Summary
Social interactions profoundly influence animal development, physiology and behavior. Yet, how sleep–a central behavioral and neurophysiological process–is modulated by social interactions is poorly understood. Here, we characterized sleep behavior and neurophysiology in freely-moving and co-living mice under different social conditions. We utilized wireless neurophysiological devices to simultaneously record multiple individuals within a group for 24 hours, alongside video acquisition. We first demonstrated that mice seek physical contact before sleep initiation and sleep while in close proximity to each other (hereafter, ‘huddling’). To determine whether huddling during sleep is a motivated behavior, we devised a novel behavioral apparatus allowing mice to choose whether to sleep in close proximity to a conspecific or in solitude, under different environmental conditions. We also applied a deep-learning-based approach to classify huddling behavior. We demonstrate that mice are willing to forgo their preferred sleep location, even under thermoneutral conditions, to gain access to social contact during sleep. This strongly suggests that the motivation for prolonged physical contact–which we term somatolonging–drives huddling behavior. We then characterized sleep architecture under different social conditions and uncovered a social-dependent modulation of sleep. We also revealed coordination in multiple neurophysiological features among co-sleeping individuals, including in the timing of falling asleep and waking up and NREM sleep intensity. Notably, the timing of REM sleep was synchronized among co-sleeping male siblings but not co-sleeping female or unfamiliar mice. Our findings provide novel insights into the motivation for physical contact and the extent of social-dependent plasticity in sleep.
Keywords: Social contact, NREM sleep, REM sleep, Huddling behavior, Wireless technology, Neurophysiological synchronization, somatolonging, Immersive social interactions assay
Sotelo et al. demonstrate that mice seek to huddle prior to sleep initiation and that sleep huddling is a motivated behavior. They further reveal social-context-dependent synchronization in multiple neurophysiological features among co-sleeping individuals, including sleep and wakefulness onset times, NREMS intensity, and REMS timing.
Graphical Abstract
Introduction
Social interactions play a crucial role in animal life, significantly affecting the development, physiology, and behavior of individuals1. Consequently, social isolation in various species is associated with negative health outcomes, including compromised immunity and mood dysregulation2–5. However, the extent to which social contact modulates sleep–a central behavioral and neurophysiological process6–remains unclear. This is mainly since sleep has traditionally been studied in experimental subjects that are individually isolated and deprived of their natural ecological and social milieu7,8.
Studies have suggested that sleep quality and need are socially-modulated in diverse species9,10. For instance, honeybees and fruit flies sleep more following an awake period with social interactions than following an awake period without social interactions11,12. In baboons, 3D acceleration tracking suggests that the size of the social group, valence of previous social interactions, and social status modulate sleep need and quality12,13. Electroencephalographic (EEG) and electromyographic (EMG) recordings from single individuals within a group or across fragments of the light/dark cycle in several mammalian species further suggest that sleep architecture and quality can be modulated by social-living and dominance hierarchy. For instance, the presence of non-sibling conspecifics affects sleep architecture and composition in the rock hyrax14 and laboratory male mice15,16.
Although previous studies strongly suggest that sleep physiology is modulated by the social environment, the extent to which the social milieu modulates sleep need, quality, and cortical oscillatory patterns during sleep remains undetermined. This is because previous studies lacked neurophysiological qualification of sleep, were based on short recordings, or originated from a single individual within the social group. Developments in biologging technologies, which permit continuous neurophysiological monitoring across days from multiple untethered individuals in semi-natural and wild conditions, have emerged as a breakthrough in the capacity to study sleep in naturalistic environments7. Indeed, the recent use of these technologies has revealed that sleep neurophysiology and behavior are plastic and modulated by internal and ecological demands17,18.
Various species, including humans19, baboons13, gulls20, and laboratory mice16, exhibit coordination in the timing of sleep and wake episodes between co-living individuals. Recent studies have also uncovered coordination in brain oscillatory patterns between awake interacting individuals in humans21, bats22, and mice23. However, there is still a significant knowledge gap regarding the extent of this coordination to brain oscillatory patterns during sleep.
Here, we utilized innovative wireless neurophysiological recording devices to investigate the sleep behavior and neurophysiology of freely-moving mice living together under various social conditions. We reveal that prior to sleep initiation, mice actively seek physical contact and sleep while in close proximity to each other, a behavior we refer to as huddling. Employing a novel preference assay and deep-learning-based huddling classification, we demonstrate that huddling during sleep is a motivated behavior. We also uncover social-dependent changes in sleep architecture and coordination in sleep oscillatory patterns among co-sleeping individuals. We reveal that while social sleeping fragments non-rapid eye movement sleep (NREMS), it synchronizes rapid eye movement sleep (REMS) episodes. Notably, REMS synchronization was specific to male sibling mice and not observed in female or unfamiliar male mice, suggesting a dependence on both sex and social familiarity. Our findings further suggest that the internal state of an animal plays a crucial role in coordinating REMS and oscillatory neurophysiological activity, and expand current understanding of the extent to which social factors can modulate sleep.
Results
Mice huddle prior to sleep initiation
To examine the influence of the presence of conspecifics on behavioral preparation for sleep, we recorded same-sex triplet sibling adult wild-type (CD-1) mice in their home-cage environment before and after transitioning into the light phase (Figure 1A). We identified eight behavioral states (‘in-motion,’ ‘transiently-socially interacting,’ ‘eating/drinking,’ ‘allo-grooming,’ ‘self-grooming,’ ‘nesting,’ ‘resting’ and ‘sleeping’) and noted whether these behaviors were displayed while in close physical proximity to conspecifics–hereafter referred to as huddling–or not (Figure 1B). We compared the amount of time spent in each behavior within two time windows: 40 to 20 minutes and 20 to 0 minutes before sleep onset (Figure 1C–D), informed by our recent identification of the 20-minute period preceding sleep initiation as a pre-sleep transitional phase24. We performed a second-by-second annotation of the videos for one individual from eight triplets (n = 4 females and 4 males). Four behavioral states (‘self-grooming,’ ‘nesting,’ ‘resting’ and ‘sleeping’) were displayed both while huddling and not-huddling during the 40-minute period prior to sleep onset. While not-huddling, mice spent significantly less time ‘in-motion’ and ‘transiently-socially interacting’ as sleep approached yet did not alter time spent ‘eating/drinking,’ ‘self-grooming,’ ‘nesting,’ and ‘resting’ (Figure 1C; data not shown for eating/drinking). Conversely, while huddling, mice spent significantly more time ‘self-grooming,’ ‘nesting,’ and ‘resting’ as sleep approached, but not ‘allo-grooming’ (Figure 1D). Overall, the proportion of behaviors performed while not-huddling decreased, and those performed while huddling increased, as sleep onset approached (Figure 1E). These findings imply that mice seek physical contact in the pre-sleep phase and engage in pre-sleep behaviors such as nesting and grooming24, while huddling.
Figure 1. Mice seek physical contact in the pre-sleep phase and engage in pre-sleep behavior while huddling.
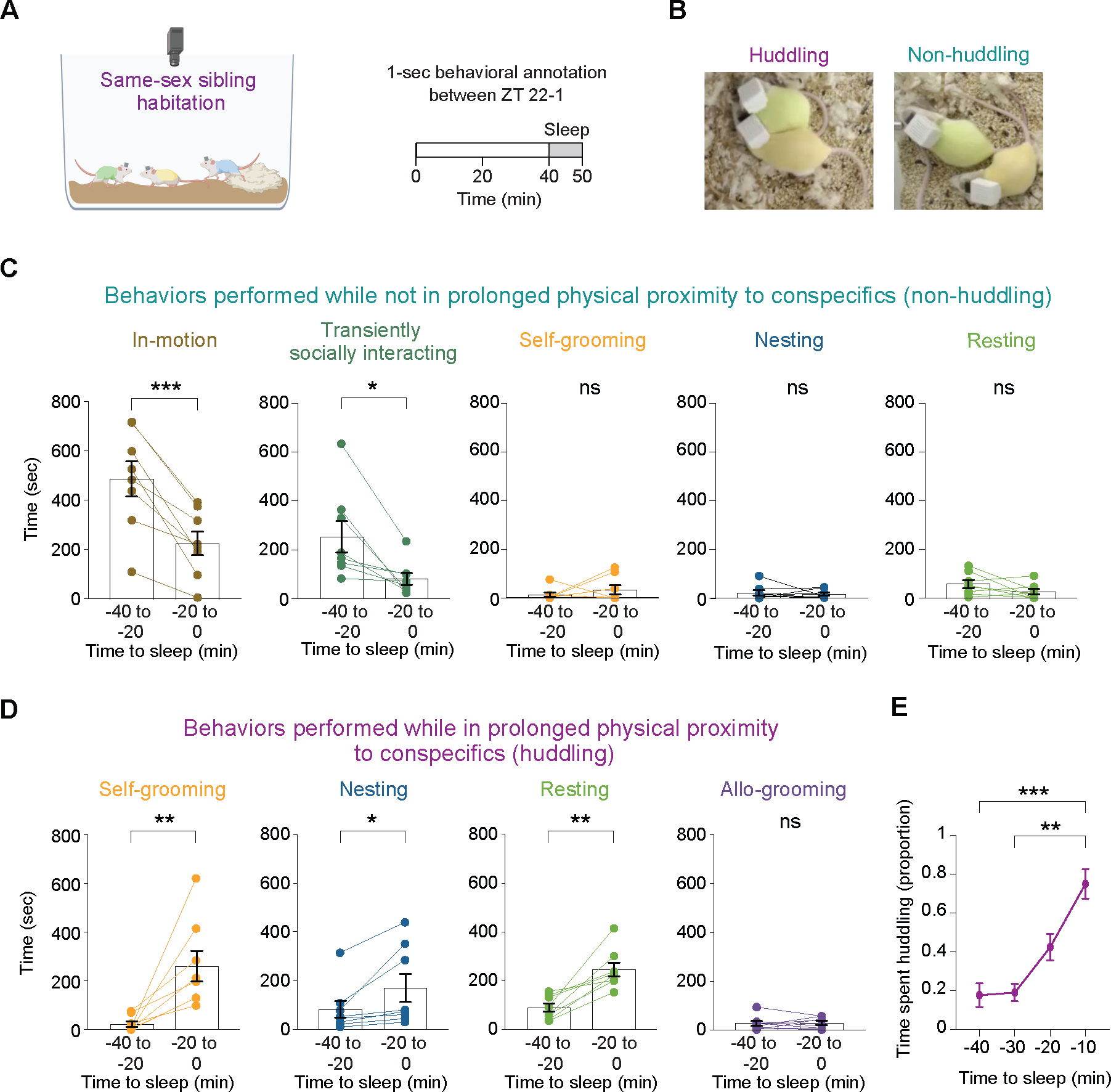
(A) Schematic representation of the home-cage environment, and the time windows for behavioral analysis. (B) Images providing a representative illustration of huddling (left) and non-huddling (right) mice, as determined through manual behavioral annotation. Huddling was classified for incidents in which two mice were in bodily contact, resulting in no visible background between them, and this contact lasted for more than a second. Mice bodies included the abdomen and thorax but not the head, legs, paws, and tail. (C) Time devoted to different behaviors prior to sleep onset in a non-huddling state. n = 8 female and male mice. Paired t-tests, two tailed. ns, p > 0.05; *, 0.01 < p < 0.05; ***, 0.001 < p < 0.01. (D) Time devoted to different behaviors prior to sleep onset in a huddling state. n = 8 female and male mice. Paired t-tests, two tailed. ns, p > 0.05; *, 0.01 < p < 0.05; **, 0.001 < p < 0.01. (E) Proportion of time spent in huddling state during the 40-minute period preceding sleep onset. n = 8 female and male mice. Friedman test (p = 0.0002) followed by multiple comparisons: **, 0.001 < p < 0.01; ***, 0.0001 < p < 0.001.
Huddling before and during sleep is driven by somatolonging–a motivation for physical contact
We next aimed to examine whether the observed increase in huddling prior to sleep was driven by a motivation for physical contact or due to difficulty sleeping alone in the shared home-cage environment. To facilitate a high throughput analysis of huddling behavior, we employed a deep-learning-based approach for huddling classification (Figure 2A and Figure S1A–G). First, we performed pose estimation via DeepLabCut25,26 to identify and track mice. We annotated two back keypoints for each mouse, which remained predominantly visible even when most of the mouse’s body was occluded by a conspecific or nesting material (Figure S1A and Figure 2A). Next, we computed the minimum Euclidean distance between the mice’s keypoints (Figure 2A). Finally, we classified huddling instances when the minimum distance fell below a pre-set threshold indicating physical contact (5 cm; Figure S1F and Table 1) and which lasted over a pre-set duration threshold for huddling (1 second, excluding transient social interactions) (Figure 2A and Figure S1G).
Figure 2. Huddling behavior is an expression of a motivation for physical contact.
