Abstract
CtIP interacts with a group of tumor suppressor proteins including RB (retinoblastoma protein), BRCA1, Ikaros, and CtBP, which regulate cell cycle progression through transcriptional repression as well as chromatin remodeling. However, how CtIP exerts its biological function in cell cycle progression remains elusive. To address this issue, we generated an inactivated Ctip allele in mice by inserting a neo gene into exon 5. The corresponding Ctip−/− embryos died at embryonic day 4.0 (E4.0), and the blastocysts failed to enter S phase but accumulated in G1, leading to a slightly elevated cell death. Mouse NIH 3T3 cells depleted of Ctip were arrested at G1 with the concomitant increase in hypophosphorylated Rb and Cdk inhibitors, p21. However, depletion of Ctip failed to arrest Rb−/− mouse embryonic fibroblasts (MEF) or human osteosarcoma Saos-2 cells at G1, suggesting that this arrest is RB dependent. Importantly, the life span of Ctip+/− heterozygotes was shortened by the development of multiple types of tumors, predominantly, large lymphomas. The wild-type Ctip allele and protein remained detectable in these tumors, suggesting that haploid insufficiency of Ctip leads to tumorigenesis. Taken together, this finding uncovers a novel G1/S regulation in that CtIP counteracts Rb-mediated G1 restraint. Deregulation of this function leads to a defect in early embryogenesis and contributes, in part, to tumor formation.
CtIP is a yet poorly understood protein that was initially discovered as a cofactor of transcriptional corepressor CtBP by yeast two-hybrid screening (34). Subsequent efforts have demonstrated that CtIP serves as an interacting partner for two retinoblastoma tumor suppressor family proteins, Rb and p130 (8, 27), and breast tumor suppressor BRCA1 (22, 38, 42), as well as members of the Ikaros family encoding Kruppel-like zinc finger proteins that regulate early hemolymphopoiesis (19).
CtIP is an 897-amino-acid protein that is ubiquitously expressed in different types of cells (8, 42). Apart from specific motifs used to bind Rb (LECEE) and CtBP (PLDLS), two coiled-coil domains of CtIP are also recognized. The one at the N terminus is used for homodimerization of CtIP (7). Interestingly, judging by sequence similarity, the CtIP gene so far has only been found in higher eukaryotic organisms, including Homo sapiens, Mus musculus, and Rattus norvegicus, with significant sequence divergence, implying a late participation during evolution.
The interaction between CtIP and CtBP is believed to play an important role in the CtIP-mediated and CtBP-dependent transcriptional repression. CtBP, identified as a C-terminal binding protein of human adenovirus E1A protein (2, 33), could function as a transcriptional corepressor in a histone deacetylase (HDAC)-dependent or -independent manner and may play critical roles during the early development and oncogenesis (3). Like CtBP, CtIP also possesses transcriptional repression activity when artificially fused to GAL4 DNA binding domain and coexpressed with reporter gene driven by a simian virus 40 promoter with upstream GAL4 binding sites (27). On the other hand, Rb family proteins bind CtIP at the LECEE motif, and mutation of this site disrupts their interaction. Using the same transcription reporter assay, mutations of the binding domain of Rb prevent the association of CtIP and fail to repress transcription (27). However, it remains unclear whether binding of CtIP to Rb has a role other than transcriptional repression.
In addition to Rb family proteins, BRCA1 can also physically and functionally interact with CtIP/CtBP complex. The N-terminal 133 to 369 amino acids of CtIP mediate its interaction with the C-terminal repeats (BRCT) of BRCA1 (42, 45). Significantly, tumor-associated mutations in BRCT domains abolish this specific interaction (22, 42, 46). DNA damage agents such as ionizing irradiation induce the hyperphosphorylation of CtIP at serine 664 and 745 by ATM kinase (23). And this event is suggested to modulate BRCA1 for the transcriptional activation of p21 and GADD45 upon DNA damage (22, 23), although the detailed mechanism remains to be clarified.
Ikaros family proteins are transcriptional repressors, which can interact with the HDAC-recruiting factors, Sin3 and Mi-2b, and exhibit HDAC-dependent repression activity (20). In addition, Ikaros can also directly bind to CtBP, Rb, and CtIP, which can interact with the basal transcription factor TFIIB (19). That may explain how Ikaros repression works through an HDAC-independent pathway.
Those observations deduced from studies of other proteins suggest that CtIP serves as a corepressor for transcriptional repression. However, the actual biological function of CtIP remains unclear. Especially, systematic study of CtIP loss of function has not been reported yet. In this communication, we show that the Ctip−/− embryos die at embryonic day 4.0 (E4.0) as blastocysts fail to enter S phase and accumulate in G1, leading to an elevated cell death. Rb+/+ cells, but not Rb−/−, depleted of Ctip are arrested at G1, suggesting that this arrest is RB dependent. Importantly, the life spans of Ctip+/− heterozygotes are shortened by the development of multiple types of tumors, predominantly, large lymphomas. This finding uncovers that CtIP is a key protein in early embryogenesis, and the loss of one allele contributes, in part, to tumor formation.
MATERIALS AND METHODS
Abbreviations.
Histone deacetylase; FACS, fluorescence-activated cell sorting; MEF, mouse embryonic fibroblast; E, embryonic day; BudR, bromodeoxyuridine; siRNA, small interfering RNA; TUNEL, terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling; GFP, green fluorescent protein; MOI, multiplicity of infection; DAPI, 4′,6-diamidino-2-phenylindole.
Cell lines and tissue culture.
NIH 3T3, Rb+/+, and Rb−/− mouse embryonic fibroblasts (MEF), and Saos-2 and SR5 human osteosarcoma cells were cultured in high-glucose Dulbecco's modified Eagle medium containing 10% fetal bovine serum (FBS) at 37°C in an incubator with 10% CO2.
Construction of targeting vectors and establishing the knockout mouse.
The Ctip gene was isolated by screening a λDASH mouse genomic library derived from the 129/Sv mouse strain (kindly provided by Tom Doetschman, University of Cincinnati), using a 2.7-kb cDNA fragment of human CtIP as probe. A 9.8-kb BamHI fragment of the mouse Ctip gene containing exon 5 was subcloned into the pBluescript SK vector. A SalI site was generated in the middle of exon 5 by site-directed mutagenesis for inserting 1.6 kb of pgkneopA cassette flanked by XhoI sites in antisense orientation. This construct was used to generate ES clones for producing chimeric mice according to procedures described previously (21).
