Abstract
The activation function AF2 in the ligand-binding domain of estrogen receptors ERα and ERβ signals through the recruitment of nuclear receptor coactivators. Recent evidence indicates that coactivators, such as the transcription intermediary factor TIF2, also bind to and transactivate the N-terminal AF1 function of the two ERs. We have generated TIF2 mutant proteins that are deficient in either AF1 or AF2 interaction and use these mutants to investigate the relative contribution of both AFs to TIF2 recruitment and transactivation. We observe that TIF2 is capable of interacting simultaneously with both the isolated N- and C-terminus of ERα in transfected mammalian cells and in vitro, indicating that TIF2 can bridge both receptor domains. The concomitant interaction of TIF2 with both AFs results in synergistic activation of transcription. Thus, synergy between ERα AF1 and AF2 is a result of the cooperative recruitment of TIF2 and/or other members of the p160 coactivator family.
INTRODUCTION
Nuclear receptors are ligand-dependent transcription factors that have evolved from an ancestral orphan receptor into a highly diverse family present throughout the entire animal kingdom and encompassing receptors for steroid and non-steroid hormones, vitamins and metabolic intermediates. They regulate the expression of a wide variety of target genes by binding either directly as mono-, homo- or heterodimers to cognate DNA-responsive elements, or indirectly by acting through poorly understood ‘crosstalk’ phenomena. Nuclear receptors are composed of five to six independent modules. These regions encode specific functions, which include transcriptional activation and repression, DNA and ligand binding, cellular compartmentalization and dimerization. Ligand binding induces major conformational changes in the ligand-binding domain (LBD), thus generating the cognate binding surfaces for nuclear receptor (NR) coactivators (CoAs) while concomitantly destroying the binding surface for corepressors (CoRs), which exist for some, but not all NRs. Direct transcriptional repression by some NRs is mediated by CoR complexes that are associated with the unliganded receptor (or with receptors bound to certain antagonists) due to condensation of chromatin in the promoter environment through histone deacetylation. Upon agonist binding, CoRs dissociate from the receptor and CoA complexes are recruited. CoAs possess one or several LxxLL NR boxes, which are critical constituents of the LBD–CoA interface (Darimont et al., 1998; McInerney et al., 1998; Nolte et al., 1998), while the so-called ‘CoRNR’ box of CoRs is the sequence that is critical for the interaction with an NR LBD surface, which is topographically closely related to that involved in CoA binding (Hu and Lazar, 1999; Nagy et al., 1999; Perissi et al., 1999) (for recent reviews on NRs and their coregulators see Moras and Gronemeyer, 1998; McKenna et al., 1999; Minucci and Pelicci, 1999; Glass and Rosenfeld, 2000). While CoA and CoR complexes are believed to modulate the chromatin structure, agonist binding results in the establishment of a second type of NR–CoA complex, referred to as SMCC complex, which establishes the links to the basal machinery (Ito et al., 1999; Rachez et al., 1999; Chiba et al., 2000).
Recent evidence indicates that some CoAs are also capable of interacting with the AF1 of several receptors. Both ERα and ERβ have been shown to recruit functionally SRC-1 or its ortholog TIF2 (transcription intermediary factor 2) via their N-terminal AF1 harboring AB domains (Webb et al., 1998; Tremblay et al., 1999; our unpublished results), and a novel ERα AF1-selective CoA has been described (Endoh et al., 1999). In the case of ERβ this interaction was shown to depend on phosphorylation of conserved serine residues in this receptor region (Tremblay et al., 1999). A similar mechanism has been demonstrated for the nuclear orphan receptor SF1, where MAP kinase signaling triggers phosphorylation of Ser203 in the AB region and concomitant recruitment of CoAs (Hammer et al., 1999). Similarly, the peroxisome proliferator-activated receptor PPARγ has been shown to stimulate transcription via the association of its AF1 with the general mediator CBP (CREB-binding protein) (Gelman et al., 1999), and the glucocorticoid receptor τ1 AF acts via binding to the transcription mediator ADA2 and associated ADA complexes and the recruitment of DRIP150 (Almlof et al., 1998; Hittelman et al., 1999). Finally, both known (Alen et al., 1999; He et al., 1999) and novel (Hsiao and Chang, 1999) CoAs have been shown to mediate the AF1 function of the androgen receptor.
