Abstract
To further understand how the mitogen-activated protein kinase (MAPK) signaling pathways regulate AP-1 activity, we have elucidated the physiological role of these cascades in the regulation of c-jun gene expression. c-Jun is a crucial component of AP-1 complexes and has been shown in vitro to be a point of integration of numerous signals that can differentially affect its expression as well as its transcriptional activity. Our strategy was based on the use of (i) genetically modified fibroblasts deficient in components of the MAPK cascades and (ii) pharmacological reagents. The results demonstrate that c-Jun NH2-terminal protein kinase (JNK) is essential for a basal level of c-Jun expression and for c-Jun phosphorylation in response to stress. In addition to JNK, p38 MAPK or ERK1/2 and ERK5 are required for mediating UV radiation- or epidermal growth factor (EGF)-induced c-Jun expression, respectively. Further studies indicate that p38 MAPK inhibits the activation of JNK in response to EGF, causing a down-regulation of c-Jun. Overall, these data provide important insights into the mechanisms that ultimately determine the function of c-Jun as a regulator of cell fate.
c-Jun is a critical component of the AP-1 transcription factors that consist of homo- or heterodimers of basic region-leucine zipper proteins that belong to the Jun, Fos, ATF, and Maf subfamilies (17). Basic region-leucine zipper dimers recognize either 12-O-tetradecanoylphorbol-13-acetate response elements (TRE) or cyclic AMP response elements in the promoter regions of numerous genes. The variety of dimeric complexes and the dual roles of AP-1 as a transcriptional activator and repressor of genes may explain how c-Jun regulates so many different, and sometimes opposing, cellular processes (37). For example, consistent with its role in cell proliferation, c-Jun is induced by the transient expression of oncogenes and is required for the transformation of fibroblasts by activated Ha-Ras (3, 16, 36). Conversely, genetically modified cells have provided evidence that c-Jun is required for mediating the apoptotic response of neurons to stress (5, 32, 37). The role of c-Jun in regulating apoptosis of fibroblasts following UV radiation (UV) remains controversial (5, 42).
As c-Jun is a typical immediate early gene, the induction of c-Jun expression following the stimulation of cells is rapid, large, and transient. Regulation occurs at two levels: (i) the stabilization of the protein via a reduction in c-Jun ubiquitination and degradation (10, 27, 40) and (ii) the control of gene transcription (2). The c-jun promoter region contains several regulatory elements, including TRE and myocyte enhancer factor 2 (MEF2) binding sites (12, 13). The TRE binds dimers of c-Jun and ATF factors, indicating that c-Jun regulates its own expression (2). MEF2A and MEF2D seem to be the predominant factors that bind to the MEF2 site (13, 29). The transcriptional activities of c-Jun, ATFs, and MEF2 are regulated upon phosphorylation by various protein kinases, including the mitogen-activated protein kinases (MAPK), which have been implicated in vitro in the transcriptional regulation of c-jun (18, 23).
At least four MAPK subfamilies have been identified: extracellular-regulated protein kinases 1 and 2 (ERK1/2), ERK5, c-Jun NH2-terminal protein kinase (JNK), and p38 MAPK. ERK1/2 and JNK are capable of phosphorylating c-Jun (25, 33). JNK, p38 MAPK, and ERK1/2 phosphorylate ATF2 (11, 26, 31, 34). However, c-Jun and ATF2 activities appear to be regulated in vivo, primarily by JNK (25, 26). MEF2A activity is controlled by both p38 MAPK and ERK5, whereas MEF2D is a specific substrate of ERK5 (19, 30, 43).
Together, these studies clearly establish the regulation of c-Jun as a point of integration of numerous signals transduced by MAPK pathways. However, the data are based on the use of different cell lines and are not directly comparable. Here, we have performed a comprehensive analysis to unravel the respective roles of each of the MAPKs in regulating c-jun gene expression in response to apoptotic stress (UV) and after mitogenic stimulation (epidermal growth factor [EGF]). Our results provide clear evidence that the regulation of c-Jun expression by UV involves both JNK and p38 MAPK, while JNK, ERK1/2, and ERK5 are required to mediate the effect of EGF. In addition, p38 MAPK negatively affects EGF-dependent regulation of c-Jun by its ability to inhibit JNK activity.
MATERIALS AND METHODS
Tissue culture and preparation of lysates.
Mouse embryonic fibroblasts (MEFs) were established from wild-type, jnk1−/− jnk2−/− (jnk−/−), or erk5−/− embryonic day 13 mouse embryos (39; Xin Wang, unpublished data). The cells were placed in 1% serum for 24 h prior to being treated with UV (60 J/m2) or EGF (50 ng/ml; Sigma). Where appropriate, the cells were pretreated for 30 min to 1 h with inhibitors as follows: SP600125, 25 μM (Calbiochem); SB203580, 10 μM (Promega); and PD184352, 2 μM. Agonists and inhibitors were added directly to the cell culture medium. Transfection of fibroblasts was carried out using the calcium phosphate method.
Proteins were extracted from cells in triton lysis buffer (20 mM Tris, pH 7.4, 137 mM NaCl, 2 mM EDTA, 1% Triton X-100, 25 mM β-glycerophosphate, 10% glycerol, 1 mM orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 10 μg/ml aprotinin). Extracts were clarified by centrifugation (14,000 × g for 10 min at 4°C). The concentration of soluble proteins in the supernatants was quantified by the Bradford method (Bio-Rad).