Embryo collection and PCR genotyping.
Ctip heterozygotes were used for timed pregnancy experiments. For embryos older than E8.5, the visceral yolk sac was collected and subjected to PCR genotyping. For embryos between E4.5 and E8.5, embryos inside the uterus were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned. Embryonic tissues stained with Mayer's hematoxylin and eosin (H&E) were microdissected from slides for genotyping. The E3.5 blastocysts were flushed from maternal uterus for analysis. PCR genotyping was performed using the primer A (5′-ACA GGT TAA GAG CAG GAT TGT GTG A-3′) and B (5′-ATA TGA AAG GGA ACT CAC TCA GCT C-3′), which flank the exon 5 and generate a 144-bp fragment from the wild-type Ctip, while primer C (5′-TGC ACG AGA CTA GTG AGA CGT GCT A-3′), which resides at PGK promoter, and primer A together generate a 388-bp fragment from the targeted Ctip allele.
In vitro blastocyst culture, BrdU incorporation, immunostaining, and TUNEL assay.
Blastocysts were isolated from pregnant females at E3.5, and an outgrowth assay was performed as previously described (24).
For BrdU incorporation, blastocysts were cultured for 1 day and then labeled with BrdU in medium for 6 h. Blastocysts were then fixed and incubated with anti-BrdU antibody (Amersham Biosciences) or rabbit polyclonal anti-CtIP antibody (C21) at 4°C overnight. After three washes in PBS, blastocysts were incubated with fluorescence-conjugated secondary antibody at room temperature for 30 min, followed by DAPI staining. Blastocysts were mounted on slides and examined under fluorescence microscopy (Zeiss Axioplan 2 imaging). Slides were demounted later, and blastocysts were collected and lysed for PCR genotyping.
For phospho-histone H3 immunostaining, blastocysts were collected, fixed, and then incubated with anti-phospho-histone H3 (ser10) antibody (6G3; Cell Signaling Technology). A TUNEL assay (24) was performed on freshly collected and fixed blastocysts, using an in situ cell death detection kit (Roche Applied Science) according to the manufacturer's instructions.
Ctip siRNA construction.
A Ctip siRNA expression cassette was constructed by inserting a DNA fragment derived from two annealed synthetic oligonucleotides (5′-GGC GTC ATC TCC GGT ATT TGC TCA AGA GAC AAA TAC CGG AGA TGA CGC CCT TTT TA-3′ and 5′-AGC TTA AAA AGG GCG TCA TCT CCG GTA TTT GTC TCT TGA GCA AAT ACC GGA GAT GAC GCC-3′) into pBS-U6 vector (37). The BamHI fragment containing the U6-siRNA construct was released and subcloned into pAdTrack vector, which carries a GFP-expressing cassette, resulting in pAd-Ctip-siRNA. The adenovirus was then produced following the protocol as described previously (13). Human CtIP siRNA construct was generated under the regulation of U6 promoter by inserting an oligonucleotide sequence from 2275 to 2295 (GGG AGC AGA CCT TTC TCA GTA) of human CtIP into U6 RNAi vector to form pBS/U6-CtIPi. The RNAi expression cassette was then inserted into a pATG vector (28), which directs GFP expression, and that was named pCtIP-RNAi.
Immunoblotting and FACS analysis.
Antibodies against p21 (F5 from Santa Cruz Biotechnology), Rb (Rb245), and CtIP (19E8) were used for Western blotting. Fluorescence-activated cell sorting (FACS) analysis for NIH 3T3 cells infected with CtIP-siRNA or GFP adenovirus was carried out using FACScalibur as instructed by the manufacturer (BD Biosciences).
Histology and immunohistochemistry.
Collected tissues were fixed in 4% paraformaldehyde and processed through paraffin embedding following standard procedures. Sections were stained with H&E for histopathological evaluation. Immunostaining was performed following the protocol described in a Vectastain Elite ABC kit (Vector Laboratories, Burlingame, CA). For antigen retrieval, slides were heated for 20 min in 10 mM citrate buffer (pH 6.0) in a microwave oven. The antibodies used were CD45R/B220 (BD Biosciences, San Diego, CA), Mac2 (Cadarlane, Ontario, Canada), and CD3 (DAKO, Carpinteria, CA).
RESULTS
Absence of wild-type Ctip alleles leads to early embryonic lethality.
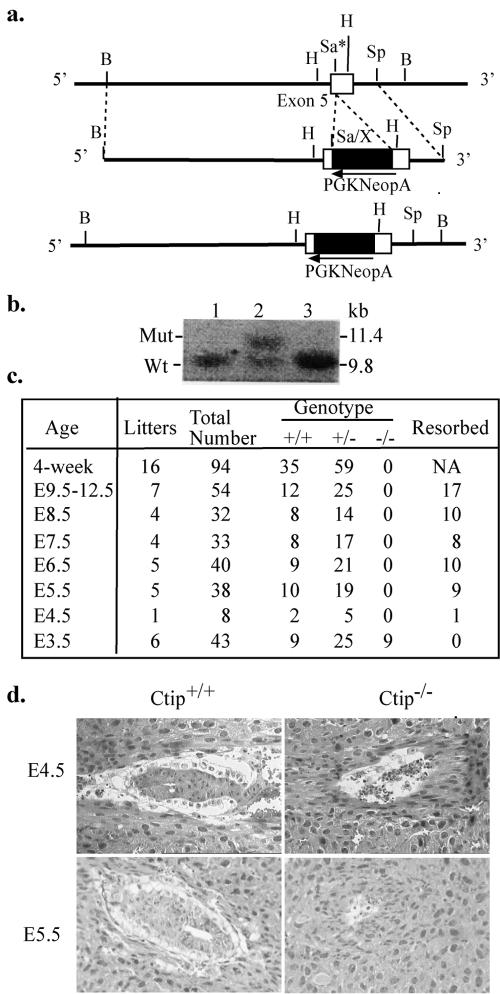
The biological function of CtIP is yet poorly understood except that it can interact with several tumor suppressor proteins. To address its roles in mouse development, we generated an inactivated Ctip allele in embryonic stem (ES) cells by inserting a pgkneopA cassette at the position equivalent to amino acid residue 109 in the human CtIP protein (Fig. 1a). Recombinant ES cell clones were identified by Southern blotting (Fig. 1b), and the correct clone was used to generate the chimeric mice and, subsequently, heterozygous mice.
FIG. 1.
