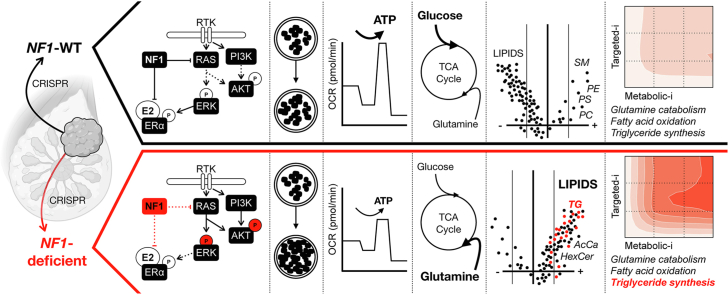
Abstract
Objective
NF1 is a tumor suppressor gene and its protein product, neurofibromin, is a negative regulator of the RAS pathway. NF1 is one of the top driver mutations in sporadic breast cancer such that 27 % of breast cancers exhibit damaging NF1 alterations. NF1 loss-of-function is a frequent event in the genomic evolution of estrogen receptor (ER)+ breast cancer metastasis and endocrine resistance. Individuals with Neurofibromatosis type 1 (NF) – a disorder caused by germline NF1 mutations – have an increased risk of dying from breast cancer [1–4]. NF-related breast cancers are associated with decreased overall survival compared to sporadic breast cancer. Despite numerous studies interrogating the role of RAS mutations in tumor metabolism, no study has comprehensively profiled the NF1-deficient breast cancer metabolome to define patterns of energetic and metabolic reprogramming. The goals of this investigation were (1) to define the role of NF1 deficiency in estrogen receptor-positive (ER+) breast cancer metabolic reprogramming and (2) to identify potential targeted pathway and metabolic inhibitor combination therapies for NF1-deficient ER + breast cancer.
Methods
We employed two ER+ NF1-deficient breast cancer models: (1) an NF1-deficient MCF7 breast cancer cell line to model sporadic breast cancer, and (2) three distinct, Nf1-deficient rat models to model NF-related breast cancer [1]. IncuCyte proliferation analysis was used to measure the effect of NF1 deficiency on cell proliferation and drug response. Protein quantity was assessed by Western Blot analysis. We then used RNAseq to investigate the transcriptional effect of NF1 deficiency on global and metabolism-related transcription. We measured cellular energetics using Agilent Seahorse XF-96 Glyco Stress Test and Mito Stress Test assays. We performed stable isotope labeling and measured [U–13C]-glucose and [U–13C]-glutamine metabolite incorporation and measured total metabolite pools using mass spectrometry. Lastly, we used a Bliss synergy model to investigate NF1-driven changes in targeted and metabolic inhibitor synergy.
Results
Our results revealed that NF1 deficiency enhanced cell proliferation, altered neurofibromin expression, and increased RAS and PI3K/AKT pathway signaling while constraining oxidative ATP production and restricting energetic flexibility. Neurofibromin deficiency also increased glutamine influx into TCA intermediates and dramatically increased lipid pools, especially triglycerides (TG). Lastly, NF1 deficiency alters the synergy between metabolic inhibitors and traditional targeted inhibitors. This includes increased synergy with inhibitors targeting glycolysis, glutamine metabolism, mitochondrial fatty acid transport, and TG synthesis.
Conclusions
NF1 deficiency drives metabolic reprogramming in ER+ breast cancer. This reprogramming is characterized by oxidative ATP constraints, glutamine TCA influx, and lipid pool expansion, and these metabolic changes introduce novel metabolic-to-targeted inhibitor synergies.
Keywords: NF1, Breast cancer, Metabolic reprogramming, Metabolic inhibition, Neurofibromatosis Type 1
Graphical abstract
Highlights
-
•
NF1 deficiency drives metabolic reprogramming in ER + breast cancer.
-
•
NF1-driven metabolic reprogramming is characterized by OXPHOS constraints, glutamine TCA influx, and lipid pool expansion.
-
•
NF1-deficient ER + breast cancer cells have increased sensitivity to combined RAS and triglyceride synthesis inhibitors.
