Abstract
Myc is a transcription factor with diverse biological effects ranging from the control of cellular proliferation and growth to the induction of apoptosis. Here we present a comprehensive analysis of the transcriptional targets of the sole Myc ortholog in Drosophila melanogaster, dMyc. We show that the genes that are down-regulated in response to dmyc inhibition are largely identical to those that are up-regulated after dMyc overexpression and that many of them play a role in growth control. The promoter regions of these targets are characterized by the presence of the E-box sequence CACGTG, a known dMyc binding site. Surprisingly, a large subgroup of (functionally related) dMyc targets contains a single E-box located within the first 100 nucleotides after the transcription start site. The relevance of this E-box and its position was confirmed by a mutational analysis of a selected dMyc target and by the observation of its evolutionary conservation in a different Drosophila species, Drosophila pseudoobscura. These observations raise the possibility that a subset of Myc targets share a distinct regulatory mechanism.
Myc proteins play a crucial role in the control of cellular proliferation and growth during normal development and in disease (34). In as much as 70% of human cancers, the expression of Myc is found to be deregulated, which places the myc genes among the most medically important human proto-oncogenes (30). Our current molecular understanding of Myc's functions is founded on the identification of the Max protein as an obligatory interaction partner for Myc (16). Myc-Max complexes bind DNA at E-boxes (CACGTG and variants thereof) and activate the transcription of nearby genes. Several mechanisms have been proposed for this activation (1, 16, 41): recruitment of histone acetylases [Tip60 complex, S(T)AGA complex, CBP], recruitment of chromatin remodeling complexes (hBrm), interactions with the TATA box binding protein (23, 27), and binding to kinases of the RNA polymerase II C-terminal domain (15, 26). The relative importance of these different pathways in vivo and for individual Myc-Max target genes is still the subject of debate. The activation of Myc's targets is opposed by complexes of Max with a transcriptional repressor of the Mad/Mnt family (4, 44). Mad-Max heterodimers also bind to E-boxes but then recruit histone deacetylases and repress the expression of nearby genes. In addition, Myc (most likely in association with Max) also functions as a transcriptional repressor on a different set of target genes by binding to, and inhibiting, other transcriptional activators such as Miz-1 (42). This repression by Myc is not mediated by E-boxes but frequently involves a loosely defined sequence motif flanking the transcription initiation site, a so-called initiator element.
Ever since Myc was recognized as a transcription factor, the quest has been on for the transcriptional targets that can explain some or all of Myc's biological functions (11). In recent years, the use of high-throughput methods has dramatically accelerated the pace of target identification, and currently more than 1,000 genes are listed as potential Myc targets (43). These putative Myc targets fall into different functional categories and, as is consistent with Myc's biological role, a large number of activated genes encode proteins involved in cell growth and cell cycle regulation, whereas many Myc-repressed genes affect cell adhesion. Despite this abundance of proposed Myc targets, only three studies have systematically addressed the sequence determinants of Myc binding sites in vivo. Fernandez et al. (17) used chromatin immunoprecipitation (ChIP) assays to analyze 533 selected E-box-containing promoters in established human cell lines; a majority of these promoters were found to bind to c-Myc, in particular when their E-boxes were located close to CpG islands. Orian et al. (33) overexpressed Drosophila Myc (dMyc) together with Drosophila Max in Kc167 cells and found 287 promoters that were able to bind to dMyc (of about half the Drosophila genome that was assayed); 40% of these promoters contain an E-box (33; also our analysis). In addition, 544 genes were found to be induced by dMyc overexpression in vivo, and their promoters also showed a significant association with E-boxes. Neither study found any additional characteristics of Myc-binding sites. A recent ChIP analysis of human chromosomes 21 and 22 found 756 c-Myc binding regions, one-third of which contained at least one E-box. Only one-quarter of these c-Myc binding regions were located close to CpG islands, and many of them were situated far away from known promoter regions (7). Importantly, all these studies sampled only a fraction of the genome, and none of them systematically assayed the importance of physiological levels of Myc for the expression of these putative targets. In contrast, O'Connell and colleagues covered a large fraction of the genome in their search for genes that were misregulated in a rat cell line in which c-Myc had been knocked out, but the promoter sequences of these targets were not systematically analyzed (31). Thus, it is not clear at present which criteria, in addition to the CACGTG sequence, are required to define a Myc-binding site, and to what extent binding to a certain promoter predicts a role for Myc in the regulation of the corresponding gene.
To address these issues, we have set out to characterize the promoters of transcriptional targets of Myc in Drosophila melanogaster D. melanogaster encodes a single Myc homolog, dMyc, with molecular functions very similar to those of its vertebrate counterparts; dMyc and vertebrate Myc can even largely substitute for each other in vivo (21, 25, 38, 40; C. Benassayag, L. Montero, N. Colombié, P. Gallant, D. Cribbs, and D. Morello, submitted for publication). To identify direct transcriptional dMyc targets and to avoid adaptive responses that could possibly be caused by prolonged proliferation of cells in the absence of dMyc, we acutely down- or up-regulated dMyc in vivo and in Schneider 2 (S2) cells and assayed the ensuing effects on the entire transcriptome by using Affymetrix whole-genome microarrays. The availability of the detailed annotation of the Drosophila melanogaster genome sequence (8, 32), as well as the recently published genome sequence of a related species, Drosophila pseudoobscura (Baylor College of Medicine Human Genome Sequencing Center), allowed us to perform an extensive analysis of dMyc-responsive promoters. This analysis revealed the existence of a functionally related subset of dMyc targets that are characterized by the presence of an E-box within the first 100 nucleotides following the transcription start site. The importance of this E-box was further demonstrated by the mutational analysis of selected dMyc targets.
MATERIALS AND METHODS
Molecular biology.
Double-stranded RNA (dsRNA) was transcribed in vitro from PCR fragments of approximately 600 bp, amplified from the gene of interest. Target sequences were subjected to BLAST analysis to ensure minimal homology with unrelated transcripts. dsRNA was produced by Megascript IVT (Ambion). Site-directed mutagenesis was carried out using the GeneEditor system (Promega). Promoter elements used in luciferase reporter expression analyses were cloned into the pGL3-basic vector (Promega). For ChIP, untransfected S2 cells or S2 cells stably transfected with hemagglutinin (HA) epitope-tagged dMyc under the control of the hsp70 promoter (12) were subjected to a heat shock at 37°C; 2 h later, triplicate samples of 8 × 106 cells each were processed for ChIP analysis using 0.6 μg of rat anti-HA monoclonal antisera (Roche) as described previously (19, 20). Sequences for PCR primers used for in vitro synthesis of dsRNA, mutagenesis, and ChIP are listed in the supplemental material.
Cell culture.
S2 Drosophila cells (37) were propagated in 1× Schneider's Drosophila medium (Gibco/BRL), supplemented with 10% fetal bovine serum, at 24°C. RNA interference (RNAi) experiments were performed by incubation of 3 × 106 cells in a six-well tissue culture plate with 15 μg of dsRNA as previously described (10). Cells were harvested for fluorescence-activated cell sorting (FACS) or RNA extraction at the time points indicated.
