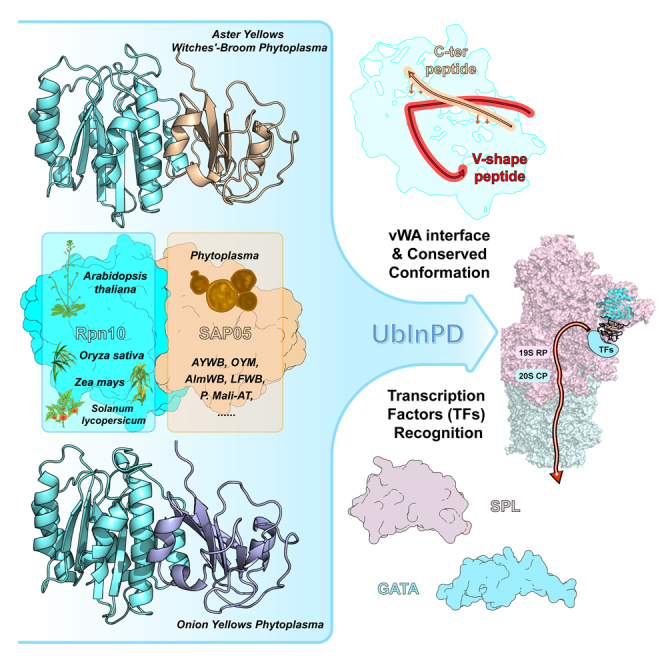
Summary
Besides traditional ubiquitin-dependent proteasome degradation, thousands of eukaryotic proteins more than previously appreciated could undergo ubiquitin-independent proteasomal degradation (UbInPD). A pathogen-encoded effector protein SAP05 secreted by phytoplasma, could hijack hostage Rpn10 subunit of proteasome derived from Arabidopsis thaliana and target the degradation of GATA BINDING FACTOR (GATA) or SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factors (TFs) without ubiquitin or additional proteasome shuttle factors. To explain how could SAP05 target the degradation bypassing the ubiquitin-dependent pathway, we have determined the crystal structure of the complex between Arabidopsis thaliana Rpn10 and Aster Yellows witches'-broom phytoplasma SAP05 or onion yellow phytoplasma SAP05, which showed a previously unknown recognition interface. Sequence alignment and structural biological evidence showed that this interaction is highly conserved in various SAP05 homologs, suggesting a general mode in plant infection. After docking the complex structure to the plant proteasome, SAP05 was near to the adenosine triphosphatase (ATPase) central pore and enough to submit substrate to degradation process, which suggested a molecular glue-like role to bridge TFs close to the ATPase central pore of proteasomes for the direct degradation.
Subject areas: Bioinformatics, Plant bioinformatics, Plant biology
Graphical abstract
Highlights
-
•
Crystal structures of At RPn10 complexed with phytoplasma SAP05 of two species
-
•
SAP05-Rpn10 complexes share conserved recognition mode across species
-
•
SAP05 may orient the substrate directly to proteasome to trigger UbInPD
Bioinformatics; Plant bioinformatics; Plant biology
Introduction
Controlled protein degradation in eukaryotic cells is crucial for protein homeostasis and primarily mediated by the ubiquitin-proteasome system (UPS). Abnormal proteins including misfolded or damaged proteins were labeled with ubiquitin as a degradation tag, followed by the recognition and degradation by the 26S proteasome.1 As the core protein machine in the pathway, the 26S proteasome is composed of a 20S catalytic core particle (CP) and one or two 19-subunit regulatory particles (RP).2 Ubiquitinated substrate is recruited by RP subunits Rpn10, Rpn13, and Rpn1, and then the substrate can be pulled into the central pore of CP and degraded into short peptides with the assistance of ATPase domains.3,4 Dysregulation of the UPS is associated with a wide range of human diseases such as cancers and degenerative diseases, and several core enzymes such as 26S proteasome are attractive therapeutic targets. Moreover, targeted protein degradation that hijacks the endogenous UPS to degrade disease-related proteins, has become a therapeutic approach of great interest in the biomedical field.5,6
Interestingly, a subset of eukaryotic proteins have been shown to undergo ubiquitin-independent proteasomal degradation (UbInPD). These proteins such as ornithine decarboxylase, p53 and its downstream apoptosis regulators BIMEL and NADPH oxidase activator (NOXA) as well as cyclin-dependent kinase inhibitor p21, can be directly degraded by the 26S proteasome in vivo and in vitro despite that all lysine sites are blocked from ubiquitination.7,8,9,10 Recently, the global protein stability (GPS)-peptidome technology has been applied to identify novel full-length protein substrates that are degraded without ubiquitin.11 Thousands of UbInPD substrates were identified, some of which required C-terminal degradation sequence (C-degron) and shuttling factors of the Ubiquilin family for proteasome recognition. Besides, a new midnolin-proteasome pathway was found to rapidly degrade immediate-early genes (IEGs) coded proteins without a ubiquitin tag.12 These studies suggest the process that UbInPD regulates substrate turnover in vivo is more common than previously appreciated.
In addition to eukaryotic endogenous homeostasis, pathogen-encoded effector proteins derived from plant bacteria hijack some hostage proteins for direct proteasomal degradation through ubiquitin-independent pathways. An insect-vectored parasitic phytoplasma Aster Yellows phytoplasma strain Witches’ Broom (AYWB) infects the plant host Arabidopsis thaliana (At) and secretes the effector protein SAP05,13 which induces almost all extended developmental phenotypes of the phytoplasma, such as plant dwarfism, prolonged growth cycle, increased number of leaves and lateral branches, and infertility of flowers, etc. A recent study found that SAP05 could bind the host proteasome Rpn10 subunit and plant GATA BINDING FACTOR (GATA) or SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factors (TFs), which results in targeted TFs degradation without ubiquitination. Therefore, concurrent destabilization of SPLs and GATAs by SAP05 effectors decouples plant developmental transitions. Detailed evidence demonstrated that SAP05 forms a bridge between the SPL5 Zinc-finger (ZnF) domain and Rpn10 N-terminal von Willebrand factor A (vWA) domain to generate a ternary complex, and ubiquitination of lysine residues within SPL5 may not be required for SAP05-mediated degradation. Moreover, although the overall structure of plant 26S proteasome is similar to the mammalians', SAP05-induced Rpn10 interaction is not conserved in Homo sapiens.14,15,16 This unique mode of action given that targeted protein degradation has been applied to engineer plant Rpn10 conferring resistance to SAP05. Besides SAP05, other effectors phytoplasma secreted have evolved to interact and degrade other families of TFs like plant MADS-domain transcription factors (MTFs).17,18,19,20 Due to the wide range of phytoplasma’s targets and the strong spreading ability assisted by insect hosts, which seriously affects crop yields including grain and causes incalculable economic losses, structural information about the molecular mechanism of phytoplasma effector protein induced directly proteasome-targeted degradation will be helpful to more accurately guide the transgenic operation of crop pest resistance, and also facilitate the development of novel chemical inducers of degradation (CIDEs) that directly target proteasome.21,22
Here, we resolved the crystal structure of phytoplasma AYWB_SAP05 protein in complex with Arabidopsis thaliana Rpn10vWA domain. The structure reveals that AYWB_SAP05 recognizes AtRpn10 through a stapled V-shape peptide and its C-terminus folded over the V-shape peptide, different from all known recognition with ubiquitin or ubiquitin-like (UBL) modified proteins. Sequence alignment showed that this recognition pattern is conserved among various SAP05 homologs. Moreover, the crystal structure of the complex between onion yellow phytoplasma SAP05 (OY_SAP05) and Rpn10vWA still showed consistency in the recognition pattern even though its poor sequence identity (42.11%) against the AYWB_SAP05. Therefore, our work suggests that phytoplasmas may perform a general mode of infecting plants by attaching proteasome subunits to destabilize development-associated TFs, thereby affecting plant phenotypes. Moreover, docking the SAP05-Rpn10 complex to the spinach 26S proteasome found no significant change in the structure of Rpn10vWA, and we thus hypothesized that SAP05 may play a molecular glue-like role to bridge TFs close to the ATPase central pore of proteasomes for the direct degradation.
