Abstract
CKIP-1 is a pleckstrin homology domain-containing protein that interacts with protein kinase CK2. To elucidate the functions of CKIP-1, we generated human osteosarcoma cell lines with tetracycline-regulated expression of Flag-CKIP-1. Flag-CKIP-1 expression resulted in distinct changes in cellular morphology. Therefore, we examined the actin profile by immunofluorescence, quantitative measurement of phalloidin binding, and immunoblot analysis. These studies demonstrate that Flag-CKIP-1 expression resulted in increases in F-actin staining and protein levels of β-actin. To elucidate the mechanisms behind the observed phenotype, we utilized tandem affinity purification to isolate CKIP-1 interacting proteins. Mass spectrometry analysis led to the identification of the actin capping protein subunits, CPα and CPβ, as novel CKIP-1 interaction partners. Interactions were confirmed by coimmunoprecipitation and by colocalization. Furthermore, we demonstrate that Ser9 of CPα is phosphorylated by protein kinase CK2 in vitro, that CPα is phosphorylated in vivo, and that treatment with a CK2-specific inhibitor results in a decrease in CPα phosphorylation. Finally, we demonstrate that CKIP-1 and CK2 inhibit the activity of actin capping protein at the barbed ends of actin filaments. Overall, our results are consistent with CKIP-1 playing a role in the regulation of the actin cytoskeleton through its interactions with actin capping protein.
The phosphorylation and dephosphorylation of proteins that is mediated by a complex network of protein kinases is a key mechanism intimately associated with the control of various aspects of cellular regulation (reviewed in references 23 and 34). One component of these regulatory kinase networks is protein kinase CK2. Protein kinase CK2 (formerly casein kinase II) is a ubiquitous and highly conserved protein serine/threonine kinase that is essential for viability in eukaryotic organisms (7, 26, 52). Although its precise functions remain poorly defined, there is mounting evidence to suggest that CK2 plays an important role in control of cell proliferation and transformation (reviewed in references 31 and 37). A number of studies have shown alteration in the expression of CK2 in a variety of tumor or leukemic cells (11, 17, 53, 59). Furthermore, the targeted overexpression of CK2α in the T cells of transgenic mice results in lymphocyte transformation. In these mice, there is evidence for collaboration between the dysregulated expression of CK2α and the c-Myc and Tal-1 oncogenes in lymphoma development (25, 51). Taken together, these studies demonstrate a role for CK2 as a component of the kinase networks that regulate the growth and division of cells. Despite the importance of CK2 in various aspects of cell regulation and its role in transformation, many questions regarding its regulation in cells remain.
An emerging paradigm in signal transduction is the importance of the clustering and targeting of signaling molecules. A notable example of this targeting is the interaction between the pleiotropic cAMP-dependent protein kinase (PKA) and the A-kinase anchoring proteins (AKAPs) (reviewed in references 1, 8, 14, and 44). Like PKA, protein kinase CK2 has a broad array of potential physiological substrates and exists in numerous subpopulations within a cell. Although it is evident that CK2 has substrates in a variety of subcellular compartments, it is unknown how distinct populations of protein kinase CK2 are targeted to appropriate substrates. In an effort to determine whether interacting proteins participate in the regulation of CK2, we previously undertook a yeast two-hybrid screen to identify novel CK2-interacting proteins. These studies led to the identification of CKIP-1 (3).
The CKIP-1 cDNA encodes a 409-amino-acid protein with an amino-terminal pleckstrin homology (PH) domain and a leucine-rich region within its carboxy terminus. Contained within CKIP-1 are five PXXP motifs, two of which match the consensus for cyclin-dependent kinases and mitogen-activated kinases. We have previously shown that CKIP-1 localizes to the plasma membrane and that the PH domain is required for binding to phospholipids in cells and in vitro (3, 40). Moreover, we have demonstrated that the pleckstrin homology domain of CKIP-1 is required for interactions with CK2 and for the recruitment of CK2 to the plasma membrane (40). It was also recently reported that CKIP-1 is induced during muscle differentiation and that it plays a role in phosphatidylinositol 3-kinase-regulated differentiation in C2C12 cells (46). On the basis of these observations, we hypothesized that CKIP-1 functions to target CK2 to specific cellular compartments thereby facilitating phosphorylation of particular substrates and perhaps at the same time limiting access to nonappropriate targets in an analogous way to the regulation of cAMP-dependent protein kinase by the AKAPs.
To understand the functions of CKIP-1 and its potential role in regulating protein kinase CK2, we generated cell lines with tetracycline-regulated expression of Flag-CKIP-1. Our objectives were to examine the phenotype associated with CKIP-1 expression and to elucidate the role, if any, that protein kinase CK2 plays in this phenotype. Here we report that CKIP-1 functions in the regulation of the actin cytoskeleton and cell morphology and forms stable interactions with the heterodimeric actin-capping protein. Furthermore, we show that the α-subunit of the actin capping protein (CPα) is phosphorylated by protein kinase CK2 on Ser9 in vitro, that CPα is phosphorylated in osteosarcoma cells, and that treatment with a CK2-specific inhibitor results in a decrease in CPα phosphate incorporation. Finally, we show that CKIP-1 and protein kinase CK2 inhibit the activity of CP at the barbed ends of actin filaments.
MATERIALS AND METHODS
Antibodies.
Monoclonal antibodies against the Flag epitope and polyclonal antibodies directed against β-actin were purchased from Sigma-Aldrich (St. Louis, Mo.). Monoclonal antibodies directed at the HA epitope (clone 12CA5) and anti-HA-biotin antibodies were purchased from Roche Applied Science (Indianapolis, Ind.). Polyclonal antibodies directed against green fluorescent protein (GFP) were purchased from Clontech (Palo Alto, Calif.). Antibodies against the CPα (MAb 5B12.3) and CPβ (MAb 3F2.3) subunits of the actin capping protein (49) were obtained from the Iowa Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biological Sciences, University of Iowa (Iowa City, Iowa). Monoclonal antibodies against β-tubulin were a generous gift from Lina Dagnino (Department of Pharmacology, University of Western Ontario). Antibodies directed against CKIP-1 were raised in rabbits against glutathione S-transferase (GST)-CKIP (positions 308 to 409) (3).
Plasmid constructs.
Flag-CKIP was generated by PCR amplification using the forward primer 5′ GAT AAG CTT GCC ACC ATG GAC TAC AAA GAC GAT GAC GAC AAG GAA TTC ATG ATG AAG AAG AAC 3′ and the reverse primer 5′ GCC CGG AAT TAG CTT GGC TGC AGG TGC GGC CGC CGC TGC CCTCAC ATC 3′. The resulting 1,300-bp fragment was subcloned into the pRc/CMV vector using the HindIII and NotI sites. For generation of a cell line with tetracycline-regulated expression, Flag-CKIP-1 was cut out of pRc/CMV with HindIII and NotI, extended with Klenow, and ligated into the BamHI sites of the vector pTRE. Deletion constructs Flag-CKIP-1(133-346), Flag-CKIP-1(133-308), and Flag-CKIP-1(133-250) were generated by PCR amplification using the forward primer 5′ GCC GAA TTC CGA GCC AAG AAC CGT ATC TTG GAT GAG GTC 3′ and the reverse primers 5′ AGC AGG CGG CCG CCG TTA AGA ATC CGG CGG AGA CCG AGG GGG GTC 3′, 5′ CTT GTG CGG CCG CCG TTA GAT CCG GGA CAG CTG CCC CGG GTT GGG 3′, and 5′ CTG GGG CGG CCG CCG TTA CCC TTT GTC TGT TTT TTC CCA AGG TCG 3′, respectively. Similarly, deletion constructs Flag-CKIP-1(193-346) and Flag-CKIP-1(235-346) were generated using the forward primers 5′ ATC GAA TTC GAA GAC CCT TCC CCT GAG GAA CCA ACC TCT 3′ and 5′ CAG GAA TTC GCA GAC CGG GCA AGC AGT CTC TTC CGA CCT 3′, respectively, and the reverse primer 5′ AGC AGG CGG CCG CCG TTA AGA ATC CGG CGG AGA CCG AGG GGG GTC 3′. The resulting fragments were subcloned into pRc/CMV vector by using the HindIII and NotI sites. TAP-CKIP was generated by PCR amplification using the forward primer 5′ G AGA TCT GAA TTC ATG ATG AAG AAG AAC AAT 3′ and the reverse primer 5′ CAG GTC GAC CCC AGC GGC CGC TCA CAT CAG 3′. Vectors expressing the TAP tag were a generous gift from Kathleen L. Gould, Vanderbilt University School of Medicine, Nashville, Tenn. (60). The TAP tag was amplified out of pFA6 using the forward primer 5′ GGG GCC ACC AAG CTT ATG AAA GCT GAT GCG CAA CAA AAT AAC TTC 3′ and the reverse primer 5′ TTC AGA TCT CCC ATA ATC AAG TGC CCC GGA GGA 3′. TAP-CKIP was assembled by a second round of PCR using the forward primer 5′ GGG GCC ACC AAG CTT ATG AAA GCT GAT GCG CAA CAA AAT AAC TTC 3′ and the reverse primer 5′ CAG GTC GAC CCC AGC GGC CGC TCA CAT CAG 3′. The resulting 1,800-bp fragment was subcloned into pRc/CMV using the HindIII and NotI sites. CP constructs for expression of HA-tagged CPα and HA-tagged CPαS9A were generated by PCR amplification using the forward primers 5′ AAA AGC GGC CGC ATG GCC GAC TTC GAT GAT CGT GTG TCG GAT GAG 3′ and 5′ AAA AGC GGC CGC ATG GCC GAC TTC GAT GAT CGT GTG GCG GAT GAG GAG AAG GTA CGC 3′, respectively, and the reverse primer 5′ AAA AGC GGC CGC TTA AGC ATT CTG CAT TTC TTT GCC AAT CTT 3′. The resulting 875-bp fragment was subcloned into the NotI sites of pRc/CMV immediately downstream of the HA-tag. Similarly, Myc-tagged CPβ was generated using the forward primer 5′ AAA AGC GGC CGC ATG AGC GAT CAG CAG CTG GAC TGC GCC TTG GAC 3′ and the reverse primer 5′ AAA AGC GGC CGC TCA ACA CTG CTG CTT TCT CTT CAA GGC CTC 3′. The resulting 800-bp fragment was subcloned into the NotI sites of pRc/CMV immediately downstream of the Myc tag. All constructs were verified by sequencing.
shRNA generation and transfection.
