Abstract
Decoding UGA as selenocysteine requires a unique tRNA, a specialized elongation factor, and specific secondary structures in the mRNA, termed SECIS elements. Eukaryotic SECIS elements are found in the 3′ untranslated region of selenoprotein mRNAs while those in prokaryotes occur immediately downstream of UGA. Consequently, a single eukaryotic SECIS element can serve multiple UGA codons, whereas prokaryotic SECIS elements only function for the adjacent UGA, suggesting distinct mechanisms for recoding in the two kingdoms. We have identified and characterized the first eukaryotic selenocysteyl-tRNA-specific elongation factor. This factor forms a complex with mammalian SECIS binding protein 2, and these two components function together in selenocysteine incorporation in mammalian cells. Expression of the two functional domains of the bacterial elongation factor–SECIS binding protein as two separate proteins in eukaryotes suggests a mechanism for rapid exchange of charged for uncharged selenocysteyl-tRNA–elongation factor complex, allowing a single SECIS element to serve multiple UGA codons.
INTRODUCTION
The fidelity of translation is dependent upon delivery of aminoacyl-tRNAs to their cognate codons in the ribosomal A site by the elongation factors, EF1 or eEF1. Sense codons are distinguished from termination codons by the tRNA–EF complexes decoding the former and release factor recognition of the latter (Ito et al., 2000). An exception to this is the incorporation of selenocysteine (Sec), in which a tRNA–EF complex is delivered to a codon that would normally be read as stop. The mechanism of Sec incorporation in bacteria has been characterized in considerable detail (Böck, 2000). Central to the process is SELB, a bifunctional protein consisting of an N-terminal Sec-tRNA-specific EF domain and a C-terminal SECIS RNA-binding domain (Forchhammer et al., 1989; Kromayer et al., 1996). Binding to the SECIS element recruits the EF to the ribosome as it approaches the UGA codon. Details of the corresponding mechanism in eukaryotes have lagged behind. A mammalian SECIS binding protein, SBP2, has recently been shown to be required for Sec incorporation in rabbit reticulocyte lysates, but this factor lacks EF homology or activity (Copeland et al., 2000). Here we describe the missing link, the murine Sec-specific EF, termed eEFsec. The purified recombinant factor specifically binds Sec-tRNA[Ser]Sec, but not seryl-tRNA[Ser]Sec. We show in vivo association of the factor with SBP2 and in vitro association with both SBP2 and the SECIS element. Finally, in transiently transfected mammalian cells, the EF is shown to function together with SBP2 in selenoprotein synthesis.
RESULTS
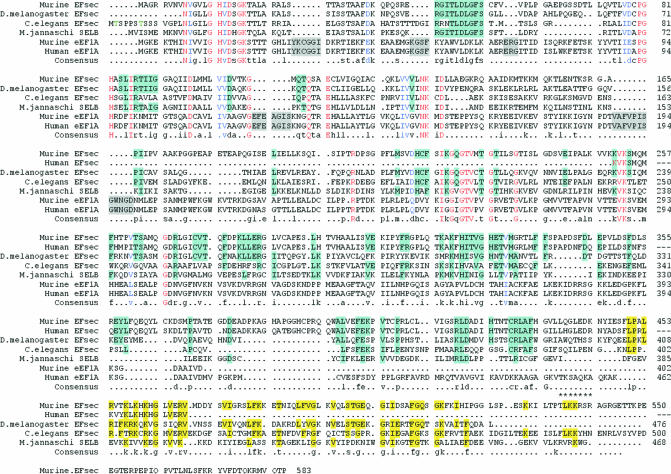
Sequential homology searches of databases of increasing complexity, progressing from the archaeon, Methanococcus jannaschii, to Caenorhabditis elegans, Drosophila melanogaster, and finally the mammalian expressed sequence tag (EST) databases resulted in identification of lower eukaryotic, murine and human Sec-specific EF candidates. The sequence of the murine EST predicts a protein of 583 amino acids, as compared with 462 for eEF1A. Alignment of the eEFsec sequences from these species along with those of murine and human eEF1A reveals high homology in the EF domain, but also regions unique to the eEFsec family (Figure 1). Insertions are present in the eEF1A sequences that are not found in eEFsec, as previously reported for the bacterial SELB family (Kromayer et al., 1996). In bacterial EF-Tu, these regions confer interaction with the GTP exchange factor, EF-Ts. Their absence in SELB is explained by a lower affinity for GDP than in EF-Tu, circumventing the need for a separate exchange factor (Hilgenfeld et al., 1996). The absence of these regions in the eEFsec family indicates that the same is likely to be true here (see below). Significant homology is also present in the C-terminal extensions of the eEFsec family, being in some regions restricted to the eukarya, but in other regions also including the archaeal protein.
