Abstract
The most frequent DNA lesions in mammalian genomes are removed by the base excision repair (BER) via multiple pathways that involve the replacement of one or more nucleotides at the lesion site. The biological consequences of a BER defect are at present largely unknown. We report here that mouse cells defective in the main BER DNA polymerase β (Pol β) display a decreased rate of DNA single-strand breaks (ssb) rejoining after methyl methanesulfonate damage when compared with wild-type cells. In contrast, Pol β seems to be dispensable for hydrogen peroxide-induced DNA ssb repair, which is equally efficient in normal and defective cells. By using an in vitro repair assay on single abasic site-containing circular duplex molecules, we show that the long-patch BER is the predominant repair route in Pol β-null cell extract. Our results strongly suggest that the Pol β-mediated single nucleotide BER is the favorite pathway for repair of N-methylpurines while oxidation-induced ssb, likely arising from oxidized abasic sites, are the substrate for long-patch BER.
INTRODUCTION
DNA apurinic/apyrimidinic (AP) sites are ubiquitous lesions that arise either from spontaneous depurination or by removal of damaged bases produced by physical and chemical treatment. Because of their ‘non instructional’ character these lesions are likely to be both cytotoxic and mutagenic. Prokaryotes and eukaryotes have evolved a defense mechanism to repair this kind of DNA damage and to guarantee the integrity of the genome, known as base excision repair (BER). For many years it has been believed that the biochemistry of the BER was completely understood. Recent data have triggered again the interest in this excision repair process by showing the existence of alternative BER pathways which involve the replacement of one (short patch BER) or more nucleotides (long patch BER) at the lesion site (for a review see 1). Both BER pathways have been reconstituted in vitro by using purified mammalian proteins (2–5).
The DNA polymerase of election for the short patch BER is DNA polymerase β (Pol β). This enzyme is able to release the remnant 5′ deoxyribosephosphate formed by the major mammalian AP endonuclease HAP1 and to fill in the gap (for a review see 6). Conversely, in the long-patch BER either Pol β (7,8) or Pol δ/ɛ (9,10) are able to perform the resynthesis step. Besides Pol δ/ɛ, other replication factors are specifically involved in this pathway: the structure-specific endonuclease FEN1, the proliferating cell nuclear antigen (PCNA) and the replication factor C responsible for the loading of PCNA onto the DNA template.
Notwithstanding the progress made in the biochemical characterization of BER, the biological significance of this repair process is largely unknown. Since no human BER mutants are available, our knowledge relies upon mice lacking a specific DNA repair activity. Whereas HAP1 endonuclease, Pol β, DNA ligase I and XRCCI gene disruptions lead to lethality during embryogenesis, the DNA glycosylase-deficient embryos (uracil DNA glycosylase, 3-methyladenine DNA glycosylase and 7,8-dihydro-8-oxoguanine-DNA glycosylase) are viable and no obvious phenotype is observed (for a review see 11). Null cells can be derived from embryos of mice carrying lethal mutations. Embryonic fibroblast cell lines homozygous for a deletion mutation in the Pol β gene have been established (12) from Pol β-knock out mice.
In this paper we show that a defect in Pol β leads to hypersensitivity to the cytotoxic effects of both methyl methanesulfonate (MMS) and hydrogen peroxide (H2O2) and to the accumulation of MMS-induced DNA single-strand breaks (ssb). In contrast, Pol β is not required for DNA ssb repair induced by H2O2. Biochemical studies clarify that the long patch BER is the predominant AP site repair route in the Pol β-null extracts. These data strongly suggest that long-patch BER is the predominant repair route for ssb arising from oxidative damage.
MATERIALS AND METHODS
Chemicals
MMS (Merck, White House Station, NJ) was dissolved in dimethyl sulfoxide (DMSO) shortly before use and quickly diluted in serum-free medium to the required concentrations (final concentration of DMSO <0.5%). H2O2 (Sigma-Aldrich, St Louis, MO) was diluted in water.
[α-32P]dCTP (3000 Ci/mmol) and [α-32P]dTTP (3000 Ci/mmol) were from Amersham Pharmacia Biotech (Buckinghamshire, UK) and unlabeled dNTPs from Roche Molecular Biochemicals (Mannheim, Germany). All other reagents were of analytical grade and purchased from Merck or Fluka-Sigma-Aldrich (St Louis, MO).
Cell culture and cytotoxicity assay
SV40 transformed wild-type and Pol β-deficient mouse embryonic fibroblasts (a gift from Dr S.H. Wilson, NIEHS, Research Triangle Park, NC) (12) were cultured in DMEM with glutamax-1 (Gibco BRL, Gaithersburg, MD) supplemented with 10% fetal calf serum, penicillin (100 U/ml), streptomycin (100 µg/ml) and hygromycin (80 µg/ml) at 34°C in a 10% CO2 incubator. For cytotoxicity studies cells were seeded at a density of 1 × 105 cells/well in six-well dishes (triplicate wells for each experimental point). After 18 h from seeding, they were exposed for 1 h to a range of concentrations of MMS or H2O2 in serum-free medium. Cell cultures were then washed, fed with complete medium and 5 days later harvested and counted microscopically. Results were expressed as the number of cells in drug-treated wells relative to cells in control wells (% control growth). Results were the average of three independent experiments.