Two small interfering RNA sequences were selected from the open reading frame of CKIP-1: TTTGACCTGAGTGACTATG (nucleotides 208 to 226) and AAATTCTGCGGGAAAGGGATTTT (nucleotides 85 to 107) (46). Sequences were verified to target only CKIP-1 by BLAST analysis. As a control, a scrambled sequence (ACTGTACATCGCACTTCTG) with similar G+C content that generated no significant BLAST results was constructed. Sense and antisense primers were obtained, annealed, and subcloned into pSuper vector (a generous gift of Reuven Agami) using the BglII and HindIII sites (4). Constructed shRNAs were verified by sequencing. For mammalian cell expression, pSuper alone, scrambled sequence, or a combination of CKIP-1 shRNA were transfected using Fugene 6 (Roche) as described by the manufacturer. The vector pEGFP-C3 was included as a transfection marker, using a ratio of small hairpin RNA (shRNA) to pEGFP-C3 of 9:1.
Generation of cell lines.
UTA6 cells were derived from the human osteosarcoma cell line U2-OS and express the tetracycline-regulated transcriptional activator protein (a generous gift from Christoph Englert, Forschungszentrum, Germany [15]). Cell lines with tetracycline-regulated expression of Flag-CKIP-1 were generated by transiently transfecting UTA6 cells with Flag-CKIP-1/pTRE and pTK-Hyg in the presence of tetracycline (1.5 μg/ml). Drug selection with hygromycin (500 μg/ml) and G418 (460 μg/ml) began 4 days after transfection. Once stably transfected colonies were formed, they were picked and transferred to 96-well plates. Individual colonies of Flag-CKIP-1-expressing cells were grown up and screened for tight inducible expression by Western blot analysis.
Cell labeling and TBB treatment.
DC1.4 cells induced for expression of Flag-CKIP-1 were labeled with carrier-free [γ-32P]orthophosphate (ICN) by washing cells three times with phosphate-free Dulbecco's modified Eagle's medium and then culturing cells in the phosphate-free medium supplemented with 0.33mCi [γ-32P]orthophosphate for 6 hours. TBB (4,5,6,7-tetrabromo-2-azabenzimidazole) treatment (28, 45, 47, 50, 54) was performed by incubating cells with 75 μM TBB or dimethyl sulfoxide (DMSO) carrier for 18 h prior to cell labeling. Subsequently, cells were labeled for 6 hs as described above in the presence of 75 μM TBB or carrier alone.
Immunofluorescence.
Cells (∼150,000) grown on sterile coverslips were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Gibco/BRL) at 37°C in an atmosphere of 5% CO2. The coverslips were washed three times with phosphate-buffered saline (PBS) and fixed for 30 min in 3.7% paraformaldehyde at 37°C. The cells were permeabilized for 5 min with 0.1% Triton X-100 in PBS and then treated with for 5 min with 0.1 M glycine in PBS. The coverslips were washed with PBS and then incubated with primary antibody (anti-FLAG at 1:250 or anti-CPβ at 1:100) for 1 h at 37°C. Following washing with PBS, secondary antibodies conjugated to tetramethylrhodamine-5-isothiocyanate (TRITC) or fluorescein isothiocyanate (FITC) (both at 1:1,000) were added for 1 h in the dark. The coverslips were washed, mounted with AirVol (Air Products and Chemicals Inc., Allentown, Pa.), and visualized using a Zeiss LSM 410 confocal microscope. Where noted, cells were prepermeabilized for 30 s with buffer containing 0.5% Triton X-100, 50 mM NaCl, 3 mM MgCl2, 0.3 M sucrose, and 10 mM HEPES (pH 6.9) prior to paraformaldehyde fixation.
Phalloidin staining, extraction, and quantitation.
For TRITC-phalloidin staining of cells, coverslips were washed three times with PBS and fixed for 30 min in 3.7% paraformaldehyde at 37°C. The coverslips were stained with TRITC-phalloidin (1 μg/ml) for 40 min in the dark, washed with PBS, and mounted with Airvol. Slides were visualized using a Zeiss LSM 410 confocal microscope. Actin quantitation experiments were performed by the method of Machesky et al. (33). Briefly, triplicate plates of cells were washed twice with PBS and scraped into 400 μl of extraction buffer (10 mM PIPES, 190 mM K2PO4 plus 10 mM KH2PO4, 5 mM EGTA, 2 mM MgCl2, 0.1% Triton X-100) with 3.7% paraformaldehyde and 2 μM TRITC-phalloidin and rocked for 1 h at room temperature. The cells were pelleted by brief centrifigation and washed twice with 200 μl of 0.1% saponin-20 mM KPO4-10mM PIPES-5 mM EGTA-2 mM MgCl2. The bound phalloidin was extracted in 1 ml of methanol by rocking for 1 h at room temperature, and TRITC levels were determined by reading the emission at 563 nm following excitation at 542 nm. Values were normalized to the overall protein concentration and represent the average for three 10-cm plates and the standard deviation from two independent experiments.
Immunoprecipitations.
For immunoprecipitations, 10-cm plates of U2-OS cells were lysed on ice in 0.5 ml of NP-40 lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% NP-40, 1% aprotinin, 0.1 mM phenylmethylsulfonyl fluoride [PMSF]). The lysates were sonicated on ice with three 10-s bursts and then centrifuged at 55,000 rpm in a Beckman TL100.2 rotor for 15 min at 4°C. The cleared lysates were subjected to immunoprecipitation using anti-Flag M2 antibodies bound to protein G-Sepharose or anti-GFP or anti-HA antibodies bound to protein A-Sepharose. Following incubation for 1 h at 4°C, the lysates were centrifuged briefly and the protein A/G-Sepharose beads were washed four times with lysis buffer. Following the last wash, bound proteins were eluted from the beads by the addition of sample buffer. For large-scale Flag immunoprecipitations, four 15-cm plates were used. For immunokinase experiments, the protein A/G-Sepharose beads were washed three times with 50 mM Tris (pH 7.5)-120 mM NaCl-10 mM MgCl2 and phosphorylated with recombinant GST-CK2α′ for 30 min at 30°C in the presence of [γ-32P]ATP. Subsequently, the beads were washed four times with lysis buffer and bound proteins were eluted from the beads by the addition of sample buffer.
Immunoblot analysis.
Samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12% polyacrylamide) using the method of Laemmli (29). Proteins were transferred to polyvinylidene difluoride (PVDF) membranes for 1 h at 15 V and 0.3 A using semidry transfer apparatus (Bio-Rad, Hercules, Calif.). Anti-Flag blotting was performed as specified by the manufacturer, using anti-Flag M2 at 1:2,500 and goat anti-mouse secondary antibody (1:20,000) conjugated to horseradish peroxidase. Anti-β-actin blots were performed as specified by the manufacturer, using anti-β-actin at 1:2,000 and goat anti-rabbit secondary antibody (1:25,000) conjugated to horseradish peroxidase. Immunoblotting with anti-β-tubulin antibodies was performed by blocking for 1 h in 5% skim milk powder followed by incubation with primary antibody (1:100). Immune complexes were detected using goat anti-mouse secondary antibodies (1:5,000) conjugated to horseradish peroxidase. Anti-CPα and anti-CPβ blotting was performed by blocking membranes for 1 h in 3% bovine serum albumin followed by incubation with primary antibodies at dilutions of 1:200 and 1:600, respectively. In both cases, secondary antibody was used at a dilution of 1:5,000. Anti-HA-biotin blotting was performed by blocking for 1 h in 3% bovine serum albumin followed by incubation with primary antibody (1:500). Immune complexes were detected using peroxidase-conjugated anti-biotin (1:10,000). Where indicated, membranes were stripped with 0.1 M NaOH for 30 min.
Tandem affinity purification.
U2-OS cells (∼108 cells) were either nontransfected or transiently transfected with tandem affinity purification (TAP)-tagged (42) CKIP-1 using the calcium phosphate method. Cells were scraped off plates, and the resulting cell pellet was lysed in NP-40 lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% NP-40, 1% aprotinin, 0.1 mM PMSF) for 30 min on ice. The lysates were sonicated on ice with five 10-s bursts and then centrifuged at 55,000 rpm in a Beckman TL100.2 rotor for 25 min at 4°C. The cleared lysate was adjusted to 10 mM Tris (pH 7.5)-150 mM NaCl-0.2% NP-40-0.5 mM dithiothreitol DTT and bound to 200 μl of immunoglobulin G IgG-Sepharose beads (Amersham Biosciences, Piscataway, N.J.) overnight at 4°C. Following extensive washing in the above buffer plus 0.5 mM EDTA, CKIP-1 and associated proteins were cleaved from the beads by treatment with tobacco etch virus (Tev) protease for 2 h. Eluate (1 ml) was collected, adjusted to 2 mM CaCl2, and incubated with 200 μl of calmodulin beads (Stratagene, La Jolla, Calif.) in the presence of Ca2+ for 2 h. The calmodulin beads were washed extensively in the presence of Ca2+ and eluted with boiling sample buffer. CKIP-1 and associated proteins were separated by SDS-PAGE and visualized using the colloidal Coomassie blue stain GelCode Blue (Pierce, Rockford, Ill.).
Phosphorylation of CPα by protein kinase CK2.
TAP purification was performed as described above with the following additions: CKIP-1 and associated proteins were bound to calmodulin beads (100 μl) in the presence of Ca2+ for 2 h. Following extensive washing with buffer containing Ca2+, beads were resuspended in 50 mM KCl-20 mM Tris-HCl (pH 7.5)-10 mM MgCl2-2 mM CaCl2 with 20 μM [γ-32P]ATP and phosphorylated with 250 U of protein kinase CK2 (Calbiochem, San Diego, Calif.) for 30 min at 30°C. The beads were washed extensively in buffer containing Ca2+ and eluted with boiling sample buffer. Following separation by SDS-PAGE, the gel was dried and radiolabeled proteins were visualized using a PhosphorImager (Molecular Dynamics).
In-gel tryptic digests, mass spectrometry, and analysis.
Bands were excised from gels, cut into 1-mm cubes, and washed in 100 μl of double-distilled ddH2O for 15 mins. The stain was removed by repeatedly washing the gel pieces with 200 μl of 50:50 acetonitrile:ddH2O for 15 min with shaking. The gel pieces were dehydrated and rehydrated with 40 μl of acetonitrile and 40 μl of 100 mM ammonium bicarbonate, respectively, and evaporated to dryness with a Speedvac. Digests were performed overnight at 37°C using 1 μg of non-self-cleaving trypsin (Promega, Madison, Wis.) in 30 μl of 25 mM ammonium bicarbonate-2.5 mM CaCl2. Peptides were extracted five times with 30 μl of acetonitrile and 30 μl of 5% formic acid by vortexing and sonication. Samples were evaporated to dryness with a Speedvac and resuspended in 5μl of 0.1% trifluoroacetic acid prior to mass spectrometry analysis. Matrix-assisted laser desorption ionization (MALDI) analyses were performed on a Micromass (Manchester, United Kingdom) Reflectron MALDI-time-of-flight (TOF) instrument. Tandem mass spectrometry-mass spectrometry (MS/MS) analyses were performed on a quadrupole-time-of-flight mass spectrometer (Q-TOF 2; Micromass). Peptide mapping and database analyses were performed online using the search engine Mascot (www.matrixscience.com). All peptide sequences were manually verified.
