Abstract
Myocyte enhancer factor 2 (MEF2) family proteins are key transcription factors controlling gene expression in myocytes, lymphocytes, and neurons. MEF2 proteins are known to be regulated by phosphorylation. We now provide evidence showing that MEF2C is acetylated by p300 both in vitro and in vivo. In C2C12 myogenic cells, MEF2 is preferentially acetylated in differentiating myocytes but not in undifferentiated myoblasts. Several major acetylation sites are mapped to the transactivation domain of MEF2C, some of which are fully conserved in other MEF2 members from several different species. Mutation of these lysines affects MEF2 DNA binding and transcriptional activity, as well as its synergistic effect with myogenin in myogenic conversion assays. When introduced into C2C12 myoblasts, the nonacetylatable MEF2C inhibits myogenic differentiation. Thus, in addition to phosphorylation, MEF2 activity is also critically regulated by acetylation during myogenesis.
Myocyte enhancer factor 2 (MEF2) family proteins consist of four members encoded by different genes: MEF2A, MEF2B, MEF2C, and MEF2D (1). MEF2 proteins belong to the MADS box superfamily (named after the first four members identified to contain this motif: MCM1, Agamous, Deficiens, and serum response factor) due to the presence of a highly conserved 59-amino-acid (59-aa) MADS box at their amino termini (Fig. 1A). In addition, all MEF2 proteins contain a conserved 26-aa region carboxyl to the MADS box, termed the MEF2 domain, which is unique to MEF2 members (Fig. 1A). Apart from the conserved MADS box and MEF2 domain, MEF2 proteins differ significantly in their central regions and carboxyl termini, which harbor transactivation domains (Fig. 1A) (1). MEF2 preferentially binds to the MEF2 site, an AT-rich consensus sequence [(C/T)TA(A/T)4TA(A/G)], which is found in the promoters and enhancers of many genes (1).
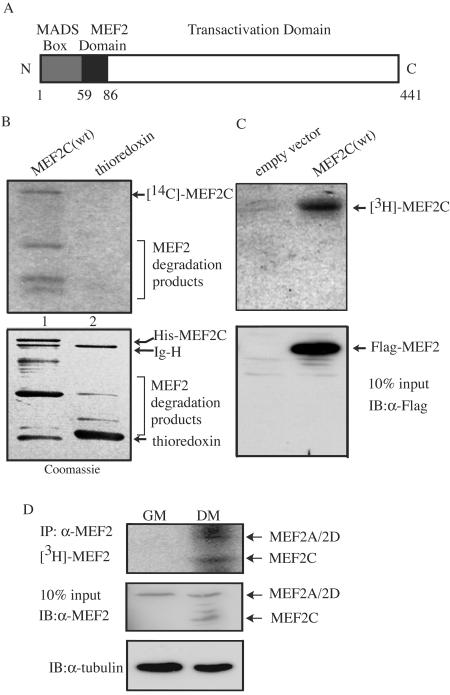
FIG. 1.
MEF2 is acetylated in vitro and in vivo. (A) Schematic representation of human MEF2C protein used in this study. The numbers give the positions of amino acids marking the boundaries of domains. (B) Purified His-thioredoxin-MEF2C (lane 1) and His-thioredoxin (lane 2) were subjected to in vitro acetylation assays with the immunoprecipitated Flag-p300. The dried gel was first subjected to fluorography (top panel), followed by rehydration and Coomassie blue staining (bottom panel). Ig-H, immunoglobulin heavy chain. (C) 293T cells were transfected with the plasmid encoding Flag-MEF2C, followed by in vivo sodium [3H]acetate labeling. The immunoprecipitated Flag-MEF2 was resolved by SDS-PAGE, and the gel was subjected to fluorography (top panel). The expression level of Flag-MEF2C was detected by immunoblotting (IB) (bottom panel). (D) Proliferating (grown in GM) and differentiating (grown in DM) C2C12 cells were labeled in vivo with sodium [3H]acetate. The endogenous MEF2s were separately immunoprecipitated (IP) from GM and DM cell lysates (twice as much GM lysate was used, because the MEF2 level is lower in GM) and resolved by SDS-PAGE. The gel was subjected to fluorography (top panel). The expression levels of MEF2s (center panel) and β-tubulin (bottom panel) present in 10% of cell lysates used in the above immunoprecipitation were revealed by immunoblotting.
MEF2 plays important roles in various biological processes. During myogenesis, many skeletal-muscle-specific genes (e.g., muscle creatine kinase, myogenin, and desmin) contain indispensable MEF2 sites in their promoters or enhancers (1). In the mouse myogenin promoter, a key proximal MEF2 site controls the expression level and tissue expression pattern of myogenin (5, 7, 38). MEF2 is also capable of directly interacting with members of the myogenic regulatory factors (MRFs) (i.e., MyoD, Myf5, myogenin, and MRF4) to synergistically activate many muscle-specific genes (22). In addition, MEF2 also plays a decisive role in cardiogenesis. It controls the expression of dHand, which is indispensable in heart development (17). Knockout of the MEF2C gene in mice is embryonically lethal due to a failure in heart development (17), whereas loss of MEF2A in mice leads to deficiency in cardiac mitochondria and sudden death due to cardiac abnormality (25). Loss of a single D-mef2 gene in Drosophila melanogaster is also embryonically lethal, due to aborted differentiation of muscle cells in somites and heart (16). Mutations in MEF2A also lead to a form of severe coronary artery disease in humans (35). In lymphocyte development, expression of the Nur77 gene, an immediate-early gene involved in apoptosis of autoreactive T cells in response to T-cell-receptor signaling, is critically regulated by MEF2 (39). In neuronal cells, MEF2 is implicated in neuronal-activity-dependent cell survival (19). Recently, MEF2 has also been found to play a role in epithelial cell survival and maintenance of blood vessel integrity (12).
In cells, MEF2 activity is regulated by diverse signaling pathways. The p38 mitogen-activated protein kinase- and BMK1/ERK5-mediated signaling pathways directly phosphorylate MEF2A and MEF2C in their transactivation domains and enhance their transcriptional activity (11, 13, 36, 41). In addition, the calcium/calmodulin-dependent protein kinase (CaMK) and phosphatase (calcineurin) also activate the transcriptional activity of MEF2. Whereas it remains unclear how calcineurin activates MEF2, the mechanism by which CaMK activates MEF2 has recently been elucidated. MEF2 interacts directly with the class II histone deacetylases (HDACs) via the MADS/MEF2 domain (20, 21). These class II HDACs directly bind and repress MEF2 in the nuclei of undifferentiated myoblasts. Upon myogenic differentiation in response to activating signals mediated by CaMK, HDACs dissociate from MEF2, translocate to the cytoplasm, and are retained by 14-3-3 proteins. Interestingly, MEF2 is also known to interact directly with p300, a histone acetyltransferase (HAT) that antagonizes the action of HDACs (30).
