Abstract
The 26S proteasome, composed of the 20S core and the 19S regulatory complex, plays a central role in ubiquitin-dependent proteolysis by catalyzing degradation of polyubiquitinated proteins. In a search for proteins involved in regulation of the proteasome, we affinity purified the 19S regulatory complex from HeLa cells and identified a novel protein of 43 kDa in size as an associated protein. Immunoprecipitation analyses suggested that this protein specifically interacted with the proteasomal ATPases. Hence the protein was named proteasomal ATPase-associated factor 1 (PAAF1). Immunoaffinity purification of PAAF1 confirmed its interaction with the 19S regulatory complex and further showed that the 19S regulatory complex bound with PAAF1 was not stably associated with the 20S core. Overexpression of PAAF1 in HeLa cells decreased the level of the 20S core associated with the 19S complex in a dose-dependent fashion, suggesting that PAAF1 binding to proteasomal ATPases inhibited the assembly of the 26S proteasome. Proteasomal degradation assays using reporters based on green fluorescent protein revealed that overexpression of PAAF1 inhibited the proteasome activity in vivo. Furthermore, the suppression of PAAF1 expression that is mediated by small inhibitory RNA enhanced the proteasome activity. These results suggest that PAAF1 functions as a negative regulator of the proteasome by controlling the assembly/disassembly of the proteasome.
The ubiquitin-dependent proteolysis regulates various physiological processes, such as cell cycle progression and signal transduction (8, 12). The 26S proteasome, the major proteolytic enzyme found in eukaryotic cells, plays a key role in the ubiquitin-dependent proteolysis by degrading proteins conjugated to ubiquitin. The 26S proteasome consists of a 20S proteolytic core particle and 19S regulatory complexes (also known as PA700), which bind to the ends of the 20S core (24, 33). The 20S particle has a barrel-shaped structure composed of two outer α rings and two inner β rings, each of which contains seven homologous subunits (10). The α subunits are catalytically inactive, whereas three of the seven β subunits are catalytically active with the active sites sequestered within the central chamber (24, 33). The α rings provide attachment sites for the regulatory complexes, such as 19S particle and 11S activator, and control the access of substrates to the core particle's catalytic chamber by functioning as a gated channel (9, 34).
The 20S core particle alone can degrade small peptides and fully denatured small proteins in an ATP-independent fashion. In contrast, degradation of ubiquitinated proteins is ATP dependent and requires the 19S regulatory particle in addition to the 20S core. The 19S regulatory particle is presumed to recognize polyubiquitin-linked proteins, remove the ubiquitin chain from the substrate, unfold the attached substrate, and translocate the substrate into the 20S core particle's catalytic chamber (8, 24). Recent biochemical and genetic studies have begun to identify specific subunits that carry out different functions of the 19S particle. For instance, Rpn11 has been shown to be responsible for substrate deubiquitination (20, 31, 37), while S6′/Rpt5 has been reported to function in ATP-modulated polyubiquitin recognition (17). The 19S particle contains six proteasomal ATPases, which are thought to assemble into a six-membered ring that directly touches the α ring of the 20S core particle. This proteasomal ATPase ring is proposed to mediate both unfolding and translocation of the substrate. Recent studies have suggested that proteasomal ATPases also function in opening the gate of the 20S core and that Rpt2 is particularly important in this process (15).
As expected from its central role in ubiquitin-dependent proteolysis, the proteasome has been reported to interact with various proteins that function in the ubiquitin-proteasome pathway, such as ubiquitin ligases (30, 36, 38), deubiquitinating enzymes (1, 18, 23), and delivery factors for ubiquitin conjugates (14, 26). Recently, affinity purification of the proteasome coupled with mass spectrometric analysis has led to the identification of novel proteasome subunits and proteasome-associated proteins in budding yeast (19, 32). In an effort to search for proteins regulating the ubiquitin-proteasome pathway, we have affinity purified the proteasome from HeLa cells and identified specifically associated proteins. In this report, we present identification of a novel protein that interacts with proteasomal ATPases and demonstrate that it negatively regulates the proteasome activity in vivo by affecting the assembly/disassembly of the 26S proteasome.
MATERIALS AND METHODS
Plasmids.
The cDNAs encoding human proteasomal ATPase-associated factor 1 (PAAF1)/FLJ11848 (accession no. BC006142), proteasome subunit β4/C7 (BC014488), S2/Rpn1 (BC002368), S11/Rpn9 (BC001100), S7/Rpt1 (D11094), S4/Rpt2 (BC000512), S6/Rpt3 (BC014488), S10b/Rpt4 (BC005390), SUG1/S8/Rpt6 (BE795619), CSN7 (BC011789), RuvB2 (BC000519) and mouse S6′/Rpt5 (BC005783) were obtained from the Medical Research Council (United Kingdom) gene service. UbG76V-GFP and Ub-R-GFP expression constructs (a kind gift from N. P. Dantuma) were previously described (5). To construct plasmids for the expression of epitope-tagged proteins, cDNAs were amplified by PCR with appropriate primers and ligated into pcDNA3.1 (Invitrogen) or pYR vectors (21).
Affinity purification of the proteasome.
Cells derived from HeLa Tet-Off (Clontech) cells stably expressing EBNA-1 were transfected with an episomal expression vector, pYR-FLAG-SUG1 or pYR-FLAG-PAAF1, that contained the gene of interest under the tetracycline-regulated promoter, oriP for episome replication, and the selection marker for hygromycin B. The cells were selected and maintained in Dulbecco modified Eagle’s medium (Life Technologies, Inc.) supplemented with 10% fetal bovine serum (Gibco BRL), 100 μg/ml G418 (Sigma), 300 μg/ml hygromycin B (Clontech), 100 units/ml penicillin, 100 μg/ml streptomycin, 1 mM l-glutamine, and 2 μg/ml tetracycline (Sigma). To induce the expression of FLAG-tagged proteins, cells were grown without tetracycline for 2 days. Nuclear extracts and cytosolic S100 extracts were prepared as described previously (6). Extracts were dialyzed against buffer BC (20 mM Tris-HCl [pH 7.9], 15% glycerol, 1 mM EDTA, 1 mM dithiothreitol, 0.2 mM phenylmethanesulfonyl fluoride, 0.05% NP-40) containing 150 mM KCl (BC150) and rotated with anti-FLAG M2 agarose (Sigma) at 4°C for 3 to 6 h. After extensive washes with BC150, proteins were eluted with 0.3 mg of FLAG peptide per ml in BC150.
Protein identification by mass spectrometry.
