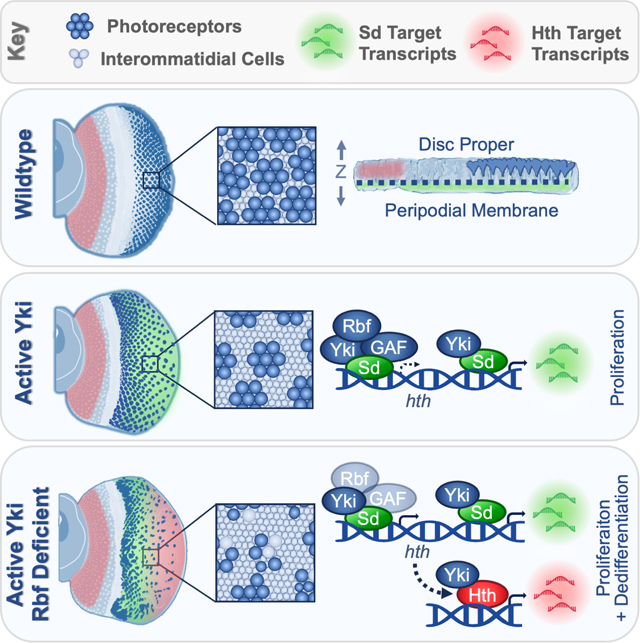
SUMMARY
The RB and Hippo pathways interact to regulate cell proliferation and differentiation. However, the mechanism of interaction is not fully understood. Drosophila photoreceptors with inactivated RB and Hippo pathways specify normally but fail to maintain their neuronal identity and dedifferentiate. We performed single-cell RNA-sequencing to elucidate the cause of dedifferentiation and to determine the fate of these cells. We find that dedifferentiated cells adopt a progenitor-like fate due to inappropriate activation of the retinal differentiation suppressor homothorax (hth) by Yki/Sd. This results in activation of a distinct Yki/Hth transcriptional program, driving photoreceptor dedifferentiation. We show that Rbf physically interacts with Yki and together with the GAGA factor, inhibits the hth expression. Thus, RB and Hippo pathways cooperate to maintain photoreceptor differentiation by preventing inappropriate expression of hth in differentiating photoreceptors. Our work highlights the importance of both RB and Hippo pathway activities for maintaining the state of terminal differentiation.
Graphical Abstract
eTOC Blurb
Rader et al report that, in Drosophila, Rbf prevents Yki/Sd from inappropriately activating hth in Hippo pathway mutant photoreceptors. Resulting Hth availability in Rbf Hippo double mutant cells enables Yki to execute a distinct, Yki/Hth progenitor-like transcriptional program, triggering photoreceptor dedifferentiation.
INTRODUCTION
Retinoblastoma (RB) tumor suppressor is important in cell fate decisions while its loss sensitizes cells to dedifferentiation1–5. However, lack of functional RB is not always sufficient for cell fate reversal. Rather, dedifferentiation is achieved through aberrant activation of developmental pathways regulating stemness and renewal of developing tissues6,7. Inappropriate activation of YAP, a Hippo pathway effector, is associated with dedifferentiation in liver8,9, pancreatic10, kidney11, and colorectal12 carcinomas, resulting in highly proliferative progenitor-like cells. Yet, the role of the Hippo pathway in maintaining differentiation of terminally specified cells, as well as its relationship with RB pathway, has not been adequately explored.
The Drosophila larval eye imaginal disc is an ideal model for studying cell differentiation. During this developmental stage, the eye is halfway complete with its transition from progenitor cells into photoreceptors (R cells). This transition occurs in the morphogenetic furrow (MF), a physical indentation that initiates in the posterior and moves towards the anterior of the eye disc. This progressive development provides a snapshot of neuronal differentiation over time, where cells that are further posterior from the MF are further differentiated.
The order in which photoreceptors are recruited is another important aspect of Drosophila eye development. Photoreceptor specification begins with the establishment of the R8 photoreceptor which uniquely expresses Senseless (Sens)13. The R8 cell recruits the remaining R1–7 cells to generate a full ommatidial cluster. As the cluster forms, every R cell expresses the late neuronal marker Elav. Since the R8 cell is absolutely required for recruitment of other R cells, each Elav-positive ommatidium always includes a Sens-positive R8 cell. Strikingly, combined inactivation of RB and Hippo pathways results in the progressive loss of photoreceptor markers in the posterior and the presence of Elav-positive ommatidia which lack a Sens-positive cell14, which are indicative of photoreceptor dedifferentiation. Identifying and characterizing the fate of dedifferentiated cells proves to be a major challenge.
Another gap in knowledge relates to how the RB and Hippo pathways prevent dedifferentiation. E2f1 transcription factor, a downstream effector of the RB pathway, has been shown to cooperate with the Drosophila YAP ortholog Yki to promote inappropriate proliferation in the eye15. Further, E2f1 was shown to prevent activation of Yki target genes in Rbf deficient wing discs16. However, E2f1 is not required for dedifferentiation in RB and Hippo pathway double mutant photoreceptors14 and the role of Yki in this process has not been fully explored. These observations highlight the importance of identifying the dedifferentiation transcriptional program. Yet, genome wide profiling using total RNA from double RB and Hippo mutant eye discs was uninformative15. This is likely due to the extensive cellular heterogeneity of the eye, which prevents extracting the dedifferentiation transcriptional signature from bulk RNA samples. Here, we employed Single-Cell RNA-Sequencing (scRNA-seq) to identify dedifferentiating photoreceptors and explore underlying transcriptional changes.
RESULTS
Genetic labeling of dedifferentiating Rbf wts double mutant photoreceptors using Flybow
The conclusion that Rbf wts double mutant photoreceptors dedifferentiate was based on progressive loss of neuronal markers14. In an alternative approach, we used Elav-GAL4 and a modified inducible mFlp5/mFRT71 from the Flybow system17 to permanently label photoreceptors by expression of a lacZ reporter. Once activated, lacZ is continually expressed regardless of photoreceptor differentiation status (Figure 1A). To test the efficiency of this system, the transgenes were combined, pupal eyes were dissected at 48 hrs after pupa formation (APF) and stained with Elav and β-Gal antibodies (Figure 1B). As expected, every β-Gal-positive cell was also positive for Elav, indicating that photoreceptors are specifically labeled. However, not every photoreceptor expressed β-Gal. On average, there were two β-Gal-positive cells per ommatidium, indicating that less than half of photoreceptors were labeled by 48 hrs APF.
Figure 1. Genetic labeling to visualize dedifferentiating Rbf wts double mutant photoreceptors.
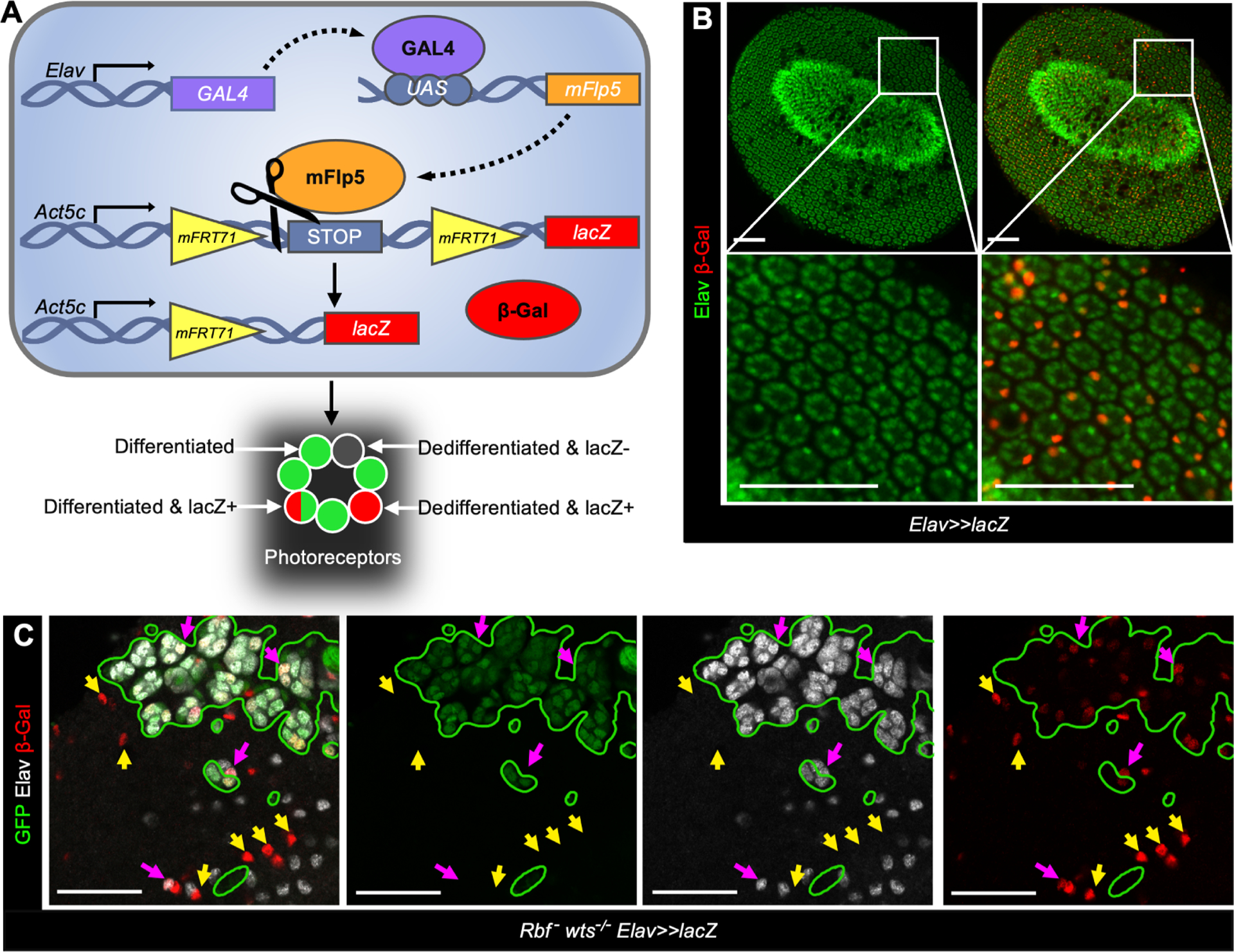
(A) A schematic of the Flybow based system to label photoreceptors.
(B-C) The expression of Elav (green) and β-Gal (red) in pupal eye at 48 hr APF in control, UASmFlp5, Act5c<stop<lacZ/+; Elav-Gal4/+ (B) and in Rbf wts double mutants, Rbf120a, ey-Flp/Y;uas-mFlp5, Act5c<stop<lacZ/+;Elav-Gal4, FRT82B, GFP /FRT82B, wtsx1 (C). Rbf mutant, wts wildtype cells are labeled with GFP, homozygous wts –/– mutant cells are GFP-negative. Elav-negative, β-Gal-positive cells visualize Rbf – wts –/– double mutant photoreceptors that fail to maintain their neuronal identity. Scale bars are 50 μm. See also Table S7.
Next, we induced clones of wts mutant cells in Rbf mutant eyes and used the aforementioned transgenes to assess lacZ expression at 48 hrs APF. In the Rbf mutant wts wildtype tissue, marked by the presence of GFP, every β-Gal positive cell maintained expression of Elav (Figure 1C). In contrast, there were multiple examples of β-Gal positive, Elav negative cells within Rbf wts double mutant tissue (marked by yellow arrows in Figure 1C). Since the presence of β-Gal indicates past Elav expression, this finding suggests that Rbf wts double mutant cells initially expressed Elav but lose expression over time, a hallmark of dedifferentiation. We note, however, that the number of dedifferentiated cells is likely severely underestimated because dedifferentiating cells that do not activate lacZ prior to losing Elav expression remain unlabeled. Given that the onset of dedifferentiation appears a few columns posterior to the MF14, this will significantly impact the number of labeled dedifferentiated cells. Nevertheless, the appearance of β-Gal positive, Elav negative cells unambiguously demonstrates that Rbf wts double mutant photoreceptors are not eliminated and indeed dedifferentiate.
scRNA-Seq reveals upregulation of hth in dedifferentiating Rbf wts double mutant cells
The inefficiency of Flybow in labeling dedifferentiating photoreceptors prompted us to seek an alternative approach. scRNA-seq has proven informative for dissecting tissue heterogeneity in the Drosophila eye and identifying mutant specific cell populations18,19. Wildtype and Rbf mutant eye discs with or without clones of homozygous wts mutant cells were processed for scRNA-seq. The Uniform Manifold Approximation and Projection (UMAP) dimensionality reduction algorithm was then used to visualize cell populations across all four genotypes. Cell populations were assigned using markers specific for each cell population (Table S1). Comparison with our eye disc cell atlas18 revealed that all previously identified cell populations were represented in the new dataset (Figure 2A). Overall, cells from each genotype contributed to every cell population and there were no significant changes in gene expression within clusters between genotypes, apart from cell cycle and Yki targets (Figure S1A).
Figure 2. Rbf wts double mutant cells inappropriately express hth in the eye posterior.