Acetylation and deacetylation have been recognized as one of the most important opposing mechanisms in controlling gene expression in eukaryotes (14). While the main target of acetylation and deacetylation is histones, more recent evidence demonstrates that many cellular proteins, especially transcription factors, are also subjected to regulation by acetylation and deacetylation. For example, p53 is acetylated at multiple sites by both p300 and PCAF, which enhances its DNA binding ability, transcriptional activity, and stability (4). In myogenic cells, MyoD is mainly acetylated by PCAF at the basic domain, and acetylation promotes MyoD to bind DNA (31).
At present, it remains unclear whether MEF2 is acetylated or not in cells. Here, we provide evidence that MEF2 is acetylated during myogenic differentiation. p300 acetylated MEF2 in vitro and in vivo. Six acetylated lysines were mapped to the transactivation domain of MEF2C. Abrogation of acetylation in MEF2 reduced its DNA binding activity and transcriptional activity and inhibited myogenic differentiation.
MATERIALS AND METHODS
Cell lines, DNA constructs, and antibodies.
10T1/2, Cos-7, and 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin in a 37°C incubator with 5% CO2. C2C12 cells were grown in DMEM with 20% fetal bovine serum and antibiotics (also called growth medium, or GM) and were induced to differentiate in DMEM containing 2% horse serum and antibiotics (also called differentiation medium, or DM) when cells were nearly confluent.
3×MEF2-luc, Flag-p300, and E1A have been described previously (27, 37). The form of MEF2C used in this work is a spliced variant of human MEF2C(1-473) without the coding sequence for aa 368 to 399. Single, double, triple, quadruple, and sixfold lysine-to-arginine (K-to-R) mutants were generated by PCR-based site-directed mutagenesis. Wild-type MEF2C(1-302) and its derivatives in pcDNA3 have the Flag epitope at their amino termini, while full-length MEF2C and its derivatives in pcDNA3.1 have the Flag tag at the carboxyl termini. The wild-type and mutant forms of MEF2C in bacterial expression vectors were constructed by inserting the PCR-amplified MEF2C cDNA fragments into the BamHI and NotI sites of pET32M (a gift from M. J. Zhang), resulting in fusion of His6-thioredoxin with MEF2C or its derivatives. All constructed clones were verified by sequencing.
Antibodies employed in this work include the mouse monoclonal antibodies to β-actin (Sigma), Flag (M2; Sigma), β-tubulin (Sigma), myosin heavy chain (MHC) (MF20; Developmental Studies Hybridoma Bank), PCAF (E-8; Santa Cruz), MEF2 (B-4; Santa Cruz), and myogenin (F5D; Santa Cruz) and the rabbit polyclonal antibodies to MEF2 (C-21; Santa Cruz) and p300 (N-15; Santa Cruz).
siRNA and plasmid transfection.
All small interfering RNAs (siRNAs) were purchased from Dharmacon Inc. (Lafayette, Colo.). Sequences were as follows: for the p300 siRNA (human, sense), 5′AACAGAGCAGUCCUGGAUUAG; for the PCAF siRNA (human, sense), 5′AAUCGCCGUGAAGAAAGCGCA. 293T cells were transfected with siRNAs twice, with a 24-h interval, using LipofectAMINE 2000 (Invitrogen), followed by a third transfection with MEF2C constructs. All plasmids were transfected into cells using LipofectAMINE PLUS reagent (Invitrogen).
In vivo acetylation assays.
Cells were labeled in the appropriate culture medium containing 0.3 mCi/ml sodium [3H]acetate (4.70 Ci/mmol; Perkin-Elmer) for 1.5 h. After removal of culture medium followed by washing with phosphate-buffered saline, cells were lysed in ice-cold radioimmunoprecipitation assay (RIPA) buffer (25 mM HEPES, pH 7.4, 1% [vol/vol] NP-40, 0.1% sodium dodecyl sulfate [SDS], 0.5% sodium deoxycholate) supplemented with phosphatase and protease inhibitors (20 mM p-nitrophenyl phosphate, 20 mM β-glycerophosphate, 50 μM sodium vanadate, 2 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 2 μg of aprotinin/ml, 0.5 μg of leupeptin/ml, 0.7 μg of pepstatin/ml). Transfected or endogenous MEF2 was immunoprecipitated from cell lysates, washed, and resolved by SDS-polyacrylamide gel electrophoresis (PAGE). The gel was soaked in Amplify solution (Amersham), dried, and subjected to fluorography at −80°C.
In vitro acetylation assays.
Flag-p300 (or Flag-PCAF) was first immunoprecipitated from 400 μg of 293T cell lysates, and the in vitro acetylation assays were reconstituted in 30 μl of reaction mixture consisting of 3 μg of His-MEF2 and 0.06 μCi of [14C]acetyl coenzyme A (60 mCi/mmol; Perkin-Elmer) in HAT buffer (50 mM Tris-HCl [pH 8.0], 0.1 mM EDTA, 10% [vol/vol] glycerol, 1 mM dithiothreitol, and 10 mM sodium butyrate). Reaction mixtures were incubated at 30°C for 1 h and stopped by the addition of SDS loading buffer. Labeled proteins were resolved by SDS-PAGE. The gel was soaked in Amplify solution, dried, and subjected to fluorography at −80°C. To visualize the amount of recombinant proteins used in each reaction, the dried gel was rehydrated and stained with Coomassie blue.
Luciferase reporter assays.
C2C12 cells were cotransfected with either 3×MEF2-luc or 5×Gal4-luc together with various plasmids. After growth in GM for 24 h, cells were shifted to DM for an additional 12 h before harvest. Cells were lysed in ice-cold lysis buffer (50 mM HEPES at pH 7.6, 1% [vol/vol] Triton X-100, 150 mM NaCl, 1 mM EGTA, 1.5 mM MgCl2, 100 mM NaF) supplemented with protease inhibitors (see above) and subjected to luciferase assays (34). Luciferase units were normalized against the total amount of protein present in each sample, determined by a protein assay reagent from Bio-Rad (Hercules, Calif.).
