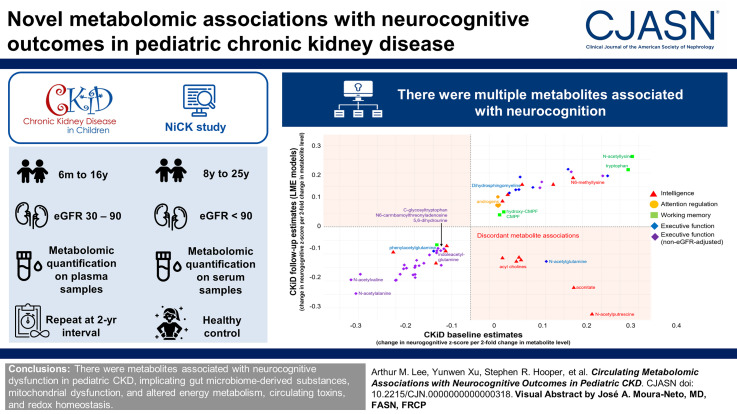
Visual Abstract
Keywords: metabolism, pediatric nephrology
Abstract
Background
Children with CKD are at risk for impaired neurocognitive functioning. We investigated metabolomic associations with neurocognition in children with CKD.
Methods
We leveraged data from the Chronic Kidney Disease in Children (CKiD) study and the Neurocognitive Assessment and Magnetic Resonance Imaging Analysis of Children and Young Adults with Chronic Kidney Disease (NiCK) study. CKiD is a multi-institutional cohort that enrolled children aged 6 months to 16 years with eGFR 30–90 ml/min per 1.73 m2 (n=569). NiCK is a single-center cross-sectional study of participants aged 8–25 years with eGFR<90 ml/min per 1.73 m2 (n=60) and matched healthy controls (n=67). Untargeted metabolomic quantification was performed on plasma (CKiD, 622 metabolites) and serum (NiCK, 825 metabolites) samples. Four neurocognitive domains were assessed: intelligence, attention regulation, working memory, and parent ratings of executive function. Repeat assessments were performed in CKiD at 2-year intervals. Linear regression and linear mixed-effects regression analyses adjusting for age, sex, delivery history, hypertension, proteinuria, CKD duration, and glomerular versus nonglomerular diagnosis were used to identify metabolites associated with neurocognitive z-scores. Analyses were performed with and without adjustment for eGFR.
Results
There were multiple metabolite associations with neurocognition observed in at least two of the analytic samples (CKiD baseline, CKiD follow-up, and NiCK CKD). Most of these metabolites were significantly elevated in children with CKD compared with healthy controls in NiCK. Notable signals included associations with parental ratings of executive function: phenylacetylglutamine, indoleacetylglutamine, and trimethylamine N-oxide—and with intelligence: γ-glutamyl amino acids and aconitate.
Conclusions
Several metabolites were associated with neurocognitive dysfunction in pediatric CKD, implicating gut microbiome–derived substances, mitochondrial dysfunction, and altered energy metabolism, circulating toxins, and redox homeostasis.
Podcast
This article contains a podcast at https://dts.podtrac.com/redirect.mp3/www.asn-online.org/media/podcast/CJASN/2023_11_17_CJN0000000000000318.mp3
Introduction
Children with CKD are at risk of neurocognitive dysfunction and poor neurodevelopmental outcomes.1–5 These neurocognitive sequelae can have lifelong consequences, including difficulties in school performance, employment, and health care self-management.6–9 Currently, the pathomechanisms of these neurocognitive sequelae are unclear but are likely mediated in part by CKD-associated dysmetabolism. Identifying metabolite associations with neurocognition may inform the development of targeted interventions for mitigating neurodevelopmental sequelae, such as improved clearance targets, dietary modifications, or novel therapies.
Metabolomics is a progressing field in nephrology research, yielding new discoveries of metabolite associations with kidney function, CKD etiology, and morbidity and mortality.10–12 Untargeted metabolomic profiling is considered an intermediate, aggregated, molecular phenotype. Circulating metabolite levels in CKD can be influenced by multiple factors, including reduced filtration, genetic variation, CKD-associated dysmetabolism, dietary modifications, intestinal dysbiosis, and pharmacologic interventions.13–18 Untargeted metabolomic profiling in pediatric CKD has identified novel metabolite associations with CKD progression and etiology.19,20 Data on metabolite associations with neurocognition in children with CKD are lacking. Metabolite signals have been associated with neuropsychiatric dysfunction in adults.21–24
This study's objective was to identify metabolite associations with neurocognition in children and adolescents with CKD. The Chronic Kidney Disease in Children (CKiD) study is the largest North American multicenter prospective cohort of pediatric CKD.5,25 Previous studies from CKiD have shown that children with CKD are at risk for lower assessment scores in intelligence, attention regulation, working memory, executive function, and academic achievement. These differences have been associated with eGFR, proteinuria, BP control, and CKD duration.26
The Neurocognitive Assessment and Magnetic Resonance Imaging Analysis of Children and Young Adults with Chronic Kidney Disease (NiCK) study is a cross-sectional study that enrolled participants with CKD and matched healthy controls.4,27 NiCK study findings are consistent with CKiD, showing children and young adults with CKD are at risk for lower assessment scores in attention regulation, executive function, and working memory. The NiCK study also showed that participants with CKD scored lower in these areas compared with matched healthy controls.1,4
This study's strength is in leveraging multiple data sources for replication analyses, a rare resource in pediatric CKD studies. Repeated longitudinal metabolomics quantification and neurocognitive phenotyping of CKiD was used for internal replication. The NiCK cohort provided a second independent sample for external replication. The NiCK healthy control population provided the opportunity to evaluate whether metabolite associations with neurocognition were unique to pediatric CKD or also present in healthy children.
Methods
CKiD and NiCK Studies
This study's CKiD participants were enrolled between January 2005 and December 2014 across 54 medical centers in the United States and Canada. Complete details of study design, methods, and cohort characteristics have been published previously.25 Children between the ages of 6 months and 16 years were enrolled if they had an eGFR of 30–90 ml/min per 1.73 m2. Exclusion criteria included history of solid organ or bone marrow transplant, dialysis within 3 years, malignancy, HIV infection within 12 months, structural cardiac disease, and genetic syndromes with central nervous system involvement. CKiD visit sample records with missing data regarding CKD duration (n=9) and proteinuria (n=42) were excluded. Participants who underwent neurocognitive assessments for both the CKiD and NiCK studies were excluded from the CKiD sample (n=11). The cross-sectional baseline CKiD data were used in the discovery analysis. The longitudinal follow-up data were leveraged for internal replication.
NiCK is a single-center, cross-sectional study based at Children's Hospital of Philadelphia that enrolled children and young adults aged 8–25 years with eGFR <90 ml/min per 1.73 m2 and healthy controls matched on age, sex, and medical insurance status between August 2011 and October 2014. Complete details of study design, methods, and cohort characteristics have been published previously.27 For this analysis, exclusion criteria included history of dialysis and kidney transplantation. There were no missing records regarding CKD duration and proteinuria. NiCK data were leveraged for external replication.
Abnormal birth history was defined as <36 weeks' gestational age or birthweight <2500 g. In the CKiD study, participant records with missing delivery history were imputed with normal birth history (n=18). Hypertension was defined on the basis of standardized study BP readings >95th percentile for age, sex, and height, or antihypertensive medication usage.
Metabolomic Profiling
Plasma (CKiD) and serum (NiCK) samples were sent to Metabolon Inc. (Durham, NC) for untargeted ultra-high–performance liquid chromatography tandem mass spectrometry-based metabolomics quantification.28–30 Plasma and serum metabolomics quantification have been shown to have good correlation and reproducibility.31–33
Metabolite quantification quality control was on the basis of the established National Institute of Diabetes and Digestive and Kidney Diseases CKD Biomarkers Consortium approaches with adaptations for longitudinal data.10,12,19,34,35 Sample records missing >50% metabolites measured, unnamed and drug metabolites, and individual metabolites with >80% missingness across all samples were excluded. For the remaining metabolites, missing values were imputed with the minimum measurement per metabolite. Metabolite levels were indexed to Metabolon standardized, pooled healthy human donor (matrix) samples included with every run.
For CKiD, Metabolon had technology platform upgrades between the baseline and follow-up sample measurements. Baseline samples were run on Metabolon's MTRX5 platform. Follow-up samples were run on the MTRX7 platform. To account for platform performance differences, conversion factors were applied to the baseline measurements on the basis of the ratio of MTRX7 median/MTRX5 median per metabolite in the matrix samples. Metabolites without conversion factors were excluded (n=220).
After natural logarithmic transformation, metabolites with variance <0.01 were excluded. Metabolite levels were log base 2 transformed for statistical analyses. Included metabolite pathways and subpathways as defined by Metabolon are shown in Supplemental Items 1 and 2.
