Abstract
Trichorhinophalangeal syndrome type 1 (TRPS1) has been reported to be a sensitive and specific immunohistochemical (IHC) marker for breast carcinomas, especially when determining primary site of origin. However, there is limited data on TRPS1 expression in prostate and bladder cancers.
A two-phase study was performed with 1) an exploratory cohort analyzing TRPS1 gene alterations in prostate, bladder, and breast carcinoma and TPRS1 mRNA expression data in prostate and bladder carcinoma; and 2) TRPS1 and GATA3 IHC in a confirmatory cohort in prostate, bladder, and breast carcinoma samples.
Gene alterations were identified in a subset of breast, bladder, and prostate carcinomas and mRNA was consistently detected. In the IHC cohort, 183/210 (87.1%) of breast, 22/69 (31.9%) of prostate, and 20/73 (27.4%) of urothelial carcinomas showed staining with TRPS1. Intermediate to high expression of TRPS1 was observed in 173/210 (82.8%) of breast, 17/69 (24.6%) of prostate, and 15/73 (20.5%) of urothelial carcinomas. Furthermore, in prostate cancer, 26.9% of pelvic lymph node metastases and 50% in sites of distant metastases showed expression. Increased TRPS1 mRNA expression (p=0.032) and IHC expression (p=0.040) correlated with worse overall survival in bladder cancer. By comparison, GATA3 IHC stained 136/210 (64.8%) of breast, 0/69 (0%) of prostate, and 63/73 (93%) of bladder carcinomas. Intermediate to high expression of GATA3 was seen in 131/210 (62.4%) of breast and 63/73 (93%) of bladder carcinomas.
This study shows there is significant staining of TRPS1 in bladder and prostate cancers. As a result, comprehensive studies are needed to establish the true specificity of TRPS1 IHC stain across various tumor types before its widespread clinical adoption.
Keywords: TRPS1, GATA3, immunohistochemistry, prostate carcinoma, urothelial carcinoma, breast carcinoma, triple-negative breast carcinoma
Introduction:
Trichorhinophalangeal syndrome 1 (TRPS1) protein is known to be a modulator in the mesenchymal to epithelial transition during the development of multiple tissue types, including cartilage, bone, kidney, and hair follicles (1–3). More recently, TRPS1 was discovered to belong to the GATA family of transcription factors, and functions as an essential regulator for growth and differentiation of normal breast epithelial cells. Importantly, TRPS1 is involved in breast cancer initiation and progression, although the exact mechanisms of its role remain unknown (4, 5).
In one of the largest initial studies, Ai et al. interrogated The Cancer Genome Atlas (TCGA) database and identified that TRPS1 shows high mRNA expression in all subtypes of breast carcinoma compared with other tumors tested. In addition, they evaluated the use of TRPS1 as an immunohistochemical (IHC) marker for breast origin and determined that TRPS1 stained 91% of breast carcinomas with 87% showing intermediate to high expression (6). Subsequent studies have supported these findings with TRPS1 expression consistently noted in >98% of estrogen receptor (ER)+/ HER2− tumors and in >90% of triple negative breast carcinomas (TNBC) (7–10). In addition to being a sensitive IHC marker for breast cancer, prior studies have indicated that TRPS1 is also specific for breast origin. Only ~15% of ovarian serous carcinomas, 24% of salivary duct carcinomas, and less than 5% of lung adenocarcinomas have been shown to stain with TRPS1 (6, 7). Furthermore, it is reported to be essentially negative in urothelial and gastrointestinal carcinomas (6, 9, 11, 12).
However, many tumor types have not been evaluated for TRPS1 staining; and therefore, additional studies are warranted before broad claims of specificity can be made. Interestingly, to date, there is no data on TRPS1 expression in prostate cancer, which is important because a subset of breast carcinomas, particularly hormone receptor positive cases in males, can be positive for the prostate marker NKX3.1 and can show overlapping morphologic features with prostate cancer (13). In addition, only a few studies provide data on TRPS1 staining in bladder cancer, which is crucial for clinical practice since both breast adenocarcinoma and bladder urothelial carcinoma are typically positive for GATA3. Furthermore, to our knowledge, TRPS1 gene level alterations have not been reported in prostate, bladder, and breast carcinomas. In addition, unlike breast carcinomas, mRNA expression data has also not been extensively reported in prostate and bladder urothelial carcinoma.
As a result, we performed a two-phase study with an exploratory cohort analyzing TRPS1 gene alterations in prostate, bladder, and breast carcinoma and TPRS1 mRNA expression data in prostate and bladder carcinoma. Next, we performed TRPS1 and GATA3 IHC in a confirmatory cohort in bladder, prostate, and breast carcinoma samples.
Methods:
This study was approved by the institutional review board at the University of Kentucky. The cBioPortal platform was utilized to gather data on the frequency of alterations, mutation types, mutation sites, and copy number alterations across all TCGA tumors (14). In the “Comparison/Survival” module, clinical prognosis data were downloaded and analyzed for all TCGA cancer types, considering the presence or absence of TRPS1 gene alterations. This encompassed information on progression-free survival (PFS), disease-specific survival (DSS), disease-free survival (DFS), and overall survival (OS).
To examine the variations in TRPS1 expression between pan-cancer and adjacent normal tissues, TIMER2.0 was employed (15). The gene expression levels were depicted on a log2 (TPM + 1) scale, with TPM representing transcripts per million. To assess the TCGA bladder tumor patients, they were categorized into high expression and low expression cohorts using cut-off values of 50%. The Kaplan-Meier plotter was used with the log-rank test to conduct survival analysis on the tumors (16).
Formalin-fixed paraffin embedded (FFPE), 4-μm thick, tissue microarray (TMA) sections with 1.5 mm diameter cores were used. Included were 210 breast carcinomas, 73 muscle-invasive high-grade urothelial carcinomas, and 69 prostate adenocarcinomas.