Results
Phytoplasma effector SAP05 recognizes proteasome subunit Rpn10 in a host species-specific manner
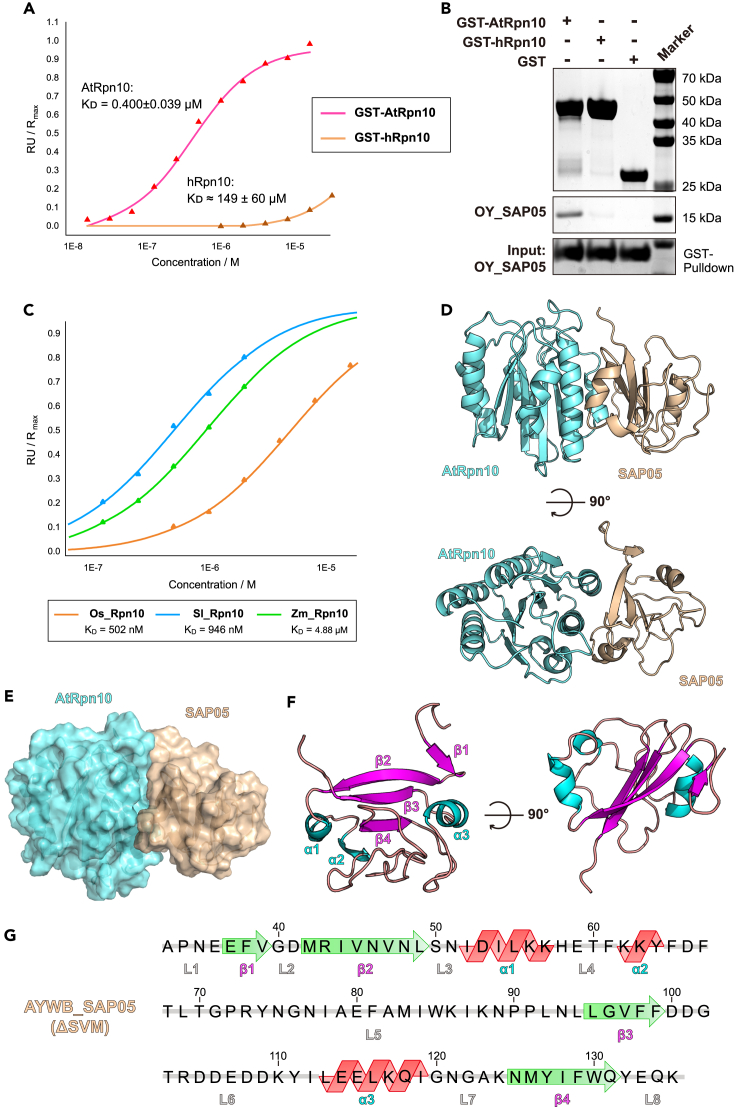
Previous data indicated that Aster Yellows witches'-broom phytoplasma SAP05 (AYWB_SAP05) mainly recognizes the N-terminal vWA domain of A. thaliana Rpn10 (AtRpn10).13 We therefore constructed and recombinantly expressed the mature SAP05 without signal peptide sequence SVM (33–135 amino acids, and just called SAP05 for convenience hereinafter) and the GST-tagged AtRpn10 vWA domain. Accordingly, surface plasmon resonance (SPR) experiments showed that the AYWB_SAP05 bound AtRpn10 with three orders of magnitude lower dissociation constant (0.4 ± 0.039 μM) compared with the Homo sapiens Rpn10 (hRpn10) vWA (Figure 1A), which is consistent with the previous finding that SAP05 cannot recognize human proteasome. We also tested another onion yellows phytoplasma OY_SAP05 that is homologous to AYWB_SAP05 with a relatively low conserved surface. It also showed a high affinity against AtRpn10 instead of hRpn10, which is similar to AYWB_SAP05 (Figure 1B). Besides, we confirmed that Rpn10 from other plant species, like Os (Oryza sativa L.), Sl (Solanum lycopersicum) as well as Zm (Zea mays) could also be recognized by AYWB_SAP05 (Figures 1C and S1A). These findings suggest a conserved recognition behavior between phytoplasma SAP05 homologs and plant host proteasome subunit Rpn10, while Rpn10 of Homo sapiens have evolved to escape this phytoplasma effector recognition.
Figure 1.
Crystal structure of AYWB_SAP05-AtRpn10 complex
(A) The curve of response value versus concentration of GST-tagged AtRpn10/hRpn10 in the SPR binding assay of Rpn10 binding to immobilized AYWB_SAP05. The dissociation constant between GST-hRpn10 and AYWB-SAP05 is far larger than GST-AtRpn10 - AYWB_SAP05 pair (Data are represented as mean ± SEM).
(B) GST-pull down of tagged AtRpn10 or hRpn10 with the appearance of analyte OY_SAP05.
(C) SPR binding assay between AYWB-SAP05 and Rpn10 of different species.
(D) Two views of the cartoon model of AYWB_SAP05-AtRpn10vWA global structure.
(E) Surface of the AYWB_SAP05 and AtRpn10 in structure.
(F and G) Secondary structures and their sequence and number in AYWB_SAP05. More details are present in Figure S1.
Structures of AYWB_SAP05-AtRpn10 complex
To gain structural insight into how AYWB_SAP05 hijacks the proteasome subunit Rpn10 in A. thaliana, we determined the crystal structure of the AYWB_SAP05-AtRpn10_vWA complex. Recombinantly expressed AYWB_SAP05 and AtRpn10 truncates were able to form a 1:1 heterodimeric complex in size exclusion chromatography coupled with multiangle light scattering (SEC-MALS) analysis (Figure S1B). The purified protein complex of AYWB_SAP05-AtRpn10 was collected and crystals were grown from sitting drop with reservoir buffer containing 0.2 M sodium malonate, 20% w/v polyethylene glycol 3,350. Utilizing the molecular replacement with Alphafold2-enabled models of two proteins (Rpn10: AF-P55034-F1, SAP05: AF-Q2NK94-F1), resulting in a final structure at 1.57 Å resolution (Figures 1C and 1D).
The AtRpn10 vWA in our structure adopts a conformation similar to that predicted by Alphafold2, which also resembles the structure of vWA in the spinach 26S proteasome15 (PDB: 8AMZ, rmsd. = 0.781 Å) (Figure S1C). And the SAP05, which previously had no homologous structure, performs to fold as a globular shape and is identical to the prediction of Alphafold2 except for the smaller amount of β-sheet secondary structure (Figure S1D). The SAP05 structure contains a four-strand β-sheet core layer and three α-helices distributed on either side (Figures 1E and 1F). There are two long and compact loops, L5, L6, between α2 and β3 and between β3 and αC, respectively, wrapping together on the opposite side of the SAP05-Rpn10 interface.
A non-classical interface of AYWB_SAP05 bound to AtRpn10
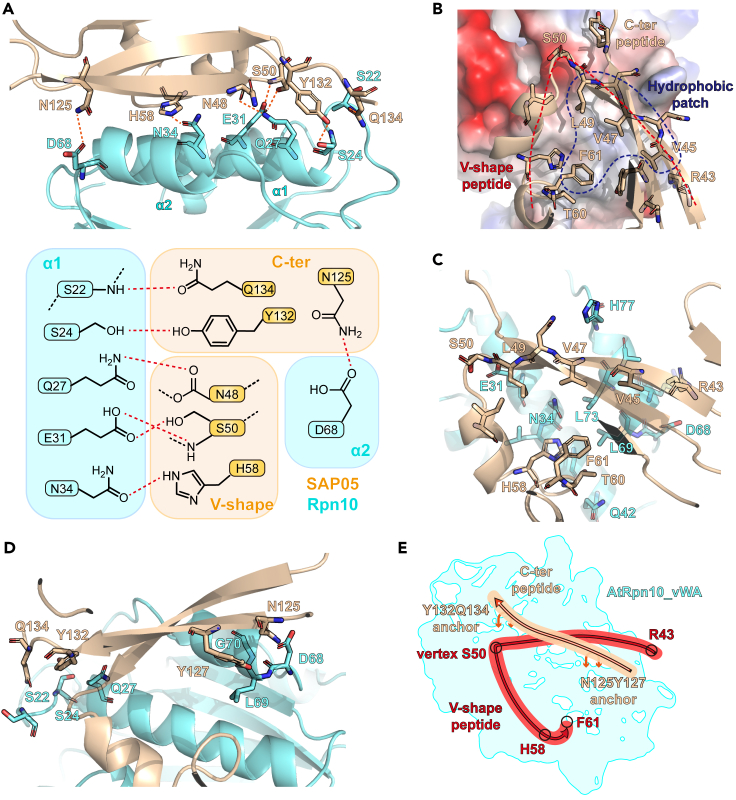
SAP05 and Rpn10 form a 1783.1 Å2 quite extensive interface, containing 6 pairs of hydrogen-bond linkages and a large area of polar contacts and hydrophobic interactions (Figures 2A and 2B). A V-shape peptide (43–61 aa.) in SAP05, which uses L3 (S50, N51) as the vertex, β2 and α1-L4 as two sides (called by β-side and α-side hereinafter, respectively), is involved in the recognition of AtRpn10 (Figure 2C). This V-shape peptide docks on the α1 and α2 of AtRpn10vWA. In detail, the S50 side chain on the vertex, the main-chain carbonyl oxygen of N48 on the β-side and H58 side chain on the α-side form three hydrogen bonds with AtRpn10 E31, Q27, and N34, respectively (Figures 2A, S2B, and S2C). SAP05 R43 and T60, which are located at the N-termini and C-termini of the V-shape peptide, have polar contacts with AtRpn10 (Figures S2A and S2C). And the V45, V47, and L49 on the β-side as well as F61 on the α-side of SAP05, contribute to the binding with hydrophobic effect simultaneously (Figures 2C, S2A, and S2C).
