Abstract
Introduction:
Mortality from preeclampsia (PE) and PE-associated morbidities are 3-to 5-fold higher in persons of African ancestry than in those of Asian and European ancestries.
Methods:
To elucidate placental contribution to worse PE outcomes in African ancestry pregnancies, we performed bulk RNA sequencing on 50 placentas from persons with severe PE (sPE) of African (n=9), Asian (n=18) and European (n=23) ancestries and 73 normotensive controls of African (n=10), Asian (n=15) and European (n=48) ancestries.
Results:
Previously described canonical preeclampsia genes, involved in metabolism and hypoxia/angiogenesis including: LEP, HK2, FSTL3, FLT1, ENG, TMEM45A, ARHGEF4 and HTRA1 were upregulated sPE versus normotensive placentas across ancestries. LTF, NPR3 and PHYHIP were higher in African vs. Asian ancestry sPE placentas. Allograft rejection/adaptive immune response genes were upregulated in placentas from African but not in Asian or European ancestry sPE patients; IL3RA was of particular interest because the patient with the highest placental IL3RA expression, a person of African ancestry with sPE, developed postpartum cardiomyopathy, and was the only patient out of 123, that developed this condition. Interestingly, the sPE patients with the highest IL3RA expression among persons of Asian and European ancestries developed unexplained tachycardia peripartum, necessitating echocardiography in the European ancestry patient. The association between elevated placental IL3RA levels and unexplained tachycardia or peripartum cardiomyopathy was found to be significant in the 50 sPE patients (p = .0005).
Discussion:
High placental upregulation of both canonical preeclampsia and allograft rejection/adaptive immune response genes may contribute to worse PE outcomes in African ancestry sPE patients.
Keywords: Preeclampsia, Placenta, African, Immune, Cardiomyopathy, IL3RA
Introduction
Preeclampsia (PE) is a hypertensive disorder of pregnancy (HDP) that the WHO lists as one of the 5 major causes of maternal mortality worldwide1. Pregnant and parturient persons of African ancestry have 1.5 to 2.5-times higher rates of PE2–5 and are 3 to 5-times more likely to die of PE than pregnant and parturient persons of Asian or European ancestry6–7. PE-associated morbidities, such as, stroke, pulmonary edema and heart failure, are also significantly higher in pregnant and parturient persons of African ancestry6–7.
Underlying comorbidities and lower socioeconomic status (SES) do not entirely explain the higher PE rates and worse prognosis in persons of African ancestry. A population-based study of 718,604 Black and White pregnant and parturient persons from a California cohort of singleton births found that, although White persons of higher SES had a lower risk of PE than White persons of lower SES, higher SES did not attenuate PE risk in Black persons8. A review of the records of 1,355 pregnant persons with HDP found similar rates of underlying chronic hypertension in Black and White pregnant persons. However, among pregnant persons with HDP without underlying chronic hypertension, Black persons were significantly more likely to be diagnosed with PE than White persons9. A retrospective cohort study of California births in 2007 found that though obesity was a risk factor for PE and low birth weight in White pregnant persons, obesity did not confer significantly increased PE risk in Black persons10.
The placenta plays a central role in the pathology of PE, as PE only occurs in the context of pregnancy, and delivery of the placenta is often curative11–13. However, how the placenta contributes to worse PE outcomes in pregnant and parturient persons of African ancestry, and which specific placental injury pathways may be dysregulated in this population (and therefore potential targets for therapy), are not well studied.
Molecular and clinical data point to multiple pathways leading to PE. Using unsupervised clustering of aggregate microarray datasets (first in 173 patient samples, including 77 with PE and 96 controls, and later in a larger dataset of 330 samples, including 157 PE and 173 non-PE samples), Leavey et al14–15 identified 3 molecular clusters/subclasses of PE which they termed maternal, canonical and immunologic. In the maternal PE group, infant birthweight and placental weight were appropriate for gestational age, and placental histology and gene expression patterns were not significantly different from normotensive controls. In the canonical PE group, birthweight and placental weight were small for gestational age, placental histology showed evidence of maternal vascular malperfusion, and genes associated with metabolism, hormone regulation and angiogenesis were overexpressed. In the immunologic PE group, birthweight and placental weight were small for gestational age, placental histology showed massive/increased perivillous fibrin deposition, and genes associated with immune and inflammatory responses were overexpressed.
Recently, our group performed RNA sequencing on placentas from PE patients with evidence of maternal vascular malperfusion and found trophoblast defects and activation of pathways associated with hypoxia, inflammation and reduced cell proliferation in placentas with maternal vascular malperfusion16, thus demonstrating that placental tissue RNA sequencing reflects pathways associated with histologic evidence of placental injury in PE.
Therefore, we sought to elucidate the cellular and molecular underpinnings of the worse PE outcomes in pregnant and parturient persons of African ancestry by performing bulk RNA sequencing on placentas from persons affected by PE with severe features (sPE) and normotensive controls of African ancestry compared to persons of Asian and European ancestries. We hypothesized that molecular PE subclasses differ in placentas from parturient persons of African ancestry versus those of Asian and European ancestries.
Materials and Methods
Study cohort selection
The study was approved by University of California, San Diego (UCSD) institutional review board, approval #181917. The study specimens were obtained from 2655 singleton parturitions at UCSD-affiliated hospitals between January 2010 and November 2020. The patients whose samples were collected gave written informed consent for accessing their electronic medical records and collection of placental tissue. Demographic, clinical and placenta histology data were collected in a REDCap-based obstetric registry. For each pregnancy, the clinical diagnosis of preeclampsia with severe features (sPE) was adjudicated by 2 Board-Certified obstetricians based on ACOG definition17. Of the 2655 parturitions, 1003 for which the maternal ethnicity was recorded as Hispanic-other were excluded, because Hispanic is a language group that can include people of diverse ancestries, ranging from African (e.g. Afro-Cubans) to North and South Americans (e.g. Mexicans and Brazilians), Asians (e.g. Spanish Filipinos) and Europeans (e.g. Spaniards). Categorization as Hispanic-other thus provided insufficient information about ancestry and had the potential to confound the study. 4 American Indian, 18 Native Hawaiian and 3 self-identified multiracial without further ancestry delineation were excluded because the numbers of patients in these categories were too few to gather generalizable information. There remained 142 parturitions with maternal African ancestry (self-identified as black, African or African American), 265 with Asian ancestry (self-identified as Asian) and 1220 with European ancestry (self-identified as white, including Hispanic and non-Hispanic). Of these, RNA sequencing was performed on 9 sPE and 9 normotensive controls of African ancestry, 18 sPE and 15 normotensive controls of Asian ancestry and 23 sPE and 49 normotensive controls of European ancestry. We then validated the self-reported maternal ancestries using a previously published method for inferring ancestry from RNAseq data,18. (Supplemental Figure 1).
Obstetric Data
The demographic data collected included maternal ancestry, maternal age at delivery, gravidity, parity, maternal body mass index (BMI), maternal history of diabetes, maternal history of chronic hypertension, gestational age at delivery, fetal sex, birth weight, mode of delivery, documented sPE, intrauterine growth restriction (IUGR), maternal pregnancy complications including renal failure, stroke, unexplained tachycardia, or peripartum cardiomyopathy.
RNA sequencing and RNA sequencing-based ancestry inference
Immunohistochemistry and Immunohistochemistry quantification
Statistical analysis
Results
Histopathologic analysis cohort
The histopathologic analysis cohort was comprised of 868 patients; 127 with sPE and 741 normotensive controls. Maternal age, gestational age at delivery, gravidity, parity, infant sex, infant birth weight and mode of delivery were evaluated in sPE patients African (n=15), Asian (n=23) and European (n=89) ancestries compared to normotensive controls of African (n=39), Asian (n=114) and European (n=588) ancestries. Gestational age at delivery and birth weight were significantly less in sPE patients than in normotensive controls across ancestries (Supplemental Table 1).
Higher incidence of maternal vascular malperfusion and perivillous fibrin deposition in placentas from patients of African ancestry with severe PE
We analyzed the 868 placentas using previously defined histopathologic criteria27–28. We found the incidence of maternal vascular malperfusion and perivillous fibrin deposition was higher in placentas from parturients of African ancestry with sPE compared to those of Asian or European ancestry, suggesting worse placental injury in the African ancestry sPE group (Supplemental Table 2).
