Abstract
Trypanosoma brucei is a flagellated protozoan with a highly polarized cellular structure. TbLRTP is a trypanosomal protein containing multiple SDS22-class leucine-rich repeats and a coiled-coil domain with high similarity to a mammalian testis-specific protein of unknown function. Homologues are present in a wide range of higher eukaryotes including zebra fish, where the gene product has been implicated in polycystic kidney disease. Western blot analysis and immunofluorescence with antibodies against recombinant TbLRTP indicate that the protein is expressed throughout the trypanosome life cycle and localizes to distal zones of the basal bodies. Overexpression and RNA interference demonstrate that TbLRTP is important for faithful basal body duplication and flagellum biogenesis. Expression of excess TbLRTP suppresses new flagellum assembly, while reduction of TbLRTP protein levels often results in the biogenesis of additional flagellar axonemes and paraflagellar rods that, most remarkably, are intracellular and fully contained within the cytoplasm. The mutant flagella are devoid of membrane and are often associated with four microtubules in an arrangement similar to that observed in the normal flagellar attachment zone. Aberrant basal body and flagellar biogenesis in TbLRTP mutants also influences cell size and cytokinesis. These findings demonstrate that TbLRTP suppresses basal body replication and subsequent flagellar biogenesis and indicate a critical role for the LRTP family of proteins in the control of the cell cycle. These data further underscore the role of aberrant flagellar biogenesis as a disease mechanism.
Eukaryotic flagella and cilia are among the most ancient of cellular organelles, and their basic architecture is conserved from protozoa to vertebrates (28). The core structure of the flagellar/ciliar axoneme comprises a membrane-bound cylinder of nine microtubule doublets plus two central singlet microtubules. Formation of the axoneme is organized by basal bodies, cylindrical organelles with walls composed of nine triplet microtubules that serve as templates for the assembly of doublet microtubules. Basal bodies and centrioles are similar in structure, and these organelles are often interchangeable in many organisms (4). The functions of flagella and cilia include roles in development, metazoan body polarity, and cell division, while many diseases involve a defect in the function of flagellar components (17).
The protozoan Trypanosoma brucei is an ideal system in which to study microtubule-mediated events, because, in addition to the flagellum, the cell is highly polarized as a consequence of an ordered array of subpellicular microtubules (8, 11). The trypanosome has a single flagellum that emerges from the flagellar pocket, an invagination of the plasma membrane at the posterior end of the cell (9, 21). The axoneme of kinetoplastid protozoa is attached to a lattice-like structure of a similar diameter, the PFR, extending along most of the axoneme (9). The flagellum is attached to the cell body throughout most of its length by the FAZ, which is composed of an electron-dense filament plus four cytoplasmic microtubules that originate from the basal body region (9).
During cell division the trypanosome must replicate and separate several single-copy organelles present in G1 cells, i.e., nucleus, kinetoplast, mitochondrion, basal body, and flagellum (36, 44). Ultrastructural studies have described a number of markers of cell cycle position and elucidated a number of discrete cell cycle phases (36). The first morphological event of the T. brucei cell cycle is the maturation and duplication of the basal body, which is followed by kinetoplast S phase and then nuclear S phase (36, 44). During flagellum morphogenesis, the new axoneme is assembled from the recently matured basal body. New probasal bodies are formed, and the elongation and emergence of the flagellum from the flagellar pocket is followed by construction of a new FAZ and PFR (14, 36, 44). The duplicated kinetoplasts are segregated by means of their attachment to the flagellar basal bodies (24, 32), and this segregation is followed by the onset of mitosis (9). Kinetoplast/basal body segregation is accompanied by increased cell length and may contribute an important element in the control of later cell cycle events such as nuclear division and cytokinesis (9, 27). A cleavage furrow originates at the anterior tip of the replicating cell and follows a helical path to the posterior end of the cell that separates the two daughter cells (36). The four FAZ-associated microtubules have been postulated to provide a structural correlate between the length and position of the new flagellum, the main growth of the cell body, and to mark the position/direction of the cleavage furrow at cytokinesis (15, 33). This well-characterized mitotic cycle provides an excellent model system by which to characterize the functions of factors involved in basal body duplication, flagellar biogenesis, and cytokinesis.
Here we describe the identification and characterization of TbLRTP, which exhibits significant similarity to a leucine-rich repeat protein most abundantly expressed in late pachytene and diplotene cells of the testes of mice and humans (46). TbLRTP also has homologues in a wide variety of eukaryotes, including zebra fish, where the respective gene, seahorse, is implicated in polycystic kidney disease (38). We demonstrate that TbLRTP is an important factor in control of basal body duplication and hence flagellar biogenesis and cytokinesis. These data uncover the function of the LRTP family of proteins and also indicate the manner in which seahorse mutants likely cause cystic disease.
MATERIALS AND METHODS
Abbreviations.
Abbreviations used in this paper are as follows: BSF, bloodstream form; FAZ, flagellar attachment zone; LRR, leucine-rich repeat; LRTP; leucine-rich testis protein; TbLRTP, T. brucei LRTP; PCF, procyclic culture form; PFR, paraflagellar rod; RNAi, double-stranded RNA interference; sds22+, a mitotic regulator of protein phosphatase 1; ORF, open reading frame; PIPES, piperazine-N,N′-bis(2-ethanesulfonic acid); DAPI, 4′,6′-diamidino-2-phenylindole; PBS, phosphate-buffered saline; PFA, paraformaldehyde; and 1K1N2BB cells, cells with one kinetoplast, one nucleus, and two basal bodies.
Cell culture.
All growth medium reagents were supplied by GIBCO-BRL Life Technologies (Paisley, United Kingdom). PCF and BSF trypanosomes, strain Lister 427, were maintained in SDM79 and HMI9 medium, respectively. TbLRTP overexpression was achieved from the pXS219 plasmid (26), and RNAi was performed using the p2T7Ti vector (16) in the T. brucei 29-13 procyclic cell line (42). Leishmania mexicana (MNYC/BZ/62/M379) promastigotes were maintained in Schneider's Drosophila medium, pH 7.5, supplemented with 10% fetal calf serum at 26°C. Metacyclogenesis was induced by passage into medium at pH 5.0; metacyclic parasites were subsequently transformed into axenic amastigotes by increasing the culture temperature to 32°C (3).