Coimmunoprecipitation.
C2C12 cells were cross-linked with 200 μg/ml dithiobis(succinimidylpropionate) (DSP; Pierce) for 8 min, followed by lysis in RIPA buffer. Four micrograms of anti-MEF2 antibody was first incubated with 300 μg of cell extracts in RIPA buffer overnight at 4°C with rotation; 30 μl of protein A-Sepharose beads was then added, and the mixture was incubated at 4°C for another hour with rotation. After extensive washing with the RIPA buffer, bound proteins were eluted out by boiling and subjected to SDS-PAGE and immunoblotting.
Electrophoretic mobility shift assays (EMSA).
Preparation of the 32P-labeled MEF2 oligonucleotide probe has been described previously (6). Increasing amounts of Cos-7 whole-cell extracts containing either Flag-MEF2C(wt) or Flag-MEF2C(6KR) were premixed with 20 μl of binding buffer containing 1 μg of poly(dI-dC), 20 μg of bovine serum albumin, 15 mM HEPES (pH 7.6), 40 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, and 5% glycerol for 30 min at 4°C. After addition of 20,000 cpm of probe, the binding reactions were carried out at room temperature for another 20 min. The samples were then loaded onto a 5% nondenaturing polyacrylamide gel, and electrophoresis was carried out at 200 V for 2 h at 4°C. The gel was subsequently dried and subjected to autoradiography.
Myogenic conversion assays.
10T1/2 cells were cotransfected with Flag-myogenin and either an empty vector (pcDNA3.1), Flag-MEF2C(wt), or Flag-MEF2C(6KR). After growth in GM for 24 h, cells were switched to DM for 5 days. Cells were then fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton in 1× phosphate-buffered saline, and subjected to MHC immunostaining. Microscopy was performed using an Olympus IX70 microscope linked to a charge-coupled device digital camera (Spot RT; Diagnostic Instruments Inc., Sterling Heights, Mich.), and the images were captured using Spot RT software, version 3.4.
Multiple sequence alignment.
Amino acid sequences of human MEF2A (NP_005578), MEF2C (NP_002388), and MEF2D (NP_005911); mouse MEF2A (NP_038625), MEF2C (AAH57650), and MEF2D (Q63943); Xenopus MEF2A (Q03414) and MEF2D (Q03413); and zebra fish MEF2A (NP_571376), MEF2C (NP_571387), and MEF2D (NP_571392) were aligned with Clustal W1.81.
RESULTS
MEF2C is acetylated both in vitro and in vivo.
Histone acetyltransferases such as p300 are known to acetylate proteins other than histones. As MEF2C was shown to interact with p300 in an in vitro glutathione S-transferase (GST)-pulldown assay (30), it is likely that MEF2 is acetylated by p300. To test this hypothesis, a purified recombinant His-thioredoxin-MEF2C fusion protein was subjected to in vitro acetylation assays using immunoprecipitated p300. As shown in Fig. 1B, MEF2C (lane 1) but not the His-thioredoxin tag (lane 2) was acetylated by p300. In addition, the PCAF immunoprecipitated from cells could also acetylate MEF2C in vitro, albeit less efficiently than p300 (unpublished data). To test whether MEF2C was acetylated in cells, we first transfected 293T cells with Flag-MEF2C and then labeled cells with sodium [3H]acetate. As shown in Fig. 1C, Flag-MEF2C was indeed acetylated in cells when overexpressed (see Discussion).
As MEF2 plays a key role in myogenesis, we next checked whether MEF2 is acetylated in myogenic cells. Both proliferating (grown in GM) and differentiating (grown in DM) myogenic C2C12 cells were subjected to in vivo labeling with sodium [3H]acetate. As shown in Fig. 1D, although MEF2A/2D were already present in GM (middle panel) (2), little acetylation signal was detected (top panel). In contrast, MEF2A/2D and MEF2C (induced only in DM) were significantly acetylated in differentiating C2C12 cells (top panel).
MEF2 interacts with and is mainly acetylated by p300 in vivo.
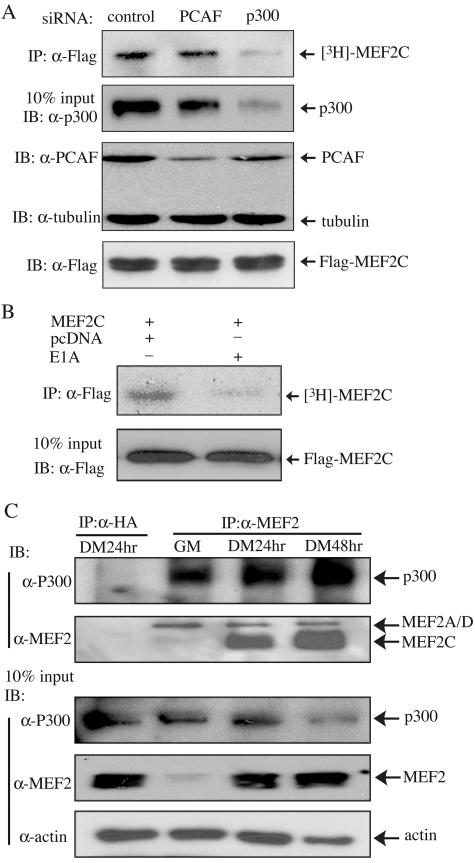
Since both p300 and PCAF acetylated MEF2C in vitro, we next sought to find out which actually acetylates MEF2C in cells. siRNA against either p300 or PCAF was transfected into 293T cells together with Flag-MEF2C, followed by in vivo labeling with sodium [3H]acetate. As shown in Fig. 2A, each siRNA specifically and effectively knocked down the expression of its target (i.e., p300 or PCAF). Importantly, only the p300-specific siRNA, but not the PCAF-specific siRNA, significantly reduced MEF2 acetylation in cells (Fig. 2A, top panel). Because adenovirus E1A protein specifically binds and inactivates p300 (8, 24), we also carried out an in vivo labeling experiment with or without E1A. Consistent with our siRNA data, we found that MEF2 acetylation was significantly reduced in the presence of E1A (Fig. 2B). Next, we wanted to find out whether and when the endogenous p300 interacts with MEF2 during myogenic differentiation. C2C12 cell lysates were subjected to coimmunoprecipitation analysis. Interestingly, we found that the endogenous p300 specifically associated with MEF2 in both proliferating myoblasts and differentiating myotubes (Fig. 2C), consistent with a recent report (21a).
