Abstract
Genes expressed in the parasitic yeast (Y) phase of the dimorphic fungal pathogen Histoplasma capsulatum which are transcriptionally silent in the mycelial (M) phase have recently been cloned and analyzed. To understand the molecular regulation of genes involved in the transition to and maintenance of the Y phase, the presumptive 5′ regulatory regions of two Y phase-specific genes (yps-3 and yps 21:E-9) were PCR amplified as labelled probes to identify nuclear DNA binding proteins which may influence phase-specific gene transcription. Protein-DNA interactions were assessed by Southwestern blot analysis in which sodium dodecyl sulfate-polyacrylamide gel electrophoresis-separated protein extracts from Y and M phases of the virulent G217B strain of H. capsulatum were visualized by their capability for in situ binding to the labelled 517-bp (G217B yps-3) or the 395-bp (G217B yps 21:E-9) putative 5′ regulatory regions. A 30-kDa nuclear protein unique to the M-phase extracts of the highly virulent G217B strain, but absent in the Y phase of the same organism, was identified. In contrast, the low-virulence, thermal-sensitive Downs strain of H. capsulatum lacked detectable p30 binding activity in either yeast- or mycelial phase extracts, regardless of the source of labelled probe (395-bp G217B yps 21:E-9 probe or 512-bp HindIII-EcoRI-labelled Downs yps21:E-9). A decanucleotide motif, TCCTTTTTTT, was identified in the upstream regulatory regions of these yps genes, as well as in the putative α-tubulin promoter, and was conserved with 70 to 100% homology. This recognition sequence was sufficient for p30M binding with 32P-labelled ligated oligonucleotides when used in the Southwestern assay. These findings describe the first nuclear DNA binding factor identified in H. capsulatum which binds to target sequences in a phase-specific manner, suggesting that p30M may govern aspects of gene transcription in this pathogenic fungus, in which a temperature-sensitive switch influences morphology and virulence.
Histoplasma capsulatum is the dimorphic fungal pathogen that is the etiologic agent of histoplasmosis, the most common systemic mycosis in the midwestern United States (23). Severe disease is rare in immunocompetent individuals in regions where the disease is endemic, but a life-threatening disseminated mycosis occurs in patients with AIDS (7) or in individuals undergoing immunosuppressive therapy (34). H. capsulatum adopts either of two morphologically distinct phases, which are controlled by environmental temperatures and other cues (19, 23, 24, 31). At 25°C, or environmental temperatures, the organism is a saprophyte in the mycelial phase, but at elevated host temperatures of 37°C it converts to a unicellular budding yeast in infected tissue (23). The yeast phase, however, is found exclusively in infected tissues (23) and is the form required for progressive infection (25, 26). Thus, genes involved in the transition to or maintenance of the parasitic yeast phase may be of prime importance in exposing the relation of morphology to virulence in this organism.
By exploiting the unique characteristics of dimorphism in H. capsulatum, several distinct genes that are expressed in the yeast phase at 37°C and that are transcriptionally silent in the mycelial form of virulent strains, such as G217B and G222B, at 25°C have been cloned by a subtraction-library strategy (16). For example, the early expression of the yps-3 gene at day 1, following a shift in temperature to 37°C of hyphal cultures of H. capsulatum, and its continued expression throughout the temperature-induced transition to the Y phase appears to correlate with virulence and temperature sensitivity in nonisogenic strains (18). The genomic nucleotide sequence of the yps-3 gene cloned from G217B and a partial cDNA isolated from the yeast phase of the same strain have been reported (17). More recently, a second yeast phase-specific gene from H. capsulatum, yps 21:E-9, has been molecularly cloned and sequenced (1, 16). In contrast to yps-3, yps 21:E-9 exhibits a late pattern of expression (day 11) when mycelia are induced to form yeast at 37°C (1). However, since neither the nucleotide sequences of yps-3 and yps 21:E-9 nor their predicted protein products are homologous to sequences currently in GenBank, the roles of the proteins in dimorphism or virulence remain obscure.
The previous reports characterizing yps-3 and yps 21:E-9 have demonstrated high levels of yeast phase gene expression in thermally tolerant, high-virulence strains of H. capsulatum (1, 16, 17, 18), including G217B. However, this same cadre of yps genes were transcriptionally silent in the low-virulence, temperature-sensitive Downs strain, although the genes and much of their putative regulatory regions were retained and even sequenced from the Downs genome (1, 17). These findings may point to a deficiency in binding or expression of a temperature-inducible regulatory protein in the low-virulence strain.
The expression of constitutive, tissue-specific, and inducible genes is regulated by specific protein factors which interact with control sequences of different genes or targets and govern their expression (22, 28) in various cell types. For example, many upstream activating sequence (UASs) have been determined, and the presence of specific binding proteins interacting with these target elements (27, 30) has been demonstrated in the yeast Saccharomyces cerevisiae. Some regulatory genes, such as GAL4 and GCN4, encode the proteins that bind specifically to the unique UASs of the GAL gene family and additional genes under general amino acid control (2, 11–14). Alternatively, some cis-regulatory elements (upstream repression sequences or silencers) repress the expression of some genes, including those controlling the yeast mating type switch (6, 10, 32). In this system, two nuclear binding factors have been detected, and one of the genes, RAP1, has been cloned. The RAP1 protein may be a regulatory control element which controls several “housekeeping genes” in yeast (20, 21), since the purified protein binds to the regulatory regions of MATα, as well as the consensus motif of the RPG box found in the ENO1 UAS and the region upstream of the ribosomal protein genes.