IHC staining for TPRS1 was performed utilizing clone PA5-84874 from ThermoFisher (Walthan MA). Antigen retrieval was performed with a citrate buffer at pH of 6.0 at 95°C for 32 minutes, followed by incubation at a 1:50 dilution for 60 minutes at 37°C. IHC staining for GATA3 IHC was performed utilizing clone L50-823 from Cell Marque (Rocklin, CA). Antigen retrieval was performed with a citrate buffer at pH 6.0 at 95°C for 48 minutes, followed by incubation in a prediluted form per manufacturer instruction for 36 minutes at room temperature. Detection for both TRPS1 and GATA3 was performed with Ventana (760-4185) anti-rabbit HQ for 20 minutes at 37°C, followed by Ventana (760-4820) anti-HQ-HRP for 20 minutes at 37°C and visualized with DAB.
IHC staining for TRPS1 and GATA3 was evaluated using a previously established expression score, which is calculated as % tumor staining (0: <1%; 1: 1-10%; 2: 11-50%; 3: 51-100%) multiplied by the average staining intensity (0-negative; 1-weak; 2-moderate; 3-strong). Scores 0-1 were considered negative, 2 = low, 3-4 = intermediate, and 6 and 9 = high expression (6, 8). Only nuclear staining was considered positive for both TRPS1 and GATA3. All cases were evaluated using visual estimation by light microscopy.
The Chi-square (and Fisher’s exact) test was used for analysis. P-values < 0.05 were considered to be statistically significant.
Results:
Exploratory Cohort
Gene alterations for TRPS1 were analyzed across 30 primary tumor sites from a dataset created from 9889 samples from the TCGA and the TCGA Pan-Cancer Atlas (https://portal.gdc.cancer.gov/). Utilizing the GISTIC algorithm, gene alterations were identified in a total of 960 samples (9.71%) and are summarized by frequency by primary site in Figure 1A.
Figure 1.
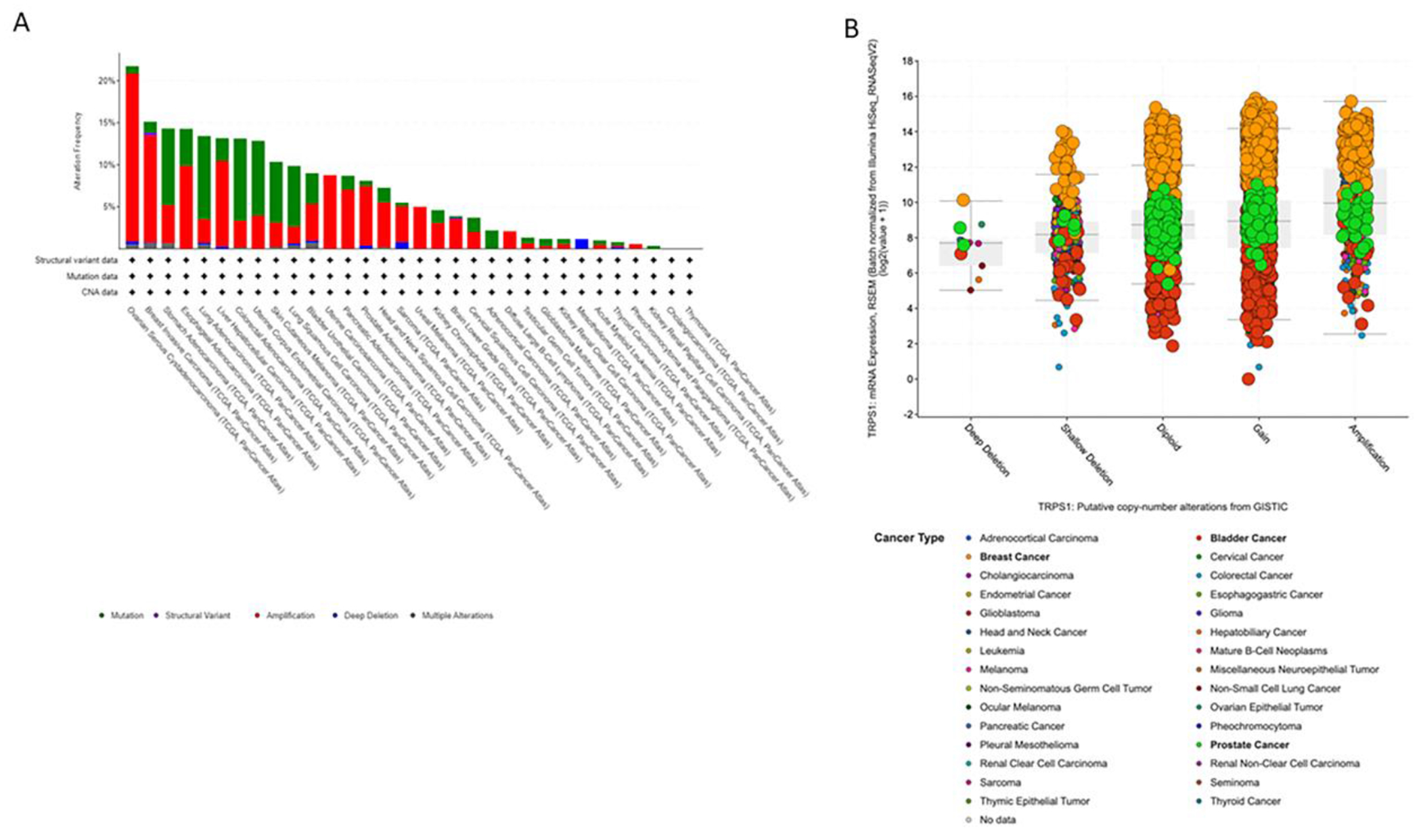
A-B. GISTIC analysis revealing frequency of TRPS1 gene alterations across multiple cancer types
Across all tumor types included, somatic TRPS1 mutations were identified in 459 (4.5%) cases, of which, 372 (81.0%) were missense mutations—most often associated with copy number gains or amplifications—while 69 (15.0%) were truncating loss of function mutations, 5 (1.1%) were inframe deletions, and 2 (0.43%) were splice site mutations. Interestingly, 11 fusions between TRPS1 and multiple gene partners were identified, 10 (90.9%) of which were found in invasive breast cancer, and were associated with gain or amplification in all cases (Supplemental Table 1) (14, 17).
