Abstract
Cells carefully modulate the rate of rRNA transcription in order to prevent an overinvestment in ribosome synthesis under less favorable nutritional conditions. In mammals, growth-dependent regulation of RNA polymerase I (Pol I) transcription is mediated by TIF-IA, an essential initiation factor that is active in extracts from growing but not starved or cycloheximide-treated mammalian cells. Here we report the molecular cloning and functional characterization of recombinant TIF-IA, which turns out to be the mammalian homolog of the yeast factor Rrn3p. We demonstrate that TIF-IA interacts with Pol I in the absence of template DNA, augments Pol I transcription in vivo and rescues transcription in extracts from growth-arrested cells in vitro.
INTRODUCTION
Alterations in cell proliferation are accompanied by profound changes in the transcription rate of rRNA genes. rRNA synthesis is down-regulated when cells approach stationary phase, are starved of an essential nutrient or are exposed to protein synthesis inhibitors. Conversely, rDNA transcription is up-regulated upon reversal of such conditions (reviewed by Grummt, 1999). Thus, by responding to changes in the cellular environment, transcription by RNA polymerase I (Pol I) ultimately determines ribosome production and the potential for cellular proliferation. One important but as yet little understood facet of rDNA transcriptional control is how information concerning the growth or proliferation state of the cell is conveyed to the transcription machinery. In mammals, growth-dependent regulation of rDNA transcription has been shown to be due to changes of the amount or activity of an essential transcription factor, termed TIF-IA. TIF-IA was initially identified as an activity that complements transcriptionally inactive extracts obtained from quiescent mouse cells (Buttgereit et al., 1985). TIF-IA is very likely to correspond to TFIC, a factor responsible for the hormonal regulation of rRNA synthesis in lymphosarcoma cells (Mahajan and Thompson, 1990) and to factor C*, a similar activity studied in cycloheximide-treated L1210 cells (Brun et al., 1994). The available data suggest that the function of factor TIF-IA/TFIC/C* (which we will collectively refer to as TIF-IA) is inherent in its tight association with Pol I. Apparently, Pol I and TIF-IA associate to form the transcriptionally active enzyme, which is capable of initiating transcription from the rDNA promoter (Schnapp et al., 1990). By this and other criteria, TIF-IA may be regarded as a functional homolog of the bacterial σ70 factor.
In Saccharomyces cerevisiae, a genetic screen for mutants defective in Pol I transcription has identified Rrn3p, which appears to be functionally analogous to TIF-IA. Like TIF-IA, Rrn3p associates with a subpopulation of Pol I to form an ‘activated’ enzyme that is capable of initiating transcription (Yamamoto et al., 1996). Significantly, extracts from stationary phase cells lack the Pol I–Rrn3p complex, suggesting that under unfavorable growth conditions the association of Rrn3p with Pol I is impaired (Milkereit and Tschochner, 1998).
The apparent functional similarities between TIF-IA and Rrn3p prompted us to clone the mammalian homolog of yeast Rrn3p. We present functional data indicating that the human Rrn3p homolog is the growth-dependent regulatory factor TIF-IA. We demonstrate that TIF-IA co-localizes with UBF in the nucleolus, interacts with Pol I, and is capable of activating Pol I transcription both in vivo and in extracts from growth-arrested cells.
RESULTS
The strategy for cloning the cDNA encoding TIF-IA was based on the functional homology between TIF-IA and Rrn3p. Starting with the sequence of Rrn3 from S. cerevisiae, homologous cDNAs were identified in Schizosaccharomyces pombe, Caenorhabditis elegans and Arabidopsis thaliana. The sequences were aligned to yield a consensus to be used for database searching. By combining expressed sequence tag (EST) walking and PCR-based cloning strategies, a human cDNA was cloned, which encodes a 651 amino acid (73.8 kDa) protein with 40% sequence similarity (27% identity) to Rrn3p (Figure 1). Consistent with an essential role in rDNA transcription, the gene is expressed ubiquitously, showing ∼4.8 kb transcripts in all tissues tested (data not shown).
Fig. 1. Amino acid sequence of human TIF-IA (H. s.) aligned with Rrn3 from S. cerevisiae (S. c.). Identical residues are shaded black and conserved residues grey. The DDBJ/EMBL/GenBank database accession No. for TIF-IA is AJ272050.
