SUMMARY
Ketamine is a multifunctional drug with clinical applications as an anesthetic, pain management medication, and a fast-acting antidepressant. However, it is also recreationally abused for its dissociative effects. Recent studies in rodents are revealing the neuronal mechanisms mediating its actions, but the impact of prolonged exposure to ketamine on brain-wide networks remains less understood. Here, we develop a sub-cellular resolution whole-brain phenotyping approach and utilize it in male mice to show that repeated ketamine administration leads to a dose-dependent decrease in dopamine neurons in midbrain regions linked to behavioral states, alongside an increase in the hypothalamus. Additionally, diverse changes are observed in long-range innervations of the prefrontal cortex, striatum, and sensory areas. Furthermore, the data support a role for post-transcriptional regulation in enabling ketamine-induced neural plasticity. Through an unbiased, high-resolution whole-brain analysis, this study provides important insights into how chronic ketamine exposure reshapes brain-wide networks.
Graphical Abstract
In brief
Datta et al. develop and use a whole-brain approach to reveal the impact of repeated ketamine exposure on the dopamine system. Findings also support post-transcriptional regulation of ketamine-induced neural network plasticity. This study provides crucial insights into how pharmacological agents like ketamine can reshape brain-wide networks in a non-uniform manner.
INTRODUCTION
Ketamine is a schedule III (US Food and Drug Administration) substance with diverse clinical applications as a dissociative anesthetic, as a pain management drug, and most recently, as a transformative fast-acting antidepressant.1-6 Its complex pharmacological actions in the brain include non-competitive antagonism of the N-methyl-D-aspartate receptor (NMDAR),7,8 blocking of hyperpolarization-activated cyclic nucleotide (HCN1)9,10 channels, and potential activation of opioid receptors.11,12 Recent studies in rodent models are unraveling the cellular and neural circuit underpinnings of ketamine’s complex action in the brain. For instance, an antidepressant acute dose of ketamine was shown to promote spinogenesis and synaptogenesis in prefrontal cortical circuits to rescue the eliminated spines in a depression mouse model.13 Another recent study showed that sub-hypnotic acute doses (50 and 100 mg/kg) of ketamine switch the spontaneous excitatory activity across the neocortex by suppressing the active neurons while activating the previously silent neurons, paralleling its dissociative property.14 Acute ketamine administration also broadly impacts the dopaminergic modulatory system (via NMDAR antagonism15-17), resulting in increased firing in the ventral tegmental area (VTA) dopamine (DA)-producing neurons and enhanced DA release in the frontal cortex, striatum, and nucleus accumbens15-17 (Figure 1A).
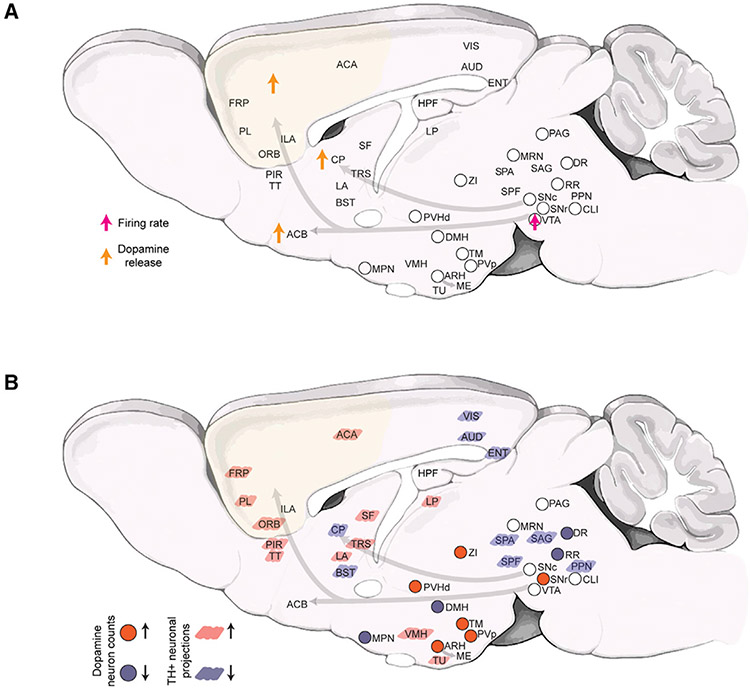
Figure 1. Brain-wide impact of ketamine exposure on the dopaminergic modulatory system.
A) Schematic summary of currently known alterations in dopaminergic neuronal activity and dopamine release after ketamine exposure.16,17
B) Schematic summary (this study) of the brain-wide alterations in the DA system after 10 days of repeated ketamine (30 and 100 mg/kg) exposure. Up and down arrows indicate increase and decrease, respectively. Note that, for the 30 mg/kg ketamine treatment, statistically significant decreases in TH+ neuron counts were only observed in DR, DMH, and MPN and increase in ZI and SNr. The TH+ neuronal projections changes are only shown for the 100 mg/kg (10 days) treatment. Abbreviations used are standard Allen Brain Atlas terms, as follows: ACA, anterior cingulate area; ACB, nucleus accumbens; ARH, arcuate hypothalamic nucleus; AUD, auditory areas; BST, bed nuclei of the stria terminalis; CLI, central linear nucleus raphe; CP, caudoputamen; DMH, dorsomedial nucleus of the hypothalamus; DR, dorsal nucleus raphe; ENT, entorhinal area; FRP, frontal pole, cerebral cortex; HPF, hippocampal formation; IF, interfascicular nucleus raphe; ILA, infralimbic area; LA, lateral amygdalar nucleus; LP, lateral posterior nucleus of the thalamus; MPN, medial preoptic nucleus; MRN, midbrain reticular nucleus; ORB, orbital area; PAG, periaqueductal gray; PIR, piriform area; PL, prelimbic area; PPN, pedunculopontine nucleus; PVH, paraventricular hypothalamic nucleus; PVp, paraventricular hypothalamic nucleus, posterior part; RR, midbrain reticular nucleus, retrorubral area; SAG, nucleus sagulum; SF, septofimbrial nucleus; SNc, substantia nigra, compact part; SPA, subparafascicular area; SPF, subparafascicular nucleus; TM, tuberomammillary nucleus; TRS, triangular nucleus of septum; TT, taenia tecta; TU, tuberal nucleus; VIS, visual areas; VMH, ventromedial hypothalamic nucleus; VTA, ventral tegmental area; ZI, zona incerta.
In contrast, the long-term impact of chronic ketamine exposure on brain networks remains much less understood, with profound scientific and clinical implications.18,19 The antidepressant effect of ketamine is known to be transient, necessitating long-term maintenance treatments,18 and recreational abuse at higher doses has been linked to cognitive and sensory impairments,20-22 as well as significant brain damage.23 Recent studies in mice have further revealed significant alterations in neocortical microcircuit synchrony after repeated exposure to ketamine.24,25 Therefore, given its broad clinical importance and increasing long-term abuse potential at higher doses, there is a considerable interest in understanding the molecular, cellular, and neural circuit adaptations caused by long-term exposure to ketamine over a wide range of doses.18,19
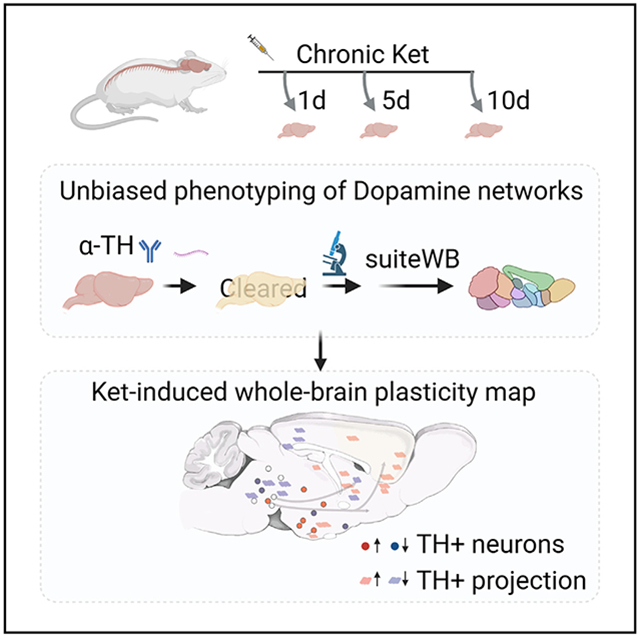
In this study, we aimed to comprehensively investigate the effects of chronic (R,S)-ketamine exposure on the entire dopaminergic system in mice. To achieve this, we utilized a range of doses (30 and 100 mg/kg),14 and we developed and employed a high-resolution whole-brain phenotyping approach to map the plasticity of the DA system in male mice. Our results reveal that chronic ketamine exposure leads to divergent and wide-spread alterations in the DA neuron populations and their long-range projections to the prefrontal cortex and sensory areas. We also uncover the role of post-transcriptional regulation mechanisms in modulating ketamine-induced structural plasticity in the dopaminergic system. Overall, our study provides a comprehensive and unbiased sub-cellular resolution mapping of the diverse effects of ketamine on the entire dopaminergic modulatory system in the male mouse brain (summarized in Figure 1B).