Although significant insights into gene regulation and control have been made by using S. cerevisiae as a model system, no companion studies have employed any of the members of the pathogenic dimorphic fungi. To explore the molecular regulation of genes involved in yeast morphology and virulence, the regulatory regions of the yps genes of H. capsulatum obtained by subtractive hybridization, and the corresponding sequences for the TUB1 (α-tubulin) gene, were PCR amplified as labelled probes to identify DNA binding proteins which may influence phase-specific gene transcription. In this study, a 30-kDa protein found in the nuclear mycelial-phase extracts of the highly virulent G217B strain but absent in the yeast phase and in the cytosol was identified. The characterization of this phase-specific, DNA binding protein represents the first report of potential regulatory factors which may govern aspects of gene transcription in this widely distributed pathogenic fungus.
MATERIALS AND METHODS
Organisms and culture conditions.
Cultures of H. capsulatum G217B (ATCC 26032) and the temperature-sensitive Downs strain (ATCC 38904) were maintained in GYE liquid medium containing 2% (wt/vol) glucose and 1% (wt/vol) yeast extract. Mycelial-phase organisms were maintained at 25°C, and yeast- phase organisms were cultured in GYE supplemented with 8.4 μg of cysteine/liter at 37°C. For the analysis of proteins during the conversion from mycelial to yeast phase, hyphal cells were induced to form yeast at 37°C in liquid culture.
Preparation of nuclear and cytoplasmic extracts from the yeast and mycelial phases of two H. capsulatum strains.
Protein extracts were prepared from the yeast phases of the G217B and Downs strains of H. capsulatum by modifications to protocols previously described for S. cerevisiae (20). Briefly, exponentially growing yeast-phase cells were pelleted, resuspended in 50 ml of TEB buffer (100 mM Tris-HCl [pH 8.0], containing 10 mM EDTA and 0.5% β-mercaptoethanol) and incubated on ice for 30 min. The cells were washed in spheroplast buffer containing 20 mM sodium phosphate buffer (pH 6.5), 1 M sorbitol, and 1 mM phenylmethylsulfonyl fluoride (PMSF). The cells, in 12 ml of spheroplast buffer, were subsequently treated with 1 mg of Zymolyase 100T (ICN) or, in some cases, the same concentration of yeast lytic enzyme (Sigma Chemical Company) for 60 min at 30°C. Spheroplasts were collected by centrifugation at 900 × g for 5 min and then lysed in breakage buffer (10 mM Tris-HCl [pH 8.0], 1.5 mM MgCl2, 15 mM KCl, 0.1 mM EDTA, 0.5 mM dithiothreitol [DTT], 1 mM PMSF, and 0.1 μg of pepstatin A/ml (20). The supernatant collected prior to lysis was retained as a cytoplasmic fraction following dialysis. Nuclei were collected from the crude lysate at 12,000 × g for 15 min, resuspended in approximately 30 ml of breakage buffer, and then lysed by the addition of 7.6 ml of 4 M ammonium sulfate. After centrifugation at 100,000 × g for 1 h, the recovered supernatant was diluted and chromatographed through a DEAE-Sephadex column containing 0.3 M KCl. Effluent proteins were collected and precipitated by the gradual addition of solid ammonium sulfate to 80%, collected by centrifugation, and then dialyzed against 50 mM Tris-HCl (pH 8.0), with 0.1 mM EDTA, 0.5 mM DTT, 50 mM ammonium sulfate, 0.1 mM PMSF, 0.1 μg of pepstatin A/ml, and 10% glycerol. The nuclear extract was aliquoted and stored at −80°C prior to use in Southwestern blot assays.
Protein extracts from the nuclear and cytosolic compartments of mycelial-phase organisms were prepared by a variation of this protocol. An exponentially growing mycelial culture maintained in GYE at 25°C (500-ml volume) was harvested by filtration. The cells were transferred to a prechilled mortar and pestle and ground under liquid nitrogen. The hyphal dust was then transferred to 50 ml of spheroplast buffer and processed as described above for the yeast-phase extracts.
Preparation of target DNA probes.
Target DNA sequences were prepared by a two-step PCR amplification of G217B or Downs genomic DNA with 5′ and 3′ primers flanking the putative regulatory regions. For the yps-3 probe, 10 pM 5′ (CGTAATGTGACGGGGGAG) and 3′ (CTCTTCCATCATTCCCATC) oligonucleotides described in Fig. 1 were annealed to 50 ng of G217B genomic DNA or to 50 ng of Downs genomic DNA with 100 μM cold deoxynucleoside triphosphates in 1× TNK buffer (10 mM Tris-HCl [pH 8.6], 50 mM KCl2, 1.5 mM MgCl2, 5 mM NH4Cl) and amplified in a Perkin-Elmer Cetus thermocycler under the following conditions: (i) 94°C, 2.5 min, 1 cycle; (ii) 94°C, 1 min; (iii) 55°C, 2 min; and (iv) 72°C, 2 min. Steps ii to iv were repeated for 30 cycles. Following amplification, the 517-bp fragment was purified on agarose gels, and with Gene Clean (BIO 101), and then 1/10 of the recovered product (approximately 5 to 10 ng) was reamplified under the same conditions in the presence of 2 μCi of [32P]dATP and 100 μM all deoxynucleoside triphosphates.
FIG. 1.
Sequences of the putative 5′ regulatory regions of the genes utilized as probes in Southwestern analysis. The flanking (5′ and 3′) primers used for probe amplification are indicated by dashed lines. (A) The previously described yeast phase-specific gene, yps-3; (B) yps 21:E-9; (C) the limited 5′ regulatory region of the α-tubulin gene (TUB1). Both yps genes are expressed in the yeast phase of the virulent strain G217B, but they are both silent in the yeast and mycelial phases of the attenuated Downs strain. α-Tubulin is expressed in both phases of the Downs and G217B strains, with fivefold-higher transcript levels detected in the mycelial phase (9). The transcriptional start sites, determined by S1 mapping of G217B poly(A)+ RNA, for each yps gene is shown (underlined), as well as the putative translational start sites (START) for all three sequences.