RNA sequencing patient demographics
Maternal age, maternal BMI, maternal history of chronic hypertension, maternal history of diabetes mellitus, gestational age at delivery, gravidity, parity, infant sex, infant birth weight, infant birth weight percentile and mode of delivery were evaluated in sPE versus normotensive control patients of African, Asian and European ancestries whose placenta samples were RNA sequenced. In comparison to normotensive controls, infant birth weight and birth weight percentiles were significantly lower in sPE parturitions across ancestries; gestational age at delivery, gravidity and parity were significantly less in African ancestry sPE parturitions; maternal BMI was significantly higher in European ancestry sPE parturitions (Supplemental Table 3).
Among the sPE patients, maternal BMI at delivery was higher in African ancestry patients than in Asian and European ancestry patients; there was no significant difference in maternal BMI at < 20 weeks of gestation, maternal age, gestational age at delivery, gravidity, parity, infant sex, birth weight and mode of delivery – across ancestries (Supplemental Table 4).
RNA sequencing-inferred ancestries and concordance with self-reported ancestries
We compared self-reported ancestry to genetic ancestry based on SNPs commonly found in different geographic regions18, 29–30 (Supplemental Figure 2A). SNPs found in self-reported Whites included those commonly found in Europe and the Americas. Of the 72 self-reported White patients, 50 (69.4%) had SNPs associated with geographic origins in Europe, 21 (29.2%) had SNPs associated with geographic origins in the Americas (including Mexico and South America) and 1 (1.4%) had SNPs associated with geographic origins in Africa. Upon delving deeper into the medical records of the 1 patient, we discovered that the reported ancestry in our electronic medical record was discrepant with that in the referral letter from the primary care facility. The ancestry in the referral letter matched the RNAseq-inferred ancestry; thus the patient’s placenta was re-assigned to the African ancestry group for subsequent analysis.
SNPs found in self-reported Asians included those commonly found in East Asia, South Asia, Europe and the Americas. Of the 33 self-reported Asian patients, 25 (75%) had SNPs associated with geographic origins in East Asia or South Asia, 6 placentas had SNPs associated with geographic origins in Europe, and 2 with geographic origins in the Americas. Review of the electronic medical records in the 8 discrepant cases revealed that discrepancy was due to paternal origins outside of Asia, but confirmed maternal origins in Asia (Korea, Japan, Cambodia, Philippines, Vietnam).
SNPs found in self-reported Blacks/African-Americans/Africans included those commonly found in Africa, Europe, East-Asia and the Americas. Of the 18 self-reported African ancestry patients, 11 (61.1%) had SNPs associated with geographic origins in Africa; 3 (16.7%) had SNPs associated with geographic origins in Europe, 2 (11.1%) had SNPs associated with geographic origins in East Asia and 2 (11.1%) had SNPs associated with geographic origins in the Americas. Review of the electronic medical records revealed 4 of the 7 discrepant cases were in African-Americans with mixed ancestry (e.g. Irish and African or Indonesian and African) and 3 of the 7 discrepant cases were in Somalis who were categorized as Europeans by RNAseq. Our reference database for generating our pipeline for inferring ancestry was the 1000 genomes project, which though diverse, did not include Somalis. The absence of Somalis in the 1000 genomes project likely explains our observed discrepancy between self-reported and RNAseq-inferred ancestry in Somalis. We thus left self-reported Somalis in the African ancestry category.
Therefore, we ended with 9 sPE and 10 normotensive controls of African ancestry, 18 sPE and 15 normotensive controls of Asian ancestry and 23 sPE and 48 normotensive controls of European ancestry (Supplemental Figure 1).
Canonical preeclampsia genes and pathways are upregulated in placentas from patients of all ancestries with sPE
We first performed principle component analysis (PCA) and found that the greatest amount of difference between these samples was between PE and the control groups as a whole (PC1: 20% and PC2: 12%) (Supplemental Figure 2B). Ancestry-dependent separation was subtle at PC10 (2%) in PE African vs. others (Supplemental figure 2C and Supplemental Table 5).
We next performed differential gene expression analysis of sPE vs. normotensive controls within each ancestry, and found 120 protein-coding genes upregulated in African-sPE, 158 in Asian-sPE, and 891 in European-sPE placentas (Figure 1A; Supplemental Tables 6–8). Among the 120 protein-coding genes upregulated in African ancestry sPE versus African ancestry normotensive controls, 67 of them were also upregulated in Asian and European sPE versus normotensive controls (Figures 1A–B).
Figure 1:
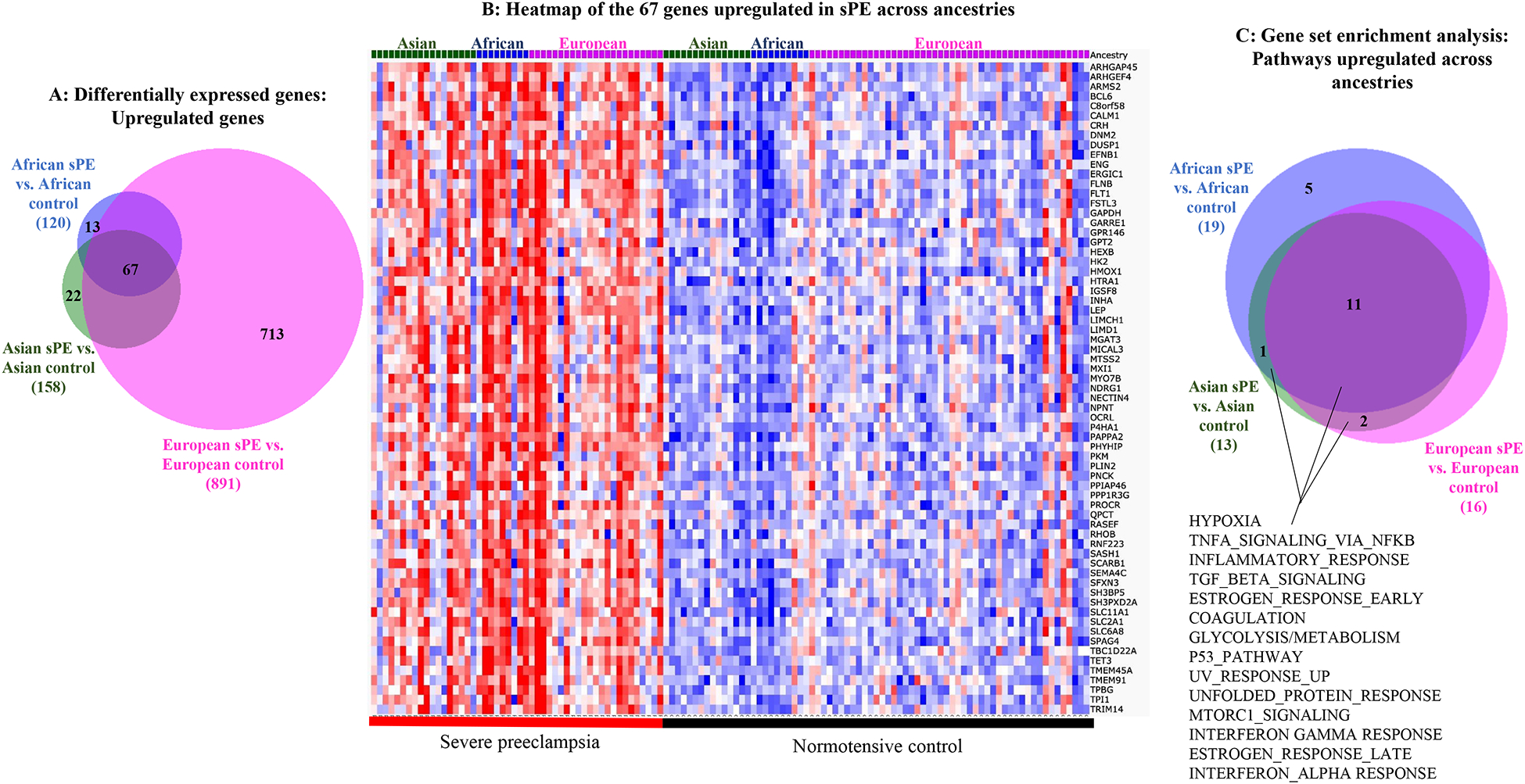
Genes and pathways upregulated in common in placentas from severe PE versus normotensive patients of African, Asian and European ancestries. A. Differential gene expression analysis of sPE vs. normotensive controls within each ancestry showing the numbers of protein-coding genes that are significantly upregulated (with DESeq2 calculated adjusted p values of <.05) in sPE versus normotensive placentas of each ancestry. B. Heat map of the 67 protein-coding genes upregulated in patients of all three ancestries with sPE. C. Results of gene set enrichment analysis showing signaling pathways upregulated in sPE versus normotensive placentas.
