Abstract
Objective
Nonalcoholic fatty liver disease (NAFLD) is an emerging public health threat as the most common chronic liver disease worldwide. However, there remains no effective medication to improve NAFLD. G protein-coupled receptors (GPCRs) are the most frequently investigated drug targets family. The Regulator of G protein signaling 14 (RGS14), as an essential negative modulator of GPCR signaling, plays important regulatory roles in liver damage and inflammatory responses. However, the role of RGS14 in NAFLD remains largely unclear.
Methods and results
In this study, we found that RGS14 was decreased in hepatocytes in NAFLD individuals in a public database. We employed genetic engineering technique to explore the function of RGS14 in NAFLD. We demonstrated that RGS14 overexpression ameliorated lipid accumulation, inflammatory response and liver fibrosis in hepatocytes in vivo and in vitro. Whereas, hepatocyte specific Rgs14-knockout (Rgs14-HKO) exacerbated high fat high cholesterol diet (HFHC) induced NASH. Further molecular experiments demonstrated that RGS14 depended on GDI activity to attenuate HFHC-feeding NASH. More importantly, RGS14 interacted with Guanine nucleotide-binding protein (Gi) alpha 1 and 3 (Giα1/3, gene named GNAI1/3), promoting the generation of cAMP and then activating the subsequent AMPK pathways. GNAI1/3 knockdown abolished the protective role of RGS14, indicating that RGS14 binding to Giα1/3 was required for prevention against hepatic steatosis.
Conclusions
RGS14 plays a protective role in the progression of NAFLD. RGS14-Giα1/3 interaction accelerated the production of cAMP and then activated cAMP-AMPK signaling. Targeting RGS14 or modulating the RGS14-Giα1/3 interaction may be a potential strategy for the treatment of NAFLD in the future.
Keywords: Lipotoxicity, Lipid accumulation, Inflammation, Steatosis, GDI, GPCR
Highlights
-
•
Hepatocyte RGS14 protects against hepatic steatosis during the progression of NAFLD.
-
•
RGS14 depends on GDI activity to ameliorate HFHC-induced NASH.
-
•
RGS14-Giα1/3 interaction promoted cAMP levels, then activated AMPK signaling pathway.
-
•
Targeting RGS14 or modulating the RGS14-Giα1/3 interaction may be a potential strategy to protect against NAFLD.
List of abbreviations
- AAV
Adeno-associated virus
- ACC
Acetyl-CoA carboxylase
- Ad
Adenovirus
- ALT
Alanine aminotransferase
- AST
Aspartate aminotransferase
- AMPK
AMP-activated protein kinase
- cAMP
Cyclic adenosine monophosphate
- BSA
Bovine serum albumin
- CD36
Cluster of differentiation 36
- CPT1a
Carnitine palmitoyltransferase 1 alpha
- CXCL10
Chemokine (C-X-C motif) ligand 10
- DGAT2
Diacylglycerol acyltransferase 2
- GAP
GTPase-activating protein
- GDI
GDP dissociation inhibitor
- GPCRs
G protein-coupled receptors
- GPAT1
Glycerol 3-phosphate acyltransferase 1
- Giα1
Guanine nucleotide-binding protein (Gi) alpha 1
- Giα3
Guanine nucleotide-binding protein (Gi) alpha 3
- HKO
Hepatocyte-specific knockout
- H&E
Hematoxylin and eosin
- HFHC
High fat high cholesterol diet
- IL1b
Interleukin 1 beta
- IL6
Interleukin 6
- NAFLD
Nonalcoholic fatty liver disease
- NASH
Nonalcoholic steatohepatitis
- NEFA
Nonestesterified fatty acid
- p-
Phosphorylated-
- PO
Plmitic acid and Oleic acid mix
- PSR
Picrate Sirius Red staining
- RBSs
Ras/Rap-binding domains
- RGS14
Regulator of G protein signaling 14
- SCD1
Stearoyl-CoA desaturase 1
- SREBP1C
Sterol regulatory element-binding proteins 1c
- TG
Triglyceride
- TC
Total cholesterol
- TNF
Tumor necrosis factor
- UCP2
Uncoupling protein 2
1. Introduction
Nonalcoholic fatty liver disease (NAFLD) is an increasingly serious health burden afflicting more than 25 % of the general population [1,2]. The dramatic increase in NAFLD prevalence and growing unmet need for health care have spurred researchers to actively explore drug targets for the treatment of NAFLD [3]. NAFLD encompasses a continuum of progressive liver abnormalities, from simple steatosis to nonalcoholic steatohepatitis (NASH) to cirrhosis [4]. Ten to twenty percent of NAFLD patients can progress to NASH, and approximately 10–15% of NASH patients progress to cirrhosis, which significantly increases the incidence of hepatocellular carcinoma [5]. To date, no effective agents have been approved for NAFLD treatment, in addition to promoting a healthy lifestyle and weight loss [6]. Therefore, it exists unmet challenges in exploring effective therapeutic targets for NAFLD.
Membrane receptors and their downstream signaling pathways modulate the functions of multicellular organelles. The largest class of membrane receptors comprises G protein-coupled receptors (GPCRs) and their downstream intracellular second messengers, such as cAMP and calcium [7,8]. GPCRs are the most frequently investigated drug targets family, which transduce chemical signals from the extracellular matrix into the cell and activate intracellular signaling [9]. RGS14 is a structurally and functionally unique subunit of the RGS family that acts as an essential negative modulator of GPCRs signaling. RGS14 contains a common RGS domain, a GoLoco/GPR motif, and Ras/Rap-binding domains (RBDs) [10,11]. GoLoco/GPR domain forms a stable complex with the inactive form of Giα1/3 (gene named GNAI1/3) and has GDP dissociation inhibitor (GDI) activity [[12], [13], [14]]. Previous studies have largely centered on the regulatory functions of RGS14 on hippocampal signaling and synaptic plasticity [15], cellular stress resistance, liver damage and inflammatory responses [16]. However, the functions of RGS14 in NAFLD have not been characterized.
In the present study, we found that RGS14 expression was significantly decreased in hepatocytes from NAFLD patients in a public database. We generated hepatocyte-specific Rgs14 knockout (Rgs14-HKO) and Rgs14-overexpressing mice in vivo to evaluate the potential roles and underlying mechanisms of RGS14 in NAFLD.
2. Materials and methods
2.1. Human liver samples
Human liver samples were obtained from individuals who had undergone liver biopsy, liver surgery or liver transplantation. Informed consent forms were signed by all donors or their families. Individuals were excluded due to viral infection (e.g., hepatitis B and C virus), autoimmune hepatitis, excessive alcohol consumption (>140 g for men or >70 g for women, per week), and drug or toxin injury. All study protocols involving human samples were approved by the Ethics Committee of Zhengzhou University People's Hospital. All procedures adhered to the principles of the Declaration of Helsinki.
2.2. Mice and treatment
All experiments including animals in our study were reviewed and approved by the Animal Experimentation Ethics Committee of Zhengzhou University and all procedures were conducted following the guidelines of the Institutional Animal Care and Use Committees of Zhengzhou University. The animal model of NASH was established by feeding high fat high cholesterol (HFHC, caloric distribution: 14 % from protein, 42 % from fat, and 44 % from carbohydrates; containing 2 % cholesterol; Trophic, Nantong, China) diet for 16 weeks. Mice that were fed a normal chow diet (NC, caloric distribution: 20.6 % from protein, 12.0 % from fat and 67.4 % from carbohydrates; Trophic) were used as normal controls.
To generate 8 weeks old hepatocyte-specific Rgs14 knockout (Rgs14-HKO) male mice, Rosa26-CAG-LSL-Cas9 knock-in male mice (Strain NO. T002249, Gempharmatech, China) were intravenously injected with AAV8-U6-sgRgs14#1-U6-sgRgs14#2-U6-sgRgs14#3-TBG-Cre adeno-associated virus (7.5x10ˆ11particles per mouse). And Rosa26-CAG-LSL-Cas9 knock-in male mice treated with AAV8-TBG-Cre Adeno-associated virus were used as normal controls. A non-viral genetic modification method based on Sleeping Beauty (SB) transposon system [17] was used to deliver with the plasmid to express Rgs14 and Rgs14 (M) (with amino acid 518 mutated from Glutamine to Glycine) in the mice liver. In brief, 8 weeks old male mice were injected with a DNA solution containing 2 ug of SB100 transposase plasmid (pCMV-SB100) together with 50 ug of pT3-Flag-Rgs14 or pT3-Flag-Rgs14 (M) via the tail vein at a volume of 10 % mouse weight within 10 s. Male mice in the blank control group were injected with the “no transposase” plasmid. The mice were housed in a specific pathogen-free environment at 22–24 °C under a 12-h light/12-h dark cycle. Primers were listed in Table 1.
Table 1.
Primers for plasmid construction.
