Abstract
Latent Epstein–Barr virus (EBV) genomes are maintained in human cells as low copy number episomes that are thought to be partitioned by attachment to the cellular mitotic chromosomes through the viral EBNA1 protein. We have identified a human protein, EBP2, which interacts with the EBNA1 sequences that govern EBV partitioning. Here we show that, in mitosis, EBP2 localizes to the condensed cellular chromosomes producing a staining pattern that is indistinguishable from that of EBNA1. The localization of EBNA1 proteins with mutations in the EBP2 binding region was also examined. An EBNA1 mutant (Δ325–376) disrupted for EBP2 binding and segregation function was nuclear but failed to attach to the cellular chromosomes in mitosis. Our results indicate that amino acids 325–376 mediate the binding of EBNA1 to mitotic chromosomes and strongly suggest that EBNA1 mediates EBV segregation by attaching to EBP2 on the cellular mitotic chromosomes.
INTRODUCTION
Epstein–Barr virus (EBV) is a human γ-herpes virus that infects >90% individuals worldwide (reviewed in Miller, 1990; Kieff, 1996; Rickinson and Kieff, 1996). After initial infection, EBV is maintained for life through latent infection of a small percentage of the B-lymphocytes. During latent infection, as many as 10 viral gene products can be expressed and the infected cells are induced to proliferate indefinitely. Latent EBV infection is associated with several malignancies including Burkitt’s lymphoma, nasopharyngeal carcinoma, Hodgkin’s disease and a variety of lymphomas in AIDS and organ transplant patients.
EBV genomes are maintained in latently infected cells as low copy number, double-stranded, circular DNA episomes. These episomes undergo one round of DNA replication per cell cycle and segregate efficiently to the daughter cells during cell division (Adams, 1987; Yates, 1996). The viral cis-acting element, oriP, has been shown to be sufficient for the replication and segregation of plasmids in human cells provided that the viral Epstein–Barr nuclear antigen 1 protein (EBNA1) is present (Yates et al., 1985). EBNA1 activates DNA replication and mediates episome partitioning by binding to specific recognition sites present in the two functional components of oriP, the family of repeats (FR) and the dyad symmetry element (DS) (Rawlins et al., 1985; Reisman et al., 1985). The DS element is the initiation site for DNA replication (Gahn and Schildkraut, 1989; Wysokenski and Yates, 1989; Niller et al., 1995; Yates et al., 2000); the FR element stimulates DNA replication and is the cis-acting segregation element of the virus (Lupton and Levine, 1985; Krysan et al., 1989; Wysokenski and Yates, 1989).
The mechanism by which EBNA1 mediates partitioning of EBV episomes and oriP-containing plasmids is thought to involve ‘piggy backing’ on the host cell mitotic chromosomes. In interphase cells, EBNA1 is present throughout the nucleus but in mitosis, EBNA1 has been shown to be associated with the condensed cellular chromosomes (Reedman and Klein, 1973; Grogan et al., 1983; Petti et al., 1990). EBV episomes and oriP-containing constructs have also been shown to associate with mitotic chromosomes in human cells expressing EBNA1 (Harris et al., 1985; Delecluse et al., 1993; Simpson et al., 1996). Hence, EBNA1 is believed to mediate episomal segregation by tethering the oriP FR element to an unknown component of the cellular mitotic chromosomes.
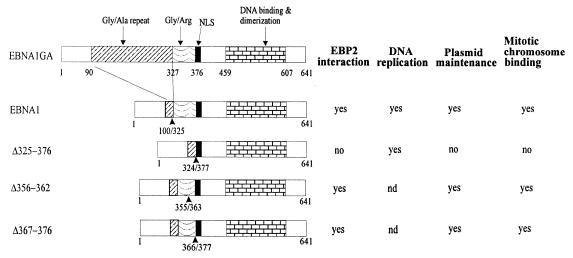
In a previous study, we identified a human protein named EBP2 (EBNA1 binding protein 2) that specifically interacts with EBNA1 (Shire et al., 1999). EBP2 is a nucleolar protein that is highly conserved in eukaryotes and whose budding yeast homologue was shown recently to provide an essential function in rRNA processing (Huber et al., 2000). EBP2 was shown to interact with a Gly-Arg rich region of EBNA1 between amino acids 325 and 376. Using EBNA1 proteins with mutations in this region, we showed a correlation between the ability of EBNA1 proteins to bind EBP2 and their ability to partition oriP-containing plasmids in human cells (Shire et al., 1999). In particular, the EBNA1 mutant that lacked sequence 325–376 (Δ325–376) was found to have wild-type DNA replication and oriP-binding activities, but did not bind EBP2 or maintain oriP plasmids in long-term culture. These observations suggested that the EBNA1–EBP2 interaction was important for the partitioning of EBV episomes during cell division.
