Abstract
We have previously shown that modification of Tcf-4, a transcription factor in the Wnt pathway, with SUMO by PIASy, a SUMO E3 ligase, enhances its transcriptional activity. Since PIASy itself was also modified with SUMO-1, we studied the role of sumoylation of PIASy in the regulation of Tcf-4. Lys35 was found to be a sumoylation site of PIASy. PIASyK35R, in which Lys35 was mutated to Arg, did not enhance sumoylation of Tcf-4, although this PIASy mutant did not lose the ligase activity of sumoylation for other proteins. Wild-type PIASy and PIASyK35R showed a distinct distribution in the nucleus, although both were colocalized with Tcf-4. Promyelocytic leukemia protein, which is involved in transcriptional regulation, was associated with PIASyK35R more frequently than wild-type PIASy in the nucleus. PIASyK35R could not stimulate the transcriptional activity of Tcf-4 under the conditions in which wild-type PIASy enhanced it. Conjugation of SUMO-1 to the amino terminus of PIASyK35R neither enhanced sumoylation of Tcf-4 nor stimulated the transcriptional activity of Tcf-4. These results suggest that sumoylation of Lys35 in PIASy determines the nuclear localization of PIASy and that it is necessary for PIASy-dependent sumoylation and transcriptional activation of Tcf-4.
Covalent interaction between SUMO and its targets is achieved by formation of an isopeptide bond between the C terminus of mature SUMO and the ɛ-amino group of a lysine in the acceptor protein (38). This reaction is ATP dependent and requires the SUMO-activating E1 enzyme Aos1/Uba2 and the SUMO-conjugating E2 enzyme Ubc9 (26, 34). Ubc9 recognizes a minimal sumoylation motif in many known targets. Although target proteins interact at least transiently with Ubc9, their efficient conjugation often requires SUMO E3 ligases (28). These factors might serve to increase the affinity of Ubc9 for a specific target, to stabilize the interaction of Ubc9 and the target protein, to help to orientate the acceptor lysine, or to contribute mechanically to conjugation.
There are three types of SUMO E3 ligases (28). The first type consists of members of the PIAS family, which were originally identified as transcriptional coregulators of the JAK-STAT pathway (25, 37). They contain a predicted RING-finger-like structure and bind to Ubc9 and selected SUMO target proteins (14). Mammals have at least five members: PIAS1, PIAS3, PIASxα, PIASxβ, and PIASy. The second type of SUMO E3 ligase is RanBP2/Nup358 (30). RanBP2 is a component of nuclear pore complexes. Although RanBP2 does not contain a RING-finger motif, it binds to Ubc9. The third is Polycomb member Pc2 (17). Pc2 has been shown to recruit the transcriptional corepressor CtBP (39) to Polycomb group (PcG) bodies that are involved in gene silencing, and to enhance sumoylation of CtBP. Known target proteins of sumoylation include RanGAP1, promyelocytic leukemia protein (PML), androgen receptor, histone H4, histone deacetylase 1, p300, IκBα, and p53 (7, 28). Sumoylation is reversible, and there are at least seven mammalian SUMO-specific proteases (10, 27, 44). Thus, sumoylation of substrates is regulated by SUMO E3 ligases and SUMO-specific proteases. Modification of target proteins by sumoylation changes their subcellular localization, transcriptional activation, and protein stability (7, 10, 14, 28). In addition to the target protein substrates, PIAS1 and PIASx themselves are also sumoylated (23). However, the roles of sumoylation of a SUMO E3 ligase in its functions are not known.
Wnt proteins constitute a large family of cysteine-rich secreted ligands that control development in organisms ranging from nematode worms to mammals (3, 42). The intracellular signaling pathway of Wnt is also conserved evolutionally and regulates various cellular functions (18). Wnt stabilizes cytoplasmic β-catenin (9, 13, 19-21), and accumulated β-catenin is translocated to the nucleus, where it binds to the transcription factor T-cell factor (Tcf) or lymphoid-enhancer factor (Lef) and thereby stimulates the expression of genes, including c-myc, fra-1, c-jun, cyclin D1, and peroxisome proliferator-activated receptor δ (12).
It has been suggested that PIASy, an E3 ligase, and Axam (SENP2), a desumoylation enzyme, are involved in the Wnt signaling pathway. Lef-1 and Tcf-4 are members of the Tcf family of proteins that function as transcription factors in the Wnt signaling pathway. PIASy interacts with Lef-1 and Tcf-4, resulting in sumoylation of these transcriptional factors (32, 43). Although PIASy does not affect the abilities of Lef-1 and Tcf-4 to bind to DNA, PIASy inhibits and enhances the transcriptional activities of Lef-1 and Tcf-4, respectively (32, 43). Axam has been identified as an Axin-binding protein (15) and regulates the Wnt signaling pathway negatively by inducing the degradation of β-catenin (16). This action of Axam requires its desumoylation activity. Thus, it is likely that sumoylation regulates the Wnt signaling pathway. In the present study we attempted to clarify the roles of sumoylation of PIASy itself in the activation of Tcf-4. We show here that PIASy is sumoylated at Lys35 and that modification of PIASy by SUMO-1 is necessary for PIASy-dependent sumoylation and transcriptional activation of Tcf-4.
MATERIALS AND METHODS
Materials and chemicals.
Glutathione S-transferase-fused Aos-1 (GST-Aos1) and His6-Uba2 purified from Spodoptera frugiperda Sf9 cells were supplied by H. Yasuda (Central Laboratory of Nippon Flour Mills Co., Ltd., Tokyo, Japan). pCMV5-Flag/PIASy, pUC/EF-1α/β-cateninSA (in which Ser33, Ser37, Thr41, and Ser45 are mutated to Ala), −163 cyclin D1-Luciferase (cyclin D1 (−163)-Luc), and pcDNAI/hTcf-4E, TOP-fos-Luc, and FOP-fos-Luc were provided by K. Shuai (University of California, San Francisco), A. Nagafuchi (Kumamoto University, Kumamoto, Japan), T. Akiyama (Tokyo University, Tokyo, Japan), and H. Clevers (University Hospital, Utrecht, The Netherlands), respectively. GST- or maltose-binding protein (MBP)-fusion proteins and His6-tagged proteins were purified from Escherichia coli according to the suppliers' instructions. COS and HEK293 cells were cultured in Dulbecco modified Eagle medium containing 10% calf serum and 10% fetal calf serum, respectively. The anti-Flag (M2) antibody, anti-Tcf-4 and anti-histone H1 antibodies, anti-α-tubulin antibody, anti-PIASy antibody, anti-SUMO-1 antibody, and anti-PML antibody were purchased from Sigma-Aldrich (Steinheim, Germany), Upstate Biotechnology (Lake Placid, N.Y.), Molecular Probes, Inc. (Eugene, Oreg.), Imgenex (San Diego, Calif.), Zymed Laboratories, Inc. (San Francisco, Calif.), and Santa Cruz (Santa Cruz, Calif.). Lipofectamine 2000 (Life Technologies, Inc.) was used for transfection of 293 and COS cells. Other materials were from commercial sources.
Plasmid construction.
pEF-BOS/hTcf-4E, pEF-BOS-HA/hTcf-4E, pEGFP-C1/hTcf-4E, pGEX-2TK/SUMO-1(GG), pCGN/SUMO-1, pCGN/SUMO-1(ΔGG), pMAL-c2/PIASy, and pCMV5-Flag/PIASyCA were constructed as described previously (16, 43). Standard recombinant DNA techniques were used to construct the following plasmids: pEGFP-C1/PIASy, pEGFP-C1/PIASyK35R,pCMV5-Flag/PIASyK35R, pMAL-c2/PIASyK35R, pCMV5-Flag/PIASyK128R, pCMV5-Flag/PIASyK35R/K128R, pCMV5-Flag/SUMO-1-(1-96)PIASyK35R,pCMV5-Flag/SUMO-1-(1-96)PIASy, pEF-BOS-HA/PIAS3, pCGN/PIASxα,pCGN/SUMO-3, and pRSETC/SUMO-1.
Sumoylation assay.
To examine the sumoylation of Tcf-4 and PIASy in intact cells, 293 cells (35-mm-diameter dish) transfected with pEF-BOS-, pCMV5-, and pCGN-derived plasmids were treated with 10% trichloroacetic acid (TCA) as described previously (43). In double immunoprecipitation (IP) analyses, 293 cells (two 35-mm-diameter dishes) transfected with pCMV5-Flag/PIASy and pCGN/SUMO-1 were lysed in 100 μl of radioimmunoprecipitation assay (RIPA) buffer (10 mM sodium phosphate [pH 7.2], 150 mM NaCl, 1% sodium deoxycholate, 1% Triton X-100, and 0.1% sodium dodecyl sulfate [SDS]) containing protease inhibitor cocktail (1 μg of aprotinin and leupeptin/ml and 10 mM phenylmethylsulfonyl fluoride [PMSF]), 1 mM NaF, 0.4 mM sodium orthovanadate, and 10 mM N-ethylmaleimide. After the lysates (200 μg of protein) were immunoprecipitated with the anti-Flag antibody, 30 μl of Laemmli sample buffer was added to the precipitates, and the samples were boiled at 100°C for 8 min. Then, 300 μl of RIPA buffer was added to each sample, followed by centrifugation at 2,300 × g for 2 min, and the supernatant was immunoprecipitated with the anti-Flag antibody again. The supernatant and precipitates of the second immunoprecipitation were probed with the anti-hemagglutinin (HA) and anti-Flag antibodies.