DNA manipulation.
The TbLRTP ORF was present in a λ-phage isolated during a genomic screen (22). PCR was performed with Pfu polymerase (Stratagene, La Jolla, Calif.), and products were subcloned using the PCR-Script Amp cloning kit (Stratagene) and sequenced with BigDye terminator chemistry, version 3.0 (Applied Biosystems, Foster City, Calif.) on a semiautomated sequencer (ABI 377; Perkin-Elmer Corp., Norwalk, Conn.). TbLRTP was PCR amplified from genomic DNA using primers CCCAAGCTTATGGGCCGCATCACTACCGACC and GGATCCCGCCCTTACCTAGTCACAGCAGTATGG and was subcloned into the trypanosomal expression vector pXS219 (26) and the RNAi vector p2T7i (16) using HindIII and BamHI sites (underlined). By the RNAit algorithm the TbLRTP ORF is not expected to cosuppress other transcripts from the trypanosome genome (30).
Antibody production.
Antiserum against TbLRTP was generated against the entire ORF expressed from the pGEX-2TK vector (Amersham Biosciences, Little Chalfont, Buckinghamshire, United Kingdom). A purified TbLRTP-glutathione S-transferase fusion protein was used to raise polyclonal antibodies in rabbits. Antibodies were affinity purified by using an Escherichia coli-expressed recombinant antigen (affinity purified on glutathione-Sepharose) immobilized on CNBr-activated Sepharose.
Cytoskeletal extraction.
Trypanosomes were harvested by centrifugation and washed in PEM buffer (100 mM PIPES [pH 6.9], 2 mM EGTA, 1 mM MgSO4) (31). Cells were resuspended in PEM buffer containing 1% Nonidet P-40. After 2 min, the mixture was centrifuged and pellets containing the cytoskeleton were processed for either biochemical or microscopic analysis. Flagellum isolation was performed using the Ca2+-resistant method (31).
Immunofluorescence.
Cells were washed in PBS, spread on poly-l-lysine-coated slides, and fixed in methanol at −20°C or in 3% PFA in PBS, pH 7.5, for 10 min (BSF) or 1 h (PCF) on ice before processing for immunofluorescence (36). For the preparation of cytoskeletons, cells were extracted prior to fixation for 2 min with cold PEM buffer (31). The following antibodies were used: TAT1 (anti-α-tubulin) (43), BBA4 (marker of the basal body) (43), L6B3 (marker of the FAZ) (14), L13D6 (marker of PFR proteins 1 and 2) (14). Primary antibodies were detected with a Texas Red-conjugated goat anti-rabbit or Alexa-conjugated goat anti-mouse secondary antibody (Molecular Probes). Cells were examined using a Nikon Eclipse E600 epifluorescence microscope equipped with a Nikon digital DMX 1200 camera (Nikon Europe B.V., AE Badhoev, The Netherlands). Digital images were captured using Nikon ACT-1 software and assembled using Adobe Photoshop (Adobe Systems, Inc., San Jose, Calif.).
Western blotting.
Protein samples were electrophoresed on 12% SDS-polyacrylamide gels and blotted onto a Hybond ECL nitrocellulose membrane (Amersham Life Science Ltd., United Kingdom) by wet transfer. For analysis of whole-cell extracts, 107 cells per lane were used. For quantitation, X-ray films after exposure with ECL reagent were scanned and analyzed by densitometry using ImageJ software (http://rsb.info.nih.gov/ij/).
Electron microscopy.
For transmission electron microscopy, cells were fixed in suspension by addition of chilled 2.5% glutaraldehyde and 4% PFA in PBS on ice for 1 h, rinsed in 0.1 M sodium cacodylate, and postfixed in 1% osmium tetroxide for 1 h. After rinsing, cells were dehydrated in an ethanol series, with 1% uranyl acetate added at the 30% stage, followed by propylene oxide, and were then embedded in Epon/Araldite. Sections were cut on a Leica Ultracut-T ultramicrotome and were contrasted with uranyl acetate and lead citrate.
Bioinformatics.
BLAST searches (2) were conducted at the National Center for Biotechnology Information site (http://www.ncbi.nlm.nih.gov/BLAST), the Department of Energy Joint Genome site (http://genome.jgi-psf.org/index.html), and the T. brucei databases at The Institute for Genomic Research (http://www.tigr.org/tdb/mdb/tbdb/index.shtml) and the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk/Projects/T_brucei/). Sequence alignments were performed using Clustal X (39). Prediction of coiled-coil domains was performed using PEPCOIL (18). Retrieval of data concerning the chromosomal environment of the TbLRTP locus was from GeneDB (http://www.genedb.org/genedb/tryp/index.jsp).
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper have been deposited at GenBank with the accession number AF152174.1. The corresponding ORFs are annotated at GeneDB as Tb03.48K5.370 and Tb03.48K5.300 on chromosome III of the T. brucei strain 927 genome.
RESULTS
A novel family of leucine-rich repeat proteins.
During a genomic screen for a T. brucei dynamin gene (22), a λ phage was isolated containing a second ORF which displayed significant similarity to LRTP (35% identity, 52% similarity), a testis-specific protein of Mus musculus and Homo sapiens; the ORF was designated TbLRTP (46). Homologues are present in Trypanosoma cruzi, Leishmania major, Giardia lamblia, Chlamydomonas reinhardtii, Drosophila melanogaster, Anopheles gambiae, Xenopus tropicalis, and Danio rerio but not Caenorhabditis elegans. The trypanosome genome carries two TbLRTP ORFs on chromosome III in a region of local tandem duplication, and no additional sequences were detected by Southern blot analysis (data not shown). It is not known if expression of both ORFs is the same, but given their identical sequences, the functions of the two proteins are likewise expected to be identical.