Immunoblot analysis.
Cell and tissue extracts (50 μg) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% polyacrylamide gel) and electrophoretically transferred to an Immobilon-P membrane (Millipore, Inc.). The membranes were incubated with 5% nonfat dry milk or 3% bovine serum albumin at 4°C overnight and then probed with polyclonal antibodies to c-Jun (Cell Signaling), MAPKAPK2 (Cell Signaling), ERK5 (41), ERK1/2 (Santa Cruz), JNK (Santa Cruz), or p38 MAPK (Santa Cruz). Immune complexes were detected by enhanced chemiluminescence (Amersham Biosciences), with rabbit or mouse immunoglobulin G coupled to horseradish peroxidase as the secondary antibody (Amersham Biosciences). Equal protein loading was monitored by detecting the levels of tubulin (Sigma) expression in the cell extracts.
Protein kinase assays.
JNK, p38 MAPK, ERK1/2, and ERK5 protein kinase activities were measured in cell lysates in the presence of [γ-32P]ATP following precipitation with glutathione S-transferase (GST)-c-Jun (34) or polyclonal antibodies to p38 MAPK (34), ERK1/2 (Santa Cruz), or ERK5 (41), respectively. GST-ATF2 (34), GST-c-Myc (1), and GST-MEF2C (28) were used as substrates for p38 MAPK, ERK1/2, and ERK5, respectively. The radioactivity incorporated into recombinant proteins was quantitated after sodium dodecyl sulfate-polyacrylamide gel electrophoresis by PhosphorImager analysis.
RNA isolation.
Total RNA was isolated from cells by using the Trizol reagent (Invitrogen) as instructed by the manufacturer. RNA was treated with DNase by using the DNA-free kit (Ambion) as instructed by the manufacturer. RNA concentrations and quality were assessed visually by ethidium bromide/agarose gel electrophoresis under UV illumination and comparison with known amounts of mouse embryonic total RNA.
Real-time quantitative PCR.
cDNA synthesis was carried out in a final volume of 20 μl of first-strand buffer containing 3 μg of total RNA, 20 U SuperScript II reverse transcriptase (Invitrogen), 0.025 μg/μl oligo(dT) (Promega), and 0.5 mM deoxynucleoside triphosphate (Roche). Primers were designed using the Primer Express software from the published mouse c-jun and β-actin sequences (GenBank accession numbers J04115 and NM-007393, respectively). These were as follows: for c-jun, forward primer, 5′-AGCAGGGACCCATGGAAGTT-3′, and reverse primer, 5′-AAAGATGACCTTTGCTTGTGCAT-3′; and for β-actin, forward primer, 5′-CCAACTTGATGTATGAAGGCTTTG-3′, and reverse primer, 5′-GCCTGTACACTGACTTGAGACCAATT-3′. These primers were to generate amplicons of 92 bp and 91 bp, respectively. Real-time quantitative PCRs were performed using the SYBR green I core kit (Eurogentec). PCR products were detected in the ABI-PRISM 7700 or 7000 sequence detection system (Applied Biosystems). Results were analyzed using the 2−ΔΔG methods (22). The level of expression of c-jun mRNA was normalized to β-actin mRNA.
Reporter gene expression assay.
The reporter plasmid TRE-Luc (35) or MEF2 Luc (13) was transiently cotransfected with or without expression vectors encoding JNK (9), ERK5, or MEKK3 (7). A pRL-Tk plasmid encoding Renilla luciferase was employed for monitoring transfection efficiency. Aliquots of cell lysates were assayed for firefly and Renilla luciferase activities according to the manufacturer's instructions (Promega).
RESULTS
JNK and p38 MAPK are both required for mediating UV-induced c-Jun expression.
We have previously provided genetic evidence for an essential role of JNK in mediating the apoptotic response of MEFs to genotoxic stress, including UV (39). To clarify the role of c-Jun in JNK-dependent cell death, we have tested the effect of deleting jnk genes on UV-dependent regulation of c-Jun expression (Fig. 1). UV transiently increased the level of c-jun mRNA, with a maximum (sixfold) reached after 1 h in wild-type cells (Fig. 1A). The absence of JNK correlated with a drastic (around 90%) decrease in the basal level of c-jun but did not prevent UV from promoting a significant induction of the transcript (16-fold after 1 h) with kinetics similar to that observed in wild-type cells (Fig. 1A). A similar decrease in c-jun mRNA was observed following the treatment of the wild-type cells with SP600125, a well-characterized inhibitor of JNK (6), but not with SB203580 or PD184352, that specifically block p38 MAPK activity (21) and ERK1/2 activation (24), respectively (Fig. 1B; see Fig. 2 and 6).
FIG. 1.
JNK is required for mediating the effect of UV on c-Jun expression. Wild-type (wt) and jnk−/− fibroblasts without treatment or pretreated with SP600125 (25 μM) were UV radiated (60 J/m2) and incubated for the times indicated. (A and B) Total RNA was extracted, and the amounts of c-jun mRNA were measured by quantitative PCR. Results are expressed as percentages of the maximums ± standard errors of duplicate samples. (C) MEF extracts were analyzed for c-Jun expression by immunoblot analysis using a specific polyclonal anti-c-Jun antibody. The detection of tubulin expression was performed to monitor protein loading. The figure is representative of three independent experiments.
