Abstract
Proteins with RNA chaperone activity are able to promote folding of RNA molecules by loosening their structure. This RNA unfolding activity is beneficial when resolving misfolded RNA conformations, but could be detrimental to RNAs with low thermodynamic stability. In order to test this idea, we constructed various RNAs with different structural stabilities derived from the thymidylate synthase (td) group I intron and measured the effect of StpA, an Escherichia coli protein with RNA chaperone activity, on their splicing activity in vivo and in vitro. While StpA promotes splicing of the wild-type td intron and of mutants with wild-type-like stability, splicing of mutants with a lower structural stability is reduced in the presence of StpA. In contrast, splicing of an intron mutant, which is not destabilized but which displays a reduced population of correctly folded RNAs, is promoted by StpA. The sensitivity of an RNA towards StpA correlates with its structural stability. By lowering the temperature to 25°C, a temperature at which the structure of these mutants becomes more stable, StpA is again able to stimulate splicing. These observations clearly suggest that the structural stability of an RNA determines whether the RNA chaperone activity of StpA is beneficial to folding.
INTRODUCTION
RNA molecules are synthesized as single-stranded coils and, with the exception of coding sequences, they have to fold into defined 3D structures in order to reach their functional states. The formation of alternative, non-native structures is very frequent due to the abundance of intramolecular sequence complementarities and results in structural promiscuity. The longevity of alternative, non-productive RNA structures significantly slows down folding of RNA molecules to the native state (1–3). As a consequence, it has been postulated that RNA folding requires the aid of proteins or of other molecules with chaperoning activity [reviewed in (4)].
Two types of proteins have been observed to promote RNA folding. Proteins with RNA chaperone activity are thought to interact rather non-specifically with RNA thereby accelerating RNA folding by resolving mispaired structures (3). They comprise a very diverse group of proteins, which have been identified by a variety of functional assays. They have initially been referred to as nucleic acid helix destabilizing proteins (5). RNA chaperone activity has been observed for the ribosomal protein, S12 (6), the HIV nucleocapsid protein, NCp7 (7–9), the RNA binding protein, hnRNP A1 (10,11), the nucleoid associated transcriptional regulators of E.coli StpA and H-NS (12–16), E.coli protein, Hfq (17–19), several proteins of the large ribosomal subunit (20) and the E.coli cold shock proteins, CspA and CspE (21–23), to mention the best studied examples.
On the other hand, there are proteins that bind specifically to a defined RNA molecule, recognizing features of its sequence or tertiary structure and leading to structural stabilization. A well-studied example of this type of protein is the tyrosyl-tRNA synthetase Cyt-18 from Neurospora crassa, which stabilizes the structure of several group I introns (24–28). Recently, a novel type of activity has been demonstrated to accelerate folding of group I introns. The DEAD box helicase Cyt-19 is recruited to the group I intron via the Cyt-18 protein and after binding, this helicase promotes splicing (29).
Heat shocks are detrimental to protein folding, but have probably no deleterious effect on RNA structure or folding, which renature rapidly and efficiently after heating. However, low temperatures are detrimental to RNA folding because the longevity of kinetic folding traps increasingly becomes a problem. Interestingly, the cold shock protein CspA from E.coli has been shown to exert RNA chaperone activity (21), suggesting that this activity is of particular importance at low temperatures. RNA folding has mainly been studied in vitro and it remains to be demonstrated to which extent RNAs misfold within cells (30).
We have previously demonstrated that the RNA chaperone activity observed in vitro is also detectable in vivo. We made use of the dependence of the thymidylate synthase (td) pre-mRNA on translation for efficient folding (31). In the absence of translation, folding of the pre-mRNA is promoted by the E.coli protein StpA (15). StpA promotes folding of the intron RNA by loosening tertiary structure elements which renders them more accessible to the methylating agent dimethyl sulfate (32). We further observed that a mutant intron with a destabilized tertiary structure was sensitive to StpA. This raised the question of whether proteins with RNA chaperone activity have different impacts on RNA folding and whether the effects depend on the structural stability of the RNA molecule. In order to address these questions, we constructed a series of group I intron variants with different structural stabilities and tested the impact of StpA on their splicing activity. We further tested the effect of StpA at 25°C, a temperature at which the RNA structures are more stable, and as a consequence kinetic traps are also more persistent. Since in in vivo assays, the observed effects can be indirect, we also measured the effect of StpA on a destabilized intron mutant in vitro.
MATERIALS AND METHODS
Plasmids, media and strains
All td mutant constructs were generated using the vector pTZ18U/tdP6Δ2 as a template for mutagenesis (33). The mutant tdC873U was previously described in (34), mutant tdC870U in (35), mutants tdU899C, tdG948A, tdG960A, tdU912C and tdG944A in (36). The plasmid encoding the RNA chaperone StpA was obtained from Marlene Belfort and is a pSU20 derivative (12). The plasmid encoding the N.crassa tyrosyl-tRNA-synthetase Cyt-18 was obtained from Alan Lambowitz and is a pACYC184 derivative (24). The plasmid used for cloning and purification of StpA and the respective mutant was pTWIN1 (no. N6951S) from the New England Biolabs IMPACT™-TWIN System. The coding sequences of StpA and G126V-StpA were cloned into the vector as a N-terminal fusion to the Mxe GyrA intein into the NdeI and SapI cloning sites. The td construct used for cis-splicing was cloned into the KpnI and XbaI cloning sites of pTZ18U. The primer sequences for cloning were 5′-GGGGTACCAACGCTCAGTAGATGTTTTC-3′ for the forward and 5′-GCTCTAGAGCATTATGTTCAGATAAGG-3′ for the reverse primer.
TBY-E was prepared from yeast extract and tryptone purchased from Sigma and Difco, respectively. LB was prepared from yeast extract and tryptone was purchased from Difco. The thymine-deficient (thyA-) variant of E.coli strain C600 (F− thr−1 leuB6 thi−1 lacY1 supE44 rfbD1 fhuA21) was used for all experiments.
Preparation of total RNA from E.coli
E.coli strain C600 thyA− was co-transformed with the respective td construct and either the vector pSU20, the StpA or Cyt-18 construct, respectively. Cells were grown in TBY-E medium at 37°C (37) supplemented with thymine to a final concentration of 52 mg/l. Ampicillin (100 μg/ml) and chloramphenicol (25 μg/ml) were added to the growth medium for double selection of plasmids. IPTG was added to a final concentration of 1 mM in order to induce the expression of the td gene and the RNA chaperones and Cyt-18. Results were reproduced for most td intron mutants using LB as a growth medium except for base triple mutants, which yield significantly lower levels of td RNA when grown in LB. For RNA preparation, bacterial growth medium was inoculated with an overnight culture in the ratio of 1:100 cells and were harvested at an OD 600 nm of 0.2–0.3, and total RNA was prepared as described previously (31). The experiments comparing splicing at different temperatures (25°C and 37°C) were performed using LB as a growth medium.