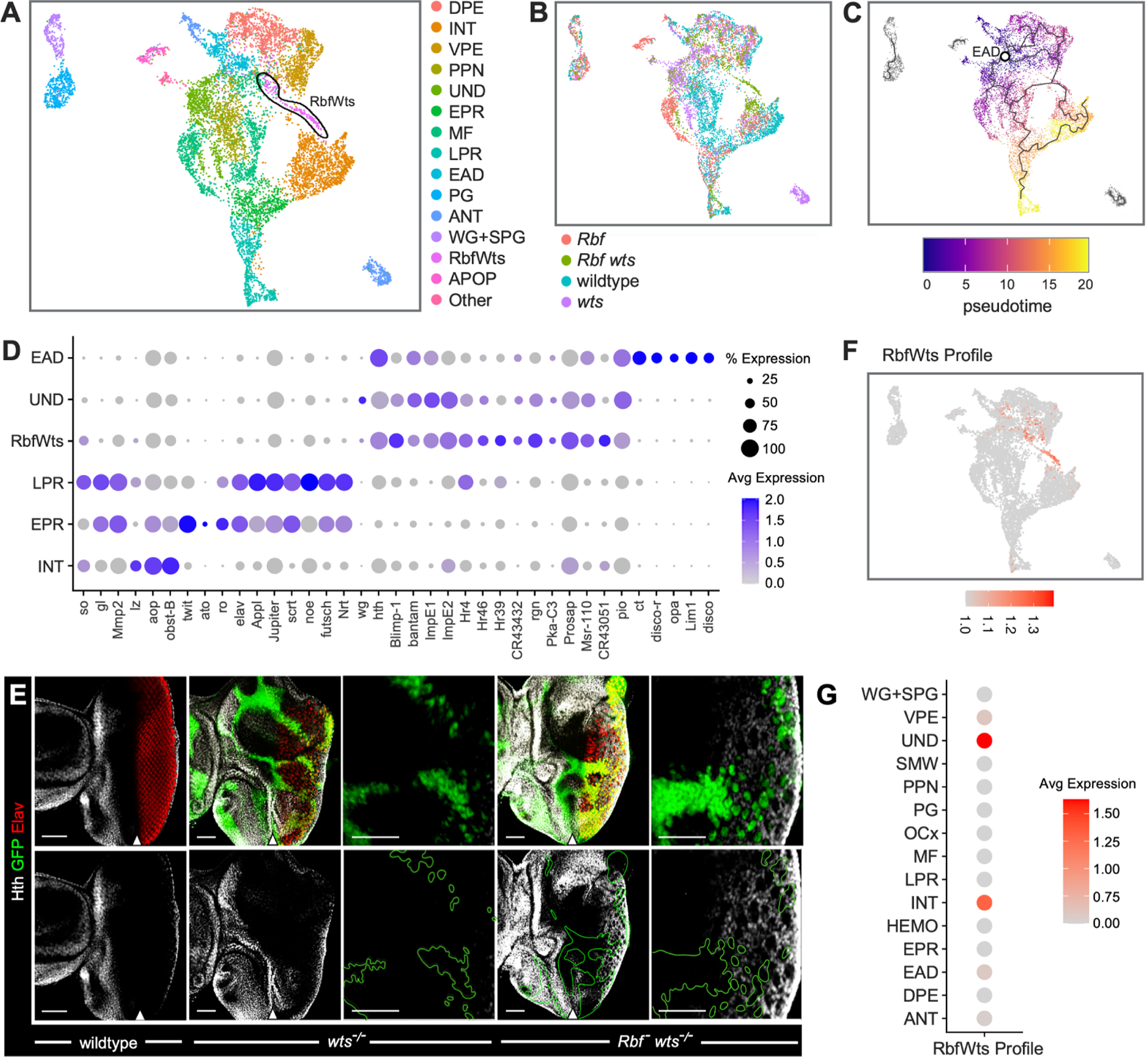
(A-B) UMAP plot of scRNA-seq data of eye discs cells of wildtype, Rbf – single mutant, wts–/– single mutant, and Rbf – wts–/– double mutants colored by assigned cell population (A) or sample genotype (B). A population specific for Rbf – wts–/– double mutants is labeled RbfWts and outlined in black.
(C) Trajectory analysis positions RbfWts cell population between anterior and posterior cells. Predicted specification from the root node (white circle) is shown with gray lines.
(D) A Dot Plot for markers of anterior (EAD, UND), RbfWts, and posterior (LPR, EPR, INT) cell populations. The percentage of cells within a population expressing a given gene is shown through dot scale and level of expression is reflected by color intensity.
(E) The expression of Elav (red) and Hth (white) in third instar larval eyes of ey-Flp/Y; FRT82B Ubi-GFP /FRT82B wtsx1 and Rbf120a ey-Flp/Y; FRT82B GFP /FRT82B wtsx1 animals. Homozygous wts−/− mutant cells are distinguished by the lack of GFP. Rbf – wts−/− double mutant cells ectopically express Hth in the posterior. Scale bars are 50 μm. The position of the MF is denoted with a white arrow.
(F) Expression of the RbfWts transcriptional profile visualized on the UMAP. The RbfWts Enrichment profile accurately and specifically identifies RbfWts cell population.
(G) Dot plot of the RbfWts transcriptional profile projected on wildtype cell populations. RbfWts most closely resembles UND and INT populations.
Since dedifferentiation only occurs in Rbf wts double mutant cells, we hypothesized that these dedifferentiating cells may comprise a distinct cell population. Indeed, the RbfWts cluster consisted entirely of Rbf wts double mutant cells (Figure 2B). We performed trajectory analysis using Monocle 3 which visualizes the position of each cell along the pseudotime differentiation axis20. Uncommitted cells from the eye-antennal domain (EAD) were designated as the earliest cell population in pseudotime. This analysis revealed the normal progression of photoreceptor differentiation in the eye from anterior compartment EAD, undifferentiated cells (UND), and preproneural (PPN) cells to posterior compartment interommatidial cells (INT), early photoreceptors (EPR), and late photoreceptors (LPR). There was a bifurcation between INT and EPR as cells exit the MF (Figure 2C). Intriguingly, RbfWts was positioned relatively early along the pseudotime neuronal differentiation axis and connected to both posterior EPR and INT clusters and anterior EAD and UND clusters.
Analysis of top markers expressed in RbfWts revealed the homeodomain transcription factor and Yki binding partner homothorax (hth), its target gene bantam21, and several nuclear hormone receptors (NHRs) which are known to respond to hth overexpression including Blimp-1 and ImpE122 (Figure 2D). This is significant since hth is a known suppressor of photoreceptor differentiation and is expressed in eye progenitor cells of the anterior compartment23,24. Accordingly, hth, bantam, Blimp-1 and ImpE1 are also highly expressed in the anterior UND and EAD clusters. However, RbfWts does not express other EAD top markers such as Lim1 or disco, suggesting that RbfWts is distinct from the EAD. Further examination of expression of posterior markers showed that RbfWts expresses posterior genes sin oculus (so) and Mmp2, but at a lower level than wildtype cells. However, RbfWts does not express the INT marker lozenge (lz) nor neuronal markers twit, Elav, or futsch. These results suggest that RbfWts is a distinct cluster with some features of both posterior and anterior transcriptional profiles.
Given that hth is highly expressed in RbfWts, we examined the pattern of Hth expression in Rbf wts double mutant mosaic eye discs to spatially localize the RbfWts cluster. Wildtype and Rbf mutant eye discs containing clones of wts mutant cells were stained with Hth antibody and examined by immunofluorescence. In the wildtype eye disc, Hth expression was restricted to anterior undifferentiated cells and the posterior margin23 (Figure 2E). In contrast, Hth was inappropriately expressed within the posterior regions of clones of wts mutant cells in Rbf mutant eye discs where photoreceptors undergo dedifferentiation (Figure 2E and Figure S1B). Importantly, ectopic hth expression in the eye posterior was specific to Rbf wts double mutants, as hth expression was normal in clones of wts single mutant cells (Figure 2E).
To further characterize the RbfWts cluster, we then generated an RbfWts transcriptional profile (Table S2) which accurately identifies RbfWts cells among other cells in UMAP plot (Figure 2F), and then calculated enrichment of the RbfWts profile across all cell populations in wildtype eye disc. The transcriptional profile of RbfWts cluster is most similar to the anterior UND cluster and, to a lesser extent, posterior INT cells (Figure 2G).
Thus, the RbfWts cluster is comprised of Rbf wts double mutant cells which are spatially localized to the posterior and transcriptionally similar to both anterior and posterior cells. As RbfWts devoid of neuronal markers, we hypothesized that RbfWts represents dedifferentiating photoreceptors.
Ectopic expression of constitutively activated Yki in Rbf mutant eye discs induces photoreceptor dedifferentiation
The finding that hth is one of the top markers of RbfWts dedifferentiating photoreceptors was unexpected because hth is normally restricted to progenitor cells of the eye disc23,24. To confirm this finding, we mimicked Hippo pathway inactivation using a constitutively active effector, YkiS111A,S168A,S250A (YkiCA), that is insensitive to Wts regulation25.
ykiCA was overexpressed in wildtype and Rbf mutant eye discs with the GMR-Gal4 driver which is active in all cells posterior to the MF. In wildtype eye, overexpression of ykiCA results in expansion of interommatidial cells, however, every ommatidium contains a Sens positive cell (Figure 3A). In contrast, ykiCA overexpression in Rbf mutant severely decreased the number of Elav positive cells toward the posterior and there were numerous examples of Elav positive clusters which lack a Sens positive cell, a hallmark of dedifferentiation14 (white arrows in Figure 3A). To quantify, we counted the number of Elav positive clusters lacking a Sens positive cell. In the wildtype, 99% of ommatidia had a Sens expressing cell (Figure 3B). Despite disruption in spacing between ommatidia in GMR>ykiCA, 97% of ommatidia contained a Sens positive cell. However, in Rbf GMR>ykiCA eye discs, in addition to severe loss of Elav in the posterior, a Sens positive cell was missing in 26% of ommatidia. Additionally, there were multiple examples of clusters with duplicated R8 cells (circled in Figure 3A), indicating that Yki prevents cell cycle exit in Rbf mutant photoreceptors, a phenotype seen in the Rbf wts double mutant14. Finally, Hth is inappropriately expressed immediately posterior to the MF in Rbf GMR>ykiCA eye discs (Figure 3C–D).
Figure 3. Ectopic ykiCA induces ectopic Hth expression and photoreceptor dedifferentiation in Rbf mutant.
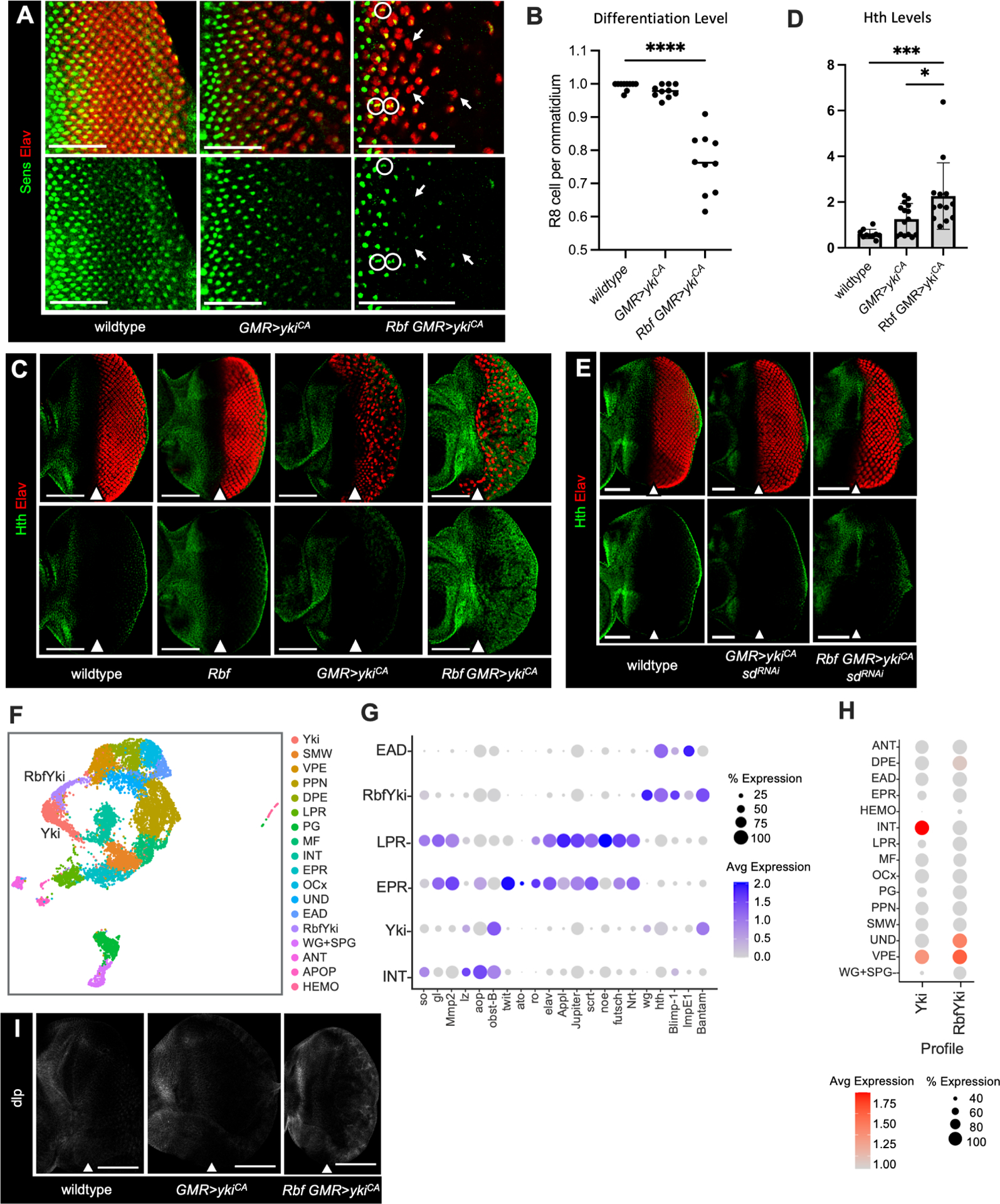
(A) Elav (red) and Sens (green) expression in wildtype, GMR>ykiCA, and Rbf GMR>ykiCA eye discs to visualize R8 cells and all photoreceptors accordingly. Photoreceptor dedifferentiation is scored by the presence of Sens-negative, Elav-positive ommatidial clusters (white arrows). Rbf GMR>ykiCA photoreceptors fail to exit the cell cycle resulting in duplicate R8 cells (white circles). Images are cropped at the morphogenetic furrow.