RESULTS
High-resolution whole-brain phenotyping of ketamine-treated animals
We established a complete pipeline for whole-brain labeling, high-resolution imaging, and comparative phenotyping of the entire dopaminergic modulatory system, and we utilized it to study the dose-dependent effects of daily (R,S)-ketamine exposure (30 and 100 mg/kg; 1, 5, and 10 days) and saline control intraperitoneal (i.p.) injections (Figure 2). To assess the cellular toxicity of ketamine exposure, we used α-activated caspase antibody staining after 10 days of 100 mg/kg daily i.p. injections. The results revealed no significant cell death in the brain (as shown in Figure S1). The locomotion of injected animals was video-recorded and quantified at 15′ and 60′ post injections (Figure S2). A dose of 30 mg/kg ketamine induced hyperlocomotion, while 100 mg/kg ketamine induced hypo-locomotion, as expected.26
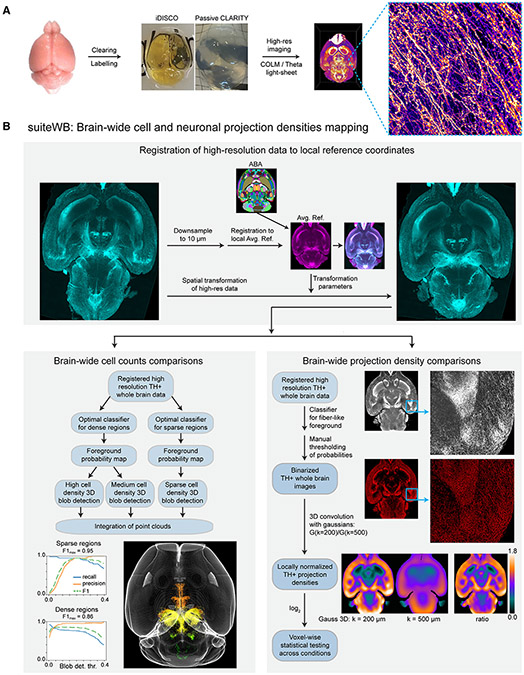
Figure 2. High-resolution whole-brain phenotyping of dopaminergic modulatory system.
A) Whole-brain mapping of TH+ neurons by optimized labeling (α-TH antibody staining, TH mRNA staining and/or TH-CreER x tdTomato signal), clearing (iDISCO+ or passive CLARITY) and high-resolution imaging with COLM or LSTM light sheet microscopy.
B) A suite of tools (suiteWB) for high-resolution comparative phenotyping of neuron counts and their brain-wide projection, including registration of high-resolution whole-brain images to a local average reference brain (with mapped ABA reference annotations), accurate brain-wide multi-model cell segmentation approach, and neuronal projections density estimation. F1 for sparse and dense region segmentations achieved are 0.95 and 0.86, respectively. See also Figures S1 and S2 and Videos S1-S4 for volume renderings of the raw data and segmentation results.
The extracted intact brains from treated animals were stained with α-tyrosine hydroxylase (TH), which is a rate-limiting enzyme for DA synthesis and a widely used marker of DA neurons27 (Figure 2, Video S1). Notably, although DA can be further converted to other catecholamines (norepinephrine and/or epinephrine) in downstream pathways, the distribution of these noradrenergic neurons is known to be localized within the hindbrain regions (pons and medulla).28,29 Nevertheless, due to their brain-wide projections (e.g., to cerebral cortex, hippocampus, amygdala, and hypothalamus), the precise identity of the TH+ neuronal projections (as DA or noradrenergic) in our datasets may only be inferred as catecholaminergic.28,29
To investigate the expression of TH mRNA, we established and utilized a whole-brain staining method based on hybridization chain reaction.30,31 Additionally, we used a well-characterized inducible TH-CreER line, crossed with a tdTomato reporter line, to indelibly label cells expressing TH mRNA (Figure S1C). The intact brain samples were cleared with either iDISCO+32 or passive CLARITY33 methods and imaged at high-resolution with COLM33 or light sheet theta microscopy (ClearScope/LSTM34) (Figure 2A).
Further, we developed a set of accurate large (~100 terabytes) data analysis methods (suiteWB, Figure 2B) for high-resolution phenotyping of the entire dopaminergic system. These methods allow for the analysis of both TH+ cell bodies and their brain-wide projections, as shown in Figure 2B. To this end, we first generated a local average reference (from seven brains), which was annotated by the registration of Allen Brain Atlas (ABA, ccfv335). Next, we developed a multi-model image segmentation approach to accurately detect the TH+ cell bodies and their brain-wide projections (Figure 2B, Video S3). Statistical comparisons were made using ANOVA and two-sided Mann-Whitney U tests with Bonferroni correction. Overall, the suiteWB methods allow for accurate high-resolution phenotyping of brain-wide structural plasticity of the dopaminergic system.
Dose-dependent divergent impact of chronic ketamine exposure on the dopamine system
We generated high-resolution whole-brain maps of TH+ neurons after 1, 5, and 10 days of daily i.p. injections of either ketamine or saline (Figures 3 and 4, Video S3). Using suiteWB, we counted TH+ neurons across all brain regions and compared the counts between treatment groups at different levels of the hierarchical ABA annotation tree (Figure 3A). We found that statistically significant alterations were only detected after 10 days of ketamine exposure for both 30 and 100 mg/kg ketamine treatment groups. The 1- and 5-day treatment datasets were not analyzed further. Additionally, we observed stronger changes in the 100 mg/kg treatment group compared to the 30 mg/kg treatment group (Figure 3).
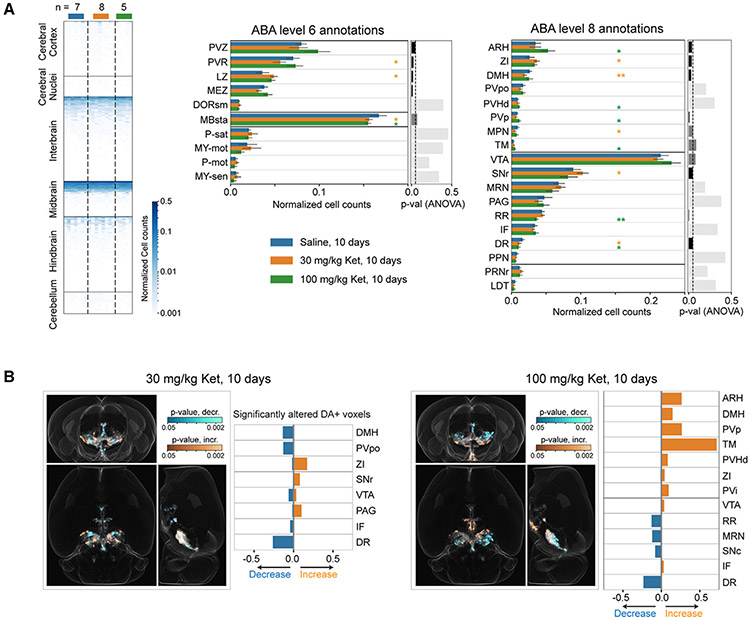
Figure 3. Repeated ketamine exposure results in divergent brain-wide changes in the dopaminergic modulatory system.
A) Brain-wide heatmap of the TH+ cell counts in ABA ROIs after 10 days ketamine (30 mg/kg and 100 mg/kg) and saline (Sal) i.p. injections. Independent biological replicates of n = 7 for Sal-, 8 for 30 mg/kg ketamine-, and 5 for 100 mg/kg ketamine-treated groups. The regional TH+ neuron counts were normalized by respective total number of TH+ neurons in brain. Orange, green, and blue bars quantify TH+ cells in 30 mg/kg ketamine-, 100 mg/kg ketamine-, and saline-treated brains, respectively. Data are represented as mean ±1.96 SD (95% confidence interval). Two-sided Mann-Whitney U tests with Bonferroni correction were performed to calculate the statistical significance. * and ** indicate p values <0.05 and <0.01, respectively. ANOVA tests p values are plotted as bar plots on the right.