More specifically, invasive breast cancers showed an altered TRPS1 gene in 164 of 1084 (15.13%) cases, including 138 with amplifications; urothelial carcinoma of the bladder showed TRPS1 gene alterations in 37 of 411 (9.0%) of cases, including 18 with amplifications; and prostate adenocarcinomas showed TRPS1 gene alterations in 40 of 494 (8.1%) cases, including 35 with amplifications. In Figure 1B, putative copy number alterations in the TRPS1 gene from GISTIC analysis are presented to highlight the three malignancies in particular that pertain to this study. Breast cancer (orange) shows frequent copy number gains and amplification in the TRPS1 gene. A subset of prostate cancers (green) and bladder cancers (red) also show gains or amplification.
In order to investigate whether TRSP1 gene alterations have any association with outcomes, we performed PFS, DFS, DSS, and OS analyses for breast, bladder, and prostate cancers from this combined cohort. In prostate cancer, patients with an altered TRPS1 gene had a statistically significant worse PFS, DFS, DSS, and OS when compared to those with an unaltered gene (Figures 2A–2D). For breast and bladder cancer, no statistically significant survival difference was seen at the gene level.
Figure 2.
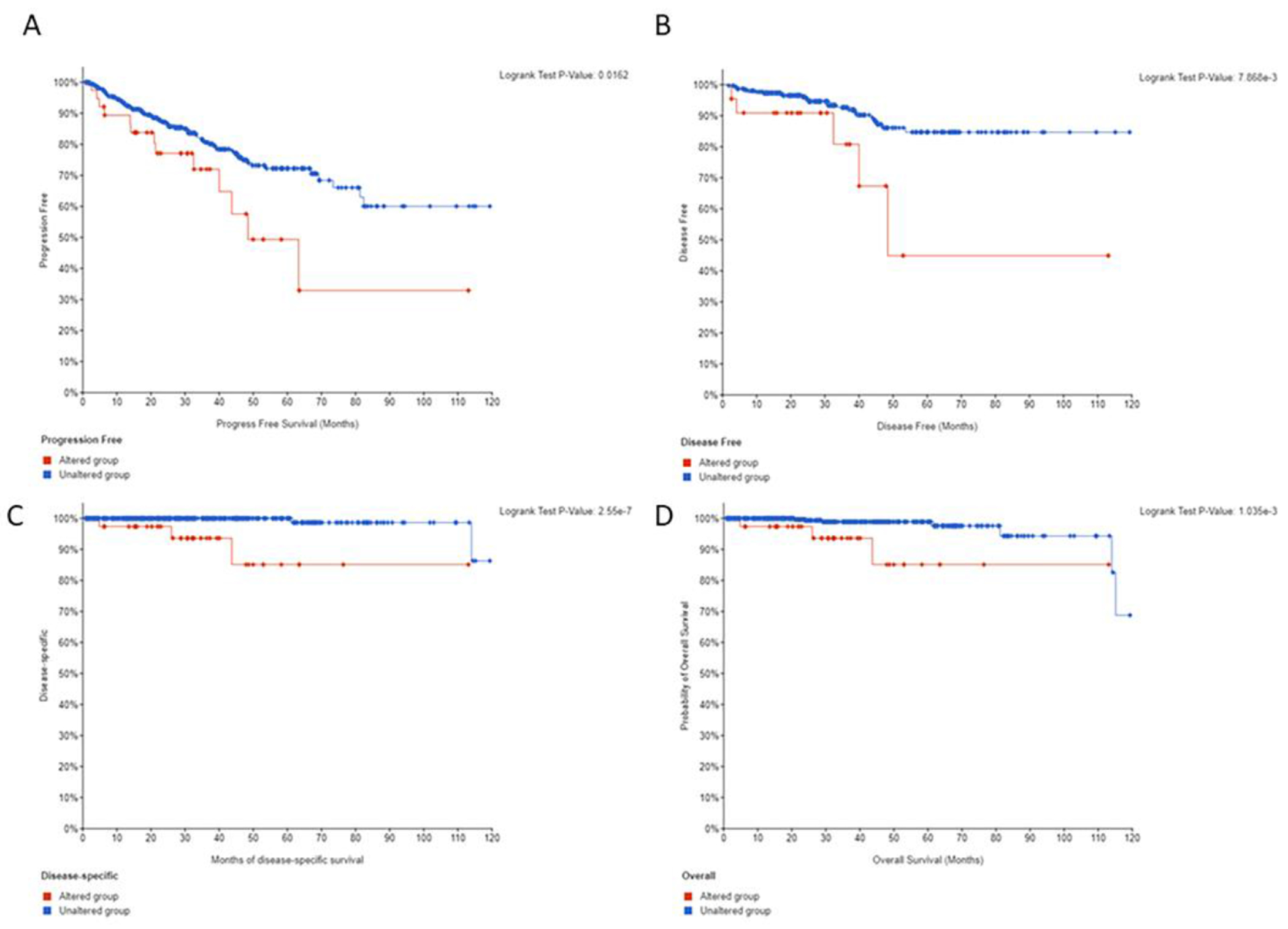
Prostate cancer outcomes for patients with TRPS1 gene alterations. A-D) In prostate cancer, patients with an altered TRPS1 gene had a statistically significant worse progression free (A), disease free (B), disease-specific (C), and overall survival (D) when compared to those with an unaltered gene in the TCGA Cohort.
