Abstract
DREAM (downstream regulatory element antagonist modulator) is a transcriptional repressor, which binds DREs (downstream response elements) in a Ca2+-regulated manner. The DREs consist of core GTCA motifs, very similar to binding motifs for non-steroid nuclear receptors. In this work, we find that DREAM stimulates basal and ligand-dependent activation of promoters containing vitamin D and retinoic acid response elements (VDREs and RAREs), consisting of direct repeats of the sequence AGT/GTCA spaced by 3 or 5 nt, respectively. Stimulation occurs when the element is located upstream, but not downstream, the transcription initiation site. Activation requires both Ca2+ binding to the EF-hands and the leucine-charged domains (LCDs), analogous to those responsible for the interaction of the nuclear receptors with coregulators. Further more, DREAM can bind both ‘in vitro’ and in chromatin immunoprecipitation assays to these elements. Importantly, ‘in vivo’ binding is only observed in vitamin D- or RA-treated cells. These results show that DREAM can function as an activator of transcription on certain promoters and demonstrate a novel role for DREAM acting as a potential modulator of genes containing binding sites for nuclear receptors.
INTRODUCTION
DREAM (downstream regulatory element antagonist modulator) is a multifunctional Ca2+-binding protein, which acts as a transcriptional repressor by binding as a tetramer to DREs (downstream regulatory elements) located downstream of the TATA box in target genes (1,2). DREs can be composed of a single copy or two inverted copies of the sequence GTCA (3). DREAM was first identified as a repressor for the prodynorphin gene in neuroblastoma cells (1) and appears to play an important role in vivo in pain modulation (4).
DREAM contains four EF-hands, of which three are responsible for binding of Ca2+ ions (1). Stimuli that increase intracellular Ca2+ or cAMP levels cause the release of DREAM from the DRE and transcriptional derepression. cAMP and Ca2+ appear to promote derepression by different mechanisms. Whereas Ca2+ modifies DREAM conformation by binding to the EF-hand motifs, leading to release of DREAM from the DRE (1,5), cAMP-dependent derepression involves a direct protein–protein interaction of DREAM with α-CREM (cAMP response element modulator) (6). The interaction with α-CREM is Ca2+-independent and is governed by leucine-charged domains (LCDs). Two LCDs in DREAM and two LCDs located in the kinase-inducible domain (KID) of α-CREM participate in a two-site interaction that results in the loss of DREAM binding to the DRE and transcriptional derepression (6). In addition, in the absence of Ca2+, DREAM binds to LCDs located in the KID domain of cAMP response element-binding protein (CREB). As a result, DREAM impairs recruitment of the coactivator CBP (CREB-binding protein) and blocks phospho-CREB-dependent transactivation (7).
In addition to the evidence indicating that DREAM is a transcription factor (8), this protein might have pleiotropic functions through the interaction with specific DNA sequences and/or with diverse proteins in different cell compartments (9–11).
Nuclear receptors are ligand-dependent transcription factors, which regulate gene expression by binding to HREs (hormone response elements) located in regulatory regions of target genes (12). In the case of the vitamin D receptor (VDR) or the retinoic acid receptor (RAR), these elements contain the consensus sequence PuGGT/GTCA, which is rather similar to the DRE. A direct repeat (DR) of this sequence spaced by 3 nt (DR3) acts as a response element for vitamin D (VDRE), whereas a DR2 or a DR5 act as retinoic acid response elements (RAREs). Heterodimerization of VDR and RAR with the retinoid X receptor (RXR) increases affinity for DNA and transcriptional activity (13).
The effects of nuclear receptors on transcription are mediated through recruitment of coregulators. In the absence of ligand, the receptors can repress target gene expression (14) because they bind corepressors, such as silencing mediator for retinoid and thyroid hormone receptors (15) or nuclear receptor corepressor (16) assembled in complexes that include histone deacetylases. Upon ligand binding corepressors are released and coactivator complexes are recruited. Coactivators cause chromatin decompactation, RNA polymerase II recruitment and transcriptional activation (12,17–19). Interaction of nuclear receptors with coactivators and corepressors is also dependent on LCDs. The LCD motif LXXLL, where X denotes any amino acid, was first described in nuclear hormone receptor p160 coactivators (20,21). Three copies of this signature motif, present in the receptor interacting domain (RID) in the coactivator proteins, mediate association with the receptors. Furthermore, LCDs in the N- and C-terminal domains of the coactivator CBP mediate interaction of this protein with the receptors and p160 proteins, respectively (21). The receptor-interacting domain (CoRNR box) of the corepressors possesses the LCD motif L/IXXV/II, which is sufficient for corepressor binding and ligand-induced release (22–24). The LCDs in α-CREM (ILNEL and LIEEL) have an antiparallel orientation compared with the motifs responsible for the association of coregulators with nuclear receptors and define a third type of LCD (6).
The homology between the DNA sequences recognized by DREAM and the nuclear receptors, as well as the presence of LCDs in DREAM, led us to examine the existence of a possible cross-talk between the receptors and this transcriptional repressor. Our results show that DREAM can bind to HREs and that, unexpectedly, can act as a transcriptional activator and increase ligand-dependent transactivation of constructs containing these motifs, when located upstream the transcription initiation site. Interestingly, the DREAM LCD appear to be required for this stimulation, suggesting that protein–protein interactions are also involved in this stimulation. Conversely, nuclear receptors can bind to DREs, although this binding is transcriptionally unproductive. These results demonstrate a novel role for DREAM acting as a modulator of nuclear receptor-mediated responses.
MATERIALS AND METHODS
Plasmids
Luciferase reporter plasmids containing different fragments of the human RARβ2 promoter have been described previously (25). R140, R90 and R37 includes fragments −124 to +14, −59 to +14 and −11 to +34 (26). In R140M3 and 140M7, the 3′ and 5′ half-sites of the RARE have been mutated, respectively. The VDRE was cloned upstream of the thymidine kinase (TK) promoter of pBL-CAT8+ to give DR3-TK (27). TK-DR3-CAT was constructed by cloning the VDRE sequence in the SalI site downstream of the TK promoter in the same vector. CAT plasmids TK-DRE and DRE-TK have been described previously (2). pHD3 contains the sequence −151 to +150 of the human prodynorphin promoter (2). A luciferase reporter plasmid containing 2.3 kb of the p21Cip1 promoter cloned in pGL3 (Promega) was also used. Expression vectors for VDR and RARα are cloned in pSG5 (27). Expression vectors for wild-type DREAM and mutants in the EF-hands and in the LCDs (L47,52V and L155V) have been described elsewhere (1,6). Constructs in which the GAL4 DNA-binding domain (DBD) is fused to the ligand-binding domain (LBD) of RAR and VDR were a kind gift from R. Evans. These constructs were transfected together with a luciferase reporter UAS plasmid containing binding sites for the GAL-DBD.
Cell culture and transfections
HeLa, GH4C1 and HL-60 cells were grown as previously described in DMEM containing 10% fetal calf serum (28,29), and SK-N-MC cells in DMEM-Ham F-12 medium with the same amount of serum (2). The cells were transfected by calcium phosphate with 5 μg of reporter plasmids and 5 μg of the expression vectors indicated in the legends for Figures 1–4 and 6–8. The amount of DNA was always kept constant by the addition of the appropriate amount of a noncoding empty vector. After transfection, the cells were incubated for 24–36 h in the presence and absence of 100 nM vitamin D (Vit.D) or 1 μM all-trans-retinoic acid (RA) in medium supplemented with 10% AG1-X8 resin and charcoal-stripped newborn calf serum. Results are shown as fold induction over the values obtained in untreated cells in the absence of transfected DREAM. All data shown are mean ± standard deviation of CAT or luciferase activities from at least five independent experiments in triplicates.
