Abstract
We have previously identified poly(A)-binding protein 1 (PABP1) as a ligand for paxillin and shown that the paxillin-PABP1 complex undergoes nucleocytoplasmic shuttling. By targeting the paxillin-binding subdomain sequences in PABP1, we have generated mutants of PABP1 that do not bind to cellular paxillin. Here we report that paxillin association is necessary for efficient nuclear export of PABP1 and that RNA interference of paxillin drives the nuclear accumulation of PABP1. Furthermore, ablation of paxillin-PABP1 association impeded a number of indices of cell motility including spreading on fibronectin, cell migration on two-dimensional matrices, and transmigration in Boyden chambers. These data indicate that PABP1 must associate with paxillin in order to be efficiently transported from the nucleus to the cytoplasm and that this event is necessary for cells to remodel their focal adhesions during cell migration.
The leading edge or lamellipodium of migrating cells is the point at which integrins engage the extracellular matrix, an interaction that leads to integrin clustering into an array of small multiprotein focal complexes (31) containing an array of structural and signaling proteins that act to establish and maintain the polarity of the migrating cell. The 68-kDa protein paxillin is an abundant component of focal complexes at the leading edge of migrating cells. In addition to binding directly to integrin cytodomains, paxillin contains numerous protein-binding modules that interact with a variety of structural and signaling proteins and is therefore classified as a molecular adaptor or scaffold protein (23). Paxillin has an N-terminal region with five leucine-rich motifs, termed LD domains, and a C-terminal portion with four tandem LIM domains. The LIM domains contain information for targeting paxillin to focal complexes and can bind directly to tubulin (9). LD domains are protein-protein interaction motifs with the consensus sequence LDXXLLXXL, and these mediate the interaction of paxillin with a number of proteins that regulate cell migration. LD domains seem to display some degree of selectivity with respect to the ligands they recruit; LD1 binds integrin-linked kinase and the actin-binding proteins vinculin and actopaxin, while LD2 associates with focal adhesion kinase (FAK) and the ARF-GAP protein, p95PKL (23). Based on mutational analysis, Turner and coworkers (23) have identified the regions of proteins that associate with the various LD domains and termed them paxillin-binding subdomains or PBSs.
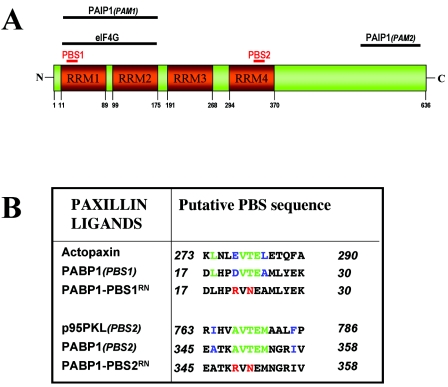
Using a proteomic approach, we have recently identified an association between paxillin and the mRNA-binding protein, PABP1. Moreover, the paxillin-PABP1 complex undergoes nucleocytoplasmic shuttling and is localized to sites of translation in the perinuclear endoplasmic reticulum and at the leading edge of migrating cells (29). PABP1 consists of an N-terminal portion that contains four tandem RNA-binding motifs (RRM) and a C-terminal region with homology to an ubiquitin E3 ligase, called HYD (Fig. 1A) (12). The RRM domains bind the mRNA poly(A) tail and also are known to interact with the eIF4G complex at the 5′ mRNA cap. This PABP1-eIF4G interaction is important for the circularization of mRNA during translation, and PABP1 is also proposed to participate in mRNA polyadenylation and nuclear export (12). Interestingly, PABP1 contains two regions of sequence with similarity to proposed PBSs. One of these is in RRM1 (PABP1-PBS1; residues 17 to 30) and has some similarity to the PBS in actopaxin, and the other is in RRM4 (PABP1-PBS2; residues 345 to 358) and has similarity to the C-terminal PBS of p95PKL (Fig. 1B). The three-dimensional structure of RRM1 and RRM2 of PABP1 cocrystallized with poly(A)-RNA (6) reveals that PABP1-PBS1 corresponds to a surface-exposed loop connecting the first β-sheet to the first α-helix of PABP1. None of the residues in this loop are directly involved with the coordination of mRNA, making it an excellent candidate for a functional paxillin-binding site.
FIG. 1.
Putative PBSs within PABP1. (A) Domain structure of PABP1, indicating the tandem RRM1 to RRM4 and a C-terminal region with homology to the hyperplastic disk protein, HYD. The positions of the putative PBS1 (residues 17 to 30) and PBS2 (residues 345 to 358) and the binding sites for elF4G and PAIP1 are indicated. (B) Sequence alignment of the putative PBSs of PABP1 with the PBSs of actopaxin and p95PKL. Residues identical to actopaxin or p95PKL are shown in green, and those with similarity are shown in blue. The amino acid substitutions introduced into the PABP1-PBS1RN and PABP1-PBS2RN mutants are shown in red.
To investigate the potential role of paxillin in PABP1 trafficking and the influence of this on cell migration, we have mutated the PBSs in PABP1. Here we report that mutation of PBS2 results in a form of PABP1 that is unable to associate with paxillin within the cell, although its ability to bind mRNA is unaffected. Mutants of PABP1 with reduced paxillin binding are inefficiently exported from the nucleus, indicating that paxillin plays a role in facilitating the nuclear export of mRNA. Moreover, expression of these mutant PABP1s markedly increases focal adhesion size and reduces cell spreading and migration.
MATERIALS AND METHODS
Materials.
Monoclonal mouse anti-paxillin and anti-Hic-5 antibodies were from Transduction Laboratories (BDBiosciences). Rabbit anti-glutathione S-transferase GST and anti-HA and mouse monoclonal anti-HA antibodies were from Santa Cruz Biotechology (Santa Cruz, Calif.). Fluorescein isothiocyanate FITC-conjugated goat anti-mouse and anti-rabbit immunoglobulins were from Southern Biotechnology (Birmingham, Ala.). Magnetic sheep anti-mouse immunoglobulin G IgG Dynabeads (Dynal, Oslo, Norway) and bovine serum albumin (BSA) were from First Link (United Kingdom) Ltd. Enhanced chemiluminescence reagents were from Pierce and Warriner Ltd. (Chester, United Kingdom). Cell culture medium was from Life Technologies (Rockville, Md.), and fetal calf serum was from Sera-Q (Tunbridge Wells, United Kingdom). Cell culture plastics including transwell inserts and chambers were from Nunc A/S (Roskilde, Denmark). Fugene 6 transfection reagent was from Roche Diagnostics GmbH (Mannheim, Germany). Nucleofector and nucleofection reagents were from Amaxa GmbH (Koeln, Germany). All other reagents were purchased from Sigma Chemical Co. (Poole, United Kingdom).