To observe sumoylation of PIASy at the endogenous level, nuclear extracts were prepared from HeLa S3 cells as described previously (43). The nuclear extracts (300 μg of protein) were incubated with His6-SUMO-1 (0.2 nmol) immobilized on nickel-agarose for 2 h at 4°C. After His6-SUMO-1 was precipitated by centrifugation, the precipitates were probed with the anti-PIASy or anti-SUMO-1 antibody.
To examine sumoylation of Tcf-4 in vitro, 0.25 μg of GST-Tcf-4 was incubated with 0.5 μg of GST-Aos1/His6-Uba2 (E1), 0.5 μg of His6-Ubc9 (E2), 10 μg of GST-SUMO-1(GG), and 0.2 μg of MBP-PIASy or MBP-PIASyK35R in 50 μl of reaction mixture (50 mM Tris-HCl [pH 7.5], 2 mM dithiothreitol, 1 mM MgCl2, and 5 mM ATP) for 30 min at 30°C (43). After the incubation, the mixtures were probed with the anti-Tcf-4 antibody.
Subcellular fractionation.
Subcellular fractionation was performed at 4°C as described previously (1) with slight modifications. Briefly, 293 cells (two 35-mm-diameter dishes) transfected with pCMV5-Flag/PIASy or PIASyK35R were suspended in 0.5 ml of buffer A (10 mM HEPES-NaOH [pH 7.9], 1.5 mM MgCl2, and 10 mM KCl) and homogenized with five strokes of a Dounce glass homogenizer, followed by centrifugation at 3,300 × g for 3 min. The precipitate was resuspended in 0.5 ml of buffer A and mixed with the supernatant. This mixture was used as total homogenate. The homogenate was centrifuged at 3,300 × g for 3 min again, and the supernatant was obtained. The precipitate was washed and suspended in 0.1 ml of buffer A and 0.3 ml of buffer B (40 mM HEPES-NaOH [pH 7.9], 0.84 M NaCl, 3 mM MgCl2 0.4 mM EDTA, 30% glycerol, 1 mM PMSF) and then incubated for 30 min on ice with occasional rocking. After centrifugation at 30,000 × g for 15 min, the supernatant was utilized as a nuclear soluble fraction. The precipitate was washed and resuspended in 0.1 ml of buffer A and 0.3 ml of buffer B and used as the nuclear insoluble fraction. The volumes of all of the fractions were adjusted to 1 ml. Aliquots (20 μl) of each fraction were probed with the anti-Flag, anti-α-tubulin, and anti-histone H1 antibodies.
Nuclear matrix preparation.
Nuclear matrices were prepared as described previously (5). After COS cells expressing green fluorescent protein-tagged Tcf-4 (GFP-Tcf-4), HA-SUMO-1, and Flag-PIASy on the slides were treated with 0.5% Triton X-100 in cytoskeleton buffer (10 mM PIPES-NaOH [pH 6.8], 100 mM KCl, 3 mM MgCl2, 1 mM EGTA, 300 mM sucrose, and 1.2 mM PMSF) for 5 min at 4°C, the cells were further treated with extraction buffer (42.5 mM Tris-HCl [pH 8.3], 8.5 mM NaCl, 2.6 mM MgCl2, 1.2 mM PMSF, 1% Tween 40, and 0.5% sodium deoxycholate) for 5 min at 4°C. After extraction, the cells were transferred to a humidified chamber, and 100 μl of digestion buffer (10 mM PIPES-NaOH [pH 6.8], 50 mM NaCl, 3 mM MgCl2, 1 mM EGTA, 300 mM sucrose, and 1.2 mM PMSF) containing 100 μg of DNase I/ml was added to the cells. The cells were incubated for 1 h at 37°C and further extracted with 0.25 M ammonium sulfate in digestion buffer for 5 min at 4°C. Then, the cells were rinsed in ice-cold phosphate-buffered saline once and fixed in methanol and acetone for 3 min each at −20°C. These samples were immunohistochemically stained with the anti-Flag or anti-HA antibody. The nuclei were colored blue by staining with DAPI (4′,6′-diamidino-2-phenylindole).
Semiquantitative RT-PCR analysis.
Total RNA was isolated from 293 cells (60-mm-diameter dish) transfected with pCGN/SUMO-1, pUC/EF-1α/β-cateninSA, and pCMV5-Flag/PIASy, or pCMV5-Flag/PIASyK35R. The RNA sample (5 μg) was subjected to reverse transcription by using murine leukemia virus reverse transcriptase (RT; PE Applied Biosystems, Foster City, Calif.) in a total volume of 20 μl. Aliquots of the reverse transcription products were amplified in a reaction mixture (20 μl) containing LightCycler FastStart DNA Master SYBR Green I, 3 mM MgCl2, and 0.5 μM primer for cyclin D1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) by using a LightCycler (Roche Diagnostic). Forward and reverse primers were as follows: cyclin D1 (5′-CATCAAGTGTGACCCGGACTG-3′ and 5′-CCTCCTCCTCAGTGGCCTTG-3′) and GAPDH (5′-CCTGTTCGACAGTCAGCCG-3′ and 5′-CGACCAAATCCGTTGACTCC-3′).
Others.
Interaction of Tcf-4 with PIASy in intact cells was examined as described previously (43). An immunofluorescence study with a confocal laser-scanning microscope (LSM510; Carl Zeiss, Jana, Germany), RNA interference analysis, and assay of Tcf-4 activity were performed as described previously (15, 43). To reduce endogenous PIASy, oligonucleotides specific for human PIASy 5′-GAGUGGACUGAAGCACGAGTT-3′ (sense) were synthesized.
RESULTS
Modification of PIASy with SUMO.
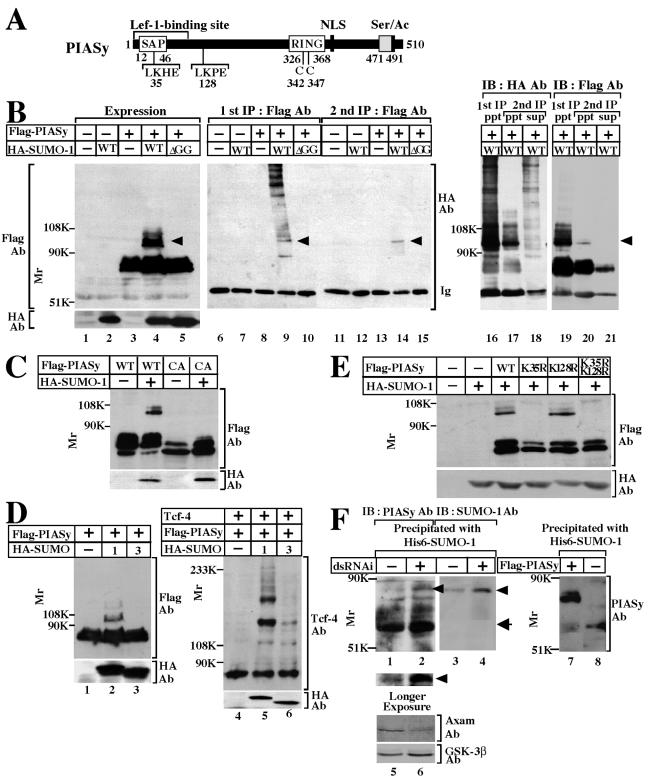
Since it has been reported that PIAS1, PIASxα, and PIASxβ are sumoylated in intact cells (23), we examined whether PIASy is also modified with SUMO. In HEK293 cells expressing Flag-PIASy and HA-SUMO-1, Flag-PIASy exhibited a mobility shift to a slowly migrating band of around 100 kDa on SDS-PAGE (Fig. 1B, lane 4). SUMO-1 precursor is processed by a C-terminal hydrolase to produce the mature form, and exposure of the C-terminal Gly-Gly residues is required for conjugation of SUMO-1 to substrates (8, 24). To examine whether the slowly migrating form of Flag-PIASy is due to modification with SUMO-1, we expressed HA-SUMO-1(ΔGG), in which the C-terminal six amino acids including Gly-Gly were deleted, in 293 cells. The higher-molecular-mass Flag-PIASy band was not observed (Fig. 1B, lane 5). When the lysates of cells expressing Flag-PIASy, together with HA-SUMO-1, were immunoprecipitated with the anti-Flag antibody, the anti-HA antibody detected the same slowly migrating band (ca. 100 kDa) as detected in cell extracts (Fig. 1B, lane 9). In addition, the anti-HA antibody revealed the presence of other SUMO-1-containing proteins at more than 150 kDa (Fig. 1B, lane 9). These high-molecular-mass proteins were assumed to represent other sumoylated proteins bound to PIASy. To examine this possibility, the immunoprecipitates obtained with the anti-Flag antibody were treated with 3.3% SDS and boiled, which would disrupt noncovalent protein-protein interactions, and the second immunoprecipitation was performed. By this treatment, the anti-HA-immunoreactive bands with high molecular mass, which were not detected by the anti-Flag antibody, were found in the supernatant of the second immunoprecipitation, whereas the anti-HA- and the anti-Flag-immunoreactive 100-kDa band was observed in the precipitates (Fig. 1B, lanes 16 to 21). Therefore, the slowly migrating band of Flag-PIASy at ca. 100 kDa indicates covalent modification with SUMO-1 and the anti-HA immunoreactive bands with high molecular mass are sumoylated proteins associated with PIASy.
