musiani P, Meyer M, Burghart E, et al. Systemic spread is an early step in breast cancer. Cancer Cell 2008;13(1):58–68 doi 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 34.Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci U S A 1992;89(22):10578–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novoa I, Zhang Y, Zeng H, Jungreis R, Harding HP, Ron D. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J 2003;22(5):1180–7 doi 10.1093/emboj/cdg112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.B'Chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, et al. The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res 2013;41(16):7683–99 doi 10.1093/nar/gkt563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim RS, Avivar-Valderas A, Estrada Y, Bragado P, Sosa MS, Aguirre-Ghiso JA, et al. Dormancy signatures and metastasis in estrogen receptor positive and negative breast cancer. PLoS One 2012;7(4):e35569 doi 10.1371/journal.pone.0035569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Axten JM, Medina JR, Feng Y, Shu A, Romeril SP, Grant SW, et al. Discovery of 7-methyl-5-(1-[3-(trifluoromethyl)phenyl]acetyl-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a potent and selective first-in-class inhibitor of protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK). J Med Chem 2012;55(16):7193–207 doi 10.1021/jm300713s. [DOI] [PubMed] [Google Scholar]
- 39.Sequeira SJ, Ranganathan AC, Adam AP, Iglesias BV, Farias EF, Aguirre-Ghiso JA. Inhibition of proliferation by PERK regulates mammary acinar morphogenesis and tumor formation. PLoS One 2007;2(7):e615 doi 10.1371/journal.pone.0000615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu Q, Zhao B, Gui J, Katlinski KV, Brice A, Gao Y, et al. Type I interferons mediate pancreatic toxicities of PERK inhibition. Proc Natl Acad Sci U S A 2015;112(50):15420–5 doi 10.1073/pnas.1516362112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Linde N, Casanova-Acebes M, Sosa MS, Mortha A, Rahman A, Farias E, et al. Macrophages orchestrate breast cancer early dissemination and metastasis. Nat Commun 2018;9(1):21 doi 10.1038/s41467-017-02481-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hosseini H, Obradovic MMS, Hoffmann M, Harper KL, Sosa MS, Werner-Klein M, et al. Early dissemination seeds metastasis in breast cancer. Nature 2016;540(7634):552–8 doi 10.1038/nature20785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun D, Singh DK, Carcamo S, Filipescu D, Khalil B, Huang X, et al. MacroH2A impedes metastatic growth by enforcing a discrete dormancy program in disseminated cancer cells. Sci Adv 2022;8(48):eabo0876 doi 10.1126/sciadv.abo0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khalil BD, Sanchez R, Rahman T, Rodriguez-Tirado C, Moritsch S, Martinez AR, et al. An NR2F1-specific agonist suppresses metastasis by inducing cancer cell dormancy. J Exp Med 2022;219(1) doi 10.1084/jem.20210836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fluegen G, Avivar-Valderas A, Wang Y, Padgen MR, Williams JK, Nobre AR, et al. Phenotypic heterogeneity of disseminated tumour cells is preset by primary tumour hypoxic microenvironments. Nat Cell Biol 2017;19(2):120–32 doi 10.1038/ncb3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sosa MS, Parikh F, Maia AG, Estrada Y, Bosch A, Bragado P, et al. NR2F1 controls tumour cell dormancy via SOX9- and RARbeta-driven quiescence programmes. Nat Commun 2015;6:6170 doi 10.1038/ncomms7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bragado P, Estrada Y, Parikh F, Krause S, Capobianco C, Farina HG, et al. TGF-beta2 dictates disseminated tumour cell fate in target organs through TGF-beta-RIII and p38alpha/beta signalling. Nat Cell Biol 2013;15(11):1351–61 doi 10.1038/ncb2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Viloria-Petit AM, David L, Jia JY, Erdemir T, Bane AL, Pinnaduwage D, et al. A role for the TGFbeta-Par6 polarity pathway in breast cancer progression. Proc Natl Acad Sci U S A 2009;106(33):14028–33 doi 10.1073/pnas.0906796106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aguirre-Ghiso JA, Ossowski L, Rosenbaum SK. Green fluorescent protein tagging of extracellular signal-regulated kinase and p38 pathways reveals novel dynamics of pathway activation during primary and metastatic growth. Cancer Res 2004;64(20):7336–45 doi 10.1158/0008-5472.CAN-04-0113. [DOI] [PubMed] [Google Scholar]
- 50.Muller WJ, Sinn E, Pattengale PK, Wallace R, Leder P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell 1988;54(1):105–15. [DOI] [PubMed] [Google Scholar]
- 51.Lu Y, Bertran S, Samuels TA, Mira-y-Lopez R, Farias EF. Mechanism of inhibition of MMTV-neu and MMTV-wnt1 induced mammary oncogenesis by RARalpha agonist AM580. Oncogene 2010;29(25):3665–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DiGiovanna MP, Lerman MA, Coffey RJ, Muller WJ, Cardiff RD, Stern DF. Active signaling by Neu in transgenic mice. Oncogene 1998;17(14):1877–84 doi 10.1038/sj.onc.1202091. [DOI] [PubMed] [Google Scholar]