In vivo splicing assay
Total RNA was subjected to reverse transcription in a primer extension reaction using the oligonucleotide NBS2 (5′-GACGCAATATTAAACGGT-3′) complementary to a td exon 2 sequence 2 nt downstream of the 3′ splice site. In addition the reaction contains dATP, dCTP and dGTP, and ddTTP, allowing discrimination between different splicing products on the basis of the extension product. The primer extension products were quantified in order to determine the splicing activity (15). The percentage of splicing was calculated in the following way: % splicing = [(mRNA + cryptic-mRNA)/(pre-mRNA + cryptic-mRNA + mRNA)] × 100.
In vitro transcription of td intron RNA for spectroscopic analysis
The td intron RNA was transcribed in vitro from PCR products (1–2 μg) amplified from plasmid tdΔP6-2 wild-type or mutant constructs with the following primers (150 pmoles): tdJ23-long: 5′-CGTAATACGACTCACTATAGAATCTATCTAAACGGGGAAC-3′ and tdP9.2-long: 5′-TGTTCAGATAAGGTCGTTAATC-3′ (T7 promoter underlined). The reaction mixture contained 5 mM of each NTP, 10 mM DTT and 150 U of T7 RNA polymerase in 100 μl of a solution of 40 mM Tris–HCl (pH 6.9), 52 mM MgCl2 and 6 mM spermidine. After incubation for 4 h at 37°C, the DNA template was digested with 10 U of RNase-free DNaseI for 30 min at 37°C. The reaction was stopped by the addition of EDTA to a final concentration of 50 mM. All transcripts were precipitated and then purified on an 8% acrylamide-urea gel. The correct band was cut out and the RNA was eluted with a buffer containing 300 mM sodium acetate 10 mM Tris (pH 7.0) and 2 mM ETDA for 1.5 h at 65°C. The filtered supernatant was precipitated with EtOHabs. The dry pellets were resuspended in water and the RNA concentrations were calculated from absorbance at 260 nm.
Purification of StpA wild-type and mutant proteins
The fusion proteins (StpA-intein, G126V-StpA-intein) were overexpressed in BL21 (DE3) E.coli cells and the lysate was loaded onto a chitin column for affinity purification. Protein splicing was induced by increasing the DTT concentration or by pH shift, respectively. All the steps of protein purification were performed according to the protocol (New England Biolabs IMPACT™-TWIN System). Finally the proteins were stored in a buffer containing 50 mM Tris–HCl (pH 7.5), 500 mM NaCl, 1 mM EDTA, 0.5 mM DTT and 12% glycerol (12).
In vitro transcription for cis-splicing assay
For the cis-splicing assay the plasmid was linearized with XbaI. Ten micrograms of the linearized plasmid were transcribed under non-splicing conditions to prevent splicing of the precursor RNA. The RNA was transcribed with 40 mM Tris–HCl (pH 7.5), 2 mM spermidine, 6 mM MgCl2, 10 mM NaCl, 20 mM DTT, 3 mM ATP, 3 mM GTP, 3 mM CTP, 1 mM UTP, 30 μCi [α-35S]UTP and 150 U of T7 RNA polymerase. The low Mg2+ concentration and the reaction temperature of 22°C were suboptimal in order to prevent splicing reactions of the precursor RNA. The reaction was performed overnight and the resulting products were purified using a 5% polyacrylamide gel.
In vitro cis-splicing assay for RNA chaperone activity
35S-labelled precursor RNA (0.5 pmol) was denatured for 1 min at 95°C and cooled to 37°C. Splicing buffer containing 50 mM Tris–HCl (pH 7.3), 0.4 mM spermidine and 5 mM MgCl2 was added followed by the respective protein with a final concentration of 1.4 μM. Splicing was induced immediately by addition of 0.5 mM GTP. Aliquots of 5 μl were taken after the following time points (15, 30 and 45 s, and 1, 2, 5, 10, 30 and 60 min). The reactions were stopped by the addition of 5 μl RNA loading buffer containing 7 M urea and 1 mM EDTA. The samples were denatured for 1 min at 95°C prior to loading onto 5% polyacrylamide gels. The gels were quantified using a PhosphoImager and the ImageQuant software. Reaction constants and graphs were calculated using the KaleidaGraph software (Synergy Software) and the data were fit to a first order equation with a double exponential: fraction pre-mRNA remaining = .
UV melting curves
The RNA probes (1 μg/μl) were denatured at 95°C and allowed to cool slowly to room temperature. The absorbance was measured at 260 nm on a Cary 100 Bio UV-Spectrophotometer in a buffer containing 50 mM sodium cacodylate, 50 mM ammonium chloride and 5 mM magnesium chloride. The heating rate was 0.5°C/min. Data were collected from 30 to 75°C at 0.5°C intervals. Derivatives were calculated from a local least square fit using a second order polynome with a program developed in the lab (available on request from N. Piganeau, N. Windbichler). For determination of the melting temperatures two Gaussian curves were fitted to the values, one for the melting of the 3D structure and the second for the transition of the secondary structure. The mean of the respective Gaussian distribution was taken as the melting temperature.
RESULTS
Splicing activity of td intron variants with mutations in tertiary structure elements
The td intron contains secondary structure elements, stems P1, P2, P4-P6, P7.1, P7.2, P8, P9, P9.1 and P9.2 and a series of tertiary interactions (Figure 1). Stems P3 and P7 form the tertiary structure, as they constitute long-range base paired elements, which fold late and are referred to as pseudoknots (38). Other important 3D structural elements, responsible for the formation of the overall tertiary structure, are the base triple interactions formed between stem P6 and joining region J3/4 or stem P4 and joining region J6/7, respectively. Peripheral long-range interactions occur between loops and stems (P5–L9, L2–P8) or loops and loops (L7.2–L9.2). It has been suggested that the internal loop E motif located between stems P7.1 and P7.2 assists in orienting stems P7 and P3 (39). Mutations introduced into the td intron in order to disturb these tertiary structure elements are shown in Figure 1, their in vivo splicing activities were measured using a primer extension assay (Figure 1B) and they are summarized in Table 1. The structural stabilities of the individual mutants were determined via UV melting and are given in Table 1. It was previously shown that, in general, the thermodynamic stability of intron mutants measured in vitro correlates well with their in vivo splicing activity (36).
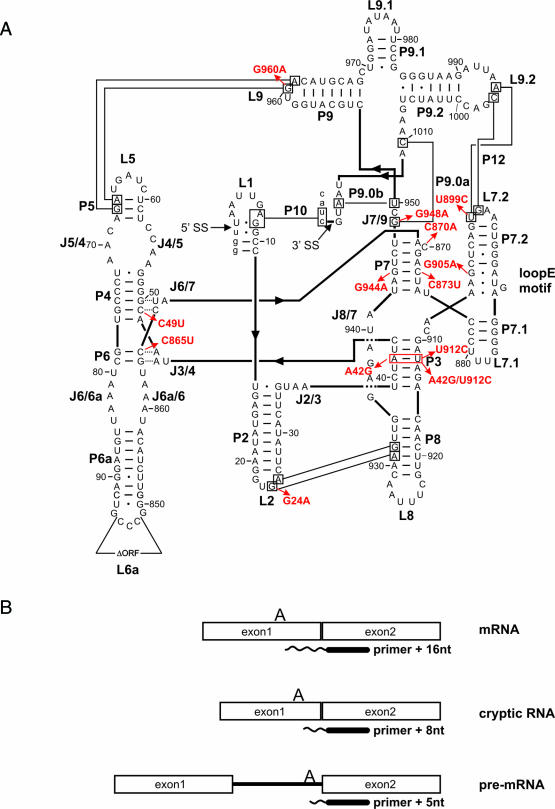
Figure 1.
