Abstract
Bioinformatics analysis of transcriptional control is guided by knowledge of the characteristics of cis-regulatory regions or enhancers. Features such as clustering of binding sites and co-occurrence of binding sites have aided enhancer identification, but quantitative predictions of enhancer function are not yet generally feasible. To facilitate the analysis of regulatory sequences in Drosophila melanogaster, we identified quantitative parameters that affect the activity of short-range transcriptional repressors, proteins that play key roles in development. In addition to the previously noted distance dependence, repression is strongly influenced by the stoichiometry, affinity, spacing, and arrangement of activator binding sites. Repression is insensitive to the type of activation domain, suggesting that short-range repression may primarily affect activators at the level of DNA binding. The activity of several short-range, but not long-range, repressors is circumscribed by the same quantitative parameters. This cis-regulatory “grammar” may aid the identification of enhancers regulated by short-range repressors and facilitate bioinformatic prediction of the functional output of transcriptional regulatory sequences.
Enhancers are cis-regulatory elements that regulate gene expression in a distance- and orientation-independent manner. Originally characterized solely as gene-activating modules, enhancers also bind transcriptional repressors to mediate activation or repression in context-dependent manners (5, 8). Combinatorial interactions of a limited suite of activators and repressors on enhancers yield a large number of distinct transcriptional outputs, providing temporal and spatial specificity (1, 15, 22). Repression is mediated by proteins that can operate in a dedicated or facultative manner to block transcription via mechanisms that include direct interaction with the basal transcriptional machinery or with chromatin-modifying machinery (2, 30). Understanding the interplay of transcriptional activators and repressors lies at the heart of deciphering gene switches. With the advent of genomic sequencing, this analysis is not limited to individual genes but encompasses entire networks of genes and regulatory elements, presenting a major challenge and opportunity.
The early Drosophila melanogaster embryo is a paradigm for developmentally regulated transcriptional control networks. Typical of higher eukaryote systems, the complex cis-regulatory elements of key patterning genes interpret maternal and early embryonic inputs to produce precisely defined transcriptional outputs (62, 70). One of the best-studied complex loci in Drosophila is the even-skipped (eve) gene, which features a series of cis-regulatory elements within a 16-kb region. Five enhancers are responsible for early expression of eve in seven regularly spaced stripes in the blastoderm embryo (29, 65, 72, 73). A key feature of the stripe enhancers is their functional autonomy; the repression of one element does not lead to the general repression of the entire locus (4, 32, 71). This autonomy is based on the properties of short-range transcriptional repressors such as Giant, Krüppel, and Knirps. These proteins block the activity of enhancers when bound within ∼100 bp of key activator sites or basal promoter elements (4, 31, 32). The magnitude of short-range repression can be modulated by the precise positioning of repressor binding sites and by utilization of C-terminal binding protein (CtBP) cofactor-dependent and -independent activities, providing additional levels of control (34, 38, 75, 79).
In contrast to the fine-tuning offered by short-range repressors, long-range repressors such as the Drosophila Hairy protein block multiple enhancers indiscriminately over distances of several kilobases (7). The molecular mechanisms of short-range and long-range repression are still poorly understood, although the short-range-long-range distinction may result from the recruitment of distinct cofactors. Short-range repressors bind the CtBP corepressor, whereas long-range repressors such as Hairy, and in some contexts Dorsal, interact with the Groucho corepressor (16). Short-range repressors, through CtBP, may mediate localized chromatin modifications, while long-range repressors, via Groucho, may generate extended transcriptionally silent chromatin structures (17, 28, 69, 74).
Traditionally, empirical tests such as analysis of transgenic reporter genes have been used to identify and analyze regulatory elements. Because of the complexity of many regulatory regions, many gene constructs must be tested to provide insights into how an expression pattern is generated. As a result, relatively few higher eukaryotic enhancers have been well characterized, and our understanding of general principles governing cis-regulatory element design remains limited. With the availability of whole genome sequences, bioinformatics methods have the promise of providing a powerful alternative route to the identification and analysis of cis-regulatory modules on a global scale. Currently used approaches include identification of clusters of putative transcription factor binding sites (11, 53, 60, 61, 66). A high local density of transcription factor binding sites has been used as a convenient signpost for computational identification of known and novel cis elements; however, not all clusters are functional enhancers. The potential for a cluster of binding sites to function as an enhancer also depends on the levels of transacting factors, which can often be inferred from gene expression or proteome data. Not only the presence of a binding site, but also the sequence context within which it is found, is also critical. This context can be considered a type of “grammar” of transcriptional code. This grammar is clearly more complex than simply the density of binding sites. Due to cooperative or antagonistic interactions between proteins and synergistic interactions with the transcriptional machinery, the activity of a given binding site can vary. Additional parameters that influence binding site activity include affinities, spacing, and positioning of transcriptional activator and repressor binding sites within cis-regulatory modules (9, 21, 25, 33, 34, 36). Previous analyses of short-range repressors on native enhancers demonstrated that these proteins can block gene expression when bound within ∼100 bp of the presumed target, either a basal promoter or activator site. In most cases, however, relevant quantitative values were not determined because short-range repression has been studied mostly in the context of complex, endogenous regulatory elements where the identity, number, relative affinities, order, and spacing of binding sites are often not known. Because of this complexity of cis-regulatory elements, the contributions of individual physical parameters to repression have been difficult to ascertain from previous empirical tests.
Evolutionary conservation of binding sites has also been used to identify enhancers (10, 27, 45, 46, 68). This approach is more likely to work with enhancers that possess rather rigid constraints on factor binding sites due to the high degree of protein-protein cooperativity, as seen with so-called “enhanceosomes” (39, 54, 55, 80). Many other enhancers that possess a more plastic structure (“billboard” or information display enhancers) are less likely to be identified by this approach, however (26, 42, 43, 47-49, 59). Even within more flexibly designed enhancers, the spacing or arrangement of activator and repressor binding sites can still be important (42, 43). In particular, spacing between short-range repressor and activator sites within the cis-regulatory element is critical for dictating repression effectiveness (4, 32, 34, 47, 79). However, there is no general understanding of how alterations in binding sites for short-range repressors or adjacent activators might affect transcription; thus, it is difficult to predict whether sequence changes introduced during evolution would affect enhancer function. Computational searches and phylogenetic comparisons that seek to identify cis-regulatory elements would be greatly facilitated by empirical determination of spatial constraints and other features of transcription factor binding sites within cis-regulatory elements.
The Drosophila segmentation network has provided an important test bed for the development of bioinformatics tools to analyze enhancers. Short-range repressors play a central role in this system; therefore, it is of particular interest to identify and quantify aspects of the cis-regulatory grammar that dictates their action. In this study, we analyze highly defined enhancer elements in which the identity, stoichiometry, and exact arrangement of activator and repressor binding sites are well defined. We show that the notion that short-range repressors block the activity of protein complexes within 100 bp is an oversimplification. By targeted alteration of these defined elements, we define contextual parameters that dictate repression effectiveness, including stoichiometry of activators and repressors, relative affinity, spacing, and position of transcription factor binding sites. We further demonstrate that the cis-regulatory logic appears to be specific to different functional classes of transcriptional regulators, indicating that identification of such class-specific rules will be critical for more detailed bioinformatics analysis.
MATERIALS AND METHODS
Gal4 activator chimeric constructs. (i) Gal4 (aa 1 to 93)-Gal4 activation domain (AD) (aa 753 to 881).