Gel mobility shift assays
Recombinant DREAM, glutathione S-transferase (GST)-fused VDR and RXR (50–100 ng) and in vitro translated RXR, VDR and RAR were used in the assays with labeled DRE, DR3 and DR5 oligonucleotides as described previously (30). Mock lysates of GST alone or unprogrammed reticulocyte lysates were used in control lanes. For competition experiments, a 30-fold excess of unlabeled oligonucleotides encompassing the DRE, DR3 or a consensus Sp-1 binding site were added to the binding reactions. For supershift experiments, a monoclonal antibody against DREAM (2 μl) (6) was added before the addition of the labeled probe. When indicated 10 μM CaCl2 was present in the incubation reaction. Analysis of coactivators recruitment by receptor heterodimers were performed with in vitro translated receptors (3 μl) and 600 ng of the RIDs of the p160 coactivators: steroid receptor coactivator 1 (SRC-1) (31), activator for thyroid and retinoid receptor (ACTR) (32) and transcription intermediary factor-2 (TIF-2) (33), fused to GST.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) assays were performed essentially according to a previously published method (34). Approximately 2 × 108 HL-60 cells, grown with charcoal-stripped serum for 12–36 h, were treated with 100 nM Vit.D or 1 μM RA for 2 h, or were left untreated. The cells were cross-linked for 10 min with 1% paraformaldehyde, nuclei were collected and the chromatin was sonicated to an average length of between 500 and 2000 bp. After preclearing with protein A/G Sepharose (Amersham), blocked with salmon sperm DNA, BSA and in the presence of preimmune rabbit IgG (Sigma), aliquots of the chromatin was immunoprecipitated with αDREAM (10 μg, affinity-purified polyclonal antibody 1013), αAcH4 (5 μl, Upstate 06-866), αSRC1 (12 μg; Santa Cruz SC-20) or, as a negative control, preimmune rabbit IgG (10 μg). Capturing of the immunecomplexes, washing and reversal of cross-linking was carried out exactly as described. Thereafter, the immunoprecipitated DNA and an aliquot of the precleared chromatin (input) were purified using Qiagen PCR purification columns (in the presence of a carrier, tRNA). DNA was subjected to semi-quantitative PCR in the linear range (22–24 cycles) in the presence of a trace amount of [α-32P]dCTP. The primers used were: for the human p21Cip1 promoter containing the VDRE (located at −788/−756) forward 5′-GGCTATGTGGGGAGTATTCAG-3′ and reverse 5′-CCAGGGAAACAGAAGAATTGGACA-3′ (156 bp), for the human p21Cip1 promoter containing the RARE (located at −1221/−1194) forward 5′-GGTCAGGGGTGTGAGGTAG-3′ and reverse 5′-GATCATGCAGCAAAGAAATGACTA-3′ (248 bp), and for the RARβ2 promoter containing the RARE (located at −37/−53) forward 5′-CTGGGAGTTGGTGATGTCAGACTA-3′ and reverse 5′-TGGCAAAGAATAGACCCTC-3′ (168 bp). As a negative control, a GAPDH promoter fragment (174 bp) was amplified with the forward 5′-AGGGTTGCCACTGGGGATCT-3′ and reverse 5′-TGCCAAAGCCTAGGGGAAGA-3′ primers. PCR fragments were analyzed by polyacrylamide gels and autoradiography. Quantification was obtained using the NHI Image software. Results obtained were corrected by the input and are expressed as fold induction of vitamin D or RA-treated versus untreated cells.
RESULTS
DREAM stimulates VDRE-mediated transcription
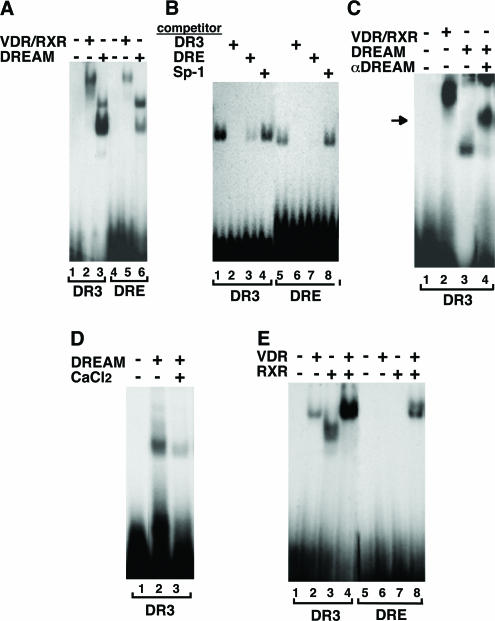
To analyze whether DREAM could bind to HREs, gel retardation assays were performed with a probe encompassing a DR3 site, a high-affinity element for RXR/VDR heterodimers. As shown in Figure 1A, recombinant DREAM bound to the DR3 with a stronger affinity than to the consensus DRE of the prodynorphin gene (compare lanes 3 and 6). Association of DREAM to both the DRE and the DR3 was competed with an excess of either unlabeled oligonucleotide, whereas a non-related competitor did not affect retardation (Figure 1B). Also showing the specificity of the binding of DREAM to the DR3 sequence, an anti-DREAM antibody caused a supershift of the DREAM-DR3 retarded band (Figure 1C). Furthermore, association of DREAM with the VDRE was sensitive to Ca2+, decreasing when CaCl2 was present in the binding buffer (Figure 1D).
Figure 1.
DREAM binds to a DR3 vitamin D response element. (A) Gel retardation assays performed with 100 ng of DREAM and 50 ng of GST-fused VDR and RXR and the DR3 and DRE oligonucleotides. (B) Binding of 50 ng of DREAM to these oligonucleotides was competed with excess of DRE, DR3 and Sp-1 oligonucleotides. (C) Binding of DREAM to the DR3 was supershifted with 2 μl of a DREAM antibody (αDREAM). Mobility of the super-retarded complex is indicated by an arrow. (D) Binding of DREAM to the DR3 was performed in the absence and in the presence of 10 μM CaCl2. (E) Binding of recombinant VDR and RXR (100 ng) alone and in combination with the DR3 and DRE oligonucleotides.
Conversely, binding of the receptor heterodimer VDR/RXR to the DRE was also observed, although with a lower affinity than to the VDRE (Figure 1A, lanes 2 versus lane 5). Figure 1E compares binding of VDR and RXR with the DR3 and the DRE. Both VDR and RXR were able to interact with the DR3 and, as expected, heterodimerization significantly increased binding. In the case of the DRE, VDR or RXR alone did not cause retardation, but a strong binding of heterodimers was found (lane 8). Therefore, although only one binding motif is present in the DRE, heterodimers but no receptor monomers associate with this element.
