Abstract
The transcriptional regulator Yin Yang 1 (YY1) controls many aspects of cell behavior and is essential for development. We analyzed the fate of YY1 during apoptosis and studied the functional consequences. We observed that this factor is rapidly translocated into the cell nucleus in response to various apoptotic stimuli, including activation of Fas, stimulation by tumor necrosis factor, and staurosporine and etoposide treatment. Furthermore, YY1 is cleaved by caspases in vitro and in vivo at two distinct sites, IATD12G and DDSD119G, resulting in the deletion of the first 119 amino acids early in the apoptotic process. This activity generates an N-terminally truncated YY1 fragment (YY1Δ119) that has lost its transactivation domain but retains its DNA binding domain. Indeed, YY1Δ119 is no longer able to stimulate gene transcription but interacts with DNA. YY1Δ119 but not the wild-type protein or the caspase-resistant mutant YY1D12A/D119A enhances Fas-induced apoptosis, suggesting that YY1 is involved in a positive feedback loop during apoptosis. Our findings provide evidence for a new mode of regulation of YY1 and define a novel aspect of the involvement of YY1 in the apoptotic process.
Yin Yang 1 (YY1) is a ubiquitously expressed Zn finger transcriptional regulator that can function as an activator, a repressor, or an initiator binding protein. YY1 is an abundant protein, and numerous potential target genes, both cellular and viral, have been identified (55, 67). It has been estimated that more than 7% of vertebrate genes contain YY1 binding sites (22). The products of some of these target genes function in the control of cell growth, development, differentiation, and tumor suppression (55, 67). By binding to DNA, YY1 controls gene expression by recruiting several different cofactors, including the histone acetyltransferases CBP and p300, various histone deacetylases, and the Arg-specific methyltransferase PRMT1 (4, 50, 74). In addition, YY1 interacts with various basal transcription factors, such as TATA binding protein, TAFs, and transcription factor IIB (4, 11, 36, 69), and other transcriptional regulators, including Sp1, c-Myc, and C/EBPβ (3, 5, 32, 53, 57). Through interplay with the above-mentioned factors and through binding to DNA response elements and to initiator sequences, YY1 can exert broad activities at target promoters. Interestingly, YY1 is also associated with polycomb group (PcG) protein complexes and is the mammalian orthologue of the Drosophila PcG protein Pleiohomeotic (1, 7, 18, 52). PcG proteins are important in maintaining repressed transcriptional states of genes and thus are important in sustaining cell fate (44). In this setting, YY1 functions as the DNA-targeting subunit of PcG complexes.
The importance of YY1 for basic biological processes is underscored by the finding that it is essential for mouse embryo development. The targeted disruption of yy1 in mice results in periimplantation lethality, a stage of development characterized by extensive proliferation and differentiation (13). Additional observations connect YY1 to the control of proliferation. YY1 interacts with the retinoblastoma tumor suppressor protein Rb, which controls the G1- to S-phase transition during the cell cycle (48). Also, the repression function of YY1 is antagonized by the adenovirus E1A protein, a potent activator of cell proliferation (33, 56). An important role of YY1 in the G1- to S-phase transition is also implied by the finding that the subcellular localization of this factor is cell cycle regulated. While YY1 is cytoplasmic in G1 phase, it becomes nuclear in early and middle S phases and returns to being primarily cytoplasmic later in S phase (45). This nuclear accumulation of YY1 near the G1- to S-phase transition coincides with both an increase in YY1 DNA binding activity and an up-regulation of histone genes (15, 24). Recent findings provide evidence for an interplay between YY1 and tumor suppressor p53. YY1 functions as a negative regulator of p53 by enhancing its MDM2-dependent ubiquitination and degradation (20, 63). Loss of YY1 results in increased apoptosis and inhibition of proliferation, possibly due to p53 activation (20, 63). YY1 and p53 antagonism occurs also at the level of DNA binding, since these two proteins have partially overlapping targeting sequences, leading to competition at some promoters (73). These findings not only support the notion that YY1 is involved in cell proliferation control but also suggest its association with apoptosis.
Apoptosis is an important mechanism for maintaining tissue homeostasis. Activation and execution of apoptosis are dependent on caspases, cysteine proteases that generally cleave at sites with an aspartic acid in an N-terminal location relative to the cleavage site (40). More than 280 caspase substrates have been described; some of these are important for specific processes that are associated with apoptosis (17). However, the function of the majority of caspase substrates during apoptosis is not yet understood.
One of the central alterations that take place in the nucleus is the cleavage of DNA during apoptosis. At an early stage of nuclear disassembly, chromatin is cleaved into large, 50- to 300-kbp fragments and later processed into oligonucleosomal fragments by several different enzymes (27, 34, 51). This process generates DNA breaks that induce a DNA repair response. The poly(ADP-ribose) polymerase PARP-1 is thought to function as a nick sensor and, by poly(ADP-ribos)ylating proteins in the vicinity of DNA nicks, to generate a favorable environment for repair processes (23, 54). YY1 is one of the few proteins that have been shown to be modified by PARP-1 in response to DNA damage (41). The activity of PARP-1 is stimulated by YY1, suggesting a role of this protein in DNA repair (19).
We analyzed the fate of YY1 in apoptotic cells. We report that YY1 is the target of rapid processing in response to apoptotic stimuli. This activity includes the efficient translocation of YY1 to the nucleus irrespective of the cell cycle status of the treated cells and the specific cleavage of YY1 at two sites within the transactivation domain by caspases. These findings suggest that YY1 possesses, in addition to its effects on proliferation and differentiation, functions during apoptosis. Indeed, the cleavage results in a YY1 protein that can still bind DNA but is deficient in transactivation. Furthermore, the apoptosis-specific YY1 fragment enhances apoptosis. Our study provides evidence for a previously nonrecognized function of YY1 during apoptosis, an additional specific aspect of cell behavior.
MATERIALS AND METHODS
Cell cultures, transient transfections, and apoptosis assays.
Jurkat T and HeLa-80Fas cells were grown in RPMI 1640 medium containing 5% fetal calf serum. HeLa, HeLa S3, HEK293, and COS7 cells were grown in Dulbecco modified Eagle medium with 10% fetal calf serum. Transient transfections with calcium phosphate were performed as described previously (60) or with ExGen500 (Fermentas) as suggested by the manufacturer. Reporter gene assays were conducted with P5 + 1-tk-luc, which contains the P5 + 1 sequence of the adeno-associated virus P5 promoter and the minimal thymidine kinase promoter (−32 to +51) (4, 56).
Apoptosis was induced in Jurkat T cells (107 cells/ml) with an anti-Fas antibody (100 ng of immunoglobulin M/ml; clone CH11; Kamiya Biomedical Co., Seattle, Wash.), Flag-FasL (100 ng/ml) in combination with anti-Flag antibody M2 (0.5 μg/ml; Sigma), or staurosporine (STS; 1 μM) in serum-free medium. For UV light induction, Jurkat T cells were washed with phosphate-buffered saline (PBS) and induced to undergo apoptosis (6 × 107 cells/2 ml of PBS per 10-cm dish) by using a Stratalinker (30 J/m2 or 120 μW/cm2 for 2.5 s). Caspase activity was inhibited by preincubation for 90 min with 100 μM z-VADfmk (Kamiya Biomedical Co.) before the induction of apoptosis.
HeLa, HeLa S3, and COS7 cells were treated with either STS at 1 μM or recombinant mouse tumor necrosis factor alpha (Sigma) at 100 ng/ml and cycloheximide at 2.5 μg/ml. HeLa-80Fas cells were preincubated with cycloheximide at 2.5 μg/ml for 30 min before the addition of an anti-Fas antibody (catalog no. 05-201; Upstate, Lake Placid, N.Y.) at 100 ng/ml (72). Etoposide (Sigma) was used at 85 μM with 2-aminopurine (Sigma) at 10 mM. At 30 min before fixation, cultures were pulse-labeled with bromodeoxyuridine (BrdU; Roche Molecular Biochemicals/Boehringer Mannheim). Cells were fixed with cold 70% ethanol-15 mM glycine (pH 2.0) for 20 to 30 min at −20°C and prepared for immunostaining.
For YY1 apoptosis assays, 2 × 104 HeLa cells were placed in each well of a 12-well plate and transfected on the next day. YY1 and YY1 mutants were coexpressed with enhanced green fluorescent protein (EGFP) and Fas to identify transfected cells and to sensitize cells for apoptosis, respectively. At 24 h after transfection, the cells were fixed with 4% paraformaldehyde at 4°C for 23 min, washed once with PBS, and then analyzed by fluorescence microscopy with Analysis software (Olympus). Transfected EGFP-expressing cells were counted, and the apoptotic subset was determined on the basis of morphological criteria (see Fig. 7).
