Summary
The erythrocyte silent Duffy blood group phenotype in Africans is thought to confer resistance to Plasmodium vivax blood-stage infection. However, recent studies report P. vivax infections across Africa in Fy-negative individuals. This suggests that the GATA-1 SNP underlying Fy-negativity does not entirely abolish Fy expression or that P. vivax has developed a Fy-independent red blood cell (RBC) invasion pathway. We show that RBCs and erythroid progenitors from in vitro differentiated CD34 cells and from bone marrow aspirates from Fy-negative samples express a functional Fy on their surface. This suggests that the GATA-1 SNP does not entirely abolish Fy expression. Given these results, we developed an in vitro culture system for P. vivax and show P. vivax can invade erythrocytes from Duffy-negative individuals. This study provides evidence that Fy is expressed in Fy-negative individuals and explains their susceptibility to P. vivax with major implications and challenges for P. vivax malaria eradication.
Keywords: Duffy protein, Duffy negative individuals, Erythroid Progenitors, Plasmodium vivax, Bone marrow
Graphical Abstract
eTOC blurb
The Duffy gene (Fy) polymorphism has historically been linked to Plasmodium vivax natural susceptibility/resistance, but clinical infections have recently been detected in Fy-negative individuals. Dechavanne and colleagues observe Fy expression during early erythropoietic stages in Fy-negative individuals, which enables P. vivax invasion and helps explain infections seen in Fy-negative individuals.
Introduction
Duffy gene (FY) polymorphism has been prominent historically. In 1968, Donahue et al. used Fya/Fyb serological inheritance patterns and human chromosome 1 centromeric staining features to make FY the first gene mapped to a human autosome1. Functionally, the Duffy blood group protein (Fy)2 also known as Duffy antigen chemokine receptor (DARC) or CD234, was the first identified member of the diverse seven-transmembrane chemokine receptor family, albeit with several interesting structural and functional differences that make its biology unique2. Fy expression on both erythroid and non-erythroid cells2 modulates homeostatic levels and gradients of chemokines between blood circulation3–7 and tissues8–12.
The decades-long observations that Africans and African Americans, who are for the most part Fy-negative, were highly resistant to Plasmodium vivax blood-stage infection13–18 suggested that interaction with the Fy protein was required for parasite invasion of human red blood cells (RBCs)19. P. vivax, selectively invades reticulocytes in a process that requires region II of Plasmodium vivax Duffy-binding protein (rPvDBPII) with DARC receptor20–26. With gene sequencing, a single nucleotide polymorphism (SNP) in the FY gene GATA-1 transcription factor binding site has been shown to inhibit FY erythroid expression27,28 and homozygosity underlies the “Fy erythrocyte silent” (herein, Fy-negative) phenotype characterized by standard serological methods27. In vitro studies using Fy-dependent P. knowlesi20,29 show that while the merozoite can orient apically to the RBC surface, the parasite failed to invade Fy-negative erythrocytes. Therefore, the onward formation of a tight mobile junction needed for infection of the RBC failed30. This background has inextricably linked Duffy blood group genetics and malaria. Recent studies now report P. vivax infections in Fy-negative individuals from many malarious African countries31,32. These observations lead to at least two hypotheses. First, constitutive low-level or periodic Fy expression may occur in Fy-negative people and facilitate occasional blood-stage infection. Second, P. vivax may not interact with Fy exclusively and has developed a Fy-independent RBC invasion pathway. Even if other P. vivax proteins appear to play a role in parasite invasion33,34 (e.g., reticulocyte-binding proteins) no alternative pathway has been so far identified.
Previous studies have demonstrated a gene dosage effect of the FY GATA-1 SNP that reduces erythrocyte surface expression of Fya and Fyb by 50% in heterozygous individuals28,35, and others have identified additional coding region SNPs associated with reduced expression of both Fya (Fya-weak)36 and Fyb (Fyb-weak)37–39; very rare nonsense SNPs and deletions in the FY gene coding region have also been reported (see text with Supplemental Information (SI) Table S1)40,41. Further, Fy is more highly expressed on reticulocytes versus older RBCs35. With this range of Fy expression phenotypes, we tested the hypothesis that the GATA-1 SNP (FYES mutation) may not entirely block Fy protein expression and here we describe how this may influence susceptibility to P. vivax blood stage infection.
Results
Fy expression on Fy-positive and Fy-negative pbRBC
To assess the presence of Fy on peripheral blood red blood cells (pbRBC), two anti-Fy6 antibodies were used: the conventional mouse monoclonal antibody 2C342,43 and the camelid single-domain antibody CA11144, which overlaps the binding site of 2C3 and possess a higher affinity45. We tested whether the anti-Fy6 antibodies recognized Fy on the surface of Fy-negative (FY*BES/*BES) and Fy-positive RBC (Figures S1, S2 and S3). Working on freshly collected pbRBC, we observed CA111 binding to pbRBC of FY*A/*A, FY*B/*B donors and FY*BES/*BES pbRBC at different time points (Figure 1A). Collection of different time points from the same donors indicated variations in CA111 binding from 74.0% to 89.4% for FY*B/*B pbRBC, 77.2% to 90.8% for FY*A/*A pbRBC and 11.5% to 47.6% for FY*BES/*BES. This latter result was markedly higher than previously observed on FY*BES/*BES donors44. Another set of experiments was performed using the conventional anti-Fy6 2C3 and the single-domain anti-Fy6 CA111 (Figure 1B) and anti-Fya and anti-Fyb on 10 other Fy-negative donors (Figure 1C). CA111 detected Fy on the pbRBC from FY*B/*B (N=7) and FY*BES/*BES donors (N=10, Figure 1B) confirming the observation of Figure 1A. Using the 2C3, Fy protein was detected on the pbRBC of FY*B/*B donors but not on the FY*BES/*BES donors. As expected, the anti-Fya antibody showed reactivity on pbRBC from FY*A/*B donor only and the anti-Fyb antibody on FY*A/*B and FY*B/*B pbRBC. Taken together, these results suggest that the Fy protein is consistently detected on the pbRBC surface of FY*BES/*BES by CA111 compared to 2C3 and the anti-Fya and anti-Fyb antibodies used for clinical serology and detection of Fy is in a reduced range in Fy-negative compared to Fy-positive individuals.
Figure 1. Binding of Fy-specific antibodies on Fy-positive and Fy-negative pbRBC.
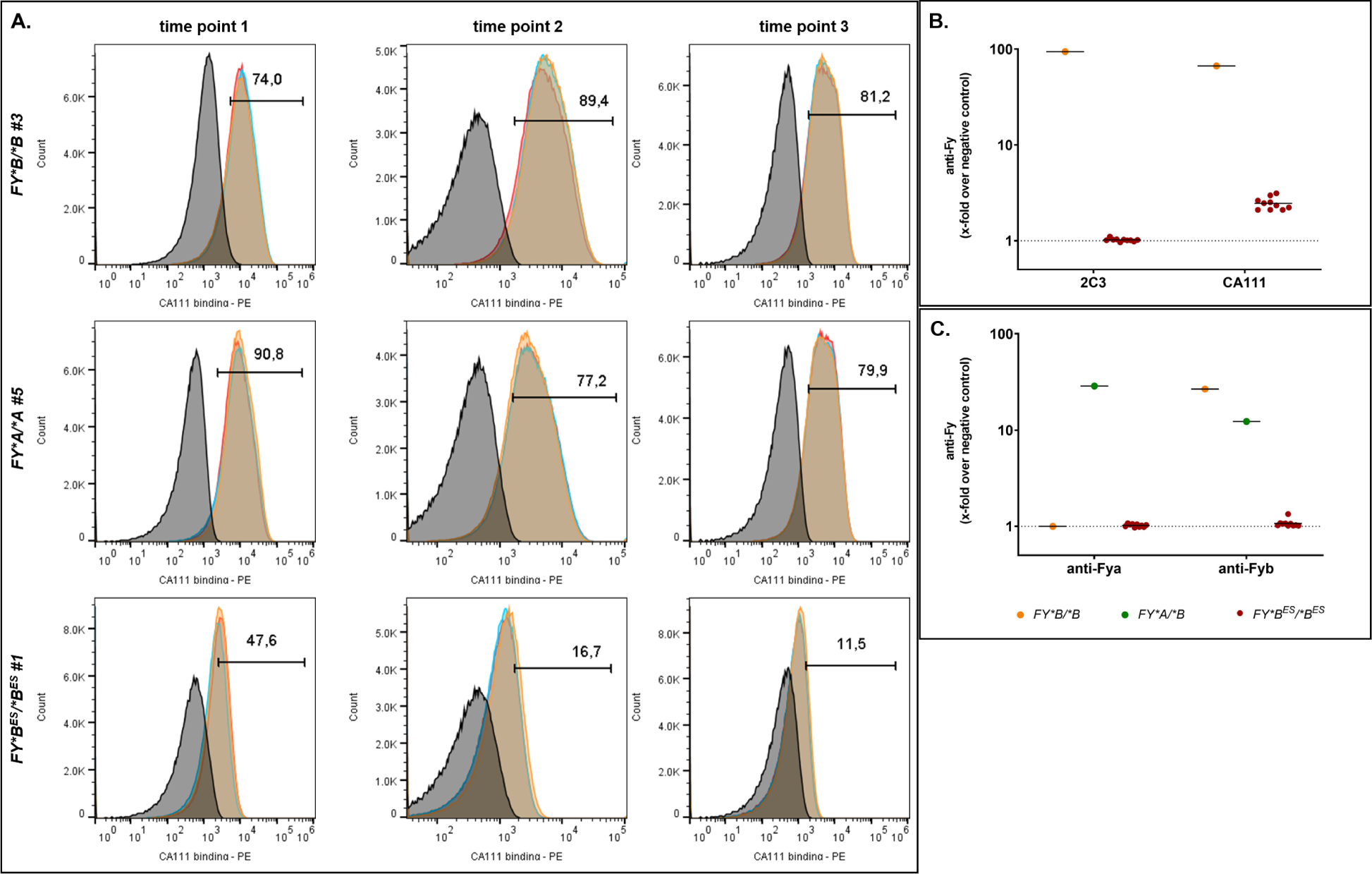
A - Direct binding of CA111 anti-Fy6 to pbRBC from FY*B/*B, FY*A/*A and FY*BES/*BES donors. Experiments were performed in triplicate (histograms in red, orange and blue in overlay) ; each donor was tested at three independent time points. Individual donor samples minus CA111 (plus anti-HA mouse antibody and goat anti-mouse PE-conjugated antibody) served as negative controls represented in each histogram in dark. The percentage of cells with CA111 binding (PE: phycoerythrin) over the background is indicated above the gate. The x-folds over negative control (geometric mean of fluorescence intensity (gmfi)) were higher than 48 for FY*B/*B #3 or FY*A/*A #5 and were equal to 6, 5 and 4 for FY*BES/*BES #1 at the three time points respectively.
B - Binding of 2C3 and CA111 anti-Fy6 to pbRBC from 1 FY*B/*B donor (orange) and 10 FY*BES/*BES donors (red). Each individual served as its own negative control: no primary antibody, only secondary antibodies (anti-HA (CA111), anti-mouse (2C3)). The x-fold is calculated using the gmfi of the binding of antibodies targeting the Fy6 epitope divided by the gmfi of the binding of the secondary antibody only.
C - Binding of anti-Fya and anti-Fyb to pbRBC from 1 FY*A/*B (green), 1 FY*B/*B donor (orange) and 10 FY*BES/*BES donors (red). Each individual served as its own negative control: no primary antibody, only secondary antibodies (anti-human-Fya or -Fyb). The x-fold is calculated using the gmfi of the binding of antibodies targeting the Fy epitope divided by the gmfi of the binding of the secondary antibody only.
rPvDBPII binding on FY*BES/*BES pbRBC
Given the unexpected interaction of the CA111 Fy6-specific antibody with FY*BES/*BES pbRBC, we tested in additional experiments the host-parasite functionality of the Fy protein by evaluating rPvDBPII binding to pbRBC. As expected, a high binding of rPvDBPII on FY*A/*B pbRBC was observed, while a much lower binding of rPvDBPII was also observed on FY*BES/*BES pbRBC (Figure 2A). We then exposed both Fy-positive and Fy-negative pbRBC to rPvDBPII, washed and eluted pbRBC bound proteins and then probed them with a polyclonal antibody to PvDBP as previously described46,47. This specific detection revealed a single protein band of approximately 35kDa - consistent with the detection of the rPvDBPII protein – from both FY*BES/*BES and FY*A/*B pbRBC on a Western blot (Figure 2B).
Figure 2. rPvDBPII binding on FY*BES /*BES pbRBC.
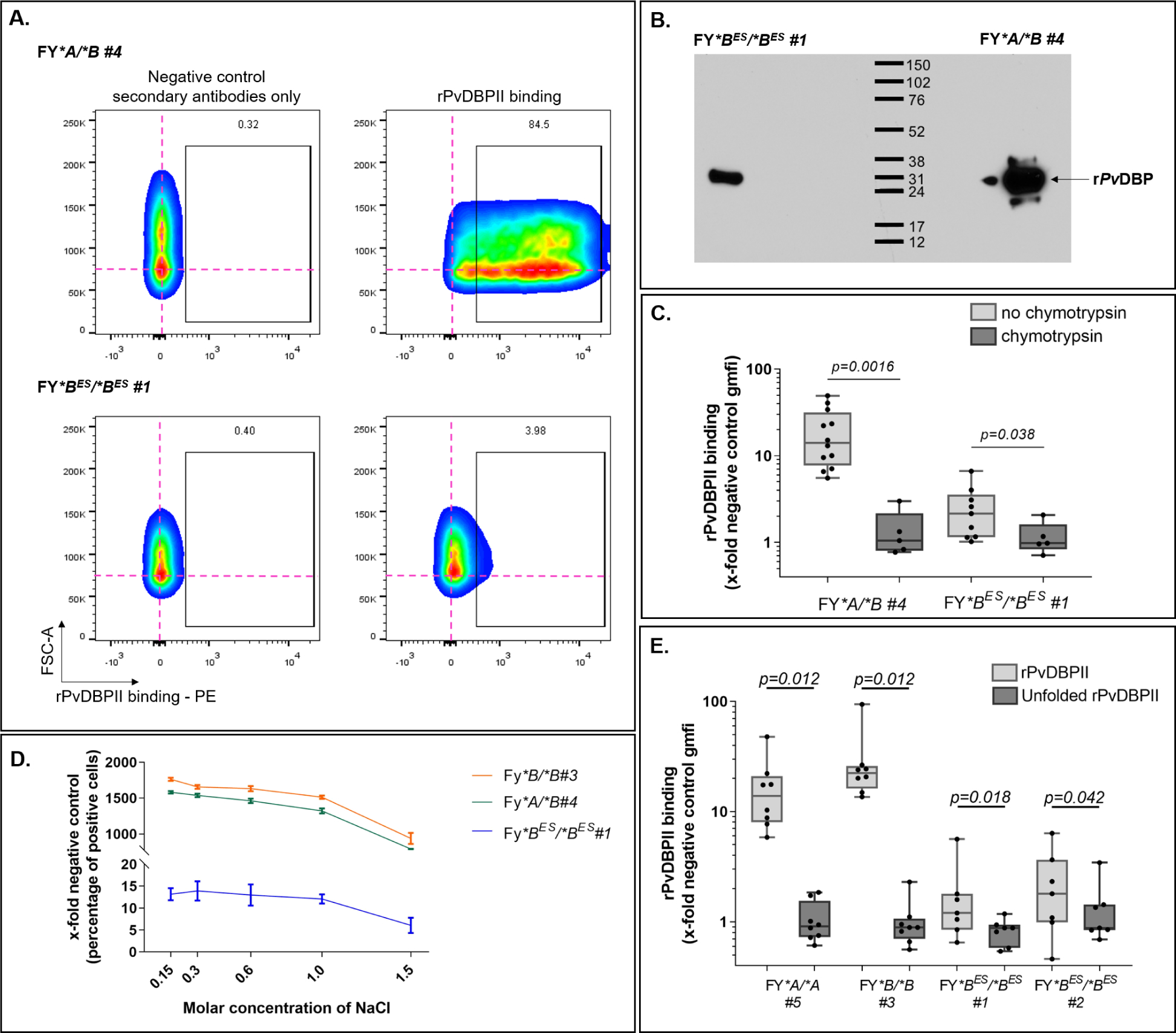
A - rPvDBPII binding on FY*A /*B and FY*BES /*BES pbRBC. The pink cross provides a point of reference for cell populations in negative controls (no rPvDBPII and secondary antibody only). The percentage of rPvDBPII binding cells over background is indicated above the gate.
