Abstract
Mixed-lineage protein kinase 3 (MLK3) is a member of the mitogen-activated protein (MAP) kinase kinase kinase group that has been implicated in multiple signaling cascades, including the NF-κB pathway and the extracellular signal-regulated kinase, c-Jun NH2-terminal kinase (JNK), and p38 MAP kinase pathways. Here, we examined the effect of targeted disruption of the murine Mlk3 gene. Mlk3−/− mice were found to be viable and healthy. Primary embryonic fibroblasts prepared from these mice exhibited no major signaling defects. However, we did find that MLK3 deficiency caused a selective reduction in tumor necrosis factor (TNF)-stimulated JNK activation. Together, these data demonstrate that MLK3 contributes to the TNF signaling pathway that activates JNK.
The mechanism of c-Jun NH2-terminal kinase (JNK) activation caused by tumor necrosis factor (TNF) is incompletely understood. It is established that JNK is activated by dual phosphorylation on the T-loop within the motif Thr-Pro-Tyr (7). This phosphorylation is mediated by the actions of two different mitogen-activated protein (MAP) kinase kinases: MKK4 and MKK7 (35). These MAP kinase kinases can be activated by MAP kinase kinase kinases (MAP3K), but the identity of the relevant TNF-stimulated MAP3K is unclear.
Three MAP3K have been implicated in the activation of JNK caused by TNF. First, apoptosis signal-regulating kinase 1 is thought to be involved in the late phase of JNK activation in response to TNF, most likely as a result of the generation of reactive oxygen species (34). The immediate activation of JNK caused by TNF may be mediated by TAK1 and/or by one or more members of the mixed-lineage protein kinase (MLK) family. MLKs may be selectively involved in TNF-stimulated JNK activation (25), while TAK1 is implicated as a common TNF-stimulated activator of JNK, p38 MAPK, and NF-κB (16, 29). The relative roles of TAK1 and MLKs in TNF signaling are unclear. In this study, we have examined the possible contribution of a MLK to TNF signaling.
There are three subgroups of MLKs (10). The MLK group (MLK1, MLK2, MLK3, and MLK4) shares similar structural domains, including an SH3 domain and a Crib motif that binds Cdc42 and Rac1. The DLK group (DLK and LZK) is structurally distinct and lacks the SH3 and Crib sequences. The third group of protein kinases consists of a single member (ZAK) that is distinctive because of the presence of a SAM domain. Many of these protein kinases are expressed in only a limited number of tissues; for example, MLK1 is expressed in epithelial cells and DLK is expressed in neurons (10). However, one member of this gene family is ubiquitously expressed, consistent with a possible role as a mediator of TNF signaling in many tissues—MLK3. This possibility is consistent with previous studies showing that TNF activates MLK3 (25), that MLK-family protein kinases (like TNF) (35) can selectively activate MKK7 (14, 18), and that a small-molecule MLK inhibitor can block TNF-stimulated JNK activation (25). However, recent RNA interference (RNAi)-based studies have suggested that MLK3 is critically required for cellular proliferation and is essential for the activation of multiple MAP kinase (MAPK) signaling pathways in response to a broad range of stimuli (5, 28). Indeed, previous studies have implicated MLK3 in extracellular signal-regulated kinase (ERK) activation (5), p38 MAPK activation (5, 22), and NF-κB activation (13). The specific role of MLK3 in signaling and the relevance of MLK3 to TNF signal transduction are therefore unclear.
The purpose of this study was to test the role of MLK3 in TNF-stimulated JNK activation. Our approach was to examine the effect of targeted disruption of the Mlk3 gene. We report that MLK3 deficiency caused a selective defect in TNF-stimulated JNK activation and that MLK3 is not required for cellular proliferation.
MATERIALS AND METHODS
Mice.
Mouse strain 129/SvJ genomic clones of the Mlk3 gene were isolated by PCR and subcloned into the vector pCRII (Invitrogen). A targeting vector designed to disrupt the Mlk3 gene (Fig. 1A) was constructed by standard techniques. Embryonic stem (ES) cells were electroporated with this vector and selected with 200 μg of G418 (Invitrogen)/ml and 2 μM gancyclovir (Syntex). Twelve Mlk3−/+ ES cell clones were identified by Southern blot analysis, and 2 were injected into C57BL/6J blastocysts to create chimeric mice that transmitted the disrupted Mlk3 allele through the germ line. The mice were backcrossed 10 generations to the C57BL/6J strain (Jackson Laboratories). Homozygous Mlk3−/− mice were obtained by crossing heterozygous Mlk3−/+ animals. The mice were housed in a facility accredited by the American Association for Laboratory Animal Care. The animal studies were approved by the Institutional Animal Care and Use Committee of the University of Massachusetts Medical School.
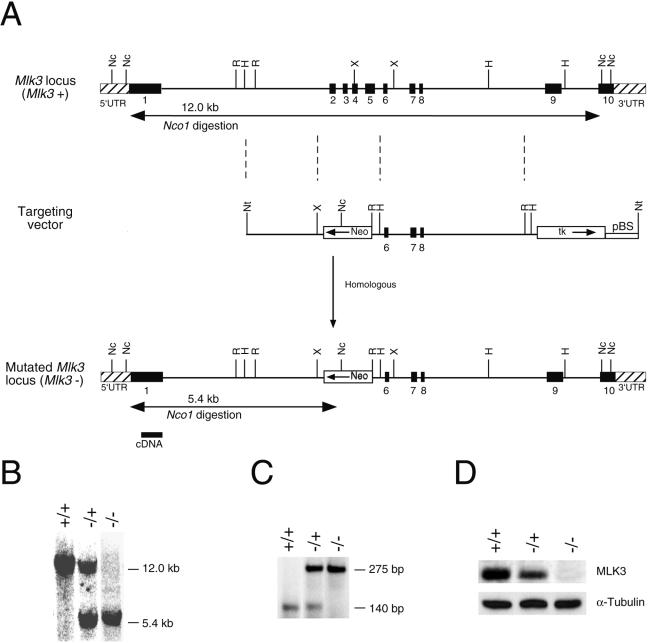
FIG. 1.
Disruption of the Mlk3 gene in mice. (A) The strategy employed to disrupt the Mlk3 gene by homologous recombination is illustrated. The structure of the Mlk3 gene, the targeting vector, and the disrupted Mlk3 gene are shown. Restriction enzyme sites are indicated: Nc (NcoI), Nt (NotI), R (EcoRI), and X (XbaI). The probe used for Southern analysis and the expected results of NcoI restriction digestion are illustrated. (B and C) Genomic DNA isolated from Mlk3+/+, Mlk3−/+, and Mlk3−/− mice was examined by Southern blot (B) and PCR (C) analysis. (D) Protein extracts prepared from MEF prepared from Mlk3+/+, Mlk3−/+, and Mlk3−/− mice were examined by immunoblot analysis using antibodies to MLK3 and α-tubulin.
Genotype analysis.