To examine further the contribution of EBP2 to EBNA1-mediated partitioning, we have examined the cellular localization of EBP2 and EBNA1 mutants. Here we show that, like EBNA1, EBP2 localizes to the condensed chromosomes in mitosis and that the EBNA1 sequences that mediate the EBP2 interaction are responsible for EBNA1 attachment to mitotic chromosomes.
RESULTS
If EBP2 plays a direct role in EBNA1-mediated DNA segregation and the ‘piggy backing’ model of EBV segregation is correct, then EBP2 should associate with the condensed chromosomes in metaphase. To address this possibility, we studied the cellular localization of EBP2 in mitotic cells. EBP2 has been shown to be predominantly nucleolar in interphase cells (Chatterjee et al., 1987), but its localization in mitosis has not been determined. To this end, we blocked Raji cells (EBV-positive human Burkitt’s lymphoma cells) in mitosis by colcemid treatment and stained the fixed cells with polyclonal antibody raised against purified EBP2. In all the mitotic cells examined, EBP2 was found to be a component of the mitotic chromosomes (Figure 1D). EBP2 immunofluorescence was coincident with DAPI staining (Figure 1F) indicating that EBP2 interaction with the chromosomes was not limited to specific sites. When the Raji cell mitotic chromosomes were stained for both EBNA1 and EBP2, the EBNA1 staining was superimposable on that of EBP2, suggesting that the two proteins co-localize on the chromosomes (Figure 1B, D and G). Identical results were obtained using an antibody that recognizes a 19 amino acid peptide of EBP2 (kindly provided by Dr Mike McAlear, Wesleyan University; data not shown).
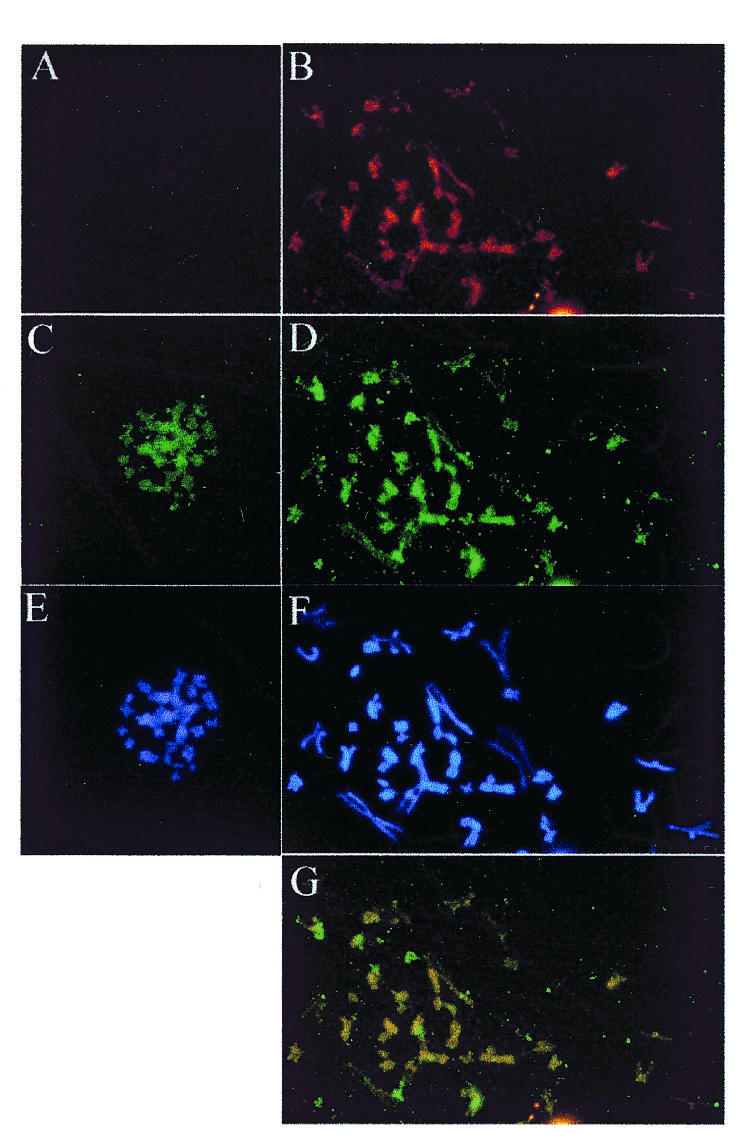
Fig. 1. Localization of EBP2 and EBNA1 on mitotic chromosomes. Mitotic chromosome spreads are shown from Raji (right panels) and BL41 (left panels) Burkitt’s lymphoma cells that do and do not express EBNA1, respectively. (A) and (B) EBNA1 staining with monoclonal antibody. (C) and (D) EBP2 staining with rabbit polyclonal antibody. (E) and (F) DAPI staining. (G) Overlay of images (B) and (D) showing co-localization (yellow). Images were captured at 630-fold magnification.