Immunopurified protein complexes were resolved on sodium dodecyl sulfate (SDS)-polyacrylamide gels with a gradient of 4 to 20% polyacrylamide (Novex). After the gels were stained first with SYPRO Ruby (Bio-Rad) and then with colloidal Coomassie blue, protein bands were excised and digested with trypsin as previously described (35). In-gel tryptic digests of proteins were analyzed by nanoelectrospray liquid chromatography tandem mass spectrometry (LC-MS-MS). High-pressure liquid chromatography separation was performed on an Ultimate instrument equipped with Famous Autosampler (LC packings) at a flow rate of 200 nl/min. The columns were constructed from fused silica capillary tubes (outer diameter of 360 μm and inner diameter of 75 μm) and packed with 300 Å C18 beads (diameter, 5 μm) (Grace Vydac) to a length of 15 cm. The outlet of the nanocolumn was in-line connected to a distal coated silica PicoTip needle (New Objective). The solvent system consisted of solvent A (0.1% formic acid, 5% acetonitrile) and solvent B (0.1% formic acid, 90% acetonitrile). The gradient was linear from 0% to 40% solvent B in 50 min followed by 90% solvent B in 5 min. Tandem mass spectra were recorded on an API QSTAR Pulsar Q-TOF mass spectrometer (Applied Biosystems) in the information-dependent acquisition mode. The MS-MS spectra were used to search the National Center for Biotechnology Information (NCBI) nonredundant and expressed sequence tag databases using the computer algorithm proID (Applied Biosystems, Framingham, MA).
Immunoprecipitations and Western blotting.
Transfection was performed using the polyethylenenimine methodology (2). After 36 h, cells were lysed in buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 0.2 mM phenylmethanesulfonyl fluoride, and 1.0% NP-40. Cell lysates were adjusted to 0.1% NP-40 and incubated with anti-FLAG or anti-HA antibody resin (Sigma) for 4 h at 4°C. The immune complexes were recovered by low-speed centrifugation, and the resin was washed extensively with the binding buffer with 0.1% NP-40 and then eluted with buffer containing 20 mM Tris-HCl (pH 8.0) and 2% SDS. Immunoprecipitated proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose membrane (Bio-Rad) and visualized by Western blotting with the enhanced chemiluminescence reagents (Amersham Pharmacia). For Western blotting, we used antibodies against FLAG (Sigma), hemagglutinin (HA) (Babco), T7 (Novagen), SUG1 (Affiniti), 20S proteasome α-core (Affiniti), 20S proteasome subunit β1 (Affiniti), 20S proteasome subunit Y/β4 (Affiniti), and green fluorescent protein (GFP) (Santa Cruz).
Flow cytometry.
To measure UbG76V-GFP and Ub-R-GFP accumulation by flow cytometry, cells were harvested by trypsinization, and fluorescence data were acquired with FACScan flow cytometer (Becton Dickinson) and analyzed with the Cellquest software. Background fluorescence and autofluorescence were determined using mock-treated cells. A total of 30,000 events per sample were considered.
RNA interference.
The target sequence for PAAF1-specific small inhibitory RNA (siRNA) was determined by the Ambion web-based criteria. PAAF1-specific siRNA was generated using the Silencer siRNA construction kit (Ambion). Two 29-mer DNA oligonucleotides (sense, 5′-AAA TCT TCA GCT GAC CAT ATC CCT GTC TC-3′; antisense, 5′-AAG ATA TGG TCA GCT GAA GAT CCT GTC TC-3′) with 21 nucleotides encoding the siRNA and 8 nucleotides complementary to the T7 promoter primer were synthesized. The two siRNA templates were hybridized to a T7 promoter primer and were extended by the Klenow DNA polymerase. The extended sense and antisense siRNA templates were then transcribed by T7 RNA polymerase and hybridized to create double-stranded siRNA following the manufacturer's instructions. The control siRNA was obtained using the two siRNA templates with a transversion at every third position of the target sequence (sense, 5′-AAG TCA TCT GCA GAT CAC ATT CCT GTC TC-3′; antisense, 5′-AAA ATT TCG TCT GCA GAG GAC CCT GTC TC-3′). Twenty picomoles of siRNA was transfected to HeLa cells grown on six-well plates with Lipofectamine (Life Technologies, Inc.). Twelve hours later, the expression construct for UbG76V-GFP was transfected into HeLa cells. After incubation for additional 48 h, reverse transcription-PCR (RT-PCR) analysis was performed to assess the effectiveness of RNA interference.
RT-PCR.
HeLa cells were lysed in Trizol reagent (Invitrogen) and total RNA was extracted according to the manufacturer's instructions by a modified version of the method of Chomczynski and Sacchi (4). RNA content was calculated by measuring A260. First-strand cDNA synthesis was performed using Superscript II RNase H− reverse transcriptase kit (Invitrogen) by the standard protocol of the supplier. The resulting cDNA was used as a template to amplify the PAAF1 and CSN2 transcripts with the following primers: PAAF1 forward, 5′-ATGGCGGCGCCTTTGA-3′; PAAF1 reverse, 5′-TCAGAGGTCAGAAAGCTG-3′; CSN2 forward, 5′-CTATGTCTGACATGGAGGAT-3′; CSN2 reverse, 5′-GAGTTAAGCCAGTTTACTGACTA-3′. PCR was subsequently performed using an optimized protocol consisting of 20 cycles. Cycling parameters were as follows: 30 s at 94°C; 30 s at 55°C; and 1 min at 72°C. The final products were separated by 0.7% agarose gel electrophoresis and visualized by ethidium bromide staining.
RESULTS
Affinity purification of the proteasome.
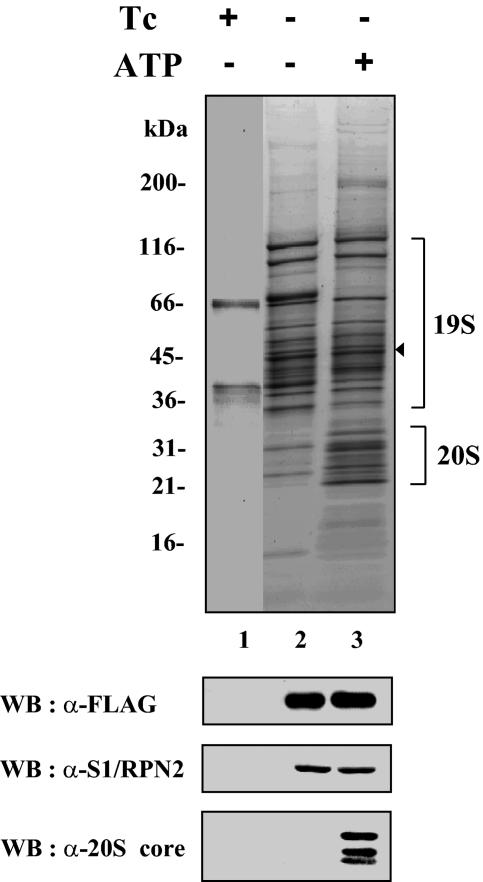
To isolate the proteasome, we established a HeLa-derived cell line that conditionally expressed FLAG-SUG1, a FLAG-tagged version of SUG1 (also known as S8 and Rpt6), an ATPase subunit of the 19S regulatory complex. FLAG-SUG1 and its associated proteins were then purified from extracts of induced and uninduced cells by an immunoaffinity purification procedure using beads immobilized by anti-FLAG antibody (21). Since ATP is required for stable interaction between the 20S core and the 19S regulatory complex, preparation of cell extracts and subsequent affinity purification were performed in the presence or absence of 2 mM ATP to obtain the 26S proteasome or the 19S complex, respectively. SDS-PAGE analysis of purified proteins and Western blotting with antibodies to 19S and 20S subunits revealed that subunits of the 19S regulatory complex were obtained with or without ATP, while 20S core subunits copurified with 19S subunits only in the presence of ATP (Fig. 1). These data indicated that depending on the ATP level, the 19S regulatory complex or the 26S proteasome could be purified from FLAG-SUG1-expressing cells.