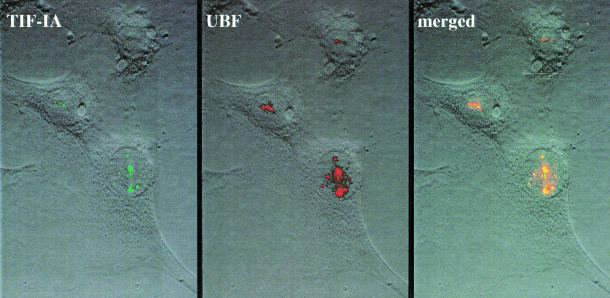
If the cloned cDNA encodes TIF-IA, then the protein should be localized within the nucleolus alongside other components of the Pol I transcription machinery. To test this, NIH 3T3 cells were transiently transfected with an expression vector encoding FLAG-tagged TIF-IA and the localization of recombinant TIF-IA was examined. As shown in Figure 2, tagged TIF-IA co-localizes with endogenous UBF in the nucleolus, indicating that TIF-IA is present at sites of active rDNA transcription.
Fig. 2. Nucleolar localization of TIF-IA. NIH 3T3 cells were transfected with CMV-FLAG-hTIF-IA, fixed in methanol for 1 min at –20°C, washed once with –20°C acetone and several times with phosphate-buffered saline. TIF-IA was visualized by indirect immunofluorescence using M2 antibodies (Sigma, 1:200) and fluorescein isothiocyanate-conjugated goat anti-mouse IgGs (Dianova, 1:300). UBF was stained with anti-UBF serum (1:500) and Texas red-conjugated goat anti-human IgGs (Dianova, 1:200).
As TIF-IA exerts a positive effect on rRNA synthesis, overexpression of the recombinant protein should augment Pol I transcription. To test this, HeLa and NIH 3T3 cells were transfected with increasing amounts of a TIF-IA expression vector together with a reporter plasmid that contains a 5′-terminal murine or human rDNA fragment fused to a 3′-terminal murine rDNA fragment containing two terminator elements. Transcripts were analyzed on northern blots using either pUC9- or CAT-specific probes that hybridize to sequences inserted between the promoter and terminators. In HeLa and NIH 3T3 cells, increasing amounts of TIF-IA stimulated transcription of the respective rDNA reporter (Figure 3), demonstrating that both the human and mouse Pol I transcription apparatuses were activated by the recombinant protein.
Fig. 3. TIF-IA augments transcription of a Pol I reporter gene. HeLa and NIH 3T3 cells were transfected with 10 µg of pHr-CBH or pMr1930-BH and increasing amounts of pCMV-FLAG-hTIF-IA. Transcripts from the reporter plasmid were visualized on northern blots. To normalize for variations in RNA loading, the filter was hybridized with a riboprobe against cytochrome c oxidase.
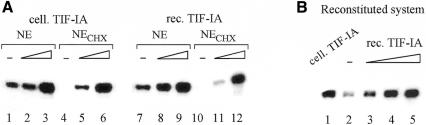
A number of previous studies have demonstrated that glucocorticoid treatment of lymphosarcoma cells, amino acid starvation or drug-induced inhibition of protein synthesis decreases the amount or activity of TIF-IA, and therefore down-regulates Pol I transcription (Buttgereit et al., 1985; Mahajan and Thompson, 1990; Brun et al., 1994). Inhibition of rDNA transcription by cycloheximide treatment of FM3A cells is illustrated in Figure 4A (lanes 4 and 10). Given that cycloheximide treatment decreases the amount or activity of TIF-IA, exogenous TIF-IA should restore transcriptional activity. Indeed, when supplemented with cellular TIF-IA, the activity of extracts from cycloheximide-treated cells was markedly stimulated (lanes 4–6), reaching levels comparable to that of the control extract (lanes 1–3). Notably, the recombinant factor exhibited the same functional properties as cellular TIF-IA, i.e. stimulated transcription of the control extract and rescued transcription in an extract from cycloheximide-treated cells (lanes 7–12). In addition, the recombinant protein was capable of activating transcription in a reconstituted TIF-IA-responsive system. The system consists of recombinant UBF, partially purified TIF-IB and Pol I, the latter containing traces of TIF-IA and saturating levels of TIF-IC, therefore exhibiting some transcriptional activity on its own (Figure 4B, lane 2). When complemented with cellular (lane 1) or recombinant TIF-IA (lanes 3–5), transcription was stimulated. Thus, recombinant TIF-IA expressed in Sf9 cells can replace cellular TIF-IA both in crude extracts and in the reconstituted transcription system.
