Abstract
Sox10 is an important transcriptional regulator in the neural crest and various neural-crest derived lineages, such as the Schwann cells of the peripheral nervous system. Recently, we identified the gene for myelin Protein zero (P0) as a transcriptional target of Sox10 in Schwann cells, allowing for the first time a detailed analysis of Sox10 responsive elements and their functional interaction with Sox10. Here we show that Sox10 functions through two different types of DNA response elements, one that allows binding of monomers, and a second that favors cooperative binding of two molecules. This dimeric binding required the presence of two heptameric Sox binding sites in a specific orientation and spacing, and was mediated by an N-terminal region of Sox10 with high conservation in the related Sox9, which also exhibited dimeric binding. This argues that the conserved region has the capacity to function as a DNA-dependent dimerization domain. The interaction between Sox10 dimers and DNA differed dramatically from that of Sox10 monomers, as it drastically reduced the protein’s off-rate and increased the protein-induced angle of DNA bending. These results indicate that functionally relevant interactions between Sox10 and DNA occur through completely different modes of binding.
INTRODUCTION
Sox proteins form a large family of transcriptional regulators characterized by possession of a high-mobility group (HMG) DNA-binding domain first identified in the mammalian sex-determining factor Sry (for reviews, see 1,2). The family is further divided into at least seven different groups on the basis of sequence relationships between the more than 20 different members identified recently. All Sox proteins are thought to bind to a 7 bp consensus DNA element (A/T)(A/T)CAA(A/T)G (3). Members of group D bind to DNA as dimers, whereas members of all other groups primarily bind to DNA as monomers (for a review, see 2). DNA-binding of Sox proteins is always accompanied by the introduction of a strong bend into the DNA (4–7). This has led to the hypothesis that Sox proteins function as architectural proteins by shaping the three-dimensional configuration of promoters and associated DNA-binding proteins in an enhanceosome-like structure (for a review, see 8). In addition to this architectural activity, Sox proteins possess the ability to transactivate. This transactivation capacity is widely variable between members and is often best observed on promoters that are activated cooperatively by Sox proteins and other transcription factors (for a review, see 2).
Several Sox proteins have proven to be important developmental regulators. One such protein is the group E protein Sox10 (9,10). Sox10 is prominently expressed in the early neural crest and in glial lineages of the peripheral and central nervous systems both during development and in the adult (9–11). Defects in Sox10 lead to multiple neural crest defects in mice (10,12) and men (13). Heterozygozity in humans always leads to Waardenburg–Hirschsprung disease which is characterized by a combination of colonic aganglionosis, hearing deficits and pigmentation abnormalities (13,14). Less frequently, patients exhibit additional signs of myelinopathies which are indicative of developmental defects in myelin-producing Schwann cells of the peripheral nervous system and oligodendrocytes of the central nervous system (15).
To understand the function of Sox10 on a molecular basis, we have started to characterize its target genes. Recently, we identified the myelin glycoprotein, Protein zero (P0), as a direct target for Sox10 both in vivo and in vitro (16). Sox10 effects were mediated by the promoter of the P0 gene. Within the promoter, a distal and a proximal region were involved. Both contributed equally to the overall stimulation and contained high-affinity binding sites for Sox10 with the functionally most important ones being site F in the distal region and sites B and C in the proximal part. The main difference between the distal and the proximal region was that the distal only functioned in the presence of the proximal, whereas the proximal region was able to function by itself.
The identification of bona fide response elements for Sox10 in one of its direct transcriptional targets offered the possibility of analyzing in detail the interaction of Sox10 with natural response elements. Here, we present the results of this study. They are unexpected in several respects as they point to a variability of binding to DNA and at the same time reveal hitherto undescribed differences of DNA binding between various Sox proteins with clear functional implications.
MATERIALS AND METHODS
Plasmids
Plasmids based on the pCMV5 backbone were used for eukaryotic expression of Sox proteins. pCMV-Sox2 and pCMV-Sox4 have been described (17,18). pCMV-Sox6 and pCMV-Sox21 contained the complete open reading frames of rat Sox6 and rat Sox21 as EcoRI fragments. A region corresponding to amino acids 1–190 of mouse Sox9 was inserted as an EcoRI/BamHI fragment into pCMV5. With the exception of pCMV5/Sox10ΔN (9), Sox10 constructs were derived from pCMV5/MIC, which expresses amino acids 1–189 of Sox10 (17). The following regions were deleted from MIC: amino acids 1–40 in pCMV5/M41, amino acids 1–60 in pCMV5/M61 and amino acids 61–102 in pCMV5/MICΔ. Bacterial pGEX expression plasmids for MIC and the isolated HMG domain (amino acids 101–180 of Sox10) were as described (16).
Luciferase reporter plasmids were derived from the previously published pTATAluc, which contains the β-globin minimal promoter, or from pTATA+Proxluc, in which the promoter consists of the β-globin minimal promoter and positions –229 to –116 of the rat P0 promoter containing the previously identified Sox10 binding sites B and C/C′ (16). Sites B, C and C′ were separately inactivated in the context of pTATA+Proxluc using site-directed mutagenesis. For determination of DNA-bending angles the region corresponding to positions –229 to –116 of the rat P0 promoter was inserted between XbaI and SalI sites of pBEND2 (19), using versions in which either site B or site C was inactivated.
Cell culture, transfections and luciferase assays
Tet-On N2A neuroblastoma capable of doxycycline-dependent induction of Sox10 expression (16) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum, 400 µg/ml G418 (Gibco BRL, Karlsruhe, Germany) and 150 µg/ml hygromycin (Roche Diagnostics, Mannheim, Germany). For luciferase assays, cells were transfected in quadruplicates on 35 mm plates with 2 µg of luciferase reporter plasmid per plate using Superfect reagent according to the manufacturer’s instructions (Qiagen, Hilden, Germany). After transfection, cells were placed back into DMEM containing 10% FCS. To induce expression of Sox10, doxycycline was added at a concentration of 2.5 µg/ml to half the plates. Cells were harvested 62 h post-transfection, and extracts were assayed for luciferase activity (20). Data are presented as reported previously (16).
COS cells were maintained in DMEM supplemented with 10% fetal calf serum. Forty-eight hours after transfection (10 µg DNA per 10 cm plate) with DEAE-dextran (500 µg/ml) and subsequent chloroquine treatment, cells were harvested and extracts were prepared as described (21).