(A) Structural model of the td intron based on phylogeny and biochemical data (39,47). Base-paired elements are termed P1 to P10, joining regions between stems are termed J2/3–J8/7, and hairpin loops are numbered L1–L9.2. Thin black lines indicate long-range tertiary interactions. The splice sites (5′ and 3′ SS) are marked with an arrow. The deletion of the intron open reading frame (ORF) is also indicated in loop L6a. The intron sequence is displayed in upper case letters and exon sequences are shown as lower case letters. Arrows indicate the td mutants investigated in this work. (B) Schematic representation of the primer extension assay: reverse transcription products obtained from mRNA, cryptic RNA and pre-mRNA. Cryptic RNA results from an alternatively folded 5′ splice site (48,49).
Table 1.
Splicing activity of td intron mutants with a destabilized tertiary structure in the absence and presence of StpA and Cyt-18
| td mutant | 3D structural element | Tm of 3D structure (°C) | % Splicing | ||
|---|---|---|---|---|---|
| No protein | +StpA | +Cyt-18 | |||
| Wt | — | 56.2 | 63.8 ± 8.7 | 64.9 ± 8.6 | — |
| C865U | Base triples | 56a | 24.2 ± 8.4 | 11.8 ± 3.7 | 61.2 ± 8.3 |
| C49U | Base triples | N.D. | 21.7 ± 5.9 | 11.1 ± 2.4 | 60.1 ± 5.3 |
| A42G | P3 (G–U base pair) | 53.5 | 64.8 ± 10.0 | 52.8 ± 5.7 | 68.9b |
| U912C | P3 (A–C mismatch) | 41.6 | 24.7 ± 6.4 | 11.7 ± 3.0 | 62.9 ± 2.5 |
| A42G/U912C | P3 (G–C base pair) | 55.1 | 70.1 ± 5.7 | 67.4 ± 9.6 | 62.1 ± 9.3 |
| C870A | P7 (bulge mutant) | N.D. | 30.3 ± 9.8 | 23.1 ± 9.5 | 61.2 ± 7.6 |
| C873U | P7 (G–U base pair) | ** | 8.0 ± 2.5 | 2.4 ± 0.9 | 22.7 ± 3.3 |
| G944Ab | P7 (A–C mismatch) | ** | 6.0 | 1.6 | 8.3 |
| G24A | L2–P8 | N.D. | 60.7 ± 13.0 | 62.8 ± 4.6 | 64.2 ± 10.7 |
| U899C | P12 | 51.2 | 37.9 ± 14.3 | 36.5 ± 13.6 | 45.6 ± 13.9 |
| G905A | Loop E | 54.6 | 22.9 ± 6.0 | 35.4 ± 7.5 | 46.9 ± 7.9 |
| G948A | P9.0a | 49.1 | 31.4 ± 11.2 | 30.2 ± 6.3 | 34.7 ± 7.1 |
| G960A | L9–P5 | 53.3 | 53.2 ± 17.7 | 59.2 ± 3.0 | 52.9 ± 2.3 |
Splicing activity is indicated as % mRNA splicing ± standard deviation (SD). **no visible transition in the UV melting profile corresponding to tertiary structure melting; N.D.: not determined; a the majority of the molecules melt between 35°C and 40°C and a very small amount melts at 56°C. b only two experiments, SD not determined. Splicing values for the wild type are mean values of all experiments.
We introduced mutations into the base triples between stem P6 and joining region J3/4 (C865U) and between stem P4 and J6/7 (C49U). These base triples are crucial for the correct orientation of the major folding domains P4–P6 and P3–P8. The mutations were chosen in such a way that GC pairs were altered to GU pairs in order to avoid disruption of stems P6 and P4. The activity of these mutants is reduced to ∼24 and 22% splicing, respectively (Table 1).
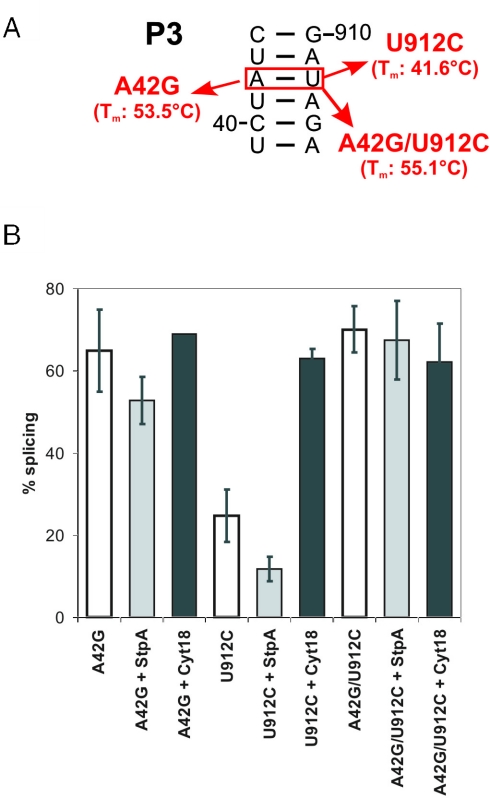
In stem P3 we introduced one mutation on either side of the helix, destabilizing the element to various extents. In the tdA42G mutant, an AU pair is altered to a GU pair, affecting splicing only marginally. The tdU912C mutation, however, results in an AC mismatch in P3 and reduces the splicing activity to 25%. The compensatory mutation tdA42G/U912C stabilizes P3 and restores wild-type splicing levels.
Mutations that disrupt stem P7 are more detrimental to splicing than mutations disturbing the base triples or disrupting stem P3, showing <10% splicing activity. The C870A mutation was used because it has an alternative pairing option with U947. The C870 is part of the guanosine binding pocket and forms a base triple with G871 (40). This mutant shows 30% splicing activity.