B - rPvDBPII binding and dissociation to Fy-positive and Fy-negative pbRBC - pbRBC were exposed to rPvDBPII (20 μg/mL, 2 hr.; room temperature) to allow protein binding. The western blot of dissociated proteins from FY*BES/*BES (left) and FY*A/*B (right) pbRBC was probed with a polyclonal anti-PvDBP. The western blot included here is from an original intact gel and blot. This experiment was repeated 3 more times on fresh RBC from 2 FY*BES/*BES donors (data not shown). Those data were complemented with flow cytometry data.
C - Chymotrypsin binding assessment - Without chymotrypsin treatment (light gray boxes); with chymotrypsin treatment (dark gray boxes); individual data points (filled black circles); FY*A/*B - without chymotrypsin (N=12); with chymotrypsin (N=5); for FY*BES/*BES - without chymotrypsin (N=9); with chymotrypsin (N=5); evaluated by non-parametric paired Wilcoxon rank-sum test. FY*A/*B (P-value = 0.0016); FY*BES/*BES (P-value = 0.038).
D - rPvDBPII dissociation assessment of non-specific binding - No significant reduction in rPvDBPII binding was observed across increasing NaCl concentrations from 0.15M to 1.0M; addition of 1.5M NaCl caused significant reduction in rPvDBPII binding (Pearson Chi2 test, p=0.014: Pearson Chi2=6.0). Each curve represents a different donor: FY*B/*B #3 (black circle), FY*A/*B #4 (white square), FY*BES/*BES #1 (black triangle).
E - Comparative binding between folded (gray boxes) and unfolded rPvDBPII (dark boxes; same amino acid sequence) to pbRBC - Non-parametric Wilcoxon matched pairs sign rank test evaluated the median differences between rPvDBPII folded vs unfolded; n=8 for each Fy-positive and n=7 for each Fy-negative donors. FY*A/*A: p=0.012; FY*B/*B: p=0.012; FY*BES/*BES #1: p=0.018; FY*BES/*BES #2: p=0.042. Unfolded rPvDBPII interaction with pbRBC from four study participants showed no significant difference (non-parametric median test Pearson Chi2: p=0.853).
Given historically consistent demonstration of the chymotrypsin-sensitive nature of the Fy protein20,48–51, we then tested the reproducibility of rPvDBPII binding to both FY*A/*B and FY*BES/*BES pbRBC after chymotrypsin treatment. We examined the chymotrypsin sensitivity of rPvDBPII binding for one FY*A/*B and one FY*BES/*BES (Donor #1) at multiple independent times. Results consistently showed that the binding of rPvDBPII to both FY*A/*B and FY*BES/*BES pbRBC were significantly higher before than after chymotrypsin treatment (Figure 2C).
To further evaluate the relative affinity of rPvDBPII interaction to Fy-positive and Fy-negative pbRBC, we tested the dissociation of PvDBPII from pbRBC with increasing NaCl concentrations (Figure 2D). While we observed that the relative binding of rPvDBPII to FY*BES/*BES pbRBC was markedly lower than that observed for FY*B/*B and FY*A/*B, dissociation of rPvDBPII from FY*B/*B and FY*A/*B versus FY*BES/*BES pbRBC had a similar pattern across increasing NaCl concentrations (0.15M, 0.3M, 0.6M and 1M NaCl). At 1.5M NaCl the pbRBC for all FY genotypes were lysing and therefore rPvDBPII signal decreased across all samples.
In a more specific test of rPvDBPII binding to Fy-positive and Fy-negative pbRBC, we compared the binding of properly folded rPvDBPII to partially denature rPvDBPII (same amino acid construct) on pbRBC (Figure 2E). Results showed that irrespective of the Duffy genotype, denatured rPvDBPII (dark boxes) compared to properly folded (light boxes) had significantly reduced binding to Fy-positive and Fy-negative pbRBC. Taken together these results show that rPvDBPII bind specifically to Fy-positive and Fy-negative pbRBC and are consistent with CA111 anti-Fy6 binding experiments.
Variable rPvDBPII binding on pbRBC over time
To observe the persistence of the interactions between rPvDBPII and pbRBC over time, we examined multiple samples from both Fy-positive and Fy-negative study participants in a time-course study (Figure 3A). The pbRBC samples were collected and examined over 282 days for one FY*B/*B donor (orange), one FY*A/*A donor (green) and two FY*BES/*BES donors (red and blue). Among the FY*BES/*BES pbRBC sampling time points (7 samples for both FY*BES/*BES #1 (red) and FY*BES/*BES #2 (blue)), five samples showed rPvDBPII binding at least 2-fold above the negative control; both FY*BES/*BES donors showed evidence of rPvDBPII binding 5-fold above the negative control. The variability observed in rPvDBPII binding among the Fy-negative donor samples was also observed for both Fy-positive donors (regardless of their genotype). Overall, the rPvDBPII binding (mean fold-increase over negative control and range) was 32.4 for FY*B/*B (1.8 –110.9), 17.2 for FY*A/*A (4.9–47.7), 1.9 for FY*BES/*BES #1 (0.7 – 5.9), 2.4 for FY*BES/*BES #2 (0.5 – 6.7). Interestingly, across this series of samples from the four pbRBC donors featured, the higher levels of Fy expression on FY*BES/*BES pbRBC were comparable to the lower level of rPvDBPII binding for the FY*A/*A pbRBC (Figure 3A). As further evidence of Fy protein expression on the FY*BES/*BES pbRBC surface, we were able to immunoprecipitate, after solubilizing the pbRBC membranes, the Fy protein using CA111 anti-Fy6 and detected it using the commercial anti-Fy [ACKR1] antibody (Figure 3B).
Figure 3. Specific rPvDBPII binding on Fy-positive and Fy-negative pbRBC over time.
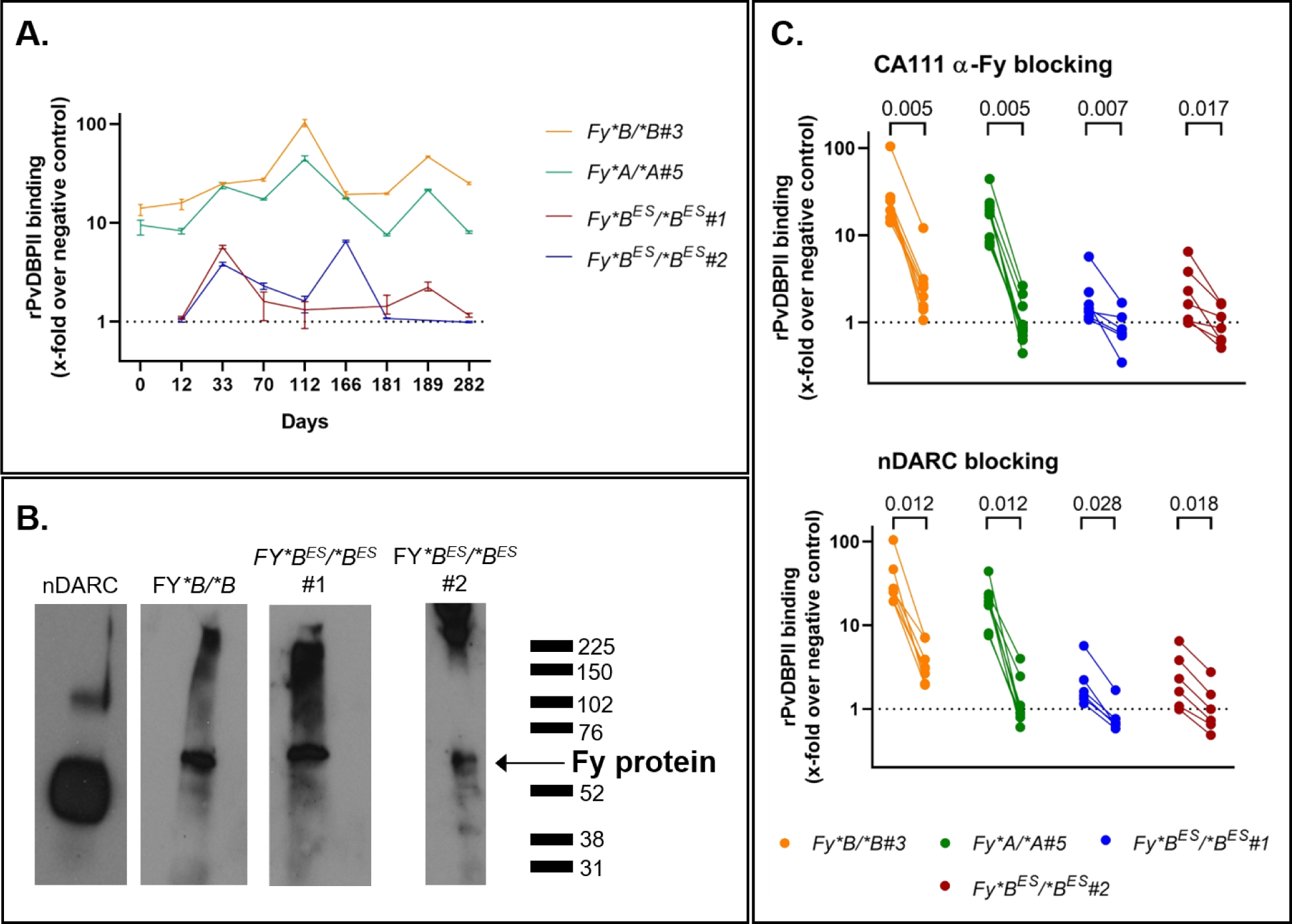
A - rPvDBPII binding on Fy-positive and Fy-negative pbRBC over time – Binding of rPvDBPII to pbRBC was followed for up to 282 days (10 months): one FY*B/*B (donor #3, orange), one FY*A/*A (donor #5, green), two FY*BES/*BES (donor #1, red; donor #2, blue); binding tested in triplicate (minimum, maximum and mean value) and represented as the x-fold increase over negative control. Those data complete the Fy-expression in the cross sectional study represented in Fig1B. Each donor served as its negative control for each time point. The same antibodies and flow cytometer (Attune NxT) were used in each experiment.
B - Specific capture and detection of the Fy protein from the surface of pbRBC.
This figure presents direct evidence of the specific capture, immunoprecipitation and detection of the Fy protein from the surface of the pbRBC of FY*B/*B and FY*BES/*BES donors. In the left panel, nDARCIg panel – the monomeric and dimeric nDARCIg species were distinguishable. Experimental panels – Fy6-specific CA111-based capture of the Fy protein from pbRBC of FY*B/*B (Donor #3; day 112), FY*BES/*BES (Donor #1; day 112) and FY*BES/*BES (Donor #2; day 189). Western blot detection of CA111-captured Fy protein was performed using polyclonal anti-ACKR1 (LS Bioscience). See SI Methods Section 7 for western blot raw data. This experiment was repeated twice on fresh RBC from 2 FY*BES/*BES donors (data not shown). Those data were complemented with flow cytometry data.
C - Blocking of the rPvDBPII binding to Fy-positive and Fy-negative pbRBC – Upper Panel – Inhibition of rPvDBPII binding with addition of the anti-Fy6-specific CA111. Lower Panel – Inhibition of rPvDBPII binding with addition of the nDARCIg. Only the experiments showing a decrease of at least two times were represented on the graphics. Experiments included in the statistical analysis: n=9 for FY*B/*B#3, n=7 for FY*A/*A#5, n=7 for FY*BES/*BES#1, n=7 for FY*BES/*BES#2 - Non-parametric Wilcoxon matched pairs sign rank test evaluated the differences of rPvDBPII binding without or with blocking for each individual. The dashed line represents equivalent binding compared to the negative controls. Each dot is the mean of 2 to 3 technical replicates. Data of rPvDBPII binding in Fig3C are also represented in Fig3A with error bars. Two connected dots refer to one experiment.
To further confirm the specificity of rPvDBPII binding to pbRBC in this time-course study, we used both CA111 and nDARCIg (a chimeric protein including Fy and IgG1-Fc region52) in competition assays to block rPvDBPII access to the RBC. The Fy-specific CA111 nanobody (Figure 3C left panels) decreased rPvDBPII binding from 25.2-fold (range, 5.8;110.9) over negative controls to 2.1-fold (0.2;12.1) for both Fy-positive donors (P-value =0.001 for FY*B/*B; P-value =0.001 for FY*A/*A). Under the same conditions, rPvDBPII binding decreased from 2.0-fold (0.5;6.4) over negative controls to 0.8-fold (0.2;1.9) for both Fy-negative donors (P-value =0.002 for FY*BES/*BES #1; P-value =0.003 for FY*BES/*BES #2). In the partner experiments (Figure 3C right panels), nDARCIg was observed to decrease rPvDBPII binding to Fy-positive pbRBC from 25.2-fold (5.8; 110.9) over negative controls to 2.81-fold (0.5;7.7) (P-value=0.005 for FY*B/*B; P-value=0.005 for FY*A/*A). For the FY*BES/*BES pbRBC, nDARC decreased rPvDBPII binding to pbRBC from 2.0-fold (0.5;6.4) over negative controls to 1.0-fold (0.3;2.8) (statistically significant P-values all <0.05 for both FY*BES/*BES samples). Thus, both CA111 and nDARCIg blocked the binding of rPvDBPII to FY*BES/*BES pbRBCs, providing evidence of Fy-specific binding of rPvDBPII to FY*BES/*BES pbRBC.