Single cell gel electrophoresis (SCGE)
Cell cultures were treated with MMS (0.5 mM) or H2O2 (50 µM) in serum free medium for 30 min. Cells were allowed to repair DNA damage by incubation in fresh medium for the indicated periods of time. DNA breaks were detected as previously described (13). Briefly, cells were embedded in agarose gel and then spread on a frosted microscope slide. Cells were lysed in 2.5 M NaCl, 10 mM Tris–HCl, 100 mM Na2EDTA, 1% Triton, 10% DMSO, pH 10, for 1 h at 4°C. After lysis, cells were preincubated for 20 min at 4°C in the electrophoresis buffer (0.3 M NaOH, 1 mM Na2EDTA, pH 13.5) and then subjected to alkaline gel electrophoresis (300 mA, 4°C, 20 min). After SCGE, cell DNA was stained with ethidium bromide and visualized by fluorescence microscopy. Slides were analyzed by computerized image analysis (Casys system, Synoptics Ltd, Cambridge, UK) to quantitate DNA damage. The tail moment, calculated by multiplying the total intensity of the comet tail by the migration distance from the center of the comet head, was used as measure of DNA damage. Fifty cells for each experimental point were scored blind from two slides.
Nucleic acid substrates
Oligonucleotides for preparing the substrate for the BER assay were purchased from M-Medical (Florence, Italy). Closed circular DNA containing a single abasic site was produced as previously described (14) by priming single-stranded (+) pGEM-3Zf DNA (Promega, Madison, WI) with a 30-fold molar excess of uracil-containing oligonucleotide and incubating with T4 DNA polymerase holoenzyme, single-stranded DNA binding protein and T4 DNA ligase (Roche Molecular Biochemicals). Closed circular DNA duplex molecules were purified by cesium chloride equilibrium centrifugation. The oligonucleotide 5′-CGGTACCCGGGGATCUTCTAGAGTCGACCTGCA-3′ was used to create plasmid molecules containing a single uracil residue. Circular closed DNA molecules were then digested with Escherichia coli uracil-DNA glycosylase (a gift from Dr S. Boiteux, Centre Energie Atomique, Fontenay aux Roses, France) to produce a single abasic site.
In vitro BER assay
Whole cell extracts from wild-type and Pol β-null mouse fibroblasts were prepared as previously described (15). The repair reactions were carried out essentially as described by Frosina et al. (14) with minor modifications. Briefly, reaction mixtures (50 µl) contained: 100 ng of plasmid DNA containing a single uracil or AP site, 40 mM HEPES–KOH, pH 7.9, 75 mM KCl, 5 mM MgCl2, 0.5 mM DTT, 50 µM of each dNTP, 2 µCi of either [α-32P]dTTP or [α-32P]dCTP as indicated (the concentration of the corresponding cold dNTP was lowered to 5 µM), 2 mM ATP, 40 mM phosphocreatine, 2.5 µg of creatine phosphokinase, 18 µg of BSA and 100 µg of whole cell extracts. After 60 min at 30°C the plasmid DNA was recovered and digested with the appropriate restriction enzymes. The digestion products were resolved on a denaturing 15% PAGE. The BER products were visualized by autoradiography and analyzed by electronic autoradiography (Instant Imager, Packard, Meriden, CT) for quantitation.
RESULTS
Pol β null cells are hypersensitive to the cytotoxic effects of MMS and H2O2
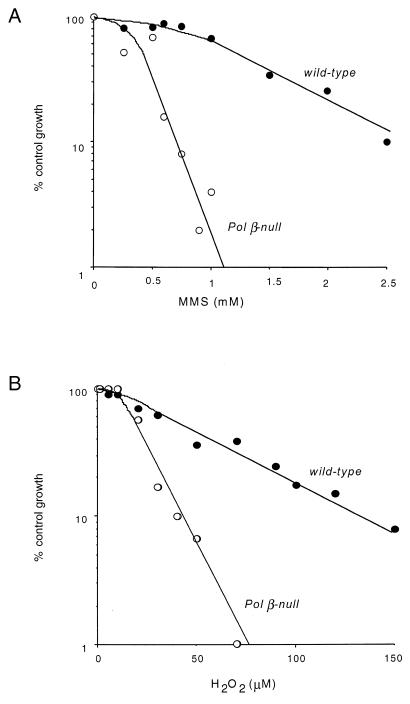
The sensitivity of Pol β-proficient and -deficient mouse cell lines to the cytotoxic effects of MMS and H2O2 was investigated. Both DNA damaging agents induce lesions, which are a substrate for BER. In particular MMS induces high levels of N-methylpurines (namely N7-methylguanine and N3-methyladenine) and H2O2 causes oxidation at purines and pyrimidines [e.g. 7,8-dihydro-8-oxoguanine (8-oxoG) and thymine glycol] and abasic sites. Pol β-deficient cells were hypersensitive to DNA damage induced by MMS (Fig. 1A) and H2O2 (Fig. 1B) although to a different extent. The sensitivity to the cytotoxic effects of MMS was increased 5-fold in Pol β-null cells (D37 = 0.3 mM MMS) as compared to the wild-type cells (D37 = 1.5 mM MMS), while the sensitivity to H2O2 was increased 2.7-fold in the defective cell line (D37 = 20 µM H2O2) as compared to the normal cells (D37 = 55 µM H2O2).