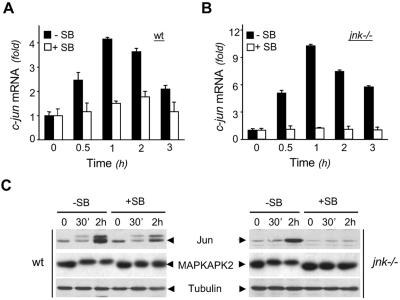
FIG. 2.
p38 MAPK contributes to the regulation of c-Jun expression in response to UV. Wild-type (wt) (A and C) and jnk−/− (B and C) MEFs without treatment (−) or pretreated (+) with SB203580 (SB; 10 μM) were UV radiated (60 J/m2) and incubated for the times indicated. (A and B) Total RNA was extracted, and the amounts of c-jun mRNA were measured by quantitative PCR. Results are expressed as increases relative to the mRNA extracted from unstimulated fibroblasts ± standard errors of duplicate samples. (C) MEF extracts were analyzed for c-Jun and MAPKAPK2 expression by immunoblot analysis using specific polyclonal anti-c-Jun and anti-MAPKAPK2 antibodies. The detection of tubulin expression was performed to monitor protein loading. The figure is representative of three independent experiments.
FIG. 6.
ERK1/2 and ERK5 are implicated in the regulation of c-Jun expression in response to EGF. Wild-type (wt), jnk−/−, and erk5−/− MEFs without treatment (−) or pretreated (+) with PD184352 (2 μM) or SP600125 (25 μM) were stimulated with EGF (50 ng/ml) or UV radiated (60 J/m2) for the times indicated. (A, B, D, and F) Total RNA was extracted, and the amounts of c-jun mRNA were measured by quantitative PCR. Results are expressed as increases relative to the mRNA extracted from unstimulated fibroblasts (A and B) or as percentages of the maximums (D and F) ± standard errors of duplicate samples. (C and E) MEF extracts were analyzed for c-Jun expression by immunoblot analysis using a specific polyclonal anti-c-Jun antibody. The detection of tubulin expression was performed to monitor protein loading. The figure is representative of three independent experiments.
Immunoblot analysis of the cell extracts confirmed that JNK was required for the basal level of c-Jun protein but was not essential for its up-regulation in response to UV (Fig. 1C). With a 90% decrease in the amount of mRNA (Fig. 1A), the jnk−/− cells displayed around three times less c-Jun protein than the wild-type cells. Despite the low level detected under basal conditions, a significant increase in c-Jun expression was observed in UV-treated jnk−/− cell extracts. In contrast, the absence of JNK completely abolished the electrophoretic mobility shift of c-Jun typical of the protein being phosphorylated in wild-type MEFs 30 min after stimulation. Pretreatment of the wild-type cells with SP600125 recapitulated the abnormal phenotype caused by jnk gene deletion (Fig. 1C). Consistent with a previous study (25), these results provide further genetic and pharmacological evidences that JNK is required for c-Jun phosphorylation in response to stress.
Since the induction of AP-1 by genotoxic stress is also mediated by the p38 MAPK cascade (17), we tested the effect of the p38 MAPK inhibitor SB203580 on the regulation of c-Jun by UV (Fig. 2). The ability of the compound to block p38 MAPK but not JNK activity is demonstrated by its specific inhibitory effect on UV-induced MAPKAPK2 but not c-Jun phosphorylation (Fig. 2C). Real-time PCR quantification and immunoblot analysis showed that SB203580 induced a partial inhibition (around 60%) of c-Jun expression in response to UV in wild-type cells (Fig. 2A and C) and a total inhibition in jnk−/− fibroblasts (Fig. 2B and C).
Altogether, the data show that JNK is essential for the basal expression of c-Jun and for c-Jun phosphorylation in response to UV. There is around 10 times less c-jun mRNA, corresponding to around 3 times less c-Jun protein in jnk−/− cells than in wild-type cells (Fig. 1). However, the up-regulation of c-Jun in response to UV stimulation requires both JNK and p38 MAPK (Fig. 2).
JNK and p38 MAPK have opposite effects on EGF-dependent regulation of c-Jun expression.
Similar to c-jun−/− MEFs, jnk−/− fibroblasts display a severe proliferation defect (16, 36, 39). This led us to investigate the role of JNK in regulating c-Jun expression in response to EGF. Compared to UV, EGF induced a more transient and less potent activation of JNK in fibroblasts, reaching a maximum at 5 min and returning to the basal level 30 min after stimulation (Fig. 3A and data not shown). However, similar to UV, EGF induced a marked and transient increase in c-jun mRNA in wild-type cells, reaching a maximum (5.5-fold) after 30 min of stimulation (Fig. 3B). With a low basal level, increased expression of c-jun in jnk−/− fibroblasts following EGF treatment was potent (11-fold after 1 h) and sustained (Fig. 3B). The sustained effect of EGF in jnk−/− cells compared to wild-type cells suggests that JNK controls a negative feedback loop resulting in a transient rather than prolonged increase in c-jun expression. Consistent with these data, EGF up-regulated the expression of c-Jun protein in jnk−/− cells to a level similar to that observed in wild-type cells 2 h following treatment (Fig. 3C).