FACS.
Cells were incubated with Hoechst 33342 (Fluka) at a final concentration of 1 ng/ml for 3 h. A suspension of 106 cells in 1 ml was analyzed in a FACStar PLUS (Becton Dickinson). Data analysis was carried out using WinMDI, version 2.8.
S2 cell microarrays.
Biologically independent triplicate samples of S2 cells were treated with experimental dsRNA and with gfp dsRNA as a control. At the indicated time points after addition of dsRNA, total RNA was extracted by using the RNeasy kit (QIAGEN). Gene expression analysis was performed with the Affymetrix (Santa Clara, Calif.) Drosophila GeneChip (36), using the methods described in the Affymetrix GeneChip expression analysis manual. Briefly, double-stranded cDNA was synthesized by using 20 μg of total RNA. Biotin-labeled cRNA was synthesized by using the BioArray high-yield RNA transcript-labeling kit (Enzo Biochem), and 20 μg of fragmented RNA was hybridized to each array. The arrays were washed by using the EukGW2 protocol on the GeneChip Fluidics Station 400 series and were scanned by using the GeneArray scanner.
Wing imaginal disk microarrays.
Flies were raised at 18°C on regular fly food supplemented with yeast. Overexpression of dMyc in vivo was performed using w1118; hs-dMyc[29]/TM3 flies (25); as a control, the isogenized w1118 line from which the transgenic line had been derived was used. Egg laying was permitted for a maximum of 12 h at 25°C; 48 h later, the flies were transferred to 18°C. Third-instar wandering larvae were subjected to a 1-h heat shock in a 37°C water bath, followed by a 1-h recovery period at 25°C. Because it was reported previously that heat shock can induce a transient cell cycle block in fly embryos (28), we monitored cell cycle progression and the levels of ectopic dMyc at different times after the heat shock (data not shown). By 1 h after the heat shock, numbers of mitotic cells had returned to normal (as assessed by phospho-histone H3 staining), while ectopic dMyc levels had dropped to 1.5-fold above background. Note that this setup differs in several aspects from that of an earlier study (33): dMyc was expressed directly, without intervening amplification by GAL4; only the fairly homogeneous wing disks were analyzed, and not whole larvae; RNA was isolated 1 h (rather than 7 h) after the onset of dMyc expression; dMyc was expressed only transiently; and only male larvae (matching the sex of S2 cells) were analyzed (this may be important, because the two sexes are known to differ by as much as 10% of their transcriptome [24, 37]).
To acutely remove dMyc function in vivo, “C(1)DX, y w / Y” females were crossed to “y w tub>FRT-dMyc-FRT>GAL4 hs-FLP / Y” control males or to “y w dmycPG45 tub>FRT-dMyc-FRT>GAL4 hs-FLP / Y” experimental males; in these flies, the lethality of the strong allele dmPG45 (6) is rescued by a dmyc cDNA expressed under the control of the ubiquitous α-tubulin promoter (12). Egg laying and growth of the larvae were carried out under the same conditions described above. Third-instar wandering larvae were subjected to a 1.5-h heat shock in a 37°C water bath, resulting in the acute loss of the dmyc cDNA in most cells of the experimental flies and uncovering the dmPG45 allele. These flies were unable to complete development and died a few hours after the heat shock, whereas similarly treated control flies developed normally to adulthood. For RNA isolation, larvae were allowed to recover for 2 h at 25°C after the heat shock, by which time the dmyc mRNA levels had dropped fivefold from those for the control (by quantitative real-time PCR [qRT-PCR]).
For both overexpression and mutant experiments, male larvae were selected and dissected in 1× phosphate-buffered saline. Approximately 120 wing disks were collected and flash-frozen in liquid N2, and RNA was isolated as described for the S2 cells. Each experimental condition and each control was represented by two biologically independent replicates.
Expression data analysis.
Data obtained from Affymetrix microarray experiments were normalized to a target signal intensity of 500. The resulting raw expression values were statistically analyzed as detailed in the supplemental material. Genes were considered to be “significantly differentially expressed” if they were expressed in all three (or two, for the in vivo data) experimental and all three (or two, for the in vivo data) control conditions, their expression differed at least 1.5-fold between control and experimental conditions, and they passed a significance cutoff (P ≤ 0.001). The same data sets were also analyzed using CyberT (3) with less stringent criteria (expression in at least three experimental or three control conditions); in this case, the numbers of significant genes were slightly higher, but the conclusions are the same (data not shown).
Promoter analysis.
Genomic sequences and sequences for open reading frames based on release 3.1 of the D. melanogaster genome were downloaded from the “Berkeley Drosophila Genome Project” (9), annotations release 3.1 from FlyBase (18), and the D. pseudoobscura genome sequence freeze 1 from the Baylor College of Medicine Human Genome Sequencing Center (http://www.hgsc.bcm.tmc.edu/projects/drosophila/). Promoter sequences were analyzed using GeneSpring (Silicon Genetics), MEME (2), the CART algorithm (see the supplemental material), and different Perl scripts. For consistency, all analyses were restricted to the 13,966 loci represented on the Affymetrix Drosophila GeneChip 1.0, corresponding to 11,810 unique loci with unique and unambiguous FlyBase gene identifiers (FBgn numbers).
For phylogenetic comparisons, first BLASTN searches were carried out with all D. melanogaster proteins to identify the corresponding D. pseudoobscura orthologs, and only orthologous gene pairs for which the protein similarity started within less than 10 amino acids of the translation start were kept. Next, all gene pairs where the translation start site in D. melanogaster fell within less than 100 nucleotides of the predicted transcription start site, or where several different transcription start sites were annotated, were discarded, resulting in 3,535 gene pairs.
Luciferase assays.
S2 cell transfections were carried out using Cellfectin (Invitrogen). nnp-1 reporter constructs were added at 1 μg per 106 cells; tubulin-Renilla luciferase control DNA and, where indicated, dsRNA were cotransfected at 0.1 μg/106 cells. Cellfectin was used at a final concentration of 6.5 μg/ml, and cells were incubated with transfection mix for 12 h. Cells were harvested 24 or 60 h posttransfection. Relative gene expression was determined using the Dual-Luciferase reporter assay (Promega) on a Wallac luminometer.
RESULTS
Identification of physiological dMyc targets in S2 cells and in vivo.
