Abstract
Chromatin insulators, or boundary elements, appear to control eukaryotic gene expression by regulating interactions between enhancers and promoters. Boundaries have been identified in the 3′ cis-regulatory region of Abd-B, which is subdivided into a series of separate iab domains. Boundary elements such as Mcp, Fab-7, and Fab-8 and adjacent silencers flank the iab domains and restrict the activity of the iab enhancers. We have identified an insulator in the 755-bp Mcp fragment that is linked to the previously characterized Polycomb response element (PRE) and silences the adjacent genes. This insulator blocks the enhancers of the yellow and white genes and protects them from PRE-mediated repression. The interaction between the Mcp elements, each containing the insulator and PRE, allows the eye enhancer to activate the white promoter over the repressed yellow domain. The same level of white activation was observed when the Mcp element combined with the insulator alone was interposed between the eye enhancer and the promoter, suggesting that the insulator is responsible for the interaction between the Mcp elements.
The discovery that eukaryotic transcriptional activators can operate over long distances in an orientation-independent manner posed several questions (3). One of these was how the long-range activation potential of eukaryotic enhancers could be restricted to the relevant target promoter. It has been proposed that eukaryotic chromatin is organized into functionally independent domains to prevent illegitimate enhancer-promoter communication. Recently, chromatin insulators, or boundary elements, have been identified. These DNA sequences are functionally characterized by two properties: they prevent enhancer-promoter interactions and buffer transgenes from the chromosomal-position effects of genomic sequences adjoining the transgene insertion site (11, 14, 22, 23, 36, 42, 43).
An idea of a probable role of insulators in the cell can be gained from data on their distribution in the bithorax complex (36, 39). The three homeotic genes of the bithorax complex, Ultrabithorax (Ubx), abdominal-A (abd-A), and Abdominal-B (Abd-B), are responsible for specifying the identity of parasegments 5 to 14 (PS5 to PS14), which form the posterior half of the thorax and all abdominal segments of an adult fly (24, 32). The PS-specific expression patterns of Ubx, abd-A, and Abd-B are determined by a complex cis-regulatory region that spans a 300-kb DNA segment (32). For example, Abd-B expression in PS10, PS11, PS12, and PS13 is controlled by the iab-5, iab-6, iab-7, and iab-8 cis-regulatory domains, respectively (4, 9, 10, 12, 19, 24, 40). Each iab domain appears to contain at least one enhancer that initiates Abd-B expression in the early embryo, as well as a PRE (Polycomb response element) silencer element that maintains the expression pattern throughout development (2, 5, 6, 17, 18, 29, 31, 33, 44, 45). It has been proposed that insulators flank each iab region and organize the Abd-B regulatory DNA into a series of separate chromatin loop domains (13, 16, 32). The recent finding that iab-7 is flanked by two insulators, Fab-7 and Fab-8, is consistent with this model (2, 17, 44, 45). The third boundary element identified, Mcp, preserves the functional autonomy of the iab-4 and iab-5 cis-regulatory domains (19, 20, 24).
It has recently been found that a core 800-bp Mcp sequence from the iab-5 regulatory region of the Abd-B gene can mediate trans-regulation between transgenes located at distant sites of the same chromosome arm or even on different arms (34). Previous data suggest that this 800-bp Mcp element functions both as a silencer and as a domain boundary element (20, 32). A silencer in the 138-bp minimal element was mapped due to its ability to maintain silencing during imaginal disk development (7). However, the predicted boundary element in the Mcp has not yet been identified (7, 34).
We describe here a PRE-linked insulator identified in the 755-bp Mcp (7). This 340-bp insulator blocks the activities of the yellow and white enhancers and the Mcp silencer. Interactions between the 755-bp Mcp elements or between the Mcp and the insulator facilitate activation of the white promoter by the eye enhancer over the repressed yellow domain, suggesting that the insulator alone is sufficient for the interaction between the Mcp elements.
MATERIALS AND METHODS
Transgenic constructs.
An 8-kb fragment containing the yellow gene was kindly provided by P. Geyer. The 5-kb BamHI-BglII fragment containing the coding region (yc) was subcloned into CaSpeR2 (C2-yc) or CaSpeR3 (C3-yc). The CaSpeR vectors carrying the white gene were kindly provided by V. Pirrotta. The 3-kb SalI-BamHI fragment containing the yellow regulatory region (yr) was subcloned into BamHI- and XhoI-cleaved pGEM7 (yr plasmid).
A 755-bp PstI-PstI fragment containing the central Mcp fragment was cloned by PCR. This fragment was subcloned between two lox sites [lox(Mcp)]. The lox(Mcp) fragment was inserted into C3-yc between the yellow and white genes [C3-lox(Mcp)-yc]. The M508 fragment was obtained by PCR amplification of the DNA fragment between 5′ GCTCAGAGTACATAAGCG 3′ and 5′ CTGCAGTCAAACGTCAC 3′. The M340 fragment was obtained by PCR amplification of the DNA fragment between primers 5′ GCTCAGAGTACATAAGCG 3′ and 5′ CCCAATCGTTGTAAGTG 3′. The M340 and M508 fragments were subcloned between two lox sites [lox(M340) and lox(M508)].
