SUMMARY
Triple-negative breast cancer (TNBC) is a heterogeneous disease with limited treatment options. To characterize TNBC heterogeneity, we defined transcriptional, epigenetic, and metabolic subtypes and subtype-driving super-enhancers and transcription factors by combining functional and molecular profiling with computational analyses. Single-cell RNA sequencing revealed relative homogeneity of the major transcriptional subtypes (luminal, basal, and mesenchymal) within samples. We found that mesenchymal TNBCs share features with mesenchymal neuroblastoma and rhabdoid tumors and that the PRRX1 transcription factor is a key driver of these tumors. PRRX1 is sufficient for inducing mesenchymal features in basal but not in luminal TNBC cells via reprogramming super-enhancer landscapes, but it is not required for mesenchymal state maintenance or for cellular viability. Our comprehensive, large-scale, multiplatform, multiomics study of both experimental and clinical TNBC is an important resource for the scientific and clinical research communities and opens venues for future investigation.
Graphical Abstract
In brief
Jovanović et al. define the cellular, molecular, and functional heterogeneity of TNBC and integrated these using multiomics factor analysis into clinically relevant subtypes. Luminal, basal, and mesenchymal gene expression and super-enhancer subtypes do not match DNA methylation, histone modification, and metabolic patterns. PRRX1 is a central transcription factor in mesenchymal TNBC.
INTRODUCTION
Triple-negative breast cancer (TNBC) is characterized by the lack of estrogen receptors (ERs) and progesterone receptors (PRs) and HER2.1 TNBC patients have worse clinical outcome with a higher 5-year recurrence rate than that of other subtypes. TNBCs are highly heterogeneous and have been further classified based on gene expression profiles.2-4 However, the transcriptional and epigenetic drivers of these TNBC subtypes and their impact on cellular phenotypes have not been defined.
We have previously described the dominant inheritance of TNBC traits over ER+ luminal features via non-genetic mechanisms.5 We also noted common super-enhancers (SEs) in ER+ luminal breast cancer cell lines and limited overlap of SEs in TNBC lines. SEs are key for cellular identity and oncogenic transcription.6,7 Thus, characterization of SEs and associated transcription factors (TFs) in TNBC may help elucidate biologically and clinically relevant subtypes.
Here we describe comprehensive molecular, metabolomic, and functional characterization of a large panel of TNBC cell lines (n = 34) and patient-derived xenografts (PDXs; n = 15) with validation of these results in The Cancer Genome Atlas (TCGA)8 and Molecular Taxonomy of Breast Cancer International Consortium (METABRIC)9 patient cohorts. By integrating our datasets using Multi-Omics Factor Analysis (MOFA),10,11 we defined TNBC heterogeneity and identified the PRRX1 TF as an orchestrator of a TF network in mesenchymal TNBC.
RESULTS
TNBC transcriptional subtypes
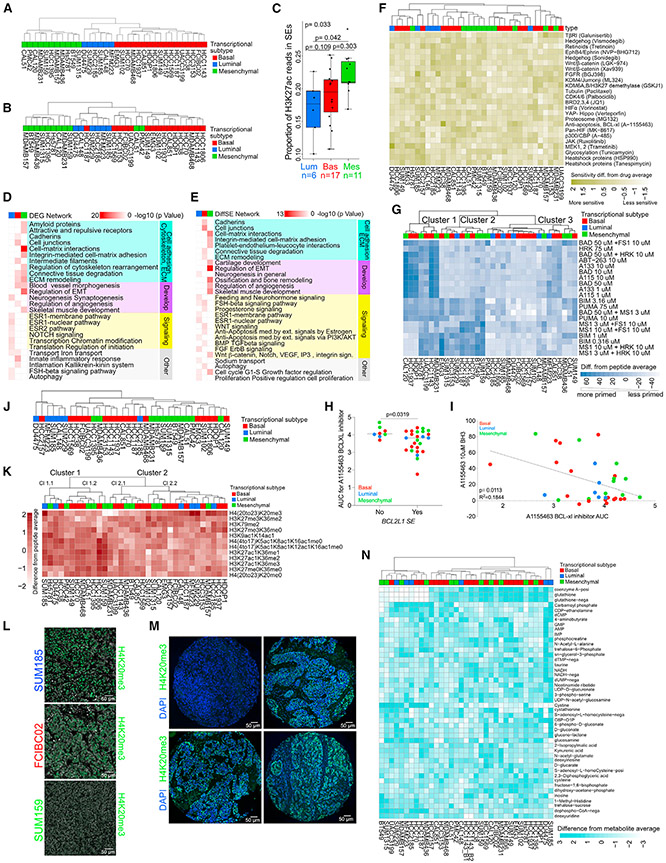
To assess TNBC transcriptional subtypes, we performed RNA sequencing (RNA-seq) and histone H3 lysine 27 acetyl (H3K27ac) chromatin immunoprecipitation sequencing (ChIP-seq) on TNBC cell lines. Based on the expression of the top 20% most variable genes, we identified three major clusters, defined as basal, luminal, and mesenchymal (Figures 1A and S1A; Table S1). Clustering of the H3K27ac ChIP-seq samples using the top 20% most variable SEs or peaks identified the same three major subtypes as RNA-seq with a few outliers (Figures 1B and S1B-S1D; Table S1). We also observed subtype-specific differences in overall H3K27ac signal in both SEs and peaks (Figures 1C and S1E). Mesenchymal TNBC had a higher proportion of H3K27ac reads in peaks compared with basal and luminal subtypes (Figure S1E) and more H3K27ac reads in SEs compared with the luminal subtype (Figure 1C). Differences in H3K27ac levels showed some associations with the number of expressed genes and variability in gene expression (Figures S1F and S1G).
Figure 1. Comprehensive molecular profiles of TNBC.
(A) Dendrogram depicting clustering of 34 TNBC cell lines based on the expression of the top 20% most variable genes. Subtype identifiers were assigned based on genes differentially expressed between the three major clusters. See also Table S1.
(B) Dendrogram depicting clustering of 33 TNBC cell lines based on H3K27ac signal in the top 20% most variable SEs.
(C) Boxplots showing the proportion of H3K27ac reads in SEs for cell lines in each TNBC subtype. Overall p value from Kruskal-Wallis test. Pairwise p values from Dunn’s test, adjusted using Holm’s method. Center lines shows medians. Hinges show interquartile ranges. Upper whiskers extend from the upper hinge to the highest value that is no further than 1.5 times the IQR from the hinge. Lower whiskerslate extends from the lower hinge to the lowest value that is nor further than 1.5 times the IQR from the hinge.
(D) Metacore networks enriched in differentially expressed genes (DEGs) among the three TNBC transcriptional subtypes. See also Table S2.
(E) Metacore networks enriched in TNBC transcriptional subtype-specific differential SEs. See also Table S2.
(F) Heatmap demonstrating TNBC cell line sensitivity to SMIs.
(G) Heatmap showing clustering of 34 TNBC cell lines based on the top 50% most variable BH3 peptides. Values shown are abundance differences from peptide average.
(H) Plot depicting sensitivity to the A1155463 BCL-xl inhibitor in TNBC lines where BCL2L1 is an SE or not. Error bars represent mean ± SEM; p value, Mann-Whitney U test.
(I) Plot depicting the correlation between BH3 profiling and drug area under the viability curve for treatment response (AUC) for the A1155463 BCL-xl inhibitor in TNBC cell lines (p = 0.0113, R2 = 0.1844, Pearson correlation).
(J) Dendrogram depicting clustering of 34 TNBC cell lines based on DNA methylation levels in the top 20% most variable SEs.
(K) Heatmap showing clustering of 34 TNBC cell lines based on the top 20% most variable histone marks determined by mass spectrometry. Average difference in mean -normalized H3K27ac, H3K27ac1K36me1, H3K27ac1K36me2, and H3K27ac1K36me3 values from cell line average = 0.032 (luminal), −0.17 (basal), and 0.25 (mesenchymal). Average difference in -normalized H4 (20–23) K20me3 value from cell line average = 1.48 (luminal), −0.063 (basal), −0.71 (mesenchymal).
(L) Immunofluorescence for H4K20me3 in SUM185 (luminal), FCIBC02 (basal), and SUM159 (mesenchymal) cell lines. Scale bars, 50 μm.
(M) Representative histone H4K20me3 immunofluorescence staining of four TNBC patient samples from the tissue microarray (TMA). Scale bars, 50 μm.
(N) Heatmap showing clustering of 34 TNBC cell lines based on the levels of the top 20% most variable metabolites. Values shown are expression differences from metabolite average. Values are capped at ±3 for the purpose of visualization.
See also Figure S1 and Tables S1-S17. Blue, red, and green colors mark luminal, basal, and mesenchymal TNBC transcriptional subtypes in all figures.
To assess the functional relevance of subtype-specific expression and SE profiles, we performed Metacore network analyses.12 Pathways related to cell matrix interactions and development showed enrichment in mesenchymal subtype-specific transcripts and SEs, while hormonal signaling- and luminal differentiation-related pathways characterized luminal-specific transcripts and SEs (Figures 1D and 1E; Table S2). Genes with basal-specific expression were enriched in cell adhesion- and inflammation-related pathways, while basal-specific SEs were enriched in cell adhesion- and stem cell-related signaling pathways.
SE-driven genes commonly drive tumorigenesis and are therapeutic targets.13 Thus, we performed a cellular viability screen in 34 TNBC lines with 24 small-molecule inhibitors (SMIs) targeting pathways corresponding to SE-associated genes or genes with known function in TNBC. Clustering of the lines based on their treatment-related decrease in cellular viability demonstrated no clear transcriptional subtype-specific pattern (Figure 1F; Table S3). Among the most differentially effective inhibitors were ones targeting MEK, BCL-xl, and FGFR. The DU4475 cell line with mutant BRAF was the most sensitive to the MEK inhibitor trametinib, while SUM185 and MFM223 luminal AR+ cell lines with FGFR amplification showed the highest sensitivity to the BGJ398 FGFR inhibitor.
To determine whether differences in drug sensitivity were driven by differences in apoptosis susceptibility, we performed BH3 profiling to determine apoptotic priming and anti-apoptotic dependencies for survival. We identified three BH3 profile clusters independent of transcriptional subtypes (Figure 1G; Table S3). Cells in cluster 1 depend on the anti-apoptotic BCL-2 family members BCL-2, BCL-xl, or BCL-w for survival, based on their response to BAD. Because of their response to the BCL-xL-specific HRK peptide and BCL-xL inhibitors A-115463 and A-1331852, this survival dependency is based mostly on BCL-xL. Cells in cluster 2 are characterized by a higher overall priming (response to BIM and PUMA) and dual dependency on the anti-apoptotic MCL-1 and BCL-xL proteins (response to MS1 and HRK peptides), while cluster 3 is overall less primed and less dependent on specific anti-apoptotic BCL2 family members for survival than the other clusters (Figure 1G). BCL2L1 (encoding BCL-xl) is one of the few recurrent SEs in TNBCs (Table S1). Sensitivity to the BCL-xl inhibitor A-115463 was significantly higher in cell lines where BCL2L1 was an SE (Figure 1H), whereas cellular sensitivity to A-1155463 only weakly correlated to the direct mitochondrion effect of the drug assessed by BH3 profiling (Figure 1I).
These data demonstrate that SEs can reveal therapeutic targets in TNBC, although the high variability among samples makes the identification of common vulnerabilities challenging.
TNBC epigenetic and metabolomic subtypes
To characterize TNBC epigenetic heterogeneity, we first assessed genome-wide DNA methylation patterns. Clustering of the samples based on variable DNA methylation in SEs, promoters, and gene bodies revealed distinct subsets that did not match the transcriptional subtypes (Figures 1J and S1H-S1J; Table S1). Integrating transcriptomic, H3K27ac, and DNA methylation data, we found that gene expression was significantly inversely correlated with DNA methylation, and it was positively correlated with H3K27ac in SEs (Figure S1K).
Next, we performed quantitative histone mass spectrometry, which revealed two main clusters driven by histone modifications associated with active (e.g., H3K27ac) and repressive (e.g., histone H3 lysine 27 trimethyl - H3K27me3) chromatin (Figure 1K; Table S3). Histone H4 lysine 20 trimethyl (H4K20me3) was the most variable histone mark and more abundant in luminal lines (Figure 1K). Genes differentially expressed between cell lines with high and low H4K20me3 levels had significant enrichment in cell adhesion, development, and inflammation-related networks (Figure S1L; Table S2). We investigated H4K20me3 in more detail by immunofluorescence in TNBC lines and primary tumors. We found subtype-specific differences with a stronger signal in luminal SUM185 than basal FCIBC02 and mesenchymal SUM159 cells (Figure 1L). Analysis of 81 primary TNBCs showed high variability for H4K20me3 signal both among and within tumors but no significant association with recurrence-free survival or luminal and basal markers (Figures 1M, S1M, and S1N).
Metabolomic profiling of TNBC lines using mass spectrometry for 228 metabolites demonstrated transcriptional subtype-independent clustering mainly driven by mutually exclusive levels of reduced glutathione (GSH) and cystine; cells with high GSH also had high coenzyme A (Figure 1N; Table S3). GSH is a major anti-oxidant, while cystine is the oxidized form of cysteine; thus, cell lines with low GSH and high cystine levels may have higher reactive oxygen species (ROS) levels compared with GSHhigh cystinelow samples.
Our multiomics profiling revealed that TNBC transcriptional subtypes correlate with SE landscape but not with other epigenetic and metabolic profiles.
Integrated evaluation of TNBC using latent factor analysis
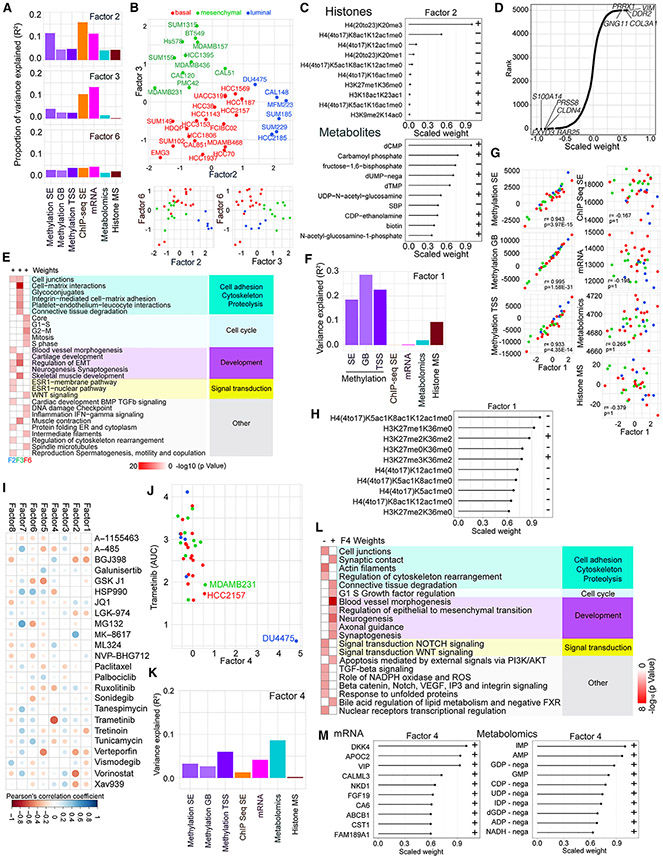
To better understand drivers of TNBC heterogeneity, we performed integrated analyses of all molecular data using MOFA.10,11 DNA methylation data were split into three datasets: promoter, gene body, and non-genic regions of SEs. The MOFA model assumes that variations from the average profile in each dataset depend linearly on the values of a few latent factors. We used MOFA to identify the latent factors active in each cell line and their molecular effects. When fitting the model to the normalized datasets, we identified eight factors based on the maximum number of factors expected to be reliably recoverable (STAR Methods; Figures S2A and S2B; Table S4). The total variance explained in each dataset, considering all MOFA factors, ranged from 19.3% (for histone mass spectrometry) to 53% (for promoter methylation). Factors 2, 3, and 6 were significantly correlated with transcriptional subtypes (Figures 2A and 2B), with the strongest correspondence observed for Factor 2 (F2) and F3. Cell lines with high F2 and F3 contribution were luminal and mesenchymal, respectively (Figure 2B). F2 explained a proportion of variance (>2%) in all datasets (Figures 2A and S2B), suggesting that biological differences between luminal and other transcriptional subtypes manifest broadly across phenotypes (Figure 2C). The top-weighted histone mark for F2 was H4K20me3 (Figure 2C), while RNA-seq for F3 showed the highest weights for genes highly expressed in mesenchymal cells (e.g., PRRX1), suggesting that these genes might regulate F3 (Figure 2D). Metacore network analysis14 of top-weighted features showed that F3 had significant enrichment in cell matrix- and extracellular matrix-related networks, while F2 had significant enrichment for a range of pathways, including ER signaling (Figure 2E). Analysis of correlations between contributions of F2 and F3 and the total signaling level (summed across all features) of each of the datasets found that F2 was negatively correlated with total signal for H3K27ac in SEs, while no correlation was observed for F3 related to the mesenchymal state (Figures S2C and S2D; Table S4).
Figure 2. Integrated analysis of the genomics data using multiomics factor analysis (MOFA).
(A) Bar graph of the proportion of variance explained in each dataset by F2, F3, and F6.
(B) Scatterplots depicting F2, F3, and F6 values across TNBC cell lines.
(C) Scaled F2 weights for the histone mark combinations with the largest absolute weights for this factor and scaled F2 weights for the metabolites with the largest absolute weights for this factor. Scaled weights for each factor in each dataset are derived from the weights for that factor in that dataset by linearly rescaling the values to lie between −1 and 1.
(D) Scaled mRNA weights for F3, with the top five negatively and positively weighted features labeled. Scaled weights for each factor in each dataset are derived from the weights for that factor in that dataset by linearly rescaling the values to lie between −1 and 1.
(E) Metacore networks for F2, F3, and F6 positive mRNA weights. See also Table S2.
(F) Bar graph showing the variance explained by F1 within each dataset.
(G) Scatterplots of total signal in each dataset against F1 scores; p values, Holm-adjusted Pearson correlation test.
(H) Scaled F1 weights for the histone mark combinations with the largest absolute weights for this factor. Scaled weights for each factor in each dataset are derived from the weights for that factor in that dataset by linearly rescaling the values to lie between −1 and 1.
(I) Correlations between MOFA F1–F8 and SMI features. Dot colors and sizes represent Pearson’s correlation coefficient values for the indicated pairs of drugs and factors.
(J) Scatterplot showing F4 scores and trametinib AUC across TNBC cell lines.
(K) Bar graph showing variance explained for F4 across each dataset.
(L) Metacore networks for F4 positive and negative mRNA weights. See also Table S2.
(M) Scaled F4 weights for the mRNA and metabolomics features with the largest absolute weights. Scaled weights for each factor in each dataset are derived from the weights for that factor in that dataset by linearly rescaling the values to lie between −1 and 1.