Neurocognitive Assessments
Details of CKiD and NiCK neurocognitive assessments have been published.1,4,26 CKiD neurocognitive phenotyping was performed at the baseline, 2-, and 4-year study visits, concurrent with samples assayed for metabolomics profiling. We analyzed four assessments of neurocognitive function: intelligence quotient as an estimate of overall intellectual abilities and three individual domains—attention regulation, working memory, and parental ratings of executive function (subsequently referred to as executive function). These four assessments were selected on the basis of previously published CKD-associated differences and alignment between CKiD and NiCK.26 Intelligence was assessed with the Wechsler Abbreviated Scales of Intelligence. Attention regulation was assessed with the Conners' Continuous Performance Test II. Working memory was assessed with the Wechsler Intelligence Scale for Children 4th Edition Integrated Digit Span for participants up to age 16. For participants aged 17 and older, working memory was assessed with the Wechsler Memory Scale 3rd Edition Digit Span and Spatial Span. Fewer CKiD participants completed working memory assessments because it was not included in CKiD's neurocognitive battery and adopted by all study sites until a later time point. Executive function was assessed by the Behavior Rating Inventory of Executive Function and Delis–Kaplan Executive Function System Tower Subtest. Each assessment was standardized using specific test normative data. For domains with multiple assessments, assessments were standardized individually, and an average z-score was calculated.
Statistical Analyses
Participants with metabolomic profiling and complete assessments for a neurocognitive domain were included in analysis of that domain. The CKiD baseline data were used for discovery analyses. CKiD participants with repeated neurocognitive assessments and metabolomic profiling at 2- and/or 4-year follow-up were evaluated for internal replication. The NiCK CKD data were used for external replication. We report metabolite associations significant in at least two of the three analytic samples on the basis of P < 0.05 and the Benjamini–Hochberg False Discovery Rate (FDR) threshold. Metabolites are nonindependent and multicollinear; for discovery analyses, multiple comparison corrections may lead to important signals being masked.36
Neurocognitive domain differences between CKD sample groups and NiCK healthy controls were assessed with Wilcoxon rank-sum tests. Differences between all sample groups were assessed with Kruskal–Wallis H tests.
We identified metabolites associated with neurocognitive z-scores using linear regression for the CKiD baseline and NiCK samples, and linear mixed-effects models with unstructured variance-covariance matrices for the CKiD follow-up samples. All models adjusted for age, sex, birth history, glomerular disease, hypertension, CKD duration, log-transformed urine protein/creatinine ratio, and log-transformed eGFR. We did not adjust for race because it is a social, nonbiologic construct.37–40 We repeated analyses without eGFR adjustment to account for potential masking of metabolite signals that are highly associated with eGFR.
Metabolite subpathways associated with neurocognition were evaluated with hypergeometric distribution tests. Linear regression assessed whether metabolite levels differed between participants with CKD and controls in NiCK. We also assessed whether these metabolites were associated with neurocognitive z-scores in the NiCK control group.
Metabolites and their related subpathways with discordant neurocognitive associations between discovery and replication analyses were selected for post hoc ratio testing. If metabolites exist within a biochemical axis connected by an enzymatic process, their ratio may provide clarifying information.41 Two metabolites A and B constituted a unique pairing, and only one of ratio A/B or B/A was tested. Fully adjusted linear regression analyses identified metabolite ratios significantly associated with neurocognitive z-scores on the basis of the p-gain statistic >10.41 R code is deposited in GitHub (Supplemental Item 6).
Results
Participant Characteristics and Neurocognitive Outcomes
The baseline CKiD sample included 569 participants. NiCK included 60 participants with CKD and 67 healthy controls. Table 1 shows participant characteristics. Follow-up CKiD participants differed from the baseline sample. CKiD participants with follow-up had higher baseline eGFR and were less likely to have glomerular disease.
Table 1.
Baseline participant characteristics of the Chronic Kidney Disease in Children and the Neurocognitive Assessment and Magnetic Resonance Imaging Analysis of Children and Young Adults with Chronic Kidney Disease studies
| Participant Characteristics | CKiD | NiCK | ||
|---|---|---|---|---|
| Overall | Subset with Follow-Up | CKD | Controls | |
| No. | 569 | 360 | 60 | 67 |
| Age, yr, median (IQR) | 12 (8–15) | 11 (8–14) | 16 (14–18) | 16 (13–18) |
| Sex, male, n (%) | 344 (60) | 220 (61) | 36 (60) | 37 (54) |
| Non-Hispanic Black race, n (%) | 111 (19) | 62 (17) | 18 (30) | 26 (38) |
| Hispanic ethnicity, n (%) | 74 (13) | 43 (12) | 3 (5) | 0 (0) |
| Abnormal delivery history, n (%) | 115 (20) | 67 (19) | 20 (33) | 20 (29) |
| eGFR, median (IQR) | 53 (40–67) | 52 (39–66) | 55 (29–73) | — |
| UPCR, median (IQR) | 0.3 (0.1–0.9) | 0.3 (0.1–0.7) | 0.7 (0.2–1.6) | — |
| CKD duration, yr, median (IQR) | 9 (4–13) | 8 (5–12) | 9 (3–15) | — |
| Glomerular diagnosis | 169 (30) | 93 (26) | 19 (32) | — |
| Hypertension | 404 (71) | 242 (67) | 31 (52) | — |
There were 569 CKiD participants with any complete neurocognitive assessment at baseline. A subset of 360 had longitudinal follow-up. NiCK participants were older than CKiD participants at baseline. NiCK participants had a larger percentage representation of non-Hispanic Black race. CKiD, Chronic Kidney Disease in Children; NiCK, Neurocognitive Assessment and Magnetic Resonance Imaging Analysis of Children and Young Adults with Chronic Kidney Disease; IQR, interquartile range; UPCR, urine protein/creatinine ratio.
Figure 1 shows neurocognitive assessment z-scores. Children with CKD had lower z-scores compared with NiCK healthy controls (full details are reported in Figure 1 and consistent with published data).1,4,26 NiCK participants scored lower in working memory compared with CKiD participants.
Figure 1.
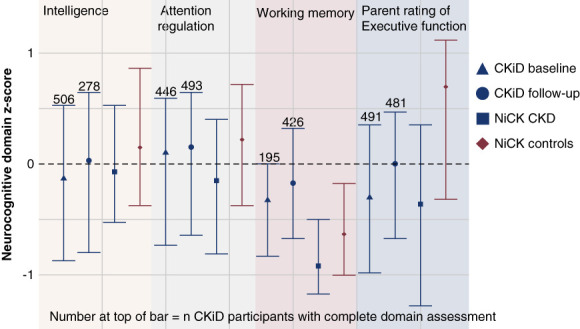
Neurocognitive assessments in CKiD and NiCK. There were slight differences in the number of CKiD participants with complete assessments per domain, denoted above each bar. Each point represents the median neurocognitive assessment z-score, and the whiskers represent the interquartile range. There were differences in neurocognitive assessment z-scores between all four sample groups on the basis of Kruskal–Wallis tests: intelligence (P = 0.027), working memory (P < 0.001), and parental rating of executive function (P < 0.001). Differences between CKD sample groups and the NiCK control group were assessed with Wilcoxon rank-sum tests. The CKiD baseline sample had lower intelligence (P = 0.0068) and executive function (P < 0.001). The CKiD follow-up sample had lower executive function (P < 0.001). The NiCK CKD sample had lower attention regulation (P = 0.028), working memory (P = 0.027), and executive function (P = 0.0012). The Wilcoxon rank-sum test showed significant difference in working memory between CKiD (baseline and follow-up) and NiCK (CKD and controls), P < 0.001. CKiD, Chronic Kidney Disease in Children; NiCK, Neurocognitive Assessment and Magnetic Resonance Imaging Analysis of Children and Young Adults with Chronic Kidney Disease.
Metabolite Associations
Six hundred and twenty-two and 825 metabolites were analyzed in CKiD and NiCK, respectively (Supplemental Items 1 and 2). Supplemental Item 3 shows 52 significant metabolite associations with neurocognition that replicated either internally or externally. In analyses not adjusted for eGFR, 29 additional metabolites associated with executive function. Two metabolites associated with neurocognitive z-scores in all three samples: aconitate with intelligence and indoleacetylglutamine with executive function. Only N6-methyllysine associated with intelligence z-score at the FDR threshold in the CKiD baseline sample. There were no significant associations at the FDR threshold in either replication sample.
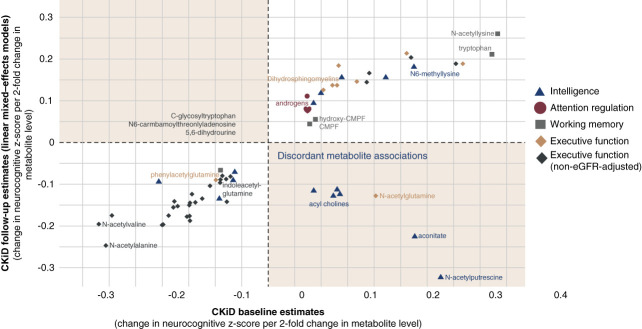
Figure 2 shows metabolite associations that internally replicated in the CKiD follow-up sample. Notably, executive function was negatively associated with eGFR-associated and gut microbiome–derived substances, including N6-carbamoylthreonyladenosine, 2,3-dihydroxy-5-methylthio-4-pentenoate, and phenylacetylglutamine.
Figure 2.
Significant metabolites in CKiD longitudinal internal replication. Associated neurocognitive domain is denoted by dot shape and color. Metabolites of interest are annotated. Notably, parental ratings of executive function associated with gut microbiome–derived phenylacetylglutamine. In analyses not adjusted for eGFR, executive function also associated with metabolites previously associated with pediatric CKD progression: N6-carbamoylthreonyladenosine, c-glycosyltryptophan, 5,6-dihydrouridine, and 2,3-dihydroxy-5-methylthio-4-pentenoate (DMTPA). CMPF, 3-carboxy-4-methyl-5-propyl-2-furanpropanoate.