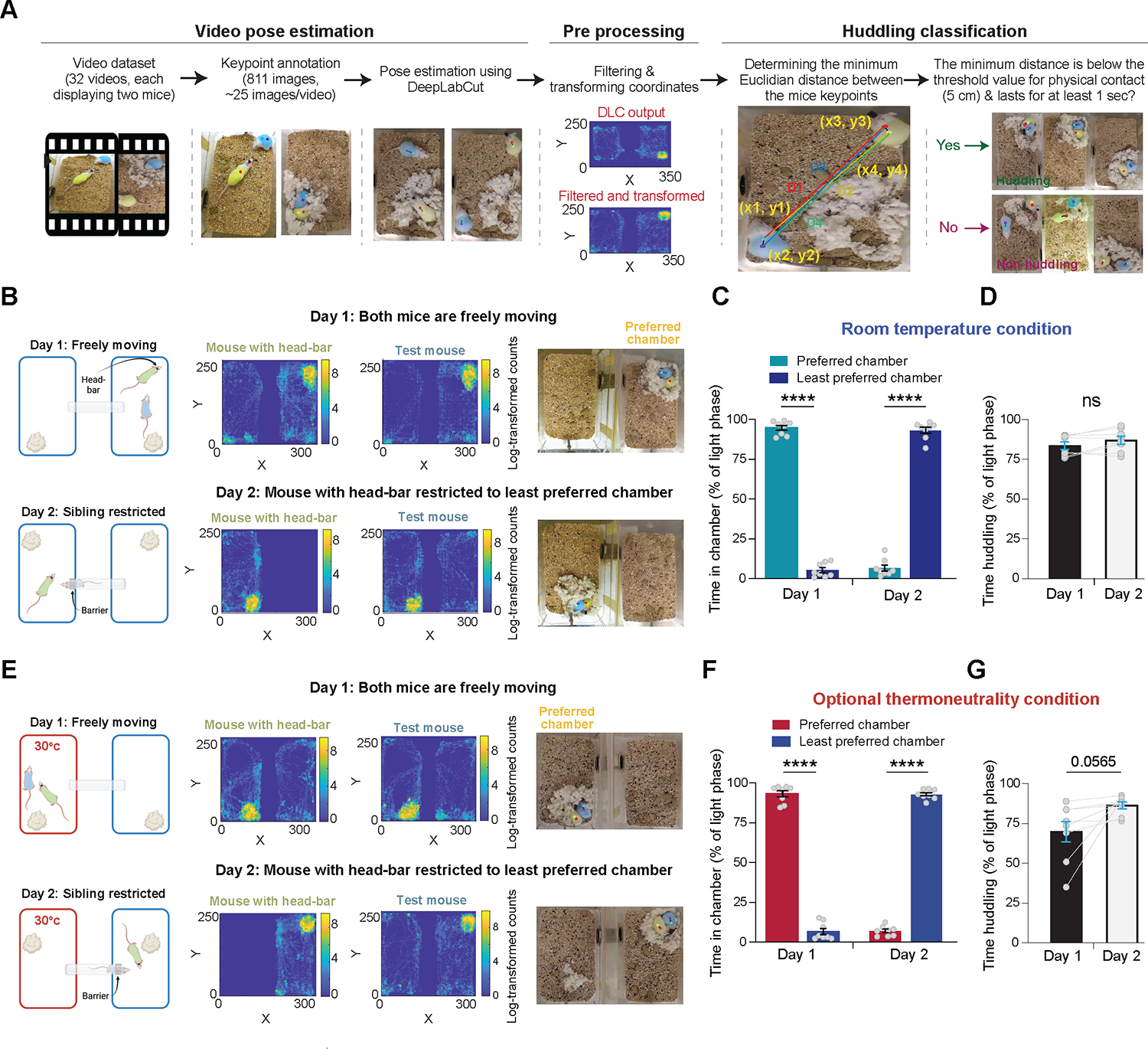
(A) Schematics illustrating the pipeline for pose estimation, coordinate data filtering, and huddling classification. (B) Schematics detailing the two-day ISIA procedure conducted under room temperature conditions, accompanied by representative data. The top row illustrates day 1 under freely moving conditions, while the bottom row details day 2 under sibling restriction conditions. Left: schematic illustrating the ISIA procedure. Middle: representative heatmaps depicting the locations of a head-bar fitted mouse and a test mouse during the 12-hour light cycle in the ISIA apparatus (log transformed histogram counts). Right: snapshots from the DLC tracking output depicting predicted key points, the mice, and their nest. Note that both the heatmaps and snapshots originate from the same recording sessions. (C) Percent time spent in each of the ISIA chambers during the light phase, measured under room temperature conditions, across the two days of the experiment. n = 8 pairs of female and male mice, RM two-way ANOVA (p(day) = 0.03, p(chamber) = 0.57, p(interaction) < 0.0001) followed by Sidak’s multiple comparisons test (****, p < 0.0001). (D) Percent time spent huddling during the light phase, measured under room temperature conditions, across the two days of the experiment. n = 8 pairs of female and male mice, Paired t-test, two-tailed, t = 2.017, df = 7, p = 0.0836. (E) As in (B), but with the ISIA procedure conducted under optional thermoneutrality conditions. This was achieved by maintaining the ambient temperature in one of the chambers at the mice’s thermoneutral zone (30°C). (F) Percent time spent in each of the ISIA chambers during the light phase, measured under optional thermoneutrality conditions, across the two days of the experiment. n = 8 pairs of female and male mice, RM two-way ANOVA (p(day) = 0.16, p(chamber) = 0.9, p(interaction) < 0.0001), followed by Sidak’s multiple comparisons test (****, p < 0.0001). (G) Percent time spent huddling during the light phase, measured under optional thermoneutrality conditions, across the two days of the experiment. n = 8 pairs of female and male mice, Paired t-test, two-tailed, t = 2.281, df = 7, p = 0.0565. See also Figure S1, Figure S2, Video S1, Video S2 and Table 1.
Table 1. Classifier performance and evaluation.
The table illustrates the confusion matrices generated for the three evaluation sets using three distinct thresholds for physical contact (4 cm, 5 cm, and 6 cm). It also presents corresponding metrics for Accuracy, Precision, Recall, Specificity, and F1 score for each of these distance thresholds. See also Figure 2, Figure S1, Video S1 and Video S2.
| 4 cm | |||
| Actual (True) values | |||
| Positive | Negative | ||
| Predictive values | Positive | 984 | 9 |
| Negative | 108 | 887 | |
| 5 cm | |||
| Actual (True) values | |||
| Positive | Negative | ||
| Predictive values | Positive | 967 | 9 |
| Negative | 8 | 1053 | |
| 6 cm | |||
| Actual (True) values | |||
| Positive | Negative | ||
| Predictive values | Positive | 934 | 31 |
| Negative | 1 | 1054 | |
| 4 cm | 5 cm | 6 cm | |
| Accuracy | 0.9411 | 0.9917 | 0.9842 |
| Precision | 0.9909 | 0.9908 | 0.9679 |
| Recall | 0.9011 | 0.9918 | 0.9989 |
| Specificity | 0.9900 | 0.9915 | 0.9714 |
| F1 score | 0.9439 | 0.9913 | 0.9832 |
Subsequently, we devised a novel behavioral assay, which we named the ‘Immersive Social Interactions Assay (ISIA)’, to investigate whether the increase in huddling prior to sleep signifies a motivation for physical contact. The ISIA apparatus consists of two home-cage chambers connected by a plastic tube–whose inner diameter can be reduced by the insertion of a plastic ring (Figure S2A). We studied same-sex pairs of adult sibling mice (n = 8 pairs). In each pair, one mouse was implanted with a 6-pin connector (hereafter referred to as ‘head-bar’), while the other underwent a sham operation. On the first day of the experiment, both mice were allowed free movement between chambers, and they consistently spent most of the ensuing light phase in one chamber, where they built their nest and slept (Figure 2B,C, Figure S2B,C and Video S1). On the second day, at the end of the dark/active phase, a plastic ring was inserted into the tube connecting the chambers to restrict the head-bar mouse to the least preferred chamber, while the test mouse remained free to move between the chambers. During the following light phase, test mice spent significantly less time in their preferred chamber and more time in their least preferred chamber (Figure 2C and Video S1), indicating that mice are willing to sacrifice their preferred sleep location for physical contact during sleep. Additionally, the proportion of time spent huddling during the light phase did not decrease under the restriction (Figure 2D), suggesting that the increase in huddling prior to sleep is driven by a motivation for physical contact rather than by difficulty sleeping alone in the shared home-cage environment.
Given that the laboratory environment falls below the thermoneutral zone for mice27, we wondered whether the observed increase in huddling behavior might be due to mice seeking physical contact as a means to mitigate the cold. We thus repeated the previous experiment, but in this instance, we modified the ISIA setup to utilize two different chambers: one kept at room temperature, henceforth referred to as the ‘cold chamber,’ and another maintained at the thermoneutral zone for mice (30°C), referred to as the ‘warm chamber’ (n = 8 pairs of sibling mice; Figure 2E and Figure S2D,E). Under baseline conditions, all mice spent significantly more time in the warm chamber during the light phase, where they constructed their nests and slept (Figure 2F, Figure S2E and Video S2). However, when the head-bar mouse was confined to the cold chamber, the test mice spent significantly less time in the warm chamber and more time in the cold chamber (Figure 2F and Video S2). Intriguingly, the time spent huddling did not decrease but rather slightly increased when the head-bar mouse was restricted to the cold chamber (Figure 2G). Together, these findings suggest that mice are willing to forgo ambient thermoneutral conditions in favor of physical contact during sleep. These results strongly suggest that huddling behavior is an expression of a motivation for prolonged physical contact, a concept we term ‘somatolonging.’
Sibling presence fragments NREMS
Next, we investigated the influence of sibling presence on the sleep/wake architecture in mice, employing wireless devices capable of monitoring neurophysiological signals in groups over extended periods (Neurologger 2A; Figure 3A). We examined the sleep/wake architecture of same-sex sibling adult mice over a 24-hour period, both in groups and isolated settings (n = 9 males and 9 females from 3 triplets per sex; Figure 3B–C). Importantly, sibling mice were recorded simultaneously, either while cohabiting the same home-cage or when housed in separate cages within the same experimental room. Sibling presence significantly altered the proportion of time spent in the different sleep/wake states in female, but not in male, mice (Figure 3D,E). Specifically, while with their siblings, females spent less time awake and more time asleep during the dark phase, compared to when individually isolated (Figure 3E). Notably, the presence of siblings significantly fragmented NREMS in both male and female mice, as evidenced by an increase in the number of NREMS episodes and a decrease in the average NREMS episode duration throughout most of the light and dark phases (Figure 3F–I and Figure S3A–D). Additionally, delta power during NREMS was significantly lower in group-housed mice compared to when individually isolated (Figure S3E). Collectively, these findings suggest that the presence of siblings adversely affects the quality of NREMS in mice.
Figure 3. Sibling presence fragments NREMs in mice.
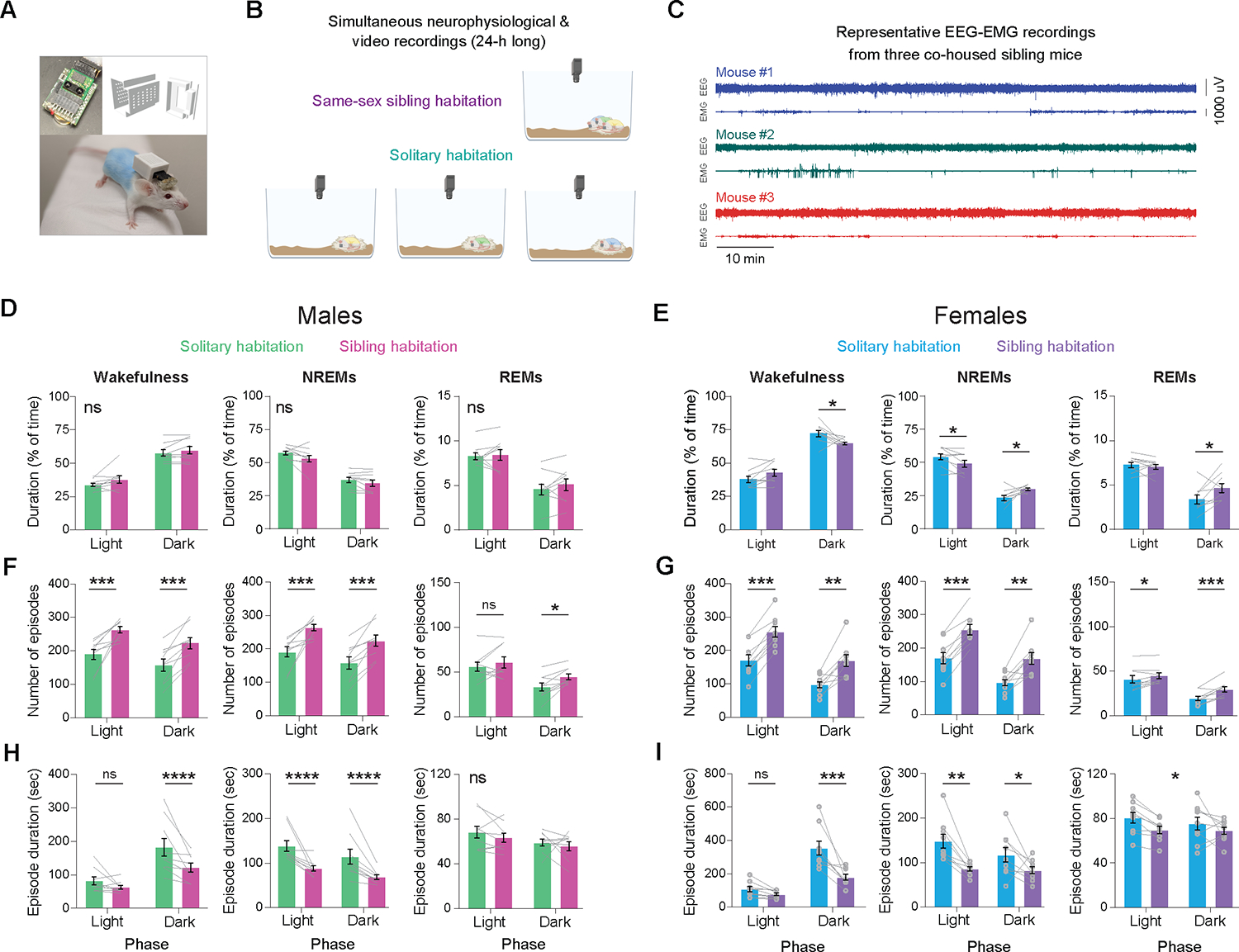
(A) Images showcasing the wireless Neurologger device, the 3D printer file of the device’s protective cover, and a CD-1 mouse with dyed fur, fitted with both the device and its cover. (B) Schematic illustrating both the same-sex sibling cohabitation and the solitary habitation conditions. (C) Representative traces of EEG-EMG signals from three simultaneously recorded, co-housed sibling mice. (D) Percentage of time that male mice spent in wakefulness, NREMs, and REMs during the light and dark phases of the day, comparing solitary habitation with sibling cohabitation. n = 9 mice, RM two-way ANOVA, p > 0.05 for both ‘social status’ and interactions for wakefulness, NREMs and REMs. (E) Same as (D) but for females. n = 9 mice, RM two-way ANOVA. Wakefulness: p(social status × phase) = 0.0022; NREMs: p(social status × phase) = 0.0021; REMs: p(social status × phase) = 0.0253. (F) Number of wake, NREMs and REMs episodes during the light and dark phases of the day for male mice in solitary habitation and sibling cohabitation. n = 9 mice, RM two-way ANOVA. Wakefulness: p(social status) = 0.0006, p(phase) = 0.037; NREMs: p(social status) = 0.0007, p(phase) = 0.036; REMs: p(social status) = 0.0291, p(phase) < 0.0001. (G) Same as (F) but for female mice. n = 9 mice, RM two-way ANOVA. Wakefulness: p(social status) and p(phase) <0.0001; NREMs: p(social status) and p(phase) <0.0001; REMs: p(social status) = 0.0069, p(phase) = 0.0002. (H) Duration of wake, NREMs and REMs episodes during the light and dark phases of the day for male mice in solitary habitation and sibling cohabitation. n = 9 mice, RM two-way ANOVA. Wakefulness: p(social status) = 0.0351, p(phase) = 0.002, p(interaction) = 0.005; NREMs: p(social status) = 0.0045, p(phase) = 0.0115; REMs: p(social status) = 0.33, p(phase) =0.0377. (I) Same as (H) but for female mice. n = 9 mice, RM two-way ANOVA. Wakefulness: p(social status) = 0.004, p(phase) < 0.0001; NREMs: p(social status) = 0.0047, p(phase) = 0.0037; REMs: p(social status) = 0.016, p(phase) =0.49. Asterisks over data points were obtained using Sidak’s multiple comparisons tests. Asterisks displayed in the center of a panel without an underlying line signify a significant main effect (no significant differences were observed in multiple comparison tests). ns, p > 0.05; *, 0.01< p < 0.05; **, 0.001 < p < 0.01; ***, 0.001 < p < 0.01; ****, p < 0.0001. See also Figure S3.