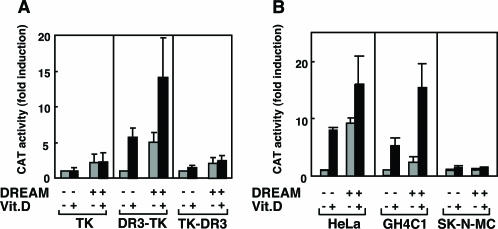
To explore the possibility that DREAM could regulate the activity of VDRE-containing promoters, we used transient transfection assays in HeLa cells with CAT reporter plasmids, in which the VDRE sequence was cloned both upstream or downstream of the TK minimal promoter. HeLa cells possess endogenous VDR, but do not express DREAM (2,28). As shown in Figure 2A, expression of DREAM did not significantly affect the activity of the empty TK reporter, confirming previous results (2). Surprisingly, DREAM, however, strongly increased the basal activity of the DR3-TK-CAT plasmid, and this activity was further enhanced in the presence of vitamin D. Therefore, in this context DREAM does not act as a repressor, but as an activator of VDRE-dependent transcription. Importantly, activation by DREAM depends on the position of the VDRE site because DREAM was unable to affect the activity of a construct in which the DR3 is located downstream of the TK promoter. This plasmid is also insensitive to vitamin D (Figure 2A).
Figure 2.
DREAM stimulates VDRE-dependent transcription. (A) HeLa cells were transfected with reporter CAT plasmids containing the DR3 element cloned upstream and downstream of the thymidine kinase promoter (DR3-TK and TK-DR3, respectively). A TK-CAT plasmid, which does not contain the response element, was also used as a control. The reporter constructs were cotransfected with an expression vector for DREAM or the corresponding empty vector, and CAT activity was determined after 36 h of treatment in the absence and presence of 100 nM Vit.D. (B) CAT activity was determined in untreated and vitamin D-treated HeLa, GH4C1 and SK-N-MC cells cotransfected with the DR3-TK reporter and expression vectors for wild-type DREAM.
To further investigate a relationship between vitamin D responsiveness and the activation of VDRE-dependent transcription by DREAM, we performed DR3-TK-CAT reporter assays in pituitary and neuroblastoma cells, which in contrast with HeLa cells express DREAM (2,35). In addition to expressing DREAM, pituitary somatolactotrop cells contain endogenous VDR and are responsive to vitamin D (27). Overexpression of DREAM in GH4C1 cells caused an increase in basal reporter activity, though less pronounced than that observed in HeLa cells that lack this factor, and significantly enhanced the ligand-dependent response (Figure 2B). These results indicate that the endogenous DREAM levels are not sufficient to produce a maximal stimulation of the VDRE-containing promoter. Interestingly, in neuroblastoma SK-N-MC cells, which express high DREAM levels (2), overexpression of DREAM failed to increase basal activity of the DR3-TK-CAT reporter (Figure 2B). We have previously shown that endogenous DREAM activity in SK-N-MC cells failed to modify the activity of a DRE-TK-CAT reporter while repressed a TK-DRE-CAT reporter (2). In these cells, vitamin D did not increase the reporter activity, demonstrating that VDR levels are not sufficient to elicit a significant transcriptional response.
DREAM activates RARE-dependent transcription
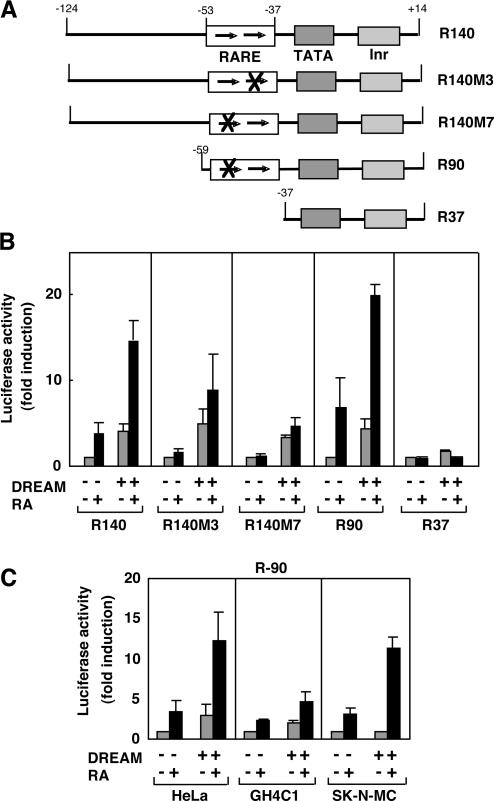
To analyze whether DREAM could also influence RARE-dependent transcription, HeLa cells were transfected with reporter plasmids containing different fragments of the RARβ2 promoter. This promoter contains a well-characterized RARE, configured as a DR5 between nucleotides −37 and −53 (Figure 3A), which binds RXR/RAR heterodimers (26). As illustrated in Figure 3B, DREAM significantly stimulated the activity of the constructs R140 and R90, which contain the RARE, and also potentiated the response to RA in HeLa cells. Interestingly, mutation of either the 5′ (R140M3) or the 3′ (R140M7) motifs of the DR5 was sufficient to strongly reduce RA-dependent transactivation but did not block stimulation by DREAM. Nevertheless, the RARE mediates the effect of DREAM on the promoter, since reporter plasmid R37, in which this element has been deleted, was not significantly stimulated by DREAM. This suggests that the requirements for DREAM and RXR/RAR heterodimer binding to these sequences are different and that whereas both hemisites of the response element are required for stimulation by RA, only one hemisite is sufficient for stimulation by DREAM.
Figure 3.
DREAM stimulates RARE-dependent transcription. (A) Schematic representation of mutations and deletions in the RARβ2 promoter, showing the positions of the RARE, the TATA box and the Initiator (Inr). The hemisites in the RARE are indicated by arrows. In R140M3 and R140M7, the 5′ and 3′ motifs, respectively, of the RARE have been mutated and mutations are shown as X. (B) HeLa cells were transfected with the RARβ2 luciferase reporter constructs indicated and an expression vector for DREAM or the noncoding vector. (C) HeLa, GH4C1 and SK-N-MC cells were transfected with the R90 construct and the expression vector for DREAM. Reporter activity was determined in cells incubated in the presence and absence of 1 μM all-trans-RA.
Figure 3C compares the influence of DREAM on the activity of the RARβ2 promoter in HeLa, GH4C1 and SK-N-MC cells. These cells are known to contain RA receptors and are responsive to the retinoid. As occurred with the DR3-containing promoter, DREAM was able to increase the activity of the R90 construct in HeLa and GH4C1 cells, but not in the neuroblastoma cells, that already express high endogenous DREAM levels. In addition, RA stimulated the activity of the RARβ2 promoter to a similar extent in HeLa and SK-N-MC cells and less strongly in GH4C1 cells, but DREAM was able to enhance RA-dependent activation in all cell types. Similar results were obtained with the R140 construct (not illustrated). Therefore, in several cellular contexts, DREAM appears to act as an activator of reporters containing DR5 and DR3 elements upstream of the TATA box.