FIG. 2.
p300 is mainly responsible for acetylating MEF2 in vivo. (A and B) 293T cells were either transfected twice with various siRNAs as indicated and once with Flag-MEF2C (A) or cotransfected with Flag-MEF2C with or without E1A (B). Cells were labeled with sodium [3H]acetate for 1.5 h before harvest. The immunoprecipitated (IP) Flag-MEF2C was resolved by SDS-PAGE, and the gels were subjected to fluorography (A and B, top panels). The levels of the endogenous p300 and PCAF were revealed by immunoblotting (IB) (A, two center panels). The level of Flag-MEF2C in 10% of the lysates used in the above IP was detected by IB (A and B, bottom panels). (C) DSP cross-linked C2C12 cell lysates were subjected to IP with antibodies against either MEF2 or hemagglutinin (HA), followed by IB with antibodies against either p300 or MEF2 (top two panels). Ten percent of the total lysates used in the above IP assays were subjected to IB with various antibodies as indicated (bottom three panels). hr, hours.
Mapping of the acetylation sites in MEF2C in vitro and in vivo.
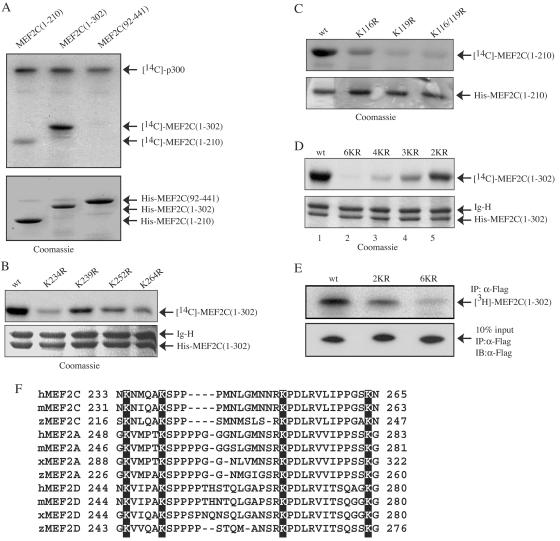
To identify lysine residues in MEF2C that are acetylated by p300, we first generated different MEF2 truncation mutants and subjected them to in vitro acetylation assays with p300. As shown in Fig. 3A, whereas MEF2C(92-441) was not acetylated, MEF2C(1-210) was weakly acetylated and MEF2C(1-302) was strongly acetylated. The failure of p300 to acetylate MEF2C(92-441) was expected, because the MADS/MEF2 domain, which is missing in MEF2C(92-441), is required for MEF2 binding by p300 (30). In addition, we found that MEF2C(1-114) was not significantly acetylated (unpublished data). This suggests that the main acetylation sites lie between aa 211 and 302 and the minor sites lie between aa 115 and 210. In human MEF2C, there are two lysines between aa 115 and 210 (i.e., K116 and 119) and four lysines between aa 211 and 302 (i.e., K234, K239, K252, and K264). We generated both single and multiple lysine mutants and subjected them to in vitro acetylation assays with p300. Whereas the single mutation of any of the three lysines (K234, K252, and K264) between aa 211 and 302 significantly decreased MEF2 acetylation by p300, the single mutation of K239 also partially reduced MEF2 acetylation (Fig. 3B). Similarly, mutation of either of the two lysines between aa 115 and 210 (K116 and K119) also partially reduced MEF2 acetylation, with that of K119 displaying a more pronounced effect (Fig. 3C). Simultaneous mutation of both lysines resulted in a near-complete loss of acetylation in MEF2C(1-210) (Fig. 3C). For the compound lysine mutants in the context of MEF2C(1-302), we found that the acetylation of the quadruple lysine mutant (4KR, with K234/239/252/264 mutated) was greatly reduced compared to that of the double lysine mutant (2KR, with K116 and K119 mutated) (Fig. 3D). This was consistent with our previous notion that K234, K239, K252, and K264 are major acetylation sites while K116 and K119 are minor acetylation sites. The fact that the acetylation of the quadruple mutant (K234/239/252/264) was further reduced relative to that of the triple mutant (K234/252/264) confirmed that K239 was also acetylated (Fig. 3D, lanes 3 and 4). Most importantly, the mutant with all six lysines between aa 115 and 302 mutated (6KR) almost completely lost the acetylation signal (Fig. 3D, lane 2). Our data revealed that K116, K119, K234, K239, K252, and K264 of MEF2C are acetylated by p300 in vitro.
FIG. 3.
Mapping of the p300-acetylated lysines in MEF2C in vitro and in vivo. (A to D) Similar amounts of the purified His-MEF2C and its mutants were subjected to in vitro acetylation assays using immunoprecipitated p300 and were resolved by SDS-PAGE. The gel was first subjected to fluorography (top panels). The dried gel was then rehydrated and stained with Coomassie blue (bottom panels). wt, wild type; 2KR, K116/119R; 3KR, K234/252/264R; 4KR, K234/239/252/264R; 6KR, K116/119/234/239/252/264R. (E) 293T cells were separately transfected with wild-type Flag-MEF2C and mutant Flag-MEF2C(1-302). Cells were labeled with sodium [3H]acetate for 1.5 h before harvest. The immunoprecipitated (IP) Flag-MEF2C was separated by SDS-PAGE, and the gel was subjected to fluorography (top panel). The expression levels of Flag-MEF2C and its mutants were revealed by IP followed by immunoblotting (IB) (bottom panel). (F) Different MEF2 isoforms from several vertebrates were aligned to show the four conserved lysines (equivalent to K234, K239, K252, and K264 in human MEF2C) using Clustal W1.81. h, human; m, mouse; x, Xenopus; z, zebra fish.
To find out whether the six acetylatable lysines we mapped in vitro are actually acetylated in vivo, we transfected 293T cells with wild-type MEF2C(1-302), the double lysine mutant (2KR), or the mutant with all six lysines mutated (6KR, with K116/119/234/239/252/264 mutated) and labeled cells with sodium [3H]acetate. As shown in Fig. 3E, whereas mutation of K116/119 partially decreased MEF2 acetylation, mutation of all six lysines between aa 115 and 302 almost completely abolished MEF2 acetylation in vivo.