Actin depolymerization assay.
F-actin at 5 μM (20% pyrene-labeled) was mixed with CP (1 nM), incubated for 5 min at room temperature, and diluted 50-fold into G buffer (2 mM Tris-HCl [pH 8.0], 0.2 mM ATP, 0.1 mM DTT, 0.2 mM CaCl2) at 25°C in a fluorometer cuvette with a stirring bar (57). The decrease in pyrene fluorescence intensity accompanying actin depolymerization was monitored for 400 s after dilution. To test the effect of CKIP-1 and protein kinase CK2, 1 nM CP was incubated for 5 min with different concentrations of proteins before addition to F-actin.
Spectrin-F-actin seeded actin polymerization assay.
Actin polymerization assays were performed essentially as described previously (57). Briefly, actin polymerization from spectrin F-actin seeds (SAS) was assayed using 1 μM actin (5% pyrene labeled), 4 nM CP, 0.4 to 0.5 nM SAS, and different concentrations of CKIP-1 and protein kinase CK2 in ATP-G-buffer (10 mM imidazole [pH 7.0], 0.2 mM ATP, 0.5 mM DTT, 0.2 mM CaCl2). The pyrene fluorescence (excitation, 368 nm; emission, 386 nm) was monitored for 500 s at 25°C.
Actin uncapping assays.
Actin uncapping assays were performed as described previously (49). Briefly, actin polymerization was initiated by addition of SAS and CP to monomeric actin (1.5 μM, 5% pyrene labeled). CKIP-1 (80 nM), protein kinase CK2α (250 nM), and phosphatidylinositol-4,5-bisphosphate PIP2 (40 μM) were added to the polymerization reaction mixture at 250 s, and the fluorescence was monitored for a total of 500 s.
RESULTS
Inducible expression of Flag-CKIP-1.
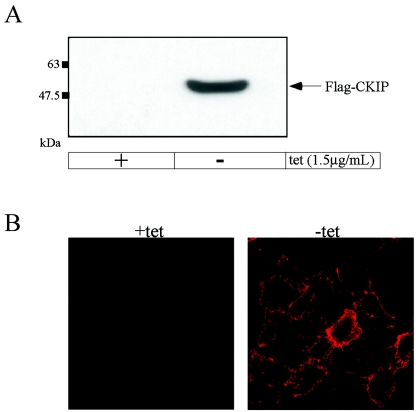
As a first step toward elucidation of the cellular functions of CKIP-1, we generated human osteosarcoma (U2-OS) cell lines, previously shown to express endogenous CKIP-1 (3), with tetracycline-regulated expression of Flag-tagged CKIP-1. For these experiments, Flag-CKIP-1 was ligated into the pTRE vector immediately downstream of the tetracycline-responsive promoter. Following transfection and drug-selection, stably transfected colonies were picked and screened for highly regulated inducible expression of Flag-CKIP-1 in the absence of tetracycline. Cells grown in the presence of tetracycline or induced for 24 h were harvested, and the lysates derived from these cells were subjected to immunoblotting with anti-Flag M2 antibody. As shown, the cell line DC1.4 exhibits strong induction and tight regulation (Fig. 1A). As a complementary approach, DC1.4 cells grown in the presence or absence of tetracycline were fixed, labeled with anti-Flag M2 antibodies, and visualized by confocal microscopy (Fig. 1B). Again, induction of Flag-CKIP-1 is tightly regulated. Furthermore, this analysis demonstrates that all cells express Flag-CKIP-1 and that it is localized predominantly at the plasma membrane in accordance with our previous results (40). Although the precise level of induction of CKIP-1 that occurs during muscle differentiation was not reported, the level of CKIP-1 induction achieved following tetracycline removal (>10-fold by immunoblot analysis) appears to be within the range of induction of CKIP-1 that was previously observed (46).
FIG. 1.
Tetracycline-regulated expression of Flag-tagged CKIP-1. Flag-tagged CKIP-1 is expressed in the cell line DC1.4 under the tight control of tetracycline (tet) as shown by immunoblotting (A) and immunofluorescence (B). (A) DC1.4 cells were grown in the presence or absence of tetracycline for 24 h to induce the expression of Flag-CKIP-1. Extracts were prepared, and 35 μg of total-cell lysate was separated by SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and immunoblotted with anti-Flag M2 antibodies. (B) DC1.4 cells grown in the presence or absence of tetracycline were fixed, permeabilized, and stained with anti-Flag M2 antibody and TRITC-goat anti-rabbit antibody.
Expression of Flag-CKIP-1 induces changes in cell morphology.
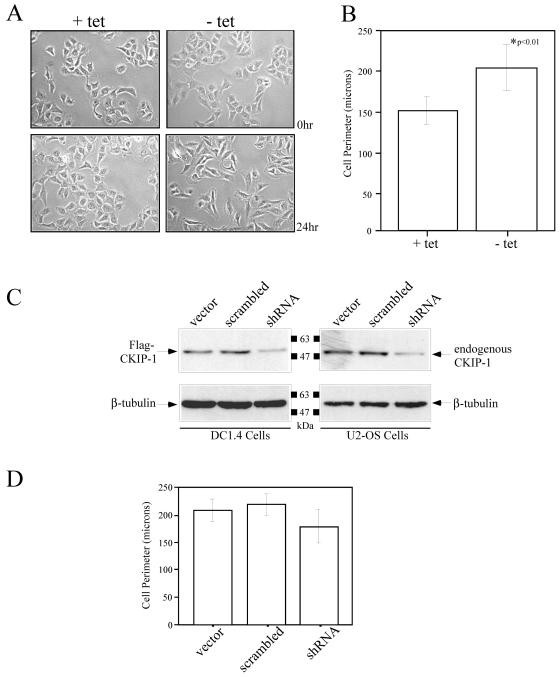
To characterize the effects of Flag-CKIP-1 expression in U2-OS cells, we examined microscopic images of DC1.4 cells grown in the presence or absence of tetracycline for 24 h (Fig. 2A). As shown, DC1.4 cells grown in the presence of tetracycline exhibit a consistent cellular morphology identical to that observed with the parent U2-OS cell line. However, after induction of Flag-CKIP-1 expression by the removal of tetracycline from the growth medium for 24 h, DC1.4 cells were consistently more elongated and exhibited morphology consistent with fibroblastic cells. To quantify this observation, we performed morphometric analysis of cells grown in the presence or absence of tetracycline by measuring the perimeter of ∼100 cells from numerous fields of view (Fig. 2B). The results of this experiment clearly show that cells expressing Flag-CKIP-1 have a significantly larger perimeter than do cells grown in the presence of tetracycline.
FIG. 2.
Expression of Flag-CKIP-1 induces changes in cell morphology that are partially blocked by treatment with shRNA. (A and B) Expression of Flag-CKIP-1 induced changes in cell morphology as shown by light microscopy (A) and Morphometric analysis (B). (A) DC1.4 cells were grown in the presence or absence of tetracycline (tet), and microscopic images were captured at 0 and 24 h. (B) The perimeter of these cells were measured and quantitated using Northern Elite software. Results shown represent the average of ∼100 cells in six fields; error bars indicate standard deviation. (C and D) Treatment of DC1.4 and parental U2-OS cells with CKIP-1 shRNA reduced the levels of Flag-CKIP-1 and endogenous CKIP-1, respectively, and partially blocked morphology changes in DC1.4 cells. (C) Transfection of DC1.4 (left panels) and U2-OS (right panels) cells with CKIP-1-targeting shRNA reduced the expression of Flag-CKIP-1 and endogenous CKIP-1, respectively, compared to that of empty vector or a scrambled sequence. To ensure equal loading, the membrane was stripped and reprobed with anti-β-tubulin. (D) DC1.4 cells cotransfected with pEGFP-C3 along with empty vector, scrambled shRNA, or CKIP-1 shRNA were induced for expression of Flag-CKIP-1. Cell perimeter measurements of GFP-positive cells were obtained 24 hs later, as described for panel B.
To extend these studies, we tested whether targeting Flag-tagged and endogenous CKIP-1 with shRNA could block the change in cell morphology. DC1.4 cells grown in the presence of tetracycline were transfected with empty vector or vector containing a scrambled shRNA or a combination of shRNA sequences directed against CKIP-1 (Fig. 2C, left panel). Flag-CKIP-1 expression was induced 18 h later by the removal of tetracycline from the growth medium, and lysates derived from these cells were subjected to immunoblotting with anti-Flag M2 antibodies. To ensure equal loading, the membrane was stripped and reprobed with antibodies against β-tubulin. Similarly, parental U2-OS cells were transfected with empty vector, scrambled shRNA, or a combination of shRNA directed against CKIP-1 (Fig. 2C, right panel), and lysates derived from these cells were analyzed by immunoblotting with anti-CKIP-1 and anti-β-tubulin antibodies. These data demonstrate that treatment with shRNA results in a partial knockdown of Flag-CKIP-1 and endogenous CKIP-1 compared to vector alone or a scrambled sequence.
To test the ability of shRNA to block the morphology change associated with Flag-CKIP-1 expression, DC1.4 cells grown in the presence of tetracycline were transfected with empty vector, scrambled shRNA, or a combination of CKIP-1 shRNA along with pEGFP-C3 as a transfection marker. Flag-CKIP-1 expression was induced 18 h later by the removal of tetracycline from the growth medium, and morphometric analysis of GFP-positive cells was performed. Treatment with CKIP-1 shRNA, but not empty vector or a scrambled sequence, was able to partially block the morphology change associated with Flag-CKIP-1 expression. These results are consistent with the partial knockdown of Flag-CKIP-1 that is observed and suggest that the change of morphology in DC1.4 cells is a direct result of Flag-CKIP-1 expression. At the level of resolution of the morphometric analyses that were performed, the partial knockdown of endogenous CKIP-1 in U2-OS cells did not elicit a significant quantifiable alteration in cell morphology (data not shown). However, more detailed investigation beyond the scope of the present investigation is required to rigorously examine subtle changes in morphology and/or the actin cytoskeleton that may result from a partial knockdown of endogenous CKIP-1.
Flag-CKIP-1 expression increases phalloidin binding and cellular β-actin levels.
Based on microscopic examination, it is evident that expression of Flag-CKIP-1 leads to changes in overall cell morphology. In order to further characterize this phenotype, we examined the F-actin profile of these cells using fluorescently-labeled phalloidin. DC1.4 cells grown in the presence (+) or absence (−) of tetracycline for 24 hours were fixed, stained with TRITC-phalloidin and visualized by confocal microscopy (Fig. 3A). When grown in the presence of tetracycline, DC1.4 cells exhibit predominantly peripheral actin stress fibers. However, DC1.4 cells expressing Flag-CKIP-1 exhibit stronger overall staining and show an increase in the number of transverse actin stress fibers. To confirm that cells expressing Flag-CKIP-1 have increased overall phalloidin binding, we performed quantitative measurements of TRITC-phalloidin binding (Fig. 3B). The data, normalized to extract protein, clearly show that DC1.4 cells exhibit increased phalloidin binding by 1.5-fold. Taken together, these data demonstrate that expression of Flag-CKIP-1 results in an increase in the overall amount of cellular F-actin.