FIG. 1.
Modification of PIASy with SUMO. (A) Schematic representation of PIASy used in the present study. (B) Modification of PIASy with SUMO-1. 293 cells expressing the indicated proteins were incubated with 10% TCA to precipitate the proteins. The resulting precipitates were dissolved in Laemmli sample buffer and probed with the anti-Flag and anti-HA antibodies (lanes 1 to 5). Lysates (200 μg of protein) prepared by extraction with RIPA buffer were immunoprecipitated with the anti-Flag antibody, and the immunoprecipitates were probed with the anti-HA antibody (lanes 6 to 10). The immunoprecipitates were further treated with SDS and immunoprecipitated with the anti-Flag antibody again. The second immunoprecipitates were also probed with the anti-HA antibody (lanes 11 to 15), and some of the supernatant (sup) and precipitates (ppt) were probed with the anti-HA and anti-Flag antibodies (lanes 16 to 21). The reason why immunoglobulin (lanes 18 and 21) and Flag-PIASy (lane 21) were detected in the supernatant of the second immunoprecipitation is that the immunoprecipitation efficiency by the anti-Flag antibody isca. 70%. Arrowheads indicate Flag-PIASy modified with HA-SUMO-1. Ab, antibody; IP, immunoprecipitation; IB, immunoblotting; WT, wild type; ΔGG, SUMO-1-(1-95). (C) 293 cells expressing the indicated proteins were incubated with TCA to precipitate the proteins, and the precipitates were probed with the anti-Flag and anti-HA antibodies. CA, a PIASy RING mutant in which Cys342 and Cys347 are changed to Ala. (D) Effects of SUMO-1 and SUMO-3 on sumoylation of PIASy and Tcf-4. Precipitates were prepared from 293 cells expressing the indicated proteins by adding TCA and probed with the anti-Flag and anti-HA antibodies (lanes 1 to 3) or the anti-Tcf-4 and anti-HA antibodies (lanes 4 to 6). (E) Identification of sumoylation site of PIASy. Precipitates were prepared from 293 cells expressing the indicated proteins by adding TCA and probed with the anti-Flag and anti-HA antibodies. K35R, a PIASy mutant in which Lys35 is changed to Arg; K128R, a PIASy mutant in which Lys128 is changed to Arg. K35R/K128R, a PIASy mutant in which both Lys35 and Lys128 are changed to Arg. (F) Modification of endogenous PIASy with endogenous SUMO-1. HeLa S3 cells were transfected with (lanes 2, 4, and 6) or without (lanes 1, 3, and 5) a dsRNAi oligonucleotide for Axam. The nuclear extracts (300 μg of protein) were incubated with His6-SUMO-1 immobilized on nickel-agarose. After His6-SUMO-1 was precipitated by centrifugation, the precipitates were probed with the anti-PIASy (lanes 1 and 2) or anti-SUMO-1 (lanes 3 and 4) antibody. Arrowheads indicate PIASy modified with SUMO-1, and the arrow indicates PIASy. Bottom panel shows a longer exposure of the same immunoblot. The lysates (20 μg of protein) were directly probed with the anti-Axam and anti-GSK-3β antibodies (lanes 5 and 6). To compare the molecular masses of endogenous PIASy and Flag-PIASy, the nuclear extracts expressing Flag-PIASy (10 μg of protein) (lane 7) and the nuclear extracts without expression of Flag-PIASy (50 μg of protein) (lane 8) were incubated with His6-SUMO-1 immobilized on nickel-agarose. After His6-SUMO-1 was precipitated by centrifugation, the precipitates were probed with the anti-PIASy antibody. The results shown are representative of three independent experiments.
PIASy has the RING domain, and a PIASy RING mutant (PIASyCA), in which Cys342 and Cys347 were mutated to Ala, does not bind to Ubc9 (43). Expression of Flag-PIASyCA and HA-SUMO-1 did not result in a mobility shift of Flag-PIASy, suggesting that PIASy is sumoylated by binding to Ubc9 (Fig. 1C). SUMO-3 shares 46% amino acid identity with SUMO-1, but expression of HA-SUMO-3 did not induce a mobility shift of Flag-PIASy (Fig. 1D). Furthermore, PIASy enhanced modification of Tcf-4 with SUMO-3 less than with SUMO-1 (Fig. 1D). Therefore, PIASy and Tcf-4 appear to prefer SUMO-1 to SUMO-3 for their sumoylation.
It has been proposed that ψKXD/E (ψ is a hydrophobic residue and X is any amino acid) is a consensus sequence for sumoylation (36). PIASy contains two of these typical consensus sequences for sumoylation: LK35HE and LK128PE (Fig. 1A). We mutated the lysine residues at amino acid positions 35 and 128 to arginine (PIASyK35R, PIASyK128R, and PIASyK35R/K128R) and expressed the lysine mutants of Flag-PIASy with HA-SUMO-1. The slowly migrating band of Flag-PIASy, corresponding to SUMO-modified PIASy, disappeared when K35 was mutated (Fig. 1E). However, when K128 was mutated, a sumoylation band was still detected (Fig. 1E). These results imply that K35 is a major sumoylation site of PIASy.
To determine whether PIASy is subject to SUMO-1 modification at the endogenous expression level, we precipitated endogenous PIASy from nuclear extracts of HeLa S3 cells by using His6-SUMO-1 because it was shown that yeast SUMO/Smt3 interacts with Nfi1, a yeast SUMO E3 ligase (40) and that PIAS family proteins interact with SUMO-1 (23). The difference between the molecular mass of Flag-PIASy and that of endogenous PIASy was ca. 15 to 20 kDa (compare Fig. 1F, lanes 7 and 8). Since the amino acid numbers of the Flag epitope are eight, the difference is larger than expected. Although we do not know the exact reason, the fusion of Flag to PIASy may affect its migration on an SDS-polyacrylamide gel. A slowly migrating band recognized by the anti-PIASy antibody was observed faintly with the unmodified form of PIASy (Fig. 1F, lane 1). The band was also recognized by the anti-SUMO-1 antibody (Fig. 1F, lane 3). The reason why sumoylation of PIASy at the endogenous level was hard to detect might be due to the presence of the desumoylation enzymes in the nuclear extracts. To examine this possibility, we reduced the protein level of Axam (SENP2) by double-stranded RNA interference (dsRNAi) (Fig. 1F, lanes 5 and 6). Knockdown of Axam by a dsRNAi oligonucleotide increased the amount of a slowly migrating band of PIASy, which was detected by the anti-SUMO-1 antibody strongly (Fig. 1F, lanes 2 and 4). These results indicate that endogenous PIASy is modified with endogenous SUMO-1.
Inability of PIASyK35R to enhance sumoylation of Tcf-4 in intact cells.
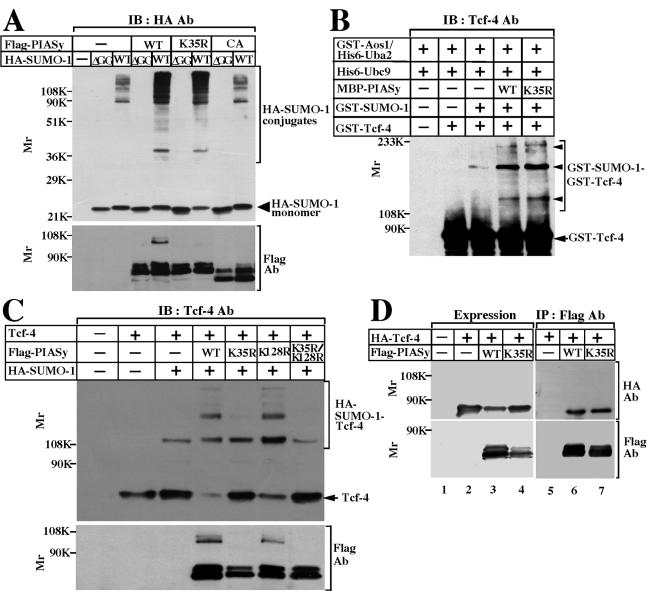
To examine whether PIASyK35R acts as an E3 ligase, this PIASy mutant was expressed with HA-SUMO-1 in 293 cells. Various high-molecular-mass forms of HA-SUMO-1 were detected even in the absence of Flag-PIASy (Fig. 2A). Flag-PIASyK35R enhanced sumoylation to the same degree as Flag-PIASy, but Flag-PIASyCA did not (Fig. 2A). There was no high-molecular-mass form of SUMO-1 when HA-SUMO-1(ΔGG) was expressed (Fig. 2A). Wild-type PIASy and its mutants were expressed almost equally (Fig. 2A, bottom panel). These results indicate that PIASyK35R does not lose the ligase activity for sumoylation.
FIG. 2.