Fig. 4. TIF-IA activates transcription in vitro. (A) Complementation of transcriptional activity in extracts from cycloheximide-treated cells. Transcription assays contained nuclear extract (40 µg protein) from mouse cells that were cultured either in the absence of cycloheximide (NE) or for 2 h in the presence of 0.1 mg/ml of cycloheximide (NECHX). Reactions were supplemented with 0.5 (lanes 2 and 5) and 1 µl (lanes 3 and 6) of cellular TIF-IA (PL-650 fraction) or 2.5 (lanes 8 and 11) and 5 ng (lanes 9 and 12) of recombinant TIF-IA. (B) Transcription in a reconstituted system. The assays contained 20 ng of template DNA, 4 µl of Pol I (H-400 fraction), 3 µl of TIF-IB (CM-400 fraction) and 2 ng of recombinant UBF. The reactions were complemented with 1 µl of partially purified cellular TIF-IA (PL-650 fraction, lane 1) or increasing amounts (0.5, 1.5 and 3 ng) of recombinant TIF-IA (lanes 3–5).
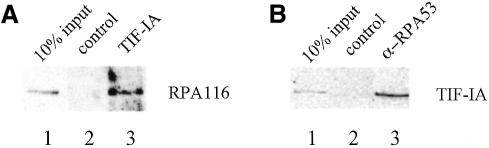
As cellular TIF-IA is associated with a subpopulation of cellular Pol I (Tower and Sollner-Webb, 1987; Schnapp et al., 1993), we wondered whether recombinant TIF-IA is capable of interacting with Pol I. To test this, TIF-IA was immobilized on agarose beads, incubated with purified Pol I, and bound Pol I was monitored on immunoblots using antibodies against RPA116, the second largest subunit of Pol I. Pol I was retained on the TIF-IA affinity resin but not on control beads (Figure 5A). Association of recombinant TIF-IA with Pol I was also demonstrated in a co-immunoprecipitation experiment using radiolabeled TIF-IA and purified Pol I. As shown in Figure 5B, in the presence of Pol I >20% of TIF-IA could be co-precipitated with anti-Pol I serum (Figure 5B).
Fig. 5. TIF-IA interacts with Pol I. (A) Pull-down assay. Immobilized TIF-IA or control beads were incubated with Pol I (MonoS-250) and bead-bound Pol I was detected on immunoblots with antibodies against RPA116. (B) Co-immunoprecipitation of Pol I and TIF-IA. Pol I was incubated with 35S-labeled TIF-IA and immunoprecipitated with either pre-immune (lane 2) or anti-RPA53 serum (lane 3).
To provide definitive evidence that we have cloned TIF-IA rather than a novel stimulatory activity that is homologous to yeast Rrn3, monoclonal antibodies against recombinant TIF-IA were used to detect cellular TIF-IA in Pol I-containing fractions. Pol I elutes in a broad peak from a MonoS column with a maximum in fractions 22–25 (Figure 6A). Antibodies against the recombinant protein recognized a 74 kDa protein in fractions 18–22, i.e. the early eluting fraction of Pol I, which is known to be transcriptionally active. Consistent with this subpopulation of Pol I containing TIF-IA, fractions 18–22 but not ‘bulk’ Pol I were capable of activating transcription in extracts from cycloheximide-treated cells (Figure 6B). This result unambiguously proves that TIF-IA is the mammalian homolog of Rrn3p.
Fig. 6. Antibodies against the recombinant protein recognize cellular TIF-IA. (A) Western blot. Fractions from a MonoS column were assayed on immunoblots for the distribution of Pol I and TIF-IA with antibodies against RPA116 and TIF-IA. One nanogram of recombinant TIF-IA is shown in the first lane (+). (B) Transcriptional activity. Individual fractions were assayed for TIF-IA activity by testing their capability to restore transcription of extracts from cycloheximide-treated cells. As a positive control, the extract was complemented with 3 ng of TIF-IA (+).
DISCUSSION
Regulation of rRNA synthesis in response to external signals that affect the physiology and metabolic state of cells has been well documented, yet the molecular basis of transcriptional regulation remains inadequately understood. It was recognized early that growth-dependent control of rDNA transcription initiation is mediated by TIF-IA, a Pol I-associated factor that is inactivated in starved or cycloheximide-treated cells. Purification of TIF-IA was based on its ability to ‘rescue’ inactive extracts from growth-arrested cells (Buttgereit et al., 1985; Schnapp et al., 1990, 1993; Brun et al., 1994). TIF-IA has been difficult to handle biochemically, and this has greatly hampered the analysis of its mode of action. While this study was submitted for publication, a paper by Moorefield et al. (2000) was published describing cloning of a human cDNA that is related to Rrn3 and is identical to that described here. Significantly, despite 64 C-terminal amino acids of hRrn3 being missing, the human protein was capable of rescuing a lethal yeast strain carrying a disruption of the Rrn3 gene, demonstrating that the factor mediating growth-dependent regulation of eukaryotic Pol I transcription is functionally conserved.