FIG. 7.
Enhancement of apoptosis by YY1Δ119. (A) HeLa cells were cotransfected with pEGFP-C1 (0.05 μg) in order to detect the transfected cells, an expression plasmid for the death receptor Fas (pCR3-Fas; 0.1 μg), and plasmids expressing YY1-T7, YY1Δ119-T7, and YY1D12A/D119A-T7 (1.2 μg each). (Left panels) GFP-expressing cells were scored for morphological signs of apoptosis at 24 h after transfection. (Right panel) Summary of two independent experiments with several slides and with scoring of at least 15 fields with at least 20 cells. Statistical values obtained by two-tailed Student's t test analysis are given. Error bars indicate standard deviations. (B) Experiments were carried out essentially as described for panel A. In addition, cells were transfected with pBabePuro and selected with puromycin after transfection. Apoptosis was induced by treatment with agonistic anti-Fas antibodies (80 ng/ml) for 3 h. Caspase 3 or caspase 7 activities in whole-cell extracts were determined. The activities measured in control samples, i.e., without anti-Fas antibody treatment, were subtracted. The means and standard deviations from two experiments performed in duplicate are shown. (C) Equal amounts of plasmids expressing the indicated proteins were expressed transiently in HeLa cells. The expression of the proteins was analyzed by Western blotting with a T7-specific antibody. (D) HEK293 cells were transfected and analyzed as described for panel A. Two independent experiments with several slides and with scoring of at least 10 fields with more than 20 cells are summarized. Statistical values obtained by two-tailed Student's t test analysis are given. Error bars indicate standard deviations.
For caspase 3 or caspase 7 assays, 4 × 105 HeLa or HEK293 cells were placed in 6-cm dishes and transfected as described above. In these transfections, pBabePuro (1.5 μg) was included. At 18 h after transfection, the cells were selected with puromycin (2 μg/ml) for 24 h. Approximately 90% of the surviving cells stained positively for coexpressed EGFP. Subsequently, the cells were allowed to recover in the absence of puromycin for 24 h before the induction of apoptosis with agonistic anti-Fas antibodies (80 ng/ml) for 3 h. Caspase-Glo 3/7 reagent (Promega) was added to the cells, and luminescence was measured after 60 min.
Plasmids and site-directed mutagenesis.
The pCB6+-YY1-T7 expression construct was generated by mutagenizing the stop codon of YY1 and inserting an oligonucleotide encoding the T7 epitope. YY1 point and deletion mutants were generated by standard PCR-based mutagenesis with specific primers and were verified by sequencing. The Fas-expressing plasmid (pCR3-Fas) was obtained from J. Tschopp. pEGFP-C1 was purchased from Clontech.
Western blotting and antibodies.
In general, lysates derived from equal numbers of cells were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), and Western blot analysis was performed as described previously (4). YY1 was detected with either a polyclonal rabbit serum (263, either affinity-purified or whole serum) against human YY1 (4) or an antibody recognizing C-terminal amino acids 395 to 414 (C20; Santa Cruz Biotechnology). Antibodies specific for Max (C-17), for USF (C-20; sc-229), and for cyclin D1 (72-13G; sc-450) were obtained from Santa Cruz Biotechnology. Antibodies specific for PARP-1 (polyclonal rabbit serum), for tubulin (Ab-4; clones DM1A and DM1B), and for the T7 tag (69522-4; Novagen) were purchased from Boehringer, from NeoMarkers, and from Calbiochem, respectively. Antibodies specific for the α subunit of CK2 were a kind gift from D. Litchfield (35). Mnt-specific antibodies were obtained from P. Hurlin (61).
In vitro caspase cleavage assays and peptide sequencing.
His-tagged YY1 (His-YY1) was purified under denaturation and renaturation conditions as described previously (4). Caspases 1 to 8 were expressed as N-terminal poly(His) fusion proteins and purified to approximately 90% purity as described previously (65). Enzymes were preactivated for 10 min at 30°C in caspase buffer (100 mM HEPES [pH 7.5], 20% glycerol, 0.5 mM EDTA, 10 mM dithiothreitol). Cleavage reaction mixtures with His-YY1 were incubated in caspase buffer in the presence of 0.5 μg of bovine serum albumin (BSA) in a reaction volume of 20 to 30 μl at 30°C for 1 h. The reactions were stopped in SDS sample buffer, and the proteins were analyzed by Western blotting.
For sequencing of the cleavage products, 4 to 5 μg of recombinant His-YY1 was incubated with preactivated caspase 6 (225 ng) or caspase 7 (450 ng) at 30°C for 15 h. The caspase 6-generated cleavage products were processed and sequenced on an Applied Biosystems (Foster City, Calif.) 477A protein sequencer with a 120-A online high-pressure liquid chromatography system (66). The fragments generated with caspase 7 were separated by SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and stained with Coomassie blue. Bands of interest were washed with 0.1% trifluoroacetic acid-60% acetonitrile and analyzed by using a Procise protein sequencer (model 492A; Applied Biosystems).
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed as described previously (4). Control or Fas-induced cells (2 × 106 cells/100 μl) were lysed in F-buffer (10 mM HEPES [pH 7.5], 50 mM NaCl, 30 mM sodium pyrophosphate, 50 mM NaF, 100 μM Na3VO4, 0.2% Triton X-100, 5 μM ZnCl2, 1 mM phenylmethylsulfonyl fluoride, 2.5 U of pepstatin A/ml, 2.5 U of leupeptin/ml, 0.15 mM benzamidine, 2.8 μg of aprotinin/ml) (60). As probes, oligonucleotides with one YY1 binding site of the immunoglobulin κ3′ enhancer (5′-GATCCTACCCCACCTCCATCTTGTTTGAAGATC) (46) or the P5 + 1 binding site (4) were used. The E-box oligonucleotide has been described elsewhere (60). Cell lysates (5 μl) were incubated with 1 μg of poly(dI) · poly(dC) and radioactively labeled oligonucleotides (50,000 cpm) in GS buffer (30 mM HEPES [pH 7.9], 25% glycerol, 25 mM MgCl2, 300 mM KCl, 5 mM 2-mercaptoethanol, 250 μg of BSA/ml, 0.25% Nonidet P-40, 10 mM dithiothreitol) at 30°C for 20 min. DNA-protein complexes were separated by 5% PAGE at 400 V and 4°C.
Immunofluorescence.
For immunocytochemistry studies, all incubations were done at 37°C. Fixed coverslips were blocked with 1 mg of immunoglobulin G-free BSA/ml and 5% normal goat serum (both from Jackson ImmunoResearch Laboratories) in PBS for 1 h. The solution was aspirated, and the coverslips were washed three times with PBS for 10 min each time on a shaker before addition of the primary antibodies (1:30 dilution of an anti-BrdU antibody and 0.4 μg of anti-YY1 antibody C20/ml; Santa Cruz Biotechnology) and incubation for 40 min. Cells were washed again and incubated with the secondary antibodies (from a BrdU kit and 2 μg of Alexa-546-goat anti-rabbit antisera/ml; Molecular Probes). Before the slides were mounted in Vectashield (Vector Laboratories), the cells were counterstained with 2 μg of 4′,6′-diamidino-2-phenylindole dihydrochloride (DAPI)/ml in PBS for 10 min, followed by a 10-min wash with PBS. Fluorescence images were captured by using a confocal laser scanning microscope (Zeiss LSM-510) with a ×40 objective lens. Differential interference contrast (DIC), Nomarski-type images were obtained with a III Plan Neofluar DIC slide. Brightness and contrast adjustments were made with Adobe Photoshop 6.0.
For quantitation, cells were scored as positive for BrdU if any BrdU signal was observed. However, it was noted that the intensity of BrdU staining in treated cells was different from that in untreated ones. Two patterns of BrdU incorporation were observed in cells treated with apoptosis-inducing agents, i.e., either diffuse or localized to distinct foci within the nucleus. This finding may have been due to replicative incorporation versus repair synthesis, but for the purpose of this study, we did not distinguish between these patterns when scoring cells. A minimum of 100 cells were counted for each time point and for every experimental treatment.
RESULTS
Caspase-dependent cleavage of YY1 in apoptotic cells.