Figure 1.
Effect of Pol β deletion on MMS- and H2O2-induced cytotoxicity. Wild-type (closed circles) and Pol β-null (open circles) cells were exposed to increasing concentrations of MMS (A) or H2O2 (B) and the cell growth rate was evaluated 5 days after treatment. Data are the averages of three independent experiments.
These data suggest that the lack of Pol β exerts its cytotoxic effect via the persistence of a cytotoxic repair intermediate that is formed during the processing of methylated and oxidized DNA bases or abasic sites.
MMS but not H2O2-induced DNA ssb are rejoined at a slower rate in Pol β-defective cells as compared with normal cells
Pol β-deficient and -proficient cell lines were treated with MMS (0.5 mM) or H2O2 (50 µM) for 30 min and then allowed to repair for different periods of time. The extent of induced or residual DNA damage was measured by alkaline SCGE. Under these conditions both DNA ssb and alkali-labile sites (i.e. abasic sites) arising from BER are detectable. The number of DNA ssb measured reflects the balance between breaks arising from BER and breaks resealed by the same repair process. The presence of breaks allows supercoiled loops of DNA to relax and migrate to form a tail, and the fraction of DNA in the tail reflects the frequency of breaks.
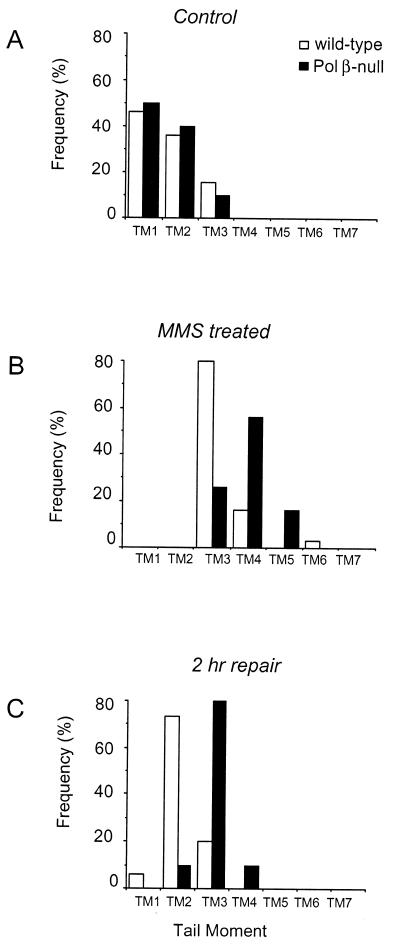
As shown in Figure 2B, the defective cells present a higher level of ssb as compared with wild-type cells immediately after 30 min incubation with MMS (Mann–Whitney test, P < 0.0001). Since MMS is a direct-acting mutagen this is unlikely to reflect differential amounts of induced DNA damage but rather identifies a defect in ssb rejoining at early times after damage. After 2 h repair incubation in drug-free medium (Fig. 2C) the mutant cells still showed an accumulation of DNA ssb as compared to the wild-type cells (Mann–Whitney test, P < 0.0001) although both cell lines were able to repair the majority of base damage (90% for the wild-type cells and 70% for the defective cells).
Figure 2.
Effect of Pol β deletion on MMS-induced DNA ssb repair. Wild-type (open bars) and Pol β-null (closed bars) cells were exposed to 0.5 mM MMS for 30 min and then incubated in drug-free medium. At different time intervals samples were taken for SCGE analysis. The tail moment (TM) of 50 comets per experimental point was measured by computerized image analysis and the comets were classified in different classes (TM1-7) according to tail moment values. (A) Heterogeneity in DNA repair in untreated cells, (B) in MMS-treated cells and (C) after 2 h repair.
These data confirm that Pol β is the polymerase of election for BER induced by alkylation damage but also provide convincing evidence that in vivo an alternative, although kinetically slower, BER system is active on base damage.
