Abstract
Human epigenetic variation is associated with both environmental exposures and allergic diseases and can potentially serve as a biomarker connecting climate change with allergy and airway diseases. In this narrative review, we summarize recent human epigenetic studies examining exposure to temperature, precipitation, extreme weather events, and malnutrition to discuss findings as they relate to allergic and airway diseases. Temperature has been the most widely studied exposure, implicating both short-term and long-term exposure with epigenetic alterations and epigenetic aging. Few studies have examined natural disasters or extreme weather events. The studies available have reported differential DNA methylation of multiple genes and pathways, some previously associated with asthma or allergy. Few studies have integrated climate-related events, epigenetic biomarkers, and allergic disease together. Prospective longitudinal studies are needed along with the collection of target tissues beyond blood samples, such as nasal and skin cells. Finally, global collaboration to increase diverse representation of study participants, particularly those most affected by climate injustice, and strengthen replication, validation, and harmonization of measurements will be needed to elucidate the impact of climate change on the human epigenome.
Keywords: Climate Change, Epigenetics, Epigenomics, DNA Methylation, Temperature, Precipitation, Extreme Weather, Malnutrition, Epigenetic Clocks
Introduction
The climate crisis is one of the greatest threats to humanity, with significant impacts on allergic, immunologic, and respiratory health.(1-5) The primary driver of climate change is the global production and consumption of fossil fuels—coal, crude oil, and natural gas—which emit greenhouse gasses like carbon monoxide and methane.(1,6) The average surface temperature of the planet has risen since preindustrial times largely due to fossil fuel use, and it is predicted to continue increasing unless immediate global efforts take place to reduce fossil fuel use. As a result, there have been changing precipitation patterns, increased frequency of natural disasters, such as wildfires and hurricanes, and alterations in land use patterns.(6-8) Global warming and the burning of fossil fuels are also associated with increased air and water pollution and increased ultraviolet (UV) radiation exposure.(9-11) These climatic and environmental changes to our planet have been linked to increases in risk for a wide range of human health outcomes, including heat stroke, vector-borne illnesses, cardiopulmonary diseases, malnutrition, and psychiatric conditions.(12,13) Relevant to allergy, changing atmospheric conditions in some regions, particularly increased temperature and precipitation, may lead to the spread of new pollen sources and increased intensity and duration of pollen generation.(4,14,15) Therefore, more extreme climatic conditions could lead to an increase in the burden of allergic diseases.
The epigenome lies at the intersection between environmental risk factors and human diseases. Epigenetic alterations refer to changes in gene expression that are not directly related to variation in the underlying genetic code. They allow us to better characterize early biomarkers of impact from environmental exposures and their connection to human health.(16,17) Epigenetic marks are tissue and cell-type specific, which can yield insights into underlying changes and responses in specific organs. In epidemiological studies, once DNA is extracted often from a mixed population of cells in a human subject, methylation can be measured after bisulfite treatment using sequencing or micro-arrays—for example, the 450K or 850K/EPIC Illumina arrays. The most commonly studied epigenetic modification in human studies is DNA methylation of cytosine nucleotides (CpG sites).(16,17) Analytic strategies can include epigenome-wide analysis, candidate gene approaches, testing of epigenetic biomarkers of aging, immune or cell type composition estimation, and identification of exposure biomarkers (e.g., smoking) and epigenetic biomarkers of health.(18) These methodologies have been used in studies on allergic diseases, highlighting potential biological pathways of disease development for environmental risk factors(19-22) and immunological components of biological aging.(23,24)
Harnessing the power of epigenetic biomarkers in the field of allergy and immunology can provide insights into how changing and extreme climatic conditions affect human health. While temperature, humidity, and precipitation have been less commonly examined in human epigenetic studies compared to air pollution,(19,25) research in plants and animal models have shown that these climatic factors affect DNA methylation and gene expression in multiple ways.(26-30) The most recent systematic review regarding climate and human epigenetic modifications was conducted in 2020 and identified 15 genes with methylation status associated with temperature, including genes associated with asthma (e.g., TLR2 and NOS2).(31) However, it did not examine other climatic factors. The objective of this review is to provide the first review of studies investigating epigenetic mechanisms related to multiple climatic factors, including temperature, humidity, and precipitation, and climate change-associated exposures, such as natural disasters and malnutrition, and how they may relate to allergic diseases. We also discuss strengths, challenges, and opportunities for future research on this topic.
Methods
For this narrative review, we conducted an online search for original research articles published on the topic of epigenetics and climatic exposures. This search included multiple epigenetic mechanisms, including DNA methylation, histone modifications, and non-coding RNAs, and various environmental exposures related to climate and climate change, such as temperature, precipitation, and natural disasters. The search was performed on 3 databases—PubMed, Web of Science, and EMBASE—for articles published between January 1, 2000 to August 1, 2023 using a combination of MeSH terms that are listed in Table 1. The findings from relevant papers from this search are discussed in the following sections organized by environmental exposure, and we provide interpretation regarding their connections to allergic and immunologic outcomes. Data on epigenetic markers and/or associated gene annotations related to climate exposures were extracted from studies, when available, and included in Supplementary Table S1. For the section on temperature, we focused on recently published studies that were not evaluated in the most recent systematic review on this topic published in 2020.(31) While air pollution is a major cause of climate change, studies regarding direct associations of air pollution and DNA methylation were excluded from this review, since they have been previously extensively reviewed.(25,32,33)
Table 1.
Description of Strategy Used to Search for Recent Articles
| Parameters | Details |
|---|---|
| Dates of Search | January 1, 2000 to August 1, 2023 |
| Databases | PubMed, EMBASE, and Web of Science |
| Search Terms | “epigenetics,” “epigenome,” “methylome,” “DNA methylation,” “histone,” “non-coding RNA,” “temperature,” “thermal,” “global warming,” “climate,” “radiation,” “humidity,” “hurricane,” “flooding,” “heat,” “wildfires,” “extreme weather,” “extreme temperature,” “heat wave,” “blizzard,” “thunderstorm,” “tornado,” “malnutrition,” “famine” |
Background on Environmental Epigenetics and Allergy
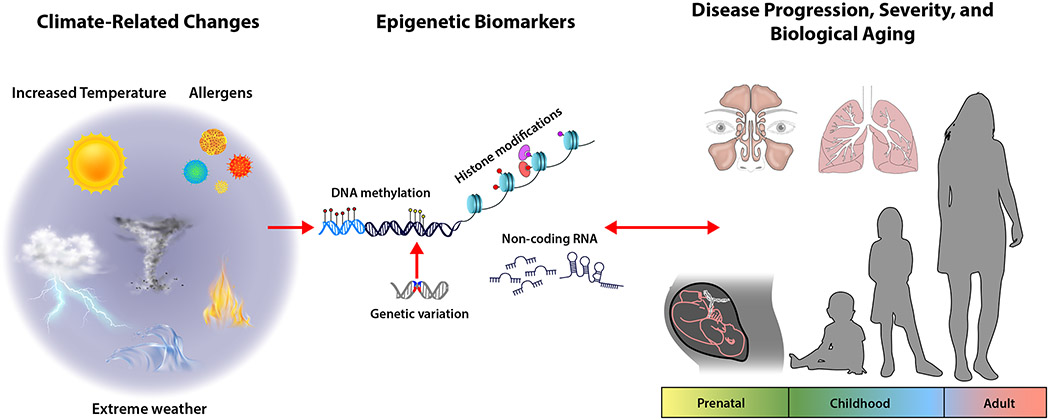
The goal of environmental epigenetic studies is to investigate how and when environmental exposures contribute to epigenetic variation, which can impact the prevalence, incidence, and severity of human diseases. It is important to note that environmental exposures, including climate-related factors, at different stages of life, such as in utero, early childhood, and adulthood, can differentially impact the epigenome and that genetic variation can also plays a role in impacting DNA methylation variability.(34) Particularly, in utero exposure is a susceptible window, as epigenetic programming of cells, organs, and tissues is established during this period.(35) Additionally, ancestry-specific methylation quantitative trait loci (meQTL) support the inclusion of ancestrally diverse and multiethnic populations in epigenetic studies. Epigenetic modifications can also reflect disease progression, so the longitudinal collection of samples is key to distinguish biomarkers of exposure from those of disease that might be characterized in cross-sectional studies (Figure 1).
Figure 1:
Conceptual framework for human epigenetic studies related to climate change, extreme weather events, and events aggravated by extreme weather and their relationship with allergic disease.