While the above results provide a mechanistic insight into the basis of NR AF1 or AF2 action, it is not yet clear how the overall transcription activity of the entire receptor is generated from its individual AFs. In this respect, the observations that both AFs of an NR can synergize and that the synergy between the two ERα AFs depends on the cell and promoter context are of particular importance. We demonstrate here that TIF2 is able to bind to the isolated AF1 and AF2 simultaneously using distinct interfaces and that this simultaneous interaction can lead to synergistic activation of transcription.
RESULTS AND DISCUSSION
Two distinct regions of TIF2 mediate the activities of ERα AF1 and AF2
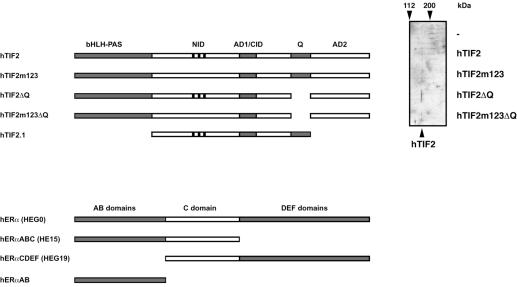
TIF2 has been shown to act as a CoA for both AF1 and AF2 of ERα and ERβ (see Introduction for references). Mapping studies in vitro have indicated that binding to AF1 is mediated by residues in the Q-rich domain of TIF2 (Webb et al., 1998; our unpublished results; Figure 1 schematizes the structures of TIF2 and ERα). Efficient interaction with AF2 requires at least two of the three previously characterized LxxLL NR boxes (Le Douarin et al., 1996) in the NR-interacting domain (NID) of TIF2 (Voegel et al., 1998). Thus, the requirements for interaction with both AFs of the ERs are different. In a previous study we reported the construction of a TIF2 mutant (hTIF2m123) that carries point mutations in all three NR boxes and is thus not capable of mediating the activity of AF2 of NRs (Voegel et al., 1998). To study the relative contribution of both ER interfaces to TIF2 activity we introduced deletions in the sequence encoding the Q-rich domain of either TIF2 wild-type (generating hTIF2ΔQ) or TIF2m123 (generating hTIF2m123ΔQ) cDNAs (illustrated in Figure 1). The immunoblot of extracts from transiently transfected COS-1 cells establishes that all these mutant proteins are expressed at similar levels to the wild-type homolog (Figure 1).
Fig. 1. Constructs used in this study. A schematic drawing of the major TIF2 and ERα constructs employed in this study is given. Note that domains are not depicted to scale. On the right a western blot confirming espression of the TIF2 mutant proteins in cDNA transfected cells at levels comparable to wild-type TIF2 is shown. The position of molecular weight markers is indicated on top of the 90° rotated image. Abbreviations used in this study are: hTIF2, human transcription intermediary factor 2 (Voegel et al., 1996); hTIF2m123, a mutant of TIF2 that contains alanine substitutions for critical leucines in all three LxxLL boxes (Voegel et al., 1998); hTIF2ΔQ, a deletion mutant of TIF2 lacking the Q-rich domain (this study); hTIF2m123ΔQ, a double mutant of TIF2 combining the LxxLL to LxxAA mutations with the Q-rich deletion mutant (this study); hERα, human estrogen receptor α; upper case letters denote the different domains of hERα present in the corresponding construct; in parentheses are the names of the different constructs used in previous studies. Note that the expression of hTIF2ΔQ is slightly higher compared with the other constructs (equal amounts of protein have been loaded for western blot analysis).