Stimulation of apoptosis results in the caspase-dependent cleavage of a significant number of intracellular proteins (14, 17). We tested whether YY1 was altered during apoptosis. Human Jurkat T cells were treated with agonistic Fas-specific antibodies, and whole-cell lysates were analyzed for YY1 by immunoblotting. By 4 and 6 h after treatment, the signal for full-length YY1 decreased, and at least two fragments of lower apparent molecular weights appeared (Fig. 1A). The formation of the two fragments was blocked by z-VADfmk, a pan-caspase inhibitor, but not by calpain inhibitor II, indicating that caspases are involved in processing YY1 (Fig. 1A and data not shown). In addition, the appearance of the YY1 fragments was not affected by the addition of z-VADfmk to the lysis buffer, demonstrating that cleavage occurred in cells before lysis (data not shown). Fragments of YY1 with identical mobilities on SDS-PAGE were also generated in response to STS or UV treatment (Fig. 1B and C). The two YY1 fragments were detected with an antibody specific for the C terminus of YY1, suggesting that N-terminal sequences were lost in response to the apoptotic signals (Fig. 1). No additional fragments could be detected when a polyclonal serum that was raised against full-length YY1 was used (4) (data not shown). In parallel, we determined the number of apoptotic cells (Fig. 1A). As a control, we analyzed the cleavage of PARP-1, a well-known caspase substrate (26, 31). PARP-1 was processed with kinetics similar to those of YY1 (Fig. 1). In addition, staining for the catalytic subunit of protein kinase CK2 served as a negative control (Fig. 1B). CK2 was not cleaved during apoptosis, as shown previously (28). Cleavage of YY1 was not restricted to Jurkat T cells. In response to anti-Fas antibodies and to STS, YY1 cleavage was observed in HeLa and COS7 cells, generating similar fragments with similar kinetics (Fig. 1D and data not shown). These findings demonstrate rapid and specific cleavage of YY1 in response to various apoptosis-inducing conditions.
FIG. 1.
Apoptosis-associated cleavage of YY1. (A) Jurkat T cells were preincubated with 100 μM z-VADfmk or dimethyl sulfoxide for 30 min and then treated with Fas-specific antibodies (100 ng/ml) for the indicated times before being harvested in SDS sample buffer. PARP-1 and YY1 (antibody C20) were detected by Western blot analysis. In parallel, cells were fixed, stained with acridine orange, and analyzed microscopically for characteristic morphological changes associated with apoptosis, i.e., chromatin condensation and nuclear fragmentation. The numbers of apoptotic cells are indicated at the bottom. (B) Jurkat T cells were treated with anti-Fas antibodies as described for panel A or with 1 μM STS. Cell lysates were generated in SDS sample buffer, and Western blotting was performed as described for panel A. In addition, the catalytic subunit of protein kinase CK2 was analyzed with specific antibodies. (C) Jurkat T cells were UV treated and then harvested in SDS sample buffer at the indicated times. The analysis of PARP-1 and YY1 was performed as described for panel A. (D) HeLa cells were treated with STS or anti-Fas antibodies for the indicated times. PARP-1 and YY1 were analyzed as described for panel A. The two YY1 fragments commonly observed are indicated by black arrowheads. Upon UV treatment, an additional, smaller YY1 fragment was detected (white arrowhead). The positions of molecular mass markers (given in kilodaltons) run on the same gels are indicated on the left.
Identification of caspase cleavage sites in YY1.
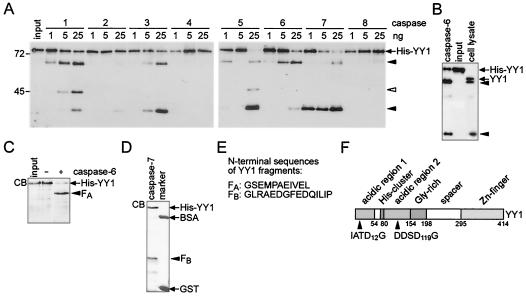
In the next set of experiments, we were interested in determining whether YY1 was a direct substrate for caspases. Therefore, we tested several recombinant caspases for their ability to hydrolyze bacterially expressed His-YY1. Cleavage with caspases 1 to 6 resulted predominantly in a larger YY1 fragment, and that with caspases 3, 5, and 7 resulted in a smaller YY1 fragment (Fig. 2A). Both fragments were detected with an antibody specific for the C terminus of YY1, suggesting that N-terminal sequences were lost. The mobility of these two fragments was identical to that of fragments generated within cells (Fig. 2B). An additional intermediate YY1 fragment was observed in some in vitro reactions (Fig. 2A). This fragment was not considered for further analysis, since it did not match any form of YY1 seen in cells (compare Fig. 1 and Fig. 2). In contrast to the caspases mentioned above, caspases 2, 4, and 8 were unable to cleave His-YY1 (Fig. 2A), although all three enzymes showed catalytic activity toward other substrates (data not shown) (28, 65).
FIG. 2.
Cleavage of His-YY1 by recombinant caspases in vitro. (A) Recombinant His-YY1 (20 ng) was treated with caspases 1 to 8 (1 to 25 ng each) at 30°C for 90 min and analyzed by Western blotting with YY1-specific antibody C20. Black arrowheads identify the YY1 fragments that possess identical apparent molecular weights as the two fragments observed in Fig. 1. An additional fragment is indicated by a white arrowhead. This fragment does not correspond to a form of YY1 observed in apoptotic cells. The positions of molecular mass markers (given in kilodaltons) run on the same gels are indicated on the left. (B) Recombinant His-YY1 was treated with caspase 6 as described for panel A. The resulting proteins were analyzed by SDS-PAGE and compared to the YY1 fragments generated in Jurkat T cells upon treatment with an agonistic anti-Fas antibody for 4 h. The proteins were visualized on Western blots with YY1-specific antibody C20. (C) His-YY1 was incubated in the presence or in the absence of recombinant caspase 6. The proteins were analyzed by SDS-PAGE and Coomassie blue staining (CB). The cleavage product is indicated (FA). (D) His-YY1 was treated with recombinant caspase 7. The resulting proteins were analyzed by SDS-PAGE and Coomassie blue staining (CB). The cleavage product is indicated (FB). The positions of the marker proteins BSA and glutathione S-transferase (GST) are indicated. (E) The N-terminal amino acid sequences of YY1 fragments FA and FB were determined by Edman degradation. (F) Schematic representation of YY1 indicating the major caspase 6 and caspase 7 cleavage sites in relation to the functional domains of YY1 (4). Caspase 6 and caspase 7 cleaved C terminal of Asp12 and Asp119, respectively.
To identify the sites of caspase cleavage in YY1, large-scale cleavage assays were performed with caspases 6 and 7 to generate the two predominant fragments of YY1 (Fig. 2C and D). Cleaved His-YY1 was detected by Coomassie blue staining, the protein fragments (FA in Fig. 2C and FB in Fig. 2D) were extracted, and the N-terminal amino acid sequences were determined as previously described (28) (Fig. 2E). The two sites identified contained an aspartic acid in an N-terminal location relative to the cleavage site, as do most caspase substrates (14, 40) (Fig. 2F). As expected from the analysis of YY1 cleavage in cells, the in vitro sites were located in the N-terminal portion of the protein. This region of YY1 functions as a transactivation domain (4).
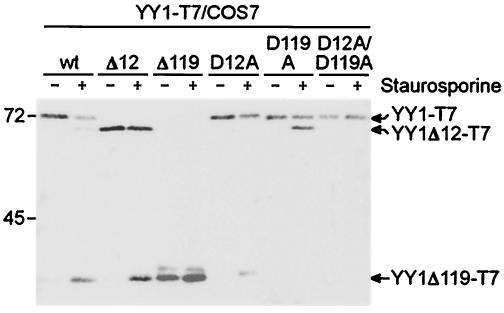
The cleavage sites defined in vitro are used in vivo.
The identified sites were evaluated by analyzing the patterns of cleavage of YY1 mutant proteins expressed in COS7 cells in response to STS-induced apoptosis. Mutants were generated in which Asp12 and/or Asp119 were changed to Ala (YY1D12A-T7, YY1D119A-T7, and YY1D12A/D119A-T7). Furthermore, the portions cleaved away during apoptosis were deleted to generate YY1Δ12-T7 and YY1Δ119-T7. The T7 tag was used to distinguish the expressed proteins from endogenous YY1. The two expected fragments were generated from YY1-T7 (Fig. 3). The apparent molecular weights of these two fragments, of YY1Δ12-T7, and of YY1Δ119-T7 were identical, further supporting the identities of the two cleavage sites. During apoptosis, YY1Δ12-T7 was cleaved, generating a fragment that comigrated with the small fragment of YY1 and with YY1Δ119-T7 (Fig. 3). Apoptosis-induced cleavage of YY1D12A-T7 and of YY1D119A-T7 resulted in the smaller and the larger fragments, respectively. The double mutant YY1D12A/D119A-T7 was no longer cleaved (Fig. 3). Together, these findings demonstrate that hydrolysis of YY1 in cells occurred at the sites mapped in the in vitro assays, thus confirming cleavage at Asp12 and Asp119 in response to apoptotic stimuli.