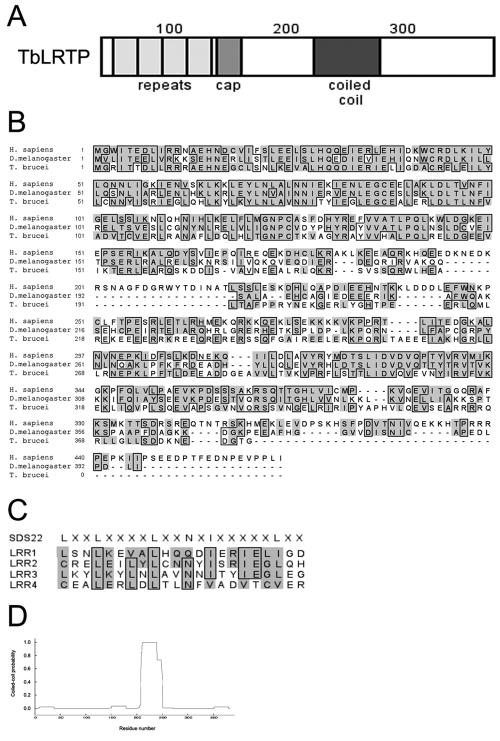
The domain structure of TbLRTP and an alignment against the D. melanogaster and H. sapiens homologues are shown in Fig. 1A and B, respectively. TbLRTP is a protein of 383 amino acids with a predicted molecular mass of 43.3 kDa and a pI of 5.8; it is composed of four N-terminal LRRs (residues 20 to 41, 43 to 64, 65 to 86, and 87 to 108), followed by an incomplete LRR (residues 109 to 130) and an LRR cap domain (residues 131 to 146). LRR motifs are involved in the formation of protein-protein interactions (13); the TbLRTP LRRs are homologous to the SDS22+ subclass (Fig. 1C), although the second and fourth repeats start with Cys, a nonconsensus alteration. SDS22+ is an inhibitory subunit of protein phosphatase 1 required in mitotic metaphase/anaphase transition (6, 25). LRTP proteins also contain several Glu-rich acidic regions near the C terminus (Fig. 1B), and PEPCOIL (18) predicts that TbLRTP will form a coiled-coil (residues 200 to 250) (Fig. 1D). Outside of the N-terminal LRRs, TbLRTP and its homologues contain no similarity to other sequences currently in the databases.
FIG. 1.
Structure and evolutionary relationship of TbLRTP. (A) Schematic representation of TbLRTP showing the locations of leucine-rich repeats (lightly shaded boxes; residues 20 to 41, 43 to 64, 65 to 86, and 87 to 108, and incomplete-LRR residues 109 to 130), the leucine-rich repeat cap (medium shaded box; residues 131 to 146), and the coiled-coil domain (dark box; residues 210 to 239). (B) The predicted amino acid sequence of TbLRTP is highly conserved between trypanosomes, arthropods, and humans. Alignments of predicted amino acids encoded by the trypanosomal TbLRTP gene, the D. melanogaster CG14620 gene product (DmLRXP), and the human testis-specific leucine-rich repeat protein (HsLRTP). Accession numbers: TbLRTP, AF152174.1; CG14620 gene product, AAF50954; HsLRTP, U60666.1. Shaded residues are conserved; boxed residues are identical. (C) Alignment of the four leucine repeats of TbLRTP. The SDS22+ consensus is shown above the alignment. Unlike the SDS22+ family, the second and fourth repeats of TbLRTP start with cysteine and the final position of the leucine in the third repeat is occupied by a valine. (D) Predicted coiled-coil formation by TbLRTP. PEPCOIL output: probabilities of the TbLRTP polypeptide forming helical coiled-coils as predicted from the algorithm of Lupas et al. (18), using a 28-residue window. The horizontal axis is the position within the protein, and the NH2 terminus is at the origin; the vertical axis is the probability of coiled-coil formation.
TbLRTP is associated with the basal body.
To investigate TbLRTP function, we raised affinity-purified antibodies against the recombinant protein (see the supplemental material). Differential NP-40 extraction, a standard technique for analysis of the trypanosome microtubular cytoskeleton, was used to probe the cytoskeletal association of TbLRTP (12, 31-33). To confirm separation of the microtubular cytoskeleton and soluble proteins, immunoblot analysis of the solubilized versus cytoskeletal/particulate fraction was performed. TbBiP, a soluble endoplasmic reticulum-resident protein, was fully extracted into the soluble fraction, while the vast majority of α-tubulin was retained within the cytoskeleton as expected (Fig. 2A). Approximately 40% of TbLRTP was associated with the particulate fraction, suggesting that a proportion is cytoskeletally associated. Immunofluorescence of NP-40-extracted trypanosomes using antibody to BBA4 antigen, which locates to the proximal end of both the mature basal body and the probasal body (45) (Fig. 2B), confirmed the close localization of TbLRTP to both the mature basal body and the probasal body, which is maintained throughout the cell cycle. In 1K1N cells, TbLRTP is juxtaposed distally to BBA4 (Fig. 2B, top). Upon basal body duplication, the TbLRTP signal was observed as a doublet with each unit in juxtaposition to a BBA4 signal (Fig. 2B and C). As the kinetoplast divides, the TbLRTP signal maintains its position relative to the BBA4 antigen (Fig. 2B and C). Further, detergent extraction removed the TbLRTP fraction that extends from the kinetoplast toward the nucleus. Therefore, TbLRTP is present both at the basal body region, where it is firmly anchored to the cytoskeleton, and in more nucleus-proximal regions, where it is less strongly cytoskeletally associated (see Fig. S1 in the supplemental material).
FIG. 2.
TbLRTP is associated with the basal body. (A) Intact trypanosomal cytoskeletons were prepared by extraction with 1% Nonidet P-40 as described in Materials and Methods. Fractions that were soluble in 1% Nonidet P-40 (S) and insoluble fractions (cytoskeletons) (P) were immunoblotted with anti-TbLRTP, anti-TbBiP, or anti-tubulin antibodies. (B) Immunofluorescence analysis of Nonidet P-40-extracted and methanol-fixed PCF cells showing a close association of TbLRTP signal (red) and the basal body (BB) revealed using BBA4 antibodies (green). (C) Immunolocalization in isolated flagellum preparations from wild-type PCF trypanosomes of TbLRTP (red; left panels), BBA4 (green; middle panel, top), and PFR (green; middle panel, top) and merged images (right panels) show that TbLRTP is localized distally to the basal body within the flagellum. Phase-contrast images are overlaid in all panels. Bars, 5 μm.
