Summary
Background
Bedaquiline is a life-saving tuberculosis drug undergoing global scale-up. People at risk of weak tuberculosis drug regimens are a priority for novel drug access despite the potential source of Mycobacterium tuberculosis-resistant strains. We aimed to characterise bedaquiline resistance in individuals who had sustained culture positivity during bedaquiline-based treatment.
Methods
We did a retrospective longitudinal cohort study of adults (aged ≥18 years) with culture-positive pulmonary tuberculosis who received at least 4 months of a bedaquiline-containing regimen from 12 drug-resistant tuberculosis treatment facilities in Cape Town, South Africa, between Jan 20, 2016, and Nov 20, 2017. Sputum was programmatically collected at baseline (ie, before bedaquiline initiation) and each month to monitor treatment response per the national algorithm. The last available isolate from the sputum collected at or after 4 months of bedaquiline was designated the follow-up isolate. Phenotypic drug susceptibility testing for bedaquiline was done on baseline and follow-up isolates in MGIT960 media (WHO-recommended critical concentration of 1 μg/mL). Targeted deep sequencing for Rv0678, atpE, and pepQ, as well as whole-genome sequencing were also done.
Findings
In total, 40 (31%) of 129 patients from an estimated pool were eligible for this study. Overall, three (8%) of 38 patients assessable by phenotypic drug susceptibility testing for bedaquiline had primary resistance, 18 (47%) gained resistance (acquired or reinfection), and 17 (45%) were susceptible at both baseline and follow-up. Several Rv0678 and pepQ single-nucleotide polymorphisms and indels were associated with resistance. Although variants occurred in Rv0676c and Rv1979c, these variants were not associated with resistance. Targeted deep sequencing detected low-level variants undetected by whole-genome sequencing; however, none were in genes without variants already detected by whole-genome sequencing. Patients with baseline fluoroquinolone resistance, clofazimine exposure, and four or less effective drugs were more likely to have bedaquiline-resistant gain. Resistance gain was primarily due to acquisition; however, some reinfection by resistant strains occurred.
Interpretation
Bedaquiline-resistance gain, for which we identified risk factors, was common in these programmatically treated patients with sustained culture positivity. Our study highlights risks associated with implementing life-saving new drugs and shows evidence of bedaquiline-resistance transmission. Routine drug susceptibility testing should urgently accompany scale-up of new drugs; however, rapid drug susceptibility testing for bedaquiline remains challenging given the diversity of variants observed.
Funding
Doris Duke Charitable Foundation, US National Institute of Allergy and Infectious Diseases, South African Medical Research Council, National Research Foundation, Research Foundation Flanders, Stellenbosch University Faculty of Medicine Health Sciences, South African National Research Foundation, Swiss National Science Foundation, and Wellcome Trust.
Introduction
Tuberculosis is an ongoing health crisis. Drug-resistant tuberculosis is difficult to diagnose and treat.1 Bedaquiline, a novel WHO-endorsed tuberculosis drug introduced for compassionate use in South Africa in 2012 because of few other treatment options,2 is now a part of all standard drug-resistant tuberculosis regimens globally.
Phenotypic drug susceptibility testing for bedaquiline should be done before patients are started on a bedaquiline-containing regimen to monitor resistance emergence.3 However, programmatic implementation of bedaquiline drug susceptibility testing has lagged because of weak laboratory infrastructure and a lack of technical expertise and easily implementable assays. This delay is alarming given the occurrence of variants in bedaquiline candidate resistance genes existing before bedaquiline’s availability,4 bedaquiline’s initial prioritisation for patients with high levels of resistance to other drugs,5 and bedaquiline-resistance emergence post-treatment cessation.6
Unsurprisingly, programmatic bedaquiline resistance has been documented in Germany, China, and Pakistan, with acquisition at rates ranging between 3% and 17%.7,8 Furthermore, in South Africa, a study of a nationally representative sample of 3005 isolates from patients receiving bedaquiline found 199 (7%) to have phenotypic resistance, far exceeding rates in clinical trials.9,10
Reduced bedaquiline susceptibility might be due to variants in the candidate resistance genes Rv0678, atpE, and pepQ. Mutations in Rv0678 are associated with clofazimine resistance,11 and some isolates with elevated minimal inhibitory concentrations to bedaquiline have not shown resistance-associated variants in any of these genes. The consequence of other bacterial genes such as Rv0676c (MmpL5), Rv0677c (MmpS5), and Rv1979c is unknown.12,13 Our understanding of genotypic mechanisms of resistance is, however, far from complete.
Not only are more data for variants associated with phenotypic resistance needed, but current data are scarce for bedaquiline-resistance emergence during treatment in which multiple isolates are analysed from the same individuals. This concern is especially important in people in whom resistance is more likely to emerge (eg, in a programmatic rather than a clinical trial context). Such individuals could include those with resistance to many second-line drugs who, despite being, on a population-level, a minority receiving bedaquiline (most patients do not receive bedaquiline), could inadvertently serve as a source of population-level bedaquiline-resistance transmission.11 These patients do not always rapidly clinically or bacteriologically respond positively: programmatic data from inpatients with second-line resistance in Cape Town, South Africa, show that 30% receiving bedaquiline as part of their regimens remain culture positive after 4 months of treatment.14 This study population is similar to the population in our current study in terms of patients who were in a hospital specialising in drug resistance in Cape Town (where many of our patients originated from), the study period overlapped somewhat (2014–19), and importantly the eligibility criteria of who received bedaquiline was the same or similar. In this study, we aimed to characterise bedaquiline resistance in people who had sustained culture positivity during bedaquiline-based treatment.
Methods
Study population and patient data collection
In this retrospective longitudinal cohort study, we retrospectively identified adults (aged ≥18 years) with culture-positive pulmonary tuberculosis who received at least 4 months of a bedaquiline-containing regimen from 12 drug-resistant tuberculosis treatment facilities (as inpatients at Brooklyn Chest Hospital or Khayelitsha Day Hospital or outpatients from surrounding facilities) in Cape Town, South Africa, between Jan 20, 2016, and Nov 20, 2017 (figure 1A). Sputum was programmatically collected at baseline and each month to monitor treatment response per the national algorithm.15 The last available isolate from the sputum collected at or after 4 months of bedaquiline was designated the follow-up isolate.
Figure 1: Study profile and bedaquiline DST results at baseline and follow-up.
