Abstract
The mating-type switching of the fission yeast, Schizosaccharomyces pombe, is highly regulated. Two consecutive asymmetric divisions are required to produce one mating-type switched cell among the four progeny. Using DNA density-gradient centrifugation we demonstrate that one-fourth of the mat1 DNA is not replicated by the conventional semi-conservative mode, but instead both DNA strands are synthesized de novo. Our data are consistent with a gene conversion event, initiated by a site- and strand-specific DNA break (SSB). We further demonstrate that the virgin switched mat1-containing chromatid no longer contained the nick, while it is reintroduced during the lagging strand synthesis of the mat1 locus on the sister chromatid. This finding establishes at the molecular level a firm experimental link between the phenotype and genotype in the process of asymmetric mating-type switching during mitotic divisions.
INTRODUCTION
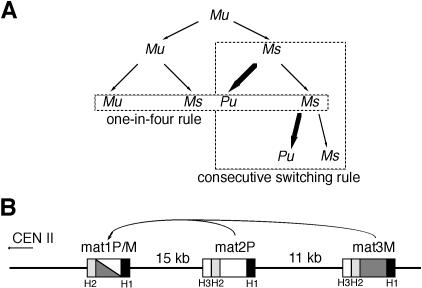
Haploid cells of the fission yeast, Schizosaccharomyces pombe, exhibit a homothallic life cycle, in which the mating-type of the cell mitotically alternates (Figure 1A) producing a population of P (+) and M (–) cells (Leupold, 1950; Egel, 1977). The mating-type region contains three loci in the right arm of chromosome II: the expressed mat1 locus and the two silent mat2P and mat3M loci (Egel and Gutz, 1981; Beach, 1983; Beach and Klar, 1984; Kelly et al., 1988). In a process similar to gene conversion, the two silent mat2P and mat3M loci can donate their genetic information to mat1 (Figure 1B). The efficiency of this process leads to a clonal haploid cell population containing roughly the same proportion of P and M cell types.
Fig. 1. Mating-type pedigree and loci of fission yeast. (A) P and M indicate the mating-type of the cell, the suffix u or s represents the unswitchable and switchable potential of the cell, respectively. For simplicity, only the M lineage is shown. The one-in-four and the consecutive switching rules are indicated. (B) The mating-type region on chromosome II. The mat1 locus contains either the P (white box) or the M (grey box) alleles, and mat2P and mat3M are donors of genetic information. The H1 and H2 homology sequences are common to all cassettes whereas the H3 sequence is common to only the silent mat2P and mat3M loci (Kelly et al., 1988).
During vegetative growth, the mating-type locus is not expressed. Consequently the cells are genotypically different at the mating-type locus (either mat1P or mat1M) but phenotypically identical (formally sterile). When cells experience starvation, they arrest in the G1 phase and the mating-type alleles (P or M) present in mat1 are transcriptionally induced (Kelly et al., 1988), allowing conjugation between cells of opposite mating-type followed by sporulation. The pattern of mating-type switching in the mitotic lineage has been determined in conditions approaching nitrogen starvation. These studies show (Figure 1A) that two consecutive asymmetric divisions are required, generating one switched and three unswitched cells (Miyata and Miyata, 1981; Egel, 1984). Furthermore, the sister of the switched cell is competent for switching during the next division cycle, forming a recurrent chain of asymmetric switching (Egel and Eie, 1987; Klar, 1987, 1990).
It was proposed that this switching pattern results from an imprinting event, marking one of the sister chromatids in a strand-specific manner at the mat1 locus (Klar, 1987). Recent molecular experiments revealed the presence of a site- and strand DNA-specific break (Arcangioli, 1998) or DNA modification (Dalgaard and Klar, 1999) at the mat1 locus, which prepares the switching event during the next round of DNA replication. The involvement of the orientation of the replication fork (Arcangioli, 1998; Dalgaard and Klar, 1999), together with the presence of a transitory gene conversion intermediate during S-phase, strongly supports the notion that DNA replication and recombination at mat1 are coupled (Arcangioli and de Lahondès, 2000). This process restricts mating-type switching to only one of the two sister chromatids during mat1 DNA replication.