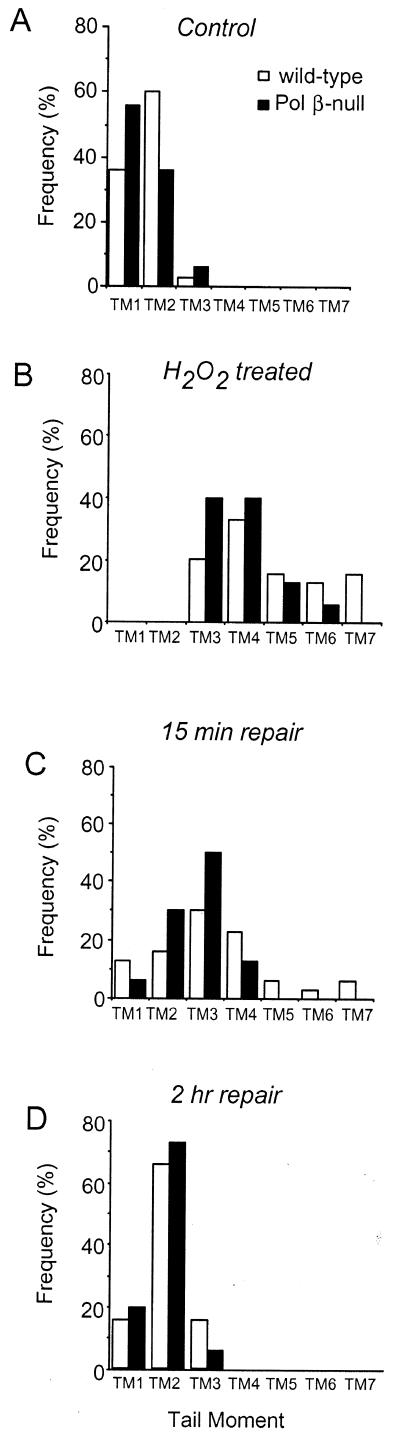
DNA strand breakage after H2O2 treatment (50 µM) was also investigated. In this case the distribution of cells according to the tail moment was very similar in normal and Pol β-null cells (Mann–Whitney test, P = 0.03, Fig. 3B) indicating that Pol β does not play a major role in oxidation-induced early strand breakage. A striking similarity between the two cell lines was also observed when the repair kinetics was investigated. Both cell lines repaired a large portion (almost 65%) of DNA ssb within 15 min incubation in drug-free medium (Fig. 3C) and in 2 h the rejoining process was complete (Fig. 3D). These data are compatible with the hypothesis that the oxidation-induced DNA ssb detected by SCGE are directly rejoined and/or do not require Pol β for their rejoining.
Figure 3.
Effect of Pol β deletion on H2O2-induced DNA ssb repair. Wild-type (open bars) and Pol β-null (closed bars) cells were exposed to 50 µM H2O2 for 30 min and then incubated in drug-free medium. At different time intervals samples were taken for SCGE analysis. The tail moment (TM) of 50 comets per experimental point was measured by computerized image analysis and the comets were classified in different classes (TM1-7) according to tail moment values. (A) Heterogeneity in DNA repair in untreated cells, (B) in H2O2-treated cells, (C) after 15 min repair and (D) after 2 h repair.
Pol β-defective cell extracts repair abasic sites predominantly via long-patch BER
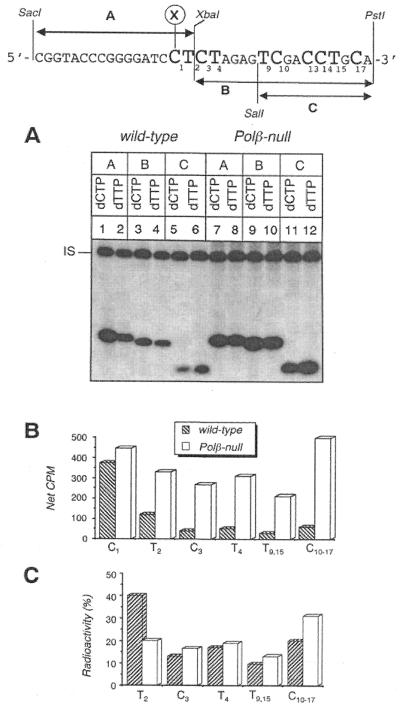
To gain insight into the repair specificity of wild-type and Pol β-null cells, whole cell extracts were tested in an in vitro BER assay using as substrate circular duplex plasmid molecules containing a single AP site. AP sites are the common repair intermediate after damage with BER-inducing agents. The duplex plasmid DNA was constructed to allow mapping of the repair patches at nucleotide resolution by combining specific restriction digestions with the use of different labeled dNTPs in the repair reaction. In particular (see scheme in Fig. 4), the incorporation of dTMP in the A, B and C restriction fragments allows quantification of long patch BER up to the T residue 15 nt 3′ to the lesion. The incorporation of dCMP in the same fragments allows identification of the contribution of both one gap-filling reactions (by subtracting the radioactivity in the B fragment from that in the A fragment) and long patch BER (B and C fragments) up to the C residue 17 nt 3′ to the lesion.
Figure 4.