DREAM is recruited in vivo to HRE-containing promoters in a ligand-dependent manner
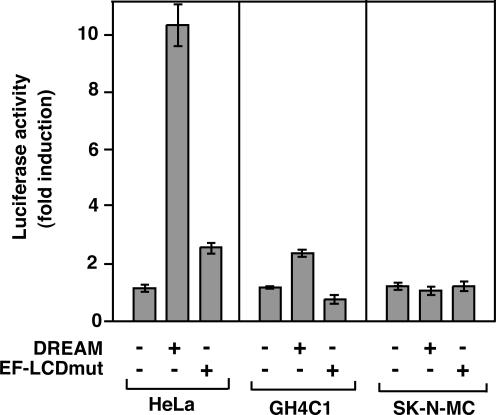
It has been shown that p21Cip1 expression is stimulated by both vitamin D and RA in hematopoietic cells (36,37), and therefore this gene constitutes a good model for analyzing the effects of DREAM. To study the response of the p21Cip1 promoter to DREAM, a reporter plasmid containing the 5′-flanking region of the p21Cip1 gene was transfected into HL-60, HeLa, pituitary and neuroblastoma cells in the presence and absence of DREAM. In addition, the effect of a version of DREAM (EF-LCDmut), in which both the Ca2+-binding domain and the LCDs were mutated (3), was also examined. The difficulty of transfection of hematopoietic cells did not allow the study of the effect of DREAM on the p21Cip1 promoter in HL-60 cells. However, as shown in Figure 4, native DREAM, but not the mutated protein, strongly stimulated the p21Cip1 promoter in HeLa cells. In agreement with the data obtained with the RARβ2 and the DR3-containing promoters, DREAM was less effective in GH4C1 cells and again did not stimulate the promoter activity in SK-N-MC cells that contain high endogenous DREAM levels. The EF-LCDmut was also uneffective in pituitary and neuroblastoma cells. In the tested cell types, we did not observe activation of the p21Cip1 promoter by vitamin D or RA (data not shown).
Figure 4.
DREAM activates the p21cip1promoter. A luciferase reporter plasmid containing 2.3 kb of the 5′-flanking region of the p21cip1 gene was transfected into HeLa, GH4C1 and SK-N-MC. Reporter activity was determined in cells transfected with an empty vector or with expression vectors for wild-type DREAM or for a DREAM protein, in which both the functional EF-hands and the LCDs have been mutated (EF-LCDmut).
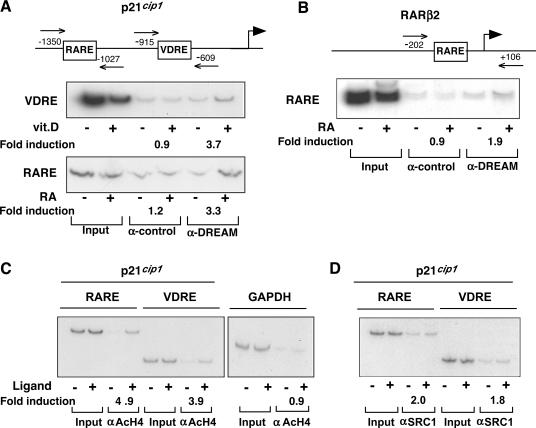
The p21Cip1 promoter contains a VDRE and a RARE at positions −770 and −1200, respectively. To analyze binding of DREAM to these sites ‘in vivo’, we carried out ChIP assays with primers amplifying the p21Cip1 promoter fragments that include the VDRE or the RARE motifs. The distance between both elements allows the study of recruitment of DREAM to these elements separately. For these studies, we used hematopoietic HL-60 cells that contain high levels of DREAM (not illustrated), and in which incubation with vitamin D or RA increases p21Cip1 expression and induces differentiation (36,37). Binding of DREAM to the proximal RARβ2 promoter was also determined by ChIP assay in these cells. Fragments encompassing the VDRE and RARE (Figure 5A) of the p21Cip1 promoter were not amplified above the levels obtained with a non-specific antibody from chromatin of untreated cells after immunoprecipitation with an anti-DREAM antibody. However, DREAM was recruited to the VDRE promoter region in cells incubated with vitamin D and to the RARE promoter region in RA-treated cells (compare lanes 5 and 6). Similarly, ‘in vivo’ binding of DREAM to the fragment encompassing the RARE of the RARβ2 gene was only detected upon incubation with the retinoid (Figure 5B). As shown in Figure 5C, incubation of HL-60 cells with either RA or vitamin D also caused the recruitment of acetylated histone H4 to the p21Cip1 promoter fragments containing the RARE and the VDRE, respectively. In contrast, acetylated histone H4 was not found after vitamin D treatment in the promoter of the GAPDH gene, used as a negative control. An antibody against the p160 coactivator SRC-1 was also used in ChIP experiments. In parallel with the increase in acetylated H4 histone and DREAM, RA and vitamin D also caused the recruitment of the coactivator to the RARE or VDRE p21Cip1 promoter regions (Figure 5D).
Figure 5.
DREAM binds in vivo to the p21cip1 promoter. (A) Schematic representation of the 5′-flanking region of the p21cip1 gene showing the location of the primers used to amplify the fragments that contain the VDRE and the RARE used in the ChIP assays. The assays were performed in untreated HL-60 cells and in cells treated with vitamin D or RA for 2 h. Representative assays for both regions performed with control and anti-DREAM antibodies are shown in the lower panels. The inputs of the assays for untreated and treated cells are shown in lanes 1 and 2, respectively. (B) The upper panel shows the position of the RARE of the RARβ2 promoter and the region amplified in the ChIP assays in control and RA-treated cells is shown in the lower panel. In (C), ChIP assays were performed with an antibody against acetylated histone H4 (αAcH4). In the left panel are shown the results obtained with the regions amplifying the RARE and the VDRE of the p21cip1 gene in cells treated with RA and vitamin D, respectively. The right panel illustrates the amplification of a GAPDH promoter fragment, used as a negative control, in control and vitamin-D treated cells. (D) Recruitment of the p160 coactivator SRC-1 to the RARE and VDRE regions of the p21cip1 promoter in control cells and after treatment with RA or vitamin D, respectively. In all panels, relative recruitment in untreated versus ligand-treated cells was quantified and is shown as fold induction.
The LCDs are required for DREAM-dependent stimulation of the DR3 and DR5-containing plasmids
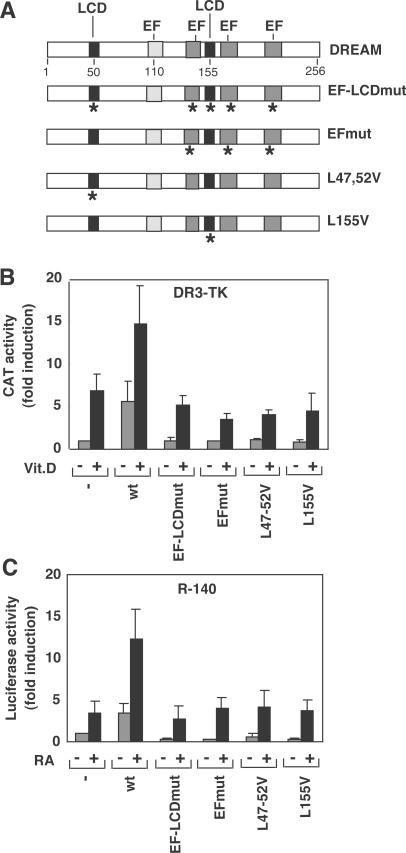
Since binding of DREAM to DREs and repression is blocked in the presence of Ca2+ (1), we examined the influence of a DREAM mutated protein (EFmutDREAM) in which the three functional EF-hands of DREAM were modified. EFmutDREAM binds to DREs and like wild-type DREAM is regulated by cAMP (3). EFmutDREAM failed to activate the basal expression of the DR3TK reporter (Figure 6B) and even had an inhibitory effect on the basal activity of the RARβ2 promoter construct (Figure 6C). Furthermore, the mutant also failed to potentiate ligand-dependent transactivation of either reporter by vitamin D or RA, respectively. The fact that DREAM and EFmutDREAM show different effects on the HRE-dependent transcription suggest that not only protein–DNA but also protein–protein interactions could be involved in the complex regulation by DREAM of VDRE- and RARE-mediated gene expression. To test the importance of these interactions in the transactivating effect of DREAM on hormone receptor mediated transcription, we used DREAM with mutations at the LCDs (Figure 6A) and assessed their activity on the DR3-TK and R-140 reporters in HeLa cells. Figure 6 shows that although wild-type DREAM causes activation of the DR3- and DR5-containing promoters, DREAM mutants bearing the mutations L47,52V in the first LCD or L155V in the second LCD did not activate basal or ligand-dependent expression of the DR3 and RARβ2 promoters. Mutation of the Ca2+-binding motifs in combination with both LCDs (EF-LCDmut) did not further repress ligand-independent or -dependent transcription. Therefore, any of the single mutations alone is enough to preclude the effect of DREAM on transcriptional activity mediated by the HREs. These results further substantiate the idea that protein–protein interactions that are disturbed upon changes in the DREAM protein could be involved in DREAM-dependent transactivation of HRE-mediated transcription.