Figure 2.
Detailed interaction between AYWB_SAP05 and AtRpn10
(A) Hydrogen bonds formed between AYWB_SAP05 and AtRpn10. The amino acids involved in the hydrogen bonds formation are shown as sticks in the upper figure. The lower diagram summarizes the hydrogen-bond interactions between different regions of two proteins. The hydrogen bond was represented by red dashed lines.
(B) Components of AYWB_SAP05-AtRpn10vWA interface. The vacuum electrostatic potential surface of AtRpn10 and two key binding regions on SAP05: V-shape peptide and C-ter peptide are shown as the cartoon model.
(C) Interactions between V-shape peptide and AtRpn10. The amino acids involved in the interaction are shown as sticks.
(D) Interactions between N-ter peptide and AtRpn10.
(E) Cartoon diagram of how AYWB_SAP05 interacts with AtRpn10vWA domain, vWA surface is colored by cyan. V-shape peptide and C-ter peptide are indicated in red and wheat lines, respectively. See also Figure S2.
Interestingly, SAP05’s C-terminal peptide (C-ter peptide) containing the β4 and L8 region, straddles the β-side of the V-shape peptide and “staples” itself onto the surface of AtRpn10, forming a two-point anchored conformation (Figure 2D). On the side of SAP05 C-termini, Y132 forms a hydrogen bond with AtRpn10 S24 while Q134 interacts with the main chain of the N-terminal of AtRpn10 L1 (Figure S2D). On the other side, SAP05 N125 and AtRpn10 D68 also take a hydrogen-bond contact (Figure S2E). In summary, SAP05-Rpn10 recognition is mediated by the cooperation of polar contact including a large number of hydrogen bonds, and hydrophobic interactions (Figure 2E).
Proteasome subunit Rpn10 is a well-known “ubiquitin receptor” which comprises the quantity different C-terminal ubiquitin interacting motifs (UIMs) to mediate the direct recognition of ubiquitin, UBL domain or UBL proteins (ATG8,23 FAT1024). Previously, shuttle factors such as the Ubiquilin family, DSK2, and RAD23 were reported to interact with Rpn10-UIM to transport ubiquitinated substrates or induce the UbInPD directly. And even the vWA of Rpn10 is able to act as an atypic region bound to FAT10 and NUB1L UBL domain. Here, we found Rpn10 vWA domain recognizes the special exogenous shuttle factor AYWB_SAP05 which has no similarity to ubiquitin (i.e., no UBL domain) through a previously unknown atypical interface. Our structure reveals that this atypical recognition depends on a stapled V-shape peptide forming extensive hydrogen bonds and hydrophobic interactions with the association of the Rpn10 vWA domain.
Key residues of SAP05 contributing to the conserved recognition of AtRpn10
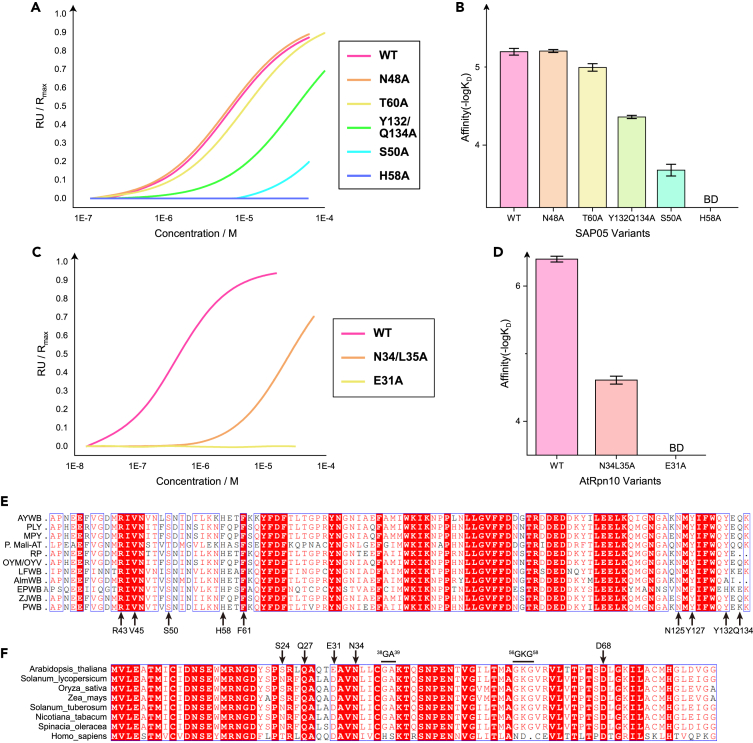
To validate the interface of the crystal structure, we introduced single or double Ala mutations to key interface residues of recombinant SAP05 or Rpn10 (Figures S3A and S3B) and tested the binding affinity by the SPR measurement (Figure S3C). We found whether residues of V-shape peptide (S50A, H58A) or C-ter peptide (Y132/Q134A) whose side chains were involved in the hydrogen bonds formation, their Ala mutations greatly weakened the interaction (Figures 3A, 3B, and S3D). The binding affinity also decreased when the T60 was turned to Ala, which diminished the polar contacts between the two proteins. Expectedly, the N48A mutation did not affect the interaction between AYWB_SAP05 and AtRpn10, since N48 uses main-chain carbonyl oxygen rather than side-chain as the hydrogen-bond acceptor (Figures 3A and 3B). Consistently, mutations of the key residues of interaction on AtRpn10 also abrogated the SAP05 binding (Figures 3C and S3E). Altogether, the residues we observed at the interface are indeed involved in the interaction between AYWB_SAP05 and AtRpn10.
Figure 3.
Conserved sequence and binding mode of SAP05-Rpn10 recognition of different species
(A) The curve of response value versus concentration of AYWB_SAP05 mutants in the SPR binding assay of SAP05 proteins binding to immobilized AtRpn10.
(B) The histogram of the dissociation constant between WT AtRpn10 and different AYWB_SAP05 mutants measured by SPR. The error bars represent the stand error of -logKD. BD: below the detectable limit.
(C) The curve of response value versus concentration of GST-AtRpn10 mutants in the SPR binding assay of Rpn10 proteins binding to immobilized AYWB_SAP05.
(D) The histogram of the dissociation constant between WT AYWB_SAP05 and different AtRpn10 mutants.
(E) Multiple sequence alignment of SAP05 homologs from different phytoplasma species. Amino acids which are involved in interaction and highly conserved are pointed with arrows at the bottom.
(F) Multiple sequence alignment of Rpn10 homologs from different plant species. Amino acids which are involved in interaction and highly conserved are pointed with arrows on the top. Two key mutation sites that plant Rpn10 distinct from the mammalian Rpn10 (PSMD4) are also shown on the top. See also Figure S3.