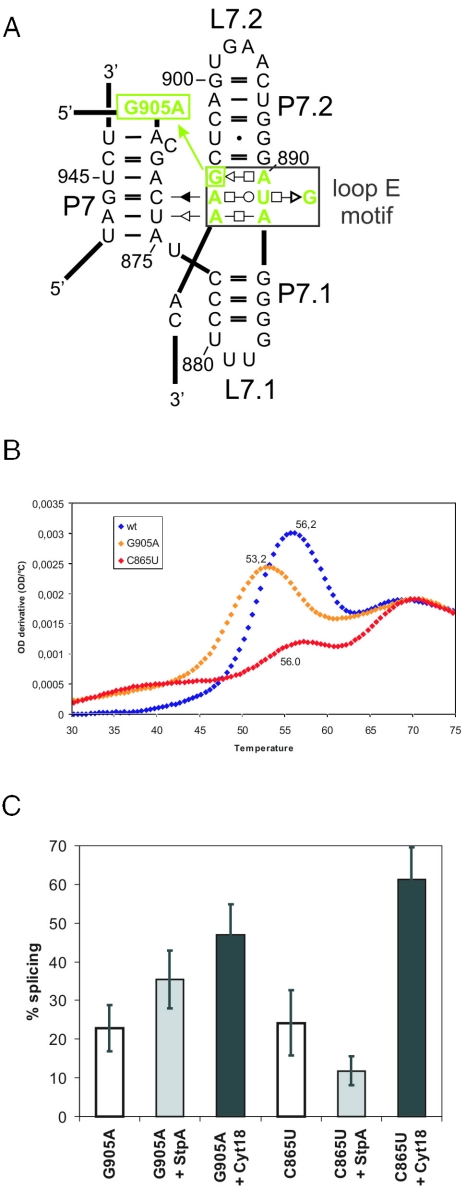
Peripheral long-range tertiary interactions were disrupted in mutants tdG24A, tdU899C, td948A and tdG960A resulting in partially destabilized tertiary structures. Their effect on splicing is moderate (Table 1). Finally, a mutation was introduced to perturb the loop E motif in the peripheral extension P7.1/P7.2, tdG905A, which has 23% splicing activity.
The impact of StpA on splicing of td intron mutants depends on their structural stability
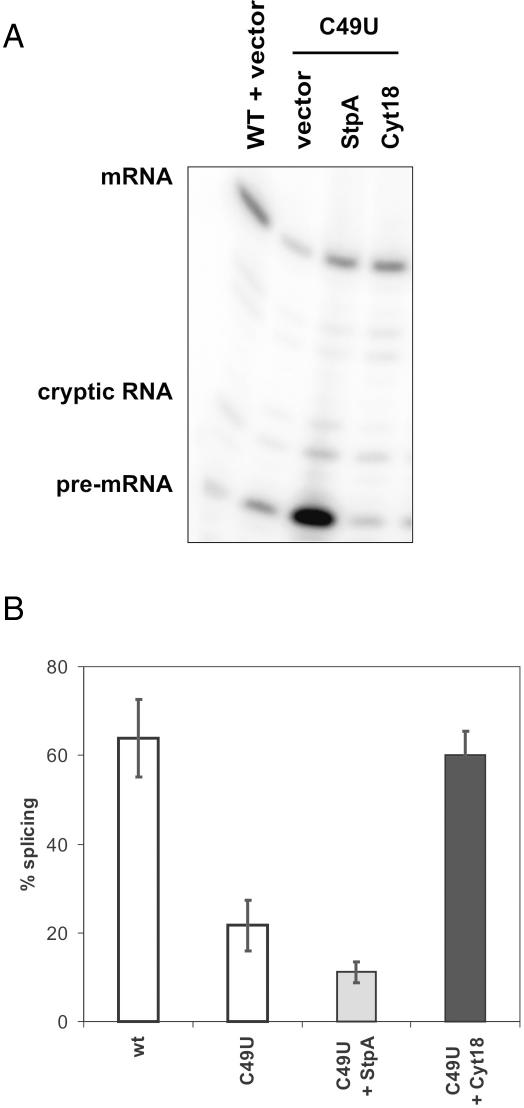
In order to investigate the impact of the RNA chaperone activity of StpA on splicing of td intron mutants with a destabilized tertiary structure, the td mutants were co-expressed with StpA, an empty vector control or the group I intron specific splicing factor, Cyt-18. Representative experiments are depicted in Figures 2–4 and the results are summarized in Table 1. As had been observed previously, the tdC865U mutant, which weakens the base triple interactions between P6 and J3/4, is sensitive to StpA but rescued by Cyt-18 (32). The same holds true for the analogous mutant tdC49U, which weakens an equivalent interaction between stem P4 and joining region J6/7, suggesting that both base triple interactions contribute equally to the structure of the intron and that the StpA sensitivity is not limited to the tdC865U mutation (Figure 2).
Figure 2.
Splicing of the base triple mutant tdC49U is sensitive to StpA. (A) Splicing of the base triple mutant tdC49U in the absence and presence of StpA and Cyt-18, respectively. Gel sample showing reverse transcription products corresponding to mRNA, cryptic RNA and pre-mRNA. (B) Quantification of four independent experiments. Values are given as (%) mRNA splicing and were calculated as described in Materials and Methods.
Figure 4.
Splicing of a loop E mutant is increased in the presence of StpA. (A) Schematic representation of the loop E motif using the base pair annotation described in (50). The loop E mutant G905A is indicated. (B) First derivative of a UV absorbance profile for melting of RNA tertiary and secondary structures due to increasing temperature is shown for wild-type td intron (in blue), the tdG905A mutant in the loop E motif (in yellow) and the tdC865U mutant in P6 (in red). (C) Splicing of the loop E mutants tdG905A and tdC865U in the absence and presence of StpA and Cyt-18, respectively. Quantification of four independent experiments.
Nucleotide changes which weaken stems P3 and P7 result in td mutants, whose splicing activity is also reduced in the presence of StpA. This sensitivity correlates with the structural stability of the mutants as determined by UV melting experiments (Table 1). For example, the tdU912C mutant in P3, whose melting temperature (Tm) is 14°C lower than that of wt, shows a splicing activity of ∼25%, which is reduced more than 2-fold in the presence of StpA. The tdA42G mutant, which is only slightly affected in splicing and whose Tm is 53.5°C compared to the wild-type's Tm of 55.6°C, is only weakly sensitive to StpA. The tdA42G/U912C double mutant displays wild-type activity and is not sensitive to StpA (Figure 3). Mutations disrupting P7 have a strong effect on splicing and they are also highly sensitive to StpA. The splicing activity of the tdC873U mutant is reduced 3-fold and the tdC870A mutant is reduced from 31 to 23% in the presence of StpA (Table 1).
Figure 3.
Splicing of mutants with a destabilized stem P3 is sensitive to StpA. (A) Schematic representation of investigated mutants. Notice that the mutation tdU912C causes a mismatch-destabilizing stem P3, whereas the double mutant tdA42G/U912C results in a stabilized P3 element which is also indicated by the Tm of the intron structure. (B) Splicing of mutants in stem P3 was assayed in the absence and presence of StpA and Cyt-18, respectively. Quantification of three independent experiments.
Mutants with weakened long-range tertiary interactions, tdG24A, tdU899C and tdG960A, do not display sensitivity to StpA, probably because the loss of a single long-range pairing does not considerably destabilize the tertiary structure. These mutants are also not efficiently rescued by Cyt-18 suggesting that the lost interaction cannot be compensated for by the binding of Cyt-18. These long-range interactions have previously been shown to exert cooperative effects on the structural stability (41,42), and these regions are not part of the primary Cyt-18 binding site (43).