The genotype at the Mlk3 locus was examined by Southern blot analysis of NcoI-restricted genomic DNA by probing with a random-primed 32P-labeled probe (529 bp) that was isolated by PCR with an Mlk3 cDNA as the template and the amplimers 5′-CTCCGAAGGCAACAGCAGCTTATGCCA-3′ and 5′-CACACGCCGACCAGCCAGGGCCCGGCT-3′. The wild-type (140-bp) and disrupted (275-bp) alleles of Mlk3 were also detected by PCR amplification of genomic DNA using the primers 5′-AAGCGGAGCAAACTCCGAGCAAG-3′, 5′-AAAGGCTAAACCAGAACTCAAG-3′, and 5′-GTAGAAGGTGGCGCGAAGGG-3′.
Histology.
Tissue was fixed in 10% formalin for 24 h, dehydrated, and embedded in paraffin. Sections (each, 4 μm thick) were cut and stained with Harris hematoxylin (Sigma) and eosin (Sigma).
Cell culture.
Primary murine embryo fibroblasts (MEF) were isolated and cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen). All experiments were performed using MEF between passage 2 and passage 5. Similar data were obtained in experiments using independently isolated MEF cultures. Proliferation assays were performed by staining with crystal violet (36). Adipocyte differentiation assays (11, 21) were performed by culturing 2-day postconfluent cells in medium supplemented with 10-μg/ml insulin (Sigma), 0.5 mM isomethylbutyl-1-xanthine (Sigma), and 1-μg/ml dexamethasone (Sigma). The medium was replaced after 72 h with fresh medium supplemented with 10-μg/ml insulin and 1 μM troglitizone (Calbiochem). The accumulation of fat droplets within the cytoplasm was detected by staining the cells with Oil-Red-O (VWR).
Migration and invasion assays.
Boyden chamber assays were performed using 2.5 × 104 cells in 0.5 ml of Dulbecco's modified Eagle's medium placed in each insert of a 24-well multiwell plate (BIOCOAT; Becton Dickinson). Migration and invasion assays were performed without and with Matrigel, respectively, by incubating the cells at 37°C (16 h). The inserts were placed in methanol (−20°C) and stained with 4′-6′-diamino-2-phenylindole (DAPI; Vector Laboratories). The cells were visualized with an Axioplan 2 microscope with a MicroImager CCD camera (Carl Zeiss).
Immunoblot analysis.
Cell extracts were prepared with Triton lysis buffer (20 mM Tris [pH 7.4], 1% Triton X-100, 10% glycerol, 137 mM NaCl, 2 mM EDTA, 25 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, and a 10-μg/ml concentration of aprotinin and leupeptin). Extracts (each, 50 μg of protein) were examined by protein immunoblot analysis by probing with antibodies to JNK (Pharmingen), phospho-JNK (Cell Signaling), p38 MAPK (Santa Cruz), phospho-p38 MAPK (Cell Signaling), ERK1/ERK2 (Santa Cruz), MLK3 (Cell Signaling), IκBα (Cell Signaling), CCAAT/enhancer binding protein α (C/EBPα) (Santa Cruz), phospho-Thr-222/226-C/EBPα (Cell Signaling), C/EBPβ (Santa Cruz), phospho-Thr-235-C/EBPβ (Cell Signaling), and α-tubulin (Sigma). Immune complexes were detected by enhanced chemiluminescence (NEN).
Protein kinase assays.
The activity of JNK, ERK, and p38 MAPK was measured by in vitro kinase assays with the substrates cJun, myelin basic protein, and ATF2, respectively (39).
RNase protection assays.
Total RNA (5 μg) was examined using the Multi-Probe RNase Protection Assay (Pharmingen) with the template sets mFos/Jun, mCR-4, and mCK-3b, following the manufacturer's recommendations. The products were separated on a 5% sequencing gel, detected by autoradiography, and quantitated by Phosphorimager analysis (Molecular Dynamics).
Immunofluorescence analysis.
Cells were grown on coverslips and fixed at −20°C in methanol (5 min) and in acetone (2 min). The coverslips were washed in phosphate-buffered saline, incubated (30 min at 22°C) in BPT buffer (3% bovine serum albumin-phosphate-buffered saline-0.2% Tween-20), and stained with a mouse monoclonal antibody to α-tubulin (Sigma) in BPT buffer (60 min at 22°C). Immune complexes were visualized with a fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin secondary antibody (Jackson ImmunoResearch). Slides were mounted with Vectashield mounting medium with DAPI (Vector Laboratories, Inc). Fluorescence microscopy was performed with a Zeiss inverted microscope (Axiovert M200) with a 10× objective (numerical aperture, 0.30), a charge-coupled device camera (Zeiss Axiocam), and image acquisition software (Zeiss Axiovision).
RESULTS
Targeted disruption of the Mlk3 gene.
We constructed a targeting vector to disrupt the Mlk3 gene. The vector was designed to replace exons 2 to 6 with a NeoR cassette (Fig. 1A). This region of the Mlk3 gene includes the kinase domain. The vector was linearized and electroporated into ES cells to obtain homologous recombination within the Mlk3 gene. Twelve ES cell clones with the correctly targeted Mlk3 gene were identified by Southern blot analysis. Two of these clones were injected into C57BL/6J blastocysts to create male chimeric mice that were bred to obtain germ line transmission of the disrupted Mlk3 allele. The mice were backcrossed to the C57BL/6J strain background. Genomic DNA isolated from the progeny obtained from crossing Mlk3−/+ mice was examined by PCR and Southern blot analysis to identify wild-type, Mlk3−/+, and Mlk3−/− littermates (Fig. 1B and C). The number of wild-type, heterozygous, and homozygous knockout mice obtained from these crosses conformed to the expected Mendelian inheritance. Immunoblot analysis demonstrated that the level of MLK3 expression was reduced in Mlk3−/+ mice and was absent in Mlk3−/− mice (Fig. 1D).
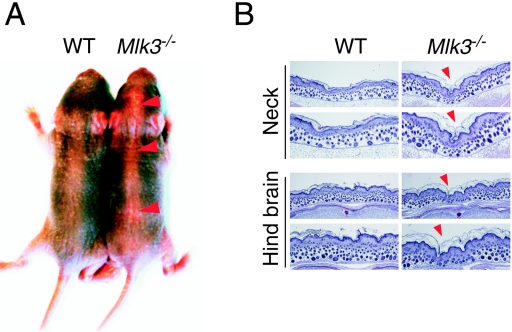
MLK3-deficient mice were found to be viable, had a normal life span, and were not found to be tumor prone compared with wild-type mice. The Mlk3−/− mice appeared to be morphologically normal, but these mice did display a minor defect along the dorsal midline (Fig. 2A). Histological analysis indicated that MLK3 deficiency reduced the thickness of the dorsal epidermal tissue (Fig. 2B). The cause of this epidermal defect has not been established. However, it is interesting that defects in the dorsal epidermis are observed in JNK-deficient mice (due to neural tube closure defects) and in JNK-deficient Drosophila melanogaster (due to dorsal closure defects) (6, 38). Furthermore, the Drosophila MLK isoform slipper is required for dorsal closure during embryogenesis (26, 27). Thus, the observation that Mlk3−/− mice exhibit a dorsal midline defect is not unexpected, but the mechanism that causes the specific defect observed in Mlk3−/− mice is unclear.