FIG. 3.
Verification of in vitro YY1 cleavage sites in vivo. T7-tagged YY1 and YY1 mutants were expressed transiently in COS7 cells. The cells then were treated with STS, and the YY1 proteins were detected by immunoblotting with an antibody specific for the tag. Note that the cleaved YY1 fragments comigrated with YY1Δ12 and YY1Δ119. The positions of molecular mass markers (given in kilodaltons) run on the same gels are indicated on the left. wt, wild type.
Reduced solubility of YY1 in response to apoptotic stimuli.
Since the two cleavage sites are localized in the N-terminal acidic domains of YY1, we predicted that the two fragments would bind to DNA in EMSAs with either the immunoglobulin κ 3′ enhancer or the P5 + 1 binding site as a probe (with both probes giving comparable results). Therefore, we prepared low-stringency F-buffer whole-cell extracts from Jurkat T cells pretreated with anti-Fas antibodies. Surprisingly, we observed a strong reduction in YY1-specific DNA binding activity over time (Fig. 4A). Within 2 h of treatment, about 50% of DNA binding was lost, and by 4 and 6 h, most was lost. At later time points, an additional weak complex appeared and was supershifted with YY1-specific antibodies; this complex most likely represented the smaller YY1 fragment (Fig. 4A). However, we did not convincingly detect an EMSA complex that would correspond to YY1Δ12. To address the DNA binding properties in more detail, we expressed YY1-T7, YY1Δ12-T7, and YY1Δ119-T7 in HeLa cells. In addition, cells expressing YY1-T7 were treated with STS to induce apoptosis. Extracts from these cells were used in EMSA experiments (Fig. 4B). The three YY1 proteins generated specific complexes with the P5 + 1 oligonucleotide; these complexes were supershifted with T7-specific but not with cyclin D1-specific antibodies used as controls. These results indicated that all three proteins were capable of binding to DNA. In response to STS, a complex with mobility identical to that seen with YY1Δ119 was observed and was also supershifted specifically (Fig. 4B). No specific complexes with a mutated P5 + 1 probe were observed with any of the proteins analyzed (data not shown). Thus, YY1Δ119 generated in response to apoptotic signals was present in the extracts and was able to bind specifically to DNA. In addition, YY1Δ12 was able to bind to DNA. Comparable findings were also obtained with HEK293 cells (data not shown). To further verify these findings, His-YY1 was treated with small amounts of caspase 6 and tested for DNA binding. We detected an additional, faster-migrating complex that was supershifted in response to YY1-specific antibodies (Fig. 4C). Its appearance was blocked by z-VADfmk, providing additional evidence that YY1Δ12 can bind to DNA. Collectively, these studies indicate that the two caspase-derived YY1 fragments, YY1Δ12 and YY1Δ119, can bind to DNA in in vitro binding assays.
FIG. 4.
YY1 DNA binding activity in extracts from apoptotic cells. (A) F-buffer lysates of Jurkat T cells were analyzed by EMSAs with the labeled κ3′ enhancer as a probe. Supershift experiments were performed with antibodies specific for YY1 (polyclonal serum 263). YY1 and supershifted (S) complexes are indicated. An additional complex appearing in response to apoptotic signals is marked by an arrowhead. The asterisk denotes nonspecific complexes; F, free probe. (B) HeLa cells were transiently transfected with expression plasmids encoding the indicated YY1 proteins. F-buffer lysates were analyzed by EMSAs with the labeled P5 + 1 oligonucleotide as a probe. Supershift experiments were performed with antibodies specific for the T7 tag or for cyclin D1. STS (1 μM) treatments were performed for 4 h. The various YY1 complexes are indicated. S, supershifted complexes; F, free probe; wt, wild type. (C) Recombinant His-YY1 (15 ng) was incubated with or without recombinant caspase 6 (10 ng) in the presence or in the absence of z-VADfmk for the indicated times. Binding of the resulting proteins to the labeled κ3′ enhancer probe was analyzed by EMSAs. A complex with the specific caspase 6 cleavage product is marked by an arrowhead. S, supershifted complexes; F, free probe. (D) F-buffer lysates of Jurkat T cells were analyzed by EMSAs with a labeled E-box oligonucleotide as a probe. Supershift experiments were performed with antibodies specific for USF, Max, or Mnt. The specific complexes are indicated. The free probe was run off the gel to increase separation of the various complexes. (E) Jurkat T cells were stimulated with Fas-specific antibodies in the presence or in the absence of z-VADfmk as described in the legend to Fig. 1A. At the indicated times, half of the cells were lysed in SDS sample buffer, and the other half were lysed in F-buffer. Equal portions were analyzed by Western blotting for YY1 cleavage as described in the legend to Fig. 1A. The arrowheads identify cleavage products YY1Δ12 and YY1Δ119. As controls, the same samples were analyzed for PARP1 and tubulin. The positions of molecular mass markers (given in kilodaltons) are indicated on the left. (F) The relative extraction of YY1, YY1Δ12, and YY1Δ119 in F-buffer versus SDS sample buffer was determined by scanning and quantifying the corresponding Western blots. The numbers are the mean percentages from two independent experiments.
We noted that the reduction in DNA binding in response to apoptotic stimuli was faster than the processing of full-length YY1 by caspases revealed by Western blot analysis (compare Fig. 1 and Fig. 4A). Also, the nonspecific complexes, defined by their insensitivity to both YY1-specific antibodies (Fig. 4A) and specific competition (data not shown), decreased in intensity during the time course. To exclude a general loss of DNA binding, control EMSA experiments with an E-box probe were performed with the same extracts. Specific complexes, as determined by antibody-induced supershift experiments, were detected for Max-Max, Mnt-Max, and USF-USF (Fig. 4D) (28, 60, 61). No major loss of USF-USF or Mnt-Max binding was seen 6 h after Fas stimulation, while Max-containing complexes showed the previously described mobility shift (28). Thus, the loss of YY1 binding activity does not reflect a general loss of DNA binding activity of transcriptional regulators. However, one difference between the analysis of YY1 by EMSA and the analysis of YY1 by Western blotting is that for the former, the extracts were generated under mild conditions (F-buffer), while for the latter, stringent conditions (SDS sample buffer) were used. Therefore, one possibility was that YY1 was degraded in F-buffer extracts of apoptotic cells after cell lysis. However, a number of control experiments with a broad range of protease inhibitors, including z-VADfmk, indicated that this possibility was highly unlikely (data not shown). Next, we considered the possibility that YY1 changed its association or distribution in cells upon the induction of apoptosis. Therefore, Jurkat T cells were stimulated with anti-Fas antibodies, and YY1 proteins were extracted either in SDS sample buffer or in F-buffer. We observed that significantly less YY1 was extracted from apoptotic cells under mild conditions than under stringent conditions (Fig. 4E and F). This finding was particularly obvious for YY1Δ12 and, to a lesser degree, for full-length YY1 and for YY1Δ119, in agreement with the reduced complex formation seen in gel shift experiments (Fig. 4A, E, and F). Comparable observations were made with HeLa cells (data not shown). When caspases were blocked by z-VADfmk in anti-Fas antibody-treated cells, no difference in solubility was seen. These findings suggested that in addition to being cleaved, full-length YY1 and YY1Δ12 showed altered solubility after stimulation of apoptosis.
Rapid uptake of YY1 into the nucleus in response to apoptotic stimuli.