A similar two-step amplification was utilized to prepare 32P-labelled DNA from the 5′ regulatory region of yps 21:E-9 from H. capsulatum G217B, as seen in Fig. 1, using the 5′ primer (CTGAATCAATCTAA) and the 3′ primer (CTTCAAACTTGGCTTGACGTC). In the case of the corresponding Downs yps 21:E-9 region, several nucleotide changes were observed in the 3′ primer sequence (Fig. 1), which prevented amplification with the primers designed for the upstream regulatory region of G217B yps 21:E-9. As a result, the HindIII site of p2.0SH, a pUC18 clone containing a 2.0-kb HindIII-SstI insert representing the 5′ region of the yps 21:E-9 gene and potential regulatory sequences from the Downs strain (1), was end labelled with T4 DNA polymerase. Following digestion of the labelled plasmid with EcoRI, a 500-bp HindIII-EcoRI fragment containing the 5′ regulatory regions of the Downs yps 21:E-9 gene was isolated from 1.2% agarose gels, purified with Gene Clean, and used as labelled target DNA.
The probe for the 5′ regulatory region of the TUB1 (α-tubulin) gene was prepared with the primers derived by the methods of Harris and coworkers (8), as indicated in Fig. 1, with 50 ng of G217B genomic DNA as the template.
Southwestern blot analysis of DNA binding proteins from H. capsulatum protein extracts. (i) SDS-polyacrylamide gel electrophoresis and electroblot transfer conditions.
Protein extracts (40 μg) prepared from the yeast and mycelial phases of H. capsulatum strains were solubilized in 12.5% glycerol, 1.25% sodium dodecyl sulfate (SDS), 178 mM β-mercaptoethanol, 0.005% bromophenol blue, and 62.5% Tris-HCl (pH 6.8) and electrophoresed with prestained protein markers (Amersham) in discontinuous Laemmli polyacrylamide gels with an SDS–10% polyacrylamide separating gel (1). When required, portions of the gel were stained with 0.01% Coomassie brilliant blue in an aqueous solution with 50% methanol and 1% acetic acid, followed by destaining in 50% methanol–1% acetic acid, and photographed. For membrane preparation, the unstained portions of the gel were transferred to 0.45-μm-pore-size nitrocellulose (Schleicher and Schuell) by electroblotting in 25 mM Tris-HCl (pH 8.3), 192 mM glycine, and 20% (vol/vol) methanol at 4°C in a Hoeffer Transphore apparatus overnight at 0.3 mA. Prestained protein markers were transferred simultaneously for molecular mass estimation in Southwestern blots.
(ii) Conditions for in situ protein-DNA interactions.
Blots were incubated with shaking in blocking buffer (10 mM Na–HEPES [pH 7.5], 70 mM NaCl, 10 mM MgCl2, 1 mM DTT, 0.1 mM EDTA) containing 0.25% low-fat skim milk at room temperature for 2 h with 5 μg of nonspecific competitor poly(dI-dC) DNA (Sigma)/ml. Labelled target DNA (105 cpm/ml) was added to the blocking buffer, and incubation was continued for 1 h. The blot was processed by washing three times in 25 ml of blocking buffer lacking skim milk, exposed to XAR X-ray film, and developed.
(iii) Localization of sequence-specific protein binding in the 5′ regulatory region of yps 21:E-9.
The 395-bp target fragment obtained by amplification of G217B genomic DNA with the 5′ and 3′ primers of yps 21:E-9 was labelled by the inclusion of 2 μCi of [32P]dATP in the PCR mixture. The amplified fragment was digested with DdeI (Promega) as recommended by the manufacturer, and the subfragments were resolved on 8% polyacrylamide gels. The 109-, 150-, and 136-bp labelled fragments were processed by electroelution and used in Southwestern blots.
(iv) Preparation and ligation of oligonucleotide probes and conditions for competition analysis.
The 10-mer oligonucleotides (TCCTTTTTTT and its complement, AAAAAAGGA; 20 μg of each) were heated together in 44 μl of distilled H2O to 95°C for 2 min and then allowed to cool to room temperature to anneal the complementary oligonucleotides. The annealed material was ligated by the addition of 5 μl of 10× ligase buffer and 20 U of T4 DNA ligase (Boehringer Mannheim) at room temperature overnight to concatenate the blunted double-stranded oligonucleotides. Ligation was subsequently assessed by electrophoresis on 1.2% agarose gels with molecular size standards (a kilobase ladder from Bio-Rad Laboratories); concatemers were 150 to 350 bp in size.
Prepared Southwestern blots were permitted to interact overnight with 105 cpm (approximately 0.5 μg) of 32P-labelled DNA target/ml prepared from the 5′ regulatory region of yps 21:E-9 (the 365-bp probe from G217B) in the absence or presence of increasing amounts (1×, 0.5 μg; 5×, 2.5 μg; and 25×, 12.5 μg) of the ligated DNA site probe.
Nucleotide sequence accession numbers.
Accession numbers for the G217B yps 21:E-9 DNA and the protein and the Downs yps 21:E-9 upstream regulatory sequence are U83168 and U83193, respectively.
RESULTS
Identification of a 30-kDa nuclear protein from H. capsulatum G217B mycelial extracts which binds to upstream regulatory regions of yps-3 (G217B probe).