FIG. 2.
MLK3 deficiency causes epidermal defects at the dorsal midline. (A) Heterozygous Mlk3 mice were crossed, and two littermates (wild type [WT] and Mlk3−/−) were examined 4 days after birth. The Mlk3−/− mice appear morphologically normal but exhibit a dorsal midline defect (red arrowheads). (B) Histological examination of WT and Mlk3−/− mice. Sections of the dorsal epidermis and underlying tissue were stained with hematoxylin and eosin (H&E). Images of low and high magnification are shown. The red arrowheads indicate the loss of epidermal tissue along the dorsal midline of the Mlk3−/− mice.
MLK3 is not essential for cellular proliferation.
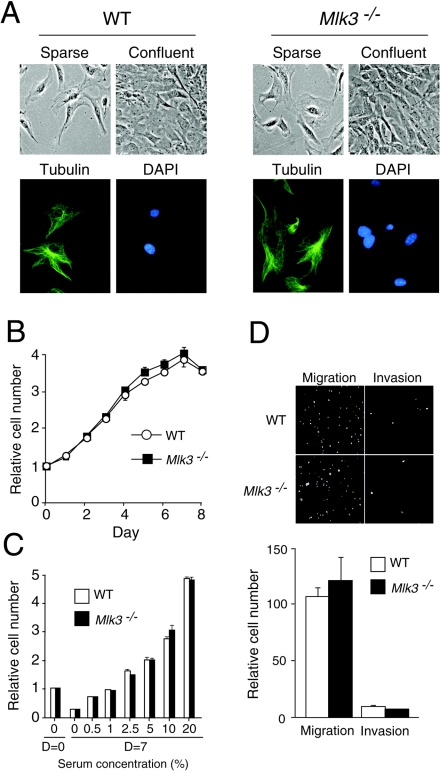
It has been established in previous RNAi-based studies that MLK3 is essential for serum-stimulated cell proliferation (5). This conclusion appears to be inconsistent with the finding that Mlk3−/− mice are viable. To directly investigate whether MLK3 is required for proliferation, we isolated primary fibroblasts (MEF) from wild-type and Mlk3−/− embryonic day 13.5 (E13.5) embryos. Phase-contrast microscopy indicated that the morphology of the wild-type and Mlk3−/− MEF was similar in sparse cultures, and both groups of MEF exhibited contact growth inhibition in confluent cultures (Fig. 3A). Measurement of proliferation during culture in medium supplemented with 10% fetal bovine serum for 8 days demonstrated no differences between wild-type and Mlk3−/− MEF (Fig. 3B). Similarly, no differences between wild-type and Mlk3−/− MEF were detected in experiments with serum concentrations ranging from 0 to 20% (Fig. 3C). The regulated expression of members of the AP-1 family of transcription factors is implicated in cell proliferation and these genes represent potential targets of MLK3 signaling; however, comparison of wild-type and Mlk3−/− MEF indicated no differences in AP-1 mRNA expression (Fig. 4A). Together, these data indicate that MLK3 is not essential for MEF proliferation.
FIG. 3.
Characterization of primary MEF isolated from wild-type and Mlk3−/− mice. (A) The morphology of sparse and confluent cultures of WT and Mlk3−/− MEF was examined by phase-contrast microscopy (top). The presence of microtubules (green) in WT and Mlk3−/− MEF was examined by immunofluorescence microscopy (bottom). DNA in the nucleus was stained with DAPI (blue). (B) Cells (2.5 × 104) were incubated in 11-mm wells with medium supplemented with 10% fetal calf serum. Relative cell numbers were measured by staining with crystal violet. The proliferation of WT and Mlk3−/− MEF during culture for 8 days was examined. The data shown represent the means ± standard deviation (SD) for three experiments. (C) The effect of serum concentration on the saturation growth density of WT and Mlk3−/− MEF was examined. Cells (2.5 × 104) were incubated in 11-mm wells with medium supplemented with different concentrations of serum for 7 days. Relative cell numbers were measured by staining with crystal violet. The data shown represent the mean ± SD for three experiments. (D) Boyden chamber assays were performed to measure chemotaxis (cell migration) from serum-free medium to medium supplemented with 10% serum. Invasion assays were performed in similar experiments using Matrigel. The cells were stained with DAPI and visualized by fluorescence microscopy. The relative cell migration and invasion were quantitated by counting the number of cells (mean ± SD for three experiments). The migration and invasion by WT and Mlk3−/− MEF are shown.
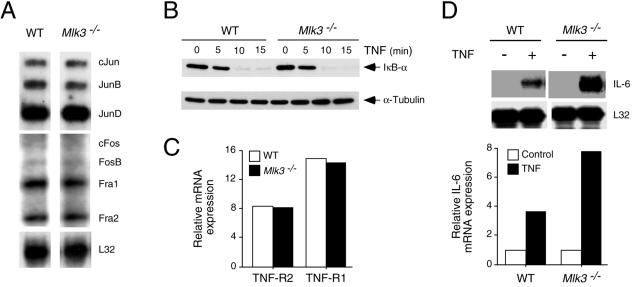
FIG. 4.
Effect of MLK3 deficiency on AP-1 and NF-κB. (A) The expression of several genes that encode proteins that can form AP-1 transcription factors by exponentially growing WT and Mlk3−/− MEF was examined by a RNase protection assay to detect mRNA. The expression of ribosomal protein L32 mRNA was examined in control experiments. (B) WT and Mlk3−/− MEF were incubated with TNF (10 ng/ml), and the degradation of IκBα was examined by immunoblot analysis. Control experiments were performed by probing immunoblots with an antibody to α-tubulin. (C) The expression of TNF-R1 and TNF-R2 was examined by an RNase protection assay to measure the amount of TNF receptor mRNA in WT and Mlk3−/− MEF. Control experiments were performed by measuring the amount of ribosomal protein L32 mRNA. The data were quantitated by phosphorimager analysis, and the relative TNF receptor expression was calculated as the ratio of TNF receptor mRNA to L32 mRNA. (D) WT and Mlk3−/− MEF were incubated with TNF (10 ng/ml) for 24 h, and the amount of IL-6 and L32 mRNA was examined by an RNase protection assay. The data were quantitated by phosphorimager analysis, and the relative IL-6 mRNA expression was calculated as the ratio of IL-6 mRNA to L32 mRNA.
A previous RNAi-based study has indicated that MLK3 is essential for the regulation of microtubule dynamics (28). Defective microtubule regulation would be expected to cause defects in mitosis and cell migration (28). Comparison of wild-type and Mlk3−/− MEF did not indicate a requirement for MLK3 in proliferation (Fig. 3B and C). We therefore investigated potential defects in cell migration caused by MLK3 deficiency. Boyden chamber assays with an imposed serum gradient (0 to 10%) demonstrated no differences in serum-induced chemotaxis between wild-type and Mlk3−/− MEF (Fig. 3D). Similarly, no differences between the ability of wild-type and Mlk3−/− MEF to invade a Matrigel layer were observed (Fig. 3D). These data suggest that MLK3 deficiency does not cause major microtubule defects. Indeed, immunofluorescence microscopy with a monoclonal antibody to α-tubulin indicated the presence of similar microtubule networks in wild-type and Mlk3−/− MEF (Fig. 3A).