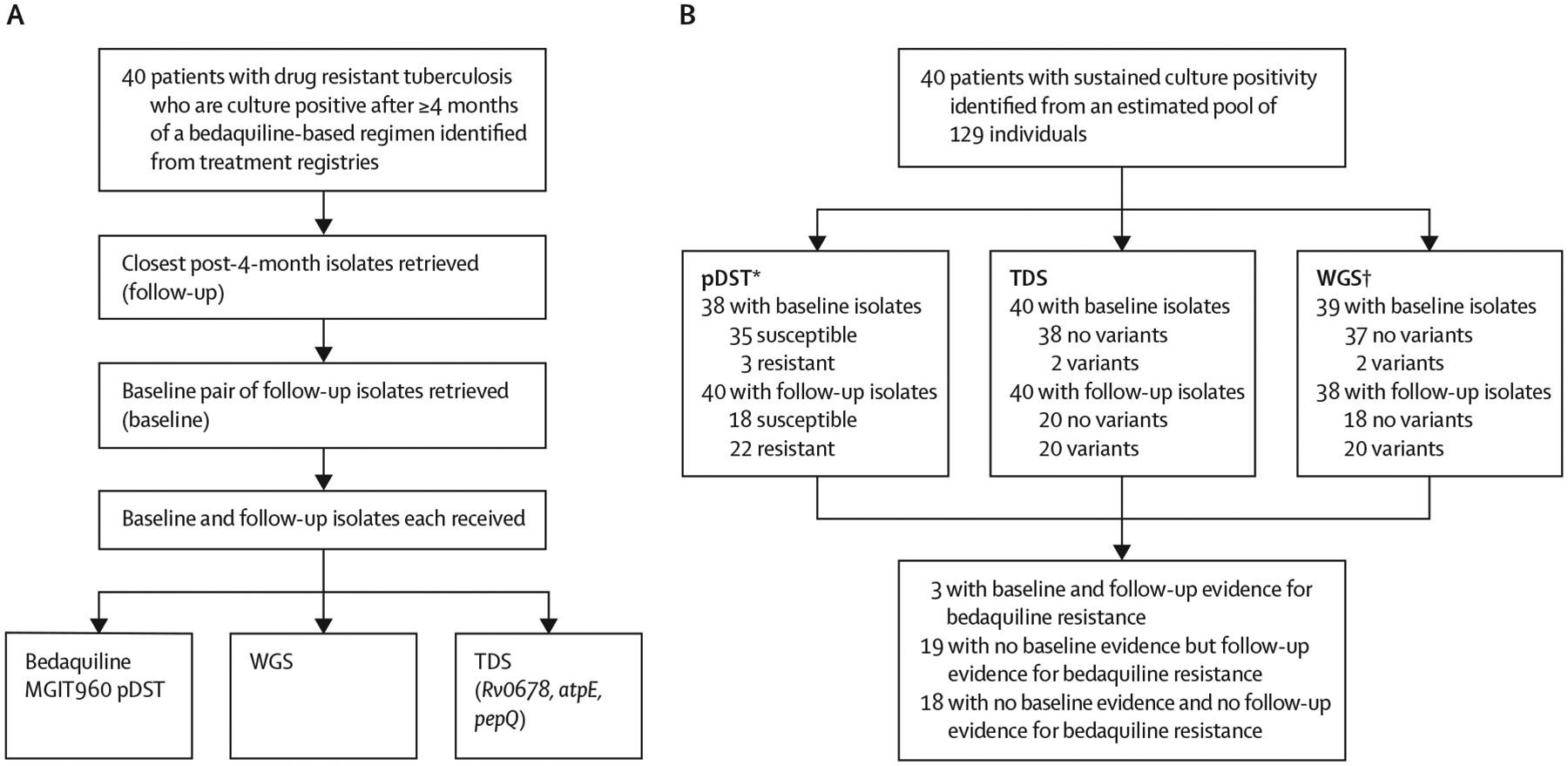
(A) Patients with pulmonary drug-resistant tuberculosis and who were culture positive after 4 months or more of a bedaquiline-based regimen had their baseline (close to bedaquiline treatment initiation) and follow-up isolate (≥4 months) retrieved and used for phenotypic and genotypic (TDS and WGS) DST. (B) This approach identified a large proportion of patients whose Mycobacterium tuberculosis isolates gained resistance. DST=drug susceptibility testing. MGIT=mycobacterial growth indicator tube. pDST=phenotypic drug susceptibility testing. TDS=targeted deep sequencing. WGS=whole-genome sequencing. *Missing data for pDST results due to non-culturable isolates. †WGS results unavailable.
We included patients who had both a post-4-month isolate and a pre-bedaquiline treatment initiation (ie, baseline) isolate stored in a biobank. We excluded patients without such isolates, not on a bedaquiline regimen, or minors (aged <18 years). Our study took place in the period before all oral regimens were being recommended for patients with drug-resistant tuberculosis.
This study was approved by the Stellenbosch University Health Research Ethics Committee (N09/11/296; N16/04/045), Western Cape Health Research Committee (WC_2016RP18_637), University of California San Francisco Human Research Protection Program (14–15090), and the University of Cape Town Human Research Ethics Committee (416/2014).
Procedures
We extracted patient demographic and clinical data, including previous and current tuberculosis treatment regimen and follow-up, from clinical records. Treatment outcome data were reported using WHO definitions for drug-resistant tuberculosis.16
Phenotypic drug susceptibility testing for bedaquiline was done on baseline and follow-up isolates in mycobacterial growth indicator tube (MGIT960) media as previously described.17 The WHO-recommended critical concentration of 1 μg/mL bedaquiline fumarate (Janssen Pharmaceuticals via the National Institute of Allergy and Infectious Diseases/National Institutes of Health AIDS Reagent Program; Bethesda, USA) was used. Mycobacterium tuberculosis H37Rv was used as a susceptible control and BCCM/ITM 121749 (Institute of Tropical Medicine; Antwerp, Belgium) as a resistant control.18
Sequencing was done on crude or purified isolate DNA.19 Genomic libraries for whole-genome sequencing were prepared using the DNA Prep kit (Illumina; San Diego, USA) or NEBNext Ultra II kit (NEB; Ipswich, USA), and sequenced using NextSeq500 (Illumina; San Diego, USA) with V2, paired-end chemistry. Reads are deposited in the European Nucleotide Archive (PRJEB47429) and were analysed as described to detect non-synonymous variants or indels in Rv0678, atpE, pepQ, Rv0676c, Rv0677c, and Rv1979c.20 TB profiler (version 3.0.4) was used for drug susceptibility testing (allele frequency of ≥10% for resistance calls). De novo assembly to detect Rv0678 structural variants for one isolate with discrepant whole-genome sequencing and targeted deep sequencing results was done using UGAP and svTyper.
Targeted deep sequencing for Rv0678, atpE, and pepQ were each analysed using multiple tiled amplicons as previously described6 (appendix 1 p 8), but were not done for other loci as primers were unavailable. Samples were pooled and sequenced using an Illumina MiSeq (Illumina; San Diego, California, USA) with V3 paired-end chemistry with a targeted coverage of 30 000 reads per amplicon. Reads are in NCBI Bioproject (PRJNA767896) and were analysed by Amplicon Sequencing Analysis Pipeline. Minority variants were counted if variants occurred at 1% or more with five or more paired reads. Mitigations steps, such as positive reference controls, were included to reduce false-positive results. For haplotype analysis, four Rv0678 amplicons were individually analysed. Haplotype analysis was not done on isolates that had a single variant or genes from phenotypic resistant patients without variants reported.
Statistical analysis
We did the statistical analyses using Stata (version 15) and GraphPad Prism (version 8.0.1) using two-sided tests (α=0·05). McNemar’s test was used to calculate differences in paired data. We estimated the number of people from which our cohort was derived using data from a contemporaneous clinical study with similar eligibility criteria that shared a facility. For drugs other than bedaquiline, whole-genome sequencing of drug susceptibility testing was used to classify a drug as likely effective. Methods for clustering, phylogeny, and reinfection analyses are summarised in appendix 1 (pp 2–3). Information about the whole-genome sequencing and targeted deep sequencing bioinformatics analysis are summarised in appendix 1 (pp 2–3).