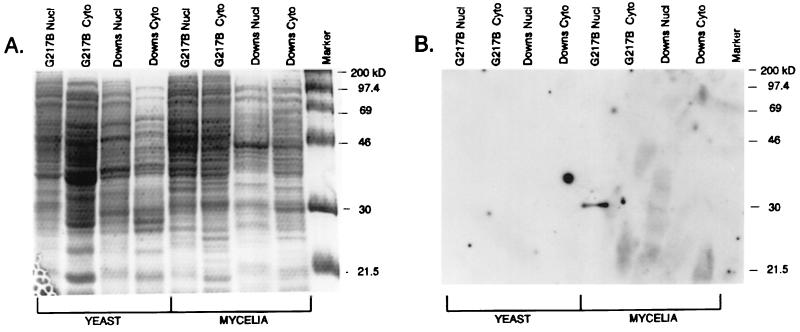
To evaluate protein-DNA interactions which potentially influence transcription of yeast phase-specific genes in H. capsulatum, protein extracts from both nuclear and cytoplasmic compartments of the yeast and mycelial phases of G217B (class 2) and Downs (class 1) strains were prepared, electrophoresed on duplicate SDS–8% polyacrylamide gels, and examined by staining (Fig. 2A) and Southwestern blot analysis (Fig. 2B), using the 5′ regions of 32P-labelled yps-3 DNA amplified from the virulent G217B strain. As seen in Fig. 2A, a broad array of proteins with apparent molecular masses in the 21.5- to 200-kDa range were found in all prepared extracts. Some visible differences between the staining patterns of the nuclear and cytoplasmic extracts of the same strain were evident in the apparent 21.5- to 40-kDa range (Fig. 2A); differences in the patterns were also noted in the same size range when yeast and mycelial nuclear extracts obtained from the G217B or Downs strain were compared in this one-dimensional analysis. As seen in Fig. 2B, however, a discrete DNA binding activity was identified exclusively in mycelial phase extracts prepared from G217B; it was absent in nuclear extracts from the yeast or mycelial phases of the hypovirulent Downs strain. This in situ DNA binding activity with an apparent molecular mass of 30 kDa was detected only in nuclear extracts from mycelia of virulent strains of H. capsulatum, including G217B (Fig. 2B) and G222B and G186B (data not shown), and it was designated p30M. Given the constraints of the Southwestern assay on proteins under denaturing conditions, it is likely that p30M contacts the 5′ regulatory region of yps-3 (the G217B probe) as a monomer or multimer of a single polypeptide with a 30-kDa molecular mass.
FIG. 2.
Southwestern blot analysis of extracts prepared from the yeast and mycelial phases of H. capsulatum strains. (A) Nuclear (Nucl) and cytoplasmic (Cyto) extracts (40 μg) prepared from the yeast and mycelial phases of the Downs and G217B strains were electrophoresed on SDS–10% polyacrylamide gels, fixed, and stained with Coomassie brilliant blue. (B) A second gel, electrophoresed in parallel under the same conditions, was transferred by electroblotting, blocked, and treated with 105 cpm of the PCR-amplified target probe (395 bp of the 5′ regulatory region of yps 21:E-9 in G217B)/ml for 2 h. The blot was washed and autoradiographed to demonstrate a protein (designated p30M) of an apparent molecular mass of 30 kDa from the mycelial phase nuclear extracts of G217B which interacts with the 32P-labelled DNA target in situ.
Attenuated Downs strain of H. capsulatum is deficient in the production of biologically active p30M protein.
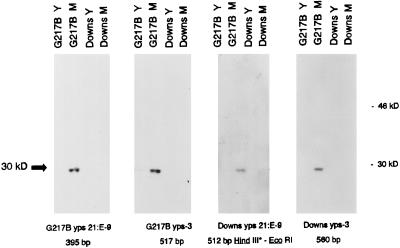
The ability of yeast- and mycelial phase nuclear extracts from the Downs and G217B strains to interact in cross-binding reactions with target probes prepared from the same or heterologous DNA sources was evaluated by Southwestern analysis. As seen in Fig. 3, a 30-kDa protein found exclusively in mycelial phase extracts of G217B (p30M) bound to target probes prepared from homologous DNA (393-bp G217B yps 21:E-9 and 517-bp G217B yps-3 probes) as well as labelled probes prepared from the same upstream regions from the Downs strain (the end-labelled 512-bp HindIII-EcoRI probe and the 560-bp Downs yps-3 probe). However, extracts from the Downs strain failed to demonstrate p30M binding in assays performed with heterologous (G217B) or homologous probes amplified from Downs genomic DNA. The findings pointed to a p30M deficiency in the low-virulence Downs strain, since no biologically active protein capable of the protein-DNA target interaction was observed.
FIG. 3.
Southwestern blot analysis of p30M binding with 32P-labelled target probes from various sources. Nuclear extract (40 μg) prepared from the yeast (Y) and mycelial (M) phases of the G217B and Downs strains of H. capsulatum were electrophoresed on SDS–10% polyacrylamide gels, transferred, and allowed to interact with the indicated DNA target probes. All probes were prepared by PCR amplification, except for the Downs yps 21:E-9 probe, which required end labelling of the 512-bp HindIII-EcoRI fragment at the HindIII site.
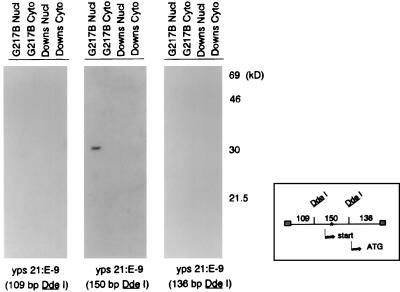
p30M DNA binding activity localizes to a 150-bp DdeI fragment in the upstream regulatory region of yps 21:E-9.
The probes used in the previous experiment (Fig. 3) ranged in size from 395 to 560 bp and might harbor single or multiple interaction sites for the p30M protein. To dissect the molecular relationship of this interaction, the 32P-labelled 395-bp yps 21:E-9 upstream regulatory region probe amplified from G217B was digested with DdeI to provide three subfragments, which were used as probes in the Southwestern assay. As seen in Fig. 4, the 136-bp DdeI 3′ primer fragment contains the putative ATG start codon while the central 150-bp fragment bordered by DdeI sites contains the transcriptional start site. The nuclear p30M protein from G217B extracts reacted exclusively with the internal 150-bp DdeI probe from the yps 21:E-9 regulatory region.
FIG. 4.