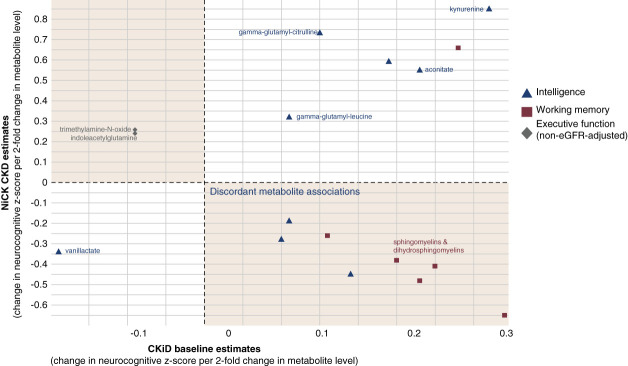
Figure 3 shows metabolite associations that externally replicated in the NiCK CKD data. Notably, intelligence was positively associated with kynurenine, γ-glutamylcitrulline, and γ-glutamylleucine.
Figure 3.
Significant metabolites in NiCK CKD external replication. Associated neurocognitive domain is denoted by dot shape and color. Metabolites of interest are annotated. Kynurenine and γ-glutamyl-amino acids positively associated with intelligence z-score.
Table 2 reports metabolite subpathway associations with neurocognition. Tryptophan and γ-glutamyl amino acid metabolite associations were externally replicated.
Table 2.
Pathway enrichment analysis
| Subpathway | Domain | Hypergeometric P Value | ||
|---|---|---|---|---|
| Baseline | Longitudinal | NiCK | ||
| Acyl cholines | Intelligence | 0 | 0 | — |
| γ-Glutamyl amino acids | 0.008 | — | <0.001 | |
| Androgenic steroids | Attention regulation | 0.002 | <0.001 | — |
| Tryptophan | 0.02 | — | 0.05 | |
| Dicarboxylates | Working memory | 0.002 | 0.01 | — |
| Glycine, serine, and threonines | 0.01 | — | 0.02 | |
| Lysines | — | 0.01 | 0.02 | |
| Tryptophan | 0.005 | 0.02 | — | |
| Dicarboxylates | Executive function | — | 0.03 | 0.05 |
| Dihydrosphingomyelins | <0.001 | <0.001 | — | |
| Vitamin A | Executive function, non–eGFR-adjusted | 0.01 | — | 0.003 |
| Acetylated peptides | 0.003 | 0.003 | — | |
| Alanine and aspartate | 0.03 | 0.02 | — | |
| Aminosugars | 0.03 | 0.002 | — | |
| Polyamines | 0.03 | <0.001 | — | |
| Purine and adenines | 0.05 | 0.04 | — | |
| Sterols | 0.05 | 0.04 | — | |
Hypergeometric distribution tests identified metabolite subpathways associated with neurocognitive domain z-scores in at least two of three analytic groups. Tryptophan and γ-glutamyl amino acid metabolite associations were externally replicated in NiCK. “—” denotes subpathways that did not replicate in the sample. NiCK, Neurocognitive Assessment and Magnetic Resonance Imaging Analysis of Children and Young Adults with Chronic Kidney Disease.
Metabolite Analyses in NiCK Healthy Controls
Table 3 shows that 56 of the 81 metabolites associated with neurocognition z-scores (from Table 2) were different between participants with CKD and healthy controls in NiCK. Levels of 55 metabolites were higher in children with CKD. Only tryptophan had lower levels in children with CKD.
Table 3.
Metabolite-level differences between CKD and control participants in Neurocognitive Assessment and Magnetic Resonance Imaging Analysis of Children and Young Adults with Chronic Kidney Disease
| Metabolite | Estimate | 95% CI | P Value |
|---|---|---|---|
| 4-Hydroxyphenylacetylglutamine | 2.75 | (2.22 to 3.27) | <0.001 |
| Orotidine | 2.36 | (1.81 to 2.9) | <0.001 |
| γ-CEHC glucuronide | 2.32 | (1.84 to 2.8) | <0.001 |
| Indoleacetylglutamine | 1.85 | (1.36 to 2.35) | <0.001 |
| Pimeloylcarnitine/3-methyladipoylcarnitine (C7-DC) | 1.83 | (1.38 to 2.28) | <0.001 |
| N6-succinyladenosine | 1.79 | (1.44 to 2.13) | <0.001 |
| Vanillactate | 1.65 | (1.27 to 2.03) | <0.001 |
| Adipoylcarnitine (C6-DC) | 1.53 | (1.12 to 1.95) | <0.001 |
| Suberoylcarnitine (C8-DC) | 1.52 | (0.99 to 2.04) | <0.001 |
| 1-Ribosyl-imidazoleacetate | 1.41 | (1.12 to 1.69) | <0.001 |
| C-glycosyltryptophan | 1.24 | (0.98 to 1.49) | <0.001 |
| Arabitol/xylitol | 1.22 | (0.99 to 1.45) | <0.001 |
| Hydroxyasparagine | 1.21 | (0.99 to 1.42) | <0.001 |
| 4-Hydroxyphenylacetate | 1.13 | (0.54 to 1.72) | <0.001 |
| N6-carbamoylthreonyladenosine | 1.11 | (0.88 to 1.34) | <0.001 |
| 5,6-Dihydrouridine | 1.06 | (0.84 to 1.29) | <0.001 |
| N-acetylneuraminate | 1.06 | (0.84 to 1.29) | <0.001 |
| Acisoga | 1.01 | (0.73 to 1.28) | <0.001 |
| N-acetylglutamine | 0.95 | (0.72 to 1.19) | <0.001 |
| Phenylacetylglutamine | 0.95 | (0.59 to 1.31) | <0.001 |
| Glutarylcarnitine (C5-DC) | 0.94 | (0.7 to 1.19) | <0.001 |
| N-acetyl-isoputreanine | 0.93 | (0.67 to 1.18) | <0.001 |
| 3-Hydroxy-3-methylglutarate | 0.92 | (0.68 to 1.16) | <0.001 |
| 3-Carboxy-4-methyl-5-propyl-2-furanpropanoate (CMPF) | 0.89 | (0.25 to 1.53) | <0.001 |
| Pro-hydroxy-pro | 0.77 | (0.37 to 1.18) | <0.001 |
| N-acetyltaurine | 0.77 | (0.57 to 0.97) | <0.001 |
| N-acetylalanine | 0.75 | (0.61 to 0.88) | <0.001 |
| N-acetylvaline | 0.74 | (0.58 to 0.9) | <0.001 |
| Trimethylamine N-oxide | 0.73 | (0.29 to 1.18) | <0.001 |
| Aconitate [cis or trans] | 0.72 | (0.5 to 0.93) | <0.001 |
| Retinol (vitamin A) | 0.71 | (0.55 to 0.86) | <0.001 |
| 3-Carboxy-4-methyl-5-pentyl-2-furanpropionate (3-Cmpfp) | 0.70 | (0.45 to 0.96) | <0.001 |
| 1-Stearoyl-2-oleoyl-GPE (18:0/18:1) | 0.70 | (0.41 to 0.99) | <0.001 |
| Kynurenine | 0.60 | (0.45 to 0.75) | <0.001 |
| N6-acetyllysine | 0.56 | (0.4 to 0.71) | <0.001 |
| N-acetylputrescine | 0.54 | (0.35 to 0.73) | <0.001 |
| Behenoyl dihydrosphingomyelin (d18:0/22:0) | 0.45 | (0.18 to 0.73) | <0.001 |
| Glycosyl-N-(2-hydroxynervonoyl)-sphingosine (d18:1/24:1(2OH)) | 0.44 | (0.02 to 0.86) | 0.044 |
| γ-Glutamylleucine | 0.38 | (0.25 to 0.51) | <0.001 |
| Indoleacetate | 0.38 | (0.15 to 0.61) | 0.002 |
| Tricosanoyl sphingomyelin (d18:1/23:0) | 0.35 | (0.19 to 0.52) | <0.001 |
| Campesterol | 0.35 | (0.04 to 0.66) | 0.03 |
| Cholesterol | 0.35 | (0.23 to 0.47) | <0.001 |
| Sphingomyelin (d17:1/14:0, d16:1/15:0) | 0.34 | (0.14 to 0.55) | 0.001 |
| Lactosyl-N-nervonoyl-sphingosine (d18:1/24:1) | 0.32 | (0.16 to 0.48) | <0.001 |
| Lignoceroyl sphingomyelin (d18:1/24:0) | 0.30 | (0.16 to 0.44) | <0.001 |
| Sphingomyelin (d18:0/20:0, d16:0/22:0) | 0.30 | (0.04 to 0.56) | 0.03 |
| trans-4-Hydroxyproline | 0.29 | (0.1 to 0.49) | 0.003 |
| γ-Glutamylcitrulline | 0.27 | (0.06 to 0.48) | 0.02 |
| Sphingomyelin (d18:1/21:0, d17:1/22:0, d16:1/23:0) | 0.27 | (0.11 to 0.43) | 0.001 |
| 1-Arachidonoyl-GPE (20:4n6) | 0.24 | (0.12 to 0.35) | <0.001 |
| Sphingomyelin (d18:1/25:0, d19:0/24:1, d20:1/23:0, d19:1/24:0) | 0.23 | (0.03 to 0.42) | 0.02 |
| Glycosyl ceramide (d18:1/20:0, d16:1/22:0) | 0.22 | (0.06 to 0.38) | 0.008 |
| Behenoyl sphingomyelin (d18:1/22:0) | 0.20 | (0.09 to 0.3) | <0.001 |
| Myristoyl dihydrosphingomyelin (d18:0/14:0) | 0.18 | (0.03 to 0.33) | 0.02 |
| Tryptophan | −0.32 | (−0.43 to −0.21) | <0.001 |
| Arachidonoylcholine | 0.21 | (−0.01 to 0.43) | 0.07 |
| l-Urobilin | −0.27 | (−0.71 to 0.17) | 0.24 |
| Linoleoylcholine | 0.12 | (−0.08 to 0.32) | 0.24 |
| Palmitoylcholine | 0.11 | (−0.09 to 0.31) | 0.30 |
| Oleoylcholine | 0.12 | (−0.11 to 0.34) | 0.32 |
| 1-Linoleoyl-GPE (18:2) | 0.10 | (−0.1 to 0.3) | 0.32 |
| Hydroxy-CMPF | −0.24 | (−0.86 to 0.37) | 0.44 |
| 5-Methyluridine (ribothymidine) | 0.05 | (−0.12 to 0.22) | 0.57 |
| Epiandrosterone sulfate | −0.10 | (−0.67 to 0.47) | 0.74 |
| Pregnenediol disulfate (C21H34O8S2) | −0.06 | (−0.46 to 0.33) | 0.75 |
| 5α-Androstan-3α,17β-diol monosulfate (1) | 0.10 | (−0.53 to 0.73) | 0.76 |
| Isoursodeoxycholate | 0.06 | (−0.53 to 0.66) | 0.83 |
| Androsterone sulfate | −0.05 | (−0.56 to 0.46) | 0.84 |
| 5α-Pregnan-diol disulfate | −0.06 | (−0.69 to 0.58) | 0.86 |
| 5α-Androstan-3α,17β-diol monosulfate (2) | 0.05 | (−0.56 to 0.65) | 0.87 |
There were 71 metabolites significantly associated with neurocognitive z-scores reported in Table 2. Regression analyses identified that 56 of these had significant differences on the basis of CKD status in NiCK. Estimates are reported as for the difference between having CKD versus being a control participant and the number of two-fold unit differences in the metabolite level. Only tryptophan was measured at lower levels in children with CKD. CI, confidence interval; NiCK, Neurocognitive Assessment and Magnetic Resonance Imaging Analysis of Children and Young Adults with Chronic Kidney Disease.