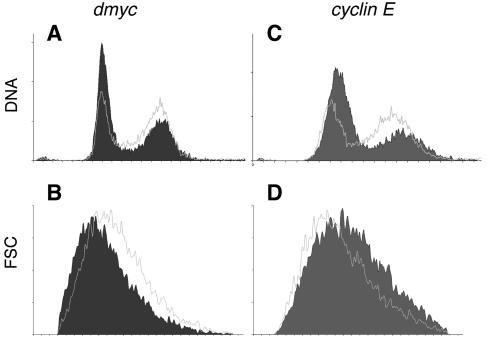
In order to characterize the Myc-responsive cis-acting regulatory sequences, we first identified transcriptional targets of dMyc in cultured Drosophila S2 cells. dMyc was acutely down-regulated by RNAi in exponentially proliferating S2 cells. As indicated by control experiments, close to 100% of the cells take up dsRNA (data not shown), and within 6 h of addition of the dmyc dsRNA, dmyc levels are reduced to 39% of those in control cells incubated with gfp dsRNA (because the available antibodies did not recognize the endogenous dMyc protein in our experiments, we measured transcript levels, either by qRT-PCR [at 48 h] or by microarrays [at the other time points]); by 48 h, dmyc levels have fallen to 19%. Thus, dMyc activity is impaired to a greater extent in these experiments than in experiments with the hypomorphic allele dmP0, which was characterized for its strong growth defects in vivo (36% of control levels as measured by qRT-PCR [25]), suggesting that relevant downstream targets of dMyc will be affected in S2 cells by the RNAi treatment. Indeed, this impairment of dMyc is accompanied by a slowing down in G1 phase, comparable to that observed after RNAi against the cell cycle regulator cyclin E (Fig. 1). Furthermore, cells with reduced dmyc levels show a decrease in cell size in all phases of the cell cycle, consistent with dMyc's essential role for cellular growth (25), whereas the growth of cells treated with cyclin E dsRNA is unaffected (Fig. 1).
FIG. 1.
FACS analysis of S2 cells treated with dsRNA against gfp, dmyc, or cyclin E. Each panel shows a single cytometric profile of S2 cells 48 h after addition of the indicated dsRNA. The data shown are representative of three independent experiments (each performed in duplicate, with similar results). Outlined traces show control cells treated with gfp dsRNAi; filled diagrams show cells treated with dmyc RNAi (A and B) or cyclin E RNAi (C and D). (A and C) Cell cycle distribution; (B and D) forward scatter (FSC) indicative of cell size.
The effects of dmyc reduction on target gene expression were assayed by Affymetrix whole-genome microarrays at 6, 12, and 48 h after addition of dsRNA. A total of 489 genes were down-regulated and 55 genes were up-regulated at at least one time point (corresponding to 12 and 1%, respectively, of the 4,101 genes that were expressed in all experiments in S2 cells [see Table S1 in the supplemental material]). The number of affected genes is largest at 6 h, raising the possibility that other proteins might progressively compensate for the loss of dMyc, e.g., other transcription factors of the basic helix-loop-helix-leucine zipper family with a DNA binding specificity similar to that of dMyc. Although none of these proteins changes dramatically at the level of mRNA abundance during our experiments (data not shown), we cannot exclude compensatory alterations at the level of protein abundance or activity. Alternatively, the experimental manipulation (which includes a short incubation in serum-free medium followed by addition of complete medium) might induce a partial serum response, accompanied by the induction of a large number of genes, which is blunted in the dmyc RNAi-treated cells. Because at present we cannot rule out either possibility and we are most interested in the direct transcriptional targets of dMyc, we focused our subsequent analysis on those genes that are down-regulated both at 6 h and at a later time point. This selection covers the genes requiring physiological dMyc levels for their steady-state expression (139 genes shared between the 6- and 12-h time points; 30 genes shared between all three time points). The up-regulated genes showed no overlap between different time points and were not examined further.
The majority of these 30 down-regulated genes play a role in ribosome biogenesis and protein synthesis, consistent with dMyc's role in cellular growth and with the types of targets that have been identified in vertebrate studies (see Tables S2 and S3 in the supplemental material; the latter gives a full list of all genes that are significantly affected by altered dMyc levels in at least one situation, many of which are involved in processes such as signaling, transcription, protein modification, transport, metabolism, cytoskeleton dynamics, cell cycle control, and RNA processing). Importantly, the dMyc targets do not overlap the genes affected by cyclin E RNAi, indicating that their misexpression is not an indirect consequence of the cell cycle effects of dmyc RNAi (see Table S1 in the supplemental material).
To confirm the generality of these dMyc targets, we also analyzed the genes controlled by dMyc in imaginal wing disks in vivo. To avoid potential long-term adaptive responses, we sampled wing disks 1 h after dMyc overexpression and 2 h after reduction of dmyc function, respectively (these time points were chosen to minimize nonspecific effects of the heat shock; see Materials and Methods). Only 12 genes were significantly down-regulated under these dmyc mutant conditions (possibly because dMyc-activated mRNAs have not sufficiently decayed in the 2 h following the heat shock), but they showed a high degree of overlap with the dMyc targets in S2 cells: of the 8 genes that were also expressed in S2 cells, 3 were down-regulated at all time points in S2 cells, and all 8 were significantly down-regulated at the 6-h time point. The 19 up-regulated genes did not overlap significantly with genes in any of the other lists, as was the case for the genes that were down-regulated in response to overexpressed dMyc. In contrast, dMyc overexpression activated 165 genes, of which 88 were down-regulated at at least one time point in S2 cells (60% of the 147 genes that are expressed in S2 cells); these genes fell into the same functional categories as the dMyc targets in S2 cells (see Table S1 in the supplemental material). We also observed good agreement with an earlier publication describing genes activated by overexpressed dMyc (33): 50 genes were shared by both studies, corresponding to 47% of the 107 genes that were represented on both microarrays (the remaining differences between the two studies are most likely due to differences in experimental setup, see Materials and Methods). Thus, very similar sets of genes are controlled by dMyc in different cell types, and the ectopic activation of dMyc (under the conditions used here) largely targets the same genes that are controlled by dMyc during normal development.
dMyc targets are characterized by the presence of a positionally conserved E-box.