Targeted disruption of Ctip mouse locus leads to embryonic lethality. a, Knockout strategy. A SalI site in exon 5 was introduced by site-directed mutagenesis, and a pgkneopA cassette was inserted into this site. Only partial genomic DNA is shown. b, Identification of the recombinant ES clones by digesting the genomic DNA with BamHI and probing with the 0.5-kb SpeI-BamHI fragment. Lane 2 indicated a correct recombinant ES cell clone. c, Genotype analysis of the progenies from Ctip+/− heterozygous intercrosses. d, Histological sections of wild-type and Ctip−/− mutant embryos grown in utero. The embryos inside the uterus were fixed, sectioned, and stained with H&E. Left panels, wild-type mouse embryos at E4.5 (top) and E5.5 (bottom) stage; right panels, Ctip−/− embryos at E4.5 (top) and E5.5 (bottom) stage. At E4.5, the Ctip−/− embryo is mostly resorbed, while at E5.5 day, the Ctip−/− embryo is nearly completely resorbed.
While examining the in vivo effect of homozygous mutations, we found no Ctip−/− mice in 94 offspring produced by the intercross of Ctip+/− heterozygotes, indicating that they died in uterus. To define the time of death, Ctip+/− females conceived by intercrosses were sacrificed, and the fetuses at different gestation times from E4.5 to E12.5 were examined. At all the time points, there were no normal conceptuses with the Ctip−/− genotype, but, instead, many resorbed embryos were observed (Fig. 1c). To precisely pinpoint the differences between the wild-type and Ctip-null embryos, intact decidual swellings from Ctip+/− intercrosses obtained between E4.5 and E5.5 were examined. Both the wild-type and heterozygous embryos showed normal growth and elongation of the egg cylinder, while Ctip−/− embryos failed to form egg cylinders and degenerated in the uterus (Fig. 1d), suggesting that Ctip is essential for very early embryonic development.
Inner stem cells of Ctip null blastocysts arrest at G1.
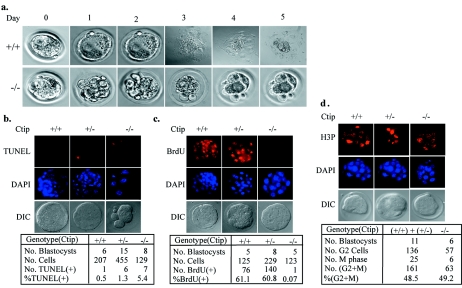
To explore the reason why Ctip−/− embryos die so early, we employed blastocyst outgrowth in culture as an alternative method to study early developmental event. While 41 cultured Ctip+/+ and Ctip+/− blastocysts gave rise to adherent sheets of trophoblastic giant cells and outgrowth of the inner cell mass, 14 Ctip−/− blastocysts showed no outgrowth but retained intact zona pellucida (an example is shown in Fig. 2a), suggesting that Ctip−/− blastocysts failed to hatch from the eggshell. We then tested whether the inner cell mass underwent apoptosis by a TUNEL assay (24). On average, the total number of cells per Ctip−/− blastocyst was slightly lower than that of the wild type or heterozygotes, whereas the number of dead cells was slightly higher in Ctip−/− blastocysts than that of the wild type or heterozygotes. These differences were subtle and may not entirely account for the dramatic phenotype seen in the in vitro blastocyst culture (Fig. 2b). To seek for an additional explanation for the phenotype, we analyzed cell cycle profiles for inner stem cells in blastocysts by BrdU uptake at S phase and by immunostaining with anti-phospho-histone H3 (H3P) antibody at G2 or M phase. Essentially, all the Ctip−/− blastocysts failed to incorporate BrdU, while 61% of inner stem cells of the wild type or heterozygotes were labeled with BrdU, suggesting that Ctip−/− blastocysts failed to enter S phase (Fig. 2c). However, Ctip−/− blastocysts contained numbers of cells at G2 and M phases similar to those of the wild-type or heterozygotes (Fig. 2d). Taken together, these results suggest that Ctip is essential for the S-phase entry in inner stem cells. Without Ctip, these cells are arrested at G1, which may eventually lead to cell death. This phenotype is much severer than those observed in mice with null genotype of Ctbp (14), Brca1 (12, 24), Ikaros (9), or Rb (21), all of which interact with Ctip.
FIG. 2.
Ctip−/− blastocysts failed to hatch in vitro and arrested in G1. a, Ctip+/+ blastocysts showed normal outgrowth and hatched during in vitro culture (top panels), while Ctip−/− blastocysts showed no outgrowth but retained the intact outer membranes (bottom panels). b, Blastocysts cultured for 1 day were subjected to a TUNEL assay by labeling with BrdU. Inner embryonic stem cells were stained with anti-BrdU antibodies and DAPI for nuclei DNA. The results of the TUNEL assay were summarized in the table. The same blastocysts as described for panel b were subjected to the BrdU uptake assay (c) or immunostained with anti-phospho-histone H3 (H3P) to determine the status of G2/M phases (d). The results of these two assays were summarized in each table following the photos.
Ctip siRNA knockdown leads to G1 arrest in mouse fibroblasts.