A KpnI-XbaI fragment from pSCTEV Gal4 (amino acids [aa] 1 to 93)-Gal4 (67) with the Gal4 activation domain (residues 753 to 881) was cloned into KpnI-XbaI-cut pTwiggy (3), which has the twist enhancer (2xPEe-Et), the twist basal promoter, and the Gal4 DNA binding domain (residues 1 to 93). To generate other Gal4 activator genes, the following fragments were cloned into this vector. For Gal4-VP16, the activation domain of herpes simplex virus VP16 from residues 412 to 490 was amplified from pRevTet off (64) using DA410 (5′-GGG TCG GTA CCG CAA CGG CCC CCC CGA CCG ATG TC-3′) and DA411 (5′-GGG GAA TCT AGA CTA ACT AAT TAC TAC CCA CCG TAC TCG TCA AT-3′). For Gal4-Sp1, a KpnI-XbaI fragment from pSCTEV Gal4 (residues 1 to 93)-Sp1:Q1 (67) encoding residues 132 to 243 of the human transcription factor Sp1 was isolated. For Gal4-human TATA binding protein (hTBP) (aa 1 to 339), the following oligonucleotides were used to amplify full-length hTBP (aa 1 to 339): DA162 (5′-GGG TCG GTA CCG CAG CCG CAA TGG ATC AGA ACA ACA GCC TG-3′) and DA164 (5′-GGG GAA TCT AGA CTA ACT AAT TAC TAC GTC GTC TTC CTG AAT CCC TT-3′).
(ii) Insulated Gal4-Gal4 AD (aa 753 to 881).
A 420-bp fragment of DNA with the gypsy insulator with 12 Su(Hw) sites was amplified from Green Pelican green fluorescent protein vector (6) using DA639 (5′-CGG AAT TCC GAA TTG TAA GCG TTA ATG ACT-3′) and DA640 (5′ CGG AAT TCC GAT ACA TAC TAG AAT TGA TCG 3′).
The fragment was inserted into pTwiggy at the EcoRI site between the twist regulatory elements and the w gene to prevent twist activation of w, allowing us to assay Gal4 activation of w in Fig. 5. The Gal4 AD (aa 753 to 881) KpnI-XbaI fragment was then inserted into this vector.
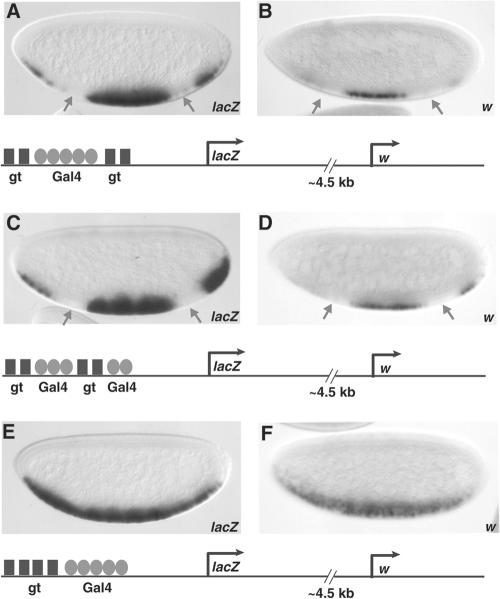
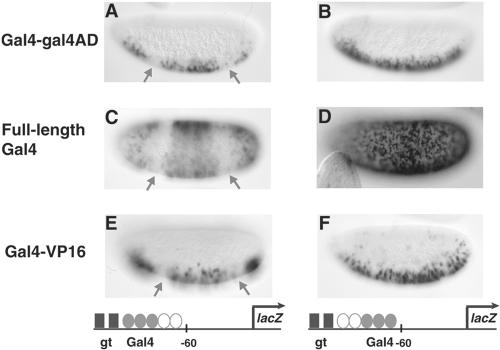
FIG. 5.
The arrangement of short-range repressor binding sites is critical in dictating repression effectiveness. Reporter genes are shown below the corresponding embryos. (A and B) Giant (gt) is able to repress transcription from both the proximal hsp70 lacZ gene and the distal w promoter, which is located 4.5 kb 3′, on a gene in which the repressor sites flank the Gal4 binding sites. (C and D) Giant also represses effectively when binding sites are interspersed between the Gal4 sites. (E and F) Giant does not repress when repressor sites are situated 5′ of the activator sites, although activators are within 100 bp of the most proximal Giant site. A minimal Gal4 activator was expressed in ventral regions under the control of the twist promoter. In order to distinguish transcription of the white gene 3′ of the lacZ reporter from transcription of the white gene present on the Gal4 driver, insulator sequences were inserted between the twist regulatory element and the white gene in the Gal4 driver to prevent direct activation of white, as described in Materials and Methods. Expression patterns were visualized in 2- to 4-h embryos by in situ hybridization with antisense lacZ probes.
Fly stocks.
Flies carrying a mutation in the giant gene gtA8/FM7c (stock number 1004.1) and gtX11/FM6 (stock number 1529) were obtained from the Bloomington Stock Center. To analyze reporter gene expression in a giant mutant background, males carrying the reporter and the Gal4 activator transgenes were crossed to females carrying the giant mutation. Half of the male progeny will be hemizygous null for giant.
Flies expressing the full-length yeast transcriptional activator Gal4 ubiquitously throughout the embryo under the actin 5C enhancer act5cGAL4/CyO (stock number 4414) were also obtained from Bloomington. To obtain ubiquitous activation of the lacZ reporter gene in the early embryo, act5cGAL4/CyO females were crossed to males carrying the reporter transgene.
Reporter genes.
The stripe 2/2x UAS/eve-lacZ vector (3) containing two Gal4 binding sites and the minimal eve basal promoter driving lacZ expression was modified to include two Giant (14) binding sites (DA127/128 [5′-AAT TCG CAT GCT ATG ACG CAA GAA GAC CCA GAT CTT TTT ATG ACG CAA GAG CAT GCG-3′; the Giant binding sites are underlined]) using EcoRI-BssH2, 5′ of the Gal4 sites. Three additional Gal4 binding sites were inserted (DA139/140 [5′ TCG GAT TAG AAG CCG CCG TCG CTA GAG GAA GAC TCT CCT CCG ACG TGA ACG CAG GAC ACT CCT GC GCT GCA-3′; the Gal4 binding sites are underlined]) at the PstI site 3′ of the existing Gal4 sites. Oligonucleotides with a 50-bp spacer (DA125 [5′-TCG CTA GAC GTG AAT CTC GTA GCT TCC GTA CCA AAT GCG TAT CAG CTG CA-3′] and DA126 [5′-GCT GAT ACG CAT TTG GTA CGG AAG CTA CGA GAT TCA CGT CTA GCG ATG CA-3′]) were introduced at the PstI 3′ site, yielding H2g5u-50 (Fig. 1C and D) with two Giant binding sites, five tandemly arrayed Gal4 binding sites, a 50-bp spacer, and minimal eve basal promoter driving lacZ expression.
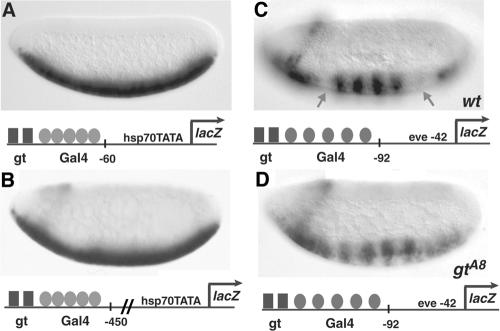
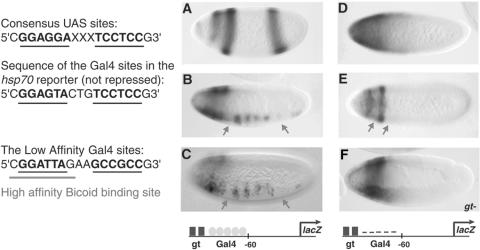
FIG. 1.
Context dependence of short-range repression. Giant is dependent on gene context for repression of Gal4 activators. (A) The hsp70 lacZ gene, activated by a cluster of five high-affinity Gal4 binding sites, is not repressed by Giant (gt). (B) lacZ expression is not repressed when the repressor-activator cluster is situated 400 bp further 5′ of the basal promoter. (C) Giant effectively represses a cluster of five Gal4 binding sites 5′ of the eve basal promoter. Arrows indicate regions of Giant expression in the anterior and posterior domains of the embryo. Striping is thought to be caused by the binding of an unidentified pair-rule regulator. wt, wild type. (D) Repression is abolished in the giant (gtA8) mutant embryo. A minimal Gal4 activator (residues 1 to 93, DNA binding, fused to residues 753 to 881, activation domain) was expressed in ventral regions under the control of the twist promoter. Expression patterns were visualized in 2- to 4-h embryos by in situ hybridization with antisense lacZ probes. In this figure and later figures, embryos are oriented anterior to the left and dorsal up. The structure of the reporter gene is shown below the corresponding embryos.