Fig. 1. Alignment of deduced amino acid sequences of eEF and eEF1A family members. Alignment was generated using Multalign software (Corpet, 1988). Red lettering indicates conserved amino acids. Blue lettering indicates similar amino acids. Light blue highlighting indicates regions conserved amongst the eEFsec family but differing from eEF1A. Gray highlighting indicates insertions in eEF1A relative to EFsec. Yellow highlighting indicates conserved or similar regions in the C-terminal eEFsec extensions. Asterisks indicate the predicted nuclear localization signal. Homology between murine eEFsec and D. melanogaster eEFsec is 41%, C.elegans is 30% and M. jannaschii is 33%.
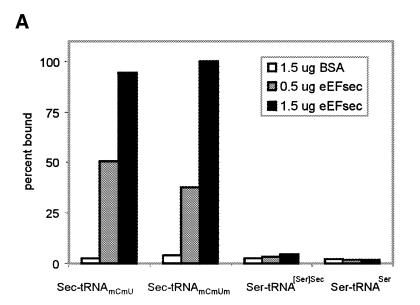
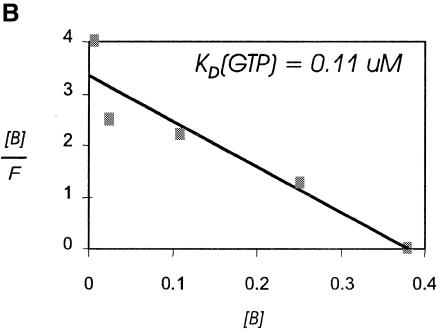
The tRNA specificity of the murine factor was determined in a nitrocellulose filter binding assay using purified recombinant protein and purified radiolabeled mammalian tRNAs. Sec-tRNA[Ser]Sec exists as two isoforms, which differ by the presence or absence of a 2′-O-ribose methylation at U34, the anticodon wobble base (Diamond et al., 1993). Binding by the factor was highly specific for the two isoforms of Sec-tRNA[Ser]Sec, whereas binding to either the serylated precursor, seryl-tRNA[Ser]Sec, or to the standard tRNA, seryl-tRNASer, occurred at background levels (Figure 2A). Binding to methionyl-tRNAMet or to the only other non-standard tRNA, initiator methionyl-tRNAMetI, also represented background levels (data not shown). GTP binding by the recombinant factor was assessed by Scatchard analysis, indicating a Kd of 0.11 µM (Figure 2B). Competition with unlabeled GDP indicated an ~3-fold higher Kd, consistent with previous findings for SELB and indicative of a lack of requirement for a GTP exchange factor. GTP binding and specificity of the factor for the two Sec-tRNA[Ser]Sec isoforms, but not the seryl-tRNA[Ser]Sec precursors, satisfy the criteria for a Sec-tRNA-specific EF. We therefore suggest the designation eEFsec for this protein to reflect this specificity and function.
Fig. 2. tRNA and guanine nucleotide binding specificity of murine eEFsec. (A) Binding of radiolabeled aminoacyl-tRNAs by the indicated proteins is given as percent total input counts retained on nitrocellulose filters. Seryl-tRNA[Ser]Sec was tested as either the individual mcmU and mcmUm isoacceptors or as a mixture of the two, with the same results. (B) Scatchard analysis of GTP binding to eEFsec. Binding was performed as described in Methods.