Figure 6.
Both the EF-hands and the LCDs are required for DREAM stimulation of VDRE and RARE-containing promoters. (A) Schematic representation of the DREAM protein showing the position of the two LCDs and the four EF-hands. EF-LCDmut has mutations in the three functional EF-hands and in both LCDs, EFmut is a mutant in the EF-hands. L47, 52V and L155V are mutants in the first and second LCD motifs in DREAM, respectively. Mutations are indicated by asterisks below the corresponding domains. DR3-TK (B) and R140 (C) reporter constructs were transfected into HeLa cells together with expression vectors for wild-type (wt) and mutated versions of DREAM. Reporter activity was determined after 36 h incubation in control cells and in cells treated with vitamin D or RA.
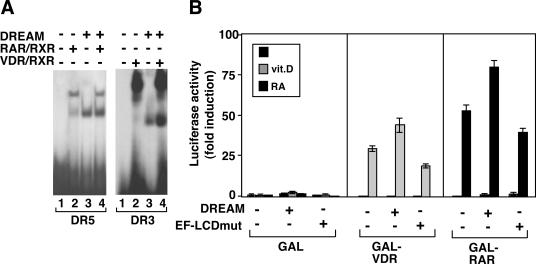
Since the LCDs in DREAM are similar to those present in several nuclear receptor coactivators, we tested the possibility that DREAM could form a complex with the receptor heterodimers when bound to DNA. However, as shown in Figure 7A, the receptors and DREAM appear to bind independently to the DR3 or DR5 elements, since neither the formation of larger complexes by occupancy of the same DNA molecule nor the appearance of complexes with intermediate mobilities due to the interaction of DREAM with the receptors was detected. In addition, binding of in vitro translated DREAM with VDR or RAR fused to GST was analyzed in pull-down assays. No direct interaction of the receptors with DREAM was observed, whereas in the same assays hormone receptors interacted in a ligand-dependent manner with a p160 coactivator used as a positive control (data not shown). Thus, DREAM can cooperate with vitamin D and RA to increase HRE-dependent transcription without a direct interaction with their receptors.
Figure 7.
DREAM does not interact directly with the receptors. (A) Gel retardation assays performed with the DR5 and DR3 oligonucleotides. DREAM (50 ng) was incubated with the probes alone or in combination with in vitro translated RAR and RXR (4 μl) or with recombinant VDR and RXR (100 ng), as indicated. (B) The GAL-DBD alone or fused with the VDR or RAR LBDs were transfected into HeLa cells together with expression vectors for wild-type or EF-LCDmut DREAM and the UAS reporter plasmid. Luciferase activity was determined after 24 h in untreated cells and in cells treated with vitamin D or RA, as indicated.
To test directly whether DREAM could affect ligand-dependent and -independent transactivation without binding of the receptor heterodimers to their recognition sites on DNA, we performed transfection studies in which fusions of the DBD of the yeast transcription factor GAL4 with the LBDs of VDR and RAR were cotransfected with expression vectors for wild-type and mutant DREAM and a reporter UAS plasmid. As shown in Figure 7B, expression of neither native DREAM nor the EF-LCDmut affected significantly the activity of GAL-VDR or GAL-RAR in the absence of ligand. In addition, DREAM caused a weak increase of the ligand-dependent response, which was absent when the EF-LCDmut DREAM was used. These data further demonstrate that the presence of the HRE plays an important role in the transactivating effects of DREAM.
Nuclear receptors do not affect DRE-dependent transcription
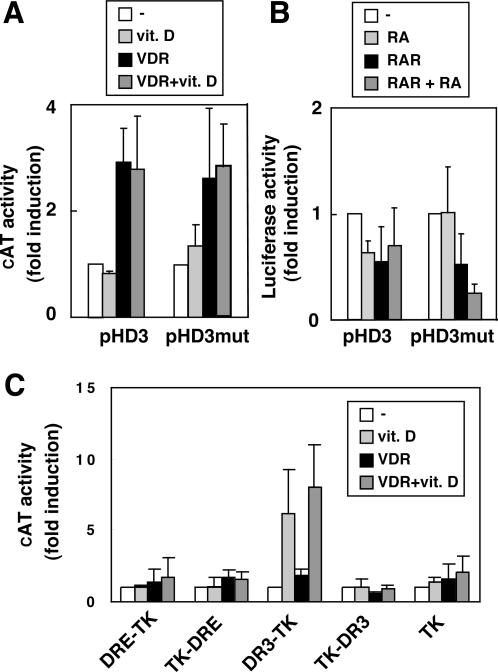
Since RXR/VDR heterodimers bind the DRE as shown in Figure 1, we next examined the possibility that vitamin D could influence the activity of constructs containing this element. For this purpose, HeLa cells were transfected with the CAT reporter plasmid HD3. This construct contains the prodynorphin promoter fragment −151 to +150 with the DRE at position +40. As illustrated in Figure 8A, vitamin D did not alter the activity of the prodynorphin promoter. Since the affinity of binding of the receptor to the DRE was lower than to a DR3, the CAT activity was also examined after the overexpression of VDR. Expression of the receptor caused some ligand-dependent increase of pHD3 activity. However, this effect appears to be independent of the DRE, as mutation of this element did not alter regulation by VDR. Similar results were obtained after treatment with RA that did not alter the activity of the prodynorphin promoter even after the overexpression of RAR (Figure 8B).
Figure 8.
Vitamin D and retinoic acid receptors do not affect DRE-mediated transcription. HeLa cells were transfected with the CAT reporter construct pHD3 containing sequences −151/+150 of the human prodynorphin promoter, or pHD3mut mutated in the DRE site. The reporter plasmids were cotransfected with expression vectors for VDR (A) or RAR (B), and CAT activity determined after incubation with vitamin D or RA as indicated. (C) Cells were cotransfected with plasmids containing the DRE and DR3 motifs upstream and downstream of the TK promoter and with an expression vector for VDR. CAT activity was determined in untreated and vitamin D-treated cells.