Proteins, cell extracts and western blots
MIC and the HMG domain of rat Sox10 were produced in bacteria as glutathione-S-transferase fusion proteins and purified according to standard procedures (22). The glutathione-S-transferase moiety was removed by thrombin cleavage. Extracts from COS cells were prepared and analyzed in western blots using the ECL detection system as described (23). Polyclonal rabbit antisera directed against Sox10 (1:3000 dilution) served as primary antibodies, horseradish-peroxidase coupled protein A as secondary detection reagent (9).
Coupled in vitro transcription/translation and glutaraldehyde cross-linking
35S-labeled proteins were produced in reticulocyte lysates using the TNT system according to the manufacturer’s instructions (Promega, Mannheim, Germany). For cross-linking experiments, reticulocyte lysates were incubated in 0.01% glutaraldehyde for 10 min at room temperature. Following size separation of lysates on SDS–polyacrylamide gels, radiolabeled proteins were analyzed by autoradiography.
Electrophoretic mobility shift assay
In general, 0.5 ng of 32P-labeled probe (for oligonucleotides, see Figs 2, 5 and 6; for restriction fragments from pBEND2, see Fig. 8) were incubated with recombinant protein or COS cell extract for 20 min on ice in a 20 µl reaction mixture containing 10 mM HEPES (pH 8.0), 5% glycerol, 50 mM NaCl, 5 mM MgCl2, 2 mM DTT, 0.1 mM EDTA, 4 µg of bovine serum albumin and 2 µg of poly(dG·dC) as unspecific competitor. Samples were loaded onto native 4% polyacrylamide gels and electrophoresed in 0.5× TBE (45 mM Tris, 45 mM boric acid, 1 mM EDTA, pH 8.3) at 120 V for 1.5 h. Gels were dried and exposed for autoradiography. For off-rate experiments, the reaction was scaled up to 80 µl. After incubation of protein and probe under the indicated conditions, 500-fold excess of unlabeled homologous competitor was added. Aliquots (10 µl) were removed after varying times from the reaction and immediately loaded onto the gel.
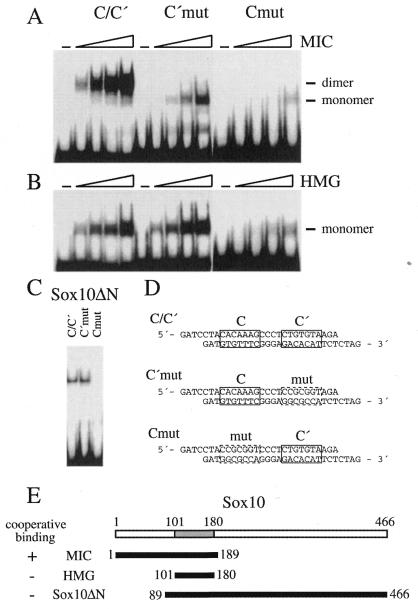
Figure 2.
The N-terminal region of Sox10 is required for cooperative binding to C/C′. Increasing amounts (2.5–50 ng) of purified MIC protein (A) and isolated Sox10 HMG domain (B), or 400 ng of extract from transiently transfected COS cells expressing Sox10ΔN (C) were analyzed in electrophoretic mobility shift assays for their ability to bind to radiolabeled oligonucleotide probes containing site C/C′ in wild-type (C/C′) or mutant versions (C′mut and Cmut). –, no protein added. The sequence of oligonucleotides is shown in (D) with the exact location of sites C and C′ indicated by boxes. The proteins used in (A–C) are depicted schematically in (E). Numbers mark the first and last amino acids present in the protein. The HMG domain (gray box) is localized between amino acids 101 and 180 of Sox10. The ability of each protein to bind cooperatively to site C/C′ is indicated by + or –.
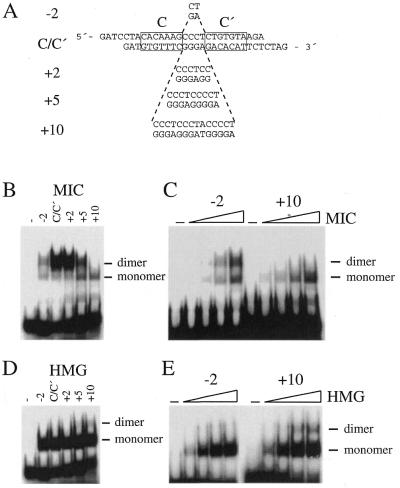
Figure 5.
Spacing requirements for cooperative binding of Sox10. (A) Sequence of oligonucleotide probes with the exact location of sites C and C′ indicated by boxes. Sequences between sites C and C′ were deleted or inserted relative to the original sequence present in the P0 promoter (C/C′) as indicated. (B and D) Purified MIC protein (50 ng) or isolated Sox10 HMG domain (50 ng) were analyzed in electrophoretic mobility shift assays for their ability to bind to the radiolabeled oligonucleotide probes shown in (A). (C and E) Increasing amounts (2.5–50 ng) of MIC protein and Sox10 HMG domain were incubated with the radiolabeled oligonucleotide probes in which the spacer region between sites C and C′ was shortened by 2 bp (–2) or increased by 10 bp (+10), respectively. Position of bound monomers and dimers are indicated.
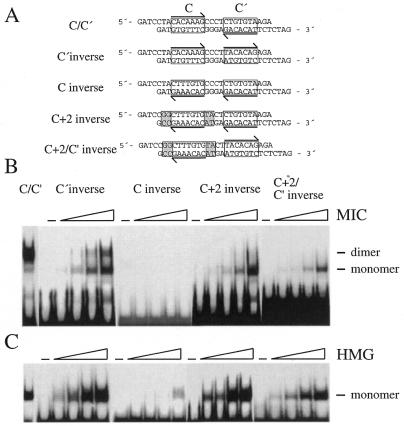
Figure 6.
Orientation requirements for cooperative binding of Sox10. (A) Sequence of oligonucleotide probes with the exact location of sites C and C′ indicated by boxes and their orientation indicated by arrows. Sites C and C′ were inverted separately (C′ inverse, C inverse, C+2 inverse) or together (C+2/C′ inverse), with or without flanking sequences as indicated, relative to the original sequence present in the P0 promoter (C/C′). Inverted flanking base pairs are on a gray background. (B and C) Increasing amounts (2.5–50 ng) of purified MIC protein and isolated Sox10 HMG domain were analyzed in electrophoretic mobility shift assays for their ability to bind to the radiolabeled oligonucleotide probes shown in (A). Twenty nanograms of protein were used for probe C/C′. Positions of bound monomers and dimers are indicated.