| Gene | Vector | Primer sequence (5′-3′) |
|---|---|---|
| 1.1 Primer sequences for overexpression plasmids | ||
| RGS14-human | phage-Flag | F: TCGGGTTTAAACGGATCCATGCCAGGGAAGCC CAAGC |
| R: GGGCCCTCTAGACTCGAGTCAGAGGGCTGAG TCGGTG | ||
| GNAI1-human | phage-HA | F: TCGGGTTTAAACGGATCCATGGGCTGCACGCT GAGCGC |
| R: GGGCCCTCTAGACTCGAGTTAAAAGAGACCA CAATCTT | ||
| GNAI3-human | phage-HA | F: TCGGGTTTAAACGGATCCATGGGCTGCACGTT GAGCGC |
| R: GGGCCCTCTAGACTCGAGTCAATAAAGTCCA CATTCCT | ||
| RGS14(1-202aa)-human | phage-Flag | F: TCGGGTTTAAACGGATCCATGCCAGGGAAGCC CAAGC |
| R: GGGCCCTCTAGACTCGAGTCAGCCGAGGCGC GAGGAGCCA | ||
| RGS14(1-373aa)-human | phage-Flag | F: TCGGGTTTAAACGGATCCATGCCAGGGAAGCC CAAGC |
| R: GGGCCCTCTAGACTCGAGTCAGATCCTGTTTT CCAGCCGC | ||
| RGS14(203-489aa)-human | phage-Flag | F: TCGGGTTTAAACGGATCCAGCCCTGACGCCAC GAGGA |
| R: GGGCCCTCTAGACTCGAGTCAGGCACTACTG GGCACCTTC | ||
| RGS14(374-566aa)-human | phage-Flag | F: TCGGGTTTAAACGGATCCACCTTCGAGCTGGA GCTGA |
| R: GGGCCCTCTAGACTCGAGTCAGAGGGCTGAG TCGGTG | ||
| RGS14(E92A/N93A)-human | phage-Flag | F: GCCTTCCAGAAAGTCACGGCTGCCGCGCTGAA CTCCTTCT |
| R: AGAAGGAGTTCAGCGCGGCAGCCGTGACTTT CTGGAAGGC | ||
| RGS14(R335L)-human | phage-Flag | F: GGTAGAGAGAGGCCTAGTTTCTCACAGATCCC |
| R: GGGATCTGTGAGAAACTAGGCCTCTCTCTACC | ||
| RGS14(Q516A)-human | phage-Flag | F: CAGAAGGCCCCTCGCGTCGTGGGCCCCG |
| R: CGGGGCCCACGACGCGAGGGGCCTTCTG | ||
| Rgs14-mice | pENTRY-CMV-Flag | F: GGCTAGCGATATCGGATCCGCCACCATGCCAG GGAAGCCCAAGC |
| R: CGTCCTTGTAATCACTAGTTGGTGGAGCCTCC CGAGAGC | ||
| Rgs14(E92A/N93A)-mice | pENTRY-CMV-Flag | F: GAAGAAGGAATTCAGCGCAGCGGCCGTAACT TTCTGGAAAGCCT |
| R: CAGGCTTTCCAGAAAGTTACGGCCGCTGCGCT GAATTCCTTCTTC | ||
| Rgs14(R336L)-mice | pENTRY-CMV-Flag | F: CAGGCAGAGAGAGGCCTAACTTCTCACAGAT GCCTGT |
| R: CAGGCATCTGTGAGAAGTTAGGCCTCTCTCTG CCTG | ||
| Rgs14(Q518A)-mice | pENTRY-CMV-Flag | F: GAAGAAGTCCTCTCGCATCGTGGGCCCCGCT |
| R: AGCGGGGCCCACGATGCGAGAGGACTTCTTC | ||
| Gnai1-mice | pENTRY-CMV-Flag | F: GCTAGCGATATCGGATCCGCCACCATGGGCTG CACATTGAGCGC |
| R: GTCCTTGTAATCACTAGTGAAGAGACCACAGT CTTTTAG | ||
| Gnai3-mice | pENTRY-CMV-Flag | F: GCTAGCGATATCGGATCCGCCACCATGGGCTG CACGTTGAGCGCC |
| R: GTCCTTGTAATCACTAGTATAAAGCCCACATT CCTTTAA | ||
| GNAI1-human | pET-21b | F: GCAAATGGGTCGGGATCCATGGGCTGCACGC TGAGCGC |
| R: GAGTGCGGCCGCAAGCTTAAAGAGACCACAA TCTTTT | ||
| GNAI3-human | pET-21b | F: CAAATGGGTCGGGATCCATGGGCTGCACGTTG AGCG |
| R: GAGTGCGGCCGCAAGCTTATAAAGTCCACATT CCTTT | ||
| RGS14-human | pGEX-6p-1 | F: CAGGGGCCCCTGGGATCCATGCCAGGGAAGC CCAAGC |
| R: ACGATGCGGCCGCTCGAGTCAGAGGGCTGAG TCGGTG | ||
| Rgs14-mice | pT3-Flag | F: ACGCGTTTAAACGGATCCATGCCAGGGAAGC CCAAGCA |
| R: TCTGCGGCCGCACTAGTCTATGGTGGAGCCTC CCGAG | ||
| Rgs14(Q518A)-mice | pT3-Flag | F: GAAGAAGTCCTCTCGCATCGTGGGCCCCGCT |
| R: AGCGGGGCCCACGATGCGAGAGGACTTCTTC | ||
| Prime name | Species | Sequence (5′-3′) |
| 1.2 sgRNA or shRNA target sequences for adeno-associated virus8 (AAV8) and Adenovirus (Ad) construction | ||
| sgRgs14#1 | Mice | TGAAGTAAGCCAGACCCCGT |
| sgRgs14#2 | Mice | GATCTGCTGGAAACGTTCGC |
| sgRgs14#3 | Mice | CCACGAGTTCCTATCCAGCC |
| ShRgs14#1 | Mice | GAAGATGCCAGTAGTTCTATT |
| ShRgs14#2 | Mice | GCAGCTTCAGATCTTCAATTT |
| ShGnai1#1 | Mice | GAAGGACCTCTTCGAAGAATT |
| ShGnai1#2 | Mice | GAATAGCACAGCCAAATTATT |
| ShGnai3#1 | Mice | GTAGTTGTCTACAGCAATATT |
| ShGnai3#2 | Mice | CAATCTGTTATCCAGAATATT |
2.3. Serum metabolites and liver function assays
Serum triglyceride (TG), total cholesterol (TC), serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were measured using an Thermo Scientific™ Indiko™ Clinical Chemistry Analyzer (Thermo Fisher Scientific, Waltham, USA) according to the manufacturer's instructions.
2.4. Histopathology analysis
Liver samples were embedded in paraffin, sectioned and stained with hematoxylin and eosin (H&E) to analyze lipid accumulation and inflammatory cell infiltration. Lipid droplets in the liver were visualized using Oil Red O (0625, Sigma, CT, USA) staining of OCT-embedded frozen liver tissues. Liver fibrosis was analyzed by Picrate Sirius Red staining (PSR) in paraffin sections of liver tissues. Immunohistochemical staining was used to analyze the expression of Rgs14 and CD11b in liver tissue. The histological features of the tissues were observed and imaged using a light microscope.
2.5. Cell lines and primary hepatocytes
Human cell lines (HEK293T cells, HEK293A cells and L02 hepatocytes) were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China), and cultured in DMEM with 10 % fetal bovine serum and 1 % penicillin-streptomycin.
Primary mouse hepatocytes were isolated from 6 to 8 weeks C57BL/6J male mice via collagenase type IV perfusion protocol [17]. In Brief, mice were anesthetized and fixed, and the liver was digested by perfusion of the solution containing 0.5 % collagenase type IV (17104-019, Thermo Fisher Scientific) through the inferior vena cava. Then take the digested live out and minced gently in cold DMEM medium, and filtered through nylon Cell Strainer (352350, BD Biosciences). The filtered solution was Centrifuge 500 rpm for 1 min at 4 °C to collect hepatocytes. Cell viability and density were determined by trypan blue. Primary hepatocytes were cultured in DMEM supplemented with 10 % FBS and 1 % penicillin-streptomycin. The L02 cells and Primary hepatocytes were stimulated with PO mixture (0.5 mM palmitic acid, P0500, Sigma Aldrich, St Louis, MO, USA and 1.0 mM oleic acid, O108485, Aladdin, China), then hepatocytes were harvested.
2.6. Nile red staining
Lipid accumulation in hepatocytes was measured by Nile red staining. In brief, cells were fixed with 4 % paraformaldehyde for 30 min after challenged with PO (0.5 mM PA and 1.0 mM OA) for 24 h. Cells were then incubated with Nile red (N121291, Aladdin, China) at room temperature for 10 min. Nuclei were visualized with DAPI. Images of lipid accumulation were acquired by a confocal laser scanning microscope (TCS SP8X, Leica).
2.7. Hepatic and cellular lipid detection
The hepatocytes or liver tissues lipid (TG, TC and nonestesterified fatty acid (NEFA)) were extracted as described [18]. In brief, the liver tissues or hepatocytes were homogenized and broken by adding an appropriate amount of PBS, and then chloroform: methanol (2:1) solution was added to extract triglyceride. Lipid levels were assayed using a commercial kit (290-63701 for TG, 294-65801 for TC, 294-63601 for NEFA, Wako, Tokyo, Japan) according to the manufacturer's instructions.
2.8. Plasmid constructs and adenovirus packaging
Plasmids encoding full-length, mutants and truncated human RGS14, GNAI1and GNAI3 were constructed by cloning the indicated coding regions of human RGS14, GNAI1and GNAI3 cDNA into phage-Flag or phage-HA vectors. The full-length, mutants and truncated mice Rgs14, Gnai1and Gnai3 were inserted into pENTRY-CMV-Flag vectors; The ShRgs14, ShGnai1and ShGnai3 sequences were synthesized (Sangon Biotech, shanghai, China) and inserted into the pEntry-U6 vectors, then subjected homologous recombination to the pAD/PL-DEST vectors. After digested with PacI restriction enzyme, the linearized fragments were transfected into 293A cells for packaging and amplification to obtain the relevant adenovirus. Adenoviruses were constructed and used to infect murine primary hepatocytes. Primers were listed in Table 1.
2.9. Cellular and hepatic tissue cAMP assay
Cellular cAMP assay was performed as previously described [19]. In brief, cultured primary hepatocytes (5 × 106) were collected and resuspended with 200 μL PBS, then hepatocytes were broken using an ultrasound crusher and were centrifuged at 3000g for 15 min at 4 °C. For hepatic tissue, the samples were accurately weighted, added with 10X volume of PBS, homogenized on ice using a Tissue Homogenizer, and then centrifuged at 3000g for 15 min at 4 °C. The cellular and hepatic tissue supernatants were measured for cyclic adenosine monophosphate (cAMP) levels using the cAMP ELISA Kit (Elabscience, Bethesda, USA) according to the manufacturer's instructions.
2.10. RNA isolation and qPCR
Total RNA from hepatocytes and liver tissues was extracted using with RNAiso Plus (9108; TAKARA; Japan) according to the reagent instructions. Then the extracted RNA was reverse transcribed into complementary DNA (cDNA) using the HiScript III RT SuperMix for qPCR kit (R323-01, Vazyme, Nanjing, China). The amplification products were quantified by Taq Pro Universal SYBR qPCR Master Mix (Q712-02, Vazyme, Nanjing, China) and analyzed on ABI QuantStudio5 Q5 (ABI, Carlsbad, USA). The targeting gene expression was normalized to the housekeeping gene -Actin. The Primers for qPCR in this study are listed in Table 2.
Table 2.
The primers for qPCR.
| Gene | Species | Primer sequence (5′-3′) |
|---|---|---|
| Rgs14 | Mice | F: GGCTTACTTCACTGAGTTCCTG |
| R: ACTCGTGGTAGATGTTGTGGG | ||
| Tnfa | Mice | F: GGTGATCGGTCCCCAAAGGGATGA |
| R: TGGTTTGCTACGACGTGGGCT | ||
| Il6 | Mice | F: TCTGCAAGAGACTTCCATCCAGTTGC |
| R: AGCCTCCGACTTGTGAAGTGGT | ||
| Il1b | Mice | F: CGGACCCCAAAAGATGAAGGGCTG |
| R: AGCTGCCACAGCTTCTCCACA | ||
| Mcp1 | Mice | F: AACGCCCCACTCACCTGCTG |
| R: GCTTCTTTGGGACACCTGCTGCT | ||
| Cxcl10 | Mice | F: ATCATCCCTGCGAGCCTATCCT |
| R: GACCTTTTTTGGCTAAACGCTTTC | ||
| Cd36 | Mice | F: AGATGACGTGGCAAAGAACAG |
| R: CCTTGGCTAGATAACGAACTCTG | ||
| Fatp5 | Mice | F: TTTCTGGGGTTGGCCAAGTT |
| R: CTCCTGGAGGTCTGGATCCA | ||
| Acc1 | Mice | F: AATGAACGTGCAATCCGATTTG |
| R: ACTCCACATTTGCGTAATTGTTG | ||
| Fasn | Mice | F: AAGTTGCCCGAGTCAGAGAA |
| R: CGTCGAACTTGGAGAGATCC | ||
| Scd1 | Mice | F: CAAACACCCGGCTGTCAAAG |
| R: TGAAGCACATCAGCAGGAGG | ||
| Gpat1 | Mice | F: ATGAAACGCACACAAGGCAC |
| R: TTGCCTCTTGGACTCTGCTG | ||
| Dgat2 | Mice | F: GCGCTACTTCCGAGACTACTT |
| R: GGGCCTTATGCCAGGAAACT | ||
| Srebp1c | Mice | F: GACCCTACGAAGTGCACACA |
| R: TGTCGGGCTCAGAGTCACTA | ||
| Pparg | Mice | F: GGGGATGTCTCACAATGCCA |
| R: CAGACTCTGGGTTCAGCTGG | ||
| Ppara | Mice | F: AACCTGAGGAAGCCGTTCTG |
| R: TCTGCAGGTGGAGCTTAAGC | ||
| Cpt1a | Mice | F: ACAATTCCCCTCTGCTCTGC |
| R: TACACGACAATGTGCCTGCT | ||
| Acox1 | Mice | F: TAACTTCCTCACTCGAAGCCA |
| R: AGTTCCATGACCCATCTCTGTC | ||
| Ucp2 | Mice | F: CAGCGCCAGATGAGCTTTG |
| R: GGAAGCGGACCTTTACCACA | ||
| Tgfb1 | Mice | F: CTTCAATACGTCAGACATTCGGG |
| R: GTAACGCCAGGAATTGTTGCTA | ||
| Timp1 | Mice | F: TACACCCCAGTCATGGAAAGC |
| R: CGGCCCGTGATGAGAAACT | ||
| Ctgf | Mice | F: GGGCCTCTTCTGCGATTTC |
| R: ATCCAGGCAAGTGCATTGGTA | ||
| Col1a1 | Mice | F: CTGGCGGTTCAGGTCCAAT |
| R: TTCCAGGCAATCCACGAGC | ||
| β-Actin | Mice | F: TACTGCTCTGGCTCCTAGCA |
| R: CGGACTCATCGTACTCCTGC |
2.11. Western blot assay
Western blot analysis was performed as previously described [17]. In brief, tissue or cell samples were lysed in RIPA lysis buffer (P0013K, Beyotime, Shanghai, China) With protease inhibitor cocktail (P1005, Beyotime). Total protein was quantified using a BCA Protein Assay Kit (P0010, Beyotime, Shanghai, China). Proteins were separated on 10 % SDS–PAGE gels and transferred to PVDF membranes (IPVH00010; Merck Millipore, Darmstadt, Germany). After blocking with 5 % skim milk in Tris-buffered saline/Tween 20 (TBST) for 1 h, membranes were incubated first with primary antibodies overnight at 4 °C and then with the corresponding secondary antibodies at room temperature for 1 h. Signals were visualized with a Chemiluminescence image analyzer (Tanon5200, Tanon, shanghai, China). β-Actin was used as a loading control. Primary antibodies were listed in Table 3.