The plasmid UAS-lacZ (12) was modified to contain two Giant sites (DA321/322 [5′-GGC CGC TAT GAC GCA AGA AGA CCC AGA TCT TTT TAT GAC GCA AGA GA-3′; the Giant sites are underlined), two Knirps sites (DA319/320 [5′-GGC CGC ATC TGA TCT AGT TTG TAC TAG ACA TCT GAT CTA GTT TCA-3′; the Knirps sites are underlined]), two Krüppel sites (DA694/695 [5′-GGC CGC AAA ACG GGT TAA GCG ACC CAA AAC GGG TTA AGC A-3′; the Krüppel sites are underlined]), or two Hairy sites (DA604/605 [5′-GGC CGC GCG GCA CGC GAC ATG ACC CGC GGC ACG CGA CAT A-3′; the Hairy sites are underlined]) 20 nucleotides 5′ of the five Gal4 binding sites (4, 14, 32, 57). The resulting vectors M2g5u-lacZ/M2k5u-lacZ/M2kr5u-lacZ/M2h5u-lacZ, respectively, have two Giant, Knirps, Krüppel, or Hairy sites, five Gal4 binding sites, and the hsp70 TATA box and transcriptional start driving lacZ expression (Fig. 1A and see Fig. 2A and B, 4, and 7B, D, F, and H).
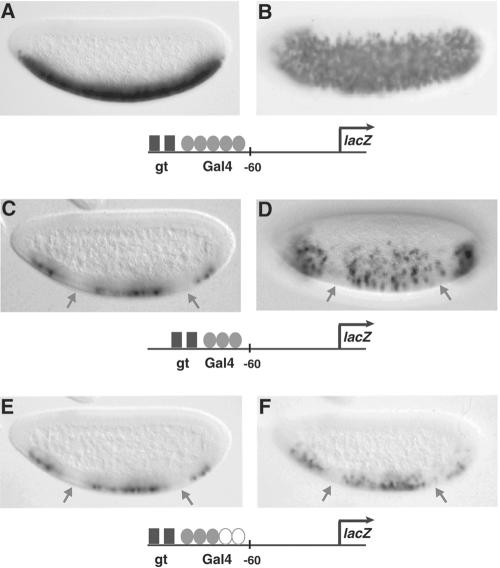
FIG. 2.
Stoichiometry of activators to repressors influences repression effectiveness. (A and B) Activity mediated by five Gal4 binding sites 5′ of the hsp70 basal promoter elements is not repressed by Giant (gt). (C to F) Reducing the number of activator sites from five to three by deletion or replacement with a neutral spacer permits repression by Giant (arrows). Ventrolateral views are shown (B and D). A minimal Gal4 activator was expressed in ventral regions under the control of the twist promoter. Expression patterns were visualized in 2- to 4-h embryos by in situ hybridization with antisense lacZ probes.
FIG. 4.

Weaker activation domains are not more susceptible to repression by Giant. Chimeric Gal4 activators used to drive expression from the reporter gene are indicated to the left of the embryos. The structure of the reporter gene is shown below. (A and B) Neither the more potent Gal4 and Gal4-VP16 activators or the weaker Gal4-Sp1 and Gal4-hTBP activators (C and D) were repressed by Giant (gt) on a lacZ reporter containing five high-affinity Gal4 binding sites. Activators were expressed in ventral regions under the control of the twist promoter. Expression patterns were visualized in 2- to 4-h embryos by in situ hybridization with antisense lacZ probes.
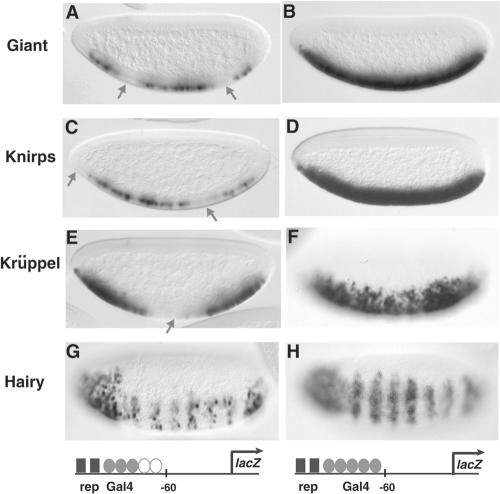
FIG. 7.
Short-range repressors exhibit similar functional limits, in contrast to a long-range repressor. The reporter genes contain two binding sites for Giant, Knirps, Krüppel, or Hairy and three or five high-affinity Gal4 binding sites. (A, C, E, and G) All repressors (rep) were able to repress the minimal Gal4 activator on genes containing three Gal4 binding sites. Repression in A, C, and E is indicated by arrows and corresponds to the pattern of expression of the repressor proteins. Hairy is expressed in seven stripes at this stage. (B, D, F, and H) None of the short-range repressors repressed a reporter containing five Gal4 binding sites; however, Hairy induces a pronounced striped pattern, indicating effective repression. To drive expression, a minimal Gal4 activator was expressed in ventral regions under the control of the twist promoter. Expression patterns were visualized in 2- to 4-h embryos by in situ hybridization with antisense lacZ probes.
The vector M2g5u-lacZ was modified by introducing oligonucleotides containing a 55-bp neutral spacer (DA65/66 [5′-TCC ATG ATA AAC GCG TGC TAG ACT ATT GCA GGT ACT GAT CGA ATG CCT CTG CAT G-3′]) at the SphI site downstream of the Gal4 binding sites. The vector was further modified by introducing a 340-bp fragment of the Knirps open reading frame amplified by using DA572/573 (DA572, 5′-ACA TGC ATG CAA CCG CTT TAG TCC CGC CAG-3′; DA573, 5′-ACA TGC ATG CTG TGC ACG GAG CTC CGC GAG-3′) from Gal4-kniF1 (38), resulting in the spaced construct M2g5u-55-340-bpkni ORF-lacZ (Fig. 1B).
M2g5u-lacZ was modified to replace the five Gal4 sites with HindIII-SphI oligonucleotides containing three high-affinity Gal4 (12) sites (DA469/470 [5′-AGC TTG CCT GCA GGT CGG AGT ACT GTC CTC CGA GCG GAG TAC TGT CCT CCG AGC GGA GTA CTG TCC TCC GAG GCA TG-3′; the Gal4 sites are underlined]) to give M2g3u-lacZ (Fig. 2C and D). This was further modified by introducing SphI spacer oligonucleotides (DA471 [5′-TCA TAC AAC TGG TCA GTG AGC ATA CAA CTG GTC AGT GAG CAT G-3′] and DA472 [5′-CTC ACT GAC CAG TTG TAT GCT CAC TGA CCA GTT GTA TGA CAT G-3′]) equal to the length of two Gal4 sites, resulting in M2g3u2x-lacZ (Fig. 2E and F and see Fig. 6A, C, and E). The two Giant binding sites in M2g3u2x-lacZ were replaced by two Knirps sites (DA319/320), two Krüppel sites(DA694/695), or two Hairy sites (DA604/605) 20 nucleotides 5′ of the three Gal4 binding sites. The resulting vectors named M2k3u2x-lacZ/M2kr3u2x-lacZ/M2h3u2x-lacZ (see Fig. 7C, E, and G) consist of two Knirps, two Krüppel, and two Hairy binding sites, respectively, three tandemly arrayed Gal4 binding sites, and a spacer followed by the hsp70 TATA box and transcriptional start driving lacZ expression.
FIG. 6.
Additional distance dependence of permissive repressor-activator stoichiometries. The reporter gene structure is shown below and chimeric Gal4 activators used to drive expression from the reporter gene are indicated to the left of the embryos. (A, C, and E) Full-length Gal4 protein was expressed ubiquitously, while the minimal Gal4 activator and a Gal4-VP16 activator were expressed in ventral regions under the twist promoter. Giant (gt) represses all three activators when three Gal4 sites are present. (B, D, and F) Giant repression activity is absent when the three Gal4 sites are moved 37 nucleotides from the Giant sites, although still less than 100 bp from the Giant sites. Expression patterns were visualized in 2- to 4-h embryos by in situ hybridization with antisense lacZ probes.