Considering that our mice were fitted with wireless devices, we wondered whether the observed sleep fragmentation is due to the devices themselves rather than a genuine fragmentation induced by conspecifics. We thus compared the average duration of sleep bouts in mice with and without wireless devices. We first validated that video-based sleep scoring yielded a similar average duration of sleep episodes as EEG-EMG based scoring in mice equipped with wireless devices, recorded under both social and individual housing conditions (n = 6 mice; Figure S3F). Subsequently, employing video-based scoring, we compared the average sleep episode in mice with and without wireless devices under both individual and social conditions (Figure S3G). We found that the social context (isolation vs social housing), but not the presence of wireless devices, significantly influenced sleep episode duration (n = 6 mice per group; Figure S3G). This finding further supports our premise that the presence of conspecifics, rather than the wireless devices, negatively affects sleep quality.
To quantify the proportion of huddling during sleep under group-housed conditions, we employed the deep-learning-based approach for huddling classification described above (Figure 2A, Figure S1 and Figure S3H), and aligned the huddling data with the sleep-state data. Both female and male mice huddled with their siblings during the majority of their NREM and REMS epochs during the light phase (n = 9 mice per sex; Figure S3I). These findings suggest that while mice actively seek physical contact prior to sleep and continue huddling throughout, such close physical proximity to conspecifics leads to a fragmentation of NREMS.
Co-sleeping siblings show coordination of multiple neurophysiological features
To investigate the effects of group housing on brain states and oscillatory brain activity synchronization between animals (n = 18 male pairs and 14 female pairs), we first assessed the synchrony of sleep and wake onset times between pairs of mice, either cohabiting in the same home-cage or housed separately within the same experimental room. Both female and male mice displayed a significant degree of synchronization in the timing of sleep/wake onsets in group-housed conditions as compared to isolated settings (Figure 4A–I). Notably, we found a positive correlation between the degree of synchronization in the timing of falling-asleep and the degree of synchronization in the timing of waking-up across mouse pairs, suggesting that pairs who typically wake-up together also tend to fall-asleep together (Figure 4J). Further, we detected a strong synchrony in the overall sleep and wake times when mice were cohabiting in the same home-cage as compared to isolated settings and shuffled data (Figure 4K,M). Our cross-correlation analysis further revealed a significant coordination in sleep/wake state timing with a peak at zero lag time in group-housed but not in isolated settings (Figure 4L,N). Together, these findings suggest that sibling mice demonstrate strong coordination, with high temporal precision, of sleep and wake timing.
Figure 4: Sibling mice show coordination of multiple neurophysiological features while co-sleeping.
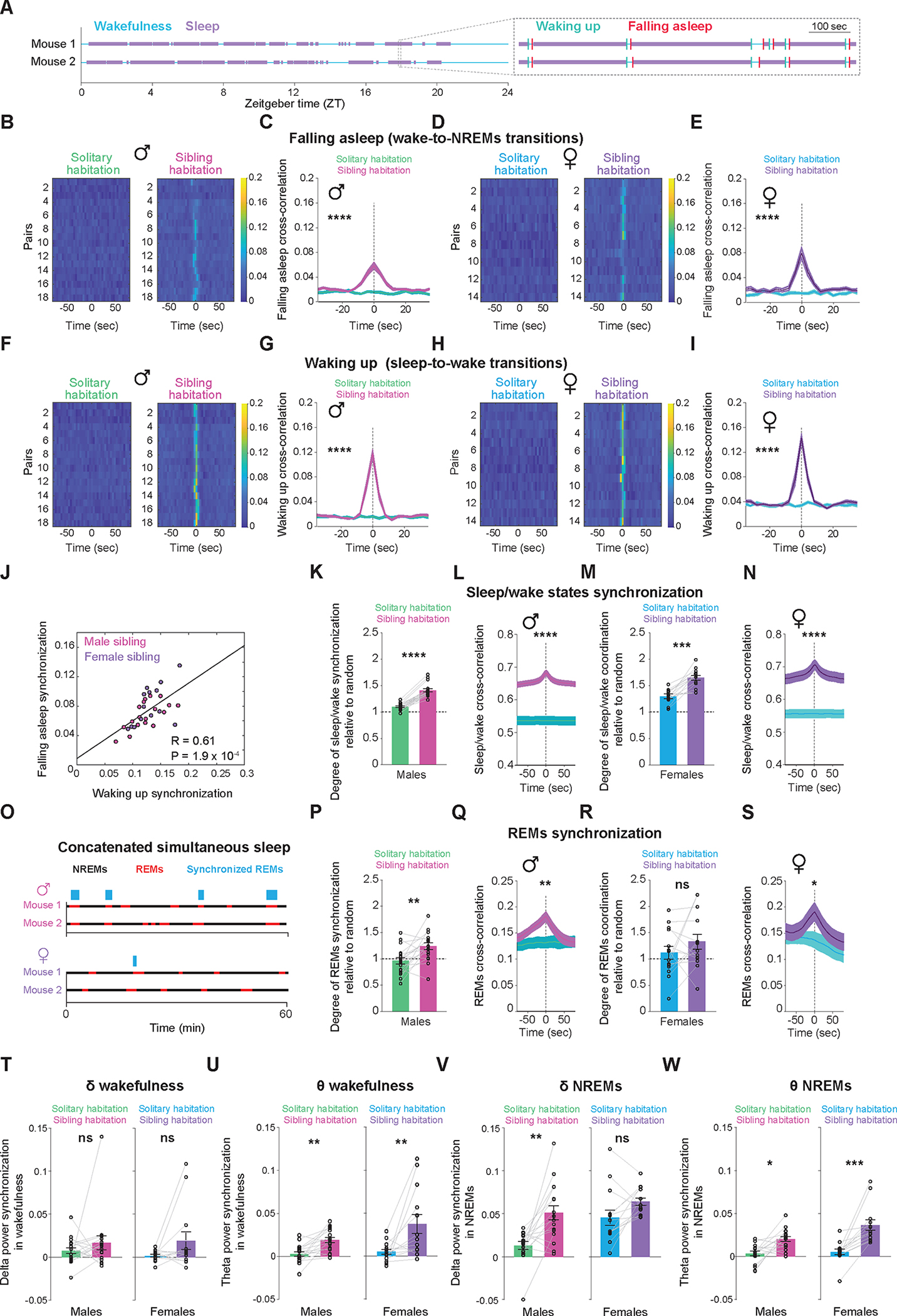
(A) On the left, binary vectors representing wake (depicted in blue) and sleep (depicted in purple) states over a 24-hour period for a pair of sibling mice. On the right, a magnified section of the vectors reveals the exact moments of wake onset (depicted in green) and sleep onset (depicted in red) for the two mice. (B-I) Synchrony of sleep and wake onset times between pairs of mice, either cohabiting in the same home-cage or housed separately within the same experimental room. (B) Heatmap representation of cross-correlation of sleep onset times for each pair of male sibling mice, whether housed together (right) or separately (left). (C) Average cross-correlations of sleep onset times from all male pairs housed separately or together (mean ± S.E.). Shuffle test, p < 0.0001. (D) Same as (B) but for female mice. (E) Same as (C) but for female mice. Shuffle test, p < 0.0001. (F) Heatmap representation of cross-correlation of wake onset times for each pair of male sibling mice, whether housed together (right) or separately (left). (G) Average cross-correlations of wake onset times from all male pairs housed together or separately (mean ± S.E.). Shuffle test, p < 0.0001. (H) Same as (F) but for female mice. (I) Same as (G) but for female mice. Shuffle test, p < 0.0001. (J) Correlation between the degree of synchronization in the timing of falling asleep and the degree of synchronization in the timing of waking up across both male (red) and female (purple) sibling mouse pairs. (K) Synchrony in the overall sleep and wake times between pairs of male sibling mice, either cohabiting in the same home-cage or housed separately within the same experimental room. Wilcoxon rank-sum test, p = 6.6 × 10−6. (L) Average cross-correlations of overall sleep and wake times from all male pairs housed together or separately (mean ± S.E.). Shuffle test, p < 0.0001. (M) Same as (K), but for females. Wilcoxon rank-sum test, p = 0.00012. (N) Same as (L), but for females. Shuffle test, p < 0.0001. (O) Binary vectors depicting NREMs (shown in black) and REMs (shown in red) states for a pair of sibling mice, with the top representing males and the bottom representing females. Blue rectangles highlight synchronized timepoints of REMs. Notably, bins in which either animal was awake were omitted from both vectors prior to the REMs synchronization quantification. Thus, this analysis specifically quantifies REMs synchronization within the sleep period. (P) Synchrony in the timing of REMs between pairs of male sibling mice, either cohabiting in the same home-cage or housed separately within the same experimental room. Wilcoxon rank-sum test, p = 0.004. (Q) Average cross-correlations of REMs time from all male pairs housed together or separately (mean ± S.E.). Shuffle test, p = 0.00124. (R) Same as (P), but for females. Wilcoxon rank-sum test, p = 0.3258. (S) Same as (Q), but for females. Shuffle test, p =0.2112. (T) Synchrony in the delta (1–4 Hz) band power during wakefulness between pairs of male (left) and female (right) sibling mice, either cohabiting in the same home-cage or housed separately within the same experimental room. Wilcoxon rank-sum test, p(males) = 0.46, p(females) = 0.19. (U) Same as (T) but for theta (5–9 Hz) band power. Wilcoxon rank-sum test, p(males) = 0.001, p(females) = 0.006. (V) Same as (T) but for NREMs. Wilcoxon rank-sum test, p(males) = 0.0017, p(females) = 0.0942. (W) Same as (U) but for NREMs. Wilcoxon rank-sum test, p(males) = 0.0109, p(females) = 0.0002.
We then examined if, beyond the overall sleep/wake synchrony, co-sleeping mice showed synchrony in their sleep states (Figure 4O). Notably, male mice demonstrated significant synchrony in the timing of REMS (Figure 4P,Q), whereas female mice exhibited minimal to no coordination (Figure 4R,S). Together, these findings suggest that social-living promotes the coordination of sleep/wake states, with REMS coordination particularly pronounced in male sibling mice.
Lastly, we examined the synchronization of oscillatory brain activity between pairs of mice. Co-habiting male and female mice exhibited synchrony in the theta band power, but not the delta band, of the EEG during wakefulness (Figure 4T,U). Notably, during NREMS, only male mice demonstrated synchronization in the power of the delta band, whereas female mice did not (Figure 4V); however, both males and females showed synchronization in the power of the theta band during NREMS (Figure 4W). Collectively, our data unravels a significant degree of coordination in multiple neurophysiological features among co-sleeping individuals, with the extent of coordination being particularly pronounced in male sibling mice.
