Abstract
Human immunodeficiency virus type 1 (HIV-1) gene transcription is characterized by two temporally distinct phases. While the initial phase relies solely on cellular transcription factors, the subsequent phase is activated by the viral Tat transactivator. We have previously reported that the subsequent phase of viral gene transcription can be repressed by the chicken ovalbumin upstream promoter transcription factor (COUP-TF)-interacting protein 2 (CTIP2) in human microglial cells [O. Rohr, D. Lecestre, S. Chasserot-Golaz, C. Marban, D. Avram, D. Aunis, M. Leid and E. Schaeffer (2003), J. Virol., 77, 5415–5427]. Here, we demonstrate that CTIP proteins also repress the initial phase of HIV-1 gene transcription, mainly supported by the cellular transcription factors Sp1 and COUP-TF in microglial cells. We report that CTIP2 represses Sp1- and COUP-TF-mediated activation of HIV-1 gene transcription and viral replication as a result of physical interactions with COUP-TF and Sp1 in microglial nuclei. Using laser confocal microscopy CTIP2 was found to colocalize with Sp1, COUP-TF and the heterochromatin-associated protein Hp1α, which is mainly detected in transcriptionally repressed heterochromatic region. Moreover, we describe that CTIP2 can be recruited to the HIV-1 promoter via its association with Sp1 bound to the GC-box sequences of the long terminal repeat (LTR). Since our findings demonstrate that CTIP2 interacts with the HIV-1 proximal promoter, it is likely that CTIP2 promotes HIV-1 gene silencing by forcing transcriptionally repressed heterochromatic environment to the viral LTR region.
INTRODUCTION
Infection of the central nervous system (CNS) by the human immunodeficiency virus type 1 (HIV-1) is associated with a spectrum of neurological damage, ranging from acute encephalopathies to AIDS dementia (1,2). The CNS constitutes a sanctuary and a reservoir for the virus despite antiretroviral therapies (3). Microglial cells, the CNS resident macrophages, may be particularly suitable for this purpose because they are long lived, HIV productive for several weeks and relatively immune to virus-induced cytopathology [reviewed in (4)]. Since microglial cells are the primary targets of HIV-1 productive infection within the CNS (5,6), investigation on HIV-1 repression in these cells are important for the global understanding of HIV pathogenesis and for the development of anti-HIV strategies.
HIV-1 gene transcription is a key step in the control of the virus life cycle [reviewed in (7)]. Viral gene transcription is characterized by two temporally distinct phases. The initial phase occurs immediately after integration and relies solely on cellular transcription factors. Most of the transcripts cannot elongate efficiently and terminate rapidly after initiation. However, a few transcripts elongate throughout the genome, resulting in transcription and expression of the viral transactivator Tat. The subsequent phase of transcription occurs when enough Tat protein has accumulated. Tat binds to TAR, recruits pTEFb complex and dramatically stimulates HIV-1 gene transcription [reviewed in (8,9)].
We have previously studied some of the cellular transcription factors that impact HIV-1 gene transcription in microglial cells. Chicken ovalbumin upstream promoter transcription factor I (COUP-TFI)/Ear3, Arp1/COUP-TFII and Ear2/COUP-TFIII proteins are the three members of the COUP-TF orphan nuclear receptors superfamily. We have described that the orphan nuclear receptor COUP-TFI/Ear3 (10,11) is expressed in numerous human CNS cell lineages, including microglial cells. COUP-TFI is an activator of HIV-1 gene transcription in oligodendrocytes (12), microglia (13) and T lymphocytes (14). Moreover, we have revealed the importance of functional interactions between the nuclear factor for interleukin 6 (NF-Il6), Sp1 and COUP-TFI in the regulation of the initial phase of HIV-1 gene transcription in brain cells (15). The transcription factor Sp1 is one of the crucial cellular proteins for efficient HIV-1 gene transcription (16–18). A number of studies have reported that Sp1 can serve as an anchor for indirect binding of other transcription factors to the HIV-1 long terminal repeat (LTR) region. In all cell types, an interaction between Sp1 and Tat is required for optimal HIV-enhancer activation (19). Moreover, Sp1 is able to recruit the Cyclin T1 subunit of P-TEFb to the LTR, which helps to bypass a requirement for TAR/Tat and promotes processive transcription without Tat (20). In microglial cells, the main HIV-1 target cells in the brain (6), Sp1 anchors COUP-TF (13) and NF-IL6 (15) to the three GC-boxes adjacent to the viral CATA box. The CATA box is used instead of the TATAA box for optimal HIV-1 gene transcription and replication (21). We have shown that Sp1 and COUP-TF interact and cooperate in the transcriptional activation of HIV-1 gene transcription (13). Moreover, COUP-TF can substitute to Sp1 association with the viral Tat protein to restore Tat function and HIV-1 replication in microglial cells (22). These results highlight the key roles of Sp1 and COUP-TF proteins in HIV-1 expression in microglial cells.
Members of the COUP-TF family have been shown to bind related zinc finger proteins named COUP-TF-interacting protein 1 (CTIP1) and 2 (CTIP2) that are highly expressed in brain and immune systems (23,24). Recent results support the selective contribution of these proteins in the development and function of the nervous and immune systems (24). We recently described that the nuclear cofactor CTIP2 inhibits the subsequent phase of HIV-1 gene transcription and viral replication by relocating the viral transactivator Tat protein to transcriptionally inactive regions of chromatin via Hp1α (25). In the present work, CTIP2 overexpression leads to the repression of HIV-1 replication. In contrast, CTIP1 was unable to affect Tat function. Using confocal microscopy to visualize Tat subcellular distribution in the presence of each CTIP proteins, we found that CTIP2, but not CTIP1, leads to the disruption of Tat nuclear localization and to its recruitment within CTIP2-induced nuclear ball-like structures. In addition, we showed that CTIP2 and Hp1α associate with Tat to form a three-protein complex. These findings suggest that the inhibition of HIV-1 expression by CTIP2 correlates with the recruitment of Tat within CTIP2-induced structures and its relocalization within inactive regions of chromatin.
On line with this study, we noticed that CTIP proteins also affect HIV-1 gene transcription in the absence of Tat, which means that CTIP proteins may also impair endogenous cellular transcription factor functions of importance for the initial phase of HIV-1 gene transcription.
Here, we report that CTIP proteins repress HIV-1 gene transcription via the proximal LTR region which binds Sp1 and COUP-TF transactivators. Since COUP-TF and Sp1 cellular transcription factors are two of the major contributors to HIV-1 gene transcription in microglial cells, we have postulated that Sp1- and COUP-TF-mediated LTR-driven activation may be impaired by CTIP1 and CTIP2. We report here that both CTIP1 and CTIP2 repress Sp1- and COUP-TF-mediated stimulation of HIV-1 gene expression. Since the CTIP2 repressive activity is much stronger than the CTIP1-mediated inhibition of HIV-1 replication, we focused our study on CTIP2. CTIP2-mediated repression of Sp1 and COUP-TF functions results from direct physical interactions of these cellular transcription factors with CTIP2. Association with CTIP2 forces Sp1 and COUP-TF relocation to CTIP2-induced subnuclear structures containing the heterochromatin-associated protein Hp1α. Moreover, CTIP2 can be recruited to the HIV-1 promoter by a physical association with Sp1 bound to the GC-box regions of the LTR. Since Sp1 or COUP-TF transcription factors are necessary for the Tat function in microglial cells (22), CTIP2-mediated repression of COUP-TF and Sp1 functions may largely contribute to the previously described CTIP2-mediated impairment of Tat activity and viral replication in microglial cells (25).
MATERIALS AND METHODS
Plasmids
Most of the constructs used in our assays were described previously: HIV-1 (LAI) 5′ LTR chloramphenicol acetyltransferase (pLTR-CAT) (22), pGC-WAP-CAT (26), pLTR-LUC 287–535 (13), pLTR-CAT mutGC and pLTR-CAT ΔGC (22), pcDNA3, full-length constructs HA-CTIP1, Flag-CTIP2 and GST-CTIP2 (23), deletions constructs Flag-CTIP2 (25), RSV-COUP-TF, CMV-Sp1, GST-COUP-TF1, GST-COUP-TF2 and GST-COUP-TF3 (13).
Several plasmids were kindly provided by the following investigators: GST-Sp1 constructs were provided by H. Rotheneder (Vienna, Austria) (27) and pLTR-LUC by C. Van Lint (Gosselies, Belgium). pNL4-3 (28) was obtained through the AIDS Research and Reference Reagent Program, division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), NIH.