In an attempt to correlate the phenotypic switching pattern with the fate of the mat1 DNA strands during growth, we used density-gradient centrifugation (Meselson and Stahl, 1958). We compared the density distribution of the mat1 locus from switchable (h90) and non-switchable strains. We also determined the density distribution of mat1 DNA during mitotic divisions and examined the distribution of the broken mat1 DNA strand. The results are interpreted in molecular terms that reveal the inheritance of mat1 DNA strands during asymmetric mating-type switching.
RESULTS AND DISCUSSION
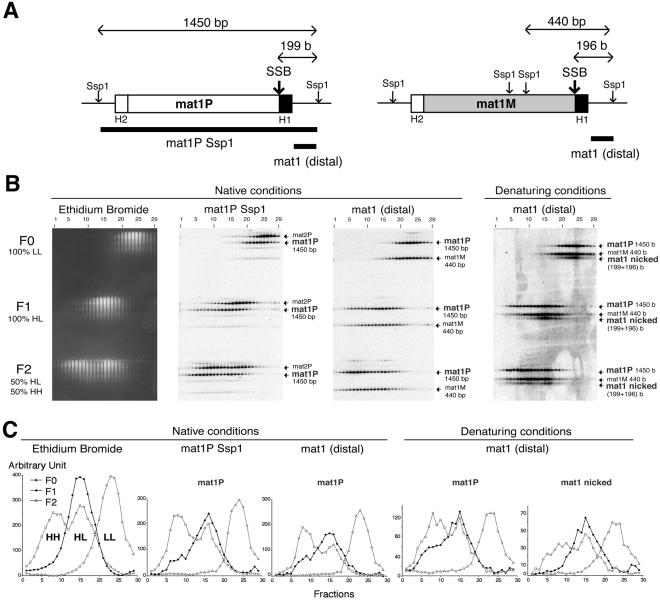
In the current study, we followed the inheritance of mat1 DNA strands during mitotic divisions by performing Meselson–Stahl type experiments (Meselson and Stahl, 1958). A homothallic wild-type cell population grown in normal (light) minimum medium was enriched in small G2 cells (see Methods). The partially synchronized cells were allowed to resume growth in an isotopically dense medium. Samples of cells were taken at time 0 and after one or two generations (F0, F1, F2). Figure 2B (left panel) shows the density distribution of the SspI-digested genomic DNA in the CsCl-eluted fractions after electrophoresis in an agarose gel. DNA quantification (Figure 2C, left panel) is typical of the semi-conservative genomic DNA replication mode.
Fig. 2. Density distribution of the mat1 locus. (A) The location of the SspI restriction sites at the mat1 locus and their respective sizes are indicated. The position of the site- and strand-specific break (SSB) is shown by an arrow. 199 and 196 bases are the sizes of the mat1P- and mat1M-distal upper DNA strand, following SspI digestion. The H1 and H2 sequences are indicated and the two DNA probes used are shown at the bottom. (B) Distribution of the mat1 locus analysed by DNA–DNA hybridization. DNA samples from the CsCl-eluted fractions were electrophoresed in native or denaturing conditions, as indicated. F0, F1, F2, LL, HL and HH indicate the number of generations in heavy media and the double-stranded DNA density, respectively. The probes used (A) are double stranded and are indicated at the top of the autoradiogram. Note that the same probe, mat1 (distal), is used for the Southern blots performed in native and denaturing conditions (right panel). The names and sizes of the DNA fragments are indicated by small arrows. The weak hybridization signals are due to the H1 and H2 sequences present at the three loci. (C) Quantification of the blots is shown above using the same nomenclature. The corresponding restriction DNA fragments are indicated in bold. The density distribution of the mat1M DNA fragments in denaturing conditions behaves similarly to mat1P and is not shown.
