Highlights
-
•
Chronic aerobic exercise decreases serum BDNF and increases serum IL-6 levels.
-
•
DNA 5-hmC undergoes significant alterations in a rodent model of TLE.
-
•
Chronic aerobic exercise reinstates DNA 5-hmC levels in TLE rats.
-
•
TET1 emerges as the central enzyme affected by chronic aerobic exercise in TLE.
-
•
Exercise-driven DNA 5-hmC changes in TLE are neuron-specific.
Keywords: Epigenetics, Neurons, Astrocytes, 5-hydroxymethylcytosine, Ten-eleven translocation, Chronic exercise, Acute exercise
Abstract
The therapeutic potential of aerobic exercise in mitigating seizures and cognitive issues in temporal lobe epilepsy (TLE) is recognized, yet the underlying mechanisms are not well understood. Using a rodent TLE model induced by Kainic acid (KA), we investigated the impact of a single bout of exercise (i.e., acute) or 4 weeks of aerobic exercise (i.e., chronic). Blood was processed for epilepsy-associated serum markers, and DNA methylation (DNAme), and hippocampal area CA3 was assessed for gene expression levels for DNAme-associated enzymes. While acute aerobic exercise did not alter serum Brain-Derived Neurotrophic Factor (BDNF) or Interleukin-6 (IL-6), chronic exercise resulted in an exercise-specific decrease in serum BDNF and an increase in serum IL-6 levels in epileptic rats. Additionally, whole blood DNAme levels, specifically 5-hydroxymethylcytosine (5-hmC), decreased in epileptic animals following chronic exercise. Hippocampal CA3 5-hmC levels and ten-eleven translocation protein (TET1) expression mirrored these changes. Furthermore, immunohistochemistry analysis revealed that most 5-hmC changes in response to chronic exercise were neuron-specific within area CA3 of the hippocampus. Together, these findings suggest that DNAme mechanisms in the rodent model of TLE are responsive to chronic aerobic exercise, with emphasis on neuronal 5-hmC DNAme in the epileptic hippocampus.
Introduction
Epilepsy is one of the most common neurological diseases characterized by unprovoked, synchronous seizure activity. Epilepsy is a complex disease with different subtypes and presentation of symptoms depending on the area of seizure origin [1]. Temporal lobe epilepsy (TLE) is the most common form of acquired epilepsy, with seizures largely originating from the hippocampus, and it is characterized by hippocampal sclerosis and cell loss [2]. The main therapeutic option for people with epilepsy continues to be anti-seizure medications (ASMs) that may require multiple tryouts and combinations before seizure regulation is achieved. More importantly, ASMs primarily help treat seizure occurrence but provide little to no relief for the associated epileptic comorbidities such as memory and other cognitive impairments, depression and other mood disorders, and sleep disorders [3]. Additional alternative treatment options that can be used in combination with ASMs are often necessary to improve the overall quality of life for people with epilepsy.
Recently, physical activity and exercise have emerged as potential options to help alleviate comorbid conditions in epilepsy [4]. Exercise is known to offer a wide range of benefits to healthy individuals, and scientists have begun to explore its potential benefits in people suffering from diseases and/or disorders [5]. Aerobic exercise promotes overall cardiovascular health and increases blood flow and oxygen to the brain in older adults [6]. A study has shown that adults with epilepsy who underwent supervised training that included aerobic exercise showed overall improved memory and cognitive performance and either improved or no change in seizure control [7], [8]. Similar effects have also been shown using rodent models of epilepsy [9]. However, how exercise induces molecular changes to impart these positive effects, specifically with epilepsy, is still unclear. One potential mechanism involves epigenetic modifications [10].
Epigenetic regulation includes DNA methylation, histone modifications, and non-coding RNAs. These modifications alter chromatin structure and function, leading to changes in gene expression [11]. There is increasing evidence in the literature indicating the importance of epigenetic mechanisms in epilepsy [12] and how they can influence gene expression [13]. Although previously believed to be static changes, epigenetic mechanisms are dynamic and active throughout a lifetime in the human brain [14]. DNA methylation is a major epigenetic mark involving a covalent addition of a methyl group at the 5-carbon cytosine, creating 5-methylcytosine (5-mC). DNA methylation at 5-mC can be oxidized to 5-hydroxymethylcytosine (5-hmC), and then further oxidized to formylcytosine (5-fC) and 5-carboxycytosine (5-caC) [15]. DNA methylation is an enzymatic process mediated by DNA methyltransferases (DNMTs) and the ten-eleven translocation (TET) enzymes [16]. Although 5-hmC was previously described as a transient epigenetic mark, we now know that it is a major regulatory epigenetic mark with enriched levels of 5-hmC found specifically in the brain [17].
The DNA methylation hypothesis in epileptogenesis suggests that DNA methylation changes contribute to the progression of the disease [12]. In our previous work, we have shown that DNA methylation at the Bdnf gene is impacted with epilepsy-related cognitive deficits, and supplementation with methionine to increase DNA methylation rescued memory deficits and methylation at the Bdnf gene [18]. Additionally, others have shown DNA methylation changes with exercise can be gene-specific, such as with Bdnf [19], [20]. Therefore, DNA methylation is likely to play an important role in how exercise exerts positive benefits with epilepsy. Unfortunately, these previous studies were limited in that they did not afford us the opportunity to independently measure the two major forms of DNA methylation in the brain, 5-mC and 5-hmC. Since then, advancements in the field have allowed us the opportunity to employ approaches to distinguish between these two marks, 5-mC and 5-hmC, and the impact of exercise on epilepsy.
In the present study, we first wanted to determine which exercise modality would be most effective to explore the effects of exercise in our experimental model of TLE. Here, we used peripheral blood markers to compare acute aerobic exercise versus chronic aerobic exercise 24 h post each intervention. There has been growing interest in monitoring blood markers in response to different exercise modalities in addition to how they can be used for better clinical relevance [21]. We chose to measure two known serum markers for epilepsy: BDNF and IL-6 [22], [23]. We then explored which of the two major forms of DNA methylation is most affected by aerobic exercise in DNA from whole blood and in the epileptic hippocampus. Furthermore, we looked at which of the different DNA methylation enzymes are driving this change. We used both ELISA and immunohistochemistry to measure bulk levels of DNA methylation in hippocampal area CA3 and then further explored the cell type specificity using neuron and astrocyte-specific labeling.
Materials and methods
Animals
2-month-old male Fisher-344 (F344) rats received from Harlan weighing 160–180 g at the time of arrival were used for these experiments. Animals were double housed in pairs in plastic cages and had access to water and NIH-31 lab rat diet ad libitum. Upon arrival, animals were put on a reverse light–dark cycle with lights off at 12:00p.m. and lights on at 12:00 a.m. Animals adapted to the new light cycle for 3 weeks prior to any experimental procedures and were handled 3–4 times a week by investigators. During the animal’s dark cycle, a red light was used to provide adequate lighting for the investigators to perform routine housing tasks, animal handling, and training. All procedures were approved by the University of Alabama at Birmingham Institutional Animal Care and Use Committee and done in accordance with the National Institute of Health and ethical guidelines.
Treadmill familiarization and pre-selection protocol
All treadmill activities were performed on the Panlab 5-lane rat treadmill (Harvard Instruments, model). Treadmill familiarization and training protocol was adapted from the NIH common fund MoTrPAC animal protocol [24]. In short, after 3 weeks of light–dark cycle adaptation, animals underwent a 12-day familiarization protocol to get accustomed to the treadmill and to identify any non-compliant runners. Day 1–2, animals were placed on a non-moving treadmill at 0 m/min for 10 min while shock grids were blocked to avoid animals sitting on the shock grid. Day 3–5, the shock area remained blocked, animals ran at 6 m/min for 10 min, and a pen was used to gently probe animals forward on the treadmill. Day 6–9, the shock area remained blocked, and animals ran at 10 m/min for 10 mins while continuing to use a pen to probe the animals forward. During days 10–12, a light shock was used to encourage animals to run at 10 m/min for 10 min. On day 11, after each 10-minute familiarization, the treadmill grade was increased to 10 degrees and speed to 12 m/min for an additional 2 min. Day 12 was evaluation day; rats ran on the treadmill for 5 min at 10 m/min. The final evaluation happened at an increased grade of 10 degrees at 12 m/min for 5 mins. Animals were scored based on their ability to complete the exercise protocol, with a score of 1 indicating the animal was non-compliant and did not complete the activity session. A score of 2 indicated the animal required assistance for more than 25 % of the activity. A score of 3 indicated the animal required some assistance for less than 25 % of the activity, and a score of 4 indicated the animal required minimal to no assistance to complete the activity. Once treadmill familiarization was complete and all animals were scored, animals were sorted into groups with the compliant runners (a score of 3–4) included in the exercise groups.
Temporal lobe epilepsy model
Forty-eight hours after completing treadmill familiarization, half of the animals were subcutaneously (s.c.) injected with saline (SA) (control), and the other half with 9 mg/kg of KA (0222, Tocris, Minneapolis, MN, USA). Behavioral seizure severity was measured using the Racine scale [25]: 1 - mouth and face clonus and head nodding; 2 - clonic jerks of one forelimb; 3 - bilateral forelimb clonus; 4 - forelimb clonus and rearing; 5 - forelimb clonus with rearing and falling. The onset of status epilepticus (SE) was defined as the time from KA injection to the start of continuous seizure activity with a score of 5 on the Racine scale until seizure activity stopped [26]. Once KA-injected animals reached 1 h and 30 min in SE, SE was terminated with an intraperitoneal injection (i.p.) of Midazolam 10 mg/kg. Control saline animals also received the Midazolam injection at 10 mg/kg to reduce any confounding factors between groups. Additionally, all animals received 3–5 ml of saline injection at about 8 h post-KA/SA injection to avoid dehydration. This F344 protocol has been characterized, and animals start to spontaneously seize at 1–2 weeks post-SE induction and develop full epileptic pathology by 28 days post-SE [27], [28].