The defects in cell proliferation and migration predicted by RNAi-based studies (5, 28) were not observed in Mlk3−/− MEF (Fig. 3). The absence of the expected phenotype could be caused by compensatory increases in the expression of another member of the MAP3K group. To test this hypothesis, we examined MAP3K gene expression in wild-type and Mlk3−/− MEF by microarray analysis of isolated mRNA (Affymetrix). This analysis did not demonstrate differences in MAP3K gene expression (other than Mlk3) between wild-type and Mlk3−/− MEF (data not shown). We conclude that either MLK3 is not required for MEF proliferation and migration or that the loss of MLK3 is compensated by increased function (but not expression) of another member of the MAP3K group in Mlk3−/− MEF.
MLK3 deficiency causes a selective defect in TNF-stimulated JNK activation.
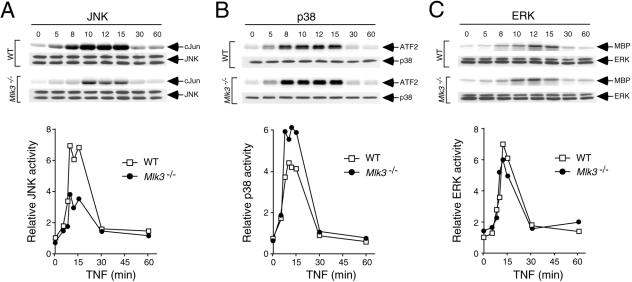
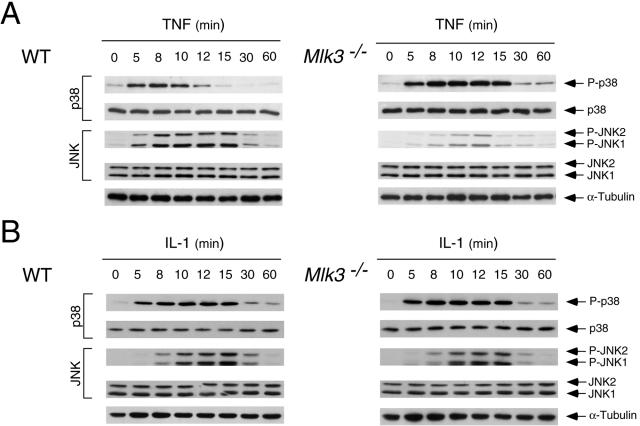
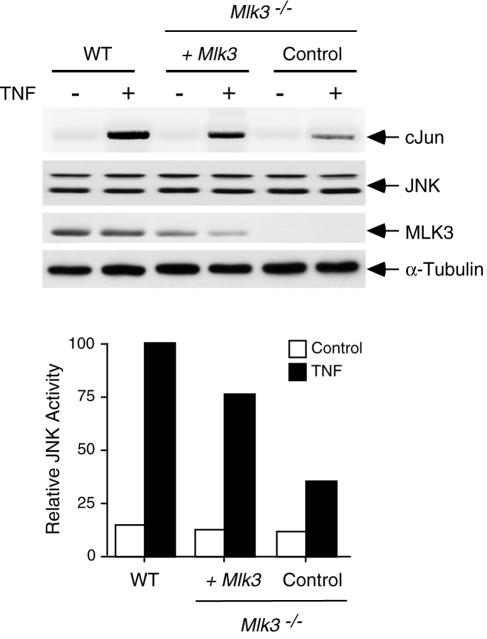
It is established that the MLK3 protein kinase is activated when cells are exposed to TNF, and pharmacological studies using the drug CEP-11004 have implicated MLK3 in TNF-stimulated JNK activation (25). We therefore examined the ability of TNF to activate JNK in wild-type and Mlk3−/− MEF with an in vitro protein kinase assay using c-Jun as the substrate. This analysis demonstrated that TNF caused a rapid and transient increase in JNK activity in wild-type MEF and that the extent of JNK activation was suppressed (but not eliminated) in Mlk3−/− MEF (Fig. 5A). The effect of MLK3 deficiency to suppress JNK regulation by TNF was also observed in experiments in which the JNK activation state was monitored by immunoblot analysis with an antibody that binds (Thr and Tyr) phosphorylated JNK (Fig. 6A). The defect in JNK activation was not a consequence of decreased TNF receptor expression because wild-type and Mlk3−/− MEF expressed similar amounts of TNF-R1 and TNF-R2 (Fig. 4C). To confirm that the reduced TNF-stimulated JNK activation observed in Mlk3−/− MEF was caused by MLK3 deficiency, we performed complementation analyses (Fig. 7). Together, these data demonstrate that MLK3 contributes to TNF-stimulated JNK activation. However, MLK3 was not essential for TNF-stimulated JNK activity.
FIG. 5.
MLK3 is required for maximal TNF-stimulated JNK activation. (A) WT and Mlk3−/− MEF were incubated with TNF (10 ng/ml) for different periods of time before the cells were harvested and protein extracts were prepared. JNK activity was measured by an in vitro kinase assay with the substrate c-Jun. The phosphorylation of c-Jun was detected by autoradiography and was quantitated by phosphorimager analysis. The amount of JNK in each assay was examined by immunoblot analysis. (B) The TNF-stimulated activation of p38 MAPK was examined by an in vitro kinase assay with the substrate ATF2. (C) The TNF-stimulated activation of ERK was examined in an in vitro kinase assay with myelin basic protein (MBP) as the substrate.
FIG. 6.
Maximal TNF-stimulated JNK phosphorylation on Thr and Tyr requires MLK3. (A) WT and Mlk3−/− MEF were incubated with TNF (10 ng/ml), and the activation of JNK and p38 MAPK was examined by immunoblot analysis with antibodies to phospho-JNK and phospho-p38. The amount of MAPK in each assay was examined by probing immunoblots with antibodies to JNK and p38 MAPK. (B) The effect of IL-1 (10 ng/ml) on JNK and p38 MAPK activation in WT and Mlk3−/− MEF was examined by immunoblot analysis.
FIG. 7.
Complementation analysis demonstrated that the expression of MLK3 increases TNF-stimulated JNK activation in MLK3-deficient MEF. WT and Mlk3−/− MEF were incubated with 10-ng/ml TNF (10 min). The effect of MLK3 expression in the Mlk3−/− MEF was examined. Protein extracts were investigated by probing immunoblots with antibodies to JNK, MLK3, and tubulin. JNK activity was measured by an immune complex kinase assay with cJun as the substrate. The incorporation of [32P]phosphate into the substrate was detected by autoradiography and was quantitated by phosphorimager analysis.