The white regulatory sequences from bp −1084 to −1465 relative to the transcription start site (Ee) were cloned between two frt sites [frt(Ee)]. These sequences contained the testis and eye enhancers. The frt(Ee) fragment was then inserted between the body and wing enhancers at position −1868 from the yellow transcription start site [yr-frt(Ee)].
To construct Ey(Mcp)YW and Ey(McpR)YW, the lox(Mcp) was inserted in both orientations into the yr plasmid cleaved by Eco47III at position −893 from the yellow transcription start site [yr-lox(Mcp)]. The yr-lox(Mcp) fragments were ligated into C3-yc cleaved by XbaI and BamHI.
To construct Ey(M508R)YW, the lox(M508) fragment was inserted in the inverse orientation into the yr plasmid cleaved by Eco47III at position −893 from the yellow transcription start site [yr-lox(M508R)]. The yr-lox(M508R) fragment was ligated into C3-yc cleaved by XbaI and BamHI.
To construct Eye(M340)YW, the lox(M340) fragment was inserted into yr-Ee cleaved by Eco47III at position −893 from the yellow transcription start site [yr-Ee-lox(M340)]. The yr-Ee-lox(M340) fragment was ligated into C3-yc cleaved by XbaI and BamHI.
To construct Ey(e)Mcp/M340YW, the Mcp fragment was inserted in the direct orientation into the yr-frt(Ee) plasmid cleaved by Eco47III at position −893 from the yellow transcription start site [yr-frt(Ee)-Mcp]. The M340 fragment was inserted into yr-frt(Ee)-Mcp cleaved by KpnI. The resulting yr-frt(Ee)-Mcp-M340 fragment was subcloned into C3-yc cleaved by XbaI and BamHI.
To construct (M340)EyeYW, the M340 fragment was inserted into the yr-Ee plasmid cleaved by NcoI (yr-M340-Ee). The yr-M340-Ee fragment was subcloned into C3-yc cleaved by XbaI and BamHI.
To construct Ey(e)Y(McpR)W, the yr-frt(Ee) fragment was ligated into C3-lox(McpR)-yc cleaved by XbaI and BamHI.
To construct Ey(e)McpY(McpR)W and Ey(e)McpRY(McpR)W, the Mcp fragment was inserted in the inverse orientation into the yr-frt(Ee) plasmid cleaved by Eco47III at position −893 from the yellow transcription start site [yr-frt(Ee)-McpR]. The resulting yr-frt(Ee)-McpR fragment and the previously described yr-frt(Ee)-Mcp fragment were subcloned into C3-lox(McpR)-yc cleaved by XbaI and BamHI.
To construct McpEy(e)Y(McpR)W, the Mcp fragment was inserted in the direct orientation into the yr-frt(Ee) plasmid cleaved by NcoI [yr-Mcp-frt(Ee)]. The yr-Mcp-frt(Ee) fragment was subcloned into C3-lox(McpR)-yc cleaved by XbaI and BamHI.
To construct Ey(e)M340Y(McpR)W, the yr-frt(Ee)-M340 fragment was subcloned into C3-lox(McpR)-yc cleaved by XbaI and BamHI.
Generation and analysis of the transgenic lines.
The construct and P25.7wc, a P element with defective inverted repeats used as a transposase source (21), were injected into yacw1118 preblastoderm embryos as described previously (38, 41). The resulting flies were crossed with yacw1118 flies, and the transgenic progeny were identified by their eye color. Chromosomal locations of various transgene insertions were determined by crossing the transformants with the yacw1118 balancer stock containing dominant markers: In(2RL),CyO for chromosome 2 and In(3LR)TM3,Sb for chromosome 3.
The lines with Mcp or eye enhancer excisions were obtained by crossing the flies bearing the transposons with lines expressing Flp (w1118; S2CyO, hsFLP, ISA/Sco;+) or Cre (y1, w1; Cyo, P[w+,cre]/Sco; +) recombinase. The Cre recombinase induces 100% excisions in the next generation. A high level of FLP recombinase (almost 100% efficiency) was produced by heat shock treatment for 2 h on the second and third days after hatching. All excisions were confirmed by PCR analysis with pairs of primers flanking the −893 insertion site (5′ ATCCAGTTGATTTTCAGGGACCA 3′ and 5′ TTGGCAGGTGATTTTGAGCATAC 3′) or the −343 insertion site (5′ TAGATCGTCAAATAAAGTCCCTA 3′ and 5′ GTTTGGTATGATTTTTGGCCTTC 3′) relative to the yellow transcription start site and the insertion site between the yellow and white genes (5′ TTTTCTTGAGCGGAAAAAGCGGA 3′ and 5′ ATCTACATTCTCCAAAAAAGGGT 3′). Details of the crosses used for genetic analysis and the excision of functional elements are available upon request.