DNA from the agarose gel was transferred to a filter, and the density distribution of the mating-type loci was determined by quantitative hybridization. The mat1P SspI probe (Figure 2A) mostly detects the mat1P and mat2P loci (Figure 2B, second panel). The intensity of the mat2P signal is about twice the intensity of the mat1P since only 50% of mat1 locus contains the P allele, while the other half contains the M allele. The density distribution of the mat2P DNA fragment in the three samples resembles the distribution of the genomic DNA (Figure 2B). However, the density distribution of the mat1P DNA fragment diverged from the mat2P and the total genomic DNA in several respects. In F0, mat1P is slightly shifted compared with mat2P (Figure 2B). A similar difference in density was observed with SspI DNA fragments purified from Escherichia coli (data not shown) and reflects the intrinsic density imposed by the nucleotide sequence composition flanking the mating-type cassettes. In F1, the mat1P DNA fragment is not only confined to the HL density fractions, but also spreads into the HH density fractions (Figure 2B, second panel in fractions 5–10). This demonstrates that both DNA strands have been synthesized de novo, in one generation, in a fraction of the cells. In F2, mat1P is more abundant in the HH than in the HL density (Figure 2B). A very similar distribution pattern is observed when the membrane is hybridized with the mat1 (distal) probe (Figure 2A), which detects both mat1P and mat1M (Figure 2B and C, third panels).
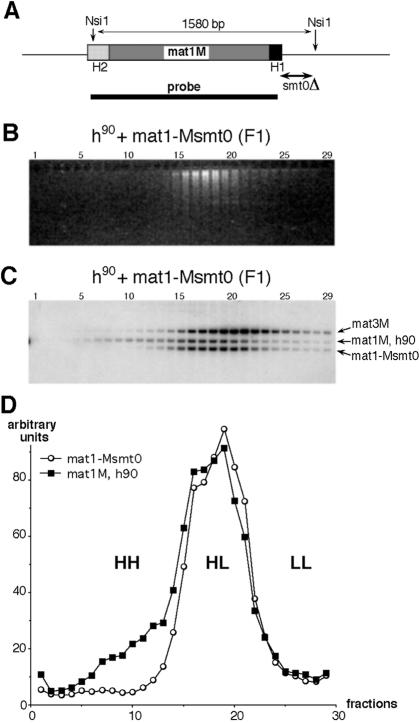
In order to confirm the intriguing density distribution of the mating-type locus we directly compared the mat1 density distribution in F1 of the wild-type h90 and the non-switchable, mat1-Msmt0 mutant, strains. The mat1-Msmt0 mutant contains a deletion of 262 bp (Figure 3A) next to the mat1M locus, abolishing mating-type switching (Arcangioli and Klar, 1991; Styrkarsdottir et al., 1993). Wild-type and mat1-Msmt0 mutant mixed culture grown in light media were shifted to heavy media [glucose (13C) and 15NH4Cl] for one generation. The genomic DNA from both strains was purified, digested with the NsiI restriction enzyme and submitted to CsCl gradient centrifugation. Genomic DNA from the CsCl-eluted fractions was separated on an agarose gel (Figure 3B) and the density distribution of the mat1 locus was analysed by DNA–DNA hybridization. Three DNA fragments (Figure 3C) are observed after hybridization with the mat1M probe (Figure 3A). The relative intensity of the signals corresponds to the number of loci. The mat3M locus is present in both strains. The mat1M signal from the wild-type strain is roughly half the level of the mat1-Msmt0 signal, since half of the wild-type mat1 locus contains the P allele and the other half contains the M allele. The density distribution of the mat1M locus clearly spreads into the HH density fractions (Figure 3C, fractions 6–13), which contrasts with the mat1-Msmt0 and mat3M loci. The quantification indicates that 20–25% (Figure 3D) of the mat1M locus have both DNA strands newly replicated in one generation. Finally, the observation that the copy transposition products are separated from the bulk of genomic DNA in F1 by density centrifugation indicates that this approach can be used to isolate known or unknown recombinant sequences in a wide range of biological systems.