Expression plasmids.
The human sequence (residues 1 to 636) of PABP1, the indicated PABP1 mutants, and enhanced green fluorescent protein (EGFP)-tagged PABP1 (generated by PCR) were cloned into the pcDNA3 vector. EGFP-paxillin-α and -β were in the pEGFP-C1 vector as described previously (13). The mU6pro vector was used for the expression of RNA duplexes (21). A gene-specific sequence exclusive to murine paxillin was targeted (5′-CCCTGACGAAAGAGAAGCCTA-3′). All plasmids were purified by CsCl banding prior to transfection.
Cell culture and transfection.
Swiss and NIH 3T3 mouse fibroblasts were grown in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal calf serum as described previously (19) and transfected with Fugene 6 as specified by the manufacturer. The ratio of Fugene 6 to DNA was maintained at 3 μl of Fugene 6 to 1 μg of DNA. Where indicated, transfections were carried out using the Amaxa Nucleofector system. Briefly, cells were grown to 80% confluence, detached by trypsinization, washed in phosphate-buffered saline, and resuspended in Amaxa solution R together with 5 μg of DNA. Following electroporation (in the Nucleofector [program T-20]), the cells were replated and all experiments were carried out 24 h posttransfection. The efficiency of transfection in the Nucleofector was determined by immunofluorescence to be >90%.
Expression and purification of paxillin and PABP1 fusion proteins.
The human sequence of PABP1 (residues 1 to 636) and the indicated PABP1 mutants were cloned into BamHI and SacI sites of the His-tagged bacterial expression vector pET15 (Qiagen). PET15-PABP1 was transformed into Escherichia coli strain BL-21, grown to a density of 0.3 (optical density at 600 nm) at 37°C, and then induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for a further 2 h at 22°C. E. coli was lysed in a French press in a buffer containing 20 mM morpholinepropanesulfonic acid (MOPS) buffer (pH 7.4), 0.5 M NaCl, 20 mM imidazole, 2 mM benzamidine, 30 μg of leupeptin per ml, 15 μg of aprotinin per ml, and 1 mM 4-(2-aminoethyl)benzynesulfonyl fluoride (AEBSF). Then 1.0% Igepal CA-630 was added, the lysates were clarified by centrifugation, and His-PABP1 was recovered by incubation with 50% nitrilotriacetate-agarose beads for 1 h at 4°C. His-PABP1 was eluted with a buffer containing 100 mM EDTA at 4°C and dialyzed against phosphate-buffered saline (pH 7.4). The construct encoding GST-tagged human paxillin (residues 1 to 591) was expressed and purified as described previously (29).
Immunoprecipitations and pull-down assays.
Cells were grown to 60% confluence, washed twice in ice-cold phosphate-buffered saline, and lysed in a buffer containing 200 mM NaCl, 75 mM Tris-HCl (pH 7.0), 15 mM NaF, 1.5 mM Na3VO4, 7.5 mM EDTA, 7.5 mM EGTA, 1.5% (vol/vol) Triton X-100, 0.75% (vol/vol) Igepal CA-630, 50 μg of leupeptin per ml, 50 μg of aprotinin per ml, and 1 mM AEBSF (1.14 μl/cm2 [culture area]). The cells were scraped from the dish with a rubber policeman, and lysates were passed three times through a 27-gauge needle and centrifuged at 10,000 × g for 10 min at 4°C. For immunoprecipitations, magnetic beads conjugated to sheep anti-mouse IgG were blocked in phosphate-buffered saline containing 0.1% (wt/vol) BSA and then bound to mouse anti-paxillin, anti-HA or the control IgG1 antibody, MOPC. For pull-down assays, anti-mouse-conjugated magnetic beads were bound to mouse anti-rabbit IgG followed by rabbit anti-GST and finally GST or GST-paxillin (residues 1 to 591). Magnetic beads that were coated with antibodies or GST fusion proteins were incubated with lysates for 2 h at 4°C with constant rotation. Unbound proteins were removed by extensive washing in lysis buffer, and coimmunoprecipitating material was analyzed by Western blotting as described previously (20).
Immunofluorescence.
For immunofluorescence analysis, cells were fixed in 4% (wt/vol) paraformaldehyde in phosphate-buffered saline for 20 min at room temperature and permeabilized with 0.2% (vol/vol) Triton X-100 in phosphate-buffered saline for 5 min, and nonspecific binding sites were blocked for 1 h with phosphate-buffered saline containing 1% (wt/vol) BSA. The cells were incubated with the primary antibodies at room temperature for 1 h. Detection was with FITC- or Texas red-conjugated secondary antibodies. The actin cytoskeleton was visualized by counterstaining with FITC- or Texas red-conjugated phalloidin in phosphate-buffered saline for 10 min at room temperature. Coverslips were mounted in Profade antifade mountant (Molecular Probes) and viewed on a Leica confocal laser-scanning microscope, with EGFP fluorescence being collected into the fluorescein channel.
Cell-spreading assays.
Tissue culture plates (24 wells) were coated overnight at 4°C with fibronectin (20 μg/ml) and then blocked with 2% (wt/vol) BSA. Cells were transfected with PABP1 or mutant PABP1s in conjunction with a β-galactosidase expression construct, and 24 h later the cells were harvested by trypsinization and collected by centrifugation in the presence of 20 μg of soyabean trypsin inhibitor per ml. The cell suspensions were added immediately to fibronectin-coated wells in serum-free DMEM containing 10 ng of PDGF-BB per ml. The cells were allowed to attach for 60 min, and nonadherent cells were removed by six washes with phosphate-buffered saline. Attached cells were fixed for 1 min in 0.2% glutaraldehyde containing 5 mM EGTA, and β-galactosidase-expressing cells were visualized by incubation with 5 mM potassium ferricyanide and 1 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) per ml overnight at 37°C. To obtain an index of cell spreading, the area of cells expressing β-galactosidase was determined by delineation of the cell envelope, using NIH Image software (21).