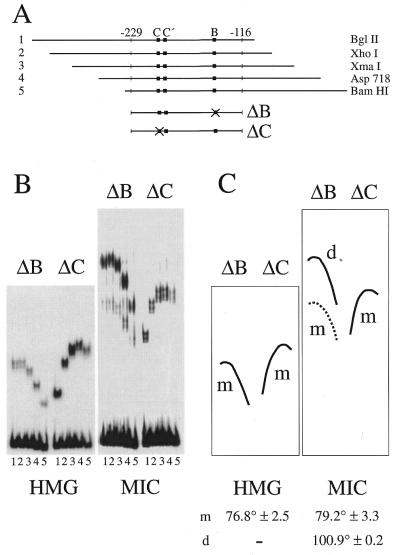
Figure 8.
Bending properties of Sox10. (A) Sequences from position –229 to –116 of the rat P0 promoter were inserted into the multiple cloning site of the pBEND2 vector and retrieved with flanking sequences using the indicated restriction enzymes (fragments 1–5). The ΔB series contained only the wild-type site C/C′; the ΔC series contained only the wild-type site B. (B) Electrophoretic mobility shift analyses of isolated Sox10 HMG domain or MIC protein with the ΔB and ΔC series of fragments 1–5 shown in (A). (C) Schematic representation of the positions of protein–DNA complexes obtained for each fragment after monomer (m) or dimer (d) binding. Bending angles (SEM) were determined in three separate experiments and are listed at the bottom.
RESULTS
Functional dissection of the proximal P0 promoter
We have previously shown that the proximal region of the P0 promoter contains several binding sites for Sox10 with two sites, designated site B and site C, exhibiting the highest affinity (16). Interestingly, sites B and C differ dramatically in their binding to Sox10. Site B fully conforms to the heptameric consensus for Sox binding sites and binds the protein in its monomeric form. Site C, on the other hand, does not conform to the consensus. It actually consists of two adjacent sites with single mismatches, called site C proper and site C′. To be able to differentiate between these two components, we will henceforth refer to this composite site as C/C′. It is bound by two Sox10 molecules in a cooperative manner (16) and both of its components are fully conserved between mouse, rat and human P0 promoters.
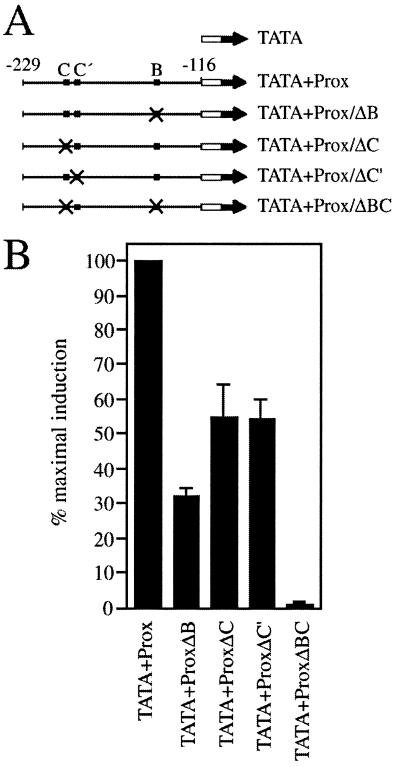
To confirm the importance of these binding sites we performed site-directed mutagenesis on the Sox10-responsive proximal region of the P0 promoter (positions –229 to –116 relative to the transcriptional start site; Fig. 1A). All introduced changes led to a complete loss of Sox10 binding to the mutated sites as judged by mobility shift experiments [for C and C′, see Fig. 2; for B, see Peirano et al. (16)]. Using Tet-On Neuro 2A cells that allowed the doxycycline-dependent induction of Sox10, this fragment from the P0 promoter had previously been shown in transient transfections to confer Sox10 responsiveness to the β-globin minimal promoter (16). Here we again observed, on average, an 8-fold induction of reporter gene expression for the intact proximal region of the P0 promoter (Fig. 1B). Mutation of site B reduced the effect by two-thirds, whereas mutation of site C halved activation rates. Both mutants, however, retained some level of Sox10 responsiveness, arguing that Sox10 worked independently through either site B or site C/C′. The Sox10 response was lost completely only upon simultaneous mutation of sites B and C (Fig. 1B). Interestingly, reporter gene induction was not only lowered by mutation of site C proper, but to a similar extent by mutation of the adjacent site C′ thus corroborating the functional relevance of dimer binding to site C/C′.
Figure 1.
The proximal fragment of the P0 promoter relies on both site B and site C/C′ to mediate Sox10-dependent transcriptional activation. (A) Schematic representation of the reporter genes used in (B). The luciferase reporter is depicted as a filled arrow, the minimal β-globin promoter as an open box. Sequences from position –229 to –116 of the rat P0 promoter were included in the construct and contained the Sox10 binding sites B and C/C′ in wild-type (filled box) or mutant (crossed box) version. (B) Tet-On N2A cells capable of expressing Sox10 [S10 according to Peirano et al. (16)] were transfected in quadruplicates with the luciferase reporters shown in (A). From each transfected quadruplicate, one duplicate was left untreated, while the second one was treated with doxycycline to induce Sox10 expression. Luciferase activities in extracts from transfected cells were determined in three independent experiments. Data were normalized as described (16) and are presented as percentages of the maximal induction of promoter activity by Sox10.
Dimeric binding requires the N-terminal region of Sox10
Previous electrophoretic mobility shift experiments on site C/C′ had used the naturally occuring Sox10 mutant MIC (16) which consists of amino acids 1–189 only (17). This protein bound with high avidity to site C/C′ (Fig. 2A). Mutation of site C′ reduced dimeric binding to monomeric binding with a concomitant reduction of the protein’s affinity to the site (C′mut in Fig. 2A). Similar alterations from dimer to monomer binding were also observed upon mutation of site C proper (Cmut in Fig. 2A). Site C′, however, is such a low-affinity site, that monomeric Sox10 binding could only be detected at the highest concentration used in the absence of site C.