The vector M2g3u-lacZ was cut with HindIII to introduce spacer oligonucleotides (DA473 [5′-AGC TTC ATA CAA CTG GTC AGT GAG CAT ACA ACT GGT CAG TG-3′] and DA474 [5′-AGC TCA CTG ACC AGT TGT ATG CTC ACT GAC CAG TTG TAT GA-3′]) equal to the length of two Gal4 sites between the Giant and three Gal4 sites to result in M2g2x3u-lacZ (see Fig. 6B, D, and F). The five high-affinity Gal4 binding sites in M2g5u-lacZ were replaced with five low-affinity Gal4 sites (13, 37) by sequentially cloning in HindIII-SphI oligonucleotides containing three low-affinity Gal4 sites (DA600/601 [5′-AGC TTG CCT GCA GGT CGG ATT AGA AGC CGC CGA GCG GAT TAG AAG CCG CCGAGC GGA TTA GAA GCC GCC GCA TG-3′; the low-affinity Gal4 sites are underlined]) followed by SphI oligonucleotides containing two low-affinity Gal4 sites (DA602/603 [5′-TCG GAT TAG AAG CCG CCG AGC GGA TTA GAA GCC GCC GCA TG 3′; the low-affinity Gal4 sites are underlined]), resulting in the vector M2g5u (low affinity)-lacZ (Fig. 3).
FIG. 3.
Effectiveness of repression correlates with the affinity of Gal4 activator binding sites. Sequences of Gal4 binding sites are shown (left). The high-affinity Gal4 sites were used in reporters shown in Fig. 1A and B and 2, and low-affinity Gal4 binding sites were used here. The fortuitous Bicoid binding site is underlined in gray. (A) For reference, giant expression in the early blastoderm embryo, visualized by in situ hybridization, refines into two stripes anteriorly and one stripe posteriorly. (B and C) Giant (gt) represses lacZ expression driven by a minimal Gal4 activator in ventral regions acting on a cluster of five low-affinity Gal4 binding sites (arrows). These sequences also appear to bind to unidentified pair-rule repressors which confer an overall striped expression pattern on the reporter gene. This pattern made analysis of the Giant repression pattern more difficult; however, lacZ expression was consistently reduced in regions of Giant expression. (D and E) Giant represses Bicoid-mediated activation of the hsp70 lacZ reporter (arrows). Even in the absence of the Gal4 activator, lacZ expression is activated by the transcription factor Bicoid in the anterior region of the embryo from five high-affinity Bicoid sites that overlap the Gal4 sites. Bicoid-mediated activation is refined into two stripes of expression as the embryo develops, in regions where giant is not expressed. (F) lacZ expression in a giant mutant shows unrepressed expression mediated by Bicoid. lacZ expression is no longer refined into a two-stripe pattern in the giant mutant background. The embryo shown in F is of an age comparable to that shown in E. A minimal Gal4 activator was expressed in ventral regions under the control of the twist promoter. Expression patterns were visualized in 2- to 4-h embryos by in situ hybridization with antisense lacZ probes. UAS, upstream activation sequence.
Two additional Giant binding sites were introduced either at the SphI site (DA50/51) in the M2g5u-lacZ vector between the five Gal4 binding sites and the hsp70 TATA box, resulting in M2g5u2g-lacZ (see Fig. 5A and B), or at the NotI site (DA637/638) in the M2g5u-lacZ vector upstream of the two Giant sites, resulting in M4g5u-lacZ (see Fig. 5E and F). Two additional binding sites for Giant (DA50/51) as well as two high-affinity Gal4 sites (DA598/599) were introduced sequentially in the M2g3u-lacZ vector at the SphI site, resulting in M2g3u2g2u-lacZ (see Fig. 5C and D).
P-element transformation, crosses to reporter genes, and in situ hybridizations.
P-element transformation vectors were introduced into the Drosophila germ line by injection of yw67 embryos as described previously (72). Embryos were collected either directly from each transgenic reporter line or from a cross between a reporter line and a line expressing the Gal4 activator chimeric proteins in the ventral regions or ubiquitously throughout the embryo. The embryos were fixed and stained using digoxigenin-UTP-labeled antisense RNA probes to either lacZ or w as described previously (72). Embryos shown are generally representative of at least 90% of scored embryos of the relevant age, except as noted otherwise.
RESULTS
Context dependence of short-range repression.
The activity of short-range transcriptional repressors has been studied mostly in the context of complex natural enhancers (3, 20, 32, 41, 72, 75). To analyze cis-acting element activity in a setting in which activator-repressor composition, stoichiometry, and spacing can be exactly defined, we constructed chromosomally integrated, compact regulatory modules containing binding sites for the endogenous short-range repressor Giant and chimeric Gal4 activators. The space between repressor and activator sites on these elements is less than 100 bp, a distance over which short-range repressors have been previously shown to be effective. The activity of the chimeric Gal4 activator is localized to the ventral regions of the embryo, where it is expressed under the control of ventral-specific enhancer elements. Strikingly, Giant was unable to repress the activity of a minimal Gal4 activator protein on a reporter gene in which Giant binding sites were located immediately 5′ of five high-affinity Gal4 sites (Fig. 1A). This lack of repression activity reveals a hitherto unknown limitation of short-range repressors. Giant represses adjacent Dorsal and Twist activators on similar reporter genes, indicating that Giant can bind such a reporter gene and that the hsp70 basal promoter is not inherently resistant to repression (43). The close proximity of the Gal4 activators to the hsp70 basal promoter may prevent Giant from mediating repression on this reporter; therefore, a neutral 400-bp spacer sequence was introduced between the Gal4 binding sites and the transcriptional start site. However, Giant was also unable to repress in this context (Fig. 1B). The inability of Giant to repress is not due to an inherent resistance of the Gal4 activation domain, for Giant was able to repress the activity of the Gal4 activator on a gene containing a cluster of five Gal4 binding sites 5′ of the eve basal promoter (Fig. 1C). The repression in anterior and posterior regions is relieved when this transgene is assayed in giant mutant embryos (Fig. 1D), confirming that the observed repression is mediated by Giant.
These results indicate that the simple notion that short-range repressors block the activity of all protein complexes within 100 bp is an oversimplification. Clearly, mere proximity is not the only determinant affecting repression by Giant. We set out to systematically define other factors that dictate repression effectiveness to uncover a potential cis-regulatory grammar of short-range repression. The repressed and nonrepressed reporter genes (Fig. 1, compare A and C) differ in the sequence of the activator sites, nature of the basal promoters, and repressor position with respect to the transcriptional start site. Activator binding site affinity or spacing seems likely to be a more important factor, because Giant has been previously shown to be able to repress genes with both types of basal promoter, and the relative spacing of the repressors to +1 should in fact favor repression as shown in Fig. 1A, in which the Giant repressor sites are closer to the start site of transcription. We first tested whether activator binding sites played a role.
Repression sensitivity correlated to the number of activator binding sites or strength of the activating signal.
Studies of the hairy gene in Drosophila led to the suggestion that the overall stoichiometry, rather than the absolute number, of activators and repressors may be critical in dictating enhancer output (44). To test whether the stoichiometry of activators to repressors is a critical factor in determining short-range repression levels by Giant, we reduced the number of Gal4 activator binding sites on the hsp70-lacZ reporter from five (Fig. 2A and B) to three (Fig. 2C and D). As anticipated, the levels of transcriptional activation by the minimal Gal4 activator were lower in the transgene containing three Gal4 sites (Fig. 2C and D), leading to a less robust ventral staining pattern. In this context, Giant was able to block transcription of the lacZ gene (Fig. 2C and D). However, the removal of two Gal4 sites also positions the repressors closer to the start of transcription, which may facilitate repression of the basal promoter (“direct repression”). Therefore, to maintain the distance between Giant binding sites and the start of transcription, a neutral spacer was placed downstream of the three Gal4 sites (Fig. 2E and F). Again, Giant was also able to repress the minimal Gal4 activator. These results demonstrate that repression is critically dependent on the number of activator binding sites but do not explicitly differentiate between the overall level of transcriptional activation and binding site number, an issue addressed in Fig. 4. These results are also consistent with previous analyses of the eve stripe 2 element, where the insertion of additional Bicoid binding sites in an otherwise normal stripe 2 enhancer causes a slight anterior expansion of its expression pattern, suggesting that an excess of Bicoid activators can “overwhelm” the Giant repressor (3).