We next attended to investigate the conservation of the interaction between SAP05 and proteasome Rpn10. Comparing the sequence of SAP05 in various phytoplasma species, R43, V45 on the β-side, S50 at the vertex, H58 on the α-side, and N125, Y127, Y132, Q134 on the C-ter peptide of SAP05 are highly conserved involving in the interaction with AYWB_SAP05-AtRpn10 (Figure 3E). Most of these residues participate in the hydrogen bond formation. On the other hand, residue N34, D68, and S24 of AtRpn10, which are the corresponding binding partners of H58, N125, and Y132 of SAP05, respectively, are also conserved among different plant species (Figure 3F). Especially, E31 of AtRpn10 is replaced by Asp in most other plant species, which may mediate distinct polar contact to stabilize the V-shape peptide anchoring (Figure 3F). These sequence alignment results indicate that most SAP05 homologs may use similar contacts to interact with plant Rpn10.
Structural conservation of SAP05 homologs bound to AtRpn10
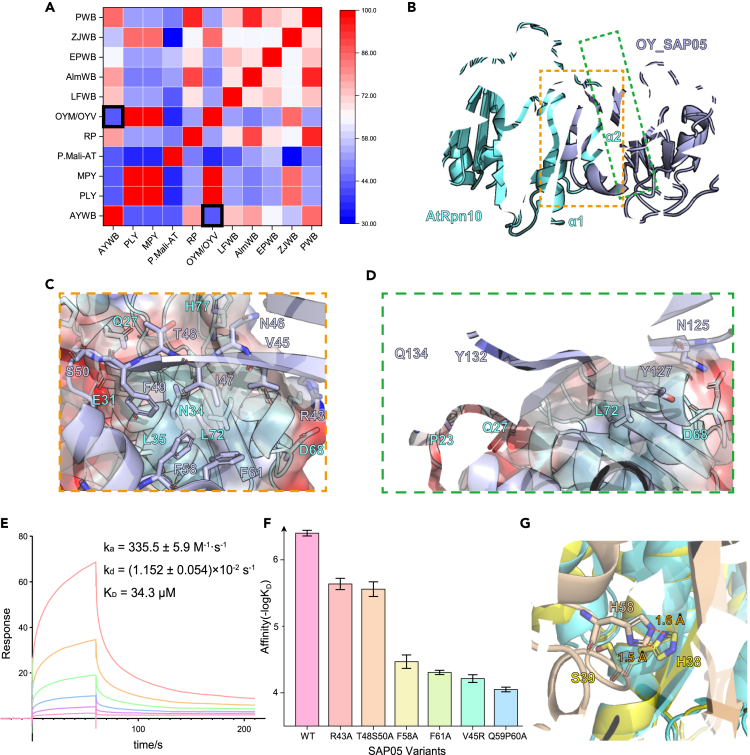
We further noticed the onion yellows phytoplasma SAP05 (OY_SAP05) whose V-shape region has only 42.11% identity against the AYWB_SAP05 interface (Figure 4A), however, its KD value (0.398 ± 0.039 μM) bound to AtRpn10 vWA domain is similar to AYWB_SAP05 (Figures 4F, S4A, S4B). Especially, some vital residues whose mutations lead to the obstacle of SAP05-Rpn10 interaction like H58 and T60, are not conserved in OY_SAP05. To further illustrate the recognition mechanism, we co-crystallized the OY_SAP05 with AtRpn10vWA and resolved the 1.78 Å structure of the AtRpn10_vWA-OY_SAP05 1:1 stoichiometric complex (Figure 4B). Compared to the AYWB_SAP05-AtRpn10 complex structure, both SAP05 and vWA in this homologous structure adopt identical conformations, except that OY_SAP05 forms a triangular β-sheet core layer harboring 6 β-strands, which is larger than AYWB_SAP05 (Figures S5A and S5B).
Figure 4.
Crystal structure and the conserved conformation of OY_SAP05-AtRpn10 complex
(A) Percent identity matrix of V-shape region of SAP05 homologs in different phytoplasma species. The blocks representing the similarity between AYWB and OY_SAP05 are highlighted.
(B) Cartoon model of OY_SAP05-AtRpn10vWA complex global structure.
(C) Interactions between OY_SAP05 V-shape peptide and AtRpn10.
(D) Interactions between OY_SAP05 N-ter peptide and AtRpn10.
(E) Response curve of V-shape-GSGS-C-ter peptide binding to AtRpn10 measured by SPR.
(F) The histogram of the dissociation constant between WT AtRpn10 and different OY_SAP05 mutants measured by SPR (Data are represented as mean ± SEM).
(G) Alignment of our crystal structure to the human Rpn10 (PSMD4 in PDB: 6MSB) vWA domain structure. The clash between aligned SAP05 with human Rpn10 was shown in sticks. See also Figures S4–S6.
Although the sequence similarity between AYWB_SAP05 and OY_SAP05 is relatively low, their recognition manners we found remain conserved (Figure 4B). The OY_SAP05 located above the α1 and α2 of Rpn10_vWA and the V-shape peptide (Figure 4C, 43-61 aa.) and two anchors (N125/Y127 & Y132/Q134) on the C-ter peptide (Figure 4D) contribute the interaction. Moreover, conserved vertex S50 of the V-shape peptide associating with adjacent T48 forms two hydrogen bonds with AtRpn10 E31, Q27, anchoring the peptide onto the vWA surface (Figure S5C), despite differences in amino acid pairing compared to AYWB_SAP05-AtRpn10 complex. The V45, I47, and F49 on the β-side and F61 on the α-side of SAP05 still have hydrophobic interactions with AtRpn10 (Figure S5D). And the conserved R43, which is involved in polar contacts in the AYWB_SAP05 structure, is linked to D68 directly by a hydrogen bond (Figure S5E). Phe-mediated strong hydrophobic interaction covers the contribution of the hydrogen bond involving key residue H58 in the AYWB_SAP05 interface (Figure S5F). In summary, the recognizing conformation of SAP05 remains conserved despite the low sequence similarity.
We also synthesized the possible shortest recognition region of SAP05 which directly fused the V-shape and C-ter peptide by the GSGS linker (Figure S6A). However, the peptide bound to the AtRpn10 with a KD value of about 34.3 μM (Figure 4E), was much lower than that of the total AYWB_SAP05 bound to AtRpn10. This phenomenon suggests that conformation recognition rather than previously reported sequence-specific degron recognition is more necessary for SAP05 transporting TFs and inducing direct proteasomal degradation.
The authenticity of this homologous structure is also verified by SPR binding assay involving wild type (WT) and mutated SAP05. The affinity binding to AtRpn10 with all OY_SAP05 mutants is reduced by one or two orders of magnitude (Figures 4F and S6B). It is delightful that although we failed to prepare V45, F61 mutated recombinant AYWB_SAP05 due to the formation of aggregates, we successfully obtained the corresponding mutants of OY_SAP05. And their performance proved the important role of hydrophobic interaction in the SAP05-Rpn10 recognition. Therefore, our data indicated that the phytoplasma SAP05 family may adopt highly similar conformations for recognizing plant Rpn10.
Changes in interface abrogate the binding of SAP05 to hRpn10
In 2021, Huang et al. reported that AWYB_SAP05 cannot bind with Homo sapiens Rpn10 homolog PSMD4.13 They found two key polymorphistic sites in Rpn10/PSMD4 sequence that prevents hRpn10 from interacting with SAP05: 38GA39 (A. thaliana) to 38HS39 (H. sapiens), and 56GKG58 (A. thaliana) to 56ND57 (H. sapiens). Superposing hRpn10_vWA (from PDB: 6MSB) to the AtRpn10_vWA in our structure shows that the hRpn10 38HS39 mutation clashes with the SAP05 H58, thus obstructing the SAP05 recognition (Figure 4G). Similar clashes also occurred between OY_SAP05 F58 and hRpn10 (Figure S5H). It is proved by SPR that a 38GA39 to 38HS39 AtRpn10 mutant (AtRpn10-m1) considerably decreased the binding of SAP05 (Figure S5I). 56GKG58 to 56ND57 mutation may prevent SAP05 binding by abolishing the β3 secondary structure and subsequently skewing the important interface α2 (Figure S5G). Our data are consistent with the previous finding and provide clues for the species selectivity of this hijacking process.