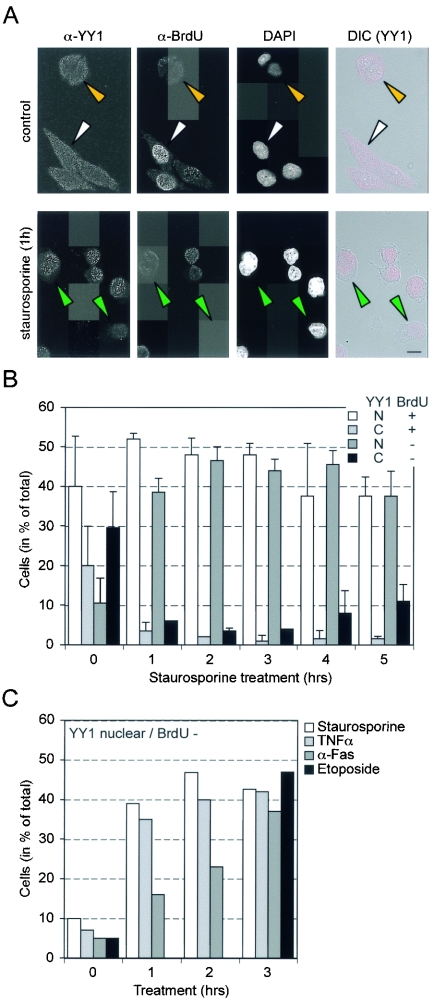
Recent work has indicated that the subcellular distribution of endogenous YY1 is regulated during the cell cycle. YY1 becomes nuclear at the G1/S boundary, when DNA synthesis commences, and is transported into the cytoplasm late in S phase (45). This fact, together with the reduced solubility of YY1, led us to address whether the subcellular localization of YY1 in HeLa cells is altered in response to apoptotic stimuli. HeLa cells were chosen for these experiments because the subcellular localization of endogenous YY1 can be analyzed (45). In addition, these cells also show fragmentation of YY1 during apoptosis (Fig. 1D). Nuclear YY1 was detected in about half of the control cells. Most of these were also in S phase, as shown by BrdU uptake (Fig. 5A and B). Within 1 h of stimulation of apoptosis with STS, most cells showed nuclear YY1 (Fig. 5A and B). Unlike the results obtained for control cells, this effect was most prominent in cells that were BrdU negative. Twenty-five percent of untreated BrdU-negative cells showed nuclear YY1, whereas upon STS treatment, 90 to 95% showed nuclear YY1 (Fig. 5A and B). Similarly, the percentage of BrdU-positive cells showing nuclear YY1 increased from about 67% to more than 90% upon induction of apoptosis (Fig. 5A and B). Tumor necrosis factor, anti-Fas antibody, and etoposide treatments also induced the nuclear accumulation of YY1 in BrdU-negative cells, albeit with somewhat different kinetics (Fig. 5C). Interestingly, nuclear uptake was not inhibited by z-VADfmk, suggesting that the relevant signal was not caspase dependent and that YY1 fragmentation was not a prerequisite (data not shown). Together, these findings indicate that various apoptotic stimuli activate the unscheduled uptake of cytoplasmic YY1 into the nucleus.
FIG. 5.
YY1 translocation to the nucleus in response to apoptosis-inducing agents. (A) Asynchronously growing HeLa S3 cells were fixed, immunostained, and imaged with a confocal microscope. (Top panels) Field of untreated control cells. Some cells display cytoplasmic YY1 and stain positively for BrdU (white arrowhead), and others show only cytoplasmic YY1 staining (yellow arrowhead). (Bottom panels) Cells treated with STS (1 μM). Some of these cells show nuclear staining of YY1 in BrdU-negative cells (green arrowheads). Images of immunodetectable YY1 (α-YY1; C20) and BrdU (α-BrdU) and DAPI images are shown separately in gray scale. DIC, Nomarski-type images showing only YY1 staining (red). Images were collected in optical sections of 1 μm at one Airy unit. Scale bar, 10 μm. (B) HeLa S3 cells were treated with STS (1 μM) for the indicated times and subjected to the same immunocytochemical analysis as that described for panel A. Cells were scored in four subcategories: both nuclear YY1 staining and BrdU-positive staining, nuclear YY1 staining without BrdU staining, and no nuclear YY1 staining with or without BrdU staining. Theexperiment was repeated three times, and for each time point indicated at the bottom of the graph, a minimum of 100 cells were scored. N, nuclear; C, cytoplasmic. Error bars indicate standard deviations. (C) HeLa S3 cells were treated with the indicated apoptosis inducers. The experiment was performed as described for panel A. Only the numbers of cells that possessed nuclear YY1 and that were BrdU negative are shown. The data displayed for STS are the mean values from panel B and are presented for comparison with the other treatments. TNFα, tumor necrosis factor alpha.
YY1Δ119 is defective for transactivation and stimulates apoptosis.
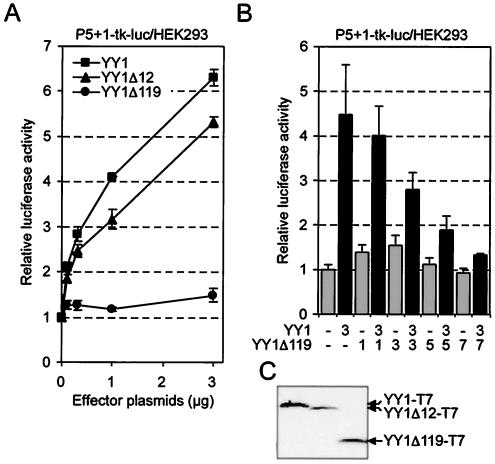
The two YY1 fragments generated upon induction of apoptosis have lost portions of the N-terminal transactivation domain (4). In particular, cleavage at Asp119 would be expected to significantly impair the ability of YY1 to transactivate. Therefore, we tested whether YY1Δ12-T7 or YY1Δ119-T7 had an altered transactivation potential. YY1Δ12-T7 stimulated a reporter gene construct to levels comparable to those obtained with YY1, considering the expression of these proteins (Fig. 6A and C). In contrast, YY1Δ119-T7 showed no transactivation activity, suggesting that during apoptosis, the transactivating function of YY1 is impaired (Fig. 6A). None of the three proteins showed activity toward a reporter construct lacking the P5 + 1 binding sites (data not shown). YY1Δ119-T7 has lost most but not all of the transactivation domain (2). Since this mutant binds to DNA in vitro (Fig. 4B), we were interested in determining whether this protein also interacts with DNA in cells. To this end, we performed competition experiments with YY1-T7 and YY1Δ119-T7 and observed that the latter competed with YY1-dependent activation of the reporter gene (Fig. 6B). This finding supported the conclusion that YY1Δ119-T7 binds to DNA but does not possess transactivation activity.
FIG. 6.
Loss of transactivation upon caspase-dependent cleavage of YY1. (A) HEK293 cells were transiently cotransfected with the YY1 reporter plasmid P5 + 1-tk-luc, a plasmid expressing β-galactosidase for standardization, and the indicated amounts of plasmids encoding YY1-T7, YY1Δ12-T7, and YY1Δ119-T7. The experiments were performed in triplicate; means and standard deviations from a representative experiment of four are shown. (B) Transient transfections were performed as described for panel A. Increasing amounts of expression plasmid for YY1Δ119-T7 were titrated with a fixed amount of expression plasmid for YY1-T7. Means and standard deviations from two independent experiments performed in triplicate are shown. (C) Equal amounts of expression plasmids for YY1-T7, YY1Δ12-T7, and YY1Δ119-T7 were transfected into HEK293 cells. The expression of the three proteins was analyzed by Western blotting with a T7-specific antibody.
The alterations to YY1 occurring during apoptosis and the broad functional role played by this protein suggested that YY1 might affect the apoptotic process. To evaluate whether YY1 affects apoptosis, we transiently expressed the short cleavage product YY1Δ119-T7 or the noncleavable mutant YY1D12A/D119A-T7 together with Fas in HeLa cells. We observed that YY1 or YY1D12A/D119A-T7 did not alter the number of apoptotic cells that appeared as a result of Fas expression (Fig. 7A). In contrast, YY1Δ119-T7 significantly enhanced the extent of apoptosis. This was confirmed by measuring the activity of caspases 3 and 7. YY1Δ119-T7 but not wt YY1-T7 or YY1D12A/D119A-T7 enhanced caspase activity upon stimulation with anti-Fas antibodies (Fig. 7B). In addition to the findings in HeLa cells, YY1Δ119-T7 also enhanced Fas-induced apoptosis in HEK293 cells (Fig. 7D). These findings suggest that YY1Δ119-T7 sensitizes cells for Fas-induced apoptosis.
DISCUSSION
In this study, we analyzed the fate of YY1, a transcriptional regulator of PcG proteins, during apoptosis. We found that the induction of apoptosis results in the rapid redistribution of YY1 from the cytoplasm to the cell nucleus. In addition, YY1 is a substrate of caspases, the intracellular executioners of apoptosis. We mapped in the transactivation domain two distinct cleavage sites that result in truncated YY1 in apoptotic cells that is deficient in transactivation and that enhances Fas-induced apoptosis. These findings defined novel regulatory mechanisms affecting YY1 function and suggested that this transcriptional regulator is somehow associated with the control of apoptosis.