FIG. 3.
Flag-CKIP-1 expression increases phalloidin binding and cellular β-actin levels. Cells expressing Flag-CKIP-1 have increased F-actin levels as shown by immunofluorescence (A) and quantitation of extracted TRITC-phalloidin (B). (A) DC1.4 cells grown in the presence of tetracycline (tet) or induced for Flag-CKIP-1 expression were fixed and stained with TRITC-phalloidin. (B) DC1.4 cells grown in the presence or absence of tetracycline for 48 h were scraped off plates, permeabilized, and labeled with TRITC-phalloidin as described in Materials and Methods. Bound phalloidin was extracted with methanol and quantitated by fluorescence emission of TRITC at 563 nm following excitation at 542 nm. Data represent the average of two independent experiments, each using three 100-mm plates of DC1.4 cells normalized for protein concentration (error bars indicate standard deviation). (C) Flag-CKIP-1 expression causes an increase in β-actin levels. DC1.4 cells were grown in the presence or absence of tetracycline for 24 h. Lysates were prepared by extraction with boiling sample buffer, and proteins were separated by SDS-PAGE, transferred to a PVFD membrane, and immunoblotted with anti-β-actin antibodies. To ensure equal loading, the membrane was stripped and reprobed with anti-β-tubulin. (D) CKIP-1 colocalizes with F-actin in DC1.4 cells. GFP-actin was transfected into DC1.4 cells expressing Flag-CKIP-1. Cells were pre-permeabilized with Triton X-100-containing buffer, fixed, and stained with anti-Flag M2 antibody as described in Materials and Methods.
Increased phalloidin binding in DC1.4 cells could result from either increased polymerization or increased amounts of actin. To determine if the overall cellular levels of actin are affected by Flag-CKIP-1, DC1.4 cells grown in the presence or absence of tetracycline were harvested with boiling sample buffer after a 24-h induction. As shown in Fig. 3C, expression of Flag-CKIP-1 clearly results in an increase in the cellular levels of β-actin. To ensure equal protein loading on the gel, the membrane was stripped and reprobed with anti-β-tubulin antibodies (Fig. 3C). Quantification of β-actin levels by densitometry and normalization to β-tubulin levels illustrated that β-actin levels are increased by approximately 60% in cells expressing Flag-CKIP-1, an increase that corresponds well to the increase in the level of F-actin. To extend these studies, we were interested in examining if CKIP-1 colocalizes with the actin cytoskeleton in mammalian cells (Fig. 3D). For these experiments, a construct expressing GFP-actin was transiently transfected into DC1.4 cells grown in the absence of tetracycline. Prior to fixing and immunofluorescence, these cells were pre-permeabilized with buffer containing Triton X-100 to remove all noncytoskeletal proteins. Immunofluorescence with anti-Flag M2 antibodies was performed, and the coverslips were mounted and visualized by fluorescence microscopy. With pre-permeabilization of the cells, Flag-CKIP-1 colocalizes with transverse actin stress fibers, as shown in the image overlaying Flag-CKIP with GFP-actin. This colocalization was not apparent when the cells were not pre-permeabilized, indicating that the majority of CKIP-1 localizes to the plasma membrane. Taken together with the results from the previous figure, these data suggest direct association of Flag-CKIP-1 with microfilaments in mammalian cells.
Identification of CKIP-1-associated proteins by tandem affinity purification of CKIP-1 and large-scale immunoprecipitations.
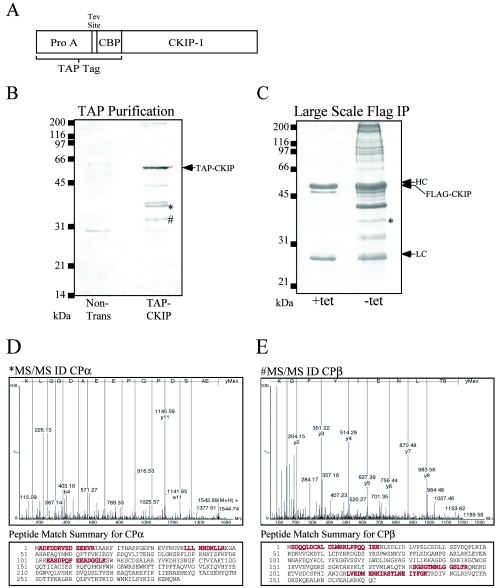
To understand the mechanistic basis for the phenotype associated with Flag-CKIP-1 expression, we utilized TAP (42) and large-scale immunoprecipitations to identify CKIP-1 interaction partners. A schematic illustration of CKIP with an amino-terminal TAP tag composed of protein A domains and a calmodulin-binding peptide is shown in Fig. 4A. Briefly, U2-OS cells (∼108) were either nontransfected or transiently transfected with TAP-CKIP by using the calcium phosphate method. The cells were harvested, and the lysates derived from them were purified on IgG-Sepharose beads, cleaved with Tev protease, and then bound to calmodulin beads in the presence of Ca2+. Following extensive washing, proteins were eluted from the calmodulin beads with boiling sample buffer. For large-scale immunoprecipitations, DC1.4 cells grown in the presence or absence of tetracycline for 24 h were harvested and the lysates were subjected to immunoprecipitation with anti-Flag M2 antibody. For both experiments, CKIP-1 and associated proteins were eluted, separated by SDS-PAGE, and stained with Gel Code Blue. Representative examples of TAP and large-scale immunoprecipitations are shown (Fig. 4B and C, respectively). Bands corresponding to CPα and CPβ were excised and subjected to in-gel trypic digestion in preparation for MS. Importantly, none of these bands were isolated when cells were transfected with TAP constructs lacking the CKIP-1 fusion (data not shown) or in the absence of Flag-CKIP-1, indicating that the presence of these bands is dependent on CKIP-1. The band labeled TAP-CKIP was positively identified using MALDI MS (data not shown). The two remaining bands of approximately 35 and 30 kDa were identified by tandem MS/MS as the heterodimeric actin capping protein subunits CPα and CPβ, respectively (Fig. 4D and E). For each protein, sequence data for one peptide and a peptide match summary highlighting three peptides that were sequenced for each protein are shown. Our results demonstrate that the heterodimeric actin capping protein is a novel CKIP-1-interacting protein. In a similar respect, it is noteworthy that yeast two-hybrid studies to identify CK2-interacting proteins previously led to the identification of CP as a potential interacting partner of protein kinase CK2 (19).
FIG. 4.
Identification of CKIP-1-associated proteins by TAP and large-scale immunoprecipitations. Identification of the heterodimeric actin-capping proteins, CPα and CPβ, by tandem mass spectrometry is shown. (A) Schematic representation of CKIP-1 with an amino-terminal TAP tag that is composed of protein A and a calmodulin binding peptide separated by a TEV cleavage site. (B) U2-OS cells (∼108 cells) were untreated (Non-trans) or transfected with TAP-CKIP-1. The cells were harvested and subjected to tandem affinity purification as described in Materials and Methods. (C) For large-scale immunoprecipitations (IP), DC1.4 cells grown in the presence or absence of tetracycline (tet) for 24 h were harvested, and the lysates were subjected to immunoprecipitation with anti-Flag M2 antibody. For both experiments, CKIP-1 and associated proteins were eluted, separated by SDS-PAGE, and stained with Gel Code Blue. Bands corresponding to CPα and CPβ, marked with * and #, respectively, were excised, processed, and analyzed by tandem MS/MS and MALDI MS (data not shown). (D) Sequence data of one representative peptide and peptide match summary for CPα. (E) Sequence data of one representative peptide and peptide match summary for CPβ. Matched peptides and sequence coverage are shown below in red.
Coimmunoprecipitation and colocalization of CKIP-1 and actin capping protein.
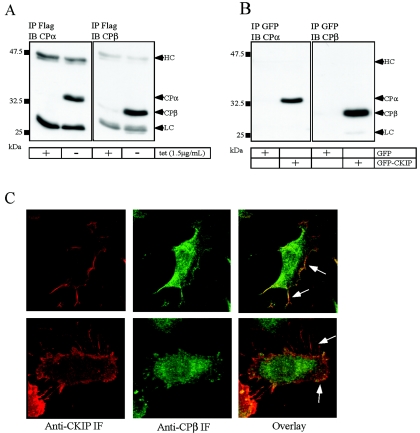
To further confirm interactions between CKIP-1 and actin capping protein, we performed coimmunoprecipitation and colocalization studies. DC1.4 cells grown in the presence or absence of tetracycline for 24 h were harvested, and the resulting lysates were subjected to immunoprecipitation with anti-Flag M2 antibodies. Proteins were separated by SDS-PAGE and immunoblotted with antibodies against CPα and CPβ (Fig. 5A). As expected on the basis of results illustrated in Fig. 4, both CPα and CPβ are found in complexes with Flag-CKIP-1 in the absence of tetracycline. Likewise, GFP-CKIP interacts with capping protein (Fig. 5B). U2-OS cells transiently transfected with GFP or GFP-CKIP-1 were harvested, and the derived lysates were immunoprecipitated with anti-GFP antibodies. Following separation by SDS-PAGE, immunoblotting was performed with antibodies against CPα and CPβ. In agreement with the anti-Flag immunoprecipitations, both CPα and CPβ interact specifically with GFP-CKIP but fail to interact with GFP alone.
FIG. 5.
Confirmation of CPα and CPβ as bona fide CKIP-1-interacting proteins. CKIP-1 interacts with the heterodimeric actin capping proteins as shown by coimmunoprecipitations (A and B) and colocalization (C). (A) DC1.4 cells grown in the presence or absence of tetracycline (tet) for 24 h were harvested, and the lysates were subjected to immunoprecipitation (IP) with anti-Flag M2 antibody. Proteins were separated by SDS-PAGE, transferred to a PVDF membrane, and immunoblotted (IB) with anti-CPα or anti-CPβ antibodies as indicated. (B) U2-0S cells were transfected with GFP or GFP-CKIP-1 as indicated. Extracts were prepared from the transfected cells, and immunoprecipitations were performed with anti-GFP antibodies. Immunoprecipitates were separated by SDS-PAGE and transferred to PVDF membranes for immunoblotting with anti-CPα or anti-CPβ antibodies. The positions of CPα, CPβ, and the heavy and light chains (HC and LC) of IgG are indicated. (C) DC1.4 cells grown in the absence of tetracycline were fixed, permeabilized, and costained with anti-CKIP-1 antibodies and anti-CPβ antibodies. The localization of Flag-CKIP and CPβ are shown separately and overlaid to show the extent of colocalization. Arrows indicate regions of colocalization. IF, immunofluorescence.