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
In total, 40 (31%) of 129 patients from an estimated pool were eligible for this study (figure 1). Of these 40 patients, 26 (65%) were inpatients and 14 (35%) were outpatients at surrounding facilities. 28 (70%) of 40 patients were initiated on bedaquiline15 when substituted for a second-line injectable because of toxicity or intolerance, or within an individualised regimen where resistance to fluoroquinolones or second-line injectables was noted. 12 (30%) of 40 patients were treated with bedaquiline-containing salvage regimens in the setting of extensive drug resistance (eight [67%] of 12 had resistance to both fluoroquinolones and second-line injectables, and four [33%] had a history of second-line treatment failure).21
Demographics and clinical characteristics, including treatment histories and outcomes are summarised in the table. Overall, 18 (46%) of 39 patients (one result unavailable) had baseline whole-genome sequencing-detected fluoroquinolone resistance, and this finding was more frequent in patients with bedaquiline resistance (at baseline or follow-up) than in those who were bedaquiline susceptible (14 [67%] of 21 vs four [22%] of 18; p=0·01). 29 (73%) of 40 patients had a history of clofazimine exposure and 20 (50%) had concurrent clofazimine and bedaquiline treatment. At follow-up, patients received a median of seven drugs (IQR 6–9). When antibiograms of baseline versus follow-up isolates were compared, the frequency of fluoroquinolone resistance (18 [46%] of 39 patients vs 28 [76%] of 37; p=0·01) and clofazimine resistance (three [8%] of 39 vs 21 [57%] of 37; p<0·0001) increased (appendix 1 p 11). In patients who had bedaquiline resistance at follow-up, the number of likely effective drugs were fewer than in patients with bedaquiline-susceptible tuberculosis (three [IQR 3–4] vs five [4–6]; p<0·0001). The odds ratio (OR) for bedaquiline resistance at follow-up for baseline fluoroquinolone resistance was 7 (95% CI 2–29; p<0·0001), clofazimine exposure was 5 (1–23; p=0·03), and four or less likely effective drugs was 12 (2–61; p<0·0001). When patients with baseline bedaquiline resistance and reinfection were omitted, the four or less likely effective drugs was the only variable with a significant OR (appendix 1 p 12). Levofloxacin, pyrazinamide, and clofazimine were each more likely to be ineffective in bedaquiline-resistant patients than in bedaquiline-susceptible patients (table; appendix 1 pp 9–10). Patients who had phenotypically bedaquiline-resistant isolates at follow-up had, at specimen collection, received bedaquiline for a similar period as susceptible patients. 25 (63%) of 40 patients had an unfavourable outcome, and all but one (patient 37-B08) with a bedaquiline resistance-associated variant had an unfavourable outcome.
Table:
Patient characteristics according to phenotypic bedaquiline resistance profile at follow-up
| Overall (n=40) | Bedaquiline phenotype at follow-up | p value | ||
|---|---|---|---|---|
| Susceptible (n=18) | Resistant (n=22) | |||
| Demographics | ||||
| Age at diagnosis of current episode (years) | 36 (27–45) | 34 (27–43) | 36 (27–45) | 0·75 |
| Sex | ||||
| Female | 11 (28%) | 5 (28%) | 6 (27%) | 0·97 |
| Male | 29 (73%) | 13 (72%) | 16 (73%) | 0·96 |
| Clinical characteristics | ||||
| HIV positive | 21 (53%) | 7 (39%) | 14 (64%) | 0·12 |
| CD4 count (× 109 cells per L)* | 168 (61–394) | 235 (28–631) | 157 (66–314) | 0·70 |
| Antiretroviral therapy | 18/21 (86%) | 5/7 (71%) | 13/14 (93%) | 0·49 |
| Treatment history | ||||
| Previous tuberculosis | 32 (80%) | 13 (72%) | 19 (86%) | 0·27 |
| Previous drug-resistant tuberculosis | 20/32 (63%) | 8/13 (62%) | 12/19 (63%) | 0·93 |
| Days between baseline and follow-up isolate | 429 (291–564) | 369 (252–592) | 439 (365–578) | 0·44 |
| Any clofazimine exposure (prior or concurrent) | 29 (73%) | 10 (56%) | 19 (86%) | 0·03 |
| Prior clofazimine† | 7/26 (27%) | 2/8 (25%) | 5/18 (28%) | 0·88 |
| Concurrent with bedaquiline† | 20/26 (77%) | 6/8 (75%) | 14/18 (78%) | 0·88 |
| Treatment and drug resistance | ||||
| Bedaquiline treatment duration (days) | 185 (168–265) | 181 (168–267) | 193 (168–266) | 0·80 |
| Baseline fluoroquinolone resistance‡ | 18/39 (46%) | 4 (22%) | 14/21 (67%) | 0·01 |
| Overall drug resistance patient categorisation | ||||
| Pre-multidrug-resistant or rifampicin mono-resistant tuberculosis | 2/36 (6%) | 2/16 (13%) | 0 | 0·10 |
| Multidrug resistant | 8/36 (22%) | 6/16 (38%) | 2/20 (10%) | 0·05 |
| Multidrug resistant plus resistance to a fluoroquinolone | 20/36 (56%) | 7/16 (44%) | 13/20 (65%) | 0·20 |
| Multidrug resistant plus resistance to a fluoroquinolone and second-line injectable | 6/36 (17%) | 1/16 (6%) | 5/20 (25%) | 0·43 |
| Treatment regimen | ||||
| Total number of drugs (excluding bedaquiline)§ | 7 (6–9) | 7 (6–12) | 8 (6–9) | 0·85 |
| Likely effective¶ | 4 (3–5) | 5 (4–6) | 3 (3–4) | <0·0001 |
| Treatment outcomes | ||||
| Favourable outcome | 8 (20%) | 7 (39%) | 1 (5%) | <0^0001 |
| Cured | 4 (10%) | 3 (17%) | 1 (5%) | 0·20 |
| Treatment completed | 4 (10%) | 4 (22%) | 0 | 0·02 |
| Unfavourable outcome | 25 (63%) | 6 (33%) | 19 (86%) | <0·0001 |
| Treatment failed | 5 (13%) | 0 | 5 (23%) | 0·03 |
| Died | 20 (50%) | 6 (33%) | 14 (64%) | 0·06 |
| Lost to follow-up | 4 (10%) | 3 (17%) | 1 (5%) | 0·20 |
| Not evaluable | 3 (8%) | 2 (11%) | 1 (5%) | 0·43 |
Data are n (%), n/N (%), or median (IQR), unless otherwise specified. Those with resistance were more likely to have baseline fluoroquinolone resistance, clofazimine exposure, fewer likely effective drugs, and an adverse treatment outcome versus susceptible patients at follow-up.
One (3%) of 40 patients had unknown CD4 count.
Three (8%) of 40 patients had a record of clofazimine treatment (two bedaquiline-susceptible patients and one bedaquiline-resistant patient) but no date.
Detected by whole-genome sequencing or programmatic line probe assay. One result unavailable.
Two (5%) of 40 patients were excluded due to unknown background tuberculosis drug regimens.
Two whole-genome sequencing results unavailable.