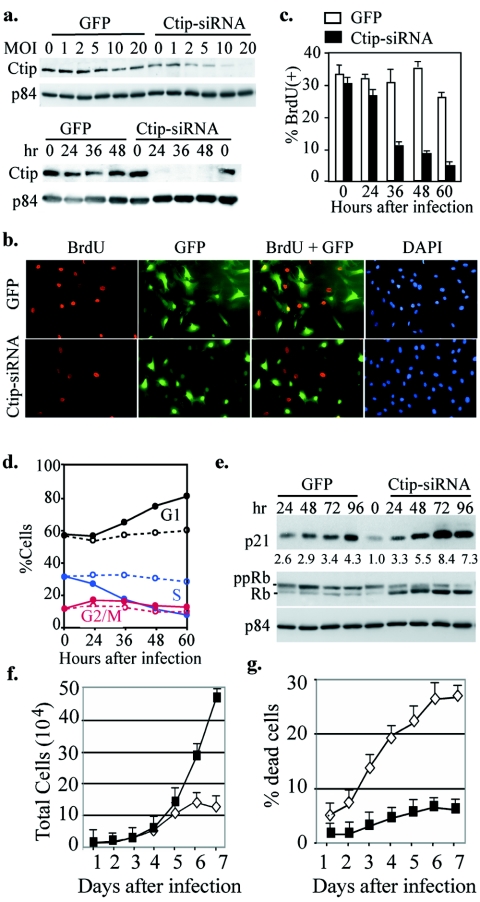
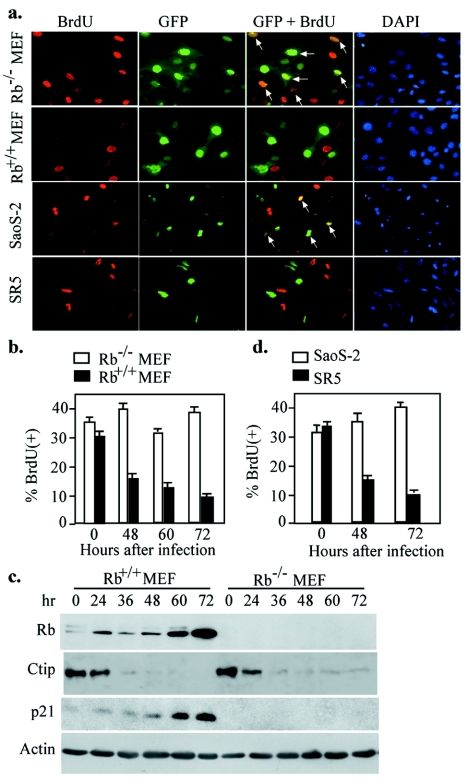
To explore the mechanism of how CtIP functions in S-phase entry, we attempted to establish Ctip−/− cells in culture for this pursuit. However, the early lethality of Ctip−/− embryo disables the establishment of mouse embryonic fibroblasts (MEF) for further study. To circumvent this difficulty, we constructed two adenoviruses carrying either GFP expression cassette only or the additional hairpin oligonucleotide for Ctip siRNA expression to deplete Ctip. Infection of NIH 3T3 cells with Ctip-siRNA adenovirus, but not control virus, at 10 multiplicities of infection (MOI) efficiently abrogated Ctip expression within 24 to 48 h (Fig. 3a). To determine whether the consequence of Ctip depletion by siRNA would be similar to that observed in vivo, we performed BrdU uptake assay in the time course after infection. Cells infected with the control adenovirus incorporated BrdU vigorously, while those infected with Ctip-siRNA virus failed to do so. The differences in BrdU incorporation between these two were obvious at 48 to 72 h postinfection (Fig. 3b and c). Similarly, by FACS analysis, we found that the population at G1, but not S or G2/M, increased significantly in Ctip-siRNA virus-infected cells compared to those infected with the control virus (Fig. 3d). Consistently, protein markers for G1 phase, namely, the cdk inhibitor p21, and hypo-phosphorylated form of Rb increased following Ctip-siRNA virus infection compared with the control (Fig. 3e). Cells depleted of Ctip eventually led to cell death after 6 days (Fig. 3f and g), further supporting the observation described for the early Ctip−/− embryos.
FIG. 3.
Depletion of Ctip by infection with adenovirus carrying Ctip siRNA lead to inhibition of S-phase entry. a, Infection with Ctip-siRNA adenovirus abrogated Ctip expression in a dose- and time-dependent manner. NIH 3T3 cells were infected with different MOI of GFP or Ctip-siRNA adenovirus for 48 h, and the corresponding cell lysates were prepared for Western analysis and probed with anti-Ctip or anti-p84, serving as internal loading control (upper panel). Similarly, NIH 3T3 cells were infected with 10 MOI and harvested at different time points for Western blotting analysis as described above (lower panel). b, Cells were labeled with BrdU for 4 h after infection with 10 MOI of GFP or Ctip-siRNA adenovirus for 48 h and stained with anti-BrdU antibody. c, Graphic quantitative summary of cells labeled with BrdU at different time points after infection with adenoviruses as described in panel b. d, Graphic summary of data from FACS analysis of cells collected at various time points after infection with GFP (dashed lines) or Ctip-siRNA viruses (solid lines). e, NIH 3T3 cells were infected with 10 MOI of GFP or Ctip-siRNA and harvested at different time points for Western blotting probed with anti-p21, Rb, or p84 antibodies. In cells depleted of Ctip, Rb became hypophosphorylated and p21 accumulated. f and g, NIH 3T3 cells were infected as indicated for panel e and cells were collected and stained with trypan blue. The numbers of both total (f) and trypan blue-positive cells were counted. The percentages of cell death are plotted in panel g.
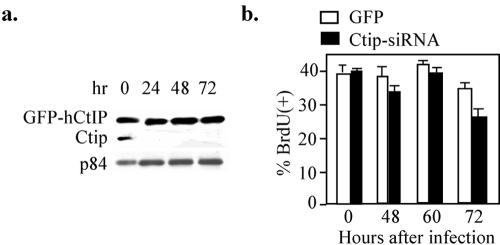
To test whether this effect was solely caused by Ctip inactivation, we ectopically expressed human CtIP, which was resistant to the mouse Ctip-siRNA due to the variation in primary sequences, in NIH 3T3 cells prior to Ctip-siRNA virus infection (Fig. 4a). As shown in Fig. 4b, expression of human CtIP rescued these cells from G1 arrest mediated by Ctip-siRNA virus infection. These results demonstrate unambiguously that Ctip-depleted cells fail to enter S phase and are arrested in G1, a phenotype similar to that observed in vivo.
FIG. 4.
Expression of the human CtIP restored proliferation of NIH 3T3 cells with mCtip knocked down. a, Human GFP-CtIP fusion protein stably expressed in NIH 3T3 cells is resistant to Ctip-siRNA depletion. A stable clone of NIH 3T3 cells expressing human GFP-CtIP fusion protein was established and infected with Ctip-siRNA adenoviruses. Cell lysates prepared from the infected cells were analyzed by Western blotting probed with anti-CtIP and anti-p84 antibodies. b. The stable clone of NIH 3T3 cells was labeled with BrdU after infection with 10 MOI of GFP or Ctip-siRNA for different time intervals, and the results of BrdU incorporation are summarized.
CtIP facilitates S entry by releasing Rb-mediated G1 restraint.