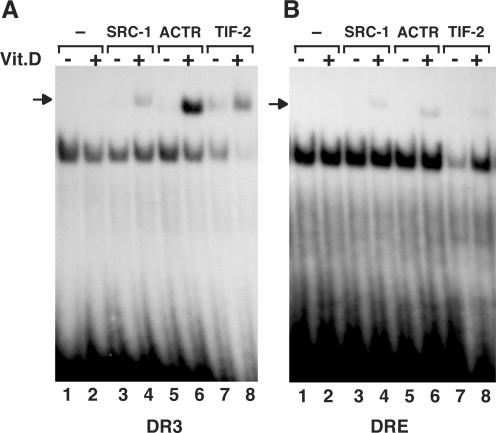
The possibility that VDR could stimulate DRE-dependent transcription when this element is located upstream of the transcription initiation site was also explored. Figure 8C compares the results obtained in cells transfected with constructs in which the DR3 and DRE elements are cloned both 5′ and 3′ of the TK promoter. Only the activity of the DR3-TK construct was stimulated by vitamin D, both in the presence and in the absence of transfected receptor. In contrast, the DRE was unable to sustain VDR-mediated transcriptional activity. Since ligand-dependent stimulation by nuclear receptors depends on recruitment of coactivators, we next examined binding of several p160 coactivators, including SRC-1, ACTR and TIF-2 to receptors bound to the DR3 and DRE motifs. As shown in Figure 9, addition of ligand caused strong recruitment of coactivators by the RXR/VDR heterodimers bound to the DR3, which was shown by the appearance of a super-retarded complex. However, intensity of this band was much less prominent when the receptors were bound to the DRE, and the supershifted bands were only detected after longer exposures.
Figure 9.
Recruitment of coactivators by the VDR/RXR heterodimer. Gel retardation assays with the DR3 (A) and the DRE (B), and in vitro translated VDR and RXR (3 μl). The assays were performed in the presence of 600 ng of GST alone or the same amount of the RIDs of p160 coactivators SRC-1, ACTR and TIF-2 fused to GST. When indicated (+), vitamin D (100 nM) was present in the binding reactions. Arrowheads, mobilities of the supershifted complexes containing the receptor heterodimer and the coactivator. Autoradiogram in the right has been subjected to a longer exposure time.
DISCUSSION
In this work, we show that the repressor DREAM can also function as a transcriptional activator of genes containing response elements for nuclear receptors, such as VDR or RAR. DREAM expression had a very strong stimulatory effect on HRE-dependent transcription in cells that do not express this factor, but was weaker in cells expressing high endogenous levels of DREAM protein. DREAM can bind in vitro to these elements, and it can also bind in vivo, in a ligand-dependent manner, to promoters of genes that contain a VDRE or a RARE. Moreover, expression of DREAM strongly potentiates ligand-dependent transcriptional regulation by VDR and RAR. Therefore, depending on the promoter context, DREAM functions as a repressor or as a transcriptional activator.
Our results show that DREAM can bind to HREs in vitro even more efficiently than to the original DRE from the prodynorphin gene (1). HREs are composed of two copies of a hexameric motif rather similar to the DRE, and the presence of two motifs probably contributes to the stabilization of DREAM binding to these sequences. Natural DREs containing two copies of the DRE motif have been found in the c-fos and CREM genes (1,3,38). Analysis of these tandem DRE sequences also showed a greater affinity for DREAM than the prodynorphin DRE (3). Moreover, our results indicate that the DRE motif can also function in a DR arrangement and show the flexibility of DREAM binding to DNA, since this factor can recognize sequences with different spacing between the binding motifs. The fact that DREAM binds to DREs as a tetramer may contribute to this flexibility. In addition to spacing, small differences in the half-site motif and in the sequence of the flanking extensions of the response elements appear to be important parameters in determining the binding efficiency both for DREAM and for nuclear receptors (3,39). Nevertheless, stimulation of HRE-mediated transcription by DREAM and the nuclear receptors appears to have distinct requirements. Thus, mutational analysis of the RARβ2 promoter shows that although stimulation by RA requires both hemisites of the response element, only one hemisite is sufficient for ligand-independent transactivation by DREAM.
The DREs of the prodynorphin and c-fos genes are located downstream of the transcription initiation site (1,2). Furthermore, the DRE sequence of the apoptotic hrk gene is located in the 3′-untranslated region (3′-UTR) (40). Here, we demonstrate that DREAM was effective in stimulating a VDRE-containing construct when the element was placed upstream but not downstream of the TK promoter. These results demonstrate that the position of the response element can determine the transcriptional outcome. This was also true for vitamin D-dependent activation that was only found when the element was located upstream of the promoter. Indeed, most positive HREs have this location. However, nuclear receptors can mediate transcriptional repression in a ligand-dependent manner through binding to the so-called negative HREs. Interestingly, a rather common finding is that these elements are positioned downstream of the transcription initiation site (12,41), or even have an unusual location at the 3′-UTR (42).
In contrast with the nuclear receptors that possess a well-characterized DBD, this functional domain has not yet been fully delineated in DREAM. DREAM appears to bind the DRE as a tetramer, and most likely, the DBD is composed upon oligomerization. Therefore, it has not been possible to analyze the role of specific mutations in DREAM that abolish binding to DNA on HRE-mediated transcriptional activation. However, we have found that DREAM proteins in which the EF-hands or the LCDs have been mutated (1) fail to stimulate HRE-dependent transcription. This is contrary to expectations based on the similar efficiency of these mutants and wild-type DREAM to bind DRE sites in the DNA (3,6,7). Furthermore, in basal conditions, these mutants are as active as the wild-type protein in repressing DRE-dependent transcription and remain bound to DNA acting as dominant negative repressors following Ca2+ or cAMP stimulation (1,3,7). Therefore, HRE-dependent transcription requires the integrity of the EF-hands.
The finding that mutation of either LCD in DREAM abolishes stimulation mediated by the HREs indicates that the interaction of DREAM with the transcriptional complex formed on the HRE site could be crucial for the transactivating effect of this factor. A prevalence of protein–protein interactions over binding of DREAM to DNA has been previously reported in the de-repression of DRE sites following cAMP stimulation, which depends on the interaction of DREAM with α- or ε-CREM (6) and in the repression by DREAM of CRE-dependent transcription (7). In both instances, the protein–protein interactions are mediated by the LCDs in DREAM and in the CREM or CREB proteins. Although DREAM binds in vitro to the HREs independently of the presence of the occupied receptors, ChIP assays demonstrated that in vivo binding could only be detected upon incubation of cells with the appropriate ligand. The ligand-dependent recruitment of DREAM parallels that of the nuclear receptor coactivators and the increase of acetylated histones to the target promoter. It is possible that the concentrations of DREAM used in the retardation assays is sufficient to detect interactions, whereas under physiological conditions DREAM could only be recruited to the promoter through a direct interaction with other proteins. One possible mechanism was that DREAM could be tethered to the HRE through interaction with the occupied receptor. However, despite the presence in DREAM of LCDs rather similar to those responsible for the association of coregulators with the nuclear receptors (20–24), we did not observe a direct interaction of the receptors with DREAM. Furthermore, we did not find a substantial increase in VDR- or RAR-mediated transactivation by DREAM when these proteins are not bound to a HRE. Therefore, recruitment to the HRE and activation of DR3- and DR5-dependent transcription by this factor does not involve a direct association with the receptors and appears to require DREAM–DNA interactions. Future studies will determine whether the association of DREAM with coactivators through the LCDs could mediate the recruitment of DREAM to the mediator complex that results in the potentiation of HRE transcription by DREAM.