There are several methodologies used in environmental epigenetics research, and we have defined commonly used terms found in this review and in the literature in Table 2. The first step involves the extraction of DNA from a population of cells from specific tissues, such as whole blood, the skin, or nasal or bronchial epithelium. Once the DNA is processed, it can be analyzed using a variety of methods including methylation arrays.(36) The 450K and 850K EPIC BeadChips have been commonly used in epigenome-wide association studies (EWASs) to systematically measure the methylation of CpG sites across the human genome in large epidemiological cohorts.(37) Then, several statistical and bioinformatic approaches are used to test for associations between methylation status of CpG sites and phenotypes and/or exposures of interest. The most common approach is to test each CpG individually across all DNA methylation measurements, often referred to as a differentially methylated position (DMP). This approach can be complemented by testing entire regions, defined by proximity or correlation structure, referred to as a differentially methylated region (DMR) with multiple CpGs. This technique has been employed to systematically assess the relationship between multiple different environmental exposures, asthma and allergic diseases.(38) A more targeted approach involves selecting certain genes a priori, for example using specific primers or an array specific for a panel of genes related to inflammation, and analyzing methylation of the relevant CpG sites. An example of this approach is the study of how farm milk exposure contributed to increased methylation of FOXP3 in regulatory T cells, which was associated with decreased atopic sensitization and asthma in childhood.(39) A newer methodology is the use of epigenetic clocks, which are DNA methylation biomarkers of biological aging found to be predictors of morbidity and mortality.(40) These biomarkers estimate an epigenetic age that is predictive of an individual’s chronological age. Deviation between both measures is referred to as epigenetic age acceleration (EAA), which is a strong risk factor for mortality and morbidity, including allergic diseases, and shown to be influenced by environmental factors. For example, one study found that cigarette smoking is associated with increased EAA among adults,(41) and another showed that greater EAA in childhood is associated with higher odds of having atopy and food allergen sensitization.(23)
Table 2.
Definition of Common Terms Used in Epigenetics Research
| Term | Definition |
|---|---|
| Epigenome-Wide Association Study (EWAS) | A comprehensive genome-wide testing of epigenetic marks and how they are associated with phenotypes or exposures of interest |
| CpG Site | DNA dinucleotides of cytosine followed by guanine in which the cytosine nucleotide may have a methyl group added by DNA methyltransferase, which can alter gene transcription |
| Differentially Methylated Region (DMR) | An entire genomic region composed of multiple CpG sites with differential DNA methylation across samples |
| Non-coding RNAs | Functional molecules of RNA that are not translated to proteins but can affect gene expression and protein functioning |
| Epigenetic Clocks | Statistical calculators that use methylation data at certain CpG sites to derive an epigenetic age, which is a marker of biological aging that is correlated with morbidity and mortality |
| Epigenetic Age Acceleration (EAA) | Deviations between epigenetic and chronological age calculated as the residuals from when each DNA methylation clock output is regressed on the chronological age of each participant |
| Methylation Euantitative Trait Loci (meQTL) | Single-nucleotide polymorphisms (SNPs) associated with DNA methylation variation of a CpG site |
Most environmental epigenetics studies on allergy and asthma have focused on cigarette smoke and air pollution as common exposures of interests. While results have been discordant, many EWASs have found that these exposures are frequently associated with differential methylation of CpG sites near or in the promoters of AHRR, FOXP3, and IL4.(33,42,43) This is relevant as DNA methylation in these regions could influence gene expression, as shown in studies of smoking and AHRR methylation.(44,45) These findings have emerged mostly from cross-sectional study designs, which are limited because the timing of exposure and disease development and the directionality of DNA methylation variability are less clear. Nonetheless, new methodological advancements in the field of environmental epigenetics show promise for elucidating the relationship between climatic factors and allergic diseases in order to improve disease prevention and treatment.
Epidemiological studies on climate change and epigenetics have focused on extreme weather events, temperature, and factors aggravated by climate change, such as famine and drought. These factors have been frequently tested for cross-sectional associations with DNA methylation isolated from leukocytes. Below, we discuss studies from our search and place their results into the context of allergic and airway diseases. We found one study that examined miRNAs as the epigenetic outcome of interest, and the rest of the studies measured DNA methylation; none investigated histone modifications.
Temperature
Mechanisms Linking Temperature to Allergy and Immunologic Diseases
High temperature exposure, both chronic and acute, are associated with increased morbidity and mortality, and various epigenetic research methodologies have been used to investigate this relationship (Table 3).(46) There might be several pathways linking temperature fluctuations to allergic disease (Table 4). For example, extreme heat might impact airway responsiveness by activating certain transient receptor potential cation channels and stimulating cholinergic reflex pathways.(47) Additionally, increased membrane fluidity and disruption of transmembrane structural proteins can increase risk for an inflammatory response. Regarding temperature-related epigenetic alterations, a systematic review of studies published before 2020 summarized results from 7 research articles and identified 15 candidate genes;(31) below, we discuss findings from more recent studies on this topic.
Table 3.
Summary of Epigenetic Studies on Temperature and Precipitation
| Study Title | Authors (year) |
Location | Population | Exposure | Epigenetic Endpoint |
Key Finding(s) |
|---|---|---|---|---|---|---|
| Temperature | ||||||
| Epigenome-wide association study of short-term temperature fluctuations based on within-sibship analyses in Australian females | Wu et al. (2023) | Australia | Adult female twins and their sisters in the Australian Mammographic Density Twins and Sisters Study (AMDTSS) | Short-term changes in temperature | DNA methylation (EWAS) of peripheral leukocytes analyzed with with the 450K BeadChip | -Temperature changes were associated with differential methylation of 14 CpGs and 70 DMRs mapping to 68 genes linked to human diseases. |
| Associations between medium- and long-term exposure to air temperature and epigenetic age acceleration | Ni et al. (2023) | Augsburg, Germany | Adults from the Cooperative Health Research in the Region of Augsburg (KORA) studies | Land surface temperature (medium-term: 4-week and 8-week; long-term: 365 days) | Epigenetic aging biomarkers: Horvath, Hannum, PhenoAge, GrimAge, and Skin and Blood clocks | -Medium-term exposures to high but not low temperature increased Horvath EAA, Hannum EAA, GrimAge EAA, and Skin-Blood EAA. -Higher annual average temperature was associated with increased Horvath EAA, Hannum EAA, Pheno EAA, GrimAge EAA, and Skin-Blood EAA. |
| Intermediate and long-term exposure to air pollution and temperature and the extracellular microRNA profile of participants in the normative aging study (NAS) | Yazdi et al. (2023) | Boston, Massachusetts, USA | Adult men living around Boston who use medical services at the Veterans Affair Hospital and are part of the Normative Aging Study (NAS) | Intermediate and long-term exposure to temperature | MicroRNAs found extracellularly in whole blood samples that were processed and sequenced | -Increased intermediate and long-term exposures to temperature were associated with levels of several extracellular miRNAs with clinical correlates to respiratory and cardiovascular diseases. |
| Short-term air pollution and temperature exposure and changes in the extracellular microRNA profile of Normative Aging Study (NAS) participants | Yazdi et al. (2023) | Boston, Massachusetts, USA | Adult men living around Boston who use medical services at the Veterans Affair Hospital and are part of the Normative Aging Study (NAS) | Short-term exposure to temperature | MicroRNAs found extracellularly in whole blood samples that were processed and sequenced | -In most cases, increased mean temperature was positively associated with extracellular microRNA levels, many of which were associated with inflammation, disease development, and fatty acid metabolism. -Longer-term changes in temperature may impact changes in microRNA profiles greater than short-term changes in temperature. |
| Weather and Birth Weight: Different Roles of Maternal and Neonatal GPR61 Promoter Methylation | Yuan et al. (2022) | Zhengzhou, China | Pregnant women who delivered in 2010-2012 and their newborns | Mean temperature and temperature range | Candidate gene methylation analysis of the GPR61 promoter in peripheral leukocytes in maternal and umbilical cord blood | -There was a positive association between daily temperature range and GPR61 methylation in maternal and cord blood, which was linked to greater birth weight -Maternal GPR61 methylation modified associations between temperature and birth weight. |
| Ambient temperature and genome-wide DNA methylation: A twin and family study in Australia | Xu (2021) | Australia | Adult female twins and their sisters in the Australian Mammographic Density Twins and Sisters Study (AMDTSS) | Mean temperature (short-, medium-, and long-term exposure) | DNA methylation (EWAS) of peripheral leukocytes analyzed with with the 450K BeadChip | -Temperature was associated with differential methylation of 31 CpGs and 82 DMRs mapping to 85 genes linked to chronic diseases, including asthma. |