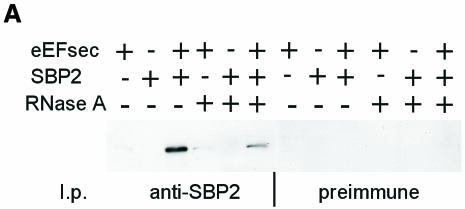
The presence of a SECIS element in the 3′ untranslated region (3′UTR) of a eukaryotic mRNA confers recoding of upstream UGA codons as Sec (Berry et al., 1991a, 1993). We hypothesize that Sec incorporation directed from the 3′UTR involves binding of SBP2 to the SECIS element, followed by recruitment of eEFsec-tRNA[Ser]Sec by the SBP2–SECIS complex. Assembly of the complex at the SECIS element, followed by delivery of eEFsec-tRNA[Ser]Sec to a UGA codon occupying the ribosomal A site, would allow translation of UGA codons at any site in the open reading frame, without strict context requirements. This model predicts the association of SBP2 and eEFsec, either through direct protein–protein interactions, protein–RNA interactions or through other interacting partners. To address these possibilities, we performed co-immunoprecipitation studies involving eEFsec and SBP2, using proteins expressed by co-transfection in a human cell line, HEK-293. eEFsec was subcloned into a mammalian expression vector with a FLAG epitope tag introduced at the N-terminus, and co-transfected with plasmids expressing SBP2 (Copeland et al., 2000) and a mammalian selenoprotein, the latter to increase selenoprotein synthesis over the endogenous level. Immunoprecipitation of transfected cell homogenates with antisera against SBP2 resulted in co-precipitation of eEFsec, detected by western blotting with FLAG antibody (Figure 3A). The amount of eEFsec in the co-precipitate increased significantly with SBP2 co-transfection, but eEFsec was also detected in the absence of co-transfected SBP2, presumably due to association with endogenous SBP2, which is present at significant levels in this cell line (data not shown). When immunoprecipitation reactions were incubated in the presence of RNase A, the amount of eEFsec recovered decreased by 75%, indicating that complex formation was enhanced by the SECIS element or other RNA ligand. However, RNase treatment did not completely abolish co-precipitation, suggesting that there is an RNA-independent interaction, which is augmented by the presence of RNA, or that association is dependent on a partially nuclease-resistant RNA.

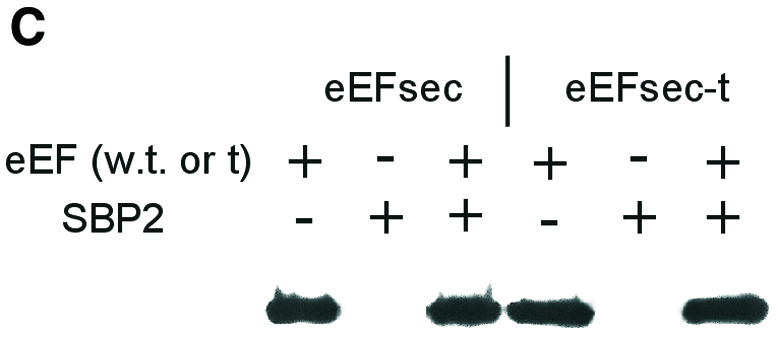
Fig. 3. Co-immunoprecipitation of murine eEFsec and SBP2 from co-transfected cells. (A) Co-immunoprecipitation of eEFsec with antisera against SBP2, followed by western blotting and detection with anti-FLAG (eEFsec) antibody. (B) Co-immunoprecipitation of truncated eEFsec-t as above. (B) was exposed ∼8-fold longer than (A). (C) Western analysis of eEFsec or eEFsec-t in total cell homogenates.
To address eEFsec binding specificity, a truncated version of eEFsec missing the last 20 amino acids, including two Lys–Arg repeats, was co-transfected with SBP2 as above. Co-precipitation of this protein, designated eEFsec-t, was greatly decreased relative to the full-length protein (Figure 3B). This is apparent by the lower amount of protein detected in Figure 3B relative to Figure 3A, despite an ∼8-fold longer exposure of the film. RNase treatment further decreased the amount of eEFsec-t recovered. Based on western blot analysis of total cell homogenates, the full-length and truncated proteins were expressed at comparable levels (Figure 3C). We were unable to assess endogenous levels of eEF-sec, due to lack of an antibody specific for this protein. However, northern blotting of a murine multi-tissue blot revealed that the mRNA is ubiquitously expressed, with highest levels in testes and liver (data not shown).