B) Whole-brain orthogonal projections visualizing voxel-by-voxel differences in TH+ neuron densities after 10 days of 30 mg/kg ketamine (n = 8) and 100 mg/kg ketamine (n = 5), compared with saline (n = 7) controls. Cyan and orange visualize p values (two-sided Mann-Whitney U test) for decreases and increases, respectively. Note that the decreases and increases in the densities are identified by comparing the sample averages of the treated and control groups. The bar plots quantify the fraction of DA neurons containing voxels (within an ABA ROI) that show significant alterations.
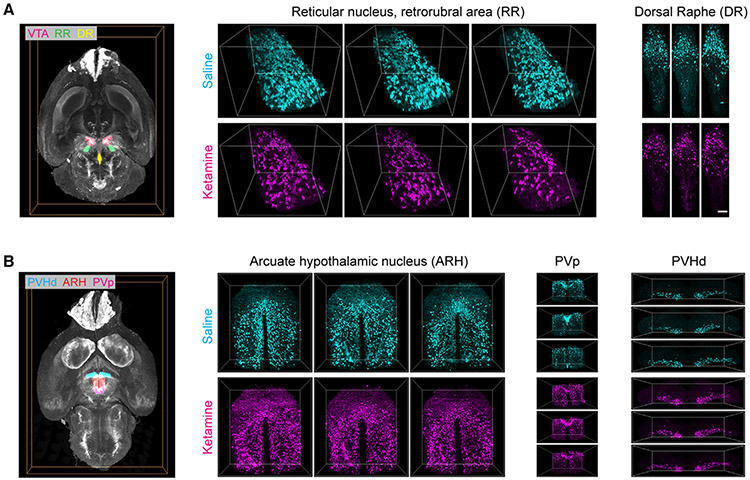
Figure 4. Examples of ketamine-induced changes in midbrain and hypothalamic dopamine domains after repeated ketamine exposure.
Representative volume renderings from three different ketamine- (100 mg/kg, 10 days) and saline-treated brains, illustrating decreases (A) in RR (reticular nucleus, retrorubral area) and DR (dorsal raphe) and increases (B) in ARH (arcuate hypothalamic nucleus), PVp (periventricular hypothalamic nucleus, posterior part), and PVHd (periventricular hypothalamic nucleus, descending division). Whole-brain renderings on the left visualize the spatial locations of these regions. Bounding boxes for RR, ARH, PVp, and PVHd are 617.5 × 500 × 595 μm3, 950 × 1,000 × 490 μm3, 910 × 470 × 630 μm3, and 1,800 × 490 × 350 μm3 respectively. Scale bar represents 100 μm.
At a higher-level annotation (i.e., larger brain regions of interest [ROIs]; level 6), we found an overall dose-dependent statistically significant decrease in TH+ neuron counts within the behavior-state-related midbrain regions (MBsta) and, conversely, an overall increase within the hypothalamic lateral zone in both the 30 and 100 mg/kg (10 days) treatment groups (Figure 3A). In addition, for the 100 mg/kg group, we observed a tendency for increase within the hypothalamic periventricular zone (PVZ) and, for the 30 mg/kg group, significant decrease within the periventricular region (PVR) (Figure 3A). Next, we compared the brains at lower-level (level 8) graph cut of the ABA annotation tree. For both 30 and 100 mg/kg treatment groups, we observed a robust decrease in the dorsal raphe (DR36) and increase in the lateral hypothalamic region zona incerta (ZI37). In addition, in the 100 mg/kg treatment group, we observed a robust decrease in the reticular nucleus retrorubral area (RR38) and increases in the arcuate hypothalamic nucleus (ARH39), the periventricular hypothalamic nucleus posterior part (PVp), tuberomammillary nucleus (TM), and the periventricular hypothalamic nucleus descending division (PVHd) (Figure 3A). Whereas, in the 30 mg/kg treatment group, we found significant increases in midbrain region SNr (substantia nigra reticular part) and a significant decrease in the dorsomedial nucleus of the hypothalamus (DMH) and the medial hypothalamic region MPN40 (preoptic nucleus). These brain-wide alterations were further validated by an independent voxels-based (20 × 20 × 20 μm3 sampling) brain parcellation approach (Figure 3B). Representative example volume renderings are shown for RR, DR, ARH, PVp, and PVHd (Figure 4, 100 mg/kg ketamine-treated group).
Overall, through an unbiased whole-brain analysis, we found that the impact of chronic ketamine exposure on TH+ neuron counts was both divergent and dosage dependent (Figure 1B). Specifically, the chronic ketamine exposure led to an increase in TH+ neuron counts in multiple hypothalamic domains, such as the ARH, which contains TH+ DA neurons that play a role in energy homeostasis.39 However, chronic ketamine exposure also led to decreases in TH+ neuron counts within midbrain regions that are associated with behavioral states, such as the DR, which contains TH+ DA neurons that modulate social isolation behaviors,36 and the RR, which contains TH+ DA neurons involved in fear and aversive signaling.38
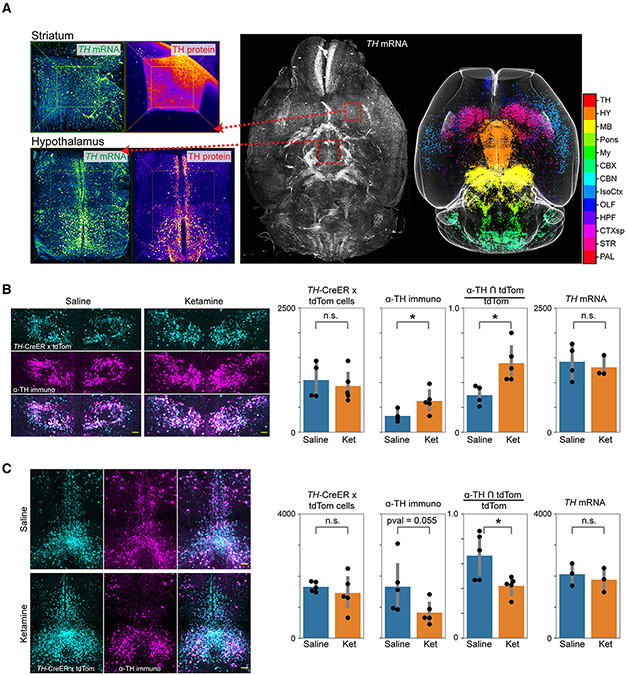
Untranslated TH mRNA+ neurons facilitate ketamine-induced cellular plasticity
We sought to further investigate the mechanistic basis of chronic ketamine-induced brain-wide cellular plasticity. Utilizing our whole-brain mRNA labeling protocol, we first mapped the expression of TH mRNA (Figure 5A, Video S4) and found that the expression was much broader than the corresponding TH protein (Video S5). This suggested the potential role of post-transcriptional mechanisms in the brain-wide modulation of the DA system. To test this hypothesis further, we utilized an inducible TH-CreER transgenic line41 (Figure S1C) crossed with a tdTomato reporter line to investigate if the newly acquired TH+ neurons originated from untranslated TH mRNA+ neurons. TH-CreER induction (by 4-OHT i.p. injections) was performed 1 week prior to the start of the ketamine exposure to permanently label the pre-treatment TH mRNA+ neurons by tdTomato. After 10 days of chronic ketamine (100 mg/kg) exposure, the brains were harvested, cleared with passive CLARITY,33 and labeled with α-TH antibody for a direct comparison of after-treatment TH+ neurons with before-treatment TH mRNA+ neurons within the same brain samples. As shown in Figure 5B, we found a significant increase in TH protein+/tdTomato+ co-labeled neurons (α-TH ∩ tdTomato) in the ketamine-treated group compared to the saline controls, while the overall number of tdTomato+ neurons remained unchanged, consistent with unchanged TH mRNA+ neurons (Figure 5B). Conversely, we observed a decrease in TH protein+/tdTomato+ co-labeled neurons in the midbrain regions (which showed reduction in TH+ DA neuron counts), while the tdTomato+ and TH mRNA+ neuron counts remain unchanged compared to the saline controls (Figure 5C). Altogether, these results suggest that the cellular plasticity in the DA system may be facilitated by the existence of much larger pools of untranslated TH mRNA+ neurons to rapidly modulate the number of available TH+ DA neurons in various brain regions.
Figure 5. Untranslated TH mRNA+ neurons facilitate adaptations in the dopamine system.
(A) Whole-brain mapping of TH mRNA, showing much broader expression than the TH protein. Color code corresponds to different ABA ROIs, as listed in adjoining color bar.