F6 was also significantly associated with transcriptional subtype and separated most of the basal cell lines from the other two types (Figure 2B). F6 explained a proportion of variance in most datasets (Figure 2A). Pathway analysis for the critical F6 genes in the RNA-seq dataset revealed an enrichment for cell-cycle-related pathways (Figure 2E). Thus, this factor might be linked to proliferation.
MOFA reveals transcription subtype-independent variation in TNBC
F1, F4, F5, F7, and F8 were not significantly correlated with transcription subtype. F1 explained substantial proportions of variance in DNA methylation (range: 18.5%-28.7%) and histone mass spectrometry data (9.4%; Figure 2F). The contribution of F1 to cell lines was strongly correlated with total signal in each of the methylation datasets (Figure 2G; Table S4). In the histone mass spectrometry data, the top positively weighted features for F1 were histone marks characteristic of repressive chromatin (H3K36me2, H3K27me2, and H3K27me3), which are associated with higher levels of DNA methylation (Figure 2H). These findings suggest that variance in DNA methylation may be an important transcription subtype-independent driver of TNBC heterogeneity.
Next, we correlated our MOFA results with SMI screen data. After correcting for multiple testing, we detected a significant association between F4 and trametinib (a MEK inhibitor), driven by the DU4475 BRAF mutant cell line (Figures 2I and 2J). Variance decomposition analysis suggested that F4 was most influential in shaping the DNA methylation, transcriptional, and metabolic landscapes (Figure 2K). Interrogation of highly weighted F4 features revealed high positive weights for metabolites linked to nucleotide synthesis and identified an enrichment for Wnt signaling-related genes among the top negatively weighted mRNA features (Figures 2L and 2M). Thus, activation of F4 in DU4475 cells might be related to the APC mutation in this cell line.15
F5 explained substantial proportions of variance in the DNA methylation and low proportions of variance in the other datasets (Figures S2E-S2G). After assessing the pathways for top-weighted genes using feature set enrichment analysis and Metacore, we found multiple pathways significantly related to sensory perception and olfactory stimulation (Figures S2H and S2I). F7 and F8 explained appreciable proportions of variance in the epigenetic and low proportions of variance in the other datasets (Figures S2J-S2O). Pathway analysis of these factors showed enrichment for RNA binding, catabolic processes, and maintenance of protein location (Figures S2I, S2L, S2O, S2P, and S2Q). F7 values were positively correlated with the total signal in H3K27ac data (Figure S2R; Table S4).
These data show that MOFA can identify features of TNBC lines not obvious in individual data types and highlight that the imprint of cellular phenotypes is discernible at multiple levels.
Clinical relevance of MOFA factors in TNBC
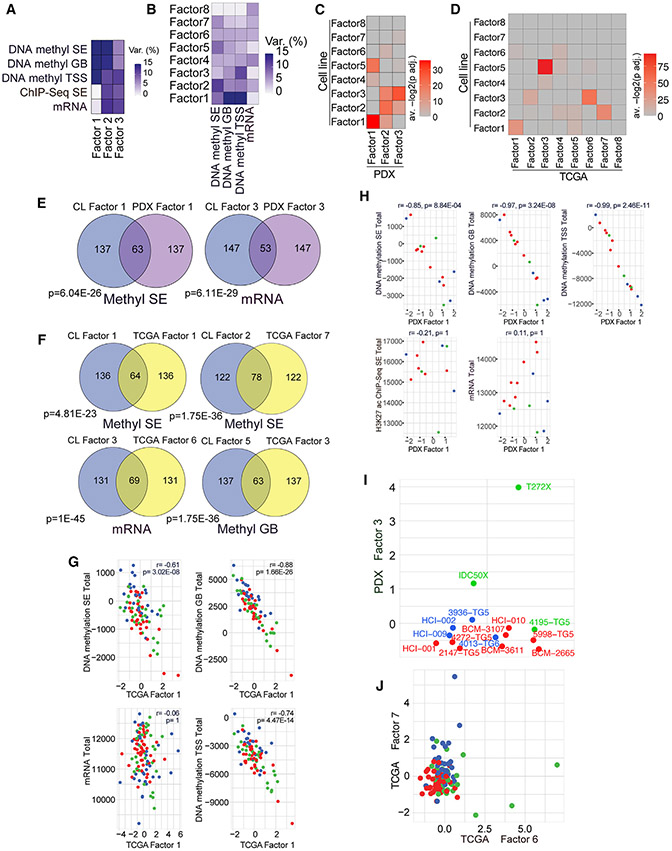
To assess the clinical relevance of these MOFA factors, we fit analogous MOFA models to data from our 15 TNBC PDXs and data for 115 TNBC TCGA samples. We used 8 factors for the TCGA cohort to match the number of factors used in the original model and 3 factors for the PDX cohort due to smaller sample size (Figures 3A-3F). We compared the top 200 most highly weighted features for each cell line MOFA factor in each dataset where the factor in question explained more than 2% of variance with the top 200 features for each of the new factors in the same dataset to assess the similarity between the original factors and the newly inferred factors. Using this approach, we found that highly weighted features from 6 (F1–F6) of 8 of the original factors significantly overlapped with highly weighted features for at least one factor in the PDX model. We also found that the same six factors had a significant overlap with at least one factor in the TCGA model (Figures 3C and 3D; Table S4). In the PDX model, the most significant overlaps for F1, F4, and F5 involved PDX F1, for F2 and F6 involved PDX F2, and for F3 involved PDX F3 (Figures 3C and 3E; Table S4). In the TCGA cohort, the most significant overlaps for F1–F3 involved TCGA F1, F7, and F6; for F4 and F5 involved TCGA F3; and for F6 involved TCGA F4 (Figures 3D and 3F).
Figure 3. Validation of MOFA factors in PDXs and clinical samples.
(A) Variance explained by each PDX MOFA factor in the PDX data. Methyl SE: n = 15,p = 2,835; methyl GB: n = 15,p = 4,992; methyl TSS: n = 15,p = 4,996; ChIP-seq SE: n = 12,p = 5,120; mRNA: n = 15, p = 5,000.
(B) Variance explained in each dataset by each TCGA TNBC MOFA factor. Methyl SE: n = 83, p = 2,462; methyl GB: n = 83, p = 4,463; methyl TSS: n = 83, p = 4,704; mRNA: n = 115,p = 4,736..
(C and D) Heatmaps showing overlaps between top features by absolute weight for each cell line MOFA factor and top features by absolute weight for each of the MOFA factors derived from PDX (C) and TCGA TNBC (D) samples. Cell colors represent the average of the negative -transformed adjusted hypergeometric test p values for tests corresponding to the pair of factors indicated by the row and column; p value adjustment by Holm’s method. Tests for overlap were performed for all datasets where the cell line factor explained at least 2% of variance in the original model. Rows indicate cell line factors, and columns indicate validation model factors.
(E) Venn diagrams of overlaps between the top 200 features by absolute weight for the indicated cell line and PDX factors in the indicated dataset. Holm-adjusted hypergeometric test p values are shown.
(F) Venn diagrams of overlaps between the top 200 features by absolute weight for the indicated cell line and TCGA factors in the indicated dataset. Holm-adjusted hypergeometric test p values are shown.
(G) Scatterplots of total signal in each dataset plotted against TCGA F1 scores. Holm-adjusted Pearson correlation test p values are shown.
(H) Scatterplots of total signal in each dataset plotted against PDX F1 scores. Holm-adjusted Pearson correlation test p values are shown.
(I) Bee swarm plot showing PDX F3 scores across PDX samples. Samples are colored by their assigned TNBC subtype based on the cell-line-defined RNA-seq signatures. Points are jittered along the horizontal axis for the purpose of visualization.
(J) Plot showing TCGA TNBC F6 and F7 scores for all TCGA TNBC samples, colored according to assigned TNBC subtype based on cell-line-derived signatures. See also Figure S2 and Table S5.
We also analyzed the correspondence between TCGA and PDX factor contributions and total signal from each dataset and TNBC subtype signatures derived from bulk RNA-seq. As observed for F1, both PDX F1 and TCGA F1 correlated strongly with total signal in each of the methylation datasets in the PDX and TCGA cohorts, respectively (Figures 3G and 3H; Table S4); similar correlations were also observed for TCGA F3 and F5 (Table S4). Additionally, PDX F3 and TCGA F6 and F7 were significantly correlated with transcriptional subtypes; however, this was also the case for TCGA F4, F5, and F8. As for F3, mesenchymal samples had high values for PDX F3 and TCGA F6 in these two datasets (Figures 3I and 3J). Like F2, luminal samples had high values for TCGA F7 (Figure 3J).
Most factors from the original model had significant overlaps with more than one factor in each validation model. For both the PDX and TCGA models, the most significant of these alternate mappings involved F3. F3 showed significant overlaps with PDX F3 and PDX F2 in the PDX model and with TCGA F2 and TCGA F6 in the TCGA model (Figures 3C and 3D; Table S4). We hypothesize that this may be due to similar but distinct biological factors active in non-tumor cells detected in the patient samples.
Taken together, these results support the conclusion that F1–F6 capture clinically relevant features that influence variability in primary TNBC.
Intra-tumor cellular heterogeneity of TNBC
To evaluate whether TNBC transcriptional subtypes are reflected in the proteome and are homogeneous within cell lines, we performed cytometry by time of flight (CyTOF)16 on 34 TNBC lines for a panel of 31 protein markers associated with luminal and mesenchymal/basal features, signaling pathways, and proliferation. CyTOF data were depicted as trees built using the X-shift method17 and a uniformly sized subset of cells from each sample. This approach allowed us to visualize the position of individual cell lines on the tree (Figure S3A). We found that each of the TNBC subtypes occupied a different general area of the tree, with limited overlap in the regions occupied by cell lines belonging to different subtypes. An exception to this was the HDQP1 basal cell line, where some cells occupied the mesenchymal region of the tree (Figure S3A), while others were more closely related to basal cell lines.
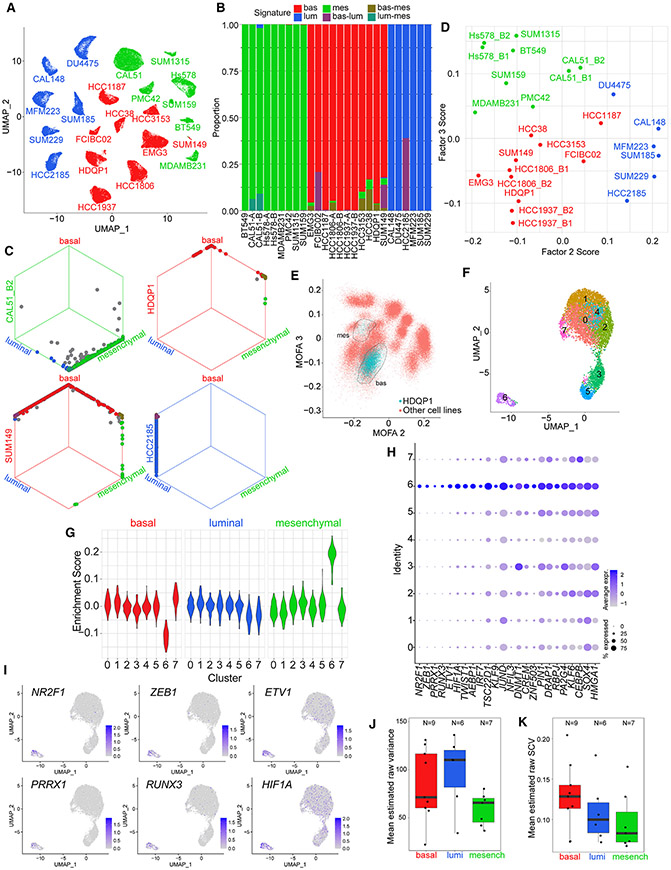
We further evaluated a subset of TNBC cell lines by single-cell RNA-seq (scRNA-seq) to better understand the extent of cellular heterogeneity. The UMAP (Uniform Manifold Approximation and Projection) plot of all cells showed cell-line-specific clustering and limited heterogeneity within individual cell lines (Figure 4A). Statistical testing of enrichment of bulk RNA-seq-derived TNBC luminal, basal, and mesenchymal signatures in single cells further suggested limited TNBC subtype heterogeneity in mesenchymal and luminal lines, with most heterogeneity present in basal lines (Figures 4B and 4C). We then calculated MOFA factor scores for the single cells of each sample. Average F2, F3, and F6 scores across single cells within each sample mirrored the factor scores observed for the same cell lines from bulk RNA-seq data (Figures 2B, 4D, S3B, and S3C). Samples with high average F2 and F3 values were luminal and mesenchymal, respectively. The HDQP1 line had two subclusters, with the majority of cells occupying the basal region of the plot and a subset closer to the mesenchymal section (Figure 4E, S3D, and S3E). The UMAP plot of HDQP1 alone showed eight cell clusters, with cluster 6 being distant from the rest of the cells and highly enriched in the mesenchymal signature (Figures 4F and 4G). To explore potential regulators of this mesenchymal subclone, we performed differential gene expression analysis for cluster 6 versus the other clusters, focusing on TFs, and identified NR2F1, PRRX1, RUNX3, ETV1, HIF1A, ZEB1, and TWIST1 as the top differentially expressed TFs (Figures 4H and 4I).
Figure 4. Intra-tumor heterogeneity assessment by single-cell analyses.
(A) UMAP visualization of scRNA-seq gene expression data from TNBC cell lines. Single cells from each cell line are colored according to assigned subtype from bulk RNA-seq.
(B) Bar plot showing significantly enriched TNBC transcriptional subtype signatures (bootstrap p <0.05) in single cells from samples belonging to each TNBC subtype.
(C) Hexagonal plots showing significantly enriched TNBC transcriptional subtype signatures for all analyzed single cells from four cell line samples. Each point represents a single cell. Cells are positioned along each axis according to bootstrap classification score (1 minus bootstrap p value) for the indicated cell identity. Cells significantly enriched for each signature are shown along the corresponding edges of the plot. Cell colors represent significantly enriched signatures; cells with no significant enrichments are shown in gray.
(D) Average MOFA F2 and F3 scores of single cells from each sample.
(E) Inferred MOFA F2 and MOFA F3 scores for HDQP1 single cells (blue) and all other single cells (red). Circled regions show two apparent HDQP1 subclusters.
(F) UMAP visualization of HDQP1 cell line scRNA-seq data.
(G) Enrichment scores of TNBC subtype signatures in single cells of HDQP1 by cluster. Scores measure the difference in TNBC subtype signature expression compared with average expression across HDQP1 cells after correcting for the differences observed for random size-match signatures.
(H) Cluster-specific expression of TFs differentially expressed in HDQP1 cluster 6.
(I) UMAP visualization of HDQP1 single cells, colored by expression of the six mostly strongly overexpressed TFs in cluster 6.
(J) Boxplot showing mean estimated raw variance across highly expressed genes in single-cell samples assigned to each subtype. For cell lines with two replicate samples, only the higher-depth replicate is shown. Bottom and top hinges of inset box plots show the 25th and 75th percentiles. Upper whiskers extend from the upper hinge to the highest value that is no further than 1.5 times the interquartile range (IQR) from the hinge. Lower whiskers extend from the lower hinge to the lowest value no further than 1.5 times the IQR from the hinge.
(K) Boxplot showing mean estimated raw SCV across highly expressed genes in single-cell samples assigned to each subtype. For cell lines with two replicate samples, only the higher-depth replicate is shown. Bottom and top hinges of inset box plots show the 25th and 75th percentiles. Upper whiskers extend from the upper hinge to the highest value that is no further than 1.5 times the interquartile range (IQR) from the hinge. Lower whiskers extend from the lower hinge to the lowest value no further than 1.5 times the IQR from the hinge.
See also Figure S3.
We then investigated whether transcriptomic heterogeneity varied by transcriptional subtype. Considering only highly expressed genes, we used the single-cell read count distribution for each gene in each sample to estimate each gene’s mean expression, biological variance, and raw squared coefficient of variation (SCV) in the sample. Using a simulation study, we confirmed that the estimates provided by our approach were expected to have low average bias across genes in each sample (Figures S3F-S3H). For these highly expressed genes, average estimated raw variance and average estimated raw SCV did not differ significantly across transcriptional subtypes, although we observed a trend toward higher raw SCV in the basal cell lines (Figures 4J and 4K). We obtained similar results when investigating the effect of TNBC type and average expression on raw variance using linear mixed-effects models (Figure S3I).
TNBC cell lines display remarkable cellular homogeneity of TNBC subtypes, implying robust regulatory mechanisms, although minor subpopulations with more unstable cell states cannot be excluded.
Clinical relevance of TNBC subtype heterogeneity
Next, to evaluate the clinical relevance of the TNBC subtypes we identified, we performed unbiased hierarchical clustering of TNBC tumors from the METABRIC9 and TCGA8 cohorts, annotated using our TNBC transcriptional subtype signatures. We found that luminal TNBC was more distinct from the other two subtypes in both datasets, while there was mixing between basal and mesenchymal subtypes (Figures S4A-S4C). In the METABRIC cohort, there were significant differences in tumor cellularity between TNBC subtypes, with mesenchymal tumors having significantly lower cellularity. In the TCGA cohort, total absolute CIBERSORTx18 scores across 22 immune cell types differed significantly between subtypes (Figure S4D), with significantly higher absolute scores in mesenchymal-classified compared with luminal-classified tumors, suggestive of higher proportions of immune cells. Thus, the apparent mixing of tumors may partly be due to stromal cells expressing mesenchymal TNBC subtype-specific genes.
To test whether the TNBC transcriptional subtypes or MOFA factors are prognostic, we calculated MOFA factor scores for samples from the TCGA and METABRIC cohorts and tested for associations between the calculated factor scores, inferred TNBC subtypes, and survival (Table S5). We first examined transcriptional subtype-independent factors (F1, F4, F5, F7, and F8). We found that high F7 scores were associated with shorter disease-specific survival (DSS) and progression-free survival (PFS) in the TCGA cohort (Figure S4E) but that neither associations remained significant when controlling for age and pathological stage (Table S5). Inferred F7 values were not correlated with survival in the METABRIC cohort (Figure S4F; Table S5). A possible explanation for these discordant results is that the F7 scores inferred using expression data only for the METABRIC cohort may be more variable than those inferred for the TCGA cohort, which make use of DNA methylation data alongside expression data.
Next, we examined associations between survival and MOFA factor scores for the three TNBC subtype-linked factors (F2, F3, and F6) as well as associations between survival and signature-based inferred TNBC subtype. High F6 scores were significantly correlated with longer PFS in the TCGA cohort (Figure S4E). We also observed significant differences in DSS by TNBC subtype in the TCGA cohort, with significantly shorter DSS in the luminal and mesenchymal subtypes (Figure S4E; Table S5). However, neither TNBC subtype nor inferred F6 scores were significantly associated with survival in the TCGA cohort after controlling for age and pathological stage (Table S5). Neither variable was associated with survival in the METABRIC cohort (Figure S4F; Table S5).