Of baseline isolates assessable by phenotypic drug susceptibility testing for bedaquiline (1 μg/mL), 35 (92%) of 38 were susceptible and three (8%) were resistant. Of the follow-up isolates, 18 (45%) of 40 patients were susceptible and 22 (55%) were resistant. Overall, three (8%) of 38 patients had primary resistance (appendix 1 pp 13–14), 18 (47%) gained resistance (acquired or reinfection; appendix 1 pp 15–20), and 17 (45%) were susceptible at both baseline and follow-up (figure 1B, 2; appendix 1 pp 21–24).
Figure 2: Timeline of bedaquiline treatment and follow-up isolate sampling per patient.
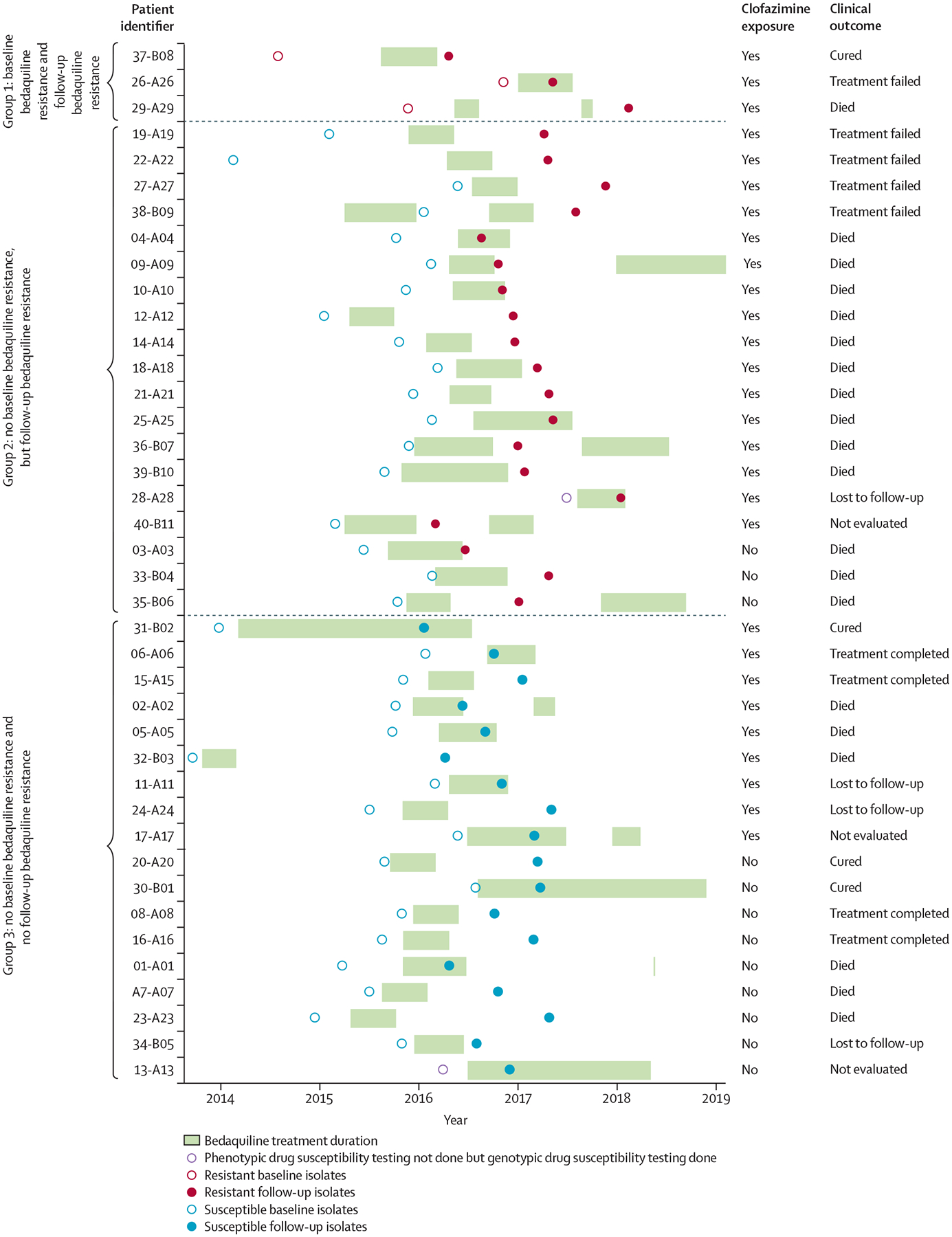
Each circle represents individual bedaquiline phenotypic drug susceptibility testing done. Three (8%) of 40 patients were phenotypically resistant at baseline and follow-up (classified as group 1), and all had clofazimine exposure (prior or concurrent with bedaquiline). 19 (48%) of 40 patients acquired bedaquiline resistance during treatment (group 2); all but three (16%) of 19 had clofazimine exposure and all with a known treatment outcome had an unfavourable outcome. 18 (45%) of 40 patients were susceptible at both timepoints (group 3).
Whole-genome sequencing data were available for 39 (98%) of 40 patients with baseline isolates and 38 (95%) with follow-up isolates. In patients whose isolates gained resistance, no baseline isolates had Rv0678, atpE, pepQ, or Rv0677c variants (excluding Rv0678-11 C/A), although five (29%) of 17 had one or more Rv1979c variant (appendix 1 pp 15–20). All isolates (resistant or susceptible) had one or more Rv0676c variant. At follow-up, the percentage of patients whose isolates gained variants associated with resistance increased to 88% (15 of 17 patients; p<0·0001 vs baseline) for Rv0678 and 24% (four patients; p=0·70 vs baseline) for Rv1979c; no atpE, pepQ, or Rv0677c variants were noted at follow-up. Notably, two (12%) of 17 patients’ isolates gained bedaquiline resistance (03-A03, 09-A09) but had no apparent variants when a genome-wide screen was done. However, after visual inspection an IS6110 insertion site was identified in patient 03-A03. By contrast, among patients whose isolates remained susceptible, new variants were noted only in pepQ (two [13%] of 16; p=0·13) and Rv1979c (five [31%] of 16; p=0·69; appendix 1 pp 21–24). When gain-or-loss variants were compared between timepoints for background drugs (excluding bedaquiline), 30 (79%) of 38 drug-resistance conferring mutations were fixed. However, for eight (21%) of 38 patients, heteroresistance was seen and this finding is detailed in appendix 1 (pp 5–6).
Targeted deep sequencing detected additional variants relative to whole-genome sequencing; 13 (33%) of 40 patient isolates with a whole-genome sequencing-detected variant had additional variants exclusively detected by targeted deep sequencing. In all patients, Rv0678 and pepQ variants were detected in two (5%) of 40 baseline isolates and 20 (50%) of 40 follow-up isolates, but none were detected in combination (no atpE variants were detected; −11C/A Rv0678 was excluded). However, no variants exclusively detected by targeted deep sequencing explained phenotypic bedaquiline resistance not otherwise explained by whole-genome sequencing-detected variants. Additionally, targeted deep sequencing identified the same majority bedaquiline-resistant associated variants as whole-genome sequencing. In those who gained phenotypic resistance, 16 (89%) of 18 patients gained one or more Rv0678 variants (–11C/A Rv0678 variants excluded; appendix 1 pp 15–20). The IS6110 insertion in the patient 03-A03 follow-up isolate was undetected by targeted deep sequencing. Haplotype analyses on phenotypically resistant isolates with targeted deep sequencing-detected variants from 17 patients showed, for Rv0678, that if more than one variant was detected 14 (82%) of 17 were on different amplicons. Three (18%) of 17 patients were on different amplicons and the same amplicon some of the time (appendix 2).