The relatively small number of differentially expressed genes did not show enrichment of ontology terms using hypergeometric statistical tests. We therefore used our ranked gene list to perform gene set enrichment analysis (GSEA) to look for enriched pathways in our dataset. We found 11 gene sets commonly enriched in PE of all three ancestries compared to their respective normotensive controls (Figure 1C).
We then performed leading edge and fold-change analysis and identified the genes driving the pathways upregulated in sPE versus normotensive controls of all ancestries; the most highly upregulated genes included genes involved in metabolism (LEP and HK2), inflammation (TPBG, ARMS2, QPCT, SH3BP5), hypoxia (TMEM45A, HTRA1, FLT1, ARHGEF4), cell migration (MYO7B), protein localization (PHYHIP), hormone synthesis/regulation (CRH, PAPPA2) and TGF-beta signaling, angiogenesis and matrix remodeling pathways (ENG, FSTL3 and INHA) (Table 1; Supplemental Tables 9–11).
Table 1:
Fold changes in genes upregulated in severe PE (versus normotensive controls) of all three ancestry groups. Listed genes include leading edge genes and genes upregulated at greater or equal to 2.5-fold in African ancestry severe PE placentas. DESeq2 adjusted p value <.05 for all listed genes.
| Leading edge genes | African ancestry fold change | Asian ancestry fold change | European ancestry fold change |
|---|---|---|---|
| LEP | 23.42 | 18.34 | 11.11 |
| FSTL3 | 14.18 | 13.28 | 3.49 |
| HK2 | 6.39 | 5.21 | 3.50 |
| TMEM45A | 4.09 | 2.26 | 3.06 |
| HTRA1 | 3.99 | 2.46 | 2.23 |
| SH3BP5 | 3.69 | 3.07 | 2.39 |
| ENG | 3.59 | 2.36 | 2.27 |
| TPBG | 3.58 | 2.30 | 1.86 |
| INHA | 3.56 | 3.35 | 2.61 |
| NDRG1 | 3.34 | 2.57 | 2.37 |
| SPAG4 | 3.18 | 2.68 | 2.94 |
| SLC6A8 | 2.79 | 2.08 | 2.18 |
| PROCR | 2.59 | 2.09 | 2.35 |
| TRIM14 | 2.58 | 1.87 | 2.03 |
| GAPDH | 2.47 | 1.82 | 1.62 |
| DUSP1 | 2.35 | 1.93 | 1.78 |
| SCARB1 | 1.82 | 1.66 | 1.64 |
| SLC2A1 | 1.79 | 1.71 | 1.61 |
| OCRL | 1.78 | 1.60 | 1.59 |
| RHOB | 1.69 | 1.59 | 1.35 |
| Non-leading edge highly upregulated genes | |||
| ARMS2 | 9.49 | 6.54 | 5.36 |
| ARHGEF4 | 6.09 | 6.31 | 3.02 |
| FLT1 | 5.55 | 4.30 | 4.14 |
| MYO7B | 5.12 | 3.29 | 3.03 |
| PHYHIP | 4.89 | 2.46 | 2.54 |
| CRH | 4.42 | 3.13 | 2.91 |
| QPCT | 4.36 | 2.54 | 2.43 |
| RNF223 | 3.98 | 4.94 | 3.35 |
| PAPPA2 | 3.72 | 4.07 | 2.97 |
| PNCK | 3.37 | 4.24 | 2.89 |
| PLIN2 | 3.29 | 2.32 | 2.09 |
| ARHGAP45 | 3.28 | 3.38 | 2.35 |
| RASEF | 3.21 | 2.32 | 2.29 |
| GPR146 | 3.18 | 1.94 | 1.74 |
| PPP1R3G | 2.86 | 2.34 | 1.57 |
| NPNT | 2.85 | 2.20 | 2.33 |
| MXI1 | 2.79 | 1.95 | 1.70 |
| TET3 | 2.76 | 1.79 | 2.08 |
| NECTIN4 | 2.75 | 2.32 | 1.97 |
| FLNB | 2.72 | 1.75 | 1.77 |
| SASH1 | 2.71 | 2.26 | 2.46 |
| GPT2 | 2.65 | 1.87 | 1.87 |
| HEXB | 2.61 | 1.86 | 1.72 |
| BCL6 | 2.54 | 2.15 | 1.91 |
| SFXN3 | 2.53 | 2.19 | 1.79 |
Allograft rejection and adaptive immune response genes and pathways are selectively upregulated in placentas from patients of African ancestry with sPE
Our GSEA identified 5 pathways, including allograft rejection and adaptive immune response (interleukin-JAK-STAT signaling) pathways, upregulated in placentas from sPE patients of African ancestry (versus African ancestry normotensive controls), but not in Asian or European ancestry sPE placentas (versus normotensive controls) (Figure 2A). We performed leading edge and fold-change analysis and identified the genes more highly upregulated in African versus Asian and European ancestry sPE placentas included genes involved in allograft rejection/immune response (IL3RA, INHBA, CXXC1, TNFAIP2, CXCL8, LTF, USP18, TREM1), hedgehog and wnt signaling (PTCH1), ion transport (CP), water transport (AQP1) and blood pressure regulation (NPR3) (Table 2).
Figure 2:
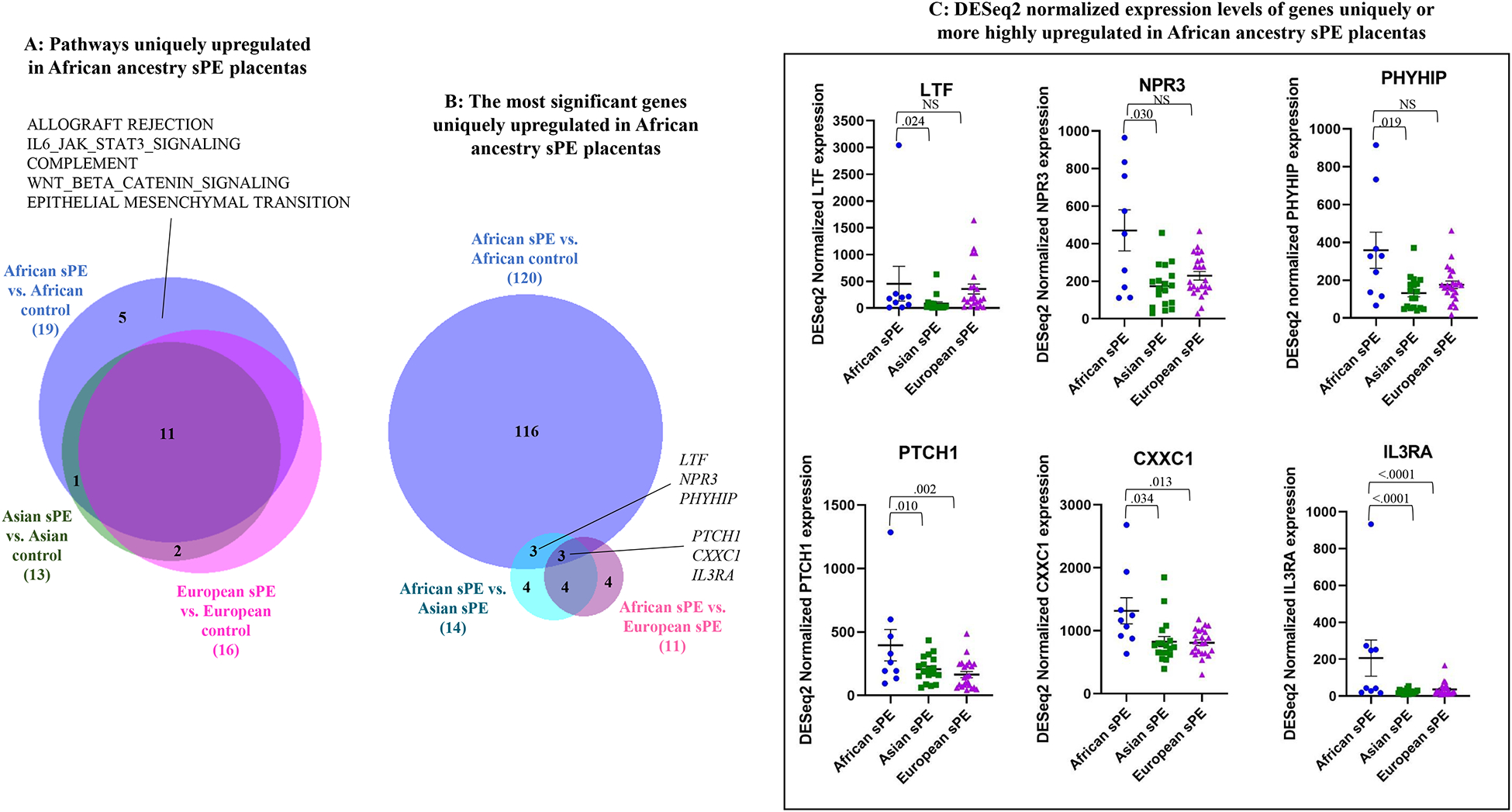
Genes and pathways selectively or more highly upregulated in African ancestry sPE placentas. A. Results of gene set enrichment analysis showing pathways upregulated in African ancestry sPE (vs. normotensive) placentas, but not in Asian or European ancestry sPE (vs. normotensive) placentas. B. Identification of the most significant protein-coding genes upregulated in African ancestry sPE placentas; differentially upregulated genes in: 1. African ancestry sPE vs. normotensive placentas; 2. African sPE vs. Asian sPE placentas and 3. African sPE vs. European sPE placentas were compared. Non-protein-coding genes were excluded. Statistical analysis was performed using DESEq2; adjusted p values <.05 were considered significant. C. Plots of DESeq2-normalized gene expression levels of the most significant genes upregulated in African ancestry sPE placentas in comparison to their levels in Asian and European ancestry sPE placentas. Statistical analysis was performed using DESEq2; adjusted p values <.05 were considered significant.