| The role of maternal methylation in the association between prenatal meteorological conditions and neonatal H19/H19-DMR methylation | Yang (2020) | Zhengzhou, China | Pregnant women who delivered in 2010-2012 and their newborns | Minimum, mean, and maximum temperature | Candidate gene methylation analysis of the H19 promoter and H19-DMR in peripheral leukocytes in maternal and umbilical cord blood | -Neonatal and maternal H19 and H19-DMR methylation were negatively associated with temperature in the first trimester and positively associated with temperature in the third trimester. |
| Precipitation | ||||||
| Epigenetic mechanisms underlying the association between maternal climate stress and child growth: characterizing severe drought and its impact on a Kenyan community engaging in a climate change-sensitive livelihood | Straight et al. (2022) | Kenya, Africa | Children of exposed women | Severe drought in 2008-2009 | DNA methylation (EWAS) from saliva analyzed with the Epic BeadChip | -16 differentially methylated CpG sites were found, and most related to immunologic and metabolic pathways. -There was an association between drought exposure and child body weight through cg03771070 methylation. |
| Weather and Birth Weight: Different Roles of Maternal and Neonatal GPR61 Promoter Methylation | Yuan et al. (2022) | Houzhai, China | Pregnant women who delivered in 2010-2012 and their newborns | 24-hour precipitation | Candidate gene methylation analysis of the GPR61 promoter in peripheral leukocytes in maternal and umbilical cord blood | -There was a positive association between precipitation and GPR61 methylation in maternal and cord blood, which was linked to greater birth weight. -Maternal GPR61 methylation modified associations between precipitation and birth weight. |
Table 4:
Biological Pathways Connecting Components of Climate Change to Risk for Allergic Diseases
| Climate Change Component | Mechanisms Related to Risk for Allergic Diseases |
|---|---|
| Increased Temperature | 1. Faster plant growth and prolonged pollen generation seasons leads to increased pollen quantity and allergenicity. 2. There is increased membrane fluidity due to redirected blood flow to the periphery and disruption of transmembrane structural proteins, which then increases risk for an inflammatory response. 3. Extreme heat can impact airway responsiveness by activating certain transient receptor potential (TRP) cation channels and stimulating cholinergic reflex pathways. |
| Increased Precipitation and Hurricanes | 1. Rainfall can cause atmospheric pollen grains to release large quantities of various small allergens that can be inhaled to exacerbate asthma and allergic rhinitis. 2. Rising air moisture after storms and flooding can increase indoor and outdoor mold growth, including allergenic ones like Alternaria, Aspergillus, and Cladosporium. |
| More Intense and Frequent Wildfires | 1. Wildfire smoke contains air pollutants that can interact with the respiratory epithelium to directly cause inflammation and bronchoconstriction. 2. These air pollutants also increase permeability of the respiratory tract and skin to facilitate penetration of allergens in people with asthma and atopic dermatitis. 3. Air pollutants can adhere to pollen grains to change their morphology and allergenic potential. |
| Disrupted Food and Water Systems | 1. Exposure to heavy metals like arsenic and other water pollutants can trigger a pro-inflammatory response. 2. Lower nutritional content of crops due to increased atmospheric carbon dioxide could alter immune system functioning. 3. Forced migration due to food and water insecurity exposes humans to new environmental allergens and infectious vectors, potentially increasing risk for food-related and other allergic reactions. |
Recent Epigenetic Studies on Temperature
More studies have been recently published to quantify temperature exposure and its impacts on the epigenome of humans. For example, an EWAS of blood cells from Australian women showed associations between whole blood methylation and short-term temperature fluctuations. A total of 14 differentially methylated CpGs and 70 differentially methylated regions (DMRs) were associated with short-term temperature fluctuations. Of note, the most statistically significant CpG that had higher DNA methylation relative to temperature was annotated to the KCNK4 gene.(48) This gene has been shown to be hypomethylated in blood samples for patients who achieved complete remission of asthma, defined as no use of asthma medications, no asthma symptoms, no airway hyperresponsiveness, and normal lung function at a recent clinic visit; however, this study did not examine the role of temperature.(49) In another study from Australia, ambient temperature ranging from the previous day to a year from sample collection was associated with differential methylation of 31 CpGs and 82 DMRs with biological pathways enriched for asthma and eczema-associated genes, such as NIPAL1 and PHF11.(50) In the Normative Aging Study of male veterans from the greater Boston Area, short-term, intermediate, and long-term temperature exposures were associated with several miRNAs derived from extracellular vesicles, including some implicated in respiratory diseases.(51,52) In a birth cohort from China employing a candidate gene approach, maternal whole blood and cord blood DNA methylation of the GPR61 gene was associated with prenatal temperature and humidity exposure, with evidence that cord blood GPR61 methylation mediated associations between prenatal exposure to temperature and humidity and birth weight.(53) Another birth cohort in China reported differential methylation of the H19 promoter in cord blood associated with prenatal temperature and humidity exposure.(54) These findings provide evidence that high temperature exposure, both chronic and short-term and at different points of the life course, influences DNA methylation in leukocytes and miRNAs, as examined in one study.
Finally, there has been one study published on temperature and epigenetic aging. A study from the Cooperative Health Research in the Region of Augsburg (KORA) in Germany reported acceleration of multiple epigenetic aging clocks (Horvath, Hannum, GrimAge and Skin-Blood) associated with medium-term exposure (4-week and 8-week) to high but not low temperature.(55) Additionally, higher average annual temperature was associated with increased EAA of those same epigenetic aging biomarkers as well as the PhenoAge epigenetic clock. These findings are relevant because acceleration of certain clocks, including PhenoAge and GrimAge, captures risk of all-cause mortality and disease morbidity, such as lower airway diseases and poor lung function.(56-58) In this study, higher annual temperature associations for select epigenetic aging markers were stronger for females, obese participants, and participants with cardiovascular disease. These findings are relevant to allergic disease, as acceleration of epigenetic aging biomarkers, particularly Horvath’s clock, has been associated with asthma and allergic sensitization in children.(23,24) Overall, among DNA methylation studies on temperature, there is heterogeneity with respect to study design, temperature range and measurements, and epigenetic approaches and biomarkers tested.
Precipitation
Mechanisms Linking Precipitation to Allergy and Immunologic Diseases
Climate change is associated with increased extreme precipitation and flooding as well as more severe drought in many parts of the world due to changes in the hydrological cycle.(59) One result of these climatic changes is increased exposure to molds, dust mite allergens, bacteria, and microbial toxins in both indoor and outdoor settings, which can trigger asthma exacerbations through both allergic and non-allergic mechanisms.(2,4) In addition, heavy precipitation and thunderstorms have been associated with severe asthma exacerbations and deaths in pollen-allergic patients.(60-63) The likely mechanism is that rainwater interacts with pollen to release a high concentration of smaller, more allergenic components that trigger asthma and allergic rhinitis, especially at the start of a storm (Table 4).(4,5)
Recent Epigenetic Studies on Precipitation
Both extremes of precipitation exposure—drought and heavy rainfall—have been shown to affect DNA methylation (Table 3). For example, in an EWAS conducted in Africa with children whose mothers were exposed to severe drought, 16 CpG sites in saliva samples were found to be differentially methylated between exposed and unexposed participants.(64) In this study, 7 CpGs were hypermethylated and 9 were hypomethylated; some were related to genes involved with metabolism and immune function, such as INFG, which encodes for the IFN-γ cytokine that likely contributes to asthma pathogenesis.(65) Another study that examined DNA methylation of the G protein-coupled receptor 61 (GPR61) promoter in newborns and their mothers found that increased precipitation exposure during pregnancy was associated with greater GPR61 methylation in maternal and cord blood samples, which affected birth weight.(53) The evidence of a relationship between precipitation exposure and DNA methylation alterations is further bolstered by an EWAS on exposure to Hurricane Maria, which is discussed in the following section.(66)
Extreme Weather Events
Climate change is associated with greater frequency and intensity of extreme weather events due to warmer temperatures, persisting drought conditions, and changes in sea level, ocean currents, and wind patterns. Examples of extreme weather events include wildfires, hurricanes, tropical cyclones, and heat waves, which have been increasing around the world, posing a global threat to public health.(8) Many of these events have been linked to allergic disease pathogenesis and other health outcomes in epidemiology studies (Table 4). For example, exposure to air pollution from wildfires has been shown to increase risk for asthma(67) and atopic dermatitis,(68,69) and extreme heat events in the U.S. increased the odds of experiencing seasonal allergic rhinitis.(70)