Aerobic exercise intervention
The treadmill aerobic exercise protocol was adapted from the NIH common fund MoTrPAC animal protocol [24]. Exercise training began at 6 weeks post-SE induction, during which all epileptic animals experienced 2 or more behavioral seizures. For the acute exercise protocol, food was removed from the cages 4 h prior to the exercise protocol. The animals ran for 30 min at 5 degrees (8.7 %) grade, at 28 cm/s, using light shock for motivation. Tissue and whole blood were collected 24 h after completion of the acute exercise protocol, which was around 6 weeks post-SE induction.
For the 4-week chronic exercise protocol, animals were exercised for 5 days per week using an incremental training protocol designed to exercise F344 rats at about 70 % VO2max. Training was conducted during the dark/active cycle and would always start 2 h after the lights were turned off. Day 1 of training started at 5 degrees grade and 13 m/min for 20 min, and day 20 concluded at 10 degrees grade and 18 m/min for 30 min. Supplementary Table 1 details the daily exercise protocol. If an animal was unable to complete the exercise activity on 3 consecutive days, it was excluded from the study and downstream analysis. Animals assigned to sedentary control groups were also exposed daily to the treadmill at 0 m/min for 10 min 5 days per week. Animals were euthanized by rapid decapitation 24 h after the last bout of exercise at about 10 weeks post-SE induction. Trunk whole blood, plasma, and serum were collected post decapitation, 1 brain hemisphere was collected and frozen on dry ice, and the other hemisphere was used to isolate the hippocampus and subdissected in ice-cold oxygenated artificial cerebral spinal fluid (ACSF). The cornu ammonis (CA) region 3 was collected and frozen on dry ice.
Serum BDNF and IL-6
Trunk whole blood was collected in silicone-coated/clot-activating tubes (BD Vacutainer) and inverted 3 times. After collection, the whole blood was left at room temperature for 1 h and then placed on ice to be kept cold, allowing clots to form. Tubes were then centrifuged 2,000 x g for 10 min in a refrigerated centrifuge for serum separation. The supernatant/serum was then collected and aliquoted into CryoTubes and then frozen at −80 °C, and later used for protein measurements. Avoiding freeze–thaw cycles, serum BDNF levels were measured using a quantikine total BDNF immunoassay (R&D Systems) according to the manufacturer’s protocols. Serum IL-6 levels were measured using an IL-6 quantikine ELISA kit (R&D Systems) according to the manufacturer’s protocol.
DNA and RNA isolation
Trunk whole blood samples collected in K2 EDTA coated tubes (BD Vacutainer) were aliquoted and frozen at −80 °C. Total DNA was isolated from frozen whole blood, and hippocampal tissue collected, area CA3, using DNeasy blood and tissue kit (QIAGEN) according to manufacturer’s protocol. DNA concentration and purity were quantified using a Nanodrop spectrometer. RNA was isolated from hippocampal tissue collected, area CA3, using RNeasy mini kit (QIAGEN) according to manufacturer protocol. RNA purity and quantity were measured using Nanodrop spectrometer.
Total 5-methylcytosine and 5-hydrocymethylcytosine
Total 5-mC and 5-hmC were measured using a colorimetric ELISA-like assay MethylFlash Methylated DNA 5-mC Quantification Kit (Epigentek) and MethylFlash Hydroxymethylated DNA 5-hmC Quantification Kit (Epigentek) according to manufacturer’s guidelines.
Quantitative real-time PCR (qRT-PCR)
100 ng of total RNA isolated was converted to cDNA using iScript cDNA synthesis kit (Bio-Rad). Real-time PCR amplification was performed on the Biorad CFX-96 Real-time system using TaqMan® Fast Advanced Master Mix and TaqMan® Gene expression assay following protocol: UNG activation at 50.0 °C for 2 min, polymerase activation at 95.0 °C for 20 s, denature at 95.0 °C for 3 s, followed by an Anneal/Extend at 60 °C for 40 cycles. Hprt1 (hypoxanthine phosphoribosyl transferase 1) expression was used to normalize gene expression. Cycle threshold (Ct) values were analyzed using the comparative Ct method to calculate differences in gene expression between samples. Primers are as specified: Hprt1 assay ID: Rn01527840_m1 VIC-MGB, Dnmt2 assay ID: Rn00709664_m1, Dnmt3 assay ID: Rn01027162_g1, Dnmt3b assay ID: Rn01536418_g1, Tet1 assay ID: Rn01428192_m1, Tet2 assay ID: Rn01522037_m1, Tet3 assay ID: Rn01425643_m1.
Immunohistochemistry, imaging, and quantification
Fresh frozen brains were embedded in Optimal Cutting Temperature compound (O.C.T., Tissue-Tek, Sakura Finetek USA), and serial coronal sections at 14 µm were collected using a Leica CM1950 cryostat. Slides were stored at −80 °C prior to immunofluorescence. Slides were fixed in 10 % Buffered Formalin for 15 min at room temperature, then washed 5 times in Phosphate-Buffered Saline (PBS) for 5 min. Sections underwent antigen retrieval by being submerged in boiling sodium citrate buffer and incubated for 30 min while the buffer cooled. Slides were then washed 5 times in PBS for 5 min and then dried using a Kimwipe. The sections were then incubated in blocking buffer for 1 h at room temperature. The blocking buffer contained 5 % normal donkey serum, 5 % normal goat serum, 0.3 % Triton-X and 10 % Bovine Serum Albumin (BSA) in PBS. Sections were incubated overnight at 4 °C with primary antibodies in blocking buffer. Primary antibodies used: NeuN (1:1000, Millipore Sigma, MAB377), GFAP (1:1000, Abcam, ab4674), 5-hmC (1:250, Active Motif, 39791). After overnight incubation, slides were washed 5 times in PBS for 5 min and then incubated in secondary antibodies at 4 °C for 2 h. Secondary antibodies used: Donkey anti-mouse Alexa fluor 647 (1:500, Invitrogen, A32787), goat anti-rabbit Alexa fluor 488 (1:500, Invitrogen, A11008), donkey anti-chicken Cy3 (1:500, Jackson Immuno Research, 703–165-155). Following secondary incubation, slides were washed 5 times for 5 min in PBS and then dried using Kimwipe. We used Invitrogen ProLong Diamond Antifade Mountant with DAPI to mount slides and allowed drying for at least 30 min prior to imaging. Images were collected using an Olympus VS200 Research Slide Scanner, the hippocampus was selected, and optical sections were acquired at 40x and collapsed to a single image for quantification. Images were imported into QuPath software for analysis, and the CA3 hippocampal subfield was traced and annotated. We used the Positive Cell detection function to measure the percentage of cells with 5-hmC (FITC) and DAPI as total cell detection by threshold measurements. Additionally, neurons were identified by NeuN (Cy5) and astrocytes by GFAP (Cy3). Cells were then identified as 5-hmC positive neurons or 5-hmC positive astrocytes. Thresholds were set using Nucleus Mean and visually confirmed for accuracy for each image. All calculations were normalized to total cell detection by DAPI and graphed as percentages of each stain compared to the total cells detected.
Protein isolation, western blot, and quantification
Protein lysates were prepared using AllPrep DNA/RNA/Protein Kit (Qiagen), according to the manufacturer’s instructions. Polyacrylamide gels were poured in 8 % concentration using 30 % acrylamide/bisacrylamide solution (Bio-Rad), sodium dodecyl sulfate (SDS) (Bio-Rad), ammonium persulfate (APS) (Bio-Rad), tetramethylethylenediamine (TEMED) (Bio-Rad), Resolving Gel Buffer (Bio-Rad) and Stacking Gel Buffer (Bio-Rad). Electrophoresis of protein lysates was performed using 10 µL of each sample, followed by Ponceau S staining. Samples were transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore). The following antibodies were used for the detection of TET1, TET3, and beta-actin: anti-TET1 in dilution of 1:1000 (Thermo Fisher, PA5-72805), anti-TET3 in dilution 1:1000 (Abcam, ab 139311) and anti-beta-actin in dilution of 1:10 000 (Cell Signaling, 4967L). All membranes were incubated with a secondary anti-rabbit IgG antibody in a dilution of 1:7500, conjugated to horseradish peroxidise (HRP) (Thermo Fisher). Protein signals were detected by chemiluminescence using Clarity Max Western ECL Substrate (Bio-Rad) and quantified with ImageLab software (Bio-Rad). TET1 and beta-actin measurements were normalized to whole-cell lysate content based on the Ponceau S staining.
Statistical analysis
Data was analyzed using one-way Analysis of Variance (ANOVA) with multiple comparisons comparing each column with the mean of every other column independently. Each comparison was analyzed as a stand-alone comparison. All data sets assumed a Gaussian distribution and equal standard deviation. Where not applicable, a Brown-Forsythe test was used and noted in the text. A repeated measures ANOVA was used to analyze the effect of the 4-week chronic exercise protocol on distance traveled and weight overtime in each experimental group. For comparisons between only 2 groups (non-epileptic vs. epileptic), we used a Two-sample t-test. Graphs are presented as a mean and standard error of the mean (SEM). Each data set was screened for technical and experimental outliers, in addition to using Grubb’s test (α = 0.05), and outliers were subsequently excluded. Statistical tests and graphs were generated in GraphPad Prism 9.5.0. For all experiments, n indicates the number of biological replicates. Significant thresholds were set as * P < 0.05, ** P < 0.01, *** P < 0.001.