To further investigate these results, we examined the distribution of MOFA F2, F3, and F6 scores in both primary data cohorts. We observed that, in both cohorts, there was a small subset of mesenchymal-assigned samples with high values of F3 (mesenchymal-high samples) (Figure S4G and S4H). Notwithstanding that the mesenchymal-high sample size was small in the TCGA cohort, we found that the mesenchymal-high group was significantly associated with shorter DSS in both cohorts (Figures S4I and S4J; Table S5). This association remained significant in the METABRIC cohort when controlling for age, tumor size, and number of positive lymph nodes (Table S5). In the METABRIC cohort, we also observed a significant association between mesenchymal-high and DSS when we used an alternate clustering-derived threshold on F3 scores to define mesenchymal-high samples (Figures S4K and S4L). However, we found no significant differences in DSS between the resulting three groups when we redefined the basal, luminal, and mesenchymal sample groups by clustering on F2, F3, and F6 scores (Figures S4M and S4N). One possible explanation for these results is that the stringent criteria used to define the mesenchymal-high groups remove false-positive mesenchymal samples from our signature-based calls and that this more stringent set of calls enables us to detect an underlying association between the mesenchymal subtype and poor outcome in primary TNBC in both cohorts that would otherwise be undetectable in the METABRIC cohort. Nevertheless, a larger sample set for the mesenchymal-high subtype is needed to follow up on these results.
TNBC subtype-specific TFs
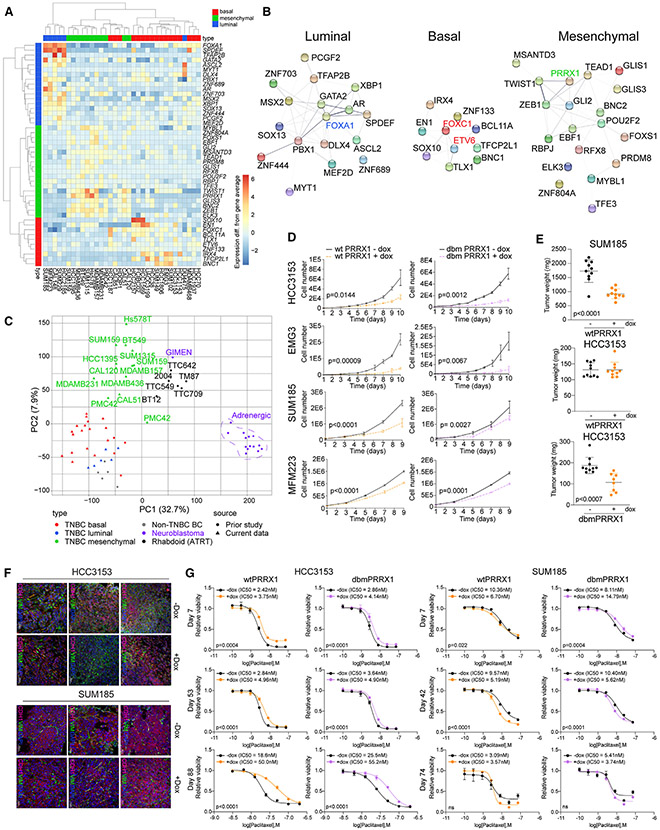
To identify TFs regulating TNBC subtypes, we integrated our RNA-seq and H3K27ac ChIP-seq data and identified 46 TFs expressed in a subtype-specific manner and associated with subtype-specific SEs (Figure 5A). Luminal TFs included many genes with well-established roles in luminal differentiation (e.g., FOXA1) and several less characterized ones (e.g., ASCL2). Mesenchymal TFs consist of some well-known EMT-inducers (e.g., ZEB1) and the PRDM8 transcriptional repressor.19 Several of the basal TFs have known roles in mammary stem cells (e.g., BCL11A)20 or are known oncogenes (e.g., ETV6).21
Figure 5. PRRX1 is a mesenchymal subtype-specific TF.
(A) Heatmap of mRNA expression of TNBC subtype-specific TFs. Differences in -normalized expression from the gene average are shown for each gene.
(B) STRING-based protein-protein interaction network for TNBC subtype-specific TFs. Selected factors discussed in the text are highlighted for emphasis.
(C) Scatterplot of cell line RNA-seq data by principal components 1 and 2. The percentages of variance explained by principal components 1 and 2 are shown in brackets.
(D) Viable cell numbers after expression of dox-inducible WT or dbm PRRX1 in the indicated cell lines. Error bars represent mean ± SEM, n = 3 replicates, p values by two-tailed unpaired t test.
(E) Plot depicting weights of xenografts derived from SUM185 and HCC3153 cell lines expressing WT or dbm PRRX1 from mice with and without dox in the diet. Error bars represent mean ± SEM, n = 10 tumors, p values by two-tailed unpaired t test.
(F) E-cadherin and vimentin immunofluorescence staining of xenografts derived from SUM185 and HCC3153 cell lines expressing WT PRRX1 from mice with and without dox in the diet. Scale bars, 50 μm and 100 μm. Multiple representative images are shown from different xenografts to illustrate intra-tumor heterogeneity.
(G) Plots depicting viable cell numbers of HCC3153 and SUM185 cells following paclitaxel treatment and induction of WT or dbm PRRX1 expression by dox for the indicated days. Error bars represent mean ± SEM, p values by nonlinear fit test, n = 3 replicates.
See also Figure S5.
To assess TF networks, we performed STRING protein interaction network analysis22 for these 46 TFs. We found highly connected TF interaction networks in both luminal and mesenchymal TNBC, while basal TFs were less interconnected except for FOXC1 (Figure 5B). The TF with the most connections in the luminal subtype was FOXA1, a known luminal pioneering factor,23 while in the mesenchymal subtype, multiple TFs (e.g., TWIST1, ZEB1, and PRRX1) were well connected, implying a cross-regulatory network. In neuroblastomas, PRRX1 acts as a switch driving adrenergic cells toward a mesenchymal state by reprogramming the SE and mRNA landscapes.24,25 To test the relatedness of PRRX1-driven mesenchymal tumors from different organs, we compared the gene expression profiles of TNBC, non-TNBC breast cancer, neuroblastomas, and atypical teratoid rhabdoid tumors (ATRTs). ATRTs are poorly differentiated pediatric tumors that are also divided into mesenchymal and neurogenic epigenetic subtypes.26 Mesenchymal TNBC lines clustered more closely with ATRT and mesenchymal neuroblastoma (e.g., GIMEN) cell lines expressing PRRX1 than with luminal and basal TNBC (Figure 5C). These data highlight the loss of organ-specific features in poorly differentiated tumors and identify PRRX1 as a candidate driver of the mesenchymal tumor subtype regardless of tissue of origin.
The functional relevance of PRRX1 in TNBC
To determine whether PRRX1 is a dependency in mesenchymal TNBC, we expressed three independent tetracycline (TET)-inducible short hairpin RNAs (shRNAs) targeting PRRX1 and a non-targeting control in Hs578T mesenchymal TNBC and TTC642 rhabdoid cell lines (Figures S5A and S5B). Downregulation of PRRX1 had no significant impact on cellular viability, cell migration, invasion, or adhesion in either cell line (Figures S5C and S5D). Downregulation of PRRX1 also did not affect Hs578T xenograft growth and histology (Figures S5E and S5F). Thus, PRRX1 is not required for the growth of mesenchymal cancer cells, although incomplete knockdown by shRNA cannot be excluded.
To investigate whether PRRX1 is sufficient to induce the mesenchymal subtype, we expressed wild-type (WT) PRRX1 and a DNA binding mutant (dbm) with reduced DNA binding affinity25 in basal (EMG3 and HCC3153) and luminal (SUM185 and MFM223) TNBC lines in a TET-inducible manner (Figure S5G). Exogenous expression of both WT and dbm PRRX1 significantly reduced cellular proliferation in all four cell lines (Figure 5D). We also observed a significant decrease in xenograft tumor growth upon WT PRRX1 expression in SUM185 luminal tumors and upon dbm PRRX1 expression in HCC3153 basal tumor (Figures 5E and S5H). Histologic analysis of the xenografts demonstrated more mesenchymal features after PRRX1 overexpression (Figure S5I). To further investigate this observation, we analyzed the expression of luminal and mesenchymal markers in our RNA-seq data. We found high variability among cell lines, with some showing a decrease in some luminal markers (e.g., GATA3 in SUM185 cells and KRT18 in EMG3), while others had an increase in mesenchymal genes (e.g., VIM in MFM223) (Figure S5J). Multicolor immunofluorescence for E-cadherin luminal and vimentin mesenchymal markers also demonstrated highly heterogeneous patterns regardless of exogenous PRRX1 expression (Figure 5F). Overexpression of PRRX1 (WT and dbm) significantly decreased cell migration in EMG3 and HCC3153 cells, while invasion had no significant differences (Figure S5K). The SUM185 cell line was neither migratory nor invasive.
To determine whether PRRX1 expression alters response to paclitaxel, we assessed IC50 (half-maximal inhibitory concentration) at different time points after induction of PRRX1 (WT and dbm) in cells. We found that, in the basal HCC3153 cell line, prolonged expression of both WT and dbm PRRX1 increased resistance to paclitaxel, while the opposite was observed in luminal SUM185 cells (Figure 5G), and no change was detected in EMG3 or MFM223 cells (Figure S5L). These data suggest that PRRX1 expression in TNBC can either positively or negatively affect response to paclitaxel, depending on cellular context.
Overall, these data show that PRRX1 is sufficient to induce certain mesenchymal features but that it is not essential for the maintenance of mesenchymal tumor growth and phenotypes.
Transcriptional and genomic targets of PRRX1
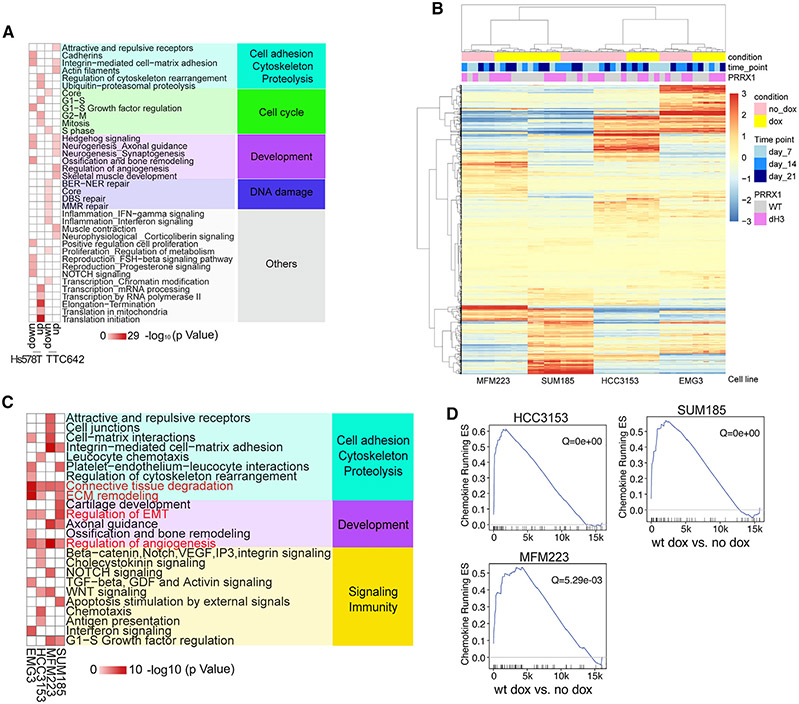
To investigate mechanisms by which PRRX1 exerts its function, we performed RNA-seq at different time points (days 5, 28, and 56) following its downregulation by shRNA in Hs578T and TTC642 cell lines and 7, 14, and 21 days following exogenous overexpression of WT or dbm PRRX1 in basal (HCC3153 and EMG3) and luminal (SUM185 and MM223) TNBC lines (Table S6). Metacore analysis of differentially expressed genes following PRRX1 downregulation revealed limited overlap between TNBC and rhabdoid cell lines, with Hs578T cells showing enrichment for transcription and translation-related networks, while in TTC642 cells, there was upregulation of neurogenesis-related processes and downregulation of immune-related pathways, including interferon signaling (Figure 6A; Table S2). Over-expression of WT PRRX1 induced common transcriptional changes characterized by enrichment for EMT, transforming growth factor (TGF-), WNT, and NOTCH signaling and immune-related functions (Figures 6B and 6C; Table S2). GSEA using immune gene signatures following WT PRRX1 overexpression also showed several significant enrichments, with the most pronounced being the chemokine gene set observed in 3 of 4 cell lines (Figure 6D; Table S5). The transcriptional changes induced by downregulation or overexpression of PRRX1 imply that PRRX1 perturbs cellular differentiation, promotes more stem cell-like states, and modulates the immune environment through both cell-autonomous and non-cell-autonomous mechanisms.
Figure 6. PRRX1 transcriptional targets.
(A) Metacore network analysis for Hs578T and TTC642 cell line DEGs following PRRX1 downregulation using TET-inducible shRNA at the 5-day time point.
(B) Heatmap showing clustering of basal (EMG3 and HCC3153) and luminal (SUM185 and MFM223) cell lines overexpressing WT or dH3 mutant PRRX1 based on expression of the union of DEGs (lfc > 1) in each cell line following PRRX1 induction by doxycycline (dox).
(C) Metacore network analyses for upregulated DEGs in basal (EMG3 and HCC3153) and luminal (MFM223 and SUM185) lines overexpressing WT PRRX1 (network gene list shown in Table S2; Figure 6C). For each cell line, dox-treated PRRX1-overexpressing samples (at three time points) were compared with untreated samples (corresponding to the same three time points) to identify DEGs.
(D) GSEA of the chemokine gene set in WT cell lines overexpressing PRRX1.
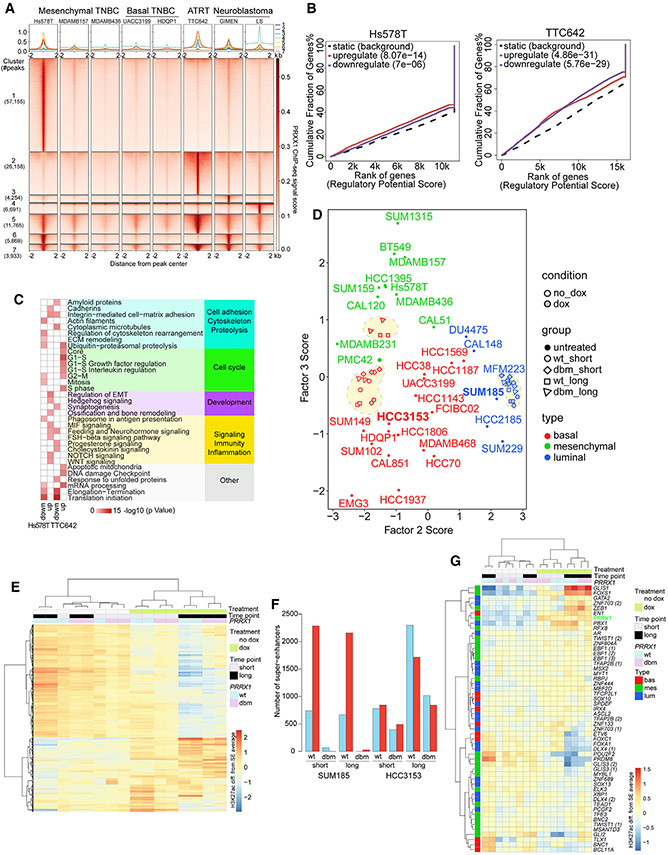
Next, we performed ChIP-seq for PRRX1 to identify its direct genomic targets in mesenchymal TNBC, ATRT, and neuroblastoma lines and in basal TNBC as a control. We found that a subset of PRRX1 binding sites was shared among all lines, but the largest subsets of peaks were unique to Hs578T or TTC642, the two cell lines with the highest endogenous PRRX1 levels (Figures 7A, S6A, and S6B; Table S6). Metacore analysis showed that gene sets 1 and 2, representing peaks unique to Hs578T and TT642, respectively, had common enrichments for EMT and WNT signaling. Common peaks between these two cell lines (set 5) were enriched for the cell cycle (Figure S6C; Table S2). These data suggest that PRRX1 has a tissue-dependent role in regulating proliferation and stem cell pathways.
Figure 7. PRRX1 genomic targets.
(A) PRRX1 ChIP-seq peaks in the indicated cell lines from experiments with the larger number of peaks. All combinations represented in more than 1% of peaks are shown.
(B) Cumulative fraction of genes up- or downregulated by PRRX1, plotted against rank of the regulatory potential score, from BETA of association between gene expression changes after PRRX1 downregulation and PRRX1 chromatin occupancy for Hs578T and TTC642.
(C) Metacore network analysis for Hs578T and TTC642 for PRRX1 ChIP-seq-based BETA targets representing enrichments for up- and downregulated PRRX1 targets (network gene list shown in Table S2; Figure 7C).
(D) Scatterplot depicting MOFA F2 and F3 scores for each sample, calculated based on SE H3K27ac signal. Red- and blue-outlined shapes within dotted lines represent HCC3153 and SUM185 samples from the PRRX1 overexpression H3K27ac experiment, respectively.
(E) Heatmap showing clustering of HCC3153 PRRX1-overexpressing samples and corresponding controls based on H3K27ac signal in the top 20% most variable SEs.
(F) Bar plot showing counts of differentially acetylated SE regions under WT and dbm PRRX1 overexpression in SUM185 and HCC3153 at short and long time points.
(G) Heatmap of SE H3K27ac signal for TNBC subtype-specific TFs in HCC3153 PRRX1 overexpression samples and corresponding controls.
To investigate whether PRRX1 functions as a transcriptional repressor or activator, we performed binding and expression target analysis (BETA).27 We integrated PRRX1 ChIP-seq with genes differentially expressed 5 days after shPRRX1 expression in Hs578T and TTC642 cells. We found that PRRX1 may both activate and repress transcripts in both cell lines, possibly due to the late time point chosen for RNA-seq (Figure 7B). Networks for the PRRX1 targets showed that BETA-inferred positive PRRX1 targets were enriched in mesenchymal genes in Hs578T cells and cell-cycle-related genes in TTC642 (Figure 7C).
We also analyzed what fractions of TNBC subtype-specifically expressed genes were direct PRRX1 targets in Hs578T cells (Table S6). We found that the proportion of overlapping genes differed significantly across subtypes, with a significantly greater proportion of mesenchymal subtype-specific genes being direct PRRX1 targets (86%) compared with basal (72%) and luminal (73%) subsets, suggesting that PRRX1 regulates mesenchymal subtype-specific genes (Figure S6D).