FIG. 3.
JNK is required for mediating the effect of EGF on c-Jun expression. Wild-type (wt) and jnk−/− fibroblasts were stimulated with EGF (50 ng/ml) for the times indicated. (A) Endogenous JNK activity was measured by protein kinase assay. (B) Total RNA was extracted, and the amounts of c-jun mRNA were measured by quantitative PCR. Results are expressed as percentages of the maximums ± standard errors of duplicate samples. (C) MEF extracts were analyzed for c-Jun expression by immunoblot analysis using a specific polyclonal anti-c-Jun antibody. The detection of tubulin expression was performed to monitor protein loading. The figure is representative of three independent experiments.
Since EGF was able to induce a transient increase in p38 MAPK activity in fibroblasts (Fig. 4A), we tested whether the p38 MAPK cascade was responsible for the JNK-independent regulation of c-Jun expression in response to EGF. Surprisingly, the pretreatment of the wild-type MEFs with SB203580 markedly potentiated and delayed the effect of EGF on c-Jun expression (Fig. 4B and D). The level of c-jun mRNA in wild-type cells exposed to both SB203580 and EGF was approximately fourfold higher than that in cells exposed only to EGF. This phenomenon was reported previously and did not implicate the destabilization of the transcript by p38 MAPK (14). In contrast, SB203580 did not have any effect on increased c-Jun expression in EGF-treated jnk−/− fibroblasts (Fig. 4C and D), indicating that p38 MAPK-induced down-regulation of c-Jun is JNK dependent. Consistent with this hypothesis, fibroblasts pretreated with SB203580 displayed higher JNK activity following EGF treatment (Fig. 4E). Based on these results, we concluded that p38 MAPK represses the EGF-dependent increase in c-Jun expression by inhibiting JNK activity. Next, we investigated whether ERKs were involved, in addition to JNK, in controlling c-jun gene expression in response to a mitogenic signal.
FIG. 4.


p38 MAPK antagonizes the effect of JNK on the transcriptional regulation of c-jun following EGF treatment. Wild-type (wt) (A, B, D, and E) and jnk−/− (C and D) MEFs without treatment (−) or pretreated (+) with SB203580 (SB; 10 μM) were incubated with EGF (50 ng/ml) for the times indicated. The activities of p38 MAPK (A) or JNK (E) were measured by protein kinase assay. (B and C) Total RNA was extracted, and the amounts of c-jun mRNA were measured by quantitative PCR. Results are expressed as increases relative to the mRNA extracted from unstimulated fibroblasts ± standard errors of duplicate samples. (D) MEF extracts were analyzed for c-Jun expression by immunoblot analysis using a specific polyclonal anti-c-Jun antibody. The detection of tubulin expression was performed to monitor protein loading. The figure is representative of three independent experiments.
Inhibition of ERK activities in fibroblasts.
The ability of PD98059 to block the activation of both ERK1/2 and ERK5 (24) has led to controversial data about the specific role of ERK1/2 and ERK5 in regulating cell function. To circumvent this problem, we employed PD184352, a more specific inhibitor of the ERK1/2 signaling pathway. Consistent with a previous study (24), PD184352 blocked EGF-dependent stimulation of ERK1/2 (Fig. 5A). At this concentration (2 μM), the drug did not inhibit ERK5 activity but consistently delayed the peak of activation of ERK5 by EGF (Fig. 5B).
FIG. 5.
Inhibition of ERK1/2 and ERK5 in fibroblasts. MEFs without treatment (−) or pretreated (+) with PD184352 (PD; 2 μM) were incubated with EGF (50 ng/ml) for the times indicated. The activities of ERK1/2 (A and E) and ERK5 (B and D) were measured by protein kinase assay. (C) Wild-type (wt) and erk5−/− MEF extracts were analyzed for ERK5, ERK1/2, JNK, and p38 MAPK expression by immunoblot analysis using specific polyclonal antibodies. The detection of tubulin expression was performed to monitor protein loading. The figure is representative of two independent experiments.
Since no specific ERK5 inhibitor exists, we used a novel model of erk5-null fibroblasts (Xin Wang, unpublished). Disruption of the erk5 gene did not alter the expression of ERK1/2, JNK, or p38 MAPK (Fig. 5C). Furthermore, EGF treatment caused a marked increase in ERK5 activity in wild-type but not homozygous knockout erk5 fibroblasts (Fig. 5D). In contrast, no marked difference was observed in the abilities of EGF to stimulate ERK1/2 and JNK activities in wild-type and erk5−/− MEFs (Fig. 5E and data not shown). These data established that erk5−/− MEFs possess no ERK5 and represent a useful model for studies of the ERK5 signaling pathway.
Both ERK1/2 and ERK5 contribute to mediating EGF-induced c-Jun expression.
Inhibition of ERK1/2 activation impaired the early increase in c-jun mRNA by EGF and subsequently delayed the induction of the protein in the wild-type fibroblasts (Fig. 6A and C). The ability of PD184352 to interfere with EGF signaling was more pronounced in jnk−/− MEFs for which the drug inhibited the response of the cells to the treatment throughout the time course of induction (Fig. 6B and C). These results indicate that ERK1/2 is required for the early increase in c-Jun expression by EGF but can contribute to the late phase of induction in cells that do not express JNK.