Social factors modulate sleep/wake architecture
We examined the impact of non-familial social interactions on the sleep/wake architecture in mice by introducing intruder mice (female or male) into a resident male’s territory, studying both male-female and male-male pairings (Figure 5A). Anticipating significant immediate behavioral changes in both resident and intruder mice, we conducted neurophysiological analysis on the fourth day post-introduction–when sexual and agonistic behaviors had significantly subsided (n = 8 male-female and 8 male-male dyads; Figure 5B and Figure S4A,B). We first investigated the propensity of non-familial mice to huddle during sleep. Not only male-female dyads but also male-male dyads huddled for most of their sleep epochs on the fourth day of cohabitation (Figure 5C), even when the home-cages contained two shelters (Figure S4C). When analyzing the sleep/wake architecture of resident and intruder mice (n = 9 male-female and 9 male-male dyads; Figure 5D–I and Figure S5A–F), we compared, for each mouse, data from non-familial dyadic experiments to data collected when the mouse was with its siblings. This comparison enabled us to pinpoint changes attributable specifically to non-familial interactions, rather than to the mere presence of conspecifics. Male residents, when in the presence of a male intruder, displayed minimal alterations in their sleep/wake architecture (Figure 5D–I and Figure S5A–F). The primary change observed was a consolidation of NREMS during the light phase, marked by fewer yet lengthier NREMS bouts compared to when they were with their siblings (Figure S5B). Conversely, when a female intruder was present, male residents exhibited an increase in wakefulness and a decrease in REMS during the light phase compared to when with their siblings (Figure 5D,F). Notably, the proportion of REMS in male residents while in the presence of a female matched that of their female counterparts (Figure S5G). The surge in wakefulness in male residents was accompanied with its consolidation, characterized by fewer but longer episodes (Figure S5A), while the decrease in REMS was primarily attributed to fewer episodes, rather than alteration in the duration of REMS bouts (Figure S5C). However, during the dark phase when in the presence of a female intruder, the male residents showed no significant alterations in their sleep/wake architecture (Figure 5G–I and Figure S5D–F).
Figure 5. Sex and social-context dependent modulation of sleep.
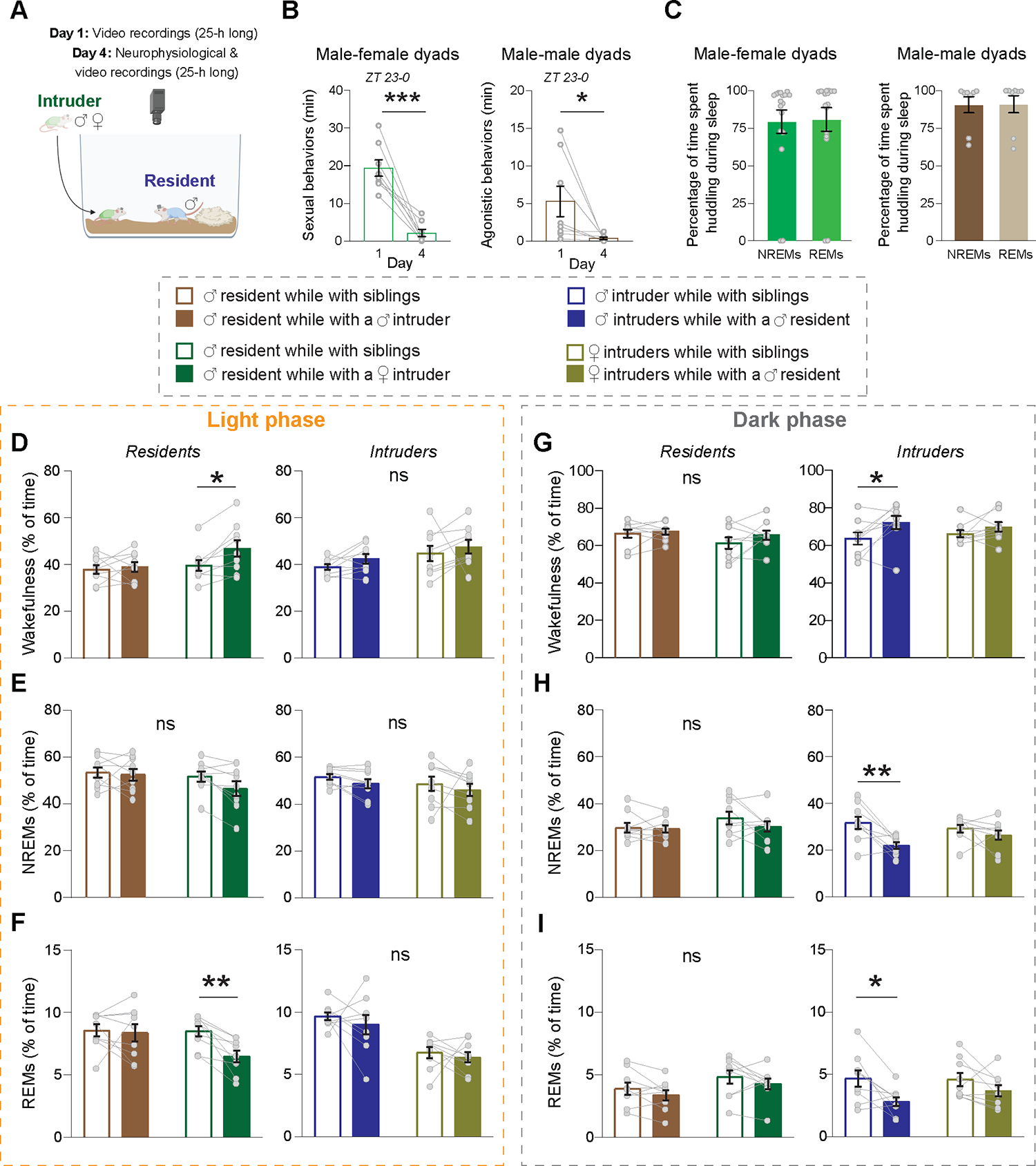
(A) Schematic illustrating experimental paradigm. (B) Total time dedicated to sexual behaviors in male-female dyads (left; n = 8 mice) and agonistic behaviors in male-male dyads (right; n = 8 mice) during ZT 23–0 immediately following the placement of an intruder mouse into the home-cage of a resident mouse on day 1 and four days later. Sexual behaviors include ‘Seeking,’ ‘Mounting without lordosis’ and ‘Mounting with lordosis.’ Agonistic behaviors include ‘Mounting,’ ‘Attacking,’ ‘Chasing,’ ‘Aggressive grooming’ and ‘Fleeing.’ Behavioral data was obtained from resident mice. Paired t-tests, two-tailed; p(sexual behaviors) = 0.0003; p(agonistic behaviors) = 0.048. (C) Percentage of time spent huddling during NREM and REM sleep in the light phase on the fourth day following cohabitation for male-female dyads (left) and male-male dyads (right). n = 9 mice per group. (D-I) Sleep/wake architecture across the light (left) and dark (right) cycle in Resident and Intruder mice compared to the architecture of the same mice while cohabiting with their siblings. (D) Percentage of time awake during the light phase for Resident mice (left) and Intruder mice (right) relative to the duration they were awake when cohabiting with their siblings. n = 9 mice per group, RM two-way ANOVA’s. Residents, p(social status) = 0.025; Intruders, p > 0.05 for both ‘social status’ and interaction. (E) Percentage of time spent in NREMs during the light phase for Resident mice (left) and Intruder mice (right) relative to the duration they were in NREMs when cohabiting with their siblings. n = 9 mice per group, RM two-way ANOVA’s. Residents and Intruders, p > 0.05 for both ‘social status’ and interaction. (F) Percentage of time spent in REMs during the light phase for Resident mice (left) and Intruder mice (right) relative to the duration they were in REMs when cohabiting with their siblings. n = 9 mice per group, RM two-way ANOVA’s Residents, p(social status) = 0.0189, p(interaction) = 0.0458; Intruders, p > 0.05 for both ‘social status’ and interaction. (G) Same as (D) but for the dark phase. n = 9 mice per group, RM two-way ANOVA’s. Residents, p > 0.05 for both ‘social status’ and interaction; Intruders, p(social status) = 0.0176. (H) Same as (E) but for the dark phase. n = 9 mice per group, RM two-way ANOVA’s. Residents, p > 0.05 for both ‘social status’ and interaction; Intruders, p(social status) = 0.0042. (I) Same as (F) but for the dark phase. n = 9 mice per group, RM two-way ANOVA’s. Residents, p > 0.05 for both ‘social status’ and interaction; Intruders, p(social status) = 0.0042. Asterisks over data points were obtained using Sidak’s multiple comparisons tests. ns, p > 0.05; *, 0.01< p < 0.05; **, 0.001 < p < 0.01; ***, 0.001 < p < 0.01; ****, p < 0.0001. See also Figure S4 and S5.
Unlike resident mice, male intruders displayed significant sleep/wake architecture changes during the dark, but not the light, phase (Figure 5D–I and Figure S5A–F). Male intruders spent more time awake and less time asleep during the dark phase compared to when with siblings (Figure 5G–I). Wake episodes were more consolidated (Figure S5D), while sleep bouts were not shorter, but less frequent (Figure S5E,F). Notably, female intruders showed minimal to no alterations in their sleep/wake architecture when housed with a male resident, compared to when with siblings (Figure 5D–I and Figure S5A–F). Together, our results suggest that for females, the presence of a social partner influences sleep more than its familiarity or sex. In contrast, for males, the prevailing social context plays a pivotal role. Our findings highlight the nuanced interplay between sex and context in social modulations of sleep.
Social context modulates the degree of neurophysiological coordination among mice
Finally, we explored the impact of non-familial social interactions on the synchronization of brain states and oscillatory brain activity among mouse pairs (n = 9 male-female and 10 male-male). We compared synchrony in mice cohabiting the same home-cage with that of the same mice when housed separately (Figure 6). We discovered a significant sleep and wake onset synchronization among both male-female and male-male dyads, compared to while individually housed (Figure 6A–H). Moreover, dyads waking up together often fell asleep together (Figure 6I). Notably, the degree of synchronization in waking up and falling asleep times among dyads surpassed that observed in sibling mice, particularly among males (Figure 6J,K). Furthermore, we identified robust synchrony in overall sleep and wake times among dyads compared to solitary housing and shuffled data (Figure 6L,N). Our cross-correlation analysis further revealed a significant coordination in sleep/wake state timing with a peak at zero lag time among dyads, a finding not observed in solitary housing (Figure 6M,O). Collectively, these findings provide compelling evidence of robust coordination in the timing of sleep and wakefulness among non-familial mouse dyads.
Figure 6: Social context modulates the degree and nature of neurophysiological coordination among mice.
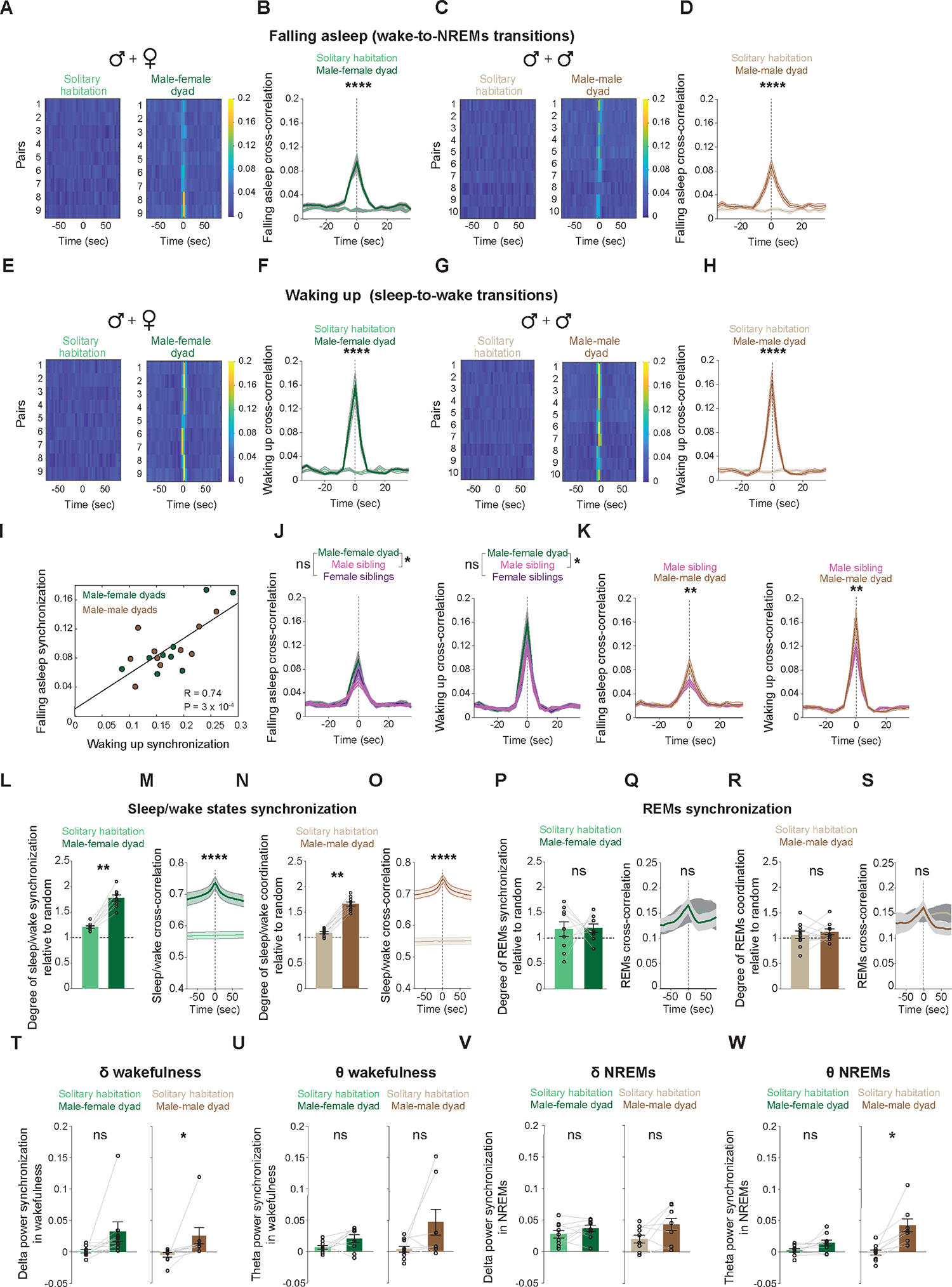
(A-H) Synchrony of sleep and wake onset times between pairs of mice, either cohabiting in the same home-cage or housed separately. (A) Heatmap representation of the cross-correlation of sleep onset times for each male-female dyad, whether housed together (right) or separately (left). (B) Average cross-correlations of sleep onset times from all male-female dyads, either housed together or separately (mean ± S.E.). Shuffle test, p < 0.0001. (C) Same as (A) but for male-male dyads. (D) Same as (B) but for male-male dyads. Shuffle test, p < 0.0001. (E) Heatmap representation of the cross-correlation of wake onset times for each male-female dyad, whether housed together (right) or separately (left). (F) Average cross-correlations of wake onset times from all male-female dyads either housed together or separately (mean ± S.E.). Shuffle test, p < 0.0001. (G) Same as (E) but for male-male dyads. (H) Same as (F) but for male-male dyads. Shuffle test, p = 0.00002. (I) Correlation between the degree of synchronization in the timing of falling asleep and waking up across both male-female (green) and male-male (brown) dyads. (J) Average cross-correlations of sleep onset (left) and wake onset (right) times for male-female, either housed together or separately (mean ± S.E.). Shuffle test, Falling asleep: p(male sibling) = 0.0086, p(female sibling) = 0.1793; Waking up: p(male sibling) = 0.0354, p(female sibling) = 0.0706. (K) Average cross-correlations of sleep onset (left) and wake onset (right) times for male-male, either housed together or separately (mean ± S.E.). Shuffle test, Falling asleep: p = 0.0091, Waking up: p = 0.0068. (L) Synchrony in overall sleep and wake times for male-female dyads, either housed together or separately. Wilcoxon rank-sum test, p = 0.0039. (M) Average cross-correlations of overall sleep and wake times from male-female dyads, either housed together or separately (mean ± S.E.). Shuffle test, p < 0.0001. (N) Same as (L), but for male-male dyads. Wilcoxon rank-sum test, p = 0.0019. (O) Same as (M), but for male-male dyads. Shuffle test, p < 0.0001. (P) Synchrony in the timing of REMs between male-female dyads, either housed together or separately. Wilcoxon rank-sum test, p = 0.99. (Q) Average cross-correlations of REMs time for male-female dyads either housed together or separately (mean ± S.E.). Shuffle test, p = 0.3342. (R) Same as (P), but for male-male dyads. Wilcoxon rank-sum test, p = 0.49. (S) Same as (Q), but for male-male dyads. Shuffle test, p = 0.2085. (T) Synchrony in the delta (1–4 Hz) band power during wakefulness between male-female (left) and male-male (right) dyads, either housed together or separately. Wilcoxon rank-sum test, p(male-female) = 0.0546, p(male-male) = 0.0156. (U) Same as (T) but for theta (5–9 Hz) band power. Wilcoxon rank-sum test, p(male-female) = 0.0976, p(male-male) = 0.1562. (V) Same as (T) but for NREMs. Wilcoxon rank-sum test, p(male-female) = 0.3007, p(male-male) = 0.2187. (W) Same as (U) but for NREMs. Wilcoxon rank-sum test, p(male-female) = 0.0546, p(male-male) = 0.0156. n = 9 male-female dyads and 10 male-male dyads.