Table 3.
Antibodies used in the research.
| Antibodies name | Source | Cat No. |
|---|---|---|
| Anti-RGS14 | Proteintech | 16258-1-AP |
| Anti-p-PKA | Santacruz | sc-377575 |
| Anti-PKA | Santacruz | sc-28315 |
| Anti-GNAI1 | Santacruz | sc-13533 |
| Anti-GNAI3 | Santacruz | sc-365422 |
| Anti-p-AMPKa | Abclonal | AP0432 |
| Anti-AMPKa | Abclonal | A1229 |
| Anti-p-ACC | Cell Signaling Technology | 11818S |
| Anti-ACC | Proteintech | 21923-1-AP |
| Anti-SREBP1C | Santacruz | sc-13551 |
| Anti-HA | Medical Biological Laboratories | M180-3 |
| Anti-Flag | Medical Biological Laboratories | M185 |
| Anti-His | Abclonal | AE003 |
| Anti-GST | Abclonal | AE006 |
| Anti-β-Actin | Abclonal | AC026 |
| Anti-Cd11b | Boster | BM3925 |
| HRP-conjugated Affinipure Goat Anti-Mouse IgG(H + L) | Abclonal | SA00001-1 |
| HRP-conjugated Affinipure Goat Anti-Rabbit IgG(H + L) | Abclonal | SA00001-2 |
2.12. Co-immunoprecipitation (Co-IP) assay and MS assays
For Co-IP assays, cultured cells were transfected with the indicated plasmids for 24 h and then were lysed in ice-cold immunoprecipitation buffer (20 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA and 1 % Triton X-100) containing protease inhibitor cocktail (P1005, Beyotime) for 30 min. Lysates were cleared by centrifugation at 12,000g for 15 min, then incubated with the indicated antibodies and Protein G Bestarose 4FF (AA0142; BESTBOCHROM; China) at 4 °C for 3 h, and washed three times in cold immunoprecipitation buffer. The immunocomplexes were eluted in 2× loading buffer and subjected to western blotting analysis. For MS analysis, the eluate was separated via10 % SDS–PAGE and visualized with Fast Silver Stain Kit (P0017S, Beyotime, Shanghai, China). Then the bands were excised and sent for mass spectrometry (APTBIO; Shanghai; China). The obtained data were searched in the UniProt database using MaxQuant 1.6.0.1 software.
2.13. GST pull-down assay
For GST Pull-down assay, the prokaryotic cell transfected with the pGEX-6p-1-GST-RGS14 or pET-21b-His-GNAI1/3 plasmids, subsequently, GST-RGS14, His-Giα1 and His-Giα3 protein was induced by IPTG at 16 °C for overnight. Then GST-RGS14 and GST alone protein were incubated with 20 μL GST Bestarose 4FF (AA0071; BESTBOCHROM; China) at 4 °C for 3 h, then washed three times in cold PBS buffer and incubated with His-Giα1 or His-Giα3 for 4 h at 4 °C on a rotating windmill. Finally, the GST Bestarose 4FF beads were washed three times with cold PBS buffer and analyzed by western blotting.
2.14. Statistical analysis
All data were presented as the means ± s.d, and statistical analysis was performed using SPSS Statistics version 21.0. For comparisons between two groups, statistical significance was performed using a two-tailed Student's t-test when data conformed to a normal distribution, while the nonparametric test (Mann Whitney U test) was used when data with a skewed distribution. For comparisons among more than two groups, a one-way ANOVA was applied to assess simple effects and followed with the Bonferroni post hoc test for data with homogeneous variance or Tamhane's T2 analysis for heteroscedastic data. P values were specified as follows: #P < 0.05, ##P < 0.01; ∗P < 0.05, ∗∗P < 0.01; n.s. indicates no significance between the two indicated groups.
3. Results
3.1. RGS14 expression is downregulated in NAFLD model
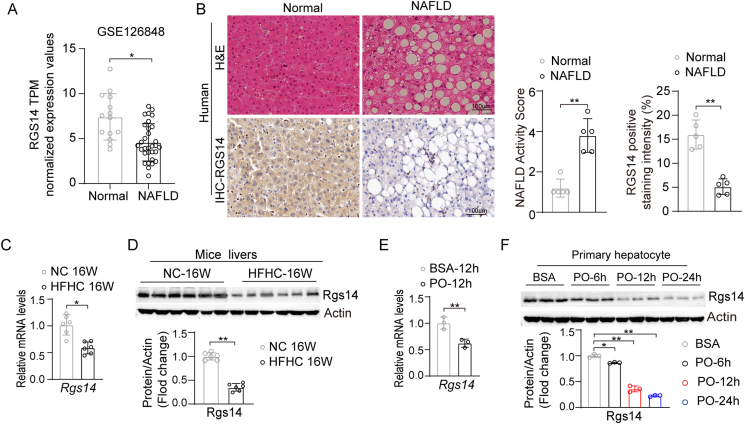
To explore whether RGS14 was implicated in nonalcoholic fatty liver disease (NAFLD), we first analyzed the expression of RGS14 both in healthy individuals and NAFLD patients in a public database (GSE126848) [20] by GEO2R online tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/) [21]. The results showed that the expression of RGS14 was strikingly lower in NAFLD patients than in healthy individuals (Figure 1A). We further confirmed that RGS14 expression in liver tissues was significantly lower in NAFLD patients than in healthy individuals by H&E staining and immunohistochemical staining (Figure 1B). Moreover, we observed a similar decrease in the high fat high cholesterol (HFHC) diet-induced NASH mice model (Figure 1C, D). To further study the expression alterations of RGS14 in hepatocytes, primary mouse hepatocytes were subjected to PO (0.5 mM palmitic acid (PA) and 1.0 mM oleic acid (OA)) for 0, 6, 12 and 24 h. Consistent with the results obtained from the livers of human and mice subjects, RGS14 expression gradually decreased in a time-dependent manner in primary mouse hepatocytes challenged by PO stimulus (Figure 1E, F). These data indicate that the downregulation of RGS14 was associated with the progression of NAFLD and that RGS14 might play a critical role in this process.
Figure 1.
RGS14 expression is down-regulated in NAFLD model. (A) mRNA levels of RGS14 in the liver samples from NAFLD and health individuals by GSEA based on the RNA-seq datasets (GSE126848). (B) Representative H&E staining and Immunohistochemical staining images of RGS14 in the individuals with or without NAFLD, (n = 5 per group). Scale bar, 100 μm. (C) The relative mRNA expressions of Rgs14 gene in the livers of 8-week-old C57BL/6 male mice subjected to normal or HFHC diet for 16 weeks. β-Actin was used as internal control, n = 6 per group. (D) Protein levels of Rgs14 in the livers of mice from the indicated groups, β-Actin was used as internal control, (n = 6 per group). (E) The relative mRNA expressions of Rgs14 gene in primary mouse hepatocytes under BSA or PO (0.5 mM Palmitic Acid and 1 mM Oleic Acid) treatment, β-Actin was used as internal control, n = 3 independent experiments. (F) Protein levels of Rgs14 in primary mouse hepatocytes from the indicated groups, β-Actin was used as internal control (n = 3 independent experiments). A two-tailed Student t test (B–E) and one-way ANOVA (F) analysis was used. ∗P < 0.05, ∗∗P < 0.01. All data are shown as the mean ± s.d. Abbreviations: NAFLD, Nonalcoholic Fatty Liver Disease; TPM, Transcripts per million; H&E, Hematoxylin and Eosin; IHC, Immunohistochemical; NC, Normal Chow diet; HFHC, High Fat High Cholesterol Diet; BSA, Bovine Serum Albumin; PO; 0.5 mM Palmitic Acid and 1 mM Oleic Acid.
3.2. RGS14 ameliorates lipid accumulation in hepatocytes
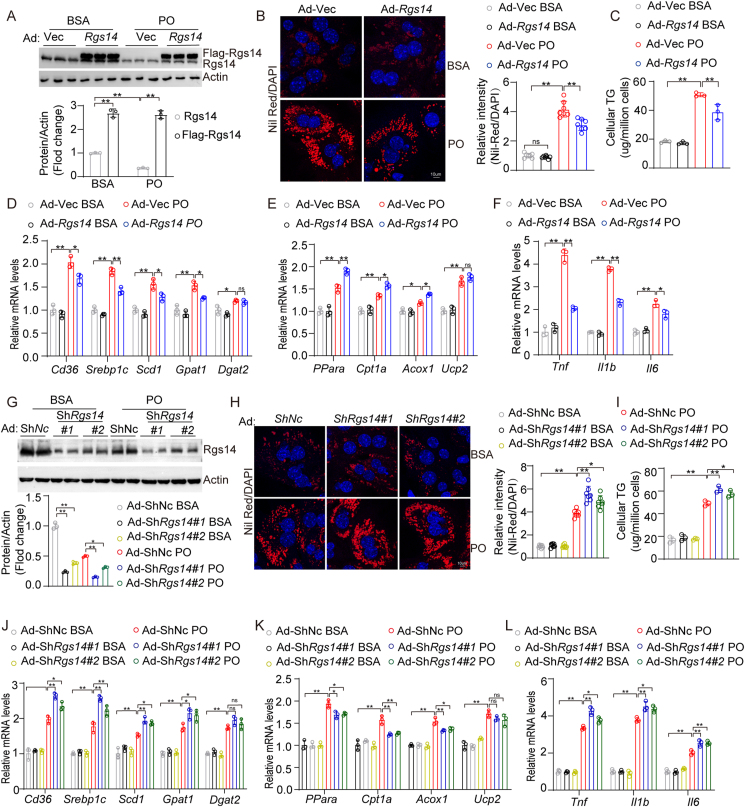
On the basis of the significant decrease of RGS14 in NAFLD, we hypothesized a crucial functional relevance of RGS14 in the progression of NAFLD. As presented in Figure 2A, we first established Rgs14-overexpressing primary mouse hepatocytes by transfected with an adenovirus vector loading the Rgs14 gene (Ad-Rgs14). Compared with those in the vector control group (Ad-Vec), the Rgs14-overexpression group sharply ameliorated lipid deposition (Figure 2B) and intracellular TG content (Figure 2C) in hepatocytes subjected to PO for 24 h. Furthermore, the overexpression of Rgs14 reduced the gene expression, including those involved in fatty acid absorption and lipogenesis (Figure 2D), inflammatory responses (Figure 2F), and accelerated the expression of genes involved in fatty acid oxidation (Figure 2E) in hepatocytes induction elicited by PO. Subsequently, we generated Rgs14-knockdown primary mouse hepatocytes by transduction with adenovirus ShRgs14 (Ad-ShRgs14) (Figure 2G). Compared to those in the vector control (Ad-ShNc), the lipid accumulation (Figure 2H) and intracellular TG content (Figure 2I) in the Rgs14-knockdown group were significantly greater. Moreover, Rgs14-knockdown markedly promoted the upregulation of fatty acid uptake and synthesis genes (Figure 2J) and the proinflammatory response (Figure 2L) and blocked the expression of fatty acid oxidation genes (Figure 2K) in primary mouse hepatocytes challenged with PO for 12 h. In conclusion, these gain- and loss-of-function data strongly suggested that RGS14 in hepatocytes is a pivotal molecule that improves liver lipid accumulation and alleviates the inflammatory response under PO treatment.