The role of PRRX1 in establishing mesenchymal SE landscapes
To determine whether PRRX1 can reprogram basal and luminal SE landscapes to a mesenchymal one, we performed H3K27ac ChIP-seq in HCC3153 basal and SUM185 luminal TNBC cell lines following expression of WT or dbm PRRX1 for 7 or 42 days and quantified H3K27ac expression in SEs previously identified in parental cells. We found that long-term (42 days) expression of both WT and dbm PRRX1 in HCC3153 cells was sufficient to induce a more mesenchymal cell state, defined based on MOFA F3 scores, while no changes in F2, F3, or F6 scores were detected in luminal SUM185 cells (Figures 7D and S6E).
To explore PRRX1-induced chromatin changes in more detail, we assessed the H3K27ac signal in the topmost variable SEs among samples. In the HCC3153 cell line, the most pronounced difference was between PRRX1 overexpressing (+doxycycline [dox]) and non-expressing (−dox) samples, with lesser variability observed between long and short time points and between WT and dbm PRRX1 (Figure 7E). In SUM185 cells, only WT PRRX1 induced distinct changes in SE H3K27ac signal, especially at longer time points (Figure S6F). Quantification of these SE changes further highlighted marked differences between the two cell lines; in SUM185 cells, only WT PRRX1 induced changes, and there was no difference between short- and long-term treatment (Figures 7F and S6F; Table S7). In HCC3153 cells, the WT and dbm PRRX1 induced the same magnitude of changes and more pronounced changes at later time points (Figures 7E and 7F). These observations suggest that PRRX1 can function both as an activator and repressor, which follows our BETA (Figure 7B); that it may modulate transcription and SE patterns via both direct and indirect DNA binding; and that PRRX1-induced changes in luminal SUM185 cells require direct PRRX1 DNA binding, possibly due to the lack of certain PRRX1-interacting TFs.
Analysis of the H3K27ac signal in SEs associated with TNBC subtype-specific TFs showed a pronounced gain in mesenchymal TFs in basal HCC3153 cells, with a minimal increase in luminal SUM185 cells (Figures 7G and S6G). PRRX1 was among the mesenchymal TFs that gained signal in PRRX1-overexpressing HCC3153 cells, implying that PRRX1 may positively autoregulate itself (Figure 7G). We also investigated overlaps between significantly gained and lost SEs in both cell lines and identified shared and cell-line-specific long-term gained and lost regions (Figure S6H). We defined long-term gains and losses induced by WT PRRX1 in HCC3153 cells as indirect when they were also observed in HCC3153 cells expressing dbm PRRX1, while changes specific to WT PRRX1 were classified as direct. We found that long-term WT PRRX1-induced indirect changes in HCC3153 cells were significantly more likely to be observed in SUM185 cell than direct changes (Figure S6H). Given the dependence of PRRX1-induced SE changes in SUM185 on WT PRRX1 (Figure 7F), this implies that some changes gain DNA dependence in SUM185 cells. By comparing DNA sequences of indirect and direct regions in HCC3153 cells, we identified putative PRRX1 co-binding TFs in HCC3153 long-term gained and lost regions. We found that, as a group, putative co-binding TFs identified from gained regions had lower expression in SUM185 compared with HCC3153 (Figure S6I). The lower expression of these TFs, which include FOXI1, ETS2, and TWIST1, may play a role in the muted changes observed in dbm-expressing SUM185 cells. These data highlight the importance of cell-type-specific TFs in establishing cellular states and epigenetic landscapes.
Clinical relevance of PRRX1 in TNBC
Last, we investigated the clinical relevance of PRRX1 activity in primary TNBC samples. In both the METABRIC and TCGA datasets, PRRX1 expression differs among basal, luminal, and mesenchymal TNBCs and is higher in mesenchymal TNBC than in the other two subtypes (Figure S7A). We also analyzed the expression of a mesenchymal RNA target (MRT) signature derived from our integrated analysis of Hs578T PRRX1-ChIP-seq and RNA-seq data (STAR Methods; Table S6). We found that correlations between PRRX1 expression and expression of its putative positive targets were higher than correlations between PRRX1 expression and expression of its putative negative targets in the TCGA cohort (Figure S7B), and we observed the same, although non-significant, trend in the METABRIC data (Figure S7B). We also investigated the Hs578T-specific RNA targets (HsRT) and noted the same patterns in both cohorts (Figure S7C). We also found that the expression of the MRT target gene set varied between TNBC subtypes, assigned to samples using cell line-derived signatures, in the METABRIC datasets, with significantly higher expression in mesenchymal than in luminal samples in both cohorts (Figure S7D). However, refined clustering of the samples showed that PRRX1 or MRT target expression did not differ significantly between mesenchymal low and high samples (Figure S7E). These results support our hypothesis that PRRX1 acts as a clinically relevant transcriptional regulator in mesenchymal TNBC.
To further investigate the clinical relevance of our findings, we tested for associations between TNBC subtype and PRRX1 and immune-related gene expression signatures28 (Table S5). We found that 8 of 24 signatures were significantly positively associated with PRRX1 expression in at least one of the two cohorts, with an association that remained significant in the other cohort (Figure S7F and S7G; Table S5). In the TCGA cohort, a single signature was significantly differentially expressed between TNBC types and significantly more highly expressed in the mesenchymal subtype compared with the other two subtypes. In the METABRIC cohort, 10 such signatures were significantly overexpressed in mesenchymal TNBC after accounting for multiple testing, including 7 of 8 signatures with a validated significant association with PRRX1 expression and an additional three signatures (Figure S7H; Table S5). For 3 of 10 of these mesenchymal-specific immune signatures in the METABRIC cohort, the same trends were statistically significant in the TCGA cohort, but the effect sizes were rather small in both cohorts (Figure S7H; Table S5). Of these three signatures, only TGF- family members had significantly different expression between mesenchymal-high and mesenchymal-low samples in TCGA or METABRIC data, and this signature was more highly expressed in mesenchymal-high samples for both METABRIC and TCGA (Figure S7I). Taken together, our results suggest that there is a link between PRRX1 expression and immune activity that may contribute to shaping the immune microenvironment of mesenchymal TNBC.
DISCUSSION
Characterization of mechanisms underlying TNBC heterogeneity may guide the design of more effective therapies. Here, we performed comprehensive multiomics and phenotypic characterization of both inter- and intra-tumor heterogeneity in TNBC to identify key regulators of disease processes that may reveal therapeutic targets and markers for patient stratification.
RNA-seq confirmed luminal, basal, and mesenchymal subtypes that have been described previously, highlighting the robustness of transcriptional differences in TNBC.2-4,29 Similar to prior studies,2-4,29 we noted that, even within basal and mesenchymal subtypes, there are further subclusters (e.g., BL1 and BL2), but we did not investigate these in more detail. SE analyses based on H3K27ac ChIP-seq data largely correlated with the three main transcriptional subtypes, in line with SEs playing key roles in cell-type-specific transcriptional patterns.7,30 Several recent studies have reported the identification of TNBC-specific SEs (e.g., FOXC1, MET, and BAMBI) and have shown that some of these SE-associated genes reflect dependencies in TNBC.31,32 FOXC1 and MET were also among the top basal and mesenchymal subtype-specific SEs in our dataset, while BAMBI was among the top variably expressed genes but did not show significant TNBC subtype specificity.
Quantitative analyses of histone modification profiles revealed a distinct pattern of variability largely driven by H3K27ac, H3K27me3, and H4K20me3, and H4 acetylation marks associated with active (H3K27ac) and repressive (H3K27me3) chromatin. The roles of H3K27ac and H3K27me3 in cellular differentiation and epigenetic states have been extensively characterized both during normal development and cancer.33 Modifiers of these histone marks, including histone H3 acetyl transferases (e.g., P300) and deacetylases (HDACs), and H3K27me3 transferase (EZH2) and demethylase (KDM6A) and readers of H3K27ac (BET bromodomain proteins) have been explored as therapeutic targets in breast and other cancer types.34 The sources and consequences of histone H4 modifications are poorly defined, even though they account for nearly half of all histone modification events.35 Our finding that H4K20me3 is the topmost variable histone mark both among and within TNBC tumors highlights a potential role of histone H4 in TNBC biology that is worth further investigations.
Using MOFA, we defined the landscape of TNBC cell line epigenetic and metabolic heterogeneity. We identified 8 biological factors and associated multiomics signatures (3 factors linked to transcriptional subtype [F2, F3, and F6] and 5 subtype-independent factors [F1, F4, F5, F7, and F8]) and found that 6 of the 8 factors (F1–F6) were validated in two independent primary TNBC cohorts. Further interrogation of the mechanisms underpinning these factors will be an important area for future work.
A main goal of our study was to identify key drivers of TNBC subtypes. Because TFs orchestrate transcriptional and SE landscapes, we focused on TFs associated with subtype-specific expression and SEs. In addition to confirming the expression of known luminal, basal, and mesenchymal TFs, we identified several previously uncharacterized TFs in each subtype. Mesenchymal TNBC represents the least differentiated subtype, suggesting that these tumors may originate from an early stem/progenitor cell or lost epithelial features during tumorigenesis. Our finding that mesenchymal TNBC is more similar to ATRT and mesenchymal neuroblastoma than to other breast cancer subtypes supports this hypothesis. We found that the PRRX1 TF is a shared driver of these mesenchymal tumors regardless of tissue of origin, highlighting the importance of TFs in establishing cell states. Based on our data, PRRX1 appears to be a trigger of mesenchymal state but not to be required for its maintenance or tumor growth. The PRRX1-associated TF interaction network, including several positive feedback loops, might maintain mesenchymal programs even without PRRX1.
Overall, our study is an excellent resource because it provides highly usable data for both scientific and clinical communities, thus providing opportunities for follow-up studies as a future direction.
Limitations of the study
The multiomics profiling was performed on TNBC cell lines but validated in the TCGA and METABRIC TNBC cohorts. While the proportion of variance explained by the MOFA model was greater than 50% for some datasets, the proportion of variance explained by the model was lower for the metabolomics and histone mass spectrometry datasets. While some of the unexplained variance may be due to factors unrelated to TNBC biology, including measurement error, we cannot rule out the possibility that some of this unexplained variability relates to TNBC biology. Applying similar approaches to larger multiomics datasets is expected to shed light on this possibility and is an area for future work. Additionally, experimental functional validation will be important to confirm the roles of the putative PRRX1 co-binding TFs identified using computational analyses.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Kornelia Polyak, Dana-Farber Cancer Institute, 450 Brookline Ave., SM1070B, Boston, MA 02215, USA. kornelia_polyak@dfci.harvard.edu; tel: 617-632-2106.
Materials availability
Hs578T, SUM185, EMG3, and MFM223, and TTC642 cell line derivatives generated using Tet-doxycycline inducible system will be made available upon request and following the execution of an MTA.
Data and code availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplemental Information. All raw genomic data was deposited to GEO under accession number: GSE202776. Processed histone mass spectrometry, metabolomics, drug screen, and BH3 profiling data is provided as supplementary information. Additional metadata files and processed data files used in the original code have been deposited at (https://figshare.com/s/2f077f7838fb5f6e8d35). All code used to analyze genomics data and produce the corresponding figures is available on the GitHub repository https://github.com/daniel-temko/TNBCEpiHet (https://doi.org/10.5281/zenodo.10139754). The raw mass spectrometry data have been deposited in the public proteomics repository MassIVE (http://massive.ucsd.edu) using the identifier: MSV000091071.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Human breast tumor samples
Human breast cancer samples were collected using protocol #93-085 approved by the DF/HCC Institutional Review Board, informed consent was obtained from all patients, and samples were de-identified prior to transport to the lab. Tumor tissues were dissociated to single cells by mechanical chopping with razor blades followed by digestion at 37°C in DMEM/F12 with 2 mg/mL bovine serum albumin (BSA), 2 mg/mL collagenase type IV, and 2 mg/mL hyaluronidase while stirring for 3–4 h. Cells were filtered sequentially through 500, 100, and 70-μm mesh, washed in DMEM/F12 with 5% fetal bovie serum (FBS), frozen in DMEM/F12 with 5% FBS and 10% DMSO, and stored in liquid nitrogen for subsequent xenograft studies. Tissue microarray of human TNBC (HTMA 240) was generated from tumors collected using tissue banking protocol #93-085 approved by the DF/HCC Institutional Review Board, informed consent was obtained from all patients.
Breast cancer cell lines
Breast cancer cell lines were obtained from ATCC, DMZE, or generously provided by multiple principal investigators (please see key resources table for details) and cultured following the provider’s recommendations. The identity of the cell lines was confirmed based on STR and exome-seq analyses. Cells were regularly tested for mycoplasma.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies (ChIP, WB, IHC and CYTOF-metal) | ||
| Rabbit polyclonal anti-H3K27Ac | Diagenode | Cat#C15410196 RRID: AB_2637079 |
| Rabbit polyclonal anti-PRRX1 | Sigma | Cat#HPA051084 RRID: AB_2681338 |
| Mouse monoclonal anti-beta-Actin | Sigma-Aldrich | Cat# A2228; RRID: AB_476697 |
| Anti-vimentin, clone D21H3 | Cell Signaling Technology | Cat#5741; RRID: AB_10695459 |
| Anti-E-cadherin, clone. 4A2 | Cell Signaling Technology | Cat#14472S; RRID AB_2728770 |
| Anti-smooth muscle actin, clone. 1A4 | Thermo Fisher Scientific | Cat#MA5-11547; RRID: AB_10979529 |
| Rabbit monoclonal anti-PR a/b (141Pr) | Cell Signaling Technology | Cat# 8757 RRID: AB_2797144 |
| Mouse monoclonal anti-CD10 (142ND) | BD Biosciences | Cat# 555373; RRID: AB_395775 |
| Rat monoclonal anti-CD44 (143ND) | Biolegend | Cat# 103002; RRID: AB_312953 |
| Mouse monoclonal anti-cyclin D3 (144ND) | Abcam | Cat# ab28283; RRID: AB_2070798 |
| Mouse monoclonal anti-Muc1 (145ND) | Biolegend | Cat# 355602; RRID: AB_2561642 |
| Mouse monoclonal anti-Lamp2 (146ND) | Biolegend | Cat# 354302; RRID: AB_11204245 |
| Mouse monoclonal anti-CDK4 (147Sm) | BD Biosciences | Cat# 559677; RRID: AB_397299 |
| Rabbit monoclonal anti-PTEN (148ND) | Cell Signaling Technology | Cat# 9559; RRID: AB_390810 |
| Rabbit monoclonal anti-E-Cadherin (149Sm) | Cell Signaling Technology | Cat# 3195; RRID: AB_2291471 |
| Mouse monoclonal anti-Epcam (150ND) | Biolegend | Cat# 324202; RRID: AB_756076 |
| Mouse monoclonal anti-Her2 (151Eu) | BD Biosciences | Cat# 554299; RRID: AB_395352 |
| Rabbit polyclonal anti-CK5 (152Sm) | Abcam | Cat# ab53121; RRID: AB_869889 |
| Mouse monoclonal anti-CD24 (153Eu) | Biolegend | Cat# 311102; RRID: AB_314851 |
| Mouse monoclonal anti-CDK1 (154Sm) | Biolegend | Cat# 626901; RRID: AB_2074779 |
| Rabbit monoclonal anti-CDK6 (155Gd) | Cell Signaling Technology | Cat# 13331; RRID:AB_2721897 |
| Rabbit monoclonal anti-p63 (158Gd) | Abcam | Cat# ab124762; RRID: AB_10971840 |
| Rabbit monoclonal anti-TCF7 (159Tb) | Cell Signaling Technology | Cat# 2203; RRID: AB_2199302 |
| Rabbit monoclonal anti-AR (160Gd) | Cell Signaling Technology | Cat# 5153; RRID: AB_10691711 |
| Mouse monoclonal anti-Cyclin A (161Dy) | BD Biosciences | Cat# 554175; RRID: AB_395286 |
| Mouse monoclonal anti-Ki-67 (162Dy) | BD Biosciences | Cat# 550609; RRID: AB_393778 |
| Mouse monoclonal anti-SMA (163Dy) | Thermo Fisher Scientific | Cat# 14-9760-82; RRID: AB_2572996 |
| Mouse monoclonal anti-cPARP (164Dy) | BD Biosciences | Cat# 552596; RRID: AB_394437 |
| Rabbit monoclonal anti-Vimentin (165Ho) | Cell Signaling Technology | Cat# 5741; RRID: AB_10695459 |
| Rat monoclonal anti-GATA-3 (166Er) | eBioscience | Cat# 14-9966-80; RRID: AB_1210520 |
| Rabbit monoclonal anti-p21 (167Er) | Cell Signaling Technology | Cat# 2947; RRID: AB_823586 |
| Rabbit monoclonal anti-phospho-AKT | Cell Signaling Technology | Cat# 4060; RRID: AB_2315049 |
| Rabbit monoclonal anti-phospho-STAT3 | Cell Signaling Technology | Cat# 9145; RRID: AB_2491009 |
| Rabbit monoclonal anti-EGFR (170Er) | Cell Signaling Technology | Cat# 4267; RRID: AB_2246311 |
