Abstract
Transcription factors are crucial to regulate gene expression in immune cells and in other cell types. In lymphocytes, there are a large number of different transcription factors that are known to contribute to cell differentiation and the balance between quiescence and activation. One such transcription factor is E26 Oncogene Homolog 1 (Ets1). Ets1 expression is high in quiescent B and T lymphocytes and its levels are decreased upon activation. The human ETS1 gene has been identified as a susceptibility locus for many autoimmune and inflammatory diseases. In accord with this, gene knockout of Ets1 in mice leads to development of a lupus-like autoimmune disease, with enhanced activation and differentiation of both B cells and T cells. Prior reviews have summarized functional roles for Ets1 based on studies of Ets1 knockout mice. In recent years, numerous additional studies have been published that further validate ETS1 as a susceptibility locus for human diseases where immune dysregulation plays a causative role. In this update, new information that further links Ets1 to human autoimmune diseases is organized and collated to serve as a resource. This update also describes recent studies that seek to understand molecularly how Ets1 regulates immune cell activation, either using human cells and tissues or mouse models. This resource is expected to be useful to investigators seeking to understand how Ets1 may regulate the human immune response, particularly in terms of its roles in autoimmunity and inflammation.
Graphical Abstract
Summary of known roles for Ets1 in regulating B and T cell function. Ets1 regulates B and T cell differentiation to limit excessive activation and development of autoimmunity.
1. INTRODUCTION
The transcription factor Ets1 has emerged as an important regulator of B and T cell activity and is essential for maintaining normal differentiation of these cells. In mice, loss of Ets1 leads to an autoimmune disease that shares features with human lupus (Mouly et al., 2010; Wang et al., 2005). In the absence of Ets1, CD4 T cells undergo enhanced differentiation into T follicular helper type 2 (Tfh2) cells that produce high levels of IL-4 and drive autoimmune pathology (Kim et al., 2018). Ets1 deficient CD4 T cells also give rise to increased numbers of Th17 cells, but reduced numbers and function of regulatory T cells (Treg) (Moisan et al., 2007; Mouly et al., 2010). Ets1 also has important cell-intrinsic roles in preventing premature B cell activation and differentiation into plasma cells (Luo et al., 2014; Sunshine et al., 2019). The levels of Ets1 in B cells are regulated by positive and negative signaling pathways, with B cell receptor (BCR) and Toll-like receptor (TLR) signaling functioning to downregulate Ets1, while inhibitory signaling through CD22-Siglec-G, Lyn and SHP1 maintain Ets1 expression (Luo et al., 2014). Figure 1 along with Tables I and II summarize the main B and T cell phenotypes identified in Ets1 knockout mice and indicate the phenotypes that have been shown to be B cell or T cell intrinsic.
Figure 1. Summary of known roles for Ets1 in regulating B and T cell function.
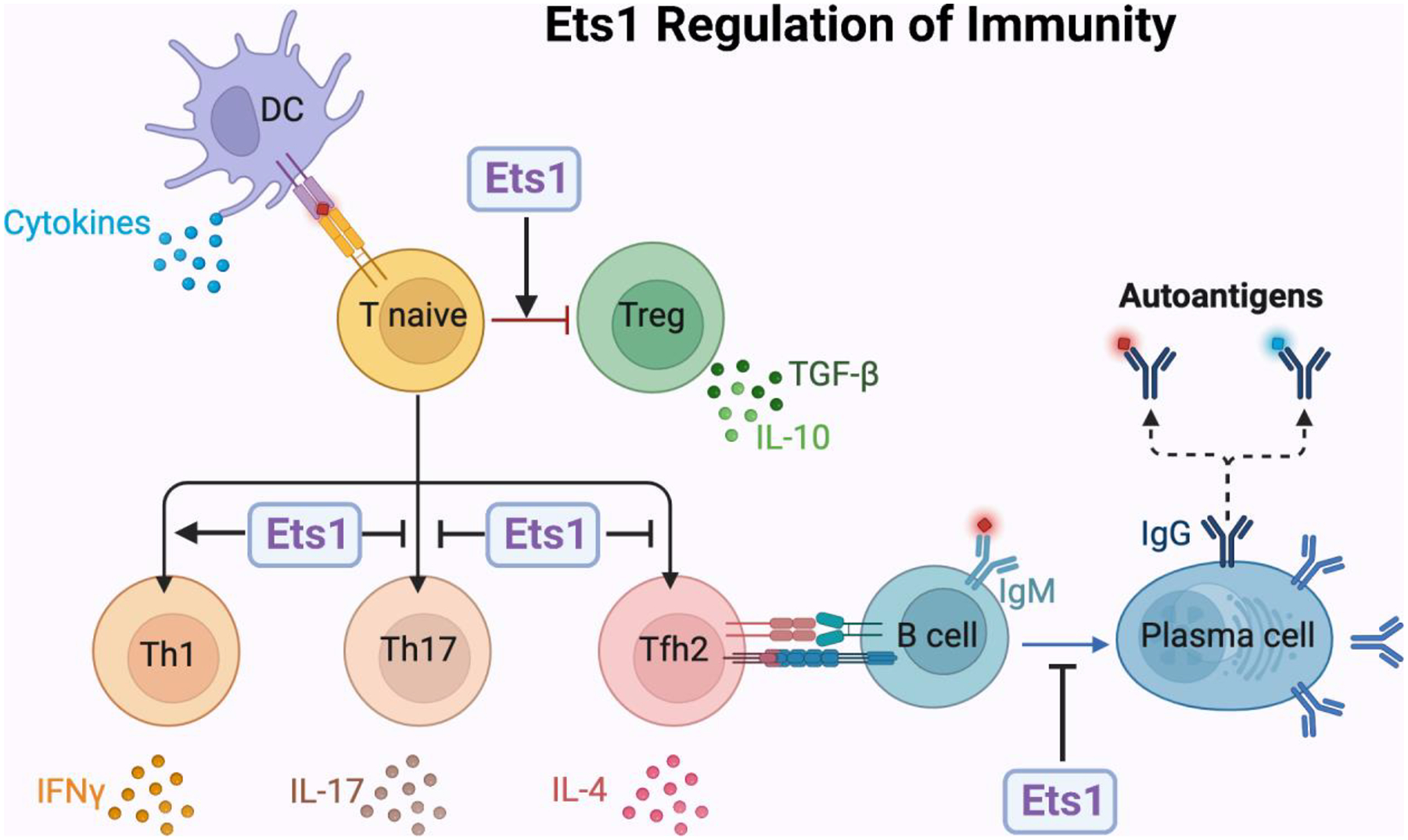
Ets1 is highly expressed in naïve quiescent B and T cells, but is downregulated upon activation. Using mouse models, the role of Ets1 in regulating the differentiation of B and T cells has been extensively studied. The figure summarizes major roles for Ets1 in these processes. In brief, Ets1 functions to inhibit T cell differentiation into T helper 17 (Th17) cells that secrete IL-17 and T follicular helper type 2 (Tfh2) cells that secrete IL-4. Ets1 also functions to inhibit B cell differentiation to antibody-secreting plasma cells. On the other hand, Ets1 promotes the development of regulatory T cells (Tregs) and T helper 1 (Th1) subsets of T cells. The combined actions of Ets1 in these different lineages is important to prevent development of autoimmune disease in Ets1 knockout mice. The functions of Ets1 in human lymphocytes is less well-studied, but current evidence suggests it has roles similar to those described in mice and summarized in the figure.
Table I –
Described Roles of Ets1 in Mature B cells
| B Cell Roles of Ets1 Described in vivo in Mice | Reference(s) |
|---|---|
| Loss of marginal zone type B cells * | (Eyquem et al., 2004; Sunshine et al., 2019; Wang et al., 2005) |
| Increased expression of activation markers | (Sunshine et al., 2019; Wang et al., 2005) |
| Increased differentiation to germinal center B cells | (Russell et al., 2015) |
| Increased differentiation to memory B cells * | (Sunshine et al., 2019) |
| Increased differentiation to plasmablasts/plasma cells * | (Barton et al., 1998; Bories et al., 1995; John et al., 2014; Luo et al., 2014; Sunshine et al., 2019) |
| Increased secretion of autoantibodies * | (Mouly et al., 2010; Sunshine et al., 2019; Wang et al., 2005) |
| Loss of peripheral tolerance to self-antigens | (Russell et al., 2015) |
| Increased secretion of IgM * | (Barton et al., 1998; Bories et al., 1995; Mouly et al., 2010; Nguyen et al., 2012; Sunshine et al., 2019) |
| Increased class-switching to IgG1 and IgE | (Mouly et al., 2010; Nguyen et al., 2012; Sunshine et al., 2019) |
| Decreased class-switching to IgG2a | (Nguyen et al., 2012) |
Functions marked by asterisks are known to be (at least in part) B cell-intrinsic
Table II –
Described Roles of Ets1 in Mature CD4+ T cells
| T Cell Roles of Ets1 Described in vivo in Mice | Reference(s) |
|---|---|
| Reduced numbers of peripheral T cells | (Bories et al., 1995; Clements et al., 2006) |
| Reduced expression of CD5, Thy1 and CD127 | (Bories et al., 1995; Clements et al., 2006; Mouly et al., 2010) |
| Reduced numbers of Tregs and reduced FoxP3 levels | (Mouly et al., 2010) |
| Skewing towards memory phenotype (CD44+) * | (Clements et al., 2006; Kim et al., 2018) |
| Decreased production of IL-2 | (Grenningloh et al., 2005; Nagaleekar et al., 2008) |
| Decreased differentiation to Th1 and Th2 cells | (Grenningloh et al., 2005) |
| Increased differentiation to Tfh2 cells * | (Kim et al., 2018) |
| Increased differentiation to Th17 cells | (Moisan et al., 2007) |
Functions marked by asterisks are known to be T cell-intrinsic
In addition to studies in mice, Ets1 has been implicated in human autoimmune diseases using a variety of different approaches (Sidebar 1). Numerous single-nucleotide polymorphisms (SNPs) in the human ETS1 gene locus have previously been associated with autoimmune and inflammatory diseases (reviewed in (Garrett-Sinha et al., 2016)). Diseases associated with ETS1 SNPs include systemic lupus erythematosus, rheumatoid arthritis, multiple sclerosis, psoriasis, uveitis, ankylosing spondylitis, celiac disease, allergy and atopic dermatitis. This is complemented by studies showing reduced Ets1 expression in PBMCs or T cells of autoimmune patients as compared to healthy controls (Y. Li et al., 2010; Shan et al., 2014; Wei et al., 2014; Wen et al., 2013; Yang et al., 2010). Together, these studies have established Ets1 as an important regulator of the immune response in humans. In this review, we focus on studies published in the past five years that further support and extend a role for Ets1 in regulation of immune cell function and in controlling autoimmune and inflammatory conditions with a focus on roles of Ets1 in human disease.
2. SYSTEMIC LUPUS ERYTHEMATOSUS (LUPUS)
By 2016 there were a large number of studies that linked polymorphisms in the human ETS1 gene with susceptibility to lupus (Bentham et al., 2015; Han et al., 2009; He et al., 2010; Leng et al., 2012; Lessard et al., 2016; Lu et al., 2015; Molineros et al., 2014; Morris et al., 2016; Sullivan et al., 2000; Wang et al., 2013; Yang et al., 2010; Zhang et al., 2013; Zhong et al., 2011). The association of ETS1 SNPs with lupus has been most thoroughly demonstrated in Asian populations, although there is also evidence that links SNPs in ETS1 to lupus susceptibility in European populations (Bentham et al., 2015; Morris et al., 2016; Wang et al., 2013). Furthermore, the levels of Ets1 mRNA have been shown to be reduced in PBMCs and T cells of lupus patients (He et al., 2010; Wen et al., 2013), suggesting that SNPs in ETS1 may lead to lower expression of the gene. Indeed, the risk allele of SNP rs6590330 located downstream of ETS1 results in increased binding of STAT1 and this is correlated with reduced expression of Ets1 mRNA (Lu et al., 2015). Another SNP, rs1128334, is in the 3’UTR of Ets1 and regulates mRNA abundance in PBMCs (Yang et al., 2010). Some SNPs are also linked with the occurrence of particular lupus clinical manifestations including age, skin rashes, arthritis, vasculitis or renal involvement (He et al., 2010; Sullivan et al., 2000; Zhong et al., 2011).