(B) A scatter plot displaying the ratio of Sens-positive R8 cells per Elav-positive ommatidial cluster. Ommatidia and R8 cell numbers were identified using ZEN software (N=10 eye discs per genotype).
(C) Elav (red) and Hth (green) expression in wildtype, Rbf, GMR>ykiCA, and Rbf GMR>ykiCA eye discs. Hth is normally restricted to the anterior compartment and posterior margin in wildtype, Rbf mutant, and GMR>ykiCA eyes. Expression of ykiCA in Rbf mutant eye discs results in aberrant Hth expression in the posterior compartment.
(D) Quantification of Hth protein levels in the posterior compartment normalized to the anterior compartment of each tissue (N≥10 eye discs per genotype).
(E) Wildtype, GMR>sdRNAi ykiCA, and Rbf GMR>sdRNAi ykiCA eye discs stained for Elav (red) and Hth (green). Knockdown of Sd limits Yki-driven interommatidial expansion and ectopic Hth expression in the posterior compartment.
(F) UMAP plot visualizing cell populations in wildtype, Rbf, GMR>ykiCA, and Rbf GMR>ykiCA samples. An Rbf GMR>ykiCA specific population, designated RbfYki, and the GMR>ykiCA specific population designated Yki are labeled.
(G) A Dot Plot of markers for posterior, anterior, and RbfYki cell populations in wildtype, Yki, and RbfYki clusters. Like RbfWts, RbfYki upregulates hth and its target genes.
(H) Enrichment of the Yki and RbfYki transcription profiles in wildtype cell populations.
(I) Expression of the Hth target gene dlp in wildtype, GMR>ykiCA, and Rbf GMR>ykiCA eye discs. dlp is normally expressed in anterior but it is inappropriately expressed in posterior of Rbf GMR>ykiCA. Scale bars are 50 μm (A, E) and 100 μm (C, I). The position of the MF is denoted with a white arrow.
Yki lacks DNA-binding activity and is recruited to DNA by Scalloped (Sd) to execute the Hippo transcriptional program26,27. To determine if inappropriate expression of Hth is dependent on Sd, we depleted Sd by RNAi in Rbf GMR>ykiCA and examined Hth expression. As has been previously shown, Sd knockdown blocks Yki driven inappropriate proliferation28, which is evident by normal spacing between ommatidia (Figure 3E). Notably, depletion of Sd fully prevents inappropriate expression of Hth in Rbf GMR>ykiCA sdRNAi (Figure 3E) suggesting that Yki induces hth in Rbf mutant eye discs in a Sd-dependent manner.
Next, we performed scRNA-seq on GMR>ykiCA and Rbf GMR>ykiCA eye discs (Table S3). These datasets were combined with previously generated wildtype and Rbf mutant eye discs to visualize cell populations across all four genotypes by UMAP (Figure 3F). To identify the dedifferentiating photoreceptors cluster, we isolated a cell population which consisted exclusively of Rbf GMR>ykiCA cells: RbfYki (Figure 3F, Figure S2A). We then generated a Dot Plot visualizing the expression of EPR, LPR, and RbfWts dedifferentiation markers in the RbfYki cluster. Strikingly, the expression pattern in RbfYki was very similar to that of RbfWts: both clusters highly expressed hth and NHRs and lacked neuronal markers (Figure 2D and 3G). Notably, RbfYki was clearly distinct from the Yki cluster, which contained only GMR>ykiCA cells (Figure 3F, Figure S2A). Unlike RbfYki, the Yki cluster did not upregulate hth and was highly similar to INT cells, indicating that Yki represents inappropriately proliferating interommatidial cells.
Finally, we generated profiles which were characteristic of RbfYki and Yki clusters (Figure S2B–C, Table S2) and projected these profiles onto wildtype cell populations (Figure 3H). Yki cells shared an expression profile similar to that of the VPE (ventral peripodial epithelium) and INT. The similarity to peripodial cells is in agreement with recent study29. Importantly, the Yki transcriptional profile most closely resembled wildtype interommatidial cells and displayed no similarity to anterior cell populations. In this respect, the RbfYki transcriptional profile was strikingly distinct from Yki. In addition to a peripodial-like signature, the RbfYki profile was remarkably akin that of the anterior UND cells (Figure 3H). Thus, only in an Rbf mutant background, ykiCA expression shifts the transcriptional signature of posterior cells towards undifferentiated progenitor cells. This is concordant with photoreceptor dedifferentiation occurring only in Rbf GMR>ykiCA but not in GMR>ykiCA eyes. To confirm this adoption of anterior identity in vivo, we performed immunostaining for anterior gene and Hth target (Figure S2D) dally-like protein (dlp)30,31 which is upregulated in the RbfYki population. Indeed, dlp was inappropriately localized to the posterior of Rbf GMR>ykiCA eye discs (Figure 3I). Thus, as with RbfWts, posterior RbfYki cells adopt an undifferentiated anterior-like phenotype.
hth is necessary and sufficient to trigger Yki-driven photoreceptor dedifferentiation
Given the known role of hth in inhibiting differentiation of eye progenitor cells24, the finding that inappropriate expression of hth is the major feature of dedifferentiating cells was intriguing. To determine the importance of hth, we used RNAi to knockdown Hth in Rbf GMR>ykiCA. As shown above, Rbf GMR>ykiCA eye discs displayed a progressive loss of Elav expression in the posterior compartment, accompanied by the presence of ommatidia without a Sens positive cell (Figure 4A–B). Strikingly, this phenotype was largely rescued by Hth knockdown as very few Elav positive clusters lacked a Sens positive cell (Figure 4A–B). Notably, the distance between ommatidia remained increased in Rbf GMR>ykiCA hthRNAi suggesting that Hth knockdown did not significantly affect the expansion of interommatidial cells, a hallmark of Yki driven proliferation. This result was confirmed by phosphor-H3 (PH3) staining to visualize mitotic cells (Figure S3A–B) although the number of PH3 positive cells was somewhat lower in Rbf GMR>ykiCA hthRNAi than in Rbf GMR>ykiCA. Additionally, there were cases of ommatidia with duplicated R8 cells (circled in white, Figure 4A) indicating that the ability of Yki to override cell cycle exit in terminally differentiated Rbf mutant cells is largely unaffected by Hth knockdown. In a reciprocal experiment, we expressed hth alone or together with ykiCA in the posterior of wildtype eye discs. Expression of hth alone had no impact on photoreceptor differentiation (Figure 4C). Strikingly, although the onset of Sens and Elav expression was unaffected, expression of both was acutely lost several columns posterior in GMR>hth ykiCA eye discs, indicative of photoreceptor dedifferentiation.
Figure 4. Hth is necessary and sufficient for Yki-driven dedifferentiation.
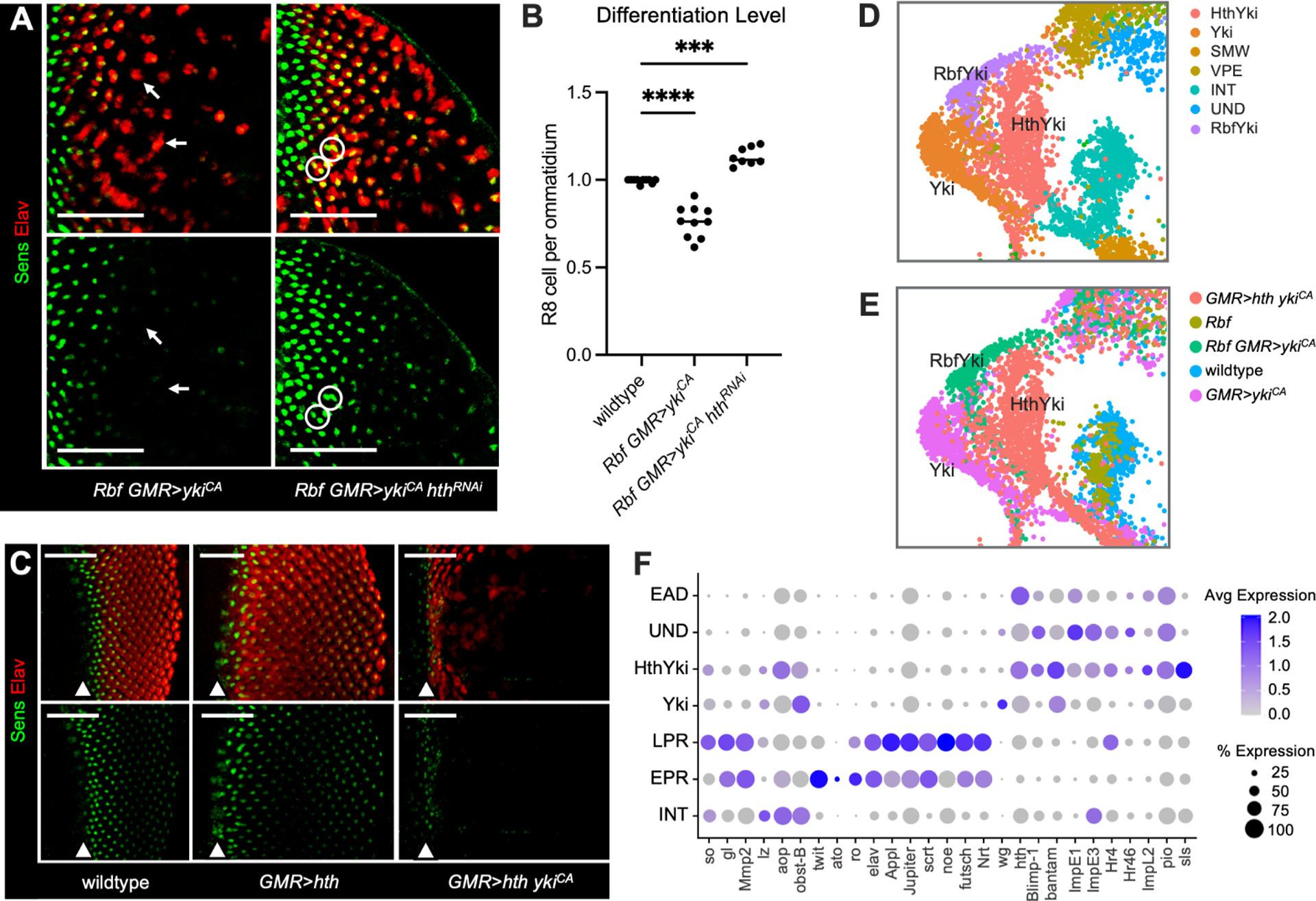
(A) Expression of Elav (red) and Sens (green) in Rbf GMR>ykiCA and Rbf GMR>ykiCA hthRNAi eye discs. Knockdown of Hth rescues photoreceptor dedifferentiation. Images are cropped at the MF.
(B) A scatter plot displaying the ratio of Sens-positive R8 cells per Elav-positive ommatidia (N≥9 eye discs per genotype) to score the extent of dedifferentiation.
(C) Expression of Elav (red) and Sens (green) in wildtype, GMR>hth and GMR>hth ykiCA eye discs. Co-expression of hth and ykiCA results in photoreceptor dedifferentiation.
(D-E) UMAP plot visualizing cell populations (D) and genotype-of-origin (E), cropped to display the GMR>hth ykiCA specific population HthYki. Genotype specific populations Yki, RbfYki, and HthYki are labeled.
(F) A Dot Plot of markers for posterior, anterior, and HthYki cell populations in wildtype, Yki, and HthYki clusters. HthYki upregulates hth and associated nuclear hormone receptors to anterior levels.
Scale bars are 50 μm. See also Figure S3, Tables S2, S4 and S7.
To characterize the effect of Hth and YkiCA co-expression, we performed scRNA-seq on GMR>hth ykiCA eye discs (Table S4) and compared it with single cell transcriptomes of wildtype, Rbf mutant, GMR>ykiCA and Rbf GMR>ykiCA cells (Figure 4D–E). Notably, the GMR>hth ykiCA specific cell population (designated HthYki) shared a number of top markers with populations of dedifferentiating cells identified above (Figure 4F). Thus, Hth is both necessary and sufficient to trigger photoreceptor dedifferentiation in the presence of YkiCA. Notably, both dedifferentiating RbfYki and HthYki populations expressed genes that are normally activated in eye progenitor cells.
Yki/Hth activate a progenitor-like transcriptional program during dedifferentiation
Hth is a Yki binding partner and the Hth-Yki interaction is important in eye progenitor cells24. The results described above raise the possibility that inappropriate expression of hth in the posterior makes Hth available for Yki to execute a Yki/Hth transcriptional program in differentiating photoreceptors which is typically restricted to eye progenitor cells. To test this idea, we generated a HthYki transcriptional profile (Figure S4A, Table S2) that was then projected onto wildtype cell populations. Notably, the HthYki profile most closely resembled undifferentiated eye progenitor cells UND (Figure 5A). In fact, the similarity to UND is the major feature shared between the HthYki, RbfWts and RbfYki profiles. Importantly, the Yki profile bore no similarity to UND, but instead resembled interommatidial (INT) and peripodial (VPE) cells. This is consistent with the observations that Yki alone is unable to trigger dedifferentiation (Figure 3A). Reciprocally, projection of the UND transcriptional profile (Table S2) revealed three populations of dedifferentiating cells: RbfWts, RbfYki and HthYki (Figure 5B). Among wildtype cells, the UND profile was specifically present in anterior UND, EAD and peripodial cells, all of which normally express hth, and was absent in the PPN and posterior cells which do not express hth.