The yellow (y) phenotype was determined from the level of pigmentation of the abdominal cuticle, wings, and bristles in 3- to 5-day-old males developing at 25°C. As a reference group, we used flies in which the y allele had been characterized previously. The level of pigmentation (i.e., of y expression) was estimated on an arbitrary five-grade scale: wild-type expression was assigned grade 5, and the absence of expression, grade 1. In the case of bristles, we used intermediate grades reflecting the level of variegation in pigmentation: weak variegation indicated that one to three bristles on the thorax and head were yellow; medium variegation, that approximately half the bristles were yellow; and extreme variegation, that only one to three bristles on the thorax and head were pigmented. The white (w) phenotype was determined from eye pigmentation in adult flies. Wild-type white expression determined bright-red eye color (R); in the absence of white expression, the eyes were white (W). Intermediate levels of white expression (in increasing order) were reflected in the eye color, ranging from pale yellow (pY) to yellow (Y), dark yellow (dY), orange (Or), dark orange (dOr), and, finally, brown (Br) or brown-red (BrR).
RESULTS
We examined the 755-bp PstI-PstI Mcp fragment (Fig. 1A) included in the 800-bp core Mcp sequence described previously (7, 34). The 755-bp Mcp fragment contains a 138-bp silencer that is sufficient to maintain silencing during imaginal disk development (7). However, the Mcp core fragment required additional sequences to act as a pairing-sensitive silencer of the white gene in adult eyes (6, 34). These additional sequences could be taken from the yellow regulatory region (34). For this reason, we used the model system containing both the yellow and white loci to map the silencer and boundary elements in the 755-bp Mcp fragment. The yellow gene is required for dark pigmentation of larval and adult cuticle and its derivatives. Two enhancers located upstream of the promoter are responsible for its activation in the body cuticle and the wing blades, while the enhancer responsible for yellow activation in bristles resides in the intron of the yellow gene (15, 28). The white gene is required for eye pigmentation and is regulated by its eye-specific enhancer (37).
FIG. 1.
(A) Diagrams of the 755-bp Mcp element and its 508-bp (M508R) and 340-bp (M340) deletion derivatives. The PRE and a putative insulator are shown in gray and black, respectively. Restriction sites for SalI and AflII are indicated. (B) Maps of the constructs. The yellow wing [E(w)], body [E(b)], and bristle enhancers [E(br)] are shown as white ovals, and the eye enhancer [E(e)] as a gray oval (not to scale). The arrows labeled FRT and LOX indicate the target sites for Flp recombinase and Cre recombinase, respectively. The 755-bp Mcp, M508R, and M340 elements are shown as white, gray, and black triangles, respectively, with the vertex of a triangle indicating orientation (normal or inverse). The yellow and white genes are shown as rectangles, with arrows indicating the direction of transcription.
The strength of repression by the 755-bp Mcp element depends on its orientation.
To test the activity of the 755-bp Mcp element in our model system, we made two constructs, Ey(Mcp)YW and Ey(McpR)YW, containing the yellow and white genes (Fig. 1B). The 755-bp Mcp fragment was inserted at −893 relative to the yellow transcription start site, between the wing and body enhancers and the yellow promoter. The Mcp fragment was flanked by loxP sites (lox) in order to permit its excision from transgenic flies by crossing the latter with flies expressing Cre recombinase. The 755-bp Mcp element was inserted in such a way that the 138-bp silencer was located closer to the yellow gene in Ey(Mcp)YW and closer to the yellow enhancers in Ey(McpR)YW (Fig. 1B).
Twelve transgenic lines containing a single copy of the Ey(Mcp)YW transposon were obtained. In 10 out of 12 transgenic lines heterozygous for Ey(Mcp)YW, bristle pigmentation was yellow (as in the y1-like null allele) or strongly variegated (Fig. 2). The location of the bristle enhancer in the yellow intron suggested that the Mcp element could repress the yellow promoter. The expression of the yellow gene in the wings and body was also markedly attenuated, which may be explained by two activities supposedly mediated by Mcp: silencing and enhancer blocking (insulation) (34). In 10 transgenic lines heterozygous for Ey(Mcp)YW, flies had eye pigmentation in the yellow-to-orange range. However, in the same lines containing 2 copies of the Ey(Mcp)YW transgene, the eye color of flies was lighter or did not change, suggesting that Mcp repressed white expression in a pairing-dependent manner, as was shown previously (34). Altogether, white expression was decreased in 11 out of 12 transgenic lines. To estimate the role of Mcp in repression, we deleted this element by inducing recombination between the lox sites (Fig. 2). The deletion of Mcp largely restored yellow expression and eliminated pairing-dependent silencing in most of the transgenic lines, suggesting that Mcp was responsible for repression.
FIG. 2.
Study of the polarity of Mcp-mediated repression based on analysis of yellow and white expression in transgenic lines heterozygous for the construct. Numbers above the rectangles show the levels of yellow pigmentation in the abdominal cuticle (Ab) (reflecting the activity of the body enhancer) and bristles (Br) on a five-grade arbitrary scale: wild-type expression was assigned grade 5 and the absence of expression, grade 1; var indicates variegated pigmentation (some bristles on the head and thorax were yellow, and some were pigmented). The number of lines with a corresponding phenotype is shown in the rectangle. In most lines, the level of pigmentation of wing blades (reflecting the activity of the wing enhancer) closely correlated with that of the abdominal cuticle. Wild-type white expression determined bright-red eye color (R); in the absence of white expression, the eyes were white (W). Intermediate levels of pigmentation, with the eye color ranging between pale yellow and white (pY), pale yellow and dark yellow (dY), dark yellow and orange (Or), or dark orange and brown-red (Br), reflect increasing levels of white expression. In the N/T ratio, N is the number of transgenic lines in which flies acquired a new y or w phenotype after the deletion of a DNA fragment flanked by either FRT or LOX sites, and T is the total number of transgenic lines examined. Numbers in parentheses show the ratio (P/P) between the lines displaying pairing-sensitive silencing of the white gene in flies homozygous for the construct and the total number of lines examined for each construct.