Seven metabolites associated with neurocognition in the NiCK control sample: intelligence with orotidine (estimate=−0.18, 95% confidence interval [−0.33 to −0.03]), l-urobilin (−0.26 [−0.41 to −0.11]), and 5-methyluridine (−0.47 [−0.92 to −0.01]); working memory with N-acetylneuraminate (−1.10 [−1.88 to −0.32]), N-acetylglutamine (−0.75 [−1.37 to −0.13]), and indoleacetate (0.57 [0.07, 1.08]); and executive function with pro-hydroxy-pro (−0.27 [−0.40 to −0.13]) (Supplemental Item 4). Estimate units represent a change in neurocognitive z-score per two-fold change in the metabolite level.
Discordant Metabolite Analysis
Replication analyses identified metabolites with discordant associations with neurocognition. Ratio analyses assessed aconitate (mitochondrial/energy metabolism) and sphingomyelins (sphingomyelin/ceramide axis) (Supplemental Item 5).
In CKiD and NiCK, there were 42 metabolites related to mitochondrial metabolism (the tricarboxylic acid cycle and acylcarnitine shuttle), generating 861 unique ratio pairings. There were 53, 5, and 5 ratios associated with intelligence in the CKiD baseline, CKiD follow-up, and NiCK CKD samples, respectively. The association of the ratio of oleoylcarnitine to stearoylcarnitine with intelligence was internally replicated. The association of the ratio of octadecanedioylcarnitine to palmitoylcarnitine with intelligence was externally replicated.
Five sphingomyelin metabolites discordantly associated with working memory. Forty-seven metabolites within the sphingomyelin and ceramide pathways generated 1081 unique ratio pairings. There were 45, 25, and 2 ratios associated with working memory in the CKiD baseline, CKiD follow-up, and NiCK CKD samples, respectively. Working memory's association with the ratios of myristoyl dihydrosphingomyelin (d18:0/14:0) to sphingomyelin (d18:2/16:0, d18:1/16:1), and sphingomyelin (d17:1/14:0, d16:1/15:0) to sphingomyelin (d18:2/16:0, d18:1/16:1) were internally replicated.
Discussion
We report novel and confirmatory metabolite associations with neurocognition in pediatric CKD. We demonstrated metabolite associations across four neurocognitive assessment domains, independent of eGFR. One of our study's key strengths is leveraging multiple samples for replication analyses. These study samples have complementary strengths. NiCK's smaller sample size but inclusion of matched, healthy controls was useful for external replication. CKiD longitudinal data provided a larger sample size for internal replication. CKiD's repeated metabolomics profiling served as an additional control for the metabolome's inherent variability. These replication samples represent a significant advance for pediatric CKD metabolomics studies because previous studies were limited by the lack of available and comparable replication cohorts.
Some of the most interesting novel signals were the positive associations of γ-glutamyl amino acids with intelligence, which were externally replicated. These γ-glutamyl amino acids were also elevated in children with CKD compared with healthy controls. γ-Glutamyl amino acids are downstream products of γ-glutamyl transpeptidase-1 in glutathione metabolism, and γ-glutamyl transpeptidase-1 is abundantly expressed in the proximal tubule.42–44 Glutathione metabolism plays an important role in cellular defense against oxidative stress.45 Glutathione and oxidation–reduction dysmetabolism have been associated with neuropsychiatric dysfunction in various conditions.46–51 Elevated levels of γ-glutamyl amino acids and their positive association with intelligence may suggest a protective metabolic adaption in children with CKD. The pharmacologic attenuation of oxidative stress is an emerging area of interest for many disease states.52–54 Our data support existing knowledge regarding increased oxidative stress and poor health outcomes and provide an exciting direction for continued study and potential therapeutic development in pediatric CKD.
A kidney–brain–gut microbiome axis in CKD is increasingly described.13,16,55–58 Impaired kidney function leads to alteration of gut membrane permeability and microbiome composition. Ultimately, gut microbiome–derived uremic toxins accumulate. These toxins may mediate biochemical pathology through inflammation, immune dysregulation, and vascular calcification.13,15,16,55
Gut microbiome–derived phenylacetylglutamine was negatively associated with executive function. Phenylacetylglutamine has gained attention for its association with cardiovascular morbidity and mortality in CKD.59–63 Our data support the theory that phenylacetylglutamine and other gut microbiome–derived metabolites may have multisystemic toxicity.64 In addition, gut microbiome–derived indoleacetylglutamine, 4-hydroxyphenylacetylglutamine, 4-hydroxyphenylacetate, indoleacetate, and trimethylamine N-oxide (TMAO) also associated with neurocognition. TMAO mediates platelet dysfunction and inflammatory signaling and can be modified by antibiotic therapy.65–69 In our study, indoleacetylglutamine and TMAO discordantly associated with neurocognition, raising question regarding their true biologic significance. All of the aforementioned gut microbiome–derived metabolite levels were increased in participants with CKD compared with healthy controls, supporting the theory that microbiome-derived toxins can accumulate in children with CKD. Our data emphasize the need for a continued study of the kidney–brain–gut axis as a potential therapeutic target in CKD.
There is growing evidence of mitochondrial dysfunction and altered energy metabolism in CKD.70,71 Internally replicated metabolites associated with neurocognition included acylcarnitines, 3-carboxy-4-methyl-5-propyl-2-furanpropionate (CMPF), and hydroxy-CMPF. Acylcarnitines are involved in the acylcarnitine/carnitine shuttle in mitochondrial energy metabolism.72 Carnitine dysmetabolism has been described in children and adults on dialysis.73 Previous studies suggest CMPF mediates neurotoxicity by inhibiting mitochondrial respiration in CKD.74,75 By contrast, our data showed CMPF and hydroxy-CMPF to be positively associated with working memory.
Aconitate associated with intelligence in all three samples. Aconitate is a mitochondrial tricarboxylic acid cycle intermediate and associated with mitochondrial DNA mutations.76 Aconitate's associations in the CKiD baseline and follow-up samples were discordant. We hypothesized that metabolite ratio testing could reveal whether aconitate was involved in different pathways affecting intelligence. Ratio testing did not identify significant associations to suggest for or against this hypothesis. Our findings highlight the need for continued investigation of mitochondrial/energy metabolism and how it may mediate complications in CKD.