Recent Epigenetic Studies on Wildfires
We found two human studies that examined the impacts of wildfires on the epigenome (Table 5). An EWAS performed with adult women in Australia assessed 3-year average wildfire and non-wildfire-related PM2.5 exposure.(71) In adjusted analyses, the researchers reported 26 CpGs that were significantly differentially methylated for wildfire-related PM2.5, most of which were hypomethylated and did not overlap with findings for non-wildfire-related PM2.5. They also found 33 significant DMRs for wildfire-related PM2.5 with no overlap for non-wildfire-related PM2.5. These epigenetic alterations for wildfire air pollution exposure mapped to 47 genes that are related to inflammation, carcinogenesis, and immune dysregulation, including HLA-DQB1, which is at a locus that is associated with asthma,(72) and LRRC43, which is associated with eczema and allergy.(73) In a smaller study of children age 7-8 years in California, researchers reported that participants exposed to wildfires compared to prescribed burns—intentional burning of land primarily for forest management and wildfire hazard reduction—had greater methylation of the promoter region of FOXP3, which is expected to be associated with decreased gene expression.(74) This result is consistent with other studies on ambient air pollution exposure(75) and suggests a mechanism by which wildfire smoke exposure affects allergic disease, as decreased Foxp3 impairs the function of regulatory T-cells that play important roles in sustaining immune tolerance to allergens. While both the studies above examined DNA methylation in blood cells, long-term differential methylation and expression of genes related to inflammation have also been found in nasal epithelium cells of rhesus macaques exposed to wildfire smoke.(76)
Table 5:
Summary of Epigenetic Studies on Extreme Weather Events and Malnutrition
| Study Title | Authors (year) |
Location | Population | Exposure | Epigenetic Endpoint |
Key Finding(s) |
|---|---|---|---|---|---|---|
| Extreme Weather Events | ||||||
| Wildfire-related PM2.5 and DNA methylation: An Australian twin and family study | Xu et al. (2023) | Australia | Adult female twins and their sisters in the Australian Mammographic Density Twins and Sisters Study (AMDTSS) | Long-term exposure to wildfire-related PM2.5 | DNA methylation of peripheral leukocytes analyzed with with the 450K BeadChip (EWAS and 7 metrics of global DNA methylation) | -Exposure was associated with differential methylation of 26 CpGs and 33 DMRs mapping to 47 genes, which did not overlap with non-wildfire-related air pollution results. -There was a negative, but not significant, association with wildfire PM2.5 and the 7 global methylation measures. |
| The Impact of Prescribed Fire versus Wildfire on the Immune and Cardiovascular Systems of Children | Prunicki et al. (2019) | Fresno, California, USA | Children aged 7-8 years | Several air pollutants comprising wildfire air pollution | Candidate gene methylation analysis of the Foxp3, IL-4, IL-10, and IFNγ genes in peripheral leukocytes | -There was increased methylation of the Foxp3 promoter region post-wildfire exposure. |
| Pre- and peri-natal hurricane exposure alters DNA methylation patterns in children | Kello et al. (2023) | Puerto Rico | Children who were prenatally exposed to Hurricane Maria or conceived within 3 months post-disaster: Project HELiOS (Hurricane Exposures and Long-term Infant Outcomes Study) | Hurricane Maria | DNA methylation of peripheral leukocytes analyzed with with the EPIC BeadChip (EWAS) | -There was differential methylation of 47 CpGs: several hypermethylated sites associated with stage of gestation at the time of hurricane impact (biggest differences at 20-25 weeks gestation). -One significant DMR was found in association with the timing of the hurricane and annotates to the LLRC39 gene. |
| Malnutrition | ||||||
| Obesity-Associated Vitamin D Deficiency Correlates with Adipose Tissue DNA Hypomethylation, Inflammation, and Vascular Dysfunction | Mirza et al. (2022) | Chicago, Illinois, USA | Premenopausal women with obesity | Vitamin D deficiency | Global DNA methylation in adipose tissue samples measured with the 5-mC DNA kit and targeted methylation analysis of 94 inflammatory genes with the EpiTect Methyl II PCR Array | -There was decreased global DNA methylation associated with Vitamin D deficiency. -More severe Vitamin D deficiency was associated with greater hypomethylation of the promoters of the majority (70%) of inflammatory genes in the array. |
| Vitamin D supplementation is associated with slower epigenetic aging | Vetter et al. (2022) | Berlin, Germany | Adults in the Berlin Aging Study II (BASE-II) and GendAge study | Vitamin D supplementation | 5 epigenetic clocks: 7-CpG, Horvath, Hannum, PhenoAge, and GrimAge | -Treatment with Vitamin D supplementation in deficient individuals was associated with 2.6 year lower 7-CpG age acceleration and 1.3 year lower Horvath age acceleration. |
| DNA methylation profile of a rural cohort exposed to early-adversity and malnutrition: An exploratory analysis | Gomez-Verjan et al. (2022) | Tlaltizapan, Mexico | Adults who experienced malnutrition and adversity early in life and healthy middle-class elders as controls | Early-life malnutrition and adversity in a rural region | DNA methylation of peripheral leukocytes analyzed with with the EPIC BeadChip (EWAS) and 4 epigenetic clocks: Horvath, Hannum, PhenoAge, and GrimAge | -Early-life malnutrition and adversity was associated with 160 hypermethylated CpG sites and 55 hypomethylated CpG sites, many of which pertain to metabolic and neurocognitive pathways. -There were differential epigenetic aging profiles based on malnutrition exposure. |
| Childhood exposure to hunger: associations with health outcomes in later life and epigenetic markers | Perna et al. (2020) | Saarland, Germany | Adults who experienced hunger during the German famine (1945-1948) earlier in life who are part of the ESTHER cohort | Early-life exposure to hunger | DNA methylation of peripheral leukocytes analyzed with with the 450K BeadChip (EWAS) | -While 12 CpGs had a raw p-value <0.05 associated with childhood hunger, no CpGs achieved epigenome-wide significance after multiple testing correction. |
| Early-life exposure to severe famine is associated with higher methylation level in the IGF2 gene and higher total cholesterol in late adulthood: the Genomic Research of the Chinese Famine (GRECF) study | Shen et al. (2019) | Anhui and Jiangxi, China | Adults in the Genomic Research of the Chinese Famine (GRECF) study who experienced the famine while an infant/fetus and those born afterwards | Chinese Great Famine in 1959-1961 | Candidate gene methylation analysis of the IGF2 gene in peripheral leukocytes in blood | -There was a positive association between famine exposure and DNA methylation of the CpG1 site of the IGF2 gene and IGF2 DMR. -Each unit increase in methylation of the CpG1 site was associated with a 1.09 unit increase in total serum cholesterol levels. |
| DNA methylation signatures link prenatal famine exposure to growth and metabolism | Tobi et al. (2014) | The Netherlands | Adults who experienced hunger during the Hunger Winter (1944-1945) earlier in life | Prenatal exposure to hunger during the Hunger Winter | Testing of differential methylation of 28 genomic annotations in whole blood samples | -181 regions were identified as prenatal malnutrition-associated DMRs, most of which were hypermethylated, occurred in gene bodies, and displayed intermediate DNA methylation levels. -There were differences in methylation patterns based on gestational timing of exposure. |
Epigenetic Studies on Storms
Regarding cyclones, a study in Puerto Rico examined the epigenetic impacts of Hurricane Maria in 2017 on children who were exposed prenatally or conceived within 3 months later (Table 5).(66) There were 47 significant differentially methylated CpGs associated with all hurricane-related variables, including stress, and 30 were associated with gestational stage at the time of the hurricane, almost all of which were hypermethylated. The researchers reported that the most biologically relevant site was the probe near the sepiapterin reductase (SPR) gene, as it is located in a CpG island and close to the gene’s transcription start site in an open chromatin region. The gene is involved in the production of tetrahydrobiopterin, a metabolite that can affect T-cell-mediated autoimmunity and allergic inflammation.(77) The greatest mean methylation level changes occurred with hurricane exposure during 20-25 weeks of gestation, suggesting a prenatal period with increased susceptibility to epigenetic alteration. Other studies have found that exposure to Superstorm Sandy was associated with alterations in placental gene expression, which may be due to epigenetic modifications.(78,79) No human epigenetic studies were found for other extreme weather events like heat waves and tornadoes.
Malnutrition
Undernutrition, obesity, and climate change are 3 conditions that affect countries worldwide and constitute a syndemic: interacting at the same time, having synergistic effects on each other, and having similar underlying drivers.(80,81) Climate change has contributed to food insecurity through several pathways, including crop failures, destruction of agricultural property due to extreme weather, increased vector-borne diseases, and civil unrest, and it is predicted to lead to the malnourishment of 25 million more children globally by 2050.(80) Malnutrition has several effects on the human body, including impaired muscle and immune function (Table 4) as well as cardiopulmonary, gastrointestinal, and psychosocial impacts.(82) While the literature on diet and epigenetic modifications is quite expansive, we focus here on discussing a diverse set of recent epigenetic studies on undernutrition and hunger (Table 5).