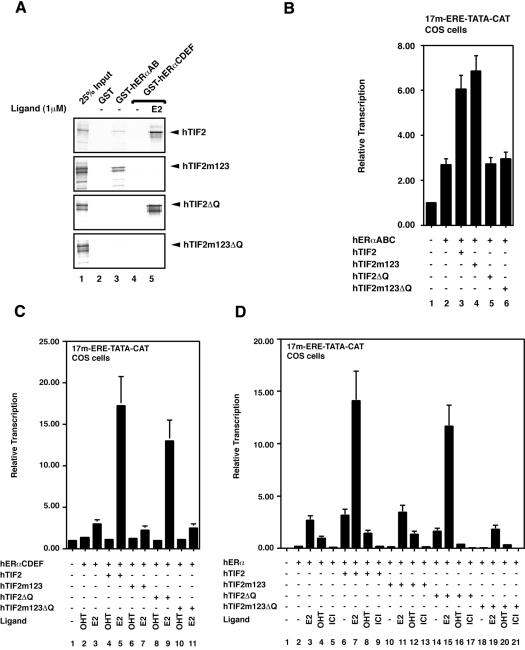
Glutathione S-transferase (GST)-based interaction assays demonstrate that the TIF2 protein lacking functional LxxLL motifs (hTIF2m123) has lost its ability to interact with the LBD harboring AF2 of ERα in a ligand-dependent manner (Figure 2A, lanes 4 and 5; Voegel et al., 1998); however, its binding to the AB region encompassing the AF1 is not affected (Figure 2A, lane 3). Inversely, hTIF2ΔQ is not able to interact with the AB domains that embody AF1 (Figure 2A, lane 3), but still binds the LBD in a ligand-dependent fashion (lanes 4 and 5). As expected, the double mutant hTIF2m123ΔQ is not able to interact with either AF. Next we analyzed the transcription properties of the different mutants on stimulating the two AFs of ERα in transient transfection experiments. To this end we transfected COS-1 cells with either expression vectors for the hERαABC domains (harboring only AF1; previously referred to as HE15; Figure 2B), hERαCDEF domains (only AF2; HEG19; Figure 2C) or the entire hERα (HEG0; Figure 2D), along with the expression vectors for TIF2 wild-type and mutant proteins. As reporter gene for monitoring the transcription activity of the different ER constructs we used an artificial minimal promoter containing binding sites for Gal4 and ER fused to the cDNA of chloramphenicol acetyltransferase (CAT; Berry et al., 1990). Use of such a promoter is advantageous as neither positive nor negative interference from other transcription factors contributes to the signal. Note, however, that due to the lack of additional core-promoter elements other than a TATA box it displays a relatively weak activity. As expected, the TIF2 mutant protein that is no longer able to interact with AF2 (hTIF2m123) still stimulates AF1 to the same extent as TIF2 wild type (Figure 2B, column 4), but fails to enhance the ligand-dependent AF2 (Figure 2C, column 7). The reverse was seen with TIF2ΔQ, which fails to stimulate AF1 (Figure 2B, column 5) but still enhances AF2 in a ligand-dependent manner (Figure 2C, column 9). The double mutant hTIF2m123ΔQ affects neither AF1 (Figure 2B, column 6) nor AF2 (Figure 2C, column 11) activity.
Fig. 2. TIF2 has two functionally independent ERα interfaces. (A) A GST interaction assay with hTIF2, hTIF2m123, hTIF2ΔQ, hTIF2m123ΔQ and purified GST–hERαAB (lane 3), as well as GST–hERαCDEF (lanes 4 and 5) in the absence and presence of estradiol (E2), is shown. The different cDNAs coding for wild-type and mutant TIF2 proteins were in vitro translated and proteins subsequently analyzed for their interaction pattern with the two ER fusion proteins. Scanned images of autoradiographs from dried gels were used to generate the figure. Note that equal loading of GST fusion proteins was confirmed by Coomassie staining (not shown). (B–D) Transient transfection experiments in COS-1 cells employing as a reporter the 17m-ERE-TATA-CAT construct, the different TIF2 cDNAs, and hERαABC (B), hERαCDEF (C) and the entire hERα (D) to reveal transcription stimulation by the different TIF2 mutants. CAT values were determined by ELISA and standardized with the aid of the activity of co-expressed β-galactosidase. Note that in (C) for the sake of clarity, values for the activity of hERαCDEF in the absence of ligand have been omitted. Hydroxy-tamoxifen (OHT) functions as complete antagonist on hERαCDEF, while being a partial agonist for hERα, which is completely blocked in its activity in the presence of ICI164-384 (ICI).
