Abstract
Biofilms have great potential for producing valuable products, and recent research has been performed on biofilms for the production of compounds with biotechnological and industrial relevance. However, the production of recombinant proteins using this system is still limited. The recombinant protein production in microbial hosts is a well-established technology and a variety of expression systems are available. Nevertheless, the production of some recombinant proteins can result in proteolyzed, insoluble, and non-functional forms, therefore it is necessary to start the exploration of non-conventional production systems that, in the future, could be helpful to produce some “difficult” proteins. Non-conventional production systems can be based on the use of alternative hosts and/or on non-conventional ways to grow recombinant cells. In this paper, the use of the Antarctic marine bacterium Pseudoalteromonas haloplanktis TAC125 grown in biofilm conditions was explored to produce two fluorescent proteins, GFP and mScarlet. The best conditions for the production were identified by working on media composition, and induction conditions, and by building a new expression vector suitable for the biofilm conditions. Results reported demonstrated that the optimized system for the recombinant protein production in biofilm, although it takes longer than planktonic production, has the same potentiality as the classical planktonic approach with additional advantages since it needs a lower concentration of the carbon sources and doesn't require antibiotic addition. Moreover, in the case of mScarlet, the production in biofilm outperforms the planktonic system in terms of a better quality of the recombinant product.
Keywords: Biofilm, Pseudoalteromonas haloplanktis TAC125, Recombinant protein production, Cold-adapted bacteria, m-Scarlet, GFP
1. Introduction
During the past few decades, considerable progress has been made in microbial platform engineering to improve the productivity and yields of recombinant proteins [[1], [2], [3]]. All advancements in this area have increased and facilitated the recombinant protein production in several organisms [4,5]. Although a variety of expression systems, vectors, and host strains are available (E.coli, Bacillus subtilis, Saccharomyces cereviae, Pichia pastoris etc …), the production of some recombinant proteins can still result in heavily proteolyzed, insoluble, and non-functional forms [1]. These phenomena can be attributed to several factors and although various strategies have been applied to overcome these limitations, there is no guarantee that every type of recombinant protein will have a high production yield or catalytic/functional activity. Therefore, it is necessary to explore and consolidate the use of non-conventional production systems that could be useful in producing some “difficult” proteins. Non-conventional production systems can be based on the use of alternative hosts and/or on non-conventional ways to grow the cells for recombinant protein production. In this paper, we explored the use of a non-conventional host, the Antarctic marine bacterium Pseudoalteromonas haloplanktis TAC125 (PhTAC125) [6], grown in biofilm conditions to produce recombinant proteins.
PhTAC125 genome was fully sequenced and annotated [7] and it is currently exploited as a new alternative expression host, its physiological features including fast growth in a wide range of temperatures and efficient protein synthesis make PhTAC125 an attractive and versatile host to produce recombinant proteins [8,9]. Several expression vectors have been developed to allow the production of several recombinant proteins either by constitutive or inducible promoters [[9], [10], [11], [12]]. The advantages in using these cold-adaptive systems in alternative to the conventional hosts were proven in the production of some “difficult proteins” [[13], [14], [15], [16]].
Most of the research on the production of recombinant proteins has been performed on cells grown in planktonic cultures. However, biofilm is the most successful and widely distributed form of life on earth [17]. It is not simply structured assemblages of cells but a dynamic complex system that evolves through a tightly controlled multistep process [18]. The first step is characterized by a loose/transient association [19], followed by robust adhesion [20] and colonization, where microorganisms are attached to the surface or interface via stronger interactions, and maturation [21]. The last stage is characterized by a return to transient motility where biofilm cells are detached, due to either intrinsic or extrinsic factors, and disseminated cells colonize other sites [22]. Biofilm provides bacteria characteristics deeply different from their planktonic counterparts [[23], [24], [25]], mainly derived by the genetic and metabolic rewiring of the biofilm-dwelling bacteria [26]. The local environmental conditions arising within the biofilm matrix, the intercellular signaling, and other phenomena, may induce cells in biofilm to modulate the expression of genes differently than in planktonic populations [25]. Moreover, biofilms are known to provide a fitness advantage under stress and harsh conditions [27], and this property could be useful to alleviate the host metabolic burden associated with heterologous protein overproduction. Furthermore, the sessile growth may allow for reaching high biomass concentration, indeed, it has been reported that biofilm reactors can retain 5-10 times more biomass per unit volume with respect to canonical one [28], and the high plasmid maintenance in biofilm [29,30] could guarantee good operation stability. All these properties could be very useful in industrial processes aimed at recombinant protein production. Although several potential drawbacks exist regarding the use of biofilm in recombinant protein production, mainly related to the control of biofilm development, this non-conventional process has several interesting potentials that justify additional studies, so much so that different studies explored this production approach in several microorganisms [31,32]. The first system for high level heterologous protein production biofilm cells was aimed at the production of eGFP in E. coli [33]. These authors demonstrated that the biofilm environment enhanced plasmid maintenance and the studies of Mergulhao and coworkers [28,32] showed that the GFP production levels were higher levels than their planktonic counterparts. Moreover, recently E. coli biofilm was successfully used to produce the human epidermal growth factor (hEGF) in a continuous process [31].
In this paper, we explored the feasibility of this alternative methodology in Pseudoalteromonas haloplanktis TAC125 (PhTAC125). This Antarctic marine bacterium is a promising unconventional host for the production of high-value proteins, thanks to its physiological features: fast growth in a wide range of temperatures and efficient protein synthesis [6,34]. Furthermore, for this bacterium an efficient gene expression technology was established [9,[35], [36], [37]] and either constitutive or inducible systems [11,38,39] were developed. The implementation of this psychrophilic system [40], the availability of engineered strains [11,41] and the formulation of suitable synthetic media [38] allowed the production of difficult-to-express proteins in an active and soluble form [42,43]. Additionally, PhTAC125 forms biofilm either when grown in rich or synthetic medium, and the quantity of biofilm produced in the synthetic medium GG was higher [44]. The analysis of PhTAC125 biofilm structure revealed the presence of different sugars which are characteristic of LPS but also, N- Acetyl-muramic acid (MurNAc, NAM), ribose, glucose, and mainly cellulose [45], this polymer is a common component of biofilms and mediates cell-cell interactions and cell adherence on surfaces [46]. Starting from the knowledge of PhTAC125 biofilm features and from the availability of different tools for recombinant protein production in this alternative host, we explored the production of recombinant proteins in biofilm. After the assessment of this option by the production of two different fluorescent proteins (GFP and mScarlet), the optimization of the production process was faced by working on media composition and by building a new expression vector suitable for the biofilm conditions.
2. Materials and methods
2.1. Bacterial strains, media, and plasmids
The strains, plasmids, and oligonucleotides used in this study are listed in Table S1. E. coli TOP10 was used for cloning purposes, while E. coli S17-1 (λpir) was employed in intergeneric conjugations as a donor strain for PhTAC125 transformations. E. coli strains were grown in Lysogen broth (LB, 10 g/L bacto-tryptone, 5 g/L yeast extract, 10 g/L NaCl) at 37 °C with 220 rpm. When required, 34 μg/mL chloramphenicol (Merck, Darmstadt, Germany) was supplemented to the medium. PhTAC125, KrPL, and KrPL LacY + strains were tested to assess the kinetic of biofilm formation. The KrPL LacY + strain was used for the expression of the GFP and mScarlet proteins. On the other hand, the resulting constitutive expression vectors, pAT-2620-mScarlet, pAT-2621-mScarlet, and pAT-2690-mScarlet, were conjugated in KrPL. The psychrophilic strains were grown in the TYP medium (16 g/L bacto-tryptone, 16 g/L yeast extract, 10 g/L NaCl) during interspecific conjugations and precultures development. The first trials of recombinant protein production in biofilm and planktonic conditions were carried out at 15 °C in the synthetic medium GG (10 g/L l-glutamic acid monosodium salt monohydrate, 10 g/L d-gluconic acid sodium salt, 10 g/L NaCl, 1 g/L NH4NO3, 1 g/L K2HPO4, 200 mg/L MgSO4·7H2O, 5 mg/L FeSO4·7H2O, 5 mg/L CaCl2) [47], and 25 μg/mL chloramphenicol. Afterwards, the recombinant production was enhanced by growing the psychrophilic strains in the GG medium with half the carbon sources (named 5/5 GG) and supplemented with 70 mg/L FeSO4.
2.2. Construction of the expression plasmids
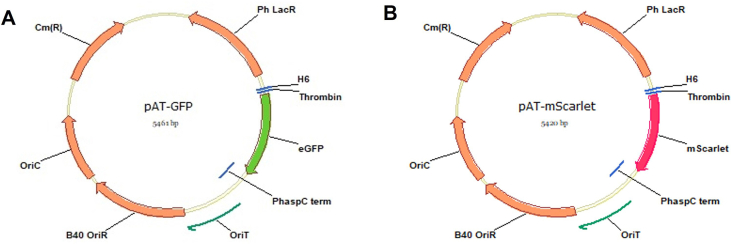
The DNA fragment containing the Shine-Dalgarno and spacing sequences of the PhTAC125 trpA gene was synthesized by Thermo Fisher Scientific and cloned into pB40-79BsC [41], a high-copy number derivative of p79C [11], using BsaI and KpnI restriction sites. The resulting vector was named pAT. The genes encoding the fluorescent proteins, GFP and mScarlet, were introduced in pAT using NdeI and SacI restriction enzymes, obtaining pAT-gfp and pAT-mScarlet (Fig. S1). Such vectors were mobilized in KrPL LacY + by conjugation [48].