To determine whether EBP2 is a natural component of mitotic chromosomes or associates with the chromosomes through EBNA1, we examined the mitotic localization of EBP2 in EBV-negative Burkitt’s lymphoma cells (BL41), which do not express EBNA1. EBP2 was found to be associated with the mitotic chromosomes in BL41 cells; as in Raji cells, the EBP2 immunofluorescence was coincident with the DAPI staining (Figure 2C and E). Hence, the binding of EBP2 to mitotic chromosomes does not require EBNA1, but, conversely, EBP2 may mediate the attachment of EBNA1 to the chromosomes.
Fig. 2. Summary of the functionalities and interactions of the EBNA1 mutants. EBNA1–EBP2 interactions and DNA replication and oriP plasmid maintenance (requiring DNA replication and segregation) activities are from Shire et al. (1999). nd, not determined.
To explore further the possibility that the attachment to EBP2 on mitotic chromosomes is necessary for EBNA1-mediated partitioning, we compared the cellular localization of the Δ325–376 EBNA1 mutant, which is defective for EBP2 binding and segregation, with that of wild-type EBNA1 (Shire et al., 1999; see Figure 2). For these experiments, human cell lines that express EBNA1 or the Δ325–376 mutant were generated from C33A cells. The expression levels of the wild-type and mutant EBNA1 proteins were shown to be similar by western blotting (data not shown). The version of EBNA1 used as wild-type in these studies is one that lacks most of the non-essential Gly-Ala repeat region but has been shown to be functional for replication and segregation activities. Immunofluorescent staining of the log-phase cells with anti-EBNA1 antibody showed that both the wild-type and the mutant proteins were expressed in at least 40% of the cells and that both proteins were nuclear (Figure 3C–F). The segregation defect associated with Δ325–376 is therefore not due to failure to enter the nucleus.
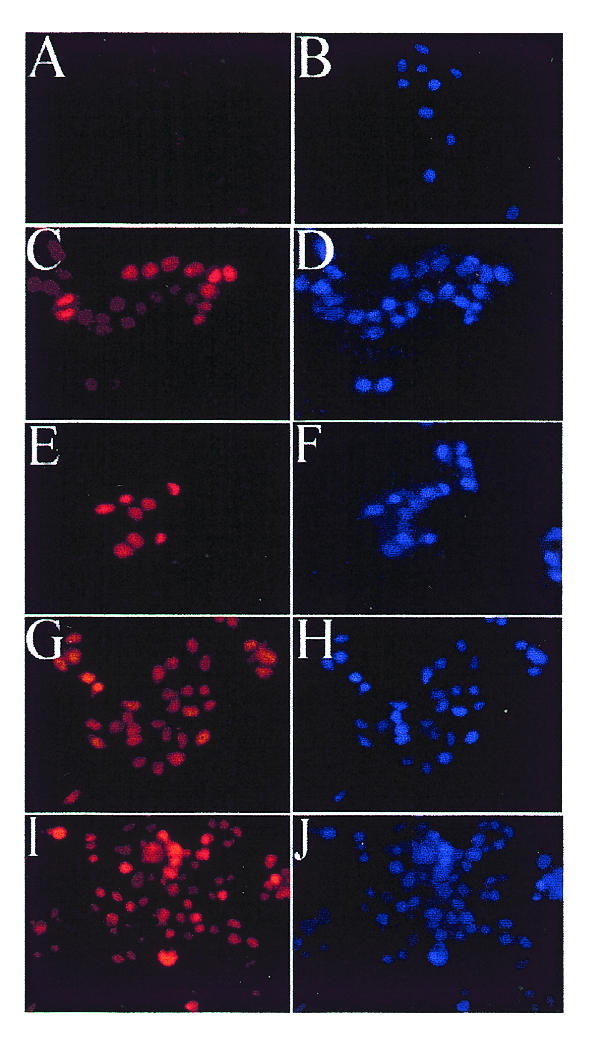
Fig. 3. Cellular localization of wild-type and mutant EBNA1 proteins in C33A cells. C33A cells that are EBNA1-negative (A and B) or that express EBNA1 (C and D), Δ325–376 (E and F), Δ356–362 (G and H) or Δ367–376 (I and J) were grown on slides then fixed for microscopy. EBNA1 proteins were detected with mouse monoclonal antibody (left panels), and DNA was detected with DAPI (right panels). Images in the left panels were captured using similar exposure times and 400-fold magnification.