A similar picture emerges when studying the transactivation properties of full-length ERα with respect to the different TIF2 mutant proteins (Figure 2D). When analyzing hERαCDEF and hERα we included synthetic anti-estrogens as ligands in the transient transfection studies. Hydroxy-tamoxifen (OHT) is an AF2 antagonist but under certain circumstances allows hERα AF1 to be active, while ICI164-384 (ICI) antagonizes both AF1 and AF2 (Berry et al., 1990). Thus, the comparison between the reporter activities in OHT- and ICI-treated cells allows us to assess AF1 activity directly in the context of full-length hERα. Indeed, in the presence of OHT the AF1 interaction-disabled TIF2ΔQ is unable to stimulate ERα activity (Figure 2D, lane 16), while TIF2m123 stimulates weakly, but significantly, ERα/OHT activity (lane 12), as does TIF2 (lane 8). No stimulation was seen in the presence of ICI or by TIF2m123ΔQ (Figure 2D). Note that the stimulation of ERα AF1 by TIF2 is marginal under the present experimental conditions (COS-1 cells, minimal promoter, compare e.g. columns 4 and 8 in Figure 2D), and that the TIF2-dependent stimulation of transcription in the absence of ligand (column 6) is likely to be due to a combined effect of residual estradiol in the culture medium and activation through AF1 (compare the antagonist effects in columns 8 and 9). In accordance with this, TIF2ΔQ displays weaker activity in the absence of ligand than TIF2, as it fails to activate AF1 (Figure 2D, column 14). In conclusion, we have generated a set of mutant TIF2 proteins that selectively transactivate either AF1 (hTIF2m123) or AF2 (hTIF2ΔQ) of ERs. Furthermore, we have ruled out the existence of interfaces other than the two described, since the double mutant hTIF2m123ΔQ is completely inactive on both AFs.
TIF2 bridges AF1 and AF2
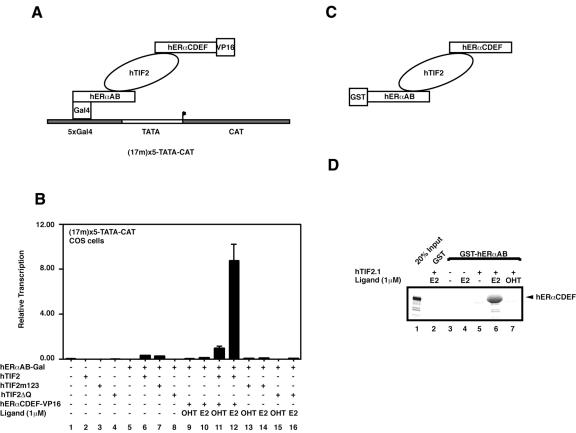
As we have established that the TIF2 binding surfaces of the two AFs of ERs operate autonomously (Figure 2), we speculated that TIF2 could be able to interact with both AFs simultaneously. To investigate the possible formation of such ternary complexes we performed in vivo bridged two-hybrid experiments. The rationale of the corresponding transient transfections (outlined in Figure 3A) is that the 17m5-TATA-CAT reporter will only display significant activity if the herpes simplex virus VP16 activation domain is recruited to the promoter, since Gal4-hERαAB is inactive on its own (Figure 3B, lane 3) and can be stimulated only weakly in this set-up (17m5-TATA-CAT, COS-1 cells) by TIF2 (lane 3). The fact that AF1 cannot stimulate transcription can be rationalized by similar arguments to those above. First, AF1 activity is cell specific and weakly active in COS-1 cells; and secondly, the absence of additional promoter and other transcription factor elements is obstructive to its activity (Berry et al., 1990). For the present experiment these conditions allow us to monitor directly the recruitment of the VP16 activation domain to the Gal4-responsive promoter by an increased transcription activity. As the ERα AB and CDEF domains do not interact under these conditions (Figure 3B, columns 9 and 10, and data not shown), such a recruitment can only occur if TIF2 binds concomitantly to both AF1 and AF2. Furthermore, transcription activation should be ligand dependent, as the binding of TIF2 or other CoAs to AF2 requires the NR LBD to be in the holo conformation. Figure 3B demonstrates that this is indeed the case. TIF2 and TIF2m123, but not TIF2ΔQ, stimulate the transcriptional activity of ERαAB to some extent (Figure 3B, compare columns 5–8). The activity of the reporter is greatly increased when ERαCDEF-VP16 is co-expressed with the AB region and wild-type TIF2 in the presence of estradiol (E2, Figure 3B, column 12), indicating that the liganded hERαCDEF-VP16 is recruited to the promoter via the Gal4-hERαAB-TIF2 bridge. As expected, this interaction depends on (i) the presence of TIF2 (Figure 3B, compare column 12 with 10); (ii) the presence of an ERα agonist (columns 11 