Fy protein expression on erythroid progenitor cells in vitro
Given a wide range of results pointing to P. vivax preference for infection of reticulocytes53–61 and the apparent low, variable expression of Fy protein on Fy*BES/*BES pbRBC (Figures 1A, 1B and 3A), we postulated that this expression pattern reflects the declining Fy expression on older pbRBC35. Indeed, recent evidence from murine studies showing that the Fy protein is expressed at higher levels on pro-erythroblasts and normoblasts, compared to mature RBC62. We initiated these in vitro studies by expanding CD34pos (expressed on megakaryocyte erythrocyte progenitors63–67) from the peripheral blood of one FY*B/*B donor and FY*BES/*BES Donors #1 and #263. At Days 11, 15 and 18 of differentiation, cells were collected to monitor erythroid lineage maturation using the surface markers CD36, CD71, Glycophorin A and Band 3 and query Fy protein expression (Figure 4A and Figure 4B). As expected, during the differentiation process, CD36 and CD71 were decreasing, whereas Glycophorin A and Band 3 increased among the most mature RBCs. No difference in the maturation stages from the different donors was observed over time (Figure 4A). Expression of the Fy protein during these differentiation time points was monitored by either flow cytometry (CA111 binding, Figure 4B) and/or Western blot (CA111 immunoprecipitation; anti-ACKR1 detection, Figure 4C and FigureS4). For the FY*B/*B donor, at Day11, 48.9% of the cell population expressed the Fy protein at the cell surface whereas from Day15 to Day18 >96% of the population expressed the Fy protein. From the two different FY*BES/*BES donors, the expression of the Fy protein was observed in vitro as early as Day11 of maturation. Consistent with our previous results on pbRBC, the Fy protein expression was lower on FY*BES/*BES cells (11.1 and 35.5%) than on FY*B/*B cells (48.9%). However, we observed that significantly higher proportions of the erythroid precursor cells from FY*BES/*BES donors tested positive for the Fy protein by Day18 (49.7% for FY*BES/*BES #1 and 83.8% for FY*BES/*BES #2) (Figure 4B) than was observed previously for pbRBC (highest proportion of Fy-positive pbRBC from Figure 3A - 6% for FY*BES/*BES #1 and 7% for FY*BES/*BES #2). Expression of the Fy protein on erythroid precursors differentiated in vitro at Day18 was further confirmed by immunoprecipitation and Western blot (Figure 4C).
Figure 4. Fy protein expression during in vitro CD34pos erythroid precursors differentiation.
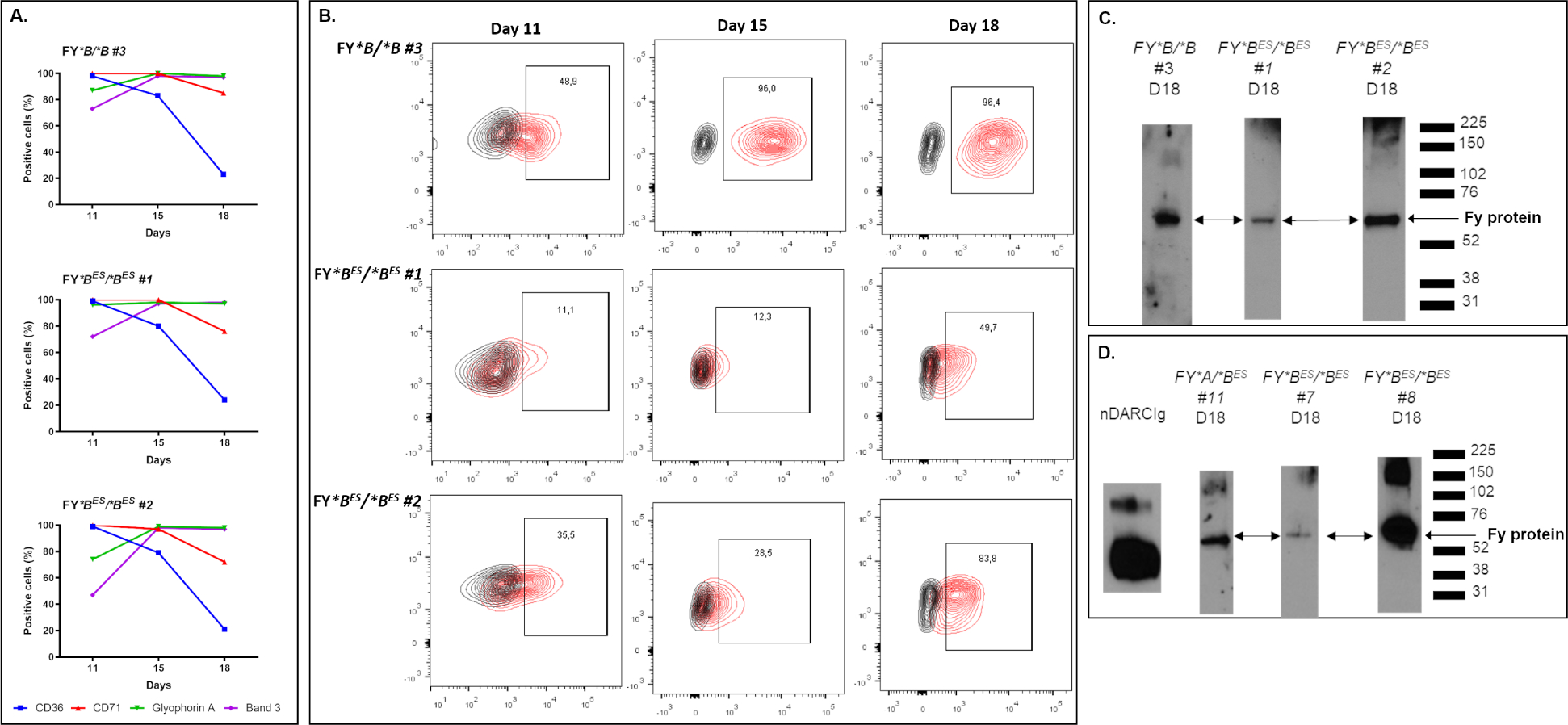
The CD34pos erythroid precursor cells were expanded and differentiated for over 21 days.
A - Monitoring of CD34 positive cells differentiation in vitro. CD34pos erythroid precursor cells were collected from the peripheral blood of 3 donors (1 Fy-positive and 2 Fy-negatives). In vitro differentiation was monitored by the presence/absence of the surface markers CD36, CD71, Glycophorin A and Band 3 on CD34pos erythroid precursor cells. As expected, during the differentiation process, the cells were losing the CD36 and CD71 signals whereas Glycophorin A and Band 3 signals were increasing. Overall, the cells from all the donors were observed to be at comparable stages over time.
B - Fy expression on differentiated CD34pos from FY*B/*B and FY*BES/*BES donors. At Day11, Day15 and Day18, the expression of Fy protein was detected by flow cytometry using CA111 anti-Fy6. For each donor, the percentage of positive cells for the binding of CA111 is annotated in the black box (population in red within the CA111-PE positive gate) in comparison to its negative control (population in black, e.g. secondary antibody only). mfi: mean of fluorescence intensity.
C, D - Expression of Fy protein in erythroid precursors isolated from peripheral blood of FY*B/*B and FY*BES/*BES donors (C) and from bone marrow of FY*A/*BES and FY*BES/*BES donors (D). Differentiated cells were harvested on Day18, membrane proteins were extracted and immunoprecipitated with CA111 anti-Fy6. The protein captured by Fy-specific CA111 were detected with a commercial polyclonal anti-ACKR1 antibody by western blot. All original western blots and re-organization of lanes to present salient results are described SI Methods Section 7. The experiments of the Fig4C were repeated at Day11 and Day18 on differentiated CD34pos from two FY*BES/*BES donors (data not shown). The experiments of the Fig4D were repeated at Day11 and Day15 on differentiated CD34pos from two FY*BES/*BES donors and Day15 on differentiated CD34pos from one other FY*BES/*BES donor (data not shown). Those data were complemented with flow cytometry data.
The same in vitro differentiation was performed on human bone marrow aspirates (HBMAs)63. From expanded CD34pos bone marrow cell populations of both Fy-positive and Fy-negative samples, we were also able to immunoprecipitate and detect the Fy protein (Figure 4D).
These studies confirm the ability of individuals with FY*BES/*BES genotype to express the Fy protein in erythroid progenitor cells cultured in vitro. Nevertheless, these results required confirmation of Fy expression in erythroid progenitor cells ex vivo.
Ex vivo Fy protein expression on bone marrow erythroid precursors.
Fy expression was evaluated ex vivo with CA111 anti-Fy6 (n=5 Fy-positive, n=2 Fy-negative) and rPvDBPII (n=2 Fy-positive, n=2 Fy-negative) binding on erythroid precursor cells from bone marrow samples (Figure 5). On the same samples, we monitored the expression of the following cell surface markers as accepted indicators of erythroid precursors: CD3463–67, CD45 (marker to differentiate erythroid from lymphoid development64), CD105 (marker of human erythrocyte progenitor (hEP)67–69), and CD71 (transferrin receptor preferentially expressed on young reticulocytes70) (Figure 5A). The cell size and expression of these markers on bone marrow subpopulations were similarly observed among all Fy phenotypes. We further assessed Fy protein expression in bone marrow erythroid precursors using CA111 and rPvDBPII recognition (Figure 5B). The highest levels of anti-Fy6 binding was observed within the CD34neg/CD45neg/CD105mid/CD71pos subpopulation (skewed toward reticulocytes; CD105 declining) for Fy-negative donors and CD34neg/CD45neg/CD105neg/CD71pos subpopulation for Fy-positive donors. Further detailed comparative analyses of bone marrow samples from individuals representing specific FY genotypes are provided in Figures S5 and S6.
Figure 5. Ex vivo expression of Fy protein among erythroid precursor sub-populations from bone marrow aspirates.
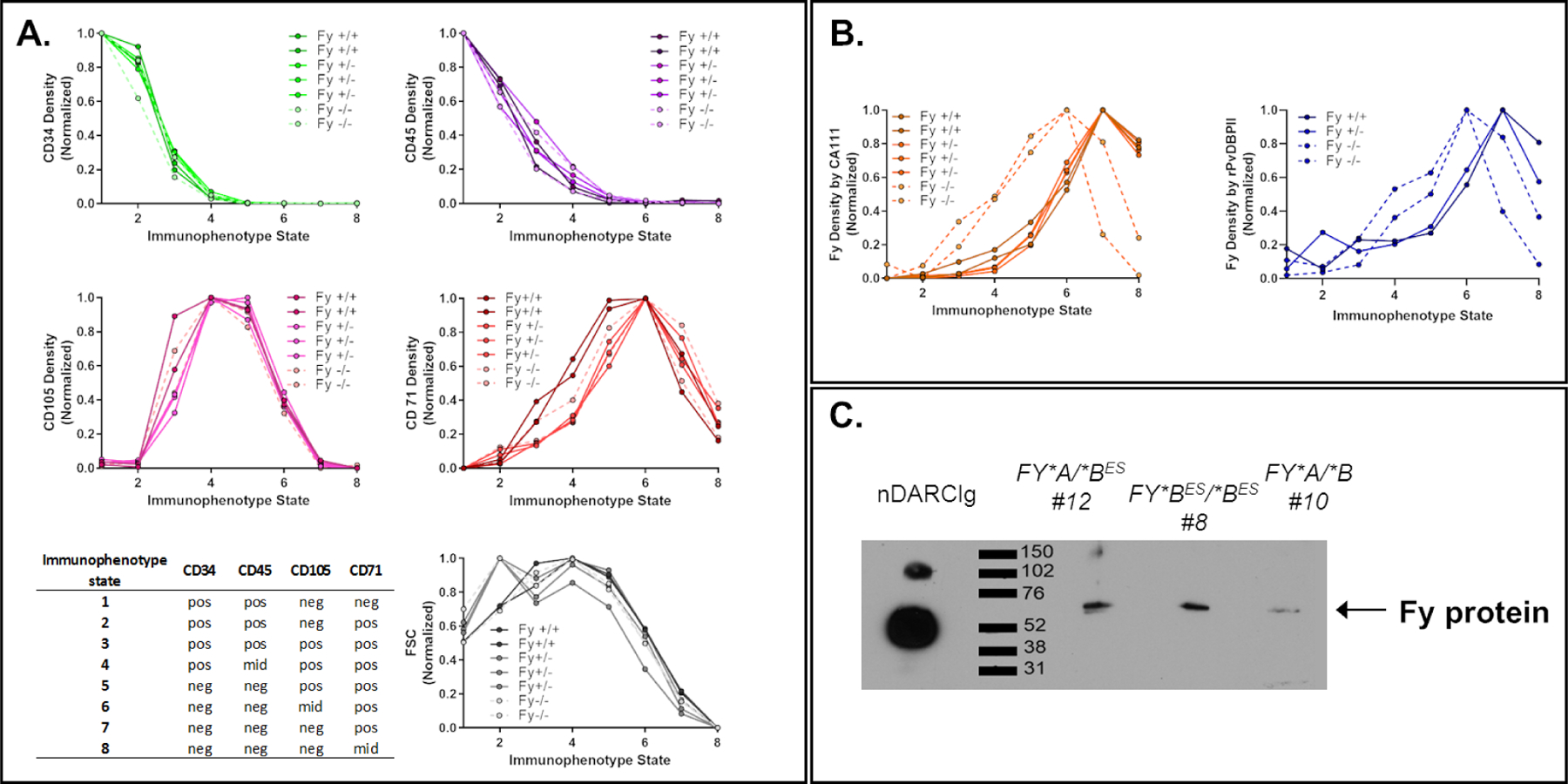
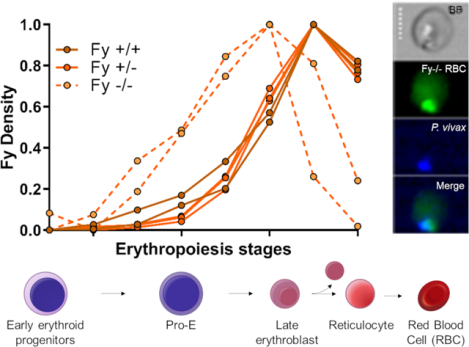
A, B - In the bone marrow, all stages of erythroid precursors are present except mature erythrocyte (that are circulating in the peripheral blood). The gating strategy is detailed in the SI and summarized in the table up left (pos: positive, mid: middle, neg: no expression). The density (mean of fluorescence intensity divided by the size of the cells) of each CD markers as well as CA111 anti-Fy6 or rPvDBPII binding to Fy protein was represented in different graphics. The Fy-negative donors were represented in dashed lines in the graphics. The profile of CD marker expression was similar between all Fy genotypes (A) whereas Fy expression appeared to be earlier during erythropoiesis in Fy negative compare to Fy positive individuals (B).
C - Expression of Fy protein in Fy*BES/*BES CD45 negative erythroid precursors from ex vivo bone marrow. CD45 negative cells (erythroid precursors in bone marrow) of three bone marrow samples were sorted by flow cytometry. The membrane proteins of the sorted cells were then immunoprecipitated with the CA111 anti-Fy6 and revealed by Western blot with a commercial polyclonal anti-ACKR1 antibody (arrow). Fy-positive and Fy-negative showed a signal for the Fy protein in erythroid precursors. The western blot included here is from an original intact gel and blot. See SI Methods Section 7 for western blot raw data. This experiment was complemented with flow cytometry data.
Finally, we confirmed expression of Fy protein by CA111-capture and commercial anti-ACKR1-detection from both Fy-negative and Fy-positive donors in the erythroid precursor bone marrow population (Figure 5C; Figure S7).