Sphingomyelins and dihydrosphingomyelins positively associated with executive function, demonstrated with internal replication. Sphingomyelins also discordantly associated with working memory, on the basis of external replication. Sphingomyelins exist in cell membranes and participate in signaling pathways. Sphingomyelin dysregulation has been associated with both neuropsychiatric and kidney pathologies.14,24,77–83
Ratio testing suggested that the discordant sphingomyelin associations with working memory may represent distinct biochemical processes in the CKiD and NiCK samples. In CKiD internal replication, a sphingomyelin/dihydrosphingomyelin ratio significantly associated with working memory, whereas in NiCK external replication, there were no common ratio associations. One possible explanation for the discordant association is the age difference between the two cohorts. In younger children, sphingomyelin metabolism is essential for myelination and brain maturation. In an older population, sphingomyelin–ceramide metabolism may be related to inflammation, atherogenesis, and cellular apoptosis. Another possibility is that the discordant sphingomyelin-working memory association reflects confounding by intrinsic racial bias within the working memory assessment itself. NiCK participants had lower working memory z-scores and greater representation of non-Hispanic Black participants than CKiD. Standardized assessments for attention regulation and working memory do not account for potential processing differences in classifying in- versus out-groups on the basis of race/ethnicity. This may result in false low assessment scores among participants belonging to minority racial/ethnic groups.84 From that perspective, the discordant NiCK sphingomyelin-working memory association may not represent a true biologic relation.
Analyses without adjustment for eGFR revealed additional metabolite associations with executive function. Notable negative associations with executive function included c-glycosyltryptophan, N6-carbamoylthreonyladenosine, DMPTA, and 5,6-dihydrouridine. These metabolites are all highly associated with eGFR. There is increasing recognition that eGFR-associated substances include both simple filtration markers and substances with independent toxic effects. These metabolites have been associated with a risk of progression to end stage kidney disease in children with CKD, independent of eGFR.11,19,85 These data support the idea that circulating filtration-associated substances can mediate neuro- and systemic toxicity.
Several N-acetylated amino acids associated with neurocognition: N-acetylglutamine, N-acetylalanine, N-acetyltaurine, and N-acetylvaline. N-acetylated amino acids are associated with genetic variation in the N-acetyltransferase-8 (NAT8) enzyme. NAT8 is highly expressed in the proximal tubule and functions in detoxification.86–89 There is population-level NAT8 genetic variation, with polymorphisms associated with higher CKD incidence and progression.88,90 Our data support the hypothesis that N-acetylated metabolite levels may represent detoxification impairment, which could mediate both neuro- and nephrotoxicity.
Polyamine metabolites associated with neurocognition, including N-acetyl-isoputreanine, N-acetyl-putrescine, and acisoga. Polyamines are spermidine derivatives, a diet-derived substance that undergoes processing by the gut microbiome.91 N-acetyl-isoputreanine associated with cognitive dysfunction in the Bogalusa Heart Study.22 N-acetyl-putrescine associated with prodromal symptoms of Parkinson disease.23 Acisoga associated with atrial fibrillation in the Atherosclerosis Risk in Communities cohort.92 Spermidine/polyamine metabolism is interesting because it is influenced by both dietary intake and the gut microbiome, representing two potential intervenable targets.
The internal replication analyses are limited by the CKiD follow-up sample representing a slightly different clinical population than the baseline sample. Participants who progressed to end stage kidney disease were also excluded from follow-up. Thus, internal replication analyses may not be representative of CKiD participants with a more advanced baseline CKD phenotype, and potentially important metabolite associations may have been missed.
Limitations exist in reconciling several discordant metabolite associations. Several notable signals included the aconitate, sphingomyelins, and the several gut microbiome–derived substances. These discordant associations could represent false-positive associations, but the extensive literature characterizing their additional clinical associations suggests otherwise. We examined metabolite pair ratios to assess whether the discordant metabolite associations may represent different biochemical processes. Metabolites often exist in complex overlapping networks, and a single metabolite's level may reflect multiple processes. Our analyses are unable to account for these complicated interactions. As more multiomics data are generated (genomics, proteomics, and microbiome characterization), their integration with metabolomics offers the opportunity for further elucidation.
Metabolomics data are highly dimensional, nonindependent, and multicollinear, raising the issue of multiple statistical comparisons. For our goals of generating pathomechanistic knowledge rather than identifying biomarkers for clinical application, the FDR might be overly restrictive and mask important signals. Conversely, a P value of < 0.05 may result in many false-positive signals. We aimed to limit false positives by reporting metabolites with both significant replication and existing published literature, supporting their biologic relation with neurocognition. We cannot infer metabolite causality on neurocognitive dysfunction. An important follow-up study could leverage CKiD genomic data for causal inference analyses.93–95
Social determinants of health have important associations with neurocognitive outcomes. How social health determinants affect the pediatric CKD metabolome has not been well characterized, limiting our ability to interpret our findings from this perspective.
We are limited in our contextual interpretation of longitudinal metabolomic signals. On the basis of our review, there are no previous published studies with repeated longitudinal metabolomic profiling in a comparable sample of children with CKD. Metabolomic variation over time in pediatric CKD may be influenced by many factors, including measurement variability, kidney function changes, metabolic adaptations, linear growth, body composition changes, and pubertal development.
We report metabolite associations with neurocognitive measures in pediatric CKD. These findings support the hypothesis of a multifactorial etiology of neurocognitive dysfunction in children with CKD, including gut microbiome alterations, mitochondrial and energy dysmetabolism, lipid-signaling changes, increased oxidative stress, and toxin accumulation. A key strength of our investigation is the use of longitudinal repeated metabolite quantification and a second independent population of children with CKD and matched healthy controls for replication analyses.
Impaired neurocognition is an important comorbidity of pediatric CKD. By identifying metabolite associations with neurocognition, our findings point to several directions for future research, including the pharmacologic modulation of oxidative stress and altered energy metabolism, improved dietary modification and clearance strategies, and gaining a better understanding of lipid signaling in pediatric CKD.
Supplementary Material
Acknowledgments
The opinions expressed in this article do not necessarily represent those of the Pennsylvania Department of Health; the National Institute of Diabetes, Digestive and Kidney Diseases; the National Institutes of Health; the Department of Health and Human Services; or the government of the United States.
Footnotes
The CKD Biomarkers Consortium members are Alison Abraham, Amanda Anderson, Shawn Ballard, Joseph Bonventre, Clary Clish, Heather Collins, Steven Coca, Josef Coresh, Rajat Deo, Michelle Denburg, Ruth Dubin, Harold I. Feldman, Bart S. Ferket, Meredith Foster, Susan Furth, Peter Ganz, Daniel Gossett, Morgan Grams, Jason Greenberg, Orlando M. Gutiérrez, Tom Hostetter, Lesley A. Inker, Joachim Ix, Paul L. Kimmel, Jon Klein, Andrew S. Levey, Joseph Massaro, Gearoid McMahon, Theodore Mifflin, Girish N. Nadkarni, Chirag Parikh, Vasan S. Ramachandran, Casey Rebholz, Eugene Rhee, Brad Rovin, Mark Sarnak, Venkata Sabbisetti, Jeffrey Schelling, Jesse Seegmiller, Michael G. Shlipak, Haochang Shou, Adriene Tin, Sushrut Waikar, Bradley Warady, Krista Whitehead, and Dawei Xie.
Contributor Information
Collaborators: Alison Abraham, Amanda Anderson, Shawn Ballard, Joseph Bonventre, Clary Clish, Heather Collins, Steven Coca, Josef Coresh, Rajat Deo, Michelle Denburg, Ruth Dubin, Harold I. Feldman, Bart S. Ferket, Meredith Foster, Susan Furth, Peter Ganz, Daniel Gossett, Morgan Grams, Jason Greenberg, Orlando M. Gutiérrez, Tom Hostetter, Lesley A. Inker, Joachim Ix, Paul L. Kimmel, Jon Klein, Andrew S. Levey, Joseph Massaro, Gearoid McMahon, Theodore Mifflin, Girish N. Nadkarni, Chirag Parikh, Vasan S. Ramachandran, Casey Rebholz, Eugene Rhee, Brad Rovin, M. Sarnak, Venkata Sabbisetti, Jeffrey Schelling, Jesse Seegmiller, Michael G. Shlipak, Haochang Shou, Adriene Tin, Sushrut Waikar, Bradley Warady, Krista Whitehead, and Dawei Xie
Disclosures
A.G. Abraham reports consultancy for Implementation Group, Inc.; research funding from NIH; honoraria from Elsevier; and advisory or leadership role as an Associate Editor of American Journal of Epidemiology and an Associate Editor of Population Health Metrics. J. Coresh reports employment with The Johns Hopkins University, Welch Center for Prevention, Epidemiology, and Clinical Research and research funding from the National Institutes of Health. M.R. Denburg reports research funding from Mallinckrodt; patents or royalties from InBore LLC; advisory or leadership role on the Editorial Board of Kidney International Reports and on KDIGO Executive Committee; and other interests or relationships with American Society of Pediatric Nephrology Research and Program Committees. M.R. Denburg's spouse reports consultancy for TriSalus Life Sciences; ownership interest in InBore LLC, Instylla, and Precision Guided Interventions LLC; and an advisory or leadership role for TriSalus Life Sciences Scientific Advisory Board. S.L. Furth reports consultancy for Genentech. E.A. Hartung reports advisory or leadership role for Polycystic Kidney Disease Foundation—Scientific Advisory Committee and PKD in Children Council/ARPKD Task Force (volunteer) and other interests or relationships as American Society of Pediatric Nephrology member and American Board of Pediatrics—Nephrology Subboard Member. P.L. Kimmel reports employment with National Institute of Diabetes and Digestive Kidney Diseases (NIDDK); royalties for co-editing Chronic Renal Disease and Psychosocial Aspects of Chronic Kidney Disease and royalties from Mayo Clinic Press for The Body's Keepers; patents or royalties from Elsevier; advisory or leadership role as an unpaid member of Board of Directors of Academy of Medicine of Washington, DC; and other interests or relationships as Co-Editor of Chronic Renal Disease (Academic Press) and Co-Editor of Psychosocial Aspects of Chronic Kidney Disease. As a Federal Employee at NIDDK, P.L. Kimmel's holdings are reviewed each year for potential conflicts of interest. At this time, P.L. Kimmel's only stock holding related in any fashion to health care is CVS and GE Healthcare. M.B. Matheson reports employment with Johns Hopkins Bloomberg School of Public Health and research funding from Toshiba Corporation. R.S. Vasan reports consultancy for NIDDK. B.A. Warady reports consultancy for Amgen, Bayer, GlaxoSmithKline, Light Line Medical, and UpToDate; research funding from Baxter Healthcare; honoraria from Amgen, Bayer, GlaxoSmithKline, and UpToDate; advisory or leadership role as Vice President of North American Pediatric Renal Trials and Collaborative Studies; and advisory or leadership roles on National Kidney Foundation Board of Directors, NTDS Board of Directors, and Midwest Transplant Network Governing Board. Y. Xu reports employment with Johns Hopkins School of Public Health. All remaining authors have nothing to disclose.