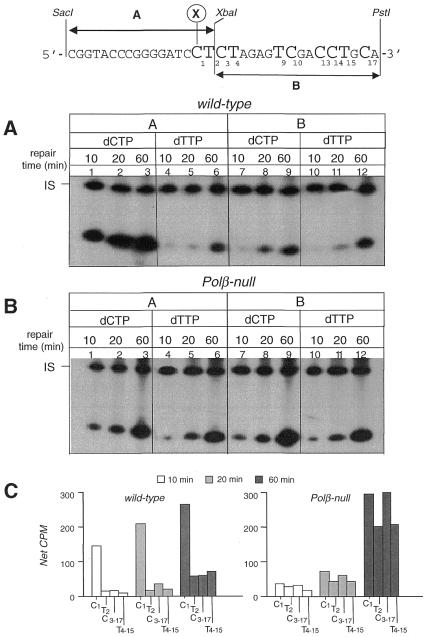
Mapping at nucleotide resolution of the repair patches at a single abasic site by wild-type and Pol β-null cell extracts. (A) Autoradiograph of a denaturing polyacrylamide gel. Whole cell extracts from Pol β-proficient and -deficient cells were used in an in vitro repair assay on circular duplex DNA molecules containing a single abasic site. Repair replication was performed for 1 h in the presence of [α-32P]dCTP or [α-32P]dTTP at 30°C. DNAs were digested with: SacI–XbaI (fragment A) to release the 17 bp fragment originally containing the lesion, XbaI–PstI (fragment B) and SalI–PstI (fragment C) to release the 16 bp and 10 bp fragments respectively, containing nucleotide residues 3′ to the AP site. IS, internal standard. (B) The levels of incorporation in the restriction fragments were measured by electronic autoradiography and corrected for DNA recovery (net c.p.m.). The incorporation at C3 and T4 was calculated by subtracting the radioactivity (dCMP or dTMP incorporation) of fragment C from that of fragment B. (C) Distribution of the radioactivity present in the long (≥2 nt) repair patches along the sequence 3′ to the AP site. Top, scheme of the restriction mapping. X, position where the AP site was originally located.
To determine and compare the repair patch profile of wild-type and Pol β-null whole cell extracts, the in vitro BER assay was performed by using either dCTP or dTTP as labeled DNA precursors in 1 h repair reaction at 30°C on a circular duplex DNA substrate containing a single abasic site (Fig. 4). Similar results were obtained when the target DNA lesion was a single uracil residue. A 5′-end labeled 60mer (indicated as internal standard, IS) was added in all reaction tubes in order to correct the repair incorporation values, as measured by electronic autoradiography, for DNA recovery. In the case of normal mouse cell extracts the level of radiolabeled dCMP incorporated in the A fragment (Fig. 4A, lane 1), which marks the resynthesis at the lesion site (C1), was well above the level of incorporated dTMP (lane 2) which marks the resynthesis including >1 nt. The long-patch BER extended beyond the T2 as shown by the presence of radioactivity in the B and C fragments (lanes 3–6) although a progressive decrease in the number of substituted nucleotides was observed at increasing distances from the lesion site. Quantitation of the radioactivity in the different restriction fragments (Fig. 4B) showed that in wild-type cell extracts 1 nt replacement events represented almost 70% of all repair reactions with the remaining 30% involving resynthesis of longer patches. Conversely, mapping of the repair patches in Pol β-null extracts showed that the level of dCMP incorporation in the A fragment (Fig. 4A, lane 7) was slightly higher (20%) than that of dTMP in the same fragment (Fig. 4A, lane 8) (Fig. 4B). Similar levels of radioactivity were detected over T2, C3 and T4 (Fig. 4B). This implies that, in the absence of Pol β, the single nucleotide replacement reactions are unfavored and the majority of the repair patches extend at least over the first 4 nt including the original lesion site. Moreover, a significant level of incorporation was detected in the C fragment (Fig. 4A, lanes 11 and 12) testifying that a significant fraction of the displaced and then resynthesized fragments are >9 nt in length.
The type of DNA polymerase involved in the BER reaction is therefore preferentially associated with a specific BER branch: Pol β with the single nucleotide BER and Pols other than Pol β, likely Pol δ/ɛ, with the long-patch BER.
When the distribution of the long (≥2 nt) repair patches in Pol β-proficient and -deficient cells was compared (Fig. 4C) a predominance of 2 nt replacement events was observed in normal cells while defective extracts were characterized by a relatively higher fraction of patches extending above T9.
The long-patch repair kinetics are similar in Pol β-proficient and -deficient cell extracts
In order to better characterize the BER process performed by normal and defective Pol β cells, the rate of nucleotide resynthesis was investigated at the lesion site and along the sequence 3′ to it. As shown in Figure 5, the single nucleotide replacement events, which are the hallmark of wild-type whole cell extracts, are fast processes that reached a plateau in 20 min (Fig. 5A, lanes 1–3; Fig. 5C). These data confirm previous reports on the higher efficiency and velocity of these reactions as compared to the long-patch BER in wild-type cell extracts (9).
Figure 5.
Mapping at nucleotide resolution of the repair patches at a single abasic site as a function of time by wild-type and Pol β-null cell extracts. (A and B) Autoradiograph of a denaturing polyacrylamide gel. Whole cell extracts from Pol β-proficient (A) and -deficient (B) cells were used in an in vitro repair assay on circular duplex DNA molecules containing a single abasic site. Repair replication was performed for the indicated periods of time (10–60 min) in the presence of [α-32P]dCTP or [α-32P)]dTTP at 30°C. DNAs were digested with: SacI–XbaI (fragment A) to release the 17 bp fragment originally containing the lesion and XbaI–PstI (fragment B) to release the 16 bp containing nucleotide residues 3′ to the AP site. IS, internal standard. (C) The levels of incorporation in the restriction fragments were measured by electronic autoradiography and corrected for DNA recovery (net c.p.m.). Top, scheme of the restriction mapping. X, position where the AP site was originally located.