P. vivax in vitro invasion into Fy-positive and Fy-negative erythroid cells.
With the Fy protein detected by Fy-specific antibodies and rPvDBPII on genotypically Fy-negative erythroid cells, we determined whether cryopreserved P. vivax field isolates (asynchronous) would interact with and/or invade host cells. Short-term invasion assays used peripheral blood of North American Fy-positive and Fy-negative participants as target cells seeded with stock P. vivax cells. In order to distinguish between P. vivax re-infection of stock culture cells and new invasion into target cells, we labeled the target cell populations with CFSE (carboxyfluorescein succinimidyl ester). Imagestream analyses were performed after 72 hours of culture. Overall assessment of the RBC invasion experiments by imaging flow cytometry provides an overview for selecting populations of cells down to assessments of individual parasite-target cell interactions (Figure 6, Figure S8, S9, S10 and S11, Table S2). Figure 6A shows separation of single cells (black data points) versus multiple (gray data points) cells by aspect ratio and area in the brightfield image. Results in Figure 6B show that strategic CSFE labeling of target RBCs in a control cell population without exposure to parasites serving as a background control. Results from the invasion assays show newly infected Fy-positive and Fy-negative target cells (CFSE+ and Hoechst+) in Figures 6C and 6D, respectively. To confirm P. vivax infected target cells, we examined individual cells of interest (circled cells) with bright field and fluorescent images (Figure 6C and 6D), with Fy-positive target cells (Figures 6E1–6 correspond to circled/numbered cells in Figure 6C) and Fy-negative target cells (Figures 6F1–6 correspond to circled/numbered cells in Figure 6D). These results show P. vivax merozoites interacting with and invading Fy-positive (Figures 6E1 to 6E4) and Fy-negative (Figures 6F1 to 6F4) target cells. Figures 6E5 & 6F5 are uninfected donor cells (stock culture) and Figures 6E6 & 6F6 are infected donor cells (stock culture). Our experiments also show that cells from FY*B/*B compared to FY*BES/*BES donors are more readily invaded by P. vivax by a factor of 2.2 (Table S2).
Figure 6. In vitro invasion of pbRBCs from Fy-positive and Fy-negative North Americans by Malagasy P. vivax isolate.
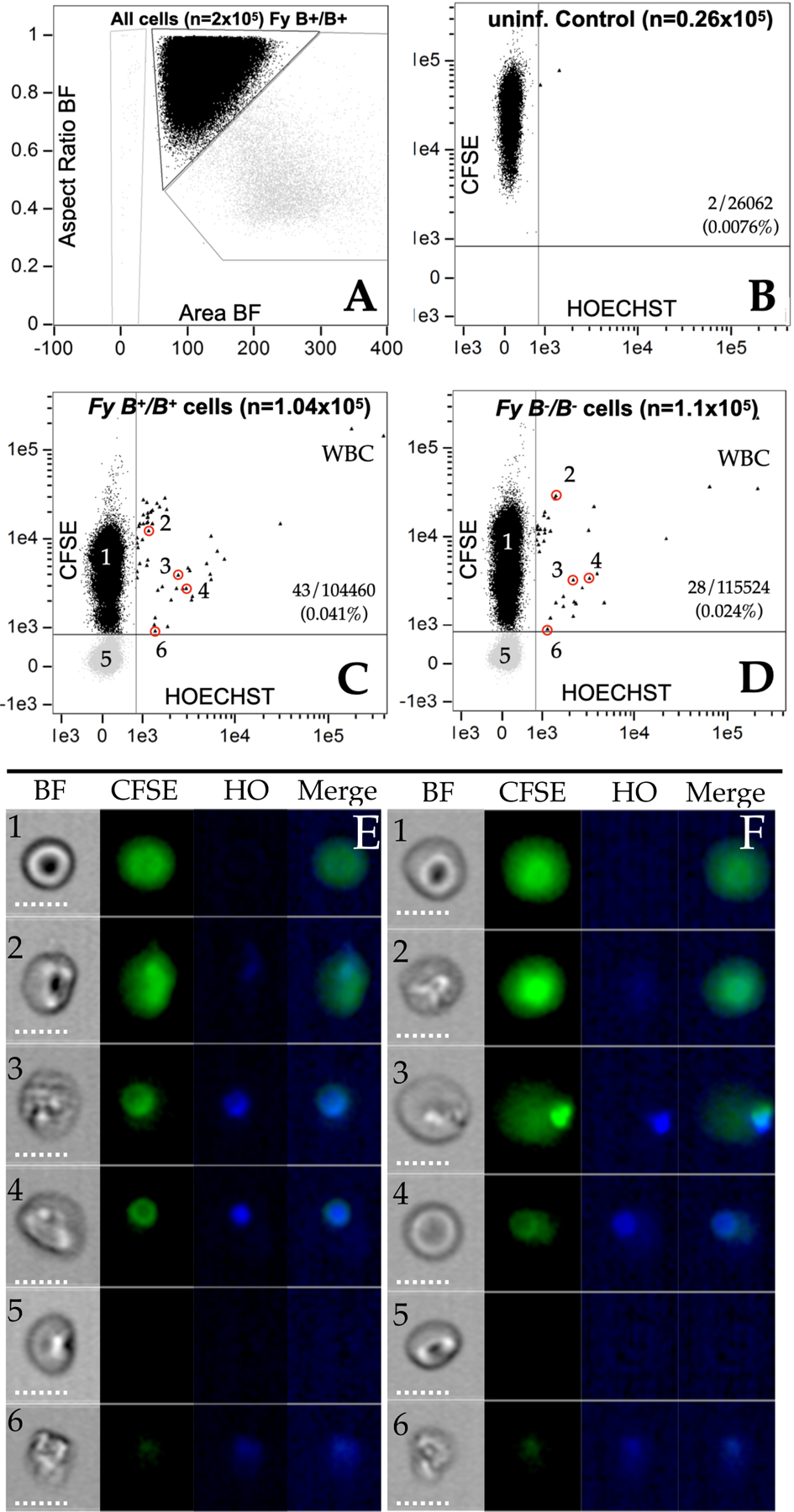
6A. single cells (black data points) versus multiple (gray data points) cells by aspect ratio and area in the brightfield image; 6B; Control image using uninfected cells previously stained with CFSE. Y-axis CFSE; X-axis Hoechst; 6C. AMP2016.14 (CFSE[-]) mixed with Fy-positive, FY*B/*B target cells (CFSE[+]); 6D. AMP2016.14 (CFSE[-]) mixed with Fy-negative, FY*BES/*BES target cells (CFSE[+]); 8E1- E6. Individual cells identified in Panel 6C from AMP2016.14 added to FY*B/*B (Fy-positive). 8F1- F6. Individual cells identified in Panel 6D from AMP2016.14 added to FY*BES/*BES (Fy-negative). Additional data and plots are provided in the SI (Figure S8, Table S2, Figure S9, Figure S10, Figure S11).
Discussion
Given our findings on P. vivax infection of Fy-negative people from Madagascar71,72 and reports from other malarious sites across Africa31,32,73, we wanted to investigate how parasite and host invasion proteins interacted with Fy-negative RBCs. We therefore assessed whether or not two different anti-Fy6 antibodies and rPvDBPII bind to the surface of different erythroid target cells (pbRBC; expanded CD34 cell populations; bone marrow). Our data show that Fy is expressed at the surface of pbRBC and this expression is variable in both Fy-negative and Fy-positive individuals. Using competitive inhibition studies, we demonstrated that adding of CA111 or nDARCIg significantly reduced rPvDBPII binding to Fy-positive pbRBC and essentially blocked rPvDBPII binding to Fy-negative pbRBC. These results suggest that rPvDBPII binding is focused on the anti-Fy6 epitope and thus limited to the Fy protein.
Our initial studies on pbRBC, showed that Fy expression on Fy-negative pbRBC was positive at some sample collection time points and that these pbRBC could interact with rPvDBPII at levels similar to RBC from FY*A/*A people. Because of P. vivax preference for infection of reticulocytes33,53–60, we studied Fy expression across a broader time frame of erythroid development. Human and murine studies show higher levels of Fy protein expression on pro-erythroblasts, normoblasts and reticulocytes compared to mature RBC35,58,62. Additionally, Malleret et al. have demonstrated that immature reticulocytes (CD71pos), mostly found in the bone marrow and matured in the spleen59 are preferentially invaded by P. vivax compared to older reticulocytes and erythrocytes (CD71neg), than in the peripheral blood. Recent studies emphasize that the bone marrow and especially the spleen, the latest accounting for more than 98% of the estimated total-body P. vivax biomass, are important reservoirs of P. vivax infection in humans and non-human primate model systems74–76. Since the spleen harbor more mature reticulocytes than in the bone marrow, the distribution of the P. vivax biomass is inconsistent with the relative paucity of invasion-receptive reticulocytes in the spleen. One possibility could be that upon P. vivax infection, the erythropoiesis within the bone marrow is impaired, resulting in extramedullary hematopoiesis (EMH)77 induction within the spleen and then allowing the expansion of parasite biomass beyond the bone marrow. With this focus on the importance of reticulocytes as P. vivax invasion-targets, interactions between reticulocyte binding proteins and CD71 and CD98 heavy chain suggest additional molecular components of the P. vivax erythrocyte invasion mechanism78.
Our studies on earlier erythroid stages focused on enriching CD34pos megakaryocyte erythrocyte progenitors that circulate in the peripheral blood, allowing them to mature in vitro under conditions favoring erythroid development63. We purposefully performed a time-course study on the same Fy-negative donors to evaluate the variability of erythroid precursors released in the peripheral circulation over time. Interestingly, results showed that significantly higher proportions of the erythroid precursor cells from FY*BES/*BES donors were positive for Fy protein detection than their pbRBC.
With the observation of Fy protein expression from the CD34 cell maturation experiments, we repeated our studies on bone marrow aspirate samples as the site for natural erythroid cell development and also an important reservoir of P. vivax. We monitored the expression of CD34, CD45, CD105 and CD71 expression as markers distinguishing erythroid from lymphoid maturation. Results from these bone marrow studies showed Fy protein expression on early-stage cells committing to erythroid development in both Fy-positive and Fy-negative people. Our observations suggest that Fy expression initiates in an early erythroid subpopulation (CD34pos/CD45pos/CD105mid/CD71neg) in both Fy-positive and Fy-negative people. Because Fy expression drops for Fy-negative individuals whereas the expression increased for Fy-positive individuals, we speculate that, before proerythroblast differentiation and transition to GATA-168,79, Fy expression may be driven by another transcription factor, such as GATA-2 and this may contribute to the results we have observed. Consistent with Tournamille et al. findings (i.e., compromised FY gene expression associated with the GATA-1 FYES SNP)27, the amount of Fy protein begins to decay earlier in Fy-negative than in Fy-positive erythroid cells. The observed decline of Fy signal in CD34neg/CD45neg/CD105mid/CD71pos cells, appears to be consistent with RBC extrusion of its nucleus, after which RBC precursors are no longer able to express their genes and the cell can only be equipped with the gene products expressed prior this important developmental event.
The potential that bone marrow could be the source of reticulocytes for in vitro cell culture is becoming an increasingly important area of P. vivax research. Fernandez-Becerra et al. have expanded CD34pos hematopoietic stem cells to generate CD71pos reticulocytes for in vitro P. vivax culture80. We followed in this direction by using human bone marrow aspirates from US donors, to provide target cells for in vitro propagation of P. vivax field isolates from the Madagascar communities where we have previously observed significant frequency of Fy-negative P. vivax infections71,72. Human bone marrow aspirates have enabled in vitro propagation of two field isolates as source parasite material for the short-term invasion assays presented here. Using these in vitro propagated P. vivax, we have shown that both Fy-positive and Fy-negative RBCs can be invaded by P. vivax.
Limitations
Although our work provides a clear demonstration that the dogma about Fy-negative individuals no longer holds and represents significant advances for understanding P. vivax malaria, we recognize limitations that future studies should address. While we have shown that P. vivax can invade Fy-negative cells in vitro, it was not possible to demonstrate that RBC invasion was Fy protein dependent and completely rule out the possibility that P. vivax can use an alternative invasion pathway. With the development of monoclonal antibodies and peptide-based inhibitors, interrupting interactions between PvDBP, PvEBP2, PvRBP and other P. vivax merozoite proteins, more specific invasion inhibition studies are warranted. For this to occur, it will be necessary to refine P. vivax in vitro invasion studies to allow more extensive studies. The demonstration of P. vivax invasion of Fy-negative cells in vitro opens potential that laboratory-adapted P. vivax strains could be used to study a range of candidate protein-protein interactions necessary for blood stage infection. Additionally, while the focus of our experiments provided assessment of Fy expression from multiple different sample types (pbRBC; expanded CD34 cell populations; bone marrow aspirates), we were not able to examine Fy-expression from splenic reticulocytes of Fy-negative and Fy-positive individuals. Emerging studies that have probed P. vivax infections outside of the peripheral blood are critically important to advance current perspectives on vivax malaria pathogenesis and evaluate Fy genotype variation in future studies of this nature.
Conclusions
Our results provide a mechanism to explain why Fy-negative people are not completely resistant to P. vivax blood-stage malaria3,4 by showing that Duffy antigen can be variably expressed in subjects with the GATA1 polymorphism associated with Fy negativity. With the peak of Fy expression during early reticulocyte stages and declining as RBCs enter peripheral circulation, our findings support observations that the bone marrow and sites of extramedullary erythropoiesis are important for P. vivax blood stage propagation. Our results are consistent with observations that P. vivax infections in Fy-negative individuals are usually detected by PCR, not blood smear microscopy. P. vivax infection in Duffy negative individuals expands human populations potentially capable of P. vivax transmission. Our results also raise questions regarding P. vivax malaria pathogenesis, especially given a wide range of clinical observations showing bone marrow involvement, from in vitro studies78,81, individual case histories82–84 and cohort studies85–87. With potential significant involvement of the bone marrow, it becomes easier to understand how considerable anemia and other markers of hematological disruption are out of proportion with low levels of observed P. vivax peripheral blood parasitemia (summarized in Baird33 and Silva-Filho et al.88). Given evidence that peripheral blood infections significantly underestimate the total body biomass of a P. vivax infection and therefore global prevalence, closer examination of P. vivax infection and disease biomarkers (lactate dehydrogenase and other circulating parasite antigens)88,89 is required to more accurately estimate the burden of vivax malaria in Fy-negative Africa and globally.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Peter A. Zimmerman (paz@case.edu).
Materials availability
Proteins generated in this study will be shared by the lead contact upon request if necessary with a Material Transfer Agreement.
Data availability
Any additional information required to reanalyze the data reported in this paper will be available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Study Participants and Sample Collection
Study protocols were approved by the University Hospitals of Cleveland Institutional Review Board (#08-03-33 and #09-90-195). All methods were carried out in accordance with guidelines and regulations included within our protocols. Informed consent was obtained from all subjects, and for subjects who were under 18, from a parent and/or legal guardian. Eleven Fy-positive (three females and eight males) and sixteen Fy-negative persons (nine females and seven males) were included (Table S1); all Fy-negative persons were homozygous for the negative GATA-1 mutation (rs2814778; FY*BES/*BES). All blood and bone marrow samples were processed within 2 hours of collection.