To construct RFP-CTIP2, the expression vector Flag-CTIP2 was digested with EcoRI and the CTIP2 insert was subcloned into the EcoRI site of the pDSRed vector (Clontech). The GFP-Sp1 was constructed by isolating the XhoI–HindIII Sp1 insert from the GST-Sp1 expression vector and subcloned into the XhoI–HindIII sites of the pEGFP-C1 vector (Clontech).
Cell culture
The human microglial (provided by M. Tardieu, Paris, France) (29), the TZM-bl (30–32) and the HEK 293T cell lines were maintained in DMEM containing 10% fetal calf serum and 100 U/ml penicillin–streptomycin.
Viral replication
Microglial cells cultured in 12-well plates were transfected using the calcium phosphate coprecipitation method with HIV-1 pNL4-3 (1.5 μg), CMV-Sp1 (1 μg) or RSV-COUP-TF (1 μg) plasmids and the indicated HA-CTIP1 or Flag-CTIP2 expression vectors (0.1, 0.5 or 1 μg). Total amounts of DNA were identical in each experiment. Compensations were made by adding the corresponding empty vectors. Each transfection was carried out in duplicate and repeated a minimum of three separate times with two different plasmid preparations. HIV-1 replication was monitored as described previously (25).
CAT assays
Microglial cells cultured in 12-well plates were transfected using the calcium phosphate coprecipitation method with pLTR-CAT (3 μg), pGC-WAP-CAT (3 μg) or pLTR-CAT ΔGC (3 μg), CMV-Sp1 (1 μg) or RSV-COUP-TF (1 μg) plasmids and the indicated HA-CTIP1 or Flag-CTIP2 (0.1, 0.5 or 1 μg) expression vectors. Total amounts of DNA were identical in each experiments. Compensations were made by adding the corresponding empty vectors. Each transfection was carried out in duplicate and repeated a minimum of three separate times with two different plasmid preparations. CAT assays were carried out using standard techniques.
Co-immunoprecipitation assays
Microglial cells cultured in 100 mm diameter dishes were transfected using Lipofectamine™ 2000 Reagent (Invitrogen) with the indicated Flag-CTIP2 (30 μg). Twenty four hours post-transfection, the cells were washed twice with cold phosphate-buffered saline, harvested and prepared for nuclear extracts (12). Nuclear proteins were first diluted in TNE buffer [50 mM Tris, pH 8, 1% Nonidet, 2 mM EDTA, protease inhibitors cocktail (Roche)], cleared with Protein A/G Plus-Agarose (Santa Cruz Biotechnology) and finally incubated overnight with primary antibodies: anti-Sp1 (Sigma), anti-COUP-TF (kindly provided by J. E. Mertz, Madison, WI) or anti-Hp1α (kindly provided by R. Losson, IGBMC, Strasbourg, France). The extracts were then immunoprecipitated by the addition of Protein A/G Plus-Agarose (Santa Cruz Biotechnology). After extensive washing with TNE buffer, the immunoprecipitates were processed for SDS–PAGE and western blot analysis.
Glutathione S-transferase (GST) pull-down assays
Production of GST fusion proteins was described previously (13) and visualized by Coomassie blue staining. The 35S-labeled proteins were prepared by in vitro transcription and translation using the TNT® T7 Coupled Wheat Germ Extract System (Promega). GST pull-down assays were performed as described previously (25).
SDS–PAGE and western blot analysis
SDS–PAGE was performed using standard techniques. Proteins were detected using antibodies directed against the Flag epitope (M2 mouse monoclonal; Sigma), against the COUP-TF protein (kindly provided by J. E. Mertz) and against the Sp1 protein (Sigma). Proteins were visualized by chemiluminescence using the Super Signal Chemiluminescence Detection System (Pierce).
Electrophoretic mobility shift assays (EMSA)
The probe used in our experiments has been described previously (13) and corresponds to the three Sp1 binding sites of the HIV-1 proximal LTR region. Once produced, GST fusion proteins were eluted in glutathione buffer (50 mM Tris, pH 8, 20 mM reduced glutathione). Purified Sp1 (Promega) and GST fusion proteins were then incubated with the 32P-labeled probe in binding buffer (20 mM HEPES, pH 7.9, 1 mM MgCl2, 60 mM KCl, 0.5 mM EDTA, 1 mM DTT and 10% glycerol) at 4°C for 15 min. For supershift experiments, GST fusion proteins were incubated with the primary antibodies: anti-COUP-TF (kindly provided by J. E. Mertz), anti-CTIP2 and anti-Sp1 (Santa Cruz Biotechnology) for ∼16 h before adding the probe. EMSA assays were performed as described previously (25).
Indirect immunofluorescence and confocal microscopy
Microglial cells cultured in 48-well plates were transfected or not using Lipofectamine™ 2000 Reagent (Invitrogen) with Flag-CTIP2, RFP-CTIP2 and/or GFP-Sp1 expression vectors. Cells were fixed and permeabilized as described previously (25). The coverslips were then incubated for 1 h at room temperature with primary antibodies directed against COUP-TF (Santa Cruz Biotechnology or kindly provided by J. E. Mertz), Sp1 (sigma) and Hp1α proteins and/or against the Flag epitope (M2 mouse monoclonal; Sigma). The primary immunocomplexes were revealed by CY2- or CY3-labeled secondary anti-species antibodies. The stained cells were analyzed by confocal microscopy using a Zeiss laser scanning microscope (model 510 invert) equipped with a Planapo oil (63×) immersion lens (numerical aperture = 1.4).
Chromatin immunoprecipitation (ChIP) assays
TZM-bl and HEK 293T cells cultured in 100 mm diameter dishes were transfected using the calcium phosphate coprecipitation method with the indicated pLTR-LUC, pLTR-CAT mutGC and Flag-CTIP2 (30 μg) expression vector. ChIP assays were performed using the ChIP Assay Kit (Upstate) 48 h post-transfection. The primary antibodies used for the ChIP were anti-Sp1 (Upstate), anti-Hp1α (Upstate), anti-COUP-TF (Santa Cruz Biotechnology) and anti-Flag M2 mouse monoclonal (Sigma). DNA was subjected to PCR amplification using a 5′ primer (5′-GATAAGGTAGAAGAGGCC-3′) corresponding to the LTR sequence located 293 nt downstream of the transcriptional start site and a 3′ primer (5′-CTAACCAGAGAGACCCAGTAC-3′) corresponding to a region just upstream of the transcriptional start site. The resulting PCR product (307 bp) was analyzed by agarose gel electrophoresis and ethidium bromide staining. Three separate experiments were performed.
RESULTS
CTIP1 and CTIP2 proteins repress HIV-1 gene transcription via the LTR proximal region
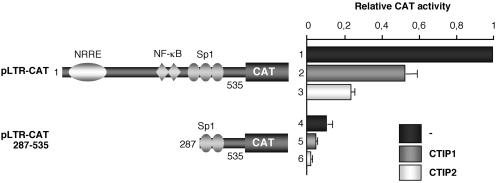
As previously shown, CTIP1 and CTIP2 proteins inhibited the LTR-driven transcription in transient transfection assays (Figure 1, lanes 2 and 3) (25). To delineate the LTR region responsible for CTIP1- and CTIP2-mediated HIV-1 gene transcriptional repression, microglial cells were transfected with a 5′ deleted pLTR-CAT reporter plasmid in the presence or absence of CTIP1 and CTIP2 expression vectors. Deletion of the 5′ region downstream of the two proximal GC-box sequences did not affect CTIP1 and CTIP2 ability to repress LTR-driven CAT activity (Figure 1, lanes 5 and 6), indicating that CTIP proteins repressive function can be mediated by the proximal region of the LTR encompassing two GC-box sequences, the CATA sequence (21) and the TAR region. We have previously observed that the cellular transcription factors, Sp1 and Sp3, are directly bound to the LTR GC-box sequences in microglial cells (13). Moreover, the orphan nuclear receptor COUP-TF is indirectly anchored to this region via its association with Sp1 (13). We have largely described that Sp1 and COUP-TF transcription factors are two of the major contributors to the initial phase of HIV-1 gene transcription in microglial cells (9,13,22). Taken together, these findings strongly suggest that CTIP repressive activity may result from impairment of endogenous Sp1 and COUP-TF protein functions.
Figure 1.
CTIP1 and CTIP2 proteins repress HIV-1 gene transcription via the proximal LTR region. Microglial cells were transfected with 3 μg of pLTR-CAT or 3 μg of pLTR-CAT (287–535) in the presence or absence of 1 μg of HA-CTIP1 or Flag-CTIP2. Two days post-transfection, CAT activities were measured and expressed relative to the CAT activity obtained with pLTR-CAT alone with the standard deviations indicated (values correspond to an average of at least three independent experiments performed in duplicate).