More recently, several additional pieces of data have added to the linkage of Ets1 to lupus. A meta-analysis combining multiple GWAS studies further confirmed the association of two ETS1 autoimmune-associated SNPs (rs1128334 and rs10893872) with lupus (Zhou et al., 2015). An additional study has also demonstrated that Ets1 expression is low in the lupus T cells as compared to T cells from healthy donors (Kim et al., 2018). In this latter study, the expression level of Ets1 in CD4+ T cells of lupus patients was shown to inversely correlate with SLEDAI scores and serum anti-dsDNA titers. In a Chinese population, Ets1 mRNA was found to be decreased more significantly in PBMCs of new onset SLE than in patients with inactive SLE (Sun et al., 2016). The large body of work associating ETS1 SNPs with lupus in many different populations around the world and from various ethnic groups makes a strong case that this gene is essential for control of human immune responses.
2.2. Roles for Ets1 in B cells and antibody production
While the prior studies described above had shown Ets1 mRNA levels to be low in total PBMCs and in T cells of lupus patients, new data show that Ets1 is also low in B cells of lupus patients (Jin et al., 2019; Zhang et al., 2019). B cells derived from patients with active SLE express the lowest levels of Ets1 mRNA, while those with inactive disease show significantly higher Ets1 mRNA expression (Jin et al., 2019). A variety of microRNAs are known to regulate Ets1 expression by binding to its mRNA transcript, resulting in degradation of the mRNA and/or impaired translation (Sidebar 2). One such miRNA is miRNA-5003 and binding of miRNA-5003 to the 3’UTR of the Ets1 transcript is increased by the presence of the “T” allele of the rs4937333 SNP. The presence of “T” at rs4937333 is associated with reduced levels of Ets1 and with development of SLE (Zhang et al., 2019). Infection of primary human B cells with lentiviruses carrying the Ets1 mRNA with either the “T” or “A” alleles of rs4937333 showed that Ets1 expression can block human B cell differentiation to plasmablasts in response to stimulation with CpG containing DNA oligos (Zhang et al., 2019). The “A” allele of the Ets1 mRNA was more effective at blocking differentiation than the “T” allele, presumably because it is less susceptible to miRNA-5003-induced effects. The Zhang et al study also showed that miR-5003 levels are higher in B cells from SLE patients than in B cells from controls. Interestingly, miR-5003 levels are higher in B cells than in T cells in SLE patients, but equivalent in B cells and T cells of controls.
The levels of Ets1 mRNA were inversely correlated with the percent of CD19+CD138+ plasmablasts among total CD19+ cells (Jin et al., 2019). The level of Ets1 mRNA was also inversely correlated with the SLEDAI score, a measure of lupus disease activity. On the other hand, Ets1 mRNA levels were negatively correlated with anti-nuclear autoantibody titers and erythrocyte sedimentation rate (ESR) (Jin et al., 2019). Ets1 levels were positively correlated with the levels of white and red blood cells, hemoglobin and complement C3 and C4.
A subset of human B cells that are CD11c+ and lack expression of CD27, IgD and CXCR5 (referred to as DN2 B cells) are expanded in lupus, particularly in African American lupus patients, and this subset has been shown to be enriched for autoreactive cells (Jenks et al., 2018). DN2 B cells are poised to differentiate into plasma cells, which can be triggered by stimulation by incubation with TLR7 ligand, IL-21 and IFNγ. DN2 cells yield increased numbers of antibody-secreting plasma cells as compared to other B cell subsets cultured under the same conditions. The DN2 subset of B cells is characterized by low expression of Ets1 as compared to activated naïve B cells (Jenks et al., 2018), which is consistent with their being primed for plasma cell differentiation. Together, these studies show important roles for Ets1 in regulating B cell activity in lupus.
2.3. Roles for Ets1 in type I interferon responses
Plasmacytoid dendritic cells are a specialized type of lymphoid dendritic cells and are the main cell type that produce type I interferons, which are known to be pathogenic in lupus (Obermoser & Pascual, 2010). As has been seen in other patient populations, Ets1 mRNA levels are low in PBMCs of Japanese lupus patients (Suzuki et al., 2022). Furthermore, the level of Ets1 mRNA was inversely correlated with the level of prednisolone treatment, suggesting that patients with low Ets1 had more severe disease requiring higher doses of immunosuppression. In this Japanese cohort, Ets1 levels were not significantly correlated with the numbers of plasmacytoid dendritic cells (pDCs) in the peripheral blood, but there was a significant positive correlation between the levels of Ets1 mRNA and the levels of mRNA for interferon-inducible genes (IFIT1, IFI44 and EIF2AK2). Since Ets1 is a transcription factor, it may play a role in regulation of interferon-inducible genes. Type I interferons activate Stat 1 signaling and Ets1 has been shown to cooperate with Stat1 to regulate gene expression (Nguyen et al., 2012; Yockell-Lelievre et al., 2009). Thus, Ets1 seems to not directly affect pDC development or survival, but may play a role in responses of target cells to type I interferons that are produced by pDC.
2.4. Roles for Ets1 in T cells and cytokine production
Ets1 has previously been implicated in regulating the differentiation programs of T cells. In humans, Sun et al showed that Tregs of SLE patients have reduced levels of Ets1, possibly due to increased expression of miR-326 (Sun et al., 2016). In another study, lupus patients that had lower levels of Ets1 in CD4 T cells had increased percentages of T follicular helper type 2 (Tfh2) cells (Kim et al., 2018), suggesting that Ets1 inhibits Tfh2 cell differentiation. Tfh2 cells are found in germinal centers, where they function to secrete IL-4, which is known to stimulate B cell proliferation and class-switching (Tangye et al., 2002).
The SNPs that have been previously linked to autoimmune diseases are relatively common variants, since genome-wide association studies (GWAS) tend to assess SNPs present at a reasonably high rate in the population. Many of these disease-associated SNPs may not be causal variants, but rather be in linkage disequilibrium with the important variants. In order to identify potential causal variants, a study by Jones et al combined analysis of DNA regulatory regions (promoters, enhancers, etc identified using Regulome DB scores) with analysis of disease-associated SNPs to identify SNPs that might be in linkage disequilibrium with causative variants in the nearby promoters and enhancers (Jones et al., 2019). Novel SNPs in these potential regulatory regions were then identified using high resolution melt (HRM) analysis. Using this technique, the authors identified a novel and rare SNP in linkage disequilibrium with lupus-associated SNP rs1128334 (Jones et al., 2019). While deletion of Ets1 from all T cells drove autoimmune responses, restricting the deletion of Ets1 to regulatory T cells by using a FoxP3-Cre line did not result in development of autoimmunity (Kim et al., 2018). This implies that Ets1 has a more important role in regulating proper effector T cell differentiation than in promoting development or function of Tregs.The rare variant identified at this locus was included within a potential FOXP3 binding motif, suggesting it might directly affect Ets1 gene regulation in Treg cells. In a luciferase reporter assay, the presence of a sequence containing the novel disease-associated variant was found to increase luciferase expression over the presence of the wild-type sequence (Jones et al., 2019). The association studies described in the sections above suggest the likely importance of Ets1 in regulating T cell responses in the context of lupus.
3. RHEUMATOID ARTHRITIS
3.1. Recent studies on SNPs and Ets1 expression in Rheumatoid Arthritis
Similar to what has been described above for lupus, SNPs in the human ETS1 locus have been associated with susceptibility to rheumatoid arthritis (RA) and with certain clinical phenotypes in RA including the disease activity score (DAS28) and the level of serum C-reactive protein level (Chatzikyriakidou et al., 2013; Chen et al., 2015; Freudenberg et al., 2011; Kim et al., 2015; Okada et al., 2012; Okada et al., 2014; Zhang et al., 2014). Many of these earlier studies have defined RA-associated SNPs in Asian or European populations. Extending the association of ETS1 and RA to other populations, Danila et al used high density genotyping to identify SNPs associated with RA in African American populations in the United States and correlate their presence with disease severity on radiographic images (Danila et al., 2017). This analysis identified SNP rs4362159 located at approximately 23 kb downstream of the ETS1 gene as being associated with severity of joint damage on radiographic images in African American subjects (Danila et al., 2017). A second linked ETS1 SNP rs7108537, about 28 kb downstream of ETS1 and located in a region rich in transcription factor binding sites, was also associated with radiographic severity (Danila et al., 2017). A third ETS1 SNP (rs73013527) was identified in a later study comparing susceptibility loci for African American subjects to those of Asian and European populations (Laufer et al., 2019). In a study of Chinese populations, another SNP in ETS1 (rs73013527) was shown to be positively correlated with serum RANKL levels (Yang et al., 2021). Elevated RANKL contributes to RA by promoting differentiation of osteoclasts that can break down bone (Komatsu & Takayanagi, 2022).
While SNPs in ETS1 have been linked to RA susceptibility and clinical phenotypes, until recently there was no direct evidence that Ets1 mRNA levels were altered in immune cells of RA patients. A recent study of Japanese patients confirmed that expression of Ets1 is decreased in PBMCs of RA patients when compared to healthy donors (Suzuki et al., 2022). Interestingly, expression of the FLI1 gene, which is located immediately adjacent to ETS1 and oriented in a head-to-head manner, also showed a decrease in PBMCs of RA patients. This suggests that these two genes may be coordinately regulated (Suzuki et al., 2022).
One of the SNPs previously associated with development of RA is rs1128334 (Zhang et al., 2014), located in the 3’UTR of Ets1. This SNP is also associated with other autoimmune diseases including lupus and ankylosing spondylitis (Lessard et al., 2016; Mansouri et al., 2016; Shan et al., 2014; Zhang et al., 2013). In silico tools were used to predict the potential effects of 152 SNPs associated with RA, including SNP rs1128334. This analysis identified rs1128334 as a potentially damaging SNP, since the risk allele is predicted to create a BRD-BOX site in the 3’ UTR (Akhtar et al., 2021). The presence of a BRD-BOX motif in a UTR is associated with reduced mRNA and protein levels (Lai & Posakony, 1997), because the BRD-BOX serves as a miRNA seed sequence binding region (Lai, 2002; Lai et al., 2005). The BRD-BOX motif generated by the “A” allele of rs1128334 is predicted to change binding sites for several miRNAs and this is predicted to potentially result in reduced stability or translation of Ets1 mRNA (Akhtar et al., 2021). Indeed, reduced levels of Ets1 mRNA containing the “A” allele at rs1128334 than Ets1 mRNA containing the “G” allele at rs1128334 in individuals heterozygous for the SNP has been noted (Yang et al., 2010). Another study has also identified rs1128334 as a SNP that might interfere with miRNA binding (Richardson et al., 2011).
3.2. Ets1 as a transcription factor relevant to RA pathogenesis
When studying CD19+ B cells from RA patients, cells from patients with a high interferon gene signature (IGS), implying high levels of type I interferon exposure, showed altered CpG methylation of DNA (Cooles et al., 2022). Examining sites of differential methylation for potential transcription factor binding indicated that Ets1 binding motifs were enriched in the methylated CpG regions of CD19+ B cells from patients with high IGS. Binding of Ets1 to DNA is known to be inhibited by methylation of its consensus binding motif (Polansky et al., 2010; Stephens & Poon, 2016). Therefore, hypermethylation of Ets1 binding sites in promoters and enhancers of target genes could lead to inhibited activation by Ets1 protein. This result suggests that Ets1 functional activity in binding to DNA may be altered in RA via changes in DNA methylation, even in patients where there may be no SNPs that affect ETS1 gene regulation.