To investigate the role of SECIS RNA in complex formation, we employed electrophoretic mobility shift assays (EMSAs) with in vitro transcribed 32P-labeled SECIS elements and purified bacterially expressed recombinant proteins. Incubation of a wild-type (w.t.) SECIS element with either SBP2 or eEFsec retarded the mobility of the SECIS element (Figure 4A). The mobility patterns of the two complexes were clearly distinguishable. Thus, the two proteins bind the SECIS element independently. Upon incubation with both proteins, an enhancement in the amount of shifted RNA was seen. Binding specificity was next tested using 32P-labeled SECIS mutants consisting of either a point mutation in the conserved essential sequence, AUGA (mutated to AUCA), or inversion of the first two nucleotides (to UAGA). Binding of the mutant SECIS elements by SBP2 was not detected in the gel shift assay (Figure 4B). In contrast, eEFsec exhibited binding to both mutants. Strikingly, when both proteins were added together, binding to the mutant SECIS elements was not seen. Thus, the presence of SBP2 prevents eEFsec binding to the SECIS mutants, conferring specificity for the w.t. sequence. This result suggests that interaction with SBP2 alters the conformation of the eEFsec RNA-binding domain. From the EMSA patterns we could not distinguish whether the shift seen in Figure 4A in the presence of both proteins was due to superimposition of the two individual complexes or a new complex consisting of the RNA and both proteins. To investigate this, the w.t. SECIS element was incubated with SBP2 in the absence or presence of eEFsec. After UV cross-linking, the RNA–protein complexes were resolved by non-denaturing PAGE and analyzed for SBP2 by western blotting (Figure 4C). A fraction of the SBP2–SECIS complex was shifted upward upon the addition of eEFsec, indicating that a new complex containing both proteins had formed on the RNA. Formation of complexes in the absence of the RNA was not detectable (not shown).
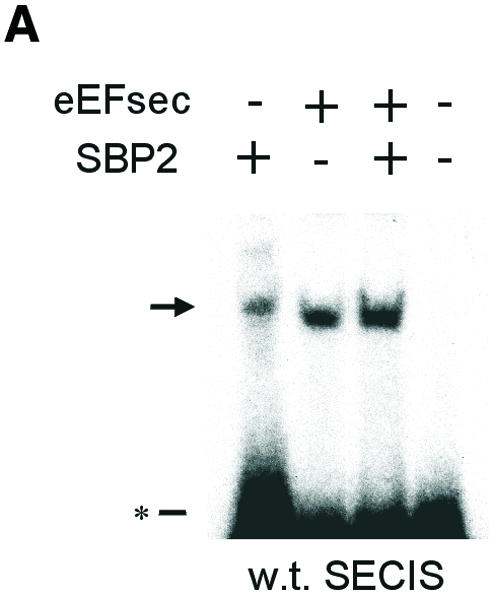
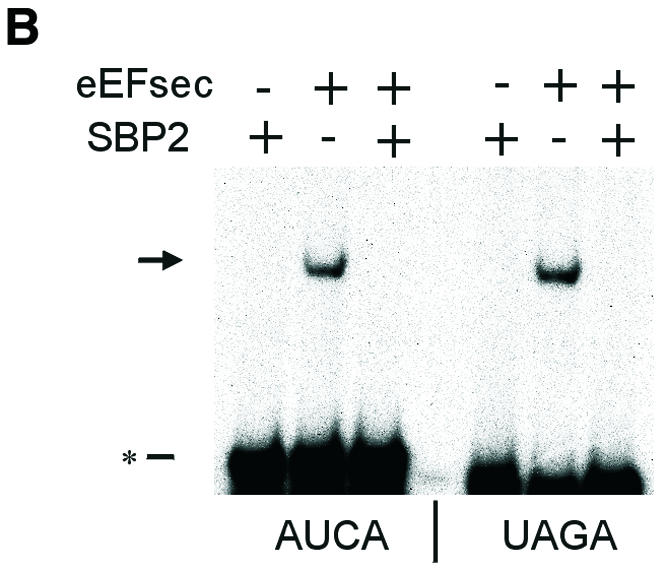
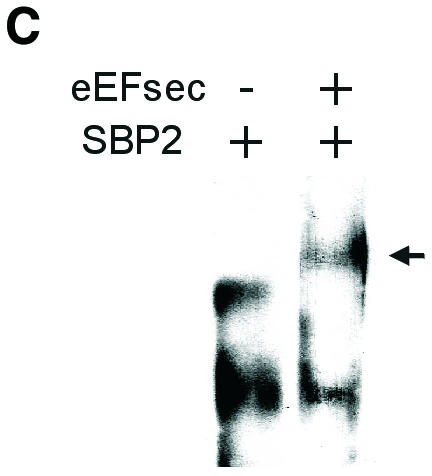
Fig. 4. Complex formation between eEFsec, SBP2 and SECIS element. (A) EMSA of w.t. type 1 deiodinase SECIS element (41 nucleotides of minimal SECIS element plus 35 nucleotides of flanking sequence). Arrows indicate shifted complexes, asterisks designate free RNA. (B) EMSA of indicated mutant SECIS elements corresponding to w.t. element above. (C) UV cross-linking of SBP2–SECIS and eEFsec–SBP2–SECIS complexes. The w.t. D1 SECIS element was incubated with the indicated proteins as for EMSA, followed by UV cross-linking, PAGE and western blotting as described in Methods. The arrow indicates the eEFsec–SBP2 complex.