To extend these results, we examined whether Flag-CKIP-1 and actin capping protein colocalize in osteosarcoma cells (Fig. 5C). DC1.4 cells grown in the absence of tetracycline for 24 h were fixed, permeabilized, and colabeled with antibodies against CKIP-1 (polyclonal) and CPβ (monoclonal). Following costaining with TRITC- and FITC-labeled secondary antibodies, coverslips were mounted and examined by confocal microscopy. As previously demonstrated, Flag-CKIP-1 is localized predominantly to the plasma membrane, with staining evident in membrane ruffles and filopodia. In U2-OS cells, both CPβ (Fig. 5C) and CPα (data not shown) are localized to the cell periphery and exhibit punctate staining surrounding the nuclei. As shown in the overlay (Fig. 5C), Flag-CKIP-1 and CPβ colocalize extensively at the cell periphery and in filopodia. Collectively, these data suggest that heterodimeric actin capping protein is a bona fide interaction partner of CKIP-1.
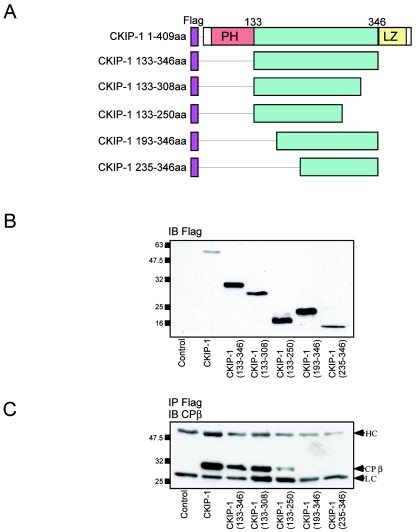
Elucidation of the domains of CKIP-1 required for interactions with CP.
We have previously reported that interactions between CKIP-1 and protein kinase CK2 are mediated by the PH domain of CKIP-1 (40). To examine if the PH domain of CKIP-1 mediates interactions with CP, we generated Flag-tagged deletions of CKIP-1 for expression in mammalian cells. These constructs are shown schematically in Fig. 6A. To verify expression, these constructs were transiently transfected into U2-OS cells and subjected to immunoblotting with anti-Flag M2 antibodies (Fig. 6B). All constructs express to the same degree as or higher than full-length Flag-CKIP-1. To test for interactions with CP, lysates expressing CKIP-1 deletion constructs were subjected to immunoprecipitation with anti-Flag M2 antibodies, separated by SDS-PAGE, and immunoblotted with anti-CPβ antibodies. As shown in Fig. 6C, interaction with CP is strongest with full-length Flag-CKIP-1. However, binding is completely abrogated only with deletions lacking amino acids 133 to 193. Similar results were obtained using GFP-tagged versions of these deletion constructs (data not shown). These data suggest that amino acids 133 to 193 of CKIP-1 are required for interactions with CP. Moreover, this implies that the domains of CKIP-1 required for binding to protein kinase CK2 and CP are distinct.
FIG. 6.
Elucidation of the domains of CKIP-1 required for interaction with CP. CKIP-1 deletions were generated and tested for interaction with CP by immunoprecipitation. (A) Schematic representation of the Flag-tagged CKIP-1 deletions generated. aa, amino acids. (B) Flag-tagged CKIP-1 deletions were transfected into U2-OS cells, and the lysates derived from these cells were separated by SDS-PAGE and transferred to PVDF membranes for immunoblotting (IB) with anti-Flag M2 antibody. (C) CKIP-1 deletions were transfected into U2-OS cells and immunoprecipitated (IP) with anti-Flag M2 antibodies. Immunoprecipitates were separated by SDS-PAGE and transferred to PVDF membranes for immunoblotting with anti-CPβ antibody. Interactions between CKIP-1 and CP require amino acids 133 to 193. HC and LC, IgG heavy and light chains, respectively.
Phosphorylation of tandem affinity-purified CPα by protein kinase CK2.
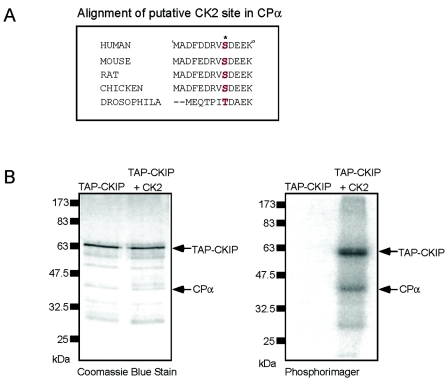
Based on the data demonstrating that protein kinase CK2 and CP appear to bind distinct regions of CKIP-1, we tested the hypothesis that CKIP-1 may function as an adaptor that targets CK2 to particular cellular compartments to facilitate the phosphorylation of proteins isolated in CKIP-1 complexes. As shown in Fig. 7A, CPα contains a highly conserved residue within its amino-terminal domain that matches the consensus sequence [(S/T)XX(D/E/Sp/Yp)] for phosphorylation by protein kinase CK2. This residue (S9) is completely conserved, with the exception of a conservative change to threonine in Drosophila melanogaster. To examine if CPα is a substrate of protein kinase CK2, we employed TAP tagging to isolate CKIP-1 and its interaction partners (Fig. 7B, left panel). Following binding to calmodulin beads, CKIP-1 complexes were phosphorylated by recombinant protein kinase CK2 in the presence of Ca2+ and [γ-32P]ATP. The calmodulin beads were washed, and the bound proteins were eluted and visualized using a PhosphorImager (Fig. 7B, right panel). As shown, three prominent bands are visible following phosphorylation by protein kinase CK2. Based on comigration with the bands stained with Coomassic Blue, we have identified two of these proteins as TAP-CKIP and CPα. At present, we do not know the identity of the third CK2 target. These data show that CPα is a target of protein kinase CK2 in vitro.
FIG. 7.
Phosphorylation of tandem affinity-purified CPα by protein kinase CK2. (A) Alignment of a conserved protein kinase CK2 consensus site in the amino terminus of CPα. The residue serine 9 is conserved in these species, with the exception of a conservation change to a threonine in D. melanogaster. (B) CKIP-1 complexes were isolated by TAP purification as in the experiment in Fig. 4. Following binding to calmodulin, beads were left untreated or phosphorylated by recombinant protein kinase CK2 in the presence of [γ-32P]ATP. The calmodulin beads were washed, and bound proteins were eluted and separated by SDS-PAGE. The gel was stained with Gel Code Blue to visualize all proteins (left panel) and dried and exposed in a PhosphorImager to visualize radiolabeled proteins (right panel). The bands identified as TAP-CKIP-1 and CPα are indicated.
Phosphorylation of CPα by protein kinase CK2 in vitro and in cells.
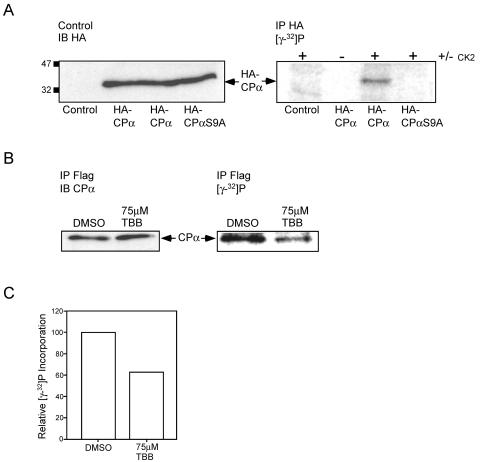
To extend these results, we generated constructs encoding HA-tagged CPα and Myc-tagged CPβ for expression of CP in mammalian cells. Additionally, the putative protein kinase CK2 phosphorylation site serine 9 was mutated to alanine in order to generate HA-CPαS9A. Importantly, we verified that HA-CPα and Myc-CPβ coimmunoprecipitate with each other and with Flag-CKIP-1 in U2-OS cell lysates (data not shown). Next, To examine if protein kinase CK2 phosphorylates CPα on Ser9 in vitro, U2-OS cells were transfected with vector alone, HA-CPα/Myc-CPβ, or HA-CPαS9A/Myc-CPβ. The lysates derived from these cells were analyzed by immunoblotting with biotinylated anti-HA antibodies to demonstrate equal loading of wild-type and mutant CPα (Fig. 8A, left panel). The derived lysates were normalized for protein concentration and subjected to immunoprecipitation with anti-HA antibodies. Bound proteins were phosphorylated with protein kinase CK2 on the protein A-Sepharose beads in the presence of [γ-32P]ATP, separated by SDS-PAGE, and visualized using a PhosphorImager (Fig. 8A, right panel). As shown, protein kinase CK2 is able to phosphorylate wild-type HA-CPα but not the mutant HA-CPαS9A. These data suggest that in vitro protein kinase CK2 phosphorylates CPα on Ser9.
FIG. 8.
Phosphorylation of CPα by protein kinase CK2 in vitro and in cells. (A) Protein kinase CK2 phosphorylates CPα on serine 9 in vitro. HA-CPα, HA-CPαS9A, and empty vector were transiently transfected into U2-OS cells. (Left panel) Lysates derived from these cells were immunoblotted (IB) with biotinylated anti-HA antibodies to demonstrate equal loading. (Right panel) The lysates were subjected to immunoprecipitation (IP) with anti-HA antibodies. Bound proteins were phosphorylated with recombinant protein kinase CK2 on the protein A-Sepharose beads in the presence of [γ-32P]ATP, separated by SDS-PAGE, and visualized using a PhosphorImager. The position of HA-CPα is indicated. (B and C) Endogenous CPα is phosphorylated in cells, and treatment with a CK2 specific inhibitor results in a decrease in phosphate incorporation. (B) DC1.4 cells expressing Flag-CKIP-1 were grown in the presence of 75 μM TBB or carrier alone for 18 h and labeled with 0.33 mCi of [γ-32P]orthophosphate for 6 h. Duplicate plates were treated identically but in the absence of 0.33 mCi of [γ-32P]orthophosphate. Lysates derived from these cells were normalized for protein concentration, and subjected to immunoprecipitation with anti-Flag M2 antibodies. (Left panel) Immunoprecipitates from nonlabeled plates were transferred to PVDF membranes and immunoblotted with anti-CPα antibodies. (Right panel) Immunoprecipitates from labeled plates were separated by SDS-PAGE, dried, and exposed in a PhosphorImager to visualize labeled proteins. The position of CPα is indicated. (C) Data in panel B were analyzed using ImageQuant software and expressed as relative phosphate incorporation.