29 (81%) of 36 patients with whole-genome sequencing data available at baseline and follow-up had seven or less variants difference between isolates (median 3·0 [IQR 2·5–3·7]), indicative of possible within-patient evolution (the number of variants difference was not associated with days-between-isolates; appendix 1 p 7). Of these 29 patients, 15 (52%) transitioned phenotypically from bedaquiline susceptible to resistant, indicating possible resistance acquisition. The other seven (19%) of 36 patients had between 39 and 1271 variants difference, suggestive of reinfection possibly from transmission. Two (29%) of seven of these patients were initially bedaquiline susceptible and hence possibly gained resistance via reinfection. Of the seven patients with possible reinfection, five (71%) who remained L2.2.1 (39 and 240 variants different, respectively) had a different sub-lineage at follow-up compared with baseline (figure 3). Patient 04-A04 had at baseline a mixed infection of two strains, both of which, based on inspection of pncA sequences, were not present at follow-up; rather, a third new strain was present.
Figure 3: Whole-genome sequencing of BL and FU isolates identified five clusters (≤12 single-nucleotide polymorphisms) and evidence of reinfection, overlayed with phenotypic bedaquiline resistance statuses.
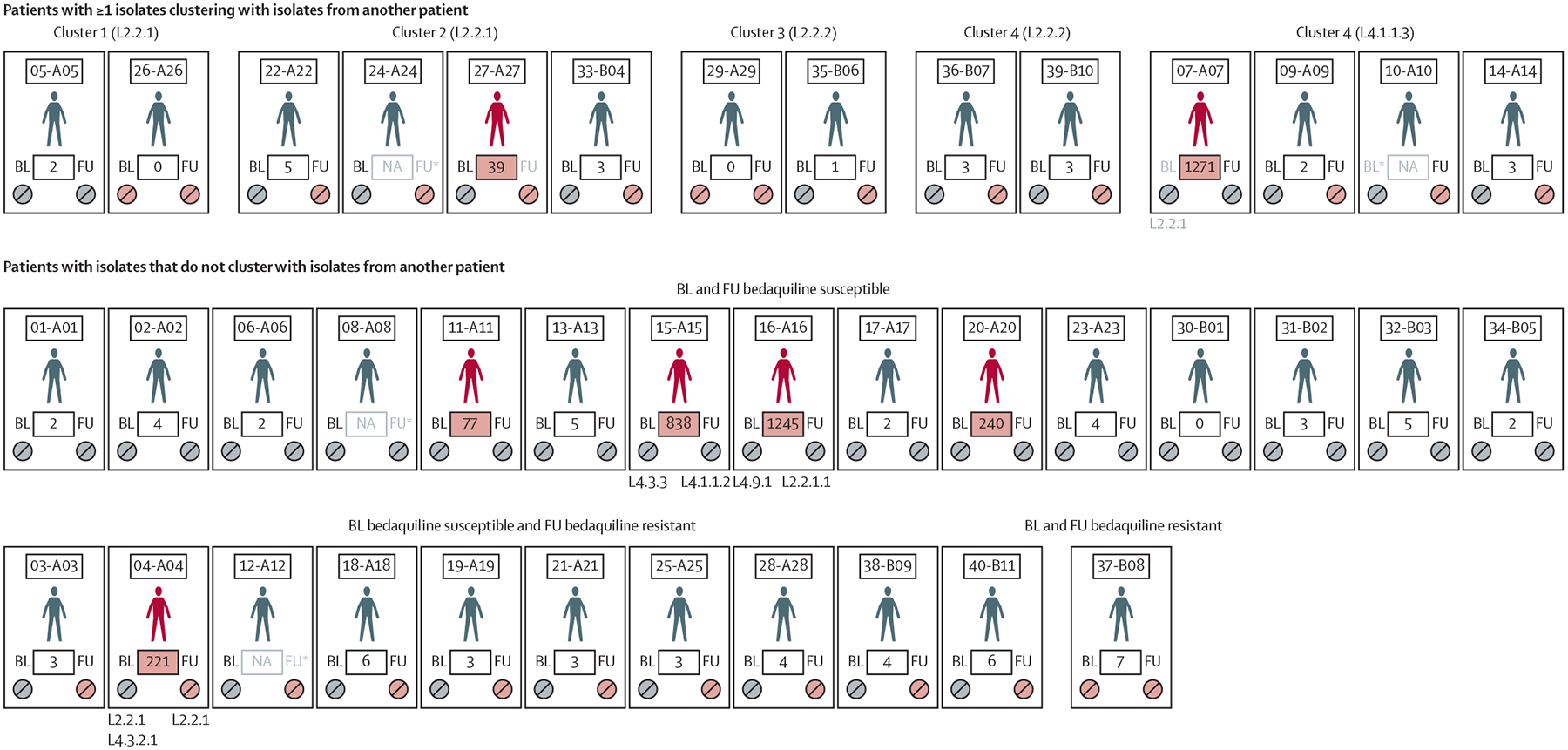
Pill colours indicate phenotypic bedaquiline susceptibility (grey) or phenotypic bedaquiline resistance (red). Greyed BL or FU labels with an asterisk indicate missing whole-genome sequencing data and those without an asterisk indicate the corresponding isolate does not cluster with its pair. Patient identifiers are shown above for each patient with variant distances between BL and FU in boxes (if distances are ≥39, reinfection is indicated by a red patient figure and red box background). Patients without isolates that cluster with other patients are grouped per baseline and follow-up phenotypic bedaquiline statuses (bottom two rows). Lineages are shown when pairs differed. BL=baseline. FU=follow-up. L=lineage. NA=not available.
At the 12 or less single-nucleotide polymorphism cutoff, five clusters totalling 14 patients with L2 or L4 strains were identified (figures 3, 4). 26 (65%) of 40 patients did not cluster with one another (the ≤5 single-nucleotide polymorphism threshold identified no clusters). All clusters except cluster 1 had at least one patient who gained phenotypic bedaquiline resistance and, in both clusters 2 and 5, one patient who gained resistance had a new follow-up strain (reinfection).
Figure 4: Cladogram showing isolate’s lineage diversity, likely reinfection, drug susceptibility, and clofazimine exposure.