To determine whether the lack of segregation activity of Δ325–376 was due to an inability to attach to the host mitotic chromosomes, we next compared the localization of EBNA1 and Δ325–376 in mitotic cells. The C33A cells expressing these proteins were treated with colcemid and the EBNA1 proteins in the mitotic cells were visualized by indirect immunofluorescence. As was observed for the EBV-infected cells, EBNA1 in the C33A cells was found to localize to the condensed chromosomes (Figure 4C and D). The Δ325–376 mutant, however, did not localize to the mitotic chromosomes but rather was diffusely distributed in the area around the chromosomes (Figure 4E–H). Localization of Δ325–376 to the mitotic chromosomes was not observed in any of the >50 cells observed that expressed this protein. Therefore, our results show that the EBNA1 region that mediates the EBP2 interaction is also responsible for chromosome binding. These results also support the contention that chromosome attachment is the mechanism of EBV segregation.
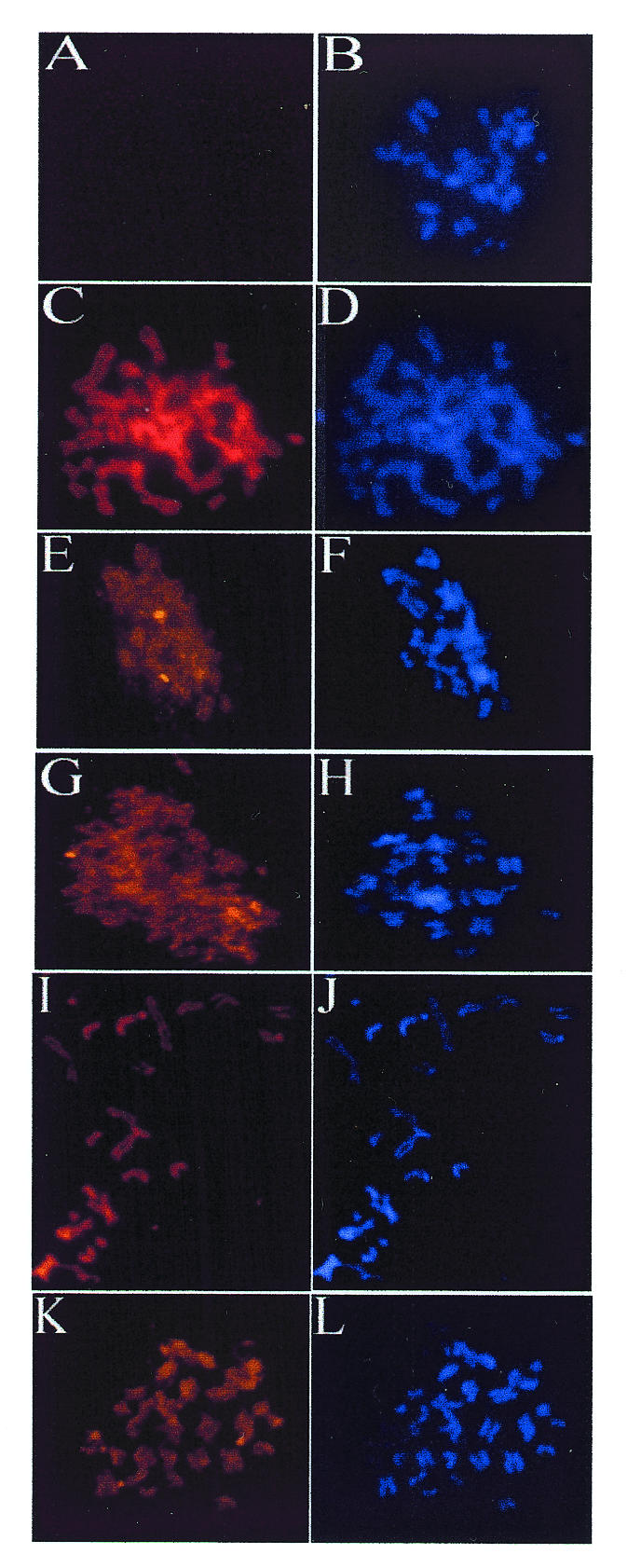
Fig. 4. Mitotic localization of EBNA1 mutants. C33A cells that are EBNA1-negative (A and B) or that express EBNA1 (C and D), Δ325–376 (E–H), Δ356–362 (I and J) or Δ367–376 (K and L) were blocked in mitosis by colcemid treatment, and chromosomes were spread for microscopy. EBNA1 proteins were detected with monoclonal antibody (left panels), and DNA was detected with DAPI (right panels). Images in left panels were captured using similar exposure times and 630-fold magnification.
To explore further the relationship between segregation activity, mitotic chromosome association and EBP2 binding, we examined the mitotic localization of two additional EBNA1 mutants with small internal deletions within the 325–376 Gly-Arg repeat (Δ356–362 and Δ367–376; Figure 2). We have shown previously that these two EBNA1 mutants interact with EBP2 and support the maintenance of oriP plasmids (Shire et al., 1999). The failure of these small deletions to disrupt