Table 2:
Fold changes in genes upregulated in African ancestry severe PE (versus normotensive control) placentas. Listed genes include leading edge genes and genes upregulated at greater or equal to 2.5-fold in African ancestry severe PE placentas. DESeq2 adjusted p value <.05, except when listed as NS (not significant).
| Leading edge genes | African ancestry fold change | Asian ancestry fold change | European ancestry fold change |
|---|---|---|---|
| LTF | 15.89 | NS | 6.22 |
| IL3RA | 3.87 | NS | NS |
| INHBA | 3.16 | NS | 2.56 |
| USP18 | 2.38 | NS | NS |
| ERO1A | 2.06 | NS | 1.73 |
| PTCH1 | 1.82 | NS | NS |
| CXXC1 | 1.75 | NS | 1.26 |
| Non-leading edge highly upregulated genes | |||
| HTRA4 | 5.83 | NS | 4.44 |
| CGB5 | 5.27 | NS | 2.86 |
| CP | 5.01 | NS | NS |
| CXCL8 | 4.44 | NS | NS |
| TREM1 | 4.28 | NS | 2.38 |
| ANKRD37 | 4.04 | NS | NS |
| PPP1R1C | 3.89 | NS | 2.42 |
| BHLHE40 | 3.32 | NS | 2.11 |
| CELSR1 | 3.28 | NS | NS |
| AQP1 | 3.15 | NS | 2.12 |
| NRIP1 | 2.96 | NS | 1.93 |
| NPR3 | 2.76 | NS | NS |
| MYO15A | 2.52 | NS | 1.67 |
| TNFAIP2 | 2.50 | NS | NS |
To narrow down the most significant of the uniquely or more highly expressed genes in placentas from African ancestry sPE patients versus Asian and European ancestry sPE patients, we performed differential gene expression analysis of placentas from African versus Asian ancestry sPE patients and African versus European ancestry sPE patients (Supplemental Tables 12–13) and then corrected for PE-independent ancestry-related differences by comparing identified upregulated genes to the 120 genes upregulated in placentas from African ancestry sPE patients versus African ancestry normotensive controls; LTF, NPR3 and PHYHIP were the most significant of the genes more highly expressed in placentas from African ancestry sPE patients versus Asian ancestry sPE patients; PTCH1 and allograft rejection/adaptive immune response genes, IL3RA and CXXC1 were the most significant of the genes more highly expressed in placentas from African ancestry sPE patients than in both Asian and European ancestry sPE patients (Figure 2B–C).
High placental upregulation of IL3RA is associated with unexplained tachycardia and peripartum cardiomyopathy
To determine the extent to which high gene expression correlated to adverse maternal outcomes, we reviewed the electronic medical records for maternal complications – renal failure, stroke, unexplained tachycardia (i.e. tachycardia in the absence of clinically explained causes such as chorioamnionitis, urinary tract infection, viral infection, post-partum hemorrhage/anemia, dehydration, sleep apnea and bulimia) and peripartum cardiomyopathy – in the patients with the highest placental expression of the 6 genes uniquely or more highly upregulated in African ancestry sPE placentas. We found that patients with the highest IL3RA expression levels for their ancestry developed either peripartum cardiomyopathy (PPCM; African ancestry) or unexplained tachycardia (UET; Asian and European ancestries) (Figure 3A).
Figure 3:
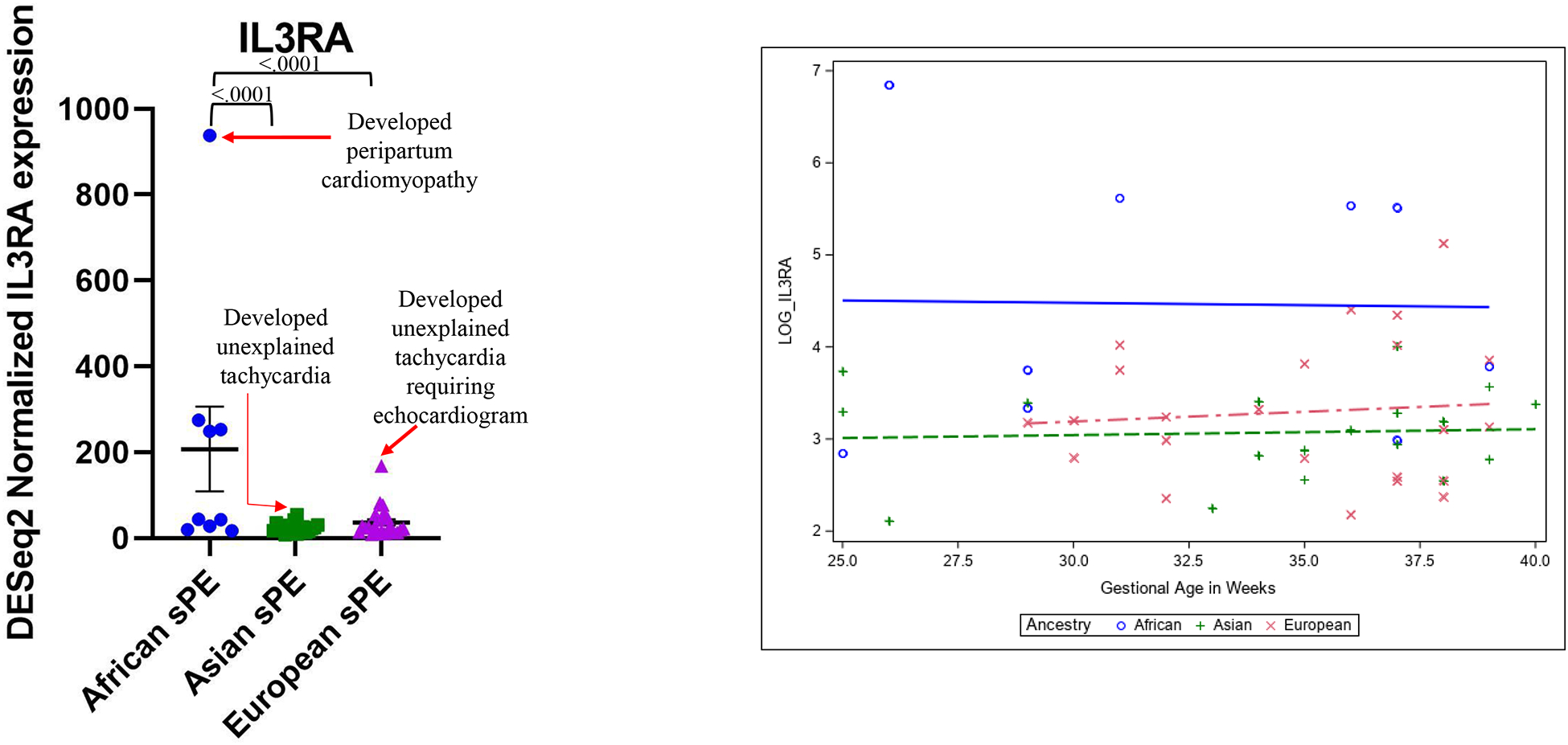
A. Patients with the highest placental DESeq2 normalized IL3RA expression levels for their ancestry developed either unexplained tachycardia or peripartum cardiomyopathy. B. Logarithmic regression analysis of DESeq2 normalized placental IL3RA expression levels in African versus Asian and European ancestry severe preeclampsia parturitions at various gestational ages.