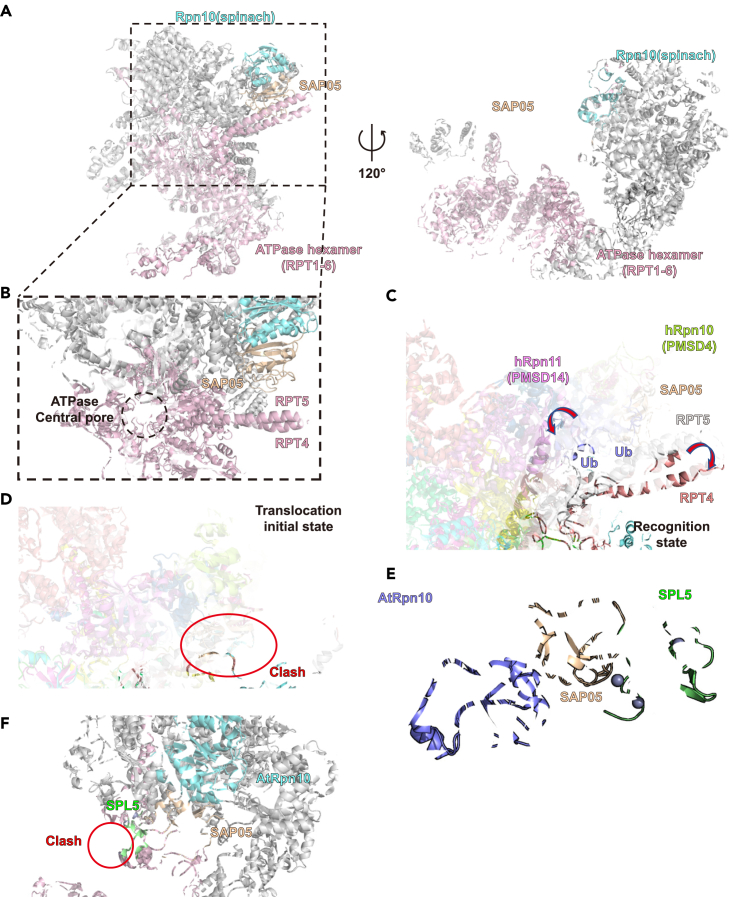
Superposing the SAP05 complex into the proteasome
Rpn10 is located at the periphery of the 19S regulatory particle, where it is in contact with lid subunits Rpn8 and Rpn9 and maintains structural integrity. The α1/α2 surface of Rpn10vWA does not interact with the other 19S subunit and is accessible for SAP05 binding. By aligning our AYWB_SAP05-AtRpn10 structure with the Rpn10 in the structure of spinach (Spinacia oleracea) proteasome 19S regulatory particle reported recently, we can observe that the SAP05 is placed proximal to the central pore of 19S ATPase OB-ring and has no clash with any constitutive subunit (Figures 5A and 5B). This binding pattern has little impact on the constitutive assembly of proteasome and recruits the TF GATA or SPL to the entrance of ATPase hexamer for the sequential substrate deposition. Moreover, SAP05 is located close to subunit RPT4-RPT5 next to Rpn10 (Figure 5C). In the reported initial recognition state of the human 26S proteasome-polyubiquitylated substrate complex, there are two ubiquitin molecules docking on the N-terminal CC (coiled-coil) domain of RPT4-RPT5 N-termini during the substrate peptide inserting to the central pore,25 which hints SAP05 may work as the substitution of ubiquitin to bring substrate to the substrate entrance of the proteasome lid.
Figure 5.
Superposing models of SAP05-Rpn10 complex in the proteasome
(A and B) Different views of superposing models of SAP05 biding with spinach proteasome 19S regulatory particle (PDB: 8AMZ).
(C and D) Superpose our AYWB_SAP05-AtRpn10 structure to the different states of substrate-engaged human 26S proteasome: recognition state (PDB: 6MSB, 6MSD, the conformation difference between these two structures are pointed out by arrows), and translocation initial state (PDB: 6MSH, the clash is marked by the red circle).
(E) Superpose the crystal structure of the AYWB_SAP05-SPL5 ZnF complex (PDB: 8PFC) to our AYWB_SAP05-AtRpn10 structure.
(F) Superpose the AtRpn10-AYWB_SAP05-SPL5 ZnF ternary complex structure model to the spinach proteasome 19S regulatory particle.
SAP05 would not collide with any subunit in the 19S regulatory particle until the substrate translocation initiates. But once the substrate-engaged proteasome entered the translocation-initial state, a conformational rotation occurred in the proteasome lid, which would make the RPT4-RPT5 N-terminal CC domain occupy the binding site of SAP05 (Figure 5D). This clash suggests that the SAP05 is supposed to dissociate from Rpn10 before the translocation initiation.
In sum, SAP05 is capable of bringing TFs proximal to the entrance of the central pore and the ubiquitin-binding region, which can initiate substrate degradation. And the SAP05 may leave from Rpn10 before translocation initiation to make the substrate unfolding process work smoothly.
Discussion
According to previous in vitro degradation assays, ∼20% of cellular proteins are able to be degraded by proteasome independent of ubiquitination.26 The plant proteasome subunit Rpn10, which is reported to recognize both ubiquitinated substrates and UbInPD-associated shuttling factors, can be hijacked by bacterial effector SAP05 to mediate ubiquitin-independent degradation of host’s TF during phytoplasma invasion.
In this work, we resolved the crystal structure of two Rpn10-SAP05 complexes and revealed the recognition mechanism between SAP05 of two different species and plant Rpn10 in this process. These SAP05 adopt similar globular conformations containing a β-sheet core layer and three α-helices. They can dock to the same Rpn10 α1/α2 interface by the collaboration of the V-shape peptide and C-terminal region, mediated by a few conserved hydrogen bonds and hydrophobic interactions, and the conserved conformation of the SAP05 interface.
At the same time that we wrote this manuscript, Liu et al. reported the crystal structure of AYWB_SAP05-AtRpn10_vWA structure same as us and a structure of SAP05 interacting with its TF target, SPL5 ZnF.27 The SPL5 ZnF binds to the loop surface consisting of L5 and L6 loops in our AYWB_SAP05 structure (Figure 5E). After superimposing their SAP05-SPL5_ZnF complex structure, our AYWB_SAP05-AtRpn10 structure and the structure of spinach proteasome 19S particle, the SPL5 will be located between SAP05 and RPT4-RPT5 CC domain (Figure 5F). The location of SPL5 has clashes with the RPT4-RPT5, which indicates the interaction between SPL5 and ATPase subunits and a conformation change during SPL5 binding.
In conclusion, our work reveals a conserved recognizing mode between phytoplasma pathogenic protein SAP05 and plant proteasome subunit Rpn10 during the phytoplasma invasion. Besides the process of SAP05 hijacking SPL and GATA TF families, another phytoplasma protein, SAP54, was reported to utilize Rad23 to mediate proteasomal degradation of MADS-box TFs. In consideration of the severe economic loss caused by phytoplasma diseases of crops, these hijack processes are worth studying to facilitate phytoplasma prevention. In addition, a heterobifunctional tool of targeted degradation via direct 26S proteasome PSMD2 subunit recruitment is reported this year.22 The SAP05/SAP5417,18,19 function of connecting host protein to the Rpn10/Rad23 to mediate ubiquitin/UBL-independent degradation can enlighten the development of similar human proteasome-targeting chimera molecules.