CTIP1 and CTIP2 cofactors inhibit Sp1- and COUP-TF-mediated activation of HIV-1 gene transcription and related viral replication
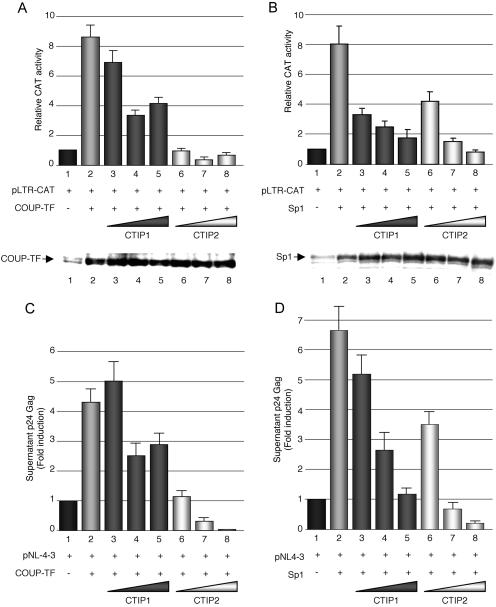
To decipher the mechanism whereby CTIP proteins affect the initial phase of HIV-1 gene transcription, we investigated the impacts of CTIP proteins on Sp1 and COUP-TF functions. Microglial cells were transfected with pLTR-CAT plasmid together with the vectors expressing COUP-TF (Figure 2A) or Sp1 (Figure 2B) proteins and with increasing amounts of HA-CTIP1 or Flag-CTIP2 expression vectors. The two proteins were able to repress COUP-TF- and Sp1-mediated stimulations of CAT activity. As a control, we performed western blot experiments, indicating that COUP-TF and Sp1 expressions were not downregulated but slightly upregulated by the cofactors. Since overexpression of COUP-TF and Sp1 stimulates HIV-1 gene expression, this modest upregulation could not be responsible for the CTIPs repressive function. To correlate CTIP1 and CTIP2 repressive activity on LTR-driven transcription to the level of HIV-1 replication, we studied their impacts on Sp1- and COUP-TF-mediated stimulations of viral replication. Cells were transfected with HIV-1 pNL4-3 and COUP-TF (Figure 2C) or Sp1 (Figure 2D) expression vectors in the presence of increasing amounts of CTIP1 or CTIP2 expression vectors. The level of viral replication was investigated by quantifying p24 Gag expression in the culture supernatants 2 days after transfection. While both CTIP1 and CTIP2 inhibited COUP-TF (Figure 2C) and Sp1 (Figure 2D) activation of HIV-1 NL4-3 replication, the strongest repression was observed with CTIP2 (Figure 2C and D, lanes 6–8), indicating that the repressing activity of CTIP2 on HIV-1 replication cannot be explained only by its previously described capacity to repress the viral Tat protein (25). These results strongly suggest that CTIP1 and CTIP2 repress the initial phase of HIV-1 gene transcription through direct or indirect interactions with endogenous Sp1 and COUP-TF proteins.
Figure 2.
CTIP1 and CTIP2 inhibit Sp1- and COUP-TF-mediated activation of HIV-1 gene transcription and related viral replication. Microglial cells were cotransfected with 3 μg of pLTR-CAT (A and B) or 3 μg of pNL4-3 (C and D), 1 μg of RSV-COUP-TF (A and C) or CMV-Sp1 (B and D) expression vectors and increasing amounts of HA-CTIP1 or Flag-CTIP2 (0.1, 0.5 or 1 μg). (A and B) Two days post-transfection, CAT activities were measured and expressed relative to the value obtained with pLTR-CAT alone with the standard deviation indicated (values correspond to an average of at least three independent experiments performed in duplicate). Western blot experiments were performed on nuclear extracts with antibodies directed against COUP-TF or Sp1 proteins as indicated. (C and D) Two days post-transfection, culture supernatants were analyzed for p24 Gag contents and expressed relative to the value obtained with pNL4-3 alone taken as 1. Depending on the cell confluency, this value varied between 500 and 5000 pg/ml. Values correspond to an average of at least three independent experiments performed in duplicate.
The HIV-1 Tat function and the subsequent phase of the HIV-1 gene transcription also depend on Tat association with Sp1 or COUP-TF in microglial cells (22). Since CTIP2-mediated repressive activities are much stronger than those observed for CTIP1 and since CTIP2, but not CTIP1, inhibits the subsequent phase of the HIV-1 gene transcription, we focused our mechanistic investigation on CTIP2.
CTIP2 interacts with COUP-TF and Sp1 in vitro by two interfaces
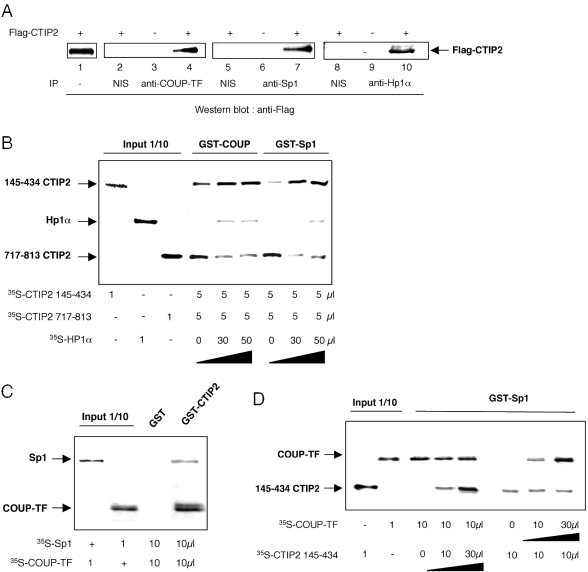
To decipher the mechanism whereby CTIP2 represses COUP-TF and Sp1 stimulatory activities, we first examined whether these proteins were able to interact in vitro. GST pull-down assays were performed with in vitro translated 35S-labeled CTIP2 and equal amounts of full-length or truncated GST-COUP-TF and GST-Sp1 fusion proteins (Figure 3A). Results show that CTIP2 bound specifically to GST-COUP-TF and GST-Sp1 (Figure 3A, lanes 3 and 8, respectively) but not to the control GST protein (Figure 3A, lanes 2 and 7, respectively). Approximately 2–5% and 1–2% of the 35S-labeled CTIP2 interacted with GST-COUP-TF and GST-Sp1, respectively. Moreover, the 49–148 region of COUP-TF (Figure 3A, lane 5) and the 622–788 region of Sp1 (Figure 3A, lane 11) were still able to mediate association with CTIP2, indicating that CTIP2 interacted with the N-terminal region of COUP-TF and the C-terminal region of Sp1, both of which include zinc finger domains.
Figure 3.
CTIP2 interacts in vitro with COUP-TF and Sp1 by two interfaces. (A) Upper panels: schematic representation of the COUP-TF and Sp1 proteins; lower panels: GST pull-down assays were performed with 35S-labeled CTIP2 incubated with GST (lanes 2 and 7) or GST fusion proteins of the indicated COUP-TF domains (lanes 3–5) or Sp1 domains (lanes 8–11). Approximately 1% of the total 35S-labeled CTIP2 obtained was loaded as input control (lanes 1 and 6). (B) GST pull-down assays were performed with 35S-labeled full-length or truncated CTIP2 proteins incubated with GST (lanes 2 and 5), GST-COUP-TF (lane 6) or GST-Sp1 (lane 3) fusion proteins. Approximately 1% of the total 35S-labeled proteins used were loaded as input control (lanes 1 and 4). Representative Coomassie stainings of GST, GST-Sp1 and GST-COUP-TF proteins were presented (B lower panels).
To precisely delineate the region of CTIP2 protein that associates with Sp1 and COUP-TF, GST pull-down assays were performed with full-length GST-COUP-TF or GST-Sp1 fusion proteins and a series of in vitro-translated 35S-labeled full-length and deletion mutants of CTIP2 (Figure 3B). The full-length as well as deletion mutants starting at position 350, 610 and 717 did mediate interaction with GST-COUP-TF (Figure 3B, lane 6) and GST-Sp1 (Figure 3B, lane 3) but not with the control GST protein (Figure 3B, lanes 2 and 5). These results show that CTIP2 is able to interact with Sp1 and COUP-TF via its C-terminal zinc finger domain region. We have previously shown that CTIP2 harbors two independent interaction domains with the viral Tat protein (25). This prompted us to determine whether an additional interaction interface was located in the N-terminal or the central region of CTIP2. To this end, GST pull-down experiments were performed with additional deletion constructs. As expected, the deletion mutant 350–716 was not able to interact with COUP-TF and Sp1, confirming that the C-terminal region of CTIP2 containing residues 717–813 mediates binding to these transcription factors. However, the 1–354 and the 145–434 constructs appeared to be able to restore interactions, suggesting that the central region of CTIP2 located between the residues 145 and 354 might be sufficient for COUP-TF and Sp1 association.