Interestingly, multiple sequence alignment revealed that the four major acetylated lysines in human MEF2C (K234, K239, K252, and K264) are also fully conserved in MEF2A, MEF2C, and MEF2D of fish, frog, mouse, and human origin (Fig. 3F).
Acetylation has a minor impact on the transactivation function of MEF2C.
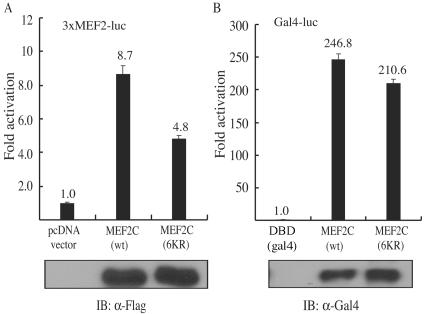
Because all six acetylated lysines fall into the transactivation domain of MEF2C, we first tested whether acetylation affects the transactivation function of MEF2C. Expression vectors encoding Flag-MEF2C(wt) or Flag-MEF2C(6KR) were separately transfected into C2C12 cells together with a MEF2-dependent reporter, 3×MEF2-luc. We found that the acetylation-defective MEF2C mutant [MEF2C(6KR)] was about 45% less active than its wild-type counterpart (Fig. 4A). Because a defect in either the DNA binding or the transactivation function of MEF2C could account for the decreased activity of Flag-MEF2C(6KR) in the reporter assays discussed above, in order to find out whether acetylation has a direct impact on the transactivation function of MEF2C, we fused full-length MEF2C with the DNA binding domain of yeast Gal4 protein. We evaluated the transcriptional activity of the fusion protein using a luciferase reporter under the control of Gal4 binding sites. In this assay, the Gal4 DNA binding domain, but not the intrinsic MEF2 DNA binding domain, was required. As shown in Fig. 4B, Gal4MEF2C(6KR) was about 15% less active than Gal4MEF2C(wt), suggesting that acetylation has a minor impact on the transactivation function of MEF2C.
FIG. 4.
MEF2 acetylation has a minor effect on its transactivation function. C2C12 cells were cotransfected with (A) 3×MEF2-luc together with either an empty vector (control), Flag-MEF2C(wt), or Flag-MEF2C(6KR) or (B) 5×Gal4-luc together with constructs expressing either Gal4DBD, Gal4MEF2C(wt), or Gal4MEF2C(6KR). After 24 h of growth in GM, cells were switched to DM for another 12 h before harvest. Whole-cell lysates were subjected to luciferase assays. The fold activation was calculated as the ratio of the normalized luciferase activity in MEF2-transfected samples to that in the control. Reporter assays were performed in duplicate three times, with similar results. Representative results showing means ± standard deviations are presented. The expression levels of the transfected Flag-MEF2C or Gal4MEF2C in the above samples were detected by immunoblotting (IB).
Acetylation of MEF2 affects its DNA binding activity.
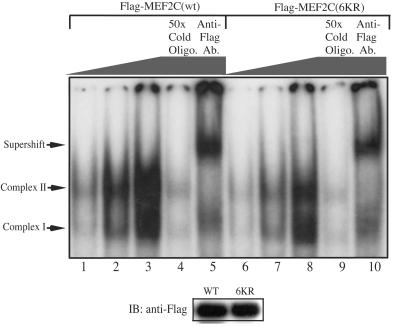
The fact that the reduction in the transcriptional activity of Gal4MEF2C(6KR) was much less obvious than that of Flag-MEF2C(6KR) compared to their respective wild-type counterparts suggested that acetylation mainly affects MEF2 DNA binding. To confirm that, increasing amounts of Cos-7 cell lysates containing either Flag-MEF2C(wt) or Flag-MEF2C(6KR) were mixed with an excess of radiolabeled oligonucleotide probe containing a consensus MEF2 site, and the mixtures were analyzed by EMSA. Two MEF2-specific complexes were detected: one containing mainly the endogenous MEF2 (complex I) and one containing mainly the exogenously transfected Flag-MEF2C (complex II). The specificity of these two MEF2-containing complexes was verified both by competition with a 50-fold excess of cold probe (Fig. 5, lanes 4 and 9) and by supershifting of the complexes (mainly complex II) using the anti-Flag antibody (lanes 5 and 10). Importantly, we found that Flag-MEF2C(6KR) had significantly lower DNA binding activity than its acetylated wild-type counterpart (Fig. 5).
FIG. 5.
The acetylation-defective MEF2 mutant has reduced DNA binding activity. Portions (1, 4, and 16 μg) of Cos-7 whole-cell extracts containing either Flag-MEF2C(wt) or Flag-MEF2C(6KR) (both expressed at similar levels, as revealed by the immunoblot below the EMSA gel) were mixed with an excess of radiolabeled MEF2 probe in an appropriate binding buffer and analyzed by EMSA. Ab., antibody; Oligo., oligonucleotide; WT, wild-type Flag-MEF2C; 6KR, Flag-MEF2C(6KR).
Acetylation of MEF2 affects its interaction with MRF.
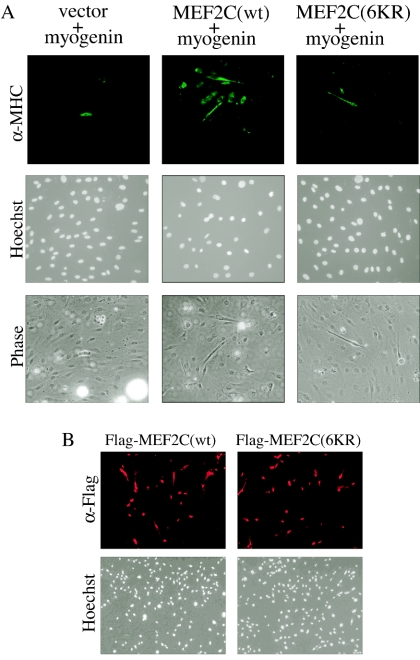
Since MEF2 is known to interact with MRF (22), we next asked whether acetylation of MEF2 affects its interaction with MRF in a myogenic conversion assay. It has been well established that any one of the four MRFs is capable of converting several nonmuscle cells into muscle lineage in cell cultures (23, 40). Coexpression of an exogenous MEF2 with MRF is known to further enhance MRF-mediated myogenic conversion (22). When 10T1/2 fibroblasts were cotransfected with a myogenin expression vector together with either an empty vector, Flag-MEF2C(wt), or Flag-MEF2C(6KR), we found that MEF2C(6KR) did not show any obvious synergistic effect (Fig. 6A, righthand panels). In contrast, MEF2C(wt) significantly enhanced myogenin-mediated myogenic conversion (Fig. 6A, middle panels). As a control, we showed that both MEF2C(wt) and MEF2C(6KR) were transfected into 10T1/2 cells at similar efficiencies (Fig. 6B).