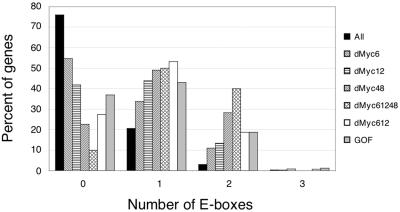
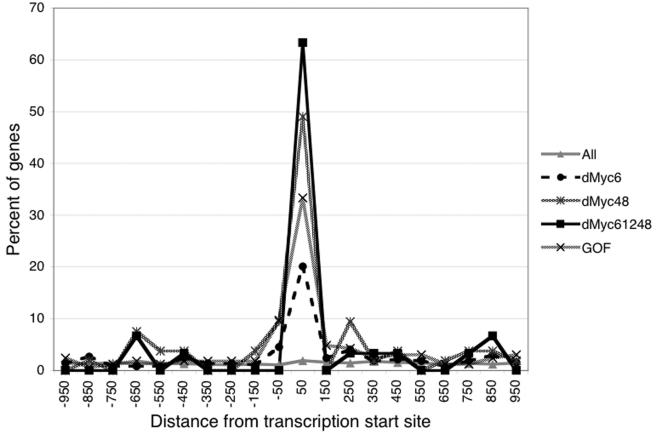
The promoter sequences of dMyc target genes, extending 1,000 bp in either direction from the predicted transcription start site, were scanned for an enrichment of sequence motifs relative to those in a random list of unaffected genes. The most common sequence found to be associated with dMyc targets in all experiments was the canonical E-box (Fig. 2); 27 of the 30 genes down-regulated at all time points contained at least one E-box (90%), with 12 containing two and none containing more. E-boxes are also highly represented in the promoters of the genes down-regulated at 6 h (169 of 373 genes [45%]) or at 12 h (143 of 246 genes [58%]), in the genes shared between these two time points (101 of 139 genes [73%]), and in the genes up-regulated in response to dMyc overexpression in vivo (104 of 165 genes [63%]). In contrast, only 2,832 out of all 11,810 genes represented on the microarrays contained an E-box within 1,000 nucleotides of the transcription start site (24%). Those E-boxes found within the promoters of dMyc targets showed a strong positional bias. A graphical representation of the positions of these E-boxes relative to the transcription start site reveals that the majority of dMyc targets changed at all time points contained one E-box within the 100 nucleotides following the transcription start site (19 of 30 genes [63%] [Fig. 3]). A similar positional bias was also seen for the other sublists of dMyc targets, whereas the distribution of E-boxes in the promoter sequences of non-dMyc targets was random (Fig. 3). The position of a second E-box, when present in a dMyc target, showed no preference (data not shown). Furthermore, the consensus sequence for the dMyc-dependent regulatory element seems to extend beyond the core sequence CACGTG. As shown in Table 1, only 11 of the 136 possible decameric sequences are found among the dMyc targets. Many of these sequences conform to the nonpalindromic consensus AACACGTG(C/T)(A/G); the motif found most frequently is AACACGTGCG. This distribution of decameric sequences is clearly different in non-dMyc targets (Table 1).
FIG. 2.
Frequencies of E-boxes in the promoter regions. Each bar shows the percentage of genes on the indicated list with 0, 1, 2, or 3 E-boxes located between nucleotides −1000 and +1000 relative to the transcription start site. Gene list abbreviations: All, all 11,810 genes included in our analysis; dMyc6, dMyc12, and dMyc48, genes that were downregulated at 6, 12, and 48 h, respectively, after dmyc RNAi in S2 cells; dMyc612, genes downregulated at both 6 and 12 h; dMyc61248, genes downregulated at all time points tested; GOF, genes up-regulated after dMyc overexpression in vivo.
FIG. 3.
Distribution of E-boxes relative to the transcription start site. The x axis indicates the center of the 100-bp window for which the frequency of E-boxes was determined (in nucleotides from the transcription start site). Gene list abbreviations are explained in the legend to Fig. 2.
TABLE 1.
Most commonly occurring extended E-box sequences
| Decameric motifa | Frequencyb of the indicated decameric motifc in the following gene listd and promoter segmente:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Not down
|
dMcy6 (+1 to +100) | dMyc12 (+1 to +100) | dMyc48 (+1 to +100) | dMyc61248
|
dMyc612
|
GOF
|
Dpse (not down) (+1 to +100) | Dpse (dMyc612) (+1 to +100) | |||||
| +1 to +100 | −1000 to +1000 | +1 to +100 | −1000 to +1000 | +1 to +100 | −1000 to +1000 | +1 to +100 | −1000 to +1000 | ||||||
| AA CACGTG CG | 0 | 2 | 25 | 21 | 27 | 32 | 26 | 28 | 16 | 23 | 13 | 3 | 14 |
| AA CACGTG TG | 0 | 2 | 5 | 7 | 12 | 16 | 13 | 7 | 7 | 5 | 8 | 0 | 7 |
| AA CACGTG CA | 3 | 2 | 7 | 8 | 12 | 11 | 5 | 9 | 5 | 4 | 3 | 0 | 21 |
| CA CACGTG CA | 3 | 4 | 9 | 7 | 4 | 5 | 3 | 9 | 5 | 7 | 5 | 3 | 7 |
| TA CACGTG CA | 0 | 1 | 5 | 4 | 4 | 5 | 3 | 5 | 5 | 7 | 4 | 0 | 0 |
| AG CACGTG TA | 3 | 1 | 3 | 3 | 4 | 5 | 3 | 3 | 2 | 5 | 4 | 3 | 0 |
| AA CACGTG CC | 0 | 1 | 1 | 3 | 4 | 5 | 3 | 2 | 1 | 2 | 1 | 0 | 0 |
| AA CACGTG CT | 0 | 1 | 1 | 1 | 4 | 5 | 3 | 2 | 2 | 2 | 1 | 0 | 14 |
| AA CACGTG TT | 0 | 0 | 1 | 1 | 4 | 5 | 3 | 2 | 1 | 2 | 1 | 0 | 0 |
| AG CACGTG CG | 3 | 1 | 3 | 1 | 4 | 5 | 3 | 2 | 2 | 2 | 1 | 3 | 0 |
| AG CACGTG GG | 0 | 1 | 1 | 1 | 4 | 5 | 3 | 2 | 1 | 2 | 1 | 0 | 0 |
| Total | 13 | 15 | 63 | 57 | 81 | 100 | 64 | 69 | 47 | 61 | 40 | 13 | 64 |
These 11 decamers are the only CACGTG-containing sequences to occur in the promoters on the dMyc61248 list.
Expressed as a percentage of all E-boxes that occur within the given promoter segment.
Combined with the reverse complement.
Not down, genes that were not down-regulated at any time after dmyc RNAi in S2 cells; dMyc6, dMyc12, and dMyc48, genes that were down-regulated at 6, 12, or 48 h, respectively; dMyc612, genes down-regulated at both 6 and 12 h; dMyc61248, genes down-regulated at all time points after dmyc RNAi in S2 cells; GOF, genes up-regulated after dMyc overexpression in vivo; Dpse (not down), D. pseudoobscura orthologs of genes in the “Not down” list; Dpse (dMyc612), D. pseudoobscura orthologs of genes in the dMyc612 list.
The region of the promoter that was sampled, comprising either nucleotides −1000 to +1000 or nucleotides +1 to +100 relative to the transcription start site.
To confirm the relevance of such downstream E-boxes for the identification of dMyc targets, we selected all Drosophila genes containing an E-box within the first 100 nucleotides following the transcription start site. Only 224 genes fulfill these criteria; 107 of these genes are expressed in all S2 microarray experiments. Thirty of these genes (28%) are not significantly changed at any time after dmyc RNAi, i.e., they correspond to false positives; 77 genes are down-regulated at at least one time point (72%), and 19 genes are down-regulated at all three time points (18%). Thus, this simple rule predicts a subset of dMyc targets with high reliability. In stark contrast, of the 1,066 genes expressed in S2 cells that simply contain an E-box anywhere in the promoter region, 875 are not affected by dmyc RNAi at any time point (corresponding to a false-positive rate of 82%). Interestingly, a large fraction of the 224 genes carrying such a downstream E-box play a role in ribosome biogenesis, RNA binding, and protein translation (44 out of the 150 genes with an annotated function), suggesting that the presence of a downstream E-box may characterize a functional subgroup of dMyc targets. This is also seen among the dMyc targets that are down-regulated at any of the three time points, where most of the genes with a downstream E-box are involved in ribosome biogenesis, RNA binding, and protein translation (29 out of 56 genes with a predicted function [52%]), as opposed to those without such an E-box (42 out of 306 genes with a predicted function [14%]).