Results
Chronic aerobic exercise decreases serum BDNF levels and increases IL-6 levels in TLE animals
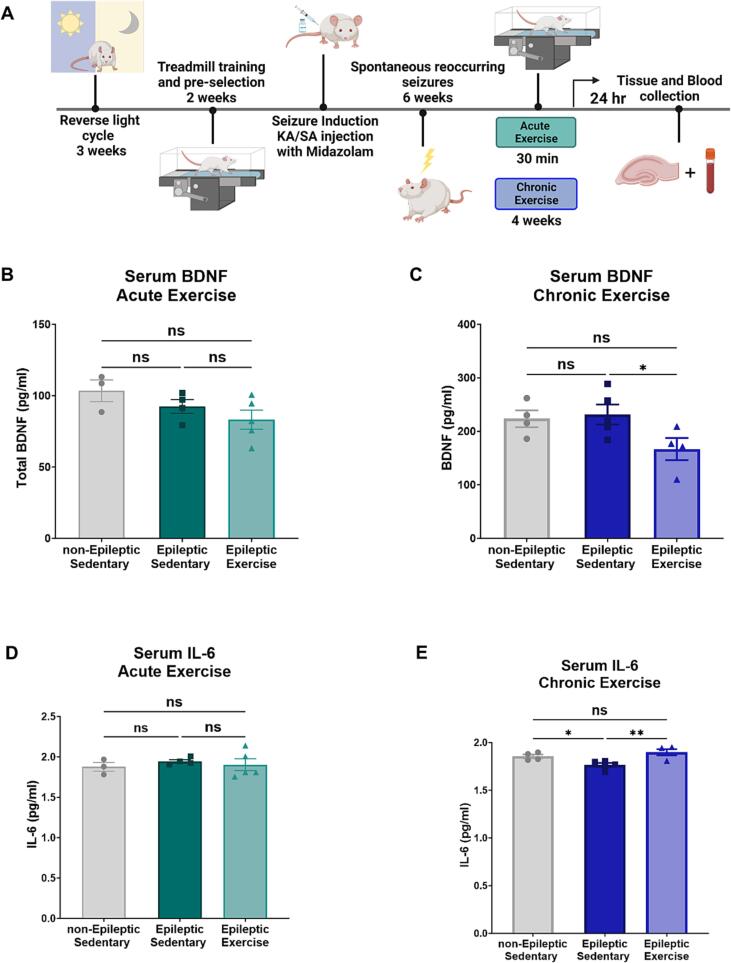
Since acute and chronic exercise can have differing effects on peripheral blood markers [21], [29], we first determined which of these two exercise protocols was sufficient to induce exercise-specific responses in our model of TLE (Fig. 1.A). To explore this, we measured two known serum markers for both epilepsy and exercise, BDNF and IL-6 [22], [23]. In our model, acute aerobic exercise did not induce any changes in serum BDNF 24 h post-exercise (Fig. 1.B) (One-way ANOVA F = 2.299, P = 0.1561). while chronic aerobic exercise (Fig. 1.C) led to a decrease in serum BDNF, specifically in epileptic exercise compared to sedentary epileptic animals (One-way ANOVA, F = 3.477, P = 0.0714, followed by multiple comparisons Uncorrected Fisher's LSD P = 0.0324 for Epileptic Sedentary vs. Epileptic Exercise). Measurement of the inflammatory marker, IL-6, showed similar results to serum BDNF with no changes in the acute aerobic exercise experiment (Fig. 1.E) (One-way ANOVA F = 0.2903, P = 0.7548). In the chronic aerobic exercise model, serum IL-6 was decreased with epilepsy and increased with the exercise intervention, similar to levels of non-epileptic controls (Fig. 1.F) (One-way ANOVA, F = 7.940, P = 0.0086, followed by multiple comparisons Uncorrected Fisher's LSD, P = 0.0257 for non-Epileptic Sedentary vs. Epileptic Sedentary and P = 0.0032 for Epileptic Sedentary vs. Epileptic Exercise). We therefore concluded that the chronic aerobic exercise protocol is more robust and effective within our outcome measures and consequently used this protocol for all other experiments.
Fig. 1.
Chronic Aerobic Exercise induces a specific response in Serum Blood markers in TLE.
Next, we confirmed the animal’s ability to complete the exercise protocol. For each experiment, we measured the distance traveled in meters for the exercise groups (Supplemental Fig. 1.A), showing no difference between epileptic and non-epileptic animals during the acute exercise (Unpaired t-test P = 0.5299). Using repeated measures ANOVA, we also compared the effect of the 4-week chronic aerobic exercise protocol on distance traveled over time between non-epileptic and epileptic animals (F (1, 8) = 1.960, P = 0.1991). Overall, epileptic animals were able to complete the exercise protocols just like their non-epileptic counterparts. We also collected weekly weight measurements for each animal for all groups. Supplemental Fig. 1.C, shows no differences in weight changes between groups during the acute exercise protocol (Repeated measure ANOVA, F (3, 28) = 0.6252, P = 0.6047). While Supplemental Fig. 1.D shows no difference in weight changes between groups during the chronic exercise protocol (Repeated measures ANOVA, F (3, 15) = 2.773, P = 0.0777). These results gave us confidence that neither distance traveled, nor weight are confounding factors in our other measurements.
Chronic aerobic exercise decreases DNA hydroxymethylation in whole blood and hippocampal area CA3 in TLE animals
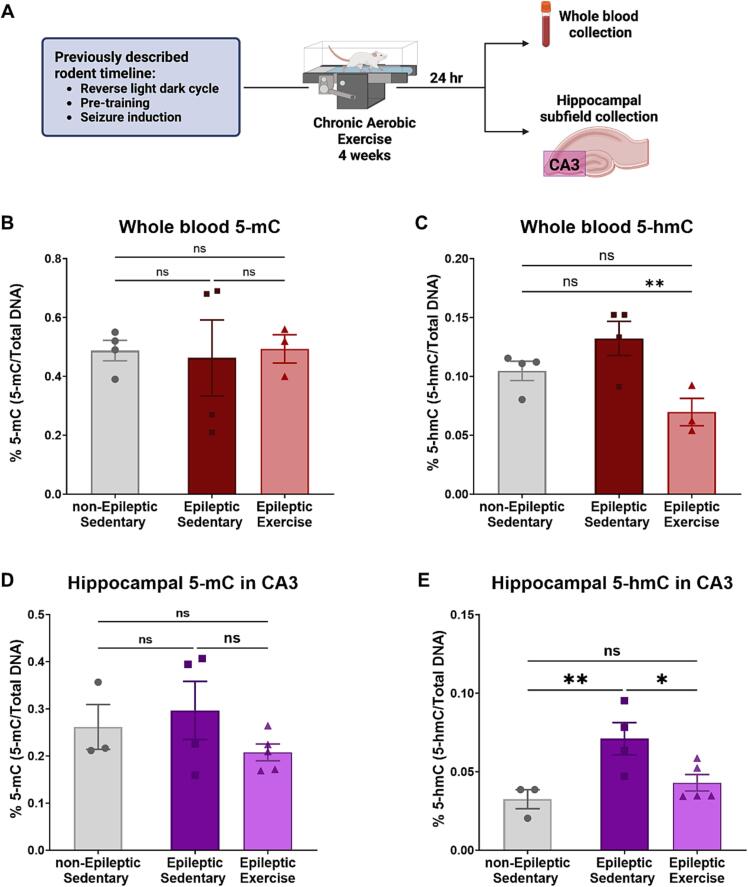
After confirming that chronic exercise is necessary to induce serum BDNF and IL-6 changes in our model of TLE, we also wanted to explore how chronic aerobic exercise impacts epigenetic markers in whole blood. Here, we investigated bulk 5-mC and 5-hmC DNA methylation in our epileptic animals in response to chronic aerobic exercise. Fig. 2.A, depicts a condensed experimental outline showing the collection of whole blood and hippocampal area CA3, 24 h after the last bout of chronic aerobic exercise (Day 20). We found that epilepsy and chronic aerobic exercise did not induce any changes in bulk 5-mC levels in whole blood DNA (Fig. 2.B, One-way ANOVA F = 0.03476, P = 0.9660). Measurement of bulk 5-hmC levels in whole blood DNA showed an increase in epileptic animals compared to non-epileptic controls, while chronic aerobic exercise led to a decrease in epileptic compared to epileptic sedentary without the exercise intervention (Fig. 2.C) (One-way ANOVA F = 6.533, P = 0.0208, followed by multiple comparisons Uncorrected Fisher's LSD, P = 0.1237 for non-Epileptic Sedentary vs. Epileptic Sedentary and P = 0.0068 for Epileptic Sedentary vs. Epileptic Exercise). Supplementary Fig. 2.A-B further confirms that these results are specific to chronic aerobic exercise, as we see no changes in 5-mC or 5-hmC in response to acute aerobic exercise.
Fig. 2.
Chronic aerobic exercise-induced changes in hippocampal area CA3 and whole blood DNA methylation.