We also show here that VDR can bind to the prodynorphin DRE, although with less affinity than to a VDRE. However, this binding appears to be transcriptionally unproductive because VDR was unable to influence DRE-mediated transcription. The same was true for RAR. Therefore, there is a discrepancy between the in vitro binding to the DRE and the lack of transcriptional activation. This situation is similar to that found with the RXR/RAR heterodimer bound to a DR1 (43), or the RXR/VDR heterodimer bound to a DR5 (44), which cannot trigger transcriptional activation through these elements. The receptor heterodimers bind to their bona fide HREs with a strict polarity. Thus, on DR3 and DR5, RXR occupies the 5′ half-site and the corresponding partner (e.g. VDR or RAR) occupies the 3′ half-site (45–47). This particular arrangement permits RAR and VDR upon ligand binding to undergo conformational changes that facilitate the recruitment of coactivator complexes. However, it has been demonstrated that in the cases of unproductive binding the heterodimer associates to DNA with a reversed or undefined polarity (43,44). The fact that the DRE is composed of a single motif could prevent anisotropic binding and this could be involved in the lack of transcriptional activation. However, a different conformation of the heterodimer bound to DRE and DRs elements could also affect interaction with coregulators, which are required for ligand-dependent transcription by nuclear receptors. This appears to be the case, because reduced recruitment of p160 coactivators by the RXR/VDR heterodimer bound to the DRE was found. Therefore, a combination of reduced binding affinity and abnormal association with coactivators could explain the lack of transcriptional activity of the RXR/VDR heterodimer upon binding to the DRE.
In summary, our results demonstrate a new function for DREAM acting as a transcriptional stimulator. The existence of a transcriptional cross-talk between DREAM and the nuclear receptor signaling, in which both protein–DNA and protein–protein interactions could participate, may have an important physiological relevance. Our results indicate that genes, such as RARβ2 or p21cip1, could be targets for DREAM. Future studies will be needed to identify additional genes in DREAM expressing tissues, which contain HRE-like DREAM binding sites and are regulated by this transcription factor.
Acknowledgments
This work was supported by Grants BMC2001-2275 and BMC2001-1431 from the MCyT. to A.A. and to J.R.N., respectively, and RGP0156/2001 from the Human Frontiers Science Program to J.R.N. Funding to pay the Open Access publication charges for this article was provided by Grant BMC2001-2275 from MCyT.
Conflict of interest statement. None declared.
REFERENCES
- 1.Carrion A.M., Link W.A., Ledo F., Mellstrom B., Naranjo J.R. DREAM is a Ca2+-regulated transcriptional repressor. Nature. 1999;398:80–84. doi: 10.1038/18044. [DOI] [PubMed] [Google Scholar]
- 2.Carrion A.M., Mellstrom B., Naranjo J.R. Protein kinase A-dependent derepression of the human prodynorphin gene via differential binding to an intragenic silencer element. Mol. Cell. Biol. 1998;18:6921–6929. doi: 10.1128/mcb.18.12.6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ledo F., Link W.A., Carrion A.M., Echeverria V., Mellstrom B., Naranjo J.R. The DREAM-DRE interaction: key nucleotides and dominant negative mutants. Biochim. Biophys. Acta. 2000;1498:162–168. doi: 10.1016/s0167-4889(00)00092-6. [DOI] [PubMed] [Google Scholar]
- 4.Cheng H.Y., Pitcher G.M., Laviolette S.R., Whishaw I.Q., Tong K.I., Kockeritz L.K., Wada T., Joza N.A., Crackower M., Goncalves J., et al. DREAM is a critical transcriptional repressor for pain modulation. Cell. 2002;108:31–43. doi: 10.1016/s0092-8674(01)00629-8. [DOI] [PubMed] [Google Scholar]
- 5.Osawa M., Tong K.I., Lilliehook C., Wasco W., Buxbaum J.D., Cheng H.Y., Penninger J.M., Ikura M., Ames J.B. Calcium-regulated DNA binding and oligomerization of the neuronal calcium-sensing protein, calsenilin/DREAM/KChIP3. J. Biol. Chem. 2001;276:41005–41013. doi: 10.1074/jbc.M105842200. [DOI] [PubMed] [Google Scholar]
- 6.Ledo F., Carrion A.M., Link W.A., Mellstrom B., Naranjo J.R. DREAM-alphaCREM interaction via leucine-charged domains derepresses downstream regulatory element-dependent transcription. Mol. Cell. Biol. 2000;20:9120–9126. doi: 10.1128/mcb.20.24.9120-9126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ledo F., Kremer L., Mellstrom B., Naranjo J.R. Ca2+-dependent block of CREB-CBP transcription by repressor DREAM. EMBO J. 2002;21:4583–4592. doi: 10.1093/emboj/cdf440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mellstrom B., Naranjo J.R. Mechanisms of Ca(2+)-dependent transcription. Curr. Opin. Neurobiol. 2001;11:312–319. doi: 10.1016/s0959-4388(00)00213-0. [DOI] [PubMed] [Google Scholar]
- 9.Buxbaum J.D., Choi E.K., Luo Y., Lilliehook C., Crowley A.C., Merriam D.E., Wasco W. Calsenilin: a calcium-binding protein that interacts with the presenilins and regulates the levels of a presenilin fragment. Nature Med. 1998;4:1177–1181. doi: 10.1038/2673. [DOI] [PubMed] [Google Scholar]
- 10.An W.F., Bowlby M.R., Betty M., Cao J., Ling H.P., Mendoza G., Hinson J.W., Mattsson K.I., Strassle B.W., Trimmer J.S., et al. Modulation of A-type potassium channels by a family of calcium sensors. Nature. 2000;403:553–556. doi: 10.1038/35000592. [DOI] [PubMed] [Google Scholar]
- 11.Ikura M., Osawa M., Ames J.B. The role of calcium-binding proteins in the control of transcription: structure to function. Bioessays. 2002;24:625–636. doi: 10.1002/bies.10105. [DOI] [PubMed] [Google Scholar]
- 12.Aranda A., Pascual A. Nuclear hormone receptors and gene expression. Physiol. Rev. 2001;81:1269–1304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- 13.Mangelsdorf D.J., Evans R.M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 14.Hu X., Lazar M.A. Transcriptional repression by nuclear hormone receptors. Trends Endocrinol. Metab. 2000;11:6–10. doi: 10.1016/s1043-2760(99)00215-5. [DOI] [PubMed] [Google Scholar]
- 15.Chen J.D., Evans R.M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 16.Horlein A.J., Naar A.M., Heinzel T., Torchia J., Gloss B., Kurokawa R., Ryan A., Kamei Y., Soderstrom M., Glass C.K., et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 17.McKenna N.J., O'Malley B.W. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 18.Rachez C., Freedman L.P. Mediator complexes and transcription. Curr. Opin. Cell Biol. 2001;13:274–280. doi: 10.1016/s0955-0674(00)00209-x. [DOI] [PubMed] [Google Scholar]
- 19.Rosenfeld M.G., Glass C.K. Coregulator codes of transcriptional regulation by nuclear receptors. J. Biol. Chem. 2001;276:36865–36868. doi: 10.1074/jbc.R100041200. [DOI] [PubMed] [Google Scholar]