Epigenetic Studies on Nutritional Status and Famine
Studies examining exposure to malnutrition associated with periods of famine or in rural communities and epigenetic alterations later in life have found mixed results. Early-life exposure to malnutrition and adversity among adult participants raised in a rural area in Mexico was associated with 160 hypermethylated CpG sites and 55 hypomethylated CpG sites in peripheral leukocytes.(83) Many of these sites annotated to pathways pertaining to biological regulation, neurocognition, and developmental processes. This study also provided evidence that EAA, which was calculated using 4 different epigenetic clocks, is affected by nutritional status, in alignment with existing studies.(84,85) On the other hand, a larger EWAS conducted with blood samples from adult participants who experienced early-life exposure to hunger during the German famine (1945-1948) found no differentially methylated CpG sites after multiple testing correction.(86) The authors suggest that this finding may be attributable to limitations of statistical power, lack of detail regarding the severity of hunger episodes, and small methylation changes that may have been reversed by adulthood. However, one targeted gene study that examined early-life exposure to the Chinese Great Famine (1959-1961) found positive associations with IGF2 gene methylation.(87) This is consistent with findings from a study on the Dutch Hunger Winter (1944-1945) that reported altered DNA methylation of IGF2 among individuals exposed prenatally to famine.(88) Of note, increased IGF2 levels has a protective effect and leads to enhanced regulatory T-cell function and IL-10 expression, so if hypermethylation of this gene due to malnutrition leads to decreased IGF2, this may increase risk for food allergy and asthma. A comprehensive testing of the epigenome among the Dutch Hunger Winter subjects prenatally exposed to famine found 181 DMRs.(89) One of the genes found to have a DMR associated with prenatal malnutrition in this study was CPT1A, which may be associated with asthma, as another study found that this gene is overexpressed in Th2 cells in patients with asthma compared to controls.(90)
Epigenetic Studies on Vitamin D
One specific example of how rising temperatures from climate change may affect nutrition is through increasing risk for Vitamin D deficiency as a result of heat-related regulation of cortisol release.(91) A few studies of Vitamin D deficiency have shown associations with DNA methylation and EAA. One study reported that Vitamin D deficiency was associated with hypomethylation in adipocytes for the majority of promoters for the 94 inflammatory genes measured within this study.(92) The existing literature suggests both that Vitamin D deficiency impacts the methylome and that the resulting epigenetic alterations reciprocally affect Vitamin D metabolism. In addition, a recent study on EAA among adults in Germany found that Vitamin D levels impacted EAA, as supplementation was associated with slower epigenetic aging: 2.6 years for a 7-CpG custom epigenetic clock and 1.3 years for the Horvath epigenetic clock.(93) Overall, more studies are needed to examine the epigenetic changes associated specifically with the ongoing and future changes in malnutrition within and across populations globally due to climate change.
Discussion and Conclusions
Currently, there is a limited body of scientific literature focusing on direct climate-related epigenetic impacts, such as from precipitation or extreme weather events. Temperature exposure has been the most widely studied weather-related variable. Other phenomena like malnutrition or famine from historical events, which are expected to worsen in some areas of the world due to climate change, have been characterized but not as a direct result of climate change. Many studies have reported multiple associations between climate factors and DNA methylation using candidate gene approaches or epigenome-wide testing of peripheral blood leukocytes. These findings provide some support for the connection between climate change-associated epigenetic changes at loci previously associated with allergic diseases, including asthma, eczema, and allergic rhinitis. Studies are limited in longitudinal follow-up and lack integration between climate-related changes, epigenetic biomarkers, and phenotyping of allergic disease. Most epigenetic studies have examined leukocyte-isolated DNA methylation, which is relevant for allergy but could miss important biological implications for other allergic diseases.
The changing climate will pose multiple health hazards, including increases in allergic disease incidence and severity in the near and distant future.(94) While immediate morbidity and mortality following an extreme weather event are concerning, long-term consequences might be larger. For example, mortality rates following Hurricane Maria in Puerto Rico remained elevated for a year afterwards.(95) Epigenetic biomarkers might serve as promising tools to survey, monitor, and evaluate the impact of climate change in global populations. While epigenetic studies have systematically surveyed allergic disease associations and found positive results, few studies have linked environmental exposures to allergic diseases prospectively.(38) Similarly, not many epigenetic studies on extreme weather events, high temperature, and climate-related dietary impacts have incorporated subsequent measures of allergic diseases within the same study design, so the current evidence remains limited. To advance the field, it will be key to leverage epigenetic biomarkers of extreme weather events in the context of allergy and atopy, ideally with large prospective epidemiological cohorts following these events incorporating biomarkers of exposure and detailed ascertainment of allergic disease. Both study design and timing of data collection are critical because epigenetic alterations can serve as both biomarkers of exposure or disease and intermediates of disease progression that can be used to target therapies or monitor progression.(38) In this context, epigenetic marks such, as DNA methylation changes, can be passengers or drivers of observed associations. Careful design of longitudinal studies along with novel causal inference methods can help elucidate relationships.
Studies related to climate change and extreme weather events with epigenetic biomarkers have been limited, and all but one investigated DNA methylation. The most widely studied meteorological condition thus far has been temperature,(31) likely reflecting the availability of spatial data to estimate exposure. However, there is a lack of studies tracking personal exposure measures that might vary substantially throughout time, day, and season. We identified a critical need to improve personal exposure measures of temperature, particle exposure, and diet associated with climatic and extreme weather events. Future work should characterize personalized measurements. This includes, for example, methods for measuring internal core temperature among some of the most vulnerable, including farm and construction workers. In this manner, epigenetic studies on climate change may help elucidate mechanisms underlying climate injustice and allergic disease prevalence based on the disproportionate exposure to harmful climatic factors across populations.(96) Among the temperature studies, differential methylation of multiple genes implicated in either asthma or inflammation have been reported. Future work should characterize epigenetic biomarkers that might respond rapidly to extreme heat, for example during a heat wave, compared to those that might be altered due to long-term exposure, for example among farm workers. This will help characterize biological pathways associated with acute and chronic exposure. While epigenetic findings might be influenced by other population characteristics, epigenetic aging biomarkers might serve as more consistent overall epigenetic indicators of the impact of climate on health given their robust link to morbidity and mortality.
Like most other epigenetic studies, those related to climate change have been limited to mostly peripheral blood cells. Epigenetic marks are tissue- and organ-specific, which poses both a challenge but also an opportunity to elucidate organ-specific effects. Increasing the collection and use of target tissues for epigenetic studies beyond leukocytes, such as through collecting nasal or skin cells, will facilitate the testing of more complex and direct hypotheses by capturing early biomarkers of allergic diseases. While it is not always feasible to collect target tissues like lung samples to study airway disease, surrogate tissues like nasal cells can provide unique biological insight compared to blood-based studies for allergic diseases.(24) Target tissues that might be accessible to study vary based on the setting. For example, skin cells are very relevant for atopic dermatitis(97) and nasal cells for airway disease and asthma.(24) These tissues are relevant to study the effects of wildfires, for example, while leukocyte-isolated DNA might serve as a common source of inflammatory cells relevant for multiple climatic exposures. It is likely that the most sensitive tissue is dependent on the type of exposure, route of exposure, and systemic or localized effects in the body. Therefore, these factors need to be carefully considered into study design.
Another important limitation of extreme weather events is the collection of data, samples, and weather information in a timely manner following these events. Very few epigenetic studies have looked at extreme weather events directly. Given that there are many indirect and direct pathways that might operate from these events on the epigenome (e.g., dietary changes, medical care access, drinking water quality, power outages, and extreme psychological stress), there is a need to better characterize specific exposure pathways related to extreme weather events. Existing frameworks have proposed multiple causal pathways for climate-related changes to impact health.(1,13) Specificity on the causal pathway associated with epigenetic changes could lead to higher reproducibility of results and the development of sensitive biomarkers. One aspect of importance is the emerging understanding of the impact of climate change on mental health,(98) especially since psychological stress plays a pathologic role in the development of allergic diseases.(99,100) This pathway should be carefully examined in epigenetic studies of extreme weather events and natural disasters.
Additionally, emerging evidence points at alterations in biological aging estimated by epigenetic clocks related to climate change events, particularly temperature and diet. The one study linking temperature to EAA also reported stronger effects for females, obese participants, and individuals with cardiovascular disease,(55) which highlights susceptible populations that might be disproportionately impacted by climate-related changes.