The absence of ERK5 significantly impaired the ability of EGF to increase c-jun mRNA expression (Fig. 6D and F). After 15 min of treatment, the level of induction in erk5−/− MEFs was half of what was observed in wild-type cells. In contrast to JNK, ERK5 did not affect the basal level of the transcript. Consistently, the induction of c-Jun protein expression by EGF was lower in erk5−/− MEFs than in wild-type MEFs (Fig. 6E). The inhibition of JNK or ERK1/2 further blocked the residual increase in c-jun mRNA in the ERK5-deficient fibroblasts treated with EGF (Fig. 6F and data not shown). Consistent with the JNK-dependent negative feedback loop, the pretreatment of the erk5−/− MEFs with SP600125 resulted in a prolonged rather than transient increase in c-jun expression in response to EGF stimulation (Fig. 6F). Control experiments showed no effect of PD184352 or of the erk5 gene deletion on UV-dependent regulation of c-Jun (Fig. 6C and E).
Altogether, these results indicate that both ERK1/2 and ERK5 are required for the regulation of c-Jun expression by EGF. The ERK1/2 signaling pathway appears to be redundant to the JNK cascade for the late induction of c-Jun.
Molecular mechanism of the transcriptional regulation of c-jun by MAPKs.
Next, we investigated the molecular mechanism by which MAPKs regulate the transcription of c-jun. Fibroblasts were transfected with the luciferase reporter plasmids containing the TRE or the MEF2 binding site present in the promoter of the c-jun gene together with constructs encoding JNK, ERK5, or MEKK3. MEKK3-induced transcriptional activity was determined by measuring luciferase activity.
MEKK3-induced activation of the TRE-Luc was enhanced following the reintroduction of JNK in the jnk−/− MEFs. This indicated that the absence of JNK impaired the ability of MEKK3 to activate transcription via AP-1 (Fig. 7A). Consistent with our in vivo data (Fig. 1 and 3), these results confirm that JNK is required but not essential for the transcriptional regulation of c-Jun. In contrast, the absence of ERK5 completely prevented MEKK3 from increasing transcriptional activity via the MEF2 binding site (Fig. 7B). Overexpression of ERK5 in the erk5−/− MEFs rescued the defect. The essential role of the ERK5 signaling pathway in regulating MEF2 activity in fibroblasts was previously observed by the inability of the p38 MAPK cascade to compensate for the deletion of the mek5 gene (41). Control experiments showed that MEKK3-induced transcriptional activation via MEF2 or TRE was not affected by the deletion of the jnk or erk5 gene, respectively (Fig. 7A and B). To investigate the roles of p38 MAPK and ERK1/2, wild-type MEFs were pretreated with SB203580 and PD184352 12 h prior to measurement of the luciferase activity, respectively (Fig. 7C). Both compounds inhibited, by around 45%, the ability of MEKK3 to activate transcription via TRE. In contrast, MEKK3-induced transcription via the MEF2 binding site was specifically blocked (80% decrease) following the inhibition of p38 MAPK activity but not ERK1/2 activation.
FIG. 7.

The targeting of c-jun mRNA by MAPK cascades via the TRE and the MEF2 binding sites. jnk−/− (A), erk5−/− (B), and wild-type (wt) (C) fibroblasts were transiently transfected as described in Materials and Methods. Levels of TRE- and MEF2-dependent transcriptional activity were measured by the dual-luciferase reporter assay system. Where indicated, the wt cells were treated with SB203580 (SB; 10 μM) or PD184352 (PD; 2 μM) 12 h prior to the performance of the assay. Firefly luciferase activity was normalized to that of Renilla luciferase and expressed as the percent increase over the control (−). The data correspond to the means ± standard errors of three independent experiments performed in duplicate.
Altogether, these results indicate that, in vitro, the promoter of the c-jun gene can be controlled by JNK, p38 MAPK, and ERK1/2 via the TRE and by p38 MAPK and ERK5 via the MEF2 binding site.
DISCUSSION
The plethora of physiological stimuli and environmental insults that regulate c-Jun function has made c-Jun a useful model to study the complexity of gene regulation via AP-1 (17). Here, we have performed a comprehensive study to elucidate how MAPK cascades control gene expression via the transcriptional regulation of c-jun. Our results demonstrate that the effect of MAPK on c-Jun expression is mediated (i) by JNK and p38 MAPK in response to UV and (ii) by JNK, ERK1/2, and ERK5 following EGF stimulation. The contribution of ERKs or p38 MAPK, together with JNK, to the transcriptional regulation of the c-jun gene may reflect the distinct function of c-Jun in the regulation of cell fate.
The small amounts of c-jun present in the jnk−/− MEFs compared to those in the wild-type cells indicate that JNK controls the basal level of the transcript (Fig. 1 and 3). JNK has previously been shown to stabilize interleukin 2 mRNA through a cis element encompassing the 5′ untranslated region and the beginning of the coding region (8). Based on this study, we examined whether JNK increased the stability of c-jun by comparing its half-life in unstimulated wild-type and jnk−/− MEFs. Consistent with the presence of an adenosine- or uridine-rich element characterized by multiple copies of the pentanucleotide AUUUA in the 3′ untranslated region of c-jun, we confirmed that c-jun is a rapidly decaying mRNA with a half-life of 20 min and that this was not significantly affected by jnk deletion (data not shown). These results indicate that JNK does not affect the stability of the c-jun transcript. Thus, the low level of c-Jun expressed in jnk-null MEFs is most likely caused by a transcriptional defect.