Thus, these findings indicate that CTIP2 interacts in vitro with the zinc finger domains of COUP-TF and Sp1 by two interfaces located in the central region (residues 145–354) and the C-terminal region (residues 717–813).
CTIP2 colocalizes with Sp1 and COUP-TF within Hp1α-associated structures
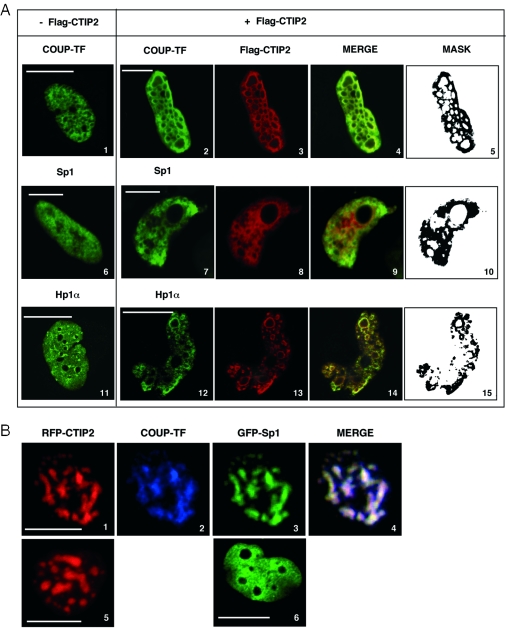
To visualize the association of CTIP2 with Sp1 and COUP-TF within the nucleus, Flag-CTIP2 transfected microglial cells were observed by immunofluorescence confocal laser microscopy (Figure 4A). As previously described (25), anti-Flag immunostaining of Flag-CTIP2 revealed the formation of CTIP2-induced nuclear ball-like structures in microglial cells nuclei (Figure 4A, images 3, 8 and 13). Flag-CTIP2 appeared as concentrated at the periphery but absent within the structures. Interestingly, observations of RFP-CTIP2 (Figure 4B, images 1 and 5) fusion protein revealed the presence of CTIP2 at the periphery but also within the previously described nuclear structures, suggesting that the Flag epitopes within these ball-like structures were probably not accessible to the anti-Flag antibodies. To control the nuclear location of RFP-CTIP2 expression, cells were transfected with RFP-CTIP2 and GFP-Lamin B expression vectors. As previously described (25), CTIP2-induced nuclear structures were located in the nucleus of microglial cells (C. Marban and O. Rohr, unpublished data). In the absence of CTIP2, endogenous COUP-TF and Sp1 proteins were expressed in the nucleoplasm with a speckled and diffused staining pattern (images 1 and 6). Moreover, in the presence of Flag-CTIP2, these transcription factors were at least partially located to CTIP2-induced nuclear structures (Figure 4A, images 2 and 7). Endogenous Sp1 (Figure 4A, images 9 and 10) and COUP-TF (Figure 4A, images 4 and 5) proteins colocalized with CTIP2 at the periphery of the immunostained structures. Up to 87 and 70% of endogenous COUP-TF and Sp1 proteins colocalized with CTIP2, respectively. No staining was observed inside the ball-like structures. We have reported that CTIP2 relocates the viral Tat transactivator to the heterochromatin-associated protein Hp1α (25). Endogenous Hp1α exhibited a diffused and speckled nuclear distribution in the absence of CTIP2 (Figure 4A, image 11). To visualize the presence of Hp1α in CTIP2-induced structures in the absence of Tat, we performed immunodetection of endogenous Hp1α in microglial cells expressing Flag-CTIP2. As shown in Figure 4A (images 14 and 15), endogenous Hp1α colocalized with CTIP2, suggesting that endogenous COUP-TF, Sp1 and Hp1α proteins are relocated to the same CTIP2-induced nuclear structures. To address whether COUP-TF and Sp1 also colocalized with CTIP2 inside the ball-like structures, we performed immunodetection of the endogenous COUP-TF protein with other different polyclonal antibodies (kindly provided by J. E. Mertz) in the presence of RFP-CTIP2 and GFP-Sp1 fusion proteins. As shown in Figure 4B (image 2), endogenous COUP-TF was observed inside the structures, confirming that the epitope accessibility to the antibodies is determinant to COUP-TF immunodetection. Since RFP-CTIP2 was expressed in ball-like structures (Figure 4B, images 1 and 5), GFP-Sp1 expressed alone was localized in the nucleoplasm with a diffused staining pattern (Figure 4B, image 6). Moreover, in the presence of RFP-CTIP2, GFP-Sp1 is relocated to CTIP2-induced structures (Figure 4B, image 3). COUP-TF and GFP-Sp1 relocation was not restricted to the periphery of the CTIP2-induced nuclear structures, since they were also detected inside the structures as shown by the presence of white staining resulting from colocalized red, blue and green staining (Figure 4B, image 4). Thus, endogenous Sp1 proteins were probably relocated within the heterochromatic structures too. However, they remained unaccessible to the currently used antibodies (Figure 4A, images 7 and 9). These results strongly suggest that the cellular transcription factors, Sp1 and COUP-TF, associate with CTIP2 in microglial cells.
Figure 4.
CTIP2 colocalizes with Sp1 and COUP-TF within Hp1α-associated structures. (A) Microglial cells were transfected or not with Flag-CTIP2 as indicated. After being treated, endogenous Sp1, COUP-TF and Hp1α proteins were immunodetected with primary anti-COUP-TF (Santa Cruz Biotechnology) (images 1 and 2), anti-Sp1 (images 6 and 7) and anti-Hp1α antibodies (images 11 and 12). Overexpressed Flag-CTIP2 was detected with antibodies directed against the Flag epitope (images 3, 8 and 13). The primary immunocomplexes were revealed by CY2- or CY3-labeled anti-species secondary antibodies (green or red staining). Mask column (images 5, 10 and 15) shows the colocalized CY2 and CY3 stainings. (B) Microglial cells expressing RFP-CTIP2 (image 1) and GFP-Sp1 (image 3) were subjected to endogenous COUP-TF immunodetection with anti-COUP-TF antibodies (kindly provided by J. E. Mertz). COUP-TF immunocomplexes were stained by CY5- (blue staining) labeled anti-species secondary antibodies (image 2). Pattern of RFP-CTIP2 and GFP-Sp1 expressed alone are presented on images 5 and 6, respectively. (A and B) Coverslips were subjected to confocal microscopy analysis. Bar, 10 μm.
CTIP2 interacts with Sp1, COUP-TF and Hp1α in microglial cells
We have observed that CTIP2 colocalizes with Sp1, COUP-TF and the heterochromatin-associated protein HP1α in microglial cells. To test whether Sp1, COUP-TF, CTIP2 and Hp1α are present in the same complex, we performed co-immunoprecipitation experiments on microglial nuclear extracts expressing or not the Flag-CTIP2 protein (Figure 5A). Anti-Flag western blot immunodetections performed on anti-COUP-TF (lanes 3 and 4), anti-Sp1 (lanes 6 and 7) and anti-Hp1α (lanes 9 and 10) immunoprecipitated nuclear extracts indicated that CTIP2 interacted with COUP-TF (lane 4), Sp1 (lane 7) and Hp1α (lane 10) in microglial cells. As a control, experiments performed on CTIP2 non-expressing extracts (lanes 3, 6 and 9) or on non-immune serum (NIS) immunoprecipitated nuclear extracts (lanes 2, 5 and 8) confirmed the specificity of the Flag immunodetection and the specificity of the COUP-TF, Sp1 and Hp1α immunoprecipitation experiments, respectively. Thus, CTIP2 may be able to recruit a protein complex, including the three proteins in the nucleus of microglial cells. To address how CTIP2 links COUP-TF and Sp1 to Hp1α, we performed competition pull-down assays (Figure 5B). GST-COUP-TF and GST-Sp1 fusion proteins were incubated with both 35S-labeled CTIP2 145–434 and CTIP2 717–813 in the presence of increasing amounts of 35S-Hp1α. Results clearly show that CTIP2 145–434 remained bound to GST-COUP-TF and GST-Sp1, while CTIP2 717–813 was displaced by Hp1α. This displacement increased the amount of GST fusion proteins available for the CTIP2 145–434 binding. Hp1α competed for COUP-TF and Sp1 binding to the C-terminal domain of CTIP2. These findings indicate that the 145–434 region of CTIP2 has a stronger binding affinity for COUP-TF and Sp1 than the 717–813 domain, which preferentially binds Hp1α. Thus, CTIP2 may link COUP-TF and Sp1 bound to the 145–434 domain and Hp1α bound to the 717–813 domain. To precisely examine the formation of a ternary complex between Sp1, COUP-TF and CTIP2, we performed GST pull-down experiments of mixed 35S-labeled-Sp1 and -COUP-TF proteins with GST-CTIP2. As shown in Figure 5C, Sp1 and COUP-TF interacted together with CTIP2. Moreover, addition of increasing amounts of 145–434 CTIP2 or COUP-TF proteins did not compete each other for Sp1 binding (Figure 5D). Thus, COUP-TF and CTIP2 do not compete for Sp1 binding in vitro. Taken together, those findings strongly suggest the formation of a ternary complex occurring between Sp1, COUP-TF and CTIP2.