We next examined if CPα found in complexes with Flag-CKIP-1 is phosphorylated in mammalian cells and if this phosphorylation can be modulated using the CK2-specific inhibitor, TBB. To achieve this objective, DC1.4 cells expressing Flag-CKIP-1 were grown with carrier (DMSO) or 75 μM TBB by using established procedures (28, 45, 47, 50, 54). Subsequently, cells were labeled for 6 h with 0.33 mCi of [γ-32P]orthophosphate in the presence of carrier or TBB. Lysates derived from labeled and nonlabeled control plates were normalized for protein concentration and subjected to immunoprecipitation with anti-Flag M2 antibodies. Immunoprecipitates from nonlabeled plates were transferred to polyvinylidene difluoride membranes and immunoblotted with anti-CPα antibodies (Fig. 8B, left panel). Importantly, treatment with 75 μM TBB had no effect on endogenous CPα protein levels. Immunoprecipitates from labeled plates were separated by SDS-PAGE, dried, and exposed in a PhosphorImager to visualize labeled proteins (Fig. 8B, right panel). As shown, treatment of DC1.4 cells with TBB results in a decrease in phosphorylation of CPα suggesting that protein kinase CK2 may phosphorylate CPα in vivo. To quantitate CPα phosphorylation, the data in Fig. 8B were analyzed using Image Quant software and expressed as relative phosphate incorporation (Fig. 8C). Treatment with 75 μM TBB results in a ∼40% decrease in CPα phosphate incorporation. At present, we do not know whether the failure of TBB to completely abolish the phosphorylation of CPα results from incomplete inhibition of CK2 or whether other kinases contribute to the phosphorylation of CPα in cells. Nevertheless, taken together, these data suggest that CPα is phosphorylated by protein kinase CK2 on Ser9 in vitro, CPα is phosphorylated in mammalian cells, and protein kinase CK2 can phosphorylate CPα in vivo.
Effects of CKIP-1 on the actin polymerization, depolymerization, and uncapping activity of CP.
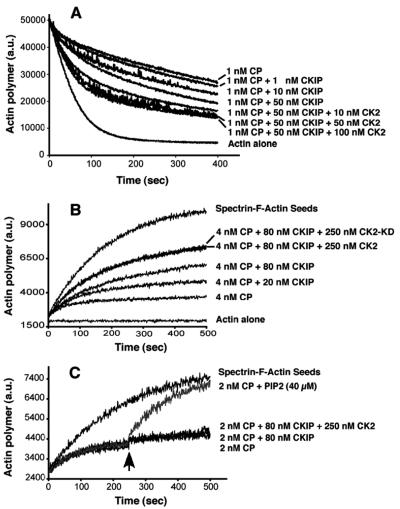
We used an actin depolymerization assay to determine whether interaction with CKIP-1 altered the capping activity of CP (Fig. 9A). CP inhibits the depolymerization of F-actin in a dose-dependent manner on dilution (57). When increasing amounts of CKIP-1 (1, 10, and 50 nM) were added to reaction mixtures with 1 nM CP, the rate of F-actin depolymerization increased (Fig. 9A). Addition of CKIP-1 at concentrations above 50 nM had no further effect (data not shown). Since protein kinase CK2 directly interacts with CKIP-1, we also asked whether CK2 might exert an additive effect on the activity of CP. In the presence of 10 to 100 nM protein kinase CK2 and 50 nM CKIP-1, the rate of actin polymerization was greater than with CKIP-1 alone (Fig. 9A). In a control experiment, 50 nM recombinant protein kinase CK2α alone with CP had no effect (data not shown).
FIG. 9.
CKIP-1 inhibits the capping activity of CP at the barbed ends of actin filaments. (A) Actin depolymerization assay. CP (1 nM) was incubated with various amounts of CKIP and CK2 for 5 min before being added to 5 μM F-actin. Pyrene-actin fluorescence is plotted against time after dilution. The concentration of CKIP and CK2 incubated with CP is indicated beside each curve. (B) Actin polymerization assay. Actin polymerization (1 μM actin) seeded by the addition of SAS was monitored by the increase in pyrene fluorescence, with CP, CKIP, protein kinase CK2α, and CK2-α-kd as indicated. (C) Uncapping assay. CKIP-1 and protein kinase CK2 do not affect the dissociation of CP from actin filaments. Polymerization of 1.5 μM actin at barbed ends was nucleated by SAS in the presence of CP. After 250 s, PIP2, CKIP-1, or protein kinase CK2α was added to reaction mixtures containing 2 nM CP.
To further analyze the effects of CKIP-1 and protein kinase CK2 on actin filament capping, actin polymerization growth assays were performed (Fig. 9B). Actin polymerization from barbed ends was initiated by SAS. Addition of 4 nM CP decreased the rate of actin polymerization. In agreement with the depolymerization assays described above, CKIP-1 (80 nM) partially inhibited capping activity. When we added protein kinase CK2α (250 nM) and CKIP-1 (80 nM) to the reaction mixture, the capping activity was further decreased. Reaction mixtures for both the depolymerization and polymerization assays contain ATP (0.2 mM), so we asked whether protein kinase CK2 phosphorylation would affect capping activity. A kinase-dead mutant of CK2α (CK2-KD) was tested in the polymerization assay (Fig. 9B) and found to have a similar effect to that of wild-type CK2. Thus, phosphorylation by protein kinase CK2 does not contribute to the effect.
Finally, we asked if CKIP-1 and protein kinase CK2 might promote the uncapping of CP from acting filaments (Fig. 9C). In a growth assay, CKIP-1 at 80 nM with or without protein kinase CK2 (250 nM) was added to a polymerization growth reaction mixture containing CP at a time when most of the actin filaments were capped (arrow at 250 s in Fig. 9C). They had no observed effect, while addition of PIP2 as a positive control induced a large increase in the rate of actin polymerization.
DISCUSSION
Functional investigation of CKIP-1.
Despite its discovery nearly 50 years ago, many details regarding the precise regulation and functions of protein kinase CK2 remain poorly defined. To test the hypothesis that interacting proteins participate in the regulation of protein kinase CK2, we previously used the yeast two-hybrid system to identify CK2-interacting proteins. These studies led to the identification of a novel CK2-interacting protein designated CKIP-1 (3). To examine the functions of this novel protein, we generated human osteosarcoma cell lines with tetracycline-regulated expression of Flag-CKIP-1. We observed that cells expressing Flag-CKIP-1 underwent a consistent morphological change that was quantitatively documented by morphometric analysis. Furthermore, we demonstrated that Flag-CKIP-1 expression results in an increase in F-actin staining in cells and an increase in cellular protein levels of β-actin. To elucidate the mechanisms behind the observed phenotype, we utilized tandem affinity purification and large-scale immunoprecipitations and identified proteins that form complexes with CKIP-1. These studies led to the identification of the α/β heterodimeric actin capping protein interacting with CKIP-1.
Recently, we reported that the PH domain of CKIP-1 is required for both localization to the plasma membrane and interactions with protein kinase CK2 (40). Here we demonstrate that CP binds to a region of CKIP-1 (amino acids 133 to 193) distinct from the PH domain, suggesting that CKIP-1 could mediate interactions between protein kinase CK2 and CP concurrently. Therefore, we examined if CKIP-1 could serve as an adaptor to facilitate the phosphorylation of CP. Our studies indicate that CPα is phosphorylated by protein kinase CK2 in vitro on Ser9. Moreover, we demonstrated that CPα is phosphorylated in osteosarcoma cells and that treatment with the specific protein kinase CK2 inhibitor TBB results in a reduction in phosphate incorporation in CPα. The use of TBB to specifically inhibit protein kinase CK2 has been reported extensively in the literature (28, 45, 47, 50, 54). Interestingly, treatment with TBB was not sufficient to completely abolish CPα phosphorylation, raising the possibility that CP is modified by other kinases in addition to protein kinase CK2. Taken together, these data suggest that one role of CKIP-1 may be to regulate the ability of CK2 to phosphorylate one of its targets.
We also examined the effect of CKIP-1 and protein kinase CK2 on the activity of CP in vitro. We observed that CKIP-1 can partially inhibit capping activity as measured in polymerization and depolymerization assays. This inhibition of CP activity was increased by the addition of protein kinase CK2. Interestingly, this effect was also seen when we used a kinase-dead mutant of protein kinase CK2. This suggests that phosphorylation of CP by protein kinase CK2 does not affect the activity of CP in vitro. In light of the study by Falck et al. (16) demonstrating that Ser9 is in the stalk domain of CP—a region that does not mediate actin binding—this result is not entirely surprising. However, it is possible that phosphorylation by protein kinase CK2 affects CP activity or mediates interaction with CP binding partners in cells. To this end, it is intriguing that the stalk region of CP containing Ser9 has been implicated in interactions between CP and the actin monomer binding protein twinfilin (16). More experimentation is required to examine if CP phosphorylation by protein kinase CK2 affects the interactions of CP with twinfilin. Finally, we found that CKIP-1 and protein kinase CK2 did not promote uncapping of CP from actin filaments.
Role of the actin capping proteins in actin filament assembly.
During actin polymerization in nonmuscle cells, actin monomers are added to the barbed (fast-growing) ends of the filament. Eventually, the fast-growing barbed ends are capped by the actin capping protein to stop filament growth in a Ca2+-independent (5, 6, 35, 36) and PIP2-dependent (2, 13, 20, 21) manner. These actin capping proteins are a family of heterodimeric proteins composed of α and β subunits of ∼35 and ∼30 kDa, respectively (9, 56). Despite having no sequence homology, the two subunits have very similar structures, giving a pseudo-twofold axis of symmetry (58). The proposed function for actin capping in the dendritic nucleation model (41) states that most barbed ends are capped so that polymerization is limited to a small set of free barbed ends that extend quickly. This model was supported by the observation that depleting Dictyostelium discoideum of capping protein caused an increase in the level of F-actin but a decrease in motility (22). Similarly, in vitro studies with Listeria monocytogenes and purified proteins have shown that when capping protein is removed, the motility of the bacterium is completely inhibited (32). Likewise, depletion of CP by shRNA in B16F1 mouse melanoma cells resulted in a large increase in filopodia, a marked decrease in lamellipodia and increased numbers of actin filaments throughout the cytoplasm (38). Finally, it was recently demonstrated that expression of CP mutants in Saccharomyces cerevisiae that fail to interact with actin led to an increase in the amount of F-actin (27). Collectively, these studies demonstrate that interference with capping protein leads to stabilization of actin filaments.
Emerging role for protein kinase CK2 in the actin cytoskeleton.