Limitations of the study
Our crystal complex structures lack a substrate, thus we cannot know how SAP05 interacts with its substrate such as SPL5, GATA18 from our work. But this part is also indispensable to understanding the mechanism of SAP05 recruiting the TF for proteasome degradation. In addition, the mechanism of SAP05 being recognized by proteasome was preliminarily explored in our study through software docking. Further cryo-EM studies can be devised to investigate the actual movie of how SAP05 induces substrate recruitment, unfolding, insertion, and degradation at the proteasomal level. At last, we suggested in our work that the SAP05-Rpn10 interaction may be conserved among many other plant-phytoplasma pairs, whether this UbInPD pathway will have impacts on the phenotypes and characteristics of other plant species can be the topic of future experiments.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Escherichia coli BL21(DE3) Chemically Competent Cell | Transgen | CD601-03 |
| Trans5α Chemically Competent Cell | Transgen | CD201-02 |
| Chemicals, peptides, and recombinant proteins | ||
| ProPlex Screen | Molecular Dimensions | MD1-38 |
| Index | Hampton | HR2-144 |
| V-shape-C-ter peptide | this study | N/A |
| AYWB_SAP05 | this study | N/A |
| AYWB_SAP05-N48A | this study | N/A |
| AYWB_SAP05-T60A | this study | N/A |
| AYWB_SAP05-Y132A/Q134A | this study | N/A |
| AYWB_SAP05-S50A | this study | N/A |
| AYWB_SAP05-H58A | this study | N/A |
| AtRpn10_vWA | this study | N/A |
| AtRpn10_vWA-N34/L35A | this study | N/A |
| AtRpn10_vWA-E31A | this study | N/A |
| hRpn10_vWA | this study | N/A |
| OsRpn10_vWA | this study | N/A |
| SlRpn10_vWA | this study | N/A |
| ZmRpn10_vWA | this study | N/A |
| OY_SAP05 | this study | N/A |
| OY_SAP05-R43A | this study | N/A |
| OY_SAP05-T48S50A | this study | N/A |
| OY_SAP05-F58A | this study | N/A |
| OY_SAP05-F61A | this study | N/A |
| OY_SAP05-V45R | this study | N/A |
| OY_SAP05-Q59P60A | this study | N/A |
| Ni NTA Beads 6FF | lablead | N30210 |
| Glutathione Beads 4FF | lablead | G10510 |
| Superdex 75 Increase 10/300 GL | Cytiva | 29148721 |
| HiLoad 16/600 Superdex 75 pg | Cytiva | 28989333 |
| Sensor Chip CM5 | Cytiva | BR100399 |
| PAGE Gel Fast Preparation Kit | epizyme | PG113 |
| Multicolor Prestained Protein Ladder | Epizyme | WJ102L |
| Ultimate XB-C18, 5μm, 21.2×150mm | Welch | 02610-21105 |
| SurePAGE™, Bis-Tris, 10x8, 4-20%, 15 wells | GenScript | M00657 |
| Recombinant DNA | ||
| pet28a(+)-His6-HRV3C-AYWB_SAP05 | GenScript | N/A |
| pet28a(+)-His6-HRV3C-OY_SAP05 | GenScript | N/A |
| pGEX-6P-1-GST-HRV3C-AtRpn10_vWA | GenScript | N/A |
| pGEX-6P-1-GST-HRV3C-hRpn10_vWA | GenScript | N/A |
| pGEX-6P-1-GST-HRV3C-OsRpn10_vWA | this study | N/A |
| pGEX-6P-1-GST-HRV3C-SlRpn10_vWA | this study | N/A |
| pGEX-6P-1-GST-HRV3C-ZmRpn10_vWA | this study | N/A |
| pet28a(+)-His6-HRV3C-AYWB_SAP05-N48A | this study | N/A |
| pet28a(+)-His6-HRV3C-AYWB_SAP05-T60A | this study | N/A |
| pet28a(+)-His6-HRV3C-AYWB_SAP05-Y132A/Q134A | this study | N/A |
| pet28a(+)-His6-HRV3C-AYWB_SAP05-S50A | this study | N/A |
| pet28a(+)-His6-HRV3C-AYWB_SAP05-H58A | this study | N/A |
| pGEX-6P-1-GST-HRV3C-AtRpn10_vWA-N34/L35A | this study | N/A |
| pGEX-6P-1-GST-HRV3C-AtRpn10_vWA-E31A | this study | N/A |
| pet28a(+)-His6-HRV3C-OY_SAP05-R43A | this study | N/A |
| pet28a(+)-His6-HRV3C-OY_SAP05-T48S50A | this study | N/A |
| pet28a(+)-His6-HRV3C-OY_SAP05-F58A | this study | N/A |
| pet28a(+)-His6-HRV3C-OY_SAP05-F61A | this study | N/A |
| pet28a(+)-His6-HRV3C-OY_SAP05-V45R | this study | N/A |
| pet28a(+)-His6-HRV3C-OY_SAP05-Q59P60A | this study | N/A |
| Software and algorithms | ||
| Origin 2023b | OriginLab Corporation | https://www.originlab.com/ |
| PHENIX | The Phenix Industrial Consortium | Phenix (phenix-online.org) |
| COOT | MRC Laboratory of Molecular Biology | Coot (cam.ac.uk) |
| Aimless | MRC Laboratory of Molecular Biology | https://www.ccp4.ac.uk/html/aimless.html |
| RCSB PDB | RCSB PDB Core Operations | https://www.rcsb.org/ |
| Image Lab | Bio-Rad Laboratories | https://www.bio-rad.com/zh-cn/product/image-lab-software?ID=KRE6P5E8Z |
| Biacore Insight Evaluation | Cytiva | https://www.cytivalifesciences.com.cn/zh/cn/support/software/biacore-downloads/Biacore-Insight-Evaluation-Software |
| GraphPad Prism | Dotmatics | https://www.graphpad-prism.cn/ |
| SnapGene | Dotmatics | https://www.snapgene.com/ |
| ASTRA 6.1 | Wyatt Technology | https://www.wyatt.com/products/software/astra.html |
| Biacore™ S200 Evaluation Software | Cytiva | https://www.cytivalifesciences.com.cn/zh/cn/support/software/biacore-downloads/Biacore-S200-Software |
Resource availability
Lead contact
For more information about experiments and reagents, please contact the lead contact, Qingyun Zheng (zheng_qy@sjtu.edu.cn).
Materials availability
Our work didn’t use any new reagents or materials.
Data and code availability
Instructions for section 1: Data
Crystal structures of the AYWB_SAP05-AtRpn10 and OY_SAP05-AtRpn10 complex have been published in the Protein Data Bank (PDB ID code 8JTK and 8JTL respectively).
Data reported in this paper will be shared by the lead contact upon request.
Instructions for section 2: Code
This paper does not report original code.
Instructions for section 3
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
Experimental model and study participant details
Escherichia coli Trans5α Chemically Competent Cells was used to confirm the target protein gene sequence and to construct mutants. These cells were plated on Luria-Bertani (LB) agar medium containing 1‰ antibiotics to screen for strains that had transformed the target gene. E. coli BL21(DE3) chemosynthesis cells were grown in LB liquid medium containing appropriate antibiotics at 37°C until OD600 ∼0.6. After cooling to 16°C, IPTG was added to induce the expression of target proteins.
Method details
Plasmids
DNA sequence of hRpn10vWA(1-193aa) was cloned from youbio PSMD4 (NM_002810) cDNA and inserted to pGEX-6p-1 vector by ClonExpress II One Step Cloning Kit (Vazyme). Other genes were synthesized and operated codon optimization for expression in E. coli by GenScript. These genes were prefixed with HRV 3C cleavage sites and cloned into the corresponding vectors between the NdeI and XhoI sites. The pet 28a (+) vector was selected for SAP05 and AtRpn10vWA used the pGEX-6p-1 vector.
Protein expression and purification
All plasmids were expressed in BL21(DE3) Chemically Competent Cell (Transgene). The antibiotic plate corresponding to the resistance gene on the vector was used to screen out the E. coli colonies importing the target gene, and the monoclonal colonies were picked and cultured in 5 mL of LB (containing 1‰ antibiotics) at 37°C overnight. The bacterial solution was transferred to 1 L LB (containing 1‰ antibiotics), and the culture was expanded in a shaker at 220 rpm 37°C for 4-5 h, until the OD600 reached 0.6-0.8. Cool the medium down to 16°C, add a final concentration of 0.4 mM IPTG into LB to induce expression of the target gene in E. coli, then adjust the shaker speed to 180 rpm and continue culture for 14-16 h.
Collect bacterial strain expressing His6-SAP05 by centrifugation and suspended in Lysis Buffer1 (20 mM HEPES pH 7.5, 150 mM NaCl, 5% glycerol, 20 mM imidazole). After sonication and ultracentrifugation, cell lysates were incubated with Ni-NTA Beads 6FF at 4°C for 1 hour. Beads were washed with Lysis buffer1 until the G250 dye did not turn blue, then changed to high salt buffer (20 mM HEPES pH 7.5, 500 mM NaCl) for 3–5 column volumes, after which they were reset to low salt buffer. The target proteins were eluted from the beads with 300 mM imidazole buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 5% glycerol, 300 mM imidazole), concentrated, and further purified by Superdex 75 Increase column (Cytiva) on a ӒKTA pure system to isolate the protein in monomeric form.
GST-Rpn10vWA was purified by glutathione affinity chromatography using Lysis Buffer 2 (20 mM HEPES pH 7.5, 150 mM NaCl, 5% glycerol, 2 mM DTT) and eluted with Lysis Buffer 2 GSH buffer containing 30 mM reduced glutathione. The elution was submitted to size exclusion chromatography using Superdex 200 Increase 10/300 GL (Cytiva). The high-purity fractions of protein samples were collected after being characterized by Coomassie-stained SDS-PAGE. Protein mutants were purified by the same method.