- 20.Heery D.M., Kalkhoven E., Hoare S., Parker M.G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 21.Torchia J., Rose D.W., Inostroza J., Kamei Y., Westin S., Glass C.K., Rosenfeld M.G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear–receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 22.Hu X., Lazar M.A. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402:93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- 23.Nagy L., Kao H.Y., Love J.D., Li C., Banayo E., Gooch J.T., Krishna V., Chatterjee K., Evans R.M., Schwabe J.W. Mechanism of corepressor binding and release from nuclear hormone receptors. Genes Dev. 1999;13:3209–3216. doi: 10.1101/gad.13.24.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perissi V., Staszewski L.M., McInerney E.M., Kurokawa R., Krones A., Rose D.W., Lambert M.H., Milburn M.V., Glass C.K., Rosenfeld M.G. Molecular determinants of nuclear receptor–corepressor interaction. Genes Dev. 1999;13:3198–3208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cosgaya J.M., Aranda A. Nerve growth factor activates the RARbeta2 promoter by a Ras-dependent mechanism. J. Neurochem. 2001;76:661–671. doi: 10.1046/j.1471-4159.2001.00078.x. [DOI] [PubMed] [Google Scholar]
- 26.Vivanco Ruiz M.M., Bugge T.H., Hirschmann P., Stunnenberg H.G. Functional characterization of a natural retinoic acid responsive element. EMBO J. 1991;10:3829–3838. doi: 10.1002/j.1460-2075.1991.tb04952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Villalba P., Jimenez-Lara A.M., Aranda A. Vitamin D interferes with transactivation of the growth hormone gene by thyroid hormone and retinoic acid. Mol. Cell. Biol. 1996;16:318–327. doi: 10.1128/mcb.16.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tolon R.M., Castillo A.I., Jimenez-Lara A.M., Aranda A. Association with Ets-1 causes ligand- and AF2-independent activation of nuclear receptors. Mol. Cell. Biol. 2000;20:8793–8802. doi: 10.1128/mcb.20.23.8793-8802.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendez-Pertuz M., Sanchez-Pacheco A., Aranda A. The thyroid hormone receptor antagonizes CREB-mediated transcription. EMBO J. 2003;22:3102–3112. doi: 10.1093/emboj/cdg295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jimenez-Lara A.M., Aranda A. Lysine 246 of the vitamin D receptor is crucial for ligand-dependent interaction with coactivators and transcriptional activity. J. Biol. Chem. 1999;274:13503–13510. doi: 10.1074/jbc.274.19.13503. [DOI] [PubMed] [Google Scholar]
- 31.Henttu P.M., Kalkhoven E., Parker M.G. AF-2 activity and recruitment of steroid receptor coactivator 1 to the estrogen receptor depend on a lysine residue conserved in nuclear receptors. Mol. Cell Biol. 1997;17:1832–1839. doi: 10.1128/mcb.17.4.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen H., Lin R.J., Schiltz R.L., Chakravarti D., Nash A., Nagy L., Privalsky M.L., Nakatani Y., Evans R.M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 33.Voegel J.J., Heine M.J., Tini M., Vivat V., Chambon P., Gronemeyer H. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takahashi Y., Rayman J.B., Dynlacht B.D. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- 35.Leclerc G.M., Leclerc G.J., Shorte S.L., Stephen Frawley L., Boockfor F.R. Cloning and mRNA expression of the Ca2+-binding DREAM protein in the pituitary. Gen. Comp. Endocrinol. 2002;129:45–55. doi: 10.1016/s0016-6480(02)00509-9. [DOI] [PubMed] [Google Scholar]
- 36.Liu M., Lee M.H., Cohen M., Bommakanti M., Freedman L.P. Transcriptional activation of the Cdk inhibitor p21 by vitamin D3 leads to the induced differentiation of the myelomonocytic cell line U937. Genes Dev. 1996;10:142–153. doi: 10.1101/gad.10.2.142. [DOI] [PubMed] [Google Scholar]
- 37.Liu M.I.A., Freedman L.P. Transcriptional activation of the human p21(WAF1/CIP1) gene by retinoic acid receptor. Correlation with retinoid induction of U937 cell differentiation. J. Biol. Chem. 1996;271:31723–31728. doi: 10.1074/jbc.271.49.31723. [DOI] [PubMed] [Google Scholar]
- 38.Mellstrom B., Naranjo J.R. Ca2+-dependent transcriptional repression and derepression: DREAM, a direct effector. Semin. Cell Dev. Biol. 2001;12:59–63. doi: 10.1006/scdb.2000.0218. [DOI] [PubMed] [Google Scholar]
- 39.Mader S., Leroy P., Chen J.Y., Chambon P. Multiple parameters control the selectivity of nuclear receptors for their response elements. Selectivity and promiscuity in response element recognition by retinoic acid receptors and retinoid X receptors. J. Biol. Chem. 1993;268:591–600. [PubMed] [Google Scholar]
- 40.Sanz C., Mellstrom B., Link W.A., Naranjo J.R., Fernandez-Luna J.L. Interleukin 3-dependent activation of DREAM is involved in transcriptional silencing of the apoptotic Hrk gene in hematopoietic progenitor cells. EMBO J. 2001;20:2286–2292. doi: 10.1093/emboj/20.9.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez-Juste G., Garcia-Silva S., Aranda A. An element in the region responsible for premature termination of transcription mediates repression of c-myc gene expression by thyroid hormone in neuroblastoma cells. J. Biol. Chem. 2000;275:1307–1314. doi: 10.1074/jbc.275.2.1307. [DOI] [PubMed] [Google Scholar]
- 42.Bigler J., Eisenman R.N. Novel location and function of a thyroid hormone response element. EMBO J. 1995;14:5710–5723. doi: 10.1002/j.1460-2075.1995.tb00258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurokawa R., Soderstrom M., Horlein A., Halachmi S., Brown M., Rosenfeld M.G., Glass C.K. Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature. 1995;377:451–454. doi: 10.1038/377451a0. [DOI] [PubMed] [Google Scholar]
- 44.Jimenez-Lara A.M., Aranda A. The vitamin D receptor binds in a transcriptionally inactive form and without a defined polarity on a retinoic acid response element. FASEB J. 1999;13:1073–1081. doi: 10.1096/fasebj.13.9.1073. [DOI] [PubMed] [Google Scholar]
- 45.Perlmann T., Rangarajan P.N., Umesono K., Evans R.M. Determinants for selective RAR and TR recognition of direct repeat HREs. Genes Dev. 1993;7:1411–1422. doi: 10.1101/gad.7.7b.1411. [DOI] [PubMed] [Google Scholar]
- 46.Kurokawa R., Yu V.C., Naar A., Kyakumoto S., Han Z., Silverman S., Rosenfeld M.G., Glass C.K. Differential orientations of the DNA-binding domain and carboxy-terminal dimerization interface regulate binding site selection by nuclear receptor heterodimers. Genes Dev. 1993;7:1423–1435. doi: 10.1101/gad.7.7b.1423. [DOI] [PubMed] [Google Scholar]
- 47.Lemon B.D., Freedman L.P. Selective effects of ligands on vitamin D3 receptor- and retinoid X receptor-mediated gene activation in vivo. Mol. Cell. Biol. 1996;16:1006–1016. doi: 10.1128/mcb.16.3.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]