4. INFLAMMATORY BOWEL DISEASE
Inflammatory bowel disease (IBD) is a generic term describing chronic inflammatory conditions of the gut and includes both ulcerative colitis (UC) and Crohn’s disease (CD). Supporting a possible role for Ets1 in inflammatory syndromes of the gut is the identification of SNPs in the human ETS1 gene locus as a susceptibility alleles for celiac disease (Dubois et al., 2010; Trynka et al., 2011). An analysis of gastrointestinal biopsy samples from patients with IBD found significantly increased protein levels of Ets1 in the colonic tissue in active ulcerative colitis compared to active Crohn’s disease, patients with non-IBD colitis or healthy controls (Konno et al., 2004). Another study comparing gene expression patterns in two bowel locations (ileum and colon) and in three disease states (control, CD or UC), also found Ets1 to be downregulated in CD as compared to control samples (Hughes, 2005). However, two more recent studies have found Ets1 to be upregulated in both CD and UC and its levels were highest in the inflamed regions of the gut as compared to less affected areas and positively correlated with disease severity (Ge et al., 2019; He et al., 2022). This is consistent with the high expression of Ets1 in lymphocytes, which are enriched at sites of inflammation. Ets1 mRNA was also found to be elevated in the peripheral blood T cells of UC and CD patients as compared to healthy donors (He et al., 2022). The relevance of this finding is unclear, since Ets1 is typically reduced in T cells upon their activation. However, it may be that peripheral blood T cells in IBD patients are not particularly activated and that only gut-resident T cells might show decreased Ets1 levels.
Treatment of healthy donor CD4 T cells with IL-1β or TNF-α increases Ets1 mRNA levels (He et al., 2022). In contrast, IBD patients that respond clinically to the anti-TNF-α drug infliximab (IFX) show a reduced expression of Ets1 mRNA in CD4 T cells after the same treatment. In another study, the Ets1 DNA binding motif was found to be enriched in the promoters and enhancers of genes that were over-expressed in intestinal biopsies of patients with active Crohn’s disease or active ulcerative colitis as compared to controls (Boyd et al., 2018). Altogether, accumulating evidence is reinforcing the link between alterations in Ets1 levels and function with development of IBD.
5. MULTIPLE SCLEROSIS
Ets1 has also been implicated in the pathogenesis of multiple sclerosis (MS). ETS1 SNPs were identified that were associated with susceptibility to multiple sclerosis development (Du et al., 2009; Lill et al., 2015). The levels of Ets1 mRNA were reported to be reduced in CD4+ T cells from multiple sclerosis patients (Du et al., 2009). A new study has confirmed reduced Ets1 mRNA expression in PBMCs of MS patients and further demonstrated that Ets1 is particularly low in patients undergoing a relapse of the disease (Li et al., 2020). The same study also showed increased levels of the miRNA miR-1–3p in PBMCs and CD4+ Th17 cells of MS patients (Li et al., 2020). miR1–3-p levels are positively correlated with expression of Th17 transcription factor RORγt and with the levels of IL-17A in serum and cerebrospinal fluid. Ets1 is known to suppress the differentiation of Th17 cells (Moisan et al., 2007) and miR1–3-p targets the 3’ UTR of Ets1 leading to degradation of the Ets1 mRNA. In keeping with this, miR1–3-p levels are inversely correlated with Ets1 levels in MS patients’ PBMCs. Further studies on the roles of Ets1 in MS would be valuable to further define the role of this gene in regulating autoimmunity in the central nervous system.
6. PSORIASIS
Psoriasis is an autoimmune disease affecting the skin and, in the case of psoriatic arthritis, also the joints. Prior GWAS studies have identified SNPs in ETS1 associated with development of psoriasis (Tsoi et al., 2012; Yin et al., 2015). Anti-TNFα therapies used in the treatment of inflammatory bowel disease (IBD) can trigger development therapy-associated psoriasis as a side effect (Vedak et al., 2016). In order to identify genetic factors that might result in susceptibility to psoriasis upon anti-TNF treatment, GWAS analysis was done for loci that have been associated with inflammatory or immune-mediated diseases (Cortes & Brown, 2011; Vedak et al., 2016). This analysis showed that the minor G allele for SNP rs3802826 in the human ETS1 gene was found at lower frequency in IBD patients who developed anti-TNF associated psoriasis than in control patients (Vedak et al., 2016). This suggests that SNP rs3802826 has a protective effect against the development of TNF-associated psoriasis. While studies in psoriasis are still at a relatively early stage, the available data indicate a likely association of Ets1 to the pathogenesis of this disease.
7. ATOPIC DERMATITIS
Atopic dermatitis (AD) is an inflammatory condition of the skin characterized by inflammation, redness, and irritation of the skin. Prior studies have linked SNPs in ETS1 to development of AD (Hinds et al., 2013; Paternoster et al., 2015). More recently, SNP rs7127307 in ETS1 has been further validated to be linked to atopic dermatitis using a bioinformatics pipeline that correlates numerous molecular properties (expression patterns and protein and DNA methylation QTL datasets in skin or immune-relevant tissues) to prioritize candidate causal variants of that disease (Sobczyk et al., 2021). In a meta-analysis of a Japanese population along with samples in the UK Biobank, SNP rs73018933 located 150 kb downstream of ETS1 was found to be associated with atopic dermatitis (Tanaka et al., 2021). The cytokine IL-22 is over-expressed in AD skin and thought to contribute to disease pathogenesis. Anti–IL22 antibody (fezakinumab) is a potential therapy for AD. To understand the genetic contributions that regulate patient outcomes in response to fezakinumab treatment, clinical disease scores were correlated with differences in gene expression between lesional and non-lesional skin of patients with AD (Brunner et al., 2019). This analysis sought to detect genes whose baseline expression levels were correlated to the outcome of fezakinumab treatment and ETS1 was one such gene. These studies implicate Ets1 as an important player in AD pathogenesis.
8. OTHER AUTOIMMUNE AND INFLAMMATORY DISEASES
8.1. Further linkage of ETS1 SNPs to autoimmune and inflammatory diseases
As described above, there are already many studies linking Ets1 to a variety of autoimmune and inflammatory conditions. Several new associations have been detected recently in additional disease states. ETS1 SNP rs1128334, previously associated with lupus and ankylosing spondylitis, is also associated with the development granulomatosis with polyangiitis (GPA, previously known as Wegener’s granulomatosis), particularly the PR3–ANCA positive sub-type of vasculitis (Kawasaki et al., 2018). A SNP previously associated with lupus (rs4937333) has recently been linked to susceptibility in primary biliary cholangitis (Xu et al., 2019). In addition, a novel ETS1 SNP rs7117932 and a SNP previously associated with lupus rs6590330 were linked with development of dermatomyositis and idiopathic inflammatory myopathies (Chen et al., 2017). Altogether, SNPs in the ETS1 locus have been linked to susceptibility to at least 12 different autoimmune and inflammatory conditions (Table III).
Table III –
Summary of Studies Linking Ets1 to Human Autoimmune and Inflammatory Diseases
| Study | Disease | Type of Analysis | Population(s) |
|---|---|---|---|
| (Sullivan et al., 2000) | Systemic Lupus Erythematosus | Candidate gene approach | U.S. population |
| (Konno et al., 2004) | Inflammatory Bowel Disease | Candidate gene approach | Japanese population |
| (Hughes, 2005) | Crohn’s Disease | Microarray for RNA expression | European population |
| (Du et al., 2009) | Multiple Sclerosis | Target of miRNA | Chinese population |
| (Han et al., 2009) | Systemic Lupus Erythematosus | GWAS | Chinese population |
| (Dubois et al., 2010) | Celiac Disease | GWAS | European population |
| (He et al., 2010) | Systemic Lupus Erythematosus | Candidate gene approach | Chinese population |
| (Y. Li et al., 2010) | Systemic Lupus Erythematosus | Candidate gene approach | Chinese population |
| (Yang et al., 2010) | Systemic Lupus Erythematosus | GWAS | Chinese and Thai populations |
| (Trynka et al., 2011) | Celiac Disease | GWAS | European and Indian populations |
| (Zhong et al., 2011) | Systemic Lupus Erythematosus | Candidate gene approach | Chinese population |
| (Freudenberg et al., 2011) | Rheumatoid Arthritis | GWAS | Korean population |
| (Leng et al., 2012) | Systemic Lupus Erythematosus | Candidate gene approach | Chinese population |
| (Okada et al., 2012) | Rheumatoid arthritis | Meta-analysis of multiple GWAS | Japanese population |
| (Tsoi et al., 2012) | Psoriasis | Meta-analysis of multiple GWAS | U.S. and European populations |
| (Chatzikyriakidou et al., 2013) | Rheumatoid Arthritis | Candidate gene approach | European population |
| (Wang et al., 2013) | Systemic Lupus Erythematosus | Candidate gene approach | U.S. and European populations |
| (Wen et al., 2013) | Systemic Lupus Erythematosus | Target of miRNA | Chinese population |
| (Hinds et al., 2013) | Atopic Dermatitis | GWAS | U.S. and UK populations |
| (Zhang et al., 2013) | Systemic Lupus Erythematosus | Candidate gene approach | Chinese population |
| (Molineros et al., 2014) | Systemic Lupus Erythematosus | GWAS | Malaysian population |
| (Okada et al., 2014) | Rheumatoid Arthritis | Meta-analysis of multiple GWAS | European and Asian populations |
| (Shan et al., 2014) | Ankylosing Spondylitis | Candidate gene approach | Chinese population |
| (Wei et al., 2014) | Pediatric Uveitis | Candidate gene approach | Chinese population |
| (Zhang et al., 2014) | Rheumatoid Arthritis | Candidate gene approach | Chinese population |
| (Bentham et al., 2015) | Systemic Lupus Erythematosus | GWAS | European and US populations |
| (Chen et al., 2015) | Rheumatoid Arthritis | Candidate gene approach | Chinese population |
| (Kim et al., 2015) | Rheumatoid Arthritis | GWAS | Korean population |
| (Lill et al., 2015) | Multiple Sclerosis | Candidate gene approach | European population |
| (Lu et al., 2015) | Systemic Lupus Erythematosus | Candidate gene approach | United States, Asian, and European populations |
| (Paternoster et al., 2015) | Atopic Dermatitis | Meta-analysis of multiple GWAS | European, African, Japanese and Latino populations |
| (Yin et al., 2015) | Psoriasis | Meta-analysis of multiple GWAS | Chinese and Caucasian populations |
| (Lessard et al., 2016) | Systemic Lupus Erythematosus | GWAS | Korean population |
| (Mansouri et al., 2016) | Systemic Lupus Erythematosus | Candidate gene approach | Iranian population |
| (Morris et al., 2016) | Systemic Lupus Erythematosus | GWAS | Chinese and European populations |
| (Na et al., 2016) | Behçet’s disease | Target of miRNA | Korean population |
| (Sun et al., 2016) | Systemic Lupus Erythematosus | Target of miRNA | Chinese population |
| (Vedak et al., 2016) | TNF-α Antagonist Associated Psoriasis in Inflammatory Bowel Diseases | GWAS | U.S. population |
| (Chen et al., 2017) | Idiopathic Inflammatory Myopathies | Candidate gene approach | Chinese population |
| (Danila et al., 2017) | Rheumatoid Arthritis | GWAS | U.S. population |
| (Boyd et al., 2018) | Inflammatory Bowel Disease | In silico predictions | N/A |
| (Jenks et al., 2018) | Systemic Lupus Erythematosus | RNA-seq | U.S. population |
| (Kawasaki et al., 2018) | Granulomatosis with Polyangiitis | Candidate gene approach | Japanese population |
| (Kim et al., 2018) | Systemic Lupus Erythematosus | Candidate gene approach | Korean population |
| (Weeding et al., 2018) | Anti-phospholipid Syndrome | CpG Methylation Assay | U.S. population |
| (Ge et al., 2019) | Inflammatory Bowel Disease | Target of miRNA | Chinese population |
| (Jin et al., 2019) | Systemic Lupus Erythematosus | Target of miRNA | Chinese population |
| (Jones et al., 2019) | Systemic Lupus Erythematosus | In silico predictions | N/A |
| (Laufer et al., 2019) | Rheumatoid Arthritis | Meta-analysis of multiple GWAS | Multiple populations |
| (Xu et al., 2019) | Primary Biliary Cholangitis | Candidate gene approach | Chinese population |
| (Zhang et al., 2019) | Systemic Lupus Erythematosus | Candidate gene approach | Chinese population |
| (Li et al., 2020) | Multiple Sclerosis | Target of miRNA | Chinese |
| (Akhtar et al., 2021) | Systemic Lupus Erythematosus | In silico predictions | N/A |
| (Sobczyk et al., 2021) | Atopic Dermatitis | In silico predictions | N/A |
| (Tanaka et al., 2021) | Atopic Dermatitis | GWAS | Japanese population |
| (Wang et al., 2021) | Asthma | Microarray for RNA expression | Chinese population |
| (Yang et al., 2021) | Rheumatoid Arthritis | Candidate gene approach | Chinese population |
| (Cooles et al., 2022) | Rheumatoid Arthritis | In silico predictions | European population |
| (He et al., 2022) | Inflammatory Bowel Disease | Candidate gene approach | Chinese population |
| (Suzuki et al., 2022) | Systemic Lupus Erythematosus | Candidate gene approach | Japanese population |
7.2. Additional regulatory pathways involved in controlling Ets1 in autoimmune and inflammatory conditions
In patients with the autoimmune disease Behçet’s syndrome CD4+ T cells show increased differentiation into Th17 cells producing the cytokine IL-17 (Na et al., 2016). This was associated with reduced expression of Ets1 protein and higher expression of microRNA miR-155, which is known to target the Ets1 mRNA for degradation (Burocchi et al., 2015; Hu et al., 2013; Wen et al., 2013). Another microRNA implicated in regulating Ets1 levels is miR-451a. PBMCs isolated from asthmatic children showed increased expression of miR-451a and decreased expression of Ets1 as compared to controls (Wang et al., 2021). Naïve human CD4+ T cells cultured under Th2 cell polarizing conditions showed decreased expression of miR-451a and increased expression of Ets1. An additional mechanism implicated in controlling Ets1 expression is CpG methylation. The human ETS1 gene was shown to be hypomethylated in the DNA of neutrophils of patients with primary anti-phospholipid syndrome (APS) compared to normal controls (Weeding et al., 2018). The hypomethylation of the ETS1 gene suggests that it may be over-expressed in neutrophils of APS patients, although this was not directly tested. In summary, many different experimental approaches have linked Ets1 to a panoply of human diseases where there is excessive activation of immune responses, indicating that Ets1 is a major regulator of the human immune system.