The constitutive plasmids pAT_2620-mScarlet, pAT_2621-mScarlet, and pAT_2690-mScarlet were constructed by replacing the regulatory elements PhTAE79 lacR-lacZ of pAT-mScarlet with the putative promoter sequences of three genes in PhTAC125 genome (PSHAa2620, PSHAa2621, and PSHAa2690). The primers (PSHAa2620_SphI Fw, PSHAa2620_NcoI Rv, PSHAa2621_SphI Fw, PSHAa2621_NcoI Rv, PSHAa2690_SphI Fw, and PSHAa2690_NcoI Rv) listed in Table S1 were used to amplify a specific genomic sequence upstream of the selected genes (682 bp for PSHAa2620 and 410 bp for both PSHAa2621 and PSHAa2690). The PCR reaction was performed in a volume of 50 μL containing 4 ng of PhTAC125 genomic DNA as template, 0.5 μM primers, 1 × HF Phusion buffer (New England Biolabs, Hitchin, UK), 200 μM of each dNTP, and 0.02 U/μL Phusion DNA Polymerase (New England Biolabs, Hitchin, UK). The PCR amplified fragments were double digested with SphI/NcoI restriction enzymes and cloned into pAT-mScarlet previously digested with the same restriction sites. The new vectors (pAT_2620-mScarlet, pAT_2621-mScarlet, and pAT_2690-mScarlet) were mobilized into KrPL by conjugation.
2.3. Growth of planktonic and biofilm cultures
To evaluate the biofilm formation, the wild-type (PhTAC125, KrPL, KrPL LacY+) and the recombinant strains (KrPL LacY + pAT-gfp, KrPL LacY + pAT-mScarlet) were grown in the GG or 5/5 GG media in static conditions. The chloramphenicol was used as a selective agent at a final concentration of 25 μg/mL when specified.
The KrPL pAT_2620-mScarlet, KrPL pAT_2621-mScarlet, and KrPL pAT_2690-mScarlet strains were grown in 5/5 GG without antibiotics and in presence of different iron sulfate concentrations (0.5 mg/L FeSO4, 5 mg/L FeSO4 the concentration used in the GG medium, 70 mg/L FeSO4) in static conditions for different times (24 h, 48 h, 72 h, 96 h).
For recombinant production in biofilm, the recombinant strains were grown by a fluidized-bed reactor using floating polystyrene supports with a total surface area of 12 cm2 [49] in the GG medium supplemented with chloramphenicol (25 μg/mL) starting from 0.2 OD600nm. The recombinant expression was induced at the beginning of growth (0.2 OD600nm) with 5 mM IPTG and incubated in a static condition at 15 °C. Afterwards, the growth conditions in the biofilm were optimized, reducing the carbon sources (5/5 GG), increasing the iron sulfate concentrations (70 mg/L FeSO4), and avoiding the use of chloramphenicol. The cells were harvested after 96 h and sonicated twice at a constant ultrasound frequency of 37 kHz for 15 min to allow biofilm detaching and biomass recovery from the polystyrene supports. After sonication, the bacterial cultures were centrifuged at 6′000 rpm for 30 min at 4 °C. Then the supernatant was discarded, and cell pellets were stored at −20 °C for the following analysis.
For planktonic cultures, the recombinant strain KrPL pAT_2620-mScarlet was grown in the 5/5 GG medium shaking at 15 °C and the culture was harvested after 72 h. The recombinant strain KrPL LacY + pAT-mScarlet was grown in the GG medium supplemented with 70 mg/L FeSO4 and chloramphenicol (25 μg/mL) and was induced during the exponential growth phase (1.0–1.5 OD600nm) with 5 mM IPTG, the culture was harvested after 72 h from the induction.
2.4. Cell lysis
To evaluate the mScarlet production in KrPL pAT_2620-mScarlet, the cell pellets (about 220 mg) were recovered from planktonic and biofilm growths after 72 h and 96 h, respectively, and resuspended in 30 mL lysis buffer (50 mM sodium phosphate pH 7.75, 500 mM NaCl, one tablet of EDTA-free Complete Ultra protease inhibitor (Roche, Mannheim, Germany). The planktonic cells were mechanically lysed by a French Press at 2 Kbar for two consecutive cycles; instead, five steps at 2.5 Kbar were used to disrupt the biofilm cells. The obtained lysates were centrifuged (6′500 rpm (4732 g) for 1 h at 4 °C) to separate the soluble and insoluble protein fractions, and then the insoluble fraction was resuspended in 30 mL PBS.
2.5. Fluorescence assay
The fluorescence intensity of GFP or mScarlet was measured using a JASCO FP-750 spectrofluorometer (Jasco Corp., Japan) at 25 °C with a 1 cm path length. Cell pellets (about 1,5 mg) were recovered at the end of the growths in planktonic or static growth conditions by centrifugation (13′000 rpm for 10 min at 4 °C), resuspended in 0.5 mL PBS, and serially diluted to achieve the best signal-to-noise ratio in fluorescence measurements. The excitation wavelength was set at 488 nm, and the intensity of emitted fluorescence of GFP at 507 nm was recorded [50]. Regarding mScarlet, the excitation and emission wavelengths were set at 569 and 690 nm, respectively [51]. Fluorescence intensities are reported in arbitrary units (AU) per biomass unit. The data were processed using the Origin 81 software (OriginLab Corporation, Northampton, MA, USA). The fluorescence of the soluble and insoluble fractions of the mScarlet protein was measured in the same conditions described previously.
2.6. SDS-PAGE and western blot analysis
To analyze the protein profile by SDS-PAGE, about 0,75 mg cell pellets recovered at the end of the growths in planktonic or static conditions by centrifugation (13′000 rpm for 20 min at 4 °C) were solubilized in 60 μL of Laemmli buffer. Then, the total cellular extracts were boiled at 95 °C for 20 min, quickly cooled on ice for 5 min, and centrifuged at 13′000 rpm for 5 min at room temperature (RT). 1 μL of samples were loaded on SDS-PAGE gel and analyzed by anti-His Western blot.
For the solubility analysis of the mScarlet protein, 45 μL of soluble or insoluble fractions were recovered and diluted in 15 μL Laemmli buffer. Then, fractions were boiled at 95 °C for different times (5 min for the soluble sample and 10 min for the insoluble fraction), quickly cooled on ice for 5 min, and finally centrifuged at 13′000 rpm for 5 min at RT. 10 μL of samples were analyzed by SDS-PAGE and anti-His Western blot. For electroblotting, the Trans-Blot Turbo Transfer System (Bio-Rad, Hercules, CA, USA) with nitrocellulose membranes was used employing the mixed molecular weight setting. After the transfer, the membrane was blocked with PBS and 5 % (w/v) milk for 1 h. Then, Monoclonal Anti-polyHistidine-Peroxidase clone HIS-1 antibody (A7058, Merck, Darmstadt, Germany) was diluted 1:2000 in PBS, 0.05 % (v/v) Tween 20, and 5 % (w/v) milk. After 1 h of incubation at RT with the antibody, the membrane was washed with PBS, 0.05 % Tween 20 three times (5 min each) and the antibody was detected with the ECL method (Cyanagen, Bologna, Italy).
2.7. Biofilm formation assay
The wells of a sterile 24-well flat-bottomed polystyrene plate were filled with 1 mL of the medium with a suitable dilution of the bacterial culture in the exponential growth phase (0.2 OD600nm) and incubated at 15 °C. After rinsing with PBS, the adherent cells were stained with 0.1 % (w/v) crystal violet, rinsed twice with double-distilled water, and thoroughly dried. Subsequently, the dye bound to the adherent cells was solubilized with 20 % (v/v) glacial acetic acid and 80 % (v/v) ethanol. After 10 min of incubation at RT, the total biofilm biomass in each well was spectrophotometrically quantified at 590 nm. Each data point was composed of three independent samples.
2.8. Confocal laser scanning microscopy
For the confocal microscopy analysis, the evaluation of fluorescent protein production in biofilm was performed on Nunc™ Lab-Tek® 8-well Chamber Slides (n° 177,445; Thermo Scientific, Ottawa, ON, Canada) used to grow the recombinant strains. All the microscopic observations and image acquisitions were performed with a confocal laser scanning microscope (CLSM) (LSM700-Zeiss, Germany) equipped with an Ar laser (488 nm), and a He–Ne laser (555 nm). The Z-stacks (XYZ isosurface) were obtained by driving the microscope to a point just out of focus on both the top and bottom of the biofilms with a step size of 1 μm. The images and z-stack were obtained using a 20X NA 0.8 objective. All images were analyzed with ZEN Black Imaging Software 3.0 (ZEISS, Jena, Germany) and recorded as a series of tif files with a file depth of 16 bits. The COMSTAT software package [52] was used to determine the biovolume (μm3/μm2). Biovolume provides an estimate of the biomass producing the fluorescent proteins for each condition, and two independent biofilm samples were used.
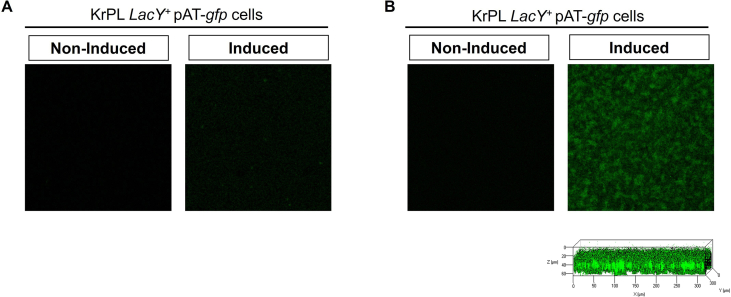
For the investigation of the effect of the induction time on GFP production, 300 μL of a suitable dilution of KrPL LacY + pAT-gfp (0.2 OD600nm) in the GG medium supplemented with chloramphenicol (25 μg/mL) was added to each well of a sterile Chamber Slide. IPTG (5 mM) was added at three different induction times: at the beginning (0 h), after 24 h, and after 48 h from the incubation. Uninduced cultures were used as a negative control. The plates were incubated for 96 h at 15 °C in static condition. Then, the plates were rinsed with filter-sterilized PBS and microscopic observations and image acquisitions were performed using an excitation/emission wavelength of 480/500 nm.
For the analyses of mScarlet production in biofilm under optimized conditions, 300 μL KrPL LacY + pAT-mScarlet (0.1 OD600nm) in the 5/5 GG medium without antibiotics and supplemented with IPTG (5 mM) were added to each well of a sterile Chamber Slide. Uninduced cultures were used as a negative control. The plates were incubated for 96 h at 15 °C in static conditions. After the incubation, the plates were rinsed with filter-sterilized PBS (as previously described) and all microscopic observations and image acquisitions were performed. The excitation/emission maxima for mScarlet are approximately 450/610 nm. The same protocol was carried out to evaluate the mScarlet production in biofilm using the recombinant strains KrPL pAT_2620-mScarlet, KrPL pAT_2621-mScarlet, and KrPL pAT_2690-mScarlet.