Binding site affinity.
Binding site affinity influences threshold responses to activator gradients in the embryo (25, 36, 76), and indeed, transcription factor binding sites of various affinities are typically found in many developmental enhancers that function during early Drosophila development. Such differences in activator site affinity might similarly influence responses to short-range repressors. We tested whether maintaining the number of activator sites but weakening their affinity would in fact change the response to repressors. We replaced the five high-affinity Gal4 binding sites in the hsp70 lacZ reporter with five copies of a site from the Saccharomyces cerevisiae Gal1-Gal10 promoter that has been characterized as a weaker Gal4 binding site (13, 37). The minimal Gal4 activator drives gene expression in a weaker, striped pattern from the lower-affinity Gal4 sites. Anterior and posterior repression by Giant is evident (Fig. 3B and C, arrows), similar to the pattern observed in Fig. 1C. As expected, later in development, when Giant protein is no longer present, lacZ is expressed in a continuous swathe (data not shown). The striped expression of the constructs is thought to be due to the binding of uncharacterized pair-rule repressors to spacer sequences in the reporter (79).
In the process of weakening the Gal4 binding sites, we inadvertently created five high-affinity binding sites for the Bicoid activator, providing an additional opportunity to assay Giant repression activity. Bicoid is maternally deposited in the anterior regions of the embryo, forming an anterior-to-posterior gradient (24). lacZ expression from the hsp70 reporter is activated even in the absence of the Gal4 activator by the Bicoid transcription factor in anterior regions (Fig. 3D and E). As the embryo develops, Giant inhibits Bicoid activation of lacZ, which is thereby progressively refined into a two-stripe pattern (Fig. 3E), in regions where giant is not expressed (Fig. 3A). Analysis of the transgene in a giant mutant background in the absence of Gal4 confirms that refinement of reporter gene expression is due to repression by Giant (Fig. 3F). These results suggest that five Bicoid binding sites are more susceptible to repression than are five high-affinity Gal4 sites, indicating that stoichiometric relationships of repressors to activators in turn may depend on either distinct DNA binding domains or the type of activation domains.
Repression not dependent on the nature of the activation domain.
The differential effectiveness of Giant against five Gal4 or five Bicoid binding sites suggests that the nature of the activation domain itself or the DNA binding domain of the transcriptional activator may play a role in dictating the response to repressors. To distinguish between those two possibilities, we compared the activities of a variety of activation domains fused to the DNA binding domain of Gal4. In addition to the Gal4 activation domain, we tested the acidic transcriptional activation domain of the herpes simplex virus activator VP16, the glutamine-rich activation domain of the mammalian transcription factor Sp1, and the hTBP, which has been shown to function as an activator when targeted to the promoter via the Gal4 DNA binding domain (50). We also sought to test the activity of Gal4-Bicoid activators (35), but unfortunately, these chimeras exhibit strong promoter specificity and are not active on the hsp70 promoter, which precluded a direct comparison (data not shown). The Gal4 chimeric proteins were used to drive expression of the hsp70 lacZ reporter from the cluster of five high-affinity Gal4 sites (Fig. 4). Giant could inhibit neither the strong Gal4 (Fig. 4A) and VP16 (Fig. 4B) activators nor the weak activation domains of Sp1 (Fig. 4C) and hTBP (Fig. 4D). These results indicate that the ability to repress does not depend on the strength of the activation domain or the activation pathway. Only those genes in which the number or affinity of Gal4 sites was reduced showed a response to Giant, suggesting that the Gal4 DNA binding domain provides a stable platform that can resist the activity of Giant. These results are consistent with a mechanism for short-range repression that involves blocking activator access to its cognate sites.
The arrangement or distribution of short-range repressor binding sites is critical in dictating repression effectiveness.
Statistical models, based on motif clustering, are only partially successful at finding novel cis-regulatory elements in the genome, perhaps because they consider only site density and relative site affinity (11, 52, 53, 66). However, it is probable that specific arrangements of binding motifs also contribute to biological function (51). We tested the effect of alternative arrangements of Giant repressor and Gal4 activator binding sites to determine if different arrangements or combinations resulted in distinct transcriptional outputs. In all reporter arrangements tested, we used four Giant binding sites and five high-affinity Gal4 binding sites, bound by the minimal Gal4 activator. Flanking the five Gal4 activator sites with two Giant sites on either side resulted in repression of the proximal hsp70 lacZ reporter gene (Fig. 5A). Interspersing the Giant repressor binding sites between the Gal4 activator sites also resulted in the inhibition of lacZ expression (Fig. 5C). However, placing all four Giant binding sites 5′ of the five Gal4 sites prevented Giant from repressing the hsp70 lacZ expression (Fig. 5E), suggesting again that promoter response cannot be calculated simply from overall activator-to-repressor stoichiometries.
The Giant binding sites in the reporter genes shown in Fig. 5A and C are in close proximity to the basal promoter; therefore, it is possible that Giant directly represses the basal promoter (4, 31, 34). To distinguish between repressor-basal promoter and repressor-activator effects, we measured transcription of the w gene, which is ∼4.5 kbp 3′ of these sites (Fig. 5B, D, and F). Again, we observed that Giant mediated repression only when flanking or interspersed with activators (Fig. 5B and D) but not when situated 5′ of the activator sites (Fig. 5F). This result suggests that Giant is acting on the activator cluster rather than only on the basal promoter element.
Previous analysis of the short-range repressor Giant demonstrated that due to the extreme distance-dependent activity of this protein, subtle changes in the spacing of Giant binding sites endowed a promoter with high or low sensitivity to repression (34). We tested whether Giant's ability to repress a smaller cluster of three Gal4 sites could be affected by small changes in spacing between the activator and repressor binding sites. Moving the smaller cluster of three Gal4 sites 37 bp away from the Giant binding sites results in the loss of repression (Fig. 6, compare A and B), suggesting that reducing the amount of activation potential does not guarantee repression by Giant in all cases, even when the activators are located within 100 bp of the repressor sites. In order to ascertain whether the spacing effects we see are specific to this particular activator protein (i.e., Gal4-Gal4 AD), we tested the ability of Giant to block transcription mediated by the full-length Gal4 protein expressed ubiquitously throughout the embryo (Fig. 6C and D) and the Gal4-VP16 fusion protein (Fig. 6E and F). As seen with the minimal Gal4 activation domain, Giant is able to repress lacZ expression mediated by the full-length Gal4 protein (Fig. 6C) and Gal4-VP16 (Fig. 6E) from three sites that are adjacent to the Giant binding sites. Moving the three sites 37 bp further away results in the loss of repression of both Gal4-mediated (Fig. 6B and D) and Gal4-VP16-mediated (Fig. 6F) activation by Giant.
Specificity of regulatory grammar.
The contextual dependencies of repression described above were characterized for the Giant repressor. To determine if similar rules applied to other types of repressors, we carried out parallel evaluations of the short-range repressors Giant, Knirps, and Krüppel. To test quantitative similarities or differences between these factors, we created reporters that would compare repressor activity on genes that represented permissive or nonpermissive contexts for the Giant protein. All three of these short-range repressors were unable to inhibit lacZ expression driven by the minimal Gal4 activator from five high-affinity Gal4 sites, indicating a similar limitation of repression on even proximally bound activators (Fig. 7B, D, and F). The Giant and Krüppel factors were active in the corresponding regions of the embryo when tested against three Gal4 sites (Fig. 7A and E). The Knirps repressor was also active in this context, although in general, the levels of repression appeared to be lower (Fig. 7C). In contrast, the long-range repressor Hairy was able to mediate repression of transgenes containing either three or five high-affinity Gal4 sites (Fig. 7G and H). Interestingly, as the embryo aged, repression by Hairy was first attenuated and then completely absent during germ band elongation (data not shown), indicating that this type of repression, though potent, is also transient. The similarity in the activity of the short-range repressors Giant, Knirps, and Krüppel, in contrast to that of Hairy, suggests that the contextual rules for repression are governed by the functional class of repressor and likely reflects mechanistic differences.