Figure 5.
CTIP2 interacts with COUP-TF, Sp1 and Hp1α in microglial cells. (A) Nuclear extracts of microglial cells expressing (lanes 2, 4, 5, 7, 8 and 10) or not (lanes 3, 6 and 9) Flag-CTIP2 were immunoprecipitated with antibodies directed against COUP-TF (lanes 3 and 4), Sp1 (lanes 6 and 7) or Hp1α (lanes 9 and 10) proteins. Proteins were separated by SDS–PAGE and western blot analysis with anti-Flag antibodies were performed. (B) GST pull-down competition assays were performed with equal amounts of 35S-labeled 145–434 and 717–813 CTIP2 proteins and increasing amounts of 35S-labeled Hp1α. Proteins were incubated with GST-COUP-TF or GST-Sp1 fusion proteins as indicated. (C) GST pull-down assays were performed with equal amounts of 35S-labeled Sp1 and COUP-TF proteins and GST or GST-CTIP2 fusion proteins as indicated. (D) GST pull-down competition assays were performed with the indicated amounts of 35S-labeled 145–434 CTIP2 and COUP-TF proteins and GST-Sp1. (A–D) Approximately 1% of the total 35S-labeled proteins used were loaded as input control.
CTIP2 is anchored to the HIV-1 LTR by direct interactions with Sp1 bound to the LTR proximal region
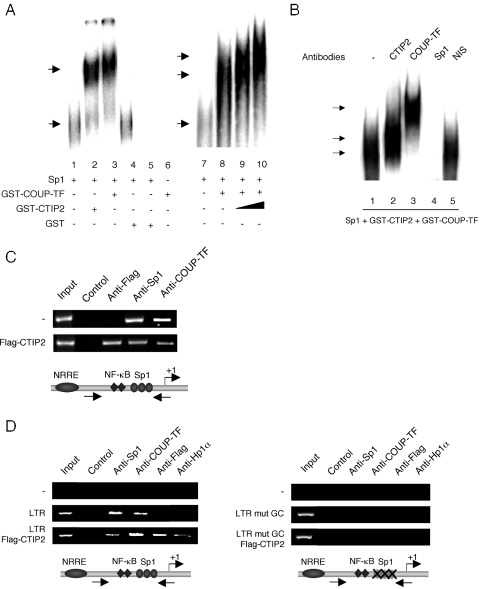
CTIP proteins are DNA-binding proteins, which bind a consensus sequence related to the canonical GC-box (33). Moreover, we have previously described that COUP-TF activates HIV-1 transcription by direct protein–protein interactions with Sp1 bound to the proximal region of HIV-1 LTR (13). The association of CTIP2 with Sp1 and COUP-TF (Figure 5) led us to examine how CTIP2 interacts with the HIV-1 LTR to promote viral repression. To do that, we performed EMSA experiments with pure Sp1 proteins and bacterially produced GST-CTIP2 and GST-COUP-TF proteins that were incubated with a 32P-labeled probe corresponding to the three Sp1 binding sites (GC-box) of the HIV-1 LTR proximal region. As expected, Sp1 bound to the probe to form a shifted nucleoproteic complex (Figure 6A, lanes 1 and 7). Interestingly, addition of GST-CTIP2 (lane 2) or GST-COUP (lane 3) but not GST (lane 4) separately promoted the formation of supershifted complexes, indicating that, as previously reported for COUP-TF (13), CTIP2 was anchored to the LTR by a physical interaction with Sp1. As a control, GST-COUP-TF and GST-CTIP2 proteins, incubated alone, were unable to directly interact with the probe (lanes 5 and 6). To confirm whether COUP-TF, CTIP2 and Sp1 form a ternary complex, we performed additional gel shift experiments in which increasing amounts of CTIP2 proteins were added to the binary Sp1/COUP-TF complex (lanes 9 and 10). As shown, the shifted complex formed by COUP-TF and Sp1 bound to the template (lane 8) was supershifted by increasing the amount of CTIP2 in a dose-dependent manner. Moreover, the intensity of the supershifted complexes also increased with CTIP2 quantity. Those findings are in favor of the formation of a ternary complex anchored to the HIV-1 Sp1 binding sites. To precisely examine this complex, the mixed three proteins were incubated with their respective antibody. EMSA experiments demonstrated that the complex observed in the presence of the three proteins (Figure 6A, lane 10 and Figure 6B, lane 1) was supershifted by the antibodies directed against CTIP2 (Figure 6B, lane 2) and COUP-TF (Figure 6B, lane 3) but not by non-immune serum (Figure 6B, lane 5) confirming the presence of the two proteins in the complex. Moreover, incubation of the mixed three proteins with the anti-Sp1 antibodies (Figure 6B, lane 4) inhibited the formation of the nucleoprotein complex. Thus, impairing Sp1 binding to the proxixmal region of the HIV-1 LTR results in the total abolition of the CTIP2 and COUP-TF ability to bind this region of the viral promoter. These results confirm that both CTIP2 and COUP-TF interact with Sp1 bound to the LTR in vitro. Previously published EMSA experiments performed with nuclear extracts of microglial cells have revealed that this region of the viral promoter binds the cellular transcription factors, Sp1 and Sp3, in microglial cells (13). EMSA experiments performed with nuclear extracts of microglial cells expressing CTIP2 could not allow us to observe CTIP2 indirect binding to the LTR (C. Marban and O. Rohr, unpublished data). To bypass this technical issue and to address whether CTIP2 interacts with the viral promoter in vivo, ChIP assays were conducted on TZM-bl cells, which contain a chromatin-integrated LTR, and on transiently transfected HEK 293T cells. Although the nature and/or the extent of chromatin formation on transiently transfected templates likely differ from that of chromosomal genes, previous reports have validated ChIP studies in transiently transfected cells (34,35). The amplified proximal LTR region includes the three Sp1 binding sites but not the previously described COUP-TF binding site (12). As expected, in the absence of CTIP2, both endogenous Sp1 and COUP-TF interact with the proximal LTR region in chromatin-integrated (Figure 6C) and transiently transfected contexts (Figure 6D, left panel). Moreover, the presence of CTIP2 did not abolish Sp1 and COUP-TF recruitment (Figure 6C, row 2 and Figure 6D, left panel row 3) in agreement with the in vitro results described above. However, in the presence of CTIP2, a lesser extent of template was immunoprecipitated with the antibodies directed against Sp1 and COUP-TF in the context of the integrated LTR (Figure 6C, row 2). The same observations were made with the antibodies directed against Sp1 in the transiently transfected LTR context (Figure 6D, left panel). As shown in Figure 6C and D (left panel), CTIP2 interacted with the proximal region of the viral promoter. In addition, mutation of the Sp1 binding sites abolished the interaction of Sp1, COUP-TF and CTIP2 with this region of the viral LTR (Figure 6D, right panel). Taken together, our in vitro and in vivo findings suggest that the Sp1-mediated recruitment of CTIP2 to the LTR occurs in a cellular context. We could not detect Hp1α interactions with the proximal region of the LTR in the absence of CTIP2, suggesting that CTIP2 significantly increase the recruitment of Hp1α to the HIV-1 proximal promoter (Figure 6D, left panel). In addition, mutation of the Sp1 binding sites also abolished CTIP2-mediated recruitment of Hp1α (Figure 6D, right panel). Since Hp1α associates with heterochromatic regions, it is likely that CTIP2 promotes transcriptionally repressed heterochromatic environment to the viral LTR region.
Figure 6.