Most other studies on Sox proteins in the past had resorted to the use of the isolated HMG domain in mobility shift assays. When we repeated our analysis on site C/C′ with the Sox10 HMG domain instead of MIC, we were surprised. Even with both site C and site C′ intact, there was only monomeric DNA binding (Fig. 2B). Mutation of site C′ did not alter the mobility shift pattern significantly, whereas mutation of site C led to a dramatic reduction of Sox10 HMG domain binding to DNA. In case of the isolated Sox10 HMG domain only one molecule thus bound to C/C′. By far the most molecules were bound by site C proper rather than site C′ in accord with the different affinities of these sites. Non-cooperative binding of two Sox10 HMG-domain molecules to site C/C′ was not even observed at the highest concentration used (Fig. 2B), arguing that for the Sox10 HMG domain, binding to site C decreased the likelihood of site C′ occupancy on the same molecule of DNA. The different behavior of MIC and Sox10 HMG domain also indicated that binding of Sox10 to site C/C′ is not only dependent on the HMG domain, but is influenced significantly by regions outside the HMG domain. Given the fact that the protein used in the original study (i.e. MIC) consisted of the HMG domain of Sox10 and all sequences N-terminal to it (16), we conclude that the region that modifies the DNA-binding properties of the HMG domain must be localized in the N-terminal part of Sox10.
To analyze whether regions C-terminal of the HMG domain of Sox10 additionally influenced its DNA binding properties, we also tested binding of a protein consisting of HMG domain and all C-terminal sequences to site C/C′ (Sox10ΔN in Fig. 2D). This protein showed binding characteristics very similar to the isolated Sox10 HMG domain and bound to site C/C′ only as a monomer (Fig. 2C).
Fine-mapping the region responsible for cooperative binding
The region N-terminal to the HMG domain of Sox10 consists of approximately 100 amino acids. To determine in greater detail which part of this region was involved in mediating the dimeric binding, we deleted increasing numbers of amino acids from the N-terminal end of Sox10 in the context of the MIC mutant (Fig. 3A). All deletion mutants were expressed at similar levels in transfected cells as evident from western blot analysis of nuclear extracts (Fig. 3B), thus allowing us to test the mutants in electrophoretic mobility shift analyses. In addition to site C/C′, a probe with mutant C′ site (C′mut) was used to determine the mobility expected for the complex between DNA and monomer.
Figure 3.
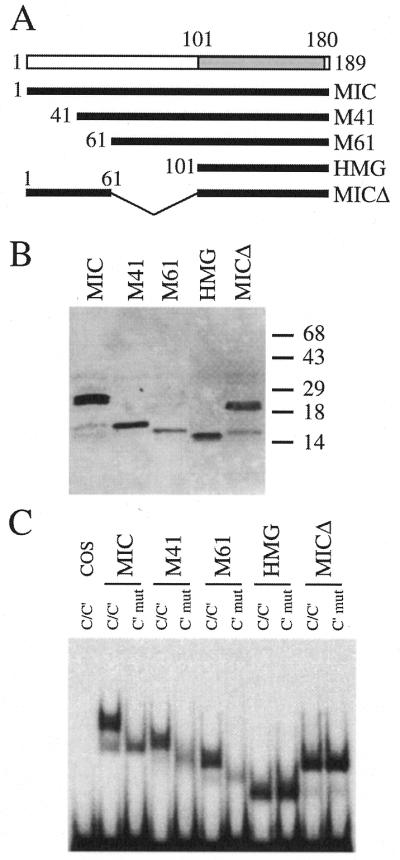
Fine-mapping of the region responsible for cooperative binding to C/C′ in the N-terminal part of Sox10. (A) Summary of Sox10-based N-terminal deletion constructs. The Sox10 regions present in each protein are indicated by the first and last amino acid. The HMG domain (amino acids 101–180) is in gray. (B) Expression of all proteins was verified by western blots of nuclear extracts from transfected COS cells with a polyclonal antiserum against Sox10. Numbers indicate size of molecular weight markers in kDa. (C) Electrophoretic mobility shift assays with the proteins shown in (A). The nuclear extracts from transfected COS cells shown in (B) served as protein source. Oligonucleotides C/C′ and C′mut (for sequence see Fig. 2) were used as probes as indicated above the lanes.
When tested for their ability to bind to site C/C′, mutants that lacked amino acids 1–40 or 1–60 behaved very similarly to wild-type MIC. They all bound as dimers (Fig. 3C). Mutation of C′ resulted in loss of dimer binding. A complex characteristic of a Sox10 monomer appeared instead. The only mutant which failed to show dimer binding similar to the isolated Sox10 HMG domain was a MIC mutant with an internal deletion of amino acids 61–100 (MICΔ in Fig. 3). We have to conclude from these results that amino acids 1–60 are dispensable for dimer binding, whereas amino acids 61–100 which immediately precede the HMG domain are essential for this property of Sox10.
The region responsible for cooperative binding is a DNA-dependent dimerization domain conserved between Sox10 and Sox9
Amino acids 61–100 of Sox10 correspond to a region found in all group E Sox proteins (i.e. Sox10, Sox9 and Sox8), but not in others (9). In agreement with the strong conservation of this region among group E proteins, Sox9 exhibited a binding pattern very similar to Sox10. It displayed cooperative, dimeric binding to wild-type site C/C′ and monomeric binding after mutation of C′ (Fig. 4A). In contrast, the majority of Sox proteins that belong to different groups and exhibit no sequence similarity to amino acids 61–100 of Sox10, failed to show cooperative binding, as exemplified by the group B proteins Sox2 and Sox21 or the group C proteins Sox4 and Sox11. They bound as monomers to site C/C′ (Fig. 4A and data not shown).
Figure 4.