EBP2 binding is likely to be due to the repetitive nature of the Gly-Arg region, which contains six octameric repeats with overlapping functions (Laine and Frappier, 1995). C33A-based cell lines were constructed that expressed either Δ356–362 or Δ367–376. By indirect immunofluorescence, we found that both Δ356–362 and Δ367–376 were nuclear in interphase (Figure 3G–J) and bound to the condensed chromosomes in mitosis with a staining pattern indistinguishable from wild-type EBNA1 (Figure 4I–L). Thus, small deletions within the Gly-Arg repeat that are insufficient to disrupt EBP2 binding and segregation function also do not abrogate binding to mitotic chromosomes.
DISCUSSION
We have shown that, like EBNA1, EBP2 localizes to the condensed cellular chromosomes during mitosis, and that an EBNA1 mutation that disrupts EBP2 binding also disrupts chromosome attachment. Thus the data suggest that the chromosomal component to which EBNA1 attaches in mitosis is EBP2. Additional proof of the importance of EBP2 for EBNA1-mediated segregation comes from our recent experiments that show that the expression of both EBNA1 and human EBP2 are necessary for efficient segregation of FR-containing plasmids in yeast (P. Kapoor and L. Frappier, in preparation). EBP2 is the only EBNA1-interacting protein identified to date that appears to mediate the DNA partitioning activity of EBNA1 activity. The other EBNA1-binding proteins that have been identified are P32/TAP (Wang et al., 1997), replication protein A (RPA) (Zhang et al., 1998) and karyopherin α1 and α2 (Fischer et al., 1997; Kim et al., 1997; Ito et al., 2000). Based on their known cellular functions, the interaction of EBNA1 with karyopherins may be important for nuclear import and the interaction with RPA may be important for DNA replication. The function of P32/TAP is undefined but its interaction with EBNA1 is not likely to mediate DNA partitioning because P32/TAP interacts with the Δ325–376 segregation mutant (M. Holowaty and L. Frappier, unpublished).