Most interesting is the observation that the only mutant that is stimulated by StpA is the tdG905A mutant in the loop E motif, which joins stems P7.1 and P7.2 (Figure 4). The loop E motif is often found in complex RNA structures and is thought to be involved in the higher order structural organization of domains (39). In the td intron, the loop E motif contacts stem P7 and is responsible for a late step in the folding pathway (44). Figure 4B shows the first derivative of the UV melting profile of the loop E motif mutant compared to those of the wild-type and the base triple mutant tdC865U. The wild-type td intron displays two melting transitions, a low temperature transition at 56.2°C corresponding to the melting of the tertiary structure and a high temperature transition corresponding to the melting of the secondary structure (36,42). The melting transition of the tertiary structure in the tdG905A mutant is at 53.2°C, almost as stable as the wild type, but the reduced height of the peak is indicative of a lower population of correctly folded molecules. This suggests that the structural phenotype of the tdG905A mutant might be a folding defect rather than a structural destabilization (C. Waldsich and R. Schroeder, unpublished work). In contrast, the tdC865U mutant displays a profile characteristic of molecules with destabilized structures, because the tertiary structure already melts between 35 and 40°C. A small peak is detectable at 56°C, which has not been assigned to a specific structural transition.
When comparing the effect of StpA on the splicing activity of different mutant td introns, it is clear that the structural stability of the mutants determines whether the RNA chaperone activities aid or impede the folding process. The sensitivity towards StpA correlates well with changes in Tm, thus with the structural destabilization. Splicing of mutants with strongly destabilized tertiary structures is further reduced in the presence of StpA, whereas splicing of the wild-type and mutant introns with stable structures is inert or further promoted by StpA.
StpA promotes splicing of destabilized mutants at 25°C
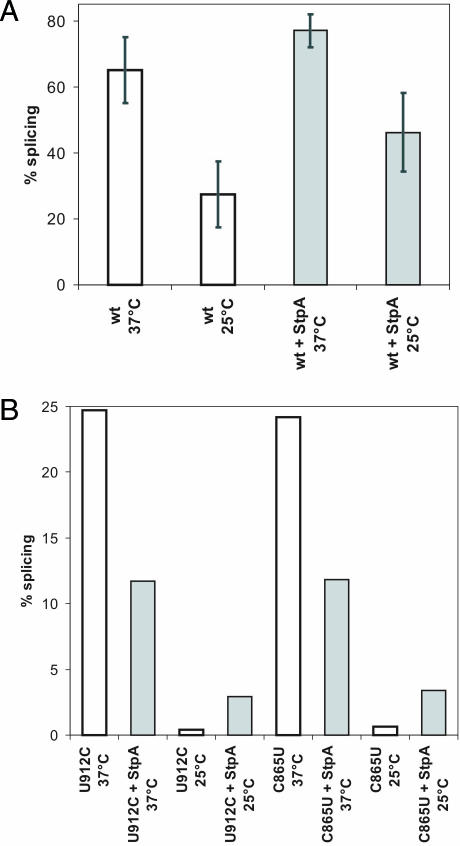
The wild-type intron construct splices to ∼70% at optimal conditions when expressed from a plasmid and is further stimulated by the presence of StpA to ∼80%. We rationalized that by lowering the temperature, folding might be slower due to the structural stabilization of non-productive intermediates and that proteins with RNA chaperone activity might be of increasing importance for RNA folding in the cold. Figure 5A shows the splicing activity of a wild-type td construct at 37 and 25°C. The drop in temperature has a strong effect on the splicing activity, which decreases from 77 to 32%. This reduction might be due to a decrease in the folding rate and to a slowing down of the chemical step. We asked whether StpA improves wild-type td splicing at 25°C. For this purpose we co-expressed the td gene with StpA at 25 and 37°C. StpA was indeed able to promote splicing of the wild-type td intron at 25°C, suggesting that RNA chaperones can counteract RNA misfolding at low temperatures.
Figure 5.
StpA can act as RNA chaperone and rescue destabilized td intron mutants at low temperature. (A) Splicing of wild-type td intron at either 37°C or 25°C in the absence or presence of StpA. Quantification of eight independent experiments. (B) Splicing activity of mutants U912C and C865U in the absence or presence of StpA at 25°C compared to the activity at 37°C. Quantification of two independent experiments.
Given that the structural stability determines whether an RNA chaperone is beneficial or detrimental to folding, we rationalized that by lowering the temperature and thereby stabilizing the RNA structure, the RNA chaperone activity of StpA could become more beneficial and less detrimental to structurally compromised intron mutants. In order to test this idea, we measured the effect of StpA on the splicing activity of two structurally destabilized mutants, tdC865U in stem P6 and tdU912C in stem P3 at 25°C. Both mutants are highly sensitive to the presence of StpA at 37°C. Figure 5B shows that the splicing activity of these mutants is very low at 25°C and that StpA indeed promotes rather than decrease splicing as it does at 37°C. These results provide further evidence that the structural stability of the RNA influences the effect of StpA on folding.
In vitro splicing of the C865U mutant is sensitive to an StpA mutant with increased RNA chaperone activity
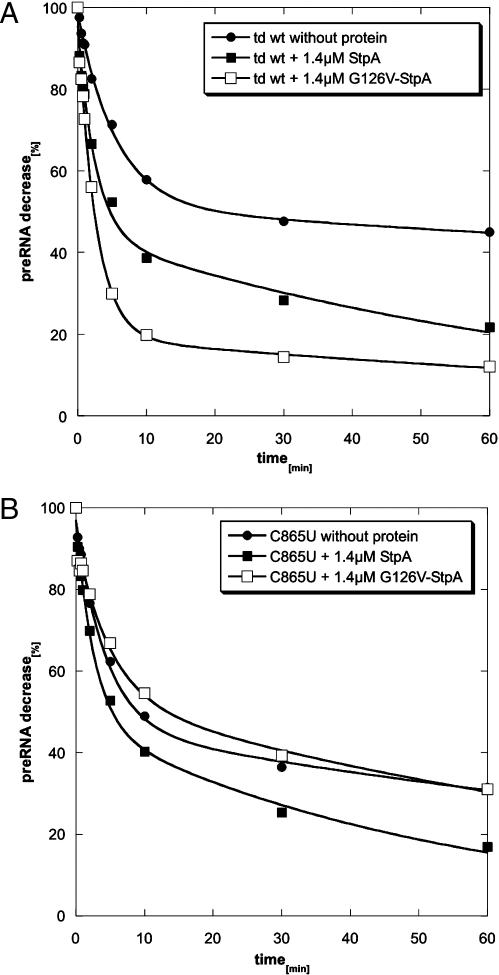
All the above effects of StpA on the various td intron mutants were measured in vivo. In vivo effects can be indirect and caused by other factors, which might be influenced by StpA. In order to ensure that the effects we observed are really due to the RNA chaperone activity of StpA, we tested its effect on the wild-type and the C865U mutant in vitro. A cis-splicing assay was used that was established to enable quantitative assessment of RNA chaperone activity (16). For this purpose a pre-RNA construct was used, which is highly impaired in folding. This construct contains 27 nt of exon 1 and only 2 nt of exon 2. When incubated for 1 min at 95°C and with subsequent folding at 37°C in the presence of 5 mM MgCl2, only 40–50% of the population of RNA molecules reach their native structure and splice rapidly with a kobs of 0.2 min−1. 50–60% of the population fold slowly displaying a kobs of 0.2 × 10−2 min−1 (Figure 6A, Table 2). Addition of StpA to this reaction promotes folding of the wild type such that 60% reach the native state and splice with a kobs of 0.4 min−1 (Figure 6A, Table 2). The tdC865U mutant in the short exon context, splices similarly to the wild type under these conditions, but is less stimulated by StpA (Figure 6B, Table 2). We also used a variant of StpA, which contains a point mutation in the nucleic acid binding domain, StpA-G126V, which has a higher RNA chaperone activity than wild-type StpA (O. Mayer, R. Grossberger and R. Schroeder, manuscript in preparation). While this StpA variant strongly stimulates splicing of the wild-type td intron, it has a slightly negative effect on splicing of the tdC865U mutant (Figure 6, Table 2). As mentioned before, the tdC865U mutant has a reduced structural stability. These results confirm our in vivo observations and clearly demonstrate that the effect we observe in vivo is reproducible in vitro and that RNA molecules with reduced structural stability are inhibited in their folding process by the RNA chaperone activity of StpA.