Due to the relationship of DNA methylation in whole blood and brain in different pathologies [30], [31], [32], we also measured 5-mC and 5-hmC in the hippocampus. For this study, we focused specially on the hippocampal CA3 aera. Hippocampal area CA3 has been shown to be important within epilepsy. Specifically, in the KA-induced model of TLE [33], excitatory outputs from CA3 are required for the generation of epileptiform activity, while the F344 model of TLE exhibits CA3 damage similar to patients with TLE [27], [28]. Here, we showed that bulk 5-mC levels in the CA3 area remain unchanged in epileptic animals and in response to exercise (Fig. 2.D) (One-way ANOVA F = 1.240, P = 0.3345). However, bulk hippocampal 5-hmC in area CA3 is increased in TLE while the exercise intervention resulted in a decrease of 5-hmC in the epileptic animals similar to the levels of non-epileptic controls (Fig. 2.E) (One-way ANOVA F = 6.476, P = 0.0181, followed by multiple comparisons Uncorrected Fisher's LSD, P = 0.0085, for non-Epileptic Sedentary vs. Epileptic Sedentary and P = 0.0214 for Epileptic Sedentary vs. Epileptic Exercise).
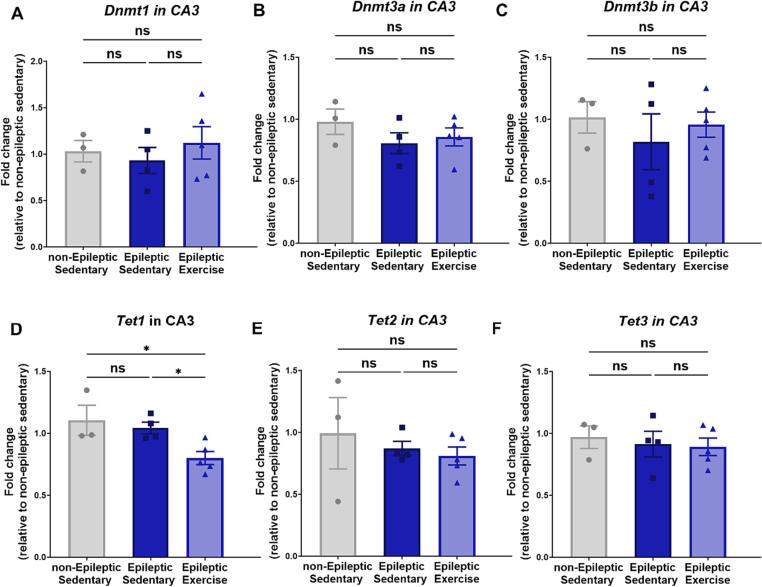
Chronic aerobic exercise decreases Tet1 expression in TLE animals
Based on the observed bulk DNA methylation changes in response to chronic aerobic exercise in TLE, we sought to determine its impact on DNA methylation enzymes. Here, we used qRT-PCR analysis to measure the expression of 3 different types of DNA DNMTs, Dnmt1, Dnmt3a, and Dnmt3b, in addition to Tet1, Tet2, and Tet3. We observed, in support of our 5-mC results, that in hippocampal area CA3, there are no changes in any of the three DNMT expressions (Fig. 3.A-C) in TLE or in response to exercise. However, upon measuring Tet genes expression, we observed an exercise-specific response in TLE with Tet1 levels (Fig. 3.D) (One-way ANOVA F = 5.747, P = 0.0247, followed by multiple comparisons Uncorrected Fisher's LSD, P = 0.0273 for Epileptic Sedentary vs. Epileptic Exercise). We observed no changes with Tet2 expression (Fig. 3.E) (One-way ANOVA F = 0.4410, P = 0.6566) in addition to Tet3 expression that also showed no changes (Fig. 3.F) (One-way ANOVA F = 0.1839, P = 0.8350).
Fig. 3.
qRT-PCR analysis in hippocampal area CA3 of DNA methylation enzymes.
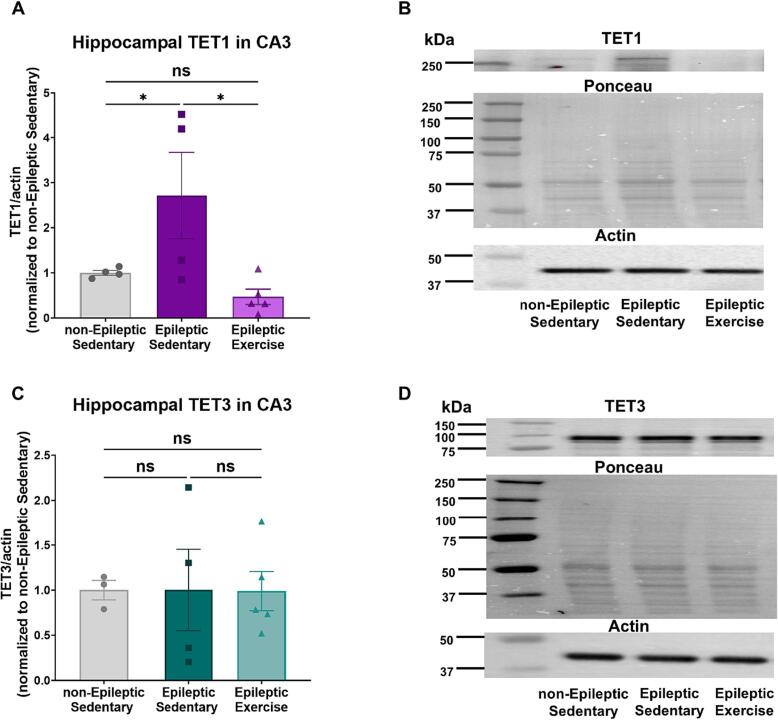
Chronic aerobic exercise reduces protein expression of TET1 in hippocampal area CA3 in epileptic animals
To analyze TET protein levels, we used western blot analysis to measure TET1 and TET3 protein expression in subdissected hippocampal area CA3. In Fig. 4.A, we observed an increase in TET1 protein expression in epileptic animals followed by a decrease in response to chronic aerobic exercise intervention (One-way ANOVA F = 5.080, P = 0.0300, followed by multiple comparisons Uncorrected Fisher's LSD, P = 0.0484 for non-Epileptic Sedentary vs. Epileptic Sedentary and P = 0.0112 for Epileptic Sedentary vs. Epileptic Exercise). This decrease resulted in similar levels as the non-epileptic control animals. Fig. 4.B, indicates representative western blot images for each group. We also measured TET3 protein expression in hippocampal area CA3. Fig. 4.C, shows no changes in TET3 protein expression in TLE or in response to exercise (One-way ANOVA F = 0.0005829, P = 0.9994). Fig. 4.D, represents western blot images for each experimental group.
Fig. 4.
Western blot analysis of protein expression TET1 and TET3 in hippocampal area CA3.
Chronic aerobic exercise-driven DNA hydroxymethylation changes in TLE hippocampus area CA3 are primarily observed in neurons
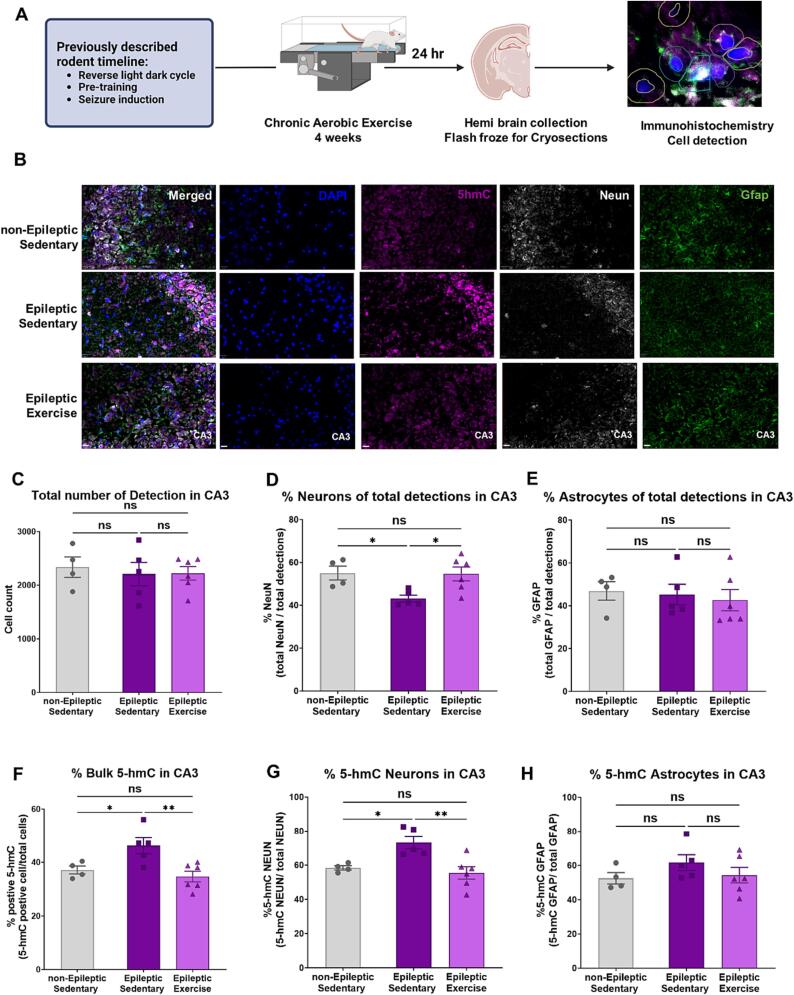
The hippocampus is a well-studied brain region, but as we discover more about cell type diversity, hippocampus heterogeneity becomes apparent [34]. This fact is even more relevant in the context of the epileptic brain and its accompanying neuronal loss [35], [36]. We therefore sought to measure DNA 5-hmC specifically in neurons and astrocytes. Fig. 5.A, depicts an abbreviated outline of the experiment, and Fig. 5.B, shows representative images for each individual group. We used DAPI stain to normalize overall cell counts as seen in Fig. 5.C, there were no differences in the total number of cells identified by DAPI stain across all groups (One-way ANOVA F = 0.1428, P = 0.8684). We also measured the percentage of neurons in Fig. 5.D, and observed that in TLE there is a decrease of cells identified as neurons by NeuN stain, compared to the non-epileptic controls while chronic aerobic exercise increased the percentage of cells identified as neurons to levels similar to non-epileptic controls (One-way ANOVA F = 5.567, P = 0.0195, followed by multiple comparisons Uncorrected Fisher's LSD, P = 0.0168 for non-Epileptic Sedentary vs. Epileptic Sedentary and P = 0.0115 for Epileptic Sedentary vs. Epileptic Exercise). We also measured the percentage of cells identified as astrocytes by GFAP stain but saw no changes between any of the experimental groups shown in Fig. 5.E, One-way ANOVA F = 0.1957, P = 0.8248). Fig. 5.F, shows the percentage of bulk 5-hmC cells compared to total cells. Here, we see as observed by our previous DNA measurements (Fig. 2), that in TLE there is an increase in bulk 5-hmC, and once the chronic aerobic exercise intervention is introduced, the bulk 5-hmC levels are decreased to levels similar to non-epileptic controls (One-way ANOVA F = 7.121, P = 0.0091, followed by multiple comparisons Uncorrected Fisher's LSD, P = 0.0231 for non-Epileptic Sedentary vs. Epileptic Sedentary and P = 0.0033 for Epileptic Sedentary vs. Epileptic Exercise). In Fig. 5.G, we see that regardless of neuronal loss observed in Fig. 5.D, the % 5-hmC cells are primarily observed in neurons (One-way ANOVA F = 8.410, p = 0.0052, followed by multiple comparisons Uncorrected Fisher's LSD, P = 0.0119 for non-Epileptic Sedentary vs. Epileptic Sedentary and P = 0.0020 for Epileptic Sedentary vs. Epileptic Exercise). No changes were observed in the percentage of astrocytes expressing 5-hmC shown in Fig. 5.H (One-way ANOVA F = 1.165, P = 0.3449).