We thus searched for UET and PPCM in the medical records of all 50 sPE study patients and found elevated IL3RA levels to be significantly associated with UET or PPCM (Fisher’s exact test p value .0005) (Table 3). We performed regression analysis of IL3RA levels in placentas of sPE patients of African, Asian and European ancestries delivered at various gestational ages and found IL3RA levels to be elevated in African ancestry sPE placentas, regardless of gestational age at delivery (Figure 3B). In African ancestry parturients, individual placentas showed upregulation of both canonical (exemplified by FLT1 and LEP) and allograft rejection/ immune response genes (exemplified by IL3RA) (Supplemental Figure 3).
Table 3:
Frequency of unexplained tachycardia or peripartum cardiomyopathy and their relationship with IL3RA levels in parturitions complicated by preeclampsia with severe features (sPE) per ancestry (N=50). Comparison placental IL3RA levels were set to 2-fold greater than average of the levels in sPE patients per ancestry.
| IL3RA | PPCM or unexplained tachycardia | No PPCM or unexplained tachycardia |
|---|---|---|
| IL3RA 2-fold greater than PE average for ancestry | African – 1 | African – 0 |
| Asian – 1 | Asian – 0 | |
| European – 1 | European – 0 | |
| Total – 3 | Total – 0 | |
| IL3RA less than 2-fold of PE average for ancestry | African – 0 | African – 8 |
| Asian – 0 | Asian – 17 | |
| European – 2 | European – 20 | |
| Total – 2 | Total – 45 |
Fisher’s exact test p-value .0005
Placental syncytiotrophoblast CD123 expression by immunohistochemistry correlates with placental DEseq2 normalized IL3RA expression levels
To confirm protein expression and determine the cell-specific placental localization of IL3RA, we evaluated the localization and expression pattern of CD123, the protein product of IL3RA by immunohistochemistry on formalin-fixed paraffin-embedded (FFPE) sections from sPE patient placentas with the highest and lowest placental IL3RA levels for their ancestry (i.e. the 2 highest and 2 lowest IL3RA-expressing sPE placentas of each ancestry; N =12). There was variable CD123 expression in chorionic plate vessel endothelial cells, stem villous vessel endothelial cells, decidual vessel endothelial cells and syncytiotrophoblasts (Figure 4A–D). CD123 expression in syncytiotrophoblasts correlated with DEseq2 normalized IL3RA expression levels (simple linear regression p = .0027), but expression in stem villous vessels did not (p = .1782) (Figure 4E–F).
Figure 4:
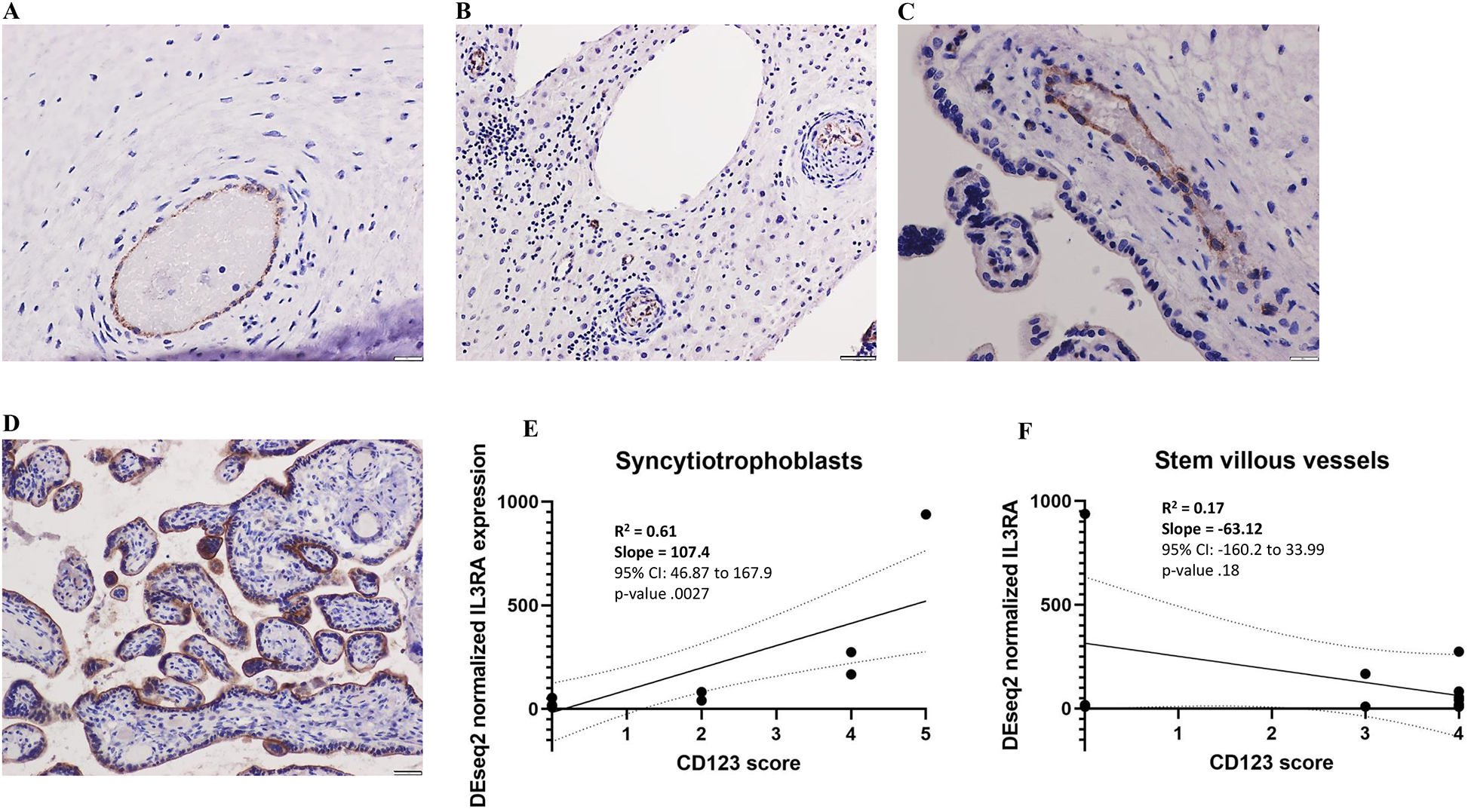
CD123 localization in placenta sections; in placentas with CD123 expression, variable expression was seen in chorionic plate vessels (A), decidual vessels (B), stem villous vessels (C) and syncytiotrophoblasts (D). CD123 expression in syncytiotrophoblasts correlated better with DESeq2 normalized IL3RA expression levels than expression in stem villous vessels (E and F).
Discussion
Factors underlying increased PE risk and worse PE outcomes in African ancestry pregnancies and parturitions are incompletely understood. In their seminal papers Leavey et al.14–15 identified molecular subtypes of preeclampsia, including canonical, associated with increased expression of genes like FLT1, LEP and FSTL3 involved in angiogenesis and metabolism and immunologic, associated with increased expression of allograft rejection-associated genes. In the current study, we evaluated placental gene expression by RNA sequencing in patients with sPE of African, Asian and European ancestries and found upregulation of canonical PE genes in sPE patients of all ancestries. Of the canonical PE genes, LEP, FSTL3, HK2 and FLT1 were among the most highly upregulated.