Ten transgenic lines carrying single copies of the Ey(McpR)YW construct were established (Fig. 2). The flies of all these lines exhibited a wild-type level of bristle pigmentation or weak variegation, indicating that Mcp weakly repressed yellow expression in bristles (Fig. 2). In Ey(McpR)YW lines, yellow expression in the body and wings was repressed to the same degree as in the transgenic Ey(Mcp)YW lines. In 5 out of 10 transgenic lines, the eyes did not become darker when there were 2 copies of the transgene, suggesting that the Mcp element located in the inverse orientation still repressed white expression. In the absence of Mcp, however, white expression in two transgenic lines was weaker in flies homozygous for the transposon than in heterozygous flies. Thus, only in 3 out of 10 transgenic lines was Mcp responsible for white repression in the case of homozygous Ey(McpR)YW. These results indicated that the ability of Mcp to repress yellow and white expression decreased considerably when the 138-bp silencer was separated from the yellow and white promoters by other Mcp sequences. Thus, the 755-bp Mcp element proved to have a polar repression activity.
The755-bp Mcp element contains an insulator.
The polarity of Mcp action could be explained by the presence of a boundary element in the 755-bp Mcp element. To map this putative boundary element, we deleted 247 bp from the end of the Mcp element that was opposite the silencer (Fig. 1A). In the EyM508RYW construct (Fig. 1B), the resulting 508-bp fragment, named M508, was flanked by the lox sites and inserted at −893 relative to the yellow transcription start site in such a way that the 138-bp silencer was located closer to the yellow enhancers.
In all nine transgenic Ey(M508R)YW lines obtained, the flies had a wild-type level of bristle pigmentation, but wing blades and body cuticle were entirely yellow (Fig. 2). In two transgenic lines displaying variegation of bristle pigmentation, the deletion of M508R had no effect on yellow expression, suggesting that M508R was not responsible for the repression of yellow in bristles. Comparing eye color in flies homozygous or heterozygous for the transgene in the presence or absence of M508R, we found that M508R induced pairing-sensitive repression of white in only one transgenic line. Thus, M508R failed to effectively repress the yellow and white genes when the silencer was separated from the yellow and white promoters by other sequences of the M508 element. This fact suggested that the 508-bp Mcp fragment still contained the putative boundary element that partially blocked the 138-bp silencer.
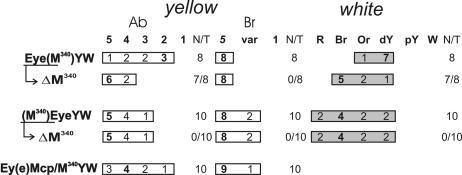
To test the putative boundary element in M508 for enhancer-blocking activity, we deleted the silencer from M508. The resulting 340-bp fragment (without the silencer), named M340 (Fig. 1A), was tested in the enhancer-blocking assay using the enhancers of the yellow and white genes. The eye enhancer was inserted between the body and wing enhancers. In the Eye(M340)YW construct, the M340 fragment flanked by the lox sites was inserted at −893 between the enhancers and promoters of the yellow and white genes (Fig. 1B). In seven out of eight transgenic Eye(M340)YW lines, wing and body pigmentation was reduced. The eye color was in the range characteristic of flies carrying the transgene without the white enhancer (Fig. 3). Deletion of the M340 fragment in the Ey(e)(ΔM340)YW derivatives increased yellow and white expression. At the same time, in all transgenic lines, flies displayed a wild-type level of bristle pigmentation and eye color became darker in the presence of 2 copies of the transgene, suggesting that the M340 element has no silencer activity, in contrast to the Mcp element.
FIG. 3.
Identification of an insulator in the Mcp element by analysis of yellow and white expression in transgenic lines. The phenotypes were examined in heterozygous flies. For designations, see the legend to Fig. 2.
If the M340 fragment functions as an insulator, it should not interfere with the enhancer-driven expression of the yellow and white genes when it is in the upstream position. To test this property, the M340 fragment flanked by the lox sites was inserted upstream of the yellow and white enhancers in the (M340)EyeYW construct (Fig. 1B). In 10 resulting transgenic lines, the deletion of the M340 fragment located upstream of the enhancers did not change yellow or white expression (Fig. 3). Thus, the M340 fragment in this location did not affect the activity of enhancers and, therefore, acted as an insulator.