and 12); and (iii) the integrity of both ER interaction motifs (LxxLL and the Q-rich domain) in TIF2 (compare column 12 with 14 and 16). Note that there is a statistically significant difference between columns 6 and 11, indicating that some hERαCDEF-VP16 is recruited to the Gal4-hERαAB by TIF2 even in the presence of OHT. We believe that the overexpression of TIF2 might to some extent shift the equilibrium of the LBD of ERα towards its active conformation even in the presence of an antagonist. Such effects have previously been observed under certain conditions (Voegel et al., 1998; our unpublished observation). Nevertheless, these data clearly demonstrate that TIF2 can bridge both AFs in vivo. To confirm these results and furthermore to rule out the possibility that other, as yet unidentified endogenous factors could contribute to this bridging phenomenon, we performed similar studies in vitro. Figure 3C gives the rationale of the experimental set-up, while Figure 3D shows the result of the interaction assay. Please note that due to the fact that hTIF2 cannot be efficiently produced in Escherichia coli we expressed the truncated hTIF2.1 (Figure 1; Voegel et al., 1996), which has the ability to interact with the two AFs of the ERs (our unpublished observation). Similar to the transfection experiment, recruitment of the 35S-labeled hERαCDEF to the GST-tagged hERαAB depends on both the presence of hTIF2.1 (Figure 3D, lanes 3 and 4) and an ER agonist (lanes 5 and 7 versus 6). Note that due to the fact that GST–hERαAB and [35S]hERαCDEF co-migrate on the polyacrylamide gel, the band in lane 6 appears broad and smeary. Together, the above results demonstrate that TIF2 is able to interact simultaneously with the two isolated AFs of ERα.
Fig. 3. TIF2 can bridge both ER AFs. (A) Set-up for the experiment shown in (B). Transient transfections similar to those in Figure 2 were performed. However, the reporter chosen [5× Gal4-binding sites in front of a TATA element directing expression of chloramphenicol acetyltransferase (17m5-TATA-CAT)] is inactive for Gal4-hERαAB. Again, activity of the system was compared in the presence or absence of TIF2 and its mutants, VP16-hERαCDEF and ligands as indicated underneath the columns. (C) Schematic for the set-up of the experiment shown in (D). A GST-based interaction assay similar to Figure 2A is shown. The retention of in vitro translated, 35S-labeled hERαCDEF on GST–hERαAB beads is determined in the absence and presence of TIF2.1 protein (see Figure 1) and ligands. An autoradiograph of a dried gel is shown.
TIF2 can mediate synergy between AF1 and AF2
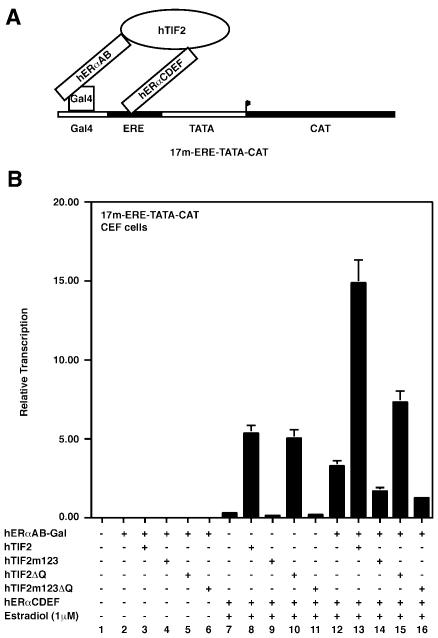
Previously, synergy between both AFs of ERα has been observed (Tasset et al., 1990; Kraus et al., 1995) but the molecular basis has remained elusive. Given the above results, we speculated that TIF2 could mediate such synergy as it is able to interact independently with the two corresponding domains (Figure 2) and can bridge both AFs (Figure 3). In order to investigate this possibility we made use of a hybrid reporter that contains one Gal4-binding site juxtaposed to an estrogen-responsive element (ERE), as well as a Gal4-hERαAB hybrid and the hERαCDEF construct. The rationale of these experiments is outlined in Figure 4A. Note that for simplicity of illustration only one molecule of hERαCDEF is depicted, whereas efficient binding to the canonical palindromic ER ERE involves ER homodimers. When such a system is implemented in chicken embryo fibroblast (CEF) cells, AF1 displays almost undetectable activity (Figure 4B, column 2), in agreement with previous studies (minimal promoter). AF2 is slightly more active in the presence of estradiol (Figure 4B, column 7; Berry et al., 1990); however, when expressed together, AF1 and AF2 strongly cooperate (Figure 4B, column 12). Together, the above conditions are optimal for the analysis of the synergistic effects between ERα AF1 and AF2 in the presence of TIF2 and confirm that the aspect of synergy can also be studied efficiently in another