The following results support our conclusion that the mammalian homolog of yeast Rrn3p is TIF-IA. The open reading frame of TIF-IA encodes a 74 kDa protein, similar to the 75 kDa polypeptide that was observed in highly purified cellular TIF-IA preparations (Schnapp et al., 1993). The recombinant protein localizes within the nucleolus, stimulates transcription of a rRNA reporter gene and is capable of restoring transcription in extracts from quiescent cells and in a TIF-IA-responsive reconstituted transcription system. Both TIF-IA and Rrn3p are required for the formation of the first phosphodiester bonds and serve a stoichiometric rather than a catalytic function in initiation. Finally, active yeast Pol I is complexed with Rrn3p (Milkereit and Tschochner, 1998), and initiation-competent mammalian Pol I is associated with TIF-IA (Tower and Sollner-Webb, 1987; Schnapp et al., 1990). Significantly, although extracts from stationary phase yeast cells contained substantial amounts of Rrn3p and Pol I, they lacked the Pol I–Rrn3p complex and were inactive in promoter-dependent transcription (Milkereit and Tschochner, 1998). This result suggests that growth-dependent association of Rrn3p/TIF-IA with Pol I may link rDNA transcription and cell proliferation.
Almost 30 years ago, Yu and Feigelson (1972) reported that down-regulation of cellular rRNA synthesis by nutrient starvation or drug-induced inhibition of protein synthesis is due to a rapidly turning over protein. Subsequent studies demonstrated that growth-dependent transcriptional regulation could be reproduced in vitro. Extracts from exponentially growing cells exhibited high transcriptional activity, whereas extracts from starved or cycloheximide-treated cells were virtually inactive (Buttgereit et al., 1985). In these early experiments, partially purified TIF-IA was used, and therefore protein(s) which co-purifies with TIF-IA could contribute to transcriptional activation. The fact that recombinant TIF-IA activates transcription in extracts from quiescent cells to levels comparable to those in extracts from exponentially growing cells demonstrates that TIF-IA is the principal target for repression of Pol I transcription by inhibitors of protein synthesis. The mechanism by which TIF-IA activity is regulated in response to cell growth is still elusive. Preliminary data indicate that TIF-IA interacts not only with Pol I but also with the TBP-containing factor TIF-IB. This finding suggests that by interaction with DNA-bound TIF-IB, the Pol I–TIF-IA complex, i.e. the initiation-competent subpopulation of Pol I, is recruited to the rDNA promoter. The availability of recombinant TIF-IA will open the path to analyzing the detailed molecular mechanism of a central component of growth control in mammalian cells.
METHODS
Plasmids. To clone the cDNA encoding human TIF-IA, Rrn3 from S. cerevisiae was aligned with homologous cDNAs from S. pombe, C. elegans and A. thaliana to yield a consensus sequence that was used for database searching, EST walking and PCR-based cloning strategies (details of the cloning procedure are available on request). The sequence for TIF-IA has been deposited in the DDBJ/EMBL/GenBank database (accession No. AJ272050). pMr600 contains 5′-terminal murine rDNA sequences from –324 to +292. pMr1930-BH (Budde and Grummt, 1999) and pHr-CBH are fusions between a 5′-terminal human or mouse rDNA fragment and a 3′-terminal BamHI–HindIII murine rDNA fragment separated by pUC sequences in pMr1930-BH, or part of the bacterial chloramphenicol acetyltransferase gene (nucleotides 4751–5001 from pSV2-cat) in pHr-CBH.
Purification of transcription factors and Pol I. Pol I and transcription factors were prepared from cultured mouse cells as described before (Schnapp and Grummt, 1996). Recombinant TIF-IA was expressed as N-terminally FLAG-tagged protein in Sf9 cells and affinity purified by binding to anti-FLAG antibodies followed by elution with 400 µg/ml FLAG epitope peptide.
In vitro transcription assays. Standard reactions (25 µl) contained 40 ng of template pMr600/EcoRI, 12 mM Tris–HCl pH 7.9, 0.1 mM EDTA, 0.5 mM dithiothreitol, 5 mM MgCl2, 80 mM KCl, 12% glycerol, 0.66 mM each ATP, CTP and GTP, 0.01 mM UTP, 1 µCi of [α-32P]UTP and 50 µg of nuclear extract proteins prepared from either exponentially growing or cycloheximide-treated (0.1 mg/ml for 2 h) FM3A cells. Alternatively, TIF-IA activity was assayed in a reconstituted transcription system containing partially purified Pol I (H-400 fraction; Schnapp and Grummt, 1996), TIF-IB (CM-400 fraction), TIF-IA/TIF-IC (PL-650 fraction) and 2–5 ng of recombinant UBF.