Figure 2.
RGS14 ameliorates lipid accumulation in hepatocytes. (A) Protein levels of Rgs14 in primary mouse hepatocytes transfected with adenovirus loading Rgs14 (Ad-Rgs14) or vector (Ad-Vec), n = 3 per group. (B) Representative images (n = 3 independent experiments) and quantification (n = 7 high power fields per group) of Nile Red staining of Rgs14-overexpressed primary mouse hepatocytes under the stimulation of PO or BSA treatment for 24 h. Scale bar, 10 um. (C) TG contents in Rgs14-overexpressed primary hepatocytes under PO treatment (n = 3 independent experiments). (D–F) mRNA levels of fatty acid uptake and synthesis genes (D), fatty acid oxidation genes (E) and inflammation genes (F) in Rgs14-overexpressing primary mouse hepatocytes treated with PO at 12 h, β-Actin was used as internal control, n = 3 independent experiments. (G) Protein expression of Rgs14 protein expression in primary mouse hepatocytes transfected with each indicated adenovirus (Ad-ShNc or Ad-ShRgs14), β-Actin was served as control, n = 3 independent experiments. (H) Representative images (n = 3 independent experiments) and quantification (n = 7 high power fields per group) of Nile Red staining of Rgs14 knockdown (ShRgs14) in primary mouse hepatocytes under PO treatment for 24 h. Scale bar, 10 um. (I) Intracellular TG content in the primary mouse hepatocytes of Rgs14 knockdown after PO treatment 24 h (n = 3 independent experiments). (J–L) The gene expression of fatty acid uptake and synthesis genes (J), fatty acid oxidation genes (K) and inflammation genes (L) in Rgs14 knock-down primary mouse hepatocytes under PO treatment for 12 h (n = 3 independent experiments). One-way ANOVA analysis was used. ∗P < 0.05, ∗∗P < 0.01, n.s. indicates no significance between the two indicated groups. All data are shown as the mean ± s.d. Abbreviations: BSA, Bovine Serum Albumin; PO, 0.5 mM Palmitic Acid and 1 mM Oleic Acid. Ad, Adenovirus; TG, Triglyceride.
3.3. Hepatocyte specific Rgs14 knockout exacerbates HFHC-induced NASH
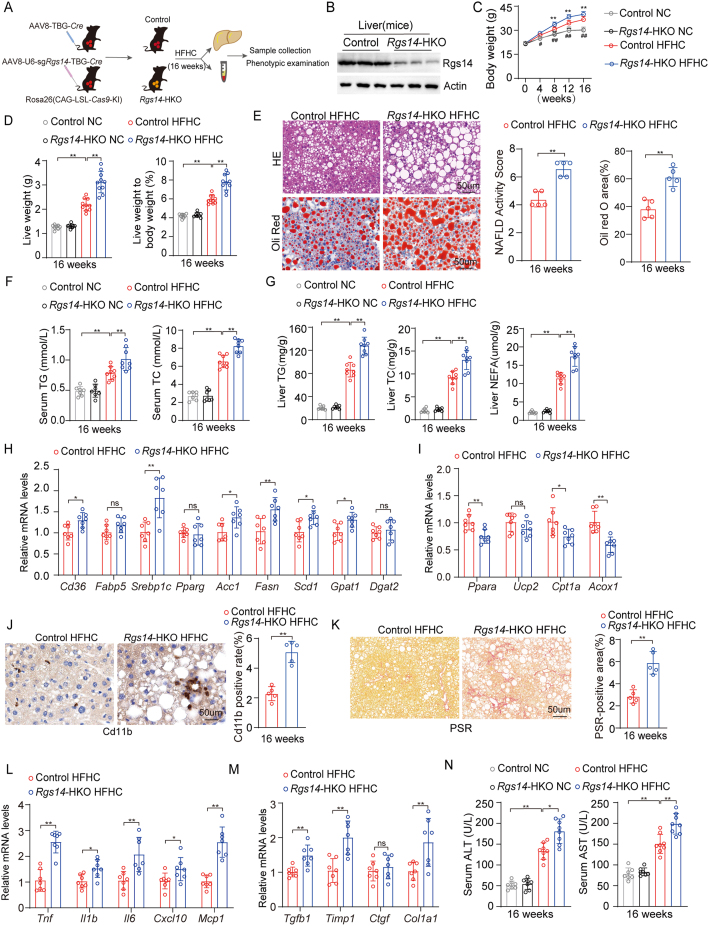
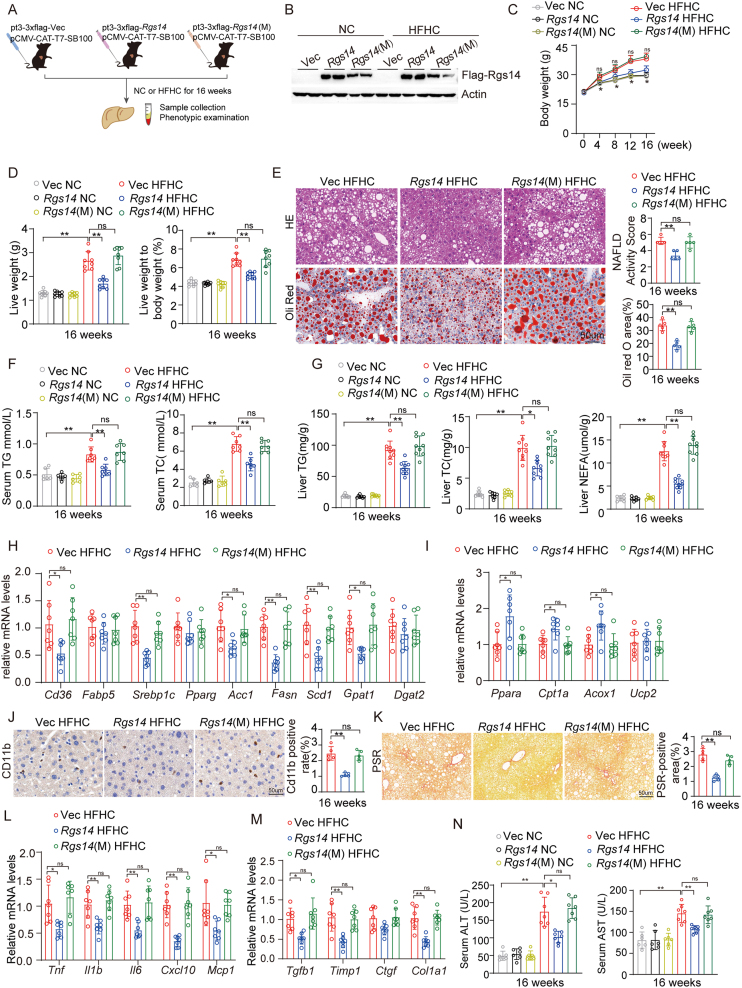
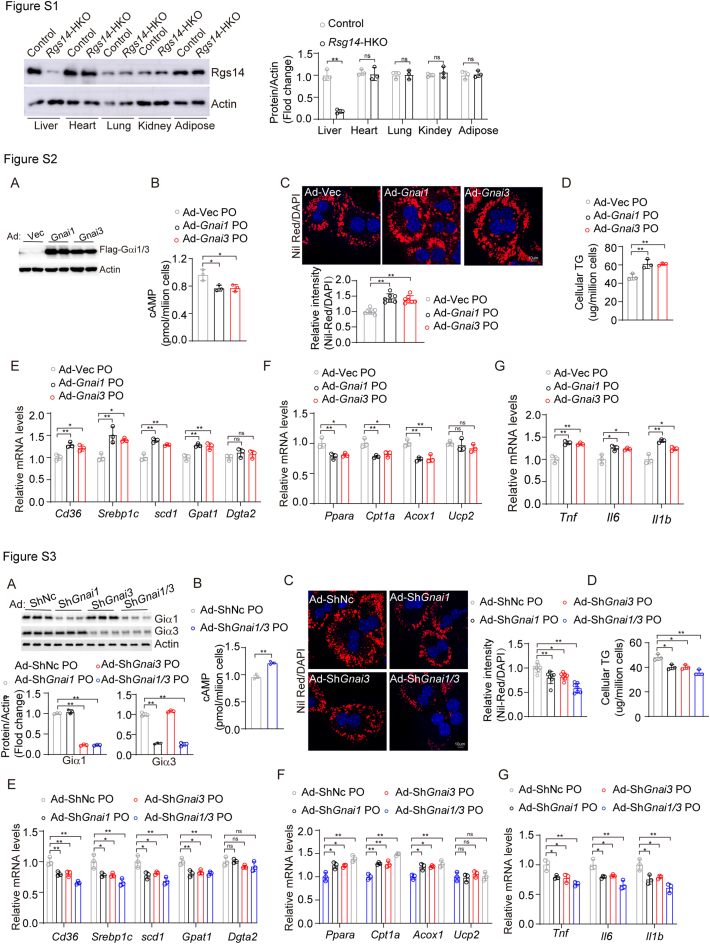
To confirm the functions of hepatic Rgs14 in vivo, hepatocyte specific Rgs14-knockout (Rgs14-HKO) male mice were generated using male Rosa26-CAG-LSL-Cas9 mice combined with AAV8-U6-sgRgs14-TBG-Cre infection and administrated to HFHC diet for 16 weeks in parallel with their parallel controls (Figure 3A). We have quantified protein expression of Rgs14 in liver, heart, lung, kidney and adipose of Control and Rgs14-HKO mice, and founded that Rgs14 was deleted in liver tissue of Rgs14-HKO mice (Figure 3B), but not in heart, lung, kidney or adipose (Fig. S1). Compared with the control mice, Rgs14-HKO mice exhibited marked increases in body weight (Figure 3C), liver weight and liver weight-to-body weight ratio (Figure 3D) after 16 weeks HFHC diet feeding. H&E and Oil Red O staining revealed that hepatic steatosis was further exacerbated in Rgs14-HKO mice fed a HFHC diet compared to control mice (Figure 3E). Moreover, the serum TG and TC concentrations were significantly greater than those in the control mice after 16 weeks of HFHC feeding (Figure 3F). Compared with control mice, Rgs14-HKO mice fed a HFHC diet for 16 weeks exhibited higher liver TG, TC and NEFA levels (Figure 3G). Consistent with these findings, the mRNA expression of genes involved in fatty acid uptake and synthesis was upregulated (Figure 3H), and the expression of genes related to fatty acid utilization and oxidation was downregulated (Figure 3I) in Rgs14-HKO mice compared to control mice after HFHC diet feeding.
Figure 3.