To assess binding specificity further, competition studies were performed with unlabeled SECIS RNAs. Addition of a 50-fold excess of unlabeled w.t. SECIS element to the labeled w.t. element prior to incubation with eEFsec resulted in 70% inhibition of binding of the labeled RNA, as assessed by nitrocellulose filter binding assay (not shown). Similarly, competition for binding by SBP2 or by the combination of the two proteins was inhibited by 60 and 67%, respectively (not shown). A 50-fold excess of the w.t. element inhibited binding of eEFsec to the labeled AUCA mutant by ∼70%, and to the UAGA mutant by >90%. The same molar excess of unlabeled AUCA SECIS mutant decreased binding of labeled w.t. SECIS by eEFsec by only ∼20%, indicating a lower affinity of eEFsec for the mutant than for the w.t. element. A 50-fold excess of unlabeled AUCA mutant competed binding of eEFsec to the labeled AUCA mutant by 70% and to the UAGA mutant by 85%. Yeast tRNA decreased binding to the w.t. and AUCA mutant elements by <10%, but inhibited binding to the UAGA mutant by ∼70%, indicating a weak or relatively non-specific interaction with the latter.
Finally, the functions of the two proteins were assessed in vivo by co-transfection in HEK-293 cells with a cDNA encoding selenoprotein P from the zebrafish, Danio rerio. This cDNA contains 17 Sec codons and two SECIS elements, making the expressed protein a sensitive indicator of effects on selenoprotein synthesis. Previous studies of selenoprotein P purified from rat plasma identified, in addition to the full-length protein, heterogeneous products resulting from premature termination at the UGA codons (Himeno et al., 1996). We have observed a similar phenomenon with transiently expressed rat and zebrafish selenoprotein P (Tujebajeva et al., 2000 and R.M. Tujebajeva, J.W. Harney and M.J. Berry, unpublished). Sec incorporation was assessed following transfection by addition of 75Se to the medium, and analysis of the resultant labeled selenoprotein P. In the absence of added factors, a diffuse band representing full-length and UGA-terminated species is seen (Figure 5, lane 1). Transfection of eEFsec alone resulted in a shift upward, indicating more full-length protein (lane 2). Transfection of SBP2 resulted in a dramatic decrease in the ratio of full-length protein to premature termination product (lane 3). Co-transfection of both factors reversed the SBP2 effect, shifting the balance towards full-length protein (lane 4). One possible explanation for this result is that overexpression of SBP2 may perturb the balance of factors required for Sec incorporation, resulting in more efficient translation at some UGA codons, but perhaps less so at others. Co-expression of eEFsec apparently restores this balance.
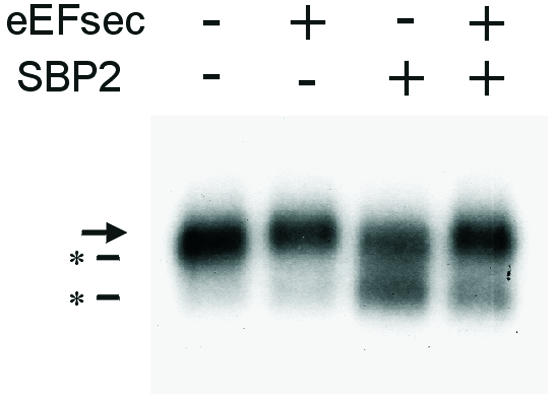
Fig. 5. Effects of eEFsec and SBP2 on selenoprotein synthesis in vivo. Danio rerio selenoprotein P cDNA in pUHD10-3 vector was transfected into HEK-293 cells either alone or with plasmids expressing eEFsec, SBP2 or both as described in Methods. Twenty-four hours after transfection, [75Se]sodium selenite was added to the medium. Media were harvested the following day and analyzed by SDS–PAGE, followed by autoradiography. The arrow indicates the position of the full-length selenoprotein. Asterisks indicate truncated forms.