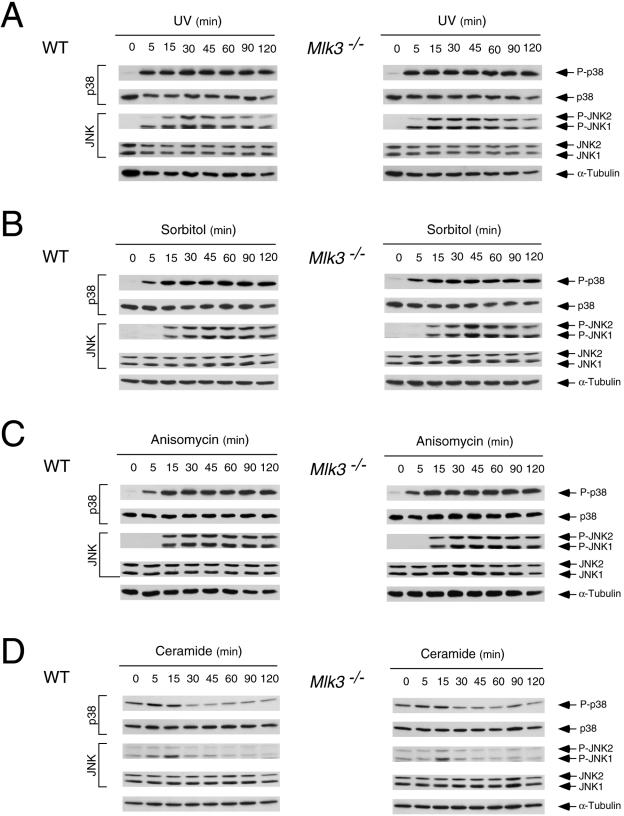
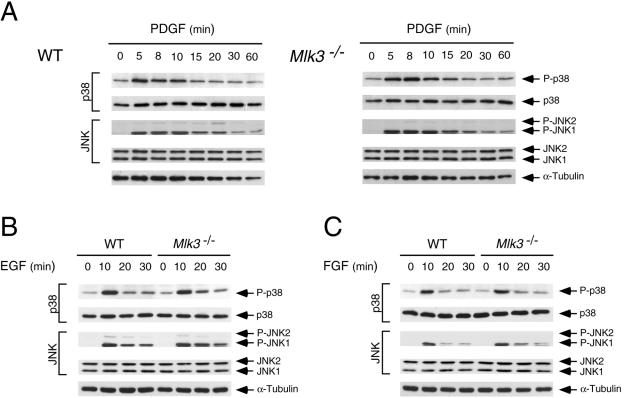
The requirement of MLK3 for maximal JNK activation appears to be selective for TNF. Thus, no defect in interleukin-1 (IL-1)-stimulated JNK activation was detected in Mlk3−/− MEF (Fig. 6B). Similarly, MLK3 deficiency did not cause defects in JNK activation when the MEF were exposed to several environmental stresses, including UV radiation, osmotic stress, anisomycin, or ceramide activation (Fig. 8). Furthermore, MLK3 deficiency did not cause defects in growth factor-stimulated JNK activation, including epidermal growth factor, platelet-derived growth factor, and fibroblast growth factor activation (Fig. 9). The selective (and partial) defect in TNF-stimulated JNK activation observed in MLK3-deficient fibroblasts markedly contrasted with previous RNAi-based studies that have implicated an essential role of MLK3 in JNK activation caused by multiple stimuli (5).
FIG. 8.
MLK3 is not required for JNK or p38 MAPK activation caused by stress. (A) WT and Mlk3−/− MEF were exposed to UV (50 J/m2), and the activation of JNK and p38 MAPK was examined by immunoblot analysis with antibodies to phospho-JNK and phospho-p38. The amount of MAPK in each assay was examined by probing immunoblots with anibodies to JNK and p38 MAPK. (B) The effect of osmotic stress (300 mM sorbitol) on JNK and p38 MAPK activation in WT and Mlk3−/− MEF was examined by immunoblot analysis. (C) The effect of anisomycin (1 μg/ml) on JNK and p38 MAPK activation in WT and Mlk3−/− MEF was examined by immunoblot analysis. (D) The effect of ceramide (10 μM) on JNK and p38 MAPK activation in WT and Mlk3−/− MEF was examined by immunoblot analysis.
FIG. 9.
MLK3 is not required for JNK or p38 MAPK activation caused by growth factors. (A) WT and Mlk3−/− MEF were exposed to platelet-derived growth factor (PDGF) (20 ng/ml), and the activation of JNK and p38 MAPK was examined by immunoblot analysis with antibodies to phospho-JNK and phospho-p38. The amount of MAPK in each assay was examined by probing immunoblots with antibodies to JNK and p38 MAPK. (B) The effect of epidermal growth factor (EGF) (50 ng/ml) on JNK and p38 MAPK activation in WT and Mlk3−/− MEF was examined by immunoblot analysis. (C) The effect of fibroblast growth factor (FGF) (25 ng/ml) on JNK and p38 MAPK activation in WT and Mlk3−/− MEF was examined by immunoblot analysis.
MLK3-mediated JNK activation negatively regulates p38 MAPK.
TNF causes the activation of several signal transduction pathways, including JNK, ERK, p38 MAPK, and NF-κB. Gene disruption experiments demonstrate that MLK3 is required for maximal TNF-stimulated JNK activation (Fig. 5 and 6). It is possible that MLK3 may also play a role in the TNF-stimulated activation of ERK, p38 MAPK, and NF-κB. Indeed, previous studies have implicated MLK3 in ERK activation (5), p38 MAPK activation (5, 22), and NF-κB activation (13). Control studies demonstrated that TNF-stimulated IκBα degradation and the activation of ERK and p38 MAPK were not reduced in MLK3-deficient MEF (Fig. 4B and 5B and C). An unexpected discovery was that TNF-stimulated p38 MAPK activity was increased in Mlk3−/− MEF (Fig. 5B). This increased TNF-stimulated p38 MAPK activation was confirmed by immunoblot analysis with an antibody to phospho-p38 MAPK (Fig. 6A) and was not observed in cells treated with IL-1 (Fig. 6B). Since JNK can suppress p38 MAPK activation and JNK-deficient cells exhibit increased TNF-stimulated p38 MAPK signaling (19), it is likely that the increased TNF-stimulated p38 MAPK activation is related to the observed decrease in TNF-stimulated JNK activation (Fig. 6A). This increased TNF-stimulated p38 MAPK activation is consistent with the finding that MLK3-deficient MEF exhibit greater TNF-stimulated expression of IL-6, a p38 MAPK target gene (2, 41), than wild-type MEF (Fig. 4D).
Altered MAPK regulation causes increased adipogenic potential.
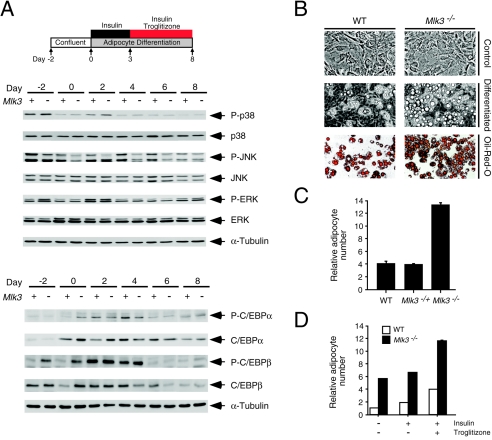
The increased p38 MAPK activation and decreased JNK activation observed in Mlk3−/− MEF is likely to be physiologically significant. It has been reported that JNK activation inhibits adipogenesis by phosphorylating and inactivating the proadipogenic transcription factor gamma peroxisome proliferator-activated receptor (PPARγ) (3). Conversely, p38 MAPK activation promotes adipogenesis by increasing the expression and phosphorylation of C/EBP transcription factors (9, 43). We therefore investigated whether altered MAPK activation might contribute to increased adipogenic differentiation of Mlk3−/− MEF in vitro. The expression and phosphorylation of JNK, p38 MAPK, and ERK during adipogenic differentiation of wild-type and Mlk3−/− MEF was examined by immunoblot analysis (Fig. 10A). During differentiation, JNK phosphorylation was slightly decreased in the Mlk3−/− MEF compared with wild-type MEF. Conversely, p38 MAPK phosphorylation was slightly increased in the Mlk3−/− MEF. No differences in the expression or phosphorylation of ERK were observed.