8. MOUSE STUDIES THAT SUPPORT RESULTS IN HUMANS
The information discussed above focuses on roles for Ets1 in regulating immune function in the context of human autoimmune and inflammatory diseases. The functions of Ets1 have also been extensively analyzed using mouse models, particularly knockout mice lacking Ets1. These animal studies support many of the observations in humans and hence strengthen the link of Ets1 to autoimmune phenomena. In addition, certain mechanistic studies can be done in mice that are not possible in human patients and these studies can provide additional insight into the functions of Ets1 in regulating immunity.
8.1. Ets1 control of immune cell differentiation
Deletion of the mouse Ets1 gene leads to development of autoimmune disease, with similarities to human SLE (Kim et al., 2018; Mouly et al., 2010; Sunshine et al., 2019; Wang et al., 2005). Both B cells and T cells in Ets1 knockout mice show evidence of hyper-activation and indeed Ets1 has cell-type intrinsic functions in both B cells and T cells (Kim et al., 2018; Sunshine et al., 2019). In B cells, Ets1 is able to block differentiation into plasma cells and the secretion of antibodies (John et al., 2014; John et al., 2008; Luo et al., 2014). Ets1-deficient mice lack marginal zone B cells and B-1a B cells (Eyquem et al., 2004; Wang et al., 2005). These two types of B cells are poised to become plasma cells and differentiate more readily than conventional follicular B cells (Genestier et al., 2007; Martin et al., 2001). Thus, their loss in Ets1−/− mice could potentially be due to their rapid differentiation into antibody-secreting cells. Using transgenic mouse models, Ets1 was shown to be essential for peripheral B cell tolerance to self-antigens, although it is dispensable for clonal deletion of highly autoreactive clones in the bone marrow (Russell et al., 2015).
Within the T cell compartment, Ets1 regulates T cell differentiation into different CD4+ T helper (Th) subsets. In particular, Ets1-deficient CD4 T cells isolated from mice failed to properly differentiate into Th1 cells when cultured under appropriate skewing conditions, but showed enhanced differentiation into Th17 cells (Grenningloh et al., 2005; Moisan et al., 2007; Strempel et al., 2010). Ets1 knockout mice also show alterations in the numbers and functions of Tregs (Mouly et al., 2010). In terms of Th2 cells, prior studies were confusing in that naïve CD4+ T cells isolated from Ets1 knockout mice failed to differentiate into IL-4 secreting cells when cultured in vitro under standard Th2 conditions (Grenningloh et al., 2005; Strempel et al., 2010), while freshly isolated CD4+ T cells from Ets1 knockout mice showed strongly enhanced production of IL-4 and other Th2 cytokines (Mouly et al., 2010). This discrepancy was resolved when it was shown that T cells lacking Ets1 (from T cell-specific Ets1 knockout mice) differentiate robustly to T follicular helper cells that secrete IL-4 (Tfh2 cells) (Kim et al., 2018). Hence, Tfh2 but not Th2 cells seem to be the main source of IL-4 in vivo in Ets1-deficient mice. Transfer of Tfh2 cells from Ets1 knockout mice to wild-type hosts drives development of autoimmune disease in the host mice (Kim et al., 2018). While deletion of Ets1 from all T cells resulted in autoimmune disease, restricting the deletion of Ets1 to regulatory T cells by using a FoxP3-Cre line did not result in development of autoimmunity (Kim et al., 2018). This implies that Ets1 has a more important role in regulating proper effector T cell differentiation than in promoting development or function of Tregs.
8.2. Molecular mechanisms of Ets1 action in B and T cell differentiation
There are several proposed mechanisms for Ets1 action in regulating immune cell differentiation. In B cells, Ets1 has been shown to block differentiation to antibody-secreting cells by interfering with the activity of plasma cell transcription factor Blimp1, while maintaining expression of the B cell transcription factor Pax-5 (John et al., 2014; John et al., 2008). In addition, Ets1 also binds to regulatory regions of a panoply of target genes that function in B cell differentiation and regulates their expression levels (Saelee et al., 2017). Ets1-deficient B cells have a defect in switching to IgG2a and this is due to a role for Ets1 in inducing expression of the transcription factor T-bet (Nguyen et al., 2012). T-bet is induced by interferon-gamma signaling and is required in B cells for switching to IgG2a. Together, these data show that Ets1 functions both by directly binding to and regulating target genes, but also by forming protein-protein interactions with other transcription factors to modulate their activity.
In T cells, Ets1 also regulates important target genes involved in T cell differentiation and function. Th1 differentiation is impaired in Ets1 knockout mice and this is due for a role of Ets1 in cooperating with T-bet to induce expression of the interferon-gamma gene (Grenningloh et al., 2005). Ets1 also seems to play a role in regulating expression of Th2 family cytokines. Naïve CD4 cells lacking Ets1 that are cultured in Th2 conditions fail to express IL-4, IL-5 or IL-13 (Grenningloh et al., 2005; Strempel et al., 2010). These three cytokine genes are clustered in a Th2-cytokine gene locus (Lee et al., 2006) and Ets1 was shown to bind to multiple sites scattered in this locus suggesting Ets1 may directly regulate their expression (Strempel et al., 2010). In contrast to impaired generation of Th1 and Th2 cells, generation of Th17 cells is enhanced in the absence of Ets1 (Moisan et al., 2007). IL-2 is known to inhibit Th17 development (Laurence et al., 2007) and Ets1-deficient T cells have reduced production of IL-2. In addition, Th cells from Ets1 knockout mice show a defect in the ability of IL-2 to suppress Th17 differentiation (Moisan et al., 2007). In terms of understanding how Ets1 controls IL-2 production, Tsao et al showed that Ets1 functions to help release the transcription factor NFAT from repressors the cytoplasm allowing it to enter the nucleus upon calcium signaling (Tsao et al., 2013). Another potential mechanism influencing Th17 differentiation in the absence of Ets1 is the observation that Ets1 knockout CD4 T cells express increased levels of gp130, a signaling component of the IL-6 receptor, and have increased phospho-STAT3 (Lee et al., 2019). Indeed, Ets1 binds to the promoter of the gp130 gene and can presumably directly induce gp130 expression. Enhanced IL-6 signaling in Ets1 knockout T cells seems to promote Th17 differentiation since inhibiting gp130 signaling with a specific inhibitor results in reduced IL-17A and IL-22 production (Lee et al., 2019).
In regulatory T cells, Ets1 has been shown to induce FoxP3 expression by binding to the CNS2 regulatory enhancer element of the gene (Mouly et al., 2010; Polansky et al., 2010). T follicular helper (Tfh) cells are also increased in mice with a T cell-specific knockout of Ets1 and in particular there is an increase in Tfh cells that secrete IL-4 (Thf2 cells) (Kim et al., 2018). This is associated with enhanced expression of the IL-4 receptor alpha chain and upregulated downstream signaling as shown by increased phospho-Stat6. Blocking IL-4 in these knockout mice led to reduced Tfh2 generation.
More recently, genome-wide epigenetic and transcriptomic studies have been used to refine mechanisms of Ets1 action in T cells. When studying the functions of transcription factor Batf in Th17 and Tfh differentiation, Pham et al showed that Batf controls the binding of the architectural protein CTCF to regulatory sites in the genome (Pham et al., 2019). CTCF is involved in generating chromatin loops that divide the genome into topologically-associating domains (TADs) and these TADs are important for proper cell-type and differentiation stage specific expression of genes. Unexpectedly, it was found that the effects of Batf on CTCF binding were dependent on Ets1, which is required for Batf to recruit CTCF to particular genomic sites.