Fig. 5.
Chronic aerobic exercise-induced changes in 5-hmC DNA methylation are primarily expressed in neurons.
Discussion
While several studies have revealed potential therapeutic effects of exercise with epilepsy [7], [9], [37], [38], [39], [40], [41], [42], [43], our understanding of the fundamental molecular mechanisms remains limited. DNA methylation is a universal epigenetic mechanism responsible for regulating gene expression in multiple diseases [44], [45], with strong evidence of its role in epilepsy [10], [12]. In this study, we found that DNA methylation, specifically 5-hmC hydroxymethylation, was altered after a 4-week chronic aerobic exercise protocol in a KA-induced model of TLE.
To study exercise-specific changes in our rodent model of TLE, we used the MoTrPAC protocol that was developed specifically for Fisher 344 rats, in parallel with a human protocol, to exercise the subjects at about 70 % VO2max [46]. This is critical as the field of exercise science is growing, and there is a need for uniform protocols that can be translated from pre-clinical studies to clinical studies. Additionally, exploring potential peripheral blood biomarkers is also of importance for this growing field. With the limitations of clinical studies in collecting tissue types, finding peripheral blood markers in addition to tissue markers in rodents that can also be measured in the clinical setting is crucial. Different exercise modalities can have varying effects on peripheral factors in blood, muscle, and brain [29], [47].
In this study, we demonstrate using the F344 model of TLE that acute exercise was not sufficient to induce peripheral blood changes in serum BDNF protein levels or serum IL-6 protein levels. In contrast, chronic exercise led to a decrease in serum BDNF protein levels in addition to an increase in serum IL-6 protein levels in TLE. We chose to use BDNF and IL-6, two well-known serum blood markers in epilepsy, to explore the response to acute exercise versus chronic exercise. BDNF has strongly been linked to epilepsy during epileptogenesis as well as influencing excitability and connectivity in the adult brain [48], [49], [50]. Additionally, BDNF has been shown to increase in response to exercise [20], [51], while studies have also shown that BDNF expression can vary depending on the exercise intensity and type [21], [52]. Serum IL-6 is a pro-inflammatory cytokine typically present at low levels in the brain but can be increased with seizures [53]. Additionally, IL-6 has been well characterized in response to exercise, with its levels also depending on multiple factors such as exercise duration and intensity [47]. In this study, we observed no changes in response to acute exercise in serum IL-6, while we observed an increase in serum IL-6 with chronic aerobic exercise. While this differs from the prior studies mentioned, we must note that there is no uniformity across studies in the exercise modality used, the model of epilepsy, or at what time point post-exercise serum was collected. Regardless of these discrepancies, we concluded from our studies that the acute aerobic exercise was not sufficient to induce measurable changes in serum BDNF and IL-6, specifically in our TLE model, and that the 4-week chronic aerobic exercise intervention did induce certain exercise-specific responses in our model of TLE. We recognize the limitation that we only measured two markers in this study, while current studies have identified other potential biomarkers that should also be explored, such as S100 calcium-binding protein B (S100B), IL-1β, and Tumor necrosis factor-alpha (TNF-α) [54]. Our ability to detect a change in levels of serum BDNF and IL-6 in response to chronic exercise provided support for the 4-week chronic aerobic exercise protocol over the acute exercise protocol. Therefore, we focused on chronic aerobic exercise protocol for all subsequent studies.
Epigenetic changes in response to exercise [20], [55], [56], [57] and how exercise can have positive effects on epilepsy [4], [7], [9], [38], [58], [59], [60] have been independently previously explored; however, how epigenetic mechanisms are impacted by exercise with epilepsy is unknown. Previous studies have shown an association between epigenetic mechanisms and epilepsy [10], [12], [61]. Specifically, DNA methylation is known to be impacted with epilepsy [18], [62], [63], [64], [65]. We found that in our model of TLE, DNA methylation, specifically 5-hmC, was increased in TLE. How DNA methylation is impacted during epilepsy is dependent on the model and during what time point these measures are taken. We have previously reported that during SE, 5-mC DNA methylation levels remained unchanged in all hippocampal regions measured, but 5-hmC was altered during SE in the hippocampal area CA3 [66]. We know from characterization studies using the F344 model of TLE, that area CA3 is impacted by neuronal loss and shows the closest relationship to the human TLE condition [27], [28]. Studies have reported that in response to exercise, DNA methylation changes occur in muscle at a gene-specific level [67], [68], [69]. Other studies specifically investigating the hippocampus also showed altered DNA methylation levels in response to exercise [57], [70], [71], [72], [73]. Our study carefully quantified global DNA methylation levels, and we show that in our model of TLE, exercise reduces 5-hmC DNA methylation to levels of non-epileptic controls. The ability of chronic aerobic exercise to alter DNA methylation, specifically 5-hmC levels in TLE, is novel and provides evidence of its therapeutic potential.
Our study also aimed to shed light on the enzyme activity responsible for these 5-hmC changes. The TET family of enzymes, including TET1, TET2, and TET3, play a pivotal role in regulating DNA methylation through the conversion of 5-mC to 5-hmC, influencing gene expression and potentially contributing to the pathogenesis of neurological disorders like epilepsy [44]. Each TET enzyme exhibits unique preferences and functions. TET1 is often associated with gene regulatory regions known as CpG islands and is primarily involved in the demethylation of promoter regions, contributing to gene activation [74]. TET2 is believed to have a broader genomic distribution and is implicated in regulating enhancer regions [75], while TET2 and TET3 depletion has been shown to increase 5-hmC levels [76]. Collectively, the TET enzymes are crucial for dynamic and context-specific regulation of DNA methylation patterns. Here, we provided evidence that TET1 is the major contributor to the 5-hmC DNA methylation changes we observed. Measured by both gene expression and protein expression, we show that TET1 is the only DNA methylation enzyme actively changing in response exercise in TLE. In accordance with our results, recent studies have also suggested a potential link between aberrant DNA methylation and epileptogenesis, highlighting TET1 as a key player in this process [65], [77].
To date, studies have reported the beneficial effects of exercise in disease models such as Alzheimer’s disease, spinal cord injury, and aging [55], [78], [79], [80]. Specifically, some studies focused on the ability of exercise to lead to functional restoration and memory improvements [78], [80], [81]. DNA methylation also plays an important role in synaptic plasticity and memory [18], [82], [83], [84], [85], [86]. However, little is known about the cell type specificity related to DNA methylation in epilepsy or with exercise. Here, we observed that there was a significant decrease in total cells identified as neurons in TLE, while the aerobic exercise intervention resulted in an increase to levels similar to non-epileptic controls. This corresponds well with what we previously know about TLE and neuronal loss [87], [88], [89]. Notably, upon measuring 5-hmC-specific changes on a cell type-specific level, we discovered that the major changes measured in 5-hmC were neuron-specific and did not correspond to astrocyte expression. Studies have begun to show the importance of cell type specificity in relation to DNA methylation [15], [90], [91]; however, to our knowledge, this is the first study to connect 5-hmC-specific DNA methylation to neurons. These measures only cover bulk DNA changes, and further gene-specific analysis and exploration of additional cell types are necessary for better understanding.