FIG. 10.
MLK3 deficiency increases adipocytic differentiation in vitro. (A) The expression and phosphorylation of JNK, ERK, and p38 MAPKs during in vitro differentiation of MEF to adipocytes were examined by immunoblot analysis. The expression and phosphorylation of C/EBPα (Ser-222/Ser-226) and C/EBPβ (Thr-235) during differentiation were also examined. (B) The morphology of WT and Mlk3−/− MEF before differentiation (Control) and following adipocyte differentiation (8 days) was examined by phase-contrast microscopy and by staining accumulated fat droplets with Oil-Red-O. (C) The number of adipocytes observed following in vitro differentiation of WT, Mlk3−/+, and Mlk3−/− MEF was counted. The data are presented as the relative number of adipocytes (mean ± SD for the results of three experiments). (D) The effect of the presence or absence of insulin and troglitizone during the final 5 days of differentiation of WT and Mlk3−/− MEF was examined. The data are presented as the relative number of adipocytes (mean ± SD for the results of three experiments).
It is established that MAPKs target transcription factors involved in adipogenesis (3, 9, 15). We therefore examined the expression and phosphorylation of C/EBPα and C/EBPβ during adipose differentiation (Fig. 10A). The expression and phosphorylation of C/EBPα and C/EBPβ were increased in Mlk3−/− MEF at early times during differentiation. These data indicate that MLK3 deficiency can alter MAPK activation and transcription factor phosphorylation during adipogenesis. Thus, MLK3 deficiency may increase adipose differentiation in vitro.
To directly investigate whether MLK3 deficiency can increase adipogenic differentiation, we examined the morphology of differentiated wild-type MEF and Mlk3−/− MEF. Morphological examination by phase-contrast microscopy indicated increased adipogenic differentiation of the Mlk3−/− MEF compared with wild-type MEF (Fig. 10B). This conclusion was confirmed by staining accumulated fat droplets with Oil-Red-O (Fig. 10B). This increase in adipogenic differentation was suppressed by expression of MLK3 (Fig. 10C), indicating that the altered differentiation of Mlk3−/− MEF was caused by MLK3 deficiency. Interestingly, the increased adipogenic differentiation potential of Mlk3−/− MEF was observed in cultures with and without the PPARγ ligand troglitizone (Fig. 10D). Together, these data demonstrate that MLK3 deficiency increases the adipogenic potential of primary MEF in vitro.
DISCUSSION
Our analysis of Mlk3−/− mice indicates that MLK3 contributes to TNF-stimulated JNK activation in primary MEF. Previous studies using RNAi-mediated gene suppression have suggested a more general role of MLK3 as an essential protein kinase for the activation of multiple MAPK signaling pathways in response to a broad array of extracellular stimuli (5). In addition, RNAi-based studies have suggested that MLK3 is critical for cell proliferation (5) and normal microtubule function (28). Our analysis of Mlk3−/− MEF does not support these conclusions drawn from RNAi-based studies (5, 28). Since our biochemical analysis was restricted to MEF, it is possible that the divergent conclusions between these studies reflect differences in the role of MLK3 between cell types. Nevertheless, it is difficult to reconcile the conclusion that MLK3 is critical for proliferation and has multiple essential signaling roles with the finding that Mlk3−/− mice are viable and healthy. A second possible explanation of the more restricted phenotype observed in our study is that a greater degree of compensation may have occurred in the MLK3 knockout mice than in vitro studies using RNAi. A number of other potential explanations also exist (20), including the possibility that the phenotype caused by a reduction of MLK3 expression is greater than that caused by the elimination of MLK3 expression.
A consensus conclusion from our MLK3 knockout analysis (this study) and previously reported RNAi-based studies (5, 28) is that the limited role of MLK3 that we observe in TNF signaling represents a nonredundant function of MLK3. Our analysis does not exclude the possibility that MLK3 may also have additional functions that are redundant with other members of the MAP3K family in primary MEF.
MLK3 contributes to the TNF signaling pathway that activates JNK.
TNF binds to the receptor TNF-R1 and activates several signaling pathways that mediate a balance between life and death (37). Thus, TNF can activate caspase 8 to promote apoptosis and can increase cell survival by activating NF-κB. TNF also activates the ERK, JNK, and p38 MAPK signaling pathways. Here, we show that the MAP3K isoform MLK3 plays a role in the response of MEF to TNF. Studies of MLK3-deficient MEF demonstrate that MLK3 partially contributes to the immediate activation of JNK in response to TNF (Fig. 5 and 6). However, MLK3 is not required for TNF-stimulated activation of ERK and p38 MAPK or TNF-stimulated NF-κB activation (Fig. 4 to 6). MLK3 is also not required for TNF-stimulated activation of caspase 8 or apoptosis (unpublished observations). These data indicate that MLK3 selectively contributes to TNF-stimulated JNK activity.
MLK3-deficient MEF exhibit a selective and partial reduction in TNF-stimulated JNK activation. Nevertheless, robust TNF-induced JNK activation was observed in Mlk3−/− MEF (Fig. 5 and 6). This observation suggests that MLK3 acts redundantly with at least one other MAP3K in TNF-stimulated JNK activation. A key question for future studies is the identity of the MAP3K(s) that mediates the effects of TNF on JNK activation in MLK3-deficient cells. A plausible hypothesis is that this protein kinase activity represents another member of the MLK group, but other MAP3K may also contribute to TNF-stimulated JNK activation. A second key question relates to the mechanism by which TNF activates MLK3. The adapter protein TRAF2 is required for TNF-stimulated JNK activation (42), but whether TRAF2 might regulate MLK3 directly or indirectly is unclear. Analysis of the regulatory relationship between TRAF2 and MLK3 represents a critical goal for future experiments.
Interestingly, TNF-stimulated p38 MAPK activation is increased in Mlk3−/− MEF (Fig. 5 and 6). Recent studies of JNK-deficient cells demonstrate increased p38 MAPK activation in response to TNF (19). Conversely, decreased p38 MAPK activity can increase JNK activation (12, 40). These data suggest that JNK and p38 MAPK are negatively regulated by cross talk between these signaling pathways. The mechanism of this cross talk may be mediated by MAPK phosphatases (4, 32), including MKP5 and MKP7 (17, 30, 31, 33, 44). Further studies are required to define the mechanism that alters TNF-stimulated p38 MAPK activation in Mlk3−/− MEF.