It is known that CtIP interacts with RB (8, 27), an essential regulator for G1/S progression (4, 11). Therefore, Ctip may be required for removing G1 restraint mediated by Rb. To test this possibility, we performed Ctip siRNA knockdown using a pair of isogenic Rb−/− and Rb+/+ MEFs. Infection of Rb−/−, but not Rb+/+, MEF with Ctip-siRNA adenoviruses had little influence on S-phase entry, suggesting that depletion of Ctip expression in Rb−/− cells failed to arrest cells at G1 (Fig. 5a and b). Instead, infection of both MEFs with GFP adenoviruses did not show any difference in S-phase entry (data not shown). Consistently, G1 protein markers, p21, and hypophosphorylated form of Rb were accumulated following Ctip-siRNA virus infection in Rb+/+ MEFs, while no Rb or p21 expression was detected in Rb−/− MEFs (Fig. 5c). Similar phenomena were observed in a pair of human osteosarcoma cells, RB-deficient Saos-2 and RB-reintroduced SR5 (11), transfected with plasmids expressing human CtIP siRNA (Fig. 5a and d). These results, taken together, suggest that CtIP is required to remove RB-mediated G1 restraint for the progression from G1 to S phase.
FIG. 5.
Ctip-mediated G1 control is Rb dependent. a, Immunofluorescence staining of BrdU uptake in the pair of Rb+/+ and Rb−/− MEFs (×400) infected with Ctip-siRNA adenovirus or in the other pair of human osteosarcoma cells (×200), Saos-2, and SR5, transiently transfected with pCtIP-RNAi plasmids. Cells were labeled with BrdU 48 h after infection or transfection. b, Summary of BrdU incorporation of Rb+/+ and Rb−/− MEFs at 48, 60, and 72 h after infection. c, The infected Rb+/+ and Rb−/− MEFs were collected for Western blotting probed with antibodies against CtIP, Rb, p21, and actin. d, Summary of the BrdU-positive cells of Saos-2 and SR5, after transfection with either pCtIP-RNAi plasmid for 48 or 72 h.
The life span of Ctip+/− heterozygotes is shortened by multiple tumor formation.
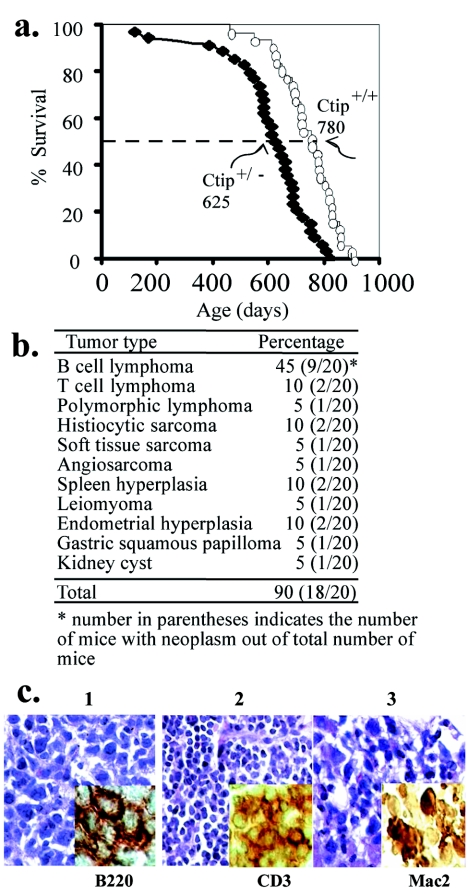
The essential role of Ctip in regulating S-phase entry allows us to speculate that deregulation of Ctip will have a significant impact on mouse development. To investigate this possibility, a cohort of more than 30 Ctip+/− mice was studied longitudinally. Ctip+/− mice died around day 625, while the life span of homozygous Ctip+/+ mice from similar genetic crosses was about 780 days (Fig. 6a). These mice, once in the moribund state, had succumbed to multiple tumors, predominantly, large lymphomas of both B and T cells (Fig. 6b and c), suggesting that Ctip is a bona fide tumor susceptibility gene.
FIG. 6.
Life span and tumor spectra of Ctip heterozygous mice. a. Loss of one Ctip allele shortened the life span of Ctip+/− compared with Ctip+/+ mice. b. Tumor incidence and spectra of Ctip+/− mice. c. Lymphatic malignancies developed in Ctip+/− mice. H&E staining was used to visualize tumor histology. In addition, antibodies specific for the distinctive tumor antigens were used (see inset). For example, B-cell lymphoma (1), T-cell lymphoma (2), and histiocytic sarcoma (3) were detected with anti-B220, anti-CD3, and anti-Mac2, respectively.
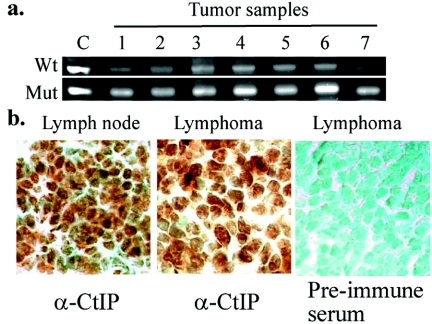
To test whether CtIP contributes to tumorigenesis following the established recessive mechanism (18, 31), we then performed microdissection to isolate those tumor cells and analyzed the Ctip genotype by PCR. In seven out of seven tumor samples, they retained the wild-type allele of Ctip (Fig. 7a). Similarly, 10 out of 10 tumors, in addition to 10 normal tissue specimens, were stained positively with anti-Ctip antibodies. A pair of normal lymph node and lymphoma cells are shown in Fig. 7b, indicating that the remaining wild-type allele is expressed. Thus, the tumor formation in these mice may be attributed to haploid insufficiency of Ctip.
FIG. 7.
Tumors derived from the Ctip heterozygotes contain an active wild-type allele of Ctip. a. PCR analysis of microdissected tumor samples showed the presence of the wild-type Ctip allele. b. Positive immunostaining with anti-Ctip antibodies, but not with preimmune antibodies, of normal lymph node and lymphoma from Ctip+/− mice. Specimens were counterstained with methylene green (×630).
DISCUSSION
In this communication, we generated an inactivated Ctip allele in mice to address how CtIP exerts its biological function during mouse development. We showed that the Ctip−/− embryos died very early, as the blastocysts failed to enter S phase, leading to a slightly elevated cell death. Consistently, depletion of Ctip arrested Rb+/+, but not Rb−/−, MEF or human osteosarcoma Saos-2 cells at G1, suggesting that this arrest is RB dependent. Importantly, Ctip+/− heterozygotes had their life spans shortened by the generation of tumors, predominantly large lymphomas. The wild-type Ctip allele and protein remained detectable in these tumors, implicating that haploid insufficiency of Ctip leads to tumorigenesis. These findings suggest that deregulation of CtIP mediated S-phase entry leads to a defect in early embryogenesis and contributes, in part, to tumor formation.