Although exciting, the findings in this study only begin to elucidate the potential role of DNAme exercise-driven epigenetic changes in TLE. A limitation of this study is that we only measured bulk 5-mC and 5-hmC DNAme changes in blood and the epileptic hippocampus in response to exercise. Further studies exploring the gene-specific methylation levels are warranted. Specifically, the relationship between 5-hmC and transcription is a topic of interest in the field of epigenetics. Changes in whole genome levels of 5-hmC can potentially influence transcription by affecting the accessibility of the DNA to the transcriptional machinery, while higher levels of 5-hmC are frequently detected within the gene bodies of actively transcribed genes [92].
Additionally, we propose exploring gene expression levels of BDNF and IL-6 in the epileptic hippocampus and how these correlate to the changes observed in serum. One proposed mechanism by which the changes observed in peripheral epilepsy-associated biomarkers travel to changes in the epileptic hippocampus is by exosomes. Exosomes demonstrate the ability to cross the blood–brain barrier and transfer information and have therefore gained increased interest as a link between peripheral blood changes and brain [93], [94].
Conclusions
In this study, we show that DNA 5-hmC is enhanced in TLE and that a chronic aerobic exercise intervention reduces DNA 5-hmC in epileptic rats. Tet1 gene and protein expression were also impacted with exercise and epilepsy. We further show that the 5-hmC specific mediated changes observed in hippocampal area CA3 were mostly observed in neurons. We therefore conclude that DNA methylation, specifically 5-hmC is a key epigenetic player in epilepsy and exercise specifically within neurons.
CRediT authorship contribution statement
Silvienne C. Sint Jago: Writing – original draft, Visualization, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Rudhab Bahabry: Writing – review & editing, Visualization, Investigation, Conceptualization. Anna Maria Schreiber: . Julia Homola: Writing – review & editing, Investigation. Tram Ngyuen: Writing – review & editing, Investigation. Fernando Meijia: Writing – review & editing, Investigation. Jane B. Allendorfer: Writing – review & editing, Funding acquisition, Conceptualization. Farah D. Lubin: Writing – review & editing, Methodology, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by National Institute of Health (NIH) grant R21NS116937 (F.D.L.), and the Evelyn F. McKnight Brain Institute at the University of Alabama at Birmingham (F.D.L. and J.B.A.). S.S.J. and R.B., were supported in part by the NIH/NINDS graduate Neuroscience Roadmap scholar training grant R25NS089463 (F.D.L.). The treadmill and behavior equipment used in these studies were supported by the UAB Neuroscience Behavioral Assessment Core. The Olympus slide scanner used in these studies was supported by the P20AG068024 grant. Schematics were created in BioRender.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebr.2023.100642.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Sirven J.I. Epilepsy: A Spectrum Disorder. Cold Spring Harb Perspect Med. 2015;5(9) doi: 10.1101/cshperspect.a022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Curia G., Lucchi C., Vinet J., Gualtieri F., Marinelli C., Torsello A., et al. Pathophysiogenesis of mesial temporal lobe epilepsy: is prevention of damage antiepileptogenic? Curr Med Chem. 2014;21(6):663–688. doi: 10.2174/0929867320666131119152201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loscher W., Klitgaard H., Twyman R.E., Schmidt D. New avenues for anti-epileptic drug discovery and development. Nat Rev Drug Discov. 2013;12(10):757–776. doi: 10.1038/nrd4126. [DOI] [PubMed] [Google Scholar]
- 4.Allendorfer J.B., Arida R.M. Role of Physical Activity and Exercise in Alleviating Cognitive Impairment in People With Epilepsy. Clin Ther. 2018;40(1):26–34. doi: 10.1016/j.clinthera.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Warburton D.E.R. Health benefits of physical activity: the evidence. Can Med Assoc J. 2006;174(6):801–809. doi: 10.1503/cmaj.051351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guadagni V., Drogos L.L., Tyndall A.V., Davenport M.H., Anderson T.J., Eskes G.A., et al. Aerobic exercise improves cognition and cerebrovascular regulation in older adults. Neurology. 2020;94(21) doi: 10.1212/WNL.0000000000009478. e2245-e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allendorfer J.B., Brokamp G.A., Nenert R., Szaflarski J.P., Morgan C.J., Tuggle S.C., et al. A pilot study of combined endurance and resistance exercise rehabilitation for verbal memory and functional connectivity improvement in epilepsy. Epilepsy Behav. 2019;96:44–56. doi: 10.1016/j.yebeh.2019.04.020. [DOI] [PubMed] [Google Scholar]
- 8.Alexander H.B., Allendorfer J.B. The relationship between physical activity and cognitive function in people with epilepsy: A systematic review. Epilepsy Behav. 2023;142 doi: 10.1016/j.yebeh.2023.109170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Almeida A.A., Gomes da Silva S., Lopim G.M., Vannucci Campos D., Fernandes J., Cabral F.R., et al. Resistance Exercise Reduces Seizure Occurrence, Attenuates Memory Deficits and Restores BDNF Signaling in Rats with Chronic Epilepsy. Neurochem Res. 2017;42(4):1230–1239. doi: 10.1007/s11064-016-2165-9. [DOI] [PubMed] [Google Scholar]
- 10.Kobow K., Blumcke I. Epigenetics in epilepsy. Neurosci Lett. 2018;667:40–46. doi: 10.1016/j.neulet.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 11.Gibney E.R., Nolan C.M. Epigenetics and gene expression. Heredity (Edinb) 2010;105(1):4–13. doi: 10.1038/hdy.2010.54. [DOI] [PubMed] [Google Scholar]
- 12.Hauser R.M., Henshall D.C., Lubin F.D. The Epigenetics of Epilepsy and Its Progression. Neuroscientist. 2018;24(2):186–200. doi: 10.1177/1073858417705840. [DOI] [PubMed] [Google Scholar]
- 13.Ryley Parrish R., Albertson A.J., Buckingham S.C., Hablitz J.J., Mascia K.L., Davis Haselden W., et al. Status epilepticus triggers early and late alterations in brain-derived neurotrophic factor and NMDA glutamate receptor Grin2b DNA methylation levels in the hippocampus. Neuroscience. 2013;248:602–619. doi: 10.1016/j.neuroscience.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roth T.L. Epigenetic mechanisms in the development of behavior: advances, challenges, and future promises of a new field. Dev Psychopathol. 2013;25(4 Pt 2):1279–1291. doi: 10.1017/S0954579413000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi D.-Q., Ali I., Tang J., Yang W.-C. New insights into 5hmC DNA modification: generation, distribution and function. Front Genet. 2017;8:100. doi: 10.3389/fgene.2017.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasmussen K.D., Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016;30(7):733–750. doi: 10.1101/gad.276568.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun W., Zang L., Shu Q., Li X. From development to diseases: the role of 5hmC in brain. Genomics. 2014;104(5):347–351. doi: 10.1016/j.ygeno.2014.08.021. [DOI] [PubMed] [Google Scholar]
- 18.Parrish R.R., Buckingham S.C., Mascia K.L., Johnson J.J., Matyjasik M.M., Lockhart R.M., et al. Methionine increases BDNF DNA methylation and improves memory in epilepsy. Ann Clin Transl Neurol. 2015;2(4):401–416. doi: 10.1002/acn3.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mastana S.S., et al. Impact of aerobic exercise and fatty acid supplementation on global and gene-specific DNA methylation. Epigenetics. 2019 doi: 10.1080/15592294.2019.1582276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sleiman S.F., Henry J., Al-Haddad R., El Hayek L., Abou Haidar E., Stringer T., et al. Exercise promotes the expression of brain derived neurotrophic factor (BDNF) through the action of the ketone body beta-hydroxybutyrate. Elife. 2016;5 doi: 10.7554/eLife.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeon Y.K., Ha C.H. The effect of exercise intensity on brain derived neurotrophic factor and memory in adolescents. Environ Health Prev Med. 2017;22(1):27. doi: 10.1186/s12199-017-0643-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li G., Bauer S., Nowak M., Norwood B., Tackenberg B., Rosenow F., et al. Cytokines and epilepsy. Seizure. 2011;20(3):249–256. doi: 10.1016/j.seizure.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 23.Kobylarek D., Iwanowski P., Lewandowska Z., Limphaibool N., Szafranek S., Labrzycka A., et al. Advances in the Potential Biomarkers of Epilepsy. Front Neurol. 2019;10:685. doi: 10.3389/fneur.2019.00685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amar D, Gay NR, Jean Beltran PM, Adkins JN, Almagro Armenteros JJ, Ashley E, et al. Temporal dynamics of the multi-omic response to endurance exercise training across tissues. bioRxiv. 2022:2022.09.21.508770.
