Abstract
It is widely accepted that the large trinucleotide repeat expansions observed in many neurological diseases occur during replication. However, genetic recombination has emerged as a major source of instability for tandem repeats, including minisatellites, and recent studies raise the possibility that it may also be responsible for trinucleotide repeat expansions. We will review data connecting tandem repeat rearrangements and recombination in humans and in eukaryotic model organisms, and discuss the possible role of recombination in trinucleotide repeat expansions in human neurological disorders.
Introduction
Two classes of tandem repeat sequences, minisatellites and microsatellites, have gained increasing attention from the scientific community over the past decade. Microsatellites are tandem arrays of short (usually <10 bp) units, while minisatellites are tandem arrays of longer units (>10 and <100 bp). These two kinds of sequence are widespread in all eukaryotic genomes (reviewed in Charlesworth et al., 1994) from yeast to mammals. A growing number of neurological disorders have been found to result from the expansion of a particular class of microsatellites, called trinucleotide repeats (reviewed in Cummings and Zoghbi, 2000).
In contrast to other forms of microsatellite instability such as that found in mismatch repair (MMR) mutants, trinucleotide repeat expansions observed in neurological diseases are often quite large (several times the initial number of repeats). The occurrence of these expansions during the lifetime is not precisely known; they might be meiotic, pre- or post-zygotic. Trinucleotide repeats are not the only class of tandem repeats prone to expansions. Three minisatellites are known to share this property. Expansion of one of them is associated with a human disease (Lalioti et al., 1997). The others, two AT-rich repeats, are responsible for fragile sites (Yu et al., 1997; Hewett et al., 1998).
Two types of mechanism have been proposed to be involved in tandem repeat instability: replication and recombination. The genome-wide microsatellite instability observed in MMR mutant cells (Simpkins et al., 1999 and references therein) strongly supports a replicative mechanism, and the somatic instability of trinucleotide repeats is generally viewed as a consequence of replication (reviewed in McMurray, 1999; Richard et al., 1999a). However, several recent results argue that recombination might also play a role in microsatellite expansions. The aim of this paper is to review the data connecting tandem repeat rearrangements with recombination, and to discuss the possible involvement of recombination in trinucleotide repeat expansions.
Gene conversion as a major source of tandem repeat instability
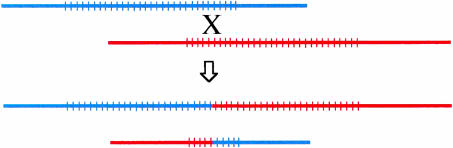
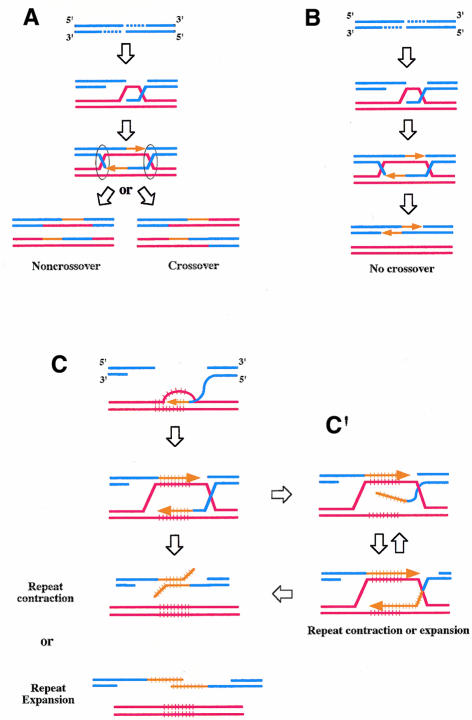
Initially, when people were thinking of tandem repeat rearrangements in terms of recombination, they were thinking of crossovers (Figure 1), i.e. a reciprocal transfer of genetic information. However, gene conversion (non-reciprocal transfer of information) has recently emerged as a major cause of tandem repeat instability. Gene conversion and crossover are often but not always associated, and a series of models, culminating in the model proposed by Szostak et al. (1983) (see Figure 2A), summarizes this relationship. However, it is mostly gene conversion without crossover that destabilizes tandem repeats.