Localization of DNA binding activity to a 150-bp DdeI fragment in the upstream regulatory region of yps 21:E-9. Nuclear (Nucl) and cytoplasmic (Cyto) extracts (40 μg) from the mycelial phases of G217B and Downs were evaluated by Southwestern analysis with 32P-labelled targets derived from the 395-bp PCR-amplified yps 21:E-9 regulatory region from G217B. The amplified probe was digested with DdeI to liberate labelled fragments of 109, 150, and 136 bp. As indicated in the diagram, the 135-bp fragment contains the ATG codon while the transcription start site mapped by S1 analysis is within the 150-bp fragment flanked by DdeI sites. The asterisk denotes the location of the single TCCTTTTTTT motif observed in this DNA ligand.
Binding sites for p30M were also mapped to at least two DdeI-generated subfragments of the 517-bp G217B yps-3 regulatory sequence (data not shown) and prompted a comparison of sequences within these regions for any consensus motifs. The decanucleotide motif TCCTTTTTTT (TCC motif) was found to be conserved at 80 to 100% homology when the yps 21:E-9 sequence from G217B was compared with the G217B yps-3 sequence (Table 1). The TCC motif was only found once within the 150-bp DdeI fragment of yps 21:E-9, consistent with the mapping results shown in Fig. 4, at 13 nucleotides downstream from the transcriptional start site and 130 nucleotides upstream of the ATG start codon for the gene (Fig. 1 and Table 1). The same motif was conserved with respect to homology (100%) and location (between the transcriptional start site and the ATG codon) in the G217B yps-3 sequence. Additional TCC consensus motifs were found upstream of the transcriptional start site mapped by S1 analysis in the yps-3 gene (Fig. 1 and Table 1). Since similar regions amplified from Downs genomic DNA mediated binding to p30M, as seen in Fig. 3, it was significant that the motif was conserved with respect to position and homology (80%) in the Downs sequences of yps-3 and yps 21:E-9 (Fig. 1 and Table 1), even though the genes are not transcribed in this strain.
TABLE 1.
Conservation of the TCC motif within yps and housekeeping genes in H. capsulatum
| Gene | Strain | Sequencea | Position |
|---|---|---|---|
| yps 21:E-9 | G217B | T C C T T T T T T T | +13 (S1 start site); ATG-130 |
| yps 21:E-9 | Downs | - - G - - G - - - - | Same position; gene is not transcribed |
| yps-3 | G217B | - - - - - - - - - - | +46 (S1 start site); ATG-114 |
| yps-3 | Downs | - T A - - - - - - C | Same position; gene is not transcribed |
| yps-3 | G217B | - - - - - - - - - A | −9 (S1 start site) |
| yps-3 | G217B | - - - - C - C - - - | −33 (S1 start site) |
| TUB1 | G217B | - A - - - - C - - - | ATG-160 |
| TUB1 | G217B | - - - A G G - - - - | ATG-110 |
| TUB2 | G217B | - T - - G - - - C - | ATG-145 |
| TUB2 | G217B | - - T - - - C A - - | ATG-28 |
Dashes indicate identity.
p30 recognizes the TCC decanucleotide motif.
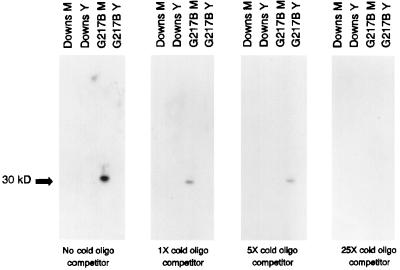
To examine the molecular basis for the interaction between p30M protein and DNA target sequences, the putative recognition motif, TCCTTTTTT, and its complement were annealed and ligated to form concatemers ranging in size from 100 to 350 bp. The excess, cold, ligated material was used as a target to challenge the binding of the 365-bp G217B yps 21:E-9 target DNA probe in Southwestern blots with yeast- and mycelial phase nuclear extracts prepared from G217B and Downs. As seen in Fig. 5, only p30M from the virulent G217B mycelial phase nuclear extract bound the probe in the absence of competitor oligonucleotides: the competition analysis with excess unlabelled double-stranded oligonucleotide demonstrated dose-response-dependent inhibition of probe binding. Experiments with DNA ligand prepared by individually end labelling the complementary oligonucleotides which form the putative motif and ligating the labelled reaction products demonstrated direct binding of the target to the 30-kDa protein (data not shown), confirming that the TCC motif is recognized by the p30M DNA binding protein.
FIG. 5.
Competition of the p30M interaction with the 32P-labelled 395-bp G217B yps 21:E-9 target by increasing amounts of unlabelled decanucleotide. Extracts from the mycelial (M) and yeast (Y) phases of the Downs and G217B strains were electrophoresed, blotted, and then evaluated for DNA binding to labelled probe in the absence and presence of the cold, ligated oligonucleotide (oligo) probe. The cold DNA ligand represented concatemers of the TCCTTTTTTT sequence and its complement and were ligated to an average range of 150 to 350 bp, as assessed by agarose gel electrophoresis.
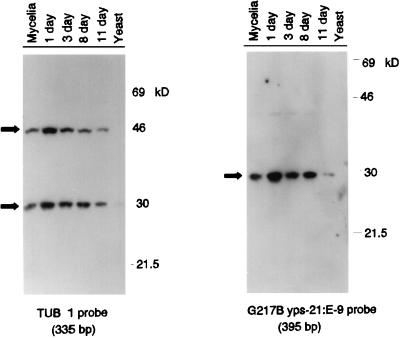
Steady-state levels of biologically active p30M diminish during the mycelial phase-yeast phase transition.