(B and C) Co-labeling of TH-CreER x tdTomato signal induced with 4-OHT injections 1 week before the start of ketamine i.p. injections and α-TH immunostaining signal capturing TH protein expression post-treatment (10 days). Left to right, bar plots compare the number of tdTomato+ neurons, α-TH immunostained neurons, their intersection, and the number of TH mRNA+ neurons. (B) and (C) show field of views that include paraventricular hypothalamic nucleus and dorsal raphe regions, respectively. Data are represented as mean ±1.96 SD (95% confidence interval). Two-sided Mann-Whitney U tests were performed for statistical significance calculations. * indicates <0.05 p value. All scale bars represent 100 μm. See also Videos S4 and S5 for whole-brain volumetric renderings.
Altered long-range TH+ projections after chronic ketamine exposure
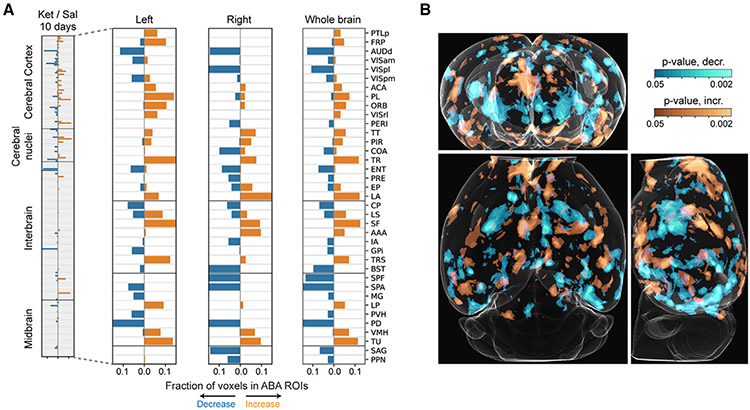
Taking advantage of the high-resolution of our datasets, we sought to map the brain-wide changes in TH+ neuronal projections in response to chronic ketamine (100 mg/kg, 10 days) exposure. TH+ neuronal projections may be dopaminergic or noradrenergic, but they can only be inferred as catecholaminergic28,29 in this study. We used the suiteWB pipeline to estimate the TH+ neuronal projection densities in the ketamine-treated and saline control whole-brain datasets and performed voxel-by-voxel (at 25 × 25 × 25 μm3 sampling) statistical comparisons between the groups. As shown in Figures 6 and 7 and Video S6, chronic ketamine exposure resulted in robust brain-wide changes in TH+ projections. Specifically, we observed increased densities in multiple associative cortical regions including the prefrontal cortex (PFC)-related prelimbic area (PL, Video S7, Figures 6 and 7), orbital area (ORB, Video S8, Figures 6 and 7), frontal pole (FRP, Figure 6A), and anterior cingulate area (ACA, Figure 6A), as well as the posterior parietal association area (PTLp, Figure 6A). The lateral amygdala (LA, Video S9, Figure 7A), which is involved in processing of threatening stimuli and fear behavior, and specific regions of the lateral septal complex (septofimbrial nucleus [SF], involved in reward and reinforcement) and the triangular nucleus of septum (TRS) also showed increased projection densities (Figure 6A). Brain regions receiving olfactory inputs, including the taenia tecta (TT) and piriform area (PIR), and regions of the thalamus (lateral posterior nucleus of the thalamus [LP]) and hypothalamus (VMH and TU) also showed increases in TH+ neuronal innervations.
Figure 6. Brain-wide changes in TH+ neuronal projections after repeated ketamine exposure.
(A) Stacked barplots of fractions of ABA ROI voxels with significantly increased (orange) and decreased (blue) TH+ neuronal projection densities after 10 days of ketamine (100 mg/kg) exposure. Independent biological replicates are as follows: n (Ket, 10 days) = 5, n (Sal, 10 days) = 7. (B) Whole-brain orthogonal projections visualizing voxel-by-voxel differences (independent of ABA) in TH+ neuronal projection densities. Orange and cyan represent significant increases and decreases, respectively. See also Video S6 for rendering of heatmaps.
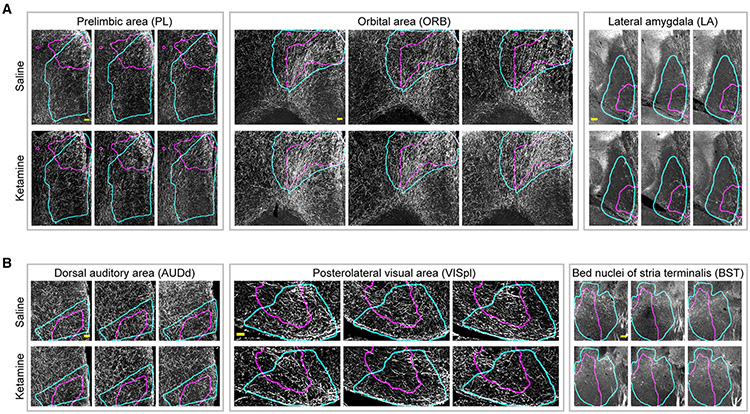
Figure 7. Examples of ketamine-induced alterations in TH+ projections after repeated ketamine exposure.
Representative examples of ABA ROIs showing increases (A) and decreases (B) in TH+ neuronal projections after 10 days of daily ketamine (100 mg/kg) exposure. Cyan and magenta curves outline the spatial location of ABA ROIs and areas with significantly changed projection densities respectively. Images shown are representative single optical planes. See also Videos S7-S12 for complete volumes. All scale bars represent 100 μm.
In contrast, certain regions exhibited decreased TH+ projection densities in response to chronic ketamine exposure, including the dorsal auditory area (AUDd, Figures 6A and 7B, Video S10), postero-lateral visual area (VISpl, Figure 7B, Video S11), entorhinal area (ENT), and presubiculum (PRE, involved in spatial information processing; Figure 6A). Striatum sub-regions caudoputamen (Cp) and bed nuclei of the stria terminalis (BST/BNST, Figures 6A and 7B, Video S12) showed reductions, consistent with the loss of TH+ neurons in DR, RR, and SNc (Figure 3). Further, thalamic regions subparafascicular area (SPA) and subparafascicular nucleus (SPF) and midbrain regions nucleus sagulum (SAG) and pedunculopontine nucleus (PPN) also showed decreased projections.
Overall, consistent with the divergent changes in TH+ neuron counts in the midbrain and hypothalamic dopaminergic domains (Figures 3 and 4), chronic ketamine exposure resulted in increased TH+ neuronal projection densities in the associative brain centers, including PFC-related regions, and decreased innervations in regions involved in visual, auditory, and spatial information processing (Figures 6 and 7).
DISCUSSION
We used an unbiased, high-resolution whole-brain mapping approach to systematically investigate the adaptability of the DA system to repeated ketamine exposure (summarized in Figure 1B). We found that chronic ketamine exposure leads to a dosage-dependent decrease in DA neurons counts within midbrain regions related to behavior state and increase within hypothalamic domains, along with altered long-range innervation of the association and sensory areas by TH+ neuronal projections. Such structural plasticity of brain-wide modulatory system may facilitate significant reconfiguration of the neuronal networks to eventually result in long-lasting cognitive behavioral changes. Note that, although DA may further get processed in the noradrenergic neurons, the distribution of these neurons is well known to be restricted to specific domains in hindbrain regions.28,29 However, the TH+ neuronal projections may still only be precisely identified as DA or noradrenergic by TH immunolabeling.28,29 In addition, the loss of TH protein in DA neurons may not result in a permanent loss of DA neurons; nevertheless, it does indicate the loss of DA-producing capacity due to the required role of TH in the L-DOPA (DA precursor) biosynthesis.
We also show that the observed adaptability of the DA system is facilitated by a large pool of neurons that stably maintain translationally suppressed TH mRNA (Figure 5). These findings were validated by a direct comparison of the newly recruited or lost TH+ DA neurons with the before-exposure TH mRNA expressing neuron populations by utilizing an inducible TH-CreER transgenic mouse line (Figure 5). Mechanistically, these observed changes in DA system may, in part, be the result of repeated ketamine-induced activation/deactivation (indirect, via NMDAR antagonism) of the DA neurons due to the heterogeneous neuronal inputs. Intriguingly, the ketamine-induced cellular plasticity is distinct from the previously reported neurotransmitter phenotypic plasticity of the hypothalamic DA neurons after changed day/night durations in rodents and included loss of TH mRNA expression.42 Overall, our data suggest that the maintenance of translationally suppressed mRNA, even though an energetically costly process, may allow for faster brain-wide adaptations to various external stresses.