Fig. 1. Tandem repeat rearrangement by unequal crossover between sister chromatids. Reciprocal exchange leads to simultaneous contraction of one of the repeats and expansion of the other.
Fig. 2. Recombination models. In all models, gene conversion is initiated by a DSB, and strand invasion of the homologous template is initiated by a resected 3′ end. Arrows represent DNA synthesis associated with DSB-repair. The broken molecule (‘donor’) is shown in blue, the template molecule (‘recipient’) is shown in red, newly synthesized DNA is shown in orange. Tandem repeats are represented by hatched lines. (A) Szostak et al. (1983) model. Resolution of the two four-stranded structures called ‘Holliday junctions’ (circled in black) results in gene conversion associated with crossovers in 50% of the cases. (B) SDSA model. Resolution is achieved by unwinding the two newly synthesized strands from the template and annealing them. No crossover is expected. (C) SDSA model of tandem repeat rearrangements. The newly synthesized strands both contain repeats and their out-of-frame annealing can lead to contraction or expansion of the tandem repeat. (C′) Alternatively, contraction and expansion can result from unwinding and out-of-frame re-invasion of one (or both) the newly synthesized strand(s). Only one kind of event (expansion) is shown here. Several rounds of unwinding/re-invasion could take place during a single gene conversion, leading to successive rounds of DNA synthesis.
Welch et al. (1990) were the first to establish a correlation between non-crossover gene conversion and tandem repeat rearrangements, when they observed meiotic expansions and contractions of the yeast CUP1 locus. Tandem repeat rearrangements that were likely to have resulted from gene conversion were also observed in Drosophila (Pâques and Wegnez, 1993; Thompson-Stewart et al., 1994; Delattre et al., 1995; Pâques et al., 1996) and provided the basis for a new model for tandem repeat rearrangements. These events were thought to result from the excision of a Drosophila P transposon. Such excisions are repaired by gene conversion, which is not usually associated with crossovers. A second series of models, termed SDSA, for synthesis-dependent strand annealing (reviewed in Pâques and Haber, 1999), accounts for non-crossover gene conversions. In SDSA models, the two newly synthesized strands unwind from their template and anneal with each other (Figure 2B). Pâques and Wegnez (1993) postulated that out-of-frame re-annealing of tandem repeats would explain the observed rearrangements detected after excision of a P transposon (Figure 2C).
Meiotic recombination in human minisatellites
Since the discovery of the first minisatellites in the human genome (Jeffreys et al., 1985), mechanisms leading to high spontaneous mutation rates of these hypervariable DNA sequences have been thoroughly studied. Jeffreys et al. (1988) proposed that small mutations involving gain or loss of 4–10 repeats could occur by a mitotic replication slippage mechanism, while large mutation events involving gain or loss of up to 200 repeats were more compatible with a meiotic recombinational process.
In further analyses, minisatellite mutations found in the germline were precisely mapped (Buard and Vergnaud, 1994; Jeffreys et al., 1994), and typical features of the mutation process were observed. First, there was a polarity in the mutational process, i.e. repeats were gained or lost preferentially at one end of the repeat array. Secondly, complex rearrangement events suggested that the new mutant allele gained genetic information from both the sister chromatid (intra-allelic event) and the homologous chromosome (inter-allelic event). Thirdly, exchange of flanking markers was rare, indicating that recombination occurred with few associated crossovers.
Again, these results were explained in terms of SDSA (Buard and Jeffreys, 1997). This mechanism provided a simple explanation for the complex events observed: both templates were used by the two ends of a double-strand break (DSB) to copy information, and the two newly synthesized strands were assembled in the recipient molecule by annealing. SDSA could also explain the low crossover rate (Figure 2B). Related SDSA models, where DNA synthesis occurred at only one end of the DSB, may also account for other features of minisatellite rearrangements observed in yeast or humans (Pâques et al., 1998; Debrauwère et al., 1999).