Fig. 3. Density distribution of the mat1 locus from homothallic (h90) and heterothallic strains in F1. (A) Schematic representation of the mat1M locus on chromosome II. The NsiI restriction sites flanking mat1M are indicated. The position of the deletions in the mat1-Msmt0 mutant strain is shown as well as the probe used. (B) The co-culture of wild-type (h90) and mat1-Msmt0 strains (as indicated) was shifted from light to heavy medium and cells were collected after one generation (F1). DNA samples from the CsCl-eluted fractions were separated by electrophoresis on agarose gel and stained with ethidium bromide. (C) Density distributions of the mating-type loci. DNA from the agarose gel was analysed by DNA–DNA hybridization. The names of the DNA fragments revealed by the mat1M probe (see above) are indicated. (D) Quantification of the hybridization signals. The mat1M signal from the wild-type strain (h90) was multiplied by 1.8 for comparison with the mat1-Msmt0 signal. HH, HL and LL indicate the density of the DNA fragments in the CsCl gradient.
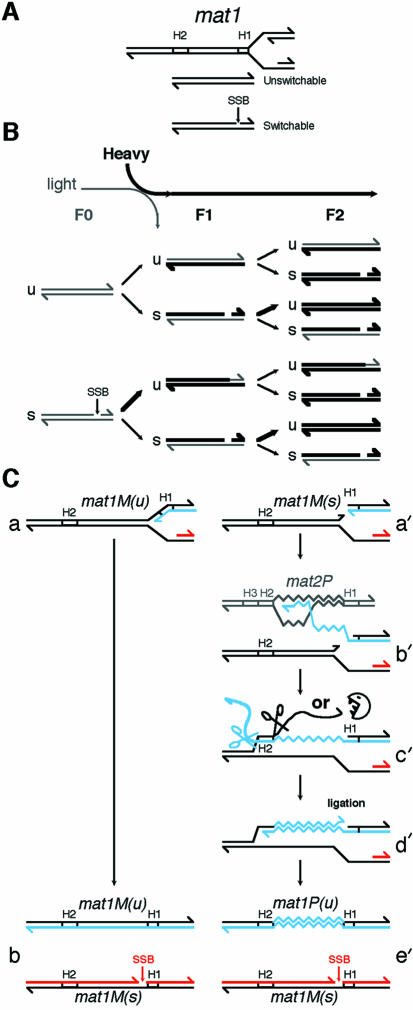
Quantification of three independent experiments shows that 20–25% of mat1 is present in the HH density in F1, and 60–65% in F2. It is assumed that during mitotic growth, half of the cell population is competent for mating-type switching (Figure 4, the switchable chromatids). Since this gene conversion is restricted to only one of the two sister chromatids, it follows that 1/4 (25%) of the mat1 locus should undergo gene conversion (Figure 4B, F0 to F1). This will replace the two mat1 parental DNA strands by copying the opposite mating-type allele during DNA replication (Arcangioli and de Lahondès, 2000). A simple extrapolation of this rule to the next generation produces an HH density proportion of 5/8 (62.5%) in F2 (Figure 4B). This result, together with the mating-type switching pedigree pattern, strongly supports the model shown in Figure 4B, which is an extension of the strand segregation model (Klar, 1990). Furthermore, this process preserves the nucleotide integrity of the donor of genetic information, mat2P (Figure 2B) and mat3M (Figure 3C), since their DNA strands followed the same density distribution as the genomic DNA. Consistently, the mat1 DNA fragment reveals an informative shoulder at the HH side of the density profile after one generation, with no deviation in the LL side of the pattern (see Figures 2 and 3). This process, proposed more than 60 years ago by Belling (1933), is known as the copy-choice recombination model (for review see Kogoma, 1997; Haber, 1999).