cellular system. In agreement with the data presented in Figure 2, both the TIF2 wild type and the TIF2ΔQ mutants significantly stimulate the transcriptional activity of the isolated AF2 also in CEF cells (Figure 4B, columns 8 and 10), whereas both TIF2m123 and TIF2m123ΔQ fail to do so (due to the absence of functional LxxLL motifs, columns 9 and 11). The activities exerted on AF1 are due to the low activity of AF1 in this system, which is not quantifiable (Figure 4B, columns 2–6). Importantly, when TIF2 wild-type cDNA is co-transfected with both the Gal4-hERαAB and the hERαCDEF constructs, a very strong stimulation of reporter activity is observed in the presence of estradiol (Figure 4B, column 13). This activity is significantly higher than the added activities in the presence of estradiol and either the AF2 interaction-deficient mutant hTIF2m123 (Figure 4B, column 14) or the AF1 interaction-deficient mutant hTIF2ΔQ (column 15). Furthermore, the overall activity of the system in the presence of TIF2 (Figure 4A, column 13) is also higher than the added activities of Gal4-hERαAB and hERαCDEF in the presence of TIF2 (columns 3 and 8). In conclusion, under these experimental conditions both AFs contribute to reporter gene activity in a synergistic fashion only when bridged by TIF2. The observed synergy between AF1 and AF2 in the absence of TIF2 (Figure 4B, column 12) is most likely due to endogenous p160 CoAs.
Fig. 4. TIF2 can mediate synergy between AF1 and AF2. (A) Experimental set-up for transient transfections in CEF cells is shown. (B) Actual experiment using indicated reporter and expression vectors, similar to Figure 2B–D.
In summary we demonstrate here that binding and transactivation requirements for TIF2 to both AFs are independent, as suggested by earlier studies, and can be separated. In keeping with this, we demonstrate that a TIF2 mutant protein with disabled LxxLL NR boxes (AF2-binding motifs) and Q-rich AF1-binding regions (hTIF2m123ΔQ) does not display any CoA activity for the ER anymore. Hence TIF2 does not possess any additional ERα-binding interfaces. ERα had previously been reported to exert synergy between both AFs (Tasset et al., 1990; Kraus et al., 1995), implying a functional link between the two transcription AFs AF1 and AF2. Briefly, our data establish that TIF2, and most likely related CoAs from the p160 family, through concomitant interaction with both AFs, can mediate the synergistic effects between ERα AF1 and AF2 on estrogen-induced target gene transactivation.
These results may have an impact on the rationale of ligand design for the treatment of NR-linked diseases. For example, OHT is widely used in the endocrine therapy of breast cancer, but frequently patients become resistant to the therapy, an effect that correlates with a loss of tamoxifen binding efficiency in the tumor cells. It is possible that enhanced expression of CoAs or recruitment of TIF2-like CoAs to AF1 is at the base of this decline in potential, and it will therefore be of great importance to understand the structural details of CoA–ER AF1 interaction, which may lead to the development of AF1 inhibitors.
METHODS
Plasmid constructions. Plasmid constructions were carried out according to standard protocols. All constructs with PCR-amplified fragments were sequenced. Details are available upon request.
Protein purification. Recombinant pGEX-based bacterial expression vectors were propagated in E. coli strains BL21 or XL1-blue. Expression of recombinant proteins was induced by treatment of exponential cultures with 0.5 mM isopropyl-β-d-thiogalactopyranoside for 2–3 h at ambient temperature. Cells were harvested, resuspended in bacterial lysis buffer [50 mM Tris pH 8.0, 100 mM KCl, 0.1 mM dithiothreitol (DTT)] containing 100 mg/ml lysozyme and proteinase inhibitors, incubated on ice for 30 min followed by sonication and centrifugation at 30 000 r.p.m. for 30 min. GST fusion proteins were purified by binding to glutathione–Sepharose beads according to the manufacturer’s recommendations (Pharmacia).