Next, we investigated whether TRPS1 mRNA levels have any association with outcomes in prostate and bladder cancer in a TCGA cohort. We analyzed mRNA expression of TRPS1 in 296 prostate cancer samples and 52 normal prostate controls. Expression was fairly similar across Gleason scores; however, interestingly, expression was most elevated in the highest Gleason Score (Figure 3A). Though, it is worth noting that the sample size for Grade Group 5 disease is incredibly low (n=4). Contrary to the worse OS observed at the gene level, there was no significant difference in survival outcomes in prostate cancer at the mRNA expression level. While these data do not support that a quantitative mRNA level of TRPS1 is predictive of outcomes, they do show consistently detectable levels of transcribed TRPS1 mRNA across a spectrum of prostate cancer samples, which may indicate that TRPS1 protein expression could be detectable by IHC in a subset of prostate cancers.
Figure 3:
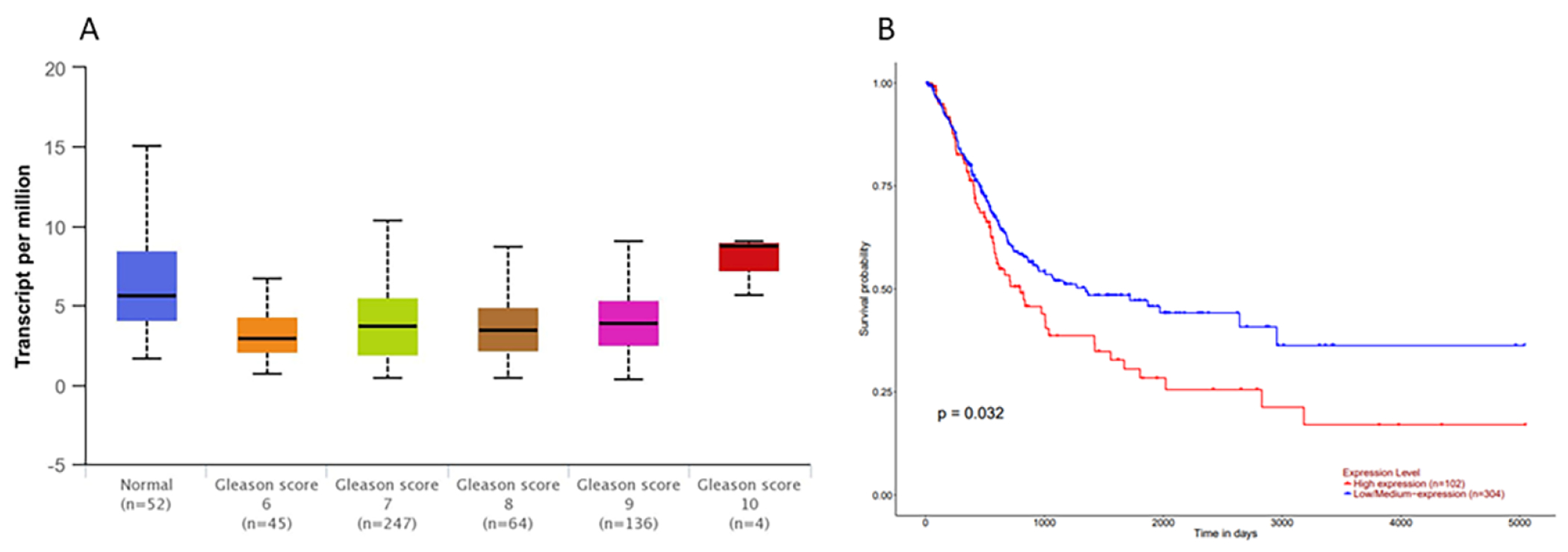
TRPS1 mRNA Expression in Prostate and Bladder Cancer. A) TRPS1 mRNA across Gleason scores in 296 prostate cancer and 52 normal samples in the TCGA Cohort. Although the highest expression is seeing in Gleason Score 10, note the small sample size and the largely uniform expression level. B) mRNA expression of TRPS1 in 406 bladder cancer samples from the TCGA cohort revealing high expression is correlated with a worse overall survival.
Next, we analyzed mRNA expression of TRPS1 in 406 bladder cancer samples from a TCGA dataset (18). High mRNA expression of TRPS1 was associated with a worse OS (Figure 3B).
Although it is difficult to make definitive conclusions from the gene level and mRNA level analyses in this data, it does provide a rational basis for studying whether or not detectable protein expression can be found in a subset of bladder and prostate cancer by IHC in clinical FFPE samples.
Tissue Microarray Cohorts:
Bladder:
A total of 73 patients with muscle invasive high-grade urothelial carcinoma on transurethral resection of bladder tumor (TURBT) were included in a TMA cohort. TRPS1 expression was observed in 20 cases (27%), with 15 of them (21%) demonstrating intermediate to high expression scores (Figure 4). All patients underwent cystectomy, and there was no statistically significant correlation in expression of TRPS1 on TURBT with final pathologic T or N stages (Supplemental Table 2). However, low expression, intermediate expression, and high expression of TRPS1 by IHC was associated with a statistically significant increasingly worse OS (p=0.040) (Supplemental Figure 1). Finally, GATA3 expression was observed in 68 of the same bladder cancer specimens (93%), with all demonstrating intermediate to high expression scores. Similar survival and stage analysis of GATA3 could not be performed due to insufficient number of cases with negative expression.
Figure 4.