To confirm that the M340 insulator blocked Mcp-mediated repression, we inserted M340 into the Ey(e)Mcp/M340YW construct at −343, between the yellow gene and the Mcp element (Fig. 1B). As shown above (Fig. 2), Mcp inserted at −893 in the direct orientation repressed yellow expression in most transgenic lines. By contrast, 9 out of 10 transgenic Ey(e)Mcp/M340YW lines had wild-type bristle pigmentation, suggesting that M340 blocked Mcp-mediated repression (Fig. 3). Interestingly, the flies of transgenic Ey(e)Mcp/M340YW lines had the same or darker wing and body pigmentation as the flies of transgenic lines carrying either Mcp or M340. This is indicative of the interaction between the M340 insulators that partially neutralizes the blocking of the yellow enhancers.
Interaction between two Mcp elements allows the eye enhancer to activate the white promoter over the repressed yellow domain.
At the next stage, we studied pairing interactions between two copies of the 755-bp Mcp element by using eye enhancer-white promoter communication as a model system. The Ey(e)McpY(McpR)W construct contained a 5′-Mcp copy at position −893 relative to the yellow transcription start site and a 3′-Mcp copy flanked by the lox sites between the yellow gene and the mini-white promoter. The 138-bp silencers of both Mcp elements were directed toward the yellow gene. The control Ey(e)Y(McpR)W construct contained only one 3′-Mcp copy (Fig. 1B). In 9 out of 10 transgenic Ey(e)McpY(McpR)W lines, yellow expression was almost completely repressed, while in Ey(e)Y(McpR)W lines it was affected only moderately (Fig. 4). Deletion of the 3′-Mcp in Ey(e)MY(McpR)W lines partially restored yellow expression, suggesting that the two copies of Mcp acted cooperatively (Fig. 4).
FIG. 4.
Analysis of transgenic lines for the interaction between two Mcp elements. The phenotypes were examined in flies heterozygous for the construct. For designations, see the legend to Fig. 2.
All 10 transgenic Ey(e)McpY(McpR)W lines had high levels of white expression (Fig. 4 and 5). The deletion of the eye enhancer markedly diminished eye pigmentation, indicating that the enhancer could activate the white promoter despite the presence of two intervening Mcp elements. Eye pigmentation was considerably weaker in transgenic lines with one Mcp element interposed between the eye enhancer and the white promoter [Ey(e)McpY(ΔMcpR)W and Ey(e)Y(McpR)W] or without such an Mcp [Ey(e)Y(ΔMcpR)W] (Fig. 4 and 5). Thus, the interaction between two Mcp elements in Ey(e)McpY(McpR)W lines proved to result in a repressed yellow domain and to facilitate enhancer-promoter communication over this domain.
FIG. 5.
Eye phenotypes in the transgenic lines. The photographs show the eyes of flies heterozygous for the constructs shown on the left (+) and eye phenotypes obtained after deletion of either Mcp (ΔMcp) or the eye enhancer (Δe). For eye color designations, see the legend to Fig. 2.
These results provided additional evidence that the Mcp element contained the boundary element that blocked the spread of the repressive chromatin beyond the yellow domain. To demonstrate the role of the boundary element in limiting the repression, we inserted Mcp into the Ey(e)McpRY(McpR)W construct between the yellow enhancers and promoter in the inverse orientation (Fig. 1B). The flies of 14 transgenic lines heterozygous for the construct had less-pigmented eyes than Ey(e)McpY(McpR)W flies (Fig. 4 and 5). Deletion of the eye enhancer led to reductions in eye pigmentation in 8 out of 11 lines tested, suggesting that the eye enhancer could activate the white promoter in most of the Ey(e)McpRY(McpR)W insertion sites (Fig. 4 and 5). Thus, the interaction between unidirectional Mcp elements still allows white activation by the eye enhancer. The lower levels of eye pigmentation in the Ey(e)McpRY(McpR)W lines can be explained by partial repression of the eye enhancer or the white promoter by the Mcp silencers. At the same time, the yellow gene in Ey(e) McpRY(McpR)W flies was less repressed than that in Ey(e)McpY(McpR)W flies, suggesting that Mcp inversion partially disrupted the synergistic interaction of Mcp silencers.
In the McpEy(e)Y(McpR)W construct, Mcp was inserted upstream of the yellow and eye enhancers (Fig. 1B). In 7 out of 11 transgenic McpEy(e)Y(McpR)W lines, flies displayed strong yellow repression in the wing blades and abdominal cuticle, which indicated that the Mcp silencers strongly repressed yellow expression (Fig. 4). In three lines, heterozygous McpEy(e)Y(McpR)W males displayed strongly variegated pigmentation of adjacent cell groups in the abdominal segments (data not shown). In some cuticle cells, the body enhancer was active, resulting in normal expression of the yellow gene; in other cells, the Mcp silencers repressed the body enhancer. Deletion of the downstream Mcp considerably restored yellow expression, which confirmed our conclusion that the Mcp silencers cooperatively repressed yellow. Deletion of the eye enhancer did not modify the eye color in 10 out of 11 transgenic McpEy(e)Y(McpR)W lines tested (Fig. 4 and 5). This result suggested that either the eye enhancer was completely inactive or the interacting Mcp elements efficiently blocked white activation by the eye enhancer.