Hepatocyte-specific Rgs14 KO exacerbates HFHC-induced NASH. (A) The strategy for the generation of 8 weeks old male hepatocyte-specific Rgs14-knockout (Rgs14-HKO) or Control mice by using male Rosa26 (CAG-LSL-Cas9-KI) mice combined with AAV8-U6-sgRgs14-TBG-Cre or AAV8-TBG-Cre infection. (B) Protein expression of Rgs14 in liver tissues of Rgs14-HKO and Control mice, n = 3 mice per group. (C) Body weight in Rgs14-HKO and Control mice fed with NC (n = 8 mice per group) and HFHC (n = 10 mice per group) diet from 0 to 16 weeks. (D) Liver weight and Liver weight to body weight ratio of Rgs14-HKO and Control mice fed NC (n = 8 mice per group) and HFHC (n = 10 mice per group) for 16 weeks. (E) Representative images of H&E and Oil red O staining and their NAFLD Activity Score/Oil red O-positive area of liver sections in Rgs14-HKO and Control mice at 16 weeks after NC and HFHC treatment. Scale bar, 100 um, n = 5 mice per group. (F) Serum TG and TC contents of Rgs14-HKO and Control mice after 16 weeks feeding with NC (n = 7 mice per group) or HFHC (n = 8 mice per group). (G) TG, TC and NEFA contents of liver tissues in Rgs14-HKO and Control mice after 16 weeks feeding with NC (n = 7 mice per group) or HFHC (n = 8 mice per group). (H, I) mRNA levels of fatty acid uptake and synthesis genes (H) and fatty acid oxidation genes (I) in Rgs14-HKO HFHC and Control HFHC mice (n = 7 mice per group). (J, K) Representative images of CD11b-positive cell staining (J) and PSR (K) of liver sections in Rgs14-HKO HFHC and Control HFHC mice, n = 5 mice per group, Scale bar, 100 um. (L, M) mRNA levels of inflammation genes (L) and fibrosis genes (M) in liver tissues of Rgs14-HKO HFHC and Control HFHC mice (n = 7 mice per group). (N) AST and ALT contents of Rgs14-HKO and Control mice after 16 weeks feeding with NC (n = 7 mice per group) or HFHC (n = 8 mice per group). A two-tailed Student t test (E, H-M) and one-way ANOVA (C, D, F, G and N) analysis were used. ∗P < 0.05, ∗∗P < 0.01 (Control HFHC group compared with Rgs14-HKO HFHC group). #P < 0.05, ##P < 0.01 (Control NC group compared with Control HFHC group); n.s. indicates no significance between the two indicated groups. All data are shown as the mean ± s.d. Abbreviations: Cas9, CRISPR-associated nuclease 9; AAV8, Adeno-associated Virus 8; TBG,Thyroid-binding globulin; Cre, Cyclization recombination enzyme; Rgs14-HKO, Hepatocyte specificity Rgs14 knock-out; H&E, Hematoxylin and Eosin; TG, Triglyceride; TC, Total Cholesterol; NEFA, Non-esterified fatty acid; PSR, Picrate Sirius Red staining; ALT, Alanine aminotransferase; AST, Aspartate aminotransferase.
Histologically, NASH is characterized by steatosis, inflammation, hepatocyte ballooning and fibrosis. After 16 weeks of HFHC diet feeding, the control mice displayed mild inflammation, fibrosis and lipid deposition in the liver. Compared to control mice, Rgs14-HKO mice displayed further increased infiltration of inflammatory Cd11b+ cells into the liver (Figure 3J) and exhibited more pronounced fibrosis as evidenced by PSR staining (Figure 3K) under HFHC diet feeding. Consistently, Rgs14-HKO aggravated expression of proinflammatory genes (Figure 3L) and profibrotic gene expression in the liver (Figure 3M). Compared to the control mice, the hepatocytes Rgs14-depleted mice further promoted HFHC-induced liver injury, as indicated by the changes in the serum transaminase alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels (Figure 3N). Taken together, these data indicate that hepatocyte specific Rgs14-knockout exacerbates HFHC-induced lipid accumulation, inflammation, fibrosis and liver injury, in other words, RGS14 plays an essential protective role in hepatocytes during HFHC-induced NASH.
3.4. RGS14 depends on GDI activity to ameliorate hepatocytes lipid accumulation
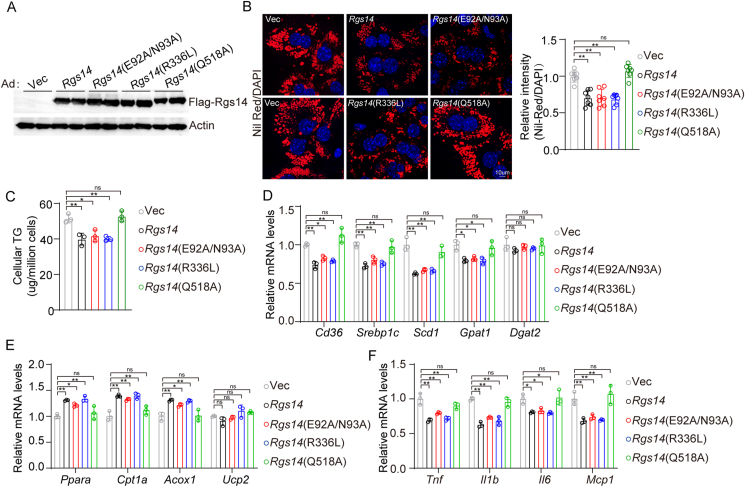
To further map the functional regions of Rgs14 that are responsible for hepatocytes lipid accumulation, we constructed and expressed a series of point mutants including E92A/N93A mutant within RGS motif which destroys GTPase-activating protein (GAP) activity of RGS14 [22], R336L mutant within the RBD motif which loses the function of binding to Ras/Raf and regulating Ras/Raf signaling pathways [11], and Q518A mutant within GoLoco/GPR motif destroying GDP dissociation inhibitor (GDI) activity of RGS14 [22]. Western blot analysis of Rgs14 and Rgs14 mutants in primary mouse hepatocytes transfected with each indicated adenovirus were performed (Figure 4A). We demonstrated Rgs14, Rgs14(E92A/N93A) and Rgs14(336L) presented decreased lipid accumulation (Figure 4B) and cellular TG contents (Figure 4C) compared with control group (Vector) in primary mouse hepatocytes under PO treatment. Furthermore, Rgs14, Rgs14(E92A/N93A) and Rgs14(R336L) reduced the mRNA expression of genes involved in fatty acid uptake and synthesis (Figure 4D) and the inflammatory response (Figure 4F), accelerated the mRNA expression of genes involved in fatty acid utilization and oxidation (Figure 4E) in primary mouse hepatocytes induction elicited by PO. The Rgs14(Q518A) mutant which dysfunctioning GDI activity, showed lost the function of inhibiting lipid accumulation and inflammatory response and was indistinguishable from the control group (Vector) in primary mouse hepatocytes under PO treatment (Figure 4B–F). Above data suggested that GDI activity of Rgs14 is responsible for hepatocytes lipid accumulation and inflammation responses under PO stimulus.
Figure 4.
RGS14 depends on GDI activity to ameliorate hepatocytes lipid accumulation. (A) Protein levels of Rgs14 and Rgs14 mutants in primary mouse hepatocytes transfected with each indicated adenovirus, n = 3 independent experiments. (B) Representative images (n = 3 independent experiments) and quantification (n = 7 high power fields per group) of Nile Red staining of Rgs14 or Rgs14 mutants overexpressed primary mouse hepatocytes under PO treatment 24 h. Scale bar, 10 um. (C) TG contents in Rgs14 or Rgs14 mutants overexpressed primary mice hepatocytes under PO treatment (n = 3 independent experiments). (D–F) mRNA levels of fatty acid uptake and synthesis genes (D), fatty acid oxidation genes (E) and inflammation genes (F) in each indicated adenovirus infected primary mouse hepatocytes stimulated by PO for 24 h (n = 3 independent experiments). One-way ANOVA analysis was used. ∗P < 0.05, ∗∗P < 0.01, n.s. indicates no significance between the two indicated groups. All data are shown as the mean ± s.d. Abbreviations: PO, 0.5 mM Palmitic Acid and 1 mM Oleic Acid; TG, Triglyceride; Ad, Adenovirus.
3.5. RGS14 depends on GDI activity to relieve HFHC-induced NASH
To further verify whether Rgs14 function in GDI activity was implicated in HFHC-induced NASH, we overexpressed Rgs14 and Rgs14 with the Q518A mutation (Rgs14(M)) in the mouse liver using the Sleeping Beauty transposon system, and Rgs14 and Rgs14(M) overexpressed mice were fed to HFHC or a normal chow diet for 16 weeks (Figure 5A,B). Remarkably, compared with the control (vector) mice, Rgs14-overexpressed effectively attenuated the HFHC-induced increase in body weight (Figure 5C), liver weight and liver weight-to-body weight ratio (Figure 5D), whereas these results were indistinguishable between Rgs14(M) and Vector-treated animals fed a HFHC diet for 16 weeks (Figure 5C,D). Hepatic steatosis was relieved by Rgs14 overexpression (Figure 5E–I), and this change was accompanied by marked reductions in proinflammatory cell infiltration, liver fibrosis, the inflammatory response, and liver injury (Figure 5J–N) compared to those in vector-treated controls under HFHC-fed diet treatment. However, Rgs14(M) lost the effect of ameliorating hepatic steatosis, inflammation, fibrosis and liver injury (Figure 5J–N). The above data demonstrated that HFHC diet–induced hepatic steatosis, inflammation, and fibrosis were suppressed by Rgs14 overexpression, but were reversed when Rgs14 was mutated in Q518. In conclusion, these findings revealed that the function of RGS14 in mediating GDI activity ameliorates HFHC-induced NASH.
Figure 5.
RGS14 depends on GDI activity to relieve HFHC-induced NASH. (A) Experimental schematic design to assess the functions of RGS14 in GDI activity on HFHC-induced NASH. (B) Protein expression of Rgs14 or Rgs14(Q518A) in male mice liver tissues that were injected with the pt3-Vec, pt3-Rgs14 and pt3-Rgs14(M) plasmids by Sleeping Beauty transposon system. (C) Body weight in Rgs14, Rgs14(M) and Vec mice fed with NC and HFHC diet from 0 to 16 weeks (n = 8 mice per group). (D) Liver weight and Liver weight to body weight ratio of Rgs14, Rgs14(M) and Vec mice that were fed with NC and HFHC for 16 weeks (n = 8 mice per group). (E) Representative images of H&E and Oil red O staining and their NAFLD Activity Score/Oil red O-positive area of liver sections in Rgs14, Rgs14(M) and Vec mice at 16 weeks after NC and HFHC treatment, n = 5 mice in each group, Scale bar, 100 um. (F) Serum TG and TC contents of Rgs14, Rgs14(M) and Vec mice fed with NC or HFHC (NC = 6 mice per group, HFHC = 7 mice per group). (G) TG, TC and NEFA contents of Rgs14, Rgs14(M) and Vec mice in hepatic samples after 16 weeks feeding with NC or HFHC (NC = 7 mice per group, HFHC = 9 mice per group). (H, I) The mRNA levels of fatty acid uptake and synthesis genes (H) and fatty acid oxidation genes (I) in liver tissues in Rgs14, Rgs14(M) and Vec HFHC mice (n = 7 mice per group). (J, K) Representative images of CD11b-positive cell staining and PSR of liver sections in Rgs14, Rgs14(M) and Vec HFHC mice (n = 5 mice per group), Scale bar, 100 um. (L, M) mRNA levels of inflammation genes (L) and fibrosis genes (M) in liver tissues in Rgs14, Rgs14(M) and Vec HFHC mice (n = 7 mice per group). (N) AST and ALT levels of Rgs14, Rgs14(M) and Vec mice after 16 weeks feeding with NC or HFHC (NC = 6 mice per group, HFHC = 7 mice per group). One-way ANOVA analysis was used. ∗P < 0.05, ∗∗P < 0.01, n.s. indicates no significance between the two indicated groups. All data are shown as the mean ± s.d. Abbreviations: GDI, GDP dissociation inhibitor; Rgs14(M), Rgs14(Q518A); H&E, Hematoxylin and Eosin; TG, Triglyceride; TC, Total Cholesterol; NEFA, Non-esterified fatty acid; PSR, Picrate Sirius Red staining; ALT, Alanine aminotransferase; AST, Aspartate aminotransferase.