In vitro studies with several recombinant caspases and analysis of YY1 and YY1 mutants in cells defined two specific cleavage sites in the transactivation domain of YY1 (Fig. 1 and 2). The two sites, Ile-Ala-Thr-Asp12↓-Gly and Asp-Asp-Ser-Asp119↓-Gly, fulfill the requirement of caspase substrates with an aspartic acid at the P1 position (17, 40). Sensitivity to the caspase inhibitor z-VADfmk also supports the notion that YY1 is cleaved by caspases in cells. The preferential cleavage of the Asp12 and Asp119 sites exemplified by caspases 6 and 7, respectively, is in agreement with the favored recognition sequence of these two enzymes with regard to the P4 position, i.e., an aliphatic amino acid (Ile in YY1) for caspase 6 and an acidic amino acid (Asp in YY1) for caspase 7 (65, 68). Cleavage at these two sites is independent of cleavage at each, since mutation of either Asp12 or Asp119 to Ala resulted in proteins that were efficiently cleaved at the nonmutated site (Fig. 3). The uptake of YY1 into the nucleus is very fast. At 1 h after induction of apoptosis, YY1 is mainly nuclear (Fig. 5). At this time, no YY1 cleavage is measurable (data not shown); thus, it is likely that caspase-dependent processing occurs in the nuclear compartment. Several caspases, including caspases 3, 6, and 7, could be relevant for the processing of YY1 in cells. These enzymes have been found in the nucleus (43, 64, 70, 75).
The fast nuclear translocation of YY1 in response to various apoptotic stimuli was unexpected (Fig. 5). Differential compartmentalization is a mechanism broadly used to regulate nuclear processes, including transcription and DNA repair (9, 59, 71). Regulation of YY1 by nucleocytoplasmic shuttling was only recently shown (45). Previously, it was demonstrated that YY1 is cytoplasmic during the early development of Xenopus embryos and accumulates in the nucleus only well beyond the midblastula transition (16). These findings suggest that endogenous YY1 is sequestered in the cytoplasm in a cell cycle-regulated and developmental stage-specific manner. In both situations, the signals that control the shuttling of YY1 are undefined. However, the regulation of nuclear transport can be inhibited by overexpression of the protein. Exogenous YY1 is localized in the cell nucleus due to a nuclear localization sequence within the Zn finger domain (4, 37). Our observation that in response to several different apoptotic stimuli, including the activation of death receptors, protein kinase C inhibitors, and DNA-damaging agents, YY1 is translocated to the nucleus suggests that these treatments interfere with the mechanism that retains YY1 in the cytoplasm in G1, late S, and G2. Since z-VADfmk is unable to inhibit translocation yet efficiently blocks caspase-dependent cleavage, we speculate that the translocation of YY1 functions as an early stress sensor, a hypothesis that needs further investigation. In support of this notion, YY1 is poly(ADP-ribos)ylated by PARP-1, a nuclear enzyme associated with the stress response (23, 42, 54).
In addition to nuclear translocation and cleavage, we also observed a reduction in YY1-DNA complex formation in vitro (Fig. 4). However, caspase-dependent cleavage did not directly affect the DNA binding domain (Fig. 1 to 4). Previous studies showed that the Zn finger domain is sufficient for specific interactions with DNA (3, 4). Reduced in vitro DNA binding correlated with decreased solubility of YY1Δ12 and, to a lesser extent, of YY1 and YY1Δ119 in the presence of apoptotic stimuli (Fig. 4). Thus, YY1 not only is translocated to but also becomes associated with the nuclear compartment in a manner that results in its resistance to mild detergent extraction. Since YY1 has been identified as a component of the nuclear matrix (6, 8, 21, 37), it will be of interest to determine whether apoptotic signals affect the distribution of YY1 in distinct subnuclear compartments.
YY1 functions as a transcriptional regulator by recruiting various cofactors. Caspase-dependent cleavage removes the transactivation domain but leaves intact the two repressor domains that are associated with the Gly-Ala-rich and Zn finger regions (55, 67). Indeed, YY1Δ119 lacks transactivation activity in vivo (Fig. 6). The expression of this truncated protein but not of wild-type YY1 or the caspase-resistant mutant YY1D12A/D119A increased the sensitivity of cells to Fas-induced apoptosis (Fig. 7). These data suggest that YY1 regulates the expression of genes associated with apoptosis. Due to the intact repressor domains and the loss of the transactivation domain, differential expression of pro- and antiapoptotic genes would be expected and could result in the observed differential sensitivity to apoptotic stimuli.
Although procaspases are ubiquitously expressed and their activation allows rapid execution of apoptotic signals in a cascade of events that are largely independent of de novo transcription and translation, several studies revealed transcriptional control of the cell death machinery (30, 49). Among the transcription factors that have been implicated in the control of apoptosis are NF-κB (25, 29), E2F factors (38, 39), the proto-oncoprotein c-Myc (47), and FOXO (10, 12, 58, 62). Our findings provide evidence that YY1 is an additional transcriptional regulator involved in the control of apoptosis. The ability of YY1Δ119 to enhance apoptosis is most likely independent of p53 binding, since the p53 binding region in YY1 is maintained (20, 63). These data suggest a YY1- or YY1Δ119-dependent mechanism, distinct from the one controlled by the interaction with p53, that will now be interesting to reveal.
Acknowledgments
We thank P. Hurlin, D. Litchfield, and J. Tschopp for reagents and Beth Alexander, Sabine Schreek, Jürgen Stahl, Sylvia Willi, and Gudrun Zimmermann for expert technical assistance.
This work was supported in part by a grant from the Deutsche Forschungsgemeinschaft (LU 466/1) to B.L.
REFERENCES
- 1.Atchison, L., A. Ghias, F. Wilkinson, N. Bonini, and M. L. Atchison. 2003. Transcription factor YY1 functions as a PcG protein in vivo. EMBO J. 22:1347-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Austen, M., C. Cerni, M. Henriksson, S. Hilfenhaus, J. M. Luscher-Firzlaff, A. Menkel, C. Seelos, A. Sommer, and B. Luscher. 1997. Regulation of cell growth by the Myc-Max-Mad network: role of Mad proteins and YY1. Curr. Top. Microbiol. Immunol. 224:123-130. [DOI] [PubMed] [Google Scholar]
- 3.Austen, M., C. Cerni, J. M. Luscher-Firzlaff, and B. Luscher. 1998. YY1 can inhibit c-Myc function through a mechanism requiring DNA binding of YY1 but neither its transactivation domain nor direct interaction with c-Myc. Oncogene 17:511-520. [DOI] [PubMed] [Google Scholar]
- 4.Austen, M., B. Luscher, and J. M. Luscher-Firzlaff. 1997. Characterization of the transcriptional regulator YY1. The bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAFII55, or cAMP-responsive element-binding protein (CPB)-binding protein. J. Biol. Chem. 272:1709-1717. [DOI] [PubMed] [Google Scholar]
- 5.Bauknecht, T., R. H. See, and Y. Shi. 1996. A novel C/EBP beta-YY1 complex controls the cell-type-specific activity of the human papillomavirus type 18 upstream regulatory region. J. Virol. 70:7695-7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bidwell, J. P., K. Torrungruang, M. Alvarez, S. J. Rhodes, R. Shah, D. R. Jones, K. Charoonpatrapong, J. M. Hock, and A. J. Watt. 2001. Involvement of the nuclear matrix in the control of skeletal genes: the NMP1 (YY1), NMP2 (Cbfa1), and NMP4 (Nmp4/CIZ) transcription factors. Crit. Rev. Eukaryot. Gene Expr. 11:279-297. [PubMed] [Google Scholar]