To examine the fate of p30M binding to target DNA probes during the phase transition, G217B mycelia were induced to transform into yeast by shifting the temperature of incubation to 37°C. Nuclear extracts were isolated at various time points during the transition, electrophoresed in 40-μg sample aliquots, and transferred to Southwestern blots for analysis with the G217B yps 21:E-9 probe (395 bp) and with a comparably sized probe (335 bp) from the putative regulatory regions of TUB1 (the α-tubulin gene) (8). The single α-tubulin gene is expressed constitutively during the temperature-induced transition, although fivefold-higher transcript levels are observed in the mycelial phase (9). Cultures were visually monitored for morphologic evidence of conversion prior to extract isolation. By day 11 following the temperature shift-up, the culture contained 90% yeast, and the transition was complete, with 100% yeast forms, at day 13. As seen in Fig. 6, a 30-kDa-molecular-mass protein recognizes the yeast phase-specific regulatory regions (in yps 21:E-9), as well as motifs within the amplified TUB1 gene region. Binding of steady-state levels of p30M in the nuclear extracts to either target appears to be maximal at day 1 of the temperature-induced phase transition, with lowered or no detectable binding observed later in the transition (day 11; yeast phase). An additional protein with an apparent molecular mass of 44 kDa interacted with the TUB1 (α-tubulin gene) regulatory region, but its role in the regulation of housekeeping gene transcription is currently unknown.
FIG. 6.
Southwestern analysis of nuclear extracts prepared from mycelial phase-to-yeast phase transforming cultures of G217B. Mycelial cells of strain G217B were induced to transform to the yeast phase at 37°C, and nuclear extracts were obtained from the terminal phases as well as 1 day, 3 days, 8 days, and 11 days following the shift-up in temperature. Proteins were electrophoresed in duplicate on SDS–10% polyacrylamide gels; half of the gel was stained to verify protein loading, and the remainder was transferred and evaluated for DNA binding. The results shown are from separate Southwestern blots with the 335-bp regulatory region of TUB1 (the α-tubulin gene) amplified from G217B and the 5′ regulatory region of yps 21:E-9 amplified from G217B.
DISCUSSION
This study describes the first DNA binding protein identified from the pathogenic dimorphic fungus H. capsulatum as a 30-kDa nuclear protein unique to the mycelial phase of the high-virulence strain G217B which binds to the 5′ regulatory regions of the yeast phase-specific genes, yps-3 and yps 21:E-9. The p30M protein from G217B binds to 32P-labelled target sequences amplified from genomic DNA prepared from the homologous G217B strain or from the temperature-sensitive low-virulence strain Downs, indicating that slight changes in a decanucleotide consensus motif fail to completely disrupt protein-DNA interaction. The Downs strain lacks detectable p30M protein in nuclear extracts prepared from yeast and mycelial phases in the Southwestern bioassay, regardless of the source of labelled target probe. The Downs strain is thus deficient in the expression of p30M protein or biologically active p30M; lack of this nuclear, phase-specific DNA binding protein may contribute to the absence of yps-3 and yps 21:E-9 transcription which is observed in this low-virulence strain.
The binding activity of p30M was localized to a 150-bp DdeI fragment in the 5′ regulatory region of yps 21:E-9. A decanucleotide motif, TCCTTTTTTT, was found in this region that was also observed with 80 to 100% homology at three sites within the putative regulatory region of yps-3. The position of the decanucleotide motif is also conserved in the genes and is localized between the transcriptional and translational start sites, where p30M binding may influence gene expression as a repressor or activator. Alterations in the TCCTTTTTTT motif found in the Downs sequence (changes at the 3rd, 6th, or 10th position) did not abolish the ability of p30M from G217B to recognize the target DNA. Cold, unlabelled oligonucleotide ligated to form 150- to 350-bp concatemers competes with 32P-labelled 365-bp yps 21:E-9 target probe amplified from G217B for binding to p30M, providing evidence that the 10-bp motif is the binding site for p30M. Moreover, recent studies have identified a 10-bp TCA motif, strikingly similar to the H. capsulatum TCC sequence, in the regulatory regions of over 20 plant genes that respond to chemical, thermal, or viral stress (5); in these cases, the motif is recognized by a 40-kDa protein which regulates the expression of essential housekeeping genes. In a promoter-GUS deletion analysis of the parsley PR-2 gene, a 60-bp region incorporating the T-rich 10-bp motif was crucial for quantitative levels of GUS expression (33).
As with other types of regulated gene expression, transcriptional induction in response to environmental cues or stress is mediated through cis DNA sequence elements which are recognized by trans-acting factors. In yeast, several families of genes, including those involved in the expression of specific mating types (27), amino acid biosynthesis (11, 12), and galactose utilization (2, 15), each contain their own common oligonucleotide sequences upstream of their respective coregulated genes. Both genetic and biochemical evidence on these yeast gene sets indicates the requirement of the oligonucleotide sequences for controlled gene expression mediated by interactions with protein products of other regulatory loci (10, 11, 22, 27, 28). However, the H. capsulatum TCCTTTTTTT motif is also found in the regulatory regions of the constitutively expressed α- and β-tubulin genes, TUB1 and TUB2 (data not shown), and is recognized by p30M protein. It is significant to note that a T-rich region, similar to the consensus p30M motif, is one of the sequences found in the tripartite upstream promoter element which is essential for expression of S. cerevisiae ribosomal protein genes (30). In yeast, this sequence may be involved in the expression of a variety of other genes, including those encoding histones (3), a negative regulator of Ty-controlled gene expression (29), and the cytoskeletal protein actin (4).