What is the efficiency of the long-patch BER in Pol β defective cells? Pol β-null extracts, which perform predominantly long-patch BER, present the same long-patch repair time course as that detected in normal extracts (Fig. 5A and B, lanes 4–12). After 10 min repair, while the one gap-filling reactions were almost at plateau in wild-type extracts (Fig. 5A, lane 1), the repair reactions of multiple nucleotide substitutions were barely detectable on the autoradiography (Fig. 5A, lanes 4, 7 and 10; Fig. 5B, lanes 1, 4, 7 and 10). In both extracts there was a time-dependent linear increase of the total yield of long patch repair products (Fig. 5C) with a 4-fold increase over 1 h repair.
The slow and comparable time courses of the long-patch BER in both cell extracts suggest that the repair machinery involved, if different, presents similar rate limiting steps, either the strand displacement reaction per se or the assembly of a multi-protein complex at the lesion site.
DISCUSSION
Pol β-dependent cytotoxicity induced by MMS and H2O2
Mouse cells defective in Pol β are hypersensitive to a variety of monofunctional alkylating agents (12,16). In this study we confirm the hypersensitivity to the cytotoxic effects of MMS and we present evidence of cross-sensitivity to H2O2. This oxidative agent causes damage to nuclear DNA by generating hydroxyl radicals in close proximity to DNA. The reaction of these reactive oxygen species (ROS) with DNA results primarily in base modifications but also in base loss and ssb due to fragmentation of the sugar. The increase in the cytotoxic effect of H2O2 associated with the loss of Pol β is lower as compared with that observed after methylation damage (compare Fig. 1B and A). This is compatible with (i) a minor role of Pol β in the processing of ROS-induced lesions; and/or (ii) with the production of repair intermediates with a lower cytototoxic potential as compared with those created during the processing of methylpurines. We (17) and others (18–20) have previously shown that Pol β is the favorite Pol for one gap filling reactions following removal by bifunctional DNA glycosylase/AP lyases of the main oxidized purines (i. e. 8-oxoG) and pyrimidines (i.e. thymine glycol) induced by ROS (for a review see 21). Pol δ/ɛ can eventually repair these gaps but at a slower rate (17). The absence of Pol β might then cause the persistence of cytotoxic strand interruptions arising from an inefficient repair of oxidized DNA bases. These strand breaks, which present a genuine 5′ nucleotide, might exert a lower lethal effect as compared with ssb with a 5′ blocked terminus as those produced during BER of methyl-purines.
Pol β-dependent DNA ssb repair induced by MMS
We report here that mammalian DNA ssb repair induced by MMS is affected by the deletion in the Pol β gene. The accumulation of DNA ssb observed after MMS treatment is likely to be responsible for the increased cytotoxicity observed in Pol β-null cells after methylation damage. A similar phenotype, i.e. increased MMS-sensitivity killing (22) and accumulation of alkylation-induced ssb (23), has been reported for XRCC1-defective cells. Interestingly, the defect in DNA ssb repair was already remarkable after 30 min incubation with MMS indicating that Pol β is required for efficient ssb repair at early times after base damage. After 2 h repair, although a higher level of ssb is still detectable in the Pol β mutant cells, the majority of breaks (70%) are resealed in these cells via a Pol β-independent process. This is in contrast with what is observed when XRCC1 is defective: the amount of ssb detected after 3 h repair following ethyl methanesulfonate (EMS) damage is even higher than the level detected immediately after DNA damage presumably reflecting continued base excision or exonuclease activity (24) in the absence of the ligation step. These data provide strong evidence that in vivo MMS damage is mainly processed via a Pol β-dependent pathway but, if Pol β is not functional, a Pol δ/ɛ-dependent back-up system can replace the main pathway limiting the hazardous effect of unrepaired ssb.
Pol β-independent DNA ssb repair induced by H2O2
We report here that DNA ssb repair induced by ROS is unaffected by Pol β mutation. What is the origin of H2O2-induced ssb detected by alkaline SCGE? In addition to frank strand breaks, which are a minor fraction of the total breaks, the majority of induced ssb present 5′ and 3′ phosphate termini implying that the release of a base has occurred (25). These abasic sites are 10% regular abasic sites and the majority are oxidized abasic sites, mainly 4′ sites (26). The contribution to the total yield of detected ssb of abasic sites arising from the processing of base damage like thymine glycol or 8-oxoG, should be limited due to the slow repair kinetics of these lesions (27). We (17) and others (18,19) have previously shown that the selection of the BER pathway is strongly affected by the structure of the termini of the abasic sites and in particular the occurrence of long-patch BER is favored by the presence of reduced or oxidized AP sites (3). In vitro the long-patch BER is performed equally well either by Pol β (3,8) or Pol δ/ɛ (10,4) . This might explain why ROS-induced DNA ssb repair is independent of a functional Pol β gene.