Figure 6.
In vitro splicing assays of wt and mutant pre-mRNA in the absence and presence of wild-type StpA and mutant G126V-StpA. Shown is the decrease of pre-mRNA with time. The indicated time points are 15, 30 and 45 s, and 1, 2, 5, 10, 30 and 60 min. The reactions were performed in the presence of 5 mM MgCl2, 0.5 mM GTP, 50 mM Tris–HCl (pH 7.3) and 0.4 mM spermidine (see Materials and Methods). (A) Splicing of the td wt intron pre-RNA without added protein (full circles), with the addition of 1.4 μM wild-type StpA (full squares) or 1.4 μM G126V-StpA mutant (empty squares). (B) Splicing of the td C865U pre-RNA intron mutant without protein (full circles), with addition of 1.4 μM wild-type StpA (full squares) or 1.4 μM G126V-StpA mutant (empty squares). The graphs were calculated according to the equation given in Materials and Methods.
Table 2.
Reaction constants gained from cis-splicing assays using td wt and tdC865U mutant RNAs in the presence and absence of StpA or StpA-G126V, respectively
| Wt short exon construct | C865U short exon construct | |||||||
|---|---|---|---|---|---|---|---|---|
| Fast reacting | Slow reacting | Fast reacting | Slow reacting | |||||
| % | kobs (min−1) | % | kobs (min−1) | % | kobs (min−1) | % | kobs (min−1) | |
| No protein | 47 ± 4 | 0.2 ± 2 × 10−2 | 53 ± 4 | 0.2 × 10−2 ± 1 × 10−3 | 50 ± 4 | 0.2 ± 3 × 10−2 | 50 ± 4 | 0.7 × 10−2 ± 2 × 10−3 |
| 1.4 μM StpA | 60 ± 4 | 0.4 ± 8 × 10−2 | 40 ± 4 | 1.0 × 10−2 ± 3 × 10−3 | 50 ± 3 | 0.3 ± 6 × 10−2 | 50 ± 4 | 1.0 × 10−2 ± 2 × 10−3 |
| 1.4 μM G126V-StpA | 77 ± 6 | 0.4 ± 3 × 10−2 | 23 ± 4 | 0.8 × 10−2 ± 4 × 10−3 | 40 ± 6 | 0.2 ± 7 × 10−2 | 60 ± 4 | 0.9 × 10−2 ± 4 × 10−3 |
The fractions of fast or slow reacting RNA molecules with the corresponding reaction rates are indicated. The reaction constants were calculated using the equation given in Materials and Methods.
DISCUSSION
Splicing of the td group I intron requires folding of the pre-mRNA into its native tertiary structure. The ribosome promotes folding of the pre-mRNA by resolving non-productive exon–intron interactions (31). In the absence of translation, folding can alternatively be aided by the group I intron specific splicing factor Cyt-18, or by StpA, a protein that exerts RNA chaperone activity (12,15,24). However, the two proteins assist intron folding in opposite ways. While Cyt-18 recognizes and binds the intron RNA specifically, thereby stabilizing its structure, StpA unfolds the intron RNA giving it another chance to fold correctly (32). The RNA unfolding activity of StpA is beneficial when resolving misfolded RNA conformations but could be detrimental to RNAs with compromised structural stability. We addressed this question by measuring the influence of StpA on the splicing activity of a series of mutant td introns with a reduced structural stability. We had previously observed that StpA increased the accessibility of bases involved in tertiary interactions, but not of those involved in secondary structure elements (32). Since the tertiary structure interactions of the td intron contribute to the overall stability of an RNA molecule and are less stable than secondary structure elements, we hypothesized that the structural stability of the target RNA dictates whether the RNA chaperone activity of StpA is beneficial or detrimental to RNA folding and in turn to splicing. Therefore we disrupted or destabilized tertiary interactions and analyzed the extent to which StpA affected the splicing activity of these mutants.
Correlation between structural stability and sensitivity towards StpA
Splicing of mutant introns, which are structurally destabilized, is sensitive to StpA. Importantly, there is a correlation between the extent to which structural stability is reduced and the extent to which StpA affects the splicing activity. For example, changing an AU to a GU base pair in stem P3 results in a very low sensitivity towards StpA, whereas introducing an AC mismatch at the same position displays a high sensitivity. A GC pair at this position, however, does not induce sensitivity. We conclude from these results that when stem P3 is only slightly destabilized, the RNA chaperone activity of StpA unfolds P3 only to some extent. However, upon destabilizing stem P3 by a mismatch, StpA further unfolds P3 and as a consequence further reduces splicing. Similar effects were observed for highly destabilized mutants with the exception of those that have weakened peripheral tertiary long-range interactions. In these mutants a single tertiary interaction is altered without significantly destabilizing the entire intron structure (42).
Notably, we are measuring the splicing activity of the intron RNA and are deducing effects on RNA folding from changes in the activity. We are confident that this is an acceptable approach because the chemical step is much faster than folding, which is rate-limiting (45). Furthermore, we had previously observed that for the td group I intron, in vivo splicing activity correlates well with structural stability (36,42). Since we measure activity and do not determine the folding state, we cannot distinguish between kinetic and thermodynamic effects of StpA on the intron RNA. StpA might equally well resolve P3 in all constructs, but since folding and, as a consequence, splicing are slowed down in the structurally destabilized mutants, the RNA chaperone activity might in those cases be faster than the splicing activity. Also, in vitro the unfolding activity of StpA on the wild-type intron RNA is slower than the chemical step (16). We further confirmed our in vivo observations by an in vitro splicing assay. At the reaction conditions (5 mM Mg2+) used, the tdC865U mutant splices as well as the wild type, because the increased magnesium rescues splicing activity. The wild-type StpA protein can still promote folding of this intron mutant, although less efficiently. The sensitivity towards the RNA chaperone activity is only observed when a variant of StpA is used which has a stronger RNA chaperone activity. We suggest that this is due to the different ionic conditions required for efficient in vitro splicing. These results nevertheless clearly demonstrate that the RNA chaperone activity of proteins can be detrimental to folding of RNAs with reduced structural stability.