The relative percentage of fluorescent cells was evaluated with Calcofluor white stain (CFW), this dye is commonly used to reveal chitin and cellulose. The bacterial culture was prepared as described above and incubated for 96 h at 15 °C. After rinsing with filter-sterilized PBS, the well of the chamber slide was filled with 300 μl of working solution of Calcofluor white stain (CFW) and incubated for 20–30 min at RT, protected from light. All excess stain was removed by rinsing gently with filter-sterilized PBS. All microscopic observations and Z-stack acquisitions were performed. The excitation/emission maxima for Calcofluor white is approximately 453/433 nm.
2.9. Statistics and reproducibility of results
Statistical analyses were performed using two-tailed Student's t-test or two-way ANOVA with either Tukey's post hoc correction for multiple comparisons. P values of ≤0.05 were considered significant. All assays were performed at least in triplicates, and all the results were reported as a mean ± standard deviation (SD). GraphPad Prism software (Version 8, GraphPad Prism Software Inc., La Jolla, California) was used for the analyses.
2.10. Bioinformatic analysis
The identification of the putative promoters upstream of the selected genes (PSHAa2620, PSHAa2621, and PSHAa2690) in the PhTAC125 genome was performed using BPROM (SoftBerry), a bacterial promoter prediction program (http://www.molquest.com).
The collected CLSM Z-stack images (saved as OME-TIFF) were analyzed with COMSTAT software package a plugin (Comstat2) to ImageJ.
3. Results
3.1. Production of the GFP and mScarlet proteins in PhTAC125 biofilm
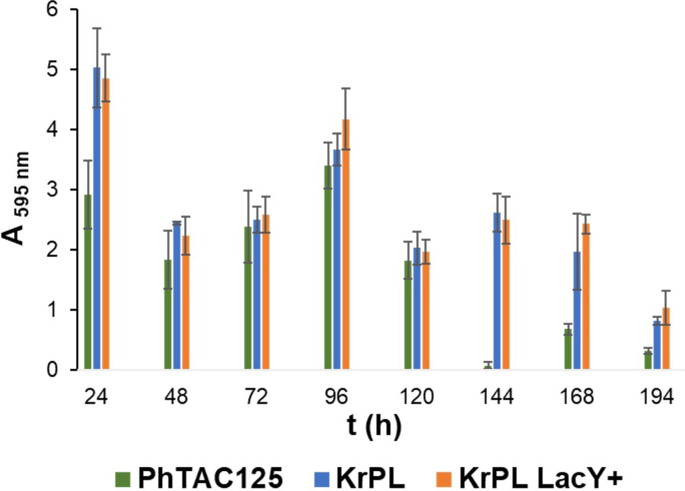
Preliminary experiments were carried out to compare the biofilm formation ability of PhTAC125 (the wild-type strain), KrPL (a PhTAC125 strain devoid of the endogenous plasmid pMtBL [11]), and KrPL LacY+ (an engineered PhTAC125 strain expressing a lactose permease to import IPTG and a truncated form of Lon protease [11]) in the GG medium at 15 °C. The tested strains showed similar biofilm formation kinetic even though a slightly higher biofilm production was observed in KrPL and KrPL LacY+ (Fig. S2). Due to KrPL LacY + well-known abilities as an unconventional host for high-quality protein production [11] it was chosen as a cell-factory to start the testing of recombinant proteins production in biofilm. The performance of this biofilm expression platform was preliminary evaluated by producing the GFP protein by the pAT vector (for details see material and methods section and Fig. S1), a derivative of the IPTG-inducible plasmid pP79 [11] consisting of a high copy replication origin, named B40-OriR [40]. This new expression plasmid conferred production yield improvements in the psychrophilic cells with respect to the pP79 vector mainly by the introduction of a high-copy number of origin replication, named B40-OriR [40].
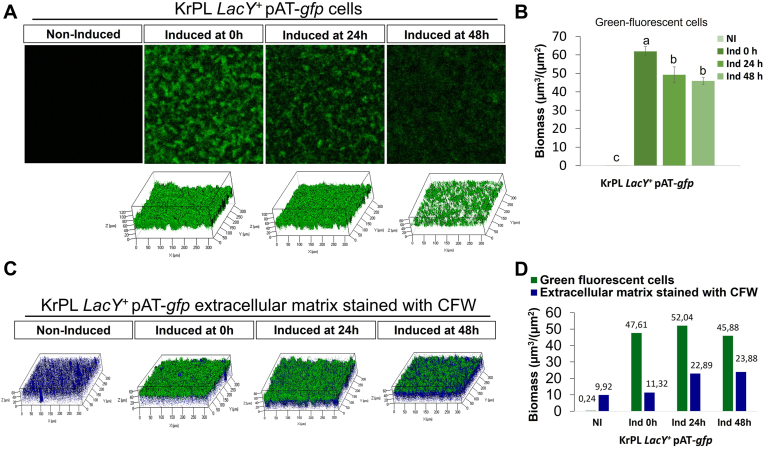
To evaluate the influence of inducer (IPTG), and antibiotic (chloramphenicol) on the biofilm formation, the first experiments were performed growing the recombinant strain, KrPL LacY + pAT-gfp, in the GG medium supplemented with chloramphenicol (25 μg/mL) and IPTG (5 mM) in static conditions at 15 °C. As shown in Fig. S3, the presence of the inducer and the selective agent didn't influence the biofilm formation of the recombinant strain compared to that of the wild-type KrPL LacY+ and higher biofilm production was observed after 24 h and 96 h (Fig. S3), these two conditions were chosen to investigate the production of the green-fluorescent protein using confocal laser scanning microscopy (CLSM). The CLSM revealed a higher fluorescence recombinant production in the biofilm cells recovered after 96 h from the inoculum (Fig. S4). Then the optimal induction time was assessed by adding 5 mM IPTG at three different times of the growth (0 h, 24 h, and 48 h) and evaluating the GFP production by CLSM on the biofilm recovered after 96 h of growth. The CLSM analysis highlighted that higher GFP fluorescence was obtained by inducing the recombinant expression at the beginning of the growth (0 h) (Fig. 1A). This data was also confirmed by determining the biomass-producing mature GFP by COMSTAT [52] image analysis (Fig. 1B). Furthermore, the CLSM analysis of biofilm structure (Fig. 1C) highlighted that the GFP fluorescence is higher in the cells exposed to the air-liquid interface (in green) than the ones present in the deep layers of the biofilm matrix (in blue). This phenomenon could be related to the higher oxygen concentration at the air-liquid interface; indeed, oxygen presence is essential for GFP maturation [53]. The COMSTAT analysis confirmed that the ratio of fluorescent cells (in green) to biofilm matrix (in blue) was highest when the cells were induced at the beginning of the growth (0 h) (Fig. 1D).
Fig. 1.
CLSM analysis of GFP protein production in biofilm at 15 °C in the GG medium for 96 h. A The recombinant strain, KrPL LacY + pAT-gfp was induced with 5 mM IPTG at three different times: at the beginning (Ind 0 h), after 24 h (Ind 24 h) or 48 h (Ind 48 h) of sessile growth and analyzed by CLSM after 96 h. Non-induced (NI) strains were used as a control. B COMSTAT quantitative analysis of biomass producing GFP in the induced cells at different times (0 h, 24 h, and 48 h from the beginning of the growth); Statistical significance was determined using two-way ANOVA followed by Tukey's post hoc test. Data with different letters (a–c) are significantly different (p value < 0.05), while those with the same letter are not significative (p value > 0.05) C Comparative analysis of EPS matrix of the biofilm (in blue) and cells producing the fluorescent protein (in green) in different conditions. Z-stack biofilm structures were obtained using the Calcofluor White Stain (CFW); D COMSTAT quantitative analysis of EPS matrix of the biofilm (stained with calcofluor) and amount of recombinant producing cells (green-fluorescent cells) at different induction times. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
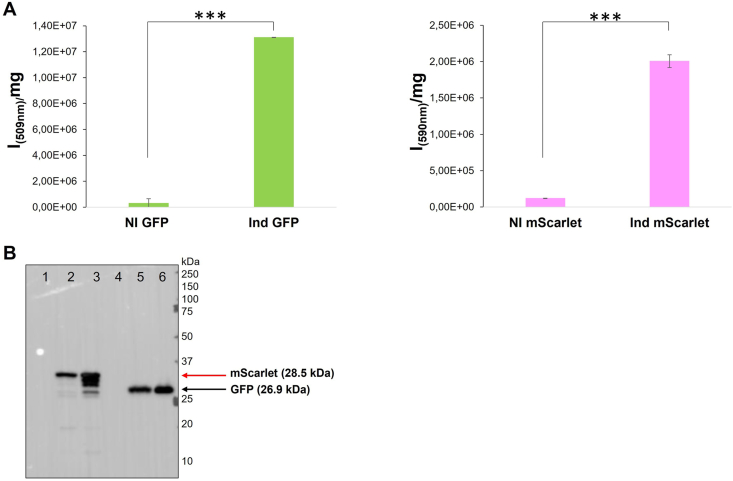
Once induction conditions in the biofilm have been optimized, the experiments were performed also evaluating the production of another fluorescent protein, mScarlet. The pAT-mScarlet vector was constructed as described in material and methods section. KrPL LacY + pAT-gfp and KrPL LacY + pAT-mScarlet recombinant cells were grown by a fluidized-bed reactor using floating polystyrene supports [54]. The recombinant expression was induced with 5 mM IPTG from the beginning of the cell growth and was performed at 15 °C in static condition for 96 h. The production of the two fluorescent proteins was evaluated by spectrofluorimetric analysis (Fig. 2A). The successful production of GFP and mScarlet was further confirmed by anti-His Western blot on total cellular extracts of the recombinant KrPL LacY + cells (Fig. 2B). As shown in Fig. 2B the protein bands assigned to the recombinant mScarlet (28.5 kDa) and GFP (26.9 kDa) produced in the biofilm are visible in lanes 2 and 5, respectively. It's interesting to note that mScarlet produced in the planktonic condition (lane 3) was more proteolyzed than that produced in biofilm (lane 2).