- 25.Racine R.J. Modification of seizure activity by electrical stimulation. II Motor seizure. Electroencephalogr Clin Neurophysiol. 1972;32(3):281–294. doi: 10.1016/0013-4694(72)90177-0. [DOI] [PubMed] [Google Scholar]
- 26.Sharma S., Puttachary S., Thippeswamy A., Kanthasamy A.G., Thippeswamy T. Status Epilepticus: Behavioral and Electroencephalography Seizure Correlates in Kainate Experimental Models. Front Neurol. 2018;9:7. doi: 10.3389/fneur.2018.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma A.K., Jordan W.H., Reams R.Y., Hall D.G., Snyder P.W. Temporal profile of clinical signs and histopathologic changes in an F-344 rat model of kainic acid-induced mesial temporal lobe epilepsy. Toxicol Pathol. 2008;36(7):932–943. doi: 10.1177/0192623308326093. [DOI] [PubMed] [Google Scholar]
- 28.Sharma A.K., Searfoss G.H., Reams R.Y., Jordan W.H., Snyder P.W., Chiang A.Y., et al. Kainic acid-induced F-344 rat model of mesial temporal lobe epilepsy: gene expression and canonical pathways. Toxicol Pathol. 2009;37(6):776–789. doi: 10.1177/0192623309344202. [DOI] [PubMed] [Google Scholar]
- 29.Pedralli M.L., Marschner R.A., Kollet D.P., Neto S.G., Eibel B., Tanaka H., et al. Different exercise training modalities produce similar endothelial function improvements in individuals with prehypertension or hypertension: a randomized clinical trial Exercise, endothelium and blood pressure. Sci Rep. 2020;10(1):7628. doi: 10.1038/s41598-020-64365-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia T., Chu C., Liu Y., van Dongen J., Papastergios E., Armstrong N.J., et al. Epigenome-wide meta-analysis of blood DNA methylation and its association with subcortical volumes: findings from the ENIGMA Epigenetics Working Group. Mol Psychiatry. 2021;26(8):3884–3895. doi: 10.1038/s41380-019-0605-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braun P.R., Tanaka-Sahker M., Chan A.C., Jellison S.S., Klisares M.J., Hing B.W., et al. Genome-wide DNA methylation investigation of glucocorticoid exposure within buccal samples. Psychiatry Clin Neurosci. 2019;73(6):323–330. doi: 10.1111/pcn.12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walton E., Hass J., Liu J., Roffman J.L., Bernardoni F., Roessner V., et al. Correspondence of DNA Methylation Between Blood and Brain Tissue and Its Application to Schizophrenia Research. Schizophr Bull. 2016;42(2):406–414. doi: 10.1093/schbul/sbv074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu L.M., Polygalov D., Wintzer M.E., Chiang M.C., McHugh T.J. CA3 Synaptic Silencing Attenuates Kainic Acid-Induced Seizures and Hippocampal Network Oscillations. eNeuro. 2016;3(1) doi: 10.1523/ENEURO.0003-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeisel A., Munoz-Manchado A.B., Codeluppi S., Lonnerberg P., La Manno G., Jureus A., et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347(6226):1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 35.Pai B., Tome-Garcia J., Cheng W.S., Nudelman G., Beaumont K.G., Ghatan S., et al. High-resolution transcriptomics informs glial pathology in human temporal lobe epilepsy. Acta Neuropathol Commun. 2022;10(1):149. doi: 10.1186/s40478-022-01453-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfisterer U., Petukhov V., Demharter S., Meichsner J., Thompson J.J., Batiuk M.Y., et al. Identification of epilepsy-associated neuronal subtypes and gene expression underlying epileptogenesis. Nat Commun. 2020;11(1):5038. doi: 10.1038/s41467-020-18752-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee J.M., Ji E.S., Kim T.W., Kim C.J., Shin M.S., Lim B.V., et al. Treadmill exercise improves memory function by inhibiting hippocampal apoptosis in pilocarpine-induced epileptic rats. J Exerc Rehabil. 2018;14(5):713–723. doi: 10.12965/jer.36394.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Almeida A.A., Gomes da Silva S., Lopim G.M., Vannucci Campos D., Fernandes J., Cabral F.R., et al. Physical exercise alters the activation of downstream proteins related to BDNF-TrkB signaling in male Wistar rats with epilepsy. J Neurosci Res. 2018;96(5):911–920. doi: 10.1002/jnr.24196. [DOI] [PubMed] [Google Scholar]
- 39.Iqbal M., Xiao X.L., Zafar S., Yang P.B., Si K.W., Han H., et al. Forced Physical Training Increases Neuronal Proliferation and Maturation with Their Integration into Normal Circuits in Pilocarpine Induced Status Epilepticus Mice. Neurochem Res. 2019;44(11):2590–2605. doi: 10.1007/s11064-019-02877-3. [DOI] [PubMed] [Google Scholar]
- 40.Lin X.Y., Cui Y., Wang L., Chen W. Chronic exercise buffers the cognitive dysfunction and decreases the susceptibility to seizures in PTZ-treated rats. Epilepsy Behav. 2019;98(Pt A):173–187. doi: 10.1016/j.yebeh.2019.07.032. [DOI] [PubMed] [Google Scholar]
- 41.Barzroodi Pour M., Bayat M., Navazesh A., Soleimani M., Karimzadeh F. Exercise Improved the Anti-Epileptic Effect of Carbamazepine through GABA Enhancement in Epileptic Rats. Neurochem Res. 2021;46(8):2112–2130. doi: 10.1007/s11064-021-03349-3. [DOI] [PubMed] [Google Scholar]
- 42.Hafele C.A., Rombaldi A.J., Feter N., Hafele V., Gervini B.L., Domingues M.R., et al. Effects of an exercise program on health of people with epilepsy: A randomized clinical trial. Epilepsy Behav. 2021;117 doi: 10.1016/j.yebeh.2021.107904. [DOI] [PubMed] [Google Scholar]
- 43.Akerlund S., Varkey E., Klecki J., Zelano J., Ben-Menachem E. Randomized controlled trial of moderate cardiovascular exercise for patients with drug-resistant epilepsy. Epilepsy Behav. 2021;124 doi: 10.1016/j.yebeh.2021.108335. [DOI] [PubMed] [Google Scholar]
- 44.Wang J., Zhang K.X., Lu G.Z., Zhao X.H. Research progress on 5hmC and TET dioxygenases in neurodevelopment and neurological diseases. Yi Chuan. 2017;39(12):1138–1149. doi: 10.16288/j.yczz.17-086. [DOI] [PubMed] [Google Scholar]
- 45.Wang K., Liu H., Hu Q., Wang L., Liu J., Zheng Z., et al. Epigenetic regulation of aging: implications for interventions of aging and diseases. Signal Transduct Target Ther. 2022;7(1):374. doi: 10.1038/s41392-022-01211-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Molecular Transducers of Physical Activity Consortium. Molecular Transducers of Physical Activity Consortium (MoTrPAC) – Adult. 2018.
- 47.Docherty S., Harley R., McAuley J.J., Crowe L.A.N., Pedret C., Kirwan P.D., et al. The effect of exercise on cytokines: implications for musculoskeletal health: a narrative review. BMC Sports Sci Med Rehabil. 2022;14(1):5. doi: 10.1186/s13102-022-00397-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Binder D.K., Croll S.D., Gall C.M., Scharfman H.E. BDNF and epilepsy: too much of a good thing? Trends Neurosci. 2001;24(1):47–53. doi: 10.1016/s0166-2236(00)01682-9. [DOI] [PubMed] [Google Scholar]
- 49.Wang X., Hu Z., Zhong K. The Role of Brain-Derived Neurotrophic Factor in Epileptogenesis: an Update. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.758232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scharfman H.E. Brain-derived neurotrophic factor and epilepsy–a missing link? Epilepsy Curr. 2005;5(3):83–88. doi: 10.1111/j.1535-7511.2005.05312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cefis M., Chaney R., Wirtz J., Meloux A., Quirie A., Leger C., et al. Molecular mechanisms underlying physical exercise-induced brain BDNF overproduction. Front Mol Neurosci. 2023;16:1275924. doi: 10.3389/fnmol.2023.1275924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodriguez-Gutierrez E., Torres-Costoso A., Saz-Lara A., Bizzozero-Peroni B., Guzman-Pavon M.J., Sanchez-Lopez M., et al. Effectiveness of high-intensity interval training on peripheral brain-derived neurotrophic factor in adults: A systematic review and network meta-analysis. Scand J Med Sci Sports. 2023 doi: 10.1111/sms.14496. [DOI] [PubMed] [Google Scholar]
- 53.Rana A., Musto A.E. The role of inflammation in the development of epilepsy. J Neuroinflammation. 2018;15(1):144. doi: 10.1186/s12974-018-1192-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Banote R.K., Akel S., Zelano J. Blood biomarkers in epilepsy. Acta Neurol Scand. 2022;146(4):362–368. doi: 10.1111/ane.13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davaa G., Hong J.Y., Kim T.U., Lee S.J., Kim S.Y., Hong K., et al. Exercise Ameliorates Spinal Cord Injury by Changing DNA Methylation. Cells. 2021;10(1) doi: 10.3390/cells10010143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fernandes J., Arida R.M., Gomez-Pinilla F. Physical exercise as an epigenetic modulator of brain plasticity and cognition. Neurosci Biobehav Rev. 2017;80:443–456. doi: 10.1016/j.neubiorev.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Solvsten C.A.E., de Paoli F., Christensen J.H., Nielsen A.L. Voluntary Physical Exercise Induces Expression and Epigenetic Remodeling of VegfA in the Rat Hippocampus. Mol Neurobiol. 2018;55(1):567–582. doi: 10.1007/s12035-016-0344-y. [DOI] [PubMed] [Google Scholar]
- 58.Arida R.M., de Almeida A.-C.-G., Cavalheiro E.A., Scorza F.A. Experimental and clinical findings from physical exercise as complementary therapy for epilepsy. Epilepsy Behav. 2013;26(3):273–278. doi: 10.1016/j.yebeh.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 59.Arida R.M., Scorza F.A., dos Santos N.F., Peres C.A., Cavalheiro E.A. Effect of physical exercise on seizure occurrence in a model of temporal lobe epilepsy in rats. Epilepsy Res. 1999;37(1):45–52. doi: 10.1016/s0920-1211(99)00032-7. [DOI] [PubMed] [Google Scholar]