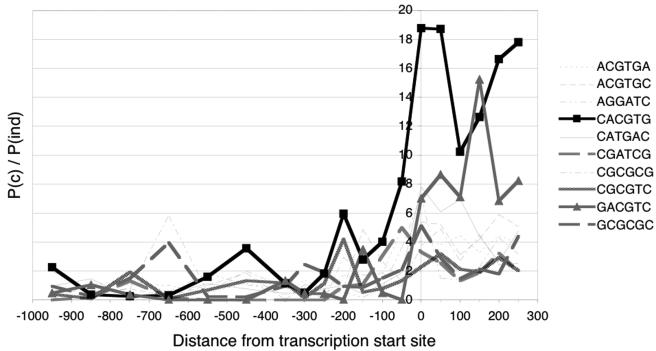
Independent confirmation of the relevance of these E-boxes was obtained by a phylogenetic comparison of promoter sequences between D. melanogaster and D. pseudoobscura. The two species diverged approximately 46 million years ago (5); hence, a conservation of sequence provides a strong indication of functional importance. We established a list of 3,535 gene pairs and used the annotated distances between transcription and translation start sites of the D. melanogaster genes to predict transcription start sites for their D. pseudoobscura orthologs (see Materials and Methods). While this procedure provides only a rough estimation of transcription start sites in D. pseudoobscura, the data presented below indicate that these estimates can be used to draw some meaningful conclusions. To identify evolutionarily conserved motifs, the orthologous promoter sequences from nucleotides −1000 to +1000 were subdivided into 100-bp segments (other segment sizes were also tested and gave qualitatively identical results). Each segment was then scanned for the occurrence of all possible hexameric sequence motifs in the D. melanogaster promoter, in the orthologous D. pseudoobscura promoter, and in both promoters simultaneously; the procedure was repeated for all 3,535 gene pairs to produce the relative frequencies of all hexameric motifs over all segments. A sequence motif with no evolutionarily conserved function would be expected to co-occur randomly in a gene pair, at a frequency that would depend on the frequency with which this motif occurs in either the D. melanogaster or the D. pseudoobscura gene. To identify evolutionarily conserved motifs, we therefore compared this frequency of random co-occurrence with the actual frequency of co-occurrence (see the supplemental material). Figure 4 shows all hexameric sequence motifs that co-occur in at least 10 gene pairs and are significantly conserved between the two species. Strikingly, the E-box is the most conserved motif, and the highest degree of conservation is seen at, and downstream of, the transcription start—where the E-box is most frequently found in dMyc-responsive genes. We notice also that the residues flanking the core E-box sequence CACGTG show some degree of conservation; 64% of the E-boxes downstream of the promoter in D. pseudoobscura correspond to one of the decameric sequences that are overrepresented among the D. melanogaster dMyc targets (Table 1).
FIG. 4.
Evolutionary comparison of hexameric sequences in the promoter regions of D. melanogaster and D. pseudoobscura. The ratio of the probability that a particular motif is conserved [P(c)] to the probability that it occurs randomly [P(ind)] is plotted (see the supplemental material) for all motifs that are conserved in at least 10 gene pairs and that are significantly conserved in at least one window of the promoter (i.e., motifs for which the ratio of the probabilities differs by at least 7 standard deviations from the average ratio of all motifs for that window). The x axis is labeled as in Fig. 3.
Experimental confirmation of E-box relevance.
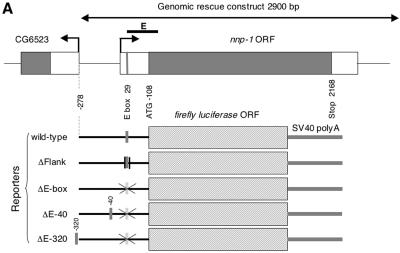
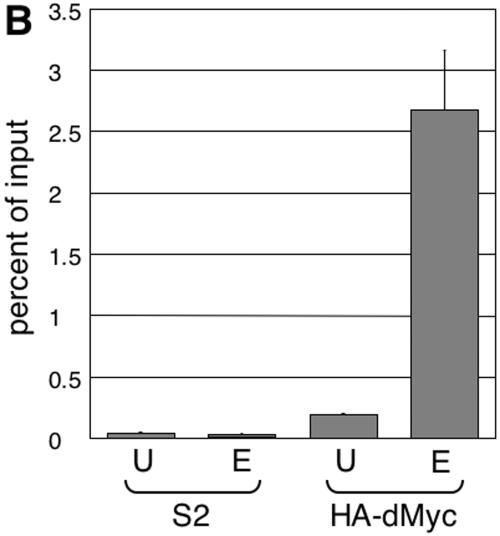
As a final demonstration of the importance of the E-box for the regulation of dMyc target genes, we experimentally analyzed a selected target, Nnp-1, a sequence homolog of the nucleolar proteins Nnp1/Nop52 (in vertebrates) and Rrp1 (in Saccharomyces cerevisiae). The nnp-1 gene is significantly down-regulated at all time points after dmyc RNAi in S2 cells and is up-regulated after dMyc overexpression in wing disks. It also contains one E-box at position +29 relative to the transcription start site (which was experimentally confirmed by rapid amplification of 5′ cDNA ends [Fig. 5A]); this E-box conforms to the extended consensus, and furthermore, it is bound by dMyc in S2 cells as demonstrated by ChIP experiments (Fig. 5B). Expression of a 2.9-kb genomic fragment partially rescues the lethality of homozygous nnp-1 mutant flies, indicating that the essential control elements of nnp-1 are located within this fragment (data not shown). To analyze the function of the nnp-1 E-box, we fused a 386-bp fragment of the nnp-1 promoter, including 108 bp downstream of the transcription start site, with the luciferase open reading frame, such that the translation of luciferase starts with the ATG of Nnp-1. In addition, we created mutant constructs (Fig. 5A) where the E-box was deleted (ΔE-box) or transposed to nucleotide −40 (ΔE-40) or −320 (ΔE-320), or where the flanking residues were altered (ΔFlank). These reporter constructs were transiently transfected into S2 cells, together with different dsRNAs and a control plasmid expressing the Renilla luciferase gene under the control of the constitutive α-tubulin promoter. The luciferase activities of the reporter and the control vector were determined at 24 h (Fig. 5C) or 60 h (Fig. 5D) after transfection.