Fig. 2.
Analysis of fluorescent proteins production in biofilm. A Analysis of the GFP and mScarlet production by spectrofluorimetry. The fluorescence indicated on the x-axis was measured on non-induced and induced cells (5 mM IPTG) after 96 h from the beginning of the growth in biofilm. Fluorescence intensities are reported in arbitrary units (I) per biomass (mg) unit. Each data point represents the mean ± SD of three independent samples and differences were considered significant since, according to Student's t-test, in all conditions was p value < 0.05 (*p < 0.05, **p < 0.01, ***p < 0.001). B Analysis of GFP and mScarlet production by anti-His Western blot performed on total extracts of the KrPL LacY + strains grown in planktonic and sessile conditions at 15 °C. The cell extracts were examined after 24 h from the induction in planktonic condition and 96 h from the inoculum in the sessile one. Lane 1, non-induced cells in sessile condition, lane 2, induced cells producing mScarlet in sessile condition, lane 3, induced cells producing mScarlet in planktonic condition, lane 4, non-induced cells in sessile condition, lane 5, induced cells producing GFP in sessile condition, lane 6, induced cells producing GFP in planktonic condition. The same quantity of biomass was analyzed in each lane. Black and red arrows on the right of the gel represent the expected molecular weights of GFP (26.9 kDa) and mScarlet (28.5 kDa). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Optimization of conditions for recombinant protein production in biofilm
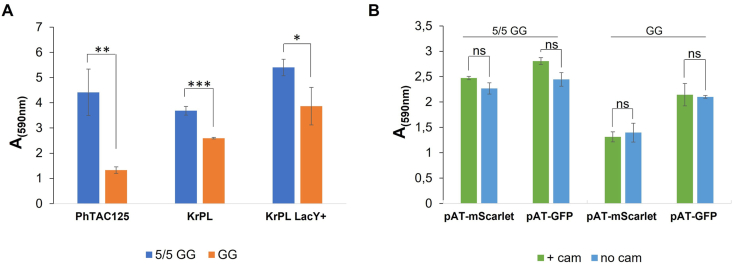
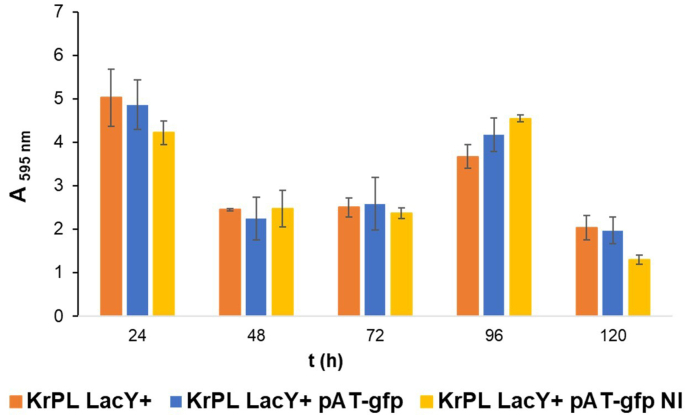
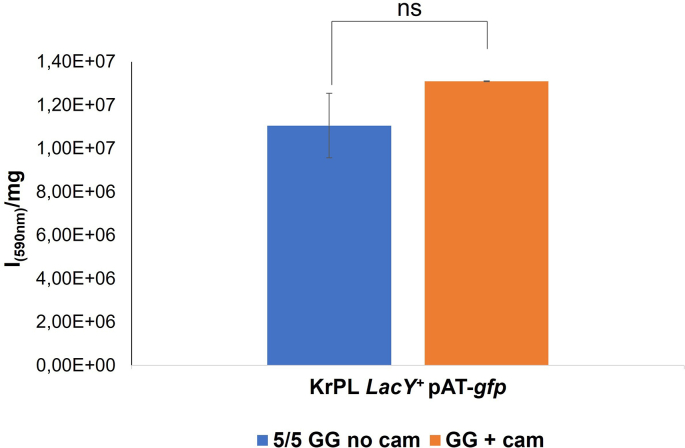
To reduce the process cost and increase biofilm production, the biofilm biomass of PhTAC125 strains was evaluated in the presence of a reduced concentration of carbon sources. PhTAC125, KrPL, and KrPL LacY+ were grown in GG with the half of carbon sources (named 5/5 GG), in static conditions for 96 h at 15 °C. The biofilm biomass was assessed by crystal violet demonstrating that the reduction of carbon sources increased the biofilm production (Fig. 3A). In addition, the influence of the antibiotic presence on biofilm biomasses produced in this new medium was explored (Fig. 3B) by growing the recombinant strains without chloramphenicol. The biofilm production resulted to be not influenced by the presence of the selective agent.
Fig. 3.
Analysis of the effect of the carbon sources concentration on biofilm formation of wild-type or antibiotics on biofilm of recombinant PhTAC125 strains. APhTAC125, KrPL, and KrPL LacY + biofilm obtained at 15 °C in 5/5 GG (GG with the half of carbon source) or GG. B KrPL LacY + pAT-mScarlet and KrPL LacY + pAT-gfp biofilm obtained at 15 °C in 5/5 GG or GG supplemented with IPTG (5 mM) in the presence (+cam) or absence (no cam) of chloramphenicol (25 μg/mL). The biofilms were analyzed after 96 h of growth with the crystal violet assay. Each data point represents the mean ± SD of three independent samples, differences were considered significant since, according to t-Student test, in all conditions was p < 0.05 (*p < 0.05, **p < 0.01, ***p < 0.001, ns: not significant). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
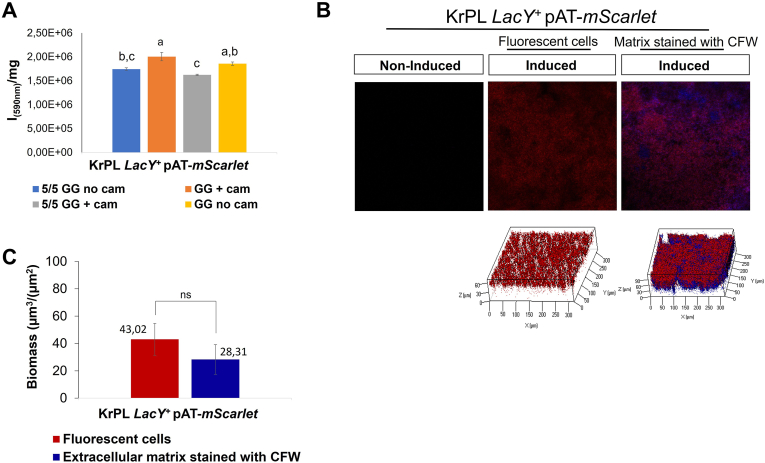
To evaluate the impact of the elimination of the selective agent on the process outcomes, the recombinant protein production of mScarlet in 5/5 GG under non-selective conditions was investigated. KrPL LacY + pAT-mScarlet cells were grown in the absence of the antibiotic by a fluidized-bed reactor as previously described, and the production was compared to that obtained in GG plus chloramphenicol. As shown in Fig. 4A, the two different growth conditions resulted in almost the same fluorescence intensity per biomass unit, an outcome substantially identical to the GFP recombinant production (Fig. S5). The mScarlet production in 5/5 GG under a non-selective condition was also investigated using confocal laser scanning microscopy (Fig. 4B). As observed previously for GFP, mScarlet fluorescence was higher in the cells exposed to the air-liquid interface (in red) than the ones present in the deep layers of the biofilm matrix (in blue). Moreover, the COMSTAT analysis revealed that the ratio between fluorescent cells (red bar) and biofilm matrix (in blue) (Fig. 4C), indicated that most of the cells embedded in the biofilm were able to produce the mature fluorescent protein.
Fig. 4.
Analysis of mScarlet production in 5/5 GG under non-selective conditions. A Analysis of the mScarlet expression by spectrofluorimetry. The fluorescence of the protein indicated on the x-axis was monitored on induced cells (5 mM IPTG) grown in 5/5 GG without chloramphenicol, GG plus chloramphenicol (25 μg/mL), 5/5 GG plus chloramphenicol (25 μg/mL) and GG without chloramphenicol, after 96 h from the beginning of the biofilm growth. Fluorescence intensities are reported in arbitrary units (I) per biomass (mg) unit. Each data point represents the mean ± SD of three independent samples. Statistical significance was determined using two-way ANOVA followed by Tukey's post hoc test. Data with different letters (a,b,c) are significantly different (p value < 0.05), while those with the same letter are not significative (p value > 0.05) B Comparative analysis of biofilm biomass (in blue) and cells producing the fluorescent protein (in red) in 5/5 GG without chloramphenicol after 96 h from the beginning of the biofilm growth. Z-stack biofilm structures were obtained using the Calcofluor White Stain (CFW); C COMSTAT quantitative analysis of EPS matrix of the biofilm (stained with Calcofluor White Stain) (blue bar) and amount of recombinant florescent cells (red bar). Each data point represents the mean ± SD of three independent samples and differences were considered significant since, according to Student's t-test, in all conditions was p value < 0.05 (*p < 0.05, **p < 0.01, ***p < 0.001, ns: not significant). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. New expression systems for the recombinant protein production in biofilm
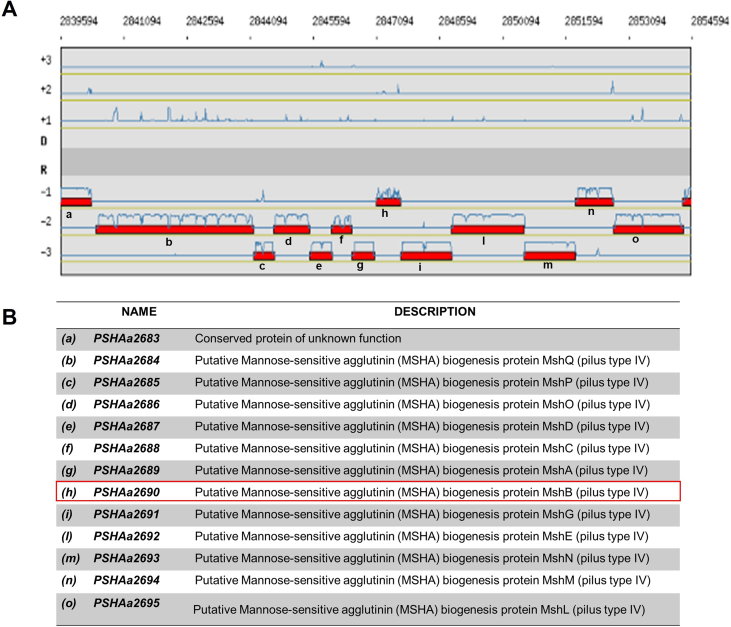
Once the carbon source concentration has been reduced and the selective agent eliminated, a further process optimization was performed by designing heterologous gene expression systems based on biofilm-related constitutive promoters. In this way, the use of the inducer was avoided. Given the limited information on molecular mechanisms involved in PhTAC125 biofilm formation, the identification of constitutive promoters was carried out by focusing on the c-di-GMP molecule [55], a second messenger that regulates the switch from planktonic to sessile growth in different bacteria [56,57]. This molecule can interact with several domains, i.e., GGDEF, EAL, or HD-GYP domains, present in many enzymes implicated in c-di-GMP synthesis or degradation [55,58]. Therefore, starting from the screening of PhTAC125 genome reported by Romling and coauthors [59], a list of genes coding for putative proteins that contain such domains was drawn up (Table S2) leading us to the selection of gene PSHAa2620 and PSHAa2690. Moreover, since the PSHAa2621 gene codifies for the putative sensor histidine kinase of the PSHAa2620-2621 two-component system (Fig. S6) it was selected too.