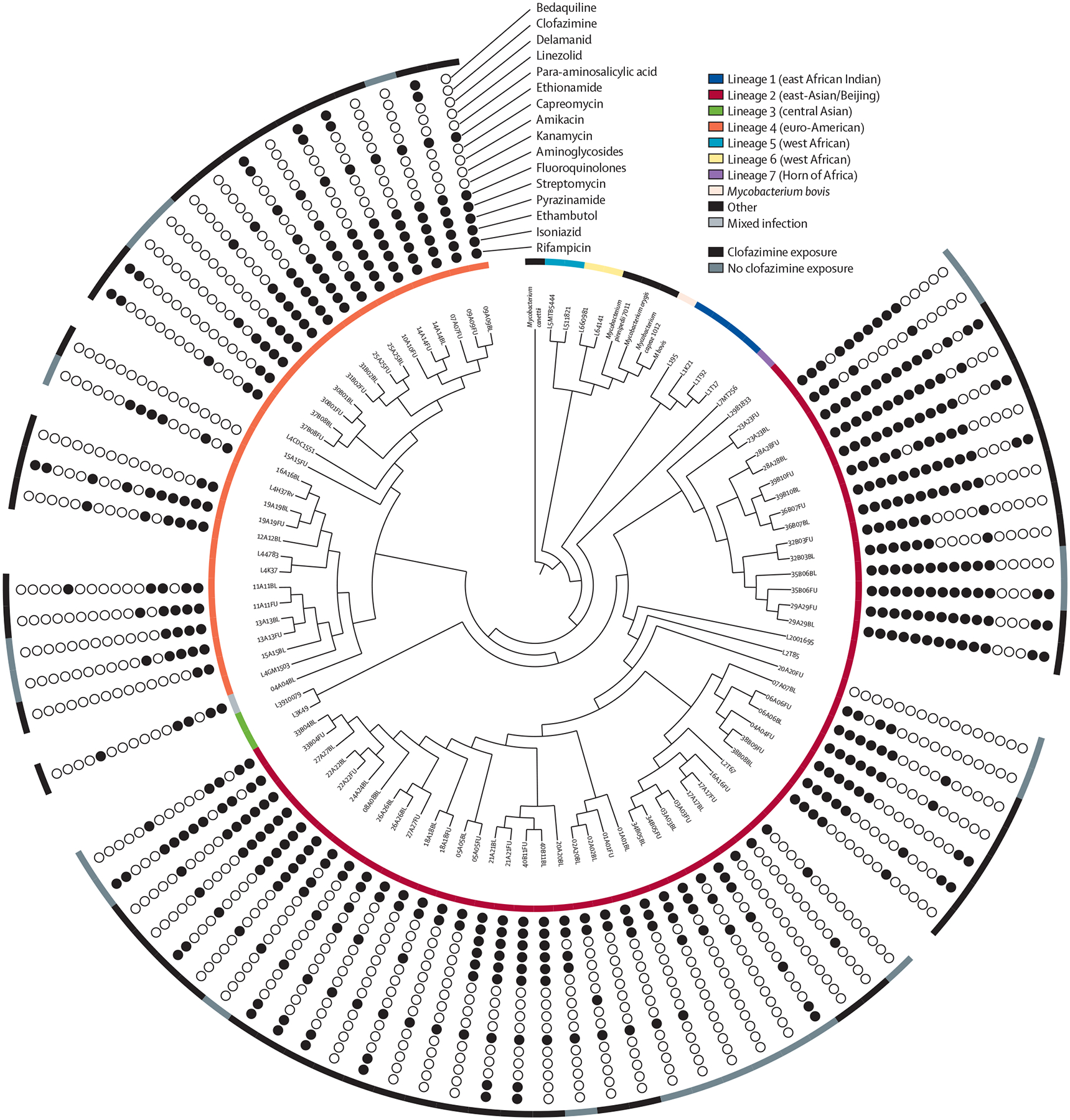
The inner ring shows the cladogram. The outer ring shows clofazimine exposure. The circles show drug susceptibility based on whole-genome sequencing excluding phenotypic drug susceptibility testing for bedaquiline. 51 (67%) of 76 isolates were in L2 (red) and 24 (32%) were in L4 (orange; one mixed with both lineages present at baseline), and isolates were resistant to many drugs. Branch lengths were ignored for visualisation and the phylogenetic analysis included 25 sequences representative of the Mycobacterium tuberculosis complex (appendix 2). BL=baseline. FU=follow-up. L=lineage.
Discussion
We evaluated phenotypic and genotypic bedaquiline resistance among programmatically treated patients with drug-resistant tuberculosis who were culture positive after 4 months of a bedaquiline-containing regimen. Key findings include the following: more than half of the patients had isolates that gained bedaquiline resistance (most due to acquisition; however, reinfection might have also occurred); diverse Rv0678 and pepQ single nucleotide variants and indels were frequently seen with phenotypic resistance, whereas isolates with Rv0676c, Rv0677c, and Rv1979c variants were seen with both susceptible and resistant isolates (suggestive of lineage markers); no atpE variants were found; and many minor variants were only detected by targeted deep sequencing, however, all isolates with exclusively targeted deep sequencing-detected variants already had other whole-genome sequencing-detected variants that were seen with phenotypic bedaquiline resistance; and bedaquiline-resistance gain at follow-up was associated with fewer likely effective drugs, clofazimine exposure, and baseline fluoroquinolone resistance. Together, these data identify a potential programmatic source of bedaquiline resistance, show how reinfection can be responsible for resistance, and highlight the complexity of associating specific variants with phenotypic bedaquiline resistance. Furthermore, these data can contribute to the WHO drug-resistant tuberculosis mutation catalogue and provide information about which patients are most at risk of gaining resistance.
This study is one of the first to report individual-level bedaquiline-resistance gain over time among patients treated in a programmatic setting with a bedaquiline-containing regimen. This rate was higher than described elsewhere,22,23 as our patient population was deliberately preselected based on elevated resistance acquisition risk, and selected for possible failure of treatment. However, these individuals reflect the type of patient originally prioritised for bedaquiline access in our setting and, should bedaquiline-resistance transmission become endemic, is one possible initial source (importantly, three patients were already resistant at bedaquiline treatment initiation). Resistance gain was primarily due to acquisition; however, in some patients it was due to reinfection.
We identified many variants in Rv0678 and pepQ alongside phenotypic bedaquiline resistance (eg, Rv0678: 343 C/T, 136 G ins, and 292 A del; pepQ: 693 A ins) hitherto undescribed.12,22 Rv0676c variants (2299 C/T, 2381 G/A, and 2842 T/C) were found in both resistant and susceptible isolates at both timepoints. In the 2021 WHO drug-resistant tuberculosis mutation catalogue, these variants are indeed classified as not associated with bedaquiline resistance13 and are suggestive of lineage markers. Interestingly, −11C/A variants have been associated with increased bedaquiline susceptibility;24 however, eight of these isolates also had additional Rv0678 variants, suggesting any hyper-susceptibility is possibly overcome.
These diverse variants represent complexity for molecular diagnostic developers that is compounded by the insertion of large elements such as IS6110 in Rv0678 that might be missed by targeted deep sequencing and whole-genome sequencing.25 This complexity is somewhat offset by the relatively low frequency of pepQ variants in the absence of Rv0678 variants (only one of these strains with only a pepQ variant was bedaquiline resistant) and a complete absence of atpE variants.