DISCUSSION
A grammar of short-range repression.
Using defined synthetic enhancer elements, we demonstrate that there is a rich set of rules or contextual grammar that influences the activity of short-range repression extending beyond the generalization that these factors block activators situated within ∼100 bp. Although distance is a critical factor in dictating repression effectiveness, it is not the only one, and in some cases, close proximity alone is not sufficient to ensure regulation by these transcriptional repressors (Fig. 1, 4, 5, and 6 and reference 43). Activators can retain function even when the binding sites are within the previously defined 100-bp effective range of short-range repression. The manipulation of these composite enhancer elements in terms of the number of activator and repressor binding sites, relative affinities, spacing, and distribution of binding sites and the type of activation domains allowed us to define other contextual parameters that dictate repression effectiveness. First, we find that the ratio of activators and repressors is an important factor; in the context of five high-affinity Gal4 sites, four Giant sites can mediate repression but two sites do not. Reducing the number of Gal4 binding sites from five to three allowed two Giant sites to repress the lacZ reporter gene. Second, although the effectiveness of repression depends on stoichiometry between the number of activators and repressors, Giant repression of a smaller cluster of activators can be attenuated by subtle changes (<40 bp) in the spacing between the repressor and activator binding sites, even when activator binding sites in this situation are within the previously defined 100-bp range of repression. Such subtle changes in spacing between Giant and activator sites may explain the internal reconfigurations in enhancer design that have been demonstrated to occur between functionally homologous even-skipped stripe 2 enhancers and presumably many other cis-regulatory elements (47). Indeed, we find that in order to mediate repression effectively, short-range repressors need to be judiciously placed, either flanking activator sites or interspersed among them, possibly to block multiple modes of activator-promoter interactions. A fourth finding is that repression effectiveness correlates with activator site affinity, and although binding affinity influences the strength of the activating signal, repression does not depend on the chemical nature of the activation domain. Although we have developed these experiments in the context of Gal4 fusion activators, it is likely that similar principles apply for repression of other activators, as repression of native activators also shows strong context dependence (3, 79). Most likely, quantitative aspects of the relationships we have identified will vary depending on the DNA binding characteristics of different factors, whose characteristics will be established by further empirical tests. Determination of such quantitative factors contributes to our understanding of enhancer design and should find application in bioinformatics analysis of novel gene regulatory sequences as well as providing insights into the evolution and biochemical activity of short-range repressors.
Computational analysis of cis-regulatory elements.
Computational approaches have focused on the identification of transcriptional regulatory regions based on patterns of binding sites and evolutionary conservation of sequences. A more ambitious objective is to identify quantitative information about enhancers, including temporal, spatial, and quantitative output of such elements. More sophisticated analytical tools might also involve identification of conserved patterns of binding site stoichiometries, arrangements, and affinities that are not readily discernible by using conventional analyses. Recently, bioinformatics analysis of number and affinity of binding sites for the Knirps and Hunchback repressors was used to successfully predict the relative sensitivity of different regulatory sequences to these factors (20). In addition to quantitating the number and affinity of factor sites, our study indicates that bioinformatics analysis should also take into account the stoichiometry of activators to repressors, the exact spacing involved, and the nature of the DNA binding domains involved. Clearly, our studies focus on the effects of one class of repressor protein; more comprehensive work will be required to elaborate parameters relevant for other types of repressors and for activators. It is unlikely that particular contextual grammars would apply to all transcription factors; however, it is encouraging that the short-range repressors tested so far show similar characteristics. It therefore appears possible to model the properties of groups of proteins without having to develop distinct cis-regulatory grammar rules for each one. Incremental improvements to current approaches, based on the identification of cis-regulatory grammars, will usefully enhance the power of computational tools and allow the extension of bioinformatics analysis to specific data sets.
Mechanisms of repression.
The contextual grammar defined in this study presents a phenomenological perspective to short-range repression, but our results also shed light on possible repression mechanisms. Three models have been presented for the action of short-range repressors. First, by binding overlapping sites, these repressors might directly compete with activators for binding to DNA, a situation that can be demonstrated experimentally (56). This mechanism has not been shown to play a role in endogenous enhancers, and where experimentally tested, the DNA binding domain of Knirps was not able to mediate repression in the embryo (75). It is in any event unlikely to be important in cases where the activator and repressor binding sites are separated, as is the case here. Second, repressors might “quench” neighboring activators, inhibiting their access to the DNA or blocking their interaction with other components of the transcriptional machinery. Third, the proteins might not affect activators but directly contact the basal transcriptional machinery. The results obtained in this study and a recent study (43) are most compatible with the second, quenching model of action. We have previously demonstrated that closely spaced factors can simultaneously mediate opposite transcriptional regulatory outputs (43), which would be hard to rationalize in the context of basal machinery interactions but is readily explainable in light of different susceptibilities of activators to chromatin remodeling. In addition, as shown in this study, the sensitivity of activators toward repression appears to be most closely linked to the DNA binding domain and affinity of the binding site rather than the activation domain, which may reflect a limited access to the DNA template under repression conditions.
The apparent lack of activator specificity demonstrated by short-range repressors also suggests that these proteins function via a general mechanism. Giant, Knirps, Krüppel, and Snail can block the activity of a number of activators such as Bicoid, Hunchback, Dorsal, Twist, and D-Stat (3, 32, 73). Many biochemical and genetic analyses suggest that at least some of these activators activate transcription via distinct pathways (40, 58, 81). Here, we have demonstrated that repression effectiveness does not depend on the nature of the activation domain but correlates instead with activator binding site affinity and placement. These findings are consistent with a mechanism that inhibits transcription by blocking access to DNA by transcriptional activators via local chromatin changes.
This model is also consistent with biochemical properties of short-range repressors. These proteins interact with CtBP, which in turn binds chromatin-modifying factors, including histone deacetylases (HDAC1 and HDAC2) and histone methyltransferases (18, 19, 69, 78). We have found that Knirps genetically and physically interacts with Rpd3, the Drosophila homolog of HDAC1 (P. Struffi, unpublished data). The Rpd3 protein in yeast is known to deacetylate histones at an extremely local level, consistent with its role in short-range repression in Drosophila (23). Knirps, Giant, and Krüppel can repress in a CtBP-independent fashion (38, 77), but this activity appears to possess similar properties to that mediated by the Drosophila CtBP-dependent activity, providing a quantitative, rather than qualitative, effect (63, 75, 79). Thus, both the Drosophila CtBP-dependent and -independent activities of the short-range repressors might work via chromatin remodeling.
Our study demonstrates that the Hairy repressor, in addition to working over a longer range, is also a more potent repressor on a local level, presumably because of its distinct biochemical mechanism for repression. By examining the nature of the promoter complexes and the chromatin state before and after repression, the defined transcriptional switch elements used in this study will facilitate further biochemical characterization of short- and long-range repressors.
Acknowledgments
We thank S. J. Triezenberg and R. W. Henry for helpful discussions and E. Fernandez-Villatoro for technical assistance.
This work was supported by NIH grant GM56976 to D.N.A.