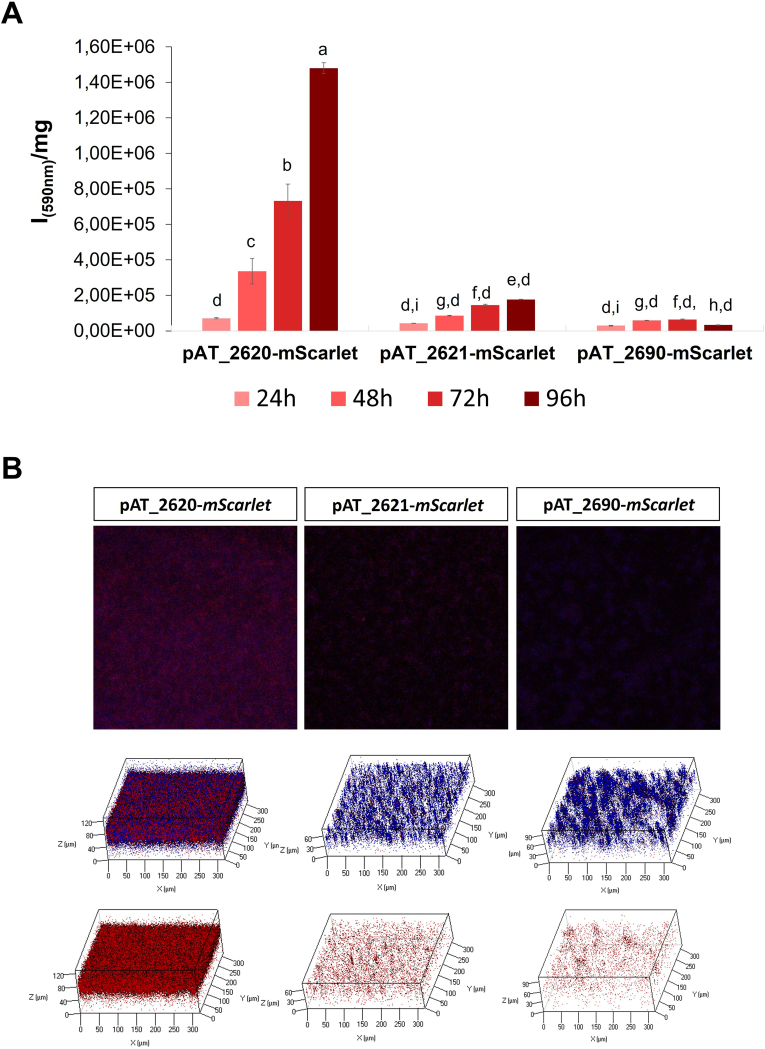
Once selected the genes, the plasmids named pAT_2620-mScarlet, pAT_2621-mScarlet, and pAT_2690-mScarlet were constructed by replacing the promoter sequence of pAT-mScarlet with the putative promoter sequences of the PSHAa2620, PSHAa2621, and PSHAa2690 genes. The performances of the new constitutive vectors were evaluated by growing the recombinant strains, KrPL pAT_2620-mScarlet, KrPL pAT_2621-mScarlet, and KrPL pAT_2690-mScarlet in the 5/5 GG medium at 15 °C in non-selective and static condition for 24 h, 48 h, 72 h, and 96 h. The spectrofluorimetric analysis per biomass unit demonstrated the ability of all the recombinant strains to produce the reporter protein during all the stages of biofilm formation, the higher mScarlet production was obtained by using pAT_2620-mScarlet vector after 96 h (Fig. 5A). This result was also confirmed by CLSM image analysis (Fig. 5B).
Fig. 5.
Analysis of mScarlet constitutive production in biofilm under the control of three putative biofilm-specific promoters. A Evaluation of the mScarlet production by spectrofluorimetry. The fluorescence of the protein indicated on the x-axis was monitored on KrPL recombinant cells after 24 h, 48 h, 72 h, and 96 h from the beginning of the growth in biofilm. Fluorescence intensities are reported in arbitrary units (I) per biomass (mg) unit. Each data point represents the mean ± SD of three independent samples. Statistical significance was determined using two-way ANOVA followed by Tukey's post hoc test. Data with different letters (a-i) are significantly different (p value < 0.05), while those with the same letter are not significative (p value > 0.05). B Comparative analysis of biofilm biomass (in blue) and cells producing the fluorescent protein (in red) in 5/5 GG after 96 h of biofilm growth. Z-stack biofilm structures were obtained using the Calcofluor White Stain (CFW) that stains the EPS matrix of the biofilm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
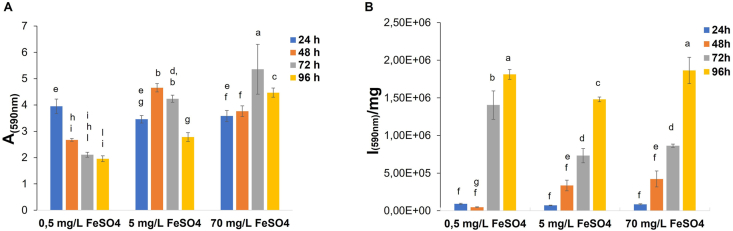
To further improve the system, the effect of iron concentration on biofilm biomass and on the overall mScarlet production was explored. To this purpose, recombinant cells KrPL pAT_2620-mScarlet, were grown in 5/5 GG with 0.5 mg/L FeSO4, 5 mg/L FeSO4 or 70 mg/L FeSO4 in previously described conditions. The biofilm biomasses were assessed by crystal violet assay (Fig. 6A) showing that the highest concentrations of iron sulfate (70 mg/L FeSO4) influenced the biofilm formation of the strain increasing the biofilm production mainly after 72 h and 96 h of growth. On the other hand, it is possible to note that the mScarlet fluorescence intensity per biomass unit was not affected by the presence of different concentrations of iron sulfate (Fig. 6B).
Fig. 6.
Effect of FeSO4 concentration on biofilm biomass and m-Scarlet production. A KrPL pAT_2620-mScarlet biofilm obtained at 15 °C in 5/5 GG supplemented with 0.5 mg/L FeSO4, 5 mg/L FeSO4 (standard concentration) or 70 mg/L FeSO4. The biofilms were analyzed after 24 h, 48 h, 72 h, and 96 h of growth with the crystal violet assay. Each data point represents the mean ± SD of three independent samples. Statistical significance was determined using two-way ANOVA followed by Tukey's post hoc test. Data with different letters (a-l) are significantly different (p value < 0.05), while those with the same letter are not significative (p value > 0.05). B Analysis of the mScarlet expression by spectrofluorimetry. The fluorescence of the protein indicated on the x-axis was monitored on KrPL recombinant cells after 24 h, 48 h, 72 h, and 96 h from the beginning of the growth in biofilm. Fluorescence intensities are reported in arbitrary units (I) per biomass (mg) unit. Each data point represents the mean ± SD of three independent samples. Statistical significance was determined using two-way ANOVA followed by Tukey's post hoc test. Data with different letters (a-g) are significantly different (p value < 0.05), while those with the same letter are not significative (p value > 0.05). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
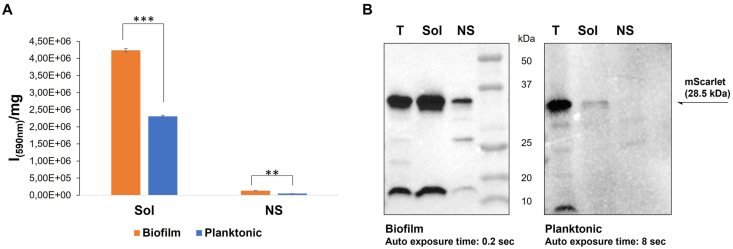
KrPL pAT_2620-mScarlet cells were grown in planktonic and optimized biofilm conditions, then recovered cells were lysed and the production of mScarlet was evaluated by spectrofluorimetric analyses (Fig. 7A). The fluorometric analysis showed that the fluorescence intensity of the soluble fraction obtained from cells grown in biofilm was higher than that gained in planktonic cells (Fig. 7A). This result was confirmed by anti-His Western blot showing that the protein band related to mScarlet in the soluble fraction of biofilm cells is more intense than one in planktonic condition (Fig. 7B). Indeed, the signal corresponding to mScarlet produced in biofilm was detected in 0.2 s, while 8 s were necessary to observe a very weak signal in case of mScarlet produced in planktonic conditions. It's interesting to note that the low quantity of recombinant protein present in the soluble fraction obtained in planktonic condition could be also due to proteolytic events. These results suggested that the specific mScarlet production was higher in biofilms than in planktonic cells. These data were confirmed by a more accurate quantitative comparison between the mScarlet production in biofilm with the production in planktonic conditions reported in supplementary section (supplementary file 1). In the supplementary file, we conducted two comparisons. The first one involved assessing mScarlet production mediated by KrPL pAT_2620-mScarlet in cells grown in biofilm or planktonic conditions and, in this case, the production obtained in biofilm is higher. In the second comparison, we evaluated biofilm production with KrPL pAT_2620-mScarlet in biofilm against planktonic production obtained by KrPL LacY + pAT-mScarlet in conditions optimized for protein production in planktonic cultures. This second assessment was aimed to compare the biofilm production against conventional planktonic production to explore the system's potential respect to established conditions for recombinant protein production, in this case, the production resulted to be equivalent in biofilm and planktonic cells (supplementary file 1).