Overall, this study sheds light on the brain-wide effects of chronic ketamine exposure on the dopamine system and provides insight into the structural plasticity that underlies these effects. The finding that ketamine exposure leads to divergent alterations in specific brain regions, rather than a uniform activating impact, is particularly intriguing and could have significant implications for the development of treatments for depression, schizophrenia, and psychosis. Moreover, such non-monolithic brain-wide impact further underscores the need for unbiased investigations of on/off-target effects of ketamine treatment at a range of doses, as well as the urgency to develop targeted pharmacological intervention approaches (e.g., focused ultra sound-based approaches43,44) for treatment of complex brain disorders.
Limitations of the study
This study also has a few limitations. Firstly, the study did not examine the progressive changes in TH expression levels over varying durations of ketamine exposure. Instead, a binary approach of presence or absence was employed. In future research, the development of strategies for whole-brain expression signal normalization will be essential to enable a more comprehensive analysis of the gradual effects of prolonged ketamine exposure on the DA system. Secondly, it’s crucial to note that TH is not exclusive to dopaminergic neurons but is also present in other catecholaminergic neurons, such as norepinephrine and epinephrine. While it is generally accepted that these neurons are primarily localized in specific regions of the hindbrain, some of the neuronal projections labeled as TH+ in this study may potentially originate from these other catecholaminergic neurons. Future studies will be necessary to precisely determine the neurotransmitter identity (whether DA or noradrenergic) of the affected neuronal projections. Thirdly, this study does not provide specific insights into the physiological mechanisms that may be mediating the impact of chronic ketamine exposure on the DA system. Fourthly, to ensure the practical feasibility of performing sub-cellular resolution comparative phenotyping, this initial whole-brain study utilized male mice to minimize potential sources of variability. Future research, building upon these initial important findings and tools, will be needed to explore any potential sex-specific impact of ketamine exposure.45 Lastly, it should be noted that the current study employed a mixture of (R,S) enantiomers of ketamine. Future follow-up studies will be necessary to understand the relative impacts of the two ketamine enantiomers.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Raju Tomer (raju.tomer@columbia.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Microscopy data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at https://doi.org/10.5281/zenodo.10067746 and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Sheep anti-Tyrosine hydroxylase | abcam | Ab113 |
| Rabbit anti-Active Caspase −3 | BD Biosciences | AB_397274 |
| DAPI (4′,6-diamidino-2-phenylindole) | VWR | 89139-054 |
| Donkey anti-Sheep Alexa Fluor 647 | ThermoFisher Scientific | A-21448 |
| Goat anti-Rabbit Alexa Fluor 647 | ThermoFisher Scientific | A-21245 |
| Chemicals, peptides, and recombinant proteins | ||
| (R,S)-Ketamine Hydrochloride | Covetrus | 071069 |
| 40% Acrylamide | Bio-Rad | 1610140 |
| 2% Bisacrylamide | Bio-Rad | 1610142 |
| VA-044 | Wako Chemicals | NC0632395 |
| Paraformaldehyde | Electron Microscopy Sciences | 15710-S |
| Boric Acid | Sigma-Aldrich | B7901 |
| Sodium dodecyl sulfate | Amresco | 0837 |
| RapiClear RI = 1.47 | SunJin lab | RC147002 |
| Methanol | Fisher Scientific | AC423950040 |
| Dichloromethane | Sigma-Aldrich | 270997 |
| Hydrogen peroxide | Sigma-Aldrich | 216763 |
| Dibenzyl ether | Sigma-Aldrich | 33630 |
| Donkey serum | Millipore Sigma | D9663 |
| Bovine serum albumin | Fisher Scientific | BP1600-100 |
| Triton X-100 | Sigma-Aldrich | T8787 |
| Dimethyl sulfoxide | Sigma-Aldrich | 472301 |
| Glycine | Sigma-Aldrich | G7126 |
| 4 – hydroxytamoxifen | Sigma-Aldrich | H7904 |
| Sodium chloride | Sigma-Aldrich | S9625 |
| Corn oil | Sigma-Aldrich | C8267 |
| 20x SSC, RNase-free | ThermoFisher Scientific | AM9765 |
| Acetic anhydride | Millipore Sigma | 320102 |
| Formamide deionized | Millipore Sigma | S4117 |
| Yeast tRNA | Fisher Scientific | AM7119 |
| Dextran sulfate | Millipore Sigma | 265152 |
| Citric acid | Sigma-Aldrich | 251275 |
| Tween 20 | Sigma-Aldrich | P9416 |
| PBS - Phosphate-Buffered Saline (10X) pH 7.4, RNase-free | ThermoFisher Scientific | AM9624 |
| Experimental models: Organisms/strains | ||
| TH-2a-CreER | David Ginty’s lab; Abraira et al.41 | TH-2a-CreER |
| B6; 129S6-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J (Ai14) | The Jackson Laboratory | Strain #:007908 |
| Oligonucleotides | ||
| HCR Amplifier: B1, Alexa 647 | Molecular Instruments | N/A |
| HCR Probes for Tyrosine Hydroxylase mRNA detection | IDT | See Table S1 |
| Software and algorithms | ||
| Python 3.7 | Python Software Foundation | https://www.python.org/ |
| ImageJ | NIH | https://imagej.net/ij/index.html |
| Terastitcher | Bria et al.46 | https://abria.github.io/TeraStitcher/ |
| Bigstitcher | David Hörl et al.47 | https://imagej.net/plugins/bigstitcher/ |
| ITK | Kitware, Inc | https://itk.org/ |
| suiteWB | This study | https://github.com/tomerlab/suiteWB |
| Napari | Chan Zuckerberg Initiative | https://napari.org/stable/index.html |
| Amira | Thermo Fisher Scientific | https://www.thermofisher.com/us/en/home/electron-microscopy/products/software-em-3d-vis/amira-software.html |
| MATLAB | Mathworks | https://www.mathworks.com/products/matlab.html |
| suiteWB | This study | https://doi.org/10.5281/zenodo.10067746 |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Animals
Male TH-2a-CreER41 mice were acquired from Dr. David Ginty’s lab and were bred with B6; 129S6-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J (Ai14; JAX Strain #:007908). All mice were group-housed in a 12:12 light:dark cycle at 22°C. All the experiments were performed with TH-CreER x Ai14 male (8–10 weeks old) mice. Food and water were provided ad libitum. (R,S)-ketamine i.p. injections were performed during the light phase. All experimental procedures were approved by the IACUC at Columbia University. Locomotor activity was recorded after 1, 5, and 10 days of treatment.
METHOD DETAILS
Drugs
4-hydroxytamoxifen (4-OHT; Sigma, H7904) was dissolved in corn oil/ethanol (90% corn oil, 10% ethanol) via the use of a vortex, ultrasonication, and 55°C heating for <15 min. Mice were intraperitoneally injected with 2 mg of 4-OHT. (R,S)-ketamine (Covetrus) was used for all ketamine exposure experiments. 3 and 10 mg/mL stocks were prepared in saline (0.9% NaCl).48 One dose of 30 mg/kg or 100 mg/kg of (R,S)-ketamine was intraperitoneally injected in a 24 h period for 1 day, 5 days, or 10 days treated animals. All mice were euthanized 24 h after the last ketamine injection.
Behavioral recording
The mice were placed within a novel home cage and were recorded (camera: GoHZQ, 1920 x 1080 pixels,30 fps transmission rate) 15 min and 1-h post-intraperitoneal ketamine injections. The total distance traveled was quantified, over 3 min duration, using the ANY-maze tracking software (ANY-maze, RRID:SCR_014289, Stoelting, Wood Dale, IL, United States).
Whole brain clearing and labeling
For iDISCO clearing, the iDISCO+ protocol was followed as previously described.32 The brains were pre-treated with methanol, placed in 66% dichloromethane/33% methanol overnight, bleached with 5% H2O2/methanol, and then rehydrated. Afterward, the whole brains were permeabilized for 2 days at 37°C in 1x phosphate buffered saline (PBS), 0.2% Triton X-100, 0.3 M glycine, and 20% dimethyl sulfoxide (DMSO). The brains were blocked with 1xPBS, 0.2% Triton X-100, 10% DMSO, 6% donkey serum for 2 days at 37°C, followed by incubation in 1xPBS, 0.1% Triton X-100, 3% donkey serum, and a 1:200-1:500 dilution of the primary antibody sheep α-TH (ab113, Abcam) for 10 days. After washing in 1xPBS/0.1% Triton X-100, the brains were placed in the secondary antibody solution containing 1xPBS/0.1% Triton X-100/3% donkey serum and a 1:1000 dilution of donkey anti-sheep 647 (A-21448, Thermofisher) for 10 days.