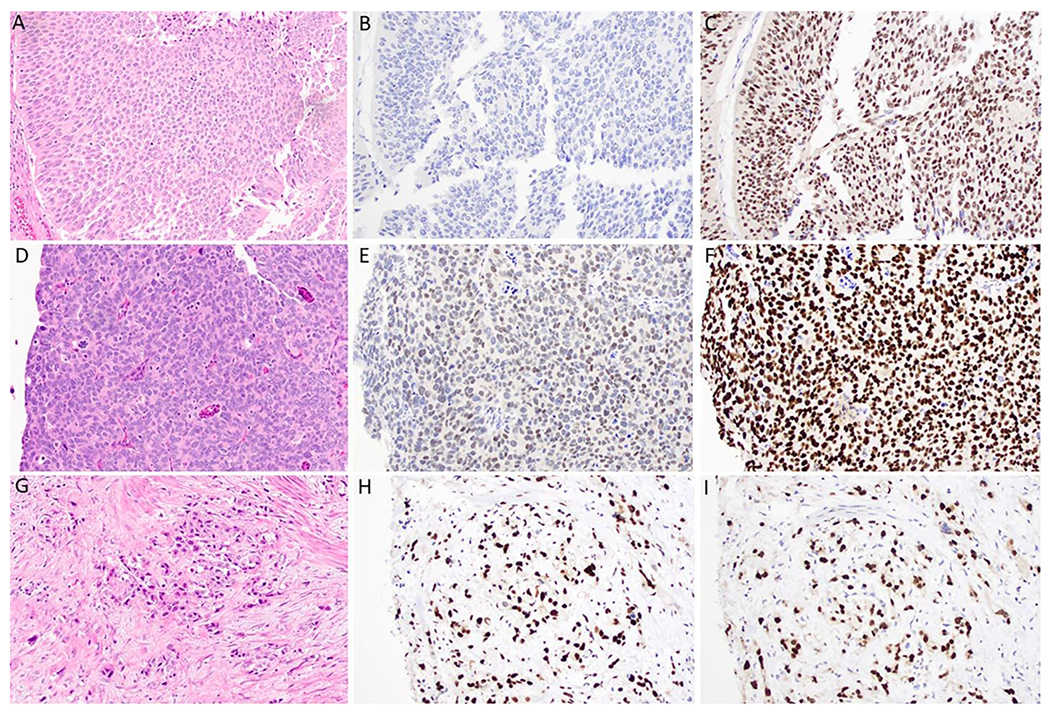
TRPS1 and GATA3 Immunohistochemical Expression in Bladder Urothelial Carcinoma. A, B, C) Urothelial carcinoma showing a lack of TRSP1 and high GATA3 expression, H&E stain, TRPS1 stain, and GATA3 stain, respectively, 20x magnification. D, E, F) Urothelial carcinoma showing intermediate TRPS1 expression and high GATA3 expression, H&E stain, TRPS1 stain, and GATA3 stain, respectively, 40x magnification. G, H, I) Urothelial carcinoma showing high TRPS1 expression and high GATA3 expression, H&E stain, TRPS1 stain, and GATA3 stain, respectively, 40x magnification.
Prostate:
A total of 69 prostatic acinar adenocarcinomas were evaluated to include 35 cases from the prostate (i.e., primary site of disease) (including Grade Groups 2-5) and 34 metastases (26 to local lymph nodes and 8 to distant sites such as lung, bone, brain, and soft tissue/subcutaneous). TRPS1 expression was observed in 22 specimens overall (31.9%), with 17 of them (24.6%) demonstrating intermediate to high expression scores (Table 1, Figure 5). In contrast, all cases were negative for GATA3. Out of the 35 primary cases evaluated, 10 (28.6%) demonstrated positive TRPS1 expression, with six (17.1%) showing intermediate or high expression scores.
Table 1.
TRPS1 expression scores in Prostate Cancer TMA Cohort with respect to Gleason grade groups and primary versus metastatic site of disease.
| Gleason Grade Group | Negative | Low | Intermediate | High |
|---|---|---|---|---|
| 2 (n=4) | 4 (100%) | 0 | 0 | 0 |
| 3 (n=6) | 6 (100%) | 0 | 0 | 0 |
| 4 (n=4) | 4 (100%) | 0 | 0 | 0 |
| 5 (n=21) | 11 (52.4%) | 4 (19.0%) | 4 (19.0%) | 2 (9.5%) |
| All Primary Prostate Cancer Total (n=35) | 25 (71.4%) | 4 (11.4%) | 4 (11.4%) | 4 (11.4%) |
| Pelvic Lymph Node Metastasis (n=26) | 18 (69.2%) | 1 (3.8%) | 2 (7.7%) | 5 (19.2%) |
| Distant Metastasis (n=8) | 4 (64.7%) | 0 | 1 (12.5%) | 3 (11.5%) |
| All Metastatic Prostate Cancer (n=34) | 22 (64.7%) | 1 (2.9%) | 3 (8.8%) | 8 (23.5%) |
| All Cases Total (n=69) | 47 (68.1%) | 5 (7.2%) | 7 (10.1%) | 10 (14.5%) |
Figure 5.
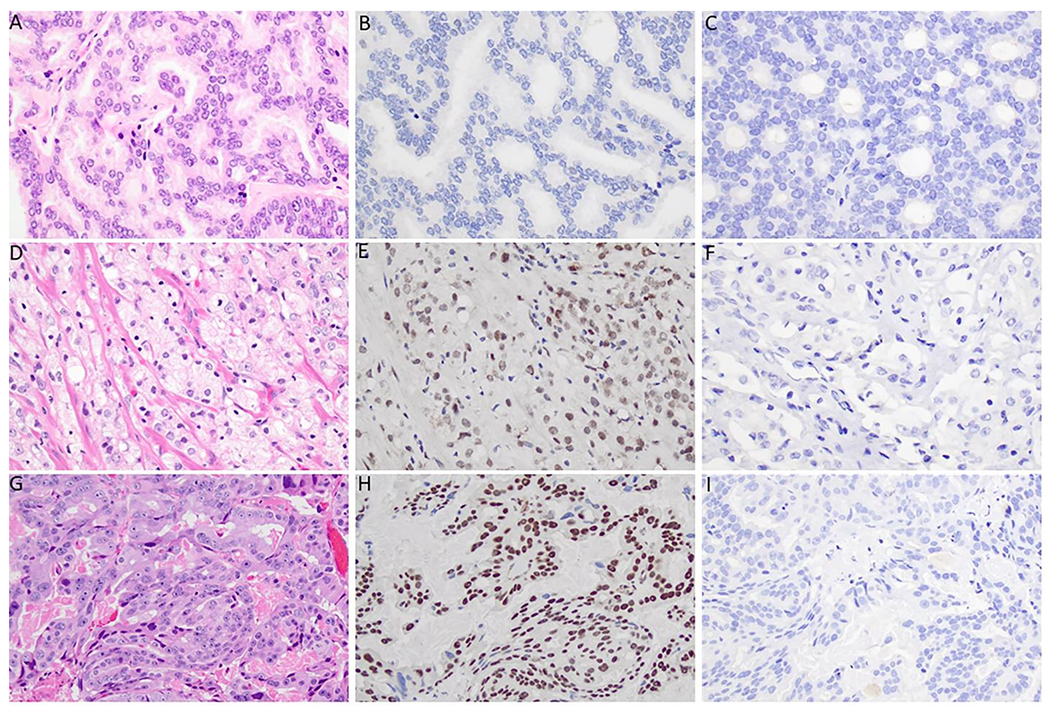
TRPS1 and GATA3 Immunohistochemical Expression in Prostate Cancer. A, B, C) Prostate cancer showing a lack of TRSP1 and GATA3 expression, H&E stain, TRPS1 stain, and GATA3 stain, respectively, 40x magnification. D, E, F) Prostate cancer showing intermediate TRPS1 expression and a lack of GATA3 expression, H&E stain, TRPS1 stain, and GATA3 stain, respectively, 40x magnification. G, H, I) Prostate cancer showing high TRPS1 expression and a lack of GATA3 expression, H&E stain, TRPS1 stain, and GATA3 stain, respectively, 40x magnification.