Ctip interacts with Ctbp (34), Brca1 (42, 46), Ikaros (19), and Rb (8, 27). The Ctip null phenotype is much severer than those observed in mice with null genotype of those interacting partners. Ctip−/− embryos die at E4 because the blastocysts fail to enter S phase, leading to an elevated cell death. On the other hand, the Ctbp1 null mice survive 20 days postnatally, while the Ctbp2-null are embryonically lethal at E10.5 (14). Brca1−/− or Rb−/− mouse embryos die at E6.5 or E14.5 (12, 21, 24), respectively, while Ikaros−/− mice are viable (9). Thus, CtIP plays a critical factor in very early embryogenesis.
The role of Ctip in G2/M checkpoint control has been reported previously (23). It was postulated that the interaction between Ctip and BRCA1 is essential for this checkpoint activity. However, whether Ctip plays roles in different cell cycle stage remains unclear. We observed that the depletion of Ctip in MEFs leads to G1 arrest, suggesting an important role of Ctip in G1 regulation. Intriguingly, our recent preliminary results revealed that depletion of Ctip in different cell types generated defects at distinct cell cycle stages, suggesting that the role of Ctip in cell cycle progression may manifest differentially depending on cell status.
On the basis of the result that depletion of Ctip arrested Rb+/+, but not Rb−/−, MEFs at G1 (Fig. 5), the Rb deficiency, at least in part, mitigates the effect of Ctip deficiency in this cell type. It was previously proposed that CtIP has an adaptor role in connecting Rb family proteins for recruiting corepressor CtBP (27). If this is the case, how this Ctip adaptor counteracts Rb in G1 restraint is intriguing. Since RB plays diverse, but concerted, roles in G1/S transition, CtIP may serve as an adaptor to recruit different factors, which counter Rb action in this context. For example, RB is mainly inactivated through hyperphosphorylation by cyclin D/CDK4/6 and cyclin E/CDK2 complexes at G1/S transition (4); however, why the presence of those kinases capable of hyperphosphorylation of RB is not sufficient for cells to enter S phase when CtIP is depleted is not known. One possible explanation is that CtIP represses the transcription of Cdk inhibitor p21 (22). In the absence of CtIP, p21 becomes derepressed and inhibits Cdk activity, leading to Rb hypophosphorylation as shown in Fig. 3 and 5. However, increased expression of p21 in Rb−/− MEF was not observed even when Ctip was depleted, suggesting that activation of p21 expression by Ctip requires Rb. Consistently, Rb has been reported to activate p21 transcription through Sp1 and Sp3 (6). When Rb is not available for transactivation, p21 is not expressed and the repression function of Ctip becomes insignificant. In this regard, Ctip may recruit a repressor to counteract the Rb-mediated coactivation of p21. However, not all the Rb-null cells fail to express p21 (35), suggesting that a much more complicated mechanism for regulating p21 exists.
In addition, active Rb has been shown to repress E2F-regulated S-phase genes, such as MCMs, DNA polymerase subunits, and deoxynucleoside triphosphate synthetic enzymes (25). If Ctip is critical for S-phase entry, such repression by Rb/E2F has to be released by the presence of Ctip. In this situation, Ctip has to recruit activators to remove the repression of Rb. However, there are very few transcriptional processes involved in early blastocyst division, albeit depletion of Ctip renders blastocysts defective in entering S phase. Thus, additional mechanism for CtIP in negatively regulating RB at the G1/S transient may exist.
In addition to the transcriptional repression function, RB also has several functions involving DNA replication and chromatin remodeling (47). DNA replication is initiated at specific chromatin elements referred to as replication origins or replicons, on which origin recognition complex could load and in turn sequentially recruit other replication components, including Cdc6, Cdt1, and Mcm2-7, to assemble the prereplication complex. Initiation of replication is then triggered by kinase activities of cyclin-dependent kinase and Cdc7/Dbf4 and by association of other replication factors at G1/S transition (1). Since RB is able to directly inhibit DNA replication by binding to DNA replication factors such as RFC, MCM7, and DNA polymerase α (29, 36, 39), CtIP may be essential for removing RB from replication origin to release RB restraint for DNA replication and S-phase entry. This possibility is under vigorous investigation.
Besides the synthesis of DNA, it has become clear that the chromatin structure maintaining the epigenetic information is critical for cell cycle progression (26). Immediately before or after replication, a subset of proteins that could modify and remodel the DNA and nucleosomes, such as DNA methyltransferase DNMT1 and chromatin assembly factor CAF1, are recruited in the vicinity of replication fork to alter chromatin domains for a variety of DNA processing, including replication, repair, and recombination (5, 15, 44). RB binds to DNMT1, which may contribute to the repression activity (32). Thus, it is likely that binding of CtIP to RB may be essential for restraining RB from the repression status, although the precise mechanism remains to be elucidated.
Interestingly, the Ctip heterozygous mice succumbed to multiple tumors, predominantly, large lymphomas of both B and T cells (Fig. 6b and c), suggesting that Ctip is a bona fide tumor susceptibility gene. In those tumor cells, they retained the wild-type allele of Ctip (Fig. 7a) and were stained positively with anti-Ctip antibodies (Fig. 7b), indicating that the remaining wild-type allele is active. Thus, the tumor formation in these mice may be attributed to haploid insufficiency of Ctip. This phenomenon has been seen in several cases, including TGFβ1 and Dmp1 (16, 40).
It is intriguing why reduction in Ctip expression predisposes mice to tumor formation. Since the interacting partners of Ctip including Rb and BRCA1 as well as Ikaros are tumor susceptibility proteins (21, 41, 43), disturbance of the delicate regulation by reducing the availability of the other significant partner may have a similar consequence in tumorigenesis. For example, reducing Ctip, which counteracts Rb in mediating G1/S progression, will increase active form of Rb and disturb Rb-mediated G1 to S progression. It was reported that expression of a hypophosphorylated form of Rb in mice leads to breast carcinoma, perhaps through reducing apoptosis, with low incidence (17). The G2/M checkpoint defect and the genomic instability resulting from the limited BRCA1/CtIP interaction could also trigger tumorigenesis in principle (23), although Brca1 heterozygous mice rarely have an increased tumor incidence (12, 24; W. H. Lee et al., unpublished). Thus, tumor predisposition in Ctip heterozygotes is not unexpected. However, it is surprising to learn that most Ctip+/− mice developed multiple tumors, predominantly lymphoma, at an age as early as 6 months. Ikaros may mediate the tissue specificity of developing large lymphoma in Ctip heterozygotes, because this transcriptional repressor is a key regulator for lymphocyte development and malignancy (9, 30, 41). Interestingly, the transcriptional repression activity of Ikaros, similar to Rb, is modulated by phosphorylation in G1 (10). CtIP may counteract RB as well as Ikaros to release the restraint at G1. Although the detailed mechanism is waiting for further elucidation, the finding described above uncovers, for the first time, a novel G1/S regulation in that CtIP directly counteracts RB restraint. Disturbing this regulation will lead to severe consequences involving embryonic lethality and tumorigenesis.