We used the passive CLARITY method as detailed previously.33 The tissue was first encapsulated in a hydrogel monomer (HM) solution consisting of 1% (w/v) acrylamide, 0.05% (w/v) bisacrylamide, 4% paraformaldehyde, 1x phosphate buffered saline (1xPBS), deionized water, and 0.25% of thermal initiator (VA-044, Fisher Scientific), followed by clearing with SBC buffer (4% (w/v) SDS, 0.2 M boric acid, pH 8.5 and deionized water) at 37°C with shaking. The SBC buffer was replaced every 2 days. After clearing, the SBC buffer was washed off with 0.2 M boric acid pH 8.5 with 0.1% Triton X-100. The cleared tissue was immunostained in 0.2 M boric acid pH 7.5 with 0.1% Triton X-100. The final refractive index matching was performed in RapiClear (SunJin lab, RI = 1.47).
For HCR-FISH, we used split-initiator DNA probes (IDT, Table S1) for detecting the tyrosine hydroxylase (Th, NM_012740.3) mRNA. During probe hybridization/detection, the brains underwent 1) equilibration with 5xSSCTw buffer, 2) acetylation with 0.25% acetic anhydride solution, 3) equilibration with probe hybridization buffer (30% formamide, 5xSSC, 0.5 mg/mL yeast tRNA, 10% dextran sulfate), and 4) probe incubation (50 nM) in probe hybridization buffer at 37C. The time/amount of solution varied depending on the thickness of the brain slices. The slices were then washed in probe wash buffer (30% formamide, 5X SSC, 9 mM citric acid, and 0.1% tween 20) at 37°C as well as two rounds of washes in 5xSSCTw. Afterward, the slices were equilibrated in amplification buffer (5xSSC, 10% dextran sulfate, and 0.1% Tween 20), and then incubated in amplification buffer with 50–150 nM of hairpins H1 and H2 conjugated with AF-647 (Molecular Instruments). The hairpin/amplification buffer mixture was then washed off with several rounds of 5xSSCTw.
For activated caspase staining, the TH-2a-CreER; Ai14 brains were sliced with a vibratome (Leica VT1000 S Vibrating blade microtome) into 50 μm sections. These sections were placed in a blocking buffer (1xPBS/0.1% bovine serum albumin/0.1% Triton X-100) for 30 min and then incubated in a 1:200-1:400 dilution of the primary antibody, rabbit anti-active caspase-3 (BD Biosciences, catalog no. 559565), with 1xPBS/0.5% bovine serum albumin/0.1% Triton X-100 overnight. After washing in 1xPBS/0.1% Triton X-100, the sections were incubated, for 2 h, in a 1:500 dilution of the secondary antibody, goat anti-rabbit 647 (ThermoFisher, Catalog #A-21245) with 1xPBS/0.5% bovine serum albumin/0.1% Triton X-100. The secondary antibody was washed off in 1xPBS/0.1% Triton X-100, and the slices were incubated in a 1:1000 dilution of a 1 mg/mL stock of DAPI with 1xPBS for 15 min. After washing off with 1xPBS/0.1% Triton X-100, the slices were mounted in 65% glycerol.
Imaging
All imaging experiments were performed with CLARITY-optimized light sheet microscopy (COLM)33 or ClearScope (MBF Biosciences)34 Olympus 10x/0.6NA/8 mmWD or ASI 16x/12mmWD detection objectives were used for most of the whole brain imaging with COLM. Olympus Macro 4×/0.28NA or Nikon 20x/1.0NA were used for ClearScope imaging.
QUANTIFICATION AND STATISTICAL ANALYSIS
suiteWB: Image registration
We first developed a fast and efficient multistep multiresolution 3D image registration tool by using Mutual Information49 as the similarity metric. The registration starts by rigid transformation (with 6 degrees of freedom for rotation and translation) of the moving image (i.e., the image being aligned) to roughly align with the reference image. Next, the moving image is affine transformed (12 degrees of freedom) to account for shearing and shrinking artifacts introduced by labeling, tissue clearing and imaging. Finally, we used a uniform grid of control points and third-order B-splines are used for non-rigid local transformations. Affine and nonrigid steps were done at three different resolutions. The algorithms were implemented with ITKv450 registration framework. All α-TH stained whole brain images were registered at 10 μm resolution to a local average reference brain (generated from 7 α-TH whole brain images), and the resulting spatial transformation parameters were applied to high-resolution datasets. ABA annotations were registered one time to the local average reference brain.
suiteWB: Multi-model segmentation pipeline
Segmentation of whole brain images presents unique challenges of high spatial variations in signal-noise ratios (SNR), object densities and varied image artifacts. Multiple open-source tools exist for cell segmentation,51-53 however, some of them are optimized for confocal images with smaller data sizes, others utilize deep learning models with pre-trained parameters that do not accurately generalize to images with different signal quality distribution and also require dense annotation training datasets. We chose to develop a semi-supervised multi-model learning approach to address the challenges of whole brain segmentation. This approach does not require dense annotated training and also allows the flexibility of using different optimal parameter sets for different regions (i.e., with dense or sparse object densities). We utilized open-source toolkit ilastik54 for semi-supervised learning of classifiers based on image features.
The high-resolution registered whole brain images were split into densely and sparsely populated regions by visual inspection, followed by pixel classification to generate probability maps. For the sparse region, a standard pixel classification workflow in ilastik was applied to generate cell probability maps. First, image features were extracted by using multiple types of filters with different kernel sizes, including intensity filters (Gaussian Smoothing), edge detection filters (Laplacian of Gaussian, Gaussian Gradient Magnitude, Difference of Gaussians), and texture detection filters (Structure Tensor Eigenvalues, Hessian of Gaussian Eigenvalues). The image features were then used to estimate the probabilities of pixels belonging to a cell or background. For denser regions, we utilized the Autocontext workflow in ilastik. In the first step, the image pixels were classified into five categories: empty space, brain background, fibers (neuronal projections), cell cytoplasm and cell nucleus. In the second step, the classification results were combined with image features (with filters described above for sparse regions) to generate probability maps of cells against background. Next, we applied blob detection on the dense and sparse probability maps separately to detect cells. This was achieved by using the difference of Gaussian (DOG) algorithm (Python scikit-image package.55 To optimize memory usage, images were processed block-wise with carefully resolved boundary conditions. Cells detected in dense and sparse regions were merged and were further tuned by using Napari viewer. F1 for sparse and dense region segmentations reached 0.95 and 0.86, with precision 0.889 and 0.976 respectively, compared to human-annotated datasets (Figure 2). For comparison, ClearMap,51 applied to immediate-early gene datasets, reported a precision of 0.83 and 0.75 compared to two human annotators.
For colocalization analysis of the multi-channel (i.e., tdTomato and α-TH immunostaining signal) CLARITY images, cell detection (as described above) was applied separately to different channels. The full cell body labels were generated by taking the union of the detected cells in the two channels that were less than 25μm apart. The cell identities (i.e., if expressing tdTomato or α-TH or both) were assigned by manually validated thresholding of probability maps of each channel.
Neuronal projection segmentation was performed by semi-supervised pixel classification by using image features (as discussed above) to generate the probability maps distinguishing projections and background. The probability threshold for binarization was determined by careful inspection throughout various regions of the brain. The binarized data was then convolved with 3D Gaussian kernels of 200 μm and 500 μm. The ratio of the two gaussians resulted in locally normalized neuronal projection densities.
Statistical comparisons between Ketamine and Saline treated samples were done with two-sided Mann Whitney U test (using Python package Scipy56) on normalized cell counts (cell count in a region divided by total cell count of that whole brain) and cell densities (estimated by 3D Gaussian kernel of size 50 μm in each dimension).
Supplementary Material
Highlights.
Whole-brain mapping of chronic ketamine-induced sub-cellular plasticity in dopamine system
Dose-dependent divergent changes in midbrain and hypothalamic dopamine domains
Divergently altered innervation of prefrontal, striatum, and sensory cortices
Data supporting post-transcriptional regulation of ketamine-induced structural plasticity
ACKNOWLEDGMENTS
We are grateful to Jordan Hamm for critically reading the manuscript and to Christine Denny, Rae Silver, and Darcy Kelley for advice throughout the project. We thank Vivek Kumar for advice on the HCR probe design and general discussions. We are grateful to Briana Chen, Christine Denny, Andre Toussaint, and Ishmail Abdus-Saboor for help with the ANY-maze software. This work was supported by NIH grant DP2MH119423 and Columbia University Arts & Sciences startup grant to R.T.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
DECLARATION OF GENERATIVE AI AND AI-ASSISTED TECHNOLOGIES IN THE WRITING PROCESS
During the preparation of this work the author(s) used ChatGPT (OpenAI) in order to proof-read the text. After using this tool/service, the author(s) reviewed and edited the content as needed and take(s) full responsibility for the content of the publication.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.113491.