Among the 34 metastatic samples assessed, 12 (35.3%) showed positive expression, of which 11 had intermediate or high scores. In the 8 cases of distant metastasis, TRPS1 expression was observed in 4 cases (50.0%), with 3 showing high expression and 1 showing intermediate expression. Seven of the 26 cases (26.9%) metastatic to pelvic lymph nodes showed intermediate to high expression with TRPS1 (5 high expression, 2 intermediate expression).
TRPS1 expression was evaluated in matched primary and metastatic prostate cancers from 13 patients. Two patients showed high TRPS1 expression scores at both sites, while seven patients were negative for TRPS1 expression at both sites. Among the remaining four patients, three had positive TRPS1 expression in the primary site but negative expression in the corresponding metastatic site, while one patient showed negative expression in the primary site but positive expression in the corresponding metastatic site. However, all four of these cases showed low to intermediate TRPS1 expression scores at both sites, with no strong expression observed.
Breast:
TRPS1 was expressed in 183 of 210 invasive breast carcinomas (87.1%), and intermediate to high expression was present in 173 of these cases (82.8%). By contrast, GATA3 was positive in only 136 cases (64.8%) with intermediate to high expression in 131 cases (62.4%) (Figure 6).
Figure 6.
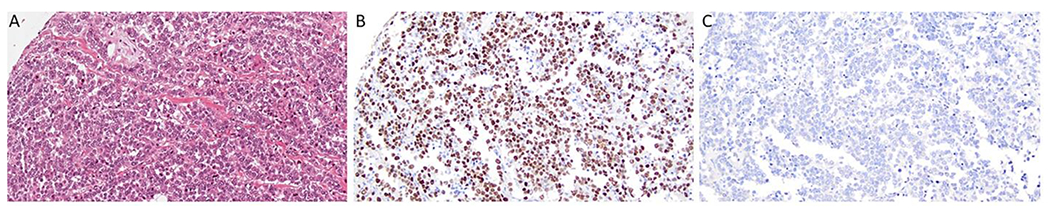
TRPS1 and GATA3 Immunohistochemical Expression in a Triple Negative Breast Cancer Case. A, B, C) Triple negative breast carcinoma showing high TRPS1 expression and a lack of GATA3 expression, H&E stain, TRPS1 stain, and GATA3 stain, respectively, 20x magnification.
TRPS1 was expressed in 86/94 (91.5%) of ER+/HER2− tumors, 29/33 (87.9%) of HER2+ tumors, and 68/83 (81.9%) of TNBC. Similarly, GATA3 was positive in 88/94 (93.6%) of ER+/HER2 negative tumors and 29/33 (87.9%) of HER2 positive cases. However, of note, GATA3 was only positive in 19/83 (22.9%) of TNBC (Supplemental Table 3). There is a statistically significant difference in TRPS1 versus GATA3 expression in TNBC (p=<0.0001); but, there is no statistically significant difference in ER+/HER2− tumors (p=0.58) or in HER2+ tumors (p=1.0).
Discussion:
This study is the first to systematically evaluate TRPS1 expression in prostate and urothelial carcinomas. In our exploratory cohort, we have shown that a significant number of urothelial and prostate carcinomas show gene alterations in TRPS1, 9.0% and 8.1% respectively. Furthermore, high mRNA expression of TRPS1 was associated with a worse OS in bladder cancer.
In our IHC cohort, TRPS1 did show some expression in urothelial tumors (27% of cases), with 21% showing intermediate to high expression. Interestingly, TRPS1 expression was also associated with increasingly worse OS in our IHC cohort. However, GATA3 stained a significantly higher proportion of these urothelial tumors (90.8% in our study), which is consistent with prior literature (19, 20). Four prior studies have evaluated TRPS1 IHC expression in urothelial carcinomas. Ai et al. included 115 urothelial carcinomas, and only 2 showed low expression scores (6). More recently, Lynn et al. included 83 urothelial carcinomas on TMA, and all were negative for TRPS1 expression (21). Two cytology-based studies also tested TRPS1 expression in urothelial carcinoma, and all cases were negative (n=6 and n=14, respectively) (11, 12).
The four aforementioned studies that included urothelial carcinomas all used the same antibody clone as in our study (PA5-84874). Indeed, only one recent study thus far, Du et al., has reported using a different antibody clone (EPR16171 from Abcam). They showed a 91.2% positivity rate in TNBC, which is similar to prior studies (10). Therefore, future studies are needed to compare the specificity of different TRPS1 antibody clones. Prior studies have used different antibody dilutions of the PA5-84874 clone, ranging from 1:50 (12), as used in our study, to 1:500 (9). The optimal dilution will vary between laboratories. As seen in figures 4–6, our staining protocol produced strong, clean nuclear expression with no significant background staining.