Migration assays.
For wound-healing assays, NIH 3T3 fibroblasts were transfected with PABP1 or mutant PABP1s by using the Amaxa Nucleofector, plated on 3-cm tissue culture plates, and allowed to grow to confluence over 24 h. Crosses were scraped into the confluent monolayer by using a plastic pipette tip. The cells were washed with phosphate-buffered saline and allowed to migrate in DMEM supplemented with 1% (vol/vol) fetal calf serum and 10 ng of PDGF-BB per ml at 37°C. Photographs of the wounds were taken at 0, 6, 14, and 28 h postwounding. Wound closure was determined by quantification of these images using NIH image software. For transmigration assays, cells were transfected with PABP1 or mutant PABP1s using the Amaxa Nucleofector and seeded into the top well of a Boyden chamber by using Transwell filter inserts that had been coated with 10 μg of fibronectin per ml on the lower surface. Migration was allowed to proceed for 3 h at 37°C with 100 ng of LPA added to the lower chamber. The number of cells that had migrated to the lower chamber was determined by staining with 1% toludine blue and expressed as a proportion of the total quantity of cells added to the assay. For time-lapse fluorescence video microscopy, cells transfected with EGFP-tagged PABP1s were plated onto coverslips coated with 3 μg of fibronectin per ml and incubated for 1 h at 37°C. The cells were then transferred to a heated microscope stage, and their migration was observed by fluorescence time-lapse video microscopy in the presence of 10 ng of PDGF-BB per ml. Images were collected at 1-min intervals for 12 min.
RESULTS
The PBS2 sequence in PABP1 is required for association with cellular paxillin.
To investigate the role of the PBS-like sequences in PABP1 in paxillin binding, we generated His-tagged PABP1 (wild type) and variants thereof with mutations in PBS1 (PABP1- PBS1RN), PBS2 (PABP1-PBS2RN), and both PBS1 and PBS2 (PABP1-PBS1/2RN) (Fig. 1B). Pull-down assays indicated that mutation of either PBS1 or PBS2 was insufficient to negate the binding of purified His-PABP1 to GST-paxillin (Fig. 2A). However, if both PBSs were disrupted (PABP1-PBS1/2RN), paxillin binding was lost (Fig. 2A) without affecting binding of the protein to poly(A)-RNA (Fig. 2B). These data indicate that regions of PABP1 within the RRM1 and RRM4 domains (Fig. 1) have the potential to bind to purified paxillin and to do so independently of one another.
FIG. 2.
In vitro association of mutant PABP1s with paxillin and poly(A). Purified recombinant His-tagged PABP1 and the indicated PABP1 mutants (see Fig. 1), were incubated with magnetic beads conjugated to GST, GST-paxillin (A), or poly(A)-RNA (B) in a buffer containing 1.5% (vol/vol) Triton X-100 and 1% (wt/vol) BSA. Coprecipitating proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by Western blotting with antibodies recognizing the hexa-His epitope (6XHis). The loading of the GST fusion proteins was confirmed by Western blotting for GST. The His-tagged 482-655 fragment of talin was included as a negative control for poly(A) binding in panel B. WT, wild type.
To assess the contribution that these sequences make to the association of PABP1 with paxillin within the cell, we expressed PABP1 and PABP1 PBS mutants as HA-tagged proteins in NIH 3T3 fibroblasts. Lysates from cells expressing these proteins were incubated with GST-paxillin or subjected to immunoprecipitation with anti-paxillin antibodies. The immobilized material was then analysed by Western blotting. In this cellular context, mutation of PBS2 alone was sufficient to abrogate the association of PABP1 with paxillin, as determined using both GST-paxillin pull-down (Fig. 3A) and paxillin immunoprecipitation (Fig. 3B) assays. To further confirm this finding, we coexpressed GFP-paxillin with HA-PABP1- PBS1RN or HA-PABP1-PBS2RN and immunoprecipitated the HA-tagged PABP1 by using either mouse monoclonal or rabbit polyclonal anti-HA antibodies. Western blotting of these immunoprecipitates indicated that GFP-tagged paxillin coimmunoprecipitated with PABP1- PBS1RN but not with PABP1-PBS2RN (Fig. 3C). Taken together, these data indicate that although both PBS1 and PBS2 sequences in PABP1 have the potential to bind purified paxillin in vitro, PBS2 is the key determinant for association of PABP1 with cellular paxillin.
FIG. 3.
Association of mutant PABP1s with paxillin in fibroblast lysates. NIH 3T3 fibroblasts were transfected with HA-tagged PABP1 and the indicated PABP1 mutants in the absence (A and B) and presence (C) of EGFP-paxillin-β. The cells were then grown to 60% confluence over 24 h, lysed in a buffer containing 1.0% (vol/vol). Triton X-100 and 0.5% (vol/vol) Igepal CA-630, and clarified by centrifugation. Lysates were incubated with magnetic beads conjugated to GST, GST-paxillin (pull-down) (A), a control IgG1 (MOPC) (B and C) anti-paxillin monoclonal antibody (anti-Pax) (B), a mouse monoclonal anti-IIA antibody [anti-HA(m)] (C), or a rabbit polyclonal antibody recognizing the HA epitope [anti-HA(r)] (C) for 2 h at 4°C. Coprecipitating proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by Western blotting with antibodies recognizing the HA epitope, paxillin, or GFP as indicated. The loading of the GST fusion proteins, paxillin, and PABP1 was confirmed by Western blotting for GST, paxillin, and anti-HA respectively (lower panels). The migration positions of paxillin, EGFP-paxillin (GFP-Pax), PABP1, and the 50-kDa IgG heavy chain (IgG-HC) are shown. The asterisk indicates the presence of a paxillin-immunoreactive band that contains ubiquitin as determined by Western blotting (not shown). IP, immunoprecipitation; WT, wild type.
Following overexpression of either wild-type PABP1 or PABP1-PBS1RN (but not PABP1-PBS2RN), an additional higher-molecular-weight form of immunoreactive paxillin was detected in the paxillin immunoprecipitates (Fig 3B; asterisk). Western blotting indicated that this may represent a ubiquitinylated form of paxillin (not shown). Ubiquitinylation of paxillin has been reported previously (7), and our data suggest that association with PABP1 may promote this post-translational modification. Furthermore, we have consistently found that overexpression of wild-type PABP1 and PABP1-PBS1RN (but not PABP1-PBS2RN or PABP1-PBS1/2RN) increased the apparent efficiency of the paxillin immunoprecipitation by approximately twofold (Fig. 3B; lower panel); this may be due to the ability of PABP1 to self-associate (14).