List of Abbreviations
- 2DG
2-Deoxy-d-glucose
- 4OHT
4-Hydroxytamoxifen or tamoxifen
- ATP
Adenosine Triphosphate
- BPTES
Bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl)ethyl sulfide
- COB
Cobimetinib
- CPT1
carnitine palmitoyl transferase 1
- CRISPR
Clustered Regularly Interspaced Short Palindromic Repeats
- DGAT1
Diacylglycerol O-Acyltransferase 1
- DGAT1/2i
Diacylglycerol O-Acyltransferase 1 and 2 Inhibitor
- DGAT2
Diacylglycerol O-Acyltransferase 2
- DMEM
Dulbecco's Modified Eagle Medium
- dFBS
Dialyzed Fetal Bovine Serum
- ECAR
Extracellular Acidification Rate
- EMT
Epithelial-Mesenchymal Transition
- ER+
Estrogen Receptor Positive
- ETC
Electron Transport Chain
- EVE
Everolimus
- EX
Etomoxir
- FA
Fatty acid
- FBS
Fetal bovine serum
- GAP
GTPase activating protein
- GDH
Glutamate dehydrogenase
- GLS
Glutaminase
- GSEA
Gene Set Enrichment Analysis
- GRD
GAP-related domain
- GFR
Growth-factor reduced
- H-ESI
Heated Electrospray Ionization
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KO
Knock-out
- LC-MS
Liquid Chromatography-Mass Spectrometry
- mRNA
Messenger RNA
- mTOR
Mechanistic Target of Rapamycin
- MEK
Mitogen-Activated Protein Kinase Kinase
- MPC
mitochondrial pyruvate carrier
- Nf1
Neurofibromin 1 (rat) gene
- NF
Neurofibromatosis Type 1
- NF1
Neurofibromin 1 (human) gene
- OCR
Oxygen Consumption Rate
- OXPHOS
Oxidative Phosphorylation
- PCA
Principal Component Analysis
- PCR
Polymerase Chain Reaction
- PG
Phosphatidylglycerol
- PI
Phosphatidylinositol
- PS
Phosphatidylserine
- PI3K-AKT
Phosphoinositide 3-kinase-AKT pathway
- RAS
Rat sarcoma viral oncogene homolog
- REACTOME
REACTOME pathway database
- RNAseq
RNA Sequencing
- SM
Sphingomyelin
- TCA
Tricarboxylic Acid Cycle
- TG
Triglyceride
- VDAC
Voltage-dependent Anion Channel
1. Introduction
Recent genomic analyses have identified that the tumor suppressor gene neurofibromin 1 (NF1) is a driver of inherited breast cancer, sporadic breast cancer, and endocrine resistance [[2], [3], [4], [5], [6], [7], [8], [9]]. The NF1 gene product, neurofibromin, is a primary human rat sarcoma viral oncogene homolog (RAS) GTPase activating protein (GAP) and negatively regulates RAS pathway signaling. Neurofibromin's GAP-related domain (GRD) binds RAS and stimulates RAS GTPase activity [1,10]. Germline NF1 mutations cause Neurofibromatosis Type 1 (NF), the most common autosomal dominant single-gene disorder, affecting 1:3000 live births [10]. Both men and women with NF have an increased risk of dying from breast cancer [[11], [12], [13], [14]]. Women <40 years old with NF have a 6.5-fold increased risk of breast cancer and a 2-fold higher lifetime risk of breast cancer compared to the general population [13]. NF-related breast cancer is associated with adverse prognostic factors and decreased overall survival compared to sporadic breast cancer [11,12,15]. Recent studies have revealed that NF1 is a genetic driver of sporadic breast cancer [2,8,9], and NF1 shallow deletions are present in up to 27 % of breast cancers [1]. Moreso, NF1 loss-of-function mutation is a frequent event in the evolution of endocrine-resistant estrogen receptor-positive (ER+) breast cancer [4,16]. RAS signaling is hyperactivated in approximately 50 % of tumors, but RAS mutations are only found in 4 % of breast cancers, suggesting that NF1 loss of function may be driving many RAS-active breast cancers [17,18]. Because mutations and indels are detected throughout the NF1 gene and all lead to variable levels of loss of function, we refer to these states of altered neurofibromin function as “NF1 deficiency” [10,19]. The results in both NF-related and sporadic breast cancer emphasize the impact of NF1 deficiency and deregulated RAS in breast cancer and in resistance to endocrine therapy.
Activated oncogenes and inactivated tumor suppressors contribute to tumorigenesis by reprogramming cellular energetics and metabolism towards macromolecular synthesis – a phenomenon termed “metabolic reprogramming” [20,21]. A canonical example of cancer metabolic reprogramming is the Warburg Effect, defined as a shift towards aerobic glycolysis to support biosynthetic processes [20,22,23]. Metabolic reprogramming can introduce novel therapeutic targets [20,[24], [25], [26], [27], [28], [29], [30], [31]]. It is well established that RAS-activating mutations drive metabolic programming and introduce energetic and metabolic dependencies that can be therapeutically exploited [[24], [25], [26], [27], [28], [29], [30], [31]]. For example, RAS-mutant cells are vulnerable to glutamine depletion and autophagy inhibition [29,[32], [33], [34]]. Targeting RAS-driven metabolic reprogramming can also sensitize cells to targeted pathway inhibitors, demonstrating therapeutic synergy between metabolic and non-metabolic inhibitors [27,29].