The findings indicate that p30M is highly expressed in the mycelial phase of H. capsulatum at 25°C and binds to yps sequences in situ. The position of the conserved pyrimidine-rich decanucleotide motifs between the transcriptional and translational start sites of the yeast phase-specific genes may suggest a potential role for the p30M protein as a transcriptional repressor (10), acting to inhibit gene expression. The inhibition of gene expression is not complete, however, even when p30M levels are presumably high, as the TUB1 and TUB2 housekeeping genes, which harbor the TCC motif, continue to be expressed at higher levels in the hyphal phase (9). It may be that the 47-kDa protein detected in Southwestern blots with the upstream regulatory sequence target of the α-tubulin gene (Fig. 6) activates housekeeping gene transcription (14, 15, 22) by directly or indirectly limiting access of p30M to target sequences in vivo. At 37°C, or during the transition of mycelia to yeast, p30M levels appear to decline, and lack of the putative repressor may be sufficient for the establishment of yeast phase-specific gene expression. In recent studies, however, a 17-kDa protein from yeast-phase extracts of H. capsulatum G217B, termed p17Y, has been detected with the concatenated oligonucleotide site probe in Southwestern blots by using the assay system described in this study (29a). Since p30M and p17Y recognize the same pyrimidine-rich motif, levels of both proteins may be important in regulating yeast phase-specific gene expression. Bimodal control of yps or tubulin gene expression may involve a consortium of repressor p30M and activators p45 and p17Y, as well as other factors not detected by the Southwestern assay system.
The results presented in this study indicate that p30M, a Histoplasma nuclear protein which binds to the TCC motif, is potentially a trans-acting transcriptional factor which may be involved in both quantitative and qualitative regulation of gene expression during the temperature-induced, yeast-to-mycelium conversion. It is likely that the TCC motif and p30M play a role in regulated expression of a number of fungal genes in response to temperature or other host signals, in concert with other elements and factors. DNA affinity-based purification of p30M and isolation of its gene will be essential in dissecting the nature and function of this novel DNA binding protein. For example, the addition and expression of the gene encoding p30M by transformation systems recently described for Histoplasma (35–37) may be sufficient to restore yeast phase-specific gene expression in the attenuated Downs strain. Alternatively, the catalog of physiologic (19, 23, 25, 26) and genetic (17, 18) differences between the Downs strain and virulent Histoplasma strains like G217B may argue that a significant number of Downs mutations prompt it to lower virulence. As an alternative approach circumventing the issue of multiple changes in the Downs genome that contribute to attenuation, construction of chimeric promoter-reporter gene fusions in transformed G217B will be informative for exploring the mechanism(s) by which this TCC motif or other sequence elements modulate gene expression in this dimorphic pathogenic fungus.
ACKNOWLEDGMENTS
We thank Judith Medoff, Robert Bolla, William Picking, and others at Saint Louis University for encouragement and discussion.
This work was supported by Public Health Service grants AI28950 and AI37540 from the National Institutes of Health to E.J.K. F.E.A. and H.R. were also supported by graduate teaching and research assistantships from the Department of Biology and the Graduate School of Saint Louis University.
REFERENCES
- 1.Abidi F E. Characterization of a yeast phase specific gene (yps 21:E-9) and identification of a DNA binding protein from the dimorphic pathogenic fungus, Histoplasma capsulatum. Ph.D thesis. University of Minnesota; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bram R J, Lue N F, Kornberg R D. A GAL family of upstream activating sequences in yeast: roles in both induction and repression of transcription. EMBO J. 1986;5:603–608. doi: 10.1002/j.1460-2075.1986.tb04253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choe J, Kolodrubetz D, Grunstein M. The two yeast histone H2A genes encode similar protein subtypes. Proc Natl Acad Sci USA. 1982;79:1484–1487. doi: 10.1073/pnas.79.5.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallwitz D, Perrin F, Seidel R. The actin gene in yeast Saccharomyces cerevisiae: 5′ and 3′ end mapping, flanking and putative regulatory sequences. Nucleic Acids Res. 1981;9:6339–6350. doi: 10.1093/nar/9.23.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldsbrough A P, Albrecht H, Stratford R. Salicylic acid-inducible binding of a tobacco nuclear protein to a 10 bp sequence which is highly conserved amongst stress-inducible genes. Plant J. 1993;3:563–571. doi: 10.1046/j.1365-313x.1993.03040563.x. [DOI] [PubMed] [Google Scholar]
- 6.Goodbourn S. Negative regulation of transcriptional initiation in eukaryotes. Biochim Biophys Acta. 1990;1032:53–77. doi: 10.1016/0304-419x(90)90012-p. [DOI] [PubMed] [Google Scholar]
- 7.Graybill J R. Histoplasmosis and AIDS. J Infect Dis. 1988;158:632–636. doi: 10.1093/infdis/158.3.623. [DOI] [PubMed] [Google Scholar]
- 8.Harris G S, Keath E J, Medoff J Z. Characterization of alpha and beta tubulin genes in the dimorphic fungus, Histoplasma capsulatum. J Gen Microbiol. 1989;135:1817–1832. doi: 10.1099/00221287-135-7-1817. [DOI] [PubMed] [Google Scholar]
- 9.Harris G S, Keath E J, Medoff J. Expression of α- and β-tubulin genes during dimorphic-phase transitions of Histoplasma capsulatum. Mol Cell Biol. 1989;9:2042–2049. doi: 10.1128/mcb.9.5.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herschbach B M, Johnston A D. Transcriptional repression in eukaryotes. Annu Rev Cell Biol. 1993;9:479–511. doi: 10.1146/annurev.cb.09.110193.002403. [DOI] [PubMed] [Google Scholar]
- 11.Hinnebush A G, Fink G R. Repeated DNA sequences upstream from HIS 1 also occur at several other co-regulated genes in Saccharomyces cerevisiae. J Biol Chem. 1983;258:5238–5247. [PubMed] [Google Scholar]