MLK3 regulates the adipogenic potential of MEF in vitro.
Adipogenesis is a physiological process that is differentially regulated by MAPKs (23, 24). For example, JNK activation inhibits the proadipogenic transcription factor PPARγ (1, 3). Conversely, p38 MAPK activation promotes adipogenesis, in part by increasing the expression and transcriptional activation of C/EBP (8, 9, 43). These observations suggest that adipogenesis is a paradigm in which reciprocal alterations in JNK and p38 MAPK activity have biological consequences. Here, we show that the decreased JNK activation and increased p38 MAPK activation observed with Mlk3−/− MEF are associated with increased expression and phosphorylation of proadipogenic transcription factors (C/EBPα and C/EBPβ) and increased adipogenic differentiation in vitro. These data suggest that MLK3 plays a role in regulating adipogenesis. Nevertheless, studies of Mlk3−/− mice maintained on a standard laboratory chow diet ad libitum did not reveal increased adiposity in vivo (unpublished observations). Further studies will be required to test whether the potential for increased adiposity in Mlk3−/− mice may be revealed if these mice are exposed to an environmental challenge (e.g., being fed a high-fat diet).
Conclusion.
We report that the MLK3 protein kinase contributes to TNF-stimulated JNK activation. This identification of a role for MLK3 fills an important gap in our understanding of the mechanism of JNK activation caused by TNF.
Acknowledgments
We thank Stephen Jones for blastocyst injections; Linda Evangelista for ES cell culture; Tammy Barrett, Judith Reilly, Vicky Benoit, and Jennifer Ayala for expert technical assistance; and Kathy Gemme for administrative assistance.
These studies were supported by a grant from the National Institutes of Health and by the Diabetes and Endocrinology Research Center (DERC) of the University of Massachusetts (NIH/NIDDK). R.A.F. and R.J.D. are investigators of the Howard Hughes Medical Institute.
REFERENCES
- 1.Adams, M., M. J. Reginato, D. Shao, M. A. Lazar, and V. K. Chatterjee. 1997. Transcriptional activation by peroxisome proliferator-activated receptor gamma is inhibited by phosphorylation at a consensus mitogen-activated protein kinase site. J. Biol. Chem. 272:5128-5132. [DOI] [PubMed] [Google Scholar]
- 2.Beyaert, R., A. Cuenda, W. Vanden Berghe, S. Plaisance, J. C. Lee, G. Haegeman, P. Cohen, and W. Fiers. 1996. The p38/RK mitogen-activated protein kinase pathway regulates interleukin-6 synthesis response to tumor necrosis factor. EMBO J. 15:1914-1923. [PMC free article] [PubMed] [Google Scholar]
- 3.Camp, H. S., S. R. Tafuri, and T. Leff. 1999. c-Jun N-terminal kinase phosphorylates peroxisome proliferator-activated receptor-gamma1 and negatively regulates its transcriptional activity. Endocrinology 140:392-397. [DOI] [PubMed] [Google Scholar]
- 4.Camps, M., A. Nichols, and S. Arkinstall. 2000. Dual specificity phosphatases: a gene family for control of MAP kinase function. FASEB J. 14:6-16. [PubMed] [Google Scholar]
- 5.Chadee, D. N., and J. M. Kyriakis. 2004. MLK3 is required for mitogen activation of B-Raf, ERK and cell proliferation. Nat. Cell Biol. 6:770-776. [DOI] [PubMed] [Google Scholar]
- 6.Davis, R. J. 2000. Signal transduction by the JNK group of MAP kinases. Cell 103:239-252. [DOI] [PubMed] [Google Scholar]
- 7.Derijard, B., M. Hibi, I. H. Wu, T. Barrett, B. Su, T. Deng, M. Karin, and R. J. Davis. 1994. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76:1025-1037. [DOI] [PubMed] [Google Scholar]
- 8.Engelman, J. A., A. H. Berg, R. Y. Lewis, A. Lin, M. P. Lisanti, and P. E. Scherer. 1999. Constitutively active mitogen-activated protein kinase kinase 6 (MKK6) or salicylate induces spontaneous 3T3-L1 adipogenesis. J. Biol. Chem. 274:35630-35638. [DOI] [PubMed] [Google Scholar]
- 9.Engelman, J. A., M. P. Lisanti, and P. E. Scherer. 1998. Specific inhibitors of p38 mitogen-activated protein kinase block 3T3-L1 adipogenesis. J. Biol. Chem. 273:32111-32120. [DOI] [PubMed] [Google Scholar]
- 10.Gallo, K. A., and G. L. Johnson. 2002. Mixed-lineage kinase control of JNK and p38 MAPK pathways. Nat. Rev. Mol. Cell Biol. 3:663-672. [DOI] [PubMed] [Google Scholar]
- 11.Green, H., and O. Kehinde. 1975. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell 5:19-27. [DOI] [PubMed] [Google Scholar]
- 12.Hall, J. P., and R. J. Davis. 2002. Inhibition of the p38 pathway upregulates macrophage JNK and ERK activities, and the ERK, JNK, and p38 MAP kinase pathways are reprogrammed during differentiation of the murine myeloid M1 cell line. J. Cell Biochem. 86:1-11. [DOI] [PubMed] [Google Scholar]
- 13.Hehner, S. P., T. G. Hofmann, A. Ushmorov, O. Dienz, I. Wing-Lan Leung, N. Lassam, C. Scheidereit, W. Droge, and M. L. Schmitz. 2000. Mixed-lineage kinase 3 delivers CD3/CD28-derived signals into the IκB kinase complex. Mol. Cell. Biol. 20:2556-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirai, S., K. Noda, T. Moriguchi, E. Nishida, A. Yamashita, T. Deyama, K. Fukuyama, and S. Ohno. 1998. Differential activation of two JNK activators, MKK7 and SEK1, by MKN28-derived nonreceptor serine/threonine kinase/mixed lineage kinase 2. J. Biol. Chem. 273:7406-7412. [DOI] [PubMed] [Google Scholar]
- 15.Hu, E., J. B. Kim, P. Sarraf, and B. M. Spiegelman. 1996. Inhibition of adipogenesis through MAP kinase-mediated phosphorylation of PPARγ. Science 274:2100-2103. [DOI] [PubMed] [Google Scholar]
- 16.Ishitani, T., G. Takaesu, J. Ninomiya-Tsuji, H. Shibuya, R. B. Gaynor, and K. Matsumoto. 2003. Role of the TAB2-related protein TAB3 in IL-1 and TNF signaling. EMBO J. 22:6277-6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuguchi, T., T. Musikacharoen, T. R. Johnson, A. S. Kraft, and Y. Yoshikai. 2001. A novel mitogen-activated protein kinase phosphatase is an important negative regulator of lipopolysaccharide-mediated c-Jun N-terminal kinase activation in mouse macrophage cell lines. Mol. Cell. Biol. 21:6999-7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merritt, S. E., M. Mata, D. Nihalani, C. Zhu, X. Hu, and L. B. Holzman. 1999. The mixed lineage kinase DLK utilizes MKK7 and not MKK4 as substrate. J. Biol. Chem. 274:10195-10202. [DOI] [PubMed] [Google Scholar]