To further define the association of TbLRTP with the trypanosome cytoskeleton, flagella were isolated using a protocol that depolymerizes the subpellicular microtubule corset (31, 32). The isolated flagella were then fixed and subjected to immunofluorescence with antibodies to TbLRTP, basal bodies (45), and PFR proteins 1 and 2 (14) (Fig. 2C). TbLRTP was localized at a distinct site at one end of the flagellum. Costaining with BBA4 and anti-PFR1 and -2 demonstrates that TbLRTP localizes to the basal body end of the flagellum, distal to the BBA4 signal but well separated from the starting point of the PFR (Fig. 2C). Hence, TbLRTP is a basal body protein. Note that the BBA4 signal is partially resolved into two spots in Fig. 2B, reflecting the basal body and the probasal body. A clear resolution of these two structures is seen in the flagellar preparation (Fig. 2C).
TbLRTP is required for the regulation of cell division.
To investigate TbLRTP function, overexpression and RNAi were used. Overexpression and RNAi of TbLRTP were confirmed by Western blotting (Fig. 3A) and achieved threefold overexpression and ∼80% suppression by RNAi. A small increase in cell growth was observed in TbLRTP overexpressers (data not shown), while suppression correlated with decreased growth (Fig. 3B), suggesting that TbLRTP is an important factor in cell proliferation. Growth restoration at 5 days following RNAi induction was accompanied by increased expression of TbLRTP, indicating escape from RNAi (1, 41).
FIG. 3.
TbLRTP is required for the correct regulation of the cell cycle. Overexpression of TbLRTP was achieved using pXS219 in wild-type (WT) PCF parasites, and RNAi was induced in procyclic p2T7Ti/TbLRTP cells by the addition of tetracycline. The parental 29-13 cell line was treated identically for 5 days. Cells were counted and diluted daily. (A) Immunoblotting of WT, TbLRTP-overexpressing, and p2T7Ti/TbLRTP cells induced using anti-TbLRTP antiserum. In TbLRTP-overexpressing cells (++), the protein was present at ∼3-fold the level in parental (WT) cells. A reduction in TbLRTP protein levels during RNAi was also seen (days 0 to 4; maximal reduction of 84% of protein at day 3). Each lane was loaded with 106 parasites, and equivalence of loading was demonstrated by reprobing with TbBiP. (B) Cumulative growth curve of p2T7Ti/TbLRTP cells with (squares) and without (diamonds) tetracycline demonstrates that PCF cells subject to RNAi exhibit a reduced growth rate relative to that of uninduced cells. (C) The effects of overexpression and RNAi-mediated depletion of TbLRTP in PCF cells were investigated using immunofluorescence microscopy of unextracted PFA-fixed cells. Cells were stained for TbLRTP (red). Note the increase in length and the nuclear aberration in the overexpresser cells and the reduction in overall size of the RNAi cells. (D) Morphometric analysis of postmitotic TbLRTP mutant cells; basic cell dimensions of WT (2K2N) and mutant (2K2N) trypanosomes were measured. AN1, anterior end to anterior nucleus; N1N2, nucleus to nucleus; N1K1, anterior nucleus to anterior kinetoplast; N2K2, posterior nucleus to posterior kinetoplast; K1K2, kinetoplast to kinetoplast; K2P, posterior kinetoplast to posterior end. Measurements are the mean parameters of 50 cells. Solid bars, RNAi-suppressed cells; open bars, TbLRTP overexpressers.
Under RNAi the amount of protein expressed by the targeted mRNA is progressively reduced. In cells under RNAi for 54 h, residual TbLRTP was found in a small pool adjacent to the kinetoplast (Fig. 3C), similar to the detergent-resistant fraction (Fig. 2B). In overexpressers, TbLRTP was extensively distributed throughout the cytoplasm (Fig. 3C, upper panels). Hence the cytoskeletal pool of TbLRTP is probably more stable than the cytoplasmic pool, and these data suggest that there is a saturable number of TbLRTP docking/binding sites at the basal body.
Cells overexpressing TbLRTP were increased in size and exhibited numerous nuclear and kinetoplastid abnormalities. By contrast, RNAi of TbLRTP resulted in reduced cell length (Fig. 3C). To determine whether these size alterations were due to modulation of a specific cellular structure, as observed for some gene products (10, 15), or an overall effect on cell volume, morphometric analysis was performed on cells in interphase, which were selected based on possessing a single nucleus and kinetoplast (see Fig. S2A for a schematic of trypanosome cellular parameters). The overall length of wild-type trypanosomes was 20.4 ± 3.7 μm, and the distance between the kinetoplast or the nucleus and the posterior end of the cell was 6.1 ± 1.4 or 9.2 ± 1.5 μm, respectively. TbLRTP overexpresser cells were longer, at 31.4 ± 8.1 μm, while the kinetoplast-posterior and nucleus-posterior distances also increased to 7.9 ± 3.6 and 12.0 ± 4.9 μm, respectively. By contrast, TbLRTP RNAi cells were shorter, at 14.1 ± 5.1 μm, and the kinetoplast-posterior and nucleus-posterior distances also decreased to 4.4 ± 2.5 and 7.4 ± 3.0 μm, respectively. We also measured flagellum and FAZ lengths in TbLRTP RNAi cells and overexpressers; these parameters correlate with the alterations in the distance between the kinetoplast and the cell posterior (data not shown). There is a direct correlation between flagellum length and cell size (15, 33, 40), and taken together, these data demonstrate that the sizes of the trypanosomes were altered globally by manipulation of TbLRTP expression without a specific change to any one region. Therefore, the mechanism by which TbLRTP affects cell size is distinct from that indicated by other reports of alterations to specific regions (10, 15, 33).
Trypanosome growth is not achieved simply by doubling of length during the cell cycle, as the positions of the nucleus and kinetoplast exhibit a high degree of coordinate movement and a relationship with overall cell extension. The inter-basal body distance (33) reflects the timing and extent of cell length, and the distances from the cell posterior to the old basal body and from the new basal body to the anterior remain constant during the cell cycle. To test if alterations in cell length in TbLRTP mutants were a result of defects arising during cytokinesis, TbLRTP mutant cell lines were analyzed by morphometry during cell division (Fig. 3D; see Fig. S2B in the supplemental material for a schematic of the cell cycle). In 2K1N cells, the distance from the cell posterior to the posterior kinetoplast was increased by overexpression of TbLRTP and was decreased by RNAi. As the cell cycle progressed, the effects of TbLRTP protein levels on cell length became more pronounced. In 2K2N cells, all of the parameters measured, with the exception of the distance between the posterior nucleus and posterior kinetoplast, were decreased in cells subjected to RNAi. By contrast, all of these parameters increased in the overexpressing mutants. The most pronounced effects are in the distance from the anterior of the cell to the anterior nucleus and the distance from the posterior kinetoplast to the posterior of the cell (Fig. 3D). This indicates that the distance from the posterior of the cell to the kinetoplast is related to TbLRTP expression levels.