Acknowledgments
We thank Houyi Wang and Andrea Nikitin for their technical contribution at the early stage of the project and Saori Furuta for her critical reading.
This work was supported by grants from the National Institutes of Health (CA 94170 to W.-H.L. and CA 85605 to P.-L.C.) and a postdoctoral fellowship from Susan G. Komen Foundation to F.L.
REFERENCES
- 1.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 2.Boyd, J. M., T. Subramanian, U. Schaeper, M. La Regina, S. Bayley, and G. Chinnadurai. 1993. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phos-phoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 12:469-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chinnadurai, G. 2002. CtBP family proteins: more than transcriptional corepressors. BioEssays 25:9-12. [DOI] [PubMed] [Google Scholar]
- 4.Classon, M., and E. Harlow. 2002. The retinoblastoma tumour suppressor in development and cancer. Nat. Rev. Cancer 2:910-917. [DOI] [PubMed] [Google Scholar]
- 5.Collins, N., R. A. Poot1, I. Kukimoto, C. García-Jiménez1, G. Dellaire, and P. D. Varga-Weisz1. 2002. An ACF1-ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat. Genet. 32:627-632. [DOI] [PubMed] [Google Scholar]
- 6.Decesse, J. T., S. Medjkane, M. B. Datto, and C. E. Cremisi. 2001. RB regulates transcription of the p21/WAF1/CIP1 gene. Oncogene 20:962-971. [DOI] [PubMed] [Google Scholar]
- 7.Dubin, M. J., P. H. Stokes, E. Y. Sum, R. S. Williams, V. A. Valova, P. J. Robinson, G. J. Lindeman, J. N. Glover, J. E. Visvader, and J. M. Matthews. 2004. Dimerization of CtIP, a BRCA1- and CtBP-interacting protein, is mediated by an N-terminal coiled-coil motif. J. Biol. Chem. 279:26932-26938. [DOI] [PubMed] [Google Scholar]
- 8.Fusco, C., A. Reymond, and A. S. Zervos. 1998. Molecular cloning and characterization of a novel retinoblastoma-binding protein. Genomics 51:351-358. [DOI] [PubMed] [Google Scholar]
- 9.Georgopoulos, K., M. Bigby, J. H. Wang, A. Molnar, P. Wu, S. Winandy, and A. Sharpe. 1994. The Ikaros gene is required for the development of all lymphoid lineages. Cell 79:143-156. [DOI] [PubMed] [Google Scholar]
- 10.Gomez-del Arco, P., K. Maki, and K. Georgopoulos. 2004. Phosphorylation controls Ikaros's ability to negatively regulate the G1-S transition. Mol. Cell. Biol. 24:2797-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodrich, D. W., N. P. Wang, Y. W. Qian, E. Y. Lee, and W. H. Lee. 1991. The retinoblastoma gene product regulates progression through the G1 phase of the cell cycle. Cell 67:293-302. [DOI] [PubMed] [Google Scholar]
- 12.Hakem, R., J. L. de la Pompa, C. Sirard, R. Mo, M. Woo, A. Hakem, A. Wakeham, J. Potter, A. Reitmair, F. Billia, E. Firpo, C. C. Hui, J. Roberts, J. Rossant, and T. W. Mak. 1996. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell 85:1009-1023. [DOI] [PubMed] [Google Scholar]
- 13.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hildebrand, J. D., and P. Soriano. 2002. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol. Cell. Biol. 22:5296-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoek, M., and B. Stillman. 2003. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc. Natl. Acad. Sci. USA 100:12183-12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue, K., F. Zindy, D. H. Randle, J. E. Rehg, and C. J. Sherr. 2001. Dmp1 is haplo-insufficient for tumor suppression and modifies the frequencies of Arf and p53 mutations in Myc-induced lymphomas. Genes Dev. 15:2934-2939.11711428 [Google Scholar]
- 17.Jiang, Z., and E. Zacksenhaus. 2002. Activation of retinoblastoma protein in mammary gland leads to ductal growth suppression, precocious differentiation, and adenocarcinoma. J. Cell Biol. 156:185-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knudson, A. G. J. 1971. Mutation and cancer: statistical study of retinoblastoma. Proc. Natl. Acad. Sci. USA 68:820-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koipally, J., and K. Georgopoulos. 2002. Ikaros-CtIP interactions do not require C-terminal binding protein and participate in a deacetylase-independent mode of repression. J. Biol. Chem. 277:23143-23149. [DOI] [PubMed] [Google Scholar]
- 20.Koipally, J., A. Renold, J. Kim, and K. Georgopoulos. 1999. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J. 18:3090-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, E. Y., C. Y. Chang, N. Hu, Y. C. Wang, C. C. Lai, K. Herrup, W. H. Lee, and A. Bradley. 1992. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature 359:288-294. [DOI] [PubMed] [Google Scholar]
- 22.Li, S., P. L. Chen, T. Subramanian, G. Chinnadurai, G. Tomlinson, C. K. Osborne, Z. D. Sharp, and W. H. Lee. 1999. Binding of CtIP to the BRCT repeats of BRCA1 involved in the transcription regulation of p21 is disrupted upon DNA damage. J. Biol. Chem. 274:11334-11338. [DOI] [PubMed] [Google Scholar]
- 23.Li, S., N. S. Ting, L. Zheng, P. L. Chen, Y. Ziv, Y. Shiloh, E. Y. Lee, and W. H. Lee. 2000. Functional link of BRCA1 and ataxia telangiectasia gene product in DNA damage response. Nature 406:210-215. [DOI] [PubMed] [Google Scholar]
- 24.Liu, C. Y., A. Flesken-Nikitin, S. Li, Y. Zeng, and W. H. Lee. 1996. Inactivation of the mouse Brca1 gene leads to failure in the morphogenesis of the egg cylinder in early postimplantation development. Genes Dev. 10:1835-1843. [DOI] [PubMed] [Google Scholar]