In addition, TRPS1 was also expressed in 31.9% of prostate carcinomas, with a substantial portion (24.6%) demonstrating intermediate to high expression. GATA3 was negative in all 69 prostate cores evaluated. Interestingly, intermediate to high TRPS1 expression was observed in 28.6% of local Grade Group 5 prostate cancer, 26.9% of pelvic lymph node metastases, and 50% in sites of distant metastases. It is important to re-emphasize that TRPS1 expression was observed in sites of metastatic disease, which is a crucial observation since these are the cases where TRPS1 staining will be used in the clinical setting. Interestingly, in the TCGA cohort, TRPS1 expression increased with higher Gleason scores. Not inconsistent with this observation, our TMA data supports a trend toward staining in higher grade group/aggressive tumors. However, case numbers are too low from either cohort to draw definitive conclusions.
Finally, TRPS1 consistently stained breast carcinomas (87.1% in our cohort) while GATA3 stained 64.8%. A statistically significant difference was seen in TRPS1 vs GATA3 expression in TNBC in our cohort, which is consistent with prior studies (6, 8, 10). While the overall rate of TRPS1 expression in breast cancer is slightly lower in our cohort than in literature published to date, our cohort was enriched for TNBC (accounting for 83 out of 210 of the breast tumors tested (39.5%)), which also explains our lower GATA3 expression rates.
While we can confirm that TRPS1 is more sensitive than GATA3 for breast cancer, our study does raise concerns about claims to its specificity, especially in regard to prostate and urothelial carcinomas (IHC results summarized in Table 2).
Table 2.
Summary of TPRS1 and GATA3 IHC expression Breast, Prostate, and Muscle Invasive Bladder Urothelial Carcinoma
| TRPS1 expression scores | ||||
|---|---|---|---|---|
| Tumor Type | Negative | Low | Intermediate | High |
| Breast (n=210) | 27 (12.9%) | 10 (4.8%) | 27 (12.9%) | 146 (69.5%) |
| Prostate (n=69) | 47 (68.1%) | 5 (7.2%) | 7 (10.1%) | 10 (14.5%) |
| Bladder (n=73) | 53 (72.6%) | 5 (6.9%) | 9 (12.3%) | 6 (8.2%) |
| GATA3 expression | ||||
| Tumor Type | Negative | Low | Intermediate | High |
| Breast (n=210) | 74 (35.2%) | 5 (2.4%) | 11 (5.2%) | 120 (57.1%) |
| Prostate (n=69) | 69 (100%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Bladder (n=73) | 5 (6.9%) | 0 (0%) | 5 (6.9%) | 63 (86.3%) |
Early studies have evaluated the use of TRPS1 as a prognostic marker in breast cancer; however, the results need to be validated in larger cohorts (22–24). Further work is needed in breast, urothelial, and prostate cancers to draw definitive conclusions about the prognostic significance of TRPS1 expression.
When evaluating metastatic tumors of unknown primary, a broad IHC panel is still needed if there is clinical concern for determining breast versus urothelial carcinoma. If the tumor is negative for ER/PR/HER2 and GATA3 positive, TRPS1 cannot provide definitive results for primary site of disease, as we have shown significant staining in bladder tumors. In this scenario, if TRPS1 is negative, one could postulate that the tumor is more likely to be of urothelial origin rather than breast, as TRPS1 stained only 27.4% of urothelial carcinomas versus 87.1% of TNBC in our study. Nevertheless, clinical and radiologic information is likely to be more informative rather than relying on TRPS1 expression alone.
However, if TRPS1 is positive in the setting of an ER/PR/HER2 negative and GATA3 positive tumor, additional markers are still required. SOX10 may be of value as it has been reported to stain 40-80% of TNBC. However, a cytokeratin stain should also be included when using SOX10, as it is a basal/myoepithelial marker rather than a specific breast carcinoma marker, and metastatic melanoma would need to be excluded (7).
In males, the scenario of a TNBC versus metastatic prostate carcinoma is rare, but the tumors can show similar glandular or cribriform architectural features. In this differential diagnosis, GATA3 remains are more useful marker for breast cancer than TRPS1, as 100% our prostate cases were GATA3 negative, versus 31.9% were positive for TRPS1.
Limitations to our study include the use of TMA sections as opposed to whole tissue sections, such that intratumoral heterogeneity may not have been accurately captured. However, if substantial heterogeneity for expression of TRPS1 does exist, the use of whole tissue sections would only have increased the rates of reported expression and further diminished its specificity.
In summary, our study confirms that TRPS1 is indeed more sensitive than GATA3 for breast cancer. Furthermore, some prostate, including a higher percentage of metastatic cancers, and muscle invasive bladder cancers show significant staining. As a result, our study raises concerns about claims to TRPS1 specificity, especially in regard to advanced prostate and muscle invasive bladder urothelial carcinomas. Furthermore, our study provides interesting insight into TRPS1 as a prognostic marker, trending toward higher expression in advanced prostate cancers and correlating with a worse OS at the mRNA and IHC level in bladder cancer. In conclusion, additional comprehensive studies are needed to help elucidate the true specificity of TRPS1 IHC stain with regards to many tumor types before it is widely adopted into clinical practice.
Supplementary Material
Highlights:
TRPS1 is not a specific immunohistochemical marker for breast cancer
A subset of prostate cancer, including metastases, and muscle invasive bladder cancer exhibit significant staining with TRPS1
TRPS1 shows higher expression in advanced prostate cancers and correlates with worse overall survival in bladder cancer at both the mRNA and IHC levels
Further comprehensive studies are needed to establish the true specificity of TRPS1 IHC stain across various tumor types before its widespread clinical adoption
Funding Statement/Acknowledgements:
This research was supported by the Biospecimen Procurement & Translational Pathology and the Cancer Research Informatics Shared Resource Facilities of the University of Kentucky Markey Cancer Center (P30CA177558). Preliminary findings in this study were presented as a platform presentation at the 112th Annual United States and Canadian Academy of Pathology Meeting in March of 2023.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement: The authors have no relevant conflicts of interest to disclose.