Despite numerous studies interrogating the role of RAS mutation in tumor metabolism, no study has investigated NF1-driven metabolic reprogramming in breast cancer. Of the limited number of NF1-related metabolic analyses, most have been done in NF1 knock-out (KO) systems. Although valuable, KO models don't account for RASGAP-independent effects. Neurofibromin's GRD accounts for only 10 % of the total neurofibromin protein, but many studies assume functional equivalence between NF1 KO and GRD inactivation. Additionally, mutations throughout the NF1 gene are NF and cancer-causal despite differential RASGAP effect. This data taken together underscores the utility of NF1 mutational analyses. NF1-KO models have been shown to decrease mitochondrial respiration [35,36], increase nutrient scavenging [37,38], increase lipid droplets [39], increase resting energy expenditure [40], and induce a switch from carbohydrate to lipid catabolism [39,40]. In non-breast cancer models, NF1-mutation has been shown to increase glutamine [41] and β-oxidation dependence [42]. Additionally, individuals affected by NF have lower fasting blood glucose, lower body mass indexes, increased lipid oxidation, and decreased carbohydrate catabolism compared to controls [[43], [44], [45]]. These analyses underscore the necessity of defining and targeting NF1-driven metabolic reprogramming in breast cancer.
The goals of this investigation were (1) to define the role of NF1 deficiency in ER + breast cancer metabolic reprogramming and (2) to identify potential targeted pathway and metabolic inhibitor combination therapies for NF1-deficient ER + breast cancer. Our results revealed that NF1 deficiency enhanced cell proliferation, altered neurofibromin isoform expression, and increased RAS and PI3K/AKT pathway signaling while constraining oxidative adenosine triphosphate (ATP) production and restricting energetic flexibility. NF1 deficiency also increased glutamine influx into TCA intermediates and dramatically increased lipid pools, particularly triglycerides (TG). Lastly, we demonstrate that NF1 deficiency is associated with a synergistic response to a combination of RAS pathway and TG synthesis inhibitors in clinically relevant breast cancer models.
2. Methods
2.1. Generation of an NF1 sporadic human breast cancer model
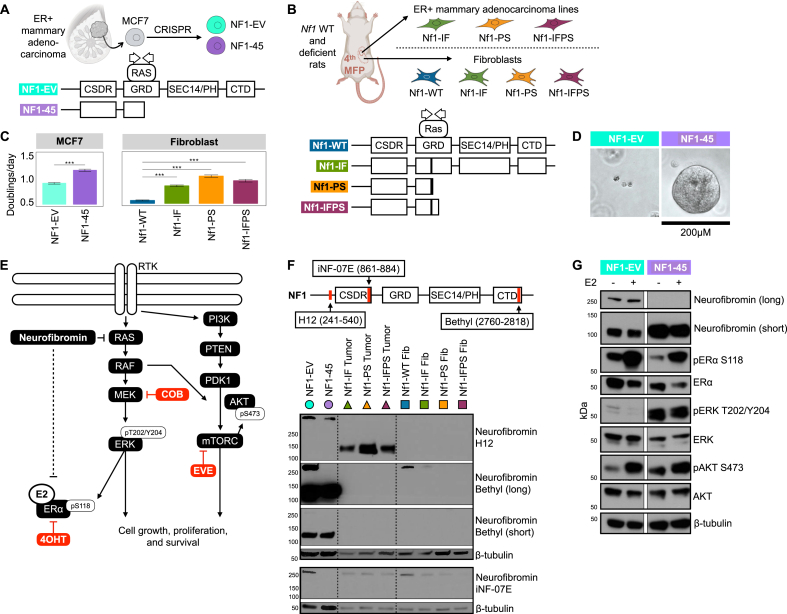
To evaluate the energetic and metabolic effect of NF1 deficiency in ER + sporadic human breast cancer, we utilized the ER + MCF7 human adenocarcinoma cell line. We targeted NF1's RASGAP GRD domain with clustered regularly interspaced short palindromic repeat (CRISPR) guides to the 21st exon and established multiple NF1-deficient cell lines of which one was chosen for this analysis, NF1-45 (Figure 1A). The NF1-45 cells have a 5-bp deletion that introduces a premature stop codon, resulting in a truncated protein product after exon 21 (Figure 1A).
Figure 1.