We have previously shown that the Δ325–376 EBNA1 mutant has wild-type DNA replication activity but is unable to maintain oriP plasmids in long term culture, indicating a specific defect in DNA partitioning (Shire et al., 1999). We have now shown that, although Δ325–376 is nuclear, it fails to associate with mitotic chromosomes. Therefore, our results provide strong support for the contention that EBNA1 governs the partitioning of latent EBV episomes by mediating an interaction with the cellular mitotic chromosomes. Although the association of EBNA1 (Reedman and Klein, 1973; Grogan et al., 1983; Petti et al., 1990), EBV episomes (Harris et al., 1985; Delecluse et al., 1993) and oriP-containing artificial chromosomes (Simpson et al., 1996) with cellular mitotic chromosomes suggested that chromosomal attachment was necessary for EBNA1-mediated segregation, it has not previously been shown that segregation-defective EBNA1 mutants fail to bind to mitotic chromosomes.
A recent study by Marechal et al. (1999) examined the contribution of EBNA1 sequences to mitotic chromosome attachment by fusing small fragments of EBNA1 to green fluorescent protein. They found that three EBNA1 regions, amino acids 8–54 (CBS-3), 72–84 (CBS-1) and 328–365 (CBS-2), could mediate chromosome binding. However, deletion of either CBS-1 or CBS-3 from the full length EBNA1 protein did not disrupt the binding of EBNA1 to mitotic chromosomes (a deletion of CBS-2 was not constructed), leading to the conclusion that the three regions perform redundant functions. Our data with the Δ325–376 mutant show that deletion of CBS-2 has a much more pronounced effect on mitotic chromosome association, and indicate that in the context of the EBNA1 protein, CBS-3 and CBS-1 are not sufficient for chromosome binding.
METHODS
Cell lines. BL41 is an EBV-negative Burkitt’s lymphoma line that was a gift from Dr Elliott Kieff. Cell lines expressing EBNA1 or the Δ325–376, Δ356–362 or Δ367–376 EBNA1 mutants were generated by transfection of human C33A cells with plasmids that contained oriP and a neomycin-resistance marker, and expressed EBNA1 or an EBNA1 mutant (Shire et al., 1999). Cells containing the EBNA1-expressing plasmids were selected by growing in medium containing 400 µg/ml G418 for at least 3 weeks. Plasmids expressing EBNA1, Δ356–362 or Δ367–376 (which support oriP plasmid maintenance) were maintained episomally at an average copy number of two copies per cell, while those expressing Δ325–376 (which is unable to maintain oriP plasmids) integrated into the cellular DNA. In all of these cell lines, EBNA1 is in large excess over oriP sequences and therefore most of the EBNA1 is not oriP-bound.
EBP2 antibodies. Full-length EBP2 was expressed in Escherichia coli as a His6 fusion (from pET15b; Novagene), and as a maltose binding protein (MBP) fusion (from pMAL-c2; New England Biolabs). The His-tagged human EBP2 was solubilized from BL21DE3 E. coli in 8 M urea and purified on a nickel column (Novagene) under denaturing conditions. The purified His-EBP2 was used to immunize rabbits. MBP-tagged EBP2 was produced in DH5α E. coli in soluble form and purified on an amylose column (New England Biolabs) according to the manufacturer’s instructions. Purified MBP-EBP2 (6 mg) was coupled to 1 ml Affi-gel 10 according to the manufacturer’s method (Bio-Rad) and used to affinity-purify anti-EBP2 antibody from the rabbit serum. The serum was diluted 1:10 in 150 mM NaCl, 16 mM Na2HPO4, 4 mM NaH2PO4 and passed through the affinity column three times. After extensive washing with phosphate-buffered saline (PBS), the bound antibody was eluted from the column with 100 mM glycine pH 2.5 and dialysed against PBS.