METHOD DETAILS
DNA Extraction and PCR-based Genotyping
All study participants were genotyped by previously described methods71. Genomic DNA was extracted from the whole blood samples using the QIAGEN QIAmp DNA Blood Kit following recommended protocols with a starting blood volume of 200 μL and an elution volume of 200 μL of Buffer AE (Valencia, CA).
PCR amplifications of Duffy blood group gene sequences were performed in reaction mixtures (28μL) with 3 μL of genomic DNA, 180 μM each dNTP, 67mM Tris-HCl (pH 8.8), 6.7 mMMgSO4, 16.6mM(NH4)2SO4, 10mM 2-mercaptoethanol, 0.1 μM each primer (Primers inclusive of promoter (rs2814778) and FY*A/FY*B (rs12075) snps - forward primer, Duffy-200up 5′-CAGGCAGTGGGCGTGGG-3′; reverse primer, Duffy +730dn 5′-CTGCTAGCTAGGATACCCAG-3′; Primers inclusive of FY*B/FY*X (rs34599082) snp - forward primer, FYBXup 5′-AGCACTGTCCTCTTCATGCTTT-3′; reverse primer, FYBXdn 5′-GCAGAGCTGCGAGTGCTAC-3′) and 2.5 units of thermostable DNA polymerase under the following conditions: 95 °C for 2min, followed by 40 cycles of 95 °C (30 s), 60 °C (30 s), and 72°C (90 s) and a final extension at 72 °C (5 min) (PCR products 912 and 1,033 bp). Following PCR amplification, products were further processed by a ligation detection reaction (LDR). The LDR was performed in a reaction mixture (15 μL) containing 20mMTris-HCl buffer (pH 7.6), 25 mM potassium acetate, 10mM magnesium acetate, 1mM NAD+, 10mM DTT, 0.1% Triton X- 100, 13nM each LDR probe,1 μL of PCR product, and 2 units of Taq DNA ligase (New England BioLabs). LDR probes consisted of six allele-specific oligonucleotides and three fluorescently labeled conserved-sequence oligonucleotides. The allele-specific probes contained an MTAG sequence for further hybridization with complementary sequence oligonucleotides bound to Luminex FlexMAP fluorescent microspheres; conserved-sequence probes were 5′ phosphorylated and 3′ biotinylated.
Sequences of the LDR probes used were as follows:
Duffy null promoter snp (rs2814778) at nucleotide −67 (t, wild-type; c, erythrocyte silent): PRO ntT MTAG_A018: 5′-ACACTTATCTTTCAATTCAATTACcattagtccttggctcttat-3′ PRO ntC MTAG_A020: 5′- CTTTCTCATACTTTCAACTAATTTtcattagtccttggctcttac-3′ PRO common: 5′-phosphate-cttggaagcacaggcgctg-biotin-3′.
Codon 42 snp (rs12075) associated with Fya and Fyb phenotypes (g, Fya; a, Fyb): FY*A ntG MTAG-A022: 5′- CAAACAAACATTCAAATATCAATCttcccagatggagactatgg-3′ FY*B ntA MTAG-A026: 5′- TACATTCAACACTCTTAAATCAAActtcccagatggagactatga-3′ Codon42 common: 5′-phosphate-tgccaacctggaagca-biotin-3′.
Codon 89 snp (rs34599082) associated with Fyb or the Fybweak phenotypes (c, Fyb; t, Fybweak): FY*B ntC MTAG-A028: 5′- CACTTAATTCATTCTAAATCTATCtgcttttcagacctctctcc-3′ FY*X ntT MTAG-A034: 5′- ACTTATTTCTTCACTACTATATCAtgcttttcagacctctctct-3′ Codon89 common: 5′-phosphate-gctggcagctctgccctggct-biotin-3′.
Protein expression
CA111 anti-Fy nanobody
Variable domain of a heavy chain only (VHH), nanobody of camelids. This nanobody includes the CA52 VHH sequence44 and an HA (human influenza virus hemagglutinin)-tag. This nanobody was kindly provided by Olivier Bertrand (National Institute of Blood Transfusion (INTS), Paris, France)44. CA111 was obtained by subcloning the CA52 VHH sequence into a Novagen pET 28-b derived plasmid allowing expression in the cytoplasm of SHuffle (C3029H) cells. This plasmid encodes (from 5’ to 3’) a polyhistidine tail, a thrombin cleavage site, the VHH, and an HA-tag; it is used for routine subcloning of other VHHs using the PstI and Eco91I sites90.
The resulting nanobody exhibits the following characteristics.
Interferes with the interleukin-8 binding to Fy on RBCs.
Interferes with P. vivax infection of red blood cells (short term culture).
Upon linkage to a solid substrate, this nanobody acts as a powerful adsorbent to purify native Fy in a single step from a detergent extract of cells.
Pepscan analysis identified 22FEDVW26 as the peptide sequence of the linear epitope.
A single dromedary was immunized with the N-terminal extracellular domain constructs of Fy expressed in E. coli (referred to as ECD1; Amino acids 6 [H] to 63 [P] – HRAELSPSTENSS QLDFEDVWNSSYGVNDSFPDGDY[G/D]ANLEAAAPCHSCNLLDDSALP)44. This domain covers the region to which Fy6 epitope, chemokines and the P. vivax Duffy binding protein (PvDBP) bind (amino acids 19 to 25 – 19QLDFEDV25) and carries the polymorphic G/D site at amino acid 42, responsible for the Fya/Fyb allotypes. From the immunized animal, a VHH library of dromedary lymphocytes was exposed to ECD1 to yield several Fy-specific VHHs. Smolarek et al. focused on one VHH, referred to as CA52. The linear epitope of CA52 exhibits capacity to recognize the glycosylated Fy protein present on human cells, although the immunogen was a non-glycosylated polypeptide.
To further validate the specificity of the CA111 antibody, we assessed its reactivity on K562 cells expressing or not the Fy antigen and Saimiri RBC91. Blood samples (2 mL) from Bolivian squirrel monkeys (S. boliviensis), collected in EDTA vacutainers, was obtained from the Biologics Production Program, Michael E. Keeling Center for Comparative Medicine and Research, The University of Texas MD Anderson Cancer Center (Protocol number: 00000451-RN01-AR001). The blood was shipped on ice overnight and used in our studies on the day of arrival.
rPvDBPII recombinant protein
Region II of Salvador I strain PvDBP (rPvDBPII; 37.6 KDa) was expressed as inclusion bodies in Escherichia coli and then refolded and purified as previously described21–23. The protein purified before refolding served as a negative control.
Duffy Antigen Receptor for Chemokine chimeric protein construct (nDARCIg Fyb)
Briefly, as previously described52, HEK293H cells were co-transfected with pCDM8-DARC-Fc (first N-terminal 60 amino acids of Fy) and pRc/CMV-TPST (human sulfotransferase).
Fy detection by flow cytometry
Erythrocyte binding assays were performed based on previously developed strategies and reagents46,47. Peripheral blood RBC (1×106) from human or Saimiri) or K562 cells92 were first blocked for 30 min with PBS 1% BSA. Anti-Fy antibodies were then incubated for 20 min at 37°C at the following concentrations: CA111 (30 μg/mL), murine monoclonal antibody, 2C3 (10 μg/mL)42,43, anti-FyA (ref: 808 186 BioRad, 1:10 dilution) or anti-FyB (ref: 808 191 BioRad, 1:10 dilution) antibodies. A mouse anti-HA antibody (Biolegend; 30 μL of a 1:1000 dilution in PBS 0.2% BSA; 20 min at 37°C) followed by a goat-anti-mouse PE-conjugated antibody (eBioscience; 40 μl of a 1:80 dilution in PBS 0.2% BSA; 20 min at 37°C in the dark) for CA111, an anti-mouse for 2C3 or an anti-human for anti-Fy antibodies were used to detect the binding. Following 2 washes in PBS 0.2% BSA, a total of 100,000 cells (all samples in triplicate) were analyzed by flow cytometry. Between each staining step, pbRBC were washed two times with PBS 0.2% BSA. For ex vivo fresh bone marrow samples, a direct anti-HA tag phycoerythrin (PE)- tagged antibody was used to reduce the background (Miltenyi, dilution 1:11). Similar procedures were followed for other Fy-specific antibody reagents. For the experiments on the maturation of CD34+ hematopoietic stem cells, the same protocol was performed and at least 50,000 cells were analyzed by flow cytometry. In evaluation of bone marrow precursors, the BSA concentration change from 1% to 2% and at least 500,000 cells were evaluated to ensure sufficient data capture for erythroid precursor subpopulations of interest.
rPvDBPII binding monitored by flow cytometry
Peripheral blood RBC (1×106) were first blocked for 30 min with PBS 1% BSA and then further incubated with rPvDBPII Sal1 (20μg/mL final concentration; 60 μL of a 1:20 dilution in PBS 0.2% BSA; 30 min at 37°C). After 2X washes with PBS 0.2%BSA, these cells were incubated with a rabbit polyclonal anti-PvDBP serum (30 μL of a 1:50 dilution in PBS 0.2% BSA; 20 min at 37°C). Cells were washed again 2X with PBS 0.2%BSA and then incubated with rabbit phycoerythrin (PE)-tagged antibody (Sigma; 40 μL of a 1:32 dilution in PBS 0.2% BSA; before 20 min at 37°C in the dark). After 2 washes in PBS 0.2% BSA and a final wash in PBS, cells were subjected to analytical flow cytometry46,47. Erythrocyte binding assays were further optimized by adjusting the blocking concentrations for BSA.
Unfolded rPvDBPII was used as the same concentration as folded rPvDBPII and the protocol is identical.
Erythrocytes were pre-treated with chymotrypsin (1 mg/mL) 30 min at 37 °C. The enzyme treatment was stopped by washing five times with PBS 0.2% BSA. A blocking step (1% BSA for an hour at 37°C) were performed before rPvDBPII binding.
For the rPvDBPII dissociation assessment, the rPvDBPII binding protocol as described above was performed and increasing NaCl concentrations from 0.15M to 1.0M were added 750 μL of each gradually. The measures were acquired on Attune NTX.
Competition assays
Blocking of rPvDBPII binding:
the competition assay was performed using pre-incubated erythrocytes with CA111 (30 μg/mL) or nDARCIg (60 μg/mL) for an hour at 37°C before rPvDBII binding following the same steps as described above.
Blocking of CA111 binding:
the competition between CA111 and 2C3 was performed on fresh FY*B/*B pbRBC, FY*BES/*BES pbRBC or enriched reticulocytes from FY*BES/*BES donor. The enrichment protocol was described elsewhere93. Briefly, after leucocyte depletion using NWF syringe filter, cells were resuspended in 3 ml high-KCl buffer (pH 7.4) and then incubated 20 minutes at room temperature on a rocker. After centrifugation, cells were resuspended in 500 μL of high-KCl buffer and layered over 300 ul 19% Nycodenz-KCl solution in a 1.1 mL microtube. Tube was centrifuged at 3,000g for 30 min without braking. Reticulocytes at the interface were harvested and washed 2 times in PBS. After a blocking step, cells were pre-incubated with increasing concentrations of CA111 (molar ratio from 0 to 25) an hour at 37°C. 2C3 was then directly added (no wash) at 5 μg/mL. The detection of the 2C3 binding was following the same steps as described above.
rPvDBPII Erythrocyte Capture Assay
This assay was previously described85. Briefly, ~106 cells were collected from packed RBC and 10 μg of recombinant PvDBPII was added. After 2 hours of incubation at room temperature, the preparation was layered on top of 500 μL of dibutylphtalate and centrifuged at 11,000 rpm for 30 sec. The pellet was collected, and the proteins bound at the RBC surface were eluted adding 20 μL of 1.5 M NaCl drop wise and shaking. After incubation (5 min), this procedure was repeated with 1.0 M of NaCl and then 0.3 M of NaCl. The preparation was centrifuged at 14,000 rpm for 2 min. The proteins present in the supernatant were separated on a Tris-Glycine SDS 4–20% gel (Biorad) in reducing conditions and then transferred onto a PVDF membrane. The membrane was blocked for one hour with TBS 0.1% tween, 5% milk, incubated with rabbit anti-PvDBPII (1: 2,000), washed, then followed by the secondary antibody anti-rabbit HRP (Thermo Scientific). Chemiluminescent signal was detected using SuperSignal (Pierce) on X-ray film.
Fy immunoprecipitation by CA111 and Western Blot detection
Solubilization and protein extraction –
Packed RBC (~5 mL); cells matured during the in vitro erythroid differentiation; and CD45 negative sorted-cells from the bone marrow were lysed by three cycles of freeze/thaw followed by sonication. After adding 5 mM Tris·HCl, pH 8.0 with protease inhibitor (Sigma Aldrich), the protein preparation was centrifuged 20 min at 16,000 × g at 4°C. The pellet was resuspended twice in 0.1 M Na2CO3 pH 11.5 with protease inhibitor and centrifuged 20 min at 16,000 × g at 4°C. To solubilize the membrane proteins from the pellet, 1mM triton X100 + protease inhibitors were added and incubated at room temperature for 30 min.
Immunoprecipitation of Fy antigen -
CA111 was covalently coupled to magnetic beads (Dynabeads M-270 Carboxylic Acid, Invitrogen) for 30 min at room temperature with slow rotation. EDC at 100mg/ml (1-ethyl-3-[3-dimethylaminopropyl] carbodiimide) was dissolved 100mM MES (2-(N-morpholino) ethanesulfonic acid)) buffer (pH 5.0) for the activation reaction and incubated for 2 hr at 4°C with slow rotation. The solubilized extract was incubated with the activated beads for 1h. Beads were resuspended in Laemmli buffer for SDS gel and western blot.
Western blot -
The beads + extract solutions were separated on a Tris-Glycine SDS 4–20% gel (Biorad) in reducing conditions and were then transferred onto a PVDF membrane. The membrane was blocked for one hour with TBS 0.1% Tween 5% milk. Polyclonal rabbit anti-ACKR1 (2ug/ml; LS Bioscience) and HRP anti-rabbit antibody (diluted 1/10,000 into 10 ml TBS 0.1% Tween 5% milk; Thermo Scientific) were used to probe Fy from the immunoprecipitated elution. The signal was detected on X-ray films using SuperSignal (Pierce).
Specificity of CA111 -
Tests of specificity were performed on recombinant proteins by direct Western blotting, or immunoprecipitating cells from patient bone marrow and subsequent Western Blot. For the direct probe of the Western Blot, 0.1 μg of nDARCIg and rCXCR2 were loaded on a gel; 0.2 μg/mL of CA111 were used to probe the Western blot and reveal the signal. For the immunoprecipitation experiment, 0.5 μg of rCXCR2 were used. The anti-CXCR2 antibody (R&D system, 0.5 μg/mL) was used to probe recCXCR2 on a Western Blot as a positive control.
Erythroid differentiation of CD34 positive cells
Isolation of CD34 positive cells was performed from peripheral blood donors (FY*B/*B and FY*BES/*BES donors; #1 and #2) and from bone marrow samples. Mononuclear cells were isolated by Ficoll Plaque Plus (GE Healthcare) and frozen in 10% DMSO. The thawed cells were positively selected for CD34 by magnetic beads (human CD34 MicroBead Kit; Miltenyi Biotech). Between 10,000 and 100,000 cells were obtained after selection and differentiated in culture over 21 days. A detailed protocol is published by Jingping Hu et al.63. The in vitro differentiation was monitored by flow cytometry with 400,000 cells per day (Day 11, Day 15, Day 18) monitoring surface markers CD36 (BD Biosciences), CD71 (BD Biosciences), Band 3 (American Research Product) and Glycophorin A (Life Technology).