By mapping these rare crossovers in the vicinity of the MS32 minisatellite, Jeffreys et al. (1998) found a meiotic hot spot (a chromosomal region where meiotic recombination is frequently observed) centered ∼200 bp upstream of MS32 and extending into the beginning of the repeat array. Their work strongly suggests that hypervariability arises in proximity to a meiotic hot spot. Their finding also explains mutational polarity, since meiotic DSBs are initiated on one side of the minisatellite and recombination propagates within the repeat array from the initiation point. The gene conversion gradient decreases from the initiation point, as observed for meiotic hot spots in yeast (Nicolas et al., 1989).
Analysis of somatic mutations at MS32 revealed simple deletions or duplications, with no detectable bias toward one extremity of the repeat array (Jeffreys and Neumann, 1997). The authors thus concluded that somatic mutational processes involved in MS32 expansions and contractions were different from those found in the germline.
The introduction of two minisatellites, MS32 and CEB1, into yeast confirmed that they are more unstable during meiosis than during mitosis. Gains and losses of repeats were again complex events involving both inter- and intra-allelic recombination (Appelgren et al., 1997; Debrauwère et al., 1999). In addition, it was shown that CEB1 meiotic instability depended on DSB formation at two well characterized meiotic hot spots, one of which lies upstream and the other downstream of the repeat array. Interestingly, instability disappeared when CEB1 was moved to a chromosomal region where no meiotic DSB could be detected.
Gene conversion-induced rearrangements of tandem repeats in yeast
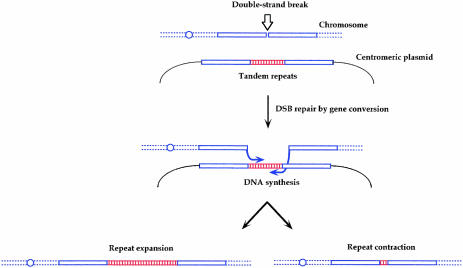
Recently, Pâques et al. (1998) designed an experimental system to study the effect of mitotic gene conversion on tandem repeat instability in Saccharomyces cerevisiae. A DSB was made on a yeast chromosome, which could be repaired using a homologous sequence containing a tandem repeat as donor (Figure 3). Both donor and recipient were recovered and analyzed for tandem repeat rearrangements.
Fig. 3. Experimental system used to study tandem repeat instability during mitotic gene conversion induced by an endonuclease in yeast. After induction of a DSB on a yeast chromosome by an inducible endonuclease, yeast cells can repair the broken molecule using a tandem repeat-containing homologous donor sequence. During DNA synthesis associated with DSB-repair, slippage of the newly synthesized strand(s) may occur, giving rise to contractions or expansions of the tandem repeat in the recipient molecule. The blue circle represents the chromosome centromere.
Different kinds of tandem repeat were studied including 5S rRNA genes, trinucleotide repeats and minisatellites (Pâques et al., 1998; Richard et al., 1999b, 2000; F. Pâques, G.-F. Richard and J.E. Haber, unpublished results), all of which rearranged with a high frequency during gene conversion (Table I). Furthermore, rearrangements were observed mainly in the recipient molecule, whereas the donor was extremely stable, as predicted by SDSA models (Figure 2C). Interestingly, the CAG39 repeat (i.e. an array of 39 CAG triplets) yielded only contractions, whereas both contractions and expansions were observed with CAG98 (Table I).
Table I. Rearrangements of tandem repeat sequences during gene conversion.
| Faithful gene conversion (%) | Gene conversion with contraction (%) | Gene conversion with expansion (%) | Total (contractions + expansions) (%) | |
|---|---|---|---|---|
| 5S rDNA genes | 63 | 23 | 14 | 37 |
| CAG39 | 84 | 16 | – | 16 |
| CAG98 |
57 |
30 |
13 |
43 |
| CAA87 | 90 | 10 | – | 10 |
These results demonstrate the destabilizing effect of gene conversion on different kinds of tandem repeat. The model proposed in Figure 2C postulates that, during gene conversion-associated DNA synthesis, slippage of the newly synthesized strands can occur, leading to addition or subtraction of a whole number of repeats. The frequency of such ‘repair-slippage’ was estimated to be 800-fold higher than that for S-phase replication-slippage (Richard et al., 1999b).