REFERENCES
- 1.Bell RF, and Kalso EA (2018). Ketamine for pain management. Pain Rep. 3, e674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman RM, Cappiello A, Anand A, Oren DA, Heninger GR, Charney DS, and Krystal JH (2000). Antidepressant effects of ketamine in depressed patients. Biol. Psychiatr 47, 351–354. [DOI] [PubMed] [Google Scholar]
- 3.Ibrahim L, DiazGranados N, Franco-Chaves J, Brutsche N, Henter ID, Kronstein P, Moaddel R, Wainer I, Luckenbaugh DA, Manji HK, and Zarate CA Jr. (2012). Course of Improvement in Depressive Symptoms to a Single Intravenous Infusion of Ketamine vs Add-on Riluzole: Results from a 4-Week, Double-Blind, Placebo-Controlled Study. Neuropsychopharmacology 37, 1526–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zarate CA, Singh JB, Carlson PJ, Brutsche NE, Ameli R, Luckenbaugh DA, Charney DS, and Manji HK (2006). A randomized trial of an N-methyl-D-aspartate antagonist in treatment-resistant major depression. Arch. Gen. Psychiatr 63, 856–864. [DOI] [PubMed] [Google Scholar]
- 5.Daly EJ, Singh JB, Fedgchin M, Cooper K, Lim P, Shelton RC, Thase ME, Winokur A, Van Nueten L, Manji H, and Drevets WC (2018). Efficacy and Safety of Intranasal Esketamine Adjunctive to Oral Antidepressant Therapy in Treatment-Resistant Depression: A Randomized Clinical Trial. JAMA Psychiatr. 75, 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zanos P, Moaddel R, Morris PJ, Georgiou P, Fischell J, Elmer GI, Alkondon M, Yuan P, Pribut HJ, Singh NS, et al. (2016). NMDAR inhibition-independent antidepressant actions of ketamine metabolites. Nature 533, 481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olney JW, Newcomer JW, and Farber NB (1999). NMDA receptor hypofunction model of schizophrenia. J. Psychiatr. Res 33, 523–533. [DOI] [PubMed] [Google Scholar]
- 8.Franks NP, and Lieb WR (1994). Molecular and cellular mechanisms of general anaesthesia. Nature 367, 607–614. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, Shu S, and Bayliss DA (2009). HCN1 Channel Subunits Are a Molecular Substrate for Hypnotic Actions of Ketamine. J. Neurosci 29, 600–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou C, Douglas JE, Kumar NN, Shu S, Bayliss DA, and Chen X. (2013). Forebrain HCN1 channels contribute to hypnotic actions of ketamine. Anesthesiology 118, 785–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sleigh J, Harvey M, Voss L, and Denny B (2014). Ketamine – More mechanisms of action than just NMDA blockade. Trends in Anaesthesia and Critical Care 4, 76–81. [Google Scholar]
- 12.Gupta A, Devi LA, and Gomes I (2011). Potentiation of μ-opioid receptor-mediated signaling by ketamine. J. Neurochem 119, 294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moda-Sava RN, Murdock MH, Parekh PK, Fetcho RN, Huang BS, Huynh TN, Witztum J, Shaver DC, Rosenthal DL, Alway EJ, et al. (2019). Sustained rescue of prefrontal circuit dysfunction by antidepressant-induced spine formation. Science 364, eaat8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cichon J, Wasilczuk AZ, Looger LL, Contreras D, Kelz MB, and Proekt A (2023). Ketamine triggers a switch in excitatory neuronal activity across neocortex. Nat. Neurosci 26, 39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iro CM, Hamati R, El Mansari M, and Blier P (2021). Repeated but Not Single Administration of Ketamine Prolongs Increases of the Firing Activity of Norepinephrine and Dopamine Neurons. Int. J. Neuropsychopharmacol 24, 570–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kokkinou M, Ashok AH, and Howes OD (2018). The effects of ketamine on dopaminergic function: meta-analysis and review of the implications for neuropsychiatric disorders. Mol. Psychiatr 23, 59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simmler LD, Li Y, Hadjas LC, Hiver A, van Zessen R, and Lüscher C (2022). Dual action of ketamine confines addiction liability. Nature 608, 368–373. [DOI] [PubMed] [Google Scholar]
- 18.Smith-Apeldoorn SY, Veraart JK, Spijker J, Kamphuis J, and Schoevers RA (2022). Maintenance ketamine treatment for depression: a systematic review of efficacy, safety, and tolerability. Lancet Psychiatr. 9, 907–921. [DOI] [PubMed] [Google Scholar]
- 19.Short B, Fong J, Galvez V, Shelker W, and Loo CK (2018). Side-effects associated with ketamine use in depression: a systematic review. Lancet Psychiatr. 5, 65–78. [DOI] [PubMed] [Google Scholar]
- 20.Morgan CJA, and Curran HV Drugs (ISCD); the IIndependent Scientific Committee on Drugs (2012). Ketamine use: a review. Addiction 107, 27–38. [DOI] [PubMed] [Google Scholar]
- 21.Ke X, Ding Y, Xu K, He H, Wang D, Deng X, Zhang X, Zhou Y, Zhou C, Liu Y, et al. (2018). The profile of cognitive impairments in chronic ketamine users. Psychiatr. Res 266, 124–131. [DOI] [PubMed] [Google Scholar]
- 22.Zhang C, Xu Y, Zhang B, Hao W, and Tang WK (2020). Cognitive impairment in chronic ketamine abusers. Psychiatr. Res 291, 113206. [DOI] [PubMed] [Google Scholar]
- 23.Strous JFM, Weeland CJ, van der Draai FA, Daams JG, Denys D, Lok A, Schoevers RA, and Figee M (2022). Brain Changes Associated With Long-Term Ketamine Abuse, A Systematic Review. Front. Neuroanat 16, 795231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Behrens MM, Ali SS, Dao DN, Lucero J, Shekhtman G, Quick KL, and Dugan LL (2007). Ketamine-Induced Loss of Phenotype of Fast-Spiking Interneurons Is Mediated by NADPH-Oxidase. Science 318, 1645–1647. [DOI] [PubMed] [Google Scholar]
- 25.Hamm JP, Peterka DS, Gogos JA, and Yuste R (2017). Altered Cortical Ensembles in Mouse Models of Schizophrenia. Neuron 94, 153–167.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayase T, Yamamoto Y, and Yamamoto K (2006). Behavioral effects of ketamine and toxic interactions with psychostimulants. BMC Neurosci. 7, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garritsen O, van Battum EY, Grossouw LM, and Pasterkamp RJ (2023). Development, wiring and function of dopamine neuron subtypes. Nat. Rev. Neurosci 24, 134–152. [DOI] [PubMed] [Google Scholar]
- 28.Hussain LS, Reddy V, and Maani CV (2023). Physiology, Noradrenergic Synapse. In StatPearls (StatPearls Publishing; ). [PubMed] [Google Scholar]
- 29.Robertson SD, Plummer NW, de Marchena J, and Jensen P (2013). Developmental origins of central norepinephrine neuron diversity. Nat. Neurosci 16, 1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dirks RM, and Pierce NA (2004). Triggered amplification by hybridization chain reaction. Proc. Natl. Acad. Sci. USA 101, 15275–15278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar V, Krolewski DM, Hebda-Bauer EK, Parsegian A, Martin B, Foltz M, Akil H, and Watson SJ (2021). Optimization and evaluation of fluorescence in situ hybridization chain reaction in cleared fresh-frozen brain tissues. Brain Struct. Funct 226, 481–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Renier N, Wu Z, Simon DJ, Yang J, Ariel P, and Tessier-Lavigne M (2014). iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910. [DOI] [PubMed] [Google Scholar]