Transient transfections. For northern blots, 5 × 105 NIH 3T3 or HeLa cells were co-transfected with 10 µg of a rDNA reporter plasmid and increasing amounts of pCMV-FLAG-hTIF-IA. Total cellular RNA was hybridized (50% formamide, 5× SSC, 50 mM sodium phosphate pH 6.5, 8× Denhardt’s, 0.5 mg/ml yeast tRNA, 0.1% SDS at 68°C) to 32P-labeled probes that are complementary to pUC or CAT sequences inserted between the rDNA promoter and terminator in pMr1930-BH and pHr-CBH.
Interaction of TIF-IA with Pol I. Fifty microliters of partially purified Pol I (MQ-250) and 5 µl (1.5 fmol) of 35S-labeled TIF-IA were incubated in buffer AM-400 for 1 h at room temperature, followed by incubation with 50 µg of α-RPA53 antibodies bound to 12.5 µl of Sepharose G. After washing with 3× 200 µl of AM-400/0.3% NP-40, bound proteins were eluted with 100 mM glycine pH 2.5, separated by SDS–PAGE, and TIF-IA was visualized by autoradiography. Alternatively, 20 µg of bead-bound TIF-IA were incubated with 50 µl of Pol I, washed with buffer AM-400/0.3% NP-40, and Pol I was analyzed on immunoblots using anti-RPA116 antibodies.
Acknowledgments
ACKNOWLEDGEMENTS
We thank W. Ross, P. Elbert and S. Haas for help and advice. This work was supported by the Deutsche Forschungsgemeinschaft, Roche Diagnostics GmbH and the Fond der Chemischen Industrie.
REFERENCES
- Brun R.P., Ryan, K. and Sollner Webb, B. (1994) Factor C*, the specific initiation component of the mouse RNA polymerase I holoenzyme, is inactivated early in the transcription process. Mol. Cell. Biol., 14, 5010–5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde A. and Grummt, I. (1999) p53 represses ribosomal gene transcription. Oncogene, 18, 1119–1124. [DOI] [PubMed] [Google Scholar]
- Buttgereit D., Pflugfelder, G. and Grummt, I. (1985) Growth-dependent regulation of rRNA synthesis is mediated by a transcription initiation factor (TIF-IA). Nucleic Acids Res., 13, 8165–8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I. (1999) Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog. Nucleic Acid Res. Mol. Biol., 62, 109–154. [DOI] [PubMed] [Google Scholar]
- Mahajan P.B. and Thompson, E.A. (1990) Hormonal regulation of transcription of rDNA. Purification and characterization of the hormone-regulated transcription factor IC. J. Biol. Chem., 265, 16225–16233. [PubMed] [Google Scholar]
- Milkereit P. and Tschochner, H. (1998) A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription. EMBO J., 17, 3692–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorefield B., Greene, E.A. and Reeder, R.H. (2000) RNA polymerase I transcription factor Rrn3 is functionally conserved between yeast and human. Proc. Natl Acad. Sci. USA, 97, 4724–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp A. and Grummt, I. (1996) Purification, assay and properties of RNA polymerase I and class I-specific transcription factors in mouse. Methods Enzymol., 273, 233–248. [DOI] [PubMed] [Google Scholar]
- Schnapp A., Pfleiderer, C., Rosenbauer, H. and Grummt, I. (1990) A growth-dependent transcription initiation factor (TIF-IA) interacting with RNA polymerase I regulates mouse ribosomal RNA synthesis. EMBO J., 9, 2857–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp A., Schnapp, G., Erny, B. and Grummt, I. (1993) Function of the growth-regulated transcription initiation factor TIF-IA in initiation complex formation at the murine ribosomal gene promoter. Mol. Cell. Biol., 13, 6723–6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J. and Sollner-Webb, B. (1987) Transcription of mouse rDNA is regulated by an activated subform of RNA polymerase I. Cell, 50, 873–883. [DOI] [PubMed] [Google Scholar]
- Yamamoto R.T., Nogi, Y., Dodd, J.A. and Nomura, M. (1996) RRN3 gene of Saccharomyces cerevisiae encodes an essential RNA polymerase I transcription factor which interacts with the polymerase independently of DNA template. EMBO J., 15, 3964–3973. [PMC free article] [PubMed] [Google Scholar]
- Yu F.L. and Feigelson, P. (1972) The rapid turnover of RNA polymerase of rat liver nucleolus and of its messenger RNA. Proc. Natl Acad. Sci. USA, 69, 2833–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]