To gain the SAP05-Rpn10vWA complex used for crystal, two types of protein elution were mixed at a molar ratio of 1.2 to 1 according to concentrations determined via a Nanodrop. After adding one percent HRV 3C protease, protein solutions were dialyzed to enzyme digestion buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 5% glycerol, 2 mM DTT) overnight. Protein solutions were incubated with glutathione beads to remove the GST tag, and the beads were washed with dialysis buffer until the G250 dye did not turn blue. Flow-through together with washing solution was diluted with Buffer A (20 mM HEPES pH 8.5) and loaded onto anion exchanger Source 15Q chromatography and eluted at around 14 mS/cm conductance by gradient against Buffer B (20 mM HEPES pH 8.5, 1 M NaCl). Peak fractions eluted at around 14 mS/cm conductance were collected and characterized by 12.5% SDS-PAGE (epizyme PAGE Gel Fast Preparation Kit) for the removal of GST-mixed complexes. Then the sample was concentrated and loaded onto Superdex 75 Increase 10/300 GL or HiLoad 16/600 Superdex 75 pg (Cytiva) equilibrated with SPR buffer (20 mM HEPES pH 7.5, 150 mM NaCl). Conduct SDS–PAGE analysis to determine the fraction of protein complex with high purity, and concentrated to about 20 mg/ml to obtain the sample for crystallization.
Crystallization, data collection and processing
20 mg/mL AYWB_SAP05-AtRpn10 or OY_SAP05-AtRpn10 complex was used for crystal initial screening with sitting-drop vapor diffusion at 291K. Each drop contained 0.5 μL protein complex solution and 0.5 μL crystallization reservoir buffer. Satisfying crystals of the AYWB_SAP05-AtRpn10 complex grew under a reservoir solution containing 0.2 M Sodium malonate pH 5.0, 20% w/v Polyethylene glycol 3,350, while the crystals of OY_SAP05-AtRpn10 complex grew under 0.1M MES pH 6.5, 15% w/v PEG 6000, 5% v/v MPD. Crystals were cryo-protected in the solution with 17 % glycerol in the respective reservoir solution.
Both the diffraction of the AYWB_SAP05-AtRpn10 and OY_SAP05-AtRpn10 complex crystal was collected under the temperature of 100 K and wavelength of 0.979183 Å at the SSRF BEAMLINE BL10U2 of Shanghai Synchrotron Radiation Facility. Raw data was auto-processed and scaled with Aimless 0.7.7.28 We used the Alphafold2-predicted model of AYWB_SAP05 (AF-Q2NK94-F1), OY_SAP05 (AF-Q6YQ57-F1), and AtRpn10 (AF-P55034-F1) for molecular replacement phasing in PHENIX 1.19.2_4158.29 The model building was visualized and operated by COOT.30 Crystallographic data collection and refinement statistics are presented in Table 1.
Table 1.
Crystallographic data collection and refinement statistics
| AYWB_SAP05-AtRpn10c (PDB 8JTK) | OY_SAP05-AtRpn10c (PDB 8JTL) | |
|---|---|---|
| Data collection | ||
| Space group | P 31 2 1 | P 2 |
| Cell dimensions | ||
| a, b, c (Å) | 49.24, 49.24, 215.154 | 52.428, 39.386, 121.952 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 93.781, 90.0 |
| Resolution range (Å) | 71.72–1.57 (1.65–1.57)a | 31.08–1.69 (1.73–1.69)a |
| No. of unique reflections | 42242 | 55704 |
| Rmerge | 0.056 (0.768)a | 0.169 (1.773)a |
| Rpim | 0.021 (0.314)a | 0.074 (0.763)a |
| Rmeas | 0.060 (0.831)a | 0.185 (1.936) |
| I/σ(I) | 18.0 (2.6)a | 5.0 (1.0)a |
| CC1/2 | 0.999 | 0.827 |
| Completeness (%) | 95.9 (100)a | 99.2 (98.6)a |
| Redundancy | 8.2 (6.9)a | 6.4 (6.4)a |
| Refinement | ||
| Resolution (Å) | 24.93–1.57 | 25.35–1.78 |
| No. reflections | 42142 | 47533 |
| Rwork/Rfree (%) | 18.70/21.54 | 18.40/21.85 |
| No. of non-hydrogen atoms | 2522 | 5062 |
| Protein | 2322 | 4652 |
| Ligand/ion | 0 | 0 |
| Water | 200 | 410 |
| B factors (Å2) | 38.0 (Average) | 35.0 (Average) |
| Proteinb | 33b | 31b |
| Solvent | 44.7 | 38.4 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.008 | 0.014 |
| Bond angles (°) | 1.033 | 1.356 |
| Clashscore | 4.0 | 10.0 |
Values in parentheses are for the highest-resolution shell.
The median values of the occupancy-weighted average B-factor per residue.
The data are collected from one crystal.
Surface plasmon resonance
SPR assay was carried out on Biacore S200 (Cytiva). At first, the ligand was diluted to 5 μg/mL by 10 mM sodium acetate (pH 4.5) and conjugated to Series S Sensor Chip CM5 (Cytiva) using the amide couple kit until the response value approached to setting level. Then use ethanolamine to block the remaining reactive sites in the channel. To continue, the analytes are diluted by SPR Buffer into a series of concentration gradients and run through the ligand binding cell one by one for Kinetics/Affinity testing. Eventually, we used the Biacore S200 Evaluation System to fit the curve and calculate the dissociation equilibrium constant.
Pull down assay
50 μg of GST-AtRpn10/hRpn10 was incubated with glutathione resin pre-equilibrated with SPR buffer (20 mM HEPES pH 7.5, 150 mM NaCl) for 2 h at 16°C. Washing beads until the G250 dye did not turn blue. And the resin was incubated with 40 μg OY_SAP05 for 2 h. Take one-tenth beads to the loading buffer after washing the unbinding protein. Then the samples were boiled at 95°C for 15 min and run on 4-20% polyacrylamide gels (GenScript).
Size exclusion chromatography coupled with multiangle light scattering (SEC-MALS) analysis
100 μL of AYWB_SAP05-AtRpn10 complex at 2.5mg/mL was injected to the loop of ӒKTA pure system connected with Superdex 75 Increase 10/300 GL pre-equilibrated with SPR buffer (20 mM HEPES pH 7.5, 150 mM NaCl). Set the system parameter by ASTRA 6.1. For experiment from method at ‘UV+LS+RI Online’ model, flow rate at 0.5 mL/min and 35min duration. Plot by elution time with calculated Malor Mass and differential refractive index on Origin.
Quantification and statistical analysis
Prior to SPR analysis, the proteins used were concentration-corrected by adjusting the volume of the target protein bands in SDS-PAGE as reported by Imagelab software. Under the condition that the concentration of the wild-type protein was used as a reference, the other mutant proteins were quantified relatively based on the fold relationship of the adjusted volume relative to the WT group. A part of SPR data (Figures 1C and S5I) from the Biacore 8K+ test was processed with the Biacore Insight Evaluation, choosing steady state affinity for the general affinity model and 1:1 binding for the kinetic model. Other SPR binding tests were performed using Biacore™ S200 Evaluation Software to fit the data and output standard deviations.
Acknowledgments
Qingyun Zheng thank the “2030” Project of Shanghai Jiao Tong University. Qian Qu thanks National Natural Science Foundation of China (23Z033004247). We thank the Tsinghua University Branch of China National Protein Science Facility, Tsinghua University, Beijing, for the device support of crystal screening (mosquito, sptlabtech) and checking (XtaLAB FR-X, Rigaku) and the guidance about the crystallization experiment from Min Li (National Protein Science Facility, Tsinghua University). The data collection was supported by the Shanghai Synchrotron Radiation Facility. We thank the Center of Pharmaceutical Technology, Tsinghua University for support of our binding assay and the experimental guidance from Ting Wang (enter of Pharmaceutical Technology, Tsinghua University).
Author contributions
L.Z., Y.D., Q.Z., Q.Q. designed research; L.Z., Y.D. performed research; All authors wrote and edited the paper.
Declaration of interests
The authors declare no competing interests.
Published: January 12, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.108892.