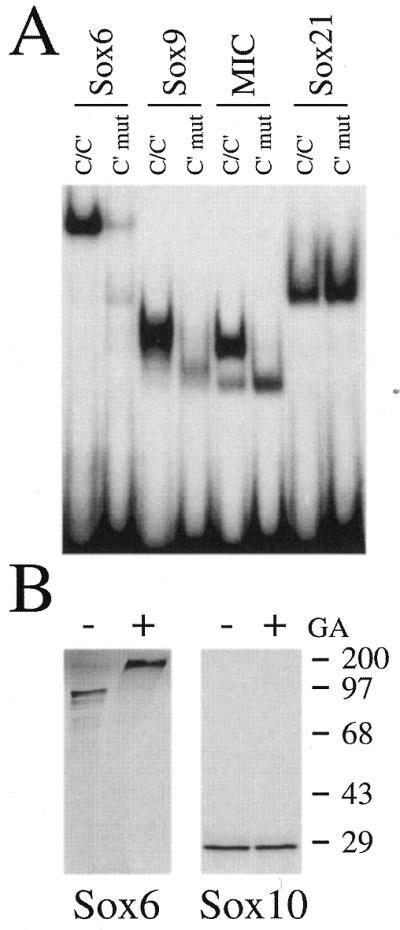
Cooperative binding and dimerization properties of Sox proteins. (A) Electrophoretic mobility shift assays with MIC (amino acids 1–189 of Sox10), Sox9 (amino acids 1–190), Sox21 and Sox6. Nuclear extracts from COS cells transfected with corresponding expression plasmids served as protein source. Oligonucleotides C/C′ and C′mut (for sequence see Fig. 2) were used as probes as indicated above the lanes. (B) Cross-linking studies with Sox10 and Sox6 proteins produced by coupled in vitro transcription/translation in the presence of [35S]methionine and incubated with (+) or without (–) glutaraldehyde (GA). Autoradiographs are shown after polypeptide separation by SDS–PAGE. Numbers indicate size of molecular weight markers in kDa.
The only exception were the group D proteins, represented by Sox6. In general, group D proteins strongly prefer dimeric binding (24). Accordingly, Sox6 also bound as a dimer to site C/C′ (Fig. 4A). After mutation of site C′, Sox6 binding was strongly reduced, but there was still as much dimer as monomer. DNA binding of group D Sox proteins is determined by a leucine zipper domain in their N-terminus which causes them to dimerize constitutively in solution with themselves or other members of group D (24–26). It is possible to visualize these dimers on SDS–polyacrylamide gels after prior treatment with the cross-linking agent glutaraldehyde as shown for Sox6 in Figure 4B. When the same procedure was employed on Sox10, dimer formation was not observed (Fig. 4B). Thus, the dimerization domain of Sox10 (and probably that of other group E proteins) differs from that of Sox6 (and other group D proteins) in that it only functions on appropriate DNA binding sites.
Sequence and spacing constraints for cooperative binding of Sox10
Site C/C′ within the P0 promoter consists of two non-consensus sites with particular spacing and orientation. The fact that cooperative binding involves non-consensus sites is not unusual and has been observed in a number of other exemplary cases of cooperative binding (27). Changing site C to the Sox consensus did not significantly alter binding pattern or affinity of either MIC or Sox 10 HMG domain, whereas changing the low-affinity site C′ to the Sox consensus resulted in a general increase of binding affinity (data not shown). Importantly, however, none of these changes altered the overall mode of DNA-binding for either MIC or the Sox10 HMG domain, showing that it is not an important determinant for cooperativity whether the sites are consensus or non-consensus binding sites.
In the P0 promoter, sites C and C′ are separated by 4 bp. The centers of both binding sites are one helix turn apart indicating that both Sox10 molecules are bound to the same side of the DNA helix. To analyze whether the exact spacing of both sites plays an important role in enabling cooperativity we changed the distance between both sites by either removing or introducing base pairs (Fig. 5A). Decreasing the spacing by 2 bp strongly reduced DNA binding of MIC. Despite this, dimer formation was still favored over monomer formation (Fig. 5B and C). Increasing the spacing by 2 bp, on the other hand, did not significantly change the binding pattern of MIC. Dimer formation was as prominent as for the wild-type C/C′ probe. When 5 bp were added to the spacer between site C and site C′, MIC dimer formation was diminished. However, as already observed for the deletion of 2 bp, the dimer was still the predominant species. This result is quite intriguing given the fact that insertion of 5 bp not only increases the distance between both sites by half a turn of the DNA helix, but in addition moves the binding sites to opposite sides of the DNA helix. Addition of 10 bp, on the other hand, moves both binding sites further apart but retains the original arrangement of both binding sites on the same side of the DNA helix. Nevertheless, insertion of 10 bp was incompatible with dimer formation of MIC (Fig. 5B and C). Thus, distance is more important for dimer formation than the spatial arrangement of both binding sites on the surface of the DNA helix.
When the isolated HMG domain of Sox10 was tested on the same set of binding sites, no significant difference in monomer formation was observed as a consequence of base pair deletion or insertion (Fig. 5D and E). There was a slightly increased frequency of two Sox10 HMG-domain molecules binding independently to the same probe following introduction of 10 bp. As mentioned previously, binding of the first Sox10 HMG-domain molecule to site C in the wild-type C/C′ probe seemed to interfere with binding of a second molecule to site C′. This interference was relieved by increased spacing between sites C and C′.
Orientation constraints for cooperative binding of Sox10
In the P0 promoter, sites C and C′ have different orientations with site C pointing towards the transcriptional start and site C′ pointing away. Thus, sites C and C′ are oriented towards each other (Fig. 6A). To determine whether the exact orientation of both sites is important, we inverted either site C or site C′ such that both sites were oriented identically, either pointing towards the transcriptional start site (C′ inverse in Fig. 6A) or away from it (C inverse in Fig. 6A). Inversion of site C′ results in a strong reduction of dimer binding (Fig. 6B). Electrophoretic mobility shift analysis with increasing amounts of MIC protein and a probe containing an inverted site C′ showed that the amount of dimer was less than the amount of monomer at any given concentration. When site C was inverted, binding was completely abolished. However, binding was not only abolished for MIC but also strongly reduced for the HMG domain of Sox10 (Fig. 6C), arguing that DNA-binding was generally compromised by the inversion of site C. To analyze whether DNA-binding depended on determinants other than the 7 bp of the core Sox binding sites, such as the exact flanking sequences, we not only inverted the 7 bp of the core Sox recognition element, but additionally included 2 bp on each side of site C. Inclusion of the flanking base pairs restored binding of the Sox10 HMG domain to levels observed for the wild-type C/C′ probe (Fig. 6C). Binding of MIC was also restored. However, instead of a dimer, we exclusively observed monomer formation. Thus, identical orientation of both sites is not compatible with dimer conformation.
Next we inverted both site C (and flanking sequences) as well as site C′ (C+2/C′ inverse in Fig. 6A). In this case, both C and C′ are differently oriented, but instead of pointing towards each other as is the case in the wild-type C/C′, both sites point away from each other. This change was also not compatible with dimer formation for MIC. Taken together, these results indicate that the exact orientation of both binding sites is crucial for obtaining cooperative binding of Sox10.