| Rabbit monoclonal anti-phospho-SMAD2 | Cell Signaling Technology | Cat# 8828; RRID: AB_2631089 |
| Rabbit monoclonal anti-ERa (172Yb) | Cell Signaling Technology | Cat# 13258; RRID: AB_2632959 |
| Rat monoclonal anti-CD49f (173Yb) | Biolegend | Cat# 313602; RRID: AB_345296 |
| Rabbit monoclonal anti-phospho-STAT5 | Cell Signaling Technology | Cat# 4322; RRID: AB_10548756 |
| Rabbit monoclonal anti-phospho-S6 | Cell Signaling Technology | Cat# 4858; RRID: AB_916156 |
| Mouse monoclonal anti-CK8/18 (176Yb) | Cell Signaling Technology | Cat# 4546; RRID: AB_2134843 |
| Rabbit polyclonal anti-histone H4K20me3 | Abcam | Cat# ab9053; RRID: AB_306969 |
| Biological samples | ||
| HCI-001 human patient-derived xenograft | Alana Welm, Huntsman Cancer Institute | N/A |
| HCI-002 human patient-derived xenograft | Alana Welm, Huntsman Cancer Institute | N/A |
| HCI-009 human patient-derived xenograft | Alana Welm, Huntsman Cancer Institute | N/A |
| HCI-010 human patient-derived xenograft | Alana Welm, Huntsman Cancer Institute | N/A |
| 2147-TG5 human patient-derived xenograft | Michael Lewis, Baylor College of Medicine | N/A |
| 3936-TG5 human patient-derived xenograft | Michael Lewis, Baylor College of Medicine | N/A |
| 4013-TG6 human patient-derived xenograft | Michael Lewis, Baylor College of Medicine | N/A |
| 4195-TG5 human patient-derived xenograft | Michael Lewis, Baylor College of Medicine | N/A |
| 4272-TG5 human patient-derived xenograft | Michael Lewis, Baylor College of Medicine | N/A |
| 5998-TG5 human patient-derived xenograft | Michael Lewis, Baylor College of Medicine | N/A |
| BCM-2665 human patient-derived xenograft | Michael Lewis, Baylor College of Medicine | N/A |
| BCM-3107 human patient-derived xenograft | Michael Lewis, Baylor College of Medicine | N/A |
| BCM-3611 human patient-derived xenograft | Michael Lewis, Baylor College of Medicine | N/A |
| T272X human patient-derived xenograft | Polyak Lab, DFCI | N/A |
| IDC50X human patient-derived xenograft | Polyak Lab, DFCI | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Galunisertib, LY2157299 (TGF-beta) | Selleckchem | S2230 |
| Xav939 (Wnt/beta-catenin) | Selleckchem | S1180 |
| LGK-974 (Wnt/beta-catenin) | Selleckchem | S7143 |
| Vismodegib (GDC-0449) (Hedgehog) | Selleckchem | S1082 |
| Sonidegib (Erismodegib, NVP-LDE225) (Hedgehog) | Selleckchem | S2151 |
| NVP-BHG712 (Ephrin) | Selleckchem | S2202 |
| BGJ398 (NVP-BGJ398) (FGFR) | Selleckchem | S2183 |
| Vorinostat (SAHA, MK0683) (HDACs1/3) | Selleckchem | S1047 |
| Tretinoin (Retinoids) | Selleckchem | S1653 |
| MK-8617 (Pan-HIF) | Selleckchem | S8443 |
| Ruxolitinib, INC018424 (JAK) | Selleckchem | S1378 |
| A-1155463 (BCL-xl) | Selleckchem | S7800 |
| ML324 (KDM4) | Selleckchem | S7296 |
| GSK J1 (KDM6A/B) | Selleckchem | S7581 |
| Verteporfin (YAP/TEAD) | Selleckchem | S1786 |
| A-485 (p300/CBP) | MedChemExpress | HY-107455 |
| Critical commercial assays | ||
| ThruPLEX DNA-seq 48S Kit | Rubicon | R400427 |
| Infinium HumanMethylation 450K BeadChIP | Illumina | WG-314-1003 |
| Deposited data | ||
| All raw genomic data | GEO | GSE202776 |
| Raw histone mass spectrometry data | MassIVE (http://massive.ucsd.edu) | MSV000091071 |
| Code associated with this manuscript | This manuscript | Zenodo: https://doi.org/10.5281/zenodo.10139754 |
| Experimental models: Cell lines | ||
| Human: BT549 cell line | ATCC | HTB-122 |
| Human: CAL120 cell line | DSMZ | ACC 459 |
| Human: CAL148 cell line | DSMZ | ACC 460 |
| Human: CAL51 cell line | DSMZ | ACC 302 |
| Human: CAL851 cell line | DSMZ | ACC 440 |
| Human: DU4475 cell line | ATCC | HTB-123 |
| Human: EMG3 cell line | Eva Matou, Czechia | N/A |
| Human: FCIBC02 cell line | Massimo Cristofanilli, Jefferson University | N/A |
| Human: GIMEN cell line | Kimberly Stegmaier, Dana-Farber Cancer Institute | N/A |
| Human: HCC1143 cell line | ATCC | CRL-2321 |
| Human: HCCC1187 cell line | ATCC | CRL-2322 |
| Human: HCC1395 cell line | ATCC | CRL-2324 |
| Human: HCC1569 cell line | ATCC | CRL-2330 |
| Human: HCC1806 cell line | ATCC | CRL-2335 |
| Human: HCC1937 cell line | ATCC | CRL-2336 |
| Human: HCC2157 cell line | ATCC | CRL-2340 |
| Human: HCC2185 cell line | Adi Gazdar, UT Southwestern | N/A |
| Human: HCC3153 cell line | Adi Gazdar, UT Southwestern | N/A |
| Human: HCC38 cell line | ATCC | CRL-2314 |
| Human: HCC70 cell line | ATCC | CRL-2315 |
| Human: HDQP1 cell line | DSMZ | ACC 494 |
| Human: Hs578 cell line | ATCC | HTB-126 |
| Human: LS cell line | DSMZ | ACC 675 |
| Human: MDAMB157 cell line | ATCC | HTB-24 |
| Human: MDAMB231 cell line | ATCC | HTB-26 |
| Human: MDAMB436 cell line | ATCC | HTB-130 |
| Human: MDAMB453 cell line | ATCC | HTB-131 |
| Human: MDAMB468 cell line | ATCC | HTB-132 |
| Human: MFM223 cell line | DSMZ | ACC 422 |
| Human: PMC42 cell line | Robert H. Whitehead, Melbourne, Australia | N/A |
| Human: SUM102 cell line | Stephen Ethier, University of Michigan | N/A |
| Human: SUM1315 cell line | Stephen Ethier, University of Michigan | N/A |
| Human: SUM149 cell line | Stephen Ethier, University of Michigan | N/A |
| Human: SUM159 cell line | Stephen Ethier, University of Michigan | N/A |
| Human: SUM185 cell line | Stephen Ethier, University of Michigan | N/A |
| Human: SUM229 cell line | Stephen Ethier, University of Michigan | N/A |
| Human: TT642 cell line | Charles Roberts, Dana-Farber Cancer Institute | N/A |
| Human: UACC3199 cell line | University of Arizona | N/A |
| Experimental models: Organisms/strains | ||
| NOG (NOD.Cg-Prkdcscid Il2rgtm1Sug/JicTac) | Taconic Biosciences | N/A |
| NOD (NOD.Cg-Prkdc< scid> Il2rg< tm1Wjl>Tg (CMV-IL3,CSF2,KITLG)1Eav/MloySzJ | Jackson Laboratory | N/A |
| Recombinant DNA | ||
| shERWOOD Lentiviral Inducible shRNA (n = 3) for PRRX1: | Transomic Technologies | cat# TLHSU2300-5396 |
| 1. ULTRA-3340261-pZIP-TRE3G-ZsGreen-Puro | Transomic Technologies | cat# TLHSU2300-5396 |
| 2. ULTRA-3340262-pZIP-TRE3G-ZsGreen-Puro | Transomic Technologies | cat# TLHSU2300-5396 |
| 3. ULTRA-3340265-pZIP-TRE3G-ZsGreen-Puro | Transomic Technologies | cat# TLHSU2300-5396 |
| PAX2 packaging plasmid | Addgene | cat#35002 |
| pMD2.G envelope plasmid | Addgene | cat#12259 |
| WT and MUT (ΔH1) PRRX1 lentiviral vectors | Sengupta et al.25 | gift from Rani George’s lab |
Animal model
For knockdown xenograft assays, female NOG (NOD.Cg-Prkdcscid Il2rgtm1Sug/JicTac) mice were purchased from Taconic at 5–6 weeks of age. For overexpression, xenograft assays female NOD (NOD.Cg-Prkdc< scid> Il2rg< tm1Wjl> Tg (CMV-IL3,CSF2,KITLG)1Eav/MloySzJ) mice at 6–7 weeks of age were purchased from Jackson Laboratories. Animal experiments were performed by B.J. and K.M. according to protocol 11–023 approved by the Dana-Farber Cancer Institute Animal Care and Use Committee. Mice were housed 5 to a cage with ad libitum access to food and water in 20°C ambient temperature, 40–50% humidity, and 12-h light/12-h dark cycle.
METHOD DETAILS
Xenograft assays
All animal experiments were performed in an AAALAC-accredited SPF rodent-only barrier facility at Dana-Farber Cancer Institute. All mice are housed in individually ventilated, solid-bottom, polysulfone 135 sq. in. microisolator cages. The cages are used in conjunction with the Optimice rack systems with integrated automatic watering. Temperature and humidity in the rodent facility is controlled at 72 ± 2°F and a target range of 35–55% relative humidity. A standard photoperiod of 12 h light/12 h dark is controlled by an automated system. All animal experiments were performed according to protocol 11–023 approved by the Dana-Farber Cancer Institute Animal Care and Use Committee. Animals were euthanized by CO2 inhalation. Maximum tumor size burden allowed for mice is 2 cm and this was not exceeded in any of the experiments. For xenograft assays using Hs578T cells expressing dox-inducible shRNAs, 5–6-weeks old female NOG (NOD.Cg-Prkdcscid Il2rgtm1Sug/JicTac) mice were purchased from Taconic. Tumors were induced by bilateral orthotopic mammary fat pad injection of 2 × 106 Hs578T (non-targeting, shPRRX1-1, 2 and 3) cells suspended in 50 μL total volume of 50% DMEM media (Corning, cat# 10-013-CV) and 50% of Matrigel (BD Biosciences, cat# 354234). After tumors became palpable, mice were randomized into two groups (+ and − doxycycline diet (625ppm)). Tumor growth was monitored weekly using caliper measurements. Mice were euthanized and tumors collected, fixed overnight in 4% formalin, stored in 70% ethanol, followed by paraffin embedding, sectioning, and hematoxylin and eosin staining by the Pathology Core of the Brigham and Women’s Hospital. For xenograft assays using PRRX1 overexpressing lines female NOD (NOD.Cg-Prkdc< scid> Il2rg< tm1Wjl> Tg (CMV-IL3,CSF2,KITLG)1Eav/MloySzJ) mice at 6–7 weeks of age were purchased from Jackson Labs. Tumors were induced by bilateral orthotopic mammary fat pad injection of 5 × 106 EMG3 WT/mutant +/− doxycycline cells, 5 × 106 SUM185 WT +/− doxycycline cells, 5 × 106 MFM223 WT/mutant +/− doxycycline cekks, and 2.625 x 106 HCC3153 WT/mutant +/− doxycycline cells suspended in 50 μL total volume of 50% DMEM-F12 media (Corning, cat# 10-090-CV) and 50% of Matrigel (BD Biosciences, cat# 354234). Plus, doxycycline cells were pretreated for 18hrs (HCC3153), 2 days (SUM185, MFM223), or 3 days (EMG3) with 3 μg/mL doxycycline before injection. Mice in the plus doxycycline group began their doxycycline diet (625ppm) starting 2 (SUM185, MFM223) or 3 (EMG3, HCC3153) days before the injection. Tumor growth was monitored weekly using caliper measurements. Mice were euthanized and tumors collected, fixed overnight in 4% formalin, stored in 70% ethanol, followed by paraffin embedding, sectioning, and hematoxylin and eosin staining by the Pathology Core of the Brigham and Women’s Hospital.
ChIP-seq
Cell lines:
H3K27ac ChIP-seq data for two samples was used from our prior publications.5,36 For histone H3K27ac ChIP-seq, 5 × 106 cells were fixed with 1% paraformaldehyde for 10 min at room temperature. For PRRX1 ChIP-seq, 1 × 107 cells were fixed with 2mM DSG (Thermo scientific 20593) for 30 min at room temperature. DSG was then removed and replaced with fixing buffer (50 mM HEPES-NaOH (pH 7.5), 100 mM NaCl, 1mM EDTA) containing 1% paraformaldehyde (Electron Microscopy Sciences, 15714) and cross-linked for 10 min at 37°C. Crosslinking was quenched by adding glycine to a final concentration of 0.125 M. The cells were washed with ice-cold PBS, harvested in PBS. The nuclear fraction was extracted by first resuspending the pellet in 1 mL of lysis buffer (50 mM HEPES-NaOH (pH 8.0), 140 mM NaCl, 1mM EDTA, 10% glycerol, 0.5% NP-40, and 0.25% Triton X-100) for 10 min at 4°C. Cells were pelleted and washed in 1 mL of wash buffer (10 mM Tris-HCL (pH 8.0), 200 mM NaCl, 1 mM EDTA) for 10 min at 4°C. Cells were then pelleted and resuspended in 1 mL of shearing buffer (10 mM Tris-HCl (pH 8), 1 mM EDTA, 0.1% SDS) and sonicated in a Covaris sonicator. Lysate were cleared by centrifugation for 5 min at 14,000 rpm. Then 100 μL of 10% Triton X-100 and 30 μL of 5M NaCl were added. The sample was then incubated with 20 μL of Dynabeads Protein G (LifeTechnologies, 10003D) for 1 h at 4°C. Primary antibodies were added to each tube, and immunoprecipitation (IP) was conducted overnight at 4°C. Cross-linked complexes were precipitated with Dynabeads Protein G for 2 h at 4°C. The beads were then washed in low salt wash buffer (20 mM Tris-HCl pH 8, 150 mM NaCl, 10 mM EDTA, and 1% SDS) for 5 min at 4°C, high salt wash buffer (50 mM Tris-HCl pH 8, 10 mM EDTA, and 1% SDS) for 5 min at 4°C and LiCl wash buffer (50 mM Tris-HCl pH 8, 10 mM EDTA, and 1% SDS) for 5 min at 4°C. DNA was eluted in elution buffer (100 mM sodium bicarbonate and 1% SDS). Cross-links were reversed overnight at 65°C. RNA and protein were digested with 0.2 mg mL–1 RNase A for 30 min at 37°C followed by 0.2 mg mL–1 Proteinase K for 1 h at 55°C. DNA was purified with phenol-chloroform extraction and isopropanol precipitation. ChIP-seq libraries were prepared using the Rubicon ThruPLEX DNA-seq Kit (cat# R400427) from 1 ng of purified ChIP DNA or input DNA according to the manufacturer’s protocol.
RNA-seq
Total RNA was extracted using the RNeasy Mini Kit (Qiagen). RNA-seq libraries were prepared using Illumina TruSeq Stranded mRNA sample preparation kits from 500 ng of purified total RNA according to the manufacturer’s protocol. The finished dsDNA libraries were quantified by Qubit fluorometer, Agilent TapeStation 2200, and RT-qPCR using the Kapa Biosystems library quantification kit according to the manufacturer’s protocols. Uniquely indexed libraries were pooled in equimolar ratios and sequenced on an Illumina NextSeq500 with single-end 75 bp reads in the Dana-Farber Cancer Institute Molecular Biology Core Facilities.
DNA methylation
Genomic DNA was extracted using the ALLPrep kit (Qiagen). DNA methylation profiling was carried out on Infinium HumanMethylation450K BeadChip 450,000 CpG site platform array (Illumina, WG-314-1003 discontinued) at the Harvard Medical School-Partners HealthCare Center for Genetics and Genomics.
Mass spectrometry analysis of histone modifications
Briefly, histones were isolated from cell nuclei using acid extraction, biochemically prepared, and analyzed by mass spectrometry against a reference of stable isotope-labeled synthetic peptide standards exactly as described.37
Metabolomic profiling
Metabolomic profiling was performed as previously described.38 Briefly, 1 × 107 cells were cultured in triplicate, and the medium was changed 2 h before metabolite extraction. After aspirating the medium completely, 4 mL of 80% (v/v) methanol which was precooled to −80°C was added to the plates on dry ice, then incubate the plates at −80°C for 20 min. The plates were scraped on dry ice with a cell scraper, and the cell lysate/methanol mixture was transferred to a 15-mL conical tube on dry ice. After centrifuging the tube at 14,000g for 5 min at 4°C to pellet the cell debris, the metabolite-containing supernatant was transferred to a 15-mL conical tube on dry ice. To collect metabolites completely, 500 μL of 80% (v/v) methanol precooled to −80°C was added to the remaining pellet in a 15-mL tube and vortex for 1 min at 4°C. The tubes were centrifuged again at 14,000g for 5 min at 4°C, and then the supernatant was transferred to a conical tube. A total of 4.5 mL of sample was divided and transferred into three 1.5-mL microcentrifuge tubes (1.5 mL in each tube), then SpeedVac/lyophilize to a pellet using no heat. Dried metabolite samples were stored at −80°.
Mass cytometry (CyTOF)
Antibodies used for mass cytometry in this study are listed in key resources table. All antibodies were purchased in carrier-free buffers from the indicated sources and conjugated with the respective lanthanide metals by the CyTOF Antibody Resource and Core at Brigham Women’s Hospital, Boston, MA, USA. Cells were treated with 50 μM IdU-127 (Fluidigm, South San Francisco, CA, USA) for 30 min and 100 μM of the intercalator-103Rh (Fluidigm) for 15 min at 37°C in their respective medium. Next, 1x106 cells of each sample were barcoded using the Cell-ID 20-Plex Pd Barcoding Kit (Fluidigm) according to the manufacturer’s instructions. Barcoded samples were pooled and stained simultaneously. Cells were fixed for 10 min with paraformaldehyde (Electron Microscopy Sciences, Hattfield, PA, USA) at a final concentration of 1.6%, followed by Fc-receptor block (Human TruStain FcX, Biolegend, San Diego, CA) for 10 min and surface antibody staining for 30 min at room temperature. Subsequently, cells were permeabilized with methanol for 10 min on ice and incubated with the antibody cocktail for intracellular epitopes for 30 min. Cells were kept at 4°C overnight in Fix, and Perm Buffer (Fluidigm) was supplemented with Intercalator-IR (Fluidigm) 1:2000. Prior to analysis, cells were washed with water, resuspended in water containing EQ Four Element Calibration Beads (Fluidigm) (1:10), and filtered through a 35 μm strainer. Samples were acquired at a CyTOF Helios instrument (Fluidigm), normalized as previously described16 and analyzed with Cytobank (Cytobank, Inc., Mountain View, CA). Cell Staining Media (PBS with 0.5% BSA, 0.02% NaN3) was used for all washes during staining.
Generation of TET-doxycycline inducible PRRX1 knockdown and overexpression cells
Hs578T and TTC642 cells were transduced with shERWOOD UltramiR Lentiviral Inducible shRNA pZIP target gene PRRX1 set of 3 shRNAs ((1) ULTRA-3340261-pZIP-TRE3G-ZsGreen-Puro, (2) ULTRA-3340262-pZIP-TRE3G-ZsGreen-Puro, (3) ULTRA-3340265-pZIP-TRE3G-ZsGreen-Puro and non-targeting control (TLNSU4300-ULTRA-NT#4-pZIP-TRE3G-ZsGreen-Puro) (Transomic Technologies Inc, cat# for the shRNA set TLHSU2300-5396). After puromycin selection, three days of doxycycline treatment induces the PRRX1 silencing. PRRX1 knockdown was confirmed both by RNA-seq and Western blot. WT or MUT (DNA-binding mutant, DH1) PRRX1 lentiviral vectors (gift from Dr. Rani George’s Lab39) were packaged in HEK293FT cells using the PAX2 packaging plasmid (Addgene plasmid, #35002), pMD2.G envelope plasmid (Addgene plasmid, #12259), and Lipofectamine 3000 transfection reagent (Life Technologies, #L3000015). The virus was collected 48 h after transfection. EMG3, HCC3153, MFM223, and SUM185 cells were transduced with 2 mL of virus and 10 μg/mL polybrene (Millipore Sigma, #TR1003G). Cells were selected with neomycin for 2–3 weeks. Five days of doxycycline treatment induces the WT and mutant PRRX1 overexpression. PRRX1 overexpression was confirmed both by RNA-seq and immunoblot.