Ets1 function in CD8 T cells is less well-studied than Ets1 function in CD4 subsets. However, some data has been obtained that help to understand how Ets1 may regulate the development of function of these cells. During CD8 T cell development in the thymus, Ets1 is required for normal maturation from CD4+CD8+ double-positive (DP) thymocytes to CD8+ single-positive (CD8 SP) thymocytes. In the absence of Ets1, MHC-I restricted cells developing to the CD8 lineage inefficiently downregulate CD4, leading to the occurrence of a subset of late-stage thymocytes and peripheral CD8 T cells that still retain CD4 expression (Zamisch et al., 2009). The failure to downregulate CD4 efficiently is due to a role for Ets1 in stimulating expression of the transcription factor Runx3, which is required for this process (Ehlers et al., 2003; Taniuchi et al., 2002; Woolf et al., 2003). In peripheral CD8 T cells, Ets1 was shown to be required for expression of the β2 subunit of the IL-12 receptor (IL-12Rβ2) (Q. Li et al., 2010). IL-12 signaling in CD8 T cells during the process of antigen stimulation promotes differentiation to IFN-gamma secreting effectors and thus loss of expression of IL-12Rβ2 impairs effector differentiation. In Ets1 knockout CD8 T cells, where IL-12Rβ2 expression is reduced, T cells fail to upregulate interferon-gamma, perforin or granzyme B upon in vitro stimulation (Q. Li et al., 2010). Furthermore, there is a decrease in cell recovery from in vitro stimulated cultures of Ets1 knockout CD8 T cells as compared with wild-type T cells. In summary, similar to the situation described in B cells, the evidence gathered from studies of Ets1 functions in T cell populations support the idea that Ets1 both directly regulates important target genes by binding to promoters and enhancers and also regulates cellular responses by engaging in protein-protein interactions with other transcription factors to modulate their activity.
8.2. Autoimmune and inflammatory phenotypes in Ets1 knockout mice
In terms of autoimmune disease, Ets1 knockout mice share certain features with human lupus patients, including high titers of anti-DNA autoantibodies and the deposition of immune complexes in the kidney glomeruli (Mouly et al., 2010; Wang et al., 2005). Immune complexes in the kidney lead to fixation and deposition of complement C3, however despite this mice lacking Ets1 do not develop proteinuria (Wang et al., 2005). On the other hand, when T follicular helper cells (Tfh) isolated from mice with a T cell-specific deletion of Ets1 are transferred into Rag1 knockout hosts, the resulting mice develop proteinuria (Kim et al., 2018). This result implies that Ets1 is required in cells other than T cells for proteinuria to develop. One possibility is that Ets1 is needed in kidney cells, such as mesangial cells, for the pathological changes that occur during glomerulonephritis. In a rat models of glomerulonephritis, Ets1 has been shown to be strongly induced in kidney cells, though the exact cell type expressing seems to depend on the model used (Naito et al., 2000; Raffetseder et al., 2004). This induction of Ets1 in resident kidney cells may be required for the proliferative and differentiation changes seen during glomerulonephritis and these changes may be required for development of proteinuria. Such a model would explain why mice lacking Ets1 in all tissues, including kidney cells, fail to develop proteinuria despite having significant deposition of immune complexes in the glomeruli coupled with complement fixation.
SNPs in human ETS1 have been linked to development of atopic dermatitis, as described above. Mice lacking Ets1 specifically in T cells develop skin dermatitis, which has similarities to human atopic dermatitis (Lee et al., 2019). This phenotype was linked to enhanced expression of gp130 on Ets1-deficient CD4 T cells, leading to enhanced IL-6 signaling in the T cells and increased differentiation into Th17 cells. Functional blocking of gp130 by an inhibitor called SC144 lead to reduced AD symptoms (Lee et al., 2019). Recently, a super-enhancer region located approximately 250 kb downstream of the mouse Ets1 gene has been identified (Chandra et al., 2023). A homologous region of the human genome contains numerous SNPs associated with allergy, asthma, and atopic dermatitis (Hinds et al., 2013; Paternoster et al., 2015), suggesting that these SNPs may affect expression of Ets1 in T cells of patients with AD and related conditions.
Ets1 is also associated with IBD in humans, as detailed above. In a mouse model of IBD involving the transfer of CD45RBhigh effector CD4+ cells into immunodeficient SCID mice, the transferred effector T cells drive an interferon-γ dependent induction of colitis in the immunodeficient hosts (Powrie et al., 1994). Transferring Ets1-deficient CD45RBhigh CD4+ cells to SCID mice failed to induce colitis, likely due to their impaired ability to produce interferon-γ (Grenningloh et al., 2005). In a transgenic mouse model that over-expresses Ets1 under the control of the CMV promoter and enhancer, IBD severity was enhanced as compared to non-transgenic controls (He et al., 2022). This was particularly dramatic in terms of inflammatory infiltration into the gut in response to dextran sodium sulfate (DSS) administration. Furthermore, these Ets1 transgenic mice developed more tumors and larger tumors in the intestine when exposed to a azomethane and DSS protocol (He et al., 2022). Bone marrow chimeras confirmed that expression of the transgenic Ets1 in bone-marrow-derived cells was required for the phenotype. The cold-inducible RNA-binding protein (CIRBP) was identified as a gene regulated by Ets1 which is involved in Th1 cell-driven immune responses in IBD (He et al., 2022). The super-enhancer described above that is located downstream of the Ets1 gene is needed for differentiation of Th1 cells (Chandra et al., 2023). As expected based on its role in Th1 cells, CD4+CD45RBhigh T cells from mice lacking this super-enhancer sequence (Ets1-SE−/− mice) cannot induce colitis when transferred to Rag1 knockout mice (Chandra et al., 2023). Together, these various studies provide substantial evidence that corroborates an important role for Ets1 in IBD.
Conclusion
There is accumulating evidence suggestive of a crucial role for transcription factor Ets1 in regulating the human immune response. Polymorphisms in the human ETS1 gene had previously been associated with susceptibility to quite a few autoimmune and inflammatory diseases, namely SLE, rheumatoid arthritis, multiple sclerosis, psoriasis, ankylosing spondylitis, uveitis, celiac disease, allergy and atopic dermatitis. New evidence suggests there are also associations with additional human diseases such as inflammatory bowel disease, idiopathic inflammatory myopathy, Behçet’s disease, granulomatosis with polyangiitis, primary biliary cirrhosis and anti-phospholipid syndrome. Furthermore, evidence has continued to accumulate indicating reduced expression of the Ets1 mRNA in immune cells from patients with these autoimmune and inflammatory conditions. To date, reduced Ets1 expression has been documented in one or more diseases, using either total PBMCs or purified subpopulations such as total B cells, total T cells and regulatory T cells. This decrease of Ets1 expression in human immune cells in autoimmune and inflammatory conditions is consistent with the known phenotype of Ets1 knockout mice that develop a lupus-like autoimmune condition, with increased B and T cell activation, increased germinal center and plasma cell responses and increased autoantibody titers. Mouse studies have demonstrated that Ets1 has both B cell-intrinsic and T cell-intrinsic roles important for normal immune regulation.
While many studies have described associations of the ETS1 gene with human autoimmune diseases, functional studies of the roles of Ets1 in humans remain fairly sparse. Much of our understanding of how Ets1 regulates immune cell responses comes from studies with Ets1 knockout mice. While these studies are valuable in understanding Ets1 functional roles, there is the complication that human and mouse immune cells do not always behave similarly (Mestas & Hughes, 2004).
Figure 2. Comparison of 3’ untranslated regions (3’UTR) of Ets1 and Ets2.
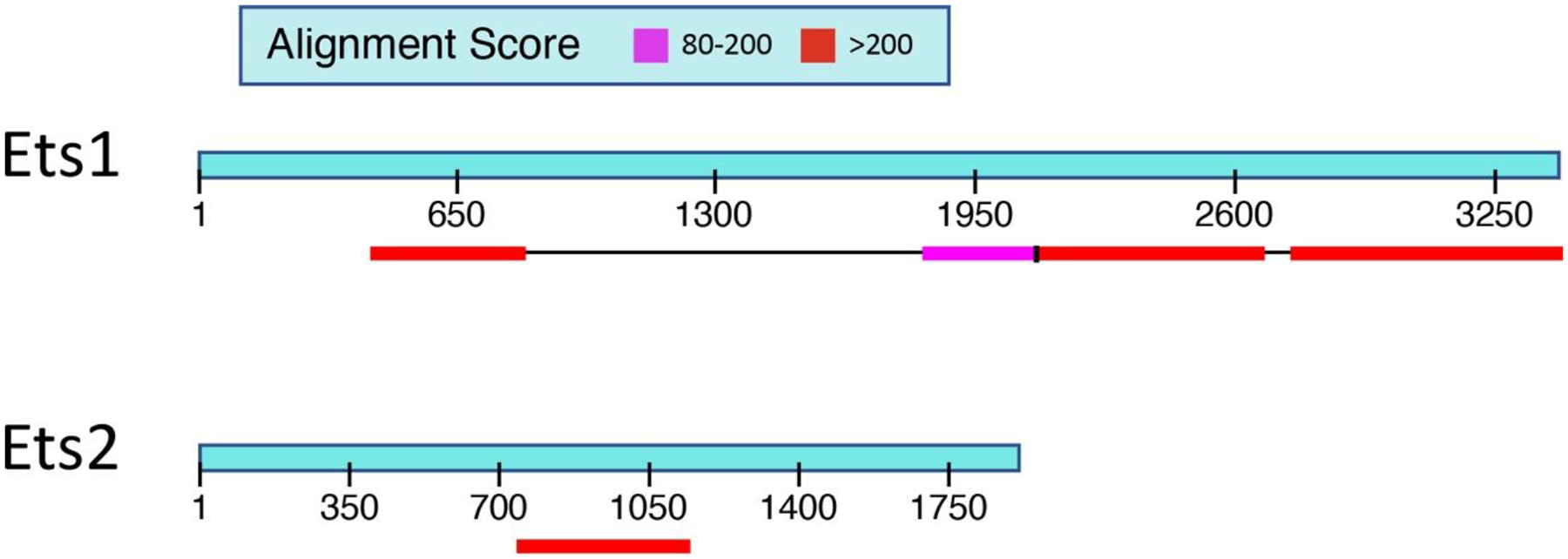
Ets1 has a very long 3’ UTR of approximately 3.6 kb and shows high levels of homology between the mouse and human mRNAs in two regions (shown in the red and pink colored boxes) that together compose almost half of the Ets1 3’ UTR. Ets2 has a shorter 3’ UTR of approximately 2 kb and shows only one region of high homology between the mouse and human mRNAs. The region of strong homology in the 3’ UTR of Ets2 comprises approximately 25% of the total UTR.
Sidebar 1: Approaches used to link Ets1 to human autoimmune and inflammatory diseases.
Numerous approaches have been used to link Ets1 to human autoimmune and inflammatory diseases (summarized in Table III). These studies may associate particular single-nucleotide polymorphisms (SNPs) in the ETS1 gene with disease (GWAS studies or meta-analyses of GWAS data that combine results from multiple GWAS studies). Other studies may show alterations in Ets1 gene expression in patients versus controls (gene expression microarrays and RNA-sequencing). Part of this change in gene expression may be due to targeting of the Ets1 mRNA by a variety of micro-RNAs (miRNAs), whose expression is changed in disease. Changes in epigenetic marks in the ETS1 locus have also been identified (CpG DNA methylation studies). In silico approaches take advantage of the large amounts of DNA genotyping, DNA sequencing, gene expression data, epigenetic data, patient clinical data and other similar information to correlate the presence of particular genetic variants to disease presence or clinical features. Given the large amount of data tying Ets1 to regulation of immune function and to aberrant immune responses, candidate gene approaches have also been used that focus on Ets1 alone or as a part of a small group of potentially-relevant genes. These candidate gene approaches can take many forms, but in general seek to further link Ets1 to disease susceptibility or clinical phenotypes or to understand the molecular function of Ets1 in regulating human immune responses.
Sidebar 2: MicroRNAs and Ets1.