NF1 deficiency increases cell proliferation and increases RAS and PI3K/AKT pathway signaling. A) Schematic of the NF1-deficient human breast cancer MCF7 model. Schematic of wild-type and mutant neurofibromin in the MCF7 NF1-EV and NF1-45 respectively. The G1 and G2 arrows within the GRD denote the CRISPR guide positioning. Non-GRD domains shown include the cysteine/serine-rich domain (CSDR), Sec14 homologous domain and pleckstrin homologous domain (SEC14/PH), and c-terminal domain (CTD). B) Schematic of Nf1-deficient rat and rat-derived fibroblast and tumor cell line models. Schematic of neurofibromin in the Nf1-WT, Nf1-IF, Nf1-PS, Nf1-IFPS rat models. The G1 and G2 arrows within the GRD denote thesCRISPR guide positioning. C) NF1 deficiency increases cell proliferation in MCF7 cells and rat mammary fibroblasts, n = 2, 24 tech reps per n. The bar represents the geometric mean, and the black, horizontal lines represent the 95 % confidence interval boundaries. D) NF1-deficient MUT-45 spheroids grow larger than NF1-EV spheroids when grown in Matrigel. (n = 3+, 1 representative technical replicate shown above, day 10 of 3D culture). E) Schematic description of the role of neurofibromin in the ER, RAS, and PI3K/AKT signaling pathways and relevant targeted inhibitors: Cobimetinib (COB), Tamoxifen (4OHT), and Everolimus (EVE). F) NF1 indels alter neurofibromin expression. Western blot analysis of total neurofibromin protein. Immunoblotting was carried out using antibodies against neurofibromin (H12, Bethyl, iNF-07E) and β-tubulin (n = 3+, 1 representative biological replicate shown). G) NF1 deficiency increases RAS and AKT/PI3K pathway signaling. Western blot analysis of ER, RAS, and PI3K/AKT phospho-signaling. Immunoblotting was carried out using antibodies against neurofibromin (Bethyl), pERα S118, ERα, pERK T202/Y204, ERK, pAKT S473, AKT, and β-tubulin (n = 3, 1 representative biological replicate shown).
2.2. NF-related rat breast cancer models
To understand the energetic and metabolic effect of Nf1 deficiency in NF-related breast cancer, we employed our previously developed and described NF-related spontaneous rat mammary adenocarcinoma models [1]. To generate these models, we targeted Nf1's RASGAP GRD domain with CRISPR guides to the 20th exon. We generated three distinct Nf1 indels in exon 20 that caused either in-frame or premature stop deletions. The Nf1 inframe indel (Nf1-IF) rats have a 54bp deletion in exon 20 (Figure 1B) [1]. The Nf1 premature stop indel (Nf1-PS) rats have an 8-bp deletion in exon 20 that introduces a premature stop codon, resulting in a truncated protein product after exon 20 (Figure 1B) [1]. The Nf1 in-frame premature stop indel (Nf1-IFPS) rats have a 57-bp deletion in exon 20 that results in a unique messenger RNA (mRNA) isoform with an additional 140-bp deletion in exon 21 [1]. The exon n21 mRNA deletion introduces a premature stop codon, resulting in a truncated protein product after exon 21 (Figure 1B) [1]. The IF, PS, and IFPS indels model “Nf1 deficiency” and induce aggressive, 100 % penetrant, multi-focal, ER/PR + mammary adenocarcinomas in both female and male rats [1]. To perform in vitro energetic and metabolic experiments using our NF-related rat breast cancer model, we utilized isolated, ex vivo rat mammary fat pad fibroblasts from each genetic background: Nf1-WT, Nf1-IF, Nf1-PS, Nf1-IFPS [46]. We also used isolated, ex vivo adenocarcinoma cell lines from each of the Nf1-deficient rat models: Nf1-IF, Nf1-PS, Nf1-IFPS [46]. Although mammary epithelial cells (MECs) are our proposed tumor cell of origin, our isolated MECs do not passage in vitro, so we used isolated fibroblasts to have a rat ex vivo Nf1-WT to Nf1-deficient control since the rat tumor cell lines do not have a Nf1-WT control.
2.3. Cell culture
Human ER + MCF7 breast cancer cells (HTB-22), were grown in Dulbecco's modified eagle medium (DMEM) no phenol red (Gibco, Cat# 21063029), 10 % fetal bovine serum (FBS) (VWR, Cat# 1300-500), 5 mL penicillin-streptomycin (Gibco, Cat# 15070063). Rat-derived fibroblast lines were grown in DMEM/F12 no phenol red (Gibco, Cat# 15070063), 10 % FBS, and 5 mL gentamycin (Gibco, Cat# 15750060). Rat adenocarcinoma-derived tumor cell lines were grown in MEC media, which is DMEM/F12 no phenol red (Gibco, Cat# 21041-025), 10 % horse serum (Gibco, Cat# 16050-122), 20 ng/mL EGF (Sigma, Cat# E9644), 0.5ug/mL hydrocortisone (Sigma, Cat# H088), 100 ng/mL cholera toxin (Sigma, Cat# C8052), and 10ug/mL insulin (Sigma, Cat# I6634). All cells were grown at 37 °C in a 5 % CO2 incubator. Mycoplasma testing (ATCC, Cat# 30-1012-K) every 6 months confirmed cells were not infected.