Paxillin association is required for efficient nuclear export of PABP1.
We used fluorescence microscopy to study the distribution of mutant PABP1s expressed in NIH 3T3 fibroblasts. Wild-type HA-tagged PABP1 presented a predominantly cytoplasmic distribution (Fig. 4A), which was indistinguishable from that of endogenous PABP1 (not shown), indicating appropriate localization of this epitope-tagged protein. Quantification of three-dimensional reconstructions of confocal image stacks indicated that less than 5% of cellular PABP1 was present in the nucleus (Fig. 4E). PABP1-PBS1RN, a mutant that associates with cellular paxillin, had an intracellular distribution similar to that of the wild-type protein (Fig. 4B) and colocalized closely with paxillin in the perinuclear region (Fig. 5A). By contrast, mutants of PABP1 that do not associate with paxillin have an altered intracellular distribution. Approximately 40% of cellular PABP1-PBS2RN and PABP1-PBS1/2RN accumulated in the nucleus (Fig. 4C to E) and had reduced colocalization with paxillin in the cytoplasm (Fig. 5B). In addition, it is notable that the size (Fig. 5C) and paxillin content (Fig. 5D) of focal adhesions was increased by the expression of PBS2 mutants of PABP1.
FIG. 4.
Subcellular localization of PABP1 with mutated PBS sequences. (A to D) NIH 3T3 fibroblasts were transfected with HA-tagged PABP1 (WT) or the indicated HA-PABP1 mutants, fixed, and stained with anti-HA followed by a Texas red-conjugated secondary antibody. Bar, 20 μm. (E) The proportion of cellular PABP1 present in the nucleus was determined by quantification of confocal images. Values are mean and standard error of the mean.
FIG. 5.
Expression of mutant PABP1s influences the size of paxillin-containing focal adhesions. (A and B) Cells were transfected with HA-PABP1-PBS1RN or HA-PABP1-PBS2RN in combination with EGFP-paxillin, fixed, and stained with anti-HA (red), with EGFP-paxillin being visualised directly (green). Images are presented as an extended focus projection of serial confocal images from a Z-stack, in which all points of the cell are seen as “in focus.” The images shown in panels A to A” and B to B” are representative examples of PABP1-PBS1RN-and PABP1-PBS2RN-expressing cells respectively. Bar, 10 μm. (C and D) NIH 3T3 fibroblasts were transfected with PABP1 (WT) or the indicated PABP1 mutants by using the Amaxa Nucleofector, fixed, and stained for paxillin by indirect immunofluorescence. The size (C) and paxillin content (D) of paxillin-containing focal adhesions were assessed by quantification of confocal images. Values are mean and standard error of the mean.
To determine whether the altered nucleocytoplasmic distribution of these mutant PABP1s was owing to their inability to associate with paxillin, we employed RNA interference (RNAi) to suppress the cellular levels of paxillin. Expression of the paxillin RNAi vector (mu6pro-PAX) reduced levels of paxillin (but not the paxillin family member Hic-5) by at least 10-fold, and a control hairpin RNA (mu6pro-CON) was ineffective in this regard (Fig. 6A). We therefore determined the effect of mu6pro-PAX on the nucleocytoplasmic distrubution of PABP1. Indeed, RNAi of paxillin strongly promoted the accumulation of PABP1 within the nucleus (Fig. 6B to D), and this could be rescued by expression of GFP-paxillin-α (Fig. 6E) and GFP-paxillin-β (Fig. 6F) but not by GFP (Fig. 6G) or an empty vector control (Fig. 6D). Quantification of confocal images indicated that the degree of nuclear accumulation of PABP1 that occurred following paxillin RNAi was similar to that found for the PBS2 mutants of PABP1 (Fig. 6H). Addition of leptomycin B (which blocks nuclear export of PABP1 and paxillin [29]) caused paxillin, PABP1, and mutants of PABP1 to accumulate in the nucleus (data not shown). These data indicate that although the nuclear import of PABP1 does not require paxillin association, PABP1 must recruit paxillin in order to be efficiently exported to the cytoplasm.
FIG. 6.
RNAi of paxillin leads to nuclear accumulation of PABP1; rescue with either the α or β isoforms of paxillin. (A) NIH 3T3 fibroblasts were transfected with mu6pro-CON or mu6pro-PAX by using the Amaxa Nucleofector. The cells were lysed 24 h following transfection, and the lysates were probed for the presence of paxillin, vinculin, Hic-5, and actin by Western blotting. (B to G) Cells were transfected with HA-PABP1 in combination with mu6pro-CON (B) or mu6pro-PAX (C) or with HA-PABP1 and mu6pro-PAX in combination with pcDNA3 (D), GFP-paxillin-α (E), GFP-paxillin-β (F), or GFP (G). The cells were fixed 24 h following transfection and stained with anti-HA followed by a Texas red-congugated secondary antibody (red). Paxillin was stained with an anti-paxillin monoclonal antibody followed by a FITC-conjugated secondary antibody (B to D; green), or GFP was visualized directly (E to G; green). The plane of focus is 2.5 μm above the level of the substratum. Bar, 20 μm. (H) The proportion of cellular PABP1 present in the nucleus was determined by quantification of confocal images. Values are mean and standard error of the mean.
Paxillin-PABP1 association is required for normal cell spreading and migration.