Misfolded molecules are rescued by StpA
In contrast to all structurally destabilized mutants, splicing of the tdG905A mutant which represents an alteration in the loop E motif that joins stems P7.1 and P7.2, is stimulated by StpA. We propose that perturbation of the loop E motif facilitates misfolding of the intron core without destabilizing structural elements. StpA can then assist in folding by resolving misfolded structures and enabling the molecules to fold correctly. This is also consistent with the view that the loop E motif serves as a structural scaffold for folding (46). These results are also in agreement with our previous observations that mutants containing non-sense codons in the upstream exon, which prevent the ribosome from translating the pre-mRNA and resolving non-productive exon–intron interactions, are impaired in splicing but, like the loop E motif mutant, rescued by StpA (15).
Furthermore, we showed that splicing of the wild-type td intron is inefficient at 25°C and that StpA also promotes splicing in vivo in the cold. The potential of StpA to rescue folding and therefore splicing at low temperatures might reflect an important functional aspect of RNA chaperones in general and supports the hypothesis that RNA chaperones could be of particular biological significance for the RNA metabolism in organisms subjected to temperature drops. These findings demonstrate that at low temperatures the RNA folding problem is of increasing significance requiring efficient RNA chaperone activity. This aspect of RNA folding needs investigation and might help clarify the general task of proteins with RNA chaperone activity.
Altogether, our data demonstrate that StpA acts by destabilizing structural elements of the intron RNA, which are partially open or not compactly folded both in vivo and in vitro. StpA and probably many other proteins with RNA chaperone activity might play a significant role in the folding of RNA molecules in the cell by acting as quality control agents for RNA conformation.
Acknowledgments
We thank N. Piganeau and N. Windbichler for help with UV melting data processing. The help of Paul Watson with the preparation of the manuscript is greatly appreciated. We thank all the members from our laboratory for constant discussions and for comments on the manuscript. This project was funded by the Austrian Science Fund FWF, grants nos. F1703 and F16026. Funding to pay the Open Access publication charges for this article was provided by FWF.
Conflict of interest statement. None declared.
REFERENCES
- 1.Treiber D.K., Williamson J.R. Beyond kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 2001;11:309–314. doi: 10.1016/s0959-440x(00)00206-2. [DOI] [PubMed] [Google Scholar]
- 2.Woodson S.A. Recent insights on RNA folding mechanisms from catalytic RNA. Cell Mol. Life Sci. 2000;57:796–808. doi: 10.1007/s000180050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herschlag D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 4.Schroeder R., Barta A., Semrad K. Strategies for RNA folding and assembly. Nat. Rev. Mol. Cell. Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- 5.Karpel R.L., Miller N.S., Fresco J.R. Mechanistic studies of ribonucleic acid renaturation by a helix-destabilizing protein. Biochemistry. 1982;21:2102–2108. doi: 10.1021/bi00538a019. [DOI] [PubMed] [Google Scholar]
- 6.Coetzee T., Herschlag D., Belfort M. Escherichia coli proteins, including ribosomal protein S12, facilitate in vitro splicing of phage T4 introns by acting as RNA chaperones. Genes Dev. 1994;8:1575–1588. doi: 10.1101/gad.8.13.1575. [DOI] [PubMed] [Google Scholar]
- 7.Tsuchihashi Z., Khosla M., Herschlag D. Protein enhancement of hammerhead ribozyme catalysis. Science. 1993;262:99–102. doi: 10.1126/science.7692597. [DOI] [PubMed] [Google Scholar]
- 8.Urbaneja M.A., Wu M., Casas-Finet J.R., Karpel R.L. HIV-1 nucleocapsid protein as a nucleic acid chaperone: spectroscopic study of its helix-destabilizing properties, structural binding specificity, and annealing activity. J. Mol. Biol. 2002;318:749–764. doi: 10.1016/S0022-2836(02)00043-8. [DOI] [PubMed] [Google Scholar]
- 9.Windbichler N., Werner M., Schroeder R. Kissing complex-mediated dimerisation of HIV-1 RNA: coupling extended duplex formation to ribozyme cleavage. Nucleic Acids Res. 2003;31:6419–6427. doi: 10.1093/nar/gkg873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pontius B.W., Berg P. Rapid assembly and disassembly of complementary DNA strands through an equilibrium intermediate state mediated by A1 hnRNP protein. J. Biol. Chem. 1992;267:13815–13818. [PubMed] [Google Scholar]
- 11.Herschlag D., Khosla M., Tsuchihashi Z., Karpel R.L. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 1994;13:2913–2924. doi: 10.1002/j.1460-2075.1994.tb06586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang A., Derbyshire V., Salvo J.L., Belfort M. Escherichia coli protein StpA stimulates self-splicing by promoting RNA assembly in vitro. RNA. 1995;1:783–793. [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang A., Rimsky S., Reaban M.E., Buc H., Belfort M. Escherichia coli protein analogs StpA and H-NS: regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J. 1996;15:1340–1349. [PMC free article] [PubMed] [Google Scholar]
- 14.Cusick M.E., Belfort M. Domain structure and RNA annealing activity of the Escherichia coli regulatory protein StpA. Mol. Microbiol. 1998;28:847–857. doi: 10.1046/j.1365-2958.1998.00848.x. [DOI] [PubMed] [Google Scholar]
- 15.Clodi E., Semrad K., Schroeder R. Assaying RNA chaperone activity in vivo using a novel RNA folding trap. EMBO J. 1999;18:3776–3782. doi: 10.1093/emboj/18.13.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayer O., Waldsich C., Grossberger R., Schroeder R. Folding of the td pre-RNA with the help of the RNA chaperone StpA. Biochem. Soc. Trans. 2002;30:1175–1180. doi: 10.1042/bst0301175. [DOI] [PubMed] [Google Scholar]
- 17.Zhang A., Wassarman K.M., Ortega J., Steven A.C., Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 18.Moller T., Franch T., Hojrup P., Keene D.R., Bachinger H.P., Brennan R.G., Valentin-Hansen P. Hfq: a bacterial Sm-like protein that mediates RNA–RNA interaction. Mol. Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 19.Moll I., Leitsch D., Steinhauser T., Blasi U. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep. 2003;4:284–289. doi: 10.1038/sj.embor.embor772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Semrad K., Green R., Schroeder R. RNA chaperone activity of large ribosomal subunit proteins from Escherichia coli. RNA. 2004;10:1855–1860. doi: 10.1261/rna.7121704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang W., Hou Y., Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 22.Phadtare S., Inouye M., Severinov K. The nucleic acid melting activity of Escherichia coli CspE is critical for transcription antitermination and cold acclimation of cells. J. Biol. Chem. 2002;277:7239–7245. doi: 10.1074/jbc.M111496200. [DOI] [PubMed] [Google Scholar]