Surprisingly, when we examined the synchrony in sleep states, we found no coordination in REMS epochs for either male-female or male-male dyads (Figure 6P–S). We also observed minimal synchronization of oscillatory brain activity between male-female and male-male dyads (Figure 6T–W). Only male-male dyads demonstrated significant synchronization in the delta power band during wakefulness (Figure 6T) and theta power band during NREMS (Figure 6W), relative to solitary housing. Taken together, these findings strongly suggest that neurophysiological coordination among mice is modulated by social context and sex.
Discussion
To assess the impact of the social environment on sleep, we employed state-of-the-art wireless neurophysiological devices, for extended monitoring of multiple freely-moving individuals within a group, alongside detailed behavioral recordings and deep-learning-based behavioral classification. We reveal that mice seek physical contact prior to sleep initiation and opt to sleep in close proximity to each other. Using a novel behavioral assay, we establish huddling during sleep as a motivated behavior. We also find that conspecific presence fragments NREMS, whereas co-sleeping animals show neurophysiological coordination across various measures. Notably, we found a synchronization in REMS timing among co-sleeping male siblings but not female or unfamiliar mice. Lastly, we highlight a social context-dependent modulation of sleep/wake architecture. Our research offers significant insights into the ethological context of sleep and furthers our understanding of sleep physiology. Considering the prevalence of sleep disturbances28 and social stress2, including loneliness, understanding the impact of social factors on sleep dynamics is essential. Gaining insights into these behavioral and neurophysiological interactions could inform strategies to mitigate sleep disturbances related to social stress, potentially enhancing human well-being.
Social factors influence both the quality and quantity of sleep across diverse species9,10. For instance, in insects, social interactions increase subsequent sleep need (based on behavioral indices)11,12, while in baboons, prior social interactions and social status modulate sleep duration and intensity (based on 3D acceleration data)13. Laboratory studies, utilizing brief recordings or focusing on single individuals within a group, further substantiate the premise that context and social status influence sleep14–16. However, until now, much remained unknown about how round-the-clock social dynamics and mating-related interactions affect sleep, as well as the extent to which social factors impact neuronal oscillatory activity during sleep. Our study provides novel insights into the complex interplay between sleep and social dynamics, through 24-hour long neurophysiological recordings from multiple subjects, while monitoring the same individuals in diverse social contexts. These contexts span from cohabitation with siblings to interactions with non-familial individuals, including mates and same-sex conspecifics.
Our study reveals that sleep/wake patterns are modulated by both sex and social context. Female mice shift their sleep timing when cohabiting with their siblings, sleeping more during the active/dark phase as compared to when alone. The presence of a male mouse does not further alter the sleep/wake states of female mice. This suggests that they are more sensitive to the presence or absence of a sleep partner, rather than its familiarity or sex. In contrast to females, male mice display sleep/wake modulation that is dependent on social context. The presence of siblings or a male intruder does not significantly alter sleep/wake duration or timing in males compared to when isolated. However, their social status plays a pivotal role. Intruding into another male’s territory increases wakefulness and reduces NREMS, especially during the dark/active phase. Mating-related interactions also alter sleep-wake states in male mice but mainly during the light/inactive phase. In the presence of a female, males stay awake longer and spend less time in REMS during the light phase, aligning their REMS duration with their female counterparts. While increased wakefulness could be due to greater opportunities to engage in motivated behaviors, the specific mechanisms and causes behind the decrease in REMS, but not NREMS, remain to be elucidated. In summary, our findings underscore the complex relationship between social factors and sleep regulation, highlighting the importance of considering both social context and sex when investigating sleep dynamics.
Our study provides novel insights into the coordination of neurobehavioral states and neurophysiological oscillatory activity during sleep among co-sleeping individuals. We identified synchronization in sleep/wake onset times, duration of sleep and wakefulness, and REMS timing. The strong synchronization in sleep/wake onset times, coupled with shorter and more frequent NREMS bouts, suggests that co-sleeping mice disturb each other’s sleep. Conspecific presence has been reported to disrupt sleep in several species. For instance, insomnia can be transferred between human bed partners29, sleep duration in baboons is reduced when in close proximity to group-mates13, and collective waves of awakenings have been observed in gulls20. Given the well-established relationship between the consolidation of sleep and its health benefits30,31, it is somewhat perplexing that animals willingly choose to sleep in situations that appear to compromise their sleep. Social living is recognized as beneficial in numerous ways1,32–35, suggesting that this type of sleep fragmentation may not be as detrimental as other forms, such as those triggered by stress. Alternatively, the benefits of group sleeping may outweigh its costs. These unresolved possibilities provide promising avenues for future research, particularly concerning any protective mechanisms that might be involved.
As opposed to the synchronization in sleep/wake onset times, the mechanisms underlying REMS synchronization remain elusive. Our data does not suggest a simple correlation between synchronized sleep initiation and REMS synchronization, as both female and male dyads exhibited strong synchronization in sleep/wake onsets, but not in REMS timing. These findings imply that while REMS timing can be synchronized among co-sleepers, this process is gated by an individual’s internal state.
While the adaptive value of REMS synchronization remains unclear, several potential benefits can be postulated. The synchronization might provide thermal benefits as thermoregulation is suppressed during REMS6 and could help mitigate the fragmentation of REMS caused by conspecific movements. Furthermore, by synchronizing a period of reduced responsiveness, animals might decrease the risk of conspecifics accessing shared resources or posing harm. Further research is needed to elucidate these potential benefits and reveal why females and unfamiliar mice do not exhibit REMS synchronization.
We also discovered synchronized oscillatory brain activity in social-living mice that was gated by the social context. Specifically, sibling mice exhibited strong theta power synchronization during both wakefulness and NREMS, while non-sibling mice showed weak to no synchronization. Synchronization in oscillatory activity during wakefulness has been reported in various species, including humans, bats and mice21–23. These studies propose that such coordination is not reliant on shared task engagement or physical contact, but is rather linked to a shared cognitive space, facilitating social communication36. Given the fundamental differences in cognitive processes between wakefulness and sleep, it is likely that diverse mechanisms oversee synchronization during these states.
Various physiological processes, including cardiac and respiratory activities, could underlie the synchronized timing of REMS and oscillatory brain activity during sleep. While responsiveness to the external environment decreases during sleep6, how external factors, as vibrations from fellow sleepers, affect sleep-regulating physiological processes and neuronal activity during sleep is still largely unknown. Notably, rocking-induced mechanosensory stimulation promote NREMS37, and an individual’s own breathing rhythm is thought to coordinate brain oscillatory activity across different regions during offline states38. Our findings significantly advance current understanding of how social factors influence sleep physiology, opening new avenues for future research. For instance, it would be of interest to identify the specific mechanisms coordinating oscillatory activity and sleep states among co-sleeping individuals and unravel the processes that gate these phenomena under different social conditions.
We demonstrated that mice actively seek physical contact prior to sleep initiation and sleep while in close proximity to each other. We demonstrate that this is a motivated behavior, as mice willingly forsake their preferred sleep location and thermoneutral conditions to huddle during sleep. Furthermore, mice choose to huddle during sleep with non-familial conspecifics, including potential same-sex rivals, even when separate sleeping structures are available. Collectively, these findings strongly suggest the existence of an inner motivation for sustained physical contact, and as there is currently no explicit word for this drive, we term it ‘somatolonging.’
Social contact plays a pivotal role in human well-being, with many studies highlighting the psychological and physiological benefits of physical touch and close relationships39–41. These benefits include improved mood, stress reduction, and a strengthened immune system4,42–44. This drive for physical contact is also observed in various animal species across different classes, including gastropod45, arthropods46, reptiles47, birds48,49 and mammals50,51. One central hypothesis for the evolution of huddling proposes it as a form of behavioral thermoregulation, providing survival advantages in harsh environments52. Our finding that mice willingly forego ambient thermoneutral conditions to huddle during sleep implies that either the thermal comfort derived from the presence of conspecifics is more desirable than ambient thermal neutrality, or that the immediate triggers for huddling differ from the thermal benefits that drove its evolutionary development.
Recently, progress has been made in identifying the neuronal circuitry of social and pleasant touch and its reinforcing properties53–57, enhancing understanding of the motivation for physical contact. Nonetheless, much remains to be revealed about the decision-making processes that initiates and maintain huddling, as well as the sensory modalities and circuitry triggering the behavior. Deepening our understanding of the behavioral-, neurophysiological-, and circuit-level facets of the motivation for social contact could provide novel insights into the regulation of natural, ethologically-relevant behaviors. Such understanding could also shed light on how social interactions influence health and well-being in humans and other species.
We have devised a novel behavioral apparatus, the ‘Immersive Social Interactions Assay,’ that presents several key features significantly enhancing current capacity to conduct comprehensive, dynamic and ethologically-relevant assessment of animal behavior within a neuroscience context (whether in its current two-chamber design or a modified three-chamber version). Firstly, our apparatus permits dynamic interactions between freely-moving individuals, enabling a full spectrum display of natural behaviors. This facet is typically missing in commonly used tests, such as the three-chamber social interaction test, which simplifies social behaviors to parameters such as proximity to other individuals and sniffing time58. Secondly, it offers animals the autonomy to either engage in or avoid social activities by choosing whether to enter or avoid a chamber. This feature introduces a new layer of complexity to the study of social behaviors like aggression and sexual behaviors, where traditionally, animals are ‘forced’ into a situation rather than given a choice to participate or not. Thirdly, in addition to allowing the placement of animals in novel testing cages, our apparatus enables the use of the animal’s home-cage. This feature may reveal behaviors not typically exhibited in a new environment, it grants animals the choice to leave their territory or not, and it eliminates the need to manipulate animals to participate in the test–eliminating the confounding effect of handling stress. Lastly, as our apparatus employs modified standard housing cages, it is accessible and cost-effective for widespread use. The integration of our apparatus with deep-learning methodologies for the detection and classification of behaviors in a social context will further support the high throughput, yet ethologically-relevant and nuanced, study of animal behavior in the lab.
STAR*Methods
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Dr. Ada Eban-Rothschild (adae@umich.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All original neurophysiological data reported in this paper will be available via Zenodo. 3D printer files will be available via github. DOIs are listed in the key resource table.