Fig. 4. Molecular pedigree of the mat1 locus during mitotic divisions. (A) The top of the figure shows the two DNA strands (upper and lower) of the mat1 locus and the direction of the replication fork. The intact and nicked (SSB) mat1 loci are called switchable and unswitchable, respectively. The position of the nick is indicated on the upper DNA strand at the junction of the mat1 specific allele (P or M) and the H1 homology box. (B) In a growing population, one half of the chromatids contain an intact mat1 locus that is not competent for switching (u for unswitchable). The other half contains a nick, restricting mating-type switching (s for switchable) to only one of the two sister chromatids during the next round of DNA replication. Since the P lineage is the mirror image of the M lineage, only the suffixes u and s are emphasized for simplicity. The density of the medium and the generations are indicated. The thickness of the DNA strands indicates the density. (See text for details). (C) The replication fork progresses from the right side of mat1M(u) (a). Following replication, the newly replicated lagging strand is broken (SSB), producing an unswitchable mat1M(u) and a switchable mat1M(s) loci (b). The replication fork progresses from the right side of mat1M(s) and is blocked by the nick (a′). Strand invasion occurs at the H1 homology box of the opposite mating-type locus (mat2P in this example) and DNA synthesis is initiated forming a migrating D-loop (Ferguson and Holloman, 1996) or a small replication bubble that coordinates leading and lagging strand synthesis (Holmes and Haber, 1999) (b′). Following strand annealing in the H2 homology region (c′) both single-stranded DNAs are digested or clipped off, presumably by swi9p/swi10p (Rad1/Rad10 in S. cerevisiae) near the H2 homology sequences, providing 3′ ends for priming leading strand and filling-in DNA synthesis and ligation (d′). Ligation and the single strand break are probably coupled. Finally a switched ligated chomatid [mat1P(u)] and an unswitched chromatid [mat1M(s)] are formed following replication in G2 (e′). The leading strand is coloured in blue and the lagging strand in red.
The density transfer approach offers a unique opportunity to address the question of the fate of the DNA modification at mat1 during growth. Genomic DNA in the CsCl-eluted fractions was denatured with formamide and formaldehyde, run on a formaldehyde gel, transferred to a filter and hybridized with the mat1 (distal) probe (Figure 2A). The denaturing conditions allow for the observation of three DNA fragments, instead of the two observed under native conditions (Figure 2B, right panel). The fast migrating additional band corresponds to the denatured mat1-distal upper strands of 199 and 196 bases (Figure 2A), melted from the complementary mat1P and mat1M SspI DNA fragments, respectively (Nielsen and Egel, 1989; Arcangioli, 1998). This result is more compatible with the presence of a nick at mat1 (Arcangioli, 1998) rather than an alkali-labile DNA modification (Dalgaard and Klar, 1999). Such a DNA strand break will stall leading strand replication restricting mating-type switching to only one chromatid (Arcangioli, 1998; Dalgaard and Klar, 1999; Arcangioli and de Lahondès, 2000; Rothstein et al., 2000).
In denaturing conditions, the additional fast migrating band is present in the HL fractions and absent in the HH fractions of the density gradient in F1 (Figure 2B and C, right panels). If a nick were also present in the recently switched mat1 (P or M) the additional band will also be observed in the HH fractions. This result demonstrates that the recently switched mat1 locus, made of two neosynthesized DNA strands, does not contain the single strand DNA break. This virgin allele is not competent for switching during its next division. On the other hand, its sister inherits the competence for switching and hence the nick. The direction of the replication fork (Arcangioli, 1998; Dalgaard and Klar, 1999), together with the nicked mat1 locus in the HL density in F1, indicates that the switchable cells inherit the intact parental lower DNA strand. It follows that the old nicked strand must be removed and synthesized de novo (Figure 4C, step c′) ruling out the possible conservation of the old nicked strand. This newly synthesized DNA strand must be cut again or remains unligated to the adjacent DNA strand. This probably occurs during lagging strand DNA synthesis (Singh and Klar, 1993) (Figure 4C) but does not exclude the participation of another replication fork arriving from the other side. The switching and cutting processes are tightly coupled in time since the steady state level of the nick appeared to be constant during S-phase (Arcangioli, 1998). However, the mechanism responsible for the nick is not coupled to switching per se since the same process occurs during replication of the unswitchable chromatid (Figure 4C, b and e’ steps) and presumably requires the cis-acting sequences next to mat1 (Arcangioli and Klar, 1991; Klar et al., 1991).
We propose that this process transfers both newly synthesized DNA strands to the mat1 locus during mating-type switching. This type of gene conversion process occurs during S-phase, is initiated at the nick in the H1 sequences and is resolved in the H2 sequences without crossing over (Arcangioli and de Lahondès, 2000). This replication/recombination coupled process is reminiscent of recombination-dependent DNA replication (RDR) in E. coli (Asai et al., 1994; Kogoma, 1997) and synthesis-dependent strand annealing (SDSA) in Drosophila, Ustilago and the yeast Saccharomyces cerevisiae (Nassif et al., 1994; Ferguson and Holloman, 1996; Haber, 1999). The present work establishes for the first time a firm experimental link between phenotype and genotype in the process of asymmetric mating-type switching in this organism. Such a link has previously been inferred indirectly according to criteria, based on mutant behaviour and cellular pedigrees, but direct positive evidence has been unavailable.