Figure 5. Yki/Hth activate a progenitor-like transcriptional program.
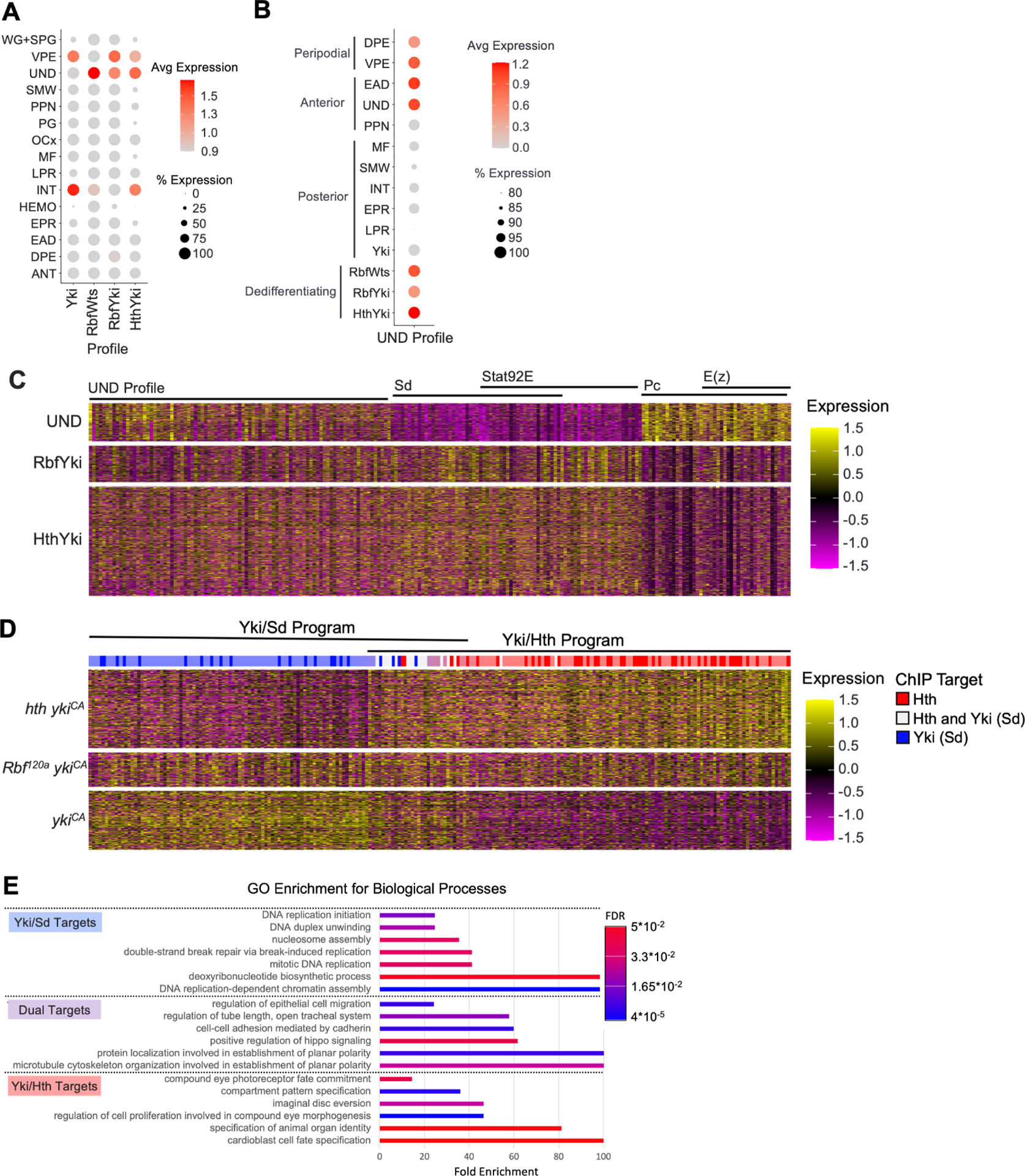
(A) Dot Plot of the Yki, RbfWts, RbfYki, and HthYki transcriptional profiles projected on wildtype cell populations. Dedifferentiating populations aberrantly activate genes expressed in UND.
(B) Dot Plot of the UND transcriptional profile projected on wildtype, Yki, and dedifferentiating cell populations. RbfWts, RbfYki, and HthYki but not Yki are enriched for the UND profile.
(C) Heatmap of differentially expressed genes in UND and HthYki.
(D) Heatmap of differentially expressed genes in GMR>ykiCA and GMR>hth ykiCA posterior cells. Genes upregulated in GMR>ykiCA cells were enriched for Sd binding (dark blue lines) while Rbf GMR>ykiCA and GMR>hth ykiCA were enriched for Hth target genes (red lines). Both Sd and Hth activate a subset of shared transcripts (white lines).
(E) Gene ontology for biological processes (GOBP) enrichment analysis on the Yki/Sd and Yki/Hth transcriptional programs.
The finding that the UND transcriptional profile is highly enriched in dedifferentiating cells raised the question of how similar dedifferentiating cells are to normal eye progenitor cells. Therefore, we identified genes that are differentially expressed between HthYki and UND cell populations (Table S5) and visualized their expression, as well as the expression of genes from the UND transcriptional profile, across UND, RbfYki and HthYki cells in a heatmap. Concordantly, a portion of genes represented in the UND transcriptional signature was expressed in UND, RbfYki and HthYki cells (Figure 5C). However, RbfYki and HthYki also expressed genes that were not expressed in UND. Interestingly, some of these genes were exclusively expressed in posterior cells but not in anterior cells. Transcription factor enrichment analysis revealed that many of these genes are Sd and Stat92E targets. Conversely, genes that were expressed only in UND were specific for anterior cells and, curiously, were enriched for Polycomb (Pc) and E(z) targets (Figure 5C). Thus, despite extensive similarities with eye progenitor cells, the transcriptional profiles of dedifferentiating cells are distinct due to differential expression of genes specific to cells of their respective compartments.
Genetic experiments suggest that Yki/Sd activity alone is insufficient to induce dedifferentiation and only Yki/Hth can activate the transcriptional signature driving transformation of photoreceptors into a progenitor-like cell type. To fully characterize the transcripts activated by Yki/Sd versus Yki/Hth we performed pseudobulk differential expression analysis on posterior compartments from GMR>ykiCA and GMR>hth ykiCA tissues. This analysis produced a list of genes that were specifically upregulated in either sample (Table S6). We then applied this gene list to generate a heatmap of gene expression in GMR>ykiCA, Rbf GMR>ykiCA, and GMR>hth ykiCA cells (Figure 5D). This identified two distinct transcriptional programs: Yki/Sd (blue bar in Figure 5D) and Yki/Hth (red bar in Figure 5D), with a small subset of genes activated by both Yki/Sd and Yki/Hth (Table S6). Reassuringly, the Yki/Hth transcriptional program was activated in both Rbf GMR>ykiCA and GMR>hth ykiCA in which dedifferentiation was observed, but not in GMR>ykiCA. We then performed motif enrichment on these expression signatures to identify transcription factors which were primarily active in each genotype32,33. As expected, cells from the GMR>ykiCA sample were exclusively enriched for Sd target genes (dark blue bars in Figure 5D), while GMR>hth ykiCA was highly enriched for Hth targets (dark red bars in Figure 5D). Genes activated in both the Yki/Sd and Yki/Hth programs contained both Sd and Hth binding sites (white bars in Figure 5D), including known Yki targets such as ex, Diap1, and kibra29 and members of the Hippo pathway ft and wts. Gene ontology of biological processes (GOBP) analysis revealed that these shared targets are primarily associated with cell polarity and cytoskeletal organization (Figure 5E). Further, gene ontology for Yki/Sd targets solely consisted of GO terms for DNA replication and cell cycle (Figure 5E), reflecting that Yki/Sd drives proliferation. Notably, Yki/Hth program was enriched for genes with GO terms related to cell differentiation and fate specification (Figure 5E).
Thus, ectopic Yki/Hth activity dedifferentiates photoreceptors by turning on the eye progenitor-like transcriptional program.
Rbf interacts with Yki and limits its ability to activate hth in transcriptional reporter assay
To determine whether Yki directly regulates hth expression, we examined the occupancy of Yki and Sd at the hth locus using publicly available genomic data34–36. Both Yki and Sd were enriched at multiple regions within the hth gene (Figure 6A). Another important factor in Yki dependent transcription is the GAGA transcription factor GAGA (GAF), which physically interacts with Yki and contributes to Yki activation34,37. To explore the role of these proteins in hth regulation, we generated a series of luciferase reporters containing different regions of the hth locus. The reporters were transfected into Drosophila S2R+ cells and their response to co-transfected V5:Yki and Flag:Sd expression plasmids was determined. Among six tested constructs, one reporter, hth-Luc-B, was robustly activated by increasing amount of V5:Yki and Flag:Sd (Figure 6B), while Rbf limited the activation (Figure 6C). Unexpectedly, although GAF was shown to potentiate Yki dependent transcription34,37, co-transfection of Flag-GAF blocked Yki/Sd activation of the hth-Luc-B reporter that was further potentiated by Rbf (Figure 6D).
Figure 6. Rbf interacts with Yki and inhibits its activation of the hth reporter in transient transfections.
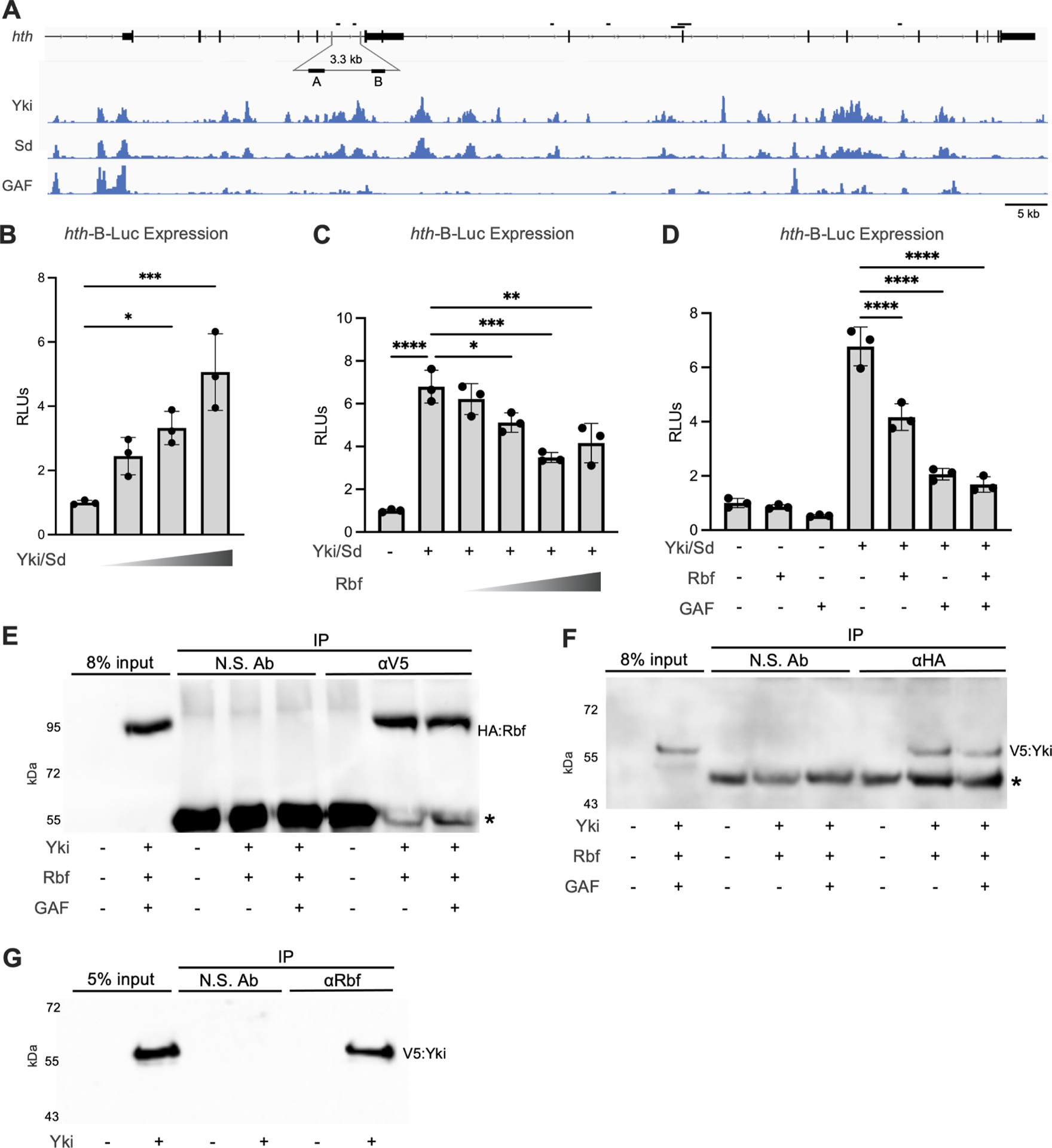
(A) ChIP-Seq data showing Yki, Sd, and GAF occupancy at the hth gene. The genomic fragments tested in luciferase reporter assays are shown.