Is DSB-repair involved in trinucleotide repeat expansions?
Trinucleotide repeats and some minisatellites are fragile sites in human (reviewed in Sutherland et al., 1998) and in yeast (Freudenreich et al., 1998). Fragile sites are loci at which chromosome breakage occurs in the presence of specific chemicals. We suggest that a mitotic or meiotic DSB could initiate the expansion process. Meiotic instability of CAG repeats in yeast has been correlated with formation of a nearby DSB in one study using long CAG tracts (Jankowski et al., 2000), but not in another using shorter CAG tracts (Moore et al., 1999). White et al. (1999) showed that CCG repeats did not exhibit a higher rate of instability during meiosis than during vegetative growth in yeast, whereas Cohen et al. (1999) found that instability of CTG repeats carried by YACs was higher during meiosis than during mitotic divisions. A very recent study showed that CA dinucleotide repeats were also destabilized during meiosis, in yeast, and that progression of strand exchange during gene conversion was inhibited by the microsatellite (Gendrel et al., 2000). In transgenic mice, CAG repeats exhibited a higher instability in the germline than in somatic cells, with a propensity towards expansion rather than contraction (Seznec et al., 2000).
In human diseases, it is common to observe expansions of more than twice the original size. Such large expansions in yeast and humans could occur by successive rounds of unwinding/re-invasion of the donor sequence by the newly synthesized strand, allowing DNA synthesis to proceed more than once within the repeats (Pâques et al., 1998; Figure 2C′).
An interesting case of large contraction of a CAG repeat was described during transmission of a myotonic dystrophy allele. O’Hoy et al. (1993) reported a large reduction of the number of CAG triplets associated with what they called a ‘discontinuous gene conversion event’. The resulting allele was a patchwork of both maternal and paternal alleles. Buard and Vergnaud (1994), Debrauwère et al. (1999) and Tamaki et al. (1999) also found complex recombination events in minisatellites. In those cases, gene conversion was inter-allelic (between homologous chromosomes), intra-allelic (between sister chromatids) or a patchwork of both. All these observations are compatible with the unwinding/re-invasion model (Figure 2C′).
Perspectives
Even though more conclusive evidence will be required to account for dramatic trinucleotide repeat expansion by a gene conversion mechanism in humans, recent data suggest that recombination can lead to large scale contractions and expansions within tandem repeat sequences at a high frequency. Experimental systems can be designed to study the effect of mitotic and meiotic recombination on tandem repeat instability. It will be particularly interesting to determine whether any kind of sequence can be expanded during gene conversion, or whether there is a sequence (or structure) specificity to this process. Moreover, even though some trans-acting factors have been shown to be involved in expansions during gene conversion (Richard et al., 2000), their precise function needs to be determined. Hopefully, further experiments in model organisms will allow us to unravel the molecular basis of tandem repeat complex rearrangements during gene conversion, and perhaps help us to understand how dramatic trinucleotide repeat expansions occur in humans.
Acknowledgments
Acknowledgements
We would like to thank Allyson Holmes and members of the Unité de Génétique Moléculaire des Levures for insightful suggestions. G.-F.R. is the recipient of a post-doctoral fellowship from the Institut Pasteur.