DISCUSSION
Insertion of Sec directed from the 3′UTR has been enigmatic until recently. Identification of SBP2 provided the first link between the SECIS element and recoding. The data presented herein identify the long sought missing link in this process, the Sec-tRNA-specific EF. Alignment of the eEFsec sequences reveals homology with the eEF1A family, but also regions of conservation unique to the eEFsec family, both in the EF domain and the C-terminal extension. As the SECIS elements are not conserved between eukarya and archaea, these regions of homology are not likely to be involved in SECIS interaction, but may function in interaction with SBP2 or its putative lower eukaryotic or archaeal counterparts. Now, for the first time a clear model can be advanced for eukaryotic selenocysteine insertion. The SECIS element, SBP2 and eEFsec form a Sec-tRNA delivery complex, with SBP2 conferring specificity on eEFsec for w.t. SECIS elements. When and where does this complex formation occur? Recruitment of factors may take place early in the life of the mRNA. eEF-1 is present in both the nucleus and the cytoplasm (Kruse et al., 1998). Interestingly, both eEFsec and SBP2 contain putative nuclear localization signals (Figure 1 and Copeland et al., 2000). Thus, SBP2 could bind to the SECIS element immediately after its transcription, in concert with or followed by eEFsec and possibly other factors. This would set the stage for Sec incorporation in the first round of translation, upon export of the mRNA to the cytoplasm and binding of the first ribosome, thus allowing the mRNA to escape nonsense-mediated decay. Unlike the situation in prokaryotes, where the SECIS elements are in the coding region, there would be no requirement for SBP2 repeatedly to dissociate from and reassociate with the SECIS element. Thus, while some aspects of the mechanisms are similar, this difference provides a solution to the question of how a SECIS element could serve multiple UGA codons. The presence of a relatively stable SECIS–SBP2 complex would allow rapid, efficient exchange of empty eEFsec for eEFsec bound to aminoacyl-tRNA. A mechanism for rapid exchange is likely to be essential for decoding the multiple Sec codons of selenoprotein P. Is termination somehow circumvented, and if so, how? It is of interest to note that SBP2 possesses homology to a yeast omnipotent suppressor of translation termination, SUP1. The role of this domain in excluding release factors has not yet been assessed. Studies by Stapulionis and Deutscher (1995), Negrutskii et al. (1999) and others have provided evidence for the existence of supramolecular translation complexes, consisting of ribosomes, mRNA, EFs and their bound aminoacyl-tRNAs, and aminoacyl-tRNA synthetases. Similar supramolecular complexes for selenoprotein mRNAs could contribute to their efficient translation. These ‘selenosome’ complexes would presumably contain SECIS-bound SBP2, eEFsec and its cognate Sec-tRNA, and perhaps also enzymes involved in Sec biosynthesis or other as yet unidentified factors. Further investigation of the components and function of the selenosome complex will certainly yield new insights into this fascinating translation mechanism.