(B) Increasing amount of Yki and Sd expression plasmids were co-transfected in S2R+ cells together with 10 ng hth-B luciferase reporter and 1 ng copia-renilla luciferase internal control plasmid. Yki/Sd activate the hth-B-Luc reporter in a does dependent manner.
(C) Increasing amount of Rbf expression plasmid was co-transfected with 100 ng Yki and 100 ng Sd expression plasmids. Rbf blocks Yki/Sd activation of hth-B-Luc in dose dependent manner.
(D) 100 ng Yki, 100 ng Sd, 200 ng GAF, and 200 ng Rbf were co-transfected either alone or in combination. Rbf and GAF repress Yki/Sd dependent activation of hth-B-Luc.
(B-D) Firefly luciferase activity was normalized to the renilla control in each transfection. Dual-luciferase assays were performed in three biological replicates. Scatter dot plots with bars, mean ± SD, One-way ANOVA by Tukey’s multiple comparisons test.
(E-F) S2R+ cells were transfected with V5:Yki, HA:Rbf, and FLAG:GAF. Lysates were immunoprecipitated with mouse anti-CD133 as a negative control (N.S. Ab) and anti-V5 to immunoprecipitate V5:Yki (E) or anti-HA to immunoprecipitate HA:Rbf (F). Associated proteins were detected by Western blot with anti-HA to detect HA:Rbf (E) or anti-V5 to detect V5:Yki (F). Asterisks denote antibody heavy chains.
(E) S2R+ cells were transfected with V5:Yki and lysates were immunoprecipitated with mouse anti-CD133 as a nonspecific control (N.S. Ab) or anti-Rbf. Associated V5:Yki and endogenous Rbf were detected by Western blot analysis with anti-V5 antibody.
See also Table S8.
Given that Rbf limited Yki/Sd activation, we tested whether Rbf and Yki physically interact. V5:Yki and HA:Rbf were transiently transfected in S2R+ cells, the complexes were immunoprecipitated with V5 antibody, separated by SDS-PAGE, and western blots were probed with HA antibody to detect HA:Rbf. As shown in Figure 6E, HA:Rbf was significantly enriched in the V5:Yki immunoprecipitates, while no HA:Rbf was immunoprecipitated with a non-specific antibody. Co-transfection of Flag:GAF did not affect the interaction between HA:Rbf and V5:Yki, although we cannot rule out that GAF and Rbf competed for Yki binding. Next, we performed a reciprocal experiment and confirmed the presence of V5:Yki in HA:Rbf immunoprecipitates (Figure 6F). Finally, cells were transfected with V5:Yki and endogenous Rbf was immunoprecipitated from cell extracts using an Rbf antibody38 which specifically recognizes endogenous Rbf39,40. The presence of V5:Yki in Rbf immunoprecipitates was detected by western blotting with V5 antibody (Figure 6G). We concluded that Yki associates with Rbf.
Knockdown of GAF leads to ectopic hth expression in GMR>ykiCA eye disc
GAF is encoded by the Trithorax-like (Trl) gene and has been shown to promote Yki/Sd transcription in the context of cell proliferation34,37. Therefore, finding that GAF limits activation of the hth-Luc-B reporter by Yki/Sd was unexpected. To explore the effect of GAF on hth expression in endogenous settings, we used Trl RNAi to deplete GAF in the posterior compartment of the GMR>ykiCA eye disc and determined Hth expression by immunofluorescence (Figure 7A). The UAS-TrlRNAi transgene has been previously validated to efficiently knockdown GAF protein in vivo, in the context of Yki dependent transcription37. Strikingly, Hth was inappropriately expressed posterior to the MF in GMR>TrlRNAi ykiCA at a level similar to Rbf GMR> ykiCA (Figure 7B) and was accompanied by progressive reduction in the number of Elav positive cells across the eye posterior (Figure 7A), indicative of dedifferentiating photoreceptors. The Hth upregulation occurred in a Yki dependent manner since its expression was unaffected in GMR>TrlRNAi (Figure S5A–B). Accordingly, staining with Elav and Sens antibodies revealed the appearance of Elav positive clusters which lack a Sens positive cell, another hallmark of dedifferentiation, in GMR>TrlRNAi ykiCA (white arrows in Figure 7C), but not in GMR>TrlRNAi. To further dissect the proliferation and dedifferentiation phenotypes, we examined mitotic cells in GMR>TrlRNAi ykiCA and found that GAF knockdown strongly reduced ykiCA driven proliferation (Figure 7D–E). Thus, although GAF is necessary for activation of the Yki/Sd pro-proliferative program it acts as a repressor at the hth gene.
Figure 7. Depletion of GAF derepresses hth expression and triggers dedifferentiation in Yki expressing photoreceptors.
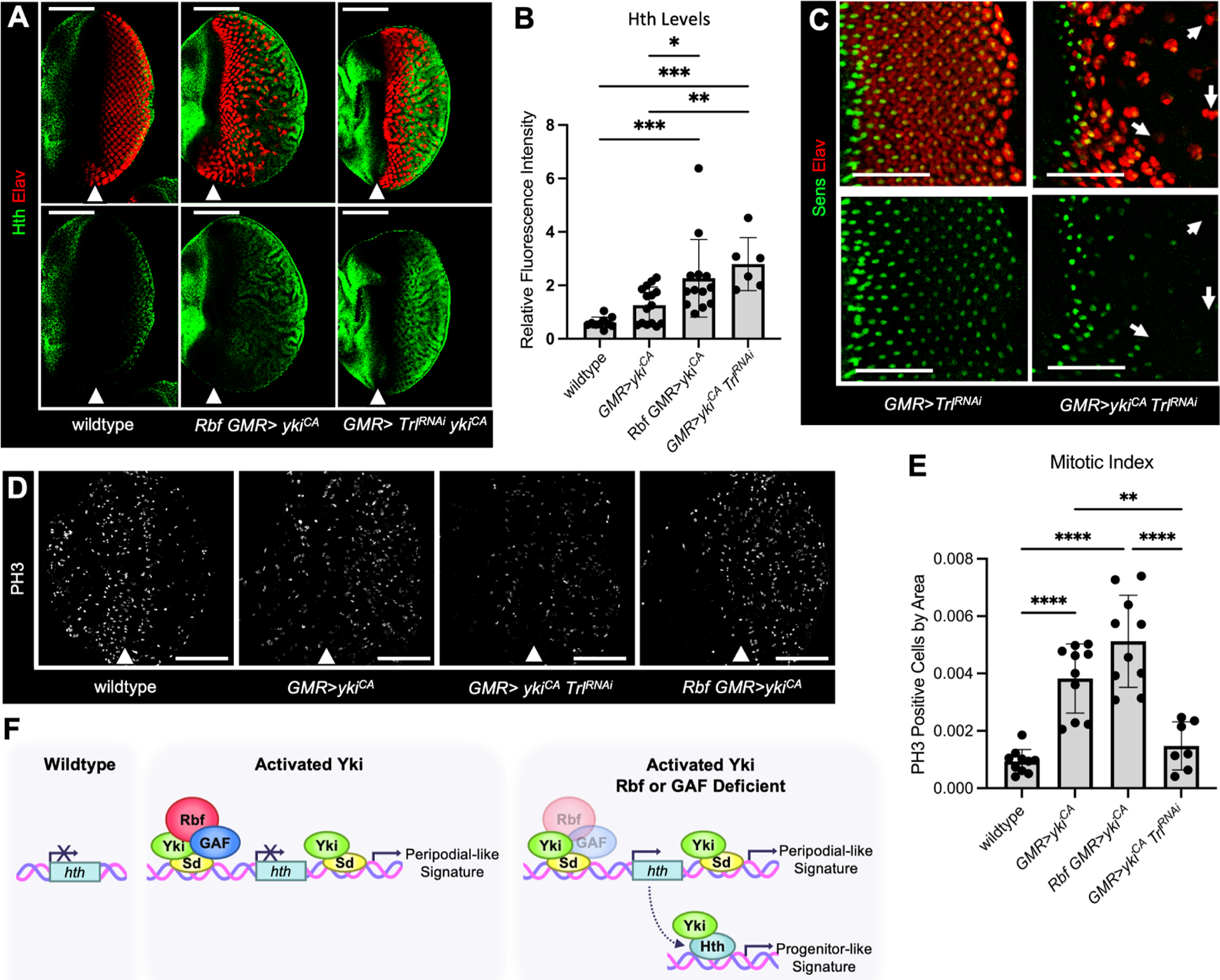
(A) Eye discs from wildtype, Rbf GMR>ykiCA, and GMR>ykiCA TrlRNAi animals were stained with Elav (red) and Hth (green) antibodies.
(B) A bar graph displaying Hth protein levels in the posterior compartment normalized to the anterior compartment of each tissue (N≥6 per genotype).
(C) GMR>TrlRNAi does not impact photoreceptor differentiation, while GMR>TrlRNAi ykiCA photoreceptors fail to maintain neuronal specification. This is visible by the occurrence of Elav-positive, Sens-negative ommatidial clusters (white arrows).
(D) Trl knockdown blocks ykiCA driven inappropriate proliferation. Eye discs from wildtype, GMR>ykiCA, GMR>ykiCA TrlRNAi, and Rbf GMR>ykiCA animals were stained for PH3 (white) to detect mitotic cells.
(E) Quantification of the number of PH3-positive cells in the posterior compartment of each genotype.
(F) A model for RB and Hippo pathways cooperation in maintenance of the differentiation state. In Rbf proficient Hippo pathway mutant cells, Rbf blocks Yki/Sd from activating hth in the eye posterior. In these cells, Yki/Sd infers a peripodial like signature. In the absence of Rbf, Yki/Sd inappropriately activates hth in differentiating photoreceptors. The availability of Hth results in activation of the Yki/Hth transcriptional program that resembles the transcriptional signature of the eye progenitor cells (UND) and leads to photoreceptors dedifferentiation. Thus, in addition to the canonical Yki/Sd signature, RB and Hippo pathways double mutant cells activate the Yki/Hth transcriptional program.
DISCUSSION
Here, we describe a mechanism by which the RB and Hippo pathways cooperate to maintain photoreceptor differentiation, a function which is distinct from the role of Hippo in cell fate specification. In Hippo pathway deficient cells with normal RB function, ectopic Yki activates a Sd-dependent program which is reminiscent of the transcriptional signature of peripodial cells29 (and this study). This results in expansion of interommatidial cells but does not impact photoreceptor identity14,29. However, combined inactivation of RB and Hippo pathways, in addition to activation of the Yki/Sd program, leads to inappropriate expression of the progenitor gene hth in differentiating photoreceptors. We suggest that availability of Hth now allows Yki to execute a distinct, Yki/Hth transcriptional program that is characteristic of eye progenitor cells (Figure 7F). This program imparts photoreceptors with an eye progenitor-like phenotype. Thus, the RB and Hippo pathways maintain photoreceptor identity by restricting Yki/Sd dependent expression of hth to inhibit reactivation of the progenitor transcriptional program in terminally differentiating cells.
In normal eye, Hth is essential for maintaining an undifferentiated state of progenitor cells while its loss results in ectopic photoreceptor differentiation23. Hth together with Teashirt (Tsh) partners with Yki to promotes progenitor cell proliferation21. Accordingly, prolonging hth expression inhibits MF initiation and differentiation23,24,41. Expression of ykiS168A in undifferentiated tissues induces wg expression which promotes hth/tsh expression to inhibit photoreceptor differentiation42. Further, in certain settings, yki can impair eye development resulting in eye tissue completely devoid of photoreceptors43,29. The common theme here is that ectopic Yki is able, in some contexts, to prevent photoreceptor differentiation by interfering with developmental cues. Therefore, these defects are explicitly distinct from the phenotype of Rbf wts double mutant cells, which initiate the neuronal differentiation, establish axonal projections, but fail to maintain a differentiated state14. Genetic tracing experiments confirm that Rbf wts double mutant photoreceptors indeed lose their neuronal identity and are not eliminated.
Our results suggest that inappropriate expression of hth is the key event in photoreceptor dedifferentiation. Hth is ectopically expressed in each of three genetic settings that are accompanied by a progressive loss of photoreceptor identity. Accordingly, depletion of Hth in Rbf mutants expressing ykiCA prevents dedifferentiation, while co-expression of hth with ykiCA in Rbf wild type eye discs triggers dedifferentiation. Thus, hth is downstream of Rbf and is necessary and sufficient for dedifferentiation of photoreceptors that express ykiCA.
Why does inappropriate upregulation of hth in the presence of activated Yki result in dedifferentiation? Our single cell data strongly argue that photoreceptor dedifferentiation is specifically accompanied by activation of a distinct transcriptional program that is most similar to that of the eye progenitor cells and is replete with Hth target genes. This program is activated in addition to the Yki/Sd program, suggesting that availability of Hth does not prevent Yki from partnering with Sd. Interestingly, several known Yki targets such as Diap1, ex, and kibra, and Hippo pathway members ft and wts are shared between the Yki/Sd and Yki/Hth programs. Gene ontology analysis for these shared targets revealed enrichment in GO terms for cytoskeletal organization and cell polarity, suggesting that regardless of binding partner Yki is highly influential in regulation of cell morphology and adhesion. Notably, overexpression of Hth alone in the posterior compartment is inconsequential and requires Yki to trigger dedifferentiation. Given that Hth and Yki physically interact and were shown to regulate Hippo pathway targets such as bantam21, this data collectively suggests that the dedifferentiation program is driven by Yki/Hth.