Consequences of cooperative binding of Sox10
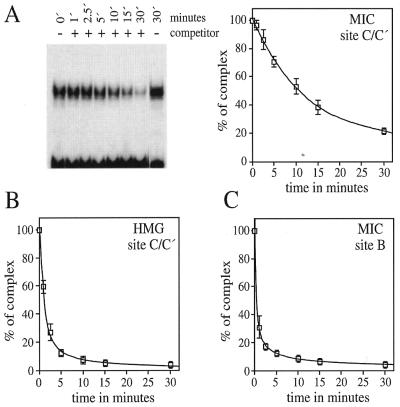
Next we analyzed whether there are any qualitative differences between Sox10 binding to DNA as a monomer or a dimer, for instance in the kinetics of complex formation. However, no difference in the on-rate was detected in electrophoretic mobility shift experiments, as complex formation on both sites was already maximal after 1 min incubation of either MIC protein or Sox10 HMG domain with excess probe under standard conditions (data not shown). Next we investigated the kinetics of decay of preformed Sox10–DNA complexes by challenging them with excess amounts of specific cold competitor DNA. When MIC was used as protein and C/C′ as probe, the protein–DNA complex was quite stable (Fig. 7A). After 10 min incubation, ∼50% of the original complex was still present (Fig. 7A). Even after a 30 min incubation with excess amounts of specific competitor, 20% of the original complex was detected. When the isolated HMG domain of Sox10 was used as protein instead, the off-rate was increased dramatically with 90% of the complex having already disappeared after a 5 min challenge period (Fig. 7B). The half-life of the protein–DNA complex was estimated to be <2 min. Even faster off-rates were observed with MIC as protein and site B as probe. Here the half-life of the protein–DNA complex was <1 min (Fig. 7C). Taken together, these results showed that Sox10 has a significantly lower off-rate on sites where it binds cooperatively as a dimer.
Figure 7.
Stability of Sox10 complexes with DNA. Preformed complexes between DNA and purified MIC protein (A and C) or isolated Sox10 HMG domain (B) were challenged with 500-fold amounts of cold competitor which was identical in sequence to the labeled probe. Aliquots were removed from the reaction after indicated times and analyzed in an electrophoretic mobility shift assay. Site C/C′ (A and B) or site B (C) served as probes. The amount of protein–DNA complex present at each time point was determined using a Phosphoimager and set in relation to the amount present in the absence of competitor, which was arbitrarily given a value of 100%. Representative experiments are shown and results from all three experiments are summarized in the respective graphs.
Sox proteins are also known to bend DNA upon binding to the minor groove (4–7). Until now, however, DNA bending properties have not been analyzed for Sox10. Thus, it was of interest to study DNA bending of Sox10 in circular permutation assays using the proximal region of the P0 promoter that contains both site B and site C/C′. For the sake of simplicity, permutation assays were performed with mutants of this region in which either site B or site C was removed. The proximal region of the P0 promoter was not intrinsically bent (Fig. 8B). When fragments were used in electrophoretic mobility shift assays in which the relative distance of the Sox binding site from the ends of the probe was varied (Fig. 8A), different mobilities of the protein–DNA complex were observed indicating that Sox10 indeed bent the DNA upon binding (Fig. 8B). The isolated HMG domain of Sox10 bound to both site B and site C/C′ as a monomer (Fig. 8B and C). Determination of the exact bending angle yielded identical angles of ∼77° for both sites (Fig. 8C). When MIC was tested on site B, to which it binds as a monomer (ΔC in Fig. 8), very similar results were obtained with average bending angles of 79° (Fig. 8C). Thus, DNA bending of a Sox protein monomer to DNA is determined by the properties of the HMG domain itself and independent of other Sox10 regions.
Next we analyzed DNA bending of MIC on site C/C′ (ΔB in Fig. 8). On this site we observed monomer as well as dimer binding with the dimer being the predominant species. Analysis of the mobility for the complexes between MIC monomers and the C/C′-containing fragments again yielded the characteristic bending angle of 79°. Bending properties were, however, different for the predominant dimer species. Here we calculated a bending angle of 101° (Fig. 8C), clearly indicating that bound monomers and dimers have distinct effects on DNA topology.
DISCUSSION
More than 20 different Sox proteins have been identified to date in mammals. They are expressed in distinct, but partially overlapping spatiotemporal patterns during development and in the adult (for reviews, see 1,2), arguing that many cells at some time express more than one Sox protein. Examples for this are the co-expression of Sox5, Sox6 and Sox9 in cells of the chondrocyte lineage (24), the parallel detection of Sox4, Sox10 and Sox11 in oligodendrocyte precursors (18), and the simultaneous presence of Sox1, Sox2, Sox3 as well as Sox14 and Sox21 in populations of developing neurons (28–30). Because of the similarity of their DNA-binding domains, all Sox proteins are believed to perform their function on DNA by binding to the heptameric consensus sequence (A/T)(A/T)CAA(A/T)G (3).
Therefore, the question arises of how specificity of target gene regulation by different Sox proteins is achieved. In some cases, there is good evidence that such specificity is not even wanted and that co-expressed Sox proteins target the exact same binding sites in the same promoters. Under these circumstances, Sox proteins function redundantly or counterbalance each other’s activity. Sox1, Sox2 and Sox3, for instance, are assumed to influence expression of the same genes that are also influenced by Sox14 and Sox21 (30,31). Whereas Sox1, Sox2 and Sox3 interchangeably work as activators, Sox14 and Sox21 work as repressors.
However, in other cases there is good indication of target gene selectivity among Sox proteins. Our analysis of the P0 promoter, for instance, revealed a strong stimulation by Sox10, but only a marginal activation by Sox11 (16). Several reasons could be envisaged for this diverging behavior. It is conceivable that other transcription factors bound to the P0 promoter determine, through protein–protein interactions, which Sox protein is compatible with them and consequently with promoter function. Evidence for such a mechanism comes from transfection analyses of synergistic actions involving Sox proteins and other transcription factors (4,18). Alternatively, target specificity of Sox proteins can also be encoded in the DNA sequence itself, if there is a greater diversity of binding site preferences among Sox proteins than appreciated so far. Then, binding sites could already restrict the number of Sox proteins eligible as potential regulators of certain targets.