Fig. 7.
Analysis of mScarlet production on cell lysates from KrPL pAT_2620-mScarlet recombinant cells in biofilm or planktonic growth conditions. A Analysis of the mScarlet expression by spectrofluorimetry. The fluorescence of the protein indicated on the x-axis was monitored on soluble and insoluble fractions obtained from KrPL pAT_2620-mScarlet cells grown in biofilm (orange bar) and planktonic (blue bar) conditions. The fluorescence intensities are reported in arbitrary units (I) per total proteins concentration. B Comparison of the soluble and insoluble fractions after the lysis of the KrPL cells producing mScarlet in planktonic and sessile conditions. The target protein in total cellular extract (T), and soluble (Sol) and insoluble fractions (NS), recovered after lysis of biofilm cells, was detected via anti-His Western blot analysis (0.2 s auto exposure time), as shown in the left panel. The right panel represents the detection of mScarlet by anti-His Western blot analysis (8 s auto exposure time) in total cellular extract (T), and soluble (Sol) and insoluble fractions (NS) obtained after lysis of planktonic cells. For Sol in biofilm and planktonic conditions, the same quantity of the protein was analyzed. The black arrow indicates the expected molecular weights of mScarlet (28.5 kDa). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
To further expand the molecular tools for recombinant protein production is helpful to explore the use of unconventional approaches. These kinds of studies, although far from a coming application, are necessary to open new perspectives and find new solutions. In this paper, we explored the possibility to produce recombinant proteins in Pseudoalteromonas haloplanktis TAC125 biofilm. The reported skills of PhTAC125 as a cell factory and as a biofilm producer inspired the idea to design an ad hoc expression system. The previously reported analyses on PhTAC125 biofilm [44] were performed on the wild-type strain, therefore it was necessary to evaluate the ability of the two PhTAC125 strains applied for the recombinant protein production to grow in biofilm. This analysis demonstrated that the capability of neither KrPL nor KrPL LacY + to grow in biofilm was affected by the antibiotic or IPTG. To evaluate the recombinant protein production in biofilm, two fluorescent proteins were chosen, GFP and mScarlet. We are aware that both the reporters require oxygen to maturate [51,53] and that they might not be the best choice, as biofilm is a matrix where the presence of an oxygen gradient is well-documented [25]. But, on the other hand, fluorescent proteins can be easily visualized by confocal laser scanning microscopy which is one of the best methodologies to investigate biofilm structure [60]. Indeed, by CLSM the best induction strategy was evaluated assessing that the addition of IPTG at time zero resulted to be the condition to obtain the best ratio between the fluorescent cells and the matrix biomass. On the contrary, the use of IPTG when the biofilm was already structured resulted in a not effective production (Fig. 1), likely this result is due to the well-known limitation in molecules diffusions within the biofilm [61] and/or a consequence of lack of oxygen required for maturation. Similar results were obtained by Gomes [62] and coworkers [63] that demonstrated that E. coli eGFP-producing biofilms were highly heterogeneous, with the cells actively producing the mature recombinant protein restricted to the top layer of the biofilm.
The production of the two fluorescent proteins in KrPL LacY + recombinant cells was investigated also by Western blot analysis (Fig. 2B). This experiment not only confirmed the production in biofilm conditions but also allowed an evaluation of recombinant protein quality. Indeed, mScarlet produced in biofilm condition resulted to be less affected by intracellular protease activity compared to the protein produced in planktonic conditions (Fig. 2B). A lot of papers report [25,64–66] the deep difference between planktonic and sessile cell physiology, this difference is demonstrated by several transcriptomic [67,68], proteomic [69], and metabolomic analysis. This deep difference is widely reported also in terms of extracellular proteases production [70–72], on the contrary, little information is available on the difference between intracellular protease in planktonic or sessile condition [73]. Further studies will be helpful to explore intracellular protease production in PhTAC125 biofilm.
After the demonstration of the feasibility of the proposed approach, the well-known features of biofilm were exploited to reduce the process cost. The use of antibiotics is necessary for the retention of plasmid-bearing cells in planktonic [33], but the cells in biofilm tend to grow more slowly than their planktonic counterparts [74] and this behaviour justifies the increased stability of the plasmids [33]. Furthermore, a recent study [30] demonstrated that biofilm can act reserve for plasmids, allowing them to persist even under non-selective conditions. Therefore, as expected, the abolishment of the antibiotic, in our experimental conditions, didn't affect protein production (Fig. 4A). A very similar results were obtained using other hosts for the recombinant protein production in biofilm conditions [32]. It's noteworthy to underline that the resistance agent could represent up to 20 % of total proteins present in a cell [75] and its synthesis subtracts energy and precursors to the production of recombinant proteins thus the elimination of the antibiotic could be very helpful in a production process.
Several studies evaluated the effect of nutrient levels on biofilm formation [25, 44, 63] and some of those suggested that a high carbon source concentration inhibits biofilm formation [76,77]. Therefore, the effect of the reduction of the carbon sources was explored. The halving of the carbon sources increased the biofilm formation without affecting the fluorescent protein production. To further reduce the cost of the process it was beneficial to abolish the use of the inducer, to attain this aim it was necessary to identify a constitutive promoter suitable for the recombinant protein production in biofilm. Since no information is available on gene expression in PhTAC125 grown in the sessile condition, the attention was focused on a second messenger, the c-di-GMP, that in most bacteria is involved in the switch from planktonic growth to the sessile biofilm lifestyle [59, 78, 79]. Responses to the intracellular concentration of c-di-GMP have been implicated in all phases of biofilm formation and its intracellular levels are regulated by diguanylate cyclase (DGC) and phosphodiesterase (PDE) enzymes that catalyze the synthesis and breakdown of this second messenger, respectively. DGC enzymes typically contain a GGDEF domain responsible for c-di-GMP synthesis [80], and a sensory domain that activates the DGC activity in response to external stimuli, such as nutrient concentrations [81], temperature [82], or phosphorylation [83]. On the other hand, PDE enzymes typically contain EAL or HD-GYP domains that catalyze the hydrolysis of c-di-GMP [55]. GGDEF, EAL, and HD-GYP domains, in addition to being widely conserved, are present in the vast majority of enzymes present in bacteria involved in c-di-GMP synthesis or degradation [84] but these domains have evolved to carry out also new functions. One of these functions may involve binding (but not processing) of the substrate [58]. Therefore, we screened the PhTAC125 genome looking for putative genes encoding for proteins that contain GGDEF, EAL, and HD-GYP domains. The literature reported that elevated concentrations of c-di-GMP enhance surface MshA pilus production [57], therefore the genes involved in the secretion and biosynthesis of MSHA in PhTAC125 were also analyzed (Fig. S7), focusing on mshB PSHAa2690 gene. Since MSHA gene locus is often organized into two operons [57], and usually, the promoter regulating the expression of the MSHA pilus structural subunits is located upstream of the mshB gene [59] we selected putative promoter the DNA region upstream of the gene upstream mshB (PSHAa2690 gene) to construct the pAT_2690 vector. This expression vector allowed a low recombinant protein production that is constant in all steps of biofilm development (Fig. 5).
Following this approach, a second gene PSHAa2620 was selected. This gene codifies a putative element of a two-component response system [85] involved in c-di-GMP production and containing the REC-GGDEF domain. Bacterial two-component system is composed of two proteins: the sensor histidine kinase (HK) and the response regulator (RR), this system serve to connect the detection of an environmental or intracellular signal to an appropriate response [85]. In particular, the RR protein is responsible for the execution of the specific cellular output in response to the input detected by the HK. The prototypical RR contains a conserved N-terminal receiver (REC) domain, which is connected to a highly variable C-terminal effector domain. The REC domain of the RR protein catalyses the transfer of the phosphoryl group from the associated HK onto itself, resulting in self-activation in a phosphorylation-dependent manner. Nearly 70 % of all classified RR contain a DNA-binding domain and are generally assumed to function as transcriptional regulators [86]. Of the classified RR, 8 % belong to a group that combines the REC domain with various enzymatic domains involved in signal transduction. Interestingly, a common enzymatic output domain in RR is involved in second messenger homeostasis such as cyclic diguanylate monophosphate (c-di-GMP) [86]. This finding led us to suppose that the PSHAa2620 (encoding for REC-GGDEF protein) and PSHAa2621 (encoding for the putative sensor histidine kinase, HK) genes could be constitutively expressed in biofilm. The vectors pAT_2620 and pAT_2621 were constructed using the upstream region of the selected genes and employed to produce mScarlet. The obtained data demonstrated that all plasmids allowed the production of the reporter protein although with a different extent, and the highest fluorescence per unit biomass was recorded after 96 h of growth (Fig. 5). Indeed, the mScarlet production is recorded in all stages of the biofilm evolution but the highest accumulation is achieved during the mature biofilm phase (Fig. 5). Moreover, the results demonstrated that pAT_2620 vector is very effective in biofilm and allows a lower production in planktonic cells. This behaviour makes pAT_2620 an ad hoc vector for biofilm production.