The ability of PABP1 to repress its own synthesis has been reported previously and is owing to its ability to associate with the 5′ untranslated region (UTR) of the PABP1 mRNA (30). Indeed, the expression of recombinant PABP1 or mutant PABP1s results in suppression of endogenous PABP1 levels (Fig. 7A) and this phenomenon has allowed us to evaluate the effect of mutant PABP1s on cellular physiology. Following transfection with PABP1-PBS1RN, NIH 3T3 fibroblasts were found to spread on fibronectin to the same degree as cells expressing the wild-type protein (Fig. 7B and C). However, spreading of cells expressing PABP1s with the PBS2 sequence mutated was suppressed by approximately 40% (Fig. 7B and C). Indeed, this suppression of cell spreading was similar to that seen following paxillin RNAi or leptomycin B treatment (Fig. 7C). To analyze this in more detail, we used immunofluorescence microscopy and determined the distribution of paxillin following engagement with the extracellular matrix. Cells expressing wild-type PABP1 (data not shown) or PABP1-PBS1RN spread rapidly on fibronectin and assembled paxillin-containing focal adhesions in a radial array (Fig. 7D), which subsequently moved centripetally to cover the underside of the cell (Fig. 7E). This indicated that focal adhesions are rapidly remodeled following matrix engagement, thus facilitating cell spreading. In cells expressing PABP1-PBS2RN (Fig. 7F) or PABP1-PBS1/2RN (data not shown), paxillin was recruited to focal contacts shortly after engagement with the matrix. However, these structures did not remodel at a speed sufficient to support rapid cell spreading (Fig. 7G), and examination of phase-contrast images indicated that cells expressing PABP1-PBS2RN spread asymmetrically and that this seemed to proceed via the extension of abnormal processes (Fig. 7B; arrowed in enlarged panels) as opposed to the assembly of a well-organized lamella.
FIG. 7.
Effect of mutant PABP1s on cell spreading. (A) NIH 3T3 fibroblasts were transfected with GFP-PABP1 or the indicated mutants of GFP-tagged PABP1 by using the Amaxa Nucleofector, which resulted in expression of tagged protein in >90% of the cells. Cells were grown to 60% confluence over 24 h, lysed, and analyzed by Western blotting with antibodies recognizing PABP1. The migration positions of endogenous PABP1 and GFP-PABP1 are indicated. These data indicate that expression of recombinant PABP1 or mutant PABP1s reduces the cellular content of endogenous PABP1. (B) Cells were transfected with PABP1 (WT), PABP1-PBS1RN, PABP1-PBS2RN, or PABP1-PBS1/2RN by using the Amaxa Nucleofector. Following trypsinization, the cells were allowed to adhere to wells coated with 20 μg of fibronectin per ml and then fixed and photographed under phase-contrast microscopy. The areas encompassed by the white boxes are enlarged four-fold in the right-hand panels. Bars, 20 μm. (C) Cells were transfected with a β-galactosidase transfection marker in combination with the indicated PABP1 and paxillin RNAi constructs. Following trypsinization, the cells were allowed to adhere to fibronectin for 1 h in the absence and presence of 10 nM leptomycin B and then fixed and stained for β-galactosidase expression. Stained cells were photographed with a digital camera, and the area of transfected cells was determined by delineation of the cell envelope using NIH image software. The data are expressed as a percentage of the cell area of wild-type PABP1 expressing cells, dimethyl sulfoxide (DMSO) control, or mu6pro-CON-expressing cells as appropriate Values are mean and standard error of the mean. (D to G) Cells were transfected with HA-PABP1-PBS1RN or HA-PABP1-PBS2RN and allowed to grow to 60% confluence over 24 h. They were trypsinized and allowed to adhere to fibronectin-coated coverslips for 15 min (D and F) or 30 min (E and G) prior to fixation. Recombinant PABP1s were visualized with anti-HA followed by a Texas red-congugated secondary antibody (red), and paxillin was stained with an anti-paxillin monoclonal antibody followed by an FITC-conjugated secondary antibody (green). For the paxillin staining, the plane of focus is at the level of the substratum, and for images showing HA-PABP1s, the plane of focus is centered 2.5 μm above the coverslip. Bar, 10 μm.
Given a requirement for paxillin-PABP1 interaction in cell spreading, we reasoned that expression of mutant PABP1s would be likely to compromise cell migration. NIH 3T3 fibroblasts were transfected with PABP1 or the PBS mutants of PABP1 by using the Amaxa Nucleofector, and confluent monolayers were wounded with a plastic pipette tip. Cells expressing PABP1 or the PBS1 mutant of PABP1 (which binds to paxillin) closed the wound by 28 h (Fig. 8A and B). In contrast, cells expressing PABP1-PBS2RN had reduced migration and were able to achieve only 30% wound closure after 28 h (Fig. 8A and B). Inability of cells to migrate into a wound may indicate either a reduction in the speed of cell translocation or a defect in the directionality of migration. For instance, appropriate localization of β-actin mRNA is not required for cells to attain normal migration speeds but is essential to the directionality and persistence of cell translocation (22). To resolve this, cells transfected with GFP-tagged PABP1 mutants were plated onto fibronectin and analyzed using time-lapse video microscopy. Cells expressing GFP-tagged PABP1-PBS1RN exhibited a typical polar migratory morphology and migrated at a speed of approximately 1.0 to 1.5 μm min−1 (Fig. 8C). In contrast, cells expressing PABP1-PBS2RN were more rounded and continuously extended and retracted plasma membrane ruffles and small lamellipodia in a disorganised fashion, while the cell body appeared to be tethered to the substratum (Fig. 8C). To test the requirement for paxillin-PABP1 interaction in transmigration, Boyden chamber assays were performed. Consistent with the wound-healing assays and time-lapse video microscopy, PABP1-PBS2RN-expressing cells migrated toward fibronectin at a significantly reduced rate (Fig. 8D). Taken together, these data indicate that association of paxillin with PABP1 is essential for cell migration.
FIG. 8.
Effect of mutant PABP1s on cell migration. (A and B) NIH 3T3 fibroblasts were transfected with PABP1 (WT), PABP1-PBS1RN, or PABP1-PBS2RN by using the Amaxa Nucleofector and allowed to grow to confluence over 24 h. Following wounding, the cells were allowed to migrate in DMEM supplemented with 1% (vol/vol) FCS and 10 ng of PDGF-BB per ml. Photographs of the wounds were taken at various times postwounding, and the degree of wound closure was determined by delineation of the wound envelope by using NIH image software (values are mean and standard error of the mean; n = 10 fields). Bar, 140 μm. (C) Cells transfected with GFP-PABP1-PBS1RN or GFP-PABP1-PBS2RN were plated onto coverslips coated with 3 μg of fibronectin per ml and incubated for 1 h at 37°C. They were then transferred to a heated microscope stage, and their migration was observed by fluorescence time-lapse video microscopy in the presence of 10 ng of PDGF-BB per ml. Images were collected at 1-min intervals for 12 min. The red mask indicates the position of the cell at the outset of the recording period. Bar, 16 μm. (D) Cells were transfected with PABP1-PBS1RN or PABP1-PBS2RN by using the Amaxa Nucleofector and seeded into the upper well of a Boyden chamber with Transwell filter inserts that had been coated with 10 μg of fibronectin per ml on the bottom surface. Migration was allowed to proceed for 3 h at 37°C, with 100 ng of lysophosphatidic acid per ml added to the lower chamber. The number of cells that had migrated to the lower chamber was determined by staining with 1% toludine blue and expressed as a proportion of the total quantity of cells added to the assay. Values are mean and standard error of the mean; n = 12.