FIG. 1.
Immunoaffinity purification of FLAG-SUG1. Cells expressing FLAG-SUG1 in a tetracycline-controlled manner were established as described in Materials and Methods. Cells were grown in the presence (+) or absence (−) of tetracycline (Tc). Immunoaffinity purification of FLAG-SUG1 was performed in the presence (+) or absence (−) of 2 mM ATP. Ten microliters each of purified FLAG-SUG1 samples was separated by a SDS-polyacrylamide gel (gradient of 4 to 20% polyacrylamide) and visualized by Sypro Ruby-Coomassie blue double staining. Protein size markers (in kDa) are indicated to the left of the gel. The location of FLAG-SUG1 is indicated by the arrowhead to the right of the gel. The presence of proteasome subunits was analyzed by immunoblotting of the purified samples with the indicated antibodies. WB, Western blotting; α-FLAG, anti-FLAG.
Identification of PAAF1 in the 19S regulatory complex.
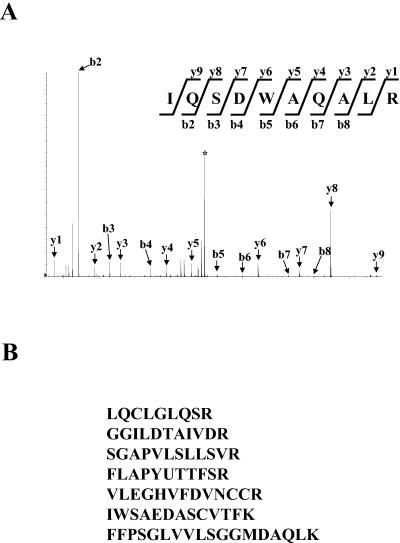
To identify proteins that copurified with FLAG-SUG1, FLAG-SUG1 preparations obtained in the presence or the absence of ATP were digested with trypsin. Peptide mixtures were then subjected to reversed-phase high-performance liquid chromatography coupled with electrospray tandem mass (MS-MS) spectrometry. Database searches with MS-MS spectra identified most subunits of the proteasome and confirmed that subunits of 19S regulatory complex were found in both preparations of FLAG-SUG1, whereas subunits of 20S core were present mainly in the preparation obtained with ATP (data not shown). In addition to peptides derived from proteasomal subunits, those from other proteins were identified by mass spectrometric analyses, including heat shock proteins, cytoskeletal proteins, proteins involved in translation, and a hypothetical protein FLJ11848 with the calculated molecular size of 43 kDa (data not shown). We named this novel protein proteasomal ATPase-associated factor 1 (PAAF1) for the reasons described below. Since we obtained the sequence of only one peptide from PAAF1 with this procedure, we repeated anti-FLAG affinity purification and analyzed purified proteins by SDS-PAGE. After the gel was stained, a gel slice that contained protein bands with the size of 40 to 45 kDa was excised, and the proteins were digested in the gel with trypsin. Subsequent analysis of resultant tryptic peptides by LC-MS-MS identified seven peptides derived from PAAF1 (Fig. 2), confirming that PAAF1 copurified with FLAG-SUG1. PAAF1 was identified from both FLAG-SUG1 preparations purified with and without ATP (data not shown). These results suggested that PAAF1 interacted with the 19S regulatory particle.
FIG. 2.
Identification of PAAF1 in the purified FLAG-SUG1 preparation. (A) MS-MS spectrum of a peptide derived from PAAF1. MS-MS analyses were performed as described in Materials and Methods. The b and y ions detected are marked with arrows on the spectra. The precursor ion is indicated by the asterisk. (B) Sequences determined by MS-MS analyses of tryptic peptides obtained from PAAF1. All peptides were identical to sequences in human PAAF1.
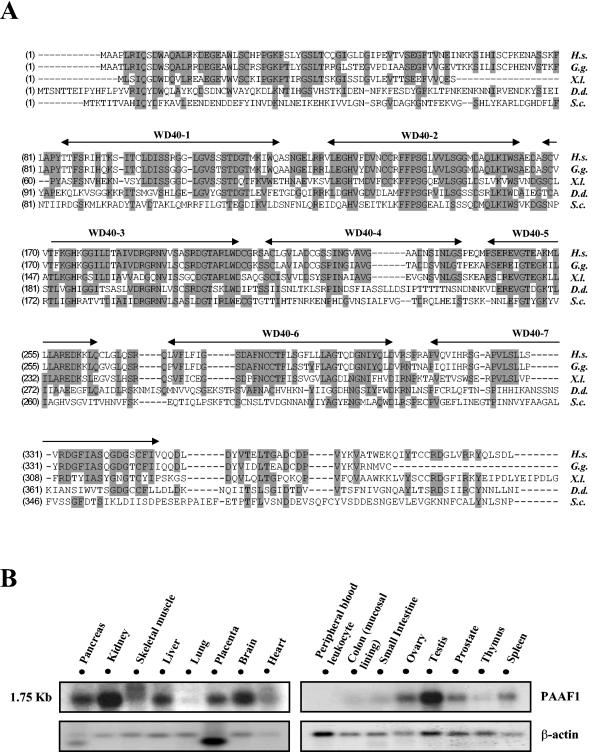
Human PAAF1 cDNA contains a gene or open reading frame encoding 392 amino acid residues (accession number BC006142). To identify functionally important regions of PAAF1, we performed a BLAST search using the amino acid sequence for human PAAF1 as a query. Figure 3A shows alignment of the human PAAF1 sequence with those from other species. There is a significant similarity between the human PAAF1 sequence and the homologous protein from Saccharomyces cerevisiae, which have 21% amino acid identity and 34% similarity. NCBI conserved domain search revealed that human PAAF1 contains seven WD repeats (Fig. 3A), which are found in a number of eukaryotic proteins involved in a variety of cellular processes. Structural studies have shown that WD repeats form a series of four-stranded antiparallel β sheets, which fold into a higher-order structure termed a β-propeller (27). WD repeats in PAAF1 are expected to function as a site for protein-protein interaction, as those in other proteins are known to do.
FIG. 3.
Sequence alignment and expression pattern of PAAF1. (A) Sequence alignment of human PAAF1 with homologous proteins from other species. Amino acid sequences of PAAF1-related proteins from different species were aligned using ClustalW. Identical residues are shaded gray. WD40 motifs are indicated. Abbreviations and accession numbers of homologous proteins are as follows: Homo sapiens (H.s.), AAH21541; Gallus gallus (G.g.), CAG31874; Xenopus laevis (X.l.), AAH70729; Dictyostelium discoideum (D.d.), AAM43783; Saccharomyces cerevisiae (S.c.), AAT92618. (B) Northern blot analysis of human PAAF1. The blots containing mRNA from the indicated tissues were probed with 32P-labeled PAAF1 cDNA (top blots), stripped, and reprobed with 32P-labeled actin cDNA (bottom blots).