Contributor Information
Qian Qu, Email: quqian22@sjtu.edu.cn.
Qingyun Zheng, Email: zheng_qy@sjtu.edu.cn.
Supplemental information
References
- 1.Goldberg A.L. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 2.Sorokin A.V., Kim E.R., Ovchinnikov L.P. Proteasome system of protein degradation and processing. Biochemistry. 2009;74:1411–1442. doi: 10.1134/s000629790913001x. [DOI] [PubMed] [Google Scholar]
- 3.Du T., Song Y., Ray A., Wan X., Yao Y., Samur M.K., Shen C., Penailillo J., Sewastianik T., Tai Y.-T., et al. Ubiquitin receptor PSMD4/Rpn10 is a novel therapeutic target in multiple myeloma. Blood. 2023;141:2599–2614. doi: 10.1182/blood.2022017897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang T.X., Zhao M., Qiu X.B. Substrate receptors of proteasomes. Biol. Rev. Camb. Philos. Soc. 2018;93:1765–1777. doi: 10.1111/brv.12419. [DOI] [PubMed] [Google Scholar]
- 5.Chirnomas D., Hornberger K.R., Crews C.M. Protein degraders enter the clinic - a new approach to cancer therapy. Nat. Rev. Clin. Oncol. 2023;20:265–278. doi: 10.1038/s41571-023-00736-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pohl C., Dikic I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science. 2019;366:818–822. doi: 10.1126/science.aax3769. [DOI] [PubMed] [Google Scholar]
- 7.Craxton A., Butterworth M., Harper N., Fairall L., Schwabe J., Ciechanover A., Cohen G.M. NOXA, a sensor of proteasome integrity, is degraded by 26S proteasomes by an ubiquitin-independent pathway that is blocked by MCL-1. Cell Death Differ. 2012;19:1424–1434. doi: 10.1038/cdd.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orlowski M., Wilk S. Ubiquitin-independent proteolytic functions of the proteasome. Arch. Biochem. Biophys. 2003;415:1–5. doi: 10.1016/s0003-9861(03)00197-8. [DOI] [PubMed] [Google Scholar]
- 9.Sheaff R.J., Singer J.D., Swanger J., Smitherman M., Roberts J.M., Clurman B.E. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol. Cell. 2000;5:403–410. doi: 10.1016/s1097-2765(00)80435-9. [DOI] [PubMed] [Google Scholar]
- 10.Tsvetkov P., Reuven N., Shaul Y. Ubiquitin-independent p53 proteasomal degradation. Cell Death Differ. 2010;17:103–108. doi: 10.1038/cdd.2009.67. [DOI] [PubMed] [Google Scholar]
- 11.Makaros Y., Raiff A., Timms R.T., Wagh A.R., Gueta M.I., Bekturova A., Guez-Haddad J., Brodsky S., Opatowsky Y., Glickman M.H., et al. Ubiquitin-independent proteasomal degradation driven by C-degron pathways. Mol. Cell. 2023;83:1921–1935.e7. doi: 10.1016/j.molcel.2023.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu X., Nardone C., Kamitaki N., Mao A., Elledge S.J., Greenberg M.E. The midnolin-proteasome pathway catches proteins for ubiquitination-independent degradation. Science. 2023;381:eadh5021. doi: 10.1126/science.adh5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang W., MacLean A.M., Sugio A., Maqbool A., Busscher M., Cho S.T., Kamoun S., Kuo C.H., Immink R.G.H., Hogenhout S.A. Parasitic modulation of host development by ubiquitin-independent protein degradation. Cell. 2021;184:5201–5214.e12. doi: 10.1016/j.cell.2021.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schrader J., Henneberg F., Mata R.A., Tittmann K., Schneider T.R., Stark H., Bourenkov G., Chari A. The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design. Science. 2016;353:594–598. doi: 10.1126/science.aaf8993. [DOI] [PubMed] [Google Scholar]
- 15.Kandolf S., Grishkovskaya I., Belačić K., Bolhuis D.L., Amann S., Foster B., Imre R., Mechtler K., Schleiffer A., Tagare H.D., et al. Cryo-EM structure of the plant 26S proteasome. Plant Commun. 2022;3 doi: 10.1016/j.xplc.2022.100310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao J., Makhija S., Zhou C., Zhang H., Wang Y., Muralidharan M., Huang B., Cheng Y. Structural insights into the human PA28-20S proteasome enabled by efficient tagging and purification of endogenous proteins. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2207200119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitazawa Y., Iwabuchi N., Maejima K., Sasano M., Matsumoto O., Koinuma H., Tokuda R., Suzuki M., Oshima K., Namba S., Yamaji Y. A phytoplasma effector acts as a ubiquitin-like mediator between floral MADS-box proteins and proteasome shuttle proteins. Plant Cell. 2022;34:1709–1723. doi: 10.1093/plcell/koac062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacLean A.M., Orlovskis Z., Kowitwanich K., Zdziarska A.M., Angenent G.C., Immink R.G.H., Hogenhout S.A. Phytoplasma effector SAP54 hijacks plant reproduction by degrading MADS-box proteins and promotes insect colonization in a RAD23-dependent manner. PLoS Biol. 2014;12 doi: 10.1371/journal.pbio.1001835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacLean A.M., Sugio A., Makarova O.V., Findlay K.C., Grieve V.M., Tóth R., Nicolaisen M., Hogenhout S.A. Phytoplasma effector SAP54 induces indeterminate leaf-like flower development in Arabidopsis plants. Plant Physiol. 2011;157:831–841. doi: 10.1104/pp.111.181586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rümpler F., Gramzow L., Theißen G., Melzer R. Did Convergent Protein Evolution Enable Phytoplasmas to Generate 'Zombie Plants. Trends Plant Sci. 2015;20:798–806. doi: 10.1016/j.tplants.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 21.Madeline B., Weijun G., Isuru M.J., Bin-Bin S., Joel T., Thomas K. Chemically Induced Degradation of Native Proteins by Direct Recruitment to the 26S Proteasome. bioRxiv. 2023 doi: 10.1101/2023.07.19.549534. [DOI] [Google Scholar]
- 22.Bashore C., Prakash S., Johnson M.C., Conrad R.J., Kekessie I.A., Scales S.J., Ishisoko N., Kleinheinz T., Liu P.S., Popovych N., et al. Targeted degradation via direct 26S proteasome recruitment. Nat. Chem. Biol. 2023;19:55–63. doi: 10.1038/s41589-022-01218-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marshall R.S., Li F., Gemperline D.C., Book A.J., Vierstra R.D. Autophagic Degradation of the 26S Proteasome Is Mediated by the Dual ATG8/Ubiquitin Receptor RPN10 in Arabidopsis. Mol. Cell. 2015;58:1053–1066. doi: 10.1016/j.molcel.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rani N., Aichem A., Schmidtke G., Kreft S.G., Groettrup M. FAT10 and NUB1L bind to the VWA domain of Rpn10 and Rpn1 to enable proteasome-mediated proteolysis. Nat. Commun. 2012;3:749. doi: 10.1038/ncomms1752. [DOI] [PubMed] [Google Scholar]
- 25.Dong Y., Zhang S., Wu Z., Li X., Wang W.L., Zhu Y., Stoilova-McPhie S., Lu Y., Finley D., Mao Y. Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature. 2019;565:49–55. doi: 10.1038/s41586-018-0736-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baugh J.M., Viktorova E.G., Pilipenko E.V. Proteasomes Can Degrade a Significant Proportion of Cellular Proteins Independent of Ubiquitination. J. Mol. Biol. 2009;386:814–827. doi: 10.1016/j.jmb.2008.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Q., Maqbool A., Mirkin F.G., Singh Y., Stevenson C.E.M., Lawson D.M., Kamoun S., Huang W., Hogenhout S.A. Bimodular architecture of bacterial effector SAP05 drives ubiquitin-independent targeted protein degradation. bioRxiv. 2023;2023 doi: 10.1073/pnas.2310664120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr. D. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liebschner D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J., et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Instructions for section 1: Data
Crystal structures of the AYWB_SAP05-AtRpn10 and OY_SAP05-AtRpn10 complex have been published in the Protein Data Bank (PDB ID code 8JTK and 8JTL respectively).
Data reported in this paper will be shared by the lead contact upon request.
Instructions for section 2: Code
This paper does not report original code.
Instructions for section 3
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.