TbLRTP is required for correct basal body replication and new flagellum biogenesis.
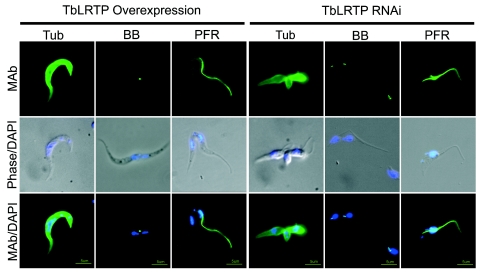
The influence of TbLRTP on components of the cytoskeleton, i.e., microtubules, the basal body, and the PFR, was also examined. In overexpressers and RNAi cells, global microtubule structure was normal by immunofluorescence (Fig. 4) and electron microscopy (Fig. 5). However, immunostaining for the basal body showed that duplication and separation of the basal body were disrupted in TbLRTP-overexpressing and RNAi cells (Fig. 4). In some of the overexpressing cells, mitosis occurred in the absence of apparent kinetoplast division and basal body duplication, whereas RNAi resulted in additional basal bodies free in the cytoplasm.
FIG. 4.
TbLRTP functions in basal body division and new flagellum biogenesis. The effect of overexpression or p2T7Ti/LRTP PCF trypanosomes induced for 3 days (RNAi) on components of the microtubule cytoskeleton in PCF cells was investigated using immunofluorescence microscopy. Cells were stained for α-tubulin (Tub), basal bodies (BB), and PFR (MAb) in green. All cells were counterstained with DAPI, which is shown in a merge with the phase-contrast image. TbLRTP overexpression resulted in increased cell size. TbLRTP RNAi knockdown resulted in smaller cells that often contained increased numbers of flagella and FAZ elements. Bars, 5 μm.
FIG. 5.
Electron microscopy of TbLRTP RNAi knockdown demonstrates aberrant flagellar axoneme, PFR, and FAZ microtubule biogenesis. TbLRTP-overexpressing cells (A) or p2T7Ti/LRTP PCF cells induced for 3 days (B-F) were fixed and processed for thin-section transmission electron microscopy. No alteration to the subpellicular microtubule array, mature basal body, or flagellar axoneme could be detected by transmission electron microscopy in the overexpressers (A) or the RNAi knockdown cells (B). However, in the cytoplasm of the p2T7Ti/LRTP PCF cells, intracellular axonemes and PFR-like structures have assembled, shown in transverse section (C, D) and longitudinal section (E, F). These elements exhibit the correct 9 + 2 flagellum axoneme and crystalline PFR but lack a delineating membrane, are surrounded by a zone of exclusion of the normal ribosome-rich cytoplasm (arrowhead, C, F), and are often associated with four microtubules which resemble those associated with the flagellum (arrowhead, D). Their assembly is from an apparently structurally normal, but ectopic, basal body (E, indicated by arrowhead). Bars, 500 nm.
To determine the effects of TbLRTP expression levels on basal body duplication and separation, both nuclear and kinetoplast contents for individual cells were determined by DAPI staining (Table 1). Perturbation of TbLRTP protein levels had a comparatively small effect on progress through the cell cycle and did not result in accumulation of cells at a specific point within the cell cycle (Table 1). However, ∼29% of overexpressers and 14% of RNAi mutant cells analyzed exhibited abnormalities in the number of nuclei (Table 1, >2N, 0N, >2K, 0K, 2N1K). Similar aberrant nuclear numbers have been demonstrated to arise as a consequence of microtubule disruption, resulting in failure to accurately complete cytokinesis (10, 27, 33).
TABLE 1.
Nuclear/kinetoplast analysis of TbLRTP mutantsa
| Strainb | % of total cell population in each strain with the following karyotypec:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1K1N | 2K1N | 2K2N | AK | >2N | 0N | >2K | 0K | 1K2N | |
| WT | 78.5 | 9.1 | 11.4 | 1.0 | 0 | 0 | 0 | 0 | 1 |
| ++ | 50.4 | 11.3 | 9.8 | 28.5 | 5.3 | 0.7 | 0.7 | 9.0 | 12.8 |
| RNAi | 57.3 | 18.3 | 10.7 | 13.7 | 3.1 | 0 | 3.7 | 0.8 | 6.1 |
Nuclear/kinetoplast classification was performed by immunofluorescence assays following DAPI staining to determine both nuclear and kinetoplast copy numbers.
WT, wild type; ++, TbLRTP overexpresser; RNAi, p2T7Ti/LRTP PCF trypanosomes induced for 3 days. More than 130 cells were analyzed for each strain.
AK, all cells with abnormal karyotypes, i.e., not 1K1N, 2K1N, or 2K2N. Percentages in the four columns on the left add up to 100%; the five rightmost columns provide a breakdown of the percentages in the AK column.
The number of basal bodies in interphase cells was also determined (Table 2). This method of analysis is used instead of cell cycle synchronization, which is not possible in trypanosomes. Cells enter mitosis with one kinetoplast, one nucleus, and two basal bodies when stained for the BBA4 antigen (45) (Fig. 2C). In wild-type cells, when the basal bodies duplicate, the cells become 1K1N4BB; following kinetoplast S phase and separation, they become 2K1N4BB; finally, following nuclear division, they become 2K2N4BB. In TbLRTP-overexpressing and RNAi cells, we observed incorrect numbers of basal bodies (reduced and elevated, respectively, relative to the numbers in the wild type) (Table 2).
TABLE 2.