- 23.Phadtare S., Tyagi S., Inouye M., Severinov K. Three amino acids in Escherichia coli CspE surface-exposed aromatic patch are critical for nucleic acid melting activity leading to transcription antitermination and cold acclimation of cells. J. Biol. Chem. 2002;277:46706–46711. doi: 10.1074/jbc.M208118200. [DOI] [PubMed] [Google Scholar]
- 24.Mohr G., Zhang A., Gianelos J.A., Belfort M., Lambowitz A.M. The neurospora CYT-18 protein suppresses defects in the phage T4 td intron by stabilizing the catalytically active structure of the intron core. Cell. 1992;69:483–494. doi: 10.1016/0092-8674(92)90449-m. [DOI] [PubMed] [Google Scholar]
- 25.Mohr G., Caprara M.G., Guo Q., Lambowitz A.M. A tyrosyl-tRNA synthetase can function similarly to an RNA structure in the tetrahymena ribozyme. Nature. 1994;370:147–150. doi: 10.1038/370147a0. [DOI] [PubMed] [Google Scholar]
- 26.Guo Q., Lambowitz A.M. A tyrosyl-tRNA synthetase binds specifically to the group I intron catalytic core. Genes Dev. 1992;6:1357–1372. doi: 10.1101/gad.6.8.1357. [DOI] [PubMed] [Google Scholar]
- 27.Myers C.A., Kuhla B., Cusack S., Lambowitz A.M. tRNA-like recognition of group I introns by a tyrosyl-tRNA synthetase. Proc. Natl Acad. Sci. USA. 2002;99:2630–2635. doi: 10.1073/pnas.052596299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caprara M.G., Lehnert V., Lambowitz A.M., Westhof E. A tyrosyl-tRNA synthetase recognizes a conserved tRNA-like structural motif in the group I intron catalytic core. Cell. 1996;87:1135–1145. doi: 10.1016/s0092-8674(00)81807-3. [DOI] [PubMed] [Google Scholar]
- 29.Mohr S., Stryker J.M., Lambowitz A.M. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell. 2002;109:769–779. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- 30.Schroeder R., Grossberger R., Pichler A., Waldsich C. RNA folding in vivo. Curr. Opin. Struct. Biol. 2002;12:296–300. doi: 10.1016/s0959-440x(02)00325-1. [DOI] [PubMed] [Google Scholar]
- 31.Semrad K., Schroeder R. A ribosomal function is necessary for efficient splicing of the T4 phage thymidylate synthase intron in vivo. Genes Dev. 1998;12:1327–1337. doi: 10.1101/gad.12.9.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waldsich C., Grossberger R., Schroeder R. RNA chaperone StpA loosens interactions of the tertiary structure in the td group I intron in vivo. Genes Dev. 2002;16:2300–2312. doi: 10.1101/gad.231302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salvo J.L., Coetzee T., Belfort M. Deletion-tolerance and trans-splicing of the bacteriophage T4 td intron. Analysis of the P6–L6a region. J. Mol. Biol. 1990;211:537–549. doi: 10.1016/0022-2836(90)90264-m. [DOI] [PubMed] [Google Scholar]
- 34.Belfort M., Chandry P.S., Pedersen-Lane J. Genetic delineation of functional components of the group I intron in the phage T4 td gene. Cold Spring Harb. Symp. Quant. Biol. 1987;52:181–192. doi: 10.1101/sqb.1987.052.01.023. [DOI] [PubMed] [Google Scholar]
- 35.Schroeder R., von Ahsen U., Belfort M. Effects of mutations of the bulged nucleotide in the conserved P7 pairing element of the phage T4 td intron on ribozyme function. Biochemistry. 1991;30:3295–3303. doi: 10.1021/bi00227a018. [DOI] [PubMed] [Google Scholar]
- 36.Brion P., Schroeder R., Michel F., Westhof E. Influence of specific mutations on the thermal stability of the td group I intron in vitro and on its splicing efficiency in vivo: a comparative study. RNA. 1999;5:947–958. doi: 10.1017/s1355838299990477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belfort M., Moelleken A., Maley G.F., Maley F. Purification and properties of T4 phage thymidylate synthetase produced by the cloned gene in an amplification vector. J. Biol. Chem. 1983;258:2045–2051. [PubMed] [Google Scholar]
- 38.Lehnert V., Jaeger L., Michel F., Westhof E. New loop–loop tertiary interactions in self-splicing introns of subgroup IC and ID: a complete 3D model of the Tetrahymena thermophila ribozyme. Chem. Biol. 1996;3:993–1009. doi: 10.1016/s1074-5521(96)90166-0. [DOI] [PubMed] [Google Scholar]
- 39.Leontis N.B., Westhof E. A common motif organizes the structure of multi-helix loops in 16S and 23S ribosomal RNAs. J. Mol. Biol. 1998;283:571–583. doi: 10.1006/jmbi.1998.2106. [DOI] [PubMed] [Google Scholar]
- 40.Adams P.L., Stahley M.R., Kosek A.B., Wang J., Strobel S.A. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- 41.Jaeger L., Westhof E., Michel F. Function of P11, a tertiary base pairing in self-splicing introns of subgroup IA. J. Mol. Biol. 1991;221:1153–1164. doi: 10.1016/0022-2836(91)90925-v. [DOI] [PubMed] [Google Scholar]
- 42.Brion P., Michel F., Schroeder R., Westhof E. Analysis of the cooperative thermal unfolding of the td intron of bacteriophage T4. Nucleic Acids Res. 1999;27:2494–2502. doi: 10.1093/nar/27.12.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caprara M.G., Myers C.A., Lambowitz A.M. Interaction of the Neurospora crassa mitochondrial tyrosyl-tRNA synthetase (CYT-18 protein) with the group I intron P4–P6 domain. Thermodynamic analysis and the role of metal ions. J. Mol. Biol. 2001;308:165–190. doi: 10.1006/jmbi.2001.4581. [DOI] [PubMed] [Google Scholar]
- 44.Waldsich C., Masquida B., Westhof E., Schroeder R. Monitoring intermediate folding states of the td group I intron in vivo. EMBO J. 2002;19:5281–5291. doi: 10.1093/emboj/cdf504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pichler A., Schroeder R. Folding problems of the 5′ splice-site containing P1 stem of the group I td intron: substrate binding inhibition in vitro and mis-splicing in vivo. J. Biol. Chem. 2002;26:26. doi: 10.1074/jbc.M111798200. [DOI] [PubMed] [Google Scholar]
- 46.Leontis N.B., Westhof E. The 5S rRNA loop E: chemical probing and phylogenetic data versus crystal structure. RNA. 1998;4:1134–1153. doi: 10.1017/s1355838298980566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cech T.R., Damberger S.H., Gutell R.R. Representation of the secondary and tertiary structure of group I introns. Nature Struct. Biol. 1994;1:273–280. doi: 10.1038/nsb0594-273. [DOI] [PubMed] [Google Scholar]
- 48.Chandry P.S., Belfort M. Activation of a cryptic 5′ splice site in the upstream exon of the phage T4 td transcript: exon context, mis-splicing, and mRNA deletion in a fidelity mutant [published erratum appears in Genes Dev 1987 Dec;1(10):1351] Genes Dev. 1987;1:1028–1037. doi: 10.1101/gad.1.9.1028. [DOI] [PubMed] [Google Scholar]
- 49.Waldsich C., Semrad K., Schroeder R. Neomycin B inhibits splicing of the td intron indirectly by interfering with translation and enhances mis-splicing in vivo. RNA. 1998;4:1653–1663. doi: 10.1017/s1355838298981444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leontis N.B., Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]