Heterogeneity of exposure assessment, timing, and measurement have precluded the replication of climate related epigenetic biomarkers across cohorts. A global collaborative network to increase diverse representation of study participants and support replication efforts and the harmonization of measurements and power will be needed to elucidate the impacts of climate change on the human epigenome. Despite these challenges, the evidence suggests that epigenetic marks are influenced by climate exposures and could serve as biomarkers of allergic disease. Additionally, with strong prospective study designs, high-quality exposure ascertainment, and precise measurement of target tissues, epigenetics research can help elucidate mechanisms through which climate change impacts atopic and airway diseases.
Supplementary Material
Sources of Funding:
This research was supported by United States National Institutes of Health grants R01ES031259 and P42ES004705. SB is supported by NIH UM1AI173380, U19AI136053, U01AI160082, and R01AI147028.
Abbreviations
- CpG site
cytosine-phosphate-guanine dinucleotide
- DMP
differentially methylated position
- DMR
differentially methylated region
- EAA
epigenetic age acceleration
- EWAS
epigenome-wide association study
- FDR
false discovery rate
- FOXP3
Forkhead box P3
- meQTLs
methylation quantitative trait loci
- PM2.5
particulate matter less than 2.5 microns in diameter
- SNP
single-nucleotide polymorphism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest: None to declare.
References
- 1.Watts N, Amann M, Arnell N, Ayeb-Karlsson S, Belesova K, Boykoff M, et al. The 2019 report of The Lancet Countdown on health and climate change: ensuring that the health of a child born today is not defined by a changing climate. The Lancet. 2019. Nov 16;394(10211):1836–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D’Amato G, Holgate ST, Pawankar R, Ledford DK, Cecchi L, Al-Ahmad M, et al. Meteorological conditions, climate change, new emerging factors, and asthma and related allergic disorders. A statement of the World Allergy Organization. World Allergy Organ J. 2015;8(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katelaris CH, Beggs PJ. Climate change: allergens and allergic diseases. Intern Med J. 2018;48(2):129–34. [DOI] [PubMed] [Google Scholar]
- 4.D’Amato G, Chong-Neto HJ, Monge Ortega OP, Vitale C, Ansotegui I, Rosario N, et al. The effects of climate change on respiratory allergy and asthma induced by pollen and mold allergens. Allergy. 2020;75(9):2219–28. [DOI] [PubMed] [Google Scholar]
- 5.Sampath V, Aguilera J, Prunicki M, Nadeau KC. Mechanisms of climate change and related air pollution on the immune system leading to allergic disease and asthma. Semin Immunol. 2023. May;67:101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossati A. Global Warming and Its Health Impact. Int J Occup Environ Med. 2017. Jan;8(1):7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashim JH, Hashim Z. Climate Change, Extreme Weather Events, and Human Health Implications in the Asia Pacific Region. Asia Pac J Public Health. 2016. Mar;28(2 Suppl):8S–14S. [DOI] [PubMed] [Google Scholar]
- 8.Ebi KL, Vanos J, Baldwin JW, Bell JE, Hondula DM, Errett NA, et al. Extreme Weather and Climate Change: Population Health and Health System Implications. Annu Rev Public Health. 2021. Apr 1;42:293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D’Amato G, Bergmann KC, Cecchi L, Annesi-Maesano I, Sanduzzi A, Liccardi G, et al. Climate change and air pollution. Allergo J Int. 2014;23(1):17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinney PL. Climate change, air quality, and human health. Am J Prev Med. 2008. Nov;35(5):459–67. [DOI] [PubMed] [Google Scholar]
- 11.Bais AF, McKenzie RL, Bernhard G, Aucamp PJ, Ilyas M, Madronich S, et al. Ozone depletion and climate change: impacts on UV radiation. Photochem Photobiol Sci Off J Eur Photochem Assoc Eur Soc Photobiol. 2015. Jan;14(1):19–52. [DOI] [PubMed] [Google Scholar]
- 12.Rocque RJ, Beaudoin C, Ndjaboue R, Cameron L, Poirier-Bergeron L, Poulin-Rheault RA, et al. Health effects of climate change: an overview of systematic reviews. BMJ Open. 2021. Jun 1;11(6):e046333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salas RN, Solomon CG. The Climate Crisis — Health and Care Delivery. N Engl J Med. 2019. Aug 22;381(8):e13. [DOI] [PubMed] [Google Scholar]
- 14.Singh AB, Kumar P. Climate change and allergic diseases: An overview. Front Allergy. 2022. Oct 13;3:964987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luschkova D, Traidl-Hoffmann C, Ludwig A. Climate change and allergies. Allergo J Int. 2022;31(4):114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harb H, Renz H. Update on epigenetics in allergic disease. J Allergy Clin Immunol. 2015. Jan 1;135(1):15–24. [DOI] [PubMed] [Google Scholar]
- 17.Potaczek DP, Harb H, Michel S, Alhamwe BA, Renz H, Tost J. Epigenetics and allergy: from basic mechanisms to clinical applications. Epigenomics. 2017. Apr;9(4):539–71. [DOI] [PubMed] [Google Scholar]
- 18.Yousefi PD, Suderman M, Langdon R, Whitehurst O, Davey Smith G, Relton CL. DNA methylation-based predictors of health: applications and statistical considerations. Nat Rev Genet. 2022. Jun;23(6):369–83. [DOI] [PubMed] [Google Scholar]
- 19.Melén E, Koppelman GH, Vicedo-Cabrera AM, Andersen ZJ, Bunyavanich S. Allergies to food and airborne allergens in children and adolescents: role of epigenetics in a changing environment. Lancet Child Adolesc Health. 2022. Nov;6(11):810–9. [DOI] [PubMed] [Google Scholar]
- 20.Somineni HK, Zhang X, Biagini Myers JM, Kovacic MB, Ulm A, Jurcak N, et al. Ten-eleven translocation 1 (TET1) methylation is associated with childhood asthma and traffic-related air pollution. J Allergy Clin Immunol. 2016. Mar;137(3):797–805.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cardenas A, Fadadu RP, Van Der Laan L, Ward-Caviness C, Granger L, Diaz-Sanchez D, et al. Controlled human exposures to diesel exhaust: a human epigenome-wide experiment of target bronchial epithelial cells. Environ Epigenetics. 2021;7(1):dvab003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDonald-Hyman C, Tager I, Nadeau K. Epigenetic Modifications of Foxp3 Locus are Associated with Asthma in Children exposed to High Levels of Ambient Air Pollution. J Allergy Clin Immunol. 2010. Feb 1;125(2):AB356. [Google Scholar]
- 23.Peng C, Cardenas A, Rifas-Shiman SL, Hivert MF, Gold DR, Platts-Mills TA, et al. Epigenetic age acceleration is associated with allergy and asthma in children in Project Viva. J Allergy Clin Immunol. 2019. Jun 1;143(6):2263–2270.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cardenas A, Sordillo JE, Rifas-Shiman SL, Chung W, Liang L, Coull BA, et al. The nasal methylome as a biomarker of asthma and airway inflammation in children. Nat Commun. 2019. Jul 12;10(1):3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji H, Biagini Myers JM, Brandt EB, Brokamp C, Ryan PH, Khurana Hershey GK. Air pollution, epigenetics, and asthma. Allergy Asthma Clin Immunol. 2016. Oct 19;12(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gugger PF, Fitz-Gibbon S, PellEgrini M, Sork VL. Species-wide patterns of DNA methylation variation in Quercus lobata and their association with climate gradients. Mol Ecol. 2016. Apr;25(8):1665–80. [DOI] [PubMed] [Google Scholar]
- 27.Weyrich A, Benz S, Karl S, Jeschek M, Jewgenow K, Fickel J. Paternal heat exposure causes DNA methylation and gene expression changes of Stat3 in Wild guinea pig sons. Ecol Evol. 2016. May;6(9):2657–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weyrich A, Lenz D, Fickel J. Environmental Change-Dependent Inherited Epigenetic Response. Genes. 2018. Dec 21;10(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marsh AG, Pasqualone AA. DNA methylation and temperature stress in an Antarctic polychaete, Spiophanes tcherniai. Front Physiol. 2014;5:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skjærven KH, Hamre K, Penglase S, Finn RN, Olsvik PA. Thermal stress alters expression of genes involved in one carbon and DNA methylation pathways in Atlantic cod embryos. Comp Biochem Physiol A Mol Integr Physiol. 2014. Jul;173C:17–27. [DOI] [PubMed] [Google Scholar]
- 31.Xu R, Li S, Guo S, Zhao Q, Abramson MJ, Li S, et al. Environmental temperature and human epigenetic modifications: A systematic review. Environ Pollut. 2020. Apr 1;259:113840. [DOI] [PubMed] [Google Scholar]