Immunofluorescence. For indirect immunofluorescence of mitotic chromosomes, Raji, BL41 or C33A-based cells were blocked in mitosis by the addition of 0.1 µg/ml colcemid to log-phase cultures. After 16 h at 37°C, the cells were harvested and swollen in hypotonic buffer as previously described (Grogan et al., 1983). The cells were then dropped onto slides, fixed in 3% paraformaldehyde at room temperature for 30 min, permeabilized in PBS containing 0.5% Triton X-100, and blocked by a 30 min incubation in 3% BSA/0.05% Tween 20 in PBS. The permeabilized cells were then incubated with anti-EBNA1 and anti-EBP2 antibodies for 1 h at room temperature, followed by incubation with FITC (Gibco-BRL; 1:30 dilution) or Texas Red (Molecular Probe; 1:200 dilution)-conjugated secondary antibodies. EBNA1 staining was performed using the mouse monoclonal antibody, OT1x, that recognizes EBNA1 amino acids 424–448 (Chen et al., 1993; kindly supplied by Dr Jaap Middeldorp, Free University Hospital, Amsterdam). The cells were counterstained with DAPI (25 ng/ml in PBS) for 5 min, and the slides were mounted in 5 µl of anti-fade solution (Molecular Probe). Staining was visualized at 630-fold magnification and captured using either a Leica DMLB microscope and Metaview Imaging software, or a Leica DMR microscope and Openlab software. For indirect immunofluorescent staining of interphase cells, log-phase cells growing on coverslips were fixed, stained and visualized (at 400-fold magnification) as described above.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Jaap Middeldorp for EBNA1 antibodies, Dr Elliott Kieff for BL41 cells and Dr Brenda Andrews for the use of her microscope. We also thank Kathy Shire for excellent technical assistance and Dr Barbara Funnell for critical reading of the manuscript. This work was supported by a grant from the Medical Research Council (MRC) of Canada. L.F. is a MRC scientist.
REFERENCES
- Adams A. (1987) Replication of latent Epstein–Barr virus genomes. J. Virol., 61, 1743–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A., Freeman, J.W. and Busch, H. (1987) Identification and partial characterization of a Mr 40,000 nucleolar antigen associated with cell proliferation. Cancer Res., 47, 1123–1129. [PubMed] [Google Scholar]
- Chen M.-R., Middeldorp, J.M. and Hayward, S.D. (1993) Separation of the complex DNA binding domain of EBNA-1 into DNA recognition and dimerization subdomains of novel structure. J. Virol., 67, 4875–4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delecluse H.-J., Bartnizke, S., Hammerschmidt, W., Bullerdiek, J. and Bornkamm, G.W. (1993) Episomal and integrated copies of Epstein–Barr virus coexist in Burkitt’s lymphoma cell lines. J. Virol., 67, 1292–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N., Kremmer, E., Lautscham, G., Mueller-Lantzsch, N. and Grasser, F.A. (1997) Epstein–Barr virus nuclear antigen 1 forms a complex with the nuclear transporter karyopherin α2. J. Biol. Chem., 272, 3999–4005. [DOI] [PubMed] [Google Scholar]
- Gahn T.A. and Schildkraut, C.L. (1989) The Epstein–Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell, 58, 527–535. [DOI] [PubMed] [Google Scholar]
- Grogan E.A., Summers, W.P., Dowling, S., Shedd, D., Gradoville, L. and Miller, G. (1983) Two Epstein–Barr viral nuclear neoantigens distinguished by gene transfer, serology and chromosome binding. Proc. Natl Acad. Sci. USA, 80, 7650–7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A., Young, B.D. and Griffin, B.E. (1985) Random association of Epstein–Barr virus genomes with host cell metaphase chromosomes in Burkitt’s lymphoma-derived cell lines. J. Virol., 56, 328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber M., Dworet, J., Shire, K., Frappier, L. and McAlear, M. (2000) The budding yeast homolog of the human EBNA1-binding protein 2 (Ebp2p) is an essential nucleolar protein required for rRNA processing. J. Biol. Chem., 275, in press. [DOI] [PubMed] [Google Scholar]
- Ito S., Ikeda, M., Kato, N., Matsumoto, A., Ishikawa, Y., Kumakubo, S. and Yanagi, K. (2000) Epstein–Barr virus nuclear antigen-1 binds to nuclear transporter karyopherin α1/NPI-1 in addition to karyopherin α2/Rch1. Virology, 266, 110–119. [DOI] [PubMed] [Google Scholar]
- Kieff E. (1996) Epstein–Barr virus and its replication. In Fields, D.M.K.B.N., Knipe, P.M. and Howley, P.M. (eds) Epstein–Barr Virus and its Replication. Lippincott-Raven Publishers, Philadelphia, PA, pp. 2343–2396.