Several lines of evidence encountered during our analysis of the P0 promoter support the assumption that binding site preferences of Sox proteins are more complex than previously thought. It became clear, for instance, that the heptameric Sox consensus is not sufficient to satisfactorily explain site recognition and transactivation capacities of Sox proteins. For one, there is no good correlation between conformity to the consensus heptamer and affinity for Sox10 among the Sox binding sites present in the P0 promoter (16). Both site C and site C′, for instance, diverge from the consensus at a single position. Nevertheless, only site C bound Sox10 efficiently in isolation. We also observed different affinities of various Sox proteins for the same site, with Sox10 exhibiting strong, and Sox11 displaying weak binding to site C (data not shown).
Our attempt to invert the Sox binding heptamers in their natural context pointed to the fact that the exact flanking sequences are important determinants in Sox protein binding. Binding was completely obliterated by inversion of the heptamer and was only restored upon inclusion of two flanking base pairs on both sides of the heptamer during the inversion. In this regard, it is interesting to note that both high-affinity binding sites from the P0 promoter share identical flanking sequences with TA on the 5′ side and CC on the 3′ side. A role for flanking sequences had also been noticed for DNA binding of Sox9 in random oligonucleotide selection assays (32).
The most striking feature, however, was the fact that two molecules of Sox10 exhibited cooperative binding on the functionally relevant site C/C′ of the P0 promoter. Site C is the high affinity site the occupancy of which facilitates binding of a second Sox10 molecule to the adjacent low-affinity site C′. Similar configurations exist in other cases where cooperative binding has been observed (27,33). The importance of binding cooperativity was clearly confirmed in transient transfection assays as Sox10-responsiveness of the P0 promoter was similarly decreased after mutation of sites C and C′, respectively.
Cooperativity was not observed for a series of other Sox proteins including the group B Sox proteins, Sox2 and Sox21, as well as the group C Sox proteins, Sox4 and Sox11, indicating that cooperative binding—at least on the functionally relevant site C/C′—is a distinctive feature of Sox10. The necessity for cooperative binding to site C/C′ and the failure of Sox11 to do so explains to some extent the extremely weak activation of the P0 promoter by Sox11 (16).
The only other Sox proteins for which cooperative binding was detected, were Sox9 and Sox6. The similar behavior of Sox9 and Sox10 indicates that cooperative binding is not so much a property unique to Sox10 alone but rather a property unique to all group E Sox proteins including Sox9 and Sox8 (2). Cooperative binding of Sox6 was also not unexpected as it is known from the literature that Sox6 and related class D proteins dimerize constitutively in solution and therefore bind as dimers to closely spaced heptamer motifs (24,26). For the same reason, monomeric DNA binding is strongly disfavored for group D proteins. The ability to switch between monomeric and dimeric binding is thus a peculiarity of Sox10 and possibly of other class E proteins. It is essential for this property that group E proteins, in contrast to class D proteins, do not effectively dimerize in solution as shown here for Sox10 in cross-linking experiments and confirmed in a separate study on Sox9 (24). This ability of group E Sox proteins makes them one of the few Sox proteins that can regulate promoters such as the P0 promoter in which occupancy of both monomeric and dimeric sites is required for full activity.
Cooperative binding of two Sox10 molecules is subject to clear constraints on both the protein and the binding site. In case of the DNA, there are obvious spacing requirements. Thus, 4–6 bp between both heptamers of the dimer site were optimal and the degree of cooperativity decreased with any additional shortening or lengthening of the spacer. Conspicuously, no cooperativity was observed when an additional 10 bp were introduced between the two sites. This showed that physical proximity is more important for obtaining cooperativity than the exact localization of both sites on the face of the DNA.
Even stricter than the spacing requirements are those concerning the orientation of both sites relative to each other. In the composite C/C′ site both heptamers are arranged such that they face to each other. Any other orientation we tested failed to yield the same cooperative binding. It is easily imaginable that this orientation is important for the function of the site as the orientation of the heptamers is a major determinant in the sort and degree of bending that is induced into the DNA.
On the side of the Sox10 protein, cooperativity was absolutely dependent on sequences outside and N-terminal of the HMG domain. Analysis of various Sox10 deletion mutants showed that the responsible region is continuous with the HMG domain and consists of the stretch of 40 amino acids previously observed to be conserved between group E Sox proteins (9). Given this degree of conservation it might be expected (and was indeed confirmed in our study) that cooperative binding of DNA dimers is a general feature of group E Sox proteins. Our results also give some indication of the function of this conserved group E-specific region. As previously speculated (2), it is a protein–protein interaction domain. Given the fact that dimerization between two molecules of Sox10 did not occur in the absence of DNA, it is likely that this interaction domain must first be configured on DNA.
We know from our analysis of the P0 promoter that binding of Sox10 to both monomeric and dimeric sites is essential for full activation. Therefore, it was important to investigate whether monomeric and dimeric sites functioned in identical ways or differently. On-rate studies did not reveal any significant difference between dimeric and monomeric binding. Not so for off-rate studies. Cooperatively bound dimers remained bound to C/C′ significantly longer in the presence of cold competitor than Sox10 monomers. In combination, similar on-rates and substantially decreased off-rates result in an increased DNA affinity of the dimer relative to the monomer.
In addition to changes of affinity there were also changes in DNA bending. A Sox10 monomer induces a bend of ∼75–80° in good accord with bending angles previously published for other Sox proteins including Sox9 (4–7,34). When we measured the apparent bending angle for the cooperative Sox10 dimer, however, we obtained a significantly different angle close to 101° and therefore much more typical of TCF/LEF proteins (35). Thus, cooperative binding leads to cooperative bending (36). The different bending angle for monomers and dimers allows variation of Sox10-induced bending according to the needs and restraints of a particular promoter. We propose that monomer and dimer sites have unique roles in the formation of the multiprotein P0 promoter complex that permits glia-specific expression in Schwann cells.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by a grant from the Deutsche Forschungsgemeinschaft to M.W. (We1326/7-1).