DISCUSSION
To investigate the significance of paxillin-PABP1 interaction within the cell, we have now mapped two paxillin-binding sites in PABP1 and generated mutants that are defective in paxillin binding. Mutations in PBS2 that abolished the binding of PABP1 to paxillin within the cell, or RNAi of paxillin resulted in altered nucleocytoplasmic trafficking of PABP1 that accumulated in the nucleus. Expression of mutants of PABP1 that are deficient in paxillin binding compromised PDGF-induced focal adhesion remodeling and led to a significant reduction in cell spreading and migration on fibronectin, in both wound healing and Boyden chamber assays.
Nature of the paxillin-PABP1 interaction.
The sequences proposed to interact with paxillin LD motifs (termed PBSs) have been identified by mutational and deletion analysis of a number of paxillin ligands, including vinculin, FAK, actopaxin, and p95PKL (23). Therefore, identification of potential paxillin-binding sites in PABP1 was initially based on sequence alignments with these proteins, and short sequences in RRM1 and RRM4 of PABP1 were found that matched the PBSs of actopaxin and p95PKL, respectively. Interestingly, these sequences are conserved in species from Xenopus to human that express paxillin or paxillin homologues but are not present in PABP1 of yeast, an organism that lacks paxillin. Subsequent mutagenesis confirmed that these PBSs are key determinants of the interaction between PABP1 and paxillin. Nevertheless, recent nuclear magnetic resonance spectroscopy (NMR) and crystallographic studies of the interaction between paxillin and FAK (1, 10) warn against a simple model for the interaction between paxillin LD motifs and their various binding partners. The FAK-paxillin interaction involves two hydrophobic patches on opposite faces of the FAK four-helix bundle, and the residues previously proposed to comprise the PBSs are not directly involved in contacting the paxillin LD domains. Similarly, our preliminary NMR studies indicate that although a single paxillin LD domain binds PABP1, a tandem array of two paxillin LD motifs binds far more tightly (data not shown). Further studies are required to provide a more detailed account of the intermolecular contact between paxillin and PABP1.
Interestingly, the nature of the paxillin-PABP1 interaction in vitro differs somewhat from that within the cell. When the interaction between purified proteins was studied, mutation of both PBS1 and PBS2 in PABP1 was required to abolish paxillin binding. However, only PBS2 was required for the interaction of PABP1 with paxillin in NIH 3T3 cells. This raises the possibility that within the cell, the PBS1 in PABP1 is occupied by an alternative LD domain-containing protein. LD domains have been identified in proteins other than those of the paxillin family, and binding of some of these to PBS sequences in target proteins is known to be mutually exclusive. For instance, binding of the LD domain protein E6-AP ubiquitin ligase to E6 protein is blocked by paxillin (25), and it will be important to establish whether the PBS1 in PABP1 recruits an LD domain-containing protein other than paxillin. Candidate proteins include the ubiquitin E3 ligase hyperplastic disk protein (HYD), which has homology to the C-terminal region of PABP1 (12). Paxillin ubiquitinylation has been proposed to displace it from focal adhesions (7), and our evidence suggests that association of paxillin with PABP1 promotes paxillin ubiquitinylation and is also required for efficient focal adhesion turnover.
Paxillin and PABP1 nucleocytoplasmic shuttling.
PABP1 mutants that do not associate with cellular paxillin accumulate in the nucleus. Since we can demonstrate no requirement for paxillin-PABP1 association in the nucleocytoplasmic shuttling of paxillin or the nuclear import of PABP1, we conclude that paxillin associates with PABP1 in the nucleus and facilitates its export to the cytoplasm. The fact that leptomycin B traps paxillin-PABP1 within the nucleus (29) indicates that it uses the CRM1-exportin pathway to exit the nucleus (28). Proteins with Rev-like leucine-rich nuclear export signal motifs generally utilize the CRM1 pathway, but no such sequence is present in PABP1, indicating that it is likely to leave the nucleus by virtue of nuclear export information contained within one of its binding partners. There are two sequences in paxillin that conform to the consensus (L***L**L**θ) NES found in a number of proteins including c-Abl, PKI-α, PKI-β, Trip6, and LPP (26). One of these is located in paxillin LD2, and the other is near LD5. Our preliminary data indicate that the putative NES in LD2 is not on its own sufficient to mediate paxillin export, and further studies are needed to establish whether paxillin contains a functional NES.
PABP1 is thought to bind mRNAs in the nucleus, thereby determining the length of the poly(A) tail. It is also reported to facilitate the export of mature mRNAs and their associated proteins (mRNPs) to the cytoplasm (12). The fact that paxillin is involved in the nucleocytoplasmic shuttling of PABP1 therefore suggests that paxillin plays a role in exporting PABP1 and, by inference, mRNAs. We have also shown that polypyrimidine tract binding protein-associated splicing factor is associated with the paxillin-PABP1 complex (29), although whether such a complex plays a role in the splicing of specific transcripts awaits further investigation.
Paxillin and PABP1 localization in the cytoplasm.
Once PABP1 has been exported from the nucleus, it must be localized to the appropriate compartment, and it is interesting that much of the cellular pool of paxillin colocalizes with PABP1 in the endoplasmic reticulum (29). Moreover, PABP1 (but not PABP2) colocalizes with paxillin at the leading edge of cells, and in situ hybridization studies suggest that focal adhesions contain mRNAs and also ribosomes (4). Thus, paxillin may also be involved in localizing PABP1 to sites involved in protein biosynthesis.