In addition to regulating c-Jun expression, JNK is essential for mediating the phosphorylation of c-Jun in response to UV (Fig. 1C). Consistent with this result, a recent study based on the use of phospho-specific antibodies to different phosphorylated forms of c-Jun has shown that JNK is the MAPK required for stress-induced phosphorylation of c-Jun on Ser63 and Ser73 residues (25). MEFs and cortical neurons in which the c-jun gene was replaced by a mutant c-jun allele with Ser63 and Ser73 mutated to Ala displayed a stress-induced apoptotic defect (5). In light of our previous results (39), these studies suggest that JNK-dependent c-Jun phosphorylation is required for mediating apoptosis. Death genes whose expression is controlled by the JNK/c-Jun signaling pathway have already been identified (37). However, c-Jun function does not always depend on JNK activation. For example, a role for c-Jun in the regulation of G1 progression has been shown to be independent of its phosphorylation (42). Similarly, JNK-dependent phosphorylation of Ser63 and Ser73 is not essential for c-Jun to cooperate with Ha-Ras in cellular transformation (15, 20).
An intriguing issue is the ability of p38 MAPK to inhibit increased c-Jun expression in response to EGF (Fig. 4). The absence of this inhibitory effect in the JNK-deficient cells indicates that the p38 MAPK-induced down-regulation of c-Jun is JNK dependent. Consistent with this hypothesis, protein kinase assays show that p38 MAPK blocks EGF-dependent activation of JNK. Negative regulation of JNK by p38 MAPK in response to EGF has previously been reported to be mediated via the inactivation of the MAPK kinase MKK4 by the protein phosphatase-2A (4). MKK4 is a dual-specificity protein kinase that functions as a selective activator of JNK in response to extracellular stimuli (38). Altogether, these studies suggest that p38 MAPK represses the EGF-dependent increase in c-Jun expression by inhibiting JNK activity via the specific dephosphorylation of MKK4.
Regulatory elements found in the promoter region of the c-jun gene include a TRE and a MEF2 site. Luciferase reporter assays demonstrate that transcriptional regulation via the TRE can be mediated by JNK, p38 MAPK, and ERK1/2 (Fig. 7A and C). This is consistent with the ability of JNK and ERK1/2 to phosphorylate c-Jun (9, 33), while ATF2 is a substrate for JNK, ERK1/2, and p38 MAPK (11, 31, 34). However, in vivo data indicate that JNK and ERK1/2 are the physiological MAPKs involved in mediating the phosphorylation of c-Jun and ATF2 (25, 26). Therefore, the p38 MAPK is likely mediating its effect via its ability to regulate MEF2 activity (Fig. 7C) (30, 43). Consistent with our previous study (41), we show that p38 MAPK is required and ERK5 is essential for the stimulation of MEF2 activity following the overexpression of MEKK3 (Fig. 7B). In light of this knowledge, this study provide for the first time physiological evidence that the transcriptional regulation of the c-jun gene is mediated by JNK and ERK1/2 via the TRE and by p38 MAPK and ERK5 via the MEF2 binding site (Fig. 8).
FIG. 8.
Model of the regulation of the promoter of the c-jun gene by MAPKs. MAPK activators include MEK1 and MEK2 for ERK1/2, MEK5 for ERK5, MKK4 and MKK7 for JNK, and MKK3 and MKK6 for p38 MAPK. UV activates JNK and p38 MAPK, leading to the subsequent increase in c-jun expression via the TRE and the MEF2 binding sites, respectively. In contrast, EGF-induced c-jun expression is mediated via TRE by JNK and ERK1/2 and via MEF2 by ERK5. The negative regulation of c-jun by p38 MAPK in response to EGF treatment is mediated by the ability of p38 MAPK to inhibit JNK activity, most probably via a phosphatase-mediated dephosphorylation of MKK4. UV- and EGF-dependent signaling pathways are represented by solid and broken lines, respectively.
Acknowledgments
We are indebted to Roger Davis and Richard Flavell for kindly providing the jnk−/− MEFs. We thank Philip Cohen for providing the ERK1/2 inhibitor PD184352 and Eric Olson, Ron Prywes, Christian Widmann, and Alan Whitmarsh for their generosity in providing the GST-MEF2C, MEF2-Luc, MEKK3, and ERK5 constructs, respectively. We thank A. Whitmarsh for critically reviewing the manuscript.
This work was supported in part by the BBSRC, and principally by the AICR, the Royal Society, and a Lister Institute of Preventive Medicine Research Fellowship to C.T.