REFERENCES
- 1.Pevny L.H. and Lovell-Badge,R. (1997) Curr. Opin. Genet. Dev., 7, 338–344. [DOI] [PubMed] [Google Scholar]
- 2.Wegner M. (1999) Nucleic Acids Res., 27, 1409–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harley V.R., Lovell-Badge,R. and Goodfellow,P.N. (1994) Nucleic Acids Res., 22, 1500–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamachi Y., Cheah,K.S. and Kondoh,H. (1999) Mol. Cell. Biol., 19, 107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connor F., Cary,P.D., Read,C.M., Preston,N.S., Driscoll,P.C., Denny,P., Crane-Robinson,C. and Ashworth,A. (1994) Nucleic Acids Res., 22, 3339–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrari S., Harley,V., Pontiggia,A., Goodfellow,P.N., Lovell-Badge,R. and Bianchi,M.E. (1992) EMBO J., 11, 4497–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDowall S., Argentaro,A., Ranganathan,S., Weller,P., Mertin,S., Mansour,S., Tolmie,J. and Harley,V. (1999) J. Biol. Chem., 274, 24023–24030. [DOI] [PubMed] [Google Scholar]
- 8.Prior H.M. and Walter,M.A. (1996) Mol. Med., 2, 405–412. [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhlbrodt K., Herbarth,B., Sock,E., Hermans-Borgmeyer,I. and Wegner,M. (1998) J. Neurosci., 18, 237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Southard-Smith E.M., Kos,L. and Pavan,W.J. (1998) Nature Genet., 18, 60–64. [DOI] [PubMed] [Google Scholar]
- 11.Pusch C., Hustert,E., Pfeifer,D., Südbeck,P., Kist,R., Roe,B., Wang,Z., Balling,R., Blin,N. and Scherer,G. (1998) Hum. Genet., 103, 115–123. [DOI] [PubMed] [Google Scholar]
- 12.Herbarth B., Pingault,V., Bondurand,N., Kuhlbrodt,K., Hermans-Borgmeyer,I., Puliti,A., Lemort,N., Goossens,M. and Wegner,M. (1998) Proc. Natl Acad. Sci. USA, 95, 5161–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pingault V., Bondurand,N., Kuhlbrodt,K., Goerich,D.E., Prehu,M.-O., Puliti,A., Herbarth,B., Hermans-Borgmeyer,I., Legius,E., Matthijs,G., Amiel,J., Lyonnet,S., Ceccherini,I., Romeo,G., Smith,J.C., Read,A.P., Wegner,M. and Goossens,M. (1998) Nature Genet., 18, 171–173. [DOI] [PubMed] [Google Scholar]
- 14.Southard-Smith E.M., Angrist,M., Ellison,J.S., Agarwala,R., Baxevanis,A.D., Chakravarti,A. and Pavan,W.J. (1999) Genome Res., 9, 215–225. [PubMed] [Google Scholar]
- 15.Inoue K., Tanabe,Y. and Lupski,J.R. (1999) Ann. Neurol., 46, 313–318. [DOI] [PubMed] [Google Scholar]
- 16.Peirano R.I., Goerich,D.E., Riethmacher,D. and Wegner,M. (2000) Mol. Cell. Biol., 20, 3198–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhlbrodt K., Schmidt,C., Sock,E., Pingault,V., Bondurand,N., Goossens,M. and Wegner,M. (1998) J. Biol. Chem., 273, 23033–23038. [DOI] [PubMed] [Google Scholar]
- 18.Kuhlbrodt K., Herbarth,B., Sock,E., Enderich,J., Hermans-Borgmeyer,I. and Wegner,M. (1998) J. Biol. Chem., 273, 16050–16057. [DOI] [PubMed] [Google Scholar]
- 19.Kim J., Zwieb,C., Wu,C. and Adhya,S. (1989) Gene, 85, 15–23. [DOI] [PubMed] [Google Scholar]
- 20.Wegner M., Drolet,D.W. and Rosenfeld,M.G. (1993) Proc. Natl Acad. Sci. USA, 90, 4743–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sock E., Renner,K., Feist,D., Leger,H. and Wegner,M. (1996) J. Virol., 70, 1512–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Renner K., Leger,H. and Wegner,M. (1994) Proc. Natl Acad. Sci. USA, 91, 6433–6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sock E., Enderich,J., Rosenfeld,M.G. and Wegner,M. (1996) J. Biol. Chem., 271, 17512–17518. [DOI] [PubMed] [Google Scholar]
- 24.Lefebvre V., Li,P. and de Crombrugghe,B. (1998) EMBO J., 17, 5718–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamashita A., Suzuki,S., Fujitani,K., Kojima,M., Kanda,H., Ito,M., Takamatsu,N., Yamashita,S. and Shiba,T. (1998) Gene, 209, 193–200. [DOI] [PubMed] [Google Scholar]
- 26.Takamatsu N., Kanda,H., Tsuchiya,I., Yamada,S., Ito,M., Kabeno,S., Shiba,T. and Yamashita,S. (1995) Mol. Cell. Biol., 15, 3759–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kemler I., Schreiber,E., Muller,M.M., Matthias,P. and Schaffner,W. (1989) EMBO J., 8, 2001–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collignon J., Sockanathan,S., Hacker,A., Cohentannoudji,M., Norris,D., Rastan,S., Stevanovic,M., Goodfellow,P.N. and Lovell-Badge,R. (1996) Development, 122, 509–520. [DOI] [PubMed] [Google Scholar]
- 29.Pevny L.H., Sockanathan,S., Placzek,M. and Lovell-Badge,R. (1998) Development, 125, 1967–1978. [DOI] [PubMed] [Google Scholar]
- 30.Uchikawa M., Kamachi,Y. and Kondoh,H. (1999) Mech. Dev., 84, 103–120. [DOI] [PubMed] [Google Scholar]
- 31.Kamachi Y., Uchikawa,M., Collignon,J., Lovell-Badge,R. and Kondoh,H. (1998) Development, 125, 2521–2532. [DOI] [PubMed] [Google Scholar]
- 32.Mertin S., McDowall,S.G. and Harley,V.R. (1999) Nucleic Acids Res., 27, 1359–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Topol J., Ruden,D.M. and Parker,C.S. (1985) Cell, 42, 527–537. [DOI] [PubMed] [Google Scholar]
- 34.Ma Y., Niemitz,E.L., Nambu,P.A., Shan,X., Sackerson,C., Fujioka,M., Goto,T. and Nambu,J.R. (1998) Mech. Dev., 73, 169–182. [DOI] [PubMed] [Google Scholar]
- 35.Love J.J., Li,X., Case,D.A., Giese,K., Grosschedl,R. and Wright,P.E. (1995) Nature, 376, 791–795. [DOI] [PubMed] [Google Scholar]
- 36.Diebold R.J., Rajaram,N., Leonard,D.A. and Kerppola,T.K. (1998) Proc. Natl Acad. Sci. USA, 95, 7915–7920. [DOI] [PMC free article] [PubMed] [Google Scholar]