Monitoring maturation and expansion of CD34+ Hematopoietic Stem Cells (HSCs).
Mononuclear cells were isolated by Ficoll Paque Plus (GE Healthcare) and frozen in 10%DMSO. The thawed cells were positively selected for CD34 by magnetic beads (human CD34 MicroBead Kit; Miltenyi Biotech). Between 10,000 and 100,000 cells were obtained after selection and differentiated in culture over 21 days. A detailed protocol is published by Jingping Hu et al63. For the FY*B/*B donor and 2 FY*BES/*BES donors, the cells were counted by light microscopy during the in vitro differentiation at different time points (Day 0, 11, 15 and 18).
The in vitro differentiation was monitored by flow cytometry with 400,000 cells per day (Day 11, Day 15, Day 18) monitoring the presence/absence of the surface markers CD36 (BD Biosciences), CD71 (BD Biosciences), Band 3 (American Research Product) and Glycophorin A (Life Technology). As expected, during the differentiation process, the cells are losing the CD36 and CD71 expressions whereas Glycophorin A and Band 3 are increasing. Overall, the cells from all the donors were observed to be at comparable stages over time.
Bone marrow cell preparation and staining
Fresh bone marrow was collected in heparin. Erythroid precursors from fresh bone marrow samples were isolated by Ficoll Paque Plus (GE Healthcare) separation. Cells were first blocked for 30 min with FcR blocking reagent (Miltenyi – 20 μL per sample) following manufacturer’s recommendations. rPvDBPII and CA111 binding experiments were performed as described above. Sub-populations of erythroid precursors were defined using directly-labeled antibodies - anti-CD45 PerCP Cy5.5 (Becton Dickinson), anti- CD34 FITC, anti-CD105 APC, anti-CD71 APC Cy7 (all from Biolegend) and an anti-Band3 PE (American Research Product). Data were acquired with a BD Biosciences LSR II (San Jose, CA) or a Thermo Fisher Attune Nxt (Waltham, MA) operated within manufacturer’s specifications, which was tested with performance beads. To ensure sufficient relevant data, we aimed to acquire 500,000 cell events. Data were analyzed with FlowJo 10.0 (Becton Dickison & Company (BD), Ashland, OR) or WinList 9.0 (Verity Software House, Topsham, ME). Two types of analysis were performed.
Flow cytometric analysis of erythroid progenitor subpopulations in bone marrow
Forward scatter (FSC-A) and side scatter (SSC-A) plots were used to select erythroid populations then standard gating was done to select positive and negative population from orthogonal quadrants on bivariate plots or univariate histograms. For each of the samples analyzed, unstained cells were used to evaluate cell auto-fluorescence. Unstained and single antibody stained cells, or unstained or single stained antibody-binding beads, were used to set fluorescence compensation. Negative controls for CD234 (Fy) were “secondary only” with either CA111 or rPvDBPII omitted in otherwise complete assays. Secondary and tertiary reagents were PE labeled anti HA-tag for CA111and PE labeled goat anti-rabbit, respectively for rPvDBPII (with a polyclonal rabbit anti- PvDBP as secondary antibody). Results collected following comparisons with these negative controls allowed for calculating fold-increase of the geometric mean of fluorescence intensity (CA111 or rPvDBPII) over the negative control or for calculating the fraction of positive cells.
Second, for data collected on bone marrow samples, we analyzed a subset of bone marrow donors to show the level of expression of CD34, CD45, CD105, CD71, and Fy. The antibodies and staining protocol was as above. However, the sub-populations were gated multi-dimensionally using a series of bivariate plots to create a sequential series of erythroid differentiation states. The fluorescence value of each parameter was divided by a correlated measure of cell size (forward scatter) providing a measure of antigen density. The expression level of Fy was made more accurate by subtracting the fluorescence of the negative control on a subpopulation basis (median fluorescence – median background fluorescence).
Erythroid cell protein extraction and Western blot analysis
Extracting the Fy protein from the surface of erythroid cells was conducted to specifically capture evidence of FY gene expression. These experiments were performed on pbRBCs from Fy-positive and Fy-negative donors. Target cell populations included pbRBC, expanded CD34 cell populations and bone marrow. Details of protein extraction, immunoprecipitation and Western blot analyses are provided earlier in the Star Method.
Competition assay of CA111 or 2C3 binding to Fy-negative RBCs in the presence of nDARC (Fc conjugated DARC)
Blood derived from three Fy negative individuals was incubated with soluble nDARC to compete with the binding to RBC bound DARC. Samples were taken on multiple days and each time tested in quadruplicates for each nDARC concentration (range: 1000 nM to 0.48 nM, twofold serial dilutions) as well as N=12 for 1000 nM nDARC, 0 nM nDARC and 2nd antibody staining alone for higher statistical power. Samples from the three individuals were tested on three different days. Samples were collected on an Attune Next using the appropriate lasers for HO, PE and FITC as well as Forward Scatter and Side Scatter. For each sample, 200,000 events were collected.
Imagestream Analysis
Samples from invasion experiments were collected on an Imagestream IS100 instrument. Target cells were labeled with CFSE, while the donor cells were CFSE negative. Each experiment was seeded using peripheral blood of North-American Fy-positive or Fy-negative participants as target cells and stock P. vivax isolates (stock culture: AMP2016.14 and AMP2016.36) at a ratio of 80% to 20% (or 90% to 10%) respectively for a total RBCs 7.2×107. After 72h incubation the cells were stained with Höchst 33342 and then fixed with 4 % PFA in 1 × PBS. For each data file 200K events were collected, for most samples we performed four technical replicate (resulting to ~1000K events). All datasets were collected in Extended Depth of Field (EDF) mode, resulting in an extended depth of field when focusing the cells. Only focused cells were further analyzed by first separating them by Area and Aspect Ratio in the brightfield channel, allowing the distinction of single versus multiple cells in an image. Single cells were then separated by intensity in CFSE and HO staining. Five areas are distinguished in the CFSE/HO plot. Unlabeled uninfected donor cells (CFSE-/HO−), infected donor cells (CFSE-/HO+), uninfected labeled target cells (CFSE+/HO−), infected target cells (CFSE+/HO+) and white blood cells (CFSE+/HO>105). The HO positive cells detected in the donor cell population versus the uninfected donor cells reflect the parasitemia of that individual sample at the time of collection. In addition, a cutoff value was determined based on a CI95 level of the mean fluorescent intensity of uninfected cells (HO channel). Cells that were above that threshold were deemed positive.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed using STATA version 13.0 and are described on the legend of each concerned figure. Additionally, Graphpad Prism 10 was used for statistical analysis of the SPR data as well as the competition assay. Imagestream data was analyzed by the IDEAS Software version 6.2.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Recombinant proteins | ||
| rPvDBPII | Gift from Dr. Niraj Tolia | https://doi.org/10.1038/nsmb.2088 |
| unfolded rPcDBPII | Material produced at CWRU | https://doi.org/10.1038/nsmb.2089 |
| nDARCIg | Construct is a gift from Chetin Chitnis, material produced at CWRU | https://doi.org/10.1371/journal.pmed.0040337 |
| rCXCR2 | NovusBio | H00003579-P01 |
| Antibodies | ||
| conventional mouse 2C3 anti-Fy6 | Gift from Dr. Yves Colin | https://doi.org/10.1046/j.1365-2141.2003.04533.x |
| single domain camelid CA111 (HA tag) anti-Fy6 | Gift from Dr. Olivier Bertrand and Dr. Yves Colin | https://doi.org/10.1007/s00018-010-0387-6 |
| Anti-ACKR1 / DARC Antibody (aa1-63) | LS Biosciences | LS-C371451 |
| HRP anti rabbit antibody | Milipore | AP187P |
| Mouse Anti-HA (IgG1) | Bioloegend | 901502 |
| Goat anti-mouse PE | eBioscience | 12-4010-82 |
| Goat anti-mouse APC | BD Biosciences | 550826 |
| Polyclonal rabbit anti-PvDBP (Rabbit 2) | https://doi.org/10.1371/journal.pmed.0040337 | |
| Goat anti-rabbit APC | Invitrogen | 31984 |
| Goat anti-rabbit PE | Sigma | P9537 |
| CD45 MicroBeads, human | Miltenyi | 130-045-801 |
| anti-CXCR2 | R&D system | MAB331 |
| Anti-Fy a (FY1) | Biorad | 808-186 |
| Anti-Fyb (FY2) | BioRad | 808-191 |
| anti-GlycoA PE-Cy7 | BD Biosciences | 563666 |
| anti-CD36 APC | BD Biosciences | 550956 |
| anti-Band3-PE | American Research Product | IBGRL-BRIC6-PE |
| anti-CD71 APC | BD Biosciences | 551374 |
| anti-CD105 PE-Cy7 | BD Biosciences | 568753 |
| anti-CD71 FITC | BD Biosciences | 555536 |
| anti-CD34 BV421 | BD Biosciences | 562577 |
| anti-CD45 APC-Cy7 | BD Biosciences | 557833 |
| anti-CD71 BV786 | BD Biosciences | 563768 |
| mouse anti-HA tag | Miltenyi | 130-098-404 |
| mouse anti-His tag | Qiagen | 34650 |
| mouse anti-PE | Invitrogen | 12-4010-82 |
| mouse anti-PE | Jackson ImmunoResearch | 115-115-164 |
| Immunoprecipitation | ||
| Protease inhibitor | Sigma Aldrich | 4693159001 ; P8849 |
| Dynabeads M-270 Carboxylic Acid | Invitrogen | 14305D ; 10003D |
| EDC 1-(3-Dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride | ACROS organics | 171440010 |
| Trypsin | Sigma Aldrich | T6567 |
| IgG Elution Buffer | Thermo | 21004 |
| Neutralizing buffer 1 M tris Base pH 9 10X | Sigma Aldrich | 1083820100 |
| Western Blot | ||
| Amersham standard autoradiography cassette, 24 × 30 cm | Dutscher | RPN11643 |
| Amersham Hyperfilm™ ECL | Dustcher | 28-9068-37 |
| Mini-P Tetra Cell,2precast gel | Biorad | 1658005 |
| Dithiothreitol (DTT), 5 g | Biorad | 1610611 |
| Mini-PROTEAN TGX,4–20%,30μl 10 well,10 | Biorad | 4561093 |
| Precision Plus Protein All Blue Standards, 500microliter, 50 applications | Biorad | 1610373 |
| Immun-Blot PVDF Membranes, 7 × 8.4 cm, 10 | Biorad | 1620174 |
| Clarity Max ECL substrate,100ml | Biorad | 1705062 |
| Surebeads Protein G Magnetic Beads 3 ml | Biorad | 1614023 |
| Surebeads Magnetic Rack, 16 tube Holder | Biorad | 1614916 |
| Tris, 500 g | Biorad | 1610716 |
| Glycine, 1kg | Biorad | 1610718 |
| SDS, 100 g | Biorad | 1610301 |
| Bio-Safe Coomassie Stain, 1 L | Biorad | 1610786 |
| CD34+ Maturation | ||
| CD34 MicroBead Kit | Miltenyi | 130-046-702 |
| holotransferrine | Sigma Aldrich | T0665 100mg |
| Human insulin | Sigma Aldrich | I9278 5ml |
| heparin | Sigma Aldrich | 2106-10VL |
| AB serum | Sigma Aldrich | H4522-100ML |
| Hydrocortisone (dexamethasone) | Sigma Aldrich | D4902-25MG |
| Stem Cell Factor (SCF) | Peprotech | 300-07 |
| IL3 | Peprotech | 100-64 |
| EPO | Peprotech | 200-03 |
| IMDM + glutamax | Gibco Thermo Fisher Scientific | 31980030 |
| Others | ||
| Chymotrypsin | Roche Diagnostic | 11418467001 |
| Bovine Serum Albumin Fraction V | Sigma Aldrich | 810531 |
| Ficoll paque plus | Sigma Aldrich | GE17-1440-02 |
| Nycodenz | MP Biomedicals | 157750 |
| Primers | ||
| Duffy forward | Sigma Aldrich | 5′-CAGGCAGTGGGCGTGGG-3′ |
| Duffy reverse | Sigma Aldrich | 5′-CTGCTAGCTAGGATACCCAG-3′ |
| FY*B/FY*X forward | Sigma Aldrich | 5′-AGCACTGTCCTCTTCATGCTTT-3′ |
| FY*B/FY*X reverse | Sigma Aldrich | 5′-GCAGAGCTGCGAGTGCTAC-3′ |
| LDR probes | ||
| PRO ntT MTAG | Sigma Aldrich | 5′-ACACTTATCTTTCAATTCAATTACcattagt ccttggctcttat-3′ |
| PRO ntC MTAG | Sigma Aldrich | 5′-CTTTCTCATACTTTCAACTAATTTtcattagt ccttggctcttac-3′ |
| PRO common | Sigma Aldrich | 5′-cttggaagcacaggcgctg-biotin-3′ |
| FY*A ntG MTAG | Sigma Aldrich | 5′-CAAACAAACATTCAAATATCAATCttccca gatggagactatgg-3′ |
| FY*B ntA MTAG | Sigma Aldrich | 5′-TACATTCAACACTCTTAAATCAAActtccca gatggagactatga-3′ |
| Codon42 common | Sigma Aldrich | 5′-tgccaacctggaagca-biotin-3′ |
| FY*B ntC MTAG | Sigma Aldrich | 5′-CACTTAATTCATTCTAAATCTATCtgcttttc agacctctctcc-3′ |
| FY*X ntT MTAG | Sigma Aldrich | 5′-ACTTATTTCTTCACTACTATATCAtgcttttc agacctctctct-3′ |
| Codon89 common | Sigma Aldrich | 5′-gctggcagctctgccctggct-biotin-3′ |
| Surface Plasmon Resonance | ||
| CM5 Sensor chip | Cytivia | 29104988 |
| Anti-DARC [2C3] | Absolute Antibody | https://absoluteantibody.com/product/anti-darc-2c3/Ab00893-10.0_human_igg1/standard/ |
| ImageStream | ||
| CellTrace CFSE Cell Proliferation Kit | Thermo Fisher | https://www.thermofisher.com/order/catalog/product/C34554 |
| Höchst 33342 | Life Technologies | H3570 |
| Software | ||
| FlowJo version 10.0 | Becton Dickison & Company (BD) | https://www.bdbiosciences.com/en-us/products/software/flowjo-v10-software |
| BD FACSDiva™ Software version 8.0 | Becton Dickison & Company (BD) | https://www.bdbiosciences.com/en-us/products/software/instrument-software/bd-facsdiva-software |
| WinList 9.0 | Verity Software House | https://www.vsh.com/products/winlist/index.asp |
| STATA version 13.0 | StataCorp LLC | https://www.stata.com/products/ |
| Graphpad Prism version 10 | GraphPad software, Inc. | https://www.graphpad.com/features |
| IDEAS Application Version 6.2.187 | Amnis part of EMD Millipore | https://ideas.com/ |
Highlights.