In the case of Ey(e)Mcp/M340YW transgenic lines, we failed to obtain efficient and stable yellow activation across the Mcp and M340 elements (Fig. 3). Hence, we examined the interaction between the M340 insulator and Mcp by using the eye enhancer-white promoter regulatory system. To this end, the M340 insulator was inserted into the Ey(e)M340Y(McpR)W construct at −893 (Fig. 1B). In the resulting 21 transgenic lines, the eye color in flies ranged from red to brown, as in Ey(e)McpY(McpR)W flies (Fig. 4 and 5). Deletion of the eye enhancer strongly reduced eye pigmentation in all transgenic lines. These results suggested that the interaction between the M340 insulator and Mcp was sufficient for facilitating white activation by the eye enhancer. It is noteworthy that flies of the Ey(e)Mcp/M340YW transgenic lines discussed above had markedly less-pigmented eyes than Ey(e)M340Y(McpR)W flies (Fig. 4 and 5). Deletion of the eye enhancer diminished eye color in 6 out of 10 lines tested, suggesting that the eye enhancer could activate white transcription over closely spaced Mcp and M340. However, to facilitate enhancer-promoter communication, the interacting Mcp and M340 should be located close to the respective enhancer and promoter, as in the Ey(e)McpY(McpR)W and Ey(e)M340Y(McpR)W lines.
DISCUSSION
We identified the boundary element in the 340-bp DNA fragment located in the middle of the 755-bp Mcp core fragment. This element (the M340 insulator) effectively blocks the enhancers of the yellow and white genes expressing at the pupa stage. At this moment, we have no information as to how the M340 insulator blocks the embryonic enhancers. At the same time, we have found that M340 protects yellow and white expression from the PRE located in the Mcp element. Thus, the M340 insulator can function as a barrier between active and repressed chromatin domains. It is noteworthy that the M340 insulator, in contrast to Fab-7 and Fab-8, is flanked on both sides by regulatory elements that cooperatively repress white and yellow expression in transgenic lines (34).
Two 755-bp Mcp elements inserted on both sides of the yellow gene cooperatively repress transcription. At the same time, they allow the eye enhancer to activate the white promoter over the repressed yellow domain if both Mcp elements are oriented so that the 138-bp silencer is closer to the yellow gene. Thus, the M340 insulators effectively protect white transcription from repression. At the same time, the interaction of the two Mcp elements allows the eye enhancer to activate the white promoter over the repressed yellow domain. Interestingly, the M340 insulator can interact with a complete Mcp element, which indicates that the PRE-binding proteins are not required for local interaction. As was shown previously, two copies of the Su(Hw) insulator inserted between an enhancer and a promoter could neutralize each other's enhancer-blocking activity (8, 35). However, studies on the interaction of other Drosophila insulators demonstrated that neutralization of enhancer-blocking activity between insulator pairs was not typical of most insulators (22, 27). It is noteworthy that the 755-bp Mcp element allows the eye enhancer to bypass the Su(Hw) insulator (30). One possible explanation of the mutual neutralization of the insulators is that the same proteins interact with the Su(Hw) and Mcp insulators. However, mutations inactivating the Su(Hw) and Mod(mdg4) proteins did not influence the ability of the M340 insulator to block enhancers in transgenic constructs (N. Gruzdeva and P. Georgiev, unpublished data).
It has been found that the 800-bp Mcp sequence is sufficient for mediating a long-distance trans-regulatory interaction between Mcp transgenes located at distant sites of the same chromosomal arm or even on different arms (34). It is possible that the high specificity of the long-distance communication is ensured by a combination of proteins bound to the PRE and insulator in the Mcp element. The same unusual case of transvection between enhancers translocated onto different chromosomes and the Abd-B promoter is also associated with the PRE and Fab-8 boundary element (45). It has recently been found that the presence of the Fab-7 element containing PRE and an insulator leads to association of transgenes with each other or with the endogenous Fab-7, even when the transgenes are on different chromosomes (1). Thus, the regulatory elements required for long-distance communication always contain an insulator and a PRE.
We observed strong white activation only when the Mcp element was close to the promoter and the M340 insulator was close to the eye enhancer. Thus, the interaction between the Mcp insulators brings the eye enhancer and the white promoter closer to one another, facilitating communication between them. The ability of the eye enhancer to activate the white promoter over the repressed yellow domain flanked by Mcp elements provides an explanation for enhancer-promoter communication in the Abd-B gene. The interaction between the boundary elements and flanking PRE silencers may effectively protect the iab enhancers from external repressing effects and regulate enhancer-promoter communication (Fig. 6). At the moment, however, we have no experimental data confirming that the interaction between different domain boundaries permits enhancer-promoter communication over a repressed domain. Recently, a novel cis-regulatory sequence was found within iab-7 (Fig. 6). This is a promoter-targeting sequence (PTS), which facilitates long-range enhancer-promoter interactions, permits distal enhancers to overcome the blocking effects of insulators, and usually restricts the enhancer activity to a single promoter when more than one promoter is present in the same transgene (25, 26, 45). It is possible that other iab domains contain PTS-like elements that have not been identified yet. Certain interactions between the insulators might either facilitate or block enhancer-promoter communication and regulate the activity of PTS-like elements that determine highly specific, long-range enhancer-promoter communications in proper regulation of Abd-B expression.