There is mounting evidence to suggest that protein kinase CK2 is involved in regulating cellular morphology and cytoskeletal events in a variety of organisms. For example, studies with S. cerevisiae have demonstrated that interfering with one CK2 isozyme, but not the other, causes loss of cell polarity and abnormal chitin deposits in the cell membrane (43). In a related vein, protein kinase CK2 orb5 mutants of Schizosaccharomyces pombe demonstrated a role for the kinase in maintenance of cell polarity and in polarized cell growth (52). Recently, it was shown that genes encoding γ-tubulin and protein kinase CK2 were able to suppress the growth defect associated with a temperature-sensitive mutant of Arc35p, a member of the Arp2/3 complex in S. cerevisiae (48). In this study, the authors demonstrated a biochemical association between CK2 and Arc35p and proposed that this interaction may play a role in the activation of γ-tubulin via calmodulin. Additionally, it was shown that the actin capping protein toxofilin in Toxoplasma gondii is regulated by phosphorylation through the actions of a CK2-like activity and a novel parasitic type 2C phosphatase (12). In these studies, CK2 phosphorylation of toxofilin (or constructs expressing phospho-mimics) caused actin disassembly in fibroblasts infected with T. gondii. There are also indications that in mammalian cells CK2 plays a role in aspects of cytoskeletal regulation. In fact, CK2 has been shown to play a role in the formation of cell-cell adherens junctions (30) and actin isolated from muscle preparations has been shown to inhibit CK2 kinase activity in vitro (24). Finally, a recent study demonstrated that the VCA domain of WASP is phosphorylated by protein kinase CK2 in vitro and in cells (10). Phosphorylation by CK2 was shown to enhance the interaction of WASP with the Arp2/3 complex and lead to increased actin polymerization. Collectively, these studies show that protein kinase CK2 plays a clear, albeit still poorly understood, role in regulation of the actin cytoskeleton.
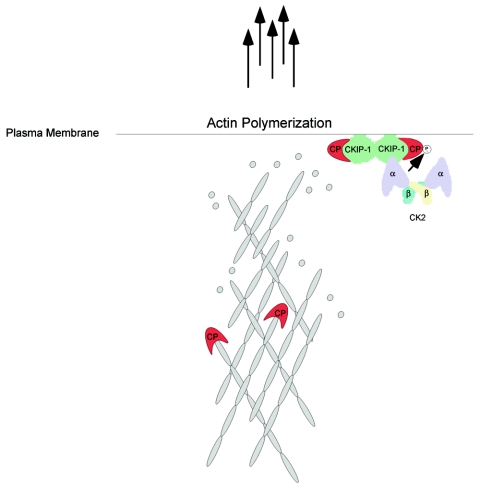
Model for the role of CKIP-1-CK2 in cytoskeletal regulation.
The data presented in this study demonstrates that CKIP-1 plays a role in the regulation of the actin cytoskeleton, interacts with the actin capping protein, and may serve as an adaptor to facilitate the phosphorylation of CP by protein kinase CK2. These results strengthen the conclusion that protein kinase CK2 is involved in regulating the actin cytoskeleton and cellular morphology. Moreover, the data demonstrating that CPα is phosphorylated by protein kinase CK2 provides an additional link between CK2 and its role in the regulation of the actin cytoskeleton. Based on these studies, we propose the hypothetical model shown in Fig. 10. We propose that when Flag-CKIP-1 is expressed, interactions between CP and CKIP-1 interfere with the binding of CP to the barbed ends of the actin filament. Under these conditions, actin polymerization at the plasma membrane would be greatly enhanced. In support of our model, we have shown that CKIP-1 and protein kinase CK2 can inhibit the activity of CP in vitro. However, there are still many questions. For instance, it is not known how interactions between CKIP-1 and the actin capping proteins lead to the increased level of F-actin and changes in morphology associated with CKIP-1 expression. However, it is worth noting that studies of D. discoideum and L. monocytogenes demonstrate that interference with capping protein leads to stabilization of actin filaments and an increase in the level of F-actin. Recently it was shown that depletion of CP by shRNA in B16F1 mouse melanoma cells resulted in a large increase in the numbers of filopodia, a marked decrease in the number of lamellipodia, and increased numbers of actin filaments throughout the cytoplasm (38). To this end, we have shown that Flag-CKIP-1 expression results in increased numbers of actin filaments and that CKIP-1 and CP colocalize at the plasma membrane and in filopodia. It is also intriguing that CKIP-1 binds to phospholipids in vivo and PIP2 in vitro (40)—a substrate known to inhibit the activity of the actin capping proteins (2, 13, 20, 21). Similarly, we do not known what function CPα phosphorylation by protein kinase CK2 might serve. More experimentation is needed to answer these questions and to determine precisely to what extent the observed effects of CKIP-1 are influenced by CK2 or to what extent functional effects of CKIP-1 are CK2 independent.
FIG. 10.
Hypothetical model illustrating the mechanistic basis for effects of CKIP-1 on cell morphology and the actin cytoskeleton. Interactions between CP and CKIP-1 at the plasma membrane inhibit the binding of CP to the barbed ends of the actin filament, leading to increased actin polymerization at the plasma membrane. As discussed in the text, CK2 phosphorylates CPα and increases the inhibitory activity of CKIP-1 toward CP in a phosphorylation-independent manner. Actin monomers are shown as grey circles. CP is shown as a monomer for simplicity. CKIP-1 is shown as a dimer, but its exact oligomeric structure is unknown.
CKIP-1: a new paradigm for regulation of CK2 and the actin cytoskeleton in cells.
An emerging model in the signaling field is the importance of the clustering and targeting of signaling molecules. One of the best examples of this is the regulation of cAMP-dependent protein kinase by the AKAP family of proteins. Clearly, there are some parallels that can be drawn between PKA-AKAP interactions and the interactions between CK2 and CKIP-1. Like PKA, protein kinase CK2 has multiple substrates localized within a variety of cellular locations. Based on these similarities, we hypothesized that CKIP-1 is a regulator that functions to target CK2 to particular cellular localizations in an analogous way to the targeting of PKA by the AKAP family of proteins. Here we provide evidence that CKIP-1 may serve to target CK2 to the plasma membrane and facilitate phosphorylation of the actin capping proteins by protein kinase CK2. This illustrates an exciting new paradigm demonstrating how phosphorylation of one specific target by CK2, a kinase proposed to have hundreds of potential cellular targets, may be regulated. In a similar respect, there are several other examples illustrating how the phosphorylation of particular CK2 subunits is regulated. These include interactions between protein kinase CK2 and Fas-associated factor FAF-1 (39), TATA binding protein (18), and the nuclear matrix (55). The present study has strengthened the evidence that protein kinase CK2 plays a role in the regulation of the actin cytoskeleton and cell morphology. Collectively, these results offer new insights regarding the regulation of one function for a protein kinase that is involved in a complex series of cellular processes.
Acknowledgments
This work was supported by operating grants from the Canadian Institutes of Health Research (MOP 37854) to D.W.L. and the National Institutes of Health (GM 38542) to J.A.C. D.A.C. is supported by a Canadian Graduate Scholarship from the Canadian Institutes of Health Research. K.K. is supported by a postdoctoral fellowship from the American Heart Association. The Biological Mass Spectrometry laboratory at the University of Western Ontario is supported by a grant from the Ontario Research and Development Challenge Fund (ORDCF).
Confocal microscopy was performed at the Imaging Center for the Faculty of Medicine and Dentistry at the University of Western Ontario. We are grateful to Eric Ball, Paul Walton, and Cunjie Zhang for helpful discussions during the course of this work, to James Duncan for generating the scrambled shRNA construct, and to Victoria Wraw for technical assistance.
REFERENCES
- 1.Alto, N., J. J. Carlisle Michel, K. L. Dodge, L. K. Langeberg, and J. D. Scott. 2002. Intracellular targeting of protein kinases and phosphatases. Diabetes 51(Suppl. 3):S385-S388. [DOI] [PubMed] [Google Scholar]
- 2.Barkalow, K., W. Witke, D. J. Kwiatkowski, and J. H. Hartwig. 1996. Coordinated regulation of platelet actin filament barbed ends by gelsolin and capping protein. J. Cell Biol. 134:389-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosc, D. G., K. C. Graham, R. B. Saulnier, C. Zhang, D. Prober, R. D. Gietz, and D. W. Litchfield. 2000. Identification and characterization of CKIP-1, a novel pleckstrin homology domain-containing protein that interacts with protein kinase CK2. J. Biol. Chem. 275:14295-14306. [DOI] [PubMed] [Google Scholar]
- 4.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 5.Caldwell, J. E., S. G. Heiss, V. Mermall, and J. A. Cooper. 1989. Effects of CapZ, an actin capping protein of muscle, on the polymerization of actin. Biochemistry 28:8506-8514. [DOI] [PubMed] [Google Scholar]
- 6.Casella, J. F., D. J. Maack, and S. Lin. 1986. Purification and initial characterization of a protein from skeletal muscle that caps the barbed ends of actin filaments. J. Biol. Chem. 261:10915-10921. [PubMed] [Google Scholar]
- 7.Chen-Wu, J. L., R. Padmanabha, and C. V. Glover. 1988. Isolation, sequencing, and disruption of the CKA1 gene encoding the alpha subunit of yeast casein kinase II. Mol. Cell. Biol. 8:4981-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colledge, M., R. A. Dean, G. K. Scott, L. K. Langeberg, R. L. Huganir, and J. D. Scott. 2000. Targeting of PKA to glutamate receptors through a MAGUK-AKAP complex. Neuron 27:107-119. [DOI] [PubMed] [Google Scholar]
- 9.Cooper, J. A., and D. A. Schafer. 2000. Control of actin assembly and disassembly at filament ends. Curr. Opin. Cell Biol. 12:97-103. [DOI] [PubMed] [Google Scholar]
- 10.Cory, G. O., R. Cramer, L. Blanchoin, and A. J. Ridley. 2003. Phosphorylation of the WASP-VCA domain increases its affinity for the Arp2/3 complex and enhances actin polymerization by WASP. Mol. Cell 11:1229-1239. [DOI] [PubMed] [Google Scholar]
- 11.Daya-Makin, M., J. S. Sanghera, T. L. Mogentale, M. Lipp, J. Parchomchuk, J. C. Hogg, and S. L. Pelech. 1994. Activation of a tumor-associated protein kinase (p40TAK) and casein kinase 2 in human squamous cell carcinomas and adenocarcinomas of the lung. Cancer Res 54:2262-2268. [PubMed] [Google Scholar]