Genome wide chromatin binding experiments revealed that hth is a direct target of Yki/Sd34,36. Accordingly, Yki/Sd activates the hth luciferase reporter and this activation is blocked by Rbf. Given that Rbf is a known transcriptional repressor that limits E2F activation through direct binding, our observation that Rbf similarly interacts with Yki provides a simple explanation for why Yki does not activate hth in Rbf proficient cells. The finding that GAF similarly inhibits Yki/Sd expression of hth was especially surprising since previous studies have shown that GAF is necessary for full activation of the Yki/Sd proliferation program37,34. Thus, GAF appears to simultaneously promote Yki/Sd driven proliferation and protect these cells from dedifferentiation through inhibition of Yki/Sd dependent activation of hth.
The idea that Rbf represses expression of genes determining the progenitor state, such as hth, is consistent with the role of mammalian pRB. pRB has been previously shown to repress transcription of key pluripotency genes Sox2 and Oct4 in somatic cells44. Further, RB protects prostate cancer cells from lineage transformation through inhibition of Sox2 and chromatin modifying protein Ezh245. Thus, cooperation between Hippo and RB pathways in maintaining a state of terminal differentiation converges on their regulation of hth, where the Hippo kinase cascade and Rbf limit Yki activity.
Limitations of the study
Although scRNA-seq approach revealed that photoreceptors dedifferentiate into progenitor-like cells, the ultimate fate of these cells remains unknown. A genetic tracing approach allowed us to positively mark dedifferentiating photoreceptors that no longer express a neuronal marker Elav. We show that such cells are present at pupal stage but we did not investigate what happens to these cells later. Our study did not explore how availability of Hth affects Yki/Sd complexes, whether Hth and Sd have different affinities for Yki and how it impacts the Yki/Hth and Yki/Sd transcriptional programs.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Maxim V. Frolov, mfrolov@uic.edu.
Materials Availability
Plasmids and Drosophila strains generated in this study are available upon request.
Data and Code Availability
The scRNA-seq datasets generated in this study have been deposited in NCBI’s Gene Expression Omnibus and is accessible through accession number GEO: GSE217380. Raw immunofluorescence images and western blots are available at DOI 10.17632/wk5c46kmr7.1. Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Drosophila cell line
Drosophila S2R+ cells were cultured in Schneider’s Drosophila Medium (Sigma S0146–500ML) supplemented with 10% heat inactivated fetal bovine serum (ThermoFisher Scientific 10082147) at 25C.
Drosophila Stocks and Maintenance
Stocks and crosses were kept at 25C in vials in an uncrowded condition on standard Bloomington formulation medium containing cornmeal, yeast, light corn syrup and agar (LabExpress; Fly Food B). w1118 was used as a wildtype stock for all single-cell RNA-Sequencing and genetic experiments. Both male and female animals were used permitting the genetic cross scheme.
METHOD DETAILS
Adapting Flybow for In vivo labeling of dedifferentiating photoreceptors.
ey-mFlp5 construct was digested with BglII and XbaI to excise mFLP5 that was then inserted into pUAST transformation vector. The embryo injections were done by BestGene Inc. (Chino Hills, CA). Ten transgenic animals with germline insertions were obtained, the insertions were mapped and one line with the insertion on the second chromosome was tested and used in this works. The P{UAS-FLPm5} was recombined with act5C-mFRT71C≥αtub 3′UTR mFRT71C≥nuclear lacZ on the second chromosome and used to generate final combinations shown in Figure 1.
Immunofluorescence
Eye imaginal discs were dissected from wandering third instar larvae, or pupa when specified, and dissected in ice-cold 1X PBS. Samples were then fixed in 4% formaldehyde (Polysciences 18814) diluted in room temperature 1X PBS for 30 minutes and subsequently permeabilized in 0.3% PBS-Triton X-100 twice for 10 minutes. Blocking was performed for one hour at room temperature in 10% normal donkey serum (Jackson ImmunoResearch 017–000-121) with 0.01% Triton X-100. Samples were then incubated in primary antibody diluted in 10% NDS, 0.01% PBS-T overnight at 4C on a nutator. The following day, samples were washed three times for 5 minutes in 0.1% PBS-Triton X-100 before incubating in fluorescent-conjugated secondary antibodies (Jackson ImmunoResearch, 1:200) for 1 hour at room temperature including 4′,6-diamidino-2-phenylindole (DAPI). Finally, samples were washed four times with 0.1% PBS-Triton X-100 for five minutes before mounting on glass slides in FluorSave reagent (MilliporeSigma 345789).
Final genotypes for each figure are shown in Table S7.
Microscopy
A Zeiss Axio Observer Z1 LSM 700 confocal microscope was used to capture fluorescent images at x20/0.8 and x100/1.45 objectives. All images were taken using 1 AU pinhole and gain settings were consistent for all images within an experiment. Resulting images were processed using ImageJ Software (1.48v, NIH, USA) unless otherwise specified. Image processing included adjustments to brightness and contrast which were consistent for all images in each experiment. Images shown are representative.
Tissue Dissection for scRNA-Seq
Third instar larvae were harvested upon wandering and eye imaginal discs were dissected in cold 1X PBS. Eye discs were then dissected from the brain and antenna by cutting the optic stalk and eye-antennal-border using a 0.15 mm microblade (Fine Science Tools, 10316–14). Discs were then deposited in ice-cold 1X Rinaldini solution. Collagenase (Sigma #C9891) was added to achieve a final concentration of 2.5 mg/mL and Trypsin (Sigma #59418C) to 1X dilution. Tubes were then attached horizontally to a shaker and agitated at 225 rpm for 10 minutes at 32C. Samples were then subjected to approximately 30 pulses of gentle mechanical digestion using a P200 pipette. The single-cell suspension was then washed twice in 0.04% BSA-PBS and resuspended in 30 uL of 0.04% BSA-PBS. The concentration of cells and their viability was assessed using a hemocytometer and Trypan blue staining.
Final genotypes subjected to single-cell RNA-sequencing include:
-w[1118]
-Rbf[120a]/Y
-yw, ey-Flp /w[*];; P{ry[+t7.2]=neoFRT}82B P{w[+mC]=Ubi-GFP.D}83/ wts[x1] P{ry[+t7.2]=neoFRT}82B
-Rbf[120]a P{ry[+t7.2]=ey-FLP.N}2/Y;; P{ry[+t7.2]=neoFRT}82B P{w[+mC]=Ubi-GFP.D}83/wts[x1] P{ry[+t7.2]=neoFRT}82B
-w[1118]/w[*]; P{GMR-GAL4.w[-]}2/+; P{y[+t7.7] w[+mC]=UAS-yki.S111A.S168A.S250A.V5}attP2/+
-Rbf[120]a/Y; P{GMR-GAL4.w[-]}2/+; P{y[+t7.7] w[+mC]=UAS-yki.S111A.S168A.S250A.V5}attP2/+
-w[1118]/w[*]; P{GMR-GAL4.w[-]}2/uas-hth; P{y[+t7.7] w[+mC]=UAS-yki.S111A.S168A.S250A.V5}attP2/+
Yki, Sd, Hth, and GAF genomic occupancy
The following publicly available datasets of genome wide occupancy of Yki, Sd, Hth, and GAF were used: Hth from the embryo30 (SRX111801), Yki and Sd from the eye-antennal disc46 (GSE54603), and Yki and GAF from the wing imaginal disc and embryo34 (GSE38594) were downloaded as BigWig files mapped to dmel6 using the ChIP-Atlas browser. BigWig files were then loaded into IGV47 (version 2.94) for visualization.
Construction of Luciferase Reporters
Selected hth gene regions were PCR amplified from w1118 genomic DNA. PCR products were then cloned into the XhoI site of a pGL2-hsp70 luciferase reporter vector including an hsp70 minimal promoter15 using NEBuilder Hifi DNA Assembly (NEB E2621). Appropriate insert sequences were confirmed by Sanger sequencing. Primers and respective gene products cloned into luciferase reporters are listed in Table S8.
Luciferase Assay
S2R+ cells were plated onto 24-well plates at 1*107 cells per well in 10% FBS-Schneider’s Drosophila Medium (Sigma S0146–500ML) and allowed to adhere for 1 hour at 25C. For each well, 10 ng of luciferase reporter and 1 ng of control copia-renilla luciferase reporter were transfected with combinations of pAc5.1-Yki-V5, pIEx-7-Sd-FLAG, pIEx-7-FLAG-GAF, and/or pIEx4-HA-Rbf. Transfection mixtures were supplemented with empty pIEx-7 vector (MilliporeSigma 71339) such that every well received the same amount of total plasmid. Mixes were then diluted in FBS-Free Schneider’s Drosophila Medium to achieve a volume of 50 μL per well and X-Tremegene HP DNA Transfection Reagent (Sigma 6366244001) was added at the ratio of 1.5 uL per 1 μg of plasmid. After incubating for 30 minutes at room temperature, transfection mix was added dropwise to the cells and incubated for 24 hours at 25C before replacing media. 48 hours after transfection cells were washed in cold PBS before lysing in 75 μL 1X Passive Lysis Buffer (Promega E1941) for 20 minutes. In triplicate, 25 μL of each lysate was loaded on a white, flat-bottom 96-well plate and subjected to the Dual-Luciferase® Reporter Assay System (Promega E1910). All luciferase assays were performed at least 3 times and representative experiments are shown.
Figure 6 Titration concentrations are as follows:
B) 0, 50, 75, and 100 ng each of pAc5.1-Yki-V5 and pIEx-7-Sd-FLAG.
C) 0, 50, 100, 150, 200 ng of pIEx4-HA-Rbf.
Immunoprecipitation-Western Blot
Drosophila S2R+ cells were plated into 6-well plates at 2*106 cells per well in 10% FBS-Schneider’s Drosophila Medium (Sigma S0146–500ML) and allowed to adhere for 1 hour at 25C. Each well was transfected with combinations of 1 μg each pAc5.1-Yki-V5, pIEx-7-FLAG-GAF, and pIE4-Rbf-HA. Transfection mixes were supplemented with empty pIEx-7 vector (MilliporeSigma 71339) such that every well received the same concentration of total plasmid. Plasmid mixes were diluted in serum-free media to achieve a volume of 100 μL per well before adding 2 μL X-Tremegene HP DNA Transfection Reagent (Sigma 6366244001). Transfection mixes were then incubated for 30 minutes at room temperature. Tranfection mix was then applied to the cells dropwise and incubated for 48 hours at 25C. Post-transfection, cells were harvested, washed with cold PBS, and resuspended in cold lysis buffer (20 mM HEPES pH 7, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol) containing 1X protease inhibitor (Sigma 11836170001), flash frozen for approximately 3 minutes, and briefly thawed at 37C. Lysates were then incubated on ice for 30 minutes before centrifugation. Supernatants were then collected for overnight incubation in primary antibody at 4C on a rotator. Mouse anti-CD133 (DSHB HB#7, 1:100) was used as a negative control, mouse anti-V5 for V5-tagged Yki (Fisher R960–25, 1:100), mouse anti-HA for HA-tagged Rbf (Fisher 26183, 1:100), and mouse anti-Rbf (DX3 and DX5, 1:10) for endogenous Rbf.
The following day, lysates were incubated in 20 μg Dynabeads Protein G and A 1:1 (Fisher 10003D and 10001D). Dynabeads were then washed four times for 10 minutes in cold lysis buffer before eluting immunoprecipitates into Laemmli buffer, boiling, and and separating by SDS-PAGE. Resulting Western Blots were blocked for 30 minutes at room temperature in 2.5% non-fat dry milk TBS with 0.1% Tween-20 before incubating in mouse anti-V5 (Fisher R960–25, 1:2000) or mouse anti-HA (Fisher 26183, 1:2000) overnight at 4C. The following day, blots were washed three times for 5 minutes with TBS-T before incubating with anti-mouse secondary conjugated to HRP (1:3000) for one hour. Samples were then washed three times for five minutes in TBS-T, incubated in SuperSignal Chemiluminescent substrate for 5 minutes (Thermo Fisher 34577), and imaged on Azure imaging system. Resulting images were processed through adjustments to brightness and contrast and horizontal cropping for clarity. Experiments were performed at least three times and representative images are shown.
scRNA-Seq Data Preprocessing
The single cell sample Fastq files were processed using the 10X Genomics CellRanger (version 6.0.0) count function with default parameters. Reads were aligned to Drosophila genome BDGP6 which was extracted from Ensemble (version 90). Features, barcodes, and matrices generated by CellRanger were then processed into Seurat objects using the Read10X and CreateSeuratObjects functions. Data was deposited to the NCBI Gene Expression Omnibus database (GEO, https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE217380.
scRNA-Seq Data Analysis
Datasets were merged using the Seurat R package48–50 and low viability cells were removed on the basis of low RNA content and enrichment for mitochondrial, heat shock, and ribosomal genes. This ultimately resulted in 4,232 wildtype, 2,643 Rbf mutant, 2,158 wts−/− mutant, 2,643 Rbf – wts−/− double mutant, 4,116 of GMR>ykiCA and 1,913 of Rbf GMR>ykiCA, and 4,004 GMR> hth ykiCA cells. Remaining cells were processed using default settings. UMAPs were created using 50 PCs before differential expression analysis with FindAllMarkers using the Wilcoxon Rank Sum. LogFC thresholds and min.pct, the minimum fraction of cells expressing a given gene, were both set to 0.25. Populations were then annotated based on top marker expression as described in Ariss et al 201818. Distinct populations were characterized by their genotype of origin. Specific transcriptional profiles were generated from differential expression analysis of the population-of-interest versus all other cells, combined with previously determined top markers. For example, the posterior compartment transcriptional profile was generated using differential expression of all posterior cells versus all anterior cells. This list was then supplemented with top marker genes for known posterior cell types which were not captured during pseudobulk differential expression analysis. Enrichment for expression of these transcriptional profiles was determined using AddModuleScore and resulting scores were visualized using the FeaturePlot, DotPlot, and DoHeatmap functions.