FIG. 6.
The acetylation-defective MEF2 mutant fails to synergize with myogenin in myogenic conversion assays. (A) 10T1/2 cells were cotransfected with Flag-myogenin and either an empty vector, Flag-MEF2C(wt), or Flag-MEF2C(6KR). After 36 h of growth in GM, cells were allowed to differentiate in DM for 5 days before fixation for immunostaining (top panels) using the anti-MHC antibody (MF20) and nuclear staining with Hoechst (middle panels). Phase, phase-contrast images (bottom panels). (B) 10T1/2 cells were transfected with vectors encoding either Flag-MEF2C(wt) or Flag-MEF2C(6KR). After 24 h of growth in GM, cells were fixed and subjected to immunostaining using the anti-Flag antibody (α-Flag). Cell nuclei were stained by Hoechst.
The acetylation-defective MEF2 mutant inhibits myogenic differentiation.
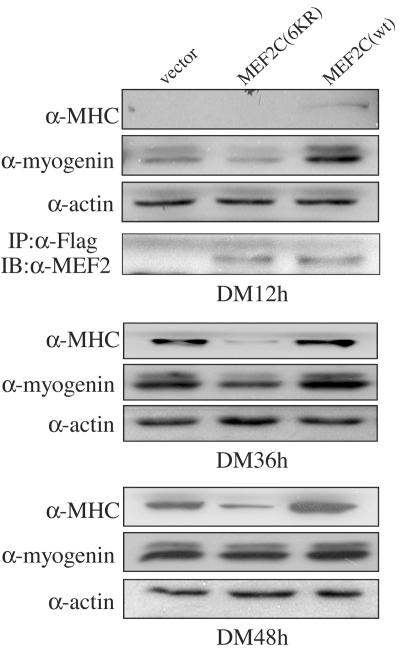
Because MEF2 is a key transcription factor controlling the expression of many muscle-specific genes, we next checked whether abrogation of MEF2 acetylation affects myogenic differentiation. C2C12 cells were separately transfected with the wild-type and the acetylation-defective (6KR) Flag-MEF2C and were induced to differentiate. Cell lysates were subjected to immunoblotting with antibodies against both myogenin and MHC, the early and late muscle differentiation markers, respectively. As shown in Fig. 7, in the early differentiation phase (DM 12 h), myogenin expression was decreased in cells transfected with MEF2C(6KR) but enhanced in cells transfected with wild-type MEF2C relative to that in the control (i.e., cells transfected with an empty expression vector only). Not much MHC was expressed at this early stage. At the late differentiation stage (DM 36 h and 48 h), MHC expression was significantly reduced in cells containing the mutant MEF2C(6KR), but enhanced in cells containing wild-type MEF2C, compared to that in the control.
FIG. 7.
Overexpression of the nonacetylatable MEF2 mutant inhibits myogenic differentiation. C2C12 cells were separately transfected with an empty vector, Flag-MEF2C(6KR), and Flag-MEF2C(wt). After 24 h of growth in GM, cells were switched to DM for either 12 h, 36 h, or 48 h before harvest. Portions (30 μg) of whole-cell extracts were subjected to immunoblotting (IB) for myogenin, MHC, and β-actin. Because the Flag-MEF2C/MEF2C(6KR) levels gradually decreased in cells grown in DM, to check the expression levels of MEF2, 100 μg of whole-cell extracts from the 12-h DM incubation was first subjected to immunoprecipitation (IP) with the anti-Flag antibody (α-Flag), followed by IB with α-MEF2.
DISCUSSION
Acetylation critically controls myogenic differentiation.
As key transcription factors involved in myogenic differentiation, both MEF2s and MRFs are known to be critically regulated by posttranslational modifications. For example, phosphorylation can either promote or inhibit myogenic differentiation, depending on distinct extracellular signals. In response to stimulation with fibroblast growth factor, protein kinase C can inhibit the activity of myogenin by phosphorylating a key threonine residue in the basic region, resulting in its decreased binding to the E box (15). At the onset of differentiation, the p38 mitogen-activated protein kinase can directly phosphorylate MEF2 at a conserved threonine in the transactivation domain, resulting in increased transcriptional activity (11, 36). Cyclin-dependent kinase has also been shown to directly phosphorylate MyoD at serine 200 and promote its fast turnover (32). In addition, ubiquitination is also known to regulate the stability or turnover of MyoD (3). Recently, acetylation and deacetylation have emerged as important posttranslational modifications modulating the activity of key myogenic factors (20, 21). MyoD was shown to be acetylated during myogenic differentiation, and acetylated MyoD displays enhanced DNA binding activity (18, 26, 31). While one group showed that p300 acetylates MyoD (26), others showed that PCAF is the specific HAT responsible for MyoD acetylation (18, 28, 31). Although p300 has been shown to enhance the transcriptional activity of MEF2 and to bind to MEF2 in an in vitro GST pulldown assay (30), the underlying mechanism was not clear. Here, we present evidence that MEF2 is acetylated by p300 upon myogenic differentiation. Abrogation of MEF2 acetylation results in decreased MEF2 DNA binding and reduced transcriptional activity. p300 is known to affect myogenesis by controlling the expression of the Myf5 and MyoD genes (29). The MEF2 acetylation we identified here represents an additional mechanism by which p300 promotes myogenesis. Although PCAF also acetylates MEF2 in vitro (unpublished data), down-regulation of PCAF by siRNA has little effect on MEF2 acetylation in vivo, indicating that PCAF is less likely the predominant HAT acetylating MEF2 in cells. It remains unclear at present which HDAC deacetylates MEF2. Considering that class II HDACs specifically bind and potently repress MEF2, it is likely that one or more of these class II HDACs antagonize the effect of p300 by deacetylating MEF2.