Sumoylation of Tcf-4 by PIASyK35R. (A) Ability of PIASyK35R to enhance sumoylation. 293 cells expressing the indicated proteins were incubated with 10% TCA, and the precipitates were probed with the anti-HA and anti-Flag antibodies. IB, immunoblotting; Ab, antibody; WT, wild type; CA, PIASy RING mutant; ΔGG, SUMO-1-(1-95). (B) Sumoylation of Tcf-4 in vitro. GST-Tcf-4 was incubated with GST-Aos1/His6-Uba2, GST-SUMO-1(GG), His6-Ubc9, and MBP-PIASy, or MBP-PIASyK35R. After the incubation, the mixtures were probed with the anti-Tcf-4 antibody. Arrowheads indicate GST-Tcf-4 modified with GST-SUMO-1, and the arrow indicates GST-Tcf-4. (C) Sumoylation of Tcf-4 in intact cells. Precipitates were prepared from 293 cells expressing the indicated proteins by adding TCA and probed with the anti-Tcf-4 and anti-Flag antibodies. The arrow indicates Tcf-4. The results shown are representative of three independent experiments. (D) Interaction of Tcf-4 with PIASy. The lysates (20 μg of protein) of COS cells expressing the indicated proteins were probed with the anti-HA and anti-Flag antibodies (lanes 1 to 4). The same lysates (200 μg of protein) were immunoprecipitated with the anti-Flag antibody, and the immunoprecipitates were probed with the anti-HA and anti-Flag antibodies (lanes 5 to 7).
An in vitro reconstituted assay system with purified proteins showed that MBP-PIASyK35R enhances sumoylation of Tcf-4 as effectively as wild-type PIASy (Fig. 2B). However, Flag-PIASyK35R did not enhance modification of Tcf-4 with HA-SUMO-1 in intact 293 cells under the conditions in which wild-type Flag-PIASy enhanced it (Fig. 2C). Flag-PIASyK128R but not Flag-PIASyK35R/K128R enhanced sumoylation of Tcf-4 to the same extent as wild-type PIASy (Fig. 2C). The expression levels of wild-type PIASy and its mutants were almost equal (Fig. 2C, bottom panel). To see the complex formation between Tcf-4 with wild-type PIASy or PIASyK35R in intact cells, HA-Tcf-4 was expressed with wild-type Flag-PIASy or Flag-PIASyK35R in COS cells (Fig. 2D). HA-Tcf-4 was precipitated with Flag-PIASyK35R to the same extent as wild-type Flag-PIASy. Therefore, sumoylation of PIASy does not affect its affinity for Tcf-4 and the interaction of PIASy with Tcf-4 is not sufficient for sumoylation of Tcf-4 in intact cells. These results collectively indicate that sumoylation of PIASy is not required for its ligase activity for sumoylation and its interaction with Tcf-4 but that PIASy is not capable of enhancing sumoylation of Tcf-4 when PIASy is not sumoylated in intact cells.
Effects of sumoylation on localization of PIASy in the nucleus.
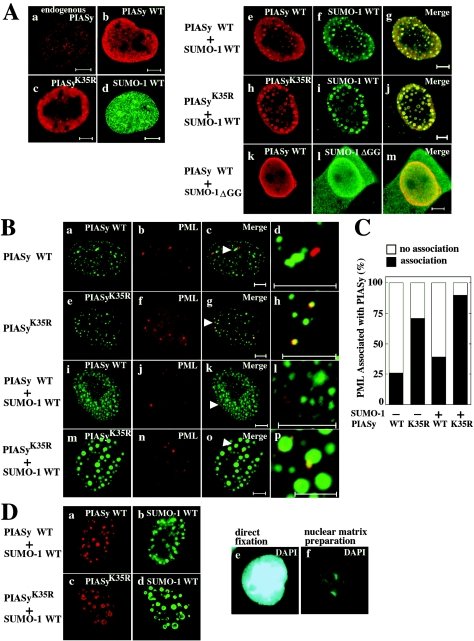
Endogenous PIASy was diffusely distributed in the nucleus with nuclear dot-like structure (Fig. 3Aa). Exogenously expressed Flag-PIASy or Flag-PIASyK35R alone displayed the nuclear diffuse distribution with a speckled pattern, similar to endogenous PIASy (Fig. 3Ab and c). HA-SUMO-1 also showed the diffuse distribution in the nucleus with nuclear dot-like structures (Fig. 3Ad). Since it is known that sumoylation determines the localization of the substrates in the nucleus (28), we examined whether sumoylation of PIASy affects its localization in the nucleus. Coexpression of Flag-PIASy with HA-SUMO-1 resulted in complete localization of SUMO-1 into small nuclear dots with PIASy (Fig. 3Ae to g). Although Flag-PIASyK35R was colocalized with HA-SUMO-1, Flag-PIASyK35R was somewhat better concentrated to large nuclear granules (Fig. 3Ah to j). When Flag-PIASy and HA-SUMO-1(ΔGG) were coexpressed, both showed diffuse nuclear distribution (Fig. 3Ak to m). Neither PIASy nor PIASyK35R was found in the cytoplasm. These results suggest that the subnuclear localization of PIASyK35R is different from that of wild-type PIASy and that sumoylation of PIASy is important for the determination of its localization in the nucleus.
FIG. 3.
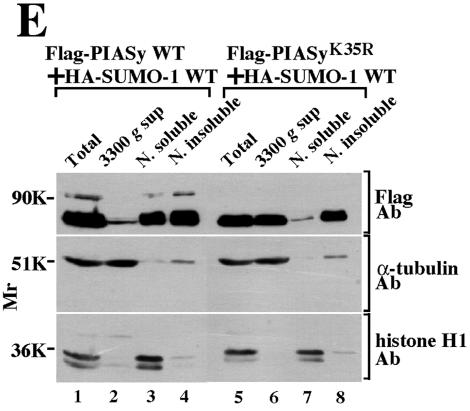
Localization of PIASy in the nucleus. (A) Sumoylation-dependent localization of PIASy. (a) Control COS cells were stained with the anti-PIASy antibody. (b to m) COS cells expressing Flag-PIASy WT alone (b), Flag-PIASyK35R alone (c), HA-SUMO-1 WT alone (d), Flag-PIASy WT and HA-SUMO-1 WT (e to g), Flag-PIASyK35R and HA-SUMO-1 WT (h to j), or Flag-PIASy WT and HA-SUMO-1(ΔGG) (k to m) were stained with the anti-Flag antibody to detect PIASy (b, c, e, h, and k) and the anti-HA antibody to detect SUMO-1 (d, f, i, and l). The merged images are shown in subpanels g, j, and m. Bars, 5 μm. (B) Localization of PIASy and PML in the nucleus. GFP-PIASy WT alone (a to d), GFP-PIASyK35R alone (e to h), GFP-PIASy WT and HA-SUMO-1 WT (i to l), or GFP-PIASyK35R and HA-SUMO-1 WT (m to p) were expressed in COS cells. The cells were viewed directly with a confocal scanning microscope to detect GFP-PIASy (a, e, i, and m) and stained with the anti-PML antibody to detect endogenous PML (b, f, j, and n). The merged images are shown in subpanels c, g, k, and o. The areas indicated by arrowheads are further enlarged (d, h, l, and p). Bars, 5 μm. (C) Percentages of PML associated with PIASy or PIASyK35R. The numbers of PML showing the colocalization or adjacent localization with PIASy or PIASyK35R were counted in the cells used in panel B. The black bars indicate the percentages of PML colocalized with or located next to PIASy or PIASyK35R. The white bars indicate the percentages of PML, which is present apart from PIASy or PIASyK35R. More than 100 cells were examined in three independent experiments. (D) Localization of PIASy in the nuclear matrix. After COS cells expressing Flag-PIASy WT and HA-SUMO-1 WT (a and b) or Flag-PIASyK35R and HA-SUMO-1 WT (c and d) were treated with DNase I, the cells were stained with the anti-Flag (a and c) and anti-HA (b and d) antibodies. The nuclei incubated with (f) or without (e) DNase I were stained blue with DAPI. (E) Biochemical subcellular fractionation. 293 cells expressing Flag-PIASy WT and HA-SUMO-1 WT (lanes 1 to 4) or Flag-PIASyK35R and HA-SUMO-1 WT (lanes 5 to 8) were fractionated into total homogenate (lanes 1 and 5), 3,300 × g supernatant fraction (3300 g sup) (lanes 2 and 6), nuclear soluble fraction (N. soluble) (lanes 3 and 7), and nuclear insoluble fraction (N. insoluble) (lanes 4 and 8). Proteins from the fractions were probed with the anti-Flag, anti-α-tubulin, and anti-histone H1 antibodies. The results shown are representative of four independent experiments. WT, wild type.
Proteins modified with SUMO-1 have been shown to localize to specific subnuclear structure, known as PML-containing nuclear bodies (NBs) (29). Endogenous PML was observed in 2 to 10 NBs (Fig. 3B). We noticed the presence of two types of PML localization with regard to the association with PIASy. The first type showed a distribution apart from PIASy. The second type was the clear colocalization or adjacent localization with PIASy. When GFP-PIASy was expressed alone, most of PML (75%) was present apart from PIASy (Fig. 3Ba to d and 3C). In contrast, when GFP-PIASyK35R was expressed alone, ca. 70% of PML was colocalized with or located next to GFP-PIASyK35R (Fig. 3Be to h and 3C). When GFP-PIASy and HA-SUMO-1 were coexpressed, 35% of PML was colocalized with or located next to GFP-PIASy (Fig. 3Bi to l and 3C). When GFP-PIASyK35R and HA-SUMO-1 were coexpressed, 85% of PML showed the colocalization with or adjacent localization with GFP-PIASy (Fig. 3Bm to p and 3C). These results suggest that PML associates with PIASyK35R more frequently than wild-type PIASy in NBs.