Cell Trajectory Analysis
Single-cell trajectory analysis was performed using Monocle 320. Seurat objects and UMAPs were converted into Monocle compatible celldataset objects and UMAP pseudotime was calculated using the branch point within the EAD cluster, the least differentiated population, as the root node.
Gene Ontology Analysis
PANTHER Overrepresentation Test for GO biological processes in Drosophila melanogaster was performed using Gene Ontology Resource51–53 (http://geneontology.org) with default settings and FDR P < 0.05. Fold enrichment and FDR values were then input to Microsoft Excel for visualization.
Motif Enrichment Analysis
Motif enrichment for transcription factor binding to differentially expressed genes was performed with i-cisTarget32,33 (database version 6.0) using the dm6 Drosophila genome. Genes were analyzed against a Transcription Factor Binding Site ChIP database containing data from 436 ChIP-seq experiments. Data was submitted with default settings: a 0.4 minimum fraction of overlap, NES threshold of 3.0, ROC threshold of 0.01, and a visualization threshold of 5000. Candidate target genes for relevant ChIP experiments, including Sd ChIP-seq on eye-antennal discs (SRX457598) and hth ChIP-seq on 8–16h embryos (SRX111801), were then extracted.
QUANTIFICATION AND STATISTICAL ANALYSIS
All data were assessed for significance in GraphPad Prism (v9.1.0) using One-way ANOVA by Tukey’s multiple comparisons test and are displayed with the mean ± SD. The number of eye imaginal discs quantified for each experiment (n) are described in corresponding figure legends. Asterisks denote p-values of: ns=p>0.05, *= p≤0.05, **= p≤0.01, ***= p≤0.001, ****p≤0.0001.
PH3 Quantification
Quantification of PH3 in Figure S3 was done as follows. Using ImageJ software, confocal images of eye tissues were cropped to the posterior compartment using Elav immunostaining. The pixel area of each posterior compartment was measured using the Measure function (Analyze->Measure). Fluorescent intensity thresholds for the PH3 channel were set to 50–255 (Image->Adjust->Threshold) before running Analyze Particles (Analyze->Analyze Particles) with size thresholds set from 5-infinity pixel units to count the number of PH3 foci. The number of PH3 positive foci was then normalized to the sampled area of the posterior compartment and averaged for each genotype (n=5–10 samples per genotype). The significance of the ratio of PH3 foci per area was assessed by ANOVA (ns=p>0.05, *= p≤0.05, **= p≤0.01, ***= p≤0.001, ****p≤0.0001). Data is presented as a bar graph with error bars representing the standard deviation of samples across each genotype.
Photoreceptor Differentiation Quantification
Quantification of Photoreceptor Differentiation was done as follows. Using ZEN Blue 2.6 software (ZEISS), Elav positive cells were used to define ommatidia into zones-of-influence (ZOI). Gaussian smoothing was then applied to better capture all photoreceptors in a single ommatidium and binary erosion was selected to separate adjacent ommatidia. R8 cells were then identified within each ommatidium by measuring the presence of Senseless signal within its corresponding ZOI. Background fluorescence was omitted by restricting the minimum area for Senseless-positive cells to 25 pixel units. Thresholds were adjusted for each image to account for variations in fluorescent intensity across samples and experiments. Data was combined to reveal the number of ommatidia and R8 cells identified per tissue. Between 9 and 10 samples were processed for each genotype and one-way ANOVA was performed to assess significance. Results are displayed as a scatter plot. Further statistical details of experiments can be found in legends for Figures 3 and 4.
Hth Protein Quantification
Hth Protein quantification was done as follows. Using ImageJ software, confocal images of eye imaginal discs were cropped to remove antennal discs before defining the anterior and posterior compartments. The DAPI channel was then used to outline the border of the tissue while Elav signal was used to define the posterior compartment. Hth protein defined the anterior compartment. In mosaic tissues, GFP signal was used to further separate wts+/+ wildtype from wts−/− mutant tissue. The average fluorescent intensity of the Hth channel was measured in the anterior and posterior compartments from 6–10 tissues across multiple experiments using the ImageJ (1.48v, NIH, USA) measure function (Analyze->Measure). The average Hth fluorescent intensity of each posterior compartment was then normalized to the anterior compartment of the same sample to account for variation across experiments. These ratios were then averaged to reflect the overall abundance of Hth in the posterior compartment for each genotype. Hth abundance ratios were then pooled and analyzed by one-way ANOVA to assess significance (ns=p>0.05, *= p≤0.05, **= p≤0.01, ***= p≤0.001, ****p≤0.0001). Data is presented as a bar graph with error bars representing the standard deviation of samples across each genotype. Further statistical details of experiments can be found in legends for Figures 3, 7, S1 and S5.
Supplementary Material
Table S1: The average expression of all genes in all annotated clusters of the Rbf wts experiment. Related to Figure 2.
Table S3: The average expression of all genes in all annotated clusters of the Rbf GMR>yki[CA] experiment. Related to Figure 3.
Table S4: The average expression of all genes in all annotated clusters of the GMR>hth yki[CA] experiment. Related to Figure 4.
Table S5: Differentially expressed genes between HthYki and wildtype UND cells. Related to Figure 5.
Table S6. Differentially expressed genes in the posterior compartments of GMR>yki[CA] and GMR>hth yki[CA] eye discs. Related to Figure 5.
Table S8: Primer sequences and gene products for luciferase constructs. Related to Figure 6.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-β-Gal (1:200), 40–1a | DSHB | RRID:AB_831022 |
| Rat anti-Elav (1:200), 7EA10 | DSHB | RRID: AB_528218 |
| Guinea Pig anti-Senseless (1:2000) | Gift from H. Bellen (Baylor College of Medicine) | RRID:AB_2567469 |
| Guinea Pig anti-hth (1:2000) | Gift from R. Mann (Columbia University) | RRID:AB_2616321 |
| Rabbit Anti-phospho-Histone H3 (1:500), 06–570 | MilliporeSigma | RRID:AB_310177 |
| Mouse Anti-dlp (1:5), 13G8 | DSHB | RRID:AB_528191 |
| Alexa Fluor® 488 AffiniPure Donkey Anti-Guinea Pig IgG (1:50) | Jackson Immunoresearch | 706–545-148 |
| Cy3 AffinoPure Goat Anti-Guinea Pig IgG (1:200) | Jackson Immunoresearch | 106–165-0030 |
| Cy5 AffiniPure Donkey Anti-Rat IgG (1:200) | Jackson Immunoresearch | 712–175-150 |
| Alexa Fluor® 488 AffiniPure Donkey Anti-Rabbit IgG (1:200) | Jackson Immunoresearch | 711–545-152 |
| Alexa Fluor® 647-conjugated AffiniPure Donkey AntiMouse IgG (1:200 | Jackson Immunoresearch | 715–605-151 |
| Mouse anti-V5 | Thermo Scientific | R960–25 |
| Mouse anti-HA, 12CA5 | Gift from N. Dyson (MGH Cancer Center) | RRID:AB_514505 |
| Mouse anti-Rbf | Gift from N. Dyson (MGH Cancer Center): Du et al.38 | DX3, DX5 |
| Goat-anti-mouse HRP secondary antibody | Azure Biosystems | AC2115 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| SuperSignal West Pico PLUS Chemiluminescent Substrate | Thermo Scientific | 34579 |
| VECTASHIELD® Antifade Mounting Medium | Vector Laboratories | H-1000–10 |
| X-tremeGENE™ HP DNA Transfection Reagent | MilliporeSigma | 6366236001 |
| Dynabeads™ Protein G for Immunoprecipitation | Invitrogen | 10003D |
| Dynabeads™ Protein A for Immunoprecipitation | Invitrogen | 10001D |
| Bacterial and virus strains | ||
| DH10B Competent Cells | Thermo Scientific | EC0113 |
| Critical commercial assays | ||
| Dual-Luciferase® Reporter Assay System | Promega | E1910 |
| NEBuilder HiFi DNA Assembly Kit | NEB | E2621 |
| Chromium Single Cell 3’ (v3.1 Chemistry Dual Index) | 10X Genomics | RRID:SCR_019326 |
| QIAGEN Plasmid Midi Kit | QIAGEN | 12143 |
| QIAGEN Gel Extraction Kit | QIAGEN | 28704 |
| Deposited data | ||
| Single-cell RNA Sequencing Data | This study | GSE217380 |
| Raw images and western blots | This study | 10.17632/wk5c46kmr7.1 |
| Experimental models: Cell lines | ||
| S2R+ | ACCC | RRID:CVCL_Z831 |
| Experimental models: Drosophila melanogaster strains | ||
| w[1118] | BDSC | RRID:BDSC_3605 |
| Rbf[120a]/FM7i, P{w[+mC]=ActGFP}JMR3 | Gift from N. Dyson (MGH Cancer Center) | N/A |
| y[d2] w[1118] P{ry[+t7.2]=ey-FLP.N}2 | BDSC | RRID:BDSC_5580 |
| P{UAS-FLPm5} | This study | N/A |
| act5C-mFRT71C≥αtub 3’UTR mFRT71C≥nuclear lacZ | Gift from Barry Dickson (Research Institute of Molecular Pathology, Vienna): Hadjieconomou et al.17 | N/A |
| P{elav-GAL4.AD}I1A1 | BDSC | RRID:BDSC_23868 |
| w[*]; P{ry[+t7.2]=neoFRT}82B P{w[+mC]=Ubi-GFP.D}83 | BDSC | RRID:BDSC_5188 |
|
Rbf[120]a P{ry[+t7.2]=ey-FLP. N}2/
FM7i,
P{w[+mC]=ActGFP}JMR3;; P{ry[+t7.2]=neoFRT}82B P{w[+mC]=Ubi-GFP.D}83/TM6B |
Gift from N. Moon (McGill University) | N/A |
|
yw, ey-Flp/FM7, GFP;;
P{ry[+t7.2]=neoFRT}82B
P{w[+mC]=Ubi-GFP.D}83/ TM6B, Tb[1] |
Gift from I. Hariharan (UC Berkeley) | N/A |
| w[*]; wts[x1] P{ry[+t7.2]=neoFRT}82B/TM6B, Tb[1] | BDSC | RRID:BDSC_44251 |
|
w[1118];
P{GMR-GAL4.w[-]}2/CyO,
P{w[+mC]=ActGFP}JMR1 |
BDSC | RRID:BDSC_9146 |
|
w[*]; P{y[+t7.7]
w[+mC]=UAS-
yki.S111A.S168A. S250A. V5}attP2 |
BDSC | RRID:BDSC_28817 |
|
w[*]; P{y[+t7.7]
w[+mC]=UAS-
yki.S168A. V5}attP2 |
BDSC | RRID:BDSC_28818 |
|
UAS-sd-RNAi
w[1118]; P{108877}101497 |
VDRC | KK-101497 |
|
UAS-hth-RNAi
w[1118]; P{108831}100630 |
VDRC | KK-100630 |
|
UAS-Trl-RNAi
w[1118]; P{112776}106433 |
VDRC | KK-106433 |
| UAS-hth | Gift from R. Mann (Columbia University) | N/A |
| Oligonucleotides | ||
| Primers for Hth Luciferase Assays | See Table S8 | |
| Recombinant DNA | ||
| pGL2-hsp70-luc Luciferase Reporter Vector with minimal hsp70 promoter | Nicolay et al.15 | N/A |
| Hth-A-Luc | This study | N/A |
| Hth-B-Luc | This study | N/A |
| Hth-E-Luc | This study | N/A |
| Hth-F-Luc | This study | N/A |
| Hth-G3-Luc | This study | N/A |
| Hth-G5-Luc | This study | N/A |
| Hth-K-Luc | This study | N/A |
| pIEx-7 Ek/LIC | MilliporeSigma | 71339 |
| pIE4-HA-Rbf | Gift from N. Dyson (MGH Cancer Center) (Stevaux et al., 2002) | N/A |
| pAc5.1-Yki-V5 | Gift from K. Irvine (Rutgers University): Oh et al.25 | N/A |
| pIEx-7- FLAG-Sd | Nicolay et al.15 | N/A |
| pIEx-7- FLAG-GAF | Bayarmagnai et al.37 | N/A |
| ey-mFlp5 | Gift from I. Salecker (MRC, London): Hadjieconomou et al.17 | N/A |
| pUAST | Gift from N. Perrimon (Harvard Medical School) | RRID:DGRC_1000 |
| Software and algorithms | ||
| ImageJ (Fiji) | NIH | RRID:SCR_003070 |
| R Studio | R | RRID:SCR_001905 |
| R Package Seurat (v 3) | Satija Lab | RRID:SCR_016341 |
| Zeiss Zen 2.6 (Blue edition) | Zeiss | RRID:SCR_013672 |
| Microsoft Excel | Microsoft | RRID:SCR_016137 |
| 10x Genomics Cell Ranger 4.0.0 and 5.0.0 | 10x Genomics | RRID:SCR_016957 |
| Integrative Genomics Viewer | Broad Institute | RRID:SCR_011793 |
| The Gene Ontology Resource | NHGRI | RRID:SCR_002811 |
| ApE | W. Davis | RRID:SCR_009122 |
| Flybase | RRID:SCR_006549 | |
| ChIP Atlas | Zou et al.54 | RRID:SCR_015511 |
| Monocle 3 | Qiu et al.55 | RRID:SCR_018685 |
Highlights.