2.4. IncuCyte proliferation analysis
Cells were grown in their respective medias (see section 2.3) and treated with 10 nM beta-estradiol (Sigma, Cat# E2758) for 48hr in a Sartorius IncuCyte. Cell proliferation was measured via cell confluency and doublings per day were calculated via the equation: Doublings per day = (final confluency/starting confluency)/assay duration (2 days).
2.5. 3-dimensional culture of cells on matrigel
MCF7 cells were plated into 8-well chamber slides (Thermo Fisher Scientific, Cat# 154534) at a density of 2 × 104 cells/well in 100uL growth-factor-reduced-Matrigel (GFR-Matrigel) (Corning, Cat# 354230). After Matrigel solidification, overlay MEC media was added (see section 2.3). Cells were fed with fresh MEC media every 3 days and imaged on Day 3, 6, and 10. Day 10 is shown.
2.6. RNAseq
RNAseq was performed on MCF7 NF1-EV and NF1-45 cell lines using 3 biological replicates. For each technical replicate, 1.2 × 105 cells were plated into a well of a 6-well plate (Corning, Cat# 3506). After 24hr. RLT buffer (Rneasy kit, Qiagen, Cat# 74104) was added to each well. The RLT buffer and cells were added to an Mpbio lysing matrix E tube (Mpbio, Cat# 6914100), and cells were homogenized for 20s in an Mpbio homogenizer (Mpbio). The supernatant was transferred to a QIAshredder column (Qiagen, Cat# 79654), and an equal volume of 70 % ethanol (Pharmco, Cat# 111000200) was added to the flow-through. The flow-through and ethanol mixture was transferred to an Rneasy column and the RNA was extracted following the manufacturer's instructions. The Van Andel Genomics Core prepared RNA libraries from 500 ng of total RNA using a KAPA mRNA Hyperprep kit (v4.17, Kapa Biosystems). RNA was sheared to 300-400bp. Before amplification, cDNA fragments were ligated to IDT for Illumina TruSeq UD Indexed adapters (Illumina Inc). Library quality was assessed using Aglient DNA High Sensitivity chip (Agilent Technologies, Inc.), QuantiFluor ® dsDNA System (Promega Corp.), and Kapa Illumina Library Quantification qPCR assays (Kapa Biosystems). Indexed libraries were combined and 100bp, paired-end sequences was performed using an Illumina NovaSeq6000 sequencer using a S4, 200-cycle sequencing kit (Illumina Inc.) to a depth of 70M reads per sample. Illumina RTA3 was used for base-calling, and NCS output was demultiplexed and converted to FastQ with Illumina Bcl2fastq v1.9.0. Trim Galore v0.6.0 was used to trim sequencing adapters from demultiplexed FASTQ files (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). GENCODE was used to map FASTQ files to GRCh38 (release 33 with STAR v2.7.8a basic two-pass mode) [47]. Raw counts were extracted from reverse strand alignments, and principal component analysis was computed using iDEP 9.1. Fold change was calculated using normalized counts per million. Msigdbr [48] and clusterProfiler [49] packages were used to perform Gene Set Enrichment Analysis (GSEA) using 10,000 permutations and genes ranked by fold change. For the metabolism-related GSEA analysis, we manually curated a list of metabolism-related Kyoto Encyclopedia of Genes and Genomes (KEGG) and REACTOME pathways from a list of all KEGG and REACTOME pathways. The data was then queried for pathways with significantly increased or decreased enrichment across 5 or more comparisons (NF1-45 vs. NF1-EV, Nf1-IF fibroblasts vs. Nf1-WT fibroblasts, Nf1-PS fibroblasts vs. Nf1-WT fibroblasts, Nf1-IFPS fibroblasts vs. Nf1-WT fibroblasts, Nf1-IF tumor vs. Nf1-WT whole mammary fat pad (MFP), Nf1-PS tumor vs. Nf1-WT whole MFP, and Nf1-IFPS tumor vs. Nf1-WT MFP).
2.7. Total protein expression Western blots
Cells were grown in their respective medias (see section 2.3) and treated with 10 nM beta-estradiol for 48hr before lysate collection. Cells were washed with ice-cold 1X PBS (Gibco, Cat# 10010023), scraped, and lysed in Rb buffer (20 mM TrisHCL, pH 7.6; 5 mM EDTA; 150 mM NaCl; 0.5 % NP-40; 50 mM NaF; 1 mM beta-glycerophosphate) supplemented with PhosSTOP (Roche, Cat# 04906837001) and protease inhibitors (Sigma, Cat# 11873580001). Samples were resolved by SDS-PAGE. Immunoblotting was carried out using antibodies against neurofibromin – Bethyl (Bethyl, Cat# A300-140A, primary 1:500, secondary 1:2000), H12 (Santa Cruz Biotechnology, Cat# sc-376886, primary 1:500, secondary 1:2000), and iNF-07E (gifted by iNFiXion Bioscience, Biointron, Cat# B21900201, primary 1:500, secondary 1:2000) – voltage-dependent anion channel (VDAC) (Invitrogen, Cat# PA1-954A, primary 1:1000, secondary 1:3000), and β-tubulin (Cell Signaling Technology, Cat#2146, primary 1:500, secondary 1:2000).