- 19.Morton, S., R. J. Davis, and P. Cohen. 2004. Signalling pathways involved in multisite phosphorylation of the transcription factor ATF-2. FEBS Lett. 572:177-183. [DOI] [PubMed] [Google Scholar]
- 20.Nature Publishing Group. 2003. Whither RNAi. Nat. Cell Biol. 5:489-490. [DOI] [PubMed] [Google Scholar]
- 21.Picard, F., M. Gehin, J. Annicotte, S. Rocchi, M. F. Champy, B. W. O'Malley, P. Chambon, and J. Auwerx. 2002. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell 111:931-941. [DOI] [PubMed] [Google Scholar]
- 22.Rana, A., K. Gallo, P. Godowski, S. Hirai, S. Ohno, L. Zon, J. M. Kyriakis, and J. Avruch. 1996. The mixed lineage kinase SPRK phosphorylates and activates the stress-activated protein kinase activator, SEK-1. J. Biol. Chem. 271:19025-19028. [DOI] [PubMed] [Google Scholar]
- 23.Rosen, E. D., and B. M. Spiegelman. 2000. Molecular regulation of adipogenesis. Annu. Rev. Cell Dev. Biol. 16:145-171. [DOI] [PubMed] [Google Scholar]
- 24.Rosen, E. D., C. J. Walkey, P. Puigserver, and B. M. Spiegelman. 2000. Transcriptional regulation of adipogenesis. Genes Dev. 14:1293-1307. [PubMed] [Google Scholar]
- 25.Sathyanarayana, P., M. K. Barthwal, C. N. Kundu, M. E. Lane, A. Bergmann, G. Tzivion, and A. Rana. 2002. Activation of the Drosophila MLK by ceramide reveals TNF-alpha and ceramide as agonists of mammalian MLK3. Mol. Cell 10:1527-1533. [DOI] [PubMed] [Google Scholar]
- 26.Sathyanarayana, P., M. K. Barthwal, M. E. Lane, S. F. Acevedo, E. M. Skoulakis, A. Bergmann, and A. Rana. 2003. Drosophila mixed lineage kinase/slipper, a missing biochemical link in Drosophila JNK signaling. Biochim. Biophys. Acta 1640:77-84. [DOI] [PubMed] [Google Scholar]
- 27.Stronach, B., and N. Perrimon. 2002. Activation of the JNK pathway during dorsal closure in Drosophila requires the mixed lineage kinase, slipper. Genes Dev. 16:377-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swenson, K. I., K. E. Winkler, and A. R. Means. 2003. A new identity for MLK3 as an NIMA-related, cell cycle-regulated kinase that is localized near centrosomes and influences microtubule organization. Mol. Biol. Cell 14:156-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takaesu, G., R. M. Surabhi, K. J. Park, J. Ninomiya-Tsuji, K. Matsumoto, and R. B. Gaynor. 2003. TAK1 is critical for IκB kinase-mediated activation of the NF-κB pathway. J. Mol. Biol. 326:105-115. [DOI] [PubMed] [Google Scholar]
- 30.Tanoue, T., T. Moriguchi, and E. Nishida. 1999. Molecular cloning and characterization of a novel dual specificity phosphatase, MKP-5. J. Biol. Chem. 274:19949-19956. [DOI] [PubMed] [Google Scholar]
- 31.Tanoue, T., T. Yamamoto, R. Maeda, and E. Nishida. 2001. A novel MAPK phosphatase MKP-7 acts preferentially on JNK/SAPK and p38 alpha and beta MAPKs. J. Biol. Chem. 276:26629-26639. [DOI] [PubMed] [Google Scholar]
- 32.Theodosiou, A., and A. Ashworth. 2002. MAP kinase phosphatases. Genome Biol 3:3009.1-3009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Theodosiou, A., A. Smith, C. Gillieron, S. Arkinstall, and A. Ashworth. 1999. MKP5, a new member of the MAP kinase phosphatase family, which selectively dephosphorylates stress-activated kinases. Oncogene 18:6981-6988. [DOI] [PubMed] [Google Scholar]
- 34.Tobiume, K., A. Matsuzawa, T. Takahashi, H. Nishitoh, K. Morita, K. Takeda, O. Minowa, K. Miyazono, T. Noda, and H. Ichijo. 2001. ASK1 is required for sustained activations of JNK/p38 MAP kinases and apoptosis. EMBO Rep. 2:222-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tournier, C., C. Dong, T. K. Turner, S. N. Jones, R. A. Flavell, and R. J. Davis. 2001. MKK7 is an essential component of the JNK signal transduction pathway activated by proinflammatory cytokines. Genes Dev. 15:1419-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tournier, C., P. Hess, D. D. Yang, J. Xu, T. K. Turner, A. Nimnual, D. Bar-Sagi, S. N. Jones, R. A. Flavell, and R. J. Davis. 2000. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science 288:870-874. [DOI] [PubMed] [Google Scholar]
- 37.Wajant, H., K. Pfizenmaier, and P. Scheurich. 2003. Tumor necrosis factor signaling. Cell Death Differ. 10:45-65. [DOI] [PubMed] [Google Scholar]
- 38.Weston, C. R., and R. J. Davis. 2002. The JNK signal transduction pathway. Curr. Opin. Genet. Dev. 12:14-21. [DOI] [PubMed] [Google Scholar]
- 39.Whitmarsh, A. J., and R. J. Davis. 2001. Analyzing JNK and p38 mitogen-activated protein kinase activity. Methods Enzymol. 332:319-336. [DOI] [PubMed] [Google Scholar]
- 40.Whitmarsh, A. J., S. H. Yang, M. S. Su, A. D. Sharrocks, and R. J. Davis. 1997. Role of p38 and JNK mitogen-activated protein kinases in the activation of ternary complex factors. Mol. Cell. Biol. 17:2360-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wysk, M., D. D. Yang, H. T. Lu, R. A. Flavell, and R. J. Davis. 1999. Requirement of mitogen-activated protein kinase kinase 3 (MKK3) for tumor necrosis factor-induced cytokine expression. Proc. Natl. Acad. Sci. USA 96:3763-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeh, W. C., A. Shahinian, D. Speiser, J. Kraunus, F. Billia, A. Wakeham, J. L. de la Pompa, D. Ferrick, B. Hum, N. Iscove, P. Ohashi, M. Rothe, D. V. Goeddel, and T. W. Mak. 1997. Early lethality, functional NF-κB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity 7:715-725. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, J. W., D. J. Klemm, C. Vinson, and M. D. Lane. 2004. Role of CREB in transcriptional regulation of CCAAT/enhancer-binding protein beta gene during adipogenesis. J. Biol. Chem. 279:4471-4478. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, Y., J. N. Blattman, N. J. Kennedy, J. Duong, T. Nguyen, Y. Wang, R. J. Davis, P. D. Greenberg, R. A. Flavell, and C. Dong. 2004. Regulation of innate and adaptive immune responses by MAP kinase phosphatase 5. Nature 430:793-797. [DOI] [PubMed] [Google Scholar]