- 32.Saenen ND, Martens DS, Neven KY, Alfano R, Bové H, Janssen BG, et al. Air pollution-induced placental alterations: an interplay of oxidative stress, epigenetics, and the aging phenotype? Clin Epigenetics. 2019. Sep 17;11(1):124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rider CF, Carlsten C. Air pollution and DNA methylation: effects of exposure in humans. Clin Epigenetics. 2019. Sep 3;11(1):131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S, Ye Z, Mather KA, Nguyen TL, Dite GS, Armstrong NJ, et al. Early life affects late-life health through determining DNA methylation across the lifespan: A twin study. EBioMedicine. 2022. Mar;77:103927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013. Oct 24;502(7472):472–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rauluseviciute I, Drabløs F, Rye MB. DNA methylation data by sequencing: experimental approaches and recommendations for tools and pipelines for data analysis. Clin Epigenetics. 2019. Dec 12;11(1):193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campagna MP, Xavier A, Lechner-Scott J, Maltby V, Scott RJ, Butzkueven H, et al. Epigenome-wide association studies: current knowledge, strategies and recommendations. Clin Epigenetics. 2021. Dec 4;13(1):214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cardenas A, Fadadu RP, Koppelman GH. Epigenome-wide association studies of allergic disease and the environment. J Allergy Clin Immunol [Internet]. 2023. Jun 6 [cited 2023 Aug 19];0(0). Available from: https://www.jacionline.org/article/S0091-6749(23)00747-9/fulltext [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lluis A, Depner M, Gaugler B, Saas P, Casaca VI, Raedler D, et al. Increased regulatory T-cell numbers are associated with farm milk exposure and lower atopic sensitization and asthma in childhood. J Allergy Clin Immunol. 2014. Feb 1;133(2):551–559.e10. [DOI] [PubMed] [Google Scholar]
- 40.Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018. Jun;19(6):371–84. [DOI] [PubMed] [Google Scholar]
- 41.Cardenas A, Ecker S, Fadadu RP, Huen K, Orozco A, McEwen LM, et al. Epigenome-wide association study and epigenetic age acceleration associated with cigarette smoking among Costa Rican adults. Sci Rep. 2022. Mar 11;12(1):4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knopik VS, Maccani MA, Francazio S, McGeary JE. The Epigenetics of Maternal Cigarette Smoking During Pregnancy and Effects on Child Development. Dev Psychopathol. 2012. Nov;24(4):1377–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukherjee S, Dasgupta S, Mishra PK, Chaudhury K. Air pollution-induced epigenetic changes: disease development and a possible link with hypersensitivity pneumonitis. Environ Sci Pollut Res Int. 2021;28(40):55981–6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reynolds LM, Wan M, Ding J, Taylor JR, Lohman K, Su D, et al. DNA Methylation of the Aryl Hydrocarbon Receptor Repressor Associations With Cigarette Smoking and Subclinical Atherosclerosis. Circ Cardiovasc Genet. 2015. Oct;8(5):707–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsai PC, Glastonbury CA, Eliot MN, Bollepalli S, Yet I, Castillo-Fernandez JE, et al. Smoking induces coordinated DNA methylation and gene expression changes in adipose tissue with consequences for metabolic health. Clin Epigenetics. 2018. Oct 20;10(1):126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ebi KL, Capon A, Berry P, Broderick C, Dear R de, Havenith G, et al. Hot weather and heat extremes: health risks. The Lancet. 2021. Aug 21;398(10301):698–708. [DOI] [PubMed] [Google Scholar]
- 47.Millqvist E. TRP channels and temperature in airway disease—clinical significance. Temperature. 2015. Jun 30;2(2):172–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu Y, Xu R, Li S, Ming Wong E, Southey MC, Hopper JL, et al. Epigenome-wide association study of short-term temperature fluctuations based on within-sibship analyses in Australian females. Environ Int. 2023. Jan 1;171:107655. [DOI] [PubMed] [Google Scholar]
- 49.Qi C, Vonk JM, van der Plaat DA, Nieuwenhuis MAE, Dijk FN, Aïssi D, et al. Epigenome-wide association study identifies DNA methylation markers for asthma remission in whole blood and nasal epithelium. Clin Transl Allergy. 2020. Dec 11;10(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu R, Li S, Li S, Wong EM, Southey MC, Hopper JL, et al. Ambient temperature and genome-wide DNA methylation: A twin and family study in Australia. Environ Pollut Barking Essex 1987. 2021. Sep 15;285:117700. [DOI] [PubMed] [Google Scholar]
- 51.Danesh Yazdi M, Nassan FL, Kosheleva A, Wang C, Xu Z, Di Q, et al. Short-term air pollution and temperature exposure and changes in the extracellular microRNA profile of Normative Aging Study (NAS) participants. Environ Int. 2023. Jan 1;171:107735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Danesh Yazdi M, Nassan FL, Kosheleva A, Wang C, Xu Z, Di Q, et al. Intermediate and long-term exposure to air pollution and temperature and the extracellular microRNA profile of participants in the normative aging study (NAS). Environ Res. 2023. Jul 15;229:115949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li ZY, Gong YX, Yang M, Chai J, Sun RJ, Li QY, et al. Weather and Birth Weight: Different Roles of Maternal and Neonatal GPR61 Promoter Methylation. Biomed Environ Sci BES. 2022. Mar 20;35(3):181–93. [DOI] [PubMed] [Google Scholar]
- 54.Yang M, He T, Jiang L, Wang H, Zhang J, Chai J, et al. The role of maternal methylation in the association between prenatal meteorological conditions and neonatal H19/H19-DMR methylation. Ecotoxicol Environ Saf. 2020. Jul 1;197:110643. [DOI] [PubMed] [Google Scholar]
- 55.Ni W, Nikolaou N, Ward-Caviness CK, Breitner S, Wolf K, Zhang S, et al. Associations between medium- and long-term exposure to air temperature and epigenetic age acceleration. Environ Int. 2023. Aug 1;178:108109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hillary RF, Stevenson AJ, McCartney DL, Campbell A, Walker RM, Howard DM, et al. Epigenetic measures of ageing predict the prevalence and incidence of leading causes of death and disease burden. Clin Epigenetics. 2020. Jul 31;12(1):115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang C, Just A, Heiss J, Coull BA, Hou L, Zheng Y, et al. Biomarkers of aging and lung function in the normative aging study. Aging. 2020. Jun 19;12(12):11942–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging. 2018. Apr 18;10(4):573–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tabari H. Climate change impact on flood and extreme precipitation increases with water availability. Sci Rep. 2020. Aug 13;10(1):13768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andrew E, Nehme Z, Bernard S, Abramson MJ, Newbigin E, Piper B, et al. Stormy weather: a retrospective analysis of demand for emergency medical services during epidemic thunderstorm asthma. BMJ. 2017. Dec 13;359:j5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marks GB, Colquhoun JR, Girgis ST, Koski MH, Treloar AB, Hansen P, et al. Thunderstorm outflows preceding epidemics of asthma during spring and summer. Thorax. 2001. Jun;56(6):468–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hew M, Lee J, Susanto NH, Prasad S, Bardin PG, Barnes S, et al. The 2016 Melbourne thunderstorm asthma epidemic: Risk factors for severe attacks requiring hospital admission. Allergy. 2019. Jan;74(1):122–30. [DOI] [PubMed] [Google Scholar]
- 63.Thien F, Beggs PJ, Csutoros D, Darvall J, Hew M, Davies JM, et al. The Melbourne epidemic thunderstorm asthma event 2016: an investigation of environmental triggers, effect on health services, and patient risk factors. Lancet Planet Health. 2018. Jun;2(6):e255–63. [DOI] [PubMed] [Google Scholar]
- 64.Straight B, Qiao X, Ngo D, Hilton CE, Olungah CO, Naugle A, et al. Epigenetic mechanisms underlying the association between maternal climate stress and child growth: characterizing severe drought and its impact on a Kenyan community engaging in a climate change-sensitive livelihood. Epigenetics. 2022. Dec;17(13):2421–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Naumov D, Gassan D, Kotova O, Sheludko E, Afanaseva E, Perelman J, et al. Role of interferon-gamma as a marker of asthma severity and control. Eur Respir J [Internet]. 2019. Sep 28 [cited 2023 Jul 11];54(suppl 63). Available from: https://erj.ersjournals.com/content/54/suppl_63/PA4378 [Google Scholar]