METHODS
Yeast growth conditions. Wild-type homothallic or mat1-Msmt0 strains were grown at 32°C in 1 l of standard minimal medium (Moreno et al., 1991) containing 1% glucose and 0.2% NH4Cl. Cells were harvested by centrifugation (5 min at 5000 r.p.m.), resuspended in minimal medium, loaded onto a lactose gradient (7–30%) and centrifuged for 8 min at 1000 r.p.m. in a Jouan CR412 centrifuge (Barbet and Carr, 1993). The smallest cells at the top of the gradients were collected (5–10% of the starting material) and pelleted by centrifugation. One half was washed in 0.2 M EDTA, 0.01% azide at 4°C (the F0 sample) and the other half was incubated in 100 ml of prewarmed minimal heavy media containing 1% glucose (13C6) and 0.2% 15NH4Cl (Advanced Research in Chemistry). In these conditions, the presence of both glucose and nitrogen is essential for cells to resume growth (data not shown). The cultures were followed by counting the cells, measuring the septation index and by flow cytometry, indicating 30–40% of cell synchrony (data not shown). Fifty milliliters of the culture were harvested after the first generation and 25 ml after the second, during the G2 phases, referred to as the F1 and F2 samples, respectively. The G2 phase is the longest cell cycle phase in S. pombe, occupying about three-quarters of the cell cycle.
DNA preparation and analysis. Genomic DNA was prepared and digested by SspI or NsiI in agarose plugs (Arcangioli, 1998), purified (Quiagene kit) and the CsCl gradient centrifugation was performed essentially as described (McCarroll and Fangman, 1988). Collected fractions (0.2 ml) were isopropanol precipitated, resuspended in TE and samples of DNA fractions were analysed in agarose gels. In denaturing conditions, the DNA was mixed with loading dye to a final concentration of 15% formamide, 2% formaldehyde, 10 mM HEPES, 2 mM EDTA, 7% glycerol pH 7.8 and incubated for 5 min at 80°C. Electrophoresis was performed in a 1.4% agarose gel containing 6% formaldehyde for 3 h at 100 V. Separated DNA was transferred onto Hybond-N+ membrane (Amersham Pharmacia), hybridized, exposed to phosphor screen (Molecular Dynamics) and quantified using ImagequantNT.
Acknowledgments
ACKNOWLEDGEMENTS
I thank B. Dujon, J. Herrick, A. Holmes, G.F. Richard and M. Yaniv for discussions or help with the manuscript, and A. Kaykov and R. de Lahondès for their various contributions to this work. This work was supported by a grant from the Association pour la Recherche sur le Cancer.