We determined the expression profile of PAAF1 in various human tissues by Northern analysis, in which a multiple tissue Northern blot was hybridized to human PAAF1 probe. These analyses reveal one major PAAF1 transcript of approximately 1.75 kb in human tissues. PAAF1 is ubiquitously expressed; it is expressed highly in kidney, brain, and testis; moderately in pancreas, skeletal muscle, liver, placenta, and heart; and at a low level in lung, thymus, colon, small intestine, and peripheral blood leukocytes (Fig. 3B).
PAAF1 stably interacts with the 19S regulatory complex but not with the 26S proteasome.
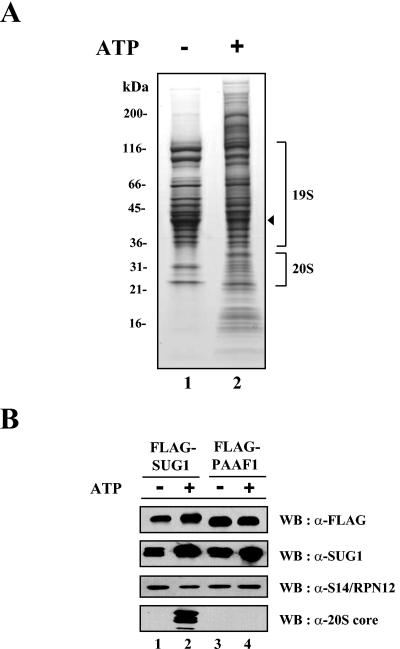
To confirm the association of PAAF1 with the 19S regulatory complex, we generated a cell line conditionally expressing FLAG-PAAF1 and immunopurified the epitope-tagged protein using anti-FLAG antibodies in the presence or absence of ATP. SDS-PAGE of purified proteins revealed that a group of proteins copurified with FLAG-PAAF1 (Fig. 4A). Mass spectrometric analyses of purified proteins and Western blotting with antibodies against 19S subunits, SUG1 and S14/Rpn12, showed that PAAF1 was stably associated with the 19S regulatory complex (Fig. 4B and data not shown). To our surprise, however, subunits of the 20S core complex were not found by Western blotting and by mass spectrometric analyses in FLAG-PAAF1 preparations obtained with or without ATP (Fig. 4B and data not shown). Since immunoaffinity purification of FLAG-SUG1 under similar experimental conditions revealed that subunits of the 20S core copurified with FLAG-SUG1 and other subunits of the 19S regulatory complex in the presence of ATP, these data suggested that the association of PAAF1 with the 19S regulatory complex might interfere with the stable interaction between the 19S complex and the 20S core. Interestingly, compared to the FLAG-PAAF1 preparations obtained in the absence of ATP, the preparations obtained in the presence of ATP consistently contained more proteins, such as the translation initiation factor eIF3 complex (Fig. 4A and data not shown). The significance of this finding was not further studied.
FIG. 4.
Immunoaffinity purification of FLAG-PAAF1. (A) SDS-PAGE analysis of purified FLAG-PAAF1. Ten microliters each of FLAG-PAAF1 samples purified in the presence (+) or absence (−) of 2 mM ATP was separated by electrophoresis on an SDS-polyacrylamide gel (gradient of 4 to 20% polyacrylamide) and visualized by Sypro Ruby-Coomassie blue double staining. Protein size markers (in kDa) are indicated to the left of the gel. The location of FLAG-PAAF1 is shown by the arrowhead to the right of the gel. (B) Western blotting (WB) analysis of purified FLAG-PAAF1. FLAG-PAAF1 and FLAG-SUG1 samples purified in the presence (+) or absence (−) of ATP were analyzed by immunoblotting with indicated antibodies. α-FLAG, anti-FLAG.
PAAF1 interacts with the proteasomal ATPases.
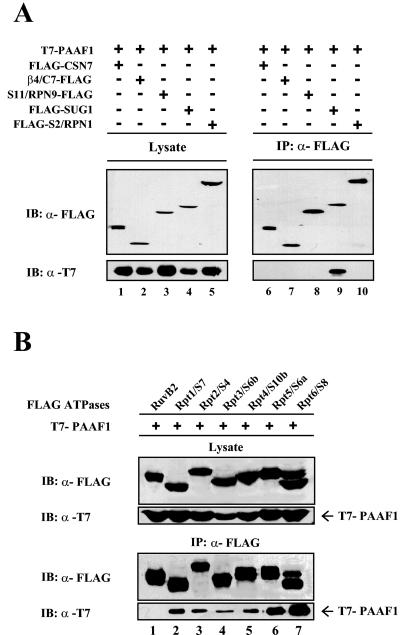
To identify the proteasome subunit that directly interacts with PAAF1, HeLa cells were cotransfected with expression constructs of T7-PAAF1 and a FLAG-tagged proteasome subunit β4/C7, S2/Rpn1, S11/Rpn9, or SUG1, which is a component of the 20S core, the base of the 19S complex, the lid of the 19S complex, or the proteasomal ATPase ring, respectively. FLAG-CSN7A, a subunit of COP9 signalosome, was included in the coimmunoprecipitation experiments as a negative control. FLAG-tagged proteins were then immunoprecipitated with anti-FLAG antibodies, and coprecipitation of T7-PAAF1 was monitored by immunoblotting with anti-T7 antibodies. As shown in Fig. 5A, PAAF1 coimmunoprecipitated with SUG1, but not with CSN7A or with the other proteasome subunits tested. Since S2/Rpn1 and S11/Rpn9 subunits of the19S regulatory complex did not interact with PAAF1, these data suggested that the association between overexpressed SUG1 and PAAF1 was direct and was not mediated by the incorporation of SUG1 into the 19S regulatory complex.
FIG. 5.
PAAF1 interacts with proteasomal ATPases. (A) Immunoprecipitations of proteasome subunits. HeLa cells were transiently transfected with plasmid vectors expressing T7-PAAF1 and a FLAG-tagged proteasome subunit or CSN7A as indicated. Cell lysates were immunoprecipitated (IP) with beads coupled to anti-FLAG antibody (α-FLAG). The immunoprecipitates were subjected to SDS-PAGE and analyzed by Western blotting with indicated antibodies. IB, immunoblotting. (B) Immunoprecipitations of proteasomal ATPases. HeLa cells were transiently transfected with expression constructs for T7-PAAF1 and a FLAG-tagged proteasomal ATPase subunit or RuvB2 as indicated. Lysates were prepared 36 h after transfection and immunoprecipitated (IP) with anti-FLAG (α-FLAG) antibodies. The precipitates were separated by SDS-PAGE and immunoblotted (IB) with the indicated antibodies.
In addition to SUG1, the 19S regulatory complex contains five other highly related ATPases, which form a subfamily among the AAA ATPase superfamily of proteins (22). To assess the specificity of the interaction between SUG1 and PAAF1, HeLa cells were cotransfected with expression constructs of T7-PAAF1 and each FLAG-tagged proteasomal ATPase subunit. The immunoprecipitation experiments were performed with anti-FLAG antibodies. Western blotting of the immunoprecipitates with anti-T7 antibodies revealed that all the proteasomal ATPases interacted appreciably with PAAF1 with the interaction of SUG1 appearing the strongest (Fig. 5B). In contrast, RuvB2, a member of the AAA ATPase superfamily, did not coprecipitate with PAAF1, indicating that the interaction of proteasomal ATPases with PAAF1 was specific.