Cellular proliferation assays
Cellular viability was assessed using the Celigo system (Nexcelom, Celigo Image Cytometer). Cells expressing doxycycline-inducible shPRRX1 or non-targeting shRNA were plated in triplicates in 24-well plates and cultured at 37°C with 5% CO2. 24hrs after plating, cells were treated with 3ug/ml doxycycline to induce shRNA expression. Culture medium was replaced every 48hrs with freshly prepared doxycycline. Cell viability was measured every 24hrs beginning at 48hrs from the start of the experiment for the duration of ten days. The viability of the EMG3, HCC3153, and MFM223 cells expressing wt or dbm PRRX1 was assessed using the Celigo system (Nexcelom, Celigo Image Cytometer). The viability of SUM185 cells was assessed using the Countess system. EMG3, HCC3153, MFM223, and SUM185 cells expressing doxycycline-inducible WT or mutant PRRX1 were plated in +/− doxycycline conditions. Each condition was plated in triplicates in 24-well plates (EMG3 and HCC3153) or 6-well plates (MFM223 and SUM185) and cultured at 37°C with 5% CO2. Cells in the + doxycycline condition were pretreated with 3ug/ml doxycycline for 30+ days to induce WT or mutant PRRX1 overexpression. Culture medium was replaced every 48–72 h with freshly prepared doxycycline. Cell viability was measured every 24hrs (EMG3 and HCC2153) or 48hrs (MFM223 and SUM185) beginning at 24hrs from the start of the experiment for the duration of nine (MFM223 and SUM185) or ten (EMG3 and HCC3153) days.
Antibodies and inhibitors
Antibodies: For immunoblotting, immunofluorescence and immunohistochemistry were anti-PRRX1 (Sigma, HPA051084), actin (Sigma, A2228), H4K20me3 (Abcam, ab9053), The antibodies used for ChIP were anti-H3K27ac (Diagenode, C15410196) and anti-PRRX1 (Sigma, HPA051084). Full list with catalog numbers available in Table S2. Inhibitors were obtained from Selleckchem: Galunisertib, LY2157299 cat#S2230, Xav939 cat#S1180, LGK-974 cat#S7143, Vismodegib (GDC-0449) cat#S1082, Sonidegib (Erismodegib, NVP-LDE225) cat#S2151, NVP-BHG712 cat#S2202, BGJ398 (NVP-BGJ398) cat#S2183, Vorinostat (SAHA, MK0683) cat#S1047, Tretinoin cat#S1653, MK-8617 cat#S8443, Ruxolitinib, INC018424 cat#S1378, A-1155463 cat#S7800, ML324 cat#S7296, GSK J1 cat#S7581, Verteporfin cat#S1786 and MedChem Express: A-485 cat#HY-107455.
Immunoblotting
Cells were lysed in RIPA buffer. Proteins were resolved in SDS-polyacrylamide gels (4–12%) and transferred to PVDF membranes by using a Tris-glycine buffer system. Membranes were blocked with 5% milk powder in 0.1% Tween 20 in TBS (TBS-T) for 1 h at room temperature, followed by incubation with primary antibodies in 5% milk TBS-T. The membranes were developed with Immobilon substrate (EMD Millipore).
Immunofluorescence staining
After deparaffinization and rehydration, slides were subjected to antigen retrieval in Tris/EDTA buffer (pH9, Dako) for 30 min in a steamer. Endogenous peroxidase was quenched after a 10 min incubation in 3% H2O2 in methanol. Blocking solution (100% goat serum) was applied for 1 h. Incubation with primary antibody 1:100 PRRX1 in PBS with 5% goat serum was held overnight at 4°C in a moist chamber. HRP-conjugated secondary antibody was applied for 1 h at room temperature. Samples were incubated with biotinylated-TSA at 1:50 in diluent solution (Akoya Biosciences) for 10 min before fluorophore-conjugated streptavidin secondary antibody was applied for 1 h at room temperature. Slides were then mounted with VectaShield HardSet Antifade Mounting Medium with DAPI (Vector Laboratories). The Dana-Farber Breast Cancer Tissue Microarray (TMA) consisted of primary untreated TNBC samples from 81 evaluable patients who underwent definitive breast surgery at Brigham and Women’s Hospital. The TMA was stained with H4K20me3 (1:100) antibody and imaged using Nikon microscope. Three images were taken per each core for 240 out of 267 cores, for the remaining 27 one or two images were taken due to tissue loss or low tumor content. The images were then analyzed using QuPath to classify cells as either H4K20me3 positive or negative based on staining intensity. H4K20me3 staining mean intensity was calculated per individual nucleus within an image. The mean intensity per image was normalized to nuclei count. Recurrence-free survival (RFS) was defined as the interval from the date of initial surgical resection to the date of recurrence (local or distant), or date of last known contact if the patient was alive and has not recurred. RFS was estimated using the Kaplan-Meier method, with hazard ratios and 95% confidence intervals from a univariate Cox proportional hazard model.
Immunohistochemistry
After deparaffinization and rehydration, slides were subjected to antigen retrieval in Tris/EDTA buffer (pH9, Dako) for 30 min in a steamer. Endogenous peroxidase was quenched after a 10 min incubation in 3% H2O2 in methanol. Blocking solution (100% goat serum) was applied for 1 h. Incubation with primary antibody 1:100 PRRX1 in PBS with 5% goat serum was held overnight at 4°C in a moist chamber. Biotinylated secondary antibody was applied for 30 min at room temperature. A Vectastain ABC peroxidase kit was applied for 30 min to conjugate secondary antibodies to HRP. The tissue was exposed with DAB (Sigma Aldrich) under a microscope until the signal was observed then the reaction was stopped with water. Samples were counter-stained using hematoxylin (Leica Biosystems, cat# 3801575) and bluing solution and dehydrated. Slides were mounted with Cytoseal 60 (Thermo Scientific).
Small molecule inhibitor screen
Using the multidrop combi microplate dispenser (Thermo Scientific, cat#D01515) 34 TNBC cell lines were seeded in quadruplicates in 50μL volume at a density of 500–2000 cells/well in 384 well plates and left to adhere for 24h. Cells were cultured at 37°C with 5% CO2. An automated liquid handling robot, JANUS (PerkinElmer), was used to deliver 100 nL of molecules from the drug panel (inhibitors listed above under ‘antibodies and inhibitors‘ section) obtained by 96-well pin-tool transfer. After 72 h, ATPlite (PerkinElmer, cat# 6016731) was performed, and luminescence was measured using a plate reader. Data was normalized to baseline (day 0). The area under the viability curve for treatment response (AUC) was calculated for each drug.
High Throughput BH3 profiling
High Throughput BH3 profiling was used to determine the apoptotic priming and anti-apoptotic dependencies of a set of 34 TNBC cell lines, as previously described.40 In brief, TNBC cell lines were seeded at a density of 500–2000 cells/well in 384 well plates and left to adhere for 24h. Then cells were washed 3 times with PBS using the BioTek 406EL plate washer (BioTek). Consequently, different BH3 peptides and BH3 mimetics at different concentrations were added via pin transfer, and cells were incubated in BH3 profiling buffer containing 0.002% digitonin for 1h. Cells were fixed in paraformaldehyde for 15 min. Afterward, the fixative was neutralized using a tris/glycine buffer. Cells were stained overnight with Hoechst33342 (Nuclei, Invitrogen) and anti-cytochrome c-Alexa Fluor 647 antibody (BioLegend). Prior to imaging, the stain solution was washed out using the BioTek 406EL plate washer. Fluorescent microscope images from the BH3 profiling plates were acquired using the IXM XLS high content widefield microscope (Molecular Devices) at the ICCB Longwood Screening Facility. The cytochrome positive cells were quantified using the Multi-Wavelength Cell Scoring module in Metamorph software. Release of cytochrome in response to the BIM and PUMA peptides indicate overall mitochondrial priming. In contrast, the release of cytochrome in response to the BAD, HRK, MS1, FS1 peptides, and BH3 mimetics indicate specific anti-apoptotic dependencies.41 Combinations of BH3 mimetics were used to test co-dependencies. Release of cytochrome in response to the BAD peptide and ABT-263 indicate BCL-2, BCL-XL, or Bcl-w dependency; to the HRK peptide, A-133, and A-115 indicate BCL-XL dependency; to ABT-199 indicates BCL-2 dependency; to the MS1 peptide and S63845 indicate MCL-1 dependency.
Single cell RNA-seq
Single-cell RNA-seq experiments were conducted in two batches. 14 TNBC cell line samples were processed using the 10x v2 kit. Briefly, cells were resuspended to a concentration of 1,000 cells/uL and 2,000 cells were targeted for recovery. 14 TNBC cell line samples were processed with the 10x v3 kit, since at this point the 10x v2 kit was discontinued. Cells were resuspended at a concentration of 1,000 cells/uL and 5,000–6,000 cells were targeted for recovery.
QUANTIFICATION AND STATISTICAL ANALYSIS
In vitro and in vivo data
Data were compiled and shown as mean ± standard error of the mean (SEM). Data were evaluated using unpaired, two-tailed t tests (95% confidence interval) or two-way analyses of variance using GraphPad Prism software version 10.1.0 (GraphPad Inc, San Diego, CA). p-values <0.05 were considered significant.
Breast cancer cohorts
METABRIC:
The “Breast (METABRIC 2016)” dataset was downloaded from www.cbioportal.org in May 2020. Microarray expression data was available for 1,904 samples. Samples annotated as “Negative” for ER_STATUS, PR_STATUS, and HER2_STATUS, from patients annotated as “Negative” for ER_IHC and as “LOSS” or “NEUTRAL” for HER2_SNP6 were considered to be TNBC. Samples that were annotated as “Positive” for ER_STATUS, PR_STATUS or HER2_STATUS, or that came from patients annotated as “Positive” for ER_IHC, or “GAIN” for HER2_SNP5 were classified as non-TNBC. Otherwise, samples were considered of indeterminate TNBC status. 227/1,904 (12%) of samples were classified as TNBC, 1,673/1,904 (88%) were classified as non-TNBC, with the remaining 4/1,904 (0%) considered indeterminate. Expression intensity data was used for downstream analysis, after removing eight genes with at least one missing value in the dataset.
TCGA:
TCGA-BRCA gene expression and DNA methylation data was downloaded from the GDC data portal in October 2021, together with TCGA-BRCA clinical supplements. For expression data, we downloaded FPKM Gene Expression Quantification data for 1,222 samples across 1,092 unique cases. We retained data for the 1,090 cases with at least one primary tumor sample and available clinical supplement. We kept the first primary tumor file for cases where there was more than one. For DNA methylation data, we downloaded Illumina Infinium HumanMethylation450 BeadChip Methylation Beta Value data for 892 samples, representing 789 unique cases. We retained data for 782 cases with at least one primary tumor sample and available clinical supplement. Out of 1,094 retained cases, both DNA methylation and RNA-seq data was available for 778 cases (71%), RNA-seq data alone was available for 312 cases (29%), and DNA methylation data alone was available 4 cases (0%).
Survival information and clinical covariate information for these samples was downloaded from the cBioPortal for Cancer Genomics in May 2020. Annotations for individual cases were obtained from patient-level annotations in the cBioPortal “Breast Invasive Carcinoma Breast (TCGA PanCan 2018)” dataset. Partial or complete annotations were available for 1,080/1,094 (99%) of TCGA-BRCA cases from this data.
TCGA patients were assigned a TNBC status based on the downloaded clinical annotation files. Cases annotated as “Negative” for “breast_carcinoma_estrogen_receptor_status”, “breast_carcinoma_progesterone_receptor_status”, and “lab_proc_her2_neu_immunohistochemistry_receptor_status”, were classified as TNBC. Patients annotated as “Positive” for at least one of these three fields were considered as non-TNBC, otherwise cases were considered indeterminate. 115/1,094 (11%) of cases were called as TNBC and 861/1,094 (79%) were called as non-TNBC, with the remaining 118/1,094 (11%) considered indeterminate. Of the 115 patients called as TNBC, 83 cases (72%) had data for both RNA-seq and methylation, and 32 (28%) had RNA-seq data only.
For RNA-seq data, Ensembl gene names were converted to HGNC symbols based on annotations downloaded from the Ensembl biomart. Genes with no matching HGNC symbol were excluded from downstream analysis. Where a gene mapped to more than one HGNC symbol, the first symbol was used. Where more than one gene mapped to the same HGNC symbol, expression data from the first gene was assigned to the gene symbol. The transformation was applied to the filtered FPKM values to obtain the -normalized expression matrix for downstream analysis.
For DNA methylation data, downloaded beta-values were converted to M-values based on the following formula39:
Loci overlapping SNP’s at the CpG site or single-base extension site were identified using the ‘SNPs.137CommonSingle’ SNP annotation from the R package “IlluminaHumanMethylation450kanno.ilmn12” and were excluded from the downstream analysis, as were CpG loci annotated as cross-reactive42,43 and loci with any missing values across the 782 retained samples. Loci were assigned to H3K27ac peaks and super-enhancers, defined based on unperturbed TNBC cell lines, based on overlap with the genomic coordinates of those regions. Loci were assigned to gene bodies and promoters based on the annotation from the R package IlluminaHumanMethylation450kanno.ilmn12.hg19. Gene body loci for each gene were defined as those loci annotated as “Body” or “3 ‘UTR” in the array annotation. Promoter loci for each gene were defined as those loci annotated as “TSS1500”, “TSS200”, “5’UTR”, or “1stExon”. Loci were assigned to the non-genic regions of super-enhancers if they fell within a super-enhancer region and were not assigned to the gene body, or promoter of any gene. Methylation levels in genomic regions of interest were quantified as the average M-value among loci in the region.
Additional cell line data
Additional RNA-seq read count data from breast cancer, neuroblastoma, and rhabdoid tumor cell lines was obtained from.44
H3K27ac ChIP-seq analysis
Cell line and PDX H3K27ac ChIP-seq:
Reads were aligned with BWA-mem v0.7.17 to hg19, and duplicates were removed with Picard MarkDuplicates v2.18.17 (REMOVE_DUPLICATES=TRUE, VALIDATION_STRINGENCY=LENIENT). Peaks for each cell line were called using MACS2 v2.1.2 (–SPMR, –B, –keep-dup=1, –extsize=146, –nomodel, –q 0.05), and super-enhancers were called for each cell line using ROSE (–t 2500) using both ranking and control bam files.6,7 Consensus super-enhancers were defined by taking the union of super-enhancers across all cell lines and were considered present in a cell line if they overlapped a super-enhancer called in that cell line. A similar approach was taken to define consensus peaks and determine their presence across cell lines. bedtools intersect v2.27.1 was used to quantify the number of reads overlapping consensus super-enhancers and peaks in each cell line and PDX sample. Read per kilobase of transcript per million mapped reads (RPKM) values for individual regions were calculated using the formula:
where is the read count for sample in region , is the length of region in base pairs, and is the total mapped reads for sample .
-normalized H3K27ac values were obtained for each sample using the following formula:
Consensus super-enhancers on canonical nuclear chromosomes were assigned to the nearest gene based on linear genomic distance.
PRRX1 over-expression experiment:
H3K27ac data was pre-processed as described above (cell line and PDX H3K27ac ChIP-seq). bedtools intersect v2.27.1 was used to quantify the number of reads overlapping consensus peak and super-enhancer regions, defined based on unperturbed cell lines, in each sample. RPKM and -normalized expression values were calculated as described above.
RNA-seq analysis
Cell lines:
RNA-seq data was aligned and preprocessed using the VIPER pipeline.45 -normalized expression values were derived from per gene FPKM values output by VIPER using the following formula:
where is the FPKM value for gene in sample .
PDXs:
RNA-seq data was initially aligned to both human hg19 and mouse mm10 genomes. Two-pass mapping was performed using the STAR RNA-seq aligner version STAR v2.5.1b (–outSAMDstrandField intronMotif, –outFilterMultimapNmax 20, – alignSJoverhangMin 8, –alignSJDBoverhangMin 1, –outFilterMismatchNmax 999, – outFilterMismatchNoverLmax 0.1, –alignIntronMin 20, –alignIntronMax 1000000, – alignMatesGapMax 1000000, -–outFilterType BySJout, –outFilterScoreMinOverLread 0.33, – outFilter-MatchNminOverLread 0.33, –limitSjdbInsertNsj 1200000, –chimSegmentMin 15, – chimJunctionOverhangMin 15, –twopassMode Basic). Reads uniquely mapped only to the hg19 genome were kept, along with uniquely mapped reads that had significantly better alignment scores in the h19 genome compared to the mm10 genome. Filtered reads were then aligned and preprocessed using the VIPER pipeline,45 and quantification was performed as described above.
PRRX1 5 day knockdown experiment:
We performed RNA-seq on 16 samples (doxycycline treated and untreated samples for each of shRNAs 1–3 and a non-targeting control for each of Hs578T and TTC642). RNA-seq data was preprocessed and quantified as described above (Cell lines).
PRRX1 28 and 56 day knockdown experiment:
The experiment comprised 24 samples (doxycycline treated and untreated samples for each of shRNAs 1 and 3 and a non-targeting control for each of two time points for each of Hs578T and TTC642). However, library preparation failed for shRNA1 for the Hs578T 8 week time point. As a result, pre-processing and downstream analysis was based on 22 samples. RNA-seq data was preprocessed and quantified as described above (Cell lines).
PRRX1 over-expression experiment:
We performed RNA-seq on 48 samples (doxycycline treated and untreated samples for each of SUM185, EMG3, MFM223, and HCC3153 for each of wt and dh3 PRRX1 for each of three time points). RNA-seq data was preprocessed and quantified as described above (Cell lines).
DNA methylation analysis
Cell lines/PDXs:
The Infinium HumanMethylation450 Beadchip (Illumina, WG-314-1003) 450,000 CpG site platform was used to generate comprehensive genome-wide profiling of DNA methylation. The cell line and PDX datasets were preprocessed using the function preprocessIllumina from the R package minfi (v.1.34.0),46 using the first annotated TNBC cell line as a common reference. Probes with detection p-value >0.01 in any sample were removed from each dataset, as were probes overlapping an annotated SNP at the CpG site, or CpG probes annotated as cross-reactive.42,43 Normalized methylation M-values for each locus in each sample were obtained using the following formula39:
Methylation levels in genomic regions of interest were quantified as the average M-value among loci in the region, as described above for TCGA data.
Mass spectrometry analysis of histone modifications
Histones were isolated from cell nuclei using acid extraction, biochemically prepared, and analyzed by mass spectrometry against a reference of stable isotope-labeled synthetic peptide standards as described.37 The experiment was performed for 35 cell lines, the 34 TNBC cell lines used in our study, and one additional cell line. Two replicate samples were analyzed for each of the 34 TNBC cell lines used for downstream analysis, with the exception of HDQP1, for which one replicate failed during preprocessing. The ratio of the intensity of each endogenous peptide to the intensity of the internal standard was calculated and normalized to the respective ratio of a mass balance peptide (termed NORM peptide). H3 and H4 marks were normalized to the H3 NORM (41–49) and H4 NORM (68–78) peptides respectively. Ratios were transformed, row- (histone mark-) median normalized, and averaged across replicates within each cell line to obtain the final -normalized peptide values. Histone marks that had missing values in any of the replicate samples for any of the 34 retained cell lines were removed from the dataset, and downstream analysis was based on the remaining 59/63 histone marks.