- 33.Tomer R, Ye L, Hsueh B, and Deisseroth K (2014). Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat. Protoc 9, 1682–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Migliori B, Datta MS, Dupre C, Apak MC, Asano S, Gao R, Boyden ES, Hermanson O, Yuste R, and Tomer R (2018). Light sheet theta microscopy for rapid high-resolution imaging of large biological samples. BMC Biol. 16, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Q, Ding S-L, Li Y, Royall J, Feng D, Lesnar P, Graddis N, Naeemi M, Facer B, Ho A, et al. (2020). The Allen Mouse Brain Common Coordinate Framework: A 3D Reference Atlas. Cell 181, 936–953.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matthews GA, Nieh EH, Vander Weele CM, Halbert SA, Pradhan RV, Yosafat AS, Glober GF, Izadmehr EM, Thomas RE, Lacy GD, et al. (2016). Dorsal Raphe Dopamine Neurons Represent the Experience of Social Isolation. Cell 164, 617–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moriya S, Yamashita A, Masukawa D, Setoyama H, Hwang Y, Yamanaka A, and Kuwaki T (2020). Involvement of A13 dopaminergic neurons located in the zona incerta in nociceptive processing: a fiber photometry study. Mol. Brain 13, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moaddab M, and McDannald MA (2021). Retrorubral field is a hub for diverse threat and aversive outcome signals. Curr. Biol 31,2099–2110.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, and van den Pol AN (2016). Hypothalamic arcuate nucleus tyrosine hydroxylase neurons play orexigenic role in energy homeostasis. Nat. Neurosci 19, 1341–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dominguez JM, and Hull EM (2005). Dopamine, the medial preoptic area, and male sexual behavior. Physiol. Behav 86, 356–368. [DOI] [PubMed] [Google Scholar]
- 41.Abraira VE, Kuehn ED, Chirila AM, Springel MW, Toliver AA, Zimmerman AL, Orefice LL, Boyle KA, Bai L, Song BJ, et al. (2017). The Cellular and Synaptic Architecture of the Mechanosensory Dorsal Horn. Cell 168, 295–310.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dulcis D, Jamshidi P, Leutgeb S, and Spitzer NC (2013). Neurotransmitter Switching in the Adult Brain Regulates Behavior. Science 340, 449–453. [DOI] [PubMed] [Google Scholar]
- 43.Ozdas MS, Shah AS, Johnson PM, Patel N, Marks M, Yasar TB, Stalder U, Bigler L, von der Behrens W, Sirsi SR, and Yanik MF (2020). Non-invasive molecularly-specific millimeter-resolution manipulation of brain circuits by ultrasound-mediated aggregation and uncaging of drug carriers. Nat. Commun 11, 4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pouliopoulos AN, Wu S-Y, Burgess MT, Karakatsani ME, Kamimura HAS, and Konofagou EE (2020). A Clinical System for Non-invasive Blood-Brain Barrier Opening Using a Neuronavigation-Guided Single-Element Focused Ultrasound Transducer. Ultrasound Med. Biol 46, 73–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen BK, Luna VM, LaGamma CT, Xu X, Deng S-X, Suckow RF, Cooper TB, Shah A, Brachman RA, Mendez-David I, et al. (2020). Sex-specific neurobiological actions of prophylactic (R,S)-ketamine, (2R,6R)-hydroxynorketamine, and (2S,6S)-hydroxynorketamine. Neuropsychopharmacology 45, 1545–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bria A, and Iannello G (2012). TeraStitcher - A tool for fast automatic 3D-stitching of teravoxel-sized microscopy images. BMC Bioinf. 13, 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hörl D, Rojas Rusak F, Preusser F, Tillberg P, Randel N, Chhetri RK, Cardona A, Keller PJ, Harz H, Leonhardt H, et al. (2019). Big-Stitcher: reconstructing high-resolution image datasets of cleared and expanded samples. Nat. Methods 16, 870–874. [DOI] [PubMed] [Google Scholar]
- 48.Brachman RA, McGowan JC, Perusini JN, Lim SC, Pham TH, Faye C, Gardier AM, Mendez-David I, David DJ, Hen R, and Denny CA (2016). Ketamine as a prophylactic against stress-induced depressive-like behavior. Biol. Psychiatr 79, 776–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viola P, and Wells III WM (1997). Alignment by Maximization of Mutual Information. Int. J. Comput. Vis 24, 137–154. [Google Scholar]
- 50.McCormick M, Liu X, Jomier J, Marion C, and Ibanez L (2014). ITK: enabling reproducible research and open science. Front. Neuroinf 8, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Renier N,Adams EL, Kirst C,Wu Z, Azevedo R, Kohl J, Autry AE, Kadiri L, Umadevi Venkataraju K, Zhou Y, et al. (2016). Mapping of Brain Activity by Automated Volume Analysis of Immediate Early Genes. Cell 165, 1789–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tyson AL, Rousseau CV, Niedworok CJ, Keshavarzi S, Tsitoura C, Cossell L, Strom M, and Margrie TW (2021).A deep learning algorithm for 3D cell detection in whole mouse brain image datasets. PLoS Comput. Biol 17, e1009074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stringer C, Wang T, Michaelos M, and Pachitariu M (2021). Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106. [DOI] [PubMed] [Google Scholar]
- 54.Berg S, Kutra D, Kroeger T, Straehle CN, Kausler BX, Haubold C, Schiegg M, Ales J, Beier T, Rudy M, et al. (2019). ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232. [DOI] [PubMed] [Google Scholar]
- 55.van der Walt S, Schönberger JL, Nunez-Iglesias J, Boulogne F, Warner JD, Yager N, Gouillart E, and Yu T; scikit-image contributors (2014). scikit-image: image processing in Python. PeerJ 2, e453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, et al. (2020). SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microscopy data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at https://doi.org/10.5281/zenodo.10067746 and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Sheep anti-Tyrosine hydroxylase | abcam | Ab113 |
| Rabbit anti-Active Caspase −3 | BD Biosciences | AB_397274 |
| DAPI (4′,6-diamidino-2-phenylindole) | VWR | 89139-054 |
| Donkey anti-Sheep Alexa Fluor 647 | ThermoFisher Scientific | A-21448 |
| Goat anti-Rabbit Alexa Fluor 647 | ThermoFisher Scientific | A-21245 |
| Chemicals, peptides, and recombinant proteins | ||
| (R,S)-Ketamine Hydrochloride | Covetrus | 071069 |
| 40% Acrylamide | Bio-Rad | 1610140 |
| 2% Bisacrylamide | Bio-Rad | 1610142 |
| VA-044 | Wako Chemicals | NC0632395 |
| Paraformaldehyde | Electron Microscopy Sciences | 15710-S |
| Boric Acid | Sigma-Aldrich | B7901 |
| Sodium dodecyl sulfate | Amresco | 0837 |
| RapiClear RI = 1.47 | SunJin lab | RC147002 |
| Methanol | Fisher Scientific | AC423950040 |
| Dichloromethane | Sigma-Aldrich | 270997 |
| Hydrogen peroxide | Sigma-Aldrich | 216763 |
| Dibenzyl ether | Sigma-Aldrich | 33630 |
| Donkey serum | Millipore Sigma | D9663 |
| Bovine serum albumin | Fisher Scientific | BP1600-100 |
| Triton X-100 | Sigma-Aldrich | T8787 |
| Dimethyl sulfoxide | Sigma-Aldrich | 472301 |
| Glycine | Sigma-Aldrich | G7126 |
| 4 – hydroxytamoxifen | Sigma-Aldrich | H7904 |
| Sodium chloride | Sigma-Aldrich | S9625 |
| Corn oil | Sigma-Aldrich | C8267 |
| 20x SSC, RNase-free | ThermoFisher Scientific | AM9765 |
| Acetic anhydride | Millipore Sigma | 320102 |
| Formamide deionized | Millipore Sigma | S4117 |
| Yeast tRNA | Fisher Scientific | AM7119 |
| Dextran sulfate | Millipore Sigma | 265152 |
| Citric acid | Sigma-Aldrich | 251275 |
| Tween 20 | Sigma-Aldrich | P9416 |
| PBS - Phosphate-Buffered Saline (10X) pH 7.4, RNase-free | ThermoFisher Scientific | AM9624 |
| Experimental models: Organisms/strains | ||
| TH-2a-CreER | David Ginty’s lab; Abraira et al.41 | TH-2a-CreER |
| B6; 129S6-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J (Ai14) | The Jackson Laboratory | Strain #:007908 |
| Oligonucleotides | ||
| HCR Amplifier: B1, Alexa 647 | Molecular Instruments | N/A |
| HCR Probes for Tyrosine Hydroxylase mRNA detection | IDT | See Table S1 |
| Software and algorithms | ||
| Python 3.7 | Python Software Foundation | https://www.python.org/ |
| ImageJ | NIH | https://imagej.net/ij/index.html |
| Terastitcher | Bria et al.46 | https://abria.github.io/TeraStitcher/ |
| Bigstitcher | David Hörl et al.47 | https://imagej.net/plugins/bigstitcher/ |
| ITK | Kitware, Inc | https://itk.org/ |
| suiteWB | This study | https://github.com/tomerlab/suiteWB |
| Napari | Chan Zuckerberg Initiative | https://napari.org/stable/index.html |
| Amira | Thermo Fisher Scientific | https://www.thermofisher.com/us/en/home/electron-microscopy/products/software-em-3d-vis/amira-software.html |
| MATLAB | Mathworks | https://www.mathworks.com/products/matlab.html |
| suiteWB | This study | https://doi.org/10.5281/zenodo.10067746 |