REFERENCES
- 1.Alvarez, E., I. C. Northwood, F. A. Gonzalez, D. A. Latour, A. Seth, C. Abate, T. Curran, and R. J. Davis. 1991. Pro-Leu-Ser/Thr-Pro is a consensus primary sequence for substrate protein phosphorylation. Characterization of the phosphorylation of c-myc and c-jun proteins by an epidermal growth factor receptor threonine 669 protein kinase. J. Biol. Chem. 266:15277-15285. [PubMed] [Google Scholar]
- 2.Angel, P., K. Hattori, T. Smeal, and M. Karin. 1988. The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell 55:875-885. [DOI] [PubMed] [Google Scholar]
- 3.Angel, P., and M. Karin. 1991. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta 1072:129-157. [DOI] [PubMed] [Google Scholar]
- 4.Avdi, N. J., K. C. Malcom, J. A. Nick, and G. S. Worthen. 2002. A role for protein phosphatase-2A in p38 mitogen-activated protein kinase-mediated regulation of c-Jun NH2-terminal kinase pathway in human neutrophils. J. Biol. Chem. 277:40687-40696. [DOI] [PubMed] [Google Scholar]
- 5.Behrens, A., M. Sibilia, and E. F. Wagner. 1999. Amino-terminal phosphorylation of c-Jun regulates stress-induced apoptosis and cellular proliferation. Nat. Genet. 21:326-329. [DOI] [PubMed] [Google Scholar]
- 6.Bennett, B. L., D. T. Sasaki, B. W. Murray, E. C. O’Leary, S. T. Sakata, W. Xu, J. C. Leisten, A. Motiwala, S. Pierce, Y. Satoh, S. S. Bhagwat, A. M. Manning, and D. W. Anderson. 2001. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl. Acad. Sci. USA 98:13681-13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonvin, C., A. Guillon, M. X. van Bemmelen, P. Gerwins, G. L. Johnson, and C. Widmann. 2002. Role of the amino-terminal domains of MEKKs in the activation of NF kappa B and MAPK pathways and in the regulation of cell proliferation and apoptosis. Cell. Signal. 14:123-131. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C.-Y., F. Del Gatto-Konczak, Z. Wu, and M. Karin. 1998. Stabilization of interleukin-2 mRNA by the c-Jun NH2-terminal kinase pathway. Science 280:1945-1949. [DOI] [PubMed] [Google Scholar]
- 9.Derijard, B., M. Hibi, I. H. Wu, T. Barrett, B. Su, T. Deng, M. Karin, and R. J. Davis. 1994. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76:1025-1037. [DOI] [PubMed] [Google Scholar]
- 10.Fuchs, S. Y., L. Dolan, R. J. Davis, and Z. Ronai. 1996. Phosphorylation-dependent targeting of c-Jun ubiquitination by Jun N-kinase. Oncogene 13:1531-1535. [PubMed] [Google Scholar]
- 11.Gupta, S., D. Campbell, B. Derijard, and R. J. Davis. 1995. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science 267:389-393. [DOI] [PubMed] [Google Scholar]
- 12.Han, T.-H., W. W. Lamph, and R. Prywes. 1992. Mapping of epidermal growth factor-, serum-, and phorbol ester-responsive sequence elements in the c-jun promoter. Mol. Cell. Biol. 12:4472-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han, T.-H., and R. Prywes. 1995. Regulatory role of MEF2D in serum induction of the c-jun promoter. Mol. Cell. Biol. 15:2907-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hazzalin, C. A., A. Cuenda, E. Cano, P. Cohen, and L. C. Mahadevan. 1997. Effects of the inhibition of p38/RK MAP kinase on induction of five fos and jun genes by diverse stimuli. Oncogene 15:2321-2331. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, R., B. Spiegelman, D. Hanahan, and R. Wisdom. 1996. Cellular transformation and malignancy induced by ras require c-jun. Mol. Cell. Biol. 16:4504-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, R. S., B. van Lingen, V. E. Papaioannou, and B. M. Spiegelman. 1993. A null mutation at the c-jun locus causes embryonic lethality and retarded cell growth in culture. Genes Dev. 7:1309-1317. [DOI] [PubMed] [Google Scholar]
- 17.Karin, M., Z.-G. Liu, and E. Zandi. 1997. AP-1 regulation and function. Curr. Opin. Cell Biol. 9:240-246. [DOI] [PubMed] [Google Scholar]
- 18.Kato, Y., V. Kravchenko, R. I. Tapping, J. Han, R. J. Ulevitch, and J.-D. Lee. 1997. BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. EMBO J. 16:7054-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato, Y., M. Zhao, A. Morikawa, T. Sugiyama, D. Chakravortty, N. Koide, T. Yoshida, R. I. Tapping, Y. Yang, T. Yokochi, and J.-D. Lee. 2000. Big mitogen-activated kinase regulates multiple members of the MEF2 protein family. J. Biol. Chem. 275:18534-18540. [DOI] [PubMed] [Google Scholar]
- 20.Kennedy, N. J., H. K. Sluss, S. N. Jones, D. Bar-Sagi, R. A. Flavell, and R. J. Davis. 2003. Suppression of Ras-stimulated transformation by the JNK signal transduction pathway. Genes Dev. 17:629-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, J. C., J. T. Laydon, P. C. McDonnell, T. F. Gallagher, S. Kumar, D. Green, D. McNulty, M. J. Blumenthal, J. R. Heys, S. W. Landvatter, J. E. Strickler, M. M. McLaughlin, I. R. Siemens, S. M. Fisher, G. P. Livi, J. R. White, J. L. Adams, and P. R. Young. 1994. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature 372:739-746. [DOI] [PubMed] [Google Scholar]