CTIP2 is anchored to the HIV-1 LTR by direct interactions with Sp1 bound to the LTR proximal region. (A and B) EMSA experiments were performed using purified Sp1, GST, GST-COUP-TF and GST-CTIP2 fusion proteins. Proteins were incubated with a 32P-labeled probe corresponding to the three Sp1 binding sites located downstream of the LTR (LAI) TATAA sequence. (A) Increasing amounts of GST-CTIP2 proteins correspond to 5 μl (lane 9) and 10 μl (lane 10) of the used GST-CTIP2 preparation. Supershift experiments performed in (B) were carried out with non-immune serum (lane 5) and with antibodies directed against CTIP2 (lane 2), COUP-TF (lane 3), and Sp1 (lane 4). The specific shifted and supershifted complexes are presented. (C) The TZM-bl cells, which contain a stably integrated LTR, were transfected or not with 30 μg of the indicated Flag-CTIP2 expression vector. (D) HEK 293T cells were transfected with 5 μg of the indicated pLTR (left panel) or pLTR mutGC (right panel) constructs and 10 μg of Flag-CTIP2 expression vector if indicated. (C and D) Input lanes correspond to positive controls conduced with a fraction of the lysates used for the immunoprecipitation. Control lanes correspond to negative controls, in which immunoprecipitation reactions were performed without antibodies. Anti-Flag, anti-Sp1, anti-COUP-TF and anti-Hp1α lanes represent amplification reactions from samples immunoprecipitated with the indicated antibodies. Results are representative of three independent experiments.
Sp1 binding sites are dominant and sufficient but not absolutely necessary for transcriptional repression mediated by CTIP2
To address the importance of the HIV-1 LTR GC-box sequence for CTIP2 repressive function, we have transfected microglial cells with the pGC-WAP-CAT reporter plasmid in the presence of Sp1 and CTIP2 expression vectors (Figure 7A). As expected, this heterologous promoter, containing three consensus binding sites for Sp1, was highly inducible by ectopic Sp1 overexpression (lane 3). Interestingly, overexpression of CTIP2 also repressed Sp1-mediated transcriptional stimulation in the context of the GC-WAP promoter (lane 4), indicating that Sp1 binding sites were sufficient for CTIP2 activity. These results also suggest that CTIP2 may be able to repress transcription of other Sp1-sensitive cellular genes.
Figure 7.
Sp1 binding sites are dominant and sufficient but not absolutely necessary for transcriptional repression mediated by CTIP2. (A) Microglial cells were transfected with 3 μg of pGC-WAP-CAT reporter vector and 1 μg of expression vectors as indicated. (B) Microglial cells were transfected with 3 μg of pLTR-CAT, pLTR-CAT mutGC or pLTR-CAT ΔGC and 1 μg of the indicated Flag-CTIP2 vector. Two days post-transfection, CAT activities were measured and expressed relative to the value obtained with the reporter plasmids pGC-WAP-CAT or pLTR-CAT alone with the standard deviations indicated (values correspond to an average of at least three independent experiments carried out in duplicate).
To decipher whether CTIP2 repressive function is exclusively mediated by the Sp1 binding sites, we have further examined the LTR sequences responsible for CTIP2-mediated transcriptional repression. To do that, we have transfected microglial cells with pLTR-CAT vectors containing mutated or deleted GC-boxes in the presence or absence of CTIP2. As previously shown, CTIP2 expression resulted in a strong repression of the basal LTR-driven CAT activity (Figure 7B, lane 2). Mutation (lane 3) or deletion (lane 5) of the Sp1 binding sites resulted in a >90% impairment of the LTR transcriptional activity, confirming the importance of endogenous Sp1 and COUP-TF proteins, which bind to this region and stimulate the initial phase of LTR-driven transcription in microglial cells (13). Surprisingly, CTIP2 was still able to repress the low remaining transcriptional activity of the mutated (lane 4) and of the deleted (lane 6) LTR, suggesting that CTIP2 may also affect HIV-1 gene transcription via association with some other cellular transcription factors yet to be characterized.
DISCUSSION
In a recent work, we have revealed the importance of the cellular transcription factors, Sp1 and COUP-TFI/Ear3, in the regulation of HIV-1 gene transcription and replication in microglial cells (13). We have shown that the orphan nuclear receptor COUP-TF associates and cooperates with Sp1 in the activation of LTR-driven transcription. CTIP1 (Bcl11a, Evi9) and CTIP2 (Bcl11b, Rit1β) are related transcriptional regulatory proteins that have been shown to bind members of the COUP-TF family (23). In a more recent work, we have described that the nuclear cofactor CTIP2 inhibits the subsequent phase of HIV-1 gene transcription by relocating the viral Tat protein to transcriptionally inactive regions of chromatin via Hp1α (25).
Here, we addressed the mechanism whereby CTIP2 impairs the initial phase (prior to Tat expression) of HIV-1 gene transcription in microglial cells.
We report that CTIP proteins inhibit Sp1- and COUP-TF-mediated activation of HIV-1 gene transcription and replication in microglial cells. To decipher the mechanism whereby CTIP2 alters COUP-TF and Sp1 functions, we examined whether these proteins interact in vitro and in cells. GST pull-down experiments revealed that CTIP2 associates with the N-terminal region of COUP-TF and with the C-terminal region of Sp1 in vitro. In this way, CTIP2 presents two interfaces for Sp1 and COUP-TF interactions. COUP-TF and Sp1 proteins bind the central and the C-terminal region of CTIP2 in vitro. These results are consistent with the previously reported interface domains of Arp1 (COUP-TFII) with CTIP1 (23). Confocal microscopy observations of COUP-TF and Sp1 location in the presence or absence of CTIP2 reveal that CTIP2 colocalizes with COUP-TF and Sp1 within nuclear ball-like structures. Moreover, the heterochromatin-associated protein Hp1α is also present in these structures. There are three Hp1 protein family members in mammals, Hp1α, Hp1β and Hp1γ. The Hp1α isoform is mainly detected in transcriptionally repressed heterochromatic region (36). Thus, the colocalization of CTIP2 with endogenous Sp1, COUP-TF and Hp1α proteins as reported here strongly suggests that these proteins are relocated to transcriptionally inactive regions of heterochromatin in microglial cells. Those CTIP2-induced structures may represent a new class of nuclear structures (25). Moreover, our observations may favor the hypothesis, suggesting that the formation of nuclear bodies inhibits HIV-1 gene transcription by sequestering a variety of factors required for transcriptional activation [reviewed in (37)].
Co-immunoprecipitation data indicate that the described subnuclear colocalization results from physical interactions between Sp1, COUP-TF, Hp1α and CTIP2 in the nucleus of microglial cells. Since Hp1α, Sp1 and COUP-TF proteins associate with CTIP2 in microglial cells, we address their respective interaction interfaces in vitro. Our findings show that the central zinc finger domain of CTIP2 may preferentially bind Sp1 and COUP-TF proteins, while the C-terminal region may bind to Hp1α. In the presence of the viral Tat transactivator, CTIP2 also associates with Hp1α via its C-terminal region. Taken together, these findings suggest a permanent association of CTIP2 with Hp1α via its C-terminal region. The central domain of CTIP2 may be reserved for the interactions with the functionally repressed transcription factors. Our observations indicate that COUP-TF and Sp1 could bind together to CTIP2 and that COUP-TF and CTIP2 do not compete for Sp1 binding. Those findings suggest the formation of a ternary complex, including COUP-TF, Sp1 and CTIP2 proteins. Thereby, CTIP2 links these transcriptional activators to transcriptionally repressed heterochromatic region. CTIP1 and CTIP2 proteins have been described as sequence-specific DNA-binding proteins (33). The core-binding site identified in this study is highly related to the canonical GC-box sequence. Since Sp1 anchors COUP-TF to the GC-box sequence of the viral LTR region (12), we addressed how CTIP2 interacts with the HIV-1 promoter. Our findings demonstrate that CTIP2 is not able to directly bind the three viral GC-box sequences in vitro. CTIP2 is recruited to the HIV-1 LTR via its association with Sp1 bound to this region. Moreover, CTIP2 is not in competition with COUP-TF for Sp1-mediated anchorage to the viral promoter, indicating that the three proteins are linked together to the LTR in vitro. The ChIP experiments conducted in a cellular context confirm that CTIP2, Sp1 and COUP-TF proteins do bind the proximal region of the HIV-1 promoter in chromatin-integrated and non-integrated contexts.
The surprising reduction of Sp1 or COUP-TF binding to the viral promoter in the presence of CTIP2 apparently does not agree with the in vitro observations. We show that Sp1 anchors CTIP2 and COUP-TF to the HIV-1 LTR and that Sp1 binding to the GC-box sequence is necessary for CTIP2 recruitment to this region. Nevertheless, since Sp1 and COUP-TF proteins are not accessible to their related antibodies in the CTIP2-induced nuclear structures, one wonders whether the observation of reduced binding to the LTR may result from protein sequestration and/or chromatin environment. ChIP experiments also indicate that CTIP2 promotes Hp1α recruitment to the LTR encompassing the Sp1 binding sequences. Mutation of the Sp1 binding sites abolished this recruitment. Taken together, these results suggest that the Sp1-mediated anchorage of CTIP2 to the HIV-1 LTR region may recruit a large complex that promotes Hp1α association, heterochromatin environment and viral gene transcriptional silencing. In Jurkat cells, CTIP2 associates with the histone deacetylase SirT1 in a large complex. CTIP2-mediated recruitment of SirT1 to promoter template results in a deacetylation of the bound histones H3 and H4 and in a transcriptional repression (35). The chromatin structure close to the HIV-1 gene promoter is involved in viral post-integration latency phenomenon (38). Further studies will be necessary to examine the CTIP2-recruited complex that promotes HIV-1 gene silencing.