References
- Appelgren H., Cederberg, H. and Rannug, U. (1997) Mutations at the human minisatellite MS32 integrated in yeast occur with high frequency in meiosis and involve complex recombination events. Mol. Gen. Genet., 256, 7–17. [DOI] [PubMed] [Google Scholar]
- Buard J. and Jeffreys, A.J. (1997) Big, bad minisatellites. Nature Genet., 15, 327–328. [DOI] [PubMed] [Google Scholar]
- Buard J. and Vergnaud, G. (1994) Complex recombination events at the hypermutable minisatellite CEB1 (D2S90). EMBO J., 13, 3203–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Sniegowski, P. and Stephan, W. (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature, 371, 215–220. [DOI] [PubMed] [Google Scholar]
- Cohen H., Sears, D.D., Zenvirth, D., Hieter, P. and Simchen, G. (1999) Increased instability of human CTG repeat tracts on yeast artificial chromosomes during gametogenesis. Mol. Cell. Biol., 19, 4153–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings C.J. and Zoghbi, H.Y. (2000) Fourteen and counting: unraveling trinucleotide repeat diseases. Hum. Mol. Genet., 9, 909–916. [DOI] [PubMed] [Google Scholar]
- Debrauwère H., Buard, J., Tessier, J., Aubert, D., Vergnaud, G. and Nicolas, A. (1999) Meiotic instability of human minisatellite CEB1 in yeast requires double-strand breaks. Nature Genet., 23, 367–371. [DOI] [PubMed] [Google Scholar]
- Delattre M., Anxolabehere, D. and Coen, D. (1995) Prevalence of localized rearrangements vs. transpositions among events induced by Drosophila P element transposase on a P transgene. Genetics, 141, 1407–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenreich C.H., Kantrow, S.M. and Zakian, V.A. (1998) Expansion and length-dependent fragility of CTG repeats in yeast. Science, 279, 853–856. [DOI] [PubMed] [Google Scholar]
- Gendrel C.-G., Boulet, A. and Dutreix, M. (2000) (CA/GT)n microsatellites affect homologous recombination during yeast meiosis. Genes Dev., 14, 1261–1268. [PMC free article] [PubMed] [Google Scholar]
- Hewett D.R. et al. (1998) FRA10B structure reveals common elements in repeat expansion and chromosomal fragile site genesis. Mol. Cell, 1, 773–781. [DOI] [PubMed] [Google Scholar]
- Jankowski C., Nasar, F. and Nag, D.K. (2000) Meiotic instability of CAG repeat tracts occurs by double-strand break repair in yeast. Proc. Natl Acad. Sci. USA, 97, 2134–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A.J. and Neumann, R. (1997) Somatic mutation processes at a human minisatellite. Hum. Mol. Genet., 6, 129–136. [DOI] [PubMed] [Google Scholar]
- Jeffreys A.J., Wilson, V. and Thein, S.L. (1985) Hypervariable ‘minisatellite’ regions in human DNA. Nature, 314, 67–73. [DOI] [PubMed] [Google Scholar]
- Jeffreys A.J., Royle, N.J., Wilson, V. and Wong, Z. (1988) Spontaneous mutation rates to new length alleles at tandem-repetitive hypervariable loci in human DNA. Nature, 332, 278–281. [DOI] [PubMed] [Google Scholar]
- Jeffreys A.J., Tamaki, K., McLeod, A., Monckton, D.G., Neil, D.L. and Armour, J.A.L. (1994) Complex gene conversion events in germline mutation at human minisatellites. Nature Genet., 6, 136–145. [DOI] [PubMed] [Google Scholar]
- Jeffreys A.J., Murray, J. and Neumann, R. (1998) High-resolution mapping of crossovers in human sperm defines a minisatellite-associated recombination hot spot. Mol. Cell, 2, 267–273. [DOI] [PubMed] [Google Scholar]
- Lalioti M.D., Scott, H.S., Buresi, C., Rossier, C., Bottani, A., Morris, M.A., Malafosse, A. and Antonarakis, S.E. (1997) Dodecamer repeat expansion in cystatin B gene in progressive myoclonus epilepsy. Nature, 386, 847–851. [DOI] [PubMed] [Google Scholar]
- McMurray C.T. (1999) DNA secondary structure: a common and causative factor for expansion in human disease. Proc. Natl Acad. Sci. USA, 96, 1823–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore H., Greenwell, P.W., Liu, C.-P., Arnheim, N. and Petes, T.D. (1999) Triplet repeats form secondary structures that escape DNA repair in yeast. Proc. Natl Acad. Sci. USA, 96, 1504–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas A., Treco, D., Schultes, N.P. and Szostak, J.W. (1989) An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature, 338, 35–39. [DOI] [PubMed] [Google Scholar]