It is interesting that both p300 and class II HDACs associate with endogenous MEF2 before myogenic differentiation (Fig. 2C). Such a scenario is not unprecedented, as Sir2, PCAF, and MyoD also form a complex in undifferentiated myoblasts (9). In the case of MyoD, the reduced expression of Sir2 and the decreased [NAD+]/[NADH] ratio at the onset of differentiation help to promote MyoD acetylation by PCAF (9). In the case of MEF2, the cytoplasmic translocation of class II HDACs in response to CaMK signaling at the beginning of differentiation should facilitate subsequent MEF2 acetylation by p300 (20, 21).
We also noticed that the overexpressed MEF2 in both proliferating myoblasts and several nonmuscle cells (e.g., 293T and Cos-7 cells) was already acetylated (Fig. 1C and Fig. 2A and B). Based on our previous studies and our present work (6), we believe that MEF2 normally complexes with both p300 and HDACs and that the numbers of MEF2s and class II HDACs in cells are well balanced, such that the endogenous MEF2s are normally bound and repressed by HDACs and are thus deacetylated. Overexpression by transfection causes MEF2 to outnumber the endogenous HDACs, resulting in enhanced MEF2 acetylation by the bound p300.
The functional role of acetylation for MEF2.
Even though the p300 acetylation sites on MEF2 are not within the N-terminal MADS/MEF2 DNA binding domain, we show that acetylation at these sites can affect MEF2 DNA binding (Fig. 5). This is reminiscent of p53 acetylation by p300, as acetylation of p53 on multiple sites at the C-terminal regulatory domain affects the function of its DNA binding domain in the middle of the protein, presumably through an altered conformation (4, 10). In addition, we also show that MEF2 acetylation affects the transactivation function of MEF2 to some extent. The exact underlying mechanism remains unclear at present. It is conceivable that acetylation can alter the conformation of MEF2 such that MEF2 efficiently recruits a transcriptional coactivator or interacts with the basal transcriptional machinery. Alternatively, acetylation at the transactivation domain of MEF2 may weaken or dissociate the binding of a transcriptional repressor (e.g., Twist) to MEF2 (33). Further investigation is needed to address this issue.
It is noteworthy that four major acetylatable lysines (K234/239/252/264) in human MEF2C are also fully conserved in MEF2A, MEF2C, and MEF2D across several different species (Fig. 3). This suggests that MEF2 acetylation by p300 at these sites is a general mechanism conserved in vertebrates. Although we have studied the acetylation of MEF2C here in the context of myogenesis, it is conceivable that the other MEF2 isoforms are similarly regulated by acetylation and that such an event may also take place and play a key role in other biological processes, including T-cell development, cardiogenesis, and neurogenesis.
Acknowledgments
We thank P. L. Puri (Burnham Institute, Calif.) and X. J. Yang (McGill University) for reagents, R. Derynck (UCSF) for helpful suggestions, L. Yu in our group for initial findings, and L. Sun and C. Wong for technical assistance.
This work was supported by grants from the Hong Kong Research Grant Council (HKUST6139/03M to G.Z. and HKUST6143/03 M to Z.W.), EHIA funding for cell signaling studies from HKUST, and the Areas of Excellence scheme established under the University Grants Committee of the Hong Kong Special Administrative Region, China (project AoE/B-15/01).
REFERENCES
- 1.Black, B. L., and E. N. Olson. 1998. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 14:167-196. [DOI] [PubMed] [Google Scholar]
- 2.Breitbart, R. E., C. S. Liang, L. B. Smoot, D. A. Laheru, V. Mahdavi, and B. Nadal-Ginard. 1993. A fourth human MEF2 transcription factor, hMEF2D, is an early marker of the myogenic lineage. Development 118:1095-1106. [DOI] [PubMed] [Google Scholar]
- 3.Breitschopf, K., E. Bengal, T. Ziv, A. Admon, and A. Ciechanover. 1998. A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 17:5964-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks, C. L., and W. Gu. 2003. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr. Opin. Cell Biol. 15:164-171. [DOI] [PubMed] [Google Scholar]
- 5.Buchberger, A., K. Ragge, and H. H. Arnold. 1994. The myogenin gene is activated during myocyte differentiation by pre-existing, not newly synthesized transcription factor MEF-2. J. Biol. Chem. 269:17289-17296. [PubMed] [Google Scholar]
- 6.Chan, J. K., L. Sun, X. J. Yang, G. Zhu, and Z. Wu. 2003. Functional characterization of an amino-terminal region of HDAC4 that possesses MEF2 binding and transcriptional repressive activity. J. Biol. Chem. 278:23515-23521. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, T. C., M. C. Wallace, J. P. Merlie, and E. N. Olson. 1993. Separable regulatory elements governing myogenin transcription in mouse embryogenesis. Science 261:215-218. [DOI] [PubMed] [Google Scholar]
- 8.Eckner, R., M. E. Ewen, D. Newsome, M. Gerdes, J. A. DeCaprio, J. B. Lawrence, and D. M. Livingston. 1994. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 8:869-884. [DOI] [PubMed] [Google Scholar]
- 9.Fulco, M., R. L. Schiltz, S. Iezzi, M. T. King, P. Zhao, Y. Kashiwaya, E. Hoffman, R. L. Veech, and V. Sartorelli. 2003. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol. Cell 12:51-62. [DOI] [PubMed] [Google Scholar]
- 10.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595-606. [DOI] [PubMed] [Google Scholar]
- 11.Han, J., Y. Jiang, Z. Li, V. V. Kravchenko, and R. J. Ulevitch. 1997. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature 386:296-299. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi, M., S. W. Kim, K. Imanaka-Yoshida, T. Yoshida, E. D. Abel, B. Eliceiri, Y. Yang, R. J. Ulevitch, and J. D. Lee. 2004. Targeted deletion of BMK1/ERK5 in adult mice perturbs vascular integrity and leads to endothelial failure. J. Clin. Investig. 113:1138-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato, Y., V. V. Kravchenko, R. I. Tapping, J. Han, R. J. Ulevitch, and J. D. Lee. 1997. BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. EMBO J. 16:7054-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuo, M. H., and C. D. Allis. 1998. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20:615-626. [DOI] [PubMed] [Google Scholar]