GST-based interaction assay. Glutathione–Sepharose beads were incubated for 4 h with bacterial extracts containing GST alone or GST fusion proteins, and subsequently washed four times with GST buffer (50 mM Tris–HCl pH 7.9, 150 mM NaCl, 5% glycerol, 0.1% NP-40, 1 mM EDTA, 1 mM DTT). For interaction assays, loaded beads were incubated with 5 µl of rabbit reticulocyte lysate containing translated protein radiolabeled with [35S]methionine (coupled transcription/translation Kit; Promega). The beads were incubated together with the proteins in a total volume of 100 µl of GST buffer for 30 min at ambient temperature. Where appropriate, the indicated ligands were added at a concentration of 10–6 M. After three to five washes with GST buffer to remove unbound material, beads were resuspended in a suitable volume of 3× SDS loading buffer and subjected to denaturing SDS–PAGE. Samples were analyzed by Coomassie staining or autoradiography of dried gels.
Transient transfections. COS-1 cells were seeded into six-well cell culture plates at 2 × 106 cells/plate in Dulbecco’s minimal essential medium supplemented with 5% fetal calf serum and antibiotics. Calcium phosphate precipitates containing 3 µg of total DNA were immediately added to the cells. After 12 h cells were washed and supplemented with fresh media. Following an additional 24 h of incubation, the cell layer was washed twice with cold phosphate-buffered saline and cells were collected and resuspended in lysis buffer (10 mM MOPS pH 6.5, 10 mM NaCl, 1 mM EGTA, 1% Triton X-100). Cellular debris was removed by centrifugation, and resulting whole cell extracts were analyzed by CAT-ELISA according to the recommendations provided (Boehringer Mannheim). In parallel, the activity of co-expressed β-galactosidase was determined according to standard procedures (Voegel et al., 1998) and used to correct for variations of the transfection efficiencies.
Immunoblots were performed according to standard procedures using a polyclonal antiserum directed against the GRIP1/TIF2 N-terminal region (kindly provided by M. Gehin).
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Astrid Pornon for help with transient transfections and Martine Gehin for the polyclonal rabbit serum against GRIP1. Work at IGBMC is supported by the CNRS, INSERM, HUS, the Collège de France and Bristol-Myers Squibb. A.B. was a recipient of Marie-Curie longterm fellowship ERBFMBICT961269 from the European Commission.
REFERENCES
- Alen P., Claessens, F., Verhoeven, G., Rombauts, W. and Peeters, B. (1999) The androgen receptor amino-terminal domain plays a key role in p160 coactivator-stimulated gene transcription. Mol. Cell. Biol., 19, 6085–6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almlof T., Wallberg, A.E., Gustafsson, J.A. and Wright, A.P. (1998) Role of important hydrophobic amino acids in the interaction between the glucocorticoid receptor τ1-core activation domain and target factors. Biochemistry, 37, 9586–9594. [DOI] [PubMed] [Google Scholar]
- Berry M., Metzger, D. and Chambon, P. (1990) Role of the two activating domains of the oestrogen receptor in the cell-type and promoter-context dependent agonistic activity of the anti-oestrogen 4-hydroxytamoxifen. EMBO J., 9, 2811–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba N., Suldan, Z., Freedman, L.P. and Parvin, J.D. (2000) Binding of liganded vitamin D receptor to the vitamin D receptor interacting protein coactivator complex induces interaction with RNA polymerase II holoenzyme. J. Biol. Chem., 275, 10719–10722. [DOI] [PubMed] [Google Scholar]
- Darimont B.D., Wagner, R.L., Apriletti, J.W., Stallcup, M.R., Kushner, P.J., Baxter, J.D., Fletterick, R.J. and Yamamoto, K.R. (1998) Structure and specificity of nuclear receptor–coactivator interactions. Genes Dev., 12, 3343–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh H. et al. (1999) Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor α. Mol. Cell. Biol., 19, 5363–5372. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gelman L., Zhou, G., Fajas, L., Raspe, E., Fruchart, J.C. and Auwerx, J. (1999) p300 interacts with the N- and C-terminal part of PPARγ2 in a ligand-independent and -dependent manner, respectively. J. Biol. Chem., 274, 7681–7688. [DOI] [PubMed] [Google Scholar]