Funding
This work is supported by NIDDK from U01DK106982 (M.R. Denburg), U01DK085689 (J. Coresh), U01DK103225 (E.P. Rhee), and K23DK109203 (E.A. Hartung); NCCIH from R21AT009752 (M.R. Denburg); Pennsylvania Department of Health from SAP4100054843 (E.A. Hartung); NCATS from KL2TR000139; Foundation for the National Institutes of Health from UL1RR024134 and UL1TR000003; and NHLBI Division of Intramural Research R38 HL143613-03 (A.M. Lee).
Author Contributions
Conceptualization: Celina Brunson, Josef Coresh, Michelle R. Denburg, Susan L. Furth, Stephen R. Hooper, Arthur M. Lee, Eugene P. Rhee, Ramachandran S. Vasan, Bradley A. Warady.
Data curation: Alison G. Abraham, Arthur M. Lee, Matthew B. Matheson, Yunwen Xu.
Formal analysis: Michelle R. Denburg, Jian Hu, Arthur M. Lee, Rui Xiao.
Funding acquisition: Josef Coresh, Michelle R. Denburg, Susan L. Furth, Erum A. Hartung, Bradley A. Warady.
Investigation: Michelle R. Denburg, Erum A. Hartung, Stephen R. Hooper, Arthur M. Lee.
Methodology: Michelle R. Denburg, Susan L. Furth, Jian Hu, Arthur M. Lee, Sarah Schrauben, Rui Xiao.
Project administration: Michelle R. Denburg, Susan L. Furth, Erum A. Hartung, Paul L. Kimmel, Sarah Schrauben, Bradley A. Warady.
Supervision: Michelle R. Denburg, Erum A. Hartung, Eugene P. Rhee, Rui Xiao.
Visualization: Arthur M. Lee.
Writing – original draft: Arthur M. Lee.
Writing – review & editing: Alison G. Abraham, Celina Brunson, Josef Coresh, Michelle R. Denburg, Susan L. Furth, Erum A. Hartung, Stephen R. Hooper, Jian Hu, Paul L. Kimmel, Arthur M. Lee, Matthew B. Matheson, Eugene P. Rhee, Sarah Schrauben, Ramachandran S. Vasan, Bradley A. Warady, Rui Xiao, Yunwen Xu.
Data Sharing Statement
Partial restrictions to the data and/or materials apply. Data will be deposited into an NIDDK repository in the future. For data inquiries, please contact Judith Jerry (jjerry@jhu.edu) at the Johns Hopkins University KIDMAC.
Supplemental Material
This article contains the following supplemental material online at http://links.lww.com/CJN/B819, http://links.lww.com/CJN/B820, http://links.lww.com/CJN/B821, http://links.lww.com/CJN/B822, http://links.lww.com/CJN/B823.
Supplemental 1. List of all metabolites included for analysis in CKiD.
Supplemental 2. List of all metabolites included for analysis in NiCK.
Supplemental 3. All metabolites associated with neurocognitive z-scores.
Supplemental 4. Metabolite associations with neurocognition in NiCK control sample.
Supplemental 5. Significant metabolite ratio associations with neurocognitive assessments.
Supplemental 6. Sample R code with relevant packages.
References
- 1.Hooper SR Laney N Radcliffe J, et al. Executive functioning in children, adolescents, and young adults with chronic kidney disease. J Dev Behav Pediatr. 2015;36(9):734–742. doi: 10.1097/DBP.0000000000000221 [DOI] [PubMed] [Google Scholar]
- 2.Gerson AC Butler R Moxey-Mims M, et al. Neurocognitive outcomes in children with chronic kidney disease: current findings and contemporary endeavors. Ment Retard Dev Disabil Res Rev. 2006;12(3):208–215. doi: 10.1002/mrdd.20116 [DOI] [PubMed] [Google Scholar]
- 3.Ruebner RL Laney N Kim JY, et al. Neurocognitive dysfunction in children, adolescents, and young adults with CKD. Am J Kidney Dis. 2016;67(4):567–575. doi: 10.1053/j.ajkd.2015.08.025 [DOI] [PubMed] [Google Scholar]
- 4.Hartung EA Kim JY Laney N, et al. Evaluation of neurocognition in youth with CKD using a novel computerized neurocognitive battery. Clin J Am Soc Nephrol. 2016;11(1):39–46. doi: 10.2215/CJN.02110215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong CJ, Moxey-Mims M, Jerry-Fluker J, Warady BA, Furth SL. CKiD (CKD in children) prospective cohort study: a review of current findings. Am J Kidney Dis. 2012;60(6):1002–1011. doi: 10.1053/j.ajkd.2012.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duquette PJ, Hooper SR, Wetherington CE, Icard PF, Gipson DS. Brief report: intellectual and academic functioning in pediatric chronic kidney disease. J Pediatr Psychol. 2007;32(8):1011–1017. doi: 10.1093/jpepsy/jsm036 [DOI] [PubMed] [Google Scholar]
- 7.Gelb SR, Shapiro RJ, Thornton WJ. Predicting medication adherence and employment status following kidney transplant: the relative utility of traditional and everyday cognitive approaches. Neuropsychology. 2010;24(4):514–526. doi: 10.1037/a0018670 [DOI] [PubMed] [Google Scholar]
- 8.Groothoff JW, Grootenhuis MA, Offringa M, Stronks K, Hutten GJ, Heymans HS. Social consequences in adult life of end-stage renal disease in childhood. J Pediatr. 2005;146(4):512–517. doi: 10.1016/j.jpeds.2004.10.060 [DOI] [PubMed] [Google Scholar]
- 9.Stilley CS, Bender CM, Dunbar-Jacob J, Sereika S, Ryan CM. The impact of cognitive function on medication management: three studies. Health Psychol. 2010;29(1):50–55. doi: 10.1037/a0016940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu JR Coresh J Inker LA, et al. Serum metabolites are associated with all-cause mortality in chronic kidney disease. Kidney Int. 2018;94(2):381–389. doi: 10.1016/j.kint.2018.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coresh J Inker LA Sang Y, et al. Metabolomic profiling to improve glomerular filtration rate estimation: a proof-of-concept study. Nephrol Dial Transplant. 2019;34(5):825–833. doi: 10.1093/ndt/gfy094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grams ME Tin A Rebholz CM, et al. Metabolomic alterations associated with cause of CKD. Clin J Am Soc Nephrol. 2017;12(11):1787–1794. doi: 10.2215/CJN.02560317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shah NB Allegretti AS Nigwekar SU, et al. Blood microbiome profile in CKD: a pilot study. Clin J Am Soc Nephrol. 2019;14(5):692–701. doi: 10.2215/CJN.12161018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitrofanova A, Drexler Y, Merscher S, Fornoni A. Role of sphingolipid signaling in glomerular diseases: focus on DKD and FSGS. J Cell Signal. 2020;1(3):56–69. doi: 10.33696/Signaling.1.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jovanovich A, Isakova T, Stubbs J. Microbiome and cardiovascular disease in CKD. Clin J Am Soc Nephrol. 2018;13(10):1598–1604. doi: 10.2215/CJN.12691117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wing MR, Patel SS, Ramezani A, Raj DS. Gut microbiome in chronic kidney disease. Exp Physiol. 2016;101(4):471–477. doi: 10.1113/EP085283 [DOI] [PubMed] [Google Scholar]
- 17.Bagheri M, Shah RD, Mosley JD, Ferguson JF. A metabolome and microbiome wide association study of healthy eating index points to the mechanisms linking dietary pattern and metabolic status. Eur J Nutr. 2021;60(8):4413–4427. doi: 10.1007/s00394-021-02599-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grams ME, Shafi T, Rhee EP. Metabolomics research in chronic kidney disease. J Am Soc Nephrol. 2018;29(6):1588–1590. doi: 10.1681/ASN.2018030256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denburg MR Xu Y Abraham AG, et al. Metabolite biomarkers of CKD progression in children. Clin J Am Soc Nephrol. 2021;16(8):1178–1189. doi: 10.2215/CJN.00220121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee AM Hu J Xu Y, et al. Using machine learning to identify metabolomic signatures of pediatric chronic kidney disease etiology. J Am Soc Nephrol. 2022;33(2):375–386. doi: 10.1681/ASN.2021040538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bressler J Yu B Mosley TH, et al. Metabolomics and cognition in African American adults in midlife: the atherosclerosis risk in communities study. Transl Psychiatry. 2017;7(7):e1173. doi: 10.1038/tp.2017.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi M Bazzano LA He J, et al. Novel serum metabolites associate with cognition phenotypes among Bogalusa Heart Study participants. Aging (Albany NY). 2019;11(14):5124–5139. doi: 10.18632/aging.102107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng K Klotz A Guven A, et al. Identification and validation of N-acetylputrescine in combination with non-canoncial clinical features as a Parkinson's disease biomarker panel. bioRxiv. 2021. doi: 10.1101/2021.07.23.453542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li D Misialek JR Boerwinkle E, et al. Prospective associations of plasma phospholipids and mild cognitive impairment/dementia among African Americans in the ARIC Neurocognitive Study. Alzheimers Dement (Amst). 2017;6:1–10. doi: 10.1016/j.dadm.2016.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furth SL Cole SR Moxey-Mims M, et al. Design and methods of the chronic kidney disease in children (CKiD) prospective cohort study. Clin J Am Soc Nephrol. 2006;1(5):1006–1015. doi: 10.2215/CJN.01941205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hooper SR Johnson RJ Gerson AC, et al. Overview of the findings and advances in the neurocognitive and psychosocial functioning of mild to moderate pediatric CKD: perspectives from the Chronic Kidney Disease in Children (CKiD) cohort study. Pediatr Nephrol. 2022;37(4):765–775. doi: 10.1007/s00467-021-05158-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartung EA Laney N Kim JY, et al. Design and methods of the NiCK study: neurocognitive assessment and magnetic resonance imaging analysis of children and young adults with chronic kidney disease. BMC Nephrol. 2015;16:66. doi: 10.1186/s12882-015-0061-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81(16):6656–6667. doi: 10.1021/ac901536h [DOI] [PubMed] [Google Scholar]