PAAF1 inhibits the association of 19S regulatory complex with the 20S core.
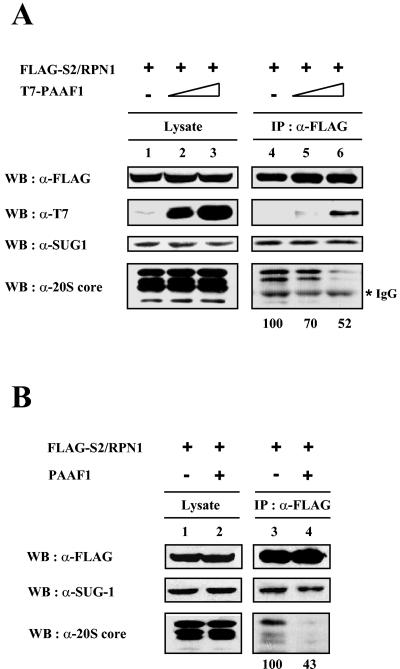
Since PAAF1 appeared to interact with SUG1 and other proteasomal ATPases, which are in direct contact with the α ring of the 20S core in the 26S proteasome, and since PAAF1 immunopurified in the presence of ATP did not contain an appreciable amount of the 20S core, we reasoned that PAAF1 might affect the binding of the proteasomal ATPases to the 20S core. To test this possibility, HeLa cells were transfected with expression constructs of T7-PAAF1 and FLAG-S2/Rpn1, a subunit of 19S regulatory particle that did not directly interact with PAAF1. FLAG-S2/Rpn1 was then immunoprecipitated with anti-FLAG antibodies in the presence of ATP, and copurified proteins were probed by immunoblotting with suitable antibodies (Fig. 6A). Overexpression of T7-PAAF1 did not affect coprecipitation of SUG1 with FLAG-S2/Rpn1. In contrast, subunits of the 20S core coprecipitated with S2/Rpn1 were reduced by the expression of exogenous PAAF1 in a dose-dependent manner, suggesting that T7-PAAF1 inhibited the association of the 19S regulatory complex with the 20S core. To exclude the possibility that this inhibition was due to the N-terminal tag of PAAF1, we repeated experiments with the expression construct of PAAF1 with no tag. As shown in Fig. 6B, overexpression of PAAF1 without any tag also reduced subunits of the 20S core in the FLAG-S2/Rpn1 immunoprecipitates. Collectively, theses data suggested that PAAF1 binding to proteasomal ATPases interfered with their association with the α ring of the 20S core, leading to inhibition of the 26S proteasome assembly.
FIG. 6.
PAAF1 interferes with the binding of 19S regulatory complex to 20S core. HeLa cells were transiently transfected with FLAG-S2/Rpn1 and T7-tagged (A) or untagged (B) PAAF1. Lysates were prepared 36 h after transfection and immunoprecipitated (IP) with anti-FLAG (α-FLAG) antibodies. The precipitates were separated by SDS-PAGE and immunoblotted with the indicated antibodies. Coprecipitated 20S α-core bands were quantified by Las-3000 luminescent image analyzer (Fuji film). WB, Western blotting; IgG, immunoglobulin G.
PAAF1 inhibits the proteasome activity in vivo.
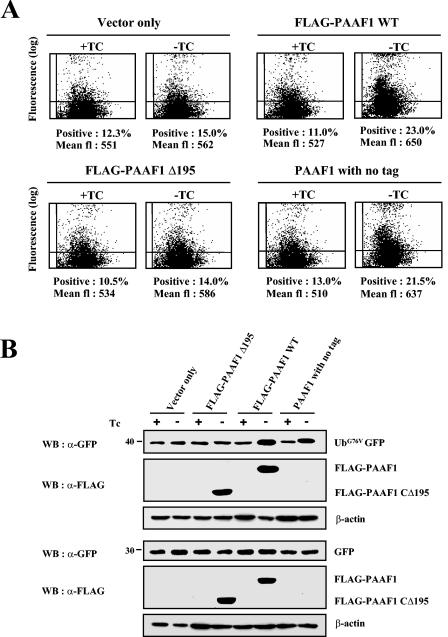
To assess the functional consequence of association of PAAF1 with proteasomal ATPases, we examined the effect of PAAF1 on the proteasome activity in vivo. Recently characterized GFP-based reporters (5), the ubiquitin fusion degradation pathway substrate (UbG76V-GFP) and the N-end rule substrate (Ub-R-GFP), were used to monitor the proteasome-dependent degradation. HeLa Tet-off cells (Clontech) were transiently cotransfected with the expression construct of UbG76V-GFP and with the expression vector for FLAG-PAAF1 under the control of a tetracycline-induced promoter. The expression level of UbG76V-GFP was then analyzed by the GFP fluorescence.
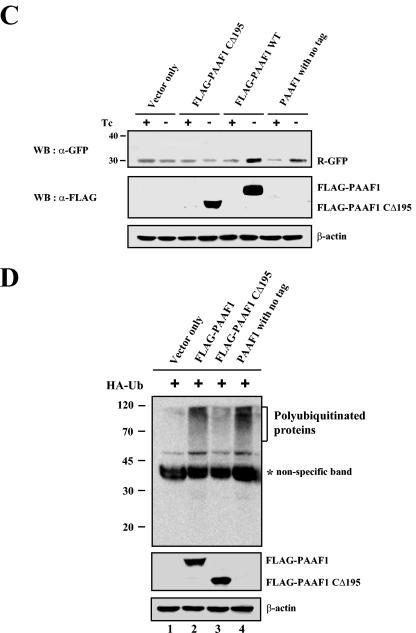
As shown in Fig. 7A, the induction of FLAG-PAAF1 by removal of tetracycline resulted in accumulation of the UbG76V-GFP, suggesting that FLAG-PAAF1 inhibited the proteasome activity. In contrast, FLAG-PAAF1CΔ195, a PAAF1 mutant with the deletion of the C-terminal 195 amino acids, which renders it incapable of binding to SUG1 (data not shown), did not increase GFP fluorescence after its induction, suggesting that the binding of PAAF1 to proteasomal ATPases was required for its inhibition of proteasome activity. Importantly, increase of GFP fluorescence was also observed with PAAF1 without any tag. These data indicated that inhibition of the proteasome activity did not result from the epitope tag of PAAF1. The expression level of UbG76V-GFP was directly monitored by Western blotting with anti-GFP antibodies, which confirmed that UbG76V-GFP was increased by FLAG-PAAF1 or untagged PAAF1, but not by FLAG-PAAF1CΔ195 (Fig. 7B). The effect of PAAF1 on UbG76V-GFP appeared specific, since the expression of GFP was not affected by PAAF1 (Fig. 7B, lower panels). When the N-end rule substrate Ub-R-GFP was used to measure the proteasome activity, FLAG-PAAF1 or untagged PAAF1 also increased both the GFP fluorescence (data not shown) and the expression level of R-GFP as determined by immunoblotting analysis (Fig. 7C). Since inhibition of proteasome activity should result in the increased level of ubiquitinated proteins, we also analyzed the effect of PAAF1 on accumulation of ubiquitinated proteins. HeLa cells were transfected with the expression constructs of HA-ubiquitin and PAAF1 derivatives, and ubiqitinated proteins were measured by immunoblotting with anti-HA antibodies. As shown in Fig. 7D, overexpression of FLAG-PAAF1 or untagged PAAF1 increased the level of ubiquitinated proteins but that of FLAG-PAAF1CΔ195 did not. These data suggested that inhibition by PAAF1 was not restricted to degradation of GFP-based proteasome substrates but degradation of ubiquitinated proteins in general was affected by PAAF1.