REFERENCES
- 1.Arnosti, D. N. 2002. Design and function of transcriptional switches in Drosophila. Insect Biochem. Mol. Biol. 32:1257-1273. [DOI] [PubMed] [Google Scholar]
- 2.Arnosti, D. N. 2004. Multiple mechanisms of transcriptional repression in eukaryotes, p. 33-67. In M. Gossen, J. Kaufmann, and S. J. Triezenberg (ed.), Handbook of experimental pharmacology: transcription factors, vol. 166. Springer, Berlin, Germany. [Google Scholar]
- 3.Arnosti, D. N., S. Barolo, M. Levine, and S. Small. 1996. The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development 122:205-214. [DOI] [PubMed] [Google Scholar]
- 4.Arnosti, D. N., S. Gray, S. Barolo, J. Zhou, and M. Levine. 1996. The gap protein knirps mediates both quenching and direct repression in the Drosophila embryo. EMBO J. 15:3659-3666. [PMC free article] [PubMed] [Google Scholar]
- 5.Banerji, J., S. Rusconi, and W. Schaffner. 1981. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 2:299-308. [DOI] [PubMed] [Google Scholar]
- 6.Barolo, S., L. A. Carver, and J. W. Posakony. 2000. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. BioTechniques 29:726, 728, 730, 732. [DOI] [PubMed] [Google Scholar]
- 7.Barolo, S., and M. Levine. 1997. hairy mediates dominant repression in the Drosophila embryo. EMBO J. 16:2883-2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barolo, S., and J. W. Posakony. 2002. Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 16:1167-1181. [DOI] [PubMed] [Google Scholar]
- 9.Beer, M. A., and S. Tavazoie. 2004. Predicting gene expression from sequence. Cell 117:185-198. [DOI] [PubMed] [Google Scholar]
- 10.Bergman, C. M., and M. Kreitman. 2001. Analysis of conserved noncoding DNA in Drosophila reveals similar constraints in intergenic and intronic sequences. Genome Res. 11:1335-1345. [DOI] [PubMed] [Google Scholar]
- 11.Berman, B. P., Y. Nibu, B. D. Pfeiffer, P. Tomancak, S. E. Celniker, M. Levine, G. M. Rubin, and M. B. Eisen. 2002. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. USA 99:757-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brand, A. H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. [DOI] [PubMed] [Google Scholar]
- 13.Burns, L. G., and C. L. Peterson. 1997. The yeast SWI-SNF complex facilitates binding of a transcriptional activator to nucleosomal sites in vivo. Mol. Cell. Biol. 17:4811-4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capovilla, M., E. D. Eldon, and V. Pirrotta. 1992. The giant gene of Drosophila encodes a b-ZIP DNA-binding protein that regulates the expression of other segmentation gap genes. Development 114:99-112. [DOI] [PubMed] [Google Scholar]
- 15.Carroll, S. B., J. K. Grenier, and S. D. Weatherbee. 2001. From DNA to diversity: molecular genetics and the evolution of animal design. Blackwell Science, Malden, Mass.
- 16.Chen, G., and A. J. Courey. 2000. Groucho/TLE family proteins and transcriptional repression. Gene 249:1-16. [DOI] [PubMed] [Google Scholar]
- 17.Chen, G., J. Fernandez, S. Mische, and A. J. Courey. 1999. A functional interaction between the histone deacetylase Rpd3 and the corepressor Groucho in Drosophila development. Genes Dev. 13:2218-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chinnadurai, G. 2002. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell 2:213-224. [DOI] [PubMed] [Google Scholar]
- 19.Chinnadurai, G. 2003. CtBP family proteins: more than transcriptional corepressors. BioEssays 25:9-12. [DOI] [PubMed] [Google Scholar]
- 20.Clyde, D. E., M. S. Corado, X. Wu, A. Paré, D. Papatsenko, and S. Small. 2003. A self-organizing system of repressor gradients establishes segmental complexity in Drosophila. Nature 426:849-853. [DOI] [PubMed] [Google Scholar]
- 21.Courey, A. J., and J. D. Huang. 1995. The establishment and interpretation of transcription factor gradients in the Drosophila embryo. Biochim. Biophys. Acta 1261:1-18. [DOI] [PubMed] [Google Scholar]
- 22.Davidson, E. H. 2001. Genomic regulatory systems: development and evolution. Academic Press, San Diego, Calif.
- 23.Deckert, J., and K. Struhl. 2001. Histone acetylation at promoters is differentially affected by specific activators and repressors. Mol. Cell. Biol. 21:2726-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Driever, W., and C. Nusslein-Volhard. 1988. A gradient of bicoid protein in Drosophila embryos. Cell 54:83-93. [DOI] [PubMed] [Google Scholar]
- 25.Driever, W., G. Thoma, and C. Nusslein-Volhard. 1989. Determination of spatial domains of zygotic gene expression in the Drosophila embryo by the affinity of binding sites for the bicoid morphogen. Nature 340:363-367. [DOI] [PubMed] [Google Scholar]
- 26.Emberly, E., N. Rajewsky, and E. D. Siggia. 2003. Conservation of regulatory elements between two species of Drosophila. BMC Bioinformatics 4:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erives, A., and M. Levine. 2004. Coordinate enhancers share common organizational features in the Drosophila genome. Proc. Natl. Acad. Sci. USA 101:3851-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flores-Saaib, R. D., and A. J. Courey. 2000. Analysis of Groucho-histone interactions suggests mechanistic similarities between Groucho- and Tup1-mediated repression. Nucleic Acids Res. 28:4189-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujioka, M., Y. Emi-Sarker, G. L. Yusibova, T. Goto, and J. B. Jaynes. 1999. Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients. Development 126:2527-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaston, K., and P. S. Jayaraman. 2003. Transcriptional repression in eukaryotes: repressors and repression mechanisms. Cell Mol. Life Sci. 60:721-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gray, S., and M. Levine. 1996. Transcriptional repression in development. Curr. Opin. Cell Biol. 8:358-364. [DOI] [PubMed] [Google Scholar]
- 32.Gray, S., P. Szymanski, and M. Levine. 1994. Short-range repression permits multiple enhancers to function autonomously within a complex promoter. Genes Dev. 8:1829-1838. [DOI] [PubMed] [Google Scholar]
- 33.Hanes, S. D., G. Riddihough, D. Ish-Horowicz, and R. Brent. 1994. Specific DNA recognition and intersite spacing are critical for action of the Bicoid morphogen. Mol. Cell. Biol. 14:3364-3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hewitt, G. F., B. S. Strunk, C. Margulies, T. Priputin, X.-D. Wang, R. Amey, B. A. Pabst, D. Kosman, J. Reinitz, and D. N. Arnosti. 1999. Transcriptional repression by the Drosophila Giant protein: cis element positioning provides an alternative means of interpreting an effector gradient. Development 126:1201-1210. [DOI] [PubMed] [Google Scholar]
- 35.Janody, F., R. Sturny, V. Schaeffer, Y. Azou, and N. Dostatni. 2001. Two distinct domains of Bicoid mediate its transcriptional downregulation by the Torso pathway. Development 128:2281-2290. [DOI] [PubMed] [Google Scholar]
- 36.Jiang, J., and M. Levine. 1993. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell 72:741-752. [DOI] [PubMed] [Google Scholar]
- 37.Johnston, M., and R. W. Davis. 1984. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol. Cell. Biol. 4:1440-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keller, S. A., Y. Mao, P. Struffi, C. Margulies, C. E. Yurk, A. R. Anderson, R. L. Amey, S. Moore, J. M. Ebels, K. Foley, M. Corado, and D. N. Arnosti. 2000. dCtBP-dependent and -independent repression activities of the Drosophila Knirps protein. Mol. Cell. Biol. 20:7247-7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim, T. K., and T. Maniatis. 1997. The mechanism of transcriptional synergy of an in vitro assembled interferon-beta enhanceosome. Mol. Cell 1:119-129. [DOI] [PubMed] [Google Scholar]
- 40.Koh, S. S., A. Z. Ansari, M. Ptashne, and R. A. Young. 1998. An activator target in the RNA polymerase II holoenzyme. Mol. Cell 1:895-904. [DOI] [PubMed] [Google Scholar]
- 41.Kosman, D., and S. Small. 1997. Concentration-dependent patterning by an ectopic expression domain of the Drosophila gap gene knirps. Development 124:1343-1354. [DOI] [PubMed] [Google Scholar]