- 12.Delorme, V., X. Cayla, G. Faure, A. Garcia, and I. Tardieux. 2003. Actin dynamics is controlled by a casein kinase II and phosphatase 2C interplay on Toxoplasma gondii toxofilin. Mol. Biol. Cell 14:1900-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiNubile, M. J., and S. Huang. 1997. High concentrations of phosphatidylinositol-4,5-bisphosphate may promote actin filament growth by three potential mechanisms: inhibiting capping by neutrophil lysates, severing actin filaments and removing capping protein-beta2 from barbed ends. Biochim. Biophys. Acta 1358:261-278. [DOI] [PubMed] [Google Scholar]
- 14.Dodge, K., and J. D. Scott. 2000. AKAP79 and the evolution of the AKAP model. FEBS Lett. 476:58-61. [DOI] [PubMed] [Google Scholar]
- 15.Englert, C., X. Hou, S. Maheswaran, P. Bennett, C. Ngwu, G. G. Re, A. J. Garvin, M. R. Rosner, and D. A. Haber. 1995. WT1 suppresses synthesis of the epidermal growth factor receptor and induces apoptosis. EMBO J. 14:4662-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falck, S., V. O. Paavilainen, M. A. Wear, J. G. Grossmann, J. A. Cooper, and P. Lappalainen. 2004. Biological role and structural mechanism of twinfilin-capping protein interaction. EMBO J. 23:3010-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faust, R. A., M. Gapany, P. Tristani, A. Davis, G. L. Adams, and K. Ahmed. 1996. Elevated protein kinase CK2 activity in chromatin of head and neck tumors: association with malignant transformation. Cancer Lett. 101:31-35. [DOI] [PubMed] [Google Scholar]
- 18.Ghavidel, A., and M. C. Schultz. 2001. TATA binding protein-associated CK2 transduces DNA damage signals to the RNA polymerase III transcriptional machinery. Cell 106:575-584. [DOI] [PubMed] [Google Scholar]
- 19.Grein, S., K. Raymond, C. Cochet, W. Pyerin, E. M. Chambaz, and O. Filhol. 1999. Searching interaction partners of protein kinase CK2beta subunit by two-hybrid screening. Mol. Cell. Biochem. 191:105-109. [PubMed] [Google Scholar]
- 20.Haus, U., H. Hartmann, P. Trommler, A. A. Noegel, and M. Schleicher. 1991. F-actin capping by cap32/34 requires heterodimeric conformation and can be inhibited with PIP2. Biochem. Biophys. Res. Commun. 181:833-839. [DOI] [PubMed] [Google Scholar]
- 21.Heiss, S. G., and J. A. Cooper. 1991. Regulation of CapZ, an actin capping protein of chicken muscle, by anionic phospholipids. Biochemistry 30:8753-8758. [DOI] [PubMed] [Google Scholar]
- 22.Hug, C., P. Y. Jay, I. Reddy, J. G. McNally, P. C. Bridgman, E. L. Elson, and J. A. Cooper. 1995. Capping protein levels influence actin assembly and cell motility in dictyostelium. Cell 81:591-600. [DOI] [PubMed] [Google Scholar]
- 23.Hunter, T. 2000. Signaling—2000 and beyond. Cell 100:113-127. [DOI] [PubMed] [Google Scholar]
- 24.Karino, A., S. Tanoue, M. Fukuda, T. Nakamura, and K. Ohtsuki. 1996. An inhibitory effect of actin on casein kinase II activity in vitro. FEBS Lett. 398:317-321. [DOI] [PubMed] [Google Scholar]
- 25.Kelliher, M. A., D. C. Seldin, and P. Leder. 1996. Tal-1 induces T cell acute lymphoblastic leukemia accelerated by casein kinase IIalpha. EMBO J. 15:5160-5166. [PMC free article] [PubMed] [Google Scholar]
- 26.Kikkawa, U., S. K. Mann, R. A. Firtel, and T. Hunter. 1992. Molecular cloning of casein kinase II alpha subunit from Dictyostelium discoideum and its expression in the life cycle. Mol. Cell. Biol. 12:5711-5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, K., A. Yamashita, M. A. Wear, Y. Maeda, and J. A. Cooper. 2004. Capping protein binding to actin in yeast: biochemical mechanism and physiological relevance. J. Cell Biol. 164:567-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulartz, M., E. Hiller, F. Kappes, L. A. Pinna, and R. Knippers. 2004. Protein kinase CK2 phosphorylates the cell cycle regulatory protein Geminin. Biochem. Biophys. Res. Commun. 315:1011-1017. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Lickert, H., A. Bauer, R. Kemler, and J. Stappert. 2000. Casein kinase II phosphorylation of E-cadherin increases E-cadherin/beta-catenin interaction and strengthens cell-cell adhesion. J. Biol. Chem. 275:5090-5095. [DOI] [PubMed] [Google Scholar]
- 31.Litchfield, D. W. 2003. Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem. J. 369:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loisel, T. P., R. Boujemaa, D. Pantaloni, and M. F. Carlier. 1999. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature 401:613-616. [DOI] [PubMed] [Google Scholar]
- 33.Machesky, L. M., and A. Hall. 1997. Role of actin polymerization and adhesion to extracellular matrix in Rac- and Rho-induced cytoskeletal reorganization. J. Cell Biol. 138:913-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manning, G., G. D. Plowman, T. Hunter, and S. Sudarsanam. 2002. Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 27:514-520. [DOI] [PubMed] [Google Scholar]
- 35.Maruyama, K., S. Kimura, T. Ishi, M. Kuroda, and K. Ohashi. 1977. beta-Actinin, a regulatory protein of muscle. Purification, characterization and function. J. Biochem. 81:215-232. [DOI] [PubMed] [Google Scholar]
- 36.Maruyama, K., H. Kurokawa, M. Oosawa, S. Shimaoka, H. Yamamoto, and M. Ito. 1990. Beta-actinin is equivalent to Cap Z protein. J. Biol. Chem. 265:8712-8715. [PubMed] [Google Scholar]
- 37.Meggio, F., and L. A. Pinna. 2003. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 17:349-368. [DOI] [PubMed] [Google Scholar]
- 38.Mejillano, M. R., S. Kojima, D. A. Applewhite, F. B. Gertler, T. M. Svitkina, and G. G. Borisy. 2004. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell 118:363-373. [DOI] [PubMed] [Google Scholar]
- 39.Olsen, B. B., V. Jessen, P. Hojrup, O. G. Issinger, and B. Boldyreff. 2003. Protein kinase CK2 phosphorylates the Fas-associated factor FAF1 in vivo and influences its transport into the nucleus. FEBS Lett. 546:218-222. [DOI] [PubMed] [Google Scholar]
- 40.Olsten, M. E., D. A. Canton, C. Zhang, P. A. Walton, and D. W. Litchfield. 2004. The Pleckstrin homology domain of CK2 interacting protein-1 is required for interactions and recruitment of protein kinase CK2 to the plasma membrane. J. Biol. Chem. 279:42114-42127. [DOI] [PubMed] [Google Scholar]
- 41.Pollard, T. D., and G. G. Borisy. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell 112:453-465. [DOI] [PubMed] [Google Scholar]
- 42.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 43.Rethinaswamy, A., M. J. Birnbaum, and C. V. Glover. 1998. Temperature-sensitive mutations of the CKA1 gene reveal a role for casein kinase II in maintenance of cell polarity in Saccharomyces cerevisiae. J. Biol. Chem. 273:5869-5877. [DOI] [PubMed] [Google Scholar]
- 44.Rubin, C. S. 1994. A kinase anchor proteins and the intracellular targeting of signals carried by cyclic AMP. Biochim. Biophys. Acta 1224:467-479. [PubMed] [Google Scholar]
- 45.Ruzzene, M., D. Penzo, and L. A. Pinna. 2002. Protein kinase CK2 inhibitor 4,5,6,7-tetrabromobenzotriazole (TBB) induces apoptosis and caspase-dependent degradation of haematopoietic lineage cell-specific protein 1 (HS1) in Jurkat cells. Biochem. J. 364:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Safi, A., M. Vandromme, S. Caussanel, L. Valdacci, D. Baas, M. Vidal, G. Brun, L. Schaeffer, and E. Goillot. 2004. Role for the pleckstrin homology domain-containing protein CKIP-1 in phosphatidylinositol 3-kinase-regulated muscle differentiation. Mol. Cell. Biol. 24:1245-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarno, S., H. Reddy, F. Meggio, M. Ruzzene, S. P. Davies, A. Donella-Deana, D. Shugar, and L. A. Pinna. 2001. Selectivity of 4,5,6,7-tetrabromobenzotriazole, an ATP site-directed inhibitor of protein kinase CK2 (‘casein kinase-2’). FEBS Lett. 496:44-48. [DOI] [PubMed] [Google Scholar]
- 48.Schaerer-Brodbeck, C., and H. Riezman. 2003. Genetic and biochemical interactions between the Arp2/3 complex, Cmd1p, casein kinase II, and Tub4p in yeast. FEMS Yeast Res. 4:37-49. [DOI] [PubMed] [Google Scholar]
- 49.Schafer, D. A., P. B. Jennings, and J. A. Cooper. 1996. Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J. Cell Biol. 135:169-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz, E. I., R. V. Intine, and R. J. Maraia. 2004. CK2 is responsible for phosphorylation of human La protein serine-366 and can modulate rpL37 5′-terminal oligopyrimidine mRNA metabolism. Mol. Cell. Biol. 24:9580-9591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seldin, D. C., and P. Leder. 1995. Casein kinase II alpha transgene-induced murine lymphoma: relation to theileriosis in cattle. Science 267:894-897. [DOI] [PubMed] [Google Scholar]
- 52.Snell, V., and P. Nurse. 1994. Genetic analysis of cell morphogenesis in fission yeast—a role for casein kinase II in the establishment of polarized growth. EMBO J. 13:2066-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stalter, G., S. Siemer, E. Becht, M. Ziegler, K. Remberger, and O. G. Issinger. 1994. Asymmetric expression of protein kinase CK2 subunits in human kidney tumors. Biochem. Biophys. Res. Commun. 202:141-147. [DOI] [PubMed] [Google Scholar]
- 54.Szyszka, R., N. Grankowski, K. Felczak, and D. Shugar. 1995. Halogenated benzimidazoles and benzotriazoles as selective inhibitors of protein kinases CK I and CK II from Saccharomyces cerevisiae and other sources. Biochem. Biophys. Res. Commun. 208:418-424. [DOI] [PubMed] [Google Scholar]
- 55.Wang, H., S. Yu, A. T. Davis, and K. Ahmed. 2003. Cell cycle dependent regulation of protein kinase CK2 signaling to the nuclear matrix. J. Cell. Biochem. 88:812-822. [DOI] [PubMed] [Google Scholar]
- 56.Wear, M. A., D. A. Schafer, and J. A. Cooper. 2000. Actin dynamics: assembly and disassembly of actin networks. Curr. Biol. 10:R891-R895. [DOI] [PubMed] [Google Scholar]
- 57.Wear, M. A., A. Yamashita, K. Kim, Y. Maeda, and J. A. Cooper. 2003. How capping protein binds the barbed end of the actin filament. Curr. Biol. 13:1531-1537. [DOI] [PubMed] [Google Scholar]
- 58.Yamashita, A., K. Maeda, and Y. Maeda. 2003. Crystal structure of CapZ: structural basis for actin filament barbed end capping. EMBO J. 22:1529-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yenice, S., A. T. Davis, S. A. Goueli, A. Akdas, C. Limas, and K. Ahmed. 1994. Nuclear casein kinase 2 (CK-2) activity in human normal, benign hyperplastic, and cancerous prostate. Prostate 24:11-16. [DOI] [PubMed] [Google Scholar]
- 60.Yoon, H. J., A. Feoktistova, B. A. Wolfe, J. L. Jennings, A. J. Link, and K. L. Gould. 2002. Proteomics analysis identifies new components of the fission and budding yeast anaphase-promoting complexes. Curr. Biol. 12:2048-2054. [DOI] [PubMed] [Google Scholar]