BER pathways in Pol β-null cell extracts
The fine mapping of the repair patches occurring at abasic sites when Pol β-defective cell extracts are used in the in vitro BER assay provided the first evidence that the predominant route of repair in the absence of Pol β is the long-patch BER. Since Pol δ/ɛ are able to perform the 1 nt polymerization step at sites of base loss (4,10) the irreplaceable function of Pol β, which is required for single nucleotide BER, should be the dRPase activity. The absence of this function and therefore the persistence of the 5′ blocked terminus is likely to be responsible for the adverse biological effects (cytotoxicity) detected in Pol β null cells treated with MMS. On the other hand, the modest (as in the case of MMS) or absent (as in the case of H2O2) effect of a defective Pol β on DNA ssb repair supports the conclusion that Pol δ/ɛ is capable of BER in vivo. Interestingly, studies performed more than one decade ago in permeable cell systems (reviewed in 28) by using DNA Pol inhibitors showed that either aphidicolin (Pol δ/ɛ inhibitor) or ddTTP (Pol β inhibitor) were able to partially inhibit alkylation damage-induced repair (29–31) while ionizing radiation-induced DNA damage was specifically inhibited by aphidicolin (32). Our data are in full agreement with these observations. The repair of damage by monofunctional alkylating agents involves preferentially Pol β but occurs, although less efficiently, also in its absence, while ROS-induced DNA ssb are efficiently repaired via Pol δ/ɛ-mediated long-patch BER.
The comparison of the long repair patch profile in wild-type versus Pol β null cell extracts showed that, although there was a significant overlapping in patch size distribution between the two extracts, the ‘short’ patches (mainly 2 nt long) were more represented in wild-type extracts while ‘long’ patches (up to 15 nt) were more frequent in Pol β-null extracts. The repair patch size distribution observed is in agreement with the different ability to perform processive gap-filling synthesis of Pol β (up to 6 nt ) (33,34) and Pol δ/ɛ (10–18 nt) (4). Pol β has been implicated in long-patch BER (7,35) and the excision of a characteristic dRP-trinucleotide was reported. This in contrast with the repair patch profile that we observed. Differences between the DNA substrates used in these studies, like the lesion sequence context, might explain this discrepancy. The time course of the repair process was similar in the two cell extracts suggesting that, if the repair machinery is different, there is a common rate-limiting step (the formation of the flap structure?). These findings are compatible with a model where both Pols are able to participate to the resynthesis step in the BER process. If DNA pols are functionally interchangeable in the repair synthesis step the challenging question is what are factors that control their participation to the BER process. Recent evidence has been provided that specific BER complexes might be acting at different stages of the cell cycle (24,36). It is tempting to speculate that the overlapping between the factors involved in DNA replication and those required for long-patch BER might favor Pol δ/ɛ in BER at post-replicative stages while the single nucleotide Pol β-mediated BER might be favored in pre-replicative stages. Future research should investigate whether the involvement of the two Pols in BER is indeed controlled by the cell cycle stage.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to M. Stucki for the biochemical characterization of the cell extracts and to U. Hubscher for helpful discussions. This work has been partially supported by the Associazione Italiana per la Ricerca sul Cancro (A.I.R.C.).
REFERENCES
- 1.Lindahl T. and Wood,R.D. (1999) Science, 286, 1897–1905. [DOI] [PubMed] [Google Scholar]
- 2.Kubota Y., Nash,R., Klungland,A., Schar,P., Barnes,D. and Lindahl,T. (1996) EMBO J., 15, 6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 3.Klungland A. and Lindahl,T. (1997) EMBO J., 16, 3341–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pascucci B., Stucki,M., Jonsson,Z.O., Dogliotti,E. and Hubscher,U. (1999) J. Biol. Chem., 274, 33696–33702. [DOI] [PubMed] [Google Scholar]