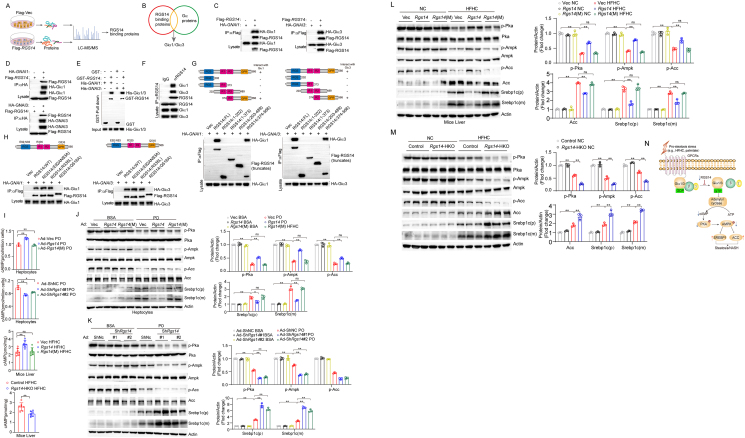
3.6. RGS14 promotes cAMP/AMPK activation by targeting Giα1/3
Based on our vivo studies, we demonstrated that the GDI activity of RGS14 is involved in the initiation and development of HFHC-induced NASH. On the basis of RGS14 binding to Gα family proteins, we performed IP-MS on RGS14-overexpressing hepatocyte line to determine the interacting target molecules (Figure 6A). We first detected the share proteins Giα1 and Giα3 (Giα1/3) between RGS14 binding proteins and Gα family proteins (Figure 6B), consistent with previous researches [23,24]. Subsequently, Co-IP assays were conducted to explore the interaction between RGS14 and Giα1/3. The results demonstrated that RGS14 interacted with Giα1/3 in human L02 hepatocytes (Figure 6C,D). Further, we performed GST pull-down assays to confirm the direct interaction between RGS14 and Giα1/3 (Figure 6E). We also determined the endogenous interaction between RGS14 and Giα1/3 in human L02 hepatocytes by Co-IP assays (Figure 6F). The domain mapping assay showed that GoLoco/GPR domain (374–566 amino acids) of RGS14 was responsible for its interaction with Giα1/3 in human L02 hepatocytes (Figure 6G). Consistently, the interaction between RGS14 and Giα1/3 in human L02 hepatocytes was terminated when Q516 was mutated in the GoLoco/GPR domain (Figure 6H). Studies have shown that Giα1 and Giα3 inhibited adenylate cyclase activity, leading to decreased intracellular cAMP levels [[25], [26], [27], [28]]. cAMP as a key second messenger, playing an important function in regulating glucose uptake/utilization and lipid metabolism in liver [29,30]. We found that Rgs14 overexpression increased cAMP contents and Rgs14(M) lost the ability to affect cAMP levels compared with the control, while Rgs14 knock-down or knockout reduced cAMP contents in vivo and vitro (Figure 6I). PKA as a cAMP-dependent protein kinase [29], also is one of the most important upstream signaling molecules of AMPK [31,32]. As a key intracellular energy sensor, AMPK can modulate lipolysis, lipogenesis, and fatty acid synthesis through phosphorylation of key substrates [32,33]. Several studies have shown that PKA/AMPK pathway attenuates hepatic steatosis and abnormal lipid metabolism [32,34]. We found that PKA/AMPK signaling was suppressed in PO-induced primary mouse hepatocytes compared to BSA control (Figure 6J,K). PKA/AMPK signaling pathway was markedly activated in Rgs14-overexpressed hepatocytes, but inhibited in Rgs14(M) hepatocytes stimulated with PO. (Figure 6J). In contrast, Rgs14 knock-down markedly inhibited PKA/AMPK signaling compared to ShNc in primary mouse hepatocytes under PO treatment (Figure 6K). In parallel with in vitro results, we found similar phenomena in liver of NAFLD mice (Figure 6L,M). Taken together, these data suggested that RGS14 improve liver steatosis through activating cAMP/PKA/AMPK signaling pathway by targeting Giα1/3 (Figure 6N).
Figure 6.
RGS14 promotes cAMP-AMPK activation by Targeting Giα1/3. (A) Schematic illustration of IP-MS experiments to explore RGS14 potential binding targets in RGS14-overexpressing hepatocytes. (B) Share proteins between RGS14 binding proteins and Gα proteins through the intersection analysis of IP-MS. (C, D) The overexpression interaction between RGS14 and Giα1/3 by Co-IP assays, Flag-tagged RGS14 and HA-tagged GNAI1or GNAI3 plasmids were cotransfected into human L02 hepatocytes. Anti-HA antibody (C) and anti-Flag antibody (D) were used for immunoprecipitation (n = 3 independent experiments). (E) The exogenous interaction of RGS14 with Giα1/3 was assayed by GST precipitation, and the purified GST protein was used as a control (n = 3 independent experiments). (F) The endogenous interaction between RGS14 and Giα1/3 in human L02 hepatocytes by Co-IP assays, and the purified IgG was used as a control (n = 3 independent experiments). (G) Domain mapping analysis of the binding domains of RGS14 to Giα1/3. Full-length RGS14 and various truncated forms of Flag-RGS14 were cotransfected into human L02 hepatocytes. An anti-Flag antibody was used for immunoprecipitation (n = 3 independent experiments). (H) Co-IP assays to analyze the interaction between WT/Mutans RGS14 and Giα1/3. Various mutants of RGS14 were cotransfected into human L02 hepatocytes. An anti-Flag antibody was used for immunoprecipitation (n = 3 independent experiments). (I) Effect of Rgs14/Rgs14(M) overexpressed, Rgs14 knockdown and Rgs14-HKO on cAMP levels in vitro (n = 3 independent experiments) and in vivo (n = 7 mice per group). (J) Expression of cAMP-PKA-AMPK signaling pathway proteins (e.g. Pka, p-Pka, Ampk, p-Ampk, Acc, p-Acc, Srebp1c-P and Srebp1c-M) in Rgs14 or Rgs14(M) overexpressed primary mouse hepatocytes after BSA or PO challenge for 12 h (n = 3 independent experiments). (K) Expression of cAMP-PKA-AMPK signaling pathway proteins in Rgs14-knockdown primary mouse hepatocytes under BSA or PO treatment 12 h. (L) The protein levels of cAMP-PKA-AMPK signaling pathway in hepatic tissue of Rgs14 or Rgs14(M) mice under NC or HFHC treatment 16 weeks, β-Actin served as an internal control (n = 3 mice per group). (M)The expression of cAMP-PKA-AMPK signaling pathway in liver tissue of Control and Rgs14-HKO mice under NC or HFHC treatment 16 weeks, β-Actin served as an internal control (n = 3 mice per group). (N) The schematic diagram shows the mechanisms of RGS14 suppressing hepatic steatosis. A two-tailed Student t test (I) and one-way ANOVA (I–M) analysis was used. ∗P < 0.05, ∗∗P < 0.01, n.s. indicates no significance between the two indicated groups. All data are shown as the mean ± s.d. Abbreviations: LC-MS, liquid chromatography-mass spectrometry; Co-IP, Co-immunoprecipitation; GST, glutathione; PO, 0.5 mM Palmitic Acid and 1 mM Oleic Acid; Ad, Adenovirus; Rgs14(M), Rgs14(Q518A); Rgs14-HKO, Hepatocyte specificity Rgs14 knockout; cAMP, cyclic adenosine monophosphate.
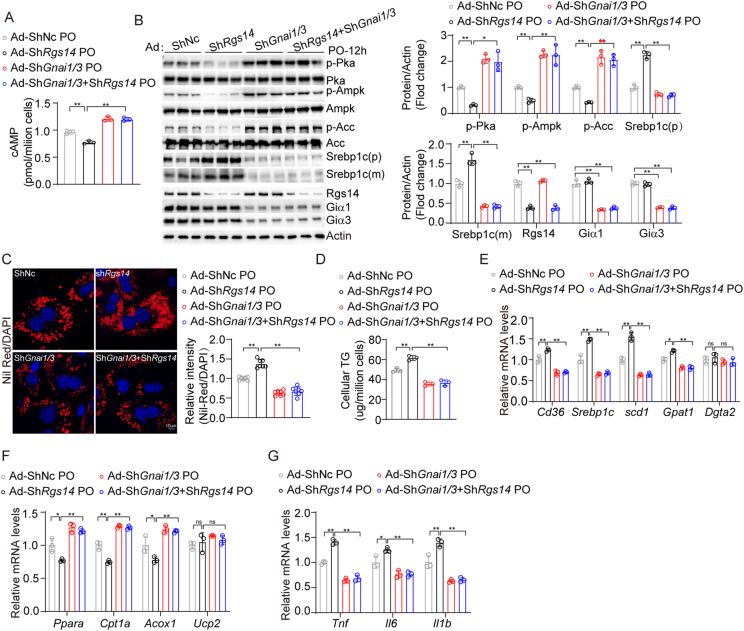
3.7. Inhibition of Giα1/3 ameliorates RGS14-knockdown induced progression of NAFLD in vitro
To further determine the contribution of Giα1/3 to RGS14-mediated regulation of NAFLD progression, we generated Rgs14-knockdown, Gnai1 and Gnai3(Gnai1/3)-knockdown and three-knockdown of primary mouse hepatocytes through transfecting with indicated adenovirus. As anticipated, knockdown of Rgs14 significantly reduced the production of cAMP, whereas Gnai1/3 knockdown largely abolished the effect of Rgs14-knockdown on cAMP production in primary hepatocytes under PO treatment (Figure 7A). Consistently, Rgs14-knockdown suppressed the activation of cAMP/PKA/AMPK pathway compared with ShNc group in primary mouse hepatocytes under PO treatment, however Gnai1/3 knockdown significantly promoted cAMP/PKA/AMPK signaling amplification in Rgs14 knockdown primary mouse hepatocytes under PO addition (Figure 7B). More remarkably, the silencing of Gnai1/3 largely abolished the exacerbating effect of shRgs14 on hepatocyte steatosis and inflammatory response after PO treatment (Figure 7C–G). Besides, we observed that the overexpression of Gnai1 or Gnai3 reduced intracellular cAMP levels, and promoted lipogenesis and the inflammatory response in primary mouse hepatocytes under PO addition (Fig. S2). It presented opposite phenomenon when Gnai1 or Gnai3 was knocked down (Fig. S3). Collectively, our findings suggest that inhibition of Giα1/3 blocked the functions of RGS14 on NAFLD, indicating the protective role of RGS14 is mainly dependent on Giα1/3.
Figure 7.
Inhibition of Giα1/3 ameliorates RGS14-deficient induced progression of NAFLD in vitro. (A) The cAMP levels in primary mouse hepatocytes transfected with indicated adenovirus with PO stimulus for 12 h (n = 3 independent experiments). (B) The protein levels of cAMP-PKA-AMPK signaling pathway in indicated groups after PO exposure, β-Actin served as an internal control (n = 3 independent experiments). (C) Representative images (n = 3 independent experiments) and quantification (n = 7 high power fields per group) of Nile Red staining of primary mouse hepatocytes transfected with indicated adenovirus under PO treatment. Scale bar, 10 um. (D) Intracellular TG content in indicated adenovirus infected primary mouse hepatocytes under the stimulation of PO (n = 3 independent experiments). (E–G) qPCR analysis of the mRNA levels of fatty acid uptake and synthesis genes (E), fatty acid oxidation genes (F) and inflammation genes (G) in infected primary mouse hepatocytes treated with PO (n = 3 independent experiments). One-way ANOVA analysis was used. ∗P < 0.05, ∗∗P < 0.01, n.s. indicates no significance between the two indicated groups. All data are shown as the mean ± s.d. Abbreviations: cAMP, cyclic adenosine monophosphate; PO, 0.5 mM Palmitic Acid and 1 mM Oleic Acid; Ad, Adenovirus; TG, Triglyceride.