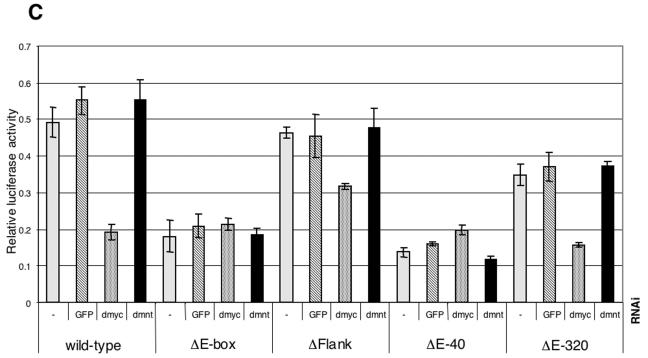
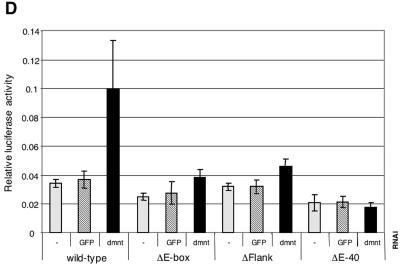
FIG. 5.
Functional analysis of the nnp-1 promoter. (A) Schematic representation of the nnp-1 promoter and the derived reporter constructs. The positions of the shifted E-boxes are indicated by vertical grey bars. (B) Binding of dMyc to the nnp-1 promoter. Chromatin recovered from naive S2 cells or from S2 cells overexpressing HA-dMyc was analyzed by ChIP using an anti-HA antiserum. DNA was amplified with primer pairs located near the E-box (E) (indicated by a thick black line in panel A) or at a distance (U) (see the supplemental material). (C and D) Normalized luciferase activities of the indicated reporter constructs at 24 h (C) or 60 h (D) after transfection. −, gfp, dmyc, and dmnt indicate the cotransfected dsRNA. Three independent experiments were performed, each in triplicate, and gave similar results. Results of a representative experiment are shown. Error bars, standard errors of the means.
The wild-type reporter accurately reflects the regulation of the endogenous nnp-1 gene; it is down-regulated to an extent similar to that of nnp-1 mRNA by dmyc RNAi but not by control RNAi. This dMyc input is entirely mediated by the E-box, since ΔE-box is unaffected by dmyc RNAi. Interestingly, the ΔE-box reporter is expressed at the same level as the wild-type reporter after dmyc RNAi, although the dMyc protein remaining after dmyc RNAi would be expected to activate the wild-type reporter to some extent; we therefore speculate that the activity of the wild-type reporter after dmyc RNAi reflects a shifted equilibrium between activation by dMyc and repression by an opposing factor, most likely dMnt (the only Drosophila member of the Mad family of Myc antagonists). Indeed, the wild-type reporter is strongly derepressed by dmnt RNAi, although this effect is visible only at later time points (perhaps due to an insufficient decrease in dMnt levels 24 h after the addition of dmnt dsRNA [Fig. 5D]).
These experiments show that an E-box positioned at −40 cannot substitute for the downstream E-box and that an E-box at −320 can do so only partially,. These observations demonstrate the relevance of the location of the E-box and suggest that, while dMyc can also function from promoter-distal positions, it does so less efficiently. In contrast, the importance of the extended consensus sequence is less clear. Under control conditions, ΔFlank is expressed at marginally lower levels than the wild-type reporter, but it is less affected by dmyc RNAi, suggesting that the mutation of the flanking residues might enable other factors to substitute for dMyc. In addition, ΔFlank and ΔE-40 are only marginally activated by dmnt RNAi, indicating that these mutations might alter the ability of dMnt to repress these reporters. Note that dmyc RNAi experiments are not included for the 60-h time point, since the dramatic effects of dmyc RNAi on cellular physiology, demonstrated above (in contrast to the marginal effects of dmnt RNAi [data not shown]) preclude any meaningful interpretation of the results.
Finally, we note that our experimental analysis has focused on a single model target of dMyc, nnp-1. To show that our findings are likely to be generalizable, we also examined the promoters of two additional dMyc targets, CG5033 and CG4364. Both confer dMyc responsiveness on a luciferase reporter, and furthermore, all of the dMyc responsiveness of CG5033 is mediated by the single downstream E-box (see Fig. S5 in the supplemental material). These observations confirm the identification of CG5033 and CG4364 as dMyc targets (and, by inference, of the other genes in Table S2 in the supplemental material as well), and they strongly suggest that the nnp-1 promoter is representative of the dMyc targets controlled by a downstream E-box.
DISCUSSION
Here, using D. melanogaster as a model system, we present the first genome-wide analysis of physiological Myc targets. Many of the dMyc targets play a role in growth-related functions, consistent with previously published Myc target gene lists, but the most important findings of this study derive from the large-scale analysis of the promoter regions of dMyc targets. We find the promoters of physiological dMyc targets to be significantly enriched in the E-box motif compared to those of non-dMyc targets; no other motifs were identified as specifically associated with dMyc targets. This is consistent with the known DNA-binding specificity of dMyc in vitro (21) and with a previous analysis of dMyc overexpression targets (33), but it should be noted that the majority of dMyc targets harbor only one such E-box. This might indicate that Drosophila Myc-Max complexes do not heterotetramerize to bind two E-boxes at the same time, as has been suggested for their vertebrate counterparts—indeed, most of the amino acids predicted to be involved in heterotetramerization in vertebrate Myc are not conserved in Drosophila (29). However, a significant number of dMyc target promoters harbor a second E-box, raising the possibility that the (independent) binding of a second dMyc-dMax dimer may increase the responsiveness of a gene to dMyc.
Most strikingly, the dMyc-responsive E-boxes are frequently located in the first 100 nucleotides following the transcription start site. This positional bias is found in all classes of dMyc-responsive genes, but it is particularly pronounced among the genes that show reduced expression at both early and late time points following addition of dmyc dsRNA (63% of these genes), suggesting that such genes are directly regulated by dMyc and that their activation cannot be appropriated by a hypothesized compensatory mechanism. The preferred location probably does not reflect a differential binding affinity of dMyc, as can be seen by comparison with published binding data (33); among the promoter regions that were found by virtue of their binding to dMyc, only those associated with differentially expressed genes also show the positional bias of the E-box (see Fig. S1 in the supplemental material). This observation also raises the possibility that dMyc may bind to some genes without affecting their expression. In agreement with such an interpretation, we have observed association of ectopically expressed dMyc with many loci on larval polytene chromosomes, but only a few of these sites colocalized with actively transcribing RNA polymerase II (see Fig. S4 in the supplemental material).