The mouse and human Ets1 genes both have very long 3’ UTRs of approximately 3.6 kb (Figure 2). When comparing the sequence of the mouse and human mRNAs, it is evident that there are two regions in the 3’UTR that show very strong sequence conservation, indicating that they likely play important roles in regulating the stability, transport or translation of the mRNA. When compared to the closely related gene Ets2, the Ets1 3’ UTR is longer and has a significantly larger percentage of the sequence that is highly conserved between mouse and human (Figure 2). There are two SNPs located in the 3’ UTR that have been associated with autoimmune diseases (rs1128334 and rs4937333). In addition, the CA repeat polymorphisms described in the Sullivan et al paper are also in the 3’ UTR (Sullivan et al., 2000). There are many microRNAs (miRNAs) that have been shown to regulate the stability and translation of the Ets1 mRNA by binding to the 3’ UTR. Five different miRNAs that regulate Ets1 in the context of autoimmunity were studied in these papers (Du et al., 2009; Ge et al., 2019; Jin et al., 2019; Li et al., 2020; Na et al., 2016; Sun et al., 2016; Wen et al., 2013). Five different miRNAs were studied in these papers, miR-1–3p, miR-125a, miR-155, miR-326 and miR-451a, which were shown to have roles in regulating Ets1 levels in various immune cell populations (total PBMC, CD4+ T cells, CD19+ B cells, Th17 cells or Treg cells). miR-1–3p, miR-125a and miR-326 are all located in the regions that are highly conserved between mouse and human. miR-155 has two potential binding sites, one of which is located in the highly conserved region. On the other hand, miR-451a does not have a binding site in the Ets1 3’UTR based on the TargetScan software prediction, so its effects on Ets1 expression would seem to be indirect.
Funding Information
This work was funded by NIH grant R01 AI122720.
Footnotes
Conflict of Interest
None
Further Reading
For further details the reader is directed to the references herein, which describe each of the studies in more detail. The role of Ets1 in autoimmune diseases has also been previously reviewed: (Gallant & Gilkeson, 2006; Garrett-Sinha, 2013; Garrett-Sinha et al., 2016; Leng et al., 2011). For further information on GWAS and autoimmune diseases, the reader is referred to these references: (Fike et al., 2019; Suzuki et al., 2021). For further information on miRNA roles in autoimmunity, the reader is referred to these references: (Xiao et al., 2020; Zhang et al., 2020).
References
- Akhtar M, Ali Y, Islam ZU, Arshad M, Rauf M, Ali M, … Jalil F (2021). Characterization of Rheumatoid Arthritis Risk-Associated SNPs and Identification of Novel Therapeutic Sites Using an In-Silico Approach. Biology (Basel), 10(6). 10.3390/biology10060501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton K, Muthusamy N, Fischer C, Ting CN, Walunas TL, Lanier LL, & Leiden JM (1998). The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity, 9(4), 555–563. 10.1016/s1074-7613(00)80638-x [DOI] [PubMed] [Google Scholar]
- Bentham J, Morris DL, Graham DSC, Pinder CL, Tombleson P, Behrens TW, … Vyse TJ (2015). Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet, 47(12), 1457–1464. 10.1038/ng.3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bories JC, Willerford DM, Grevin D, Davidson L, Camus A, Martin P, … Alt FW (1995). Increased T-cell apoptosis and terminal B-cell differentiation induced by inactivation of the Ets-1 proto-oncogene. Nature, 377(6550), 635–638. 10.1038/377635a0 [DOI] [PubMed] [Google Scholar]
- Boyd M, Thodberg M, Vitezic M, Bornholdt J, Vitting-Seerup K, Chen Y, … Sandelin A (2018). Characterization of the enhancer and promoter landscape of inflammatory bowel disease from human colon biopsies. Nat Commun, 9(1), 1661. 10.1038/s41467-018-03766-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner PM, Pavel AB, Khattri S, Leonard A, Malik K, Rose S, … Guttman-Yassky E (2019). Baseline IL-22 expression in patients with atopic dermatitis stratifies tissue responses to fezakinumab. J Allergy Clin Immunol, 143(1), 142–154. 10.1016/j.jaci.2018.07.028 [DOI] [PubMed] [Google Scholar]
- Burocchi A, Pittoni P, Tili E, Rigoni A, Costinean S, Croce CM, & Colombo MP (2015). Regulated Expression of miR-155 is Required for iNKT Cell Development. Front Immunol, 6, 140. 10.3389/fimmu.2015.00140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra A, Yoon S, Michieletto MF, Goldman N, Ferrari EK, Abedi M, … Vahedi G (2023). Quantitative control of Ets1 dosage by a multi-enhancer hub promotes Th1 cell differentiation and protects from allergic inflammation. Immunity. 10.1016/j.immuni.2023.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzikyriakidou A, Voulgari PV, Georgiou I, & Drosos AA (2013). Altered sequence of the ETS1 transcription factor may predispose to rheumatoid arthritis susceptibility. Scand J Rheumatol, 42(1), 11–14. 10.3109/03009742.2012.711367 [DOI] [PubMed] [Google Scholar]
- Chen L, Huang Z, Yang B, Cai B, Su Z, & Wang L (2015). Association of E26 Transformation Specific Sequence 1 Variants with Rheumatoid Arthritis in Chinese Han Population. PLoS One, 10(8), e0134875. 10.1371/journal.pone.0134875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wen X, Li L, Li J, Li Y, Wang Q, … Li Y (2017). Single nucleotide polymorphisms in the ETS1 gene are associated with idiopathic inflammatory myopathies in a northern Chinese Han population. Sci Rep, 7(1), 13128. 10.1038/s41598-017-13385-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements JL, John SA, & Garrett-Sinha LA (2006). Impaired generation of CD8+ thymocytes in Ets-1-deficient mice. J Immunol, 177(2), 905–912. 10.4049/jimmunol.177.2.905 [DOI] [PubMed] [Google Scholar]
- Cooles FAH, Tarn J, Lendrem DW, Naamane N, Lin CM, Millar B, … Isaacs JD (2022). Interferon-alpha-mediated therapeutic resistance in early rheumatoid arthritis implicates epigenetic reprogramming. Ann Rheum Dis. 10.1136/annrheumdis-2022-222370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes A, & Brown MA (2011). Promise and pitfalls of the Immunochip. Arthritis Res Ther, 13(1), 101. 10.1186/ar3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danila MI, Laufer VA, Reynolds RJ, Yan Q, Liu N, Gregersen PK, … Bridges SL Jr. (2017). Dense Genotyping of Immune-Related Regions Identifies Loci for Rheumatoid Arthritis Risk and Damage in African Americans. Mol Med, 23, 177–187. 10.2119/molmed.2017.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, … Pei G (2009). MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol, 10(12), 1252–1259. 10.1038/ni.1798 [DOI] [PubMed] [Google Scholar]
- Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A, … van Heel DA (2010). Multiple common variants for celiac disease influencing immune gene expression. Nat Genet, 42(4), 295–302. 10.1038/ng.543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers M, Laule-Kilian K, Petter M, Aldrian CJ, Grueter B, Wurch A, … Steimle V (2003). Morpholino antisense oligonucleotide-mediated gene knockdown during thymocyte development reveals role for Runx3 transcription factor in CD4 silencing during development of CD4-/CD8+ thymocytes. J Immunol, 171(7), 3594–3604. 10.4049/jimmunol.171.7.3594 [DOI] [PubMed] [Google Scholar]
- Eyquem S, Chemin K, Fasseu M, Chopin M, Sigaux F, Cumano A, & Bories JC (2004). The development of early and mature B cells is impaired in mice deficient for the Ets-1 transcription factor. Eur J Immunol, 34(11), 3187–3196. 10.1002/eji.200425352 [DOI] [PubMed] [Google Scholar]
- Fike AJ, Elcheva I, & Rahman ZSM (2019). The Post-GWAS Era: How to Validate the Contribution of Gene Variants in Lupus. Curr Rheumatol Rep, 21(1), 3. 10.1007/s11926-019-0801-5 [DOI] [PubMed] [Google Scholar]
- Freudenberg J, Lee HS, Han BG, Shin HD, Kang YM, Sung YK, … Bae SC (2011). Genome-wide association study of rheumatoid arthritis in Koreans: population-specific loci as well as overlap with European susceptibility loci. Arthritis Rheum, 63(4), 884–893. 10.1002/art.30235 [DOI] [PubMed] [Google Scholar]
- Gallant S, & Gilkeson G (2006). ETS transcription factors and regulation of immunity. Arch Immunol Ther Exp (Warsz), 54(3), 149–163. 10.1007/s00005-006-0017-z [DOI] [PubMed] [Google Scholar]
- Garrett-Sinha LA (2013). Review of Ets1 structure, function, and roles in immunity. Cell Mol Life Sci, 70(18), 3375–3390. 10.1007/s00018-012-1243-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett-Sinha LA, Kearly A, & Satterthwaite AB (2016). The Role of the Transcription Factor Ets1 in Lupus and Other Autoimmune Diseases. Crit Rev Immunol, 36(6), 485–510. 10.1615/CritRevImmunol.2017020284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Sun M, Wu W, Ma C, Zhang C, He C, … Liu Z (2019). MicroRNA-125a suppresses intestinal mucosal inflammation through targeting ETS-1 in patients with inflammatory bowel diseases. J Autoimmun, 101, 109–120. 10.1016/j.jaut.2019.04.014 [DOI] [PubMed] [Google Scholar]
- Genestier L, Taillardet M, Mondiere P, Gheit H, Bella C, & Defrance T (2007). TLR agonists selectively promote terminal plasma cell differentiation of B cell subsets specialized in thymus-independent responses. J Immunol, 178(12), 7779–7786. 10.4049/jimmunol.178.12.7779 [DOI] [PubMed] [Google Scholar]
- Grenningloh R, Kang BY, & Ho IC (2005). Ets-1, a functional cofactor of T-bet, is essential for Th1 inflammatory responses. J Exp Med, 201(4), 615–626. 10.1084/jem.20041330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, … Zhang XJ (2009). Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet, 41(11), 1234–1237. 10.1038/ng.472 [DOI] [PubMed] [Google Scholar]
- He CF, Liu YS, Cheng YL, Gao JP, Pan TM, Han JW, … Yang S (2010). TNIP1, SLC15A4, ETS1, RasGRP3 and IKZF1 are associated with clinical features of systemic lupus erythematosus in a Chinese Han population. Lupus, 19(10), 1181–1186. 10.1177/0961203310367918 [DOI] [PubMed] [Google Scholar]
- He Q, Gao H, Chang YL, Wu X, Lin R, Li G, … Liu Z (2022). ETS-1 facilitates Th1 cell-mediated mucosal inflammation in inflammatory bowel diseases through upregulating CIRBP. J Autoimmun, 132, 102872. 10.1016/j.jaut.2022.102872 [DOI] [PubMed] [Google Scholar]
- Hinds DA, McMahon G, Kiefer AK, Do CB, Eriksson N, Evans DM, … Tung JY (2013). A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat Genet, 45(8), 907–911. 10.1038/ng.2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Huffaker TB, Kagele DA, Runtsch MC, Bake E, Chaudhuri AA, … O’Connell RM (2013). MicroRNA-155 confers encephalogenic potential to Th17 cells by promoting effector gene expression. J Immunol, 190(12), 5972–5980. 10.4049/jimmunol.1300351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL (2005). Consistent across-tissue signatures of differential gene expression in Crohn’s disease. Immunogenetics, 57(10), 709–716. 10.1007/s00251-005-0044-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks SA, Cashman KS, Zumaquero E, Marigorta UM, Patel AV, Wang X, … Sanz I (2018). Distinct Effector B Cells Induced by Unregulated Toll-like Receptor 7 Contribute to Pathogenic Responses in Systemic Lupus Erythematosus. Immunity, 49(4), 725–739 e726. 10.1016/j.immuni.2018.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Fang X, Dai C, Xiang N, Tao J, Sun X, … Li X (2019). The potential role of Ets-1 and miR-326 in CD19(+)B cells in the pathogenesis of patients with systemic lupus erythematosus. Clin Rheumatol, 38(4), 1031–1038. 10.1007/s10067-018-4371-0 [DOI] [PubMed] [Google Scholar]