- 29.Evans AM Bridgewater B Liu Q, et al. High resolution mass spectrometry improves data quantity and wality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. Metabolomics. 2014;4:2. doi: 10.4172/2153-0769.1000132 [DOI] [Google Scholar]
- 30.Dehaven CD, Evans AM, Dai H, Lawton KA. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Cheminform. 2010;2(1):9. doi: 10.1186/1758-2946-2-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu Z Kastenmuller G He Y, et al. Differences between human plasma and serum metabolite profiles. PLoS One. 2011;6(7):e21230. doi: 10.1371/journal.pone.0021230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teahan O Gamble S Holmes E, et al. Impact of analytical bias in metabonomic studies of human blood serum and plasma. Anal Chem. 2006;78(13):4307–4318. doi: 10.1021/ac051972y [DOI] [PubMed] [Google Scholar]
- 33.Dettmer K Almstetter MF Appel IJ, et al. Comparison of serum versus plasma collection in gas chromatography—mass spectrometry-based metabolomics. Electrophoresis. 2010;31(14):2365–2373. doi: 10.1002/elps.200900778 [DOI] [PubMed] [Google Scholar]
- 34.Rhee EP Waikar SS Rebholz CM, et al. Variability of two metabolomic platforms in CKD. Clin J Am Soc Nephrol. 2019;14(1):40–48. doi: 10.2215/CJN.07070618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee AM Hu J Xu Y, et al. CKD Biomarkers Consortium. Using machine learning to identify metabolomic signatures of pediatric chronic kidney disease etiology. J Am Soc Nephrol. 2022;33(2):375–386. doi: 10.1681/ASN.2021040538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perneger TV. What's wrong with Bonferroni adjustments. BMJ. 1998;316(7139):1236–1238. doi: 10.1136/bmj.316.7139.1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delgado C Baweja M Burrows NR, et al. Reassessing the inclusion of race in diagnosing kidney diseases: an interim report from the NKF-ASN task force. Am J Kidney Dis. 2021;78(1):103–115. doi: 10.1053/j.ajkd.2021.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borrell LN Elhawary JR Fuentes-Afflick E, et al. Race and genetic ancestry in medicine - a time for reckoning with racism. N Engl J Med. 2021;384(5):474–480. doi: 10.1056/NEJMms2029562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ioannidis JPA, Powe NR, Yancy C. Recalibrating the use of race in medical research. JAMA. 2021;325(7):623–624. doi: 10.1001/jama.2021.0003 [DOI] [PubMed] [Google Scholar]
- 40.Vyas DA, Eisenstein LG, Jones DS. Hidden in plain sight—reconsidering the use of race correction in clinical algorithms. N Engl J Med. 2020;383(9):874–882. doi: 10.1056/NEJMms2004740 [DOI] [PubMed] [Google Scholar]
- 41.Petersen AK Krumsiek J Wagele B, et al. On the hypothesis-free testing of metabolite ratios in genome-wide and metabolome-wide association studies. BMC Bioinformatics. 2012;13:120. doi: 10.1186/1471-2105-13-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wickham S, West MB, Cook PF, Hanigan MH. Gamma-glutamyl compounds: substrate specificity of gamma-glutamyl transpeptidase enzymes. Anal Biochem. 2011;414(2):208–214. doi: 10.1016/j.ab.2011.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson GA, Meister A. Hydrolysis and transfer reactions catalyzed by gamma-glutamyl transpeptidase; evidence for separate substrate sites and for high affinity of L-cystine. Biochem Biophys Res Commun. 1976;71(1):32–36. doi: 10.1016/0006-291x(76)90245-x [DOI] [PubMed] [Google Scholar]
- 44.Weber EJ Chapron A Chapron BD, et al. Development of a microphysiological model of human kidney proximal tubule function. Kidney Int. 2016;90(3):627–637. doi: 10.1016/j.kint.2016.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ndrepepa G, Kastrati A. Gamma-glutamyl transferase and cardiovascular disease. Ann Transl Med. 2016;4(24):481. doi: 10.21037/atm.2016.12.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Do KQ Trabesinger AH Kirsten-Kruger M, et al. Schizophrenia: glutathione deficit in cerebrospinal fluid and prefrontal cortex in vivo. Eur J Neurosci. 2000;12(10):3721–3728. doi: 10.1046/j.1460-9568.2000.00229.x [DOI] [PubMed] [Google Scholar]
- 47.Matsuzawa D Obata T Shirayama Y, et al. Negative correlation between brain glutathione level and negative symptoms in schizophrenia: a 3T 1H-MRS study. PLoS One. 2008;3(4):e1944. doi: 10.1371/journal.pone.0001944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krull KR Bhojwani D Conklin HM, et al. Genetic mediators of neurocognitive outcomes in survivors of childhood acute lymphoblastic leukemia. J Clin Oncol. 2013;31(17):2182–2188. doi: 10.1200/JCO.2012.46.7944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang WJ, Zhang X, Chen WW. Role of oxidative stress in Alzheimer's disease. Biomed Rep. 2016;4(5):519–522. doi: 10.3892/br.2016.630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.D'Amico E, Factor-Litvak P, Santella RM, Mitsumoto H. Clinical perspective on oxidative stress in sporadic amyotrophic lateral sclerosis. Free Radic Biol Med. 2013;65:509–527. doi: 10.1016/j.freeradbiomed.2013.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Black CN, Bot M, Scheffer PG, Cuijpers P, Penninx BW. Is depression associated with increased oxidative stress? A systematic review and meta-analysis. Psychoneuroendocrinology. 2015;51:164–175. doi: 10.1016/j.psyneuen.2014.09.025 [DOI] [PubMed] [Google Scholar]
- 52.Zhang Q, Ju Y, Ma Y, Wang T. N-acetylcysteine improves oxidative stress and inflammatory response in patients with community acquired pneumonia: a randomized controlled trial. Medicine (Baltimore). 2018;97(45):e13087. doi: 10.1097/MD.0000000000013087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharifi N, Amani R, Hajiani E, Cheraghian B. Does vitamin D improve liver enzymes, oxidative stress, and inflammatory biomarkers in adults with non-alcoholic fatty liver disease? A randomized clinical trial. Endocrine. 2014;47(1):70–80. doi: 10.1007/s12020-014-0336-5 [DOI] [PubMed] [Google Scholar]
- 54.Rasmussen ST Andersen JT Nielsen TK, et al. Simvastatin and oxidative stress in humans: a randomized, double-blinded, placebo-controlled clinical trial. Redox Biol. 2016;9:32–38. doi: 10.1016/j.redox.2016.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holle J Bartolomaeus H Lober U, et al. Inflammation in children with CKD linked to gut dysbiosis and metabolite imbalance. J Am Soc Nephrol. 2022;33(12):2259–2275. doi: 10.1681/ASN.2022030378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Halverson T, Alagiakrishnan K. Gut microbes in neurocognitive and mental health disorders. Ann Med. 2020;52(8):423–443. doi: 10.1080/07853890.2020.1808239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Noble EE, Hsu TM, Kanoski SE. Gut to brain dysbiosis: mechanisms linking western diet consumption, the microbiome, and cognitive impairment. Front Behav Neurosci. 2017;11:9. doi: 10.3389/fnbeh.2017.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manderino L Carroll I Azcarate-Peril MA, et al. Preliminary evidence for an association between the composition of the gut microbiome and cognitive function in neurologically healthy older adults. J Int Neuropsychol Soc. 2017;23(8):700–705. doi: 10.1017/S1355617717000492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Niwa T, Ise M. Indoxyl sulfate, a circulating uremic toxin, stimulates the progression of glomerular sclerosis. J Lab Clin Med. 1994;124(1):96–104. doi: 10.5555/uri:pii:0022214394901600 [DOI] [PubMed] [Google Scholar]
- 60.Vanholder R De Smet R Glorieux G, et al. Review on uremic toxins: classification, concentration, and interindividual variability. Kidney Int. 2003;63(5):1934–1943. doi: 10.1046/j.1523-1755.2003.00924.x [DOI] [PubMed] [Google Scholar]