- 22.Livak, K. L., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 23.Marinissen, M. J., M. Chiariello, M. Pallante, and J. S. Gutkind. 1999. A network of mitogen-activated protein kinases links G protein-coupled receptors to the c-jun promoter: a role for c-Jun NH2-terminal kinase, p38s, and extracellular signal-regulated kinase 5. Mol. Cell. Biol. 19:4289-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mody, N., J. Leitch, C. Armstrong, J. Dixon, and P. Cohen. 2001. Effect of MAP kinase cascade inhibitors on the MKK5/ERK5 pathway. FEBS Lett. 502:21-24. [DOI] [PubMed] [Google Scholar]
- 25.Morton, S., R. J. Davis, A. McLaren, and P. Cohen. 2003. A reinvestigation of the multisite phosphorylation of the transcription factor c-Jun. EMBO J. 22:3876-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morton, S., R. J. Davis, and P. Cohen. 2004. Signalling pathways involved in multisite phosphorylation of the transcription factor ATF-2. FEBS Lett. 572:177-183. [DOI] [PubMed] [Google Scholar]
- 27.Musti, A. M., M. Treier, and D. Bohmann. 1997. Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science 275:400-402. [DOI] [PubMed] [Google Scholar]
- 28.Nicol, R. L., N. Frey, G. Pearson, M. Cobb, J. Richardson, and E. N. Olson. 2001. Activated MEK5 induces serial assembly of sarcomeres and eccentric cardiac hypertrophy. EMBO J. 20:2757-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ornatsky, O. I., and J. C. McDermott. 1996. MEF2 protein expression, DNA binding specificity and complex composition, and transcriptional activity in muscle and non-muscle cells. J. Biol. Chem. 271:24927-24933. [DOI] [PubMed] [Google Scholar]
- 30.Ornatsky, O. I., D. M. Cox, P. Tangirala, J. J. Andreucci, Z. A. Quinn, J. L. Wrana, R. Prywes, Y.-T. Yu, and J. C. McDermott. 1999. Post-translational control of the MEF2A transcriptional regulatory protein. Nucleic Acids Res. 27:2646-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Owens, D. M., N. D. de Ruiter, G. C. M. van der Zon, A. P. Carter, J. Schouten, C. van der Burgt, K. Kooistra, J. L. Bos, J. A. Maassen, and H. van Dam. 2002. Growth factors can activate ATF2 via a two-step mechanism: phosphorylation of Thr71 through the Ras-MEK-ERK pathway and of Thr69 through RalGDS-Src-p38. EMBO J. 21:3782-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmada, M., S. Kanwal, N. J. Rutkoski, C. Gufstafson-Brown, R. S. Johnson, R. Wisdom, and B. D. Carter. 2002. c-jun is essential for sympathetic neuronal death induced by NGF withdrawal but not by p75 activation. J. Cell Biol. 158:453-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pulverer, B. J., J. M. Kyriakis, J. Avruch, E. Nikolakaki, and J. R. Woodgett. 1991. Phosphorylation of c-jun mediated by MAP kinases. Nature 353:670-674. [DOI] [PubMed] [Google Scholar]
- 34.Raingeaud, J., A. J. Whitmarsh, B. Barrett, B. Derijard, and R. J. Davis. 1996. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol. Cell. Biol. 16:1247-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rincon, M., and R. A. Flavell. 1994. AP-1 transcriptional activity requires both T-cell receptor-mediated and co-stimulatory signals in primary T lymphocytes. EMBO J. 13:4370-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schreiber, M., A. Kolbus, F. Piu, A. Szabowski, U. Mohle-Steinlein, J. Tian, M. Karin, P. Angel, and E. F. Wagner. 1999. Control of cell cycle progression by c-Jun is p53 dependent. Gene Dev. 13:607-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaulian, E., and M. Karin. 2002. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4:E131-E136. [DOI] [PubMed] [Google Scholar]
- 38.Tournier, C., C. Dong, T. K. Turner, S. N. Jones, R. A. Flavell, and R. J. Davis. 2001. MKK7 is an essential component of the JNK signal transduction pathway activated by proinflammatory cytokines. Genes Dev. 15:1419-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tournier, C., P. Hess, D. D. Yang, J. Xu, T. K. Turner, A. Nimnual, D. Bar-Sagi, S. N. Jones, R. A. Flavell, and R. J. Davis. 2000. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science 288:870-874. [DOI] [PubMed] [Google Scholar]
- 40.Treier, M., L. M. Staszewski, and D. Bohmann. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78:787-798. [DOI] [PubMed] [Google Scholar]
- 41.Wang, X., A. J. Merritt, J. Seyfried, C. Guo, E. S. Papadakis, K. G. Finegan, M. Kayahara, J. Dixon, R. P. Boot-Handford, E. J. Cartwright, U. Mayer, and C. Tournier. 2005. Targeted deletion of mek5 causes early embryonic death and defects in the ERK5/MEF2 cell survival pathway. Mol. Cell. Biol. 25:336-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wisdom, R., R. S. Johnson, and C. Moore. 1999. c-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J. 18:188-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao, M., L. New, V. V. Kravchenko, Y. Kato, H. Gram, F. Di Padova, E. N. Olson, R. J. Ulevitch, and J. Han. 1999. Regulation of the MEF2 family of transcription factors by p38. Mol. Cell. Biol. 19:21-30. [DOI] [PMC free article] [PubMed] [Google Scholar]