FIG. 7.
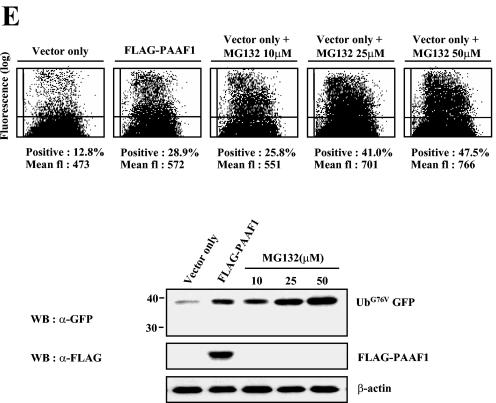
PAAF1 inhibits the proteasome activity. (A) Flow cytometric analysis of UbG76V-GFP. HeLa Tet-off cells (Clontech) were transfected with the expression construct of UbG76V-GFP and with the expression vector for PAAF1 under the control of a tetracycline-inducible promoter as indicated. Cells were incubated in the presence (+) or absence (−) of tetracycline (TC) for 48 h. The GFP fluorescence (fl) was then analyzed by flow cytometry. WT, wild type. (B) Immunoblotting analyses of UbG76V-GFP and GFP. HeLa Tet-off cells were transfected with the expression construct of UbG76V-GFP (top blots) or GFP (bottom blots) and with the expression vector for PAAF1 under the control of a tetracycline-inducible promoter as indicated. After induction of PAAF1 derivatives for 48 h, the expression levels of UbG76V-GFP and GFP were determined by immunoblotting with anti-GFP (α-GFP) antibodies. WB, Western blotting; Tc, tetracycline. (C) Immunoblotting analysis of Ub-R-GFP. HeLa Tet-off cells were transfected with the expression construct of Ub-R-GFP and with the expression vector for PAAF1 under the control of a tetracycline-inducible promoter as indicated. After induction of PAAF1 derivatives for 48 h, the expression levels of Ub-R-GFP were determined by immunoblotting with anti-GFP (α-GFP) antibodies. WB, Western blotting; Tc, tetracycline. (D) Immunoblotting analysis of polyubiquitinated proteins. HeLa cells were transfected with the expression constructs of HA-Ub and FLAG-PAAF1 as indicated. Polyubiqitinated proteins were analyzed by immunoblotting with anti-HA antibodies. The asterisk indicates a nonspecific band detected in whole-cell lysates by anti-HA antibodies. (E) Comparison of the inhibition of proteasome activity by PAAF1 overexpression with that by the proteasome inhibitor MG132. HeLa cells were transfected with the expression construct of UbG76V-GFP and with the expression vector for PAAF1 or the empty vector as indicated. Thirty-six hours after transfection, cells transfected with the empty vector were treated for 10 h with 0, 10, 25, or 50 μM MG132 as indicated. The expression level of UbG76V-GFP was then analyzed by flow cytometry and by immunoblotting with anti-GFP (α-GFP) antibodies. fl, fluorescence; WB, Western blotting.
To evaluate the level of inhibition of proteasome activity by PAAF1, we compared stabilization of UbG76V-GFP by PAAF1 with that by the proteasome inhibitor MG132. The extent of UbG76V-GFP accumulation by PAAF1 overexpression was comparable to that after treating HeLa cells with 10 μM MG132 for 10 h, as measured by the GFP fluorescence and by Western blotting with anti-GFP antibodies (Fig. 7E). The inhibition of proteasome activity by overexpression of PAAF1, however, appeared to have little effect on the cell growth for the period of transient transfection, since the rate of thymidine incorporation was not appreciably affected (data not shown).
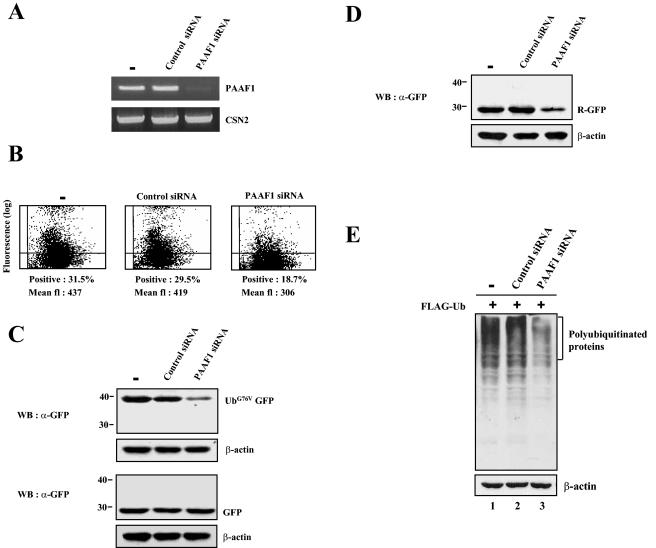
To further examine the modulation of proteasome activity by PAAF1, we used siRNA to suppress PAAF1 expression. The efficiency of siRNA-mediated PAAF1 suppression was monitored by RT-PCR. PAAF1 expression was greatly diminished only in cells transfected with PAAF1 siRNA, but not in mock-treated cells or in cells transfected with control siRNA with a transversion at every third position of PAAF1 siRNA (Fig. 8A). CSN2 expression was not affected by siRNA, indicating the specificity of PAAF1 siRNA.
FIG. 8.
Proteasome activity is enhanced by siRNA-mediated PAAF1 suppression. (A) siRNA-mediated suppression of PAAF1 mRNA. HeLa cells were transfected with control or PAAF1-specific siRNA as indicated. Total cellular RNA was isolated 48 h after transfection and subjected to RT-PCR analysis using specific primers for PAAF1 and CSN2 mRNA. (B) Flow cytometric analysis of UbG76V-GFP. HeLa cells were transfected with control or PAAF1-specific siRNA as indicated. Twelve hours later, the expression construct for UbG76V-GFP was transfected into siRNA-treated HeLa cells. After incubation for 48 h, the GFP fluorescence (fl) was analyzed by flow cytometry. (C) Immunoblotting analysis of UbG76V-GFP. Transfection experiments were performed as described in the legend to panel B. The expression levels of UbG76V-GFP were determined by immunoblotting with anti-GFP (α-GFP) antibodies. WB, Western blotting. (D) Immunoblotting analysis of Ub-R-GFP. Transfection experiments were performed with the expression construct for Ub-R-GFP as described in the legend to panel B. (E) Immunoblotting analysis of polyubiquitinated proteins. HeLa cells were transfected with control or PAAF1-specific siRNA as indicated. Twelve hours later, the expression construct for FLAG-Ub was transfected into siRNA-treated HeLa cells. After incubation for 48 h, polyubiqitinated proteins were analyzed by immunoblotting with anti-FLAG antibodies.