The functional relevance of the E-box position is further demonstrated by its evolutionary conservation and by reporter gene assays in which the E-box was deleted or transposed. The dMyc-responsive downstream E-boxes are also characterized by a nonrandom distribution of the two flanking nucleotides on either side. The molecular basis for any extended consensus is not apparent from the published structure of the Myc-Max DNA-binding domains, and no preference for flanking sites was found in the large-scale screen for genomic c-Myc binding sites (17). However, our reporter assays suggest that the flanking residues do play a role in modulating the activity of the nnp-1 reporter and its response to dmyc and dmnt levels. We consider it possible, therefore, that the extended consensus sequence reflects the responsiveness of these target promoters not only to dMyc but also to dMnt and to other transcription factors that might contact flanking nucleotides in addition to the core sequence CACGTG.
The vast majority of genes with such a downstream E-box appear to be dMyc targets. It is intriguing that these genes also fall into common functional classes, with many of them playing a role in nucleolar function and ribosome biogenesis. This suggests that these fundamental biological processes are coordinately regulated at the level of transcription, by the binding of a single transcriptional activator, Myc. The question then arises whether such a positional preference of Myc-regulated E-boxes is also found in species other than insects. No comprehensive unbiased analysis of c-Myc target promoters in vertebrates has been published, although it is generally accepted that such genes are most often regulated through Myc binding to E-boxes (1). There is anecdotal evidence that some of these E-boxes are located immediately downstream of the transcription start site (e.g., the cad gene, which is discussed in more detail below; see also recent compilations of Myc targets [22, 39]). In an unbiased survey of a small number of human Myc-responsive promoters, we found a slight preference of E-boxes for the 100 bp immediately preceding the transcription start site (see Fig. S6 in the supplemental material). While it remains to be seen whether this distribution of E-boxes (centered upstream of the transcription start) is a vertebrate manifestation of the same underlying cause as the E-box distribution in Drosophila (centered downstream of the transcription start), these observations strengthen the notion that many functional vertebrate Myc binding sites are also preferentially located close to the transcription start site.
A possible molecular basis for such a bias may be found in the analysis of the vertebrate cad gene, which contains an E-box immediately downstream of the transcription start site. It has been proposed that c-Myc is required not for bringing RNA polymerase II to the cad promoter, but rather for recruiting the P-TEFb components Cdk9 and cyclin T1, which then trigger promoter clearance and transcriptional elongation by RNA polymerase II (14, 15). Whether Myc also induces histone acetylation (via Tip60 or GCN5) at the cad promoter is still subject to debate (13, 19), but for many other target promoters this has been well demonstrated (see, e.g., reference 19). Based on these observations, it has been proposed that Myc needs to recruit both P-TEFb and histone acetyltransferases to activate its target genes but that the relative contributions of these two pathways differ for individual target promoters (14). It is tempting to speculate that the P-TEFb-dependent activation pathway requires Myc binding sites in close proximity to the transcription start site and therefore that the target genes with heavy reliance on P-TEFb for their activation make up the dMyc targets with a downstream E-box. We have therefore addressed the roles of P-TEFb and the Tip60 complex in the regulation of these genes. We found that RNAi against the Tip60 components pontin/tip49 or tra1/trrap did not affect the activity of the nnp-1 or the CG4364 luciferase reporter within 48 h after transfection of S2 cells, and RNAi against the P-TEFb component cdk9 or cyclin T led to reproducible increases rather than decreases in reporter gene activity (data not shown). These observations raise the possibility that P-TEFb and the Tip60 complex act redundantly in this process; alternatively, other cofactors might be involved in the regulation of these dMyc targets, e.g., components of the Brm complex (the vertebrate Brm homologs Brg1 and hBrm have also been shown to be recruited to the cad promoter by c-Myc and to play a role in its regulation [35]). The identification of these cofactors will undoubtedly be of major importance for an understanding of Myc function, and we believe that the target genes identified in this report as well as the reporter constructs that were established will be of great help in this endeavor.
Supplementary Material
Acknowledgments
We thank Eva Niederer for FACS analysis, Ruth Keist and Andrea Patrignani for help with microarrays, George Hausmann for S2 cells and an introduction to luciferase assays, and Ernst Hafen and Martin Eilers for critical reading of the manuscript.
This work was supported by grants from the Swiss National Science Foundation and the Zürcher Hochschulverein/FAN (to P.G.).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Amati, B., S. R. Frank, D. Donjerkovic, and S. Taubert. 2001. Function of the c-Myc oncoprotein in chromatin remodeling and transcription. Biochim. Biophys. Acta 1471:M135-M145. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28-36. [PubMed] [Google Scholar]
- 3.Baldi, P., and A. D. Long. 2001. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17:509-519. [DOI] [PubMed] [Google Scholar]
- 4.Baudino, T. A., and J. L. Cleveland. 2001. The Max network gone mad. Mol. Cell. Biol. 21:691-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergman, C. M., B. D. Pfeiffer, D. E. Rincon-Limas, R. A. Hoskins, A. Gnirke, C. J. Mungall, A. M. Wang, B. Kronmiller, J. Pacleb, S. Park, M. Stapleton, K. Wan, R. A. George, P. J. de Jong, J. Botas, G. M. Rubin, and S. E. Celniker. 2002. Assessing the impact of comparative genomic sequence data on the functional annotation of the Drosophila genome. Genome Biol. 3:RESEARCH0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourbon, H. M., G. Gonzy-Treboul, F. Peronnet, M. F. Alin, C. Ardourel, C. Benassayag, D. Cribbs, J. Deutsch, P. Ferrer, M. Haenlin, J. A. Lepesant, S. Noselli, and A. Vincent. 2002. A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech. Dev. 110:71-83. [DOI] [PubMed] [Google Scholar]
- 7.Cawley, S., S. Bekiranov, H. H. Ng, P. Kapranov, E. A. Sekinger, D. Kampa, A. Piccolboni, V. Sementchenko, J. Cheng, A. J. Williams, R. Wheeler, B. Wong, J. Drenkow, M. Yamanaka, S. Patel, S. Brubaker, H. Tammana, G. Helt, K. Struhl, and T. R. Gingeras. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116:499-509. [DOI] [PubMed] [Google Scholar]
- 8.Celniker, S. E., and G. M. Rubin. 2003. The Drosophila melanogaster genome. Annu. Rev. Genomics Hum. Genet. 4:89-117. [DOI] [PubMed] [Google Scholar]