Targeted deep sequencing detected many minor variants missed by whole-genome sequencing; however, the overall genotypic classification of resistance did not change because all variants exclusively detected by targeted deep sequencing were in loci that had whole-genome sequencing-detected variants. This finding might be because this patient population has a very low number of effective drugs and thus many variants can emerge—some of these variants to a high level (ie, non-minority heteroresistance)—at which point they are detectable to whole-genome sequencing. This finding should not rule out the use of targeted deep sequencing, as variants exclusively detected by targeted deep sequencing might affect minimum inhibitory concentrations, acquired resistance, and treatment failure. Furthermore, with bigger sample size, exclusively detected targeted deep sequencing variants might have occurred in phenotypically resistant isolates. Targeted deep sequencing is also useful (and often easier to do prospectively) for second-line genotypic drug susceptibility testing.26
Several characteristics differed at baseline in patients who later gained bedaquiline resistance versus those who did not. In addition to previously described prior or concurrent clofazimine exposure,27 a weaker background regimen (especially fluoroquinolones being possibly ineffective) puts patients at risk of resistance acquisition. This finding suggests protecting the fluoroquinolone (eg, via rapid drug susceptibility testing or enhanced dosing) is crucial for preventing bedaquiline-resistance acquisition, as well as ensuring that the number of likely effective drugs always exceeds four. Lastly, almost all patients with bedaquiline resistance had a poor clinical outcome, indicating the need for new treatment strategies in patients with bedaquiline-resistant tuberculosis.
Our study has strengths and limitations. First, we aimed to understand bedaquiline-resistance emergence in a programmatic setting where we expected it to be most likely. This finding is intentionally not representative of most patients who receive bedaquiline for drug-resistant tuberculosis, for which excellent outcomes have been observed.11 Our study took place before all oral regimens were recommended for patients with multidrug-resistant or rifampicin-resistant tuberculosis but was in a facility where novel drugs such as bedaquiline are first used before population-level scale-up. The baseline frequency of fluoroquinolone resistance is, although reflective of that in the types of patients prioritised for initial bedaquiline access in our setting, not representative of that in patients who have uncomplicated rifampicin-resistant tuberculosis who are now, per the latest guidelines, eligible for bedaquiline-based regimens (about 13% in South Africa, about 20% globally).28 Nevertheless, our study highlights the importance of fluoroquinolone drug-susceptible testing and suggests that protecting fluoroquinolone susceptibility is possibly essential to prevent emergence of bedaquiline resistance. Our aim was not to quantify population-level bedaquiline resistance, which is important. Large national or subnational molecular epidemiology studies have quantified population-level bedaquiline resistance,14,29 and one study found an overall baseline bedaquiline resistance rate of 5·0% (19 of 383) in people with drug-resistant tuberculosis in South Africa, with 3·5% (five of 142) of people gaining resistance in the longitudinal cohort.30 This difference in the magnitude of resistance gain versus our study is possibly attributable to patient profiles (all patients starting bedaquiline in these other studies vs patients with complex treatment histories and sustained culture positivity in our study). We did not quantify bedaquiline minimal inhibitory concentrations, as our objective was to characterise the proportion of people whose minimal inhibitory concentrations exceeded the critical concentration for bedaquiline; however, our isolates will contribute to future efforts to accomplish this important goal. On a technical level, a mix of crude or purified isolate DNA was used for sequencing, which might explain some discordance (eg, targeted deep sequencing missing a few whole-genome sequencing-detected variants). For targeted deep sequencing, even though positive amplification and sequencing reference controls were included, we recognise that erroneous single-nucleotide polymorphisms could be identified by chance. Our clustering analysis was a convenient secondary analysis done to rule in not rule out transmission; we therefore possibly underestimated transmission events.
In conclusion, this study highlights the existence of bedaquiline-resistant strains, for which we show transmission occurring, created under programmatic conditions in a population of patients prioritised at the beginning of bedaquiline roll-out in South Africa. These findings are most generalisable to settings where new drugs such as bedaquiline were given, even if only initially, to patients such as those in this study, where there was a high level of pre-existing drug resistance and few effective treatment options. The study illustrates high rates of resistance in people with a delayed bacteriological response, the risks of starting patients with complex tuberculosis treatment histories on a regimen containing a novel drug without routinely available drug susceptibility testing (which requires balancing with ethical considerations), provides information about novel bedaquiline resistance-associated and susceptibility-associated variants, and informs upon clinical risk factors associated with resistance gain.
Supplementary Material
Research in context.
Evidence before this study
Bedaquiline is a landmark drug. Programmatic implementation of this drug is being rapidly scaled up to treat many forms of drug-resistant tuberculosis, a life-threatening disease that affects hundreds of thousands of people globally. South Africa was one of the first countries to make bedaquiline available and prioritise the drug for people with few other treatment options; however, the use of bedaquiline was done in the absence of an ability to regularly test for resistance. We searched PubMed and Google Scholar using the search terms “bedaquiline”, “resistance”, and “tuberculosis” for primary research and reviews from database inception to Sept 5, 2022. We found publications describing bedaquiline resistance primarily in clinical trials rather than in programmatic contexts. The few publications describing programmatic data were mostly cross-sectional molecular epidemiology studies, none repeatedly sampled individuals at risk of bedaquiline resistance over time, and no descriptions of bedaquiline resistance transmission were found. We also noted a paucity of information regarding variants and risk factors associated with resistance.
Added value of this study
We uniquely analysed more than one culture isolate per patient over time and presented data from patients who had weak background regimens stemming from pre-existing advanced drug-resistant tuberculosis and were prioritised for bedaquiline access. We showed that more than half of these patients gained bedaquiline resistance and noted two individuals who gained a new bedaquiline resistant strain (probable primary resistance transmission). We reported diverse and previously undocumented Rv0678 and pepQ variants that are associated with resistance; that resistance acquisition itself is associated with baseline fluoroquinolone resistance, previous or current clofazimine exposure, and fewer drugs likely to be effective; and that bedaquiline resistance significantly increases the likelihood of a poor clinical outcome.
Implications of all the available evidence
The rapidly growing body of evidence, including this work, highlights the existence of an infectious pool of bedaquiline resistant strains that might prove to be a source of population-level resistance transmission. This finding demonstrates the challenges and risks associated with starting patients with complex treatment histories on a novel drug, for which there are several in the pipeline, without routinely available drug susceptibility testing and strong background regimens. Clinical risk factors for incident resistance and descriptions of resistance-associated variants are increasingly well known. This collective work could be used to develop guidelines to assist clinicians, simultaneously inform the development of rapid genotypic assays and their prioritisation in key at-risk populations, and advise future novel tuberculosis drug roll-out strategies.
Acknowledgments
This work was funded by Doris Duke Charitable Foundation (JM), US National Institute of Allergy and Infectious Diseases (R01AI131939; JM and DE), South African Medical Research Council, National Research Foundation, Research Foundation Flanders (G0F8316N; AVR and AD), Stellenbosch University Faculty of Medicine Health Sciences, South African National Research Foundation and Swiss National Science Foundation (107799; HC and SG), and the Wellcome Trust (099818/Z/12/Z; HC). We thank the National Health Laboratory Services, Green Point, Cape Town, South Africa. We also thank members of the TGen team, including Andrew Goedderz, Jason Agundez, and Meagan Papineau for doing targeted sequencing and data analysis support. Lastly, we thank the patients, their families, and the health-care staff who cared for them.
Footnotes
Declaration of interests
We declare no competing interests.
See Online for appendix 2
See Comment page e964
For UGAP see https://github.com/jasonsahl/UGAP
For svTyper see https://github.com/SemiQuant/svTyper.git
See Online for appendix 1
For the latest version of Amplicon Sequencing Analysis Pipeline see https://github.com/TGenNorth/ASAP
Data sharing
All data are available in the appendices. WGS reads are deposited in the European Nucleotide Archive (PRJEB47429) and TDS reads are in NCBI Bioproject (PRJNA767896).