Rbf physically interacts with Yki to prevent activation of hth
Yki/Hth induce a progenitor-like transcriptional program in photoreceptors
Yki/Hth dedifferentiate photoreceptors into progenitor-like cells
GAF limits Yki/Sd activation of hth to maintain photoreceptor differentiation
Acknowledgements
We thank Paula Zappia for advice on scRNA-seq and data analysis, James Kwon for technical help and Kieran Harvey for critically reading the manuscript. We thank Kenneth Irvine for the V5:Yki plasmid, Hugo Bellen for anti-Senseless antibody, Richard Mann for anti-Hth antibody and UAS-hth flies and the Developmental Studies Hybridoma Bank at the University of Iowa for anti-Elav, anti-β-Gal, and anti-dlp antibodies. This work was supported by NIH grant R35GM131707 to M.V.F.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- 1.Dick FA, Goodrich DW, Sage J, and Dyson NJ (2018). Non-canonical functions of the RB protein in cancer. Nat Rev Cancer 18, 442–451. 10.1038/s41568-018-0008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dyson N (2016). RB1: a prototype tumor suppressor and an enigma. Genes Dev 30, 1492–1502. 10.1101/gad.282145.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merrell AJ, and Stanger BZ (2016). Adult cell plasticity in vivo: de-differentiation and transdifferentiation are back in style. Nat Rev Mol Cell Biol 17, 413–425. 10.1038/nrm.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sage J (2012). The retinoblastoma tumor suppressor and stem cell biology. Genes Dev 26, 1409–1420. 10.1101/gad.193730.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanahan D, and Weinberg RA (2011). Hallmarks of Cancer: The Next Generation. Cell 144, 646–674. 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Jing N, Gao W-Q, and Fang Y-X (2021). Regulation of Formation, Stemness and Therapeutic Resistance of Cancer Stem Cells. Front Cell Dev Biol 9, 641498. 10.3389/fcell.2021.641498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turkel N, Sahota VK, Bolden JE, Goulding KR, Doggett K, Willoughby LF, Blanco E, Martin-Blanco E, Corominas M, Ellul J, et al. (2013). The BTB-zinc Finger Transcription Factor Abrupt Acts as an Epithelial Oncogene in Drosophila melanogaster through Maintaining a Progenitor-like Cell State. PLoS Genet. 9, e1003627. 10.1371/journal.pgen.1003627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yimlamai D, Christodoulou C, Galli GG, Yanger K, Pepe-Mooney B, Gurung B, Shrestha K, Cahan P, Stanger BZ, and Camargo FD (2014). Hippo Pathway Activity Influences Liver Cell Fate. Cell 157, 1324–1338. 10.1016/j.cell.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Zhuo S, Zhou Y, Ma L, Sun Z, Wu X, Wang XW, Gao B, and Yang Y (2022). Yap-Sox9 signaling determines hepatocyte plasticity and lineage-specific hepatocarcinogenesis. J Hepatol 76, 652–664. 10.1016/j.jhep.2021.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L, Shi H, Chen H, Gong A, Liu Y, Song L, Xu X, You T, Fan X, Wang D, et al. (2019). Dedifferentiation process driven by radiotherapy-induced HMGB1/TLR2/YAP/HIF-1α signaling enhances pancreatic cancer stemness. Cell Death Dis 10, 724. 10.1038/s41419-019-1956-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel S, Tang J, Overstreet JM, Anorga S, Lian F, Arnouk A, Goldschmeding R, Higgins PJ, and Samarakoon R (2019). Rac-GTPase promotes fibrotic TGF-β1 signaling and chronic kidney disease via EGFR, p53, and Hippo/YAP/TAZ pathways. The FASEB Journal 33, 9797–9810. 10.1096/fj.201802489RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwarzmueller L, Bril O, Vermeulen L, and Léveillé N (2020). Emerging Role and Therapeutic Potential of lncRNAs in Colorectal Cancer. Cancers 12, 3843. 10.3390/cancers12123843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frankfort BJ, and Mardon G (2002). R8 development in the Drosophila eye: a paradigm for neural selection and differentiation. Development 129, 1295–1306. 10.1242/dev.129.6.1295. [DOI] [PubMed] [Google Scholar]
- 14.Nicolay BN, Bayarmagnai B, Moon NS, Benevolenskaya EV, and Frolov MV (2010). Combined inactivation of pRB and hippo pathways induces dedifferentiation in the Drosophila retina. PLoS Genet 6, e1000918. 10.1371/journal.pgen.1000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicolay BN, Bayarmagnai B, Islam ABMMK, Lopez-Bigas N, and Frolov MV (2011). Cooperation between dE2F1 and Yki/Sd defines a distinct transcriptional program necessary to bypass cell cycle exit. Genes Dev 25, 323–335. 10.1101/gad.1999211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang P, Pei C, Wang X, Xiang J, Sun B-F, Cheng Y, Qi X, Marchetti M, Xu J-W, Sun Y-P, et al. (2017). A Balance of Yki/Sd Activator and E2F1/Sd Repressor Complexes Controls Cell Survival and Affects Organ Size. Dev Cell 43, 603–617.e5. 10.1016/j.devcel.2017.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hadjieconomou D, Rotkopf S, Alexandre C, Bell DM, Dickson BJ, and Salecker I (2011). Flybow: genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster. Nat Methods 8, 260–266. 10.1038/nmeth.1567. [DOI] [PubMed] [Google Scholar]
- 18.Ariss MM, Islam ABMMK, Critcher M, Zappia MP, and Frolov MV (2018). Single cell RNA-sequencing identifies a metabolic aspect of apoptosis in Rbf mutant. Nat Commun 9, 5024. 10.1038/s41467-018-07540-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bravo González-Blas C, Quan X, Duran-Romaña R, Taskiran II, Koldere D, Davie K, Christiaens V, Makhzami S, Hulselmans G, de Waegeneer M, et al. (2020). Identification of genomic enhancers through spatial integration of single-cell transcriptomics and epigenomics. Mol Syst Biol 16, e9438. 10.15252/msb.20209438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32, 381–386. 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng HW, Slattery M, and Mann RS (2009). Transcription factor choice in the Hippo signaling pathway: Homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev 23, 2307–2319. 10.1101/gad.1820009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neto M, Naval-Sánchez M, Potier D, Pereira PS, Geerts D, Aerts S, and Casares F (2017). Nuclear receptors connect progenitor transcription factors to cell cycle control. Sci Rep 7, 4845. 10.1038/s41598-017-04936-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pai C-YCY, Kuo T-STS, Jaw TJ, Kurant E, Chen C-TCT, Bessarab DA, Salzberg A, and Sun YH (1998). The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, Extradenticle, and suppresses eye development in Drosophila. Genes Dev 12, 435–446. 10.1101/gad.12.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopes CS, and Casares F (2010). Hth maintains the pool of eye progenitors and its downregulation by Dpp and Hh couples retinal fate acquisition with cell cycle exit. Dev Biol 339, 78–88. 10.1016/j.ydbio.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 25.Oh H, and Irvine KD (2009). In vivo analysis of Yorkie phosphorylation sites. Oncogene 28, 1916–1927. 10.1038/onc.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell S, Inamdar M, Rodrigues V, Raghavan V, Palazzolo M, and Chovnick A (1992). The scalloped gene encodes a novel, evolutionarily conserved transcription factor required for sensory organ differentiation in Drosophila. Genes Dev 6, 367–379. 10.1101/gad.6.3.367. [DOI] [PubMed] [Google Scholar]
- 27.Wu Shian, Liu Yi, Zheng Yonggang, Dong Jixin, P. D (2008). The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell 14, 388–398. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Ren F, Zhang Q, Chen Y, Wang B, and Jiang J (2008). The TEAD/TEF Family of Transcription Factor Scalloped Mediates Hippo Signaling in Organ Size Control. Dev Cell 14, 377–387. 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kowalczyk W, Romanelli L, Atkins M, Hillen H, Bravo González-Blas C, Jacobs J, Xie J, Soheily S, Verboven E, Moya IM, et al. (2022). Hippo signaling instructs ectopic but not normal organ growth. Science 378, eabg3679. 10.1126/science.abg3679. [DOI] [PubMed] [Google Scholar]
- 30.Nègre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, et al. (2011). A cis-regulatory map of the Drosophila genome. Nature 471, 527–531. 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neto M, Aguilar-Hidalgo D, and Casares F (2016). Increased avidity for Dpp/BMP2 maintains the proliferation of progenitors-like cells in the Drosophila eye. Dev Biol 418, 98–107. 10.1016/j.ydbio.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 32.Herrmann C, Van de Sande B, Potier D, and Aerts S (2012). i-cisTarget: an integrative genomics method for the prediction of regulatory features and cis-regulatory modules. Nucleic Acids Res 40, e114–e114. 10.1093/nar/gks543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imrichová H, Hulselmans G, Kalender Atak Z, Potier D, and Aerts S (2015). icisTarget 2015 update: generalized cis-regulatory enrichment analysis in human, mouse and fly. Nucleic Acids Res 43, W57–W64. 10.1093/nar/gkv395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh H, Slattery M, Ma L, Crofts A, White KP, Mann RS, and Irvine KD (2013). Genome-wide association of Yorkie with chromatin and chromatin-remodeling complexes. Cell Rep 3, 309–318. 10.1016/j.celrep.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slattery M, Voutev R, Ma L, Nègre N, White KP, and Mann RS (2013). Divergent Transcriptional Regulatory Logic at the Intersection of Tissue Growth and Developmental Patterning. PLoS Genet 9, e1003753. 10.1371/journal.pgen.1003753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pascual J, Jacobs J, Sansores-Garcia L, Natarajan M, Zeitlinger J, Aerts S, Halder G, and Hamaratoglu F (2017). Hippo Reprograms the Transcriptional Response to Ras Signaling. Dev Cell 42, 667–680.e4. 10.1016/j.devcel.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 37.Bayarmagnai B, Nicolay BN, Islam ABMMK, Lopez-Bigas N, and Frolov MV (2012). Drosophila GAGA factor is required for full activation of the dE2f1-Yki/Sd transcriptional program. Cell Cycle 11, 4191–4202. 10.4161/cc.22486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du W, Vidal M, Xie JE, and Dyson N (1996). RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes Dev 10, 1206–1218. 10.1101/gad.10.10.1206. [DOI] [PubMed] [Google Scholar]
- 39.Stevaux, et al. (2002). Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2. EMBO J 21, 4927–4937. 10.1093/emboj/cdf501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dimova DK, Stevaux O, Frolov MV, and Dyson NJ (2003). Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev 17, 2308–2320. 10.1101/gad.1116703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao H, Moberg KH, and Veraksa A (2023). Hippo pathway and Bonus control developmental cell fate decisions in the Drosophila eye. Dev Cell 58, 416–434.e12. 10.1016/j.devcel.2023.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wittkorn E, Sarkar A, Garcia K, Kango-Singh M, and Singh A (2015). The Hippo pathway effector Yki downregulates Wg signaling to promote retinal differentiation in the Drosophila eye. Development 142, 2002–2013. 10.1242/dev.117358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang T, Zhou Q, and Pignoni F (2011). Yki/YAP, Sd/TEAD and Hth/MEIS Control Tissue Specification in the Drosophila Eye Disc Epithelium. PLoS One 6, e22278. 10.1371/journal.pone.0022278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kareta MS, Gorges LL, Hafeez S, Benayoun BA, Marro S, Zmoos A-F, Cecchini MJ, Spacek D, Batista LFZ, O’Brien M, et al. (2015). Inhibition of Pluripotency Networks by the Rb Tumor Suppressor Restricts Reprogramming and Tumorigenesis. Cell Stem Cell 16, 39–50. 10.1016/j.stem.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, Goodrich MM, Labbé DP, Gomez EC, Wang J, et al. (2017). Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 355, 78–83. 10.1126/science.aah4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ikmi A, Gaertner B, Seidel C, Srivastava M, Zeitlinger J, and Gibson MC (2014). Molecular evolution of the Yap/Yorkie proto-oncogene and elucidation of its core transcriptional program. Mol Biol Evol 31, 1375–1390. 10.1093/molbev/msu071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat Biotechnol 29, 24–26. 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21. 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000). Gene Ontology: tool for the unification of biology. Nat Genet 25, 25–29. 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carbon S, Douglass E, Good BM, Unni DR, Harris NL, Mungall CJ, Basu S, Chisholm RL, Dodson RJ, Hartline E, et al. (2021). The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res 49, D325–D334. 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mi H, Muruganujan A, Ebert D, Huang X, and Thomas PD (2019). PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 47, D419–D426. 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zou Z, Ohta T, Miura F, and Oki S (2022). ChIP-Atlas 2021 update: a data-mining suite for exploring epigenomic landscapes by fully integrating ChIP-seq, ATAC-seq and Bisulfite-seq data. Nucleic Acids Res 50, W175–W182. 10.1093/nar/gkac199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14, 979–982. 10.1038/nmeth.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: The average expression of all genes in all annotated clusters of the Rbf wts experiment. Related to Figure 2.
Table S3: The average expression of all genes in all annotated clusters of the Rbf GMR>yki[CA] experiment. Related to Figure 3.
Table S4: The average expression of all genes in all annotated clusters of the GMR>hth yki[CA] experiment. Related to Figure 4.
Table S5: Differentially expressed genes between HthYki and wildtype UND cells. Related to Figure 5.
Table S6. Differentially expressed genes in the posterior compartments of GMR>yki[CA] and GMR>hth yki[CA] eye discs. Related to Figure 5.
Table S8: Primer sequences and gene products for luciferase constructs. Related to Figure 6.
Data Availability Statement
The scRNA-seq datasets generated in this study have been deposited in NCBI’s Gene Expression Omnibus and is accessible through accession number GEO: GSE217380. Raw immunofluorescence images and western blots are available at DOI 10.17632/wk5c46kmr7.1. Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.