2.8. ER, RAS, and PI3K esolute signaling Western blots
The MCF7 cells were grown in their aforementioned media (see section 2.3) and treated with 10 nM beta-estradiol in DMEM no phenol red, 2 % charcoal-stripped FBS (cFBS) (Sigma, Cat# 12676029, Lot#18A392), and 5 mL penicillin-streptomycin for 24hr before lysate collection. Cells were washed with ice-cold 1X PBS, scraped, and lysed in Rb buffer supplemented with PhosSTOP and protease inhibitors. Samples were resolved by SDS-PAGE. Immunoblotting was carried out using antibodies against neurofibromin (Bethyl), pERα S118 (Sigma, Cat# SAB450399), Erα E115 (Abcam, Cat# 32063), pERK T202/Y204 (Cell Signaling Technology, Cat#9101L), ERK (Cell Signaling Technology, Cat#4695S), pAKT S473 (Cell Signaling Technology, Cat#9271L), AKT (Cell Signaling Technology, Cat#9272S), and β-tubulin.
2.9. Seahorse XF-96 energetic analysis
Cells were grown in their respective medias (see section 2.3) and treated with 10 nM beta-estradiol for 48hr before plating for the Seahorse assay. Cells were trypsinized (Gibco, Cat# 25300-054), collected, quenched, and spun down. Cells were counted with an automated BioRad cell counter (BioRad, Cat# TC20) and plated in poly-d-lysine-coated (Gibco, Cat# A38904-01) Seahorse XF-96 assay plates (Agilent, Cat# 101085-004). Cells were plated with the goal of ∼80–90 % final confluency, so MCF7 cells were plated at 30,000 cells per well, fibroblasts at 5,000 cells per well, and tumor cells at 15,000 cells per well. Cells were spun down and allowed to fully adhere to the plates for 4hr at 37 °C in a 5 % CO2 incubator. After 4hr, growth media was gently removed by pipetting, and 180 μL Seahorse XF DMEM media (Agilent, Cat# 103575-100) was added to each well. Seahorse Mito Stress Test XF DMEM assay media contained 25 mM glucose (Fischer Scientific, Cat#D16-500), 4 mM glutamine (Sigma, Cat#25030-081), and 1 mM pyruvate (Gibco, Cat#11360-070). Seahorse Glyco Stress Test XF DMEM assay media contained 4 mM glutamine. The assay was performed according to Agilent Mito Stress Test and Glyco Stress Test instructions. Extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) shown are per 5000 cells. ATP metrics were calculated following Mookerjee et al., 2017 [50] and using the CEAS R Package [51].
2.10. Stable isotope labeling and metabolomics
For stable isotope labeling experiments with in vitro MCF7 NF1-EV and NF1-45 cells, cells were grown in DMEM no phenol red, 10 % FBS, 5 mL penicillin-streptomycin, and treated with 10 nM beta-estradiol for 48 h. Cells were washed in PBS and incubated in DMEM no phenol red, no glucose, no glutamine (Gibco, Cat# A1443001), 10 % dialyzed FBS (dFBS) (Dialyzed from VWR, Cat# 1300-500), 5 mL penicillin-streptomycin, and 10 nM beta-estradiol with either 25 mM [U–13C]-glucose (Cambridge Isotope Laboratories, Cat# CLM-1396-PK) and 4 mM [U–12C]-glutamine (Gibco, Cat# 21051-040) or 25 mM [U–12C]-glucose (Fischer Scientific, Cat#D16-500) and 4 mM [U–13C]-glutamine (Cambridge Isotope Laboratories, Cat# CLM-1822-H-PK) for 2 h. After 2hr, cells were washed 2X with ice-cold NaCl saline (Gibco Cat# S8776), snap-frozen on dry ice, and stored at −80 °C. Metabolites were extracted using a modified Bligh Dyer method [52]. This modified Bligh Dyer extraction involves the addition of ice-cold methanol (Fisher Chemical, Cat# A456-4) to the cell plate (Corning, Cat# 3506) from which cells are scraped and transferred to an Eppendorf tube containing ice-cold chloroform (Millipore Sigma, Cat# 1024441000) and water (Fisher Chemical, Cat# W6-4), with a final v/v ratio of 2:2:1.8 methanol to chloroform to water. The cell extract was then vortexed for 10 s and incubated on wet ice for 30 min. The samples were then vortexed again for 10 s and spun down at 4 °C for 10 min at 13,000rpm to achieve aqueous and organic phase separation. The aqueous and organic phases were collected and dried down in a speedvac.