- O’Hoy K.L., Tsilfidis, C., Mahadevan, M.S., Neville, C.E., Barcelo, J., Hunter, A.G. and Korneluk, R.G. (1993) Reduction in size of the myotonic dystrophy trinucleotide repeat mutation during transmission. Science, 259, 809–812. [DOI] [PubMed] [Google Scholar]
- Pâques F. and Haber, J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F. and Wegnez, M. (1993) Deletions and amplifications of tandemly arranged ribosomal 5S genes internal to a P element occur at a high rate in a dysgenic context. Genetics, 135, 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F., Bucheton, B. and Wegnez, M. (1996) Rearrangements involving repeated sequences within a P element preferentially occur between units close to the transposon extremities. Genetics, 142, 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F., Leung, W.-Y. and Haber, J.E. (1998) Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol. Cell. Biol., 18, 2045–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard G.-F., Hennequin, C., Thierry, A. and Dujon, B. (1999a) Trinucleotide repeats and other microsatellites in yeasts. Res. Microbiol., 150, 589–602. [DOI] [PubMed] [Google Scholar]
- Richard G.-F., Dujon, B. and Haber, J. (1999b) High frequency of rearrangements of short CAG/CTG trinucleotide repeats in yeast induced by double-strand break repair. Mol. Gen. Genet., 261, 871–882. [DOI] [PubMed] [Google Scholar]
- Richard G.-F., Goellner, G.M., McMurray, C.T. and Haber, J.E. (2000) Recombination-induced CAG trinucleotide repeat expansions in yeast involve the MRE11/RAD50/XRS2 complex. EMBO J., 19, 2381–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seznec H., Lia-Baldini, A.-S., Duros, C., Fouquet, C., Lacroix, C., Hofmann-Radvanyi, H., Junien, C. and Gourdon, G. (2000) Transgenic mice carrying large human genomic sequences with expanded CTG repeat mimic closely the DM CTG repeat intergenerational and somatic instability. Hum. Mol. Genet., 9, 1185–1194. [DOI] [PubMed] [Google Scholar]
- Simpkins S.B., Bocker, T., Swisher, E.M., Mutch, D.G., Gersell, D.J., Kovatich, J.P., Fishel, R. and Goodfellow, P.J. (1999) MLH1 promoter methylation and gene silencing is the primary cause of microsatellite instability in sporadic endometrial cancers. Hum. Mol. Genet., 8, 661–666. [DOI] [PubMed] [Google Scholar]
- Sutherland G.R., Baker, E. and Richards, R.I. (1998) Fragile sites still breaking. Trends Genet., 14, 501–506. [DOI] [PubMed] [Google Scholar]
- Szostak J.W., Orr-Weaver, T.L., Rothstein, R.J. and Stahl, F.W. (1983) The double-strand break repair model for recombination. Cell, 33, 25–35. [DOI] [PubMed] [Google Scholar]
- Tamaki K., May, C.A., Dubrova, Y.E. and Jeffreys, A.J. (1999) Extremely complex repeat shuffling during germline mutation at human minisatellite B6.7. Hum. Mol. Genet., 8, 879–888. [DOI] [PubMed] [Google Scholar]
- Thompson-Stewart D., Karpen, G.H. and Spradling, A.C. (1994) A transposable element can drive the concerted evolution of tandemly repetitious DNA. Proc. Natl Acad. Sci. USA, 91, 9042–9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch J.W., Maloney, D.H. and Fogel, S. (1990) Unequal crossing-over and gene conversion at the amplified CUP1 locus of yeast. Mol. Gen. Genet., 222, 304–310. [DOI] [PubMed] [Google Scholar]
- White P.J., Borts, R.H. and Hirst, M.C. (1999) Stability of the human Fragile X (CCG)n triplet repeat array in Saccharomyces cerevisiae deficient in aspects of DNA metabolism. Mol. Cell. Biol., 19, 5675–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S. et al. (1997) Human chromosomal fragile site FRA16B is an amplified AT-rich minisatellite repeat. Cell, 88, 367–374. [DOI] [PubMed] [Google Scholar]