It has been reported that PIASy binds to the nuclear matrix through its SAP domain (amino acids 12 to 46) (32). Since the SAP domain contains a sumoylation site of PIASy, we examined whether sumoylation of PIASy is necessary for its nuclear matrix association. COS cells expressing Flag-PIASy or Flag-PIASyK35R with HA-SUMO-1 were treated with DNase I and salt. Both wild-type PIASy and PIASyK35R were retained in the nuclear matrix compartment (Fig. 3Da to d). Therefore, sumoylation is not necessary for the binding of PIASy to the nuclear matrix, but wild-type PIASy and PIASyK35R appear to show different localization.
We also examined the subnuclear localization of PIASy by biochemical fractionation. α-Tubulin and histone H1 were mainly localized in the 3,300 × g supernatant and the nuclear soluble fractions, respectively (Fig. 3E). Consistent with immunohistochemical studies, both Flag-PIASy and Flag-PIASyK35R were observed in the nuclear insoluble fraction (Fig. 3E). Flag-PIASy was detected in the nuclear soluble fraction in addition to the insoluble fraction, whereas Flag-PIASyK35R was detected in the 3,300 × g supernatant fraction (Fig. 3E). Since we could not detect Flag-PIASyK35R in the cytoplasm by immunohistochemical studies, the presence of Flag-PIASyK35R in the 3,300 × g supernatant fraction by biochemical fractionation suggests that the binding affinity of PIASyK35R for nuclear components is lower than that of wild-type PIASy. Taken together, it is likely that wild-type PIASy and PIASyK35R are localized at the different places in the nucleus.
Changes of localization of Tcf-4 by sumoylation of PIASy.
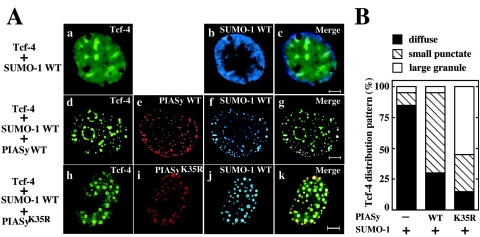
Since PIASy and Tcf-4 are colocalized in the nucleus (43), we examined whether sumoylation of PIASy is required for the colocalization of PIASy and Tcf-4. When GFP-Tcf-4 and HA-SUMO-1 were coexpressed, both proteins were diffusely present throughout the nucleus in >80% of cells expressed, and they were not colocalized with each other (Fig. 4Aa to c and B). Additional expression of Flag-PIASy resulted in a distribution of Tcf-4 into a small punctate structure that coincided with Flag-PIASy and HA-SUMO-1 in 65% of cells, and 30% of cells showed the diffused pattern (Fig. 4Ad to g and B). Although Flag-PIASyK35R also changed the nuclear distribution of GFP-Tcf-4, and these two proteins were colocalized with each other in the presence of HA-SUMO-1, they were concentrated into large nuclear granules in 55% of cells (Fig. 4Ah to k and B). The localization of this complex appeared to be different from that of the complex of wild-type PIASy and Tcf-4. Approximately 15 and 30% of the cells expressing Flag-PIASyK35R and GFP-Tcf-4 showed the diffuse pattern and the small punctate pattern, respectively (Fig. 4Ah to k and B). Taken together with the previous observations that Flag-PIASyCA showed diffuse nuclear localization and did not induce the redistribution of GFP-Tcf-4 into a small nuclear structure (43), these results suggest that sumoylation of PIASy and Tcf-4 determines their subnuclear localization.
FIG. 4.
Colocalization of PIASy and Tcf-4 in the nucleus. (A) Representative patterns of Tcf-4 distribution. COS cells expressing GFP-Tcf-4 and HA-SUMO-1 WT (a to c), GFP-Tcf-4, Flag-PIASy WT, and HA-SUMO-1 WT (d to g), or GFP-Tcf-4, Flag-PIASyK35R, and HA-SUMO-1 WT (h to k) were stained with the anti-HA antibody to detect SUMO-1 (b, f, and j), and the anti-Flag antibody to detect PIASy (e and i). Some cells were viewed directly with a confocal scanning microscope to detect GFP-Tcf-4 (a, d, and h). The merged images are shown in panels c, g, and k. Bars, 5 μm. (B) Percentages of the cells showing the different distribution of Tcf-4. The numbers of the cells showing different distribution of GFP-Tcf-4 were counted among the cells used in Fig. 4A. The black, hatched, and white bars indicate the diffuse, small punctate, and large granule patterns, respectively. More than 100 cells were examined in four independent experiments. WT, wild type.
Activation of Tcf-4 by sumoylation of PIASy.
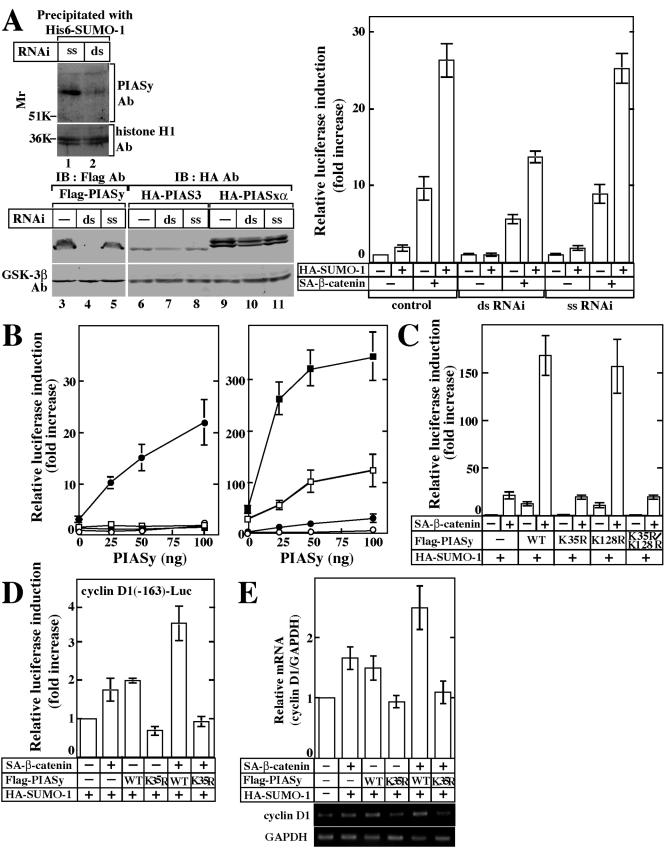
We previously showed that PIASy activates Tcf-4 and that sumoylation of Tcf-4 is necessary for the synergistic activation of Tcf-4 by PIASy and β-catenin (43). Since these results were based on overexpression of PIASy, we examined whether PIASy is involved in the transcriptional activity of Tcf-4 by the use of dsRNAi. A dsRNAi oligonucleotide for PIASy reduced the protein level of endogenous PIASy in the nuclear extracts of HeLa S3 cells (Fig. 5A, lanes 1 and 2). Since it was difficult to detect endogenous PIAS3 and PIASxα in HeLa S3 cells by their antibodies, we examined the effect of a dsRNAi oligonucleotide for PIASy on the expression of other PIAS family members by using expression plasmids. This dsRNAi oligonucleotide suppressed the expression of Flag-PIASy but little affected the expression of HA-PIAS3 and HA-PIASxα (Fig. 5A). A single-stranded sense (ss) oligonucleotide for PIASy did not affect the exogenously expressed Flag-PIASy, HA-PIAS3, and HA-PIASxα. Therefore, a dsRNAi oligonucleotide for PIASy could affect PIASy specifically among the PIAS family members.
FIG. 5.