2.11. LC-MS method (aqueous)
For liquid chromatography-mass spectrometry (LC-MS) analysis, the aqueous dry-down was resuspended in 50 μL of 50:50 acetonitrile (Fisher Scientific, Cat# A955) and water. The resuspended metabolite extracts were analyzed on an Orbitrap ID-X mass spectrometer (Thermo Fisher Scientific) with a Thermo Vanquish Horizon LC system. Each sample injection was 2 μL. The Van Andel Institute Mass Spectrometry Core performed chromatographic separation using Acquity BEH Amide (1.7 μm, 2.1 mm × 150 mm) analytical columns (Waters, Eschborn, Germany, Cat#176001909). These columns were fitted with a pre-guard column (Waters, 1.7 μm, 2.1 mm × 5 mm; Cat# 186004799) and used an elution gradient with two solvents. Solvent A was LC/MS grade water, and Solvent B was 90 % LC/MS grade acetonitrile. For negative mode analysis, both mobile phases contained 10 mM ammonium acetate (Millipore Sigma, Cat# 73594-100G-F), 0.1 % ammonium hydroxide (Fisher Scientific, Cat# A470-250), and 5 μM medronic acid. For positive mode analysis, both mobile phases included 10 mM ammonium formate and 0.1 % formic acid (Fisher Chemical, Cat# A11710X1-AMP). The 20-minute gradient at 400 μL/min consisted of the following phases: 0–1.0 min (100 % B to 90 % B), 1.0–12.5 min (90 % B to 75 % B), 12.5–19 min (75 % B to 60 % B), and 19–20 min (60 % B). After each run, the column was re-equilibrated over 20 min. The column temperature was maintained at 40 °C. The heated electrospray ionization (H-ESI) source used a spray voltage of −2500 V (negative mode) or 3500 V (positive mode).
The isotopically labeled experimental replicates were collected in full-scan mode with an MS1 scan range from 70 to 1000 m/z, a mass resolution of 120,000 FWHM, RF lens at 35 %, and automatic gain control. The unlabeled samples were used for data-dependent MS2 (ddMS2) fragmentation and were annotated using AquireX workflow. ddMS2 data was obtained with MS1 resolution set at 60,000, MS2 resolution at 30,000, an intensity threshold of 2.0 x 104, and dynamic exclusion activated after a single trigger for a duration of 10 s. MS2 fragmentation comprised HCD at 20 %, 35 %, and 50 % collision energies, which was followed by CID at 15 %, 30 %, and 45 % with an activation Q of 0.25. Both MS2 scans used standard automatic gain control and a maximum injection time of 54 ms. The total cycle time for MS1 and ddMS2 scans was 0.6 s.
2.12. LC-MS method (organic)
The organic dry-down was resuspended in 50 μL of 50:50 isopropanol (Fisher Scientific, Cat# A461-4):water. The resuspended metabolite extracts were analyzed on an Orbitrap ID-X mass spectrometer (Thermo Fisher Scientific) with a Thermo Vanquish Horizon LC system. Each sample injection was 2 μL. The Van Andel Institute Mass Spectrometry Core performed chromatographic separation using C30 (2.6 μm, 2.1 mm × 150 mm) analytical columns (Thermo Fisher Scientific, Cat# 27826–152130). These columns were fitted with a pre-guard column (1.7 μm, 2.1 mm × 5 mm; Waters, Cat# 186004799) and used an elution gradient with two solvents. Solvent A was 60 % acetonitrile, 40 % water, 0.1 % formic acid, 10 mM ammonium formate (Fisher Scientific, Cat# A11550), and Solvent B was 90 % isopropanol, 8 % acetonitrile, 2 % water, 0.1 % formic acid, 10 mM ammonium formate. The 30-minute gradient at 400 μL/min consisted of the following phases: 0–1.0 min (75%A), 1.0–3.0 min (75%A to 60%A), 3–19 min (60%A to 25%A), 19–20.5 min (25 % A to 10 % A), 20.5–28 min (10 % A to 5 % A), and 28.1–30 min (5 %–0%A). After each run, the column was re-equilibrated over 15 min. The column temperature was maintained at 50 °C. The heated electrospray ionization (H-ESI) source used a spray voltage of −2800 V (negative mode) or 3250 V (positive mode). The isotopically labeled experimental replicates were collected in full-scan mode with an MS1 scan range from 200 to 1000 m/z, a esoluteion of 240,000 FWHM, RF lens at 45 %, and automatic gain control. The unlabeled samples were used for data dependent MS3 (ddMS3) fragmentation. ddMS2 scans were collected with HCD fragmentation settings (Assisted Collision Energy Mode, Normalized HCD Collision Energies at 15 %, 30 %, 45 %, 75 %, and 110 % energy levels, Orbitrap resolution: 15,000). A m/z 184 mass trigger, indicative of phosphatidylcholines, was used for CID fragmentation (Collision energy: 35 %, activation time: 10 ms, Orbitrap resolution: 15,000). CID MS3 scans were triggered by specific acyl chain losses for detailed analysis of mono-, di-, and tri-acylglycerides (Collision energy: 35 %, activation time: 10 ms, Ion trap detection at Rapid Scan Rate).