All codes will be publicly available via github.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact, Ada Eban-Rothschild adae@umich.edu, upon request.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Raw and analyzed data | This paper | https://zenodo.org/deposit/8399441 |
| Code | This paper | https://zenodo.org/deposit/8399441 |
| Image data sets | This paper | https://github.com/AER-Lab/Social-sleep |
| 3D printed files | This paper | hhttps://github.com/AER-Lab/ISIA-apparatus |
| Experimental models: Organisms/strains | ||
| Mouse: CD-1 | Charles River Laboratories | 022 |
| Software and algorithms | ||
| Adobe Illustrator/ Premiere Pro | Adobe Creative Cloud | https://www.adobe.com/ |
| BORIS | Friard and Gamba, 2016 | https://www.boris.unito.it/ |
| iSpy | iSpy | https://www.ispyconnect.com/ |
| Prism 9.5 | Graphpad software | https://www.graphpad.com/ |
| MATLAB 2020a | MathWorks | https://www.mathworks.com/products/matlab.html |
| SleepSign for Animal 3.0 | Kissei Comtec | http://www.sleepsign.com/ |
| BioRender | BioRender | https://biorender.com |
Experimental model and subject details
We utilized CD-1 mice (Strain #: 022, Charles River Laboratories) of both sexes that were reproductively inexperienced unless otherwise specified. The mice were bred in-house and were between 7–10 weeks old at the beginning of the experimental procedures. They were housed in a controlled environment with a 12-hour light/dark cycle at a temperature of 22 ± 1°C, and were provided with nesting material consisting of 4–6 grams of compressed cotton (‘Nestlet’; Ancare, Bellmore, NY, U.S.A) and 1 gram of shredded paper (‘Enviro-Dri’; Shepherd Specialty Papers, Watertown, TN, U.S.A) as well as ad libitum access to food and water. All experiments were conducted in accordance with the guidelines set forth in the US National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by both the University of Michigan Animal Care and Use Office and the University of Michigan Unit of Laboratory Animal Medicine.
Method details
Surgery
The mice were anesthetized with a ketamine-xylazine mixture (100 and 10 mg kg-1, respectively; administered via intraperitoneal injection, IP) and received lidocaine and carprofen (4 mg kg-1 and 5 mg kg-1, respectively) prior to being placed in a stereotaxic frame (David Kopf Instruments, Tujunga, CA, U.S.A.). During the surgery, the mice were kept under isoflurane anesthesia (~1% in O2). To enable individual identification and tracking, all mice, except for two male triplets (see below), were dyed with either Electric Tiger Lily (orange), Blue Moon (blue), or Electric Lizard (green) dyes from Tish & Snooky’s Manic Panic (manicpanic.com), as in59.
EEG-EMG implantation
Mice were surgically fitted with five miniature screws. Two screws serving as reference electrodes and placed on the right hemisphere (frontal: AP = 1.5 mm, ML = −1.5 mm, parietal: AP = −3.5 mm, ML = −2.8 mm), one screw serving as a ground electrode (AP = −5.9 mm, ML = 0), and two additional screws serving as support and were placed over the left hemisphere. Two EMG electrodes were inserted between the trapezius muscles. All electrodes were pre-soldered to a 7-pin connector and secured to the skull using either C&B Metabond (Parkell) and dental cement or OptiBond Universal adhesive and Revolution flowable light-cured composite (Kerr). Mice were given a minimum of 10 days for recovery before initiating experimental habituation.
Head-bar implantation
Mice underwent surgery to secure a 6-pin connector (10 mm (L) × 2 mm (W) × 10 mm (H), referred to as ‘head-bar’) to the skull using two support screws (AP = 1.5 mm, ML = 1.5 mm and AP = −3.5 mm, ML = −2.8 mm) and either C&B Metabond (Parkell) and dental cement or OptiBond Universal adhesive and Revolution flowable light-cured composite (Kerr). Sibling mice of head-bar implanted mice underwent a sham operation in which their skin was opened at the top of the skull and closed with surgical sutures. Mice were allowed at least 5 days of recovery before experimental habituation.
Video and neurophysiological acquisition
Triplet same-sex sibling mice were implanted with EEG-EMG electrodes and placed in custom plexiglass chambers (50 cm × 25 cm × 40 cm). Mice were habituated to dummy wireless neurophysiological devices, which were of similar size and weight as the genuine devices, for over a week. Video data was collected via USB cameras (either Logitech C920 or Angetube 1080p webcams) mounted above the chambers, using iSpy software (iSpyConnect.com). Neurophysiological data was collected at 400 Hz by Neurologger 2A devices (Evolocus, Tarrytown, NY, USA) powered by two 1.45 V batteries (#312, Renata Batteries). The loggers were equipped with a 3D accelerometer and an infrared (IR)-receiver micro-board, and IR pulses (Neurologger Synchronizer, Evolocus) were used to synchronize recordings from multiple devices mounted onto different mice. To protect the loggers from damage, they were covered with parafilm and a custom 3D printed perforated plastic enclosure (Figure 2A), resulting in a final combined weight of 3 gr (which accounts for approximately 8% and 9% of the body weight for CD1 adult male and female mice, respectively). The loggers were mounted onto the experimental mice 2–4 hrs prior to the initiation of recordings. The neurophysiological recordings were performed in mice that had inhabited their chambers for at least two days, and simultaneous video and neurophysiological data were collected for 24 consecutive hours, starting from Zeitgeber time (ZT) 0 (‘Lights on’).
We recorded mice under four different sequential conditions: 1) same-sex triplet sibling, 2) individual isolation, 3) non-sibling stranger male dyads, and 4) opposite-sex dyads. Conditions 1–3 were carried out while sexually inexperienced mice were used. For Condition 2, mice were individually housed for three days before the experiment. Simultaneous 24-hour long EEG-EMG and video recordings were carried out for mice from the same triplet while they were individually housed in adjacent cages in the same room. For Conditions 3 and 4, mice were individually housed for approximately one week prior to the experiment. In Condition 3, experiments were conducted in the home-cage of a “resident” mouse, into which a male “intruder” was placed at the end of the dark phase (ZT 23). In a subset of experiments (6/9), the “resident” home-cage contained two shelters (one: 8 × 5 × 5 cm and the second: 5 × 5 × 5 cm; Figure S4C). In Condition 4, experiments were conducted in the home-cage of a male “resident” mouse into which a female “intruder” mouse at the proestrus/estrus phase of the reproduction cycle was introduced at the end of the dark phase (ZT 23). Only video data was collected on Day 1 for Conditions 3 and 4, while both video and neurophysiological data were collected on Day 4.
Estrus cycle determination
To determine the estrus cycle phase, we conducted vaginal cytology by flushing vaginal cells with saline using a pipette, collecting the liquid onto a glass slide, and examining it under an epifluorescence microscope (Zeiss Axio Imager.M2). Females were tested once a day at ZT 22 for up to 5 days until nucleated epithelial cells and/or cornified cells were identified, indicating the pro-estrus/estrus phase.
Immersive social interactions assay (ISIA)
The ISIA apparatus consisted of two chambers (each 27 (L) × 16 (w) × 25 (H) cm), which were equipped with video cameras and connected by a perforated plastic tube (Length: 13.5 mm; Inner diameter: 3.7 mm; Figure S2A). To restrict the movement of mice with head-bars, a plastic ring (Inner diameter: 2.5 cm; Length: 1.5 cm) was inserted into the tube, reducing its diameter. This modification ensured that mice with head-bars were confined to one of the chambers, while mice without head-bars could move freely between the two chambers. Ad libitum food and water, and nesting material were provided in both chambers of the apparatus. Two successive experiments were conducted, each lasting two days. In Experiment 1, both chambers were maintained at room temperature (21.4 ± 0.01°C). In Experiment 2, one chamber was kept at room temperature (23.8 ± 0.01°C) while the other was at the thermoneutral zone for mice (29.9 ± 0.01°C). To maintain a temperature of 30°C in the latter chamber, an infrared ceramic heat lamp (Wuhostam, Black bulb, 100W) was placed above it. Temperature was continuously monitored via iButton devices (one measurement / 10 min; DS1925, Maxim Integrated) placed on the inner walls of the chambers (14.5 cm above chamber floor) during the experiments.
Pairs of head-bar and sham-operated sibling WT CD-1 mice (4 pairs of females and 4 pairs of males) were placed in the apparatus one day before the experiments began. One hour prior to the recordings (at ZT 23) on each experimental day, nesting material was divided between the two chambers. On Day 1, both mice were free to move between chambers. During the following light phase, mice built their nests and slept mostly in one chamber (Figure 2B,C). On Day 2, one hour prior to the recording (at ZT 23), the plastic ring was inserted into the tube, restricting the movement of mice with head-bars to the least preferred chamber defined as the one where mice spent less than 50% of the time during the light phase on Day 1. Video recordings were made during the light phase (ZT 0–12) on all experimental days. It is important to note that only mice from litters with fewer than 9 pups were used for the experiments.
To generate a unified video that incorporated the two chambers of the ISIA, each originally captured by a separate video camera, we merged the individual recordings using Adobe Premiere Pro (Adobe Creative Cloud). The original videos were resized to standardize the chambers dimensions, temporally synchronized, and subsequently converted to a .3gp file format, at a resolution of 352 × 288 pixels.
Polysomnographic analysis
We down-sampled the neurophysiological data to 256 Hz using custom MATLAB code. We digitally filtered the EEG/EMG signal and then performed a fast Fourier transformation analysis using SleepSign 3.0 for Animal software (Kissei Comtec America). The EEG was filtered between 0.3–25 Hz and the EMG was filtered between 25–50 Hz. We classified arousal states (wakefulness, NREMs, and REMs) during 4-sec time windows. An automatic analysis was first performed based on EEG power spectra, EEG integral, and EMG integral, followed by visual inspection and correction when necessary. Wakefulness was defined as desynchronized, low-amplitude EEG signal occurring concomitantly with high tonic EMG signal with phasic bursts. NREMs was defined as synchronized, high-amplitude, low-frequency (0.5–4 Hz; delta band) EEG signal with reduced EMG activity compared to wakefulness. REMS was defined as EEG signal with a dominant theta band (6–9 Hz) with very low EMG activity. The scoring was performed by investigators (M.I.S. and T.K.) who were blind to the experimental manipulation. EEG power spectrum data were expressed as values relative to the average total power during wakefulness at ZT 12–15 of each recording. Due to EEG artifacts, we excluded the data of two, five, and four mice, respectively, from the analyses presented in Figure S3H, Figure 4, and Figure 6.
Sleep/wake synchronization
To quantify sleep/wake synchronization between pairs of mice, we transformed their sleep scoring vectors into binary vectors. These vectors indicated whether each mouse was asleep or awake in each time bin over a 24-hour period. We then counted the number of time bins where both mice were asleep. To evaluate the extent of sleep/wake synchronization, we employed a shuffle test. In this procedure, the binary sleep/wake vectors of two mice underwent circular shifting 500 times by a random quantity. With each iteration, we counted the number of time bins where both shuffled vectors indicated sleep. This process allowed us to derive a distribution of sleep/wake synchronization values based on random alterations of the original data. The synchronization ratio between two mice (Figure 4K,M and Figure 6L,N) was calculated by dividing the sleep/wake synchronization value from the original data by the average sleep/wake synchronization value derived from the shuffled data.
Cross-correlation analyses
To further explore the temporal profile of sleep/wake synchronization, we implemented a complementary approach to the one outlined above. We computed the cross-correlation of binary sleep/wake vectors for each pair of animals, normalizing the cross-correlation vectors so that autocorrelations equaled one at zero lag (Figure 4L,N and Figure 6M,O). Similarly, cross-correlations were derived for shuffled vectors as well. To determine the significance of the cross-correlation, we derived a p-value by computing the proportion of shuffled cross-correlations that exhibited a peak surpassing the peak of the original data. To test whether the peak cross-correlation of group-housed mice was higher than that of isolated mice, we utilized a shuffling procedure. We calculated the difference between the peaks of the average cross-correlations for the two groups. Next, we randomly mixed cross-correlation vectors from the group-housed and isolated pairs 100,000 times, and computed the differences between the average groups of cross-correlations. The p-value for the difference between the original mean cross-correlations was obtained by calculating the proportion of differences from the shuffled data that exceeded the difference of the original data.
To compute the cross-correlation of ‘falling asleep’ and ‘waking up’ (Figure 4C,E,G,I and Figure 6B, D,F,H), we utilized a procedure akin to that detailed above for sleep/wake cross-correlation. However, in this case, the cross-correlated vectors included ‘1’s in bins where an animal transitioned from wakefulness to sleep (for the ‘falling asleep’ analysis) and from sleep to wakefulness (for the ‘waking up’ analysis), while ‘0’s were placed in all other time bins.
For the computation of REMs synchronization and cross-correlation (Figure 4P–S), we utilized a similar method to the one outlined above for sleep/wake synchronization. The key difference was that the vectors incorporated ‘1’s for bins during which the animal was in the REMs, and ‘0’s for bins when the animal was in NREMs. Notably, bins in which either animal was awake were omitted from both vectors prior to the REMs synchronization quantification. Thus, this analysis specifically quantified REMs synchronization within the sleep period.
EEG power synchronization
The power of the EEG at the delta and theta bands (detailed above) was computed by filtering the EEG within the specific bands and taking the envelope of the squared filtered signal. To assess the synchronization of the EEG power in the delta and theta bands within a specific arousal state (Figure 4T–W and Figure 6T–W), we first identified and concatenated all time bins during which both animals were in a specific arousal state, excluding any time bins in which either animal was not in that state. Subsequently, we calculated the Pearson correlation of the EEG power within the specific band across these time periods.
Manual behavioral analysis
We annotated mice behavior in detail using the Behavioral Observation Research Interactive Software (BORIS)60 at sub-second resolution from video data.