Activation of Tcf-4 by PIASy and SUMO-1. (A) dsRNAi for PIASy. The nuclear extracts (50 μg of protein) of HeLa S3 cells transfected with a dsRNAi oligonucleotide (lane 2) or an ss oligonucleotide (lane 1) for PIASy were precipitated with His6-SUMO-1, and the precipitates were probed with the anti-PIASy antibody. The same nuclear extracts (5 μg of protein) were probed with the anti-histone H1 antibody. Lysates from HeLa S3 cells transfected with pCMV5-Flag/PIASy (lanes 4 and 5), pEF-BOS-HA/PIAS3 (lanes 7 and 8), or pCGN/PIASxα (lanes 10 and 11), and a dsRNAi oligonucleotide (lanes 4, 7, and 10) or an ss oligonucleotide (lanes 5, 8, and 11) for PIASy were probed with the anti-Flag or anti-HA and anti-GSK-3β antibodies. Lysates from HeLa S3 cells transfected with pCMV5-Flag/PIASy (lane 3), pEF-BOS-HA/PIAS3 (lane 6), or pCGN/PIASxα (lane 9) alone were used as a control. After HeLa S3 cells had been transfected with or without the indicated RNAi oligonucleotide for PIASy, the cells were further transfected with pEF-BOS/hTcf-4E (0.1 μg), TOP-fos-Luc (0.5 μg), pCGN/SUMO-1 (0.5 μg) and pUC/EF-1α/β-cateninSA (0.5 μg) (right panel). The luciferase activity was measured and expressed as the fold increase compared to the level observed in cells transfected with TOP-fos-Luc and pEF-BOS/hTcf-4E. All of the transfection experiments were performed at least five times, and the results shown are means ± the standard error. ds, double-stranded oligonucleotide; ss, single-stranded sense oligonucleotide. (B) SUMO-1-dependent activation of Tcf-4. The indicated amounts of pCMV5-Flag/PIASy, pEF-BOS/hTcf-4E (0.1 μg), and TOP-fos-Luc (0.5 μg) were transfected into 293 cells with pCGN/SUMO-1 (•), pCGN/SUMO-3 (▪), or pCGN/SUMO-1(ΔGG) (□) (0.5 μg) or without these plasmids (○) (left panel). The indicated amounts of pCMV5-Flag/PIASy, pEF-BOS/hTcf-4E (0.1 μg), and TOP-fos-Luc (0.5 μg) were transfected into 293 cells with pCGN/SUMO-1 (0.5 μg) (•), pUC/EF-1α/β-cateninSA (0.1 μg) (□), or pCGN/SUMO-1 and pUC/EF-1α/β-cateninSA (▪) or without these plasmids (○) (right panel). (C) Effects of sumoylation of PIASy on Tcf-4. pCGN/SUMO-1 (0.5 μg), pUC/EF-1α/β-cateninSA (0.1 μg), pEF-BOS/hTcf-4E (0.1 μg), and TOP-fos-Luc or FOP-fos-Luc (0.5 μg) were transfected into 293 cells with pCMV5-Flag/PIASy, pCMV5-Flag/PIASyK35R, pCMV5-Flag/PIASyK128R, or pCMV5-Flag/PIASyK35R/K128R (0.1 μg). (D) Effects of PIASy on the Tcf activity for natural Tcf-responsive promoters. Cyclin D1 (−163)-Luc (0.5 μg), pCGN/SUMO-1 (0.1 μg), pUC/EF-1α/β-cateninSA (0.5 μg), and pEF-BOS/hTcf-4E (0.1 μg) were transfected into 293 cells with pCMV5-Flag/PIASy or pCMV5-Flag/PIASyK35R (0.5 μg). The luciferase activity was measured and is expressed as the fold increase compared to the level observed in the cells transfected with cyclin D1 (−163)-Luc, pCGN/SUMO-1, and pEF-BOS/hTcf-4E. (E) RT-PCR analysis of expression of endogenous cyclin D1 mRNA. Total RNA from 293 cells transfected with pCGN/SUMO-1 (0.1 μg), pUC/EF-1α/β-cateninSA (0.5 μg), and pCMV5-Flag/PIASy or pCMV5-Flag/PIASyK35R (0.5 μg) was subjected to semiquantitative RT-PCR at 24 h after transfection. The result for cyclin D1 mRNA levels was normalized by that for GAPDH and is expressed as the fold increase compared to that of cells transfected with control vectors alone (top panel). The results shown are means ± the standard errors of the mean from four independent experiments. Middle panel, cyclin D1; bottom panel, GAPDH. WT, wild type.
The effects of a dsRNAi oligonucleotide on the endogenous PIASy-dependent activation of Tcf-4 were examined by the use of TOP-fos-Luc (22), which contains four Tcf-binding sites and a c-fos promoter sequence ligated to a luciferase gene, as a reporter gene. SUMO-1 alone did not activate Tcf-4 significantly but it enhanced β-catenin-dependent Tcf-4 activity in HeLa S3 cells (Fig. 5A). A dsRNAi oligonucleotide but not a ss oligonucleotide for PIASy suppressed SUMO-1- and β-catenin-dependent Tcf-4 transcriptional activity (Fig. 5A). These results indicate that endogenous PIASy is involved in the regulation of Tcf-4. Therefore, the effects of sumoylation of PIASy on the activation of Tcf-4 were examined by the expression of wild-type PIASy or PIASyK35R.
Under conditions in which PIASy alone (transfection with <100 ng of pCMV5-Flag/PIASy) did not activate Tcf-4 in 293 cells, coexpression with HA-SUMO-1 activated Tcf-4 (Fig. 5B, left panel). Neither HA-SUMO-1(ΔGG) nor HA-SUMO-3 affected the transcriptional activity of Tcf-4 (Fig. 5B, left panel). Flag-PIASy markedly activated Tcf-4 in a dose-dependent manner in the presence of both β-catenin and HA-SUMO-1 (Fig. 5B, right panel). Therefore, the action of PIASy on Tcf-4 is dependent on SUMO-1 under these experimental conditions.
Flag-PIASyK128R activated Tcf-4 to the same extent as Flag-PIASy, whereas Flag-PIASyK35R or Flag-PIASyK35R/K128R did not (Fig. 5C). Furthermore, Flag-PIASyK128R enhanced β-catenin-dependent activation of Tcf-4 to the same extent as Flag-PIASy, while Flag-PIASyK35R or Flag-PIASyK35R/K128R did not (Fig. 5C). We also used another reporter gene, FOP-fos-Luc, in which the Tcf-binding sites were mutated. No substantial change in this reporter activity was observed under similar conditions (data not shown). The protein expression levels of Tcf-4, Flag-PIASy, HA-β-cateninSA, and HA-SUMO-1 were almost the same in these assays (data not shown).
We confirmed the requirement of sumoylation of PIASy for gene expression by using cyclin D1 promoter fused with a luciferase gene [cyclinD1 (−163)-Luc], which is the natural Tcf-responsive promoter (41). Flag-PIASy increased the transcriptional activity of Tcf-4 toward the cyclin D1 promoter, while Flag-PIASyK35R did not activate it (Fig. 5D). Although Flag-PIASy enhanced β-catenin-dependent Tcf-4 activity, Flag-PIASyK35R rather repressed it (Fig. 5D). Consistent with these results, semiquantitative RT-PCR analyses also showed that Flag-PIASy enhances the β-catenin-dependent increase of cyclin D1 mRNA but that Flag-PIASyK35R represses it (Fig. 5E). Taken together, these results suggest that modification of PIASy with SUMO-1 is required for the PIASy-dependent activation of Tcf-4.
Effects of SUMO-fused PIASyK35R on nuclear localization and transcriptional activity of Tcf-4.
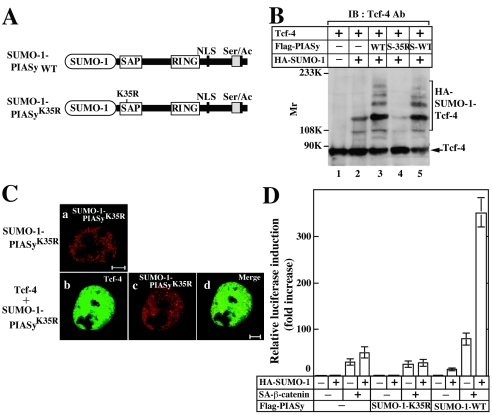
To examine whether sumoylation of PIASy is sufficient for its activities, we generated gene constructs in which SUMO-1 is fused to the N termini of PIASyK35R and wild-type PIASy (SUMO-1-PIASyK35R and SUMO-1-PIASy WT, respectively) (Fig. 6A). SUMO-1-PIASyK35R was not able to enhance sumoylation of Tcf-4 irrespective of the presence of exogenously expressed SUMO-1 (Fig. 6B). SUMO-1-PIASyK35R alone showed a speckled pattern with diffuse nuclear distribution, which was similar to the pattern of PIASyK35R alone, and this localization was distinct from that of wild-type PIASy modified with SUMO-1 at Lys35 (compare with Fig. 3A) (Fig. 6Ca). SUMO-1-PIASyK35R did not change the nuclear localization of Tcf-4 without expression of HA-SUMO-1 (Fig. 6Cb to d). Furthermore, SUMO-1-PIASyK35R neither activated basal Tcf-4 activity nor enhanced β-catenin-dependent activation of Tcf-4 even in the presence of exogenously expressed SUMO-1 (Fig. 6D). Fusion of SUMO-1 to PIASy did not impair its activity, because SUMO-1-PIASy WT enhanced the basal and β-catenin-dependent transcriptional activity of Tcf-4 in the presence of SUMO-1 (Fig. 6D). These results indicate that fusion of SUMO-1 to the N terminus of PIASyK35R does not rescue its lost abilities, suggesting that SUMO-1 must be conjugated to an appropriate site (Lys35) of PIASy to exert its functions.
FIG. 6.