- 5.Matsumoto Y., Kim,K., Hurwitz,J., Gary,R., Levin,D.S., Tomkinson,A.E. and Park,M.S. (1999) J. Biol. Chem., 274, 33703–33708. [DOI] [PubMed] [Google Scholar]
- 6.Wilson S.H. (1998) Mutat. Res., 407, 203–215. [DOI] [PubMed] [Google Scholar]
- 7.Dianov G., Prasad,R., Wilson,S. and Bohr,V. (1999) J. Biol. Chem., 274, 13741–13743. [DOI] [PubMed] [Google Scholar]
- 8.Prasad R., Dianov,G.L., Bohr,V.A. and Wilson,S.H. (2000) J. Biol. Chem., 275, 4460–4466. [DOI] [PubMed] [Google Scholar]
- 9.Fortini P., Pascucci,B., Parlanti,E., Sobol,R.W., Wilson,S.H. and Dogliotti,E. (1998) Biochemistry, 37, 3575–3580. [DOI] [PubMed] [Google Scholar]
- 10.Stucki M., Pascucci,B., Parlanti,E., Fortini,P., Wilson,S.H., Hubscher,U. and Dogliotti,E. (1998) Oncogene, 17, 835–843. [DOI] [PubMed] [Google Scholar]
- 11.Friedberg E.C. and Meira,L.B. (1999) Mutat. Res., 433, 69–87. [DOI] [PubMed] [Google Scholar]
- 12.Sobol R.W., Horton,J.K., Kuhn,R., Gu,H., Singhal,R.K., Prasad,R., Rajewsky,K. and Wilson,S.H. (1996) Nature, 379, 183–186. [DOI] [PubMed] [Google Scholar]
- 13.Fortini P., Raspaglio,G., Falchi,M. and Dogliotti,E. (1996) Mutagenesis, 11, 169–175. [DOI] [PubMed] [Google Scholar]
- 14.Frosina G., Fortini,P., Rossi,O., Carrozzino,F., Raspaglio,G., Cox,L.S., Lane,D.P., Abbondandolo,A. and Dogliotti,E. (1996) J. Biol. Chem., 271, 9573–9578. [DOI] [PubMed] [Google Scholar]
- 15.Frosina G., Fortini,P., Rossi,O., Abbondandolo,A. and Dogliotti,E. (1994) Biochem. J., 304, 699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ochs K., Sobol,R.W., Wilson,S.H. and Kaina,B. (1999) Cancer Res., 59, 1544–1551. [PubMed] [Google Scholar]
- 17.Fortini P., Parlanti,E., Sidorkina,O.M., Laval,J. and Dogliotti,E. (1999) J. Biol. Chem., 274, 15230–15236. [DOI] [PubMed] [Google Scholar]
- 18.Dianov G., Bischoff,C., Piotrowski,J. and Bohr,V.A. (1998) J. Biol. Chem., 278, 33511–33516. [Google Scholar]
- 19.Klungland A., Hoss,M., Gunz,D., Constantinou,A., Clarkson,S.G., Doetsch,P.W., Boltin,P.H., Wood,R.D. and Lindahl,T. (1999) Mol. Cell, 3, 1–20. [DOI] [PubMed] [Google Scholar]
- 20.Dianov G.L., Thybo,T., Dianova,I.I., Lipinski,L.J. and Bohr,V.A. (2000) J. Biol. Chem., 275, 11809–11813. [DOI] [PubMed] [Google Scholar]
- 21.Wallace S.S. (1998) Radiat. Res., 150, S60–S79. [PubMed] [Google Scholar]
- 22.Op het Veld C.W., Jansen,J., Zdzienicke,M.Z., Vrieling,H. and van Zeeland,A.A. (1998) Mutat. Res., 398, 83–92. [DOI] [PubMed] [Google Scholar]
- 23.Thompson L.H., Brookman,K.W., Jones,N.J., Allen,S.A. and Carr,A.V. (1990) Mol. Cell. Biol., 10, 6160–6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor R.M., Moore,D.J., Whitehouse,J., Johnson,P. and Caldecott,K.W. (2000) Mol. Cell. Biol., 20, 735–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward J.E. (1998) In Nickoloff,J.A. and Hoekstra,M.F. (eds), DNA Damage and Repair, Vol. 2. Humana Press, Totowa, New Jersey, pp. 65–84.
- 26.von Sonntag C. (1987) The Chemical Basis of Radiation Biology. Taylor and Francis, London.
- 27.Collins A.R., Dobson,V.L., Dusinska,M., Kennedy,G. and Stetina,R. (1997) Mutat. Res., 375, 183–193. [DOI] [PubMed] [Google Scholar]
- 28.Kenney S. and Linn,S. (1990) Mutat. Res., 236, 239–252. [DOI] [PubMed] [Google Scholar]
- 29.Miller M.R. and Chinault,D.N. (1982) J. Biol. Chem., 257, 46–49. [PubMed] [Google Scholar]
- 30.Hammond R.A., McClung,J.K. and Miller,M.R. (1990) Biochemistry, 29, 286–291. [DOI] [PubMed] [Google Scholar]
- 31.Hjertvik M., Erixon,K. and Ahnstrom,G. (1998) Mutat. Res., 407, 87–96. [DOI] [PubMed] [Google Scholar]
- 32.Mirzayans R., Enns,M.R., Cubitt,S., Karimian,K., Radatus,B. and Paterson,M.C. (1994) Biochim. Biophys. Acta, 1227, 92–100. [DOI] [PubMed] [Google Scholar]
- 33.Singhal R.K. and Wilson,S.H. (1993) J. Biol. Chem., 268, 15906–15911. [PubMed] [Google Scholar]
- 34.Singhal R.K., Prasad,R. and Wilson,S.H. (1995) J. Biol. Chem., 270, 949–957. [DOI] [PubMed] [Google Scholar]
- 35.Prasad R. Dianov,G.L., Bohr,V.A. and Wilson,S.H. (2000) J. Biol. Chem., 275, 4460–4466. [DOI] [PubMed] [Google Scholar]
- 36.Otterlei M., Warbrick,E., Nagelhus,T.A., Haug,T., Slupphaug,G., Akbari,M., Aas,P.A., Steinsbekk,K., Bakke,O. and Krokan,H.E. (1999) EMBO J., 18, 3834–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]