- 61.Barreto FC Barreto DV Liabeuf S, et al. Serum indoxyl sulfate is associated with vascular disease and mortality in chronic kidney disease patients. Clin J Am Soc Nephrol. 2009;4(10):1551–1558. doi: 10.2215/CJN.03980609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang CH Cheng ML Liu MH, et al. Increased p-cresyl sulfate level is independently associated with poor outcomes in patients with heart failure. Heart Vessels. 2016;31(7):1100–1108. doi: 10.1007/s00380-015-0702-0 [DOI] [PubMed] [Google Scholar]
- 63.Poesen R Claes K Evenepoel P, et al. Microbiota-derived phenylacetylglutamine associates with overall mortality and cardiovascular disease in patients with CKD. J Am Soc Nephrol. 2016;27(11):3479–3487. doi: 10.1681/ASN.2015121302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Russell WR Duncan SH Scobbie L, et al. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol Nutr Food Res. 2013;57(3):523–535. doi: 10.1002/mnfr.201200594 [DOI] [PubMed] [Google Scholar]
- 65.Kalim S, Rhee EP. An overview of renal metabolomics. Kidney Int. 2017;91(1):61–69. doi: 10.1016/j.kint.2016.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang WH Wang Z Levison BS, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368(17):1575–1584. doi: 10.1056/NEJMoa1109400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Z Klipfell E Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57–63. doi: 10.1038/nature09922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu W Gregory JC Org E, et al. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell. 2016;165(1):111–124. doi: 10.1016/j.cell.2016.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sapa H Gutierrez OM Shlipak MG, et al. Association of uremic solutes with cardiovascular death in diabetic kidney disease. Am J Kidney Dis. 2022;80(4):502–512.e1. doi: 10.1053/j.ajkd.2022.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gamboa JL Billings FT 4th Bojanowski MT, et al. Mitochondrial dysfunction and oxidative stress in patients with chronic kidney disease. Physiol Rep. 2016;4(9):e12780. doi: 10.14814/phy2.12780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roshanravan B Kestenbaum B Gamboa J, et al. CKD and muscle mitochondrial energetics. Am J Kidney Dis. 2016;68(4):658–659. doi: 10.1053/j.ajkd.2016.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Indiveri C Iacobazzi V Tonazzi A, et al. The mitochondrial carnitine/acylcarnitine carrier: function, structure and physiopathology. Mol Aspects Med. 2011;32(4-6):223–233. doi: 10.1016/j.mam.2011.10.008 [DOI] [PubMed] [Google Scholar]
- 73.Morgans HA, Chadha V, Warady BA. The role of carnitine in maintenance dialysis therapy. Pediatr Nephrol. 2021;36(8):2545–2551. doi: 10.1007/s00467-021-05101-z [DOI] [PubMed] [Google Scholar]
- 74.Costigan MG, Callaghan CA, Lindup WE. Hypothesis: is accumulation of a furan dicarboxylic acid (3-carboxy-4- methyl-5-propyl-2-furanpropanoic acid) related to the neurological abnormalities in patients with renal failure? Nephron. 1996;73(2):169–173. doi: 10.1159/000189035 [DOI] [PubMed] [Google Scholar]
- 75.Niwa T, Aiuchi T, Nakaya K, Emoto Y, Miyazaki T, Maeda K. Inhibition of mitochondrial respiration by furancarboxylic acid accumulated in uremic serum in its albumin-bound and non-dialyzable form. Clin Nephrol. 1993;39(2):92–96. PMID: 8448924 [PubMed] [Google Scholar]
- 76.Abela L Spiegel R Crowther LM, et al. Plasma metabolomics reveals a diagnostic metabolic fingerprint for mitochondrial aconitase (ACO2) deficiency. PLoS One. 2017;12(5):e0176363. doi: 10.1371/journal.pone.0176363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fornoni A Sageshima J Wei C, et al. Rituximab targets podocytes in recurrent focal segmental glomerulosclerosis. Sci Transl Med. 2011;3(85):85ra46. doi: 10.1126/scitranslmed.3002231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Watanabe S, Tsugawa K, Tsuruga K, Imaizumi T, Tanaka H. Urinary excretion of sphingomyelinase phosphodiesterase acid-like 3b in children with intractable nephrotic syndrome. Pediatr Int. 2017;59(10):1112–1115. doi: 10.1111/ped.13355 [DOI] [PubMed] [Google Scholar]
- 79.Chen SD, Yin JH, Hwang CS, Tang CM, Yang DI. Anti-apoptotic and anti-oxidative mechanisms of minocycline against sphingomyelinase/ceramide neurotoxicity: implication in Alzheimer's disease and cerebral ischemia. Free Radic Res. 2012;46(8):940–950. doi: 10.3109/10715762.2012.674640 [DOI] [PubMed] [Google Scholar]
- 80.Haughey NJ Cutler RG Tamara A, et al. Perturbation of sphingolipid metabolism and ceramide production in HIV-dementia. Ann Neurol. 2004;55(2):257–267. doi: 10.1002/ana.10828 [DOI] [PubMed] [Google Scholar]
- 81.Mallela SK, Mitrofanova A, Merscher S, Fornoni A. Regulation of the amount of ceramide-1-phosphate synthesized in differentiated human podocytes. Biochim Biophys Acta Mol Cell Biol Lipids. 2019;1864(12):158517. doi: 10.1016/j.bbalip.2019.158517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Auge N Maupas-Schwalm F Elbaz M, et al. Role for matrix metalloproteinase-2 in oxidized low-density lipoprotein-induced activation of the sphingomyelin/ceramide pathway and smooth muscle cell proliferation. Circulation. 2004;110(5):571–578. doi: 10.1161/01.CIR.0000136995.83451.1D [DOI] [PubMed] [Google Scholar]
- 83.Mielke MM, Bandaru VV, Haughey NJ, Rabins PV, Lyketsos CG, Carlson MC. Serum sphingomyelins and ceramides are early predictors of memory impairment. Neurobiol Aging. 2010;31(1):17–24. doi: 10.1016/j.neurobiolaging.2008.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gonzalez GDS, Schnyer DM. Attention and working memory biases to black and asian faces during intergroup contexts. Front Psychol. 2018;9:2743. doi: 10.3389/fpsyg.2018.02743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Solini A, Manca ML, Penno G, Pugliese G, Cobb JE, Ferrannini E. Prediction of declining renal function and albuminuria in patients with type 2 diabetes by metabolomics. J Clin Endocrinol Metab. 2016;101(2):696–704. doi: 10.1210/jc.2015-3345 [DOI] [PubMed] [Google Scholar]
- 86.Luo S Feofanova EV Tin A, et al. Genome-wide association study of serum metabolites in the African American study of kidney disease and hypertension. Kidney Int. 2021;100(2):430–439. doi: 10.1016/j.kint.2021.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Luo S Surapaneni A Zheng Z, et al. NAT8 variants, N-acetylated amino acids, and progression of CKD. Clin J Am Soc Nephrol. 2020;16(1):37–47. doi: 10.2215/CJN.08600520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chambers JC Zhang W Lord GM, et al. Genetic loci influencing kidney function and chronic kidney disease. Nat Genet. 2010;42(5):373–375. doi: 10.1038/ng.566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Veiga-da-Cunha M, Tyteca D, Stroobant V, Courtoy PJ, Opperdoes FR, Van Schaftingen E. Molecular identification of NAT8 as the enzyme that acetylates cysteine S-conjugates to mercapturic acids. J Biol Chem. 2010;285(24):18888–18898. doi: 10.1074/jbc.M110.110924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kottgen A Glazer NL Dehghan A, et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet. 2009;41(6):712–717. doi: 10.1038/ng.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Madeo F, Eisenberg T, Pietrocola F, Kroemer G. Spermidine in health and disease. Science. 2018;359(6374):eaan2788. doi: 10.1126/science.aan2788 [DOI] [PubMed] [Google Scholar]
- 92.Alonso A Yu B Sun YV, et al. Serum metabolomics and incidence of atrial fibrillation (from the atherosclerosis risk in communities study). Am J Cardiol. 2019;123(12):1955–1961. doi: 10.1016/j.amjcard.2019.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Verbitsky M Kogon AJ Matheson M, et al. Genomic disorders and neurocognitive impairment in pediatric CKD. J Am Soc Nephrol. 2017;28(8):2303–2309. doi: 10.1681/ASN.2016101108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Verbitsky M Sanna-Cherchi S Fasel DA, et al. Genomic imbalances in pediatric patients with chronic kidney disease. J Clin Invest. 2015;125(5):2171–2178. doi: 10.1172/JCI80877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Smith GD, Ebrahim S. ’Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32(1):1–22. doi: 10.1093/ije/dyg070 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Partial restrictions to the data and/or materials apply. Data will be deposited into an NIDDK repository in the future. For data inquiries, please contact Judith Jerry (jjerry@jhu.edu) at the Johns Hopkins University KIDMAC.