Fms-related receptor tyrosine kinase, FLT1 is probably the most widely recognized PE-associated gene, having been found to be upregulated in PE in multiple studies31–34. The ratio of soluble FLT-1 (sFlt-1) to placental growth factor (PIGF) has been validated as a test to predict impending PE35. Leptin, LEP plays a major role in the regulation of energy homeostasis and body weight36. Elevated LEP has been previously described in preeclampsia in multiple studies and leptin infusion in mice induces clinical characteristics of preeclampsia37–38. Longitudinal measurement of leptin/ceramide ratio outperformed sFlt-1/PIGF ratio in predicting impending PE39. Follistatin-like 3, FSTL3 is an activin antagonist that Mukherjee et al. found to be involved in glucose and fat metabolism40. In their unsupervised placental gene expression profiling, Leavey et al14–15 found FSTL3 to be the top gene associated with canonical PE. Hexokinase 2, HK2, is involved in glucose metabolism41. A number of studies have reported differential expression of HK2 in PE32, 42–43.
We observed lactotransferrin (LTF), phantonyl-CoA 2-hydroxylase interacting protein (PHYHIP) and natriuretic peptide receptor 3 (NPR3) to be more highly expressed in placentas from African ancestry sPE patients than in Asian ancestry sPE patient. LTF encodes an iron-binding protein involved in the innate immune response. Prior studies have reported upregulation of LTF in preeclampsia31. PHYHIP has been described to be involved in protein localization and has been suggested to be involved in the development of Refsum disease, a neurologic disorder caused by the toxic accumulation of phytanic acid in brain and peripheral neurons44. A few studies have identified PHYHIP as one of the placenta genes associated with early-onset preeclampsia45–46. NPR3 gene encodes one of three natriuretic peptide receptors, involved in regulation of blood volume and pressure47. To our knowledge, ours is the first study to report elevation of NPR3 in preeclamptic placentas. Interestingly however, genome-wide association studies have identified risk loci in NPR3 to be associated with preeclampsia48–49.
Another important finding in this study is that PTCH1 and genes and pathways involved in allograft rejection and adaptive immune responses –, CXXC1 and IL3RA – are selectively or more highly upregulated in placentas from parturient persons of African ancestry with sPE than in both Asian and European ancestry sPE parturients. Patched 1 (PTCH1) is a component of the hedgehog signaling pathway that also regulates WNT signaling50. Interestingly, though we observe upregulation of placental PTCH1 in African ancestry sPE patients, Takai et al.51 reported down-regulation of placental PTCH1 in a Japanese cohort of preeclamptic patients; we do not observe any significant change in PTCH1 in our cohort of Asian sPE versus normotensive patients. Cxxc finger protein 1 (CXXC1) is a CpG-binding transcriptional activator that plays a role in the survival and differentiation of T cells52–54. Interleukin 3 receptor alpha (IL3RA) is a cell surface receptor for IL3, expressed on hematopoietic progenitor cells, monocytes and B-lymphocytes55. To our knowledge, ours is the first to report the upregulation of CXXC1 and IL3RA in preeclampsia.
The most striking finding in our study is the association between high placental expression of IL3RA and unexplained tachycardia and peripartum cardiomyopathy in sPE patients. Binding of IL3 to IL3RA causes heterodimerization with the beta subunit, IL3RB, and downstream JAK2-STAT5 signaling56. IL5 and GM-CSF share the beta subunit with IL3 and can activate signal transduction upon binding57. High levels of IL3 and GM-CSF have been observed in various models of heart failure. Vistnes et al.58 found IL3 and GM-CSF among the cytokines increased upon inducing heart failure in a mouse cardiomyopathy model. IL3 caused allograft fibrosis and chronic rejection of mouse heart transplants59. IL3 impaired the cardioprotective effects of endothelial cell-derived extracellular vesicles in a rat cardiac ischemia/reperfusion model60. In humans, Oren et al.61 found CRP, GM-CSF and IL3 to be significantly upregulated in the first 12 hours of patients presenting to the emergency department with acute myocardial infarctions.
Placental endothelial cells and syncytiotrophoblasts express CD123, the protein product of IL3RA, with syncytiotrophoblast CD123 expression being a better correlate of IL3RA levels by RNA-sequencing. Syncytiotrophoblasts and decidual endothelial cells interact with the maternal systemic circulation at the intervillous space and decidua respectively62–63. CD123/IL3RA localization to these cell types suggests IL3RA may exert its adverse cardiovascular effects via interactions with the maternal systemic circulation and circulating immune cells. In support of this theory is the finding by Anzai et al.64 that T-cell derived IL3 acted on IL3RA-expressing cardiac macrophages and fibroblasts to amplify autoimmune inflammation in a mouse experimental myocarditis model. Furthermore, Pistulli et al65 found accumulation of CD123+ dendritic cells in hearts of humans with acute myocarditis, and that dendritic cell accumulation caused adverse left ventricular remodeling in murine model of experimental myocarditis. IL5, which shares the signal transducing beta subunit with IL3-IL3RA, is a contributor to the pathogenesis of eosinophilic myocarditis66–69. Altogether, the upregulation of placental IL3RA in patients with sPE who develop unexplained tachycardia and/or peripartum cardiomyopathy suggest that this marker may be a predictor and/or cause of cardiac dysfunction, perhaps via interaction with circulating immune cells that then accumulate in, and cause damage to, the heart. Studies of the crosstalk between syncytiotrophoblasts, immune cells, and cardiomyocytes may yield further mechanistic insights into how the placenta may cause cardiovascular damage in susceptible patients.
In summary, we have identified high placental expression of metabolism and angiogenesis (canonical PE) genes as well as allograft rejection/immune response (immunologic PE) genes, particularly CXXC1 and IL3RA in African ancestry sPE parturients. Notably, individual placentas from patients of African ancestry with sPE had upregulation of both canonical PE genes and immune response genes, and high placental IL3RA was associated with unexplained tachycardia and peripartum cardiomyopathy. This high expression of both canonical and immunologic PE genes likely explains the worse placental histopathology, with increased rates of maternal vascular malperfusion and perivillous fibrin deposition, we observe in African ancestry sPE parturients. Our findings suggest that the combined activation of canonical PE and allograft rejection/adaptive immune response genes may be a marker of worse placental injury and harbinger of worse PE outcomes. As pregnancy is a semi-allogeneic state and HLA diversity is higher in people of African ancestry70, the possibility that the higher upregulation of canonical and immunologic PE genes in pregnancies and parturitions with maternal African ancestry may be related to higher rates of maternal-fetal HLA mismatches in this population is an intriguing concept for future studies.
Limitations
Our study was retrospective and limited by the small number of patients with preeclampsia who developed unexplained tachycardia or peripartum cardiomyopathy peripartum, which raises the possibility that our observed association between high placental IL3RA and unexplained tachycardia and peripartum cardiomyopathy is coincidental. Nevertheless, we think IL3RA is worth further investigation in preeclampsia and peripartum cardiomyopathy for the following reasons: 1. The relationship between high IL3RA levels and cardiovascular outcomes in sPE patients appears to cut across ancestries. 2. CD123, the protein product of IL3RA is expressed by cells in contact with the maternal systemic circulation (syncytiotrophoblasts and endothelial cells) and therefore likely mediates systemic effects. 3. The ligands for IL3RA/CD123 have been shown to cause cardiovascular damage in animal models and to be increased in human patients with heart failure. Prospective studies of how expression of placental genes relate to peripartum cardiovascular complications are warranted. Another limitation of our study is that we were unable to obtain pre-pregnancy BMI in a majority of our patients due to the fact that many of our patients were transferred to UCSD to obtain higher level of care. Furthermore, we did not have enough samples in our African ancestry group to include birth weight percentile in our DESeq2 adjustment formula and maintain statistical power. Larger studies that include higher numbers of African ancestry patients are warranted.