- 25.Markey, M. P., S. P. Angus, M. W. Strobeck, S. L. Williams, R. W. Gunawardena, B. J. Aronow, and E. S. Knudsen. 2002. Unbiased analysis of RB-mediated transcriptional repression identifies novel targets and distinctions from E2F action. Cancer Res. 62:6587-6597. [PubMed] [Google Scholar]
- 26.McNairn, A. J., and D. M. Gilbert. 2003. Epigenomic replication: linking epigenetics to DNA replication. BioEssays 25:647-656. [DOI] [PubMed] [Google Scholar]
- 27.Meloni, A. R., E. J. Smith, and J. R. Nevins. 1999. A mechanism for Rb/p130-mediated transcription repression involving recruitment of the CtBP corepressor. Proc. Natl. Acad. Sci. USA 96:9574-9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng, A., and P. L. Chen. 2003. NFBD1, like 53BP1, is an early and redundant transducer mediating Chk2 phosphorylation in response to DNA damage. J. Biol. Chem. 278:8873-8876. [DOI] [PubMed] [Google Scholar]
- 29.Pennaneach, V., I. Salles-Passador, A. Munshi, H. Brickner, K. Regazzoni, F. Dick, N. Dyson, T. T. Chen, J. Y. Wang, R. Fotedar, and A. Fotedar. 2001. The large subunit of replication factor C promotes cell survival after DNA damage in an LxCxE motif- and Rb-dependent manner. Mol. Cell 7:715-727. [DOI] [PubMed] [Google Scholar]
- 30.Rebollo, A., and C. Schmitt. 2003. Ikaros, Aiolos and Helios: transcription regulators and lymphoid malignancies. Immunol. Cell Biol. 81:171-175. [DOI] [PubMed] [Google Scholar]
- 31.Riley, D. J., E. Y. Lee, and W. H. Lee. 1994. The retinoblastoma protein: more than a tumor suppressor. Annu. Rev. Cell Biol. 10:1-29. [DOI] [PubMed] [Google Scholar]
- 32.Robertson, K. D., S. Ait-Si-Ali, T. Yokochi, P. A. Wade, P. L. Jones, and A. P. Wolffe. 2000. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25:338-342. [DOI] [PubMed] [Google Scholar]
- 33.Schaeper, U., J. M. Boyd, S. Verma, E. Uhlmann, T. Subramanian, and G. Chinnadurai. 1995. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc. Natl. Acad. Sci. USA 92:10467-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaeper, U., T. Subramanian, L. Lim, J. M. Boyd, and G. Chinnadurai. 1998. Interaction between a cellular protein that binds to the C-terminal region of adenovirus E1A (CtBP) and a novel cellular protein is disrupted by E1A through a conserved PLDLS motif. J. Biol. Chem. 273:8549-8552. [DOI] [PubMed] [Google Scholar]
- 35.Sheahan, S., C. O. Bellamy, L. Treanor, D. J. Harrison, and S. Prost. 2004. Additive effect of p53, p21 and Rb deletion in triple knockout primary hepatocytes. Oncogene 23:1489-1497. [DOI] [PubMed] [Google Scholar]
- 36.Sterner, J. M., S. Dew-Knight, C. Musahl, S. Kornbluth, and J. M. Horowitz. 1998. Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with MCM7. Mol. Cell. Biol. 18:2748-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sui, G., C. Soohoo, B. Affar el, F. Gay, Y. Shi, W. C. Forrester, and Y. Shi. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 99:5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sum, E. Y., B. Peng, X. Yu, J. Chen, J. Byrne, G. J. Lindeman, and J. E. Visvader. 2002. The LIM domain protein LMO4 interacts with the cofactor CtIP and the tumor suppressor BRCA1 and inhibits BRCA1 activity. J. Biol. Chem. 277:7849-7856. [DOI] [PubMed] [Google Scholar]
- 39.Takemura, M., T. Kitagawa, S. Izuta, J. Wasa, A. Takai, T. Akiyama, and S. Yoshida. 1997. Phosphorylated retinoblastoma protein stimulates DNA polymerase alpha. Oncogene 15:2483-2492. [DOI] [PubMed] [Google Scholar]
- 40.Tang, B., E. P. Bottinger, S. B. Jakowlew, K. M. Bagnall, J. Mariano, M. R. Anver, J. J. Letterio, and L. M. Wakefield. 1998. Transforming growth factor-beta1 is a new form of tumor suppressor with true haploid insufficiency. Nat. Med. 4:802-807. [DOI] [PubMed] [Google Scholar]
- 41.Wang, J. H., A. Nichogiannopoulou, L. Wu, L. Sun, A. H. Sharpe, M. Bigby, and K. Georgopoulos. 1996. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity 5:537-549. [DOI] [PubMed] [Google Scholar]
- 42.Wong, A. K., P. A. Ormonde, R. Pero, Y. Chen, L. Lian, G. Salada, S. Berry, Q. Lawrence, P. Dayananth, P. Ha, S. V. Tavtigian, D. H. Teng, and P. L. Bartel. 1998. Characterization of a carboxy-terminal BRCA1 interacting protein. Oncogene 17:2279-2285. [DOI] [PubMed] [Google Scholar]
- 43.Xu, X., K. U. Wagner, D. Larson, Z. Weaver, C. Li, T. Ried, L. Hennighausen, A. Wynshaw-Boris, and C. X. Deng. 1999. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat. Genet. 22:37-43. [DOI] [PubMed] [Google Scholar]
- 44.Ye, X., A. A. Franco, H. Santos, D. M. Nelson, P. D. Kaufman, and P. D. Adams. 2003. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol. Cell 11:341-351. [DOI] [PubMed] [Google Scholar]
- 45.Yu, X., and R. Baer. 2000. Nuclear localization and cell cycle-specific expression of CtIP, a protein that associates with the BRCA1 tumor suppressor. J. Biol. Chem. 275:18541-18549. [DOI] [PubMed] [Google Scholar]
- 46.Yu, X., L. C. Wu, A. M. Bowcock, A. Aronheim, and R. Baer. 1998. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J. Biol. Chem. 273:25388-25392. [DOI] [PubMed] [Google Scholar]
- 47.Zheng, L., and W.-H. Lee. 2002. Retinoblastoma tumor suppressor and genome stability. Adv. Cancer Res. 85:13-50. [DOI] [PubMed] [Google Scholar]