Given the importance of iron availability in biofilm formation [87–90] the effect of iron concentration on KrPL pAT_2620-mScarlet biofilm was explored. How iron affects biofilm formation varies according to the species and strain, in the case of KrPL pAT_2620-mScarlet reported results demonstrated that the presence of a higher concentration lets an increase of biofilm biomass at 96 h (Fig. 6A). The studies on the regulatory role of iron in several bacterial biofilms have revealed the complexity of this process and thus further analyses will be dedicated to uncovering the molecular mechanism by which iron controls the PhTAC125 biofilm development. Notably, the iron concentration didn't affect the recombinant production of mScarlet, confirming that the pAT_2620 plasmid provided higher protein production at 96 h (Fig. 6B). This biofilm stage, in the presence of a higher concentration of iron, is characterized by a high biomass accumulation therefore this condition could guarantee a good total production yield. It's interesting to note that the more accurate quantitative analysis of the mScarlet production for cell unit (Supplementary file 1) revealed that the system for the recombinant protein production in biofilm has the same potentiality as the classical planktonic approach with some additional advantages. Indeed, it needs a lower concentration of carbon sources and doesn't require antibiotic and inducer addition.
5. Conclusion
In conclusion, the data reported in this paper demonstrated the feasibility of recombinant protein production in PhTAC125 biofilm and allowed us to establish an optimized production strategy working on media composition and the construction of an ad hoc expression vector. Certainly, despite the promising preliminary results, it is necessary to evaluate the performance of the new system in the production of “difficult” proteins. Moreover, further efforts will be aimed to overcome limitations mainly related to the control of PhTAC125 biofilm development and to the identification of the best processing conditions for recombinant protein production in automatic biofilm reactors.
CRediT authorship contribution statement
Marzia Calvanese: Methodology, Investigation, Data curation, Conceptualization. Caterina D'Angelo: Methodology, Investigation, Data curation, Conceptualization. Concetta Lauro: Writing – review & editing, Methodology, Data curation. Maria Luisa Tutino: Writing – review & editing, Investigation, Formal analysis. Ermenegilda Parrilli: Writing – review & editing, Writing – original draft, Supervision, Methodology, Investigation, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioflm.2024.100179.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Fig. S1.
Maps of pAT-gfp and pAT-mScarlet vectors.
Fig. S2.
Biofilm formation kinetic of PhTAC125, KrPL, and KrPL LacY + strains.
Fig. S3.
Effect of inducer and antibiotic presence on the biofilm formation.
Fig. S4.
CLSM analysis of mature GFP production in biofilm at 15 °C in the GG medium.
Fig. S5.
Analysis of GFP production in 5/5 GG under non-selective conditions.
Fig. S6.
Representation of PSHAa2620 and PSHAa2621 gene locus in the PhTAC125 genome.
Fig. S7.
Representation of the MSHA gene cluster gene in the PhTAC125 genome.
Data availability
No data was used for the research described in the article.
References
- 1.Rong Y., Jensen S.I., Lindorff-Larsen K., Nielsen A.T. Folding of heterologous proteins in bacterial cell factories: cellular mechanisms and engineering strategies. Biotechnol Adv. 2023;63 doi: 10.1016/j.biotechadv.2022.108079. [DOI] [PubMed] [Google Scholar]
- 2.Wu Y., Feng S., Sun Z., Hu Y., Jia X., Zeng B. An outlook to sophisticated technologies and novel developments for metabolic regulation in the Saccharomyces cerevisiae expression system. Front Bioeng Biotechnol. 2023;11:1–16. doi: 10.3389/fbioe.2023.1249841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gasser B., Saloheimo M., Rinas U., Dragosits M., Rodríguez-Carmona E., Baumann K., Giuliani M., Parrilli E., Branduardi P., Lang C., Porro D., Ferrer P., Tutino M., Mattanovich D., Villaverde A. Protein folding and conformational stress in microbial cells producing recombinant proteins: a host comparative overview. Microb Cell Factories. 2008;7 doi: 10.1186/1475-2859-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKenzie E.A., Abbott W.M. Expression of recombinant proteins in insect and mammalian cells. Methods. 2018;147:40–49. doi: 10.1016/j.ymeth.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Puetz J., Wurm F.M. Recombinant proteins for industrial versus pharmaceutical purposes: a review of process and pricing. Processes. 2019;7 doi: 10.3390/pr7080476. [DOI] [Google Scholar]
- 6.Medigue C., Krin E., Pascal G., Barbe V., Bernsel A., Bertin P.N., et al. Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 2005;15:1325–1335. doi: 10.1101/gr.4126905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Médigue C., Krin E., Pascal G., Barbe V., Bernsel A., Bertin P.N., Cheung F., Cruveiller S., D'Amico S., Duilio A., Fang G., Feller G., Ho C., Mangenot S., Marino G., Nilsson J., Parrilli E., Rocha E.P.C., Rouy Z., Sekowska A., Tutino M.L., Vallenet D., Von Heijne G., Danchin A. Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 2005;15:1325–1335. doi: 10.1101/gr.4126905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parrilli E., Duilio A., Tutino M.L. 2008. Heterologous protein expression in psychrophilic hosts. [DOI] [Google Scholar]
- 9.Tutino M.L., Parrilli E., Giaquinto L., Duilio A., Sannia G., Feller G., Marino G. Secretion of α-amylase from Pseudoalteromonas haloplanktis TAB23: two different pathways in different hosts. J Bacteriol. 2002;184 doi: 10.1128/JB.184.20.5814-5817.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sannino F., Giuliani M., Salvatore U., Apuzzo G.A., de Pascale D., Fani R., Fondi M., Marino G., Tutino M.L., Parrilli E. A novel synthetic medium and expression system for subzero growth and recombinant protein production in Pseudoalteromonas haloplanktis TAC125. Appl Microbiol Biotechnol. 2017;101 doi: 10.1007/s00253-016-7942-5. [DOI] [PubMed] [Google Scholar]
- 11.Colarusso A., Lauro C., Calvanese M., Parrilli E., Tutino M.L. Improvement of pseudoalteromonas haloplanktis tac125 as a cell factory: iptg-inducible plasmid construction and strain engineering. Microorganisms. 2020;8:1–24. doi: 10.3390/microorganisms8101466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giuliani M., Parrilli E., Ferrer P., Baumann K., Marino G., Tutino M.L. Process optimization for recombinant protein production in the psychrophilic bacterium Pseudoalteromonas haloplanktis. Process Biochem. 2011;46:953–959. doi: 10.1016/j.procbio.2011.01.011. [DOI] [Google Scholar]
- 13.Dragosits M., Frascotti G., Bernard-Granger L., Vázquez F., Giuliani M., Baumann K., Rodríguez-Carmona E., Tokkanen J., Parrilli E., Wiebe M.G., Kunert R., Maurer M., Gasser B., Sauer M., Branduardi P., Pakula T., Saloheimo M., Penttilä M., Ferrer P., Luisa Tutino M., Villaverde A., Porro D., Mattanovich D. Influence of growth temperature on the production of antibody Fab fragments in different microbes: a host comparative analysis. Biotechnol Prog. 2011;27 doi: 10.1002/btpr.524. [DOI] [PubMed] [Google Scholar]
- 14.Vigentini I., Merico A., Tutino M.L., Compagno C., Marino G. Optimization of recombinant human nerve growth factor production in the psychrophilic Pseudoalteromonas haloplanktis. J Biotechnol. 2006;127:141–150. doi: 10.1016/j.jbiotec.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 15.Papa R., Rippa V., Sannia G., Marino G., Duilio A. An effective cold inducible expression system developed in Pseudoalteromonas haloplanktis TAC125. J Biotechnol. 2007;127:199–210. doi: 10.1016/j.jbiotec.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 16.Colarusso A., Caterino M., Fabbri A., Fiorentini C., Vergara A., Sica F., Parrilli E., Tutino M.L. High yield purification and first structural characterization of the full-length bacterial toxin CNF1. Biotechnol Prog. 2018;34:150–159. doi: 10.1002/btpr.2574. [DOI] [PubMed] [Google Scholar]
- 17.Flemming H.C., Wuertz S. Bacteria and archaea on Earth and their abundance in biofilms. Nat Rev Microbiol. 2019;17:247–260. doi: 10.1038/s41579-019-0158-9. [DOI] [PubMed] [Google Scholar]
- 18.Zhou L., Zhang Y., Ge Y., Zhu X., Pan J. Regulatory mechanisms and promising applications of quorum sensing-inhibiting agents in control of bacterial biofilm formation. Front Microbiol. 2020;11:1–11. doi: 10.3389/fmicb.2020.589640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bos R., Van Der Mei H.C., Busscher H.J. Physico-chemistry of initial microbial adhesive interactions - its mechanisms and methods for study. FEMS Microbiol Rev. 1999;23:179–230. doi: 10.1016/S0168-6445(99)00004-2. [DOI] [PubMed] [Google Scholar]
- 20.Muhammad M.H., Idris A.L., Fan X., Guo Y., Yu Y., Jin X., Qiu J., Guan X., Huang T. Beyond risk: bacterial biofilms and their regulating approaches. Front Microbiol. 2020;11:1–20. doi: 10.3389/fmicb.2020.00928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juhas M., Eberl L., Tümmler B. Quorum sensing: the power of cooperation in the world of Pseudomonas. Environ Microbiol. 2005;7:459–471. doi: 10.1111/j.1462-2920.2005.00769.x. [DOI] [PubMed] [Google Scholar]