Supplementary Material
References
- 1.Say L, Chou D, Gemmill A, Tuncalp O, Moller AB, Daniels J, Gulmezoglu AM, Temmerman M, Alkema L (2014). Global causes of maternal death: a WHO systemic analysis. Lancet Global Health; 2(6): e323–e333. [DOI] [PubMed] [Google Scholar]
- 2.Breathett K, Muhlestein D, Foraker R, Gulati M (2014). Differences in preeclampsia rates between African American and Caucasian women: trends from the National Hospital discharge survey. J Womens Health (Larcmt). 23(11): 886–893. [DOI] [PubMed] [Google Scholar]
- 3.Ghosh G, Grewal J, Mannisto T, Mendola P, Chen Z, Xie Y, Laughon SK (2014). Racial/ethnic differences in pregnancy-related hypertensive disease in nulliparous women. Ethn Dis. 24(3): 283–289. [PMC free article] [PubMed] [Google Scholar]
- 4.Caughey AB, Stotland NE, Washington AE, Escobar GJ (2005). Maternal ethnicity, paternal ethnicity and parental ethnic discordance: predictors of preeclampsia. Obstet Gynecol 106(1): 156–161. [DOI] [PubMed] [Google Scholar]
- 5.Bibbins-Domingo K, Grossman DC, Curry SJ, Barry MJ, Davidson KW, Doubeni CA, Epling JW Jr, Kemper AR, Krist AH, Kurth AE, et al. (2017). Screening for preeclampsia: US preventive task force recommendation statement. JAMA 317(16):1661–1667. [DOI] [PubMed] [Google Scholar]
- 6.Gyamfi-Bannerman C, Pandita A, Miller EC, Boehme AK, Wright JD, Siddiq Z, D’Alton ME, Friedman AM (2020). Preeclampsia outcomes at delivery and race. J Matern Fetal Neonatal Med 33(21): 3619–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang M, Wan P, Ng K, Singh K, Cheng TH, Velickovic I, Dalloul M, Wlody D (2020). Preeclampsia among African American pregnant women: an update on prevalence, complications, etiology and biomarkers. Obstet Gynecol Surv 75(2): 111–120. [DOI] [PubMed] [Google Scholar]
- 8.Ross KM, Dunkel Schetter C, McLemore MR, Chambers BD, Paynter RA, Baer R, Feuer SK, Flowers E, Karasek D, Pantell M (2019). Socioeconomic status, preeclampsia risk and gestational length in black and white women. J Racial Ethn Health Disparities 6(6): 1182–1191. [DOI] [PubMed] [Google Scholar]
- 9.Bryant AS, Seely EW, Cohen A, Lieberman E (2005). Patterns of pregnancy-related hypertension in black and white women. Hypertens Pregnancy 24(3): 281–290. [DOI] [PubMed] [Google Scholar]
- 10.Snowden JM, Mission JF, Marshall NE, Quigley B, Main E, Gilbert WM, Chung JH, Caughey AB (2016). The impact of maternal obesity and race/ethnicity on perinatal outcomes: independent and joint effects. Obesity 7: 1590–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soto-Wright V, Bernstein M, Goldstein DP, Berkowitz RS (1995). The changing clinical presentation of complete molar pregnancy. Obstet Gynecol. 86(5):775e779. [DOI] [PubMed] [Google Scholar]
- 12.Steegers EA, Von Dadelszen P, Duvekot JJ, Pijnenborg R (2010). Pre-eclampsia. Lancet. 376(9741):631e644. [DOI] [PubMed] [Google Scholar]
- 13.August P, Sibai BM. Preeclampsia: clinical features and diagnosis. In: Lockwood CJ, ed. UpToDate. UpToDate, Inc; 2019. [Google Scholar]
- 14.Leavey K, Bainbridge SA, Cox BJ (2015). Large scale aggregate microanalysis reveals three distinct molecular subclasses of human preeclampsia. PLoS ONE 10(2): e0116508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leavey K, Benton SJ, Grynspan D, Kingdom JC, Bainbridge SA, Cox BJ (2016). Unsupervised placental gene expression profiling identifies clinically relevant subclasses of human preeclampsia. Hypertension 68: 137–147 [DOI] [PubMed] [Google Scholar]
- 16.Horii M, To C, Morey R, Jacobs MB, Li Y, Nelson KK, Meads M, Siegel BA, Pizzo D, Adami R, et al. (2023). Histopathologic and transcriptomic profiling identifies novel trophoblast defects in patients with preeclampsia and maternal vascular malperfusion. Mod Pathol. 36(2): 100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gestational Hypertension and Preeclampsia (2020): ACOG Practice Bulletin, Number 222. Obstet Gynecol 135, e237–e260. [DOI] [PubMed] [Google Scholar]
- 18.Barral-Arca R, Pardo-Seco J, Bello X, Martinon-Torres F, Salas A (2019). Ancestry patterns inferred from massive RNA-seq data. RNA 25(7): 857–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29(1):15e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Towns J, Cockerill T, Dahan M, Foster I, Gaither K, Grimshaw A, Hazlewood V, Lathrop S, Lifka D, Peterson G, et al. (2014). XSEDE: accelerating scientific discovery. Comput Sci Eng. 16(5):62e74. [Google Scholar]
- 21.Love MI, Huber W, Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durinck S, Spellman PT, Birney E, Huber W. (2009). Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 4(8):1184e1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44(W1):W90eW97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. (2015). The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 23;1(6):417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hulsen T, de Vlieg J, Alkema W. (2008). BioVenn – a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9 (1): 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alexander DH, Novembre J, Lange K (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19(9): 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khong TY, Mooney EE, Ariel I, Balmus NCM, Boyd TK, Brundler MA, Derricott H, Evans MJ, Faye-Petersen OM, Gillan JE, et al. (2016). Sampling and definitions of placental lesions: Amsterdam placental workshop group consensus statement. Arch Pathol Lab Med. 140(7):698e713. [DOI] [PubMed] [Google Scholar]
- 28.Redline RW, Ravishankar S, Bagby CM, Saab ST, Zarei S (2021). Four major patterns of placental injury: a stepwise guide for understanding and implementing the 2016 Amsterdam consensus. Mod Pathol. 34(6):1074e1092. [DOI] [PubMed] [Google Scholar]
- 29.Yang HC, Chen CW, Lin YT, Chu SK (2021). Genetic ancestry plays a central role in population pharmacogenomics. Commun Biol 4(1): DOI: 10.1038/s42003-021-01681-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The 1000 genomes project consortium (2015). A global reference for human genetic variation. Nature 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brew O, Sullivan MHF, Woodman A (2016). Comparison of normal and preeclamptic placental gene expression: a systematic review with meta-analysis. PLoS ONE 11(8): e0161504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaartokallio T, Cervera A, Kyllonen A, Laivuori K, Kere J, Laivuori H (2015). Gene expression profiling of preeclamptic placentae by RNA sequencing. Scientific reports 5: 14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varkonyi T, Nagy B, Fule T, Tarca AL, Karaszi K, Schonleber J, Hupuczi P, Mihalik N, Kovalsky I, Rigo J Jr, et al. (2011). Microarray profiling reveals that placental transcriptomes of early-onset HELLP syndrome and preeclampsia are similar. Placenta. 32: S21–S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maynard SE, Min JY, Merchan J, Lim KH, Li J, Mondal S, Libermann TA, Morgan JP, Sellke FW, Stillman IE, et al. (2003). Excess placental soluble fms-like tyrosine kinase 1 (sFlt1) may contribute to endothelial dysfunction, hypertension and proteinuria in preeclampsia. J Clin Invest 111(5): 649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeisler H, Llurba E, Chantraine F, Vatish M, Staff AC, Sennstrom M, Olovsson M, Brennecke SP, Stephan H, Allegranza D, et al. (2016). Predictive value of the sFlt-1:PIGF ratio in women with suspected preeclampsia. N Engl J Med 374(1): 13–22. [DOI] [PubMed] [Google Scholar]
- 36.Hamann A, Matthaei S (1996). Regulation of energy balance by leptin. Exp Clin Endocrinol Diabetes 104(4): 293–300. [DOI] [PubMed] [Google Scholar]