Duffy blood group protein (Fy) is naturally expressed on Fy-negative red blood cells.
Fy expression is higher on erythroid precursors than on red blood cells.
Fy protein is expressed in early erythropoietic stages in Fy-negative individuals.
P. vivax invades Fy-positive and Fy-negative erythroid cells in vitro.
Acknowledgments
We are grateful to the study donors who patiently participated over the course of this investigation in the United States and in Madagascar. We are also grateful to the Madagascar study team members, Rosalind E. Howes, Thierry Franchard, Tovonahary Angelo Rakotomanga and Brune Ramiranirina for recruitment of Malagasy study participants, collection, cryopreservation and cataloging of P. vivax-infected blood samples. We thank David N. Wald, Director of the Hematopoietic Biorepository & Cellular Therapy Core and Basabi Maitra for recruitment of bone marrow donors. Howard Meyerson, Keith E. Shults and the Comprehensive Cancer Center of Case Western Reserve University and University Hospitals of Cleveland generously contributed to the flow cytometry study design and provided technical support. We thank Chetan Chitnis for providing nDARCIg; Olivier S. Bertrand and Sylvie Cochet (INSERM/University Paris Diderot) for the gift of anti-Fy antibodies (2C3 and CA111) and objective criticisms integral to this study; Anne-Lyse Baudrier, Emmanuel Collec and the Centre National de Reference pour les Groupes Sanguins (CNRGS) for providing the anti-FyA and anti-FyB antibodies.
Funding
This study was funded by grants from the Veterans Affairs Research Service (BX001350) to CLK, NIH Grant (R01 AI064687 and R01 AI143694) to CLK and an NIH Grant (R01 AI097366 and R01 AI148469) to PAZ and the Agence Nationale de la Recherche (ANR-11-LABEX-0024-01 ParaFrap) to BG. N.H.T. and N.D.S are supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Ethics approval and consent to participate
Study protocols were approved by the University Hospitals of Cleveland Institutional Review Board (#08-03-33 and #09-90-195) and the Madagascar Ethical Committee (No. 099-MSANP/CE).
Consent for publication
All authors read and approved the final manuscript.
Competing interests
The authors have declared that no conflict of interest exists.
Declaration of Interests
The authors declare no competing interests.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- 1.Donahue RP, Bias WB, Renwick JH, and McKusick VA (1968). Probable assignment of the Duffy blood group locus to chromosome 1 in man. Proceedings of the National Academy of Sciences 61, 949–955. 10.1073/pnas.61.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachelerie F, Ben-Baruch A, Burkhardt AM, Combadiere C, Farber JM, Graham GJ, Horuk R, Sparre-Ulrich AH, Locati M, Luster AD, et al. (2014). International Union of Pharmacology. LXXXIX. Update on the Extended Family of Chemokine Receptors and Introducing a New Nomenclature for Atypical Chemokine Receptors. Pharmacol Rev 66, 1–79. 10.1124/pr.113.007724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darbonne WC, Rice GC, Mohler MA, Apple T, Hébert CA, Valente AJ, and Baker JB (1991). Red blood cells are a sink for interleukin 8, a leukocyte chemotaxin. J Clin Invest 88, 1362–1369. 10.1172/JCI115442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dawson TC, Lentsch AB, Wang Z, Cowhig JE, Rot A, Maeda N, and Peiper SC (2000). Exaggerated response to endotoxin in mice lacking the Duffy antigen/receptor for chemokines (DARC). Blood 96, 1681–1684. [PubMed] [Google Scholar]
- 5.Neote K, Darbonne W, Ogez J, Horuk R, and Schall TJ (1993). Identification of a promiscuous inflammatory peptide receptor on the surface of red blood cells. J Biol Chem 268, 12247–12249. [PubMed] [Google Scholar]
- 6.Reutershan J, Harry B, Chang D, Bagby GJ, and Ley K (2009). DARC on RBC limits lung injury by balancing compartmental distribution of CXC chemokines. Eur J Immunol 39, 1597–1607. 10.1002/eji.200839089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vergara C, Tsai YJ, Grant AV, Rafaels N, Gao L, Hand T, Stockton M, Campbell M, Mercado D, Faruque M, et al. (2008). Gene encoding Duffy antigen/receptor for chemokines is associated with asthma and IgE in three populations. Am J Respir Crit Care Med 178, 1017–1022. 10.1164/rccm.200801-182OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hub E, and Rot A (1998). Binding of RANTES, MCP-1, MCP-3, and MIP-1alpha to cells in human skin. Am J Pathol 152, 749–757. [PMC free article] [PubMed] [Google Scholar]
- 9.Middleton J, Neil S, Wintle J, Clark-Lewis I, Moore H, Lam C, Auer M, Hub E, and Rot A (1997). Transcytosis and surface presentation of IL-8 by venular endothelial cells. Cell 91, 385–395. 10.1016/s0092-8674(00)80422-5. [DOI] [PubMed] [Google Scholar]
- 10.Pruenster M, Mudde L, Bombosi P, Dimitrova S, Zsak M, Middleton J, Richmond A, Graham GJ, Segerer S, Nibbs RJB, et al. (2009). The Duffy antigen receptor for chemokines transports chemokines and supports their promigratory activity. Nat Immunol 10, 101–108. 10.1038/ni.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rot A (1992). Endothelial cell binding of NAP-1/IL-8: role in neutrophil emigration. Immunol Today 13, 291–294. 10.1016/0167-5699(92)90039-A. [DOI] [PubMed] [Google Scholar]
- 12.Zarbock A, Schmolke M, Bockhorn SG, Scharte M, Buschmann K, Ley K, and Singbartl K (2007). The Duffy antigen receptor for chemokines in acute renal failure: A facilitator of renal chemokine presentation. Crit Care Med 35, 2156–2163. 10.1097/01.ccm.0000280570.82885.32. [DOI] [PubMed] [Google Scholar]
- 13.O’Leary PA (1927). Treatment of neurosyphilis by malaria: report on the three year’s observation of the first one hundred patients treated. Journal of the American Medical Association 89, 95–100. 10.1001/jama.1927.02690020019007. [DOI] [Google Scholar]
- 14.Boyd MF, and Stratman-Thomas WK (1933). Studies on benign tertian malaria. 4. On the refractoriness of negroes to inoculation with Plasmodium vivax. American Journal of Epidemiology 18, 485–489. 10.1093/oxfordjournals.aje.a117964. [DOI] [Google Scholar]
- 15.Young MD, Ellis JM, and Stubbs TH (1946). Studies on imported malarias; transmission of foreign Plasmodium vivax by Anopheles quadrimaculatus. Am J Trop Med Hyg 26, 477–483. [PubMed] [Google Scholar]
- 16.Young MD, Eyles DE, Burgess RW, and Jeffery GM (1955). Experimental testing of the immunity of Negroes to Plasmodium vivax. J Parasitol 41, 315–318. [PubMed] [Google Scholar]
- 17.Bray RS (1957). Plasmodium ovale in Liberia. Am J Trop Med Hyg 6, 961–970. 10.4269/ajtmh.1957.6.961. [DOI] [PubMed] [Google Scholar]
- 18.Bray RS (1958). The susceptibility of Liberians to the Madagascar strain of Plasmodium vivax. J Parasitol 44, 371–373. [PubMed] [Google Scholar]
- 19.Miller LH, Mason SJ, Clyde DF, and McGinniss MH (1976). The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med 295, 302–304. 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 20.Chitnis CE, and Miller LH (1994). Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J Exp Med 180, 497–506. 10.1084/jem.180.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Batchelor JD, Malpede BM, Omattage NS, DeKoster GT, Henzler-Wildman KA, and Tolia NH (2014). Red blood cell invasion by Plasmodium vivax: structural basis for DBP engagement of DARC. PLoS Pathog 10, e1003869. 10.1371/journal.ppat.1003869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Batchelor JD, Zahm JA, and Tolia NH (2011). Dimerization of Plasmodium vivax DBP is induced upon receptor binding and drives recognition of DARC. Nat Struct Mol Biol 18, 908–914. 10.1038/nsmb.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen E, Salinas ND, Huang Y, Ntumngia F, Plasencia MD, Gross ML, Adams JH, and Tolia NH (2016). Broadly neutralizing epitopes in the Plasmodium vivax vaccine candidate Duffy Binding Protein. Proc Natl Acad Sci U S A 113, 6277–6282. 10.1073/pnas.1600488113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carias LL, Dechavanne S, Nicolete VC, Sreng S, Suon S, Amaratunga C, Fairhurst RM, Dechavanne C, Barnes S, Witkowski B, et al. (2019). Identification and Characterization of Functional Human Monoclonal Antibodies to Plasmodium vivax Duffy-Binding Protein. J Immunol 202, 2648–2660. 10.4049/jimmunol.1801631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urusova D, Carias L, Huang Y, Nicolete VC, Popovici J, Roesch C, Salinas ND, Dechavanne S, Witkowski B, Ferreira MU, et al. (2019). Structural basis for neutralization of Plasmodium vivax by naturally acquired human antibodies that target DBP. Nat Microbiol 4, 1486–1496. 10.1038/s41564-019-0461-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dickey TH, and Tolia NH (2023). Designing an effective malaria vaccine targeting Plasmodium vivax Duffy-binding protein. Trends Parasitol, S1471–4922(23)00147–2. 10.1016/j.pt.2023.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tournamille C, Colin Y, Cartron JP, and Le Van Kim C (1995). Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet 10, 224–228. 10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]
- 28.Zimmerman PA, Woolley I, Masinde GL, Miller SM, McNamara DT, Hazlett F, Mgone CS, Alpers MP, Genton B, Boatin BA, et al. (1999). Emergence of FY*A(null) in a Plasmodium vivax-endemic region of Papua New Guinea. Proc Natl Acad Sci U S A 96, 13973–13977. 10.1073/pnas.96.24.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ranjan A, and Chitnis CE (1999). Mapping regions containing binding residues within functional domains of Plasmodium vivax and Plasmodium knowlesi erythrocyte-binding proteins. Proc Natl Acad Sci U S A 96, 14067–14072. 10.1073/pnas.96.24.14067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aikawa M, Miller LH, Johnson J, and Rabbege J (1978). Erythrocyte entry by malarial parasites. A moving junction between erythrocyte and parasite. J Cell Biol 77, 72–82. 10.1083/jcb.77.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zimmerman PA (2017). Plasmodium vivax Infection in Duffy-Negative People in Africa. Am J Trop Med Hyg 97, 636–638. 10.4269/ajtmh.17-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Twohig KA, Pfeffer DA, Baird JK, Price RN, Zimmerman PA, Hay SI, Gething PW, Battle KE, and Howes RE (2019). Growing evidence of Plasmodium vivax across malaria-endemic Africa. PLoS Negl Trop Dis 13, e0007140. 10.1371/journal.pntd.0007140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baird JK (2022). African Plasmodium vivax malaria improbably rare or benign. Trends in Parasitology 38, 683–696. 10.1016/j.pt.2022.05.006. [DOI] [PubMed] [Google Scholar]
- 34.Leong YW, Russell B, Malleret B, and Rénia L (2022). Erythrocyte tropism of malarial parasites: The reticulocyte appeal. Frontiers in Microbiology 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woolley IJ, Hotmire KA, Sramkoski RM, Zimmerman PA, and Kazura JW (2000). Differential expression of the duffy antigen receptor for chemokines according to RBC age and FY genotype. Transfusion 40, 949–953. 10.1046/j.1537-2995.2000.40080949.x. [DOI] [PubMed] [Google Scholar]
- 36.Lopez GH, Condon JA, Wilson B, Martin JR, Liew Y-W, Flower RL, and Hyland CA (2015). A novel FY*A allele with the 265T and 298A SNPs formerly associated exclusively with the FY*B allele and weak Fy(b) antigen expression: implication for genotyping interpretative algorithms. Vox Sang 108, 52–57. 10.1111/vox.12185. [DOI] [PubMed] [Google Scholar]
- 37.Olsson ML, Smythe JS, Hansson C, Poole J, Mallinson G, Jones J, Avent ND, and Daniels G (1998). The Fy(x) phenotype is associated with a missense mutation in the Fy(b) allele predicting Arg89Cys in the Duffy glycoprotein. Br J Haematol 103, 1184–1191. 10.1046/j.1365-2141.1998.01083.x. [DOI] [PubMed] [Google Scholar]
- 38.Parasol N, Reid M, Rios M, Castilho L, Harari I, and Kosower NS (1998). A novel mutation in the coding sequence of the FY*B allele of the Duffy chemokine receptor gene is associated with an altered erythrocyte phenotype. Blood 92, 2237–2243. [PubMed] [Google Scholar]
- 39.Tournamille C, Le Van Kim C, Gane P, Le Pennec PY, Roubinet F, Babinet J, Cartron JP, and Colin Y (1998). Arg89Cys substitution results in very low membrane expression of the Duffy antigen/receptor for chemokines in Fy(x) individuals. Blood 92, 2147–2156. [PubMed] [Google Scholar]
- 40.Mallinson G, Soo KS, Schall TJ, Pisacka M, and Anstee DJ (1995). Mutations in the erythrocyte chemokine receptor (Duffy) gene: the molecular basis of the Fya/Fyb antigens and identification of a deletion in the Duffy gene of an apparently healthy individual with the Fy(a-b-) phenotype. Br J Haematol 90, 823–829. 10.1111/j.1365-2141.1995.tb05202.x. [DOI] [PubMed] [Google Scholar]
- 41.Rios M, Chaudhuri A, Mallinson G, Sausais L, Gomensoro-Garcia AE, Hannon J, Rosenberger S, Poole J, Burgess G, Pogo O, et al. (2000). New genotypes in Fy(a-b-) individuals: nonsense mutations (Trp to stop) in the coding sequence of either FY A or FY B. Br J Haematol 108, 448–454. 10.1046/j.1365-2141.2000.01882.x. [DOI] [PubMed] [Google Scholar]
- 42.Wasniowska K, Blanchard D, Janvier D, Wang ZX, Peiper SC, Hadley TJ, and Lisowska E (1996). Identification of the Fy6 epitope recognized by two monoclonal antibodies in the N-terminal extracellular portion of the Duffy antigen receptor for chemokines. Mol Immunol 33, 917–923. 10.1016/s0161-5890(96)00056-9. [DOI] [PubMed] [Google Scholar]
- 43.Wasniowska K, Petit-LeRoux Y, Tournamille C, Le van Kim C, Cartron JP, Colin Y, Lisowska E, and Blanchard D (2002). Structural characterization of the epitope recognized by the new anti-Fy6 monoclonal antibody NaM 185–2C3. Transfus Med 12, 205–211. 10.1046/j.1365-3148.2002.00373.x. [DOI] [PubMed] [Google Scholar]