Metabolomics analysis
Metabolomics profiling was performed in two batches (MB1 and MB2) with three replicates performed for each cell line in each batch. 34 TNBC cell lines were profiled in total; 17 cell lines were profiled in MB1 and 18 were profiled in MB2, with one cell line, HCC1143, profiled in both batches. Metabolites that had missing values in any sample were removed from the dataset, and downstream analysis was based on the remaining 228/302 metabolites. To correct for potential batch effects, we used an approach based on.47 For each metabolite, the ratio between the average raw peak area in each batch and the overall average for the metabolite was computed. The metabolite raw peak area values for each replicate were then divided by the ratio for the relevant batch to obtain batch-corrected peak areas. The batch-corrected peak area values were transformed. To correct for potential differences in cell line biomass that could lead to systematic differences in measured metabolite abundance we applied the two-step procedure described in47: An additive transformation was applied to each row (metabolite) to equalize the median value across metabolites, and subsequently an additive transformation was applied to each column (sample) to equalize the median value across samples. Processed values were averaged across replicates corresponding to the same cell line within each batch to obtain -normalized metabolite abundance values.
Drug screen data
The sensitivity of each cell line to each drug was quantified as one minus the area under the viability curve for treatment response (AUC) for the drug in the cell line.
PRRX1 ChIP-seq analysis
Cell line PRRX1 transcription factor ChIP-seq:
Cell line PRRX1 ChIP-seq experiments were performed 2–3 times in each cell line under 2–3 different sonication conditions. Downstream analysis was based on results from the two sonication conditions common to all cell lines. Reads were aligned and de-duplicated, and peaks were called as described above for H3K27ac ChIP-seq. For TTC642, one of the two input samples failed the “Per base sequence module” of the fastqc tool (www.bioinformatics.babraham.ac.uk/projects/fastqc/). In this case the passing input was used to call peaks in both replicate samples. Consensus PRRX1 peaks were defined by taking the union of PRRX1 peaks across all cell lines and sonication conditions. Consensus peaks were considered present in a cell line if they overlapped a peak called in that cell line in either sonication condition.
Hierarchical clustering of cell lines
Datasets were mean-centered for each feature prior to clustering. Clustering was performed based on -normalized data for the RNA-seq, H3K27ac ChIP-seq, histone mass spectrometry, and metabolomics datasets. For the DNA methylation, BH3 profiling and drug screen datasets clustering was based on M-values, peptide abundance values, and sensitivity values respectively. Features with zero variance across cell lines were removed from each dataset as an initial filtering step prior to selecting highly variable features. For the sequencing-based datasets, RNA-seq and ChIP-seq, we performed an additional initial filtering step, where we removed features which had FPKM >1 or RPKM >1 in fewer than two samples, respectively. The R function “hclust” with the “ward.D2” method was used to cluster both rows (features), and cell lines (columns) based on Euclidean distances.
Transcriptomic heterogeneity estimation using bulk RNA-seq
Shannon’s equitability for each sample was calculated using RNA-seq FPKM values using the following formula:
where , , and is the number of genes measured in the dataset. For the purposes of this calculation, we defined 0 log 0 to be equal to 0.
Analysis of correlations between gene expression, H3K27ac expression and DNA methylation across genes
To correlate gene expression with H3K27ac ChIP-seq expression in super-enhancers, genes were assigned to super-enhancers by linear genomic distance. Each gene was matched with the nearest super-enhancer, by linear genomic distance to the gene TSS, among all super-enhancers annotated to that gene, if any. Correlations were calculated based on -normalized gene expression data for mRNA data, average M-values for DNA methylation data, and -normalized H3K27ac expression for H3K27ac ChIP-seq data.
Differential H3K27ac and gene expression analysis
Differential H3K27ac analysis was performed based on count data using DESeq2,48 after filtering to remove regions with zero counts across all samples. The ratio of the total mapped reads in each sample to the average value of the total mapped reads across samples was used as the size factor for each sample, as described.49 Differential expression analysis was performed using DESeq2 (v.1.30.1)48 based on count data output by VIPER, after filtering to remove genes with zero counts across all samples. Regions or genes with adjusted p values <0.05 were considered differentially expressed.
MOFA data integration analysis
TNBC cell lines original model:
The R package MOFA2 (v.1.1.6)10,11 was used to fit a multi-omics factor analysis (MOFA) model to infer latent biological factors active in TNBC cell lines. The MOFA model was fit using RNA-seq data (-normalized FPKM values), DNA methylation data (average M-values), metabolomics data (-normalized metabolite abundance), histone mass spectrometry data (-normalized peptide values), and H3K27ac ChIP-seq data (-normalized RPKM values). The MOFA model assumes constant residual variance for a given feature across samples. To satisfy this assumption, for this analysis we removed the HDQP1 sample from the histone mass spectrometry dataset which, unlike the other samples in this dataset, was based on only a single replicate. The MOFA model additionally assumes uncorrelated residuals across samples. To satisfy this assumption, for this analysis we also excluded the second (MB2) replicate of the duplicated HCC1143 cell line from the metabolomics dataset. Features with zero variance across cell lines were removed from each dataset as an initial preprocessing step. And for the RNA-seq and ChIP-seq datasets we additionally removed features with FPKM <1 in fewer than two samples and RPKM <1 in fewer than two samples, respectively. Analysis was based on the top 5,000 most variable features in each dataset after filtering, or all features, where the top 5,000 features accounted for 90% or more of the remaining features. We fit a model to infer 8 factors, based on the software recommendation that the number of factors should not exceed ~8 given the number of samples. The model was fit with convergence mode set to “slow”. The software automatically inferred Gaussian likelihood models for all datasets (out of the three options Guassian, Poisson, and Bernoulli). The ontology (C5) gene set was downloaded from the Molecular Signatures Database50,51 in February 2021. Feature set enrichment analysis was run using the MOFA2 function run_enrichment.
Given the setup of the MOFA model, some factors can explain very low, but non-zero, proportions of variance in individual datasets. It can therefore be necessary to choose a threshold of variance explained which is considered non-negligible for the purposes of interpretation. Here we chose to use a threshold of 2%, in keeping with the threshold used in the original MOFA study to prune factors from the model ([PMID: 29925568]).
PDX validation model:
The MOFA model was fit using PDX RNA-seq data (-normalized FPKM values), DNA methylation data (average M-values), and H3K27ac ChIP-seq data (-normalized RPKM values). The datasets were filtered to retain only common features that were used in the original TNBC cell line model. We fit a model (convergence mode “slow”, inferred likelihood models Gaussian) to infer 3 factors, based on the software recommendation that the number of factors should not exceed ~3 given the number of samples.
TCGA validation model:
The MOFA model was fit using TCGA RNA-seq data (-normalized FPKM values), and DNA methylation data (average M-values). As for the PDX validation model, the datasets were subset to common features present in the original cell line model. We fit a model (convergence mode “slow”, inferred likelihood models Gaussian) to infer 8 factors, to match the number of factors used in the original cell line model.
MOFA scaled weights
Scaled weights were obtained from the original fitted weights for each factor in each dataset by dividing by the largest absolute value among all weights for that factor and dataset. As a result, the largest absolute value among the rescaled weights for a factor in a dataset is equal to −1 or 1, depending on whether the original fitted weight with the largest absolute value is negative or positive.
Signature-based assignment of patient samples to TNBC types
We carried out the following procedure separately for the METABRIC and TCGA cohorts: For each of the three TNBC types, we overlapped the positively and negatively differentially expressed genes from the TNBC cell line RNA-seq data with genes present in the primary patient data, and determined, n_min_overlap, the minimum size of the six sets of overlapping genes. We defined positive and negative signature genes for each TNBC type as the top n_min_overlap positively and negatively differentially expressed genes, respectively, that were also present in the primary patient data. For each sample, we calculated a signature score for each TNBC subtype as the difference between the average expression of the positive signature genes and the average expression of the negative signature genes of that type after mean-centering the expression data for each gene. Patient samples were then assigned to the TNBC type with the highest corresponding signature score.
MOFA factor scores for patient samples
For METABRIC data, multiple linear regression based on mRNA feature weights for genes with data available in the METABRIC cohort, was used to calculate scores for MOFA Factors 1 to 8, after mean-centering the METABRIC expression data for these genes. For TCGA data, multiple linear regression based on mRNA and DNA methylation features available in the TCGA cohort was used to calculate scores for MOFA Factors 1 to 8, after mean-centering the data for these features.
Alternate clustering-based assignment of METABRIC patient samples to TNBC types
We used the function Mclust from the R package ‘mclust’ (v.5.4.6), with default parameters, to cluster samples according to Factor 2, 3, and 6 scores. For each of the resulting clusters, we calculated the average Factor 2, 3, and 6 scores across samples assigned to that cluster, and identified the factor with the highest average score for the cluster. Samples were classified as basal, luminal, or mesenchymal if they were assigned to a cluster for which the factor with the highest average score was Factor 6, 2, or 3, respectively.
Classification of mesenchymal tumors into mesenchymal-high and mesenchymal-low groups
Samples considered mesenchymal using signature-based assignment were classified as mesenchymal-high if the Factor 3 score for the sample was higher than the maximum factor 3 score among basal and luminal samples in the same cohort, and were classified as mesenchymal-low otherwise. For the METABRIC cohort, we also considered an alternate clustering-based approach to define the mesenchymal-high and mesenchymal-low groups. Here, we used the function Mclust from the R package ‘mclust’ (v.5.4.6), with default parameters, to cluster samples according to Factor 3 scores. Samples considered mesenchymal using our signature-based approach were classified as mesenchymal-high if they were assigned to the cluster with the highest average factor 3 score, and were classified as mesenchymal-low otherwise.
CIBERSORTx analysis
The CIBERSORTx web portal (https://cibersortx.stanford.edu/) was used to run CIBERSORTx in “Impute Cell Fractions” mode. The tool was run using the provided LM22 signature matrix in conjunction with the TCGA TNBC FPKM data. “B-mode” batch correction was used based on the LM22 source GEP file, with quantile normalization disabled. 100 permutations were used for significance testing.
Survival analysis
Survival analysis was performed using the ‘survival’ R package. Kaplan-Meier plots were made using the ‘survminer’ (v.0.4.9) R package. For this analysis, signature scores for each sample for each putative PRRX1 target set were defined by subtracting the average expression of putative negative targets from the average expression of putative positive targets after mean-centering the expression data for each gene.
Definition of transcription factors
Genes were considered to be transcription factors if they were listed as such in the list of human transcription factors from,52 which was downloaded from http://humantfs.ccbr.utoronto.ca/download.php in October 2022.
Hierarchical clustering of cell lines by expression of subtype-specific transcription factors
Clustering of genes and samples was performed based on -normalized expression data, after mean-centering per gene, as described above (See hierarchical clustering of cell lines).
RNA-seq principal component analysis
A combined RNA-seq count matrix was formed for the combined cohort of TNBC, non-TNBC breast cancer, rhabdoid, and neuroblastoma cell lines from this study and44 including all 18,677 genes measured in both datasets. For 38 genes in common between the two datasets which were annotated to more than one row in the44 count matrix, the first expression row was retained. Only genes with a total count of at least 10 were considered for downstream analysis. DESeq248 was used to obtain size factor-corrected RNA-seq counts for the combined dataset and -normalized count values were then obtained using the formula:
where is the DESeq2 size factor-corrected count value for gene in sample
Principal component analysis was performed on the -normalized count values for the top 20% most variable genes.
Hierarchical clustering of PRRX1 over-expression RNA-seq samples
Data was gene mean-centered prior to clustering. Clustering was performed based on -normalized gene expression values. Features with zero variance across samples and features which had FPKM>1 in fewer than two samples were removed as initial filtering steps, prior to selecting highly variable features. The R function “hclust” with the “ward.D2” method was used to cluster both rows (features), and cell lines (columns) based on Euclidean distances.
Immune signature gene set enrichment analysis in PRRX1 over-expression RNA-seq data
RNA-seq count data was filtered to remove genes with total counts <2 in the conditions under consideration. Counts were normalized using DESeq2 size factors.48 GSEA analysis was run using the GSEA v4.1.0 command line tool.53 Gene set permutation was used in conjunction with 5% false discovery rate significance threshold, in line with the software recommendations for datasets with fewer than 7 samples. Results for each cell line were based on contrasting three doxycycline treated samples (corresponding to the three measured time points) with three untreated samples (corresponding to the same three time points). Immune gene signatures with 15 or fewer genes remaining in the filtered data for a cell line were excluded from testing in that cell line.
Assessment of PRRX1 expression levels in cell lines used for PRRX1 ChIP-seq
Log-normalized count values for PRRX1 were calculated as described above (See RNA-seq principal component analysis).
PRRX1 targets
Consensus PRRX1 peaks on canonical nuclear chromosomes were assigned to the nearest gene based on linear genomic distance. The Hs578T and Mes target sets were defined as genes assigned to consensus peaks that were called present in Hs578T and all mesenchymal cell lines (Hs578T, MDAMB157, and MDAMB436), respectively. The Hs578T-RNA and Mes-RNA target sets were defined by intersecting the Hs578T and Mes target sets with genes differentially expressed on PRRX1 knock-down in the Hs578T cell line at the 5 day time point. Genes that down went with PRRX1 knock-down were used to define positive targets and genes that went up with PRRX1 knock-down were used to define negative targets. BETA27 analysis was run with BETA plus v1.0.7.
ChIP-seq heatmaps
Read per million (RPM) normalized BedGraph signal track files generated by MACS2 were converted to BigWig files using bedGraph-ToBigWig v4 and Deeptools v3.3.254,55 was used to plot the heatmaps based on bigwig files.
PRRX1 target signature scores and immune signature scores for patient samples
PRRX1 target and immune signature scores were calculated as the difference between the average expression of positive signature genes and the average expression of negative signature genes after mean-centering the expression data for each gene within each patient cohort.
MOFA factor scores for PRRX1 over-expression samples
Log-normalized super-enhancer H3K27ac values for the combined cohort of PRRX1 over-expression samples, corresponding control samples, and untreated cell line samples were mean-centered by super-enhancer. Scores for MOFA Factors 1 to 8 were then calculated for each sample using multiple linear regression based on super-enhancer weights from the MOFA cell line model.
Hierarchical clustering of PRRX1 over-expression H3K27ac samples
H3K27ac super-enhancer values were mean-centered prior to clustering. Clustering was performed based on -normalized H3K27ac signal. Features with zero variance across samples and features which had RPKM>1 in fewer than two samples were removed as initial filtering steps, prior to selecting highly variable features. The R function “hclust” with the “ward.D2” method was used to cluster both rows (features), and cell lines (columns) based on Euclidean distances.
Hierarchical clustering of PRRX1 over-expression H3K27ac based on subtype-specific transcription factors
Clustering of genes and samples was performed based on -normalized H3K27ac data, after mean-centering per gene, as described above (See hierarchical clustering of cell lines).
Identification and analysis of PRRX1 putative co-binding TF’s
HOCOMOCO v11 core collection human transcription factor binding model matrices in HOMER format (p-value = 0.0001) were downloaded from (https://hocomoco11.autosome.org/downloads_v11) in November 2022, and were filtered to remove quality C motifs. Peak and super-enhancer regions that significantly gained or lost H3K27ac in HCC3153 under long-term wt and dh3 PRRX1 over-expression were overlapped to identify wild-type unique and common (wild-type and dh3) significantly gained and lost peaks and super-enhancers. Differential motif analysis was performed using the HOMER v3.12 script56 findMotifsGenome.pl (-size given –h –mknown) in conjunction with the filtered HOCOMOCO motif matrices to identify motifs enriched in common significantly gained (resp. lost) peaks within common significantly gained (resp. lost) super-enhancers compared to wild-type unique significantly gained (resp. lost) peaks within wild-type unique significantly gained (resp. lost) super-enhancers. The transcription factors corresponding to the top 10 significantly enriched (Q < 0.05) motifs found for the gained and lost groups were identified and a two-sided one-sample Mann Whitney U test was performed based on the difference in -normalized gene expression for these factors between SUM185 and HCC3153, after removing factors with zero expression in both cell lines.
CyTOF analysis
CyTOF profiling was performed in two batches (CY1 and CY2). 34 TNBC cell lines were profiled in total; 18 cell lines were profiled in CY1 and 19 were profiled in MB2, with three cell lines (CAL120, CAL51, and HCC38) profiled in both batches.
CyTOF clustering analysis
Clustering analysis of CyTOF data was performed using VorteX clustering environment (v.26).17 The CyTOF dataset was read in to VorteX using default parameter settings (arcsinh (x/5) transformation, and a maximum of 1,000 rows imported per.fcs file, to limit the size of the dataset).
Clustering was performed using the X-shift algorithm with Distance Measure “Euclidean”. K = 25 was selected by the software as the optimal number of neighbors used for density estimation, leading to 36 clusters. Visualization was performed in VorteX, using a minimum spanning tree reconstructed using Euclidean distance.
scRNA-seq analysis
Cell lines:
scRNA-seq samples were sequenced in two batches, as described above, one using the 10x v2 chemistry, the other using the 10x v3 chemistry. In addition, 10x v2 chemistry sequencing data for two additional cell lines, SUM149 and SUM159, was obtained from.57 Cell Ranger (v.3.1.0) mkfastq was used to create fastq files. UMI count matrices were obtained from fastq files using Cell Ranger count with expect-cells set to 2,000 for v2 samples and set to 5,500 for v3 samples. Four samples from the v2 sequencing batch were excluded from downstream analysis due to Cell Ranger warnings (two samples), or aberrantly high cell numbers (two samples where detected cell numbers were >2.9x the targeted cell numbers).
We applied a two-step filtering procedure within each sample to remove cells with high mitochondrial content and cells with low numbers of detected genes. First, we calculated the proportion of UMI counts from mitochondrial genes for each cell (mitochondrial_read_proportion). The function isOutlier from the R package “scater” (v.1.18.6) was used to remove cells for which the base 2 logarithm of the mitochondrial_read_proportion was greater than 4 median absolute deviations (MADs) above the median value when considering cells with mitochondrial read proportion less than 50%. Secondly, among the remaining cells, we removed cells where the base 2 logarithm of the number of detected genes was less than 4 MADs below the median.
For analyses within individual cell lines, we considered all genes detected in 10 or more cells. For each gene , in each cell , we calculated counts per 10,000, , by dividing the sample-count for the gene by the total counts across all genes and multiplying by 10,000. Log-normalized counts were then calculated as the natural logarithm of one plus the counts per 10,000 values. Clustering and UMAP visualization for individual samples was performed using Seurat (v.3.2.3), using the Seurat standard -normalization workflow (v. 3).58 The top 20 principal components were used for all cell lines. The resolution parameter was set to 0.5 to run clustering with the Seurat function FindClusters. Single cell cluster differentially expressed genes were identified using the Seurat function FindMarkers with default parameters.