To examine the effect of PAAF1 suppression on proteasome activity, we transfected siRNA-treated cells with the expression construct for UbG76V-GFP or Ub-R-GFP and analyzed its expression by GFP fluorescence (Fig. 8B and data not shown) and by immunoblotting with anti-GFP antibodies (Fig. 8C and D). The GFP fluorescence and the expression levels of UbG76V-GFP and Ub-R-GFP decreased in cells transfected with PAAF1 siRNA, but not in cells transfected with control siRNA or in mock-treated cells. In contrast, the expression of GFP was not affected by siRNA-mediated PAAF1 suppression (Fig. 8C, lower panels), indicating that the effect of PAAF1 siRNA was specific to unstable substrates of the proteasome. We also analyzed the effect of PAAF1 suppression on accumulation of ubiquitinated proteins by transfecting siRNA-treated cells with the expression construct of FLAG-ubiquitin. Immunoblotting of cell lysates with anti-FLAG antibodies revealed that the level of ubiquitinated proteins was specifically decreased in cells treated with PAAF1 siRNA (Fig. 8E). Collectively, these data strongly suggested that endogenous PAAF1 was involved in negative regulation of proteasome activity.
DISCUSSION
Recent studies on the proteasome composition using affinity purification of the proteasome from S. cerevisiae have uncovered novel subunits and interacting proteins of the proteasome (19, 28, 32). In this paper, we have applied a similar approach to human proteasomes and have found that PAAF1 is a novel regulator of the proteasome. Several lines of evidence presented here indicate that PAAF1 negatively regulates proteasome activity by interfering with the formation of functional 26S proteasomes. First, it was found by affinity purification of PAAF1 and subsequent analysis of copurified proteins by mass spectrometry and immunoblotting experiments that PAAF1 is associated with the 19S regulatory particle but not with the 20S core even in the presence of ATP, which facilitates assembly of the 26S proteasome. Second, coimmunoprecipitation analyses revealed that PAAF1 directly interacts with the proteasomal ATPases of the 19S particle, which sit on the α ring of the 20S core in the 26S proteasome. Third, overexpression of PAAF1 in HeLa cells decreases the level of the 20S core immunoprecipitated with the 19S particle in a dose-dependent fashion. Fourth, it was shown by in vivo proteasome-dependent proteolysis assays using GFP-based reporters that overexpression of PAAF1 inhibits proteasome activity. Finally, siRNA-mediated PAAF1 suppression revealed that the proteasome activity in cells is normally compromised by endogenous PAAF1. Our results illustrate the first example of a factor that affects assembly of the 26S proteasome by its binding to the 19S regulatory complex to our knowledge.
In this study, due to the unavailability of antibodies against PAAF1, we were not able to measure directly the cellular abundance of PAAF1 and to examine the possibility that PAAF1 is a stoichiometric component of the free 19S regulatory complex. However, on the basis of our findings that PAAF1 is one of the major copurified proteins with the 19S complex and that siRNA-mediated suppression of PAAF1 significantly increases the proteasome activity, we expect that the level of PAAF1 in HeLa cells is not very low compared with that of the proteasome.
Database searches indicate that the PAAF1 genes are conserved from yeast to higher eukaryotes. The regulation of PAAF1 gene expression has been reported in Xenopus laevis. Thus, the PAAF1 gene of X. laevis has been known to be one of the up-regulated genes in the tail resorption program induced by thyroid hormone (3). Since cell death and resorption without concomitant growth and differentiation are the main physiological changes in the tail resorption program, the induction of PAAF1 is expected to be related to cell death and resorption processes. Interestingly, a recent report has shown that the function of the 26S proteasome is impaired in apoptotic cells (29). Although the decrease in the proteasome activity could be explained by caspase-mediated cleavage of proteasome subunits (29), it is tempting to speculate that the induction of PAAF1 would contribute to the suppression of proteasome activity, which may facilitate apoptosis by inhibiting the proteasomal degradation of proapoptotic molecules in tail resorption of X. laevis.
The protein encoded by Saccharomyces cerevisiae open reading frame YGL004C is likely to be the ortholog of human PAAF1, as indicated by BLAST searches (Fig. 3A). In recent large-scale protein-protein interaction studies, Ygl004cp has been included as a bait to identify protein complexes interacting with it and found to be associated with subunits of the 19S regulatory complex (13). Furthermore, a large-scale bioinformatic study has predicted a role for Ygl004cp related to the proteasome based on the assumption that if two proteins share more interaction partners than random ones, they have close functional associations (25), and therefore the name RPN14 was proposed for YGL004C. Although the protein product of YGL004C/RPN14 is largely uncharacterized, these high-throughput studies suggest that it may have similar cellular functions to human PAAF1. Large-scale gene deletion analyses have indicated that YGL004C/RPN14 is not an essential gene (7). However, yeast mutants with deletions in the YGL004C/RPN14 gene show a severe growth defect on minimal culture media and a moderate growth defect after 20 or more generations on common complete media (see the Saccharomyces Genome Database [http://www.yeastgenome.org/]), suggesting its role in the growth or survival control of stressed cells.
The simplest model for inhibition of proteasome activity by PAAF1, based on our data presented here, is that PAAF1 binding to proteasomal ATPases prevents their association with the 20S core due to the steric hindrance, thereby increasing the pool of free 19S regulatory complexes. In this model, PAAF1 binds only to the free 19S regulatory complex but not to the 26S proteasome and indirectly helps 19S complexes dissociate themselves from 20S core by shifting the equilibrium between assembly and disassembly of 26S proteasome toward the latter. The exchange rate of 19S regulatory complex in the assembly-disassembly cycle of 26S proteasome might be required to be fast enough for the inhibition of proteasome by PAAF1 to be effective. Although recent studies have shown that the rate of disassembly and reassembly of 26S proteasomes is rather slow (11), the possibility still exists that a fraction of 19S complexes exchange faster than the bulk of 19S complexes. Our data are also compatible with another model, in which PAAF1 is able to bind to the proteasomal ATPases in the context of the 26S proteasome and thereby promotes the disassembly of the 26S proteasome into subcomplexes. Recent cross-linking studies on the assembly of the Drosophila 26S proteasome have indicated that it is not a simple docking of two rigid subcomplexes but a process accompanied by substantial rearrangements of their structures (16). PAAF1 binding to the proteasomal ATPases might cause them to favor the conformation in the disassembled state. The net result of PAAF1 in each model would be the decrease of the functional 26S proteasomes either by inhibition of their assembly or by promotion of their disassembly. Further studies on potential modulation of the PAAF1 activity under various physiological conditions and its mechanism in regulation of the proteasome activity will be required to clarify the role of PAAF1 in ubiquitin-dependent proteolysis.
Acknowledgments
This work was supported in part by grants from the Korea Science and Engineering Foundation through the Protein Network Research Center at Yonsei University and from the Korean Ministry of Science and Technology through the 21C Frontier Project.
We are grateful to Nico P. Dantuma for a generous gift of reagents.