- 66.Kello E, Vieira AR, Rivas-Tumanyan S, Campos-Rivera M, Martinez-Gonzalez KG, Buxó CJ, et al. Pre- and peri-natal hurricane exposure alters DNA methylation patterns in children. Sci Rep. 2023. Mar 8;13(1):3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Borchers Arriagada N, Horsley JA, Palmer AJ, Morgan GG, Tham R, Johnston FH. Association between fire smoke fine particulate matter and asthma-related outcomes: Systematic review and meta-analysis. Environ Res. 2019. Dec;179(Pt A):108777. [DOI] [PubMed] [Google Scholar]
- 68.Fadadu R, Grimes B, Jewell N, Vargo J, Young A, Abuabara K, et al. Association of Wildfire Air Pollution and Health Care Use for Atopic Dermatitis and Itch. JAMA Dermatol. 2021. Jun;157(6):658–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fadadu RP, Green M, Jewell NP, Grimes B, Vargo J, Wei ML. Association of Exposure to Wildfire Air Pollution With Exacerbations of Atopic Dermatitis and Itch Among Older Adults. JAMA Netw Open. 2022. Oct 26;5(10):e2238594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Upperman CR, Parker JD, Akinbami LJ, Jiang C, He X, Murtugudde R, et al. Exposure to extreme heat events is associated with increased hay fever prevalence among nationally representative sample of US adults: 1997–2013. J Allergy Clin Immunol Pract. 2017;5(2):435–441.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu R, Li S, Wu Y, Yue X, Wong EM, Southey MC, et al. Wildfire-related PM2.5 and DNA methylation: An Australian twin and family study. Environ Int. 2023. Jan 1;171:107704. [DOI] [PubMed] [Google Scholar]
- 72.Li X, Howard TD, Zheng SL, Haselkorn T, Peters SP, Meyers DA, et al. Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol. 2010. Feb 1;125(2):328–335.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ferreira MA, Vonk JM, Baurecht H, Marenholz I, Tian C, Hoffman JD, et al. Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nat Genet. 2017. Dec;49(12):1752–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prunicki M, Kelsey R, Lee J, Zhou X, Smith E, Haddad F, et al. The Impact of Prescribed Fire versus Wildfire on the Immune and Cardiovascular Systems of Children. Allergy. 2019. Oct;74(10):1989–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nadeau K, McDonald-Hyman C, Noth EM, Pratt B, Hammond SK, Balmes J, et al. Ambient air pollution impairs regulatory T-cell function in asthma. J Allergy Clin Immunol. 2010. Oct 1;126(4):845–852.e10. [DOI] [PubMed] [Google Scholar]
- 76.Brown AP, Cai L, Laufer BI, Miller LA, LaSalle JM, Ji H. Long-term effects of wildfire smoke exposure during early life on the nasal epigenome in rhesus macaques. Environ Int. 2022. Jan;158:106993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cronin SJF, Seehus C, Weidinger A, Talbot S, Reissig S, Seifert M, et al. The metabolite BH4 controls T cell proliferation in autoimmunity and cancer. Nature. 2018. Nov;563(7732):564–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang W, Li Q, Deyssenroth M, Lambertini L, Finik J, Ham J, et al. Timing of prenatal exposure to trauma and altered placental expressions of hypothalamic-pituitary-adrenal axis genes and genes driving neurodevelopment. J Neuroendocrinol. 2018. Apr;30(4):e12581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang W, Ham J, Li Q, Deyssenroth MA, Lambertini L, Huang Y, et al. Moderate prenatal stress may buffer the impact of Superstorm Sandy on placental genes: Stress in Pregnancy (SIP) Study. PloS One. 2020;15(1):e0226605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Swinburn BA, Kraak VI, Allender S, Atkins VJ, Baker PI, Bogard JR, et al. The Global Syndemic of Obesity, Undernutrition, and Climate Change: The Lancet Commission report. The Lancet. 2019. Feb 23;393(10173):791–846. [DOI] [PubMed] [Google Scholar]
- 81.Dietz WH. Climate change and malnutrition: we need to act now. J Clin Invest. 130(2):556–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saunders J, Smith T. Malnutrition: causes and consequences. Clin Med. 2010. Dec;10(6):624–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gomez-Verjan JC, Esparza-Aguilar M, Martín-Martín V, Salazar-Pérez C, Cadena-Trejo C, Gutiérrez-Robledo LM, et al. DNA methylation profile of a rural cohort exposed to early-adversity and malnutrition: An exploratory analysis. Exp Gerontol. 2022. Oct 1;167:111899. [DOI] [PubMed] [Google Scholar]
- 84.Kim Y, Huan T, Joehanes R, McKeown NM, Horvath S, Levy D, et al. Higher diet quality relates to decelerated epigenetic aging. Am J Clin Nutr. 2022. Jan 1;115(1):163–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Koemel NA, Skilton MR. Epigenetic Aging in Early Life: Role of Maternal and Early Childhood Nutrition. Curr Nutr Rep. 2022;11(2):318–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Perna L, Zhang Y, Wild B, Kliegel M, Ihle A, Schöttker B, et al. Childhood exposure to hunger: associations with health outcomes in later life and epigenetic markers. Epigenomics. 2020. Nov;12(21):1861–70. [DOI] [PubMed] [Google Scholar]
- 87.Shen L, Li C, Wang Z, Zhang R, Shen Y, Miles T, et al. Early-life exposure to severe famine is associated with higher methylation level in the IGF2 gene and higher total cholesterol in late adulthood: the Genomic Research of the Chinese Famine (GRECF) study. Clin Epigenetics. 2019. Jun 10;11:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008. Nov 4;105(44):17046–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tobi EW, Goeman JJ, Monajemi R, Gu H, Putter H, Zhang Y, et al. DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nat Commun. 2014. Nov 26;5:5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Seumois G, Zapardiel-Gonzalo J, White B, Singh D, Schulten V, Dillon M, et al. Transcriptional Profiling of Th2 Cells Identifies Pathogenic Features Associated with Asthma. J Immunol. 2016. Jul 15;197(2):655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Balducci M, Pruccoli L, Tarozzi A. Heat stress as a potential risk factor for vitamin D deficiency. Med Hypotheses. 2023. Jul 1;176:111085. [Google Scholar]
- 92.Mirza I, Mohamed A, Deen H, Balaji S, Elsabbahi D, Munasser A, et al. Obesity-Associated Vitamin D Deficiency Correlates with Adipose Tissue DNA Hypomethylation, Inflammation, and Vascular Dysfunction. Int J Mol Sci. 2022. Nov 19;23(22):14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vetter VM, Sommerer Y, Kalies CH, Spira D, Bertram L, Demuth I. Vitamin D supplementation is associated with slower epigenetic aging. GeroScience. 2022. Jun;44(3):1847–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pacheco SE, Guidos-Fogelbach G, Annesi-Maesano I, Pawankar R, D’ Amato G, Latour-Staffeld P, et al. Climate change and global issues in allergy and immunology. J Allergy Clin Immunol. 2021. Dec;148(6):1366–77. [DOI] [PubMed] [Google Scholar]
- 95.Kishore N, Marqués D, Mahmud A, Kiang MV, Rodriguez I, Fuller A, et al. Mortality in Puerto Rico after Hurricane Maria. N Engl J Med. 2018. Jul 12;379(2):162–70. [DOI] [PubMed] [Google Scholar]
- 96.Burbank AJ, Hernandez ML, Jefferson A, Perry TT, Phipatanakul W, Poole J, et al. Environmental justice and allergic disease: A Work Group Report of the AAAAI Environmental Exposure and Respiratory Health Committee and the Diversity, Equity and Inclusion Committee. J Allergy Clin Immunol. 2023. Mar 1;151(3):656–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Olisova OY, Kochergin NG, Kayumova LN, Zavarykina TM, Dmitriev AA, Asanov AY. Skin DNA methylation profile in atopic dermatitis patients: A case-control study. Exp Dermatol. 2020. Feb;29(2):184–9. [DOI] [PubMed] [Google Scholar]
- 98.Berry HL, Waite TD, Dear KBG, Capon AG, Murray V. The case for systems thinking about climate change and mental health. Nat Clim Change. 2018. Apr;8(4):282–90. [Google Scholar]
- 99.Chen E, Hanson MD, Paterson LQ, Griffin MJ, Walker HA, Miller GE. Socioeconomic status and inflammatory processes in childhood asthma: The role of psychological stress. J Allergy Clin Immunol. 2006. May 1;117(5):1014–20. [DOI] [PubMed] [Google Scholar]
- 100.Dave ND, Xiang L, Rehm KE, Marshall GD. Stress and Allergic Diseases. Immunol Allergy Clin North Am. 2011. Feb;31(1):55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.