For analyses involving all samples, downstream analysis was based on genes detected in at least 10 cells across all samples. Counts per 10,000 were calculated as described above. We took steps to try to remove potential artifacts due to differences in chemistry, and, where possible, differences due to batch, using four cell lines that were represented in both batches. We found the minimum number of cells among the eight samples corresponding to these four cell lines and selected a random subset of this size, without replacement, from each of the eight samples. The data were initially -normalized, as described above. We then used the function rescaleBatches from the R package ‘batchelor’ (v.1.6.3)59 to estimate a scaling difference for each gene based on the replicated cell subset, and to correct the -normalized data for v2-sequenced and v3-sequenced cells accordingly. No correction was applied for genes with zero UMI counts in cells from the replicate cell lines in either batch. Variable features were identified separately for v2 and v3 cells based on uncorrected -normalized data using the Seurat function FindVariableFeatures. These features were combined using the Seurat function SelectIntegrationFeatures to give a combined set of variable features for the dataset. The top 25 principal components were used for UMAP visualization.
MOFA factor scores for scRNA-seq data
Log-normalized expression data were mean-centered across all single cells from all samples for each gene. Multiple linear regression based on weights for all available mRNA features used in the cell line model was then used to calculate MOFA factor scores for Factors 1–8 for each single cell.
Clustering of single cells by MOFA factor scores
The function Mclust from the R package ‘mclust’ (v.5.4.6) was used to explore evidence for multiple clusters within scRNA-seq samples. We used the function to fit “VVV” models (ellipsoidal, varying volume, shape, and orientation) with between one and five clusters for each single-cell sample, and to extract the log likelihood for each fit. MOFA factors scores for Factors 2, 3, and 6 were used for clustering.
TNBC subtype signature analysis in scRNA-seq data
Statistical testing of signature enrichment in all samples:
For this analysis, data from each sample in the combined dataset containing all samples was down-sampled to the size of the smallest sample (447 cells). Equally-sized sets of positive and negative signature genes for each TNBC type were defined, as described above for bulk data. For each retained single cell, we first centered gene expression for each gene across all retained single cells from all samples. We then calculated a signature statistic as the difference between the centered -normalized expression of positive signature genes, and the centered -normalized expression of negative signature genes in the single cell. The significance of the expression statistic for each type was estimated using a previously described bootstrap procedure.57 Briefly, a bootstrap p value for each type was calculated by comparing the observed value to the same statistic for 1,000 size-matched sets of random positive and negative signature genes in the same cell.
TNBC subtype signature enrichment scores in HDQP1 single cells:
Equally-sized sets of positive and negative signature genes for each TNBC type were defined as described above. Log-normalized expression data for HDQP1 single-cells was centered across all HDQP1 cells. Statistics for each subtype signature in each HDQP1 cell were calculated as described above. The enrichment score for each signature in each cell was calculated as the average difference between the observed statistic and the same statistic for 1,000 size-matched sets of random positive and negative signature genes in the same cell.
Transcriptomic heterogeneity estimation using single-cell data
For each cell , we estimated the cell’s size factor using the fomula:
where is the count for gene in , and the sum runs over all genes measured in .
For each gene , in each sample, , we estimated the average expression level of in using the formula:
where is the total number of cells in , and the sum runs over all cells in .
Analysis was based on the 378 consistently highly expressed genes with in all samples, after excluding ribosomal proteins (gene names beginning RP). We used an approach based on that taken in60 to estimate the raw biological variance of each gene in each cell line. We estimated the biological variance for gene in sample , using the formula:
where is the sample variance of in , and is a bias correction factor given by:
We estimated the raw squared coefficient of variation (SCV) for in sample , using the formula:
A linear mixed effect model, implemented in the R package lme4 (v.1.1–26), was used to model the relationship between raw biological variance, average expression level, and TNBC type, across samples. To satisfy the model independence assumptions we excluded the replicate with lower depth for each of the four replicated cell lines, which in each case was the v3-sequenced replicate. The analysis was based on 330/378 highly expressed genes with in all samples considered. Exploratory analysis of the data suggested a model where the logarithm of biological variance is linearly related to the logarithm of average expression level for each gene (consistent with constant per-gene raw SCV), with gene-specific intercepts. Based on this analysis we chose to investigate the following model for :
where and are random intercepts corresponding to the gene and sample, respectively, and is a normally distributed random error term. Confidence intervals for the fixed effects were obtained using the R function confint.
Transcriptomic heterogeneity estimation simulations
We confirmed that the average absolute bias of , , and across genes was small under relevant parameter values using a simulation study. For each sample, we created 1,000 simulated UMI datasets for the 378 genes used in this analysis, with ground truth cell size factors, average gene expression levels, and raw gene SCV values equal to the estimated values for the cell line. The counts for each gene and cell in each dataset were simulated using a negative binomial distribution,61,62 with parameters , . Similar to,60 differences between the ground truth size factor and estimated size factors were ignored. The , , and values were estimated for each gene in each dataset as described above. The estimation bias for each parameter for each gene in each sample was estimated as the average difference between the estimate and the true value across simulated datasets.
Supplementary Material
Highlights.
Major transcriptional and super-enhancer subtypes of triple-negative breast cancer (TNBC)
Multiomics factor analysis of TNBC epigenetic, functional, and clinical heterogeneity
PRRX1 is a main transcriptional regulator of mesenchymal TNBC subtype
ACKNOWLEDGMENTS
We thank members of our laboratories for critical reading of the manuscript and discussions. We thank Rani George for providing the PRRX1 expression constructs. We thank the Dana-Farber Cancer Institute Molecular Biology Core Facility for outstanding sequencing service. This research was supported by Department of Defense Breast Cancer Research Program W81XWH-18-1-0027 (to B.J.); National Cancer Institute PSOC U54 CA193461 (to F.M. and K.P.), R35 CA197623 (to K.P.), P01 CA250959 (to K.P., M.B., P.S., H.L., and D.D.), and R01CA251599 (to K.W.W.); DF/HCC SPORE P50CA168504 (to K.P., P.S., and D.D.), and the Ludwig Center at Harvard (to K.P., F.M., and M.B). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health/NCI.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.113564.
DECLARATION OF INTERESTS
The following authors report current employment: Eli Lilly (B.J.), Shasqi, Inc (M.A.), GenieUsGenomics (A.T.), Morrison & Foerster LLP (A.G.), AstraZeneca (M.B.E. and L.E.S.), Odyssey Therapeutics (J.D.J.). K.P. serves on the Scientific Advisory Boards (SABs) of Novartis, Ideaya Biosciences, and Scorpion Therapeutics; holds equity options in Scorpion Therapeutics and Ideaya Biosciences; and receives sponsored research funding from Novartis, where she consults. F.M. is a cofounder of and has equity in Harbinger Health, has equity in Zephyr AI, and consults for Harbinger Health and Zephyr AI. She is on the board of directors of Exscientia Plc. She declares that none of these relationships are directly or indirectly related to the content of this manuscript. P.S. is a consultant for Novartis, Genovis, Guidepoint, The Planning Shop, ORIC Pharmaceuticals, Cedilla Therapeutics, Syros Pharmaceuticals, Blueprint Medicines, Curie Bio, Differentiated Therapeutics, Excientia, Ligature Therapeutics, Merck, Redesign Science, Sibylla Biotech, and Exo Therapeutics; he receives research funding from Novartis. A.G.L. serves on the SAB of Flash Therapeutics, Zentalis Pharmaceuticals, and Trueline Therapeutics and consults for AbbVie. M.B. receives research funding from Novartis, where he also serves on the SAB and acts as a consultant. He is a member of the SAB for Kronos Bio and GV20 Therapeutics and holds equity in both companies. He also serves on the SAB for FibroGen and is a consultant for Belharra Therapeutics. K.W.W. serves on the SAB of TScan Therapeutics, SQZ Biotech, Bisou Bioscience Company, DEM BioPharma, and Nextechinvest; receives sponsored research funding from Novartis; and is a co-founder, stockholder, and advisory board member of Immunitas Therapeutics. D.D. receives research support from Canon, Inc. H.W.L. receives research funding from Novartis.
REFERENCES
- 1.Garrido-Castro AC, Lin NU, and Polyak K (2019). Insights into Molecular Classifications of Triple-Negative Breast Cancer: Improving Patient Selection for Treatment. Cancer Discov. 9, 176–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, and Pietenpol JA (2011). Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest 121, 2750–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehmann BD, Jovanović B, Chen X, Estrada MV, Johnson KN, Shyr Y, Moses HL, Sanders ME, and Pietenpol JA (2016). Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS One 11, e0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lehmann BD, Colaprico A, Silva TC, Chen J, An H, Ban Y, Huang H, Wang L, James JL, Balko JM, et al. (2021). Multi-omics analysis identifies therapeutic vulnerabilities in triple-negative breast cancer subtypes. Nat. Commun 12, 6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su Y, Subedee A, Bloushtain-Qimron N, Savova V, Krzystanek M, Li L, Marusyk A, Tabassum DP, Zak A, Flacker MJ, et al. (2015). Somatic Cell Fusions Reveal Extensive Heterogeneity in Basal-like Breast Cancer. Cell Rep. 11, 1549–1563. [DOI] [PubMed] [Google Scholar]
- 6.Lovdn J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, and Young RA (2013). Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, and Young RA (2013). Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Network (2012). Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. (2012). The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Argelaguet R, Velten B, Arnol D, Dietrich S, Zenz T, Marioni JC, Buettner F, Huber W, and Stegle O (2018). Multi-Omics Factor Analysis-a framework for unsupervised integration of multi-omics data sets. Mol. Syst. Biol 14, e8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Argelaguet R, Arnol D, Bredikhin D, Deloro Y, Velten B, Marioni JC, and Stegle O (2020). MOFA+: a statistical framework for comprehensive integration of multi-modal single-cell data. Genome Biol. 21, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nikolsky Y, Ekins S, Nikolskaya T, and Bugrim A (2005). A novel method for generation of signature networks as biomarkers from complex high throughput data. Toxicol. Lett 158, 20–29. [DOI] [PubMed] [Google Scholar]
- 13.Sengupta S, and George RE(2017). Super-Enhancer-Driven Transcriptional Dependencies in Cancer. Trends Cancer 3, 269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikolsky Y, Nikolskaya T, and Bugrim A (2005). Biological networks and analysis of experimental data in drug discovery. Drug Discov. Today 10, 653–662. [DOI] [PubMed] [Google Scholar]
- 15.van de Wetering M, Barker N, Harkes IC, van der Heyden M, Dijk NJ, Hollestelle A, Klijn JG, Clevers H, and Schutte M (2001). Mutant E-cadherin breast cancer cells do not display constitutive Wnt signaling. Cancer Res. 61, 278–284. [PubMed] [Google Scholar]
- 16.Bendall SC, Simonds EF, Qiu P, Amir E.a.D, Krutzik PO, Finck R, Bruggner RV, Melamed R, rejo A, Ornatsky OI, et al. (2011). Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samusik N, Good Z, Spitzer MH, Davis KL, and Nolan GP (2016). Automated mapping of phenotype space with single-cell data. Nat. Methods 13, 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, and Alizadeh AA (2015). Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12,453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross SE, McCord AE, Jung C, Atan D, Mok SI, Hemberg M, Kim TK, Salogiannis J, Hu L, Cohen S, et al. (2012). Bhlhb5 and Prdm8 form a repressor complex involved in neuronal circuit assembly. Neuron 73, 292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khaled WT, Choon Lee S, Stingl J, Chen X, Raza Ali H, Rueda OM, Hadi F, Wang J, Yu Y, Chin SF, et al. (2015). BCL11A is a triple-negative breast cancer gene with critical functions in stem and progenitor cells. Nat. Commun 6, 5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Euhus DM, Timmons CF, and Tomlinson GE (2002). ETV6-NTRK3–Trk-ing the primary event in human secretory breast cancer. Cancer Cell 2, 347–348. [DOI] [PubMed] [Google Scholar]
- 22.Snel B, Lehmann G, Bork P, and Huynen MA (2000). STRING: a webserver to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 28, 3442–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jozwik KM, and Carroll JS (2012). Pioneer factors in hormone-dependent cancers. Nat. Rev. Cancer 12, 381–385. [DOI] [PubMed] [Google Scholar]
- 24.van Groningen T, Koster J, Valentijn LJ, Zwijnenburg DA, Akogul N, Hasselt NE, Broekmans M, Haneveld F, Nowakowska NE, Bras J, et al. (2017). Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nat. Genet 49, 1261–1266. [DOI] [PubMed] [Google Scholar]
- 25.Sengupta S, Das S, Crespo AC, Cornel AM, Patel AG, Mahadevan NR, Campisi M, Ali AK, Sharma B, Rowe JH, et al. (2022). Mesenchymal and adrenergic cell lineage states in neuroblastoma possess distinct immunogenic phenotypes. Nat. Cancer 3, 1228–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torchia J, Golbourn B, Feng S, Ho KC, Sin-Chan P, Vasiljevic A, Norman JD, Guilhamon P, Garzia L, Agamez NR, et al. (2016). Integrated (epi)-Genomic Analyses Identify Subgroup-Specific Therapeutic Targets in CNS Rhabdoid Tumors. Cancer Cell 30, 891–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S, Sun H, Ma J, Zang C, Wang C, Wang J, Tang Q, Meyer CA, Zhang Y, and Liu XS (2013). Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat. Protoc 8, 2502–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gil Del Alcazar CR, Huh SJ, Ekram MB, Trinh A, Liu LL, Beca F, Zi X, Kwak M, Bergholtz H, Su Y, et al. (2017). Immune Escape in Breast Cancer During In Situ to Invasive Carcinoma Transition. Cancer Discov. 7, 1098–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang DY, Jiang Z, Ben-David Y, Woodgett JR, and Zacksenhaus E (2019). Molecular stratification within triple-negative breast cancer subtypes. Sci. Rep 9, 19107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, and Young RA (2013). Super-enhancers in the control of cell identity and disease. Cell 155, 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang H, Hu J, Maryam A, Huang Q, Zhang Y, Ramakrishnan S, Li J, Ma H, Ma VWS, Cheuk W, et al. (2021). Defining super-enhancer landscape in triple-negative breast cancer by multiomic profiling. Nat. Commun 12, 2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raisner R, Bainer R, Haverty PM, Benedetti KL, and Gascoigne KE (2020). Super-enhancer acquisition drives oncogene expression in triple negative breast cancer. PLoS One 15, e0235343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feinberg AP, Koldobskiy MA, and Göndör A (2016). Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat. Rev. Genet 17, 284–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamdan FH, and Johnsen SA (2019). Perturbing Enhancer Activity in Cancer Therapy. Cancers 11, 634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jørgensen S, Schotta G, and Sørensen CS (2013). Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Res. 41, 2797–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM, Janiszewska M, Huh SJ, Liang Y, Ryan J, et al. (2016). Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 529, 413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Creech AL, Taylor JE, Maier VK, Wu X, Feeney CM, Udeshi ND, Peach SE, Boehm JS, Lee JT, Carr SA, and Jaffe JD (2015). Building the Connectivity Map of epigenetics: chromatin profiling by quantitative targeted mass spectrometry. Methods 72, 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan M, Breitkopf SB, Yang X, and Asara JM (2012). A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc 7, 872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, and Lin SM (2010). Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinf. 11, 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhola PD, Mar BG, Lindsley RC, Ryan JA, Hogdal LJ, Vo TT, DeAngelo DJ, Galinsky I, Ebert BL, and Letai A (2016). Functionally identifiable apoptosis-insensitive subpopulations determine chemoresistance in acute myeloid leukemia. J. Clin. Invest 126, 3827–3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Certo M, Del Gaizo Moore V, Nishino M, Wei G, Korsmeyer S, Arm-strong SA, and Letai A (2006). Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell 9, 351–365. [DOI] [PubMed] [Google Scholar]
- 42.Maksimovic J, Phipson B, and Oshlack A (2016). A cross-package Bioconductor workflow for analysing methylation array data. F1000Res. 5, 1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, Gallinger S, Hudson TJ, and Weksberg R (2013). Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 8, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Witwicki RM, Ekram MB, Qiu X, Janiszewska M, Shu S, Kwon M, Trinh A, Frias E, Ramadan N, Hoffman G, et al. (2018). TRPS1 Is a Lineage-Specific Transcriptional Dependency in Breast Cancer. Cell Rep. 25, 1255–1267.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cornwell M, Vangala M, Taing L, Herbert Z, Keister J, Li B, Sun H, Li T, Zhang J, Qiu X, et al. (2018). VIPER: Visualization Pipeline for RNA-seq, a Snakemake workflow for efficient and complete RNA-seq analysis. BMC Bioinf. 19, 135–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, and Irizarry RA (2014). Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30, 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Ning S, Ghandi M, Kryukov GV, Gopal S, Deik A, Souza A, Pierce K, Keskula P, Hernandez D, et al. (2019). The landscape of cancer cell line metabolism. Nat. Med 25, 850–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiu X, Feit AS, Feiglin A, Xie Y, Kesten N, Taing L, Perkins J, Gu S, Li Y, Cejas P, et al. (2021). CoBRA: Containerized Bioinformatics Workflow for Reproducible ChIP/ATAC-seq Analysis. Dev. Reprod. Biol 19, 652–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, and Tamayo P (2015). The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, and Mesirov JP (2011). Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, and Weirauch MT (2018). The Human Transcription Factors. Cell 172, 650–665. [DOI] [PubMed] [Google Scholar]
- 53.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kent WJ, Zweig AS, Barber G, Hinrichs AS, and Karolchik D (2010). BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26, 2204–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shu S, Wu HJ, Ge JY, Zeid R, Harris IS, Jovanović B, Murphy K, Wang B, Qiu X, Endress JE, et al. (2020). Synthetic Lethal and Resistance Interactions with BET Bromodomain Inhibitors in Triple-Negative Breast Cancer. Mol. Cell 78, 1096–1113.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Satija R, Farrell JA, Gennert D, Schier AF, and Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haghverdi L, Lun ATL, Morgan MD, and Marioni JC (2018). Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol 36, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anders S, and Huber W (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lause J, Berens P, and Kobak D (2021). Analytic Pearson residuals for normalization of single-cell RNA-seq UMI data. Genome Biol. 22, 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hafemeister C, and Satija R (2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplemental Information. All raw genomic data was deposited to GEO under accession number: GSE202776. Processed histone mass spectrometry, metabolomics, drug screen, and BH3 profiling data is provided as supplementary information. Additional metadata files and processed data files used in the original code have been deposited at (https://figshare.com/s/2f077f7838fb5f6e8d35). All code used to analyze genomics data and produce the corresponding figures is available on the GitHub repository https://github.com/daniel-temko/TNBCEpiHet (https://doi.org/10.5281/zenodo.10139754). The raw mass spectrometry data have been deposited in the public proteomics repository MassIVE (http://massive.ucsd.edu) using the identifier: MSV000091071.