- John S, Russell L, Chin SS, Luo W, Oshima R, & Garrett-Sinha LA (2014). Transcription factor Ets1, but not the closely related factor Ets2, inhibits antibody-secreting cell differentiation. Mol Cell Biol, 34(3), 522–532. 10.1128/MCB.00612-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John SA, Clements JL, Russell LM, & Garrett-Sinha LA (2008). Ets-1 regulates plasma cell differentiation by interfering with the activity of the transcription factor Blimp-1. J Biol Chem, 283(2), 951–962. 10.1074/jbc.M705262200 [DOI] [PubMed] [Google Scholar]
- Jones SA, Cantsilieris S, Fan H, Cheng Q, Russ BE, Tucker EJ, … Morand EF (2019). Rare variants in non-coding regulatory regions of the genome that affect gene expression in systemic lupus erythematosus. Sci Rep, 9(1), 15433. 10.1038/s41598-019-51864-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki A, Yamashita K, Hirano F, Sada KE, Tsukui D, Kondo Y, … Tsuchiya N (2018). Association of ETS1 polymorphism with granulomatosis with polyangiitis and proteinase 3-anti-neutrophil cytoplasmic antibody positive vasculitis in a Japanese population. J Hum Genet, 63(1), 55–62. 10.1038/s10038-017-0362-2 [DOI] [PubMed] [Google Scholar]
- Kim CJ, Lee CG, Jung JY, Ghosh A, Hasan SN, Hwang SM, … Im SH (2018). The Transcription Factor Ets1 Suppresses T Follicular Helper Type 2 Cell Differentiation to Halt the Onset of Systemic Lupus Erythematosus. Immunity, 49(6), 1034–1048 e1038. 10.1016/j.immuni.2018.10.012 [DOI] [PubMed] [Google Scholar]
- Kim K, Bang SY, Lee HS, Cho SK, Choi CB, Sung YK, … Bae SC (2015). High-density genotyping of immune loci in Koreans and Europeans identifies eight new rheumatoid arthritis risk loci. Ann Rheum Dis, 74(3), e13. 10.1136/annrheumdis-2013-204749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu N, & Takayanagi H (2022). Mechanisms of joint destruction in rheumatoid arthritis - immune cell-fibroblast-bone interactions. Nat Rev Rheumatol, 18(7), 415–429. 10.1038/s41584-022-00793-5 [DOI] [PubMed] [Google Scholar]
- Konno S, Iizuka M, Yukawa M, Sasaki K, Sato A, Horie Y, … Watanabe S (2004). Altered expression of angiogenic factors in the VEGF-Ets-1 cascades in inflammatory bowel disease. J Gastroenterol, 39(10), 931–939. 10.1007/s00535-004-1423-9 [DOI] [PubMed] [Google Scholar]
- Lai EC (2002). Micro RNAs are complementary to 3’ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet, 30(4), 363–364. 10.1038/ng865 [DOI] [PubMed] [Google Scholar]
- Lai EC, & Posakony JW (1997). The Bearded box, a novel 3’ UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split Complex gene expression. Development, 124(23), 4847–4856. 10.1242/dev.124.23.4847 [DOI] [PubMed] [Google Scholar]
- Lai EC, Tam B, & Rubin GM (2005). Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev, 19(9), 1067–1080. 10.1101/gad.1291905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer VA, Tiwari HK, Reynolds RJ, Danila MI, Wang J, Edberg JC, … Bridges SL Jr. (2019). Genetic influences on susceptibility to rheumatoid arthritis in African-Americans. Hum Mol Genet, 28(5), 858–874. 10.1093/hmg/ddy395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, … O’Shea J,J (2007). Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity, 26(3), 371–381. 10.1016/j.immuni.2007.02.009 [DOI] [PubMed] [Google Scholar]
- Lee CG, Kwon HK, Kang H, Kim Y, Nam JH, Won YH, … Im SH (2019). Ets1 suppresses atopic dermatitis by suppressing pathogenic T cell responses. JCI Insight, 4(5). 10.1172/jci.insight.124202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GR, Kim ST, Spilianakis CG, Fields PE, & Flavell RA (2006). T helper cell differentiation: regulation by cis elements and epigenetics. Immunity, 24(4), 369–379. 10.1016/j.immuni.2006.03.007 [DOI] [PubMed] [Google Scholar]
- Leng RX, Pan HF, Chen GM, Feng CC, Fan YG, Ye DQ, & Li XP (2011). The dual nature of Ets-1: focus to the pathogenesis of systemic lupus erythematosus. Autoimmun Rev, 10(8), 439–443. 10.1016/j.autrev.2011.01.007 [DOI] [PubMed] [Google Scholar]
- Leng RX, Wang W, Cen H, Zhou M, Feng CC, Zhu Y, … Ye DQ (2012). Gene-gene and gene-sex epistatic interactions of MiR146a, IRF5, IKZF1, ETS1 and IL21 in systemic lupus erythematosus. PLoS One, 7(12), e51090. 10.1371/journal.pone.0051090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard CJ, Sajuthi S, Zhao J, Kim K, Ice JA, Li H, … Tsao BP (2016). Identification of a Systemic Lupus Erythematosus Risk Locus Spanning ATG16L2, FCHSD2, and P2RY2 in Koreans. Arthritis Rheumatol, 68(5), 1197–1209. 10.1002/art.39548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ma X, Zhao YF, & Zhang C (2020). MiR-1–3p facilitates Th17 differentiation associating with multiple sclerosis via targeting ETS1. Eur Rev Med Pharmacol Sci, 24(12), 6881–6892. 10.26355/eurrev_202006_21678 [DOI] [PubMed] [Google Scholar]
- Li Q, Eppolito C, Odunsi K, & Shrikant PA (2010). Antigen-induced Erk1/2 activation regulates Ets-1-mediated sensitization of CD8+ T cells for IL-12 responses. J Leukoc Biol, 87(2), 257–263. 10.1189/jlb.0409221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sun LD, Lu WS, Hu WL, Gao JP, Cheng YL, … Yang S (2010). Expression analysis of ETS1 gene in peripheral blood mononuclear cells with systemic lupus erythematosus by real-time reverse transcription PCR. Chin Med J (Engl), 123(16), 2287–2288. https://www.ncbi.nlm.nih.gov/pubmed/20819682 [PubMed] [Google Scholar]
- Lill CM, Luessi F, Alcina A, Sokolova EA, Ugidos N, de la Hera B, … Bertram L (2015). Genome-wide significant association with seven novel multiple sclerosis risk loci. J Med Genet, 52(12), 848–855. 10.1136/jmedgenet-2015-103442 [DOI] [PubMed] [Google Scholar]
- Lu X, Zoller EE, Weirauch MT, Wu Z, Namjou B, Williams AH, … Kottyan LC (2015). Lupus Risk Variant Increases pSTAT1 Binding and Decreases ETS1 Expression. Am J Hum Genet, 96(5), 731–739. 10.1016/j.ajhg.2015.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Mayeux J, Gutierrez T, Russell L, Getahun A, Muller J, … Garrett-Sinha LA (2014). A balance between B cell receptor and inhibitory receptor signaling controls plasma cell differentiation by maintaining optimal Ets1 levels. J Immunol, 193(2), 909–920. 10.4049/jimmunol.1400666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri R, Mahmoudi M, Mirkazemi S, Mahmoudi MB, Karimizadeh E, Salimi Y, … Jamshidi AR (2016). Determination of the association of ETS1 and WDFY4 gene polymorphisms with systemic lupus erythematosus in an Iranian population. Rheumatology Research Journal, 1(1), 11–16. [Google Scholar]
- Martin F, Oliver AM, & Kearney JF (2001). Marginal zone and B1 B cells unite in the early response against T-independent blood-borne particulate antigens. Immunity, 14(5), 617–629. 10.1016/s1074-7613(01)00129-7 [DOI] [PubMed] [Google Scholar]
- Mestas J, & Hughes CC (2004). Of mice and not men: differences between mouse and human immunology. J Immunol, 172(5), 2731–2738. 10.4049/jimmunol.172.5.2731 [DOI] [PubMed] [Google Scholar]
- Moisan J, Grenningloh R, Bettelli E, Oukka M, & Ho IC (2007). Ets-1 is a negative regulator of Th17 differentiation. J Exp Med, 204(12), 2825–2835. 10.1084/jem.20070994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molineros JE, Chua KH, Sun C, Lian LH, Motghare P, Kim-Howard X, & Nath SK (2014). Evaluation of SLE Susceptibility Genes in Malaysians. Autoimmune Dis, 2014, 305436. 10.1155/2014/305436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DL, Sheng Y, Zhang Y, Wang YF, Zhu Z, Tombleson P, … Vyse TJ (2016). Genome-wide association meta-analysis in Chinese and European individuals identifies ten new loci associated with systemic lupus erythematosus. Nat Genet, 48(8), 940–946. 10.1038/ng.3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouly E, Chemin K, Nguyen HV, Chopin M, Mesnard L, Leite-de-Moraes M, … Bories JC (2010). The Ets-1 transcription factor controls the development and function of natural regulatory T cells. J Exp Med, 207(10), 2113–2125. 10.1084/jem.20092153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na SY, Park MJ, Park S, & Lee ES (2016). MicroRNA-155 regulates the Th17 immune response by targeting Ets-1 in Behcet’s disease. Clin Exp Rheumatol, 34(6 Suppl 102), S56–S63. https://www.ncbi.nlm.nih.gov/pubmed/27156371 [PubMed] [Google Scholar]
- Nagaleekar VK, Diehl SA, Juncadella I, Charland C, Muthusamy N, Eaton S, … Rincon M (2008). IP3 receptor-mediated Ca2+ release in naive CD4 T cells dictates their cytokine program. J Immunol, 181(12), 8315–8322. 10.4049/jimmunol.181.12.8315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T, Razzaque MS, Nazneen A, Liu D, Nihei H, Koji T, & Taguchi T (2000). Renal expression of the Ets-1 proto-oncogene during progression of rat crescentic glomerulonephritis. J Am Soc Nephrol, 11(12), 2243–2255. 10.1681/ASN.V11122243 [DOI] [PubMed] [Google Scholar]
- Nguyen HV, Mouly E, Chemin K, Luinaud R, Despres R, Fermand JP, … Bories JC (2012). The Ets-1 transcription factor is required for Stat1-mediated T-bet expression and IgG2a class switching in mouse B cells. Blood, 119(18), 4174–4181. 10.1182/blood-2011-09-378182 [DOI] [PubMed] [Google Scholar]
- Obermoser G, & Pascual V (2010). The interferon-alpha signature of systemic lupus erythematosus. Lupus, 19(9), 1012–1019. 10.1177/0961203310371161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Terao C, Ikari K, Kochi Y, Ohmura K, Suzuki A, … Yamamoto K (2012). Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat Genet, 44(5), 511–516. 10.1038/ng.2231 [DOI] [PubMed] [Google Scholar]
- Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, … Plenge RM (2014). Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature, 506(7488), 376–381. 10.1038/nature12873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paternoster L, Standl M, Waage J, Baurecht H, Hotze M, Strachan DP, … Weidinger S (2015). Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet, 47(12), 1449–1456. 10.1038/ng.3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham D, Moseley CE, Gao M, Savic D, Winstead CJ, Sun M, … Hatton RD (2019). Batf Pioneers the Reorganization of Chromatin in Developing Effector T Cells via Ets1-Dependent Recruitment of Ctcf. Cell Rep, 29(5), 1203–1220 e1207. 10.1016/j.celrep.2019.09.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polansky JK, Schreiber L, Thelemann C, Ludwig L, Kruger M, Baumgrass R, … Huehn J (2010). Methylation matters: binding of Ets-1 to the demethylated Foxp3 gene contributes to the stabilization of Foxp3 expression in regulatory T cells. J Mol Med (Berl), 88(10), 1029–1040. 10.1007/s00109-010-0642-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powrie F, Leach MW, Mauze S, Menon S, Caddle LB, & Coffman RL (1994). Inhibition of Th1 responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4+ T cells. Immunity, 1(7), 553–562. 10.1016/1074-7613(94)90045-0 [DOI] [PubMed] [Google Scholar]