FIG. 6.
Model for enhancer-promoter communication in the case of the best-studied iab-7 regulatory domain. An extensive region upstream (UPR) of the Abd-B gene is required for tethering the iab regulatory domains to the Abd-B promoter. Active PREs and domain boundaries interact to produce repressed chromatin and block incorrect interactions between the inactive iab enhancers and the Abd-B promoter. Fab-7 and Fab-8 boundaries protect the active iab-7 enhancer from repression. The Fab-8 boundary flanking the active iab-7 enhancer interacts with the UPR and stimulates the PTS to facilitate communication between the iab-7 enhancer and the Abd-B promoter (25, 26).
Acknowledgments
We thank Pamela Geyer and Vincenzo Pirrotta for the plasmids.
This work was supported by the Molecular and Cellular Biology Program of the Russian Academy of Sciences, the Russian Foundation for Basic Research, and the International Research Scholar Award from the Howard Hughes Medical Institute (to P.G.).
REFERENCES
- 1.Bantignies, F., C. Grimaud, S. Lavrov, M. Gabut, and G. Cavalli. 2003. Inheritance of Polycomb-dependent chromosomal interactions in Drosophila. Genes Dev. 17:2406-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barges, S., J. Mihaly, M. Galloni, K. Hagstrom, M. Muller, G. Shanower, P. Schedl, H. Gyurkovics, and F. Karch. 2000. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127:779-790. [DOI] [PubMed] [Google Scholar]
- 3.Bondarenko, V. A., Y. V. Liu, Y. I. Jiang, and V. M. Studitsky. 2003. Communication over a large distance: enhancers and insulators. Biochem. Cell. Biol. 81:241-251. [DOI] [PubMed] [Google Scholar]
- 4.Boulet, A. M., A. Lloyd, and S. Sakonju. 1991. Molecular definition of the morphogenetic and regulatory functions and the cis-regulatory elements of the Drosophila Abd-B homeotic gene. Development 111:393-405. [DOI] [PubMed] [Google Scholar]
- 5.Busturia, A., and M. Bienz. 1993. Silencers in abdominal-B, a homeotic Drosophila gene. EMBO J. 12:1415-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busturia, A., C. D. Wightman, and S. Sakonju. 1997. A silencer is required for maintenance of transcriptional repression throughout Drosophila development. Development 124:4343-4350. [DOI] [PubMed] [Google Scholar]
- 7.Busturia, A., A. Lloyd, F. Bejarano, M. Zavortink, H. Xin, and S. Sakonju. 2001. The MCP silencer of the Drosophila Abd-B gene requires both Pleiohomeotic and GAGA factor for the maintenance of repression. Development 128:2163-2173. [DOI] [PubMed] [Google Scholar]
- 8.Cai, H. N., and P. Shen. 2001. Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science 291:493-495. [DOI] [PubMed] [Google Scholar]
- 9.Celniker, S. E., S. Sharma, D. J. Keelan, and E. B. Lewis. 1990. The molecular genetics of the bithorax complex of Drosophila: cis-regulation in the Abdominal-B domain. EMBO J. 9:4227-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dejardin, J., and G. Cavalli. 2004. Chromatin inheritance upon Zeste-mediated Brahma recruitment at a minimal cellular memory module. EMBO J. 23:857-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorsett, D. 1999. Distant liaisons: long range enhancer-promoter interactions in Drosophila. Curr. Opin. Genet. 9:505-514. [DOI] [PubMed] [Google Scholar]
- 12.Duncan, I. 1987. The bithorax complex. Annu. Rev. Genet. 21:285-319. [DOI] [PubMed] [Google Scholar]
- 13.Galloni, M., H. Gyurkovics, P. Schedl, and F. Karch. 1993. The bluetail transposon: evidence for independent cis-regulatory domains and domain boundaries in the bithorax complex. EMBO J. 12:1087-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerasimova, T. I., and V. G. Corces. 2001. Chromatin insulators and boundaries: effects on transcription and nuclear organization. Annu. Rev. Genet. 35:193-208. [DOI] [PubMed] [Google Scholar]
- 15.Geyer, P. K., and V. G. Corces. 1987. Separate regulatory elements are responsible for the complex pattern of tissue-specific and developmental transcription of the yellow locus in Drosophila melanogaster. Genes Dev. 1:996-1004. [DOI] [PubMed] [Google Scholar]
- 16.Gyurkovics, H., J. Gausz, J. Kummer, and F. Karch. 1990. A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J. 9:2579-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagstrom, K., M. Muller, and P. Schedl. 1996. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 10:3202-3215. [DOI] [PubMed] [Google Scholar]
- 18.Hagstrom, K., M. Muller, and P. Schedl. 1997. A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithorax complex. Genetics 146:1365-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karch, F., B. Weiffenbach, M. Peifer, W. Bender, I. Duncan, S. Celniker, M. Crosby, and E. B. Lewis. 1985. The abdominal region of the bithorax complex. Cell 43:81-96. [DOI] [PubMed] [Google Scholar]