PABP1 mediates the circularization of mRNA during translation, and the PABP1 RRM1/2 domains associate with eIF4G of the translation complex (12). It is notable that eIF4G and eIF4E are absent from paxillin immunoprecipitates, indicating that paxillin is not part of the translation complex and suggesting that binding of paxillin and eIF4G to PABP1 is mutually exclusive. Indeed, since mRNAs are thought to be translationally repressed during transport (16), the various protein-protein interactions within mRNA transport complexes are likely to prevent association with the translational machinery, and paxillin may fulfil this role. Paxillin present in PABP1 immunoprecipitates is hypophosphorylated in comparison to total cellular paxillin (data not shown), suggesting that it is the nonphosphorylated form of paxillin that transports PABP1 out of the nucleus. Paxillin is extensively phosphorylated on both serine/threonine and tyrosine residues, and this is known to be one of the mechanisms by which the activity of the protein is regulated (24). It is therefore possible that phosphorylation of paxillin in the endoplasmic reticulum or at the lamellipodium triggers the release of PABP1, thereby activating translation.
Is paxillin involved in mRNA localization within cells?
Cells expressing PABP1-PBS2 mutants have well-defined effects on cell physiology, indicating that it is unlikely that binding of paxillin to PABP1 is required for global protein synthesis. Thus, although cell migration and focal adhesion remodeling are reduced, two-dimensional gel analysis reveals that the overall pattern of cellular protein expression is unchanged following disruption of the paxillin-PABP1 interaction (data not shown), suggesting that the paxillin-PABP1 interaction must play a more specific role within the cell. In addition to binding poly(A) tracts, PABP1 can selectively bind to a subset of mRNAs. It can bind regions in the 5′ UTR of its own mRNA thereby repressing its own translation (30), and it has also been shown to bind to sequences in certain 3′ UTRs, e.g., the dendritic localizer sequence of the vasopressin mRNA, and to be involved in the subcellular localization of this mRNA (15). It is therefore tempting to speculate that paxillin is involved in the targeting of a subset of PABP1-mRNA complexes within the cell. Given that the PABP1 RRM4 domain has a preference for sequences other than poly(A) (3), it is perhaps significant that it is this domain that associates with cellular paxillin. It will be interesting to determine whether paxillin is able to influence the mRNA species recruited to PABP1 and whether these are enriched in transcripts for proteins that are involved in cell adhesion and migration.
The concept that certain mRNAs are selectively localized within the cell is well established and is known to be crucial to development in Drosophila and Xenopus. Moreover, cytoskeletal elements are thought to be involved in this process (2). Interestingly, proteins of the VICKZ family of RNA-binding proteins appear to coordinate the transport and localization of mRNAs with cell migration (8, 17). For example, the protein ZBP1 is thought to shuttle β-actin mRNA to the leading edge of migrating fibroblasts (18), and IMP1, which binds the insulin-like growth factor II mRNA, is also localized to the leading edge in migrating cells (17). Delocalization of the β-actin mRNA either by microinjection of antisense oligonucleotides corresponding to the 3′ UTR of β-actin mRNA (22) or by overexpressing the regions of ZBP (KH domains 1 to 4) that bind to this mRNA (8) affects fibroblast migration. As a consequence, directional cell movement is compromised and the migration speed is reduced somewhat.
However, ablation of the paxillin-PABP1 interaction inhibits cell migration in a way that is more reminiscent of FAK−/− mouse embryo fibroblasts, which exhibit profoundly decreased rates of spreading and migration and more abundant and larger focal adhesions (11). These migratory defects are accounted for by a 10-fold reduction in the rate of focal adhesion disassembly (27), and we show here that cells expressing PABP1-PBS2RN have larger focal adhesions and appear to be incapable of remodeling these during cell spreading. Thus, our results support a role for the paxillin-PABP1 complex in a pathway that is required for focal adhesion remodeling. One possibility is that the paxillin-PABP1 complex targets mRNAs encoding proteins that regulate focal adhesion dynamics during cell spreading and migration. Indeed, a recent proteomic study has found PABP1 to be one of a range of mRNA-binding proteins that associate with focal adhesion components during the early stages of cell spreading (5). These workers propose these interactions to occur within a novel type of adhesive structure, termed a spreading initiation center, and report that they are rich in a number of transcripts, including rRNAs, suggesting that actively translating ribosomes are recruited to the initial points of cell contact with the extracellular matrix (5). It is possible that the paxillin-PABP1 complex is found within such a structure and that it contributes to the localization of mRNA during cell spreading and migration, but we do not rule out alternative roles for the paxillin-PABP1 interaction in focal adhesion turnover. The fact that this interaction appears to be correlated with paxillin ubiquitinylation is of interest in this regard. Whether this reveals a novel function for PABP1 or whether this, too, is linked to the well-established role of PABP1 in binding mRNAs remains to be established.
Acknowledgments
This work was generously supported by grants from the Wellcome Trust and Cancer Research U.K.
We thank Kul Sikand and Sam Wattam their invaluable assistance with the fluorescence microscopy.