Ethics Approval Statement: This study was approved by the institutional review board at the University of Kentucky (Protocol 79922).
Data availability statement:
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References:
- 1.Malik TH, Von Stechow D, Bronson RT, Shivdasani RA. Deletion of the GATA domain of TRPS1 causes an absence of facial hair and provides new insights into the bone disorder in inherited trichorhino-phalangeal syndromes. Mol Cell Biol. 2002;22(24):8592–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suemoto H, Muragaki Y, Nishioka K, Sato M, Ooshima A, Itoh S, et al. Trps1 regulates proliferation and apoptosis of chondrocytes through Stat3 signaling. Dev Biol. 2007;312(2):572–81. [DOI] [PubMed] [Google Scholar]
- 3.Gai Z, Zhou G, Itoh S, Morimoto Y, Tanishima H, Hatamura I, et al. Trps1 functions downstream of Bmp7 in kidney development. J Am Soc Nephrol. 2009;20(11):2403–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornelissen LM, Drenth AP, van der Burg E, de Bruijn R, Pritchard CEJ, Huijbers IJ, et al. TRPS1 acts as a context-dependent regulator of mammary epithelial cell growth/differentiation and breast cancer development. Genes Dev. 2020;34(3-4):179–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang J, Liu X, Huang Y, He L, Zhang W, Ren J, et al. TRPS1 drives heterochromatic origin refiring and cancer genome evolution. Cell reports. 2021;34(10):108814. [DOI] [PubMed] [Google Scholar]
- 6.Ai D, Yao J, Yang F, Huo L, Chen H, Lu W, et al. TRPS1: a highly sensitive and specific marker for breast carcinoma, especially for triple-negative breast cancer. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2021;34(4):710–9. [DOI] [PubMed] [Google Scholar]
- 7.Ding Q, Huo L, Peng Y, Yoon EC, Li Z, Sahin AA. Immunohistochemical Markers for Distinguishing Metastatic Breast Carcinoma from Other Common Malignancies: Update and Revisit. Seminars in diagnostic pathology. 2022;39(5):313–21. [DOI] [PubMed] [Google Scholar]
- 8.Yoon EC, Wang G, Parkinson B, Huo L, Peng Y, Wang J, et al. TRPS1, GATA3, and SOX10 expression in triple-negative breast carcinoma. Human pathology. 2022;125:97–107. [DOI] [PubMed] [Google Scholar]
- 9.Parkinson B, Chen W, Shen T, Parwani AV, Li Z. TRPS1 Expression in Breast Carcinomas: Focusing on Metaplastic Breast Carcinomas. The American journal of surgical pathology. 2022;46(3):415–23. [DOI] [PubMed] [Google Scholar]
- 10.Du T, Pan L, Zheng C, Chen K, Yang Y, Chen J, et al. Matrix Gla protein (MGP), GATA3, and TRPS1: a novel diagnostic panel to determine breast origin. Breast cancer research : BCR. 2022;24(1):70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang M, Stendahl K, Cai G, Adeniran A, Harigopal M, Gilani SM. Evaluation of TRPS1 Expression in Pleural Effusion Cytology Specimens With Metastatic Breast Carcinoma. Am J Clin Pathol. 2022;158(3):416–25. [DOI] [PubMed] [Google Scholar]
- 12.Chen G, Hang JF, Chen YA, Lin SJ, Chiu HM, Hsu CY, et al. The diagnostic utility of trichorhinophalangeal syndrome type 1 immunohistochemistry for metastatic breast carcinoma in effusion cytology specimens. Cancer Cytopathol. 2023;131(4):226–33. [DOI] [PubMed] [Google Scholar]
- 13.Asch-Kendrick RJ, Samols MA, Lilo MT, Subhawong AP, Sharma R, IIlei PB, et al. NKX3.1 is expressed in ER-positive and AR-positive primary breast carcinomas. J Clin Pathol. 2014;67(9):768–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–w14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lánczky A, Győrffy B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J Med Internet Res. 2021;23(7):e27633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu H, Shi J, Wilkerson ML, Lin F. Immunohistochemical evaluation of GATA3 expression in tumors and normal tissues: a useful immunomarker for breast and urothelial carcinomas. Am J Clin Pathol. 2012;138(1):57–64. [DOI] [PubMed] [Google Scholar]
- 20.Chang A, Amin A, Gabrielson E, Illei P, Roden RB, Sharma R, et al. Utility of GATA3 immunohistochemistry in differentiating urothelial carcinoma from prostate adenocarcinoma and squamous cell carcinomas of the uterine cervix, anus, and lung. Am J Surg Pathol. 2012;36(10):1472–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lynn TJ, Shi J, Liu H, Monaco SE, Prichard JW, Lin F. Trichorhinophalangeal Syndrome Type 1 Is a Highly Sensitive and Specific Marker for Diagnosing Triple-Negative Breast Carcinomas on Cytologic Samples. Archives of pathology & laboratory medicine. 2023. [DOI] [PubMed] [Google Scholar]
- 22.Chen JQ, Bao Y, Litton J, Xiao L, Zhang HZ, Warneke CL, et al. Expression and relevance of TRPS-1: a new GATA transcription factor in breast cancer. Horm Cancer. 2011;2(2):132–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen JQ, Litton J, Xiao L, Zhang HZ, Warneke CL, Wu Y, et al. Quantitative immunohistochemical analysis and prognostic significance of TRPS-1, a new GATA transcription factor family member, in breast cancer. Horm Cancer. 2010;1(1):21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen JQ, Bao Y, Lee J, Murray JL, Litton JK, Xiao L, et al. Prognostic value of the trichorhinophalangeal syndrome-1 (TRPS-1), a GATA family transcription factor, in early-stage breast cancer. Ann Oncol. 2013;24(10):2534–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