Effect of SUMO-1 conjugation to N terminus of PIASy. (A) SUMO-1 conjugated PIASy. SUMO-1 (amino acids 1 to 96) was directly fused to the N terminus of wild-type PIASy or PIASyK35R. (B) Effect of SUMO-1-PIASyK35R on sumoylation of Tcf-4 in intact cells. Precipitates from 293 cells expressing the indicated proteins were prepared by incubating with TCA and they were probed with the anti-Tcf-4 antibody. S-35R, SUMO-1-PIASyK35R; S-WT, SUMO-1-PIASy WT. (C) Subnuclear localization of SUMO-1-PIASyK35R. COS cells expressing Flag-SUMO-1-PIASyK35R (a) and GFP-Tcf-4 and Flag-SUMO-1-PIASyK35R (b to d) were stained with the anti-Flag antibody to detect SUMO-1-PIASyK35R (a and c). (b) GFP-Tcf-4 was directly viewed. The merged image is shown in subpanel d. Bars, 5 μm. (D) Effect of SUMO-1-PIASyK35R on Tcf transcriptional activity. pCGN/SUMO-1 (0.5 μg), pUC/EF-1α/β-cateninSA (0.1 μg), pEF-BOS/hTcf-4E (0.1 μg), and TOP-fos-Luc (0.5 μg) were transfected into 293 cells with pCMV5-Flag, pCMV5-Flag/SUMO-1-PIASy WT, or pCMV5-Flag/SUMO-1-PIASyK35R (0.1 μg). The luciferase activity was measured and expressed as the fold increase compared to the level observed in cells transfected with TOP-fos-Luc and pEF-BOS/hTcf-4E. All of the transfection experiments were performed five times, and the results shown are means ± the standard errors of the mean. WT, wild type.
DISCUSSION
In this study we demonstrated the modification of PIASy with SUMO-1 at the endogenous level, identified the sumoylation site of PIASy, and clarified the roles of sumoylation in the functions of PIASy. Our results provide the first evidence that sumoylation of an E3 ligase itself is necessary for the regulation of sumoylation and function of the substrate.
Roles of sumoylation of PIASy in its nuclear localization.
PIASy was colocalized with SUMO-1 in small nuclear dots, whereas PIASyK35R was colocalized with SUMO-1 in large nuclear granules. Since PIASyK35R was not sumoylated, the colocalization of PIASyK35R with SUMO-1 suggests that PIASyK35R associates noncovalently with proteins modified with SUMO-1 in the nucleus. This idea is supported by the observations that PIASy and SUMO-1(ΔGG) showed diffuse nuclear localization when both proteins were coexpressed. Since PIASy enhances sumoylation of various proteins and associates with sumoylated proteins noncovalently, PIASy may interact with sumoylated proteins at the correct place in the nucleus when it is modified with SUMO-1.
PML associated with PIASyK35R more frequently than wild-type PIASy, although we do not know the physiological significance of the difference between the colocalization and adjacent localization of PML with PIASy. It has been reported that NB formation requires PML to be modified with SUMO-1 (29). However, conjugation of SUMO-1 to PIASy may suppress the binding of PIASy to PML or other proteins modified with SUMO-1 in NBs. It has been reported that PIASy binds to the nuclear matrix through its SAP domain (amino acids 12 to 46) (32). Immunohistochemical analyses showed that sumoylation is not essential for the localization of PIASy in the nuclear matrix, although it appears that sumoylated and nonsumoylated forms of PIASy are present in the different places. Furthermore, biochemical fractionation studies also confirmed that both wild-type PIASy and PIASyK35R are localized in the nuclear insoluble fraction. They were also present in the soluble fraction but showed distinct distribution, suggesting that the binding affinity of PIASyK35R for nuclear components is lower than that of wild-type PIASy. Taken together, these results indicate the subnuclear localization of PIASyK35R is different from that of PIASy and that the subnuclear localization of PIASy is determined in a sumoylation-dependent manner.
Although PIASyK35R was colocalized with Tcf-4 in the nucleus, its localization was different from that of wild-type PIASy and Tcf-4. PIASyK35R did not lose the ligase activity that allows it to enhance sumoylation and interacted with Tcf-4 as effectively as wild-type PIASy, but this PIASy mutant was not able to enhance sumoylation of Tcf-4 in intact cells. It is possible that Tcf-4 complexed with sumoylated PIASy is efficiently modified with SUMO-1. It is intriguing to speculate that when PIASy is sumoylated and present with Tcf-4 at an appropriate place in the nucleus, desumoylation enzymes may not act on sumoylated Tcf-4.
Roles of sumoylation of PIASy in activation of Tcf-4.
Since a decrease of PIASy by dsRNAi suppressed the activity of Tcf-4, PIASy is thought to be involved in the activation of Tcf-4 through sumoylation. Therefore, it was expected that PIASyK35R would not be able to affect the activity of Tcf-4. All of the experiments using reporter genes and RT-PCR of a target gene demonstrated that PIASyK35R does not activate the transcriptional activity of Tcf-4. Although we do not know the mechanism by which sumoylation of Tcf-4 and PIASy enhances Tcf-4 activity, the recruitment of the β-catenin and Tcf-4 complex to promoters may be enhanced in a sumoylated PIASy-dependent manner. Alternatively, a coactivator may be recruited to an appropriate place where Tcf-4 and PIASy are complexed, or sumoylation of Tcf-4 and PIASy may prevent the binding of a repressor to Tcf-4. However, we could not show completely that a dsRNAi oligonucleotide for PIASy does not affect endogenous levels of other PIAS family members. Therefore, if the dsRNAi affects other PIAS family members, we cannot rule out the possibility that sumoylation events other than directly involving Tcf-4 affect the transcriptional activity of Tcf-4.
The sumoylation of PML is important for its localization in NBs. NBs are sites of protein modification, and at least one function of PML may be to target p53 to nuclear bodies where it can be acetylated, phosphorylated, and activated as a transcription factor (6, 11). PML also complexes with and modifies the activity of a wide variety of additional transcription factors, coactivators, and corepressors (35). Since PML associated with PIASyK35R more efficiently than wild-type PIASy, if the complex of PIASyK35R and PML recruits transcriptional repressors or modifies Tcf-4 negatively, this would be the reason for the inability of PIASyK35R to activate Tcf-4.
SUMO-2 and SUMO-3 have been identified as proteins related to SUMO-1 (33). SUMO-3 is very similar to SUMO-2 (95% identity) but relatively different from SUMO-1 (46% identity). However, little has been elucidated about the functional differences between SUMO-1, -2, and -3. It has been reported that topoisomerase II is modified with SUMO-2/3 but not with SUMO-1 during mitosis and that this modification is important for the topoisomerase II activity (2). We demonstrated that PIASy and Tcf-4 are exclusively modified with SUMO-1 rather than SUMO-3 and that PIASy stimulates the transcriptional activity of Tcf-4 in the presence of SUMO-1 but not SUMO-3. These results indicate discrimination among the different SUMO isoforms.
Since Lef-1, a Tcf-4 homolog, is sumoylated with SUMO-2 more efficiently than SUMO-1 (32), it is intriguing to speculate that Tcf/Lef family proteins are modified with different SUMO isoforms. It has been reported that PIASy represses the β-catenin-stimulated transcriptional activity of Lef-1 and that sumoylation of Lef-1 is not involved in the PIASy-dependent repression (32). Although we do not know the reasons for the differences of the effects of sumoylation on Lef-1 and Tcf-4, it is possible to speculate that sumoylation regulates positively or negatively the transcriptional activity of Tcf-4 and Lef-1 for their target genes, depending on the promoter architectures of the target genes. Of course, we cannot rule out the possibility that the expression levels of proteins and cell-type-specific differences may contribute to the differences between our observations and those of the other report. It has been recently reported that β-catenin-dependent Lef-1 activity is moderately reduced in mouse embryonic fibroblasts prepared from PIASy-deficient mice (31). This finding supports our results that PIASy activates Tcf-4.
Importance of sumoylation of PIASy at K35.
It has been reported that there is a correlation of sumoylation of SATB2, which binds to a nuclear matrix attachment region and enhances gene expression in pre-B cells, and its subnuclear localization (4). SATB2 is localized at the nuclear periphery and enhances gene expression, whereas the sumoylation-deficient SATB2 mutant shows diffuse nuclear localization and further increases gene expression. Covalent attachment of SUMO-1 to the N terminus of the SATB2 mutant localizes the protein into nuclear bodies and allows it to repress gene expression (4). However, fusion of SUMO-1 to the N terminus of PIASyK35R did not alter the subnuclear localization of PIASyK35R, which is different from that of wild-type PIASy modified with SUMO-1 at K35, indicating that sumoylation at site other than K35 is not involved in the correct localization of PIASy. Furthermore, SUMO-1-PIASyK35R enhanced neither sumoylation nor the transcriptional activity of Tcf-4. Therefore, these results suggest that sumoylation of PIASy at K35 is important for the nuclear localization of PIASy and its regulation of the activity of Tcf-4. Since K35 of PIASy is conserved in other PIAS family proteins (25), this lysine residue in other PIAS may be modified with SUMO and may be involved in the regulation of gene expression. Further experiments will be necessary to achieve a full understanding of the regulation of gene expression in the Wnt pathway by PIAS family proteins.
FIG. 5
Acknowledgments
We thank H. Yasuda, K. Shuai, A. Nagafuchi, T. Akiyama, and H. Clevers for donating proteins and plasmids. We thank K. Igarashi, S. Tashiro, and J. Sun (Hiroshima University) for helpful discussions.
This study was supported by Grants-in-Aid for Scientific Research and for Scientific Research on priority areas from the Ministry of Education, Science, and Culture of Japan (in 2002, 2003, and 2004) and by grants from the Yamanouchi Foundation for Research on Metabolic Disorders (in 2002 and 2003) and the Uehara Memorial Foundation (in 2003).