- 7.Brown, J. L., D. Mucci, M. Whiteley, M. L. Dirksen, and J. A. Kassis. 1998. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol. Cell 1:1057-1064. [DOI] [PubMed] [Google Scholar]
- 8.Bushmeyer, S. M., and M. L. Atchison. 1998. Identification of YY1 sequences necessary for association with the nuclear matrix and for transcriptional repression functions. J. Cell. Biochem. 68:484-499. [PubMed] [Google Scholar]
- 9.Cartwright, P., and K. Helin. 2000. Nucleocytoplasmic shuttling of transcription factors. Cell. Mol. Life Sci. 57:1193-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charvet, C., I. Alberti, F. Luciano, A. Jacquel, A. Bernard, P. Auberger, and M. Deckert. 2003. Proteolytic regulation of Forkhead transcription factor FOXO3a by caspase-3-like proteases. Oncogene 22:4557-4568. [DOI] [PubMed] [Google Scholar]
- 11.Chiang, C. M., and R. G. Roeder. 1995. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science 267:531-536. [DOI] [PubMed] [Google Scholar]
- 12.Dijkers, P. F., R. H. Medema, J. W. Lammers, L. Koenderman, and P. J. Coffer. 2000. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr. Biol. 10:1201-1204. [DOI] [PubMed] [Google Scholar]
- 13.Donohoe, M. E., X. Zhang, L. McGinnis, J. Biggers, E. Li, and Y. Shi. 1999. Targeted disruption of mouse Yin Yang 1 transcription factor results in periimplantation lethality. Mol. Cell. Biol. 19:7237-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Earnshaw, W. C., L. M. Martins, and S. H. Kaufmann. 1999. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu. Rev. Biochem. 68:383-424. [DOI] [PubMed] [Google Scholar]
- 15.Eliassen, K. A., A. Baldwin, E. M. Sikorski, and M. M. Hurt. 1998. Role for a YY1-binding element in replication-dependent mouse histone gene expression. Mol. Cell. Biol. 18:7106-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ficzycz, A., C. Eskiw, D. Meyer, K. E. Marley, M. Hurt, and N. Ovsenek. 2001. Expression, activity, and subcellular localization of the Yin Yang 1 transcription factor in Xenopus oocytes and embryos. J. Biol. Chem. 276:22819-22825. [DOI] [PubMed] [Google Scholar]
- 17.Fischer, U., R. U. Janicke, and K. Schulze-Osthoff. 2003. Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ. 10:76-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia, E., C. Marcos-Gutierrez, M. del Mar Lorente, J. C. Moreno, and M. Vidal. 1999. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J. 18:3404-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griesenbeck, J., M. Ziegler, N. Tomilin, M. Schweiger, and S. L. Oei. 1999. Stimulation of the catalytic activity of poly(ADP-ribosyl) transferase by transcription factor Yin Yang 1. FEBS Lett. 443:20-24. [DOI] [PubMed] [Google Scholar]
- 20.Gronroos, E., A. A. Terentiev, T. Punga, and J. Ericsson. 2004. YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc. Natl. Acad. Sci. USA 101:12165-12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo, B., P. R. Odgren, A. J. van Wijnen, T. J. Last, J. Nickerson, S. Penman, J. B. Lian, J. L. Stein, and G. S. Stein. 1995. The nuclear matrix protein NMP-1 is the transcription factor YY1. Proc. Natl. Acad. Sci. USA 92:10526-10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hyde-DeRuyscher, R. P., E. Jennings, and T. Shenk. 1995. DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res. 23:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeggo, P. A. 1998. DNA repair: PARP—another guardian angel? Curr. Biol. 8:R49-R51. [DOI] [PubMed] [Google Scholar]
- 24.Kaludov, N. K., T. L. Bowman, E. M. Sikorski, and M. M. Hurt. 1996. Cell cycle-regulated binding of nuclear proteins to elements within a mouse H3.2 histone gene. Proc. Natl. Acad. Sci. USA 93:4465-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karin, M., and A. Lin. 2002. NF-kappaB at the crossroads of life and death. Nat. Immunol. 3:221-227. [DOI] [PubMed] [Google Scholar]
- 26.Kaufmann, S. H., S. Desnoyers, Y. Ottaviano, N. E. Davidson, and G. G. Poirier. 1993. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res. 53:3976-3985. [PubMed] [Google Scholar]
- 27.Kovacsovics, M., F. Martinon, O. Micheau, J. L. Bodmer, K. Hofmann, and J. Tschopp. 2002. Overexpression of Helicard, a CARD-containing helicase cleaved during apoptosis, accelerates DNA degradation. Curr. Biol. 12:838-843. [DOI] [PubMed] [Google Scholar]
- 28.Krippner-Heidenreich, A., R. V. Talanian, R. Sekul, R. Kraft, H. Thole, H. Ottleben, and B. Luscher. 2001. Targeting of the transcription factor Max during apoptosis: phosphorylation-regulated cleavage by caspase-5 at an unusual glutamic acid residue in position P1. Biochem. J. 358:705-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kucharczak, J., M. J. Simmons, Y. Fan, and C. Gelinas. 2003. To be, or not to be: NF-kappaB is the answer—role of Rel/NF-kappaB in the regulation of apoptosis. Oncogene 22:8961-8982. [DOI] [PubMed] [Google Scholar]
- 30.Kumar, S., and D. Cakouros. 2004. Transcriptional control of the core cell-death machinery. Trends Biochem. Sci. 29:193-199. [DOI] [PubMed] [Google Scholar]
- 31.Lazebnik, Y. A., S. H. Kaufmann, S. Desnoyers, G. G. Poirier, and W. C. Earnshaw. 1994. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature 371:346-347. [DOI] [PubMed] [Google Scholar]
- 32.Lee, J. S., K. M. Galvin, and Y. Shi. 1993. Evidence for physical interaction between the zinc-finger transcription factors YY1 and Sp1. Proc. Natl. Acad. Sci. USA 90:6145-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, J. S., R. H. See, K. M. Galvin, J. Wang, and Y. Shi. 1995. Functional interactions between YY1 and adenovirus E1A. Nucleic Acids Res. 23:925-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, L. Y., X. Luo, and X. Wang. 2001. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature 412:95-99. [DOI] [PubMed] [Google Scholar]
- 35.Litchfield, D. W., B. Luscher, F. J. Lozeman, R. N. Eisenman, and E. G. Krebs. 1992. Phosphorylation of casein kinase II by p34cdc2 in vitro and at mitosis. J. Biol. Chem. 267:13943-13951. [PubMed] [Google Scholar]
- 36.Maldonado, E., R. Shiekhattar, M. Sheldon, H. Cho, R. Drapkin, P. Rickert, E. Lees, C. W. Anderson, S. Linn, and D. Reinberg. 1996. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature 381:86-89. [DOI] [PubMed] [Google Scholar]
- 37.McNeil, S., B. Guo, J. L. Stein, J. B. Lian, S. Bushmeyer, E. Seto, M. L. Atchison, S. Penman, A. J. van Wijnen, and G. S. Stein. 1998. Targeting of the YY1 transcription factor to the nucleolus and the nuclear matrix in situ: the C-terminus is a principal determinant for nuclear trafficking. J. Cell. Biochem. 68:500-510. [PubMed] [Google Scholar]
- 38.Moroni, M. C., E. S. Hickman, E. L. Denchi, G. Caprara, E. Colli, F. Cecconi, H. Muller, and K. Helin. 2001. Apaf-1 is a transcriptional target for E2F and p53. Nat. Cell Biol. 3:552-558. [DOI] [PubMed] [Google Scholar]
- 39.Nahle, Z., J. Polakoff, R. V. Davuluri, M. E. McCurrach, M. D. Jacobson, M. Narita, M. Q. Zhang, Y. Lazebnik, D. Bar-Sagi, and S. W. Lowe. 2002. Direct coupling of the cell cycle and cell death machinery by E2F. Nat. Cell Biol. 4:859-864. [DOI] [PubMed] [Google Scholar]
- 40.Nicholson, D. W. 1999. Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ. 6:1028-1042. [DOI] [PubMed] [Google Scholar]
- 41.Oei, S. L., and Y. Shi. 2001. Poly(ADP-ribosyl)ation of transcription factor Yin Yang 1 under conditions of DNA damage. Biochem. Biophys. Res. Commun. 285:27-31. [DOI] [PubMed] [Google Scholar]
- 42.Oei, S. L., and Y. Shi. 2001. Transcription factor Yin Yang 1 stimulates poly(ADP-ribosyl)ation and DNA repair. Biochem. Biophys. Res. Commun. 284:450-454. [DOI] [PubMed] [Google Scholar]