- 22.Liu C., Sun D., Zhu J., Liu J., Liu W. The regulation of bacterial biofilm formation by cAMP-CRP: a mini-review. Front Microbiol. 2020;11:1–7. doi: 10.3389/fmicb.2020.00802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flemming H.C., Baveye P., Neu T.R., Stoodley P., Szewzyk U., Wingender J., Wuertz S. Who put the film in biofilm? The migration of a term from wastewater engineering to medicine and beyond. Npj Biofilms Microbiomes. 2021;7:1–5. doi: 10.1038/s41522-020-00183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin W., Wang Y., Liu L., He J. Biofilms: the microbial “protective clothing” in extreme environments. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20143423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flemming H.C., Wingender J., Szewzyk U., Steinberg P., Rice S.A., Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016 doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 26.Beloin C., Ghigo J.M. Finding gene-expression patterns in bacterial biofilms. Trends Microbiol. 2005;13:16–19. doi: 10.1016/j.tim.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Parrilli E., Luisa M., Gennaro T. Biofilm as an adaptation strategy to extreme conditions. Rendiconti Lincei Sci Fis Nat. 2022 doi: 10.1007/s12210-022-01083-8. [DOI] [Google Scholar]
- 28.Carvalho F.M., Azevedo A., Ferreira M.M., Mergulhão F.J.M., Gomes L.C. Advances on bacterial and fungal biofilms for the production of added-value compounds. Biology. 2022;11:1–30. doi: 10.3390/biology11081126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madsen J.S., Burmølle M., Hansen L.H., Sørensen S.J. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol Med Microbiol. 2012;65:183–195. doi: 10.1111/j.1574-695X.2012.00960.x. [DOI] [PubMed] [Google Scholar]
- 30.Røder H.L., Trivedi U., Russel J., Kragh K.N., Herschend J., Thalsø-Madsen I., Tolker-Nielsen T., Bjarnsholt T., Burmølle M., Madsen J.S. Biofilms can act as plasmid reserves in the absence of plasmid specific selection. Npj Biofilms Microbiomes. 2021;7 doi: 10.1038/s41522-021-00249-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li M., Wang Z., Zhou M., Zhang C., Zhi K., Liu S., Sun X., Wang Z., Liu J., Liu D. Continuous production of human epidermal growth factor using Escherichia coli biofilm. Front Microbiol. 2022;13:1–11. doi: 10.3389/fmicb.2022.855059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soares A., Azevedo A., Gomes L.C., Mergulhão F.J. Recombinant protein expression in biofilms. AIMS Microbiol. 2019;5:232–250. doi: 10.3934/microbiol.2019.3.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Connell H.A., Niu C., Gilbert E.S. Enhanced high copy number plasmid maintenance and heterologous protein production in an Escherichia coli biofilm. Biotechnol Bioeng. 2007;97:439–446. doi: 10.1002/bit.21240. [DOI] [PubMed] [Google Scholar]
- 34.Parrilli E., Tedesco P., Fondi M., Tutino M.L., Lo Giudice A., de Pascale D., Fani R. The art of adapting to extreme environments: the model system Pseudoalteromonas. Phys Life Rev. 2021;36:137–161. doi: 10.1016/j.plrev.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 35.Parrilli E., Tedesco P., Fondi M., Tutino M.L., Lo Giudice A., de Pascale D., Fani R. The art of adapting to extreme environments: the model system Pseudoalteromonas. Phys Life Rev. 2021;36:137–161. doi: 10.1016/j.plrev.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 36.Cusano A.M., Parrilli E., Duilio A., Sannia G., Marino G., Tutino M.L. Secretion of psychrophilic α-amylase deletion mutants in Pseudoalteromonas haloplanktis TAC125. FEMS Microbiol Lett. 2006;258:67–71. doi: 10.1111/j.1574-6968.2006.00193.x. [DOI] [PubMed] [Google Scholar]
- 37.Tutino M.L., Duilio A., Parrilli E., Remaut E., Sannia G., Marino G. A novel replication element from an Antarctic plasmid as a tool for the expression of proteins at low temperature. Extremophiles. 2001;5 doi: 10.1007/s007920100203. [DOI] [PubMed] [Google Scholar]
- 38.Sannino F., Giuliani M., Salvatore U., Apuzzo G.A., de Pascale D., Fani R., Fondi M., Marino G., Tutino M.L., Parrilli E. A novel synthetic medium and expression system for subzero growth and recombinant protein production in Pseudoalteromonas haloplanktis TAC125. Appl Microbiol Biotechnol. 2017;101:725–734. doi: 10.1007/s00253-016-7942-5. [DOI] [PubMed] [Google Scholar]
- 39.Papa R., Rippa V., Sannia G., Marino G., Duilio A. An effective cold inducible expression system developed in Pseudoalteromonas haloplanktis TAC125. J Biotechnol. 2007;127:199–210. doi: 10.1016/j.jbiotec.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 40.Calvanese M., Balestra C., Colarusso A., Lauro C., Riccardi C., Fondi M., Parrilli E., Tutino M.L. Development of high-copy number plasmids in Pseudoalteromonas haloplanktis TAC125. Appl Microbiol Biotechnol. 2023:2469–2481. doi: 10.1007/s00253-023-12448-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lauro C., Colarusso A., Calvanese M., Parrilli E., Tutino M.L. Conditional gene silencing in the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Res Microbiol. 2022 doi: 10.1016/j.resmic.2022.103939. [DOI] [PubMed] [Google Scholar]
- 42.Calvanese M., Colarusso A., Lauro C., Parrilli E., Tutino M.L. Insoluble proteins: methods and protocols. second ed. Garcia-Fruitos E and Aris GiraltAEditors, Springer, 2021; 2022. Soluble recombinant protein production in Pseudoaltermonas haloplanktis TAC125: the case study of the full-length human CDKL5 protein; pp. 219–232. [DOI] [PubMed] [Google Scholar]
- 43.Colarusso A., Lauro C., Calvanese M., Parrilli E., Tutino M.L. Active human full-length CDKL5 produced in the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Microb Cell Factories. 2022;21:1–18. doi: 10.1186/s12934-022-01939-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ricciardelli A., Casillo A., Vergara A., Balasco N., Corsaro M.M., Tutino M.L., Parrilli E. Environmental conditions shape the biofilm of the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Microbiol Res. 2019;218:66–75. doi: 10.1016/j.micres.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 45.Ricciardelli A., Casillo A., Vergara A., Balasco N., Corsaro M.M., Tutino M.L., Parrilli E. Environmental conditions shape the biofilm of the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Microbiol Res. 2019;218 doi: 10.1016/j.micres.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 46.Römling U., Galperin M.Y. Bacterial cellulose biosynthesis: diversity of operons, subunits, products, and functions. Trends Microbiol. 2015;23:545–557. doi: 10.1016/j.tim.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sannino F., Giuliani M., Salvatore U., Apuzzo G.A., de Pascale D., Fani R., Fondi M., Marino G., Tutino M.L., Parrilli E. A novel synthetic medium and expression system for subzero growth and recombinant protein production in Pseudoalteromonas haloplanktis TAC125. Appl Microbiol Biotechnol. 2017 doi: 10.1007/s00253-016-7942-5. [DOI] [PubMed] [Google Scholar]
- 48.Tutino M.L., Duilio A., Parrilli E., Remaut E., Sannia G., Marino G. A novel replication element from an Antarctic plasmid as a tool for the expression of proteins at low temperature. Extremophiles. 2001;5:257–264. doi: 10.1007/s007920100203. [DOI] [PubMed] [Google Scholar]
- 49.Parrilli E., Ricciardelli A., Casillo A., Sannino F., Papa R., Tilotta M., Artini M., Selan L., Corsaro M.M., Tutino M.L. Large-scale biofilm cultivation of Antarctic bacterium Pseudoalteromonas haloplanktis TAC125 for physiologic studies and drug discovery. Extremophiles. 2016;20 doi: 10.1007/s00792-016-0813-2. [DOI] [PubMed] [Google Scholar]
- 50.Tsien R.Y. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 51.Bindels D.S., Haarbosch L., Van Weeren L., Postma M., Wiese K.E., Mastop M., Aumonier S., Gotthard G., Royant A., Hink M.A., Gadella T.W.J. MScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat Methods. 2016;14:53–56. doi: 10.1038/nmeth.4074. [DOI] [PubMed] [Google Scholar]
- 52.Heydorn A., Nielsen A.T., Hentzer M., Sternberg C., Givskov M., Ersboll B.K., Molin S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 53.Hebisch E., Knebel J., Landsberg J., Frey E., Leisner M. High variation of fluorescence protein maturation times in closely related Escherichia coli strains. PLoS One. 2013;8 doi: 10.1371/journal.pone.0075991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parrilli E., Ricciardelli A., Casillo A., Sannino F., Papa R., Tilotta M., Artini M., Selan L., Corsaro M.M., Tutino M.L. Large-scale biofilm cultivation of Antarctic bacterium Pseudoalteromonas haloplanktis TAC125 for physiologic studies and drug discovery. Extremophiles. 2016;20:227–234. doi: 10.1007/s00792-016-0813-2. [DOI] [PubMed] [Google Scholar]
- 55.Ryjenkov D.A., Tarutina M., Moskvin O.V., Gomelsky M. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol. 2005;187:1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumari N., Kumar S., Karmacharya M., Dubbu S., Kwon T., Singh V., Chae K.H., Kumar A., Cho Y.K., Lee I.S. Surface-textured mixed-metal-oxide nanocrystals as efficient catalysts for ROS production and biofilm eradication. Nano Lett. 2021 doi: 10.1021/acs.nanolett.0c03639. [DOI] [PubMed] [Google Scholar]
- 57.Floyd K.A., Lee C.K., Xian W., Nametalla M., Valentine A., Crair B., Zhu S., Hughes H.Q., Chlebek J.L., Wu D.C., Hwan Park J., Farhat A.M., Lomba C.J., Ellison C.K., Brun Y.V., Campos-Gomez J., Dalia A.B., Liu J., Biais N., Wong G.C.L., Yildiz F.H. c-di-GMP modulates type IV MSHA pilus retraction and surface attachment in Vibrio cholerae. Nat Commun. 2020;11:1–16. doi: 10.1038/s41467-020-15331-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lindenberg S., Klauck G., Pesavento C., Klauck E., Hengge R. The EAL domain protein YciR acts as a trigger enzyme in a c-di-GMP signalling cascade in E. coli biofilm control. EMBO J. 2013;32:2001–2014. doi: 10.1038/emboj.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Römling U., Galperin M.Y., Gomelsky M. Cyclic di-GMP: the first 25 Years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. doi: 10.1128/mmbr.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tasse J., Cara A., Saglio M., Villet R., Laurent F. A steam-based method to investigate biofilm. Sci Rep. 2018;8:1–8. doi: 10.1038/s41598-018-31437-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flemming H.C., Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.