- Kim A.L., Maher, M., Hayman, J.B., Ozer, J., Zerby, D., Yates, J.L. and Lieberman, P.M. (1997) An imperfect correlation between DNA replication activity of Epstein–Barr virus nuclear antigen 1 (EBNA1) and binding to the nuclear import receptor, Rch1/importin α. Virology, 239, 340–351. [DOI] [PubMed] [Google Scholar]
- Krysan P.J., Haase, S.B. and Calos, M.P. (1989) Isolation of human sequences that replicate autonomously in human cells. Mol. Cell. Biol., 9, 1026–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine A. and Frappier, L. (1995) Identification of Epstein–Barr nuclear antigen 1 protein domains that direct interactions at a distance between DNA-bound proteins. J. Biol. Chem., 52, 30914–30918. [DOI] [PubMed] [Google Scholar]
- Lupton S. and Levine, A.J. (1985) Mapping of genetic elements of Epstein–Barr virus that facilitate extrachromosomal persistence of Epstein–Barr virus-derived plasmids in human cells. Mol. Cell. Biol., 5, 2533–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal V., Dehee, A., Chikhi-Brachet, R., Piolot, T., Coppey-Moisan, M. and Nicolas, J. (1999) Mapping EBNA1 domains involved in binding to metaphse chromosomes. J. Virol., 73, 4385–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G. (1990) Epstein–Barr virus. In Fields, B.N. (ed.), Epstein–Barr Virus. Raven Press, New York, NY, pp. 1921–1958.
- Niller H.H., Glaser, G., Knuchel, R. and Wolf, H. (1995) Nucleoprotein complexes and DNA 5′-ends at oriP of Epstein–Barr virus. J. Biol. Chem., 270, 12864–12868. [DOI] [PubMed] [Google Scholar]
- Petti L., Sample, C. and Kieff, E. (1990) Subnuclear localization and phosphorylation or Epstein–Barr virus latent infection nuclear proteins. Virology, 176, 563–574. [DOI] [PubMed] [Google Scholar]
- Rawlins D.R., Milman, G., Hayward, S.D. and Hayward, G.S. (1985) Sequence-specific DNA binding of the Epstein–Barr virus nuclear antigen (EBNA1) to clustered sites in the plasmid maintenance region. Cell, 42, 859–868. [DOI] [PubMed] [Google Scholar]
- Reedman B.M. and Klein, G. (1973) Cellular localization of an Epstein–Barr virus (EBV)-associated complement-fixing antigen in producer and non-producer lymphoblastoid cell lines. Int. J. Cancer, 11, 499–520. [DOI] [PubMed] [Google Scholar]
- Reisman D., Yates, J. and Sugden, B. (1985) A putative origin of replication of plasmids derived from Epstein–Barr virus is composed of two cis-acting components. Mol. Cell. Biol, 5, 1822–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickinson A.B. and Kieff, E. (1996) Epstein–Barr virus. In Fields, D.M.K.B.N., Knipe, P.M. and Howley, P.M. (eds) Epstein–Barr Virus and its Replication. Lippincott-Raven Publishers, Philadelphia, PA, pp. 2397–2446.
- Shire K., Ceccarelli, D.F.J., Avolio-Hunter, T.M. and Frappier, L. (1999) EBP2, a human protein that interacts with sequences of the Epstein–Barr nuclear antigen 1 important for plasmid maintenance. J. Virol., 73, 2587–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson K., McGuigan, A. and Huxley, C. (1996) Stable episomal maintenance of yeast artificial chromosomes in human cells. Mol. Cell. Biol., 16, 5117–5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Finan, J.E., Middeldorp, J.M. and Hayward, S.D. (1997) P32/TAP, a cellular protein that interacts with EBNA-1 of Epstein–Barr virus. Virology, 236, 18–29. [DOI] [PubMed] [Google Scholar]
- Wysokenski D.A. and Yates, J.L. (1989) Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein–Barr virus. J. Virol., 63, 2657–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J.L. (1996) Epstein–Barr virus DNA replication. In M.L. DePamphilis (ed.) Epstein–Barr Virus DNA Replication. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp. 751–773.
- Yates J.L., Warren, N. and Sugden, B. (1985) Stable replication of plasmids derived from Epstein–Barr virus in various mammalian cells. Nature, 313, 812–815. [DOI] [PubMed] [Google Scholar]
- Yates J.L., Camiolo, S.M. and Bashaw, J.M. (2000) The minimal replicator of Epstein–Barr virus oriP. J. Virol. 74, 4512–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Frappier, L., Gibbs, E., Hurwitz, J. and O’Donnell, M. (1998) Human RPA (hSSB) interacts with EBNA1, the latent origin binding protein of Epstein–Barr virus. Nucleic Acids Res., 26, 631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]