- 9.Celniker, S. E., D. A. Wheeler, B. Kronmiller, J. W. Carlson, A. Halpern, S. Patel, M. Adams, M. Champe, S. P. Dugan, E. Frise, A. Hodgson, R. A. George, R. A. Hoskins, T. Laverty, D. M. Muzny, C. R. Nelson, J. M. Pacleb, S. Park, B. D. Pfeiffer, S. Richards, E. J. Sodergren, R. Svirskas, P. E. Tabor, K. Wan, M. Stapleton, G. G. Sutton, C. Venter, G. Weinstock, S. E. Scherer, E. W. Myers, R. A. Gibbs, and G. M. Rubin. 2002. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3:RESEARCH0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clemens, J. C., C. A. Worby, N. Simonson-Leff, M. Muda, T. Maehama, B. A. Hemmings, and J. E. Dixon. 2000. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. USA 97:6499-6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang, C. V. 1999. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 19:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De La Cova, C., M. Abril, P. Bellosta, P. Gallant, and L. A. Johnston. 2004. Drosophila myc regulates organ size by inducing cell competition. Cell 117:107-116. [DOI] [PubMed] [Google Scholar]
- 13.Eberhardy, S. R., C. A. D'Cunha, and P. J. Farnham. 2000. Direct examination of histone acetylation on Myc target genes using chromatin immunoprecipitation. J. Biol. Chem. 275:33798-33805. [DOI] [PubMed] [Google Scholar]
- 14.Eberhardy, S. R., and P. J. Farnham. 2001. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J. Biol. Chem. 276:48562-48571. [DOI] [PubMed] [Google Scholar]
- 15.Eberhardy, S. R., and P. J. Farnham. 2002. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J. Biol. Chem. 277:40156-40162. [DOI] [PubMed] [Google Scholar]
- 16.Eisenman, R. N. 2001. Deconstructing myc. Genes Dev. 15:2023-2030. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez, P. C., S. R. Frank, L. Wang, M. Schroeder, S. Liu, J. Greene, A. Cocito, and B. Amati. 2003. Genomic targets of the human c-Myc protein. Genes Dev. 17:1115-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.FlyBase Consortium. 2003. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 31:172-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frank, S. R., T. Parisi, S. Taubert, P. Fernandez, M. Fuchs, H. M. Chan, D. M. Livingston, and B. Amati. 2003. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 4:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frank, S. R., M. Schroeder, P. Fernandez, S. Taubert, and B. Amati. 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15:2069-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallant, P., Y. Shiio, P. F. Cheng, S. M. Parkhurst, and R. N. Eisenman. 1996. Myc and Max homologs in Drosophila. Science 274:1523-1527. [DOI] [PubMed] [Google Scholar]
- 22.Haggerty, T. J., K. I. Zeller, R. C. Osthus, D. R. Wonsey, and C. V. Dang. 2003. A strategy for identifying transcription factor binding sites reveals two classes of genomic c-Myc target sites. Proc. Natl. Acad. Sci. USA 100:5313-5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hateboer, G., H. T. Timmers, A. K. Rustgi, M. Billaud, L. J. van't Veer, and R. Bernards. 1993. TATA-binding protein and the retinoblastoma gene product bind to overlapping epitopes on c-Myc and adenovirus E1A protein. Proc. Natl. Acad. Sci. USA 90:8489-8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin, W., R. M. Riley, R. D. Wolfinger, K. P. White, G. Passador-Gurgel, and G. Gibson. 2001. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat. Genet. 29:389-395. [DOI] [PubMed] [Google Scholar]
- 25.Johnston, L. A., D. A. Prober, B. A. Edgar, R. N. Eisenman, and P. Gallant. 1999. Drosophila myc regulates cellular growth during development. Cell 98:779-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanazawa, S., L. Soucek, G. Evan, T. Okamoto, and B. M. Peterlin. 2003. c-Myc recruits P-TEFb for transcription, cellular proliferation and apoptosis. Oncogene 22:5707-5711. [DOI] [PubMed] [Google Scholar]
- 27.Maheswaran, S., H. Lee, and G. E. Sonenshein. 1994. Intracellular association of the protein product of the c-myc oncogene with the TATA-binding protein. Mol. Cell. Biol. 14:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maldonado-Codina, G., S. Llamazares, and D. M. Glover. 1993. Heat shock results in cell cycle delay and synchronisation of mitotic domains in cellularised Drosophila melanogaster embryos. J. Cell Sci. 105:711-720. [DOI] [PubMed] [Google Scholar]
- 29.Nair, S. K., and S. K. Burley. 2003. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell 112:193-205. [DOI] [PubMed] [Google Scholar]
- 30.Nilsson, J. A., and J. L. Cleveland. 2003. Myc pathways provoking cell suicide and cancer. Oncogene 22:9007-9021. [DOI] [PubMed] [Google Scholar]
- 31.O'Connell, B. C., A. F. Cheung, C. P. Simkevich, W. Tam, X. Ren, M. K. Mateyak, and J. M. Sedivy. 2003. A large scale genetic analysis of c-Myc-regulated gene expression patterns. J. Biol. Chem. 278:12563-12573. [DOI] [PubMed] [Google Scholar]
- 32.Ohler, U., G. C. Liao, H. Niemann, and G. M. Rubin. 2002. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 3:RESEARCH0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orian, A., B. Van Steensel, J. Delrow, H. J. Bussemaker, L. Li, T. Sawado, E. Williams, L. W. Loo, S. M. Cowley, C. Yost, S. Pierce, B. A. Edgar, S. M. Parkhurst, and R. N. Eisenman. 2003. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 17:1101-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oster, S. K., C. S. Ho, E. L. Soucie, and L. Z. Penn. 2002. The myc oncogene: MarvelouslY Complex. Adv. Cancer Res. 84:81-154. [DOI] [PubMed] [Google Scholar]
- 35.Pal, S., R. Yun, A. Datta, L. Lacomis, H. Erdjument-Bromage, J. Kumar, P. Tempst, and S. Sif. 2003. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol. Cell. Biol. 23:7475-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pease, A. C., D. Solas, E. J. Sullivan, M. T. Cronin, C. P. Holmes, and S. P. Fodor. 1994. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc. Natl. Acad. Sci. USA 91:5022-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider, I. 1972. Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 27:353-365. [PubMed] [Google Scholar]
- 38.Schreiber-Agus, N., D. Stein, K. Chen, J. S. Goltz, L. Stevens, and R. A. DePinho. 1997. Drosophila Myc is oncogenic in mammalian cells and plays a role in the diminutive phenotype. Proc. Natl. Acad. Sci. USA 94:1235-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuldiner, O., S. Shor, and N. Benvenisty. 2002. A computerized database-scan to identify c-MYC targets. Gene 292:91-99. [DOI] [PubMed] [Google Scholar]
- 40.Trumpp, A., Y. Refaeli, T. Oskarsson, S. Gasser, M. Murphy, G. R. Martin, and J. M. Bishop. 2001. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature 414:768-773. [DOI] [PubMed] [Google Scholar]
- 41.Vervoorts, J., J. M. Luscher-Firzlaff, S. Rottmann, R. Lilischkis, G. Walsemann, K. Dohmann, M. Austen, and B. Luscher. 2003. Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep. 4:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wanzel, M., S. Herold, and M. Eilers. 2003. Transcriptional repression by Myc. Trends Cell Biol. 13:146-150. [DOI] [PubMed] [Google Scholar]
- 43.Zeller, K. I., A. G. Jegga, B. J. Aronow, K. A. O'Donnell, and C. V. Dang. 2003. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 4:R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou, Z. Q., and P. J. Hurlin. 2001. The interplay between Mad and Myc in proliferation and differentiation. Trends Cell Biol. 11:S10-S14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.