- Raffetseder U, Wernert N, Ostendorf T, van Roeyen C, Rauen T, Behrens P, … Mertens PR (2004). Mesangial cell expression of proto-oncogene Ets-1 during progression of mesangioproliferative glomerulonephritis. Kidney Int, 66(2), 622–632. 10.1111/j.1523-1755.2004.00782.x [DOI] [PubMed] [Google Scholar]
- Richardson K, Lai CQ, Parnell LD, Lee YC, & Ordovas JM (2011). A genome-wide survey for SNPs altering microRNA seed sites identifies functional candidates in GWAS. BMC Genomics, 12, 504. 10.1186/1471-2164-12-504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell L, John S, Cullen J, Luo W, Shlomchik MJ, & Garrett-Sinha LA (2015). Requirement for Transcription Factor Ets1 in B Cell Tolerance to Self-Antigens. J Immunol, 195(8), 3574–3583. 10.4049/jimmunol.1500776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saelee P, Kearly A, Nutt SL, & Garrett-Sinha LA (2017). Genome-Wide Identification of Target Genes for the Key B Cell Transcription Factor Ets1. Front Immunol, 8, 383. 10.3389/fimmu.2017.00383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan S, Dang J, Li J, Yang Z, Zhao H, Xin Q, … Liu Q (2014). ETS1 variants confer susceptibility to ankylosing spondylitis in Han Chinese. Arthritis Res Ther, 16(2), R87. 10.1186/ar4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczyk MK, Richardson TG, Zuber V, Min JL, Gaunt TR, Paternoster L, & eQtlgen Consortium B. C. G. (2021). Triangulating Molecular Evidence to Prioritize Candidate Causal Genes at Established Atopic Dermatitis Loci. J Invest Dermatol, 141(11), 2620–2629. 10.1016/j.jid.2021.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens DC, & Poon GM (2016). Differential sensitivity to methylated DNA by ETS-family transcription factors is intrinsically encoded in their DNA-binding domains. Nucleic Acids Res, 44(18), 8671–8681. 10.1093/nar/gkw528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strempel JM, Grenningloh R, Ho IC, & Vercelli D (2010). Phylogenetic and functional analysis identifies Ets-1 as a novel regulator of the Th2 cytokine gene locus. J Immunol, 184(3), 1309–1316. 10.4049/jimmunol.0804162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KE, Piliero LM, Dharia T, Goldman D, & Petri MA (2000). 3’ polymorphisms of ETS1 are associated with different clinical phenotypes in SLE. Hum Mutat, 16(1), 49–53. [DOI] [PubMed] [Google Scholar]
- Sun XG, Tao JH, Xiang N, Li XM, Wang GS, Fang X, … Li XP (2016). Negative Correlation Between miR-326 and Ets-1 in Regulatory T Cells from new-Onset SLE Patients. Inflammation, 39(2), 822–829. 10.1007/s10753-016-0312-8 [DOI] [PubMed] [Google Scholar]
- Sunshine A, Goich D, Stith A, Sortino K, Dalton J, Metcalfe S, … Garrett-Sinha LA (2019). Ets1 Controls the Development of B Cell Autoimmune Responses in a Cell-Intrinsic Manner. Immunohorizons, 3(7), 331–340. 10.4049/immunohorizons.1900033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Guerrini MM, & Yamamoto K (2021). Functional genomics of autoimmune diseases. Ann Rheum Dis, 80(6), 689–697. 10.1136/annrheumdis-2019-216794 [DOI] [PubMed] [Google Scholar]
- Suzuki E, Zhang X, Yashiro-Furuya M, Asano T, Kanno T, Kobayashi H, … Ohira H (2022). The expression of Ets-1 and Fli-1 is associated with interferon-inducible genes in peripheral blood mononuclear cells from Japanese patients with systemic lupus erythematosus. Medicine (Baltimore), 101(45), e31522. https://doi.org/doi: 10.1097/MD.0000000000031522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Koido M, Suzuki A, Otomo N, Suetsugu H, Kochi Y, … Terao C (2021). Eight novel susceptibility loci and putative causal variants in atopic dermatitis. J Allergy Clin Immunol, 148(5), 1293–1306. 10.1016/j.jaci.2021.04.019 [DOI] [PubMed] [Google Scholar]
- Tangye SG, Ferguson A, Avery DT, Ma CS, & Hodgkin PD (2002). Isotype switching by human B cells is division-associated and regulated by cytokines. J Immunol, 169(8), 4298–4306. 10.4049/jimmunol.169.8.4298 [DOI] [PubMed] [Google Scholar]
- Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, … Littman DR (2002). Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell, 111(5), 621–633. 10.1016/s0092-8674(02)01111-x [DOI] [PubMed] [Google Scholar]
- Trynka G, Hunt KA, Bockett NA, Romanos J, Mistry V, Szperl A, … van Heel DA (2011). Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet, 43(12), 1193–1201. 10.1038/ng.998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao HW, Tai TS, Tseng W, Chang HH, Grenningloh R, Miaw SC, & Ho IC (2013). Ets-1 facilitates nuclear entry of NFAT proteins and their recruitment to the IL-2 promoter. Proc Natl Acad Sci U S A, 110(39), 15776–15781. 10.1073/pnas.1304343110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi LC, Spain SL, Knight J, Ellinghaus E, Stuart PE, Capon F, … Trembath RC (2012). Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet, 44(12), 1341–1348. 10.1038/ng.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedak P, Kroshinsky D, St John J, Xavier RJ, Yajnik V, & Ananthakrishnan AN (2016). Genetic basis of TNF-alpha antagonist associated psoriasis in inflammatory bowel diseases: a genotype-phenotype analysis. Aliment Pharmacol Ther, 43(6), 697–704. 10.1111/apt.13542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Ahlford A, Jarvinen TM, Nordmark G, Eloranta ML, Gunnarsson I, … Sandling JK (2013). Genes identified in Asian SLE GWASs are also associated with SLE in Caucasian populations. Eur J Hum Genet, 21(9), 994–999. 10.1038/ejhg.2012.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, John SA, Clements JL, Percy DH, Barton KP, & Garrett-Sinha LA (2005). Ets-1 deficiency leads to altered B cell differentiation, hyperresponsiveness to TLR9 and autoimmune disease. Int Immunol, 17(9), 1179–1191. 10.1093/intimm/dxh295 [DOI] [PubMed] [Google Scholar]
- Wang T, Zhou Q, & Shang Y (2021). Downregulation of miRNA-451a Promotes the Differentiation of CD4+ T Cells towards Th2 Cells by Upregulating ETS1 in Childhood Asthma. J Innate Immun, 13(1), 38–48. 10.1159/000509714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeding E, Coit P, Yalavarthi S, Kaplan MJ, Knight JS, & Sawalha AH (2018). Genome-wide DNA methylation analysis in primary antiphospholipid syndrome neutrophils. Clin Immunol, 196, 110–116. 10.1016/j.clim.2018.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Zhou Q, Hou S, Bai L, Liu Y, Qi J, … Yang P (2014). MicroRNA-146a and Ets-1 gene polymorphisms are associated with pediatric uveitis. PLoS One, 9(3), e91199. 10.1371/journal.pone.0091199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Xu L, Chen X, Xu W, Yin Z, Gao X, & Xiong S (2013). Autoantibody induction by DNA-containing immune complexes requires HMGB1 with the TLR2/microRNA-155 pathway. J Immunol, 190(11), 5411–5422. 10.4049/jimmunol.1203301 [DOI] [PubMed] [Google Scholar]
- Woolf E, Xiao C, Fainaru O, Lotem J, Rosen D, Negreanu V, … Groner Y (2003). Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci U S A, 100(13), 7731–7736. 10.1073/pnas.1232420100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Nemazee D, & Gonzalez-Martin A (2020). MicroRNA control of B cell tolerance, autoimmunity and cancer. Semin Cancer Biol, 64, 102–107. 10.1016/j.semcancer.2019.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Niu Q, Su Z, Wang F, Zhang J, Yang B, & Huang Z (2019). Genetic association of E26 transformation specific sequence 1 polymorphisms with the susceptibility of primary biliary cholangitis in China. Sci Rep, 9(1), 19680. 10.1038/s41598-019-56181-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Luo L, Chen L, Niu Q, Zhang J, Xu H, … Huang Z (2021). ETS1 polymorphism rs73013527 in relation to serum RANKL levels among patients with RA. Medicine (Baltimore), 100(5), e24562. 10.1097/MD.0000000000024562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Shen N, Ye DQ, Liu Q, Zhang Y, Qian XX, … Asian Lupus Genetics, C. (2010). Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet, 6(2), e1000841. 10.1371/journal.pgen.1000841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Low HQ, Wang L, Li Y, Ellinghaus E, Han J, … Liu J (2015). Genome-wide meta-analysis identifies multiple novel associations and ethnic heterogeneity of psoriasis susceptibility. Nat Commun, 6, 6916. 10.1038/ncomms7916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yockell-Lelievre J, Spriet C, Cantin P, Malenfant P, Heliot L, de Launoit Y, & Audette M (2009). Functional cooperation between Stat-1 and ets-1 to optimize icam-1 gene transcription. Biochem Cell Biol, 87(6), 905–918. 10.1139/o09-055 [DOI] [PubMed] [Google Scholar]
- Zamisch M, Tian L, Grenningloh R, Xiong Y, Wildt KF, Ehlers M, … Bosselut R (2009). The transcription factor Ets1 is important for CD4 repression and Runx3 up-regulation during CD8 T cell differentiation in the thymus. J Exp Med, 206(12), 2685–2699. 10.1084/jem.20092024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhang Y, Zhang L, Yang J, Ying D, Zeng S, … Yang W (2013). Epistatic interaction between genetic variants in susceptibility gene ETS1 correlates with IL-17 levels in SLE patients. Ann Hum Genet, 77(4), 344–350. https://doi.org/doi: 10.1111/ahg.12018 [DOI] [PubMed] [Google Scholar]
- Zhang L, Wu H, Zhao M, Chang C, & Lu Q (2020). Clinical significance of miRNAs in autoimmunity. J Autoimmun, 109, 102438. 10.1016/j.jaut.2020.102438 [DOI] [PubMed] [Google Scholar]
- Zhang R, Pan B, Li Y, & Li X (2019). SNP rs4937333 in the miRNA-5003-Binding Site of the ETS1 3’-UTR Decreases ETS1 Expression. Front Genet, 10, 581. 10.3389/fgene.2019.00581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Bo L, Zhang H, Zhuang C, & Liu R (2014). E26 transformation-specific-1 (ETS1) and WDFY family member 4 (WDFY4) polymorphisms in Chinese patients with rheumatoid arthritis. Int J Mol Sci, 15(2), 2712–2721. 10.3390/ijms15022712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Li XL, Li M, Hao LX, Chen RW, Xiang K, … Su B (2011). Replicated associations of TNFAIP3, TNIP1 and ETS1 with systemic lupus erythematosus in a southwestern Chinese population. Arthritis Res Ther, 13(6), R186. 10.1186/ar3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liu M, Li J, Hashmi F, Mao Z, Zhang N, … Li C (2015). Impact of V-ets Erythroblastosis Virus E26 Oncogene Homolog 1 Gene Polymorphisms Upon Susceptibility to Autoimmune Diseases: A Meta-Analysis. Medicine (Baltimore), 94(22), e923. 10.1097/MD.0000000000000923 [DOI] [PMC free article] [PubMed] [Google Scholar]