- 20.Karch, F., M. Galloni, L. Sipos, J. Gausz, H. Gyurkovics, and P. Schedl. 1994. Mcp and Fab-7: molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res. 22:3138-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kares, R. E., and G. M. Rubin. 1984. Analysis of P transposable element functions in Drosophila. Cell 38:135-146. [DOI] [PubMed] [Google Scholar]
- 22.Kuhn, E. J., and P. K. Geyer. 2003. Genomic insulators: connecting properties to mechanism. Curr. Opin. Cell Biol. 15:259-265. [DOI] [PubMed] [Google Scholar]
- 23.Labrador, M., and V. G. Corces. 2002. Setting the boundaries of chromatin domains and nuclear organization. Cell 111:151-154. [DOI] [PubMed] [Google Scholar]
- 24.Lewis, E. B. 1978. A gene complex controlling segmentation in Drosophila. Nature 276:565-570. [DOI] [PubMed] [Google Scholar]
- 25.Lin, Q., D. Wu, and J. Zhou. 2003. The promoter targeting sequence facilitates and restricts a distant enhancer to a single promoter in the Drosophila embryo. Development 130:519-526. [DOI] [PubMed] [Google Scholar]
- 26.Lin, Q., Q. Chen, L. Lin, and J. Zhou. 2004. The Promoter Targeting Sequence mediates epigenetically heritable transcription memory. Genes Dev. 18:2639-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Majumder, P., and H. N. Cai. 2003. The functional analysis of insulator interactions in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 100:5223-5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin, M., Y. B. Meng, and W. Chia. 1989. Regulatory elements involved in the tissue-specific expression of the yellow gene of Drosophila. Mol. Gen. Genet. 218:118-126. [DOI] [PubMed] [Google Scholar]
- 29.McCall, K., M. B. O'Connor, and W. Bender. 1994. Enhancer traps in the Drosophila bithorax complex mark parasegmental domains. Genetics 138:387-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melnikova, L., F. Juge, N. Gruzdeva, A. Mazur, G. Cavalli, and P. Georgiev. 2004. Interaction between the GAGA factor and Mod(mdg4) proteins promotes insulator bypass in Drosophila. Proc. Natl. Acad. Sci. USA 101:14806-14811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mihaly, J., I. Hogga, J. Gausz, H. Gyurkovics, and F. Karch. 1997. In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development 124:1809-1820. [DOI] [PubMed] [Google Scholar]
- 32.Mihaly, J., I. Hogga, S. Barges, M. Galloni, R. K. Mishra, K. Hagstrom, M. Muller, P. Schedl, L. Sipos, J. Gausz, H. Gyurkovics, and F. Karch. 1998. Chromatin domain boundaries in the Bithorax complex. Cell. Mol. Life Sci. 54:60-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mihaly, J., R. K. Mishra, and F. Karch. 1998. A conserved sequence motif in Polycomb-response elements. Mol. Cell 1:1065-1066. [DOI] [PubMed] [Google Scholar]
- 34.Muller, M., K. Hagstrom, H. Gyurkovics, V. Pirrotta, and P. Schedl. 1999. The Mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Genetics 153:1333-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muravyova, E., A. Golovnin, E. Gracheva, A. Parshikov, T. Belenkaya, V. Pirrotta, and P. Georgiev. 2001. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science 291:495-498. [DOI] [PubMed] [Google Scholar]
- 36.Oki, M., and R. T. Kamakaka. 2002. Blockers and barriers to transcription: competing activities? Curr. Opin. Cell Biol. 14:299-304. [DOI] [PubMed] [Google Scholar]
- 37.Qian, S., B. Varjavand, and V. Pirrotta. 1992. Molecular analysis of the zeste-white interaction reveals a promoter-proximal element essential for distant enhancer-promoter communication. Genetics 131:79-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rubin, G. M., and A. C. Spradling. 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218:348-353. [DOI] [PubMed] [Google Scholar]
- 39.Sanchez-Herrero, E., I. Vernos, R. Marco, and G. Morato. 1985. Genetic organization of Drosophila bithorax complex. Nature 313:108-113. [DOI] [PubMed] [Google Scholar]
- 40.Sanchez-Herrero, E. 1991. Control of the expression of the bithorax complex abdominal-A and abdominal-B by cis-regulatory regions in Drosophila embryos. Development 111:437-448. [DOI] [PubMed] [Google Scholar]
- 41.Spradling, A. C., and G. M. Rubin. 1982. Transposition of cloned P elements into germline chromosomes. Science 218:341-347. [DOI] [PubMed] [Google Scholar]
- 42.Udvardy, A. 1999. Dividing the empire: boundary chromatin elements delimit the territory of enhancers. EMBO J. 18:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.West, A. G., M. Gaszner, and G. Felsenfeld. 2002. Insulators: many functions, many mechanisms. Genes Dev. 16:271-288. [DOI] [PubMed] [Google Scholar]
- 44.Zhou, J., S. Barolo, P. Szymanski, and M. Levine. 1996. The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes Dev. 10:3195-3201. [DOI] [PubMed] [Google Scholar]
- 45.Zhou, J., H. Ashe, C. Burks, and M. Levine. 1999. Characterization of the transvection mediating region of the abdominal-B locus in Drosophila. Development 126:3057-3065. [DOI] [PubMed] [Google Scholar]