- 43.Orth, K., A. M. Chinnaiyan, M. Garg, C. J. Froelich, and V. M. Dixit. 1996. The CED-3/ICE-like protease Mch2 is activated during apoptosis and cleaves the death substrate lamin A. J. Biol. Chem. 271:16443-16446. [PubMed] [Google Scholar]
- 44.Otte, A. P., and T. H. Kwaks. 2003. Gene repression by Polycomb group protein complexes: a distinct complex for every occasion? Curr. Opin. Genet. Dev. 13:448-454. [DOI] [PubMed] [Google Scholar]
- 45.Palko, L., H. W. Bass, M. J. Beyrouthy, and M. M. Hurt. 2004. The Yin Yang-1 (YY1) protein undergoes a DNA-replication-associated switch in localization from the cytoplasm to the nucleus at the onset of S phase. J. Cell Sci. 117:465-476. [DOI] [PubMed] [Google Scholar]
- 46.Park, K., and M. L. Atchison. 1991. Isolation of a candidate repressor/activator, NF-E1 (YY-1, delta), that binds to the immunoglobulin kappa 3′ enhancer and the immunoglobulin heavy-chain mu E1 site. Proc. Natl. Acad. Sci. USA 88:9804-9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pelengaris, S., M. Khan, and G. Evan. 2002. c-MYC: more than just a matter of life and death. Nat. Rev. Cancer 2:764-776. [DOI] [PubMed] [Google Scholar]
- 48.Petkova, V., M. J. Romanowski, I. Sulijoadikusumo, D. Rohne, P. Kang, T. Shenk, and A. Usheva. 2001. Interaction between YY1 and the retinoblastoma protein. Regulation of cell cycle progression in differentiated cells. J. Biol. Chem. 276:7932-7936. [DOI] [PubMed] [Google Scholar]
- 49.Puthalakath, H., and A. Strasser. 2002. Keeping killers on a tight leash: transcriptional and post-translational control of the pro-apoptotic activity of BH3-only proteins. Cell Death Differ. 9:505-512. [DOI] [PubMed] [Google Scholar]
- 50.Rezai-Zadeh, N., X. Zhang, F. Namour, G. Fejer, Y. D. Wen, Y. L. Yao, I. Gyory, K. Wright, and E. Seto. 2003. Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes Dev. 17:1019-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakahira, H., M. Enari, and S. Nagata. 1998. Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature 391:96-99. [DOI] [PubMed] [Google Scholar]
- 52.Satijn, D. P., K. M. Hamer, J. den Blaauwen, and A. P. Otte. 2001. The polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol. Cell. Biol. 21:1360-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seto, E., B. Lewis, and T. Shenk. 1993. Interaction between transcription factors Sp1 and YY1. Nature 365:462-464. [DOI] [PubMed] [Google Scholar]
- 54.Shall, S., and G. de Murcia. 2000. Poly(ADP-ribose) polymerase-1: what have we learned from the deficient mouse model? Mutat. Res. 460:1-15. [DOI] [PubMed] [Google Scholar]
- 55.Shi, Y., J. S. Lee, and K. M. Galvin. 1997. Everything you have ever wanted to know about Yin Yang 1. Biochim. Biophys. Acta 1332:F49-F66. [DOI] [PubMed] [Google Scholar]
- 56.Shi, Y., E. Seto, L. S. Chang, and T. Shenk. 1991. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell 67:377-388. [DOI] [PubMed] [Google Scholar]
- 57.Shrivastava, A., S. Saleque, G. V. Kalpana, S. Artandi, S. P. Goff, and K. Calame. 1993. Inhibition of transcriptional regulator Yin-Yang-1 by association with c-Myc. Science 262:1889-1892. [DOI] [PubMed] [Google Scholar]
- 58.Skurk, C., H. Maatz, H. S. Kim, J. Yang, M. R. Abid, W. C. Aird, and K. Walsh. 2004. The Akt-regulated forkhead transcription factor FOXO3a controls endothelial cell viability through modulation of the caspase-8 inhibitor FLIP. J. Biol. Chem. 279:1513-1525. [DOI] [PubMed] [Google Scholar]
- 59.Smith, J. M., and P. A. Koopman. 2004. The ins and outs of transcriptional control: nucleocytoplasmic shuttling in development and disease. Trends Genet. 20:4-8. [DOI] [PubMed] [Google Scholar]
- 60.Sommer, A., K. Bousset, E. Kremmer, M. Austen, and B. Luscher. 1998. Identification and characterization of specific DNA-binding complexes containing members of the Myc/Max/Mad network of transcriptional regulators. J. Biol. Chem. 273:6632-6642. [DOI] [PubMed] [Google Scholar]
- 61.Sommer, A., A. Waha, J. Tonn, N. Sorensen, P. J. Hurlin, R. N. Eisenman, B. Luscher, and T. Pietsch. 1999. Analysis of the Max-binding protein MNT in human medulloblastomas. Int. J. Cancer 82:810-816. [DOI] [PubMed] [Google Scholar]
- 62.Stahl, M., P. F. Dijkers, G. J. Kops, S. M. Lens, P. J. Coffer, B. M. Burgering, and R. H. Medema. 2002. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J. Immunol. 168:5024-5031. [DOI] [PubMed] [Google Scholar]
- 63.Sui, G., B. Affar el, Y. Shi, C. Brignone, N. R. Wall, P. Yin, M. Donohoe, M. P. Luke, D. Calvo, S. R. Grossman, and Y. Shi. 2004. Yin Yang 1 is a negative regulator of p53. Cell 117:859-872. [DOI] [PubMed] [Google Scholar]
- 64.Takahashi, A., E. S. Alnemri, Y. A. Lazebnik, T. Fernandes-Alnemri, G. Litwack, R. D. Moir, R. D. Goldman, G. G. Poirier, S. H. Kaufmann, and W. C. Earnshaw. 1996. Cleavage of lamin A by Mch2 alpha but not CPP32: multiple interleukin 1 beta-converting enzyme-related proteases with distinct substrate recognition properties are active in apoptosis. Proc. Natl. Acad. Sci. USA 93:8395-8400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Talanian, R. V., C. Quinlan, S. Trautz, M. C. Hackett, J. A. Mankovich, D. Banach, T. Ghayur, K. D. Brady, and W. W. Wong. 1997. Substrate specificities of caspase family proteases. J. Biol. Chem. 272:9677-9682. [DOI] [PubMed] [Google Scholar]
- 66.Thole, H. H., I. Maschler, and P. W. Jungblut. 1995. Surface mapping of the ligand-filled C-terminal half of the porcine estradiol receptor by restricted proteolysis. Eur. J. Biochem. 231:510-516. [DOI] [PubMed] [Google Scholar]
- 67.Thomas, M. J., and E. Seto. 1999. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene 236:197-208. [DOI] [PubMed] [Google Scholar]
- 68.Thornberry, N. A., T. A. Rano, E. P. Peterson, D. M. Rasper, T. Timkey, M. Garcia-Calvo, V. M. Houtzager, P. A. Nordstrom, S. Roy, J. P. Vaillancourt, K. T. Chapman, and D. W. Nicholson. 1997. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem. 272:17907-17911. [DOI] [PubMed] [Google Scholar]
- 69.Usheva, A., and T. Shenk. 1994. TATA-binding protein-independent initiation: YY1, TFIIB, and RNA polymerase II direct basal transcription on supercoiled template DNA. Cell 76:1115-1121. [DOI] [PubMed] [Google Scholar]
- 70.van Loo, G., X. Saelens, F. Matthijssens, P. Schotte, R. Beyaert, W. Declercq, and P. Vandenabeele. 2002. Caspases are not localized in mitochondria during life or death. Cell Death Differ. 9:1207-1211. [DOI] [PubMed] [Google Scholar]
- 71.Weis, K. 2003. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 112:441-451. [DOI] [PubMed] [Google Scholar]
- 72.Weiss, T., M. Grell, K. Siemienski, F. Muhlenbeck, H. Durkop, K. Pfizenmaier, P. Scheurich, and H. Wajant. 1998. TNFR80-dependent enhancement of TNFR60-induced cell death is mediated by TNFR-associated factor 2 and is specific for TNFR60. J. Immunol. 161:3136-3142. [PubMed] [Google Scholar]
- 73.Yakovleva, T., L. Kolesnikova, V. Vukojevic, I. Gileva, K. Tan-No, M. Austen, B. Luscher, T. J. Ekstrom, L. Terenius, and G. Bakalkin. 2004. YY1 binding to a subset of p53 DNA-target sites regulates p53-dependent transcription. Biochem. Biophys. Res. Commun. 318:615-624. [DOI] [PubMed] [Google Scholar]
- 74.Yang, W. M., C. Inouye, Y. Zeng, D. Bearss, and E. Seto. 1996. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl. Acad. Sci. USA 93:12845-12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhivotovsky, B., A. Samali, A. Gahm, and S. Orrenius. 1999. Caspases: their intracellular localization and translocation during apoptosis. Cell Death Differ. 6:644-651. [DOI] [PubMed] [Google Scholar]