4. Discussion
In this study, we demonstrated that RGS14 protected against the progression of NAFLD and NASH. Hepatocyte-specific Rgs14 overexpression significantly ameliorated lipid accumulation, the inflammatory response and liver fibrosis in response to metabolic stress, while Rgs14-deficiency had the opposite effects. Mechanistically, further molecular research demonstrated that GoLoco/GPR domain of Rgs14 was responsible for binding with Giα1/3, which promoted the production of cAMP and activated the subsequent AMPK pathways. More importantly, inhibition of Giα1/3 blocked the destructive effects of Rgs14-deficiency in NAFLD, indicating that Rgs14 attenuated NAFLD by targeting Giα1/3.
Lacking pharmacological therapies for NAFLD, promising therapeutic targets for NAFLD are urgently needed [35,36]. GPCRs are the most frequently investigated drug targets family and mediate the therapeutic effects of ∼34 % of drugs [10,37]. The RGS proteins are emerging as essential negative modulators of GPCR signaling, and play important regulatory roles in stress resistance, hepatic lipid accumulation, inflammatory responses, fibrosis and NASH [38]. RGS14, which belongs to a subunit of the RGS family, contains a canonical RGS domain, a tandem (R1 and R2) Ras/Rap binding domain, and a GoLoco/GPR motif [11]. Most prior studies on RGS14 have focused on its functions on central nervous system and on hepatic ischemic-reperfusion injury [16,39]. However, to the best of our knowledge, no articles have reported the function and contribution of RGS14 to the initiation and progression of NAFLD. Therefore, we first used a public database (GSE126848) to analyze the expression of RGS14 in healthy individuals and NAFLD patients. The results indicated that RGS14 gene was down-regulated in NAFLD patients. We further observed that RGS14 expression gradually decreased after PO stimulation at both the protein and mRNA levels. Based on these results, we speculated that a decrease in RGS14 expression was associated with the pathological process of NAFLD.
It has become increasingly clear that lipotoxicity is a major initial trigger and accelerator of NASH and a significant facilitator of the transition from overloaded lipid accumulation to subsequent oxidative stress, an inflammatory response, and steatosis [6,40,41]. Using unbiased systemic analysis, we found that RGS14 significantly attenuated HFHC-induced lipid deposition and inhibited expression of genes involved in lipid metabolism and the inflammatory response, these two processes have been demonstrated to be the major mechanisms accelerating the progression of NAFLD. These findings further support our hypothesis that RGS14 plays a critical role in the progression of NAFLD.
As a critical regulator of intracellular energy homeostasis, AMPK can orchestrate lipolysis, lipogenesis, and fatty acid synthesis through phosphorylation and inhibition of key substrates, involving acetyl-CoA carboxylase (ACC), and sterol response element-binding protein 1c (SREBP1c) [32,33]. The cAMP/AMPK axis negatively regulates lipogenesis and increases the expression of genes involved in fatty acid oxidation in hepatocytes [42]. However, how RGS14 fine-tunes the activation of AMPK signaling has not been determined. In this study, we demonstrated that RGS14 augmented cAMP levels and preceded the activation of AMPK pathway signaling. Importantly, we demonstrated that RGS14 physically interacted with Giα1/3 via Q518 amino acid site of GoLoco/GPR domain and subsequently inhibited the generation of cAMP to ameliorate hepatic steatosis.
Previous studies reported that RGS domain and GoLoco/GPR domain of RGS14 mediate inhibitory interactions with inactive Giα1/3, thus temporally inhibiting Gi protein signaling [11,43]. The GoLoco/GPR motif serves as an inhibitor of GDP dissociation (GDI), and the RGS domain is responsible for their GTPase-activating protein activity [44]. However, which function is dominant in the modulation of NAFLD has not been determined. In our study, we found that the destruction of GDI activity by the point mutant Q518 blocked the beneficial effect of RGS14. Thus, we speculate GoLoco/GPR motif, rather than its RGS domain, forms intimate contacts with the Giα1/3 in NAFLD. Our present study found RGS14 interacted with Giα1/3 through Q518 amino acid site in GoLoco/GPR motif of RGS14. Furthermore, there are no published report the contribution of Giα1/3 to the modulation of RGS14 in NAFLD. In the present study, we demonstrated that RGS14 inhibited Giα1/3 to rescue the progression of NAFLD. The upregulation of RGS14 may inhibit the inhibitory effect of Giα1/3 on adenylyl cyclase, thereby augmenting Gαs-induced adenylyl cyclase activation, which results in increased cAMP accumulation, activating the subsequent AMPK signaling pathway. Overall, the RGS14-Giα1/3 interaction is essential for RGS14-modulated liver steatosis, and the RGS14-Giα1/3 axis is a promising therapeutic target for the treatment of these pathological conditions.
However, there are few studies on the direct interaction between Giα-coupled GPCRs and RGS14. It well-known that cannabinoid receptor type 1 (CB1R), as a GPCR, interacts with the Giα1 protein, increasing lipogenesis in liver by maximizing de novo lipogenesis and triglyceride accumulation and minimizing lipolysis and fatty acid oxidation via AMPK signaling [45]. Agudelo et al. reported that RGS14 inhibited the Giα-coupled receptor 35 (GPR35) in adipose tissue, stimulated cAMP signaling [43]. In the future, we will further explore which Gi-coupled GPCRs are regulated by RGS14 in hepatocytes.
In conclusion, our present study showed that hepatocyte RGS14 protected against steatosis during the progression of NAFLD. Mechanistically, we demonstrated that the RGS14-Giα1/3 interaction promoted the activation of adenylyl cyclase, resulting in an increase in cAMP and subsequent activation of the AMPK signaling pathway. Our findings provide novel insights into NAFLD and suggest that targeting RGS14 or modulating the RGS14-Giα1/3 interaction may be potential strategies for conferring protection against NAFLD.
Funding
This work was supported by the National Natural Science Foundation of China (No. 82100602).
AUTHOR CONTRIBUTIONS
Junyong Wang: Writing – review & editing, Writing – original draft, Visualization, Validation, Resources, Project administration, Methodology, Data curation, Conceptualization. Yaping Guo: Visualization, Supervision, Software, Formal analysis. Yunduan He: Visualization, Validation, Supervision, Methodology, Formal analysis, Data curation, Conceptualization. Yifan Qin: Visualization, Software, Methodology, Formal analysis, Data curation. Xiuling Li: Supervision, Project administration, Conceptualization. Ling Yang: Supervision, Investigation, Formal analysis, Conceptualization. Kangdong Liu: Writing – review & editing, Writing – original draft, Supervision, Project administration, Investigation, Formal analysis, Conceptualization. Li Xiao: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Resources, Methodology, Investigation, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2024.101882.
Contributor Information
Kangdong Liu, Email: kdliu@zzu.edu.cn.
Li Xiao, Email: xiaoli73@zzu.edu.cn, lixiao5366@163.com.
Appendix A. Supplementary data
The following is the supplementary data to this article:
figs1.
Figure S1. RGS14 protein expression in various metabolically tissues of control and Rgs14-HKO mice. Western bolt analysis of Rgs14 protein expression of Control and Rgs14-HKO mice in liver, heart, lung, kidney and adipose, β-Actin was served as internal control (n = 3 mice per group). A two-tailed Student t test analysis was used. ∗P < 0.05, ∗∗P < 0.01, n.s. indicates no significance between the two indicated groups. All data are shown as the mean ± s.d.; Figure S2. Overexpression of GNAI1/3 promotes hepatocyte steatosis and inflammation. (A) Western blot analysis of Giα1/3 expression in primary mouse hepatocytes transfected with indicated adenovirus (n = 3 independent experiments). (B) cAMP levels in Giα1 or Giα3 overexpressed primary mouse hepatocytes following PO exposure for 12 h (n = 3 independent experiments). (C) Representative images (n = 3 independent experiments) and quantification (n = 7 high power fields per group) of Nile Red staining in Giα1 or Giα3 overexpressed primary mouse hepatocytes under PO treatment. Scale bar, 10 um. (D) Intracellular TG content in in Giα1 or Giα3 overexpressed primary mouse hepatocytes under PO treatment (n = 3 independent experiments). (E–G) qPCR analysis of mRNA levels of fatty acid uptake and synthesis genes (E), fatty acid oxidation genes (F) and inflammation genes (G) in infected primary mouse hepatocytes treated with PO (n = 3 independent experiments). One-way ANOVA analysis was used. ∗P < 0.05, ∗∗P < 0.01, n.s. indicates no significance between the two indicated groups. All data are shown as the mean ± s.d. Abbreviations: Ad, Adenovirus; cAMP, cyclic adenosine monophosphate; TG, Triglyceride; PO, 0.5mM Palmitic Acid and 1mM Oleic Acid; Figure S3. Inhibition of GNAI1/3 improves hepatocyte steatosis and inflammation. (A) Western blot analysis of Giα1or Giα3 expression in primary mouse hepatocytes transfected with indicated adenovirus (n = 3 independent experiments). (B) cAMP levels in Giα1-knockdown, Giα3-knockdown or Giα1/3-knockdown primary mouse hepatocytes following PO exposure for 12 h (n = 3 independent experiments). (C) Representative images (n = 3 independent experiments) and quantification (n = 7 high power fields per group) of Nile Red staining in Giα1-knockdown, Giα3-knockdown or Giα1/3-knockdown primary mouse hepatocytes under PO treatment. Scale bar, 10 um. (D) Intracellular TG content in Giα1-knockdown, Giα3-knockdown or Giα1/3-knockdown primary mouse hepatocytes under PO treatment (n = 3 independent experiments). (E–G) qPCR analysis of the mRNA level of fatty acid uptake and synthesis genes (E), fatty acid oxidation genes (F) and inflammation genes (G) in Giα1-knockdown, Giα3-knockdown or Giα1/3-knockdown primary mouse hepatocytes under treated with PO 12 h (n = 3 independent experiments). A two-tailed Student t test (B) and one-way ANOVA (A and C–G) analysis was used. ∗P < 0.05, ∗∗P < 0.01, n.s. indicates no significance between the two indicated groups. All data are shown as the mean ± s.d. Abbreviations: Ad, Adenovirus; cAMP, cyclic adenosine monophosphate; TG, Triglyceride; PO, 0.5 mM Palmitic Acid and 1 mM Oleic Acid.
Data availability
Data will be made available on request.