METHODS
eEFsec expression, purification, and tRNA and GTP binding assays. Murine eEFsec cDNA (DDBJ/EMBL/GenBank accession No. AF283518) was subcloned into pET30 (Novagen) via a BglII site introduced at the N-terminus by PCR, and an XhoI site at the 3′ end of the EST clone in Bluescript. This resulted in its fusion with a region encoding six N-terminal His residues. The plasmid was transformed into BL21-DE3-pLysS bacteria (Novagen), grown to late log phase, and expression induced with IPTG. The culture was harvested, resuspended in phosphate-buffered saline containing 25% glycerol, 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.2 U/ml aprotinin, and cells lysed by three cycles of freeze–thawing, followed by sonication, and passage through a 27 gauge needle. Lysates were centrifugated at 4500 r.p.m. (3980 g) for 15 min. NaCl was added to the supernatant to a final concentration of 0.3 M and GTP to 0.1 mM, followed by chromatography on a Ni-NTA–agarose column (Qiagen) equilibrated in the same buffer (column buffer). The resin was washed with column buffer containing 50 mM imidazole, and protein eluted in buffer containing 250 mM imidazole. The presence of eEFsec was assessed by western blotting of column fractions and detection with an anti-His5 antibody (Qiagen). Peak fractions were concentrated and exchanged into column buffer without imidazole using Spintrap columns (VIVA Science). [75Se]Sec-tRNAs were purified from HL60 cells following labeling with [75Se]sodium selenite (1000 Ci/mmol) and the mcmU34 and mcmUm34 isoacceptors separated on an RPC-5 column (Lee et al., 1989). [3H]Seryl-tRNA[Ser]Sec, [3H]seryl-tRNASer, [35S]methionyl-tRNAMetm and [35S]methionyl-tRNAMetI were prepared with [3H]serine (36.0 Ci/mmol) or [35S]methionine (>1000 Ci/mmol) as given (Hatfield et al., 1979). The nitrocellulose filter binding assay was performed by incubation of tRNAs (10–15 pmol) with recombinant mouse eEFsec (0.5–1.5 µg) or bovine serum albumin (BSA) in binding buffer for 10 min at ambient temperature, followed by 10 min at 30°C. Binding buffer consisted of 50 mM HEPES pH 7.5, 10 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 25% glycerol and 0.1 mM GTP. Nitrocellulose filters (Millipore HAWP 025 00) were pre-wetted, placed in a vacuum manifold, binding reactions applied, vacuum slowly applied, then filters were washed three times with 0.5 ml of binding buffer at 4°C. Filters were air dried and counted by liquid scintillation (Beckman LS5801). GTP binding was performed in the nitrocellulose filter assay using 300 nCi of [γ-32P]GTP and 0.2–5.0 µM unlabeled GTP or GDP.
Transfection and co-immunoprecipitation. HEK-293 cells were transfected with cDNAs encoding type 1 iodothyronine deiodinase (all lanes; Berry et al., 1991b) and the indicated factors in mammalian expression vectors [eEFsec and eEFsec-t in pUHD10-3 (Gossen and Bujard, 1992) and SBP2 in pCR3.1 (Copeland et al., 2000)] as described previously (Berry et al., 1991a). Cells were harvested, sonicated in STN (10 mM Tris–HCl pH 7.6, 150 mM NaCl, 0.5% NP-40, 5% glycerol, 1 mM PMSF and 0.2 U/ml aprotinin), pre-cleared with normal rabbit serum plus Pansorbin (CalBiochem), and the supernatants incubated with the indicated antibodies for 1 h. Reactions were divided into two aliquots and RNase A (10 µg/ml final concentration) added to one aliquot of each, followed by further incubation for 15 min at ambient temperature. Pansorbin was added and pellets collected by centrifugation, washed three times in STN, and analyzed by SDS–PAGE, followed by western blotting and detection by enhanced chemiluminescence.
SECIS binding studies. 32P-labeled rat type 1 deiodinase w.t. or mutant SECIS elements were transcribed in vitro (Riboprobe, Promega), denatured and slowly annealed. Unlabeled competitor RNAs were generated using a MEGAshortscript T7 Kit (Ambion) according to the manufacturer’s protocol. RNAs were incubated with purified recombinant eEFsec, SBP2 (Copeland et al., 2000) or both in binding buffer for 10 min at 30°C. The linear range of binding was determined for both proteins in the nitrocellulose filter binding assay, and protein levels in the middle of that range used for subsequent assays. Reactions typically contained 105 c.p.m. of 32P-labeled SECIS RNAs, 300 ng of SBP2 and 200–400 ng of eEFsec. Following incubation, aliquots were taken for the nitrocellulose filter binding assay and for EMSA. For the latter, reactions were electrophoresed on 5 or 10% acrylamide–TBE gels (Ready gel, Bio-Rad), followed by autoradiography. UV cross-linking was performed for 30 min in a UV Stratalinker 1800 (Stratagene), followed by electrophoresis on 7.5% non-denaturing polyacrylamide gels (Ready gel, Bio-Rad) and western blotting as above.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr William Merrick for helpful suggestions. This work was supported by grants from the NIH to P.R.C., D.M.D. and M.J.B.