- 44.Smolarek D, Hattab C, Hassanzadeh-Ghassabeh G, Cochet S, Gutiérrez C, de Brevern AG, Udomsangpetch R, Picot J, Grodecka M, Wasniowska K, et al. (2010). A recombinant dromedary antibody fragment (VHH or nanobody) directed against human Duffy antigen receptor for chemokines. Cell Mol Life Sci 67, 3371–3387. 10.1007/s00018-010-0387-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smolarek D, Hattab C, Buczkowska A, Kaczmarek R, Jarząb A, Cochet S, de Brevern AG, Lukasiewicz J, Jachymek W, Niedziela T, et al. (2015). Studies of a murine monoclonal antibody directed against DARC: reappraisal of its specificity. PLoS One 10, e0116472. 10.1371/journal.pone.0116472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.King CL, Adams JH, Xianli J, Grimberg BT, McHenry AM, Greenberg LJ, Siddiqui A, Howes RE, da Silva-Nunes M, Ferreira MU, et al. (2011). Fy(a)/Fy(b) antigen polymorphism in human erythrocyte Duffy antigen affects susceptibility to Plasmodium vivax malaria. Proc Natl Acad Sci U S A 108, 20113–20118. 10.1073/pnas.1109621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tran TM, Moreno A, Yazdani SS, Chitnis CE, Barnwell JW, and Galinski MR (2005). Detection of a Plasmodium vivax erythrocyte binding protein by flow cytometry. Cytometry A 63, 59–66. 10.1002/cyto.a.20098. [DOI] [PubMed] [Google Scholar]
- 48.Haynes JD, Dalton JP, Klotz FW, McGinniss MH, Hadley TJ, Hudson DE, and Miller LH (1988). Receptor-like specificity of a Plasmodium knowlesi malarial protein that binds to Duffy antigen ligands on erythrocytes. J Exp Med 167, 1873–1881. 10.1084/jem.167.6.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horuk R (2015). The Duffy Antigen Receptor for Chemokines DARC/ACKR1. Front Immunol 6, 279. 10.3389/fimmu.2015.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller LH, Hudson D, and Haynes JD (1988). Identification of Plasmodium knowlesi erythrocyte binding proteins. Mol Biochem Parasitol 31, 217–222. 10.1016/0166-6851(88)90151-x. [DOI] [PubMed] [Google Scholar]
- 51.Wertheimer SP, and Barnwell JW (1989). Plasmodium vivax interaction with the human Duffy blood group glycoprotein: identification of a parasite receptor-like protein. Exp Parasitol 69, 340–350. 10.1016/0014-4894(89)90083-0. [DOI] [PubMed] [Google Scholar]
- 52.Choe H, Moore MJ, Owens CM, Wright PL, Vasilieva N, Li W, Singh AP, Shakri R, Chitnis CE, and Farzan M (2005). Sulphated tyrosines mediate association of chemokines and Plasmodium vivax Duffy binding protein with the Duffy antigen/receptor for chemokines (DARC). Mol Microbiol 55, 1413–1422. 10.1111/j.1365-2958.2004.04478.x. [DOI] [PubMed] [Google Scholar]
- 53.Kitchen SF (1938). The Infection of Reticulocytes by Plasmodium Vivax. The American Journal of Tropical Medicine and Hygiene s1–18, 347–359. 10.4269/ajtmh.1938.s1-18.347. [DOI] [Google Scholar]
- 54.Mons B (1990). Preferential invasion of malarial merozoites into young red blood cells. Blood Cells 16, 299–312. [PubMed] [Google Scholar]
- 55.Russell B, Suwanarusk R, Borlon C, Costa FTM, Chu CS, Rijken MJ, Sriprawat K, Warter L, Koh EGL, Malleret B, et al. (2011). A reliable ex vivo invasion assay of human reticulocytes by Plasmodium vivax. Blood 118, e74–81. 10.1182/blood-2011-04-348748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lim C, Pereira L, Saliba KS, Mascarenhas A, Maki JN, Chery L, Gomes E, Rathod PK, and Duraisingh MT (2016). Reticulocyte Preference and Stage Development of Plasmodium vivax Isolates. J Infect Dis 214, 1081–1084. 10.1093/infdis/jiw303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cho J-S, Russell B, Kosaisavee V, Zhang R, Colin Y, Bertrand O, Chandramohanadas R, Chu CS, Nosten F, Renia L, et al. (2016). Unambiguous determination of Plasmodium vivax reticulocyte invasion by flow cytometry. Int J Parasitol 46, 31–39. 10.1016/j.ijpara.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 58.Ovchynnikova E, Aglialoro F, Bentlage AEH, Vidarsson G, Salinas ND, von Lindern M, Tolia NH, and van den Akker E (2017). DARC extracellular domain remodeling in maturating reticulocytes explains Plasmodium vivax tropism. Blood 130, 1441–1444. 10.1182/blood-2017-03-774364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malleret B, Li A, Zhang R, Tan KSW, Suwanarusk R, Claser C, Cho JS, Koh EGL, Chu CS, Pukrittayakamee S, et al. (2015). Plasmodium vivax: restricted tropism and rapid remodeling of CD71-positive reticulocytes. Blood 125, 1314–1324. 10.1182/blood-2014-08-596015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gruszczyk J, Kanjee U, Chan L-J, Menant S, Malleret B, Lim NTY, Schmidt CQ, Mok Y-F, Lin K-M, Pearson RD, et al. (2018). Transferrin receptor 1 is a reticulocyte-specific receptor for Plasmodium vivax. Science 359, 48–55. 10.1126/science.aan1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garnham PCC (1966). Malaria parasites and other haemosporidia (Blackwell Scientific; ). [Google Scholar]
- 62.Duchene J, Novitzky-Basso I, Thiriot A, Casanova-Acebes M, Bianchini M, Etheridge SL, Hub E, Nitz K, Artinger K, Eller K, et al. (2017). Atypical chemokine receptor 1 on nucleated erythroid cells regulates hematopoiesis. Nat Immunol 18, 753–761. 10.1038/ni.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu J, Liu J, Xue F, Halverson G, Reid M, Guo A, Chen L, Raza A, Galili N, Jaffray J, et al. (2013). Isolation and functional characterization of human erythroblasts at distinct stages: implications for understanding of normal and disordered erythropoiesis in vivo. Blood 121, 3246–3253. 10.1182/blood-2013-01-476390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kondo M, Wagers AJ, Manz MG, Prohaska SS, Scherer DC, Beilhack GF, Shizuru JA, and Weissman IL (2003). Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol 21, 759–806. 10.1146/annurev.immunol.21.120601.141007. [DOI] [PubMed] [Google Scholar]
- 65.Attar A (2014). Changes in the Cell Surface Markers During Normal Hematopoiesis: A Guide to Cell Isolation. In Global Journal of Hematology and Blood Transfusion, pp. 20–28. 10.15379/2408-9877.2014.01.01.4. [DOI] [Google Scholar]
- 66.Sidney LE, Branch MJ, Dunphy SE, Dua HS, and Hopkinson A (2014). Concise review: evidence for CD34 as a common marker for diverse progenitors. Stem Cells 32, 1380–1389. 10.1002/stem.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mori Y, Chen JY, Pluvinage JV, Seita J, and Weissman IL (2015). Prospective isolation of human erythroid lineage-committed progenitors. Proc Natl Acad Sci U S A 112, 9638–9643. 10.1073/pnas.1512076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Machherndl-Spandl S, Suessner S, Danzer M, Proell J, Gabriel C, Lauf J, Sylie R, Klein H-U, Béné MC, Weltermann A, et al. (2013). Molecular pathways of early CD105-positive erythroid cells as compared with CD34-positive common precursor cells by flow cytometric cell-sorting and gene expression profiling. Blood Cancer J 3, e100. 10.1038/bcj.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wangen JR, Eidenschink Brodersen L, Stolk TT, Wells DA, and Loken MR (2014). Assessment of normal erythropoiesis by flow cytometry: important considerations for specimen preparation. Int J Lab Hematol 36, 184–196. 10.1111/ijlh.12151. [DOI] [PubMed] [Google Scholar]
- 70.Kono M, Kondo T, Takagi Y, Wada A, and Fujimoto K (2009). Morphological definition of CD71 positive reticulocytes by various staining techniques and electron microscopy compared to reticulocytes detected by an automated hematology analyzer. Clin Chim Acta 404, 105–110. 10.1016/j.cca.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 71.Menard D, Barnadas C, Bouchier C, Henry-Halldin C, Gray LR, Ratsimbasoa A, Thonier V, Carod J-F, Domarle O, Colin Y, et al. (2010). Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc Natl Acad Sci U S A 107, 5967–5971. 10.1073/pnas.0912496107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Howes RE, Franchard T, Rakotomanga TA, Ramiranirina B, Zikursh M, Cramer EY, Tisch DJ, Kang SY, Ramboarina S, Ratsimbasoa A, et al. (2018). Risk Factors for Malaria Infection in Central Madagascar: Insights from a Cross-Sectional Population Survey. Am J Trop Med Hyg 99, 995–1002. 10.4269/ajtmh.18-0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oboh MA, Singh US, Ndiaye D, Badiane AS, Ali NA, Bharti PK, and Das A (2020). Presence of additional Plasmodium vivax malaria in Duffy negative individuals from Southwestern Nigeria. Malar J 19, 229. 10.1186/s12936-020-03301-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Obaldia N, Meibalan E, Sa JM, Ma S, Clark MA, Mejia P, Moraes Barros RR, Otero W, Ferreira MU, Mitchell JR, et al. (2018). Bone Marrow Is a Major Parasite Reservoir in Plasmodium vivax Infection. mBio 9, e00625–18. 10.1128/mBio.00625-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kho S, Qotrunnada L, Leonardo L, Andries B, Wardani PAI, Fricot A, Henry B, Hardy D, Margyaningsih NI, Apriyanti D, et al. (2021). Evaluation of splenic accumulation and colocalization of immature reticulocytes and Plasmodium vivax in asymptomatic malaria: A prospective human splenectomy study. PLoS Med 18, e1003632. 10.1371/journal.pmed.1003632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kho S, Qotrunnada L, Leonardo L, Andries B, Wardani PAI, Fricot A, Henry B, Hardy D, Margyaningsih NI, Apriyanti D, et al. (2021). Hidden Biomass of Intact Malaria Parasites in the Human Spleen. N Engl J Med 384, 2067–2069. 10.1056/NEJMc2023884. [DOI] [PubMed] [Google Scholar]
- 77.Cenariu D, Iluta S, Zimta A-A, Petrushev B, Qian L, Dirzu N, Tomuleasa C, Bumbea H, and Zaharie F (2021). Extramedullary Hematopoiesis of the Liver and Spleen. Journal of Clinical Medicine 10, 5831. 10.3390/jcm10245831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Malleret B, El Sahili A, Tay MZ, Carissimo G, Ong ASM, Novera W, Lin J, Suwanarusk R, Kosaisavee V, Chu TTT, et al. (2021). Plasmodium vivax binds host CD98hc (SLC3A2) to enter immature red blood cells. Nat Microbiol 6, 991–999. 10.1038/s41564-021-00939-3. [DOI] [PubMed] [Google Scholar]
- 79.Ferreira R, Ohneda K, Yamamoto M, and Philipsen S (2005). GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol Cell Biol 25, 1215–1227. 10.1128/MCB.25.4.1215-1227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fernandez-Becerra C, Lelievre J, Ferrer M, Anton N, Thomson R, Peligero C, Almela MJ, Lacerda MV, Herreros E, and Del Portillo HA (2013). Red blood cells derived from peripheral blood and bone marrow CD34+ human haematopoietic stem cells are permissive to Plasmodium parasites infection. Mem Inst Oswaldo Cruz 108, 801–803. 10.1590/0074-0276108062013019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thomson-Luque R, and Bautista JM (2021). Home Sweet Home: Plasmodium vivax-Infected Reticulocytes-The Younger the Better? Front Cell Infect Microbiol 11, 675156. 10.3389/fcimb.2021.675156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aitken GJ (1943). Sternal puncture in the diagnosis of malaria. The Lancet 242, 466–468. 10.1016/S0140-6736(00)87486-3. [DOI] [Google Scholar]
- 83.Lacerda M.V.G. de, Hipólito JR, and Passos L.N. da M. (2008). Chronic Plasmodium vivax infection in a patient with splenomegaly and severe thrombocytopenia. Rev Soc Bras Med Trop 41, 522–523. 10.1590/s0037-86822008000500021. [DOI] [PubMed] [Google Scholar]
- 84.Brito MAM, Baro B, Raiol TC, Ayllon-Hermida A, Safe IP, Deroost K, Figueiredo EFG, Costa AG, Armengol MDP, Sumoy L, et al. (2022). Morphological and Transcriptional Changes in Human Bone Marrow During Natural Plasmodium vivax Malaria Infections. J Infect Dis 225, 1274–1283. 10.1093/infdis/jiaa177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wickramarachchi T, Devi YS, Mohmmed A, and Chauhan VS (2008). Identification and characterization of a novel Plasmodium falciparum merozoite apical protein involved in erythrocyte binding and invasion. PLoS One 3, e1732. 10.1371/journal.pone.0001732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rastogi N, and Rehman N (2018). Changes in bone marrow in malaria-a prospective study of 47 cases. International Journal of Research in Medical Sciences 6, 232–235. 10.18203/2320-6012.ijrms20175725. [DOI] [Google Scholar]
- 87.Jandial R, Bhagat R, and Kumar V (2019). Different Causes of Pyrexia of Unknown Origin on Bone Marrow Examination- An Institutional Experience. jmscr 7. 10.18535/jmscr/v7i7.93. [DOI] [Google Scholar]
- 88.Silva-Filho JL, Dos-Santos JC, Judice C, Beraldi D, Venugopal K, Lima D, Nakaya HI, De Paula EV, Lopes SC, Lacerda MV, et al. (2021). Total parasite biomass but not peripheral parasitaemia is associated with endothelial and haematological perturbations in Plasmodium vivax patients. Elife 10, e71351. 10.7554/eLife.71351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Barber BE, William T, Grigg MJ, Parameswaran U, Piera KA, Price RN, Yeo TW, and Anstey NM (2015). Parasite biomass-related inflammation, endothelial activation, microvascular dysfunction and disease severity in vivax malaria. PLoS Pathog. 11, e1004558. 10.1371/journal.ppat.1004558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Habib I, Smolarek D, Hattab C, Grodecka M, Hassanzadeh-Ghassabeh G, Muyldermans S, Sagan S, Gutiérrez C, Laperche S, Le-Van-Kim C, et al. (2013). V(H)H (nanobody) directed against human glycophorin A: a tool for autologous red cell agglutination assays. Anal Biochem 438, 82–89. 10.1016/j.ab.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 91.Tournamille C, Blancher A, Le Van Kim C, Gane P, Apoil PA, Nakamoto W, Cartron JP, and Colin Y (2004). Sequence, evolution and ligand binding properties of mammalian Duffy antigen/receptor for chemokines. Immunogenetics 55, 682–694. 10.1007/s00251-003-0633-2. [DOI] [PubMed] [Google Scholar]
- 92.Chaudhuri A, Zbrzezna V, Polyakova J, Pogo AO, Hesselgesser J, and Horuk R (1994). Expression of the Duffy antigen in K562 cells. Evidence that it is the human erythrocyte chemokine receptor. J Biol Chem 269, 7835–7838. [PubMed] [Google Scholar]
- 93.Roobsoong W (2015). The In Vitro Invasion Inhibition Assay (IIA) for Plasmodium vivax. In Malaria Vaccines: Methods and Protocols Methods in Molecular Biology., Vaughan A, ed. (Springer; ), pp. 187–196. 10.1007/978-1-4939-2815-6_15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Any additional information required to reanalyze the data reported in this paper will be available from the lead contact upon request.
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