REFERENCES
- Arcangioli B. (1998) A site- and strand-specific DNA break confers asymmetric switching potential in fission yeast. EMBO J., 17, 4503–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangioli B. and Klar, A.J.S. (1991) A novel switch-activating site (SAS1) and its cognate binding factor (sap1) required for efficient mat1 switching in Schizosaccharomyces pombe. EMBO J., 10, 3025–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangioli B. and de Lahondès, R. (2000) Fission yeast switches mating-type by a replication–recombination coupled process. EMBO J., 19, 1389–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Bates, D.B. and Kogoma, T. (1994) DNA replication trigger by double-stranded breaks in E. coli: dependence on chromosome recombination functions. Cell, 78, 1051–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet N.C. and Carr, A.M. (1993) Fission yeast wee1 protein kinase is not required for 5 DNA damage-dependent mitotic arrest. Nature, 364, 824–827. [DOI] [PubMed] [Google Scholar]
- Beach D.H. (1983) Cell type switching by DNA transposition in fission yeast. Nature, 305, 682–688. [Google Scholar]
- Beach D. and Klar, A.J.S. (1984). Rearrangements of the transposable mating-type cassettes of fission yeast. EMBO J., 3, 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belling J. (1933) Crossing over and gene rearrangement in flowering plants. Genetics, 18, 388–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard J.Z. and Klar, A.J.S. (1999) Orientation of DNA replication establishes mating-type switching pattern in S. pombe. Nature, 400, 181–184. [DOI] [PubMed] [Google Scholar]
- Egel R. (1977) Frequency of mating-type switching in homothallic fission yeast. Nature, 266, 172–174. [DOI] [PubMed] [Google Scholar]
- Egel R. (1984) The pedigree pattern of mating-type switching in Schizosaccharomyces pombe. Curr. Genet., 8, 205–210. [DOI] [PubMed] [Google Scholar]
- Egel R. and Eie, B. (1987) Cell lineage asymmetry in Schizosaccharomyces pombe: unilateral transmission of a high-frequency state for mating-type switching in diploid pedigrees. Curr. Genet., 12, 429–433. [Google Scholar]
- Egel R. and Gutz, H. (1981) Gene activation by copy transposition in mating-type switching of a homothallic fission yeast. Curr. Genet., 12, 5–12. [DOI] [PubMed] [Google Scholar]
- Ferguson D.O. and Holloman, W.K. (1996) Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc. Natl Acad. Sci. USA, 93, 5419–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J.E. (1999) DNA recombination: the replication connection. Trends Biochem. Sci., 24, 271–275. [DOI] [PubMed] [Google Scholar]
- Holmes A. and Haber, J. (1999) Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell, 96, 415–424. [DOI] [PubMed] [Google Scholar]
- Kelly M., Burke, J., Smith, M., Klar, A. and Beach, D. (1988) Four mating-type genes control sexual differentiation in the fission yeast. EMBO J., 7, 1537–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A.J.S. (1987) Differential parental DNA strands confer developmental asymmetry on daughter cells in fission yeast. Nature, 326, 466–470. [DOI] [PubMed] [Google Scholar]
- Klar A.J.S. (1990) The developmental fate of fission yeast cells is determined by the pattern of inheritance of parental and grandparental DNA strands. EMBO J., 9, 1407–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A.J.S., Bonaduce, M.J. and Cafferkey, R. (1991) The mechanism of fission yeast mating type interconversion: seal/replicate/cleave model of replication across the double-stranded break site at mat1. Genetics, 127, 489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T. (1997) Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev., 61, 212–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leupold U. (1950) Die vererbung von homothallie und heterothallie bei Schizosaccharomyces pombe. C.R. Lab. Carlsberg Ser. Physiol., 24, 381–480. [Google Scholar]
- McCarroll R.M., and Fangman, W.L. (1988) Time of replication of yeast centromeres and telomeres. Cell, 54, 505–513. [DOI] [PubMed] [Google Scholar]
- Meselson M. and Stahl, F.W. (1958) The replication of DNA in Escherichia coli. Proc. Natl Acad. Sci. USA, 44, 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata H. and Miyata, M. (1981) Mode of conjugation in homothallic cells of Schizosaccharomyces pombe. J. Gen. Appl. Microbiol., 27, 365–369. [Google Scholar]
- Moreno S.A., Klar, A.J.S. and Nurse, P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Nassif N., Penney, J., Pal, S., Engels, W.R. and Gloor, G.B. (1994) Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol., 14, 1613–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen O. and Egel, R. (1989) Mapping of the double-strand breaks at the mating-type locus in fission yeast by genomic sequencing. EMBO J., 8, 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R., Michel, B. and Gangloff, S. (2000) Replication fork pausing and recombination or ‘gimme a break’. Genes Dev., 14, 1–10. [PubMed] [Google Scholar]
- Singh J. and Klar, A.J.S. (1993) DNA polymerase α is essential for mating-type switching in fission yeast. Nature, 361, 271–276. [DOI] [PubMed] [Google Scholar]
- Styrkarsdottir U., Egel, R. and Nielsen, O. (1993) The smt-0 mutation which abolishes mating-type switching in fission yeast is a deletion. Curr. Genet., 23, 184–186. [DOI] [PubMed] [Google Scholar]