- 12.Hinnebush A G, Lucchini G, Fink G R. A synthetic His4 regulatory element confers general amino acid control on the cytochrome c gene (CYC1) of yeast. Proc Natl Acad Sci USA. 1985;82:498–502. doi: 10.1073/pnas.82.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hope I A, Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN 4 of yeast. Cell. 1986;46:885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- 14.Hope I A, Struhl K. GCN 4, a eukaryotic transcriptional activator protein, binds as a dimer to target DNA. EMBO J. 1987;6:2781–2784. doi: 10.1002/j.1460-2075.1987.tb02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnston M, Davis R W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keath E J. Generation of subtraction libraries for phase specific genes in the dimorphic fungus, Histoplasma capsulatum. In: Maresca B, Kobayashi G S, editors. Molecular biology of pathogenic fungi: a laboratory manual. New York, N.Y: Telos Press; 1994. pp. 391–397. [Google Scholar]
- 17.Keath E J, Abidi F E. Molecular cloning and sequence analysis of yps-3, a yeast phase specific gene in the dimorphic fungal pathogen, Histoplasma capsulatum. Microbiology. 1994;140:759–767. doi: 10.1099/00221287-140-4-759. [DOI] [PubMed] [Google Scholar]
- 18.Keath E J, Painter A A, Kobayashi G S, Medoff G. Variable expression of a yeast-phase-specific gene in Histoplasma capsulatum strains differing in thermotolerance and virulence. Infect Immun. 1989;57:1384–1390. doi: 10.1128/iai.57.5.1384-1390.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lambowitz A M, Kobayashi G S, Painter A, Medoff G. Possible relationship of morphogenesis in pathogen fungus, Histoplasma capsulatum, to heat shock response. Nature. 1983;303:806–808. doi: 10.1038/303806a0. [DOI] [PubMed] [Google Scholar]
- 20.Machida M, Jigami Y, Tanaka H. Purification and characterization of a nuclear factor which binds specifically to the upstream activating sequences of S. cerevisiae enolase 1 gene. Eur J Biochem. 1982;184:305–311. doi: 10.1111/j.1432-1033.1989.tb15020.x. [DOI] [PubMed] [Google Scholar]
- 21.Machida M, Uemora H, Jigami Y, Tanaka H. The protein factor which binds to the upstream regulatory sequence of S. cerevisiae ENO1 gene. Nucleic Acids Res. 1988;16:1407–1422. doi: 10.1093/nar/16.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maniatis T, Goodbourn S, Fischer J A. Regulation of inducible and tissue specific gene expression. Science. 1987;236:1237–1245. doi: 10.1126/science.3296191. [DOI] [PubMed] [Google Scholar]
- 23.Maresca B, Kobayashi G S. Dimorphism in Histoplasma capsulatum: a model for the study of cell differentiation in pathogenic fungi. Microbiol Rev. 1989;54:186–209. doi: 10.1128/mr.53.2.186-209.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maresca B, Lambowitz A M, Kumar B V, Grant G A, Kobayashi G S, Medoff G. Role of cysteine in regulating morphogenesis and mitochondrial activity in the dimorphic fungus Histoplasma capsulatum. Proc Natl Acad Sci USA. 1981;78:4596–4600. doi: 10.1073/pnas.78.7.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medoff G, Kobayashi G S, Painter A, Travis S. Morphogenesis and pathogenicity of Histoplasma capsulatum. Infect Immun. 1987;55:1355–1358. doi: 10.1128/iai.55.6.1355-1358.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medoff G, Maresca B, Lambowitz A M, Kobayashi G S, Painter A, Sacco M, Carratu L. Correlation between pathogenicity and temperature sensitivity in different strains of Histoplasma capsulatum. J Clin Invest. 1986;78:1638–1647. doi: 10.1172/JCI112757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller A M, MacKay V L, Nasmyth K A. Identification and comparison of two sequence elements that confer cell-type specific transcription in yeast. Nature. 1985;314:598–603. doi: 10.1038/314598a0. [DOI] [PubMed] [Google Scholar]
- 28.Mitchell P J, Tjian R. Transcriptional regulation in mammalian cells by sequence specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 29.Roeder G S, Beard C, Smith M, Keranen S. Isolation and characterization of the SPT2 gene, a negative regulator of Ty-controlled yeast gene expression. Mol Cell Biol. 1985;5:1543–1553. doi: 10.1128/mcb.5.7.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Roh, H., and E. J. Keath. Unpublished data.
- 30.Rotenberg M O, Woolford J L., Jr Tripartite upstream promoter element essential for expression of Saccharomyces cerevisiae ribosomal protein genes. Mol Cell Biol. 1986;6:674–687. doi: 10.1128/mcb.6.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sacco M, Maresca B, Kobayashi G S, Medoff G. Temperature and cyclic nucleotide induced phase transitions of Histoplasma capsulatum. J Bacteriol. 1981;146:117–120. doi: 10.1128/jb.146.1.117-120.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shore D, Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987;51:721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- 33.Van de Locht U, Meier I, Hahlbrock K, Somssich I. A 125 bp promoter fragment is sufficient for strong elicitor mediated gene activation in parsley. EMBO J. 1990;9:2945–2950. doi: 10.1002/j.1460-2075.1990.tb07486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wheat L J. Histoplasmosis: recognition and treatment. Clin Infect Dis. 1994;19:19–27. doi: 10.1093/clinids/19.supplement_1.s19. [DOI] [PubMed] [Google Scholar]
- 35.Woods J P, Goldman W E. In vivo generation of linear plasmids with addition of telomeric sequences by H. capsulatum. Mol Microbiol. 1992;6:3603–3610. doi: 10.1111/j.1365-2958.1992.tb01796.x. [DOI] [PubMed] [Google Scholar]
- 36.Woods J P, Goldman W E. Autonomous replication of foreign DNA in Histoplasma capsulatum: role of native telomeric sequences. J Bacteriol. 1993;175:636–641. doi: 10.1128/jb.175.3.636-641.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Worsham P L, Goldman W E. Development of a genetic transformation system for Histoplasma capsulatum: complementation of uracil auxotrophy. Mol Gen Genet. 1990;221:358–362. doi: 10.1007/BF00259400. [DOI] [PubMed] [Google Scholar]