- 60.Arida R.M., Scorza F.A., Gomes da Silva S., Schachter S.C., Cavalheiro E.A. The potential role of physical exercise in the treatment of epilepsy. Epilepsy Behav. 2010;17(4):432–435. doi: 10.1016/j.yebeh.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 61.Henshall D.C., Kobow K. Epigenetics and Epilepsy. Cold Spring Harb Perspect Med. 2015;5(12) doi: 10.1101/cshperspect.a022731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berger T.C., Vigeland M.D., Hjorthaug H.S., Etholm L., Nome C.G., Taubøll E., et al. Neuronal and glial DNA methylation and gene expression changes in early epileptogenesis. PLoS One. 2019;14(12):e0226575. doi: 10.1371/journal.pone.0226575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Debski K.J., Pitkanen A., Puhakka N., Bot A.M., Khurana I., Harikrishnan K.N., et al. Etiology matters - Genomic DNA Methylation Patterns in Three Rat Models of Acquired Epilepsy. Sci Rep. 2016;6:25668. doi: 10.1038/srep25668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miller-Delaney S.F., Das S., Sano T., Jimenez-Mateos E.M., Bryan K., Buckley P.G., et al. Differential DNA methylation patterns define status epilepticus and epileptic tolerance. J Neurosci. 2012;32(5):1577–1588. doi: 10.1523/JNEUROSCI.5180-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen H.Y., Weltha L., Cook J.M., Gesese R., Omi W., Baer S.B., et al. Sarcosine Suppresses Epileptogenesis in Rats With Effects on Hippocampal DNA Methylation. Front Mol Neurosci. 2020;13:97. doi: 10.3389/fnmol.2020.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parrish R.R., Albertson A.J., Buckingham S.C., Hablitz J.J., Mascia K.L., Davis Haselden W., et al. Status epilepticus triggers early and late alterations in brain-derived neurotrophic factor and NMDA glutamate receptor Grin2b DNA methylation levels in the hippocampus. Neuroscience. 2013;248:602–619. doi: 10.1016/j.neuroscience.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kanzleiter T., Jahnert M., Schulze G., Selbig J., Hallahan N., Schwenk R.W., et al. Exercise training alters DNA methylation patterns in genes related to muscle growth and differentiation in mice. Am J Physiol Endocrinol Metab. 2015;308(10):E912–E920. doi: 10.1152/ajpendo.00289.2014. [DOI] [PubMed] [Google Scholar]
- 68.Nitert M.D., Dayeh T., Volkov P., Elgzyri T., Hall E., Nilsson E., et al. Impact of an exercise intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes. 2012;61(12):3322–3332. doi: 10.2337/db11-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Swiatowy W.J., Drzewiecka H., Kliber M., Sasiadek M., Karpinski P., Plawski A., et al. Physical Activity and DNA Methylation in Humans. Int J Mol Sci. 2021;22(23) doi: 10.3390/ijms222312989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Elsner V.R., Lovatel G.A., Moyses F., Bertoldi K., Spindler C., Cechinel L.R., et al. Exercise induces age-dependent changes on epigenetic parameters in rat hippocampus: a preliminary study. Exp Gerontol. 2013;48(2):136–139. doi: 10.1016/j.exger.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jessop P., Toledo-Rodriguez M. Hippocampal TET1 and TET2 Expression and DNA Hydroxymethylation Are Affected by Physical Exercise in Aged Mice. Front Cell Dev Biol. 2018;6:45. doi: 10.3389/fcell.2018.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kashimoto R.K., Toffoli L.V., Manfredo M.H.F., Volpini V.L., Martins-Pinge M.C., Pelosi G.G., et al. Physical exercise affects the epigenetic programming of rat brain and modulates the adaptive response evoked by repeated restraint stress. Behav Brain Res. 2016;296:286–289. doi: 10.1016/j.bbr.2015.08.038. [DOI] [PubMed] [Google Scholar]
- 73.Zhang J., Li J., Zhu Y., Miao Z., Tian Y. Forced running exercise mitigates radiation-induced cognitive deficits via regulated DNA hydroxymethylation. Epigenomics. 2020;12(5):385–396. doi: 10.2217/epi-2019-0370. [DOI] [PubMed] [Google Scholar]
- 74.Williams K., Christensen J., Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2011;13(1):28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ge L., Zhang R.P., Wan F., Guo D.Y., Wang P., Xiang L.X., et al. TET2 plays an essential role in erythropoiesis by regulating lineage-specific genes via DNA oxidative demethylation in a zebrafish model. Mol Cell Biol. 2014;34(6):989–1002. doi: 10.1128/MCB.01061-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Putiri E.L., Tiedemann R.L., Thompson J.J., Liu C., Ho T., Choi J.H., et al. Distinct and overlapping control of 5-methylcytosine and 5-hydroxymethylcytosine by the TET proteins in human cancer cells. Genome Biol. 2014;15(6):R81. doi: 10.1186/gb-2014-15-6-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaas G.A., Zhong C., Eason D.E., Ross D.L., Vachhani R.V., Ming G.L., et al. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron. 2013;79(6):1086–1093. doi: 10.1016/j.neuron.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lourenco M.V., Frozza R.L., de Freitas G.B., Zhang H., Kincheski G.C., Ribeiro F.C., et al. Exercise-linked FNDC5/irisin rescues synaptic plasticity and memory defects in Alzheimer's models. Nat Med. 2019;25(1):165–175. doi: 10.1038/s41591-018-0275-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Neri MFGZPMDMPMLM. Role of physical exercise in the regulation of epigenetic mechanisms in inflammation, cancer, neurodegenerative diseases, and aging process. 2019. [DOI] [PubMed]
- 80.De Miguel Z., Khoury N., Betley M.J., Lehallier B., Willoughby D., Olsson N., et al. Exercise plasma boosts memory and dampens brain inflammation via clusterin. Nature. 2021;600(7889):494–499. doi: 10.1038/s41586-021-04183-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vaynman S., Ying Z., Gomez-Pinilla F. Hippocampal BDNF mediates the efficacy of exercise on synaptic plasticity and cognition. Eur J Neurosci. 2004;20(10):2580–2590. doi: 10.1111/j.1460-9568.2004.03720.x. [DOI] [PubMed] [Google Scholar]
- 82.Jarome T.J., Lubin F.D. Epigenetic mechanisms of memory formation and reconsolidation. Neurobiol Learn Mem. 2014;115:116–127. doi: 10.1016/j.nlm.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jarome T.J., Butler A.A., Nichols J.N., Pacheco N.L., Lubin F.D. NF-kappaB mediates Gadd45beta expression and DNA demethylation in the hippocampus during fear memory formation. Front Mol Neurosci. 2015;8:54. doi: 10.3389/fnmol.2015.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kumar D., Aggarwal M., Kaas G.A., Lewis J., Wang J., Ross D.L., et al. Tet1 Oxidase Regulates Neuronal Gene Transcription, Active DNA Hydroxy-methylation, Object Location Memory, and Threat Recognition Memory. Neuroepigenetics. 2015;4:12–27. doi: 10.1016/j.nepig.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bernstein C. DNA Methylation and Establishing Memory. Epigenet Insights. 2022;15 doi: 10.1177/25168657211072499. 25168657211072499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Campbell R.R., Wood M.A. How the epigenome integrates information and reshapes the synapse. Nat Rev Neurosci. 2019;20(3):133–147. doi: 10.1038/s41583-019-0121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang X., Cui S.S., Wallace A.E., Hannesson D.K., Schmued L.C., Saucier D.M., et al. Relations between brain pathology and temporal lobe epilepsy. J Neurosci. 2002;22(14):6052–6061. doi: 10.1523/JNEUROSCI.22-14-06052.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kapur J. Role of Neuronal Loss in the Pathogenesis of Recurrent Spontaneous Seizures. Epilepsy Curr. 2003;3(5):166–167. doi: 10.1046/j.1535-7597.2003.03506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ren E., Curia G. Synaptic Reshaping and Neuronal Outcomes in the Temporal Lobe Epilepsy. Int J Mol Sci. 2021;22(8) doi: 10.3390/ijms22083860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kozlenkov A., Roussos P., Timashpolsky A., Barbu M., Rudchenko S., Bibikova M., et al. Differences in DNA methylation between human neuronal and glial cells are concentrated in enhancers and non-CpG sites. Nucleic Acids Res. 2014;42(1):109–127. doi: 10.1093/nar/gkt838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Skvortsova K., Zotenko E., Luu P.L., Gould C.M., Nair S.S., Clark S.J., et al. Comprehensive evaluation of genome-wide 5-hydroxymethylcytosine profiling approaches in human DNA. Epigenetics Chromatin. 2017;10:16. doi: 10.1186/s13072-017-0123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shi D.Q., Ali I., Tang J., Yang W.C. New Insights into 5hmC DNA Modification: Generation. Distribution and Function Front Genet. 2017;8:100. doi: 10.3389/fgene.2017.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Han W., Zhang H., Feng L., Dang R., Wang J., Cui C., et al. The emerging role of exosomes in communication between the periphery and the central nervous system. MedComm. 2020;2023;4(6):e410 doi: 10.1002/mco2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Janas A.M., Sapon K., Janas T., Stowell M.H., Janas T. Exosomes and other extracellular vesicles in neural cells and neurodegenerative diseases. BBA. 2016;1858(6):1139–1151. doi: 10.1016/j.bbamem.2016.02.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.