REFERENCES
- Berry M.J., Banu, L., Chen, Y., Mandel, S.J., Kieffer, J.D., Harney, J.W. and Larsen, P.R. (1991a) Recognition of UGA as a selenocysteine codon in Type I deiodinase requires sequences in the 3′ untranslated region. Nature, 353, 273–276. [DOI] [PubMed] [Google Scholar]
- Berry M.J., Banu, L. and Larsen, P.R. (1991b) Type I iodothyronine deiodinase is a selenocysteine-containing enzyme. Nature, 349, 438–440. [DOI] [PubMed] [Google Scholar]
- Berry M.J., Banu, L., Harney, J.W. and Larsen, P.R. (1993) Functional characterization of the eukaryotic SECIS elements which direct selenocysteine insertion at UGA codons. EMBO J., 12, 3315–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böck A. (2000) Biosynthesis of selenoproteins—an overview. BioFactors, 11, 77–78. [DOI] [PubMed] [Google Scholar]
- Copeland P.R., Fletcher, J.E., Carlson, B.A., Hatfield, D.L. and Driscoll, D.M. (2000) A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J., 19, 306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res., 16, 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond A.M. et al. (1993) Dietary selenium affects methylation of the wobble nucleoside in the anticodon of selenocysteine tRNA. J. Biol. Chem., 268, 14215–14223. [PubMed] [Google Scholar]
- Forchhammer K., Leinfelder, W. and Böck, A. (1989) Identification of a novel translation factor necessary for the incorporation of selenocysteine into protein. Nature, 342, 453–456. [DOI] [PubMed] [Google Scholar]
- Gossen M. and Bujard, H. (1992) Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. USA, 89, 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield D.L., Matthews, C.R. and Rice, M. (1979) Aminoacyl-transfer RNA populations in mammalian cells: chromatographic profiles and patterns of codon recognition. Biochim. Biophys. Acta, 564, 414–423. [DOI] [PubMed] [Google Scholar]
- Hilgenfeld R., Bock, A. and Wilting, R. (1996) Structural model for the selenocysteine-specific elongation factor SelB. Biochimie, 78, 971–978. [DOI] [PubMed] [Google Scholar]
- Himeno S., Chittum, H.S. and Burk, R.F. (1996) Isoforms of selenoprotein P in rat plasma. J. Biol. Chem., 271, 15769–15775. [DOI] [PubMed] [Google Scholar]
- Ito K., Uno, M. and Nakamura, Y. (2000) A tripeptide ‘anticodon’ deciphers stop codons in messenger RNA. Nature, 403, 680–684. [DOI] [PubMed] [Google Scholar]
- Kromayer M., Wilting, R., Tormay, P. and Böck, A. (1996) Domain structure of the prokaryotic selenocysteine-specific elongation factor SELB. J. Mol. Biol., 262, 413–420. [DOI] [PubMed] [Google Scholar]
- Kruse C., Grunweller, A., Willkomm, D.K., Pfeiffer, T., Hartmann, R.K. and Muller, P.K. (1998) tRNA is entrapped in similar, but distinct, nuclear and cytoplasmic ribonucleoprotein complexes, both of which contain vigilin and elongation factor 1 α. Biochem. J., 329, 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.J., Worland, P.J., Davis, J.N., Stadtman, T.C. and Hatfield, D. (1989) Identification of a selenocysteyl-tRNASer in mammalian cells that recognizes the nonsense codon, UGA. J. Biol. Chem., 264, 9724–9727. [PubMed] [Google Scholar]
- Negrutskii B.S., Shalak, V.F., Kerjan, P., El’skaya, A.V. and Mirande, M. (1999) Functional interaction of mammalian valyl-tRNA synthetase with elongation factor EF-1α in the complex with EF-1H. J. Biol. Chem., 274, 4545–4550. [DOI] [PubMed] [Google Scholar]
- Stapulionis R. and Deutscher, M.P. (1995) A channeled tRNA cycle during mammalian protein synthesis. Proc. Natl Acad. Sci. USA, 92, 7158–7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tujebajeva R.M., Harney, J.W. and Berry, M.J. (2000) Selenoprotein P expression, purification and immunochemical characterization. J. Biol. Chem., 275, 6288–6294. [DOI] [PubMed] [Google Scholar]