References
- 1.Eslam M., Valenti L., Romeo S. Genetics and epigenetics of NAFLD and NASH: clinical impact. J Hepatol. 2018;68(2):268–279. doi: 10.1016/j.jhep.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Younossi Z., Anstee Q.M., Marietti M., Hardy T., Henry L., Eslam M., et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15(1):11–20. doi: 10.1038/nrgastro.2017.109. [DOI] [PubMed] [Google Scholar]
- 3.Drew L. Drug development: sprint finish. Nature. 2017;551(7681) doi: 10.1038/d41586-017-06926-1. [DOI] [PubMed] [Google Scholar]
- 4.Sheka A.C., Adeyi O., Thompson J., Hameed B., Crawford P.A., Ikramuddin S. Nonalcoholic steatohepatitis: a review. JAMA. 2020;323(12):1175–1183. doi: 10.1001/jama.2020.2298. [DOI] [PubMed] [Google Scholar]
- 5.Yang L., Roh Y.S., Song J., Zhang B., Liu C., Loomba R., et al. Transforming growth factor beta signaling in hepatocytes participates in steatohepatitis through regulation of cell death and lipid metabolism in mice. Hepatology. 2014;59(2):483–495. doi: 10.1002/hep.26698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman S.L., Neuschwander-Tetri B.A., Rinella M., Sanyal A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24(7):908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun D., Yang X., Wu B., Zhang X.J., Li H., She Z.G. Therapeutic potential of G protein-coupled receptors against nonalcoholic steatohepatitis. Hepatology. 2021;74(5):2831–2838. doi: 10.1002/hep.31852. [DOI] [PubMed] [Google Scholar]
- 8.Anton S.E., Kayser C., Maiellaro I., Nemec K., Möller J., Koschinski A., et al. Receptor-associated independent cAMP nanodomains mediate spatiotemporal specificity of GPCR signaling. Cell. 2022;185(7):1130–1142. doi: 10.1016/j.cell.2022.02.011. e11. [DOI] [PubMed] [Google Scholar]
- 9.Hauser A.S., Chavali S., Masuho I., Jahn L.J., Martemyanov K.A., Gloriam D.E., et al. Pharmacogenomics of GPCR drug targets. Cell. 2018;172(1–2):41–54. doi: 10.1016/j.cell.2017.11.033. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell A.P., Smrcka A.V. Targeting G protein-coupled receptor signalling by blocking G proteins. Nat Rev Drug Discov. 2018;17(11):789–803. doi: 10.1038/nrd.2018.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shu F.J., Ramineni S., Hepler J.R. RGS14 is a multifunctional scaffold that integrates G protein and Ras/Raf MAPkinase signalling pathways. Cell Signal. 2010;22(3):366–376. doi: 10.1016/j.cellsig.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coleman D.E., Berghuis A.M., Lee E., Linder M.E., Gilman A.G., Sprang S.R. Structures of active conformations of Gi alpha 1 and the mechanism of GTP hydrolysis. Science. 1994;265(5177):1405–1412. doi: 10.1126/science.8073283. [DOI] [PubMed] [Google Scholar]
- 13.Zhang P., Mende U. Regulators of G-protein signaling in the heart and their potential as therapeutic targets. Circ Res. 2011;109(3):320–333. doi: 10.1161/circresaha.110.231423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DerMardirossian C., Bokoch G.M. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15(7):356–363. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Vellano C.P., Lee S.E., Dudek S.M., Hepler J.R. RGS14 at the interface of hippocampal signaling and synaptic plasticity. Trends Pharmacol Sci. 2011;32(11):666–674. doi: 10.1016/j.tips.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J.K., Ding M.J., Liu H., Shi J.H., Wang Z.H., Wen P.H., et al. Regulator of G-protein signaling 14 protects the liver from ischemia-reperfusion injury by suppressing TGF-β-activated kinase 1 activation. Hepatology. 2022;75(2):338–352. doi: 10.1002/hep.32133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X.J., She Z.G., Wang J., Sun D., Shen L.J., Xiang H., et al. Multiple omics study identifies an interspecies conserved driver for nonalcoholic steatohepatitis. Sci Transl Med. 2021;13(624) doi: 10.1126/scitranslmed.abg8117. [DOI] [PubMed] [Google Scholar]
- 18.Hu Y., He W., Huang Y., Xiang H., Guo J., Che Y., et al. Fatty acid synthase-suppressor screening identifies sorting Nexin 8 as a therapeutic target for NAFLD. Hepatology. 2021;74(5):2508–2525. doi: 10.1002/hep.32045. [DOI] [PubMed] [Google Scholar]
- 19.Bai Z., Yang P., Yu F., Li Z., Yao Z., Martinez J., et al. Combining adoptive NK cell infusion with a dopamine-releasing peptide reduces senescent cells in aged mice. Cell Death Dis. 2022;13(4):305. doi: 10.1038/s41419-022-04562-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suppli M.P., Rigbolt K.T.G., Veidal S.S., Heebøll S., Eriksen P.L., Demant M., et al. Hepatic transcriptome signatures in patients with varying degrees of nonalcoholic fatty liver disease compared with healthy normal-weight individuals. Am J Physiol Gastrointest Liver Physiol. 2019;316(4):G462–G472. doi: 10.1152/ajpgi.00358.2018. [DOI] [PubMed] [Google Scholar]
- 21.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho H., Kim D.U., Kehrl J.H. RGS14 is a centrosomal and nuclear cytoplasmic shuttling protein that traffics to promyelocytic leukemia nuclear bodies following heat shock. J Biol Chem. 2005;280(1):805–814. doi: 10.1074/jbc.M408163200. [DOI] [PubMed] [Google Scholar]
- 23.Brown N.E., Goswami D., Branch M.R., Ramineni S., Ortlund E.A., Griffin P.R., et al. Integration of G protein α (Gα) signaling by the regulator of G protein signaling 14 (RGS14) J Biol Chem. 2015;290(14):9037–9049. doi: 10.1074/jbc.M114.634329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vellano C.P., Maher E.M., Hepler J.R., Blumer J.B. G protein-coupled receptors and resistance to inhibitors of cholinesterase-8A (Ric-8A) both regulate the regulator of g protein signaling 14 RGS14·Gαi1 complex in live cells. J Biol Chem. 2011;286(44):38659–38669. doi: 10.1074/jbc.M111.274928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Q., Yang S., Wei C., Lu G., Lee K., He J.C., et al. Puerarin attenuates diabetic kidney injury through interaction with Guanidine nucleotide-binding protein Gi subunit alpha-1 (Gnai1) subunit. J Cell Mol Med. 2022;26(14):3816–3827. doi: 10.1111/jcmm.17414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh M., Bergmann L., Lang A., Pexa K., Kuck F., Stibane D., et al. Gα(i3) signaling is associated with sexual dimorphic expression of the clock-controlled output gene Dbp in murine liver. Oncotarget. 2018;9(54):30213–30224. doi: 10.18632/oncotarget.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gebski E.B., Parikh V., Lam H., Kim N., Bochkov Y.A., Cao G., et al. Rhinovirus C15 attenuates relaxation and cAMP production in human airways and smooth muscle. Am J Respir Cell Mol Biol. 2023;69(2):172–181. doi: 10.1165/rcmb.2021-0526OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin S., Song R., Ma J., Liu C., Wu Z., Cao G., et al. Receptor activity-modifying protein 1 regulates mouse skin fibroblast proliferation via the Gαi3-PKA-CREB-YAP axis. Cell Commun Signal. 2022;20(1):52. doi: 10.1186/s12964-022-00852-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X., Wang K., Wang L., Kong L., Hou S., Wan Y., et al. Hepatocyte leukotriene B4 receptor 1 promotes NAFLD development in obesity. Hepatology. 2023;78(2):562–577. doi: 10.1002/hep.32708. [DOI] [PubMed] [Google Scholar]
- 30.Ravnskjaer K., Madiraju A., Montminy M. Role of the cAMP pathway in glucose and lipid metabolism. Handb Exp Pharmacol. 2016;233:29–49. doi: 10.1007/164_2015_32. [DOI] [PubMed] [Google Scholar]
- 31.Medina E.A., Oberheu K., Polusani S.R., Ortega V., Velagaleti G.V., Oyajobi B.O. PKA/AMPK signaling in relation to adiponectin's antiproliferative effect on multiple myeloma cells. Leukemia. 2014;28(10):2080–2089. doi: 10.1038/leu.2014.112. [DOI] [PubMed] [Google Scholar]
- 32.Liang C., Li Y., Bai M., Huang Y., Yang H., Liu L., et al. Hypericin attenuates nonalcoholic fatty liver disease and abnormal lipid metabolism via the PKA-mediated AMPK signaling pathway in vitro and in vivo. Pharmacol Res. 2020;153 doi: 10.1016/j.phrs.2020.104657. [DOI] [PubMed] [Google Scholar]
- 33.Jian C., Fu J., Cheng X., Shen L.J., Ji Y.X., Wang X., et al. Low-dose Sorafenib acts as a mitochondrial uncoupler and ameliorates nonalcoholic steatohepatitis. Cell Metab. 2020;31(5):892–908. doi: 10.1016/j.cmet.2020.04.011. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu H.Y., Yu L., Chen J.H., Yang L.N., Lin C., Shi X.Q., et al. Sesamol alleviates obesity-related hepatic steatosis via activating hepatic PKA pathway. Nutrients. 2020;12(2) doi: 10.3390/nu12020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Powell E.E., Wong V.W., Rinella M. Non-alcoholic fatty liver disease. Lancet. 2021;397(10290):2212–2224. doi: 10.1016/s0140-6736(20)32511-3. [DOI] [PubMed] [Google Scholar]
- 36.Younossi Z., Tacke F., Arrese M., Chander Sharma B., Mostafa I., Bugianesi E., et al. Global perspectives on nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatology. 2019;69(6):2672–2682. doi: 10.1002/hep.30251. [DOI] [PubMed] [Google Scholar]
- 37.Hauser A.S., Chavali S., Masuho I., Jahn L.J., Martemyanov K.A., Gloriam D.E., et al. Pharmacogenomics of GPCR drug targets. Cell. 2018;172(1–2):41–54. doi: 10.1016/j.cell.2017.11.033. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J., Ma J., Nie H., Zhang X.J., Zhang P., She Z.G., et al. Hepatic regulator of G protein signaling 5 ameliorates nonalcoholic fatty liver disease by suppressing transforming growth factor beta-activated kinase 1-c-Jun-N-terminal kinase/p38 signaling. Hepatology. 2021;73(1):104–125. doi: 10.1002/hep.31242. [DOI] [PubMed] [Google Scholar]
- 39.Harbin N.H., Bramlett S.N., Montanez-Miranda C., Terzioglu G., Hepler J.R. RGS14 regulation of post-synaptic signaling and spine plasticity in brain. Int J Mol Sci. 2021;22(13) doi: 10.3390/ijms22136823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuster S., Cabrera D., Arrese M., Feldstein A.E. Triggering and resolution of inflammation in NASH. Nat Rev Gastroenterol Hepatol. 2018;15(6):349–364. doi: 10.1038/s41575-018-0009-6. [DOI] [PubMed] [Google Scholar]
- 41.Mota M., Banini B.A., Cazanave S.C., Sanyal A.J. Molecular mechanisms of lipotoxicity and glucotoxicity in nonalcoholic fatty liver disease. Metabolism. 2016;65(8):1049–1061. doi: 10.1016/j.metabol.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Omar B., Zmuda-Trzebiatowska E., Manganiello V., Göransson O., Degerman E. Regulation of AMP-activated protein kinase by cAMP in adipocytes: roles for phosphodiesterases, protein kinase B, protein kinase A, Epac and lipolysis. Cell Signal. 2009;21(5):760–766. doi: 10.1016/j.cellsig.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Agudelo L.Z., Ferreira D.M.S., Cervenka I., Bryzgalova G., Dadvar S., Jannig P.R., et al. Kynurenic acid and Gpr35 regulate adipose tissue energy homeostasis and inflammation. Cell Metab. 2018;27(2):378–392. doi: 10.1016/j.cmet.2018.01.004. e5. [DOI] [PubMed] [Google Scholar]
- 44.Shu F-j, Ramineni S., Hepler J.R. RGS14 is a multifunctional scaffold that integrates G protein and Ras/Raf MAPkinase signalling pathways. Cell Signal. 2010;22(3):366–376. doi: 10.1016/j.cellsig.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J., Godlewski G., Jourdan T., Liu Z., Cinar R., Xiong K., et al. Cannabinoid-1 receptor antagonism improves glycemic control and increases energy expenditure through Sirtuin-1/mechanistic target of Rapamycin complex 2 and 5'Adenosine monophosphate-activated protein kinase signaling. Hepatology. 2019;69(4):1535–1548. doi: 10.1002/hep.30364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.