Analysis of basal body numbers in TbLRTP mutantsa
| Strain | % of cells with the following no. of BBA4 spots:
|
|||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | >5 | |
| WT | 0 | 77 | 0 | 23 | 0 | 0 |
| ++ | 46 | 35 | 14 | 5 | 0 | 0 |
| RNAi | 6 | 52 | 11 | 11 | 3 | 17 |
The number of basal bodies, as defined by BBA4 antibody staining, was counted in 1K1N cells only. This stage of the cell cycle corresponds to G1/interphase and is shown in the schematic in Fig. S2 in the supplemental material. For a normal cell in G1, BBA4 detects two closely associated spots, representing the old basal body and the probasal body. A cell that has entered into cell division will have duplicated its basal bodies as one of the first events of cell division and hence will have four BBA4 signals. This state will persist until cytokinesis is complete.
WT, wild type; ++, TbLRTP overexpresser; RNAi, p2T7Ti/LRTP PCF trypanosomes induced for 3 days. The number of cells analyzed was >100 for each strain.
Staining with antibodies against the PFR (14) also revealed significant aberrations in flagellar biogenesis. TbLRTP-overexpressing cells appear to undergo nuclear division in the absence of kinetoplast division and new flagellum biogenesis (Fig. 4). Consequently, the cells are unable to correctly complete cytokinesis, resulting in cells of increased size and a partial loss of cell polarity (Fig. 4). Counts of 50 2K2N cells showed that wild-type cells have a mean of 2.0 ± 0 flagella per cell, RNAi 2K2N cells have a mean of 2.7 ± 0.9 flagella per cell, and 2K2N overexpresser cells have 1.3 ± 0.4 flagella per cell.
Analysis of TbLRTP RNAi mutant ultrastructure shows aberrant flagellar biogenesis.
To determine the structure of the flagellar structures that accumulated in the TbLRTP mutant cells, transmission electron microscopy was performed. The subpellicular microtubule array, mature basal body, and flagellar axoneme appear normal in both the TbLRTP-overexpressing and RNAi cells. However, in the cytoplasm of the RNAi cells, intracellular axonemes and PFR-like structures have assembled (shown in transverse section in Fig. 5C and D and in longitudinal section in Fig. 5E and F). The axonemes of these structures are highly similar to those of wild-type trypanosomes, with the correct 9 + 2 flagellum axoneme and a paracrystalline PFR, but lack a delineating membrane. Hence, promiscuous flagellar initiation appears to take place in these cells, but the resulting structures are normal. A zone of exclusion of the normal ribosome-rich cytoplasm (Fig. 5C and F) surrounds the intracellular flagella, and remarkably, the flagella are also sometimes seen associated with four microtubules typical of the FAZ (Fig. 5D). Their assembly appears to occur from an apparently structurally normal basal body located within the cytoplasm (Fig. 5E).
An LRTP homologue is also present in Leishmania.
Experimentally accessible stages of T. brucei are replicative forms, but in the related organism Leishmania, it is possible to study nondividing cell populations. L. major encodes an LRTP homologue (LmjF29.2210) that is 61% identical and 77% similar to the trypanosome protein. Anti-TbLRTP sera recognized a single band in L. mexicana promastigotes (designated LmLRTP), consistent with the predicted molecular mass derived from the gene sequence (53.8 kDa). Western blot analysis demonstrated that LmLRTP was not detectable in nonproliferative metacyclic promastigotes or slowly replicating axenic amastigotes (Fig. 6). These data suggest that LRTP is expressed mainly in rapidly proliferating cells.
FIG. 6.

The TbLRTP orthologue of Leishmania is differentially expressed during the life cycle. Western blot of protein lysates from procyclic (P) (flagellated, noninfective, proliferative form), metacyclic (M) (flagellated, infective, nonproliferative form), and amastigote (nonflagellated infective, proliferative form) L. mexicana probed with antibodies against TbLRTP or, as a loading control, N-myristoyl transferase (LmNMT) (29). The positions of molecular weight markers (in thousands) are indicated. Anti-TbLRTP defines a protein in proliferative, procyclic lysates whose migration is consistent with the predicted molecular mass of 53.8 kDa, which we designate LmLRTP. Expression of LmLRTP is severely down-regulated in quiescent metacyclics and replicating, aflagellate amastigotes.
DISCUSSION
Flagella, cilia, and basal bodies are widespread structures in cells of the eukaryotic lineage, and a large number of diseases and pathologies can be caused by immotile or misassembled flagella or cilia (17, 34, 38). Hence, investigation of factors coordinating flagellar biogenesis is of substantial clinical relevance as well as of importance to basic biology. In the present work we describe the identification and characterization of TbLRTP, a trypanosome leucine-rich repeat protein that has an important role in the control of basal body function and subsequent flagellar biogenesis. The major findings here are that TbLRTP is associated with the basal body and expression is required for proliferation and completion of the cell cycle. Overexpression of TbLRTP suppressed basal body replication and new flagellum biogenesis, while an RNAi-mediated decrease in TbLRTP levels resulted in the biogenesis of additional ectopic basal bodies complete with an attached axoneme, paraflagellar rod, and FAZ-associated microtubules, all contained within the cytosol. The consequences of this abnormal basal body replication and flagellar assembly are manifest in alterations in cell size, fully consistent with a model where the flagellum controls this parameter as well as the positioning of the cytokinesis cleavage furrow (15, 33, 40). Hence, TbLRTP likely participates in a pathway that negatively regulates biogenesis of the new basal body/centriole. The presence of such a pathway has been suggested previously (19). It is significant that the LRTP orthologue in Leishmania (LmLRTP) is developmentally regulated: it is absent both in nonproliferative metacyclic forms and in slowly replicating axenic amastigotes. Hence, LRTP appears to be expressed only in actively replicating cells, i.e., those within the mitotic cycle where basal body duplication is taking place. Presumably, in cells that have dropped out of the cell cycle and entered G0, LRTP expression is not required to modulate basal body duplication, because replication has ceased. It is formally possible that TbLRTP is also involved in basal body segregation and that disruption of this process could account for some of the phenotypic features described here. However, the absence of a significant defect in basal body positioning and the close similarity to phenotypes where basal body replication has been disrupted (27) are consistent with the major function of TbLRTP being involved in basal body replication.