- Glass C.K. and Rosenfeld, M.G. (2000) The coregulator exchange in transcriptional functions. Genes Dev., 14, 121–141. [PubMed] [Google Scholar]
- Hammer G.D., Krylova, I., Zhang, Y., Darimont, B.D., Simpson, K., Weigel, N.L. and Ingraham, H.A. (1999) Phosphorylation of the nuclear receptor SF-1 modulates cofactor recruitment: integration of hormone signaling in reproduction and stress. Mol. Cell, 3, 521–526. [DOI] [PubMed] [Google Scholar]
- He B., Kemppainen, J.A., Voegel, J.J., Gronemeyer, H. and Wilson, E.M. (1999) A novel role for nuclear receptor activation function 2 in the human androgen receptor. J. Biol. Chem., 274, 37219–37225. [DOI] [PubMed] [Google Scholar]
- Hittelman A.B., Burakov, D., Iniguez-Lluhi, J.A., Freedman, L.P. and Garabedian, M.J. (1999) Differential regulation of glucocorticoid receptor transcriptional activation via AF-1 associated proteins. EMBO J., 18, 5380–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao P.W. and Chang, C. (1999) Isolation and characterization of ARA160 as the first androgen receptor N-terminal-associated coactivator in human prostate cells. J. Biol. Chem., 274, 22373–22379. [DOI] [PubMed] [Google Scholar]
- Hu X. and Lazar, M.A. (1999) The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature, 402, 93–96. [DOI] [PubMed] [Google Scholar]
- Ito M. et al. (1999) Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell, 3, 361–370. [DOI] [PubMed] [Google Scholar]
- Kraus W.L., McInerney, E.M. and Katzenellenbogen, B.S. (1995) Ligand-dependent, transcriptionally productive association of the amino- and carboxyl-terminal regions of a steroid hormone nuclear receptor. Proc. Natl Acad. Sci. USA, 92, 12314–12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin B., Nielsen, A.L., Garnier, J.M., Ichinose, H., Jeanmougin, F., Losson, R. and Chambon, P. (1996) A possible involvement of TIF1α and TIF1β in the epigenetic control of transcription by nuclear receptors. EMBO J., 15, 6701–6715. [PMC free article] [PubMed] [Google Scholar]
- McInerney E.M. et al. (1998) Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev., 12, 3357–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna N.J., Lanz, R.B. and O’Malley, B.W. (1999) Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev., 20, 321–344. [DOI] [PubMed] [Google Scholar]
- Minucci S. and Pelicci, P.G. (1999) Retinoid receptors in health and disease: co-regulators and the chromatin connection. Semin. Cell Dev. Biol., 10, 215–225. [DOI] [PubMed] [Google Scholar]
- Moras D. and Gronemeyer, H. (1998) The nuclear receptor ligand-binding domain: structure and function. Curr. Opin. Cell Biol., 10, 384–391. [DOI] [PubMed] [Google Scholar]
- Nagy L. et al. (1999) Mechanisms of corepressor binding and release from nuclear hormone receptors. Genes Dev., 13, 3209–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte R.T. et al. (1998) Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature, 395, 137–143. [DOI] [PubMed] [Google Scholar]
- Perissi V. et al. (1999) Molecular determinants of nuclear receptor–corepressor interaction. Genes Dev., 13, 3198–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachez C., Lemon, B.D., Suldan, Z., Bromleigh, V., Gamble, M., Naar, A.M., Erdjument-Bromage, H., Tempst, P. and Freedman, L.P. (1999) Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature, 398, 824–828. [DOI] [PubMed] [Google Scholar]
- Tasset D., Tora, L., Fromental, C., Scheer, E. and Chambon, P. (1990) Distinct classes of transcriptional activating domains function by different mechanisms. Cell, 62, 1177–1187. [DOI] [PubMed] [Google Scholar]
- Tremblay A., Tremblay, G.B., Labrie, F. and Giguere, V. (1999) Ligand-independent recruitment of SRC-1 to estrogen receptor β through phosphorylation of activation function AF-1. Mol. Cell, 3, 513–519. [DOI] [PubMed] [Google Scholar]
- Voegel J.J., Heine, M.J., Zechel, C., Chambon, P. and Gronemeyer, H. (1996) TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J., 15, 3667–3675. [PMC free article] [PubMed] [Google Scholar]
- Voegel J.J., Heine, M.J., Tini, M., Vivat, V., Chambon, P. and Gronemeyer, H. (1998) The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J., 17, 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb P. et al. (1998) Estrogen receptor activation function 1 works by binding p160 coactivator proteins. Mol. Endocrinol., 12, 1605–1618. [DOI] [PubMed] [Google Scholar]