- 42.Kulkarni, M. M., and D. N. Arnosti. 2005. Transcriptional enhancers: intelligent enhanceosomes or flexible billboards? J. Cell. Biochem. 94:890-898. [DOI] [PubMed] [Google Scholar]
- 43.Kulkarni, M. M., and D. N. Arnosti. 2003. Information display by transcriptional enhancers. Development 130:6569-6575. [DOI] [PubMed] [Google Scholar]
- 44.La Rosée A., T. Häder, H. Taubert, R. Rivera-Pomar, and H. Jäckle. 1997. Mechanism and Bicoid-dependent control of hairy stripe 7 expression in the posterior region of the Drosophila embryo. EMBO J. 16:4403-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lenhard, B., A. Sandelin, L. Mendoza, P. Engstrom, N. Jareborg, and W. W. Wasserman. 2003. Identification of conserved regulatory elements by comparative genome analysis. J. Biol. 2:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loots, G. G., R. M. Locksley, C. M. Blankespoor, Z. E. Wang, W. Miller, E. M. Rubin, and K. A. Frazer. 2000. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288:136-140. [DOI] [PubMed] [Google Scholar]
- 47.Ludwig, M. Z., C. Bergman, N. H. Patel, and M. Kreitman. 2000. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 403:564-567. [DOI] [PubMed] [Google Scholar]
- 48.Ludwig, M. Z., and M. Kreitman. 1995. Evolutionary dynamics of the enhancer region of even-skipped in Drosophila. Mol. Biol. Evol. 12:1002-1011. [DOI] [PubMed] [Google Scholar]
- 49.Ludwig, M. Z., N. H. Patel, and M. Kreitman. 1998. Functional analysis of eve stripe 2 enhancer evolution in Drosophila: rules governing conservation and change. Development 125:949-958. [DOI] [PubMed] [Google Scholar]
- 50.Majello, B., G. Napolitano, P. De Luca, and L. Lania. 1998. Recruitment of human TBP selectively activates RNA polymerase II TATA-dependent promoters. J. Biol. Chem. 273:16509-16516. [DOI] [PubMed] [Google Scholar]
- 51.Makeev, V. J., A. P. Lifanov, A. G. Nazina, and D. A. Papatsenko. 2003. Distance preferences in the arrangement of binding motifs and hierarchical levels in organization of transcription regulatory information. Nucleic Acids Res. 31:6016-6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Markstein, M., and M. Levine. 2002. Decoding cis-regulatory DNAs in the Drosophila genome. Curr. Opin. Genet. Dev. 12:601-606. [DOI] [PubMed] [Google Scholar]
- 53.Markstein, M., P. Markstein, V. Markstein, and M. S. Levine. 2002. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 99:763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Merika, M., and D. Thanos. 2001. Enhanceosomes. Curr. Opin. Genet. Dev. 11:205-208. [DOI] [PubMed] [Google Scholar]
- 55.Munshi, N., T. Agalioti, S. Lomvardas, M. Merika, G. Chen, and D. Thanos. 2001. Coordination of a transcriptional switch by HMGI(Y) acetylation. Science 293:1133-1136. [DOI] [PubMed] [Google Scholar]
- 56.Nibu, Y., K. Senger, and M. Levine. 2003. CtBP-independent repression in the Drosophila embryo. Mol. Cell. Biol. 23:3990-3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nibu, Y., H. Zhang, and M. Levine. 2001. Local action of long-range repressors in the Drosophila embryo. EMBO J. 20:2246-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pham, A. D., S. Muller, and F. Sauer. 1999. Mesoderm-determining transcription in Drosophila is alleviated by mutations in TAF(II)60 and TAF(II)110. Mech. Dev. 84:3-16. [DOI] [PubMed] [Google Scholar]
- 59.Piano, F., M. J. Parisi, R. Karess, and M. P. Kambysellis. 1999. Evidence for redundancy but not trans factor-cis element coevolution in the regulation of Drosophila Yp genes. Genetics 152:605-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rajewsky, N., M. Vergassola, U. Gaul, and E. D. Siggia. 2002. Computational detection of genomic cis-regulatory modules applied to body patterning in the early Drosophila embryo. BMC Bioinformatics 3:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rebeiz, M., N. L. Reeves, and J. W. Posakony. 2002. SCORE: a computational approach to the identification of cis-regulatory modules and target genes in whole-genome sequence data. Site clustering over random expectation. Proc. Natl. Acad. Sci. USA 99:9888-9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rivera-Pomar, R., and H. Jãckle. 1996. From gradients to stripes in Drosophila embryogenesis: filling in the gaps. Trends Genet. 12:478-483. [DOI] [PubMed] [Google Scholar]
- 63.Ryu, J. R., and D. N. Arnosti. 2003. Functional similarity of Knirps CtBP-dependent and CtBP-independent transcriptional repressor activities. Nucleic Acids Res. 31:4654-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ryu, J. R., L. K. Olson, and D. N. Arnosti. 2001. Cell-type specificity of short-range transcriptional repressors. Proc. Natl. Acad. Sci. USA 98:12960-12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sackerson, C., M. Fujioka, and T. Goto. 1999. The even-skipped locus is contained in a 16-kb chromatin domain. Dev. Biol. 211:39-52. [DOI] [PubMed] [Google Scholar]
- 66.Schroeder, M. D., M. Pearce, J. Fak, H. Fan, U. Unnerstall, E. Emberly, N. Rajewsky, E. D. Siggia, and U. Gaul. 2004. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2:E271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seipel, K., O. Georgiev, and W. Schaffner. 1992. Different activation domains stimulate transcription from remote (′enhancer') and proximal (′promoter') positions. EMBO J. 11:4961-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Senger, K., G. W. Armstrong, W. J. Rowell, J. M. Kwan, M. Markstein, and M. Levine. 2004. Immunity regulatory DNAs share common organizational features in Drosophila. Mol. Cell 13:19-32. [DOI] [PubMed] [Google Scholar]
- 69.Shi, Y., J.-I. Sawada, G. Sui, E. B. Affar, J. R. Whetstine, F. Lan, H. Ogawa, M. P.-S. Luke, Y. Nakatani, and Y. Shi. 2003. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422:735-738. [DOI] [PubMed] [Google Scholar]
- 70.Small, S. 1997. Mechanisms of segmental pattern formation in Drosophila melanogaster, p. 137-178. In J. R. Collier (ed.), Reproductive biology of invertebrates, vol. 7. Progress in developmental biology. John Wiley & Sons, Hoboken, N.J. [Google Scholar]
- 71.Small, S., D. N. Arnosti, and M. Levine. 1993. Spacing ensures autonomous expression of different stripe enhancers in the even-skipped promoter. Development 119:762-772. [PubMed] [Google Scholar]
- 72.Small, S., A. Blair, and M. Levine. 1992. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 11:4047-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Small, S., A. Blair, and M. Levine. 1996. Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Dev. Biol. 175:314-324. [DOI] [PubMed] [Google Scholar]
- 74.Song, H., P. Hasson, Z. Paroush, and A. J. Courey. 2004. Groucho oligomerization is required for repression in vivo. Mol. Cell. Biol. 24:4341-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Struffi, P., M. Corado, M. Kulkarni, and D. N. Arnosti. 2004. Quantitative contributions of CtBP-dependent and -independent repression activities of Knirps. Development 131:2419-2429. [DOI] [PubMed] [Google Scholar]
- 76.Struhl, G. 1989. Morphogen gradients and the control of body pattern in insect embryos. Ciba Found. Symp. 144:65-98. [DOI] [PubMed] [Google Scholar]
- 77.Strunk, B., P. Struffi, K. Wright, B. Pabst, J. Thomas, L. Qin, and D. N. Arnosti. 2001. Role of CtBP in transcriptional repression by the Drosophila giant protein. Dev. Biol. 239:229-240. [DOI] [PubMed] [Google Scholar]
- 78.Subramanian, T., and G. Chinnadurai. 2003. Association of class I histone deacetylases with transcriptional corepressor CtBP. FEBS Lett. 540:255-258. [DOI] [PubMed] [Google Scholar]
- 79.Sutrias-Grau, M., and D. N. Arnosti. 2004. CtBP contributes quantitatively to Knirps repression activity in an NAD binding-dependent manner. Mol. Cell. Biol. 24:5953-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thanos, D., and T. Maniatis. 1995. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 83:1091-1100. [DOI] [PubMed] [Google Scholar]
- 81.Zhou, J., J. Zwicker, P. Szymanski, M. Levine, and R. Tjian. 1998. TAFII mutations disrupt Dorsal activation in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 95:13483-13488. [DOI] [PMC free article] [PubMed] [Google Scholar]