Pre-sleep behavior
We began by identifying the sleep initiation time for each mouse closest to ZT 0 and preceded by a 1-hour period of wakefulness. We then analyzed 50 minutes of data for each mouse, starting 40 minutes prior to sleep initiation and ending 10 minutes after. We annotated nine different behaviors: 1) “In-motion,” where the mouse was actively moving, including walking, running, rearing, or digging; 2) “Eating/Drinking,” where the mouse was holding or consuming food pellets or feces with its forepaws or where the mouse was licking the nozzle of the water bottle spout; 3) “Transient social interactions,” where the mouse was touching another mouse with its snout or chasing/fleeing from another mouse; 4) “Self grooming,” where the mouse was licking its fur, stroking its body with its forepaws, or scratching itself with any limb; 5) “Allo-grooming,” where the mouse was grooming the body or head of another mouse or being groomed by another mouse; 6) “Nesting,” where the mouse was engaged in pulling, carrying, fraying, push-digging, sorting or fluffing of nesting material; 7) “Resting,” where the mouse was awake and alert, either lying or sitting inside the nest with its head lifted. Occasional movements were present, and it was distinguished from sleeping by the lifted head; 8) “Sleeping,” where the mouse was motionless in a sleeping posture (lying curled up or sitting with its face tucked into its body) for ≥ 8 consecutive seconds. Brief twitches (<1 second long) may be present. We also recorded whether behaviors were performed in close physical proximity to a conspecific (‘huddling’) or otherwise (‘non-huddling’). Huddling was classified for incidents during which the bodies of two mice were in contact, resulting in the lack of visible background between the two bodies, for > 1 second. Mice bodies included the abdomen and thorax but not the head, legs, paws, and tail.
Sleep/wake states
We manually classified sleep/wake states during the ZT 5–6 time window—a period during which mice are predominantly asleep—in 3 female and 3 male WT CD-1 mice per group (with and without wireless devices). We assigned the label “Wake” when a mouse was engaged in active behaviors such as eating, drinking, grooming, walking, nesting or digging, or when it was lying down with its head lifted or displaying body movement for ≥ 1 second. “Sleep” was assigned as described in the Pre-sleep behavior section.
Sexual behavior
We manually classified the behavior of resident male mice during ZT 23–0, immediately following the placement of a female intruder into their home-cage, and four days later. We annotated four different behaviors: 1) ‘Seeking’: Mouse is approaching or sniffing conspecific; 2) ‘Mounting with lordosis’: Mouse lays over the hind quarter of a conspecific with his two front paws over their back, while conspecific displays a downward arching of the back; 3) ‘Mounting without lordosis’: Mouse lays over the hind quarter of a conspecific with his two front paws over their back, while conspecific displays a straightened back; 4) ‘Other’: Mouse is not engaged in the aforementioned behaviors but may display non-sexual-related behaviors, including eating, drinking, grooming, nesting, resting and sleeping.
Agonistic behavior
We manually classified the behavior of resident male mice during ZT 23–0, immediately following the placement of a male intruder into their home-cage, and four days later. We annotated six different behaviors: 1) ‘Mounting’: mouse lays over the hind quarter of a conspecific with his two front paws over their back; 2) ‘Attacking’: mouse is running/leaping at a conspecific, and may bite; 3) ‘Chasing’: mouse is running towards a conspecific, while conspecific is running away, but not biting; 4) ‘Aggressive grooming’: mouse is allo-grooming a conspecific while the conspecific is rigid and motionless; 5) ‘Fleeing’: mouse is running away from a conspecific while the conspecific is running toward it, with no bites present; 6) ‘Other’: mouse is not engaged in the aforementioned behaviors, but may display non-aggressive-related behaviors, including eating, drinking, grooming, nesting, and sleeping.
Markerless pose estimation
We utilized DeepLabCut software25,26 (DLC; version 2.3.1) to identify and track individual mice body parts. For each mouse, we annotated two back keypoints. The first was located midway between the tail base and the ears, while the second was situated midway between the first keypoint and the ears. We selected these keypoints as they remained predominantly visible even when the majority of the mouse’s body was occluded by nesting material or a conspecific (Figure S1A). We generated and validated separate models for each of our datasets: (a) two-chambers containing two mice (ISIA data), (b) a single chamber containing three mice with wireless devices, and (c) a single chamber containing two mice with wireless devices.
For the ISIA video dataset, we annotated a total of 811 frames. Approximately 65% of these frames were randomly selected from the dataset, consisting of 32 videos across 16 mice from 8 different experiments. The remaining frames were manually chosen to represent instances that posed a tracking challenge. This includes frames depicting one or more mice in the connecting plastic tube, frames where the mice were entirely obscured, frames containing visually confusing objects outside the behavioral apparatus, and frames from video sections where a prior model underperformed (see below). For the datasets involving either two or three mice with wireless devices in a single chamber, we applied a similar strategy, utilizing both randomly selected images from the datasets and frames selected to represent challenging tracking scenarios. We annotated a total of 986 frames for the dataset with three mice and 841 frames for the dataset with two mice.
We employed a ResNet-50-based neural network with default parameters and trained each model for 250,000 iterations. To estimate the performance of our models on novel data, we validated each model using an evaluation set, which we created by randomly sampling 20% of each dataset. This evaluation procedure was repeated five times for each model, with resampled evaluation sets for each run. We computed the mean average Euclidian error (MAE, which is proportional to the average root mean square error) between manual labels and the ones predicted by DLC. For the ISIA dataset, we recorded an average (± S.E.M.) training error of 3.934 ± 0.2 pixels and an average (± S.E.M.) testing error of 5.864 ± 0.19 pixels (the size of most images was 352 × 288 pixels). The finalized models, which were used to perform markerless pose estimation, incorporated all annotated frames from each dataset. An experimenter validated the accurate identification and tracking of keypoints across all videos at 2 to 64 times the normal playback speed. We progressively increased the size of the training set, specifically incorporating instances that posed tracking challenges, until no identification switches or tracking errors were detected.
For each recording, we also annotated the edge points of the chambers, which allowed us to standardize the coordinate data across all trials (Figure S1B). This step was necessary due to varying distances between the cameras and the cages in different recordings. Accordingly, we transformed the DLC output coordinates so that a consistent real-world distance (1 mm) corresponded to the same pixel distance (1 unit) both within and across datasets (Figure S1C).
Huddling classification
We started by filtering the coordinate data, excluding frames where the coordinates either fell outside the edge points of the chambers or had a likelihood value equal to or below 0.1 (Figure 2A and S1C). The latter typically corresponded to instances where the mice were visually undetectable, due to being completely occluded by nesting material, conspecifics, or tucked away inside the tube (Figure S1D,E).
We then computed, for every frame, the closest proximity (minimum Euclidian distance) between each of the two keypoints of each mouse. We classified framed by comparing this distance to a pre-set hyper-parameter threshold value for physical contact (5 cm; see below). We first classified frames into two categories: those in which the minimum Euclidian distance was equal to or below the pre-set threshold, designated as ‘physical contact,’ and those in which the distance exceeded the threshold, designated as ‘no physical contact.’ If the ‘physical contact’ state persisted for more than a second, we classified the frames as instances of ‘huddling.’ If not, they were considered ‘non-huddling’ (Figure S1G).
To establish the hyper-parameter threshold value for physical contact, we initially generated a training set by randomly sampling frames from our entire dataset. We manually excluded frames where the outlines of the mice bodies were occluded, which hindered a clear determination of whether the mice were touching or not. Subsequently, we manually classified the images as depicting either ‘physical contact’ or ‘no physical contact,’ using the previously described criteria (see Pre-sleep behavior section), resulting in 935 ‘physical contact’ frames and 1178 ‘no physical contact’ frames. We plotted a histogram of the minimum Euclidean distances for the two classified states (Figure S1F). This visualization highlighted 5 cm as an optimal threshold value for physical contact. To assess the chosen hyper-parameter threshold, we generated three evaluation sets by randomly re-sampling our dataset (excluding images used for training), testing three distinct threshold values for physical contact: 4 cm, 5 cm, and 6 cm. Once again, we excluded frames where the outlines of the mice were obscured, yielding 1988, 2037, and 2020 frames for the 4 cm, 5 cm, and 6 cm thresholds, respectively. We computed false and true positives, along with Accuracy (number of correct predictions divided by dataset size), Precision (True Positives divided by the sum of True Positives and False Positives), Recall (True Positives divided by the sum of True Positives and False Negatives), Specificity (True Negatives divided by the sum of True Negatives and False Positives), and F1 score [2 multiplied by ((Precision multiplied by Recall) divided by the sum of Precision and Recall)] for the three distance values (Table 1). This evaluation procedure further affirmed our selection of 5 cm as the threshold value for physical contact.
Quantification and statistical analysis
Data and statistical analyses were conducted using SleepSign for Animals (3.0), MATLAB (2020a), and Prism 9.5.1 (GraphPad Software). The sample sizes were analogous to those reported in previous studies61. The data analysis was carried out blind to the experimental conditions. While we assumed data distribution was normal, this wasn’t formally tested in every instance. For comparing the data of two paired groups, we employed Wilcoxon signed-rank tests or paired t-tests. In cases involving more than two groups, we utilized the Friedman test or one-way repeated-measures (RM) ANOVA. For multiple comparisons, we used RM two-way ANOVA or, for datasets with missing values (e.g., absence of a particular sleep state at a specific time point), we utilized mixed-effects model ANOVA. Post-hoc tests were conducted using Sidak’s multiple comparisons tests. The exact P values and n values for the statistical tests are detailed in the figure legends. Adobe Illustrator 2023 (Adobe Creative Cloud) was used for figure preparation.
Supplementary Material
Video S1. Representative DLC tracking output videos for a pair of sibling mice across the ISIA assay under room temperature conditions, Related to Figure 2.
Video S2. Representative DLC tracking output videos for a pair of sibling mice across the ISIA assay under optional ambient thermoneutrality conditions, Related to Figure 2.
Mice seek physical contact before sleep initiation and huddle during their sleep
Somatolonging–a motivation for prolonged physical contact–drives huddling behavior
Co-sleeping individuals show coordination in multiple neurophysiological features
REMS timing synchronizes among male siblings, but not female or unfamiliar mice
Acknowledgments
We thank Gaurav Kaul and Dr. Matt Gaidica for 3D design, Maya Sheth and Pramith Senaratne for annotations and video processing and Gaurav Kaul and Shijie Qu for advice on huddling classification. Figures 1A, 2B and 2E, 3B and 5A were created with BioRender.com. The graphical abstract was created utilizing Adobe Firefly Generative AI. This study was supported by the Alfred P. Sloan Foundation (A.E.-R.), the Konishi Neuroethology Award (M.I.S.), the Sleep Research Society Small Grant (M.I.S.) and the Sigma-Xi Grant in Aid of Research (C.K.).