To control whether the CTIP2 repressive function is mediated by Sp1 binding sites, we have examined its ability to repress a heterologous promoter containing three consensus GC-boxes. Our findings reveal that GC-box sequences are sufficient for CTIP2-mediated repression of Sp1 stimulation, suggesting that CTIP2 may impact the transcriptional activity of other GC-box containing promoters. In agreement with these observations, CTIP2 is still able to repress LTR-driven transcriptional activity of a LTR deleted downstream of the GC-box sequences. Recruitment of CTIP1 to the template by a COUP-TF family member has been found to result in a transcriptional repression of a reporter gene harboring a COUP-TF binding site (23). In the context of the HIV-1 promoter, deletion of the previously described COUP-TF binding site did not impact CTIP2 repressive function. Moreover, EMSA and ChIP experiments demonstrate that CTIP2 interacts with the proximal region of the LTR excluding the COUP-TF binding sequence. Taken together, these findings indicate that the COUP-TF binding sequence, located downstream to the GC-box region, might not be implicated in the CTIP2 repressive functions. As widely reported, deletion of the LTR GC-box region results in a drastic (>90%) reduction of the LTR-driven transcriptional activity (13–15,22). These observations confirm the crucial contribution of Sp1 and COUP-TF transcription factors to support viral gene transcription in microglial cells. Moreover, they highlight the importance of CTIP2-mediated repression of Sp1 and COUP-TF protein functions. Most of the 80% CTIP2-mediated reduction of the basal transcriptional level may be charged on to this repressive activity. Surprisingly, CTIP2 is still able to repress the 10% remaining transcriptional activity driven by a GC-box deleted LTR, suggesting that CTIP2 may also be able to repress the expression of mutated strains of virus. This remaining activity represents <10% of the global CTIP2-mediated repressive activity.
Nevertheless, we have previously suggested that COUP-TF restores the viral Tat function in the context of a GC-box mutated LTR by anchoring Tat to the basal transcriptional machinery (22). Interestingly, we observed that CTIP2 is also able to repress this functional cooperation (C. Marban and O. Rohr, unpublished data). These results suggest that, in this context, COUP-TF may bind CTIP2 via its N-terminal region and the general transcription factor TFIIB via its C-terminal part as described previously (39). Thus, in the absence of Sp1-mediated linkage to the GC-box region of the HIV-1 LTR, CTIP2 may bypass Sp1 association by a COUP-TF-mediated linkage to the basal transcriptional machinery. Moreover, this proposed indirect linkage might be weak since it appeared undetectable by ChIP experiments. Along this line, CTIP2 might also associate to some other cellular transcription factors yet to be characterized.
These studies fully complete those reporting CTIP2-mediated repression of the viral Tat function (25). Since Sp1 or COUP-TF association is necessary for Tat function in microglial cells (22), it can be postulated that CTIP2-mediated impairment of Sp1 and COUP-TF function contributes to the previously observed repression of Tat function.
The chromatin structure at the site of provirus integration is reminiscent to post-integration latency phenomenon (38). In vitro, latent T-cell clones frequently contain HIV-1 genome integrated in heterochromatin structures. This is in contrast to a productive infection where integration in or near heterochromatin is disfavored (40). No antiretroviral drugs that are currently available can inhibit transcription of HIV RNA from the integrated HIV proviral DNA in infected cells. Nevertheless, HDAC inhibitors are able to induce quiescent provirus, suggesting that derepression of heterochromatin structures may force viral expression from latently infected reservoirs (41,42). Thus, investigation on factors that may promote heterochromatin structures to the viral gene promoter, such as CTIP2, appears crucial for understanding these phenomena and for the development of new anti-HIV strategies. At this stage, further studies will be necessary to precisely examine the CTIP2-associated enzymatic activities that promote transcriptional silencing. In addition, since our investigations were restricted to the transcriptional step of the viral life cycle, it cannot be excluded that CTIP2 also counteracts other critical steps necessary to optimal viral replication. This possibility needs further clarification.
Acknowledgments
The authors thank R. Losson for providing Hp1α vectors and anti-Hp1α antibodies, H. Rotheneder for providing GST-SP1 vectors, J. E. Mertz for providing anti-COUP-TF antibodies and Ann Dekoninck for technical support. The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: TZM-bl from Dr John C. Kappes, Dr Xiaoyun Wu and Tranzyme Inc. and pNL4-3 from Dr Malcolm Martin. This work was supported by the Institut National de la Santé et de la Recherche Médicale (INSERM), by the Agence Nationale de Recherches sur le SIDA (ANRS), by grants from the French Ministry of Research (‘ACI JC 5364’ to O.R. and grant to S.S.) and by a grant to C.M. from the Conseil Régional d'Alsace. C.V.L. is a ‘Maître de Recherches’ at the ‘Fonds National de la Recherche Scientifique’ (FNRS, Belgium) and O.R. is a ‘Maître de Conférences’ at the ‘IUT Louis Pasteur de Schiltigheim’ (University of Strasbourg I, France). Work in the Leid Laboratory was supported by a grant from NIH (GM60852) and in part by grant number P30 ES00210 from the National Institute of Environmental Health Sciences, NIH. Funding to pay the Open Access publication charges for this article was provided by INSERM.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fauci A.S. The human immunodeficiency virus: infectivity and mechanisms of pathogenesis. Science. 1988;239:617–622. doi: 10.1126/science.3277274. [DOI] [PubMed] [Google Scholar]
- 2.Portegies P., Brew B.J. Update on HIV-related neurological illness. AIDS. 1991;5(Suppl. 2):S211–S217. doi: 10.1097/00002030-199101001-00029. [DOI] [PubMed] [Google Scholar]
- 3.Pialoux G., Fournier S., Moulignier A., Poveda J.D., Clavel F., Dupont B. Central nervous system as a sanctuary for HIV-1 infection despite treatment with zidovudine, lamivudine and indinavir. AIDS. 1997;11:1302–1303. doi: 10.1097/00002030-199710001-00009. [DOI] [PubMed] [Google Scholar]
- 4.Kolson D.L., Lavi E., Gonzalez-Scarano F. The effects of human immunodeficiency virus in the central nervous system. Adv. Virus Res. 1998;50:1–47. doi: 10.1016/s0065-3527(08)60804-0. [DOI] [PubMed] [Google Scholar]
- 5.Peudenier S., Hery C., Montagnier L., Tardieu M. Human microglial cells: characterization in cerebral tissue and in primary culture, and study of their susceptibility to HIV-1 infection. Ann. Neurol. 1991;29:152–161. doi: 10.1002/ana.410290207. [DOI] [PubMed] [Google Scholar]
- 6.Perry V.H., Lawson L.J., Reid D.M. Biology of the mononuclear phagocyte system of the central nervous system and HIV infection. J. Leukoc. Biol. 1994;56:399–406. doi: 10.1002/jlb.56.3.399. [DOI] [PubMed] [Google Scholar]
- 7.Greene W.C., Peterlin B.M. Charting HIV's remarkable voyage through the cell: Basic science as a passport to future therapy. Nature Med. 2002;8:673–680. doi: 10.1038/nm0702-673. [DOI] [PubMed] [Google Scholar]
- 8.Pereira L.A., Bentley K., Peeters A., Churchill M.J., Deacon N.J. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 2000;28:663–668. doi: 10.1093/nar/28.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rohr O., Marban C., Aunis D., Schaeffer E. Regulation of HIV-1 gene transcription: from lymphocytes to microglial cells. J. Leukoc. Biol. 2003;74:736–749. doi: 10.1189/jlb.0403180. [DOI] [PubMed] [Google Scholar]
- 10.Wang L.H., Tsai S.Y., Cook R.G., Beattie W.G., Tsai M.J., O'Malley B.W. COUP transcription factor is a member of the steroid receptor superfamily. Nature. 1989;340:163–166. doi: 10.1038/340163a0. [DOI] [PubMed] [Google Scholar]
- 11.Wang L.H., Tsai S.Y., Sagami I., Tsai M.J., O'Malley B.W. Purification and characterization of chicken ovalbumin upstream promoter transcription factor from HeLa cells. J. Biol. Chem. 1987;262:16080–16086. [PubMed] [Google Scholar]