- 37.de Knegt VE, Hedley PL, Kanters JK, Thagaard IN, Krebs L, Christiansen M, Lausten-Thomsen U (2021). The role of leptin in fetal growth during preeclampsia. Int. J. Mol. Sci 22: 4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mellot E, Faulkner JL (2023). Mechanisms of leptin-induced endothelial dysfunction. Curr Opin Nephrol Hypertens. 32(2): 118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Q, Hao S, You J, Yao X, Li Z, Schilling J, Thyparambil S, Liao WL, Zhou X, Mo L, et al. (2021). Early pregnancy prediction of risk for preeclampsia using maternal blood leptin/ceramide ratio: discovery and confirmation. BMJ Open 11: e050963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murkherjee A, Sidis Y, Mahan A, Raher MJ, Xia Y, Rosen EB, Bloch KD, Thomas MK, Schneyer AL (2007). FSTL3 deletion reveals roles for TGF-beta family ligands in glucose and fat homeostasis in adults. Proc Natl Acad Sci 104 (4): 1348–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nawaz MH, Ferreira JC, Nedyalkova L, Zhu H, Carrasco-Lopez C, Kirmizialtin S, Rabeh WM (2018). The catalytic inactivation of the N-half of human hexokinase 2 and structural and biochemical characterization of its mitochondrial conformation. Biosci Rep. 38(1): BSR20171666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tong J, Zhao W, Lv H, Li WP, Chen ZJ, Zhang C (2018). Transcriptomic profiling of human decidua of severe preeclampsia detected by RNA sequencing. J Cell Biochem 119(1): 607–615. [DOI] [PubMed] [Google Scholar]
- 43.Li C, Liu W, Lao Q, Lu H, Zhao Y (2022). Placenta autophagy is closely associated with preeclampsia. Aging 19; 14. doi: 10.18632/aging.204436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koh JT, Lee ZH, Ahn KY, Kim JK, Bae CS, Kim HH, Kee JH, Kim KK (2001). Characterization of mouse brain-specific angiogenesis inhibitor 1 (BAI1) and phytonyl-CoA-alpha-hydroxylase-associated protein 1, a novel BAI1-binding protein. Brain Res Mol Brain Res 87(2): 223–237. [DOI] [PubMed] [Google Scholar]
- 45.Blair JD, Yuen RKC, Lin BK, McFadden DE, von Dadelszen P, Robinson WP (2013). Widespread DNA hypomethylation at gene enhancer regions in placentas associated with early-onset preeclampsia. Mol Hum Reprod 19(10): 697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leavey K, Wilson SL, Bainbridge SA, Robinson WP, Cox BJ (2018). Epigenetic regulation of placental gene expression in transcriptional subtypes of preeclampsia. Clin Epigenetics 10:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tokudome T, Otani K (2022). Molecular mechanism of blood pressure regulation through the atrial natriuretic peptide. Biology (Basel) 11(9): 1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tyrmi JS, Kaartokallio T, Lokki AI, Jaaskelainen T, Kortelainen E, Ruotsalainen S, Karjalainen J, Ripatti S, Kivioja A, Laisk T, et al. (2023). Genetic risk factors associated with preeclampsia and hypertensive disorders of pregnancy. JAMA Cardiol. 8(7): 674–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Honigberg MC, Truong B, Khan RR, Xiao B, Bhatta L, Vy HMT, Guerrero RF, Schuermans A, Selvaraj MS, Patel AP, et al. (2023). Polygenic prediction of preeclampsia and gestational hypertension. Nat Med 29(6): 1540–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Regan JL, Schumacher D, Staudte S, Steffen A, Haybaeck J, Keilholz U, Schweiger C, Golob-Schwarzl S, Mumberg D, Henderson D, et al. (2017). Non-canonical hedgehog signaling is a positive regulator of the WNT pathway and is required for the survival of colon cancer stem cells. Cell reports 21: 2813–2828. [DOI] [PubMed] [Google Scholar]
- 51.Takai H, Kondoh E, Mogami H, Kawasaki K, Chigusa Y, Sato M, Kawamura Y, Murakami R, Matsumura N, Konishi I, Mandai M (2019). Placental sonic hedgehog pathway regulates fetal growth via the IGF axis in preeclampsia. J Clin Endocrinol Metab 104(9): 4239–4252. [DOI] [PubMed] [Google Scholar]
- 52.Kiuchi M, Onodera A, Kokubo K, Ichikawa T, Morimoto Y, Kawakami E, Takayama N, Eto K, Koseki H, Hirahara K, Nakayama T (2021). The Cxxc1 subunit of the trithorax complex directs epigenetic licensing of CD4+ T cell differentiation. J Exp Med 218(4): e20201690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin F, Meng X, Guo Y, Cao W, Liu W, Xia Q, Hui Z, Chen J, Hong S, Zhang X, et al. (2019). Epigenetic initiation of the TH17 differentiation program is promoted by Cxxc finger protein 1. Sci Adv 5(10): eaax1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao W, Guo J, Wen X, Miao L, Lin F, Xu G, Ma R, Yin S, Hui Z, Chen T, et al. (2016). CXXC finger protein 1 is critical for T-cell intrathymic development through regulating H3K4 trimethylation. Nat Commun 7:11687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang S, Chen Z, Yu JF, Young D, Bashey A, Ho AD, Law P (1999). Correlation between IL-3 receptor expression and growth potential of human CD34+ hematopoietic cells from different tissues. Stem Cells 17(5): 265–272. [DOI] [PubMed] [Google Scholar]
- 56.Broughton SE, Hercus TR, Nero TL, Kan WL, Barry EF, Dottore M, Cheung Tung Shing KS, Morton CJ, Dhagat U, Hardy MP, et al. (2018). A dual role for the N-terminal domain of the IL-3 receptor in cell signaling. Nat. Commun 9: 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Broughton SE, et al. (2012). The GM-CSF/IL-3/IL-5 cytokine receptor family: from ligand recognition to initiation of signaling. Immunol. Rev 250: 277–302. [DOI] [PubMed] [Google Scholar]
- 58.Vistnes M, Waehre A, Nygard S, Sjaastad I, Andersson KB, Husberg C, Christensen G (2010). Circulating cytokine levels in mice with heart failure are etiology dependent. J Appl Physiol. 108: 1357–1364. [DOI] [PubMed] [Google Scholar]
- 59.Balam S, Schiechl-Brachner G, Buchtler S, Halbritter D, Schmidbauer K, Talke Y, Neumayer S, Salewski J, Winter F, Karasuyama K, et al. (2019). IL-3 triggers chronic rejection of cardiac allografts by activation of infiltrating basophils. J Immunol. 202: 3514–3523. [DOI] [PubMed] [Google Scholar]
- 60.Penna C, Femmino S, Tapparo M, Lopatina T, Espolin Fladmark K, Ravera F, Comita S, Alloatti G, Giusti I, Dolo V, et al. (2020). The inflammatory cytokine IL-3 hampers cardioprotection mediated by endothelial cell-derived extracellular vesicles possibly via their protein cargo. Cells. 10:13. doi: 10.3390/cells10010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oren H, Riza Erbay A, Balci M, Cehreli S (2007). Role of novel mediators of inflammation in left ventricular remodeling in patients with acute myocardial infarction: do they affect outcome of patients? Angiology 58: 45–54. [DOI] [PubMed] [Google Scholar]
- 62.Moffett A, Locke C (2006). Immunology of placentation in eutherian mammals. Nat. Rev. Immunol 6: 584–594. [DOI] [PubMed] [Google Scholar]
- 63.Aisagbonhi A, Morris GP (2022). Human leukocyte antigens in pregnancy and preeclampsia. Front Genet 13: 884275. doi: 10.3389/fgene.2022.884275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anzai A, Mindur JE, Halle L, Sano S, Choi JL, He S, McAlpine CS, Chan CT, Kahles F, Valet C, et al. (2019). Self-reactive CD4+ IL-3+ T cells amplify autoimmune inflammation in myocarditis by inciting monocyte chemotaxis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pistulli R, Andreas E, Konig S, Drobnik S, Kretzschmar D, Rohm I, Lichtnauer M, Heidecker B, Franz M, Mall G, et al. (2020). Characterization of dendritic cells in human and experimental myocarditis. ESC Heart Fail. 7: 2305–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yadavalli CS, Mishra A (2022). IL-5 overexpression induced blood eosinophilia associated progression of cardiac abnormalities. Int J Basic Clin Immunol. 5: 1–7. [PMC free article] [PubMed] [Google Scholar]
- 67.Hirasawa M, Ito Y, Shibata MA, Otsuki Y (2007). Mechanism of inflammation in murine eosinophilic myocarditis produced by adoptive transfer with ovalbumin challenge. Int Arch Allergy Immunol. 142: 28–39. [DOI] [PubMed] [Google Scholar]
- 68.Song T, Jones DM, Homsi Y (2017). Therapeutic effect of anti-IL-5 on eosinophilic myocarditis with large pericardial effusion. BMJ Case Rep. 2017:bcr2016218992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Desreumax P, Janin A, Dubucquoi S, Copin MC, Torpier G, Capron A, Capron M, Prin L (1993). Synthesis of interleukin-5 by activated eosinophils in patients with eosinophilic heart diseases. Blood. 82: 1553–1560. [PubMed] [Google Scholar]
- 70.Prugnolle F, Manica A, Charpentier M, Guegan JF, Guernier V, Balloux F (2005). Pathogen-driven selection and worldwide HLA class I diversity. Cur Biol 15(11): 1022–1027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