References
- 1.Laurence YV, Griffiths UK, Vassall A. Costs to health services and the patient of treating tuberculosis: a systematic literature review. Pharmacoeconomics 2015; 33: 939–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conradie F, Meintjes G, Hughes J, et al. Clinical access to bedaquiline programme for the treatment of drug-resistant tuberculosis. Afr Med J 2014; 104: 164–65. [DOI] [PubMed] [Google Scholar]
- 3.WHO. WHO consolidated guidelines on drug-resistant tuberculosis treatment 2019. Geneva: World Health Organization, 2020. [PubMed] [Google Scholar]
- 4.Klopper M, Heupink TH, Hill-Cawthorne G, et al. A landscape of genomic alterations at the root of a near-untreatable tuberculosis epidemic. BCM Med 2020; 18: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Dorp L, Nimmo C, Ortiz AT, et al. Detection of a bedaquiline/clofazimine resistance reservoir in Mycobacterium tuberculosis predating the antibiotic era. bioRxiv 2023; published online Jan 13. 10.1101/2020.10.06.328799 (preprint). [DOI] [Google Scholar]
- 6.De Vos M, Ley SD, Wiggins KB, et al. Bedaquiline microheteroresistance after cessation of tuberculosis treatment. N Engl J Med 2019; 380: 2178–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghodousi A, Rizvi AH, Baloch AQ, et al. Acquisition of cross-resistance to bedaquiline and clofazimine following treatment for tuberculosis in Pakistan. Antimicrob Agents Chemother 2019;63: e00915–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pang Y, Zong Z, Huo F, et al. In vitro drug susceptibility of bedaquiline, delamanid, linezolid, clofazimine, moxifloxacin, and gatifloxacin against extensively drug-resistant tuberculosis in Beijing, China. Antimicrob Agents Chemother 2017; 61: e00900–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Omar SV, Ismail F, Ndjeka N, Kaniga K, Ismail NA. Bedaquiline-resistant tuberculosis associated with Rv0678 mutations. N Engl J Med 2022; 386: 93–94. [DOI] [PubMed] [Google Scholar]
- 10.Conradie F, Diacon AH, Ngubane N, et al. Treatment of highly drug-resistant pulmonary tuberculosis. N Engl J Med 2020; 382: 893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nimmo C, Millard J, Van Dorp L, et al. Population-level emergence of bedaquiline and clofazimine resistance-associated variants among patients with drug-resistant tuberculosis in southern Africa: a phenotypic and phylogenetic analysis. Lancet Microbe 2020; 1: e165–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadura S, King N, Nakhoul M, et al. Systematic review of mutations associated with resistance to the new and repurposed Mycobacterium tuberculosis drugs bedaquiline, clofazimine, linezolid, delamanid and pretomanid. J Antimicrob Chemother 2020; 75: 2031–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.WHO. Catalogue of mutations in Mycobacterium tuberculosis complex and their association with drug resistance. Geneva: World Health Organization, 2021. [Google Scholar]
- 14.Olayanju O, Limberis J, Esmail A, et al. Long-term bedaquiline-related treatment outcomes in patients with extensively drug resistant tuberculosis from South Africa. Eur Respir J 2018; 51: 1800544. [DOI] [PubMed] [Google Scholar]
- 15.Mudaly V, Voget J. Clinical guidelines & standard operating procedure for the implementation of the short & long DR-TB regimens for adults, adolescents and children. 2018. https://www.westerncape.gov.za/assets/departments/health/tuberculosis_-_dr-tb_clinical_guidelines_2018.pdf (accessed Dec 23, 2022).
- 16.WHO. Definitions and reporting framework for tuberculosis—2013 revision: updated December 2014 and January 2020. Geneva: World Health Organization, 2013. [Google Scholar]
- 17.Torrea G, Coeck N, Desmaretz C, et al. Bedaquiline susceptibility testing of Mycobacterium tuberculosis in an automated liquid culture system. J Antimicrob Chemother 2015; 70: 2300–05. [DOI] [PubMed] [Google Scholar]
- 18.Andries K, Verhasselt P, Guillemont J, et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 2005; 307: 223–27. [DOI] [PubMed] [Google Scholar]
- 19.Warren R, de Kock M, Engelke E, et al. Safe Mycobacterium tuberculosis DNA extraction method that does not compromise integrity. J Clin Microbiol 2006; 44: 254–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dippenaar A, De Vos M, Marx FM, et al. Whole genome sequencing provides additional insights into recurrent tuberculosis classified as endogenous reactivation by IS6110 DNA fingerprinting. Infect Genet Evol 2019; 75: 103948. [DOI] [PubMed] [Google Scholar]
- 21.Cox H, Hughes J, Daniels J, et al. Community-based treatment of drug-resistant tuberculosis in Khayelitsha, South Africa. Int J Tuberc Lung Dis 2014; 18: 441–48. [DOI] [PubMed] [Google Scholar]
- 22.Ismail N, Rivière E, Limberis J, et al. Genetic variants and their association with phenotypic resistance to bedaquiline in Mycobacterium tuberculosis: a systematic review and individual isolate data analysis. Lancet Microbe 2021; 2: e604–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chesov E, Chesov D, Maurer FP, et al. Emergence of bedaquiline resistance in a high tuberculosis burden country. Eur Respir J 2022; 59: 2100621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villellas C, Coeck N, Meehan CJ, et al. Unexpected high prevalence of resistance-associated Rv0678 variants in MDR-TB patients without documented prior use of clofazimine or bedaquiline. J Antimicrob Chemother 2016; 72: 684–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antoine R, Gaudin C, Hartkoorn RC. Intragenic distribution of IS6110 in clinical Mycobacterium tuberculosis strains: bioinformatic evidence for gene disruption leading to underdiagnosed antibiotic resistance. Microbiol Spectr 2021; 9: e0001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engelthaler DM, Streicher EM, Kelley EJ, et al. Minority Mycobacterium tuberculosis genotypic populations as an indicator of subsequent phenotypic resistance. Am J Respir Cell Mol Biol 2019; 61: 789–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen TVA, Anthony RM, Bañuls A-L, Nguyen TVA, Vu DH, Alffenaar J-WC. Bedaquiline resistance: its emergence, mechanism, and prevention. Clin Infect Dis 2018; 66: 1625–30. [DOI] [PubMed] [Google Scholar]
- 28.Mirzayev F, Viney K, Linh NN, et al. World Health Organization recommendations on the treatment of drug-resistant tuberculosis, 2020 update. Eur Respir J 2021; 57: 2003300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ismail NA, Omar SV, Joseph L, et al. Defining bedaquiline susceptibility, resistance, cross-resistance and associated genetic determinants: a retrospective cohort study. EBioMedicine 2018; 28: 136–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pai H, Ndjeka N, Mbuagbaw L, et al. Bedaquiline safety, efficacy, utilization and emergence of resistance following treatment of multidrug-resistant tuberculosis patients in South Africa: a retrospective cohort analysis. BMC Infect Dis 2022; 22: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the appendices. WGS reads are deposited in the European Nucleotide Archive (PRJEB47429) and TDS reads are in NCBI Bioproject (PRJNA767896).