- 12.Sawaya B.E., Rohr O., Aunis D., Schaeffer E. Chicken ovalbumin upstream promoter transcription factor, a transcriptional activator of HIV-1 gene expression in human brain cells. J. Biol. Chem. 1996;271:23572–23576. doi: 10.1074/jbc.271.38.23572. [DOI] [PubMed] [Google Scholar]
- 13.Rohr O., Aunis D., Schaeffer E. COUP-TF and Sp1 interact and cooperate in the transcriptional activation of the human immunodeficiency virus type 1 long terminal repeat in human microglial cells. J. Biol. Chem. 1997;272:31149–31155. doi: 10.1074/jbc.272.49.31149. [DOI] [PubMed] [Google Scholar]
- 14.Rohr O., Schwartz C., Aunis D., Schaeffer E. CREB and COUP-TF mediate transcriptional activation of the human immunodeficiency virus type 1 genome in Jurkat T cells in response to cyclic AMP and dopamine. J. Cell. Biochem. 1999;75:404–413. [PubMed] [Google Scholar]
- 15.Schwartz C., Catez P., Rohr O., Lecestre D., Aunis D., Schaeffer E. Functional interactions between C/EBP, Sp1, and COUP-TF regulate human immunodeficiency virus type 1 gene transcription in human brain cells. J. Virol. 2000;74:65–73. doi: 10.1128/jvi.74.1.65-73.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones K.A., Kadonaga J.T., Luciw P.A., Tjian R. Activation of the AIDS retrovirus promoter by the cellular transcription factor, Sp1. Science. 1986;232:755–759. doi: 10.1126/science.3008338. [DOI] [PubMed] [Google Scholar]
- 17.Harrich D., Garcia J., Wu F., Mitsuyasu R., Gonazalez J., Gaynor R. Role of SP1-binding domains in in vivo transcriptional regulation of the human immunodeficiency virus type 1 long terminal repeat. J. Virol. 1989;63:2585–2591. doi: 10.1128/jvi.63.6.2585-2591.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sune C., Garcia-Blanco M.A. Transcriptional trans activation by human immunodeficiency virus type 1 Tat requires specific coactivators that are not basal factors. J. Virol. 1995;69:3098–3107. doi: 10.1128/jvi.69.5.3098-3107.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perkins N.D., Edwards N.L., Duckett C.S., Agranoff A.B., Schmid R.M., Nabel G.J. A cooperative interaction between NF-kappa B and Sp1 is required for HIV-1 enhancer activation. EMBO J. 1993;12:3551–3558. doi: 10.1002/j.1460-2075.1993.tb06029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yedavalli V.S., Benkirane M., Jeang K.T. Tat and trans-activation-responsive (TAR) RNA-independent induction of HIV-1 long terminal repeat by human and murine cyclin T1 requires Sp1. J. Biol. Chem. 2003;278:6404–6410. doi: 10.1074/jbc.M209162200. [DOI] [PubMed] [Google Scholar]
- 21.van Opijnen T., Kamoschinski J., Jeeninga R.E., Berkhout B. The human immunodeficiency virus type 1 promoter contains a CATA box instead of a TATA box for optimal transcription and replication. J. Virol. 2004;78:6883–6890. doi: 10.1128/JVI.78.13.6883-6890.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rohr O., Schwartz C., Hery C., Aunis D., Tardieu M., Schaeffer E. The nuclear receptor chicken ovalbumin upstream promoter transcription factor interacts with HIV-1 Tat and stimulates viral replication in human microglial cells. J. Biol. Chem. 2000;275:2654–2660. doi: 10.1074/jbc.275.4.2654. [DOI] [PubMed] [Google Scholar]
- 23.Avram D., Fields A., Pretty On Top K., Nevrivy D.J., Ishmael J.E., Leid M. Isolation of a novel family of C(2)H(2) zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (COUP-TF) orphan nuclear receptors. J. Biol. Chem. 2000;275:10315–10322. doi: 10.1074/jbc.275.14.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leid M., Ishmael J.E., Avram D., Shepherd D., Fraulob V., Dolle P. CTIP1 and CTIP2 are differentially expressed during mouse embryogenesis. Gene Expr. Patterns. 2004;4:733–739. doi: 10.1016/j.modgep.2004.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rohr O., Lecestre D., Chasserot-Golaz S., Marban C., Avram D., Aunis D., Leid M., Schaeffer E. Recruitment of Tat to heterochromatin protein HP1 via interaction with CTIP2 inhibits human immunodeficiency virus type 1 replication in microglial cells. J. Virol. 2003;77:5415–5427. doi: 10.1128/JVI.77.9.5415-5427.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohr O., Sawaya B.E., Lecestre D., Aunis D., Schaeffer E. Dopamine stimulates expression of the human immunodeficiency virus type 1 via NF-kappaB in cells of the immune system. Nucleic Acids Res. 1999;27:3291–3299. doi: 10.1093/nar/27.16.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karlseder J., Rotheneder H., Wintersberger E. Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F. Mol. Cell. Biol. 1996;16:1659–1667. doi: 10.1128/mcb.16.4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adachi A., Koenig S., Gendelman H.E., Daugherty D., Gattoni-Celli S., Fauci A.S., Martin M.A. Productive, persistent infection of human colorectal cell lines with human immunodeficiency virus. J. Virol. 1987;61:209–213. doi: 10.1128/jvi.61.1.209-213.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janabi N., Peudenier S., Heron B., Ng K.H., Tardieu M. Establishment of human microglial cell lines after transfection of primary cultures of embryonic microglial cells with the SV40 large T antigen. Neurosci. Lett. 1995;195:105–108. doi: 10.1016/0304-3940(94)11792-h. [DOI] [PubMed] [Google Scholar]
- 30.Derdeyn C.A., Decker J.M., Sfakianos J.N., Wu X., O'Brien W.A., Ratner L., Kappes J.C., Shaw G.M., Hunter E. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 2000;74:8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Platt E.J., Wehrly K., Kuhmann S.E., Chesebro B., Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei X., Decker J.M., Liu H., Zhang Z., Arani R.B., Kilby J.M., Saag M.S., Wu X., Shaw G.M., Kappes J.C. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 2002;46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Avram D., Fields A., Senawong T., Topark-Ngarm A., Leid M. COUP-TF (chicken ovalbumin upstream promoter transcription factor)-interacting protein 1 (CTIP1) is a sequence-specific DNA binding protein. Biochem. J. 2002;368:555–563. doi: 10.1042/BJ20020496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo R.X., Postigo A.A., Dean D.C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 35.Senawong T., Peterson V.J., Avram D., Shepherd D.M., Frye R.A., Minucci S., Leid M. Involvement of the histone deacetylase SIRT1 in chicken ovalbumin upstream promoter transcription factor (COUP-TF)-interacting protein 2-mediated transcriptional repression. J. Biol. Chem. 2003;278:43041–43050. doi: 10.1074/jbc.M307477200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen A.L., Oulad-Abdelghani M., Ortiz J.A., Remboutsika E., Chambon P., Losson R. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell. 2001;7:729–739. doi: 10.1016/s1097-2765(01)00218-0. [DOI] [PubMed] [Google Scholar]
- 37.Marcello A., Lusic M., Pegoraro G., Pellegrini V., Beltram F., Giacca M. Nuclear organization and the control of HIV-1 transcription. Gene. 2004;326:1–11. doi: 10.1016/j.gene.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 38.Jordan A., Defechereux P., Verdin E. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 2001;20:1726–1738. doi: 10.1093/emboj/20.7.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ing N.H., Beekman J.M., Tsai S.Y., Tsai M.J., O'Malley B.W. Members of the steroid hormone receptor superfamily interact with TFIIB (S300-II) J. Biol. Chem. 1992;267:17617–17623. [PubMed] [Google Scholar]
- 40.Jordan A., Bisgrove D., Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 2003;22:1868–1877. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ylisastigui L., Archin N.M., Lehrman G., Bosch R.J., Margolis D.M. Coaxing HIV-1 from resting CD4 T cells: histone deacetylase inhibition allows latent viral expression. AIDS. 2004;18:1101–1108. doi: 10.1097/00002030-200405210-00003. [DOI] [PubMed] [Google Scholar]
- 42.Quivy V., Adam E., Collette Y., Demonte D., Chariot A., Vanhulle C., Berkhout B., Castellano R., de Launoit Y., Burny A., et al. Synergistic activation of human immunodeficiency virus type 1 promoter activity by NF-kappaB and inhibitors of deacetylases: potential perspectives for the development of therapeutic strategies. J. Virol. 2002;76:11091–11103. doi: 10.1128/JVI.76.21.11091-11103.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]