REFERENCES
- 1.Abmayr, S. M., and J. L. Workman. 1989. Preparation of nuclear and cytoplasmic extracts from mammalian cells, p. 12.1.1-12.1.9. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 2. Wiley, New York, N.Y. [DOI] [PubMed] [Google Scholar]
- 2.Azuma, Y., A. Arnaoutov, and M. Dasso. 2003. SUMO-2/3 regulates topoisomerase II in mitosis. J. Cell Biol. 163:477-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bienz, M., and H. Clevers. 2000. Linking colorectal cancer to Wnt signaling. Cell 103:311-320. [DOI] [PubMed] [Google Scholar]
- 4.Dobreva, G., J. Dambacher, and R. Grosschedl. 2003. SUMO modification of a novel MAR-binding protein, SATB2, modulates immunoglobulin μ gene expression. Genes Dev. 17:3048-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fey, E. G., K. M. Wan, and S. Penman. 1984. Epithelial cytoskeletal framework and nuclear matrix-intermediate filament scaffold: three-dimensional organization and protein composition. J. Cell Biol. 98:1973-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fogal, V., M. Gostissa, P. Sandy, P. Zacchi, T. Sternsdorf, K. Jensen, P. P. Pandolfi, H. Will, C. Schneider, and G. Del Sal. 2000. Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J. 19:6185-6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gill, G. 2004. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 18:2046-2059. [DOI] [PubMed] [Google Scholar]
- 8.Gong, L., S. Millas, G. G. Maul, and E. T. Yeh. 2000. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J. Biol. Chem. 275:3355-3359. [DOI] [PubMed] [Google Scholar]
- 9.Hino, S.-I., T. Michiue, M. Asashima, and A. Kikuchi. 2003. Casein kinase Iɛ enhances the binding of Dvl-1 to Frat-1 and is essential for Wnt-3a-induced accumulation of β-catenin. J. Biol. Chem. 278:14066-14073. [DOI] [PubMed] [Google Scholar]
- 10.Hochstrasser, M. 2000. Evolution and function of ubiquitin-like protein-conjugation systems. Nat. Cell Biol. 2:E153-E157. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann, T. G., A. Moller, H. Sirma, H. Zentgraf, Y. Taya, W. Droge, H. Will, and M. L. Schmitz. 2002. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell Biol. 4:1-10. [DOI] [PubMed] [Google Scholar]
- 12.Hurlstone, A., and H. Clevers. 2002. T-cell factors: turn-ons and turn-offs. EMBO J. 21:2303-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeda, S., S. Kishida, H. Yamamoto, H. Murai, S. Koyama, and A. Kikuchi. 1998. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3β and β-catenin and promotes GSK-3β-dependent phosphorylation of β-catenin. EMBO J. 17:1371-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson, P. K. 2001. A new RING for SUMO: wrestling transcriptional responses into nuclear bodies with PIAS family E3 SUMO ligases. Genes Dev. 15:3053-3058. [DOI] [PubMed] [Google Scholar]
- 15.Kadoya, T., S. Kishida, A. Fukui, T. Hinoi, T. Michiue, M. Asashima, and A. Kikuchi. 2000. Inhibition of Wnt signaling pathway by a novel Axin-binding protein. J. Biol. Chem. 275:37030-37037. [DOI] [PubMed] [Google Scholar]
- 16.Kadoya, T., H. Yamamoto, T. Suzuki, A. Yukita, A. Fukui, T. Michiue, T. Asahara, K. Tanaka, M. Asashima, and A. Kikuchi. 2002. Desumoylation activity of Axam, a novel Axin-binding protein, is involved in downregulation of β-catenin. Mol. Cell. Biol. 22:3803-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kagey, M. H., T. A. Melhuish, and D. Wotton. 2003. The Polycomb protein Pc2 is a SUMO E3. Cell 113:127-137. [DOI] [PubMed] [Google Scholar]
- 18.Kikuchi, A. 1999. Roles of Axin in the Wnt signalling pathway. Cell. Signal 11:777-788. [DOI] [PubMed] [Google Scholar]
- 19.Kishida, M., S. Koyama, S. Kishida, K. Matsubara, S. Nakashima, K. Higano, R. Takada, S. Takada, and A. Kikuchi. 1999. Axin prevents Wnt-3a-induced accumulation of β-catenin. Oncogene 18:979-985. [DOI] [PubMed] [Google Scholar]
- 20.Kishida, S., H. Yamamoto, S.-I. Hino, S. Ikeda, M. Kishida, and A. Kikuchi. 1999. DIX domains of Dvl and Axin are necessary for protein interactions and their ability to regulate β-catenin stability. Mol. Cell. Biol. 19:4414-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kishida, S., H. Yamamoto, S. Ikeda, M. Kishida, I. Sakamoto, S. Koyama, and A. Kikuchi. 1998. Axin, a negative regulator of the Wnt signaling pathway, directly interacts with adenomatous polyposis coli and regulates the stabilization of β-catenin. J. Biol. Chem. 273:10823-10826. [DOI] [PubMed] [Google Scholar]
- 22.Korinek, V., N. Barker, P. J. Morin, D. van Wichen, R. de Weger, K. W. Kinzler, B. Vogelstein, and H. Clevers. 1997. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science 275:1784-1787. [DOI] [PubMed] [Google Scholar]
- 23.Kotaja, N., U. Karvonen, O. A. Janne, and J. J. Palvimo. 2002. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 22:5222-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, S. J., and M. Hochstrasser. 1999. A new protease required for cell-cycle progression in yeast. Nature 398:246-251. [DOI] [PubMed] [Google Scholar]
- 25.Liu, B., J. Liao, X. Rao, S. A. Kushner, C. D. Chung, D. D. Chang, and K. Shuai. 1998. Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl. Acad. Sci. USA 95:10626-10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matunis, M. J., E. Coutavas, and G. Blobel. 1996. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135:1457-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melchior, F., M. Schergaut, and A. Pichler. 2003. SUMO: ligases, isopeptidases, and nuclear pores. Trends Biochem. Sci. 28:612-618. [DOI] [PubMed] [Google Scholar]
- 28.Müller, S., A. Ledl, and D. Schmidt. 2004. SUMO: a regulator of gene expression and genome integrity. Oncogene 23:1998-2008. [DOI] [PubMed] [Google Scholar]
- 29.Müller, S., M. J. Matunis, and A. Dejean. 1998. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 17:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pichler, A., A. Gast, J. Seeler, A. Dejean, and F. Melchior. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108:109-120. [DOI] [PubMed] [Google Scholar]
- 31.Roth, W., C. Sustmann, M. Kieslinger, A. Gilmozzi, D. Irmer, E. Kremmer, C. Turck, and R. Grosschedl. 2004. PIASy-deficient mice display modest defects in IFN and Wnt signaling. J. Immunol. 173:6189-6199. [DOI] [PubMed] [Google Scholar]
- 32.Sachdev, S., L. Bruhn, H. Sieber, A. Pichler, F. Melchior, and R. Grosschedl. 2001. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15:3088-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saitoh, H., and J. Hinchey. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275:6252-6258. [DOI] [PubMed] [Google Scholar]
- 34.Saitoh, H., D. B. Sparrow, T. Shiomi, R. T. Pu, T. Nishimoto, T. J. Mohun, and M. Dasso. 1998. Ubc9p and the conjugation of SUMO-1 to RanGAP1 and RanBP2. Curr. Biol. 8:121-124. [DOI] [PubMed] [Google Scholar]
- 35.Salomoni, P., and P. P. Pandolfi. 2002. The role of PML in tumor suppression. Cell 108:165-170. [DOI] [PubMed] [Google Scholar]
- 36.Sampson, D. A., M. Wang, and M. J. Matunis. 2001. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276:21664-21669. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt, D., and S. Müller. 2003. PIAS/SUMO: new partners in transcriptional regulation. Cell. Mol. Life Sci. 60:2561-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwartz, D. C., and M. Hochstrasser. 2003. A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem. Sci. 28:321-328. [DOI] [PubMed] [Google Scholar]
- 39.Sewalt, R. G., M. G. Gunster, J. van der Vlag, D. P. Satijin, and A. P. Otte. 1999. C-Terminal binding protein is a transcriptional repressor that interacts with a specific class of vertebrate Polycomb proteins. Mol. Cell. Biol. 19:777-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takahashi, Y., A. Toh-E, and Y. Kikuchi. 2003. Comparative analysis of yeast PIAS-type SUMO ligases in vivo and in vitro. J. Biochem. 133:415-422. [DOI] [PubMed] [Google Scholar]
- 41.Tetsu, O., and F. McCormick. 1999. β-Catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398:422-426. [DOI] [PubMed] [Google Scholar]
- 42.Wodarz, A., and R. Nusse. 1998. Mechanisms of Wnt signaling in development. Annu. Rev. Cell Dev. Biol. 14:59-88. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto, H., M. Ihara, Y. Matsuura, and A. Kikuchi. 2003. Sumoylation is involved in β-catenin-dependent activation of Tcf-4. EMBO J. 22:2047-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeh, E. T., L. Gong, and T. Kamitani. 2000. Ubiquitin-like proteins: new wines in new bottles. Gene 248:1-14. [DOI] [PubMed] [Google Scholar]