Mutations affecting basal body number have a corresponding effect on cell length and shape in Tetrahymena (23), Paramecium (35), and Chlamydomonas (37) spp. TbLRTP RNAi generates pleiotropic effects very similar to those observed in Chlamydomonas VFL1 mutants, although the two proteins are not orthologues (37). A recent analysis of the Chlamydomonas flagellum proteome did not identify an LRTP homologue (17), but a clear orthologue is present in the Chlamydomonas genome. In both T. brucei and Chlamydomonas, there are specialized microtubules that originate near the basal bodies and extend into the cell beneath the plasma membrane: four FAZ-associated microtubules in T. brucei and four rootlet microtubules in Chlamydomonas. These microtubules are involved in the spatial positioning of cellular organelles and the cytokinesis cleavage furrow, as well as in regulation of cell volume (7, 15, 33, 37). Analysis of TbLRTP mutants supports previous data demonstrating that the correct assembly of the new flagellum/FAZ is required for the completion of cytokinesis and the maintenance of cell size/polarity (15, 20, 33, 40).
The assembly of additional axonemes, PFRs, and FAZ microtubules in TbLRTP RNAi mutants supports a model of concomitant biosynthesis and suggests that assembly occurs from a common nucleation/docking site, most likely located at the basal body region (14). IFT52 from Chlamydomonas is involved in the formation of such docking sites and has a location similar to that of TbLRTP, suggesting that these two factors, while distinct at the sequence level, may function in the same or a related pathway (5). In trypanosomes the old flagellum acts as a template for assembly and growth of the new flagellum (20), and where there is more than one new flagellum, presumably the excess copies cannot be accommodated and are assembled in a nontemplated manner. The consequence of additional basal bodies likely affects positioning of the cytokinesis cleavage furrow, resulting in nonsymmetrical cell division and production of small cells.
The TbLRTP family is highly conserved throughout evolution, and homologues are present in protozoan and metazoan systems. Significantly, homologues are absent from the genomes of fungi, nematodes, and plants; given the earlier time of divergence of the trypanosome and metazoan lineages compared to the plant/metazoan separation, this distribution suggests secondary loss of LRTP from selected lineages and hence differences in the mechanisms of basal body duplication between those lineages retaining LRTP and those where it is absent, for example, between C. elegans, trypanosomes, and humans. Most significantly, the zebra fish homologue of TbLRTP, seahorse, has been reported to be involved in ciliar function (38) and, further, was recently implicated as having a role in the development of polycystic kidney disease, although detailed functional data were not available from that analysis (38). The data reported here provide a function for the LRTP family of proteins in the modulation of basal body functions and by inference implicate that process in human disease.
Supplementary Material
Acknowledgments
We thank Ervin Goldberg (Northwestern University) and Philippe Bastin (Muséum National d'Histoire Naturelle, Paris, France) for helpful discussions and comments.
This work was funded by program grants from the Wellcome Trust (to M.C.F., D.F.S., and K.G.), and this support is gratefully acknowledged.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Allen, C. L., D. Goulding, and M. C. Field. 2003. Clathrin-mediated endocytosis is essential in Trypanosoma brucei. EMBO J. 22:4991-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Bates, P. A. 1994. Complete developmental cycle of Leishmania mexicana in axenic culture. Parasitology 108:1-9. [DOI] [PubMed] [Google Scholar]
- 4.Beisson, J., and M. Wright. 2003. Basal body/centriole assembly and continuity. Curr. Opin. Cell Biol. 15:96-104. [DOI] [PubMed] [Google Scholar]
- 5.Deane, J. A., D. G. Cole, E. S. Seeley, D. R. Diener, and J. L. Rosenbaum. 2001. Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr. Biol. 11:1586-1590. [DOI] [PubMed] [Google Scholar]
- 6.Dinischiotu, A., M. Beullens, W. Stalmans, and M. Bollen. 1997. Identification of sds22 as an inhibitory subunit of protein phosphatase 1 in rat liver nuclei. FEBS Lett. 402:141-144. [DOI] [PubMed] [Google Scholar]
- 7.Ehler, L. L., J. A. Holmes, and S. K. Dutcher. 1995. Loss of spatial control of the mitotic spindle apparatus in a Chlamydomonas reinhardtii mutant strain lacking basal bodies. Genetics 141:945-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gull, K. 1999. The cytoskeleton of trypanosomatid parasites. Annu. Rev. Microbiol. 53:629-655. [DOI] [PubMed] [Google Scholar]
- 9.Gull, K. 2003. Host-parasite interactions and trypanosome morphogenesis: a flagellar pocketful of goodies. Curr. Opin. Microbiol. 4:365-370. [DOI] [PubMed] [Google Scholar]
- 10.Hendriks, E. F., D. R. Robinson, M. Hinkins, and K. R. Matthews. 2001. A novel CCCH protein which modulates differentiation of Trypanosoma brucei to its procyclic form. EMBO J. 20:6700-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill, K. L. 2003. Biology and mechanism of trypanosome cell motility. Eukaryot. Cell 2:200-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hutchings, N. R., J. E. Donelson, and K. L. Hill. 2002. Trypanin is a cytoskeletal linker protein and is required for cell motility in African trypanosomes. J. Cell Biol. 156:867-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobe, B., and A. V. Kajava. 2001. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11:725-732. [DOI] [PubMed] [Google Scholar]
- 14.Kohl, L., T. Sherwin, and K. Gull. 1999. Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J. Eukaryot. Microbiol. 46:105-109. [DOI] [PubMed] [Google Scholar]
- 15.Kohl, L., D. Robinson, and P. Bastin. 2003. Novel roles for the flagellum in cell morphogenesis and cytokinesis of trypanosomes. EMBO J. 22:5336-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LaCount, D. J., S. Bruse, K. L. Hill, and J. E. Donelson. 2000. Double-stranded RNA interference in Trypanosoma brucei using head-to-head promoters. Mol. Biochem. Parasitol. 111:67-76. [DOI] [PubMed] [Google Scholar]