REFERENCES
- 1.Borodovsky, A., B. M. Kessler, R. Casagrande, H. S. Overkleeft, K. D. Wilkinson, and H. L. Ploegh. 2001. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 20:5187-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boussif, O., F. Lezoualc'h, M. A. Zanta, M. D. Mergny, D. Scherman, B. Demeneix, and J. P. Behr. 1995. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci USA 92:7297-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, D. D., Z. Wang, J. D. Furlow, A. Kanamori, R. A. Schwartzman, B. F. Remo, and A. Pinder. 1996. The thyroid hormone-induced tail resorption program during Xenopus laevis metamorphosis. Proc. Natl. Acad. Sci. USA 93:1924-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 5.Dantuma, N. P., K. Lindsten, R. Glas, M. Jellne, and M. G. Masucci. 2000. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat. Biotechnol. 18:538-543. [DOI] [PubMed] [Google Scholar]
- 6.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 8.Glickman, M. H., and A. Ciechanover. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82:373-428. [DOI] [PubMed] [Google Scholar]
- 9.Groll, M., M. Bajorek, A. Kohler, L. Moroder, D. M. Rubin, R. Huber, M. H. Glickman, and D. Finley. 2000. A gated channel into the proteasome core particle. Nat. Struct. Biol. 7:1062-1067. [DOI] [PubMed] [Google Scholar]
- 10.Groll, M., L. Ditzel, J. Lowe, D. Stock, M. Bochtler, H. D. Bartunik, and R. Huber. 1997. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 386:463-471. [DOI] [PubMed] [Google Scholar]
- 11.Hendil, K. B., R. Hartmann-Petersen, and K. Tanaka. 2002. 26 S proteasomes function as stable entities. J. Mol. Biol. 315:627-636. [DOI] [PubMed] [Google Scholar]
- 12.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 13.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 14.Jager, S., J. Strayle, W. Heinemeyer, and D. H. Wolf. 2001. Cic1, an adaptor protein specifically linking the 26S proteasome to its substrate, the SCF component Cdc4. EMBO J. 20:4423-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohler, A., P. Cascio, D. S. Leggett, K. M. Woo, A. L. Goldberg, and D. Finley. 2001. The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol. Cell 7:1143-1152. [DOI] [PubMed] [Google Scholar]
- 16.Kurucz, E., I. Ando, M. Sumegi, H. Holzl, B. Kapelari, W. Baumeister, and A. Udvardy. 2002. Assembly of the Drosophila 26 S proteasome is accompanied by extensive subunit rearrangements. Biochem. J. 365:527-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lam, Y. A., T. G. Lawson, M. Velayutham, J. L. Zweier, and C. M. Pickart. 2002. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature 416:763-767. [DOI] [PubMed] [Google Scholar]
- 18.Lam, Y. A., W. Xu, G. N. DeMartino, and R. E. Cohen. 1997. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature 385:737-740. [DOI] [PubMed] [Google Scholar]
- 19.Leggett, D. S., J. Hanna, A. Borodovsky, B. Crosas, M. Schmidt, R. T. Baker, T. Walz, H. Ploegh, and D. Finley. 2002. Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10:495-507. [DOI] [PubMed] [Google Scholar]
- 20.Maytal-Kivity, V., N. Reis, K. Hofmann, and M. H. Glickman. 20. September 2002, posting date. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 3:28. [Online.] http://www.biomedcentral.com/1471-2091/3/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Min, K. W., J. W. Hwang, J. S. Lee, Y. Park, T. A. Tamura, and J. B. Yoon. 2003. TIP120A associates with cullins and modulates ubiquitin ligase activity. J. Biol. Chem. 278:15905-15910. [DOI] [PubMed] [Google Scholar]
- 22.Ogura, T., and A. J. Wilkinson. 2001. AAA+ superfamily ATPases: common structure—diverse function. Genes Cells 6:575-597. [DOI] [PubMed] [Google Scholar]
- 23.Papa, F. R., A. Y. Amerik, and M. Hochstrasser. 1999. Interaction of the Doa4 deubiquitinating enzyme with the yeast 26S proteasome. Mol. Biol. Cell 10:741-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pickart, C. M., and R. E. Cohen. 2004. Proteasomes and their kin: proteases in the machine age. Nat. Rev. Mol. Cell. Biol. 5:177-187. [DOI] [PubMed] [Google Scholar]
- 25.Samanta, M. P., and S. Liang. 2003. Predicting protein functions from redundancies in large-scale protein interaction networks. Proc. Natl. Acad. Sci. USA 100:12579-12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schauber, C., L. Chen, P. Tongaonkar, I. Vega, D. Lambertson, W. Potts, and K. Madura. 1998. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature 391:715-718. [DOI] [PubMed] [Google Scholar]
- 27.Smith, T. F., C. Gaitatzes, K. Saxena, and E. J. Neer. 1999. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24:181-185. [DOI] [PubMed] [Google Scholar]
- 28.Sone, T., Y. Saeki, A. Toh-e, and H. Yokosawa. 2004. Sem1p is a novel subunit of the 26S proteasome from Saccharomyces cerevisiae. J. Biol. Chem. 279:28807-28816. [DOI] [PubMed] [Google Scholar]
- 29.Sun, X. M., M. Butterworth, M. MacFarlane, W. Dubiel, A. Ciechanover, and G. M. Cohen. 2004. Caspase activation inhibits proteasome function during apoptosis. Mol. Cell 14:81-93. [DOI] [PubMed] [Google Scholar]
- 30.Tongaonkar, P., L. Chen, D. Lambertson, B. Ko, and K. Madura. 2000. Evidence for an interaction between ubiquitin-conjugating enzymes and the 26S proteasome. Mol. Cell. Biol. 20:4691-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verma, R., L. Aravind, R. Oania, W. H. McDonald, J. R. Yates III, E. V. Koonin, and R. J. Deshaies. 2002. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298:611-615. [DOI] [PubMed] [Google Scholar]
- 32.Verma, R., S. Chen, R. Feldman, D. Schieltz, J. Yates, J. Dohmen, and R. J. Deshaies. 2000. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 11:3425-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voges, D., P. Zwickl, and W. Baumeister. 1999. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68:1015-1068. [DOI] [PubMed] [Google Scholar]
- 34.Whitby, F. G., E. I. Masters, L. Kramer, J. R. Knowlton, Y. Yao, C. C. Wang, and C. P. Hill. 2000. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature 408:115-120. [DOI] [PubMed] [Google Scholar]
- 35.Wilm, M., A. Shevchenko, T. Houthaeve, S. Breit, L. Schweigerer, T. Fotsis, and M. Mann. 1996. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379:466-469. [DOI] [PubMed] [Google Scholar]
- 36.Xie, Y., and A. Varshavsky. 2000. Physical association of ubiquitin ligases and the 26S proteasome. Proc. Natl. Acad. Sci. USA 97:2497-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao, T., and R. E. Cohen. 2002. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419:403-407. [DOI] [PubMed] [Google Scholar]
- 38.You, J., and C. M. Pickart. 2001. A HECT domain E3 enzyme assembles novel polyubiquitin chains. J. Biol. Chem. 276:19871-19878. [DOI] [PubMed] [Google Scholar]