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Footnotes
Declaration of interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wilson EO (2000). Sociobiology The New Synthesis, Twenty-Fifth Anniversary Edition (Harvard University Press; ). 10.2307/j.ctvjnrttd. [DOI] [Google Scholar]
- 2.Leigh-Hunt N, Bagguley D, Bash K, Turner V, Turnbull S, Valtorta N, and Caan W (2017). An overview of systematic reviews on the public health consequences of social isolation and loneliness. Public Health 152, 157–171. 10.1016/j.puhe.2017.07.035. [DOI] [PubMed] [Google Scholar]
- 3.Ruan H, and Wu CF (2008). Social interaction-mediated lifespan extension of Drosophila Cu/Zn superoxide dismutase mutants. Proc Natl Acad Sci U S A 105, 7506–7510. 10.1073/pnas.0711127105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hermes GL, Delgado B, Tretiakova M, Cavigelli SA, Krausz T, Conzen SD, and McClintock MK (2009). Social isolation dysregulates endocrine and behavioral stress while increasing malignant burden of spontaneous mammary tumors. Proc Natl Acad Sci U S A 106, 22393–22398. 10.1073/pnas.0910753106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanitz E, Tuchscherer M, Puppe B, Tuchscherer A, and Stabenow B (2004). Consequences of repeated early isolation in domestic piglets (Sus scrofa) on their behavioural, neuroendocrine, and immunological responses. Brain Behav Immun 18, 35–45. 10.1016/s0889-1591(03)00085-0. [DOI] [PubMed] [Google Scholar]
- 6.Sulaman BA, Wang S, Tyan J, and Eban-Rothschild A (2023). Neuro-orchestration of sleep and wakefulness. Nat Neurosci 26, 196–212. 10.1038/s41593-022-01236-w. [DOI] [PubMed] [Google Scholar]
- 7.Rattenborg NC, de la Iglesia HO, Kempenaers B, Lesku JA, Meerlo P, and Scriba MF (2017). Sleep research goes wild: new methods and approaches to investigate the ecology, evolution and functions of sleep. Philos Trans R Soc Lond B Biol Sci 372. 10.1098/rstb.2016.0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eban-Rothschild A (2022). Beyond model organisms: diversifying experimental species and ecological complexity to reveal the evolutionary history and functions of sleep. Sleep 45. 10.1093/sleep/zsac102. [DOI] [PubMed] [Google Scholar]
- 9.Eban-Rothschild A, and Bloch G (2012). Social influences on circadian rhythms and sleep in insects. Adv Genet 77, 1–32. 10.1016/b978-0-12-387687-4.00001-5. [DOI] [PubMed] [Google Scholar]
- 10.Smeltzer EA, Stead SM, Li MF, Samson D, Kumpan LT, and Teichroeb JA (2022). Social sleepers: The effects of social status on sleep in terrestrial mammals. Horm Behav 143, 105181. 10.1016/j.yhbeh.2022.105181. [DOI] [PubMed] [Google Scholar]
- 11.Eban-Rothschild A, and Bloch G (2015). The colony environment modulates sleep in honey bee workers. J Exp Biol 218, 404–411. 10.1242/jeb.110619. [DOI] [PubMed] [Google Scholar]
- 12.Ganguly-Fitzgerald I, Donlea J, and Shaw PJ (2006). Waking experience affects sleep need in Drosophila. Science 313, 1775–1781. 10.1126/science.1130408. [DOI] [PubMed] [Google Scholar]
- 13.Loftus JC, Harel R, Núñez CL, and Crofoot MC (2022). Ecological and social pressures interfere with homeostatic sleep regulation in the wild. Elife 11. 10.7554/eLife.73695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gravett N, Bhagwandin A, Lyamin OI, Siegel JM, and Manger PR (2017). Sociality Affects REM Sleep Episode Duration Under Controlled Laboratory Conditions in the Rock Hyrax, Procavia capensis. Front Neuroanat 11, 105. 10.3389/fnana.2017.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Febinger HY, George A, Priestley J, Toth LA, and Opp MR (2014). Effects of housing condition and cage change on characteristics of sleep in mice. J Am Assoc Lab Anim Sci 53, 29–37. [PMC free article] [PubMed] [Google Scholar]
- 16.Karamihalev S, Flachskamm C, Eren N, Kimura M, and Chen A (2019). Social context and dominance status contribute to sleep patterns and quality in groups of freely-moving mice. Sci Rep 9, 15190. 10.1038/s41598-019-51375-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keene AC, and Duboue ER (2018). The origins and evolution of sleep. J Exp Biol 221. 10.1242/jeb.159533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lesku JA, and Schmidt MH (2022). Energetic costs and benefits of sleep. Curr Biol 32, R656–r661. 10.1016/j.cub.2022.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Drews HJ, Wallot S, Brysch P, Berger-Johannsen H, Weinhold SL, Mitkidis P, Baier PC, Lechinger J, Roepstorff A, and Göder R (2020). Bed-Sharing in Couples Is Associated With Increased and Stabilized REM Sleep and Sleep-Stage Synchronization. Front Psychiatry 11, 583. 10.3389/fpsyt.2020.00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beauchamp G (2011). Collective Waves of Sleep in Gulls (Larus spp.). Ethology 117, 326–331. 10.1111/j.1439-0310.2011.01875.x. [DOI] [Google Scholar]
- 21.Dumas G, Nadel J, Soussignan R, Martinerie J, and Garnero L (2010). Inter-brain synchronization during social interaction. PLoS One 5, e12166. 10.1371/journal.pone.0012166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang W, and Yartsev MM (2019). Correlated Neural Activity across the Brains of Socially Interacting Bats. Cell 178, 413–428.e422. 10.1016/j.cell.2019.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kingsbury L, Huang S, Wang J, Gu K, Golshani P, Wu YE, and Hong W (2019). Correlated Neural Activity and Encoding of Behavior across Brains of Socially Interacting Animals. Cell 178, 429–446.e416. 10.1016/j.cell.2019.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sotelo MI, Tyan J, Markunas C, Sulaman BA, Horwitz L, Lee H, Morrow JG, Rothschild G, Duan B, and Eban-Rothschild A (2022). Lateral hypothalamic neuronal ensembles regulate pre-sleep nest-building behavior. Curr Biol 32, 806–822.e807. 10.1016/j.cub.2021.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathis A, Mamidanna P, Cury KM, Abe T, Murthy VN, Mathis MW, and Bethge M (2018). DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat Neurosci 21, 1281–1289. 10.1038/s41593-018-0209-y. [DOI] [PubMed] [Google Scholar]
- 26.Nath T, Mathis A, Chen AC, Patel A, Bethge M, and Mathis MW (2019). Using DeepLabCut for 3D markerless pose estimation across species and behaviors. Nat Protoc 14, 2152–2176. 10.1038/s41596-019-0176-0. [DOI] [PubMed] [Google Scholar]
- 27.Gordon CJ, Becker P, and Ali JS (1998). Behavioral thermoregulatory responses of single- and group-housed mice11This paper has been reviewed by the National Health and Environmental Effects Research Laboratory, U.S. Environmental Protection Agency, and approved for publication. Mention of trade names or commercial products does not constitute endorsement or recommendation for use. Physiology & Behavior 65, 255–262. 10.1016/S0031-9384(98)00148-6. [DOI] [PubMed] [Google Scholar]
- 28.Khurshid KA (2018). Comorbid Insomnia and Psychiatric Disorders: An Update. Innov Clin Neurosci 15, 28–32. [PMC free article] [PubMed] [Google Scholar]
- 29.Walters EM, Phillips AJK, Mellor A, Hamill K, Jenkins MM, Norton PJ, Baucom DH, and Drummond SPA (2020). Sleep and wake are shared and transmitted between individuals with insomnia and their bed-sharing partners. Sleep 43. 10.1093/sleep/zsz206. [DOI] [PubMed] [Google Scholar]
- 30.Grandner MA (2020). Sleep, Health, and Society. Sleep Med Clin 15, 319–340. 10.1016/j.jsmc.2020.02.017. [DOI] [PubMed] [Google Scholar]
- 31.Lim DC, and Pack AI (2017). Obstructive Sleep Apnea: Update and Future. Annual Review of Medicine 68, 99–112. 10.1146/annurev-med-042915-102623. [DOI] [PubMed] [Google Scholar]
- 32.Cremer S, Armitage SAO, and Schmid-Hempel P (2007). Social Immunity. Current Biology 17, R693–R702. 10.1016/j.cub.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 33.Almberg ES, Cross PC, Dobson AP, Smith DW, Metz MC, Stahler DR, and Hudson PJ (2015). Social living mitigates the costs of a chronic illness in a cooperative carnivore. Ecology Letters 18, 660–667. 10.1111/ele.12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrison I (2016). Keep Calm and Cuddle on: Social Touch as a Stress Buffer. Adaptive Human Behavior and Physiology 2, 344–362. 10.1007/s40750-016-0052-x. [DOI] [Google Scholar]
- 35.Ebensperger LA (2001). A review of the evolutionary causes of rodent group-living. Acta Theriologica 46, 115–144. 10.1007/BF03192423. [DOI] [Google Scholar]
- 36.Kingsbury L, and Hong W (2020). A Multi-Brain Framework for Social Interaction. Trends Neurosci 43, 651–666. 10.1016/j.tins.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kompotis K, Hubbard J, Emmenegger Y, Perrault A, Mühlethaler M, Schwartz S, Bayer L, and Franken P (2019). Rocking Promotes Sleep in Mice through Rhythmic Stimulation of the Vestibular System. Current Biology 29, 392–401.e394. 10.1016/j.cub.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 38.Karalis N, and Sirota A (2022). Breathing coordinates cortico-hippocampal dynamics in mice during offline states. Nat Commun 13, 467. 10.1038/s41467-022-28090-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.House JS, Landis KR, and Umberson D (1988). Social Relationships and Health. Science 241, 540–545. doi: 10.1126/science.3399889. [DOI] [PubMed] [Google Scholar]
- 40.Eisenberger NI, and Cole SW (2012). Social neuroscience and health: neurophysiological mechanisms linking social ties with physical health. Nature Neuroscience 15, 669–674. 10.1038/nn.3086. [DOI] [PubMed] [Google Scholar]
- 41.Cascio CJ, Moore D, and McGlone F (2019). Social touch and human development. Developmental Cognitive Neuroscience 35, 5–11. 10.1016/j.dcn.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holt-Lunstad J, Smith TB, and Layton JB (2010). Social relationships and mortality risk: a meta-analytic review. PLoS Med 7, e1000316. 10.1371/journal.pmed.1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berkman LF, and Syme SL (1979). Social networks, host resistance, and mortality: a nine-year follow-up study of Alameda county residents. American Journal of Epidemiology 109, 186–204. 10.1093/oxfordjournals.aje.a112674. [DOI] [PubMed] [Google Scholar]
- 44.Steptoe A, Shankar A, Demakakos P, and Wardle J (2013). Social isolation, loneliness, and all-cause mortality in older men and women. Proc Natl Acad Sci U S A 110, 5797–5801. 10.1073/pnas.1219686110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cook A (1981). Huddling and the control of water loss by the slug Limax pseudoflavus Evans. Animal Behaviour 29, 289–298. 10.1016/S0003-3472(81)80177-7. [DOI] [Google Scholar]
- 46.Hassall M, Edwards D, Carmenta R, Derhé M, and Moss A (2010). Predicting the effect of climate change on aggregation behaviour in four species of terrestrial isopods. Behaviour 147, 151–164. 10.1163/000579509X12512861455834. [DOI] [Google Scholar]
- 47.Shah B, Shine R, Hudson S, and Kearney M (2003). Sociality in Lizards: Why Do Thick-Tailed Geckos (Nephrurus milii) Aggregate? Behaviour 140, 1039–1052. [Google Scholar]
- 48.Prévost J (1961). Ecologie du manchot empereur. Expéditions polaires françaises. Hermann Press, Paris. [Google Scholar]
- 49.Rowan M (1967). A study of the colies of southern Africa. Ostrich 38, 63–115. [Google Scholar]
- 50.Eppley TM, Watzek J, Dausmann KH, Ganzhorn JU, and Donati G (2017). Huddling is more important than rest site selection for thermoregulation in southern bamboo lemurs. Animal Behaviour 127, 153–161. [Google Scholar]
- 51.Takahashi H (1997). Huddling relationships in night sleeping groups among wild Japanese macaques in Kinkazan Island during winter. Primates 38, 57–68. [Google Scholar]
- 52.Gilbert C, McCafferty D, Le Maho Y, Martrette JM, Giroud S, Blanc S, and Ancel A (2010). One for all and all for one: the energetic benefits of huddling in endotherms. Biol Rev Camb Philos Soc 85, 545–569. 10.1111/j.1469-185X.2009.00115.x. [DOI] [PubMed] [Google Scholar]
- 53.Elias LJ, Succi IK, Schaffler MD, Foster W, Gradwell MA, Bohic M, Fushiki A, Upadhyay A, Ejoh LL, Schwark R, et al. (2023). Touch neurons underlying dopaminergic pleasurable touch and sexual receptivity. Cell 186, 577–590.e516. 10.1016/j.cell.2022.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang Y, Benusiglio D, Lefevre A, Hilfiger L, Althammer F, Bludau A, Hagiwara D, Baudon A, Darbon P, Schimmer J, et al. (2020). Social touch promotes interfemale communication via activation of parvocellular oxytocin neurons. Nature Neuroscience 23, 1125–1137. 10.1038/s41593-020-0674-y. [DOI] [PubMed] [Google Scholar]
- 55.Olausson H, Lamarre Y, Backlund H, Morin C, Wallin BG, Starck G, Ekholm S, Strigo I, Worsley K, Vallbo ÅB, and Bushnell MC (2002). Unmyelinated tactile afferents signal touch and project to insular cortex. Nature Neuroscience 5, 900–904. 10.1038/nn896. [DOI] [PubMed] [Google Scholar]
- 56.Vrontou S, Wong AM, Rau KK, Koerber HR, and Anderson DJ (2013). Genetic identification of C fibres that detect massage-like stroking of hairy skin in vivo. Nature 493, 669–673. 10.1038/nature11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu H, Miao W, Ji E, Huang S, Jin S, Zhu X, Liu MZ, Sun YG, Xu F, and Yu X (2022). Social touch-like tactile stimulation activates a tachykinin 1-oxytocin pathway to promote social interactions. Neuron 110, 1051–1067.e1057. 10.1016/j.neuron.2021.12.022. [DOI] [PubMed] [Google Scholar]
- 58.Jabarin R, Netser S, and Wagner S (2022). Beyond the three-chamber test: toward a multimodal and objective assessment of social behavior in rodents. Mol Autism 13, 41. 10.1186/s13229-022-00521-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shemesh Y, Sztainberg Y, Forkosh O, Shlapobersky T, Chen A, and Schneidman E (2013). High-order social interactions in groups of mice. Elife 2, e00759. 10.7554/eLife.00759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Friard O, and Gamba M (2016). BORIS: a free, versatile open-source event-logging software for video/audio coding and live observations. Methods in Ecology and Evolution 7, 1325–1330. 10.1111/2041-210X.12584. [DOI] [Google Scholar]
- 61.Eban-Rothschild A, Rothschild G, Giardino WJ, Jones JR, and de Lecea L (2016). VTA dopaminergic neurons regulate ethologically relevant sleep-wake behaviors. Nat Neurosci 19, 1356–1366. 10.1038/nn.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Representative DLC tracking output videos for a pair of sibling mice across the ISIA assay under room temperature conditions, Related to Figure 2.
Video S2. Representative DLC tracking output videos for a pair of sibling mice across the ISIA assay under optional ambient thermoneutrality conditions, Related to Figure 2.
Data Availability Statement
All original neurophysiological data reported in this paper will be available via Zenodo. 3D printer files will be available via github. DOIs are listed in the key resource table.
All codes will be publicly available via github.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact, Ada Eban-Rothschild adae@umich.edu, upon request.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Raw and analyzed data | This paper | https://zenodo.org/deposit/8399441 |
| Code | This paper | https://zenodo.org/deposit/8399441 |
| Image data sets | This paper | https://github.com/AER-Lab/Social-sleep |
| 3D printed files | This paper | hhttps://github.com/AER-Lab/ISIA-apparatus |
| Experimental models: Organisms/strains | ||
| Mouse: CD-1 | Charles River Laboratories | 022 |
| Software and algorithms | ||
| Adobe Illustrator/ Premiere Pro | Adobe Creative Cloud | https://www.adobe.com/ |
| BORIS | Friard and Gamba, 2016 | https://www.boris.unito.it/ |
| iSpy | iSpy | https://www.ispyconnect.com/ |
| Prism 9.5 | Graphpad software | https://www.graphpad.com/ |
| MATLAB 2020a | MathWorks | https://www.mathworks.com/products/matlab.html |
| SleepSign for Animal 3.0 | Kissei Comtec | http://www.sleepsign.com/ |
| BioRender | BioRender | https://biorender.com |