- 17.Li, J. B., J. M. Gerdes, C. J. Haycraft, Y. Fan, T. M. Teslovich, H. May-Simera, H. Li, O. E. Blacque, L. Li, C. C. Leitch, R. A. Lewis, J. S. Green, P. S. Parfrey, M. R. Leroux, W. S. Davidson, P. L. Beales, L. M. Guay-Woodford, B. K. Yoder, G. D. Stormo, N. Katsanis, and S. K. Dutcher. 2004. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117:541-552. [DOI] [PubMed] [Google Scholar]
- 18.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 19.Marshall, W. F., Y. Vucica, and J. L. Rosenbaum. 2001. Kinetics and regulation of de novo centriole assembly. Implications for the mechanism of centriole duplication. Curr. Biol. 11:308-317. [DOI] [PubMed] [Google Scholar]
- 20.Moreira-Leite, F. F., T. Sherwin, L. Kohl, and K. Gull. 2001. A trypanosome structure involved in transmitting cytoplasmic information during cell division. Science 294:610-612. [DOI] [PubMed] [Google Scholar]
- 21.Morgan, G. W., B. S. Hall, P. W. Denny, M. Carrington, and M. C. Field. 2002. The endocytic apparatus of the kinetoplastida. Part I. A dynamic system for nutrition and evasion of host defences. Trends Parasitol. 18:491-496. [DOI] [PubMed] [Google Scholar]
- 22.Morgan, G. W., D. Goulding, and M. C. Field. 2004. The single dynamin-like protein of Trypanosoma brucei regulates mitochondrial division and is not required for endocytosis. J. Biol. Chem. 279:10692-10701. [DOI] [PubMed] [Google Scholar]
- 23.Nanney, D. L., and M. C. Chow. 1974. Basal body homeostasis in Tetrahymena. Am. Nat. 108:125-129. [Google Scholar]
- 24.Ogbadoyi, E. O., D. R. Robinson, and K. Gull. 2003. A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol. Biol. Cell 14:1769-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohkura, H., and M. Yanagida. 1991. S. pombe gene sds22+ essential for a mid-mitotic transition encodes a leucine-rich repeat protein that positively modulates protein phosphatase-1. Cell 64:149-157. [DOI] [PubMed] [Google Scholar]
- 26.Pal, A., B. S. Hall, D. N. Nesbeth, H. I. Field, and M. C. Field. 2002. Differential endocytic functions of Trypanosoma brucei Rab5 isoforms reveal a glycosylphosphatidylinositol-specific endosomal pathway. J. Biol. Chem. 277:9529-9539. [DOI] [PubMed] [Google Scholar]
- 27.Ploubidou, A., D. R. Robinson, R. C. Docherty, E. O. Ogbadoyi, and K. Gull. 1999. Evidence for novel cell cycle checkpoints in trypanosomes: kinetoplast segregation and cytokinesis in the absence of mitosis. J. Cell Sci. 112:4641-4650. [DOI] [PubMed] [Google Scholar]
- 28.Porter, M. E., and W. S. Sale. 2000. The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J. Cell Biol. 151:37-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price, H. P., M. R. Menon, C. Panethymitaki, D. Goulding, P. G. McKean, and D. F. Smith. 2003. Myristoyl-CoA:protein N-myristoyltransferase, an essential enzyme and potential drug target in kinetoplastid parasites. J. Biol. Chem. 278:7206-7214. [DOI] [PubMed] [Google Scholar]
- 30.Redmond, S., J. Vadivelu, and M. C. Field. 2003. RNAit: an automated web-based tool for the selection of RNAi targets in Trypanosoma brucei. Mol. Biochem. Parasitol. 128:115-118. [DOI] [PubMed] [Google Scholar]
- 31.Robinson, D., P. Beattie, T. Sherwin, and K. Gull. 1991. Microtubules, tubulin, and microtubule-associated proteins of trypanosomes. Methods Enzymol. 196:285-299. [DOI] [PubMed] [Google Scholar]
- 32.Robinson, D. R., and K. Gull. 1991. Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature 352:731-733. [DOI] [PubMed] [Google Scholar]
- 33.Robinson, D. R., T. Sherwin, A. Ploubidou, E. H. Byard, and K. Gull. 1995. Microtubule polarity and dynamics in the control of organelle positioning, segregation, and cytokinesis in the trypanosome cell cycle. J. Cell Biol. 128:1163-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenbaum, J. L., and G. B. Witman. 2002. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3:813-825. [DOI] [PubMed] [Google Scholar]
- 35.Ruiz, F., N. Garreau de Loubresse, and J. Beisson. 1987. A mutation affecting basal body duplication and cell shape in Paramecium. J. Cell Biol. 104:417-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherwin, T., and K. Gull. 1989. The cell division cycle of Trypanosoma brucei brucei: timing of event markers and cytoskeletal modulations. Philos. Trans. R. Soc. Lond. B 323:573-588. [DOI] [PubMed] [Google Scholar]
- 37.Silflow, C. D., M. LaVoie, L. W. Tam, S. Tousey, M. Sanders, W. Wu, M. Borodovsky, and P. A. Lefebvre. 2001. Vfl1 protein in Chlamydomonas localizes in a rotationally asymmetric pattern at the distal ends of the basal bodies. J. Cell Biol. 153:63-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun, Z., A. Amsterdam, G. J. Pazour, D. Cole, M. S. Miller, and N. Hopkins. 2004. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development 131:4085-4093. [DOI] [PubMed] [Google Scholar]
- 39.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tyler, K. M., K. R. Matthews, and K. Gull. 2001. Anisomorphic cell division by African trypanosomes. Protist 152:367-378. [DOI] [PubMed] [Google Scholar]
- 41.Wang, Z., J. C. Morris, M. E. Drew, and P. T. Englund. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275:40174-40179. [DOI] [PubMed] [Google Scholar]
- 42.Wirtz, E., S. Leal, C. Ochatt, and G. A. Cross. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89-101. [DOI] [PubMed] [Google Scholar]
- 43.Woods, A., T. Sherwin, R. Sasse, T. H. MacRae, A. J. Baines, and K. Gull. 1989. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93:491-500. [DOI] [PubMed] [Google Scholar]
- 44.Woodward, R., and K. Gull. 1990. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. J. Cell Sci. 95:49-57. [DOI] [PubMed] [Google Scholar]
- 45.Woodward, R., M. J. Carden, and K. Gull. 1995. Immunological characterization of cytoskeletal proteins associated with the basal body, axoneme and flagellum attachment zone of Trypanosoma brucei. Parasitology 111:77-85. [DOI] [PubMed] [Google Scholar]
- 46.Xue, J. C., and E. Goldberg. 2000. Identification of a novel testis-specific leucine-rich protein in humans and mice. Biol. Reprod. 62:1278-1284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.