- 15.Li, L., J. Zhou, G. James, R. Heller-Harrison, M. P. Czech, and E. N. Olson. 1992. FGF inactivates myogenic helix-loop-helix proteins through phosphorylation of a conserved protein kinase C site in their DNA-binding domains. Cell 71:1181-1194. [DOI] [PubMed] [Google Scholar]
- 16.Lilly, B., B. Zhao, G. Ranganayakulu, B. M. Paterson, R. A. Schulz, and E. N. Olson. 1995. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science 267:688-693. [DOI] [PubMed] [Google Scholar]
- 17.Lin, Q., J. Schwarz, C. Bucana, and E. N. Olson. 1997. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science 276:1404-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mal, A., M. Sturniolo, R. L. Schiltz, M. K. Ghosh, and M. L. Harter. 2001. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J. 20:1739-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao, Z., A. Bonni, F. Xia, M. Nadal-Vicens, and M. E. Greenberg. 1999. Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science 286:785-790. [DOI] [PubMed] [Google Scholar]
- 20.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2001. Control of muscle development by dueling HATs and HDACs. Curr. Opin. Genet. Dev. 11:497-504. [DOI] [PubMed] [Google Scholar]
- 21.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2002. Signaling chromatin to make muscle. Curr. Opin. Cell Biol. 14:763-772. [DOI] [PubMed] [Google Scholar]
- 21a.Molinari, S., F. Relaix, M. Lemonnier, B. Kirschbaum, B. Schafer, and M. Buckingham. 2004. A novel complex regulates cardiac actin gene expression through interaction of Emb, a class VI POU domain protein, MEF2D, and the histone transacetylase p300. Mol. Cell. Biol. 24:2944-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molkentin, J. D., B. L. Black, J. F. Martin, and E. N. Olson. 1995. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83:1125-1136. [DOI] [PubMed] [Google Scholar]
- 23.Molkentin, J. D., and E. N. Olson. 1996. Defining the regulatory networks for muscle development. Curr. Opin. Genet. Dev. 6:445-453. [DOI] [PubMed] [Google Scholar]
- 24.Mymryk, J. S., R. W. Lee, and S. T. Bayley. 1992. Ability of adenovirus 5 E1A proteins to suppress differentiation of BC3H1 myoblasts correlates with their binding to a 300 kDa cellular protein. Mol. Biol. Cell 3:1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naya, F. J., B. L. Black, H. Wu, R. Bassel-Duby, J. A. Richardson, J. A. Hill, and E. N. Olson. 2002. Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat. Med. 8:1303-1309. [DOI] [PubMed] [Google Scholar]
- 26.Polesskaya, A., and A. Harel-Bellan. 2001. Acetylation of MyoD by p300 requires more than its histone acetyltransferase domain. J. Biol. Chem. 276:44502-44503. [DOI] [PubMed] [Google Scholar]
- 27.Puri, P. L., M. L. Avantaggiati, C. Balsano, N. Sang, A. Graessmann, A. Giordano, and M. Levrero. 1997. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 16:369-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puri, P. L., V. Sartorelli, X. J. Yang, Y. Hamamori, V. V. Ogryzko, B. H. Howard, L. Kedes, J. Y. Wang, A. Graessmann, Y. Nakatani, and M. Levrero. 1997. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell 1:35-45. [DOI] [PubMed] [Google Scholar]
- 29.Roth, J. F., N. Shikama, C. Henzen, I. Desbaillets, W. Lutz, S. Marino, J. Wittwer, H. Schorle, M. Gassmann, and R. Eckner. 2003. Differential role of p300 and CBP acetyltransferase during myogenesis: p300 acts upstream of MyoD and Myf5. EMBO J. 22:5186-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sartorelli, V., J. Huang, Y. Hamamori, and L. Kedes. 1997. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol. Cell. Biol. 17:1010-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sartorelli, V., P. L. Puri, Y. Hamamori, V. Ogryzko, G. Chung, Y. Nakatani, J. Y. Wang, and L. Kedes. 1999. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol. Cell 4:725-734. [DOI] [PubMed] [Google Scholar]
- 32.Song, A., Q. Wang, M. G. Goebl, and M. A. Harrington. 1998. Phosphorylation of nuclear MyoD is required for its rapid degradation. Mol. Cell. Biol. 18:4994-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spicer, D. B., J. Rhee, W. L. Cheung, and A. B. Lassar. 1996. Inhibition of myogenic bHLH and MEF2 transcription factors by the bHLH protein Twist. Science 272:1476-1480. [DOI] [PubMed] [Google Scholar]
- 34.Sun, L., L. Liu, X. J. Yang, and Z. Wu. 2004. Akt binds prohibitin 2 and relieves its repression of MyoD and muscle differentiation. J. Cell Sci. 117:3021-3029. [DOI] [PubMed] [Google Scholar]
- 35.Wang, L., C. Fan, S. E. Topol, E. J. Topol, and Q. Wang. 2003. Mutation of MEF2A in an inherited disorder with features of coronary artery disease. Science 302:1578-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu, Z., P. J. Woodring, K. S. Bhakta, K. Tamura, F. Wen, J. R. Feramisco, M. Karin, J. Y. Wang, and P. L. Puri. 2000. p38 and extracellular signal-regulated kinases regulate the myogenic program at multiple steps. Mol. Cell. Biol. 20:3951-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu, Q., and Z. Wu. 2000. The insulin-like growth factor-phosphatidylinositol 3-kinase-Akt signaling pathway regulates myogenin expression in normal myogenic cells but not in rhabdomyosarcoma-derived RD cells. J. Biol. Chem. 275:36750-36757. [DOI] [PubMed] [Google Scholar]
- 38.Yee, S. P., and P. W. Rigby. 1993. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev. 7:1277-1289. [DOI] [PubMed] [Google Scholar]
- 39.Youn, H. D., L. Sun, R. Prywes, and J. O. Liu. 1999. Apoptosis of T cells mediated by Ca2+-induced release of the transcription factor MEF2. Science 286:790-793. [DOI] [PubMed] [Google Scholar]
- 40.Yun, K., and B. Wold. 1996. Skeletal muscle determination and differentiation: story of a core regulatory network and its context. Curr. Opin. Cell Biol. 8:877-889. [DOI] [PubMed] [Google Scholar]
- 41.Zhao, M., L. New, V. V. Kravchenko, Y. Kato, H. Gram, F. di Padova, E. N. Olson, R. J. Ulevitch, and J. Han. 1999. Regulation of the MEF2 family of transcription factors by p38. Mol. Cell. Biol. 19:21-30. [DOI] [PMC free article] [PubMed] [Google Scholar]