- 53.Stokes ME, Surman MD, Calvo V, Surguladze D, Li AH, Gasparek J, et al. Optimization of a Novel Mandelamide-Derived Pyrrolopyrimidine Series of PERK Inhibitors. Pharmaceutics 2022;14(10) doi 10.3390/pharmaceutics14102233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hommelgaard AM, Lerdrup M, van Deurs B. Association with membrane protrusions makes ErbB2 an internalization-resistant receptor. Mol Biol Cell 2004;15(4):1557–67 doi 10.1091/mbc.e03-08-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bertelsen V, Stang E. The Mysterious Ways of ErbB2/HER2 Trafficking. Membranes (Basel) 2014;4(3):424–46 doi 10.3390/membranes4030424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mahameed M, Boukeileh S, Obiedat A, Darawshi O, Dipta P, Rimon A, et al. Pharmacological induction of selective endoplasmic reticulum retention as a strategy for cancer therapy. Nat Commun 2020;11(1):1304 doi 10.1038/s41467-020-15067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang SS, Koh HA, Konish Y, Bullock LD, Huang JS. Differential processing and turnover of the oncogenically activated neu/erb B2 gene product and its normal cellular counterpart. J Biol Chem 1990;265(6):3340–6. [PubMed] [Google Scholar]
- 58.Sosa MS, Bragado P, Aguirre-Ghiso JA. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat Rev Cancer 2014;14(9):611–22 doi 10.1038/nrc3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chéry L, Lam H-M, Coleman I, Lakely B, Coleman R, Larson S, et al. Characterization of single disseminated prostate cancer cells reveals tumor cell heterogeneity and identifies dormancy associated pathways. Oncotarget 2014; 30;5(20):9939–51. doi: 10.18632/oncotarget.2480.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adomako A, Calvo V, Biran N, Osman K, Chari A, Paton JC, et al. Identification of markers that functionally define a quiescent multiple myeloma cell sub-population surviving bortezomib treatment. BMC Cancer 2015;15:444 doi 10.1186/s12885-015-1460-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schewe DM, Aguirre-Ghiso JA. Inhibition of eIF2alpha dephosphorylation maximizes bortezomib efficiency and eliminates quiescent multiple myeloma cells surviving proteasome inhibitor therapy. Cancer Res 2009;69(4):1545–52 doi 10.1158/0008-5472.CAN-08-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brewer JW, Hendershot LM, Sherr CJ, Diehl JA. Mammalian unfolded protein response inhibits cyclin D1 translation and cell-cycle progression. Proc Natl Acad Sci U S A 1999;96(15):8505–10 doi 10.1073/pnas.96.15.8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Borgen E, Rypdal MC, Sosa MS, Renolen A, Schlichting E, Lonning PE, et al. NR2F1 stratifies dormant disseminated tumor cells in breast cancer patients. Breast Cancer Res 2018;20(1):120 doi 10.1186/s13058-018-1049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Di Martino JS, Nobre AR, Mondal C, Taha I, Farias EF, Fertig EJ, et al. A tumor-derived type III collagen-rich ECM niche regulates tumor cell dormancy. Nat Cancer 2022;3(1):90–107 doi 10.1038/s43018-021-00291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res 2009;315(4):683–96 doi 10.1016/j.yexcr.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 66.Aguirre-Ghiso JA, Bragado P, Sosa MS. Metastasis awakening: targeting dormant cancer. Nat Med 2013;19(3):276–7 doi 10.1038/nm.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Naumov GN, Townson JL, MacDonald IC, Wilson SM, Bramwell VH, Groom AC, et al. Ineffectiveness of doxorubicin treatment on solitary dormant mammary carcinoma cells or late-developing metastases. Breast Cancer Res Treat 2003;82(3):199–206. [DOI] [PubMed] [Google Scholar]
- 68.Oshimori N, Oristian D, Fuchs E. TGF-beta promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell 2015;160(5):963–76 doi 10.1016/j.cell.2015.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carlson P, Dasgupta A, Grzelak CA, Kim J, Barrett A, Coleman IM, et al. Targeting the perivascular niche sensitizes disseminated tumour cells to chemotherapy. Nat Cell Biol 2019;21(2):238–50 doi 10.1038/s41556-018-0267-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aguirre-Ghiso JA. Translating the Science of Cancer Dormancy to the Clinic. Cancer Res 2021;81(18):4673–5 doi 10.1158/0008-5472.CAN-21-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fox DB, Garcia NMG, McKinney BJ, Lupo R, Noteware LC, Newcomb R, et al. NRF2 activation promotes the recurrence of dormant tumour cells through regulation of redox and nucleotide metabolism. Nat Metab 2020;2(4):318–34 doi 10.1038/s42255-020-0191-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Romani P, Nirchio N, Arboit M, Barbieri V, Tosi A, Michielin F, et al. Mitochondrial fission links ECM mechanotransduction to metabolic redox homeostasis and metastatic chemotherapy resistance. Nat Cell Biol 2022;24(2):168–80 doi 10.1038/s41556-022-00843-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ghajar CM, Peinado H, Mori H, Matei IR, Evason KJ, Brazier H, et al. The perivascular niche regulates breast tumour dormancy. Nat Cell Biol 2013;15(7):807–17 doi 10.1038/ncb2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nobre AR, Risson E, Singh DK, Di Martino JS, Cheung JF, Wang J, et al. Bone marrow NG2(+)/Nestin(+) mesenchymal stem cells drive DTC dormancy via TGFbeta2. Nat Cancer 2021;2(3):327–39 doi 10.1038/s43018-021-00179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Previously published RNAseq of MMTV-Neu EL and PT spheres is available under the accession code GSE165431. All data needed to evaluate the conclusions are included in the paper and Supplementary Materials. Other data supporting the findings are available from the corresponding authors on reasonable request.