REFERENCES
- 1.Arold, S. T., M. K. Hoellerer, and M. E. Noble. 2002. The structural basis of localization and signaling by the focal adhesion targeting domain. Structure 10:319-327. [DOI] [PubMed] [Google Scholar]
- 2.Bassell, G., and R. H. Singer. 1997. mRNA and cytoskeletal filaments. Curr. Opin. Cell Biol. 9:109-115. [DOI] [PubMed] [Google Scholar]
- 3.Burd, C. G., E. L. Matunis, and G. Dreyfuss. 1991. The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol. Cell. Biol. 11:3419-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chicurel, M. E., R. H. Singer, C. J. Meyer, and D. E. Ingber. 1998. Integrin binding and mechanical tension induce movement of mRNA and ribosomes to focal adhesions. Nature 392:730-733. [DOI] [PubMed] [Google Scholar]
- 5.de Hoog, C. L., L. J. Foster, and M. Mann. 2004. RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell 117:649-662. [DOI] [PubMed] [Google Scholar]
- 6.Deo, R. C., J. B. Bonanno, N. Sonenberg, and S. K. Burley. 1999. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell 98:835-845. [DOI] [PubMed] [Google Scholar]
- 7.Didier, C., L. Broday, A. Bhoumik, S. Israeli, S. Takahashi, K. Nakayama, S. M. Thomas, C. E. Turner, S. Henderson, H. Sabe, and Z. Ronai. 2003. RNF5, a RING finger protein that regulates cell motility by targeting paxillin ubiquitination and altered localization. Mol. Cell. Biol. 23:5331-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farina, K. L., S. Huttelmaier, K. Musunuru, R. Darnell, and R. H. Singer. 2003. Two ZBP1 KH domains facilitate beta-actin mRNA localization, granule formation, and cytoskeletal attachment. J. Cell Biol. 160:77-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herreros, L., J. L. Rodriguez-Fernandez, M. C. Brown, J. L. Alonso-Lebrero, C. Cabanas, F. Sanchez-Madrid, N. Longo, C. E. Turner, and P. Sanchez-Mateos. 2000. Paxillin localizes to the lymphocyte microtubule organizing center and associates with the microtubule cytoskeleton. J. Biol. Chem. 275:26436-26440. [DOI] [PubMed] [Google Scholar]
- 10.Hoellerer, M. K., M. E. Noble, G. Labesse, I. D. Campbell, J. M. Werner, and S. T. Arold. 2003. Molecular recognition of paxillin LD motifs by the focal adhesion targeting domain. Structure 11:1207-1217. [DOI] [PubMed] [Google Scholar]
- 11.Ilic, D., Y. Furuta, S. Kanazawa, N. Takeda, K. Sobue, N. Nakatsuji, S. Nomura, J. Fujimoto, M. Okada, and T. Yamamoto. 1995. Reduced cell motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature 377:539-544. [DOI] [PubMed] [Google Scholar]
- 12.Mangus, D. A., M. C. Evans, and A. Jacobson. 2003. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazaki, Y., H. Uchida, O. Hino, S. Hashimoto, and H. Sabe. 1998. Paxillin isoforms in mouse. Lack of the gamma isoform and developmentally specific beta isoform expression. J. Biol. Chem. 273:22435-22441. [DOI] [PubMed] [Google Scholar]
- 14.Melo, E. O., R. Dhalia, C. Martins de Sa, N. Standart, and O. P. de Melo Neto. 2003. Identification of a C-terminal poly(A)-binding protein (PABP)-PABP interaction domain: role in cooperative binding to poly (A) and efficient cap distal translational repression. J. Biol. Chem. 278:46357-46368. [DOI] [PubMed] [Google Scholar]
- 15.Mohr, E., N. Prakash, K. Vieluf, C. Fuhrmann, F. Buck, and D. Richter. 2001. Vasopressin mRNA localization in nerve cells: characterization of cis-acting elements and trans-acting factors. Proc. Natl. Acad. Sci. USA 98:7072-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielsen, J., J. Christiansen, J. Lykke-Andersen, A. H. Johnsen, U. M. Wewer, and F. C. Nielsen. 1999. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol. Cell. Biol. 19:1262-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nielsen, F. C., J. Nielsen, M. A. Kristensen, G. Koch, and J. Christiansen. 2002. Cytoplasmic trafficking of IGF-II mRNA-binding protein by conserved KH domains. J. Cell Sci. 115:2087-2097. [DOI] [PubMed] [Google Scholar]
- 18.Oleynikov, Y., and R. H. Singer. 1998. RNA localization: different zipcodes, same postman? Trends Cell Biol. 8:381-383. [DOI] [PubMed] [Google Scholar]
- 19.Roberts, M., S. Barry, A. Woods, P. van der Sluijs, and J. Norman. 2001. PDGF-regulated rab4-dependent recycling of alphavbeta3 integrin from early endosomes is necessary for cell adhesion and spreading. Curr. Biol. 11:1392-1402. [DOI] [PubMed] [Google Scholar]
- 20.Roberts, M. S., A. J. Woods, P. E. Shaw, and J. C. Norman. 2003. ERK1 associates with alpha(v)beta 3 integrin and regulates cell spreading on vitronectin. J. Biol. Chem. 278:1975-1985. [DOI] [PubMed] [Google Scholar]
- 21.Roberts, M. S., A. J. Woods, T. C. Dale, P. Van Der Sluijs, and J. C. Norman. 2004. Protein kinase B/Akt acts via glycogen synthase kinase 3 to regulate recycling of alpha v beta 3 and alpha 5 beta 1 integrins. Mol. Cell. Biol. 24:1505-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shestakova, E. A., R. H. Singer, and J. Condeelis. 2001. The physiological significance of beta-actin mRNA localization in determining cell polarity and directional motility. Proc. Natl. Acad. Sci. USA 98:7045-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner, C. E. 2000. Paxillin interactions. J. Cell Sci. 113:4139-4140. [DOI] [PubMed] [Google Scholar]
- 24.Turner, C. E. 2000. Paxillin and focal adhesion signalling. Nat. Cell Biol. 2:E231-E236. [DOI] [PubMed] [Google Scholar]
- 25.Vande Pol, S. B., M. C. Brown, and C. E. Turner. 1998. Association of Bovine Papillomavirus type 1 E6 oncoprotein with the focal adhesion protein paxillin through a conserved protein interaction motif. Oncogene 16:43-52. [DOI] [PubMed] [Google Scholar]
- 26.Wang, Y., and T. D. Gilmore. 2001. LIM domain protein Trip6 has a conserved nuclear export signal, nuclear targeting sequences, and multiple transactivation domains. Biochim. Biophys. Acta 1538:260-272. [DOI] [PubMed] [Google Scholar]
- 27.Webb, D. J., K. Donais, L. A. Whitmore, S. M. Thomas, C. E. Turner, J. T. Parsons, and A. F. Horwitz. 2004. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 6:154-161. [DOI] [PubMed] [Google Scholar]
- 28.Wolff, B., J. J. Sanglier, and Y. Wang. 1997. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4:139-147. [DOI] [PubMed] [Google Scholar]
- 29.Woods, A. J., M. S. Roberts, J. Choudhary, S. T. Barry, Y. Mazaki, H. Sabe, S. J. Morley, D. R. Critchley, and J. C. Norman. 2002. Paxillin associates with poly(A)-binding protein 1 at the dense endoplasmic reticulum and the leading edge of migrating cells. J. Biol. Chem. 277:6428-6437. [DOI] [PubMed] [Google Scholar]
- 30.Wu, J., and J. Bag. 1998. Negative control of the poly(A)-binding protein mRNA translation is mediated by the adenine-rich region of its 5′-untranslated region. J. Biol. Chem. 273:34535-34542. [DOI] [PubMed] [Google Scholar]
- 31.Zamir, E., and B. Geiger. 2001. Components of cell-matrix adhesions. J. Cell Sci. 114:3577-3579. [DOI] [PubMed] [Google Scholar]