2.13. LC-MS data analysis
Lipid identifications were assigned using LipidSearch (v5.0, Thermo Fisher Scientific). Data was analyzed using Compound Discoverer (v 3.2, Thermo Fisher Scientific). Compounds were identified via retention compared external standards and MS2 spectral matching using the mzCloud database (Thermo Fisher Scientific). Mass isotopologue distribution analysis was performed using isotopologue peak area data from labeled and unlabeled samples, natural abundance correction was performed using an internal VAI matrix-based algorithm like those described in Fernandez et al., 1996 and Trefely et al., 2016. PCA analysis was performed using the FactoMineR R package.
2.14. FBS dialysis
To deplete glucose and glutamine from the FBS used during stable isotope labeling, we prepared dialyzed FBS. In short, FBS was pipetted into SnakeSkin™ Dialysis Tubing, 10K MWCO, 35 mm, 10.5m (Thermo Fisher Scientific, Cat# 88245). The tubing was clamped closed using SnakeSkin Dialysis clips (Thermo Fisher Scientific, Cat# 68011). The FBS-containing dialysis tubing was suspended in 1X PBS (1:300 ratio of FBS:PBS) using a foam Eppendorf tube holder. The PBS was stirred around the FBS-containing tubing using a magnetic stir bar and stir plate for 2hr at 4 °C, after which the PBS was discarded and replaced. The PBS was then stirred around the FBS-containing tubing for 3hr at 4 °C, after which the PBS was discarded and replaced. Lastly, the PBS was stirred around the FBS-containing tubing for 18–20hr at 4 °C, after which the FBS was pipetted out of the tubing, filtered with a 0.22 μM filter (VWR, Cat# 28145-501), aliquoted, and stored at −20 °C. In total, the FBS was dialyzed against 1X PBS for approximately 24hr. Mass spectrometry was used to confirm metabolite depletion (Supplementary Fig. 2).
2.15. Targeted and metabolic inhibitor in vitro drug studies
MCF7 cells were grown in DMEM no phenol red, 10 % FBS, 5 mL penicillin-streptomycin and treated with 10 nM beta-estradiol and the targeted and/or metabolic inhibitors for 48hr in a Sartorius IncuCyte: Tamoxifen (4OHT) (Sigma, Cat# H7904), Cobimetinib (COB) (Genentech, gifted), Everolimus (EVE) (Selleckchem, Cat# S1120), 2-Deoxy-d-glucose (2DG) (Sigma, Cat# D8375), bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl)ethyl sulfide (BPTES) (Cayman Chemical Company, Cat# 19284), Etomoxir (EX) (Cayman Chemical Company, Cat# 11969), PF-04620110 (Cayman Chemical Company, Cat# 16425), PF-06424439 (Cayman Chemical Company, Cat# 17680). Cell proliferation was measured via cell confluency and doublings per day were calculated via the equation: Doublings per day = (final confluency/starting confluency)/assay duration (2 days). Synergy was calculated using the SynergyFinder R package.
3. Results
3.1. NF1 deficiency increases cell proliferation and increases RAS and PI3K/AKT pathway signaling
To evaluate the metabolic effect of NF1 mutation in ER + sporadic human breast cancer, we designed and employed a novel NF1-deficient ER + MCF-7 human adenocarcinoma cell line model, NF1-45 (Figure 1A). The NF1-45 cells have a 5-bp GRD deletion that introduces a premature stop codon, resulting in a truncated protein product after exon 21 (Figure 1A). To evaluate the metabolic effect of NF1 deficiency in an NF context, we derived ex vivo fibroblast and mammary adenocarcinoma cell lines from our previously described Nf1-deficient rat models [1]. The Nf1 in-frame indel (Nf1-IF) rats have a 54bp deletion in exon 20 (Figure 1B) [1]. The Nf1 premature stop indel (Nf1-PS) rats have an 8-bp deletion in exon 20 that introduces a premature stop codon, resulting in a truncated protein product after exon 20 (Figure 1B) [1]. The Nf1 in-frame premature stop indel (Nf1-IFPS) rats have a 57-bp deletion in exon 20 that results in a unique messenger RNA (mRNA) isoform with an additional 140-bp deletion in exon 21 [1]. The exon n21 mRNA deletion introduces a premature stop codon, resulting in a truncated protein product after exon 21 [1]. The Nf1 IF, PS, and IFPS indels model “Nf1 deficiency” and induce aggressive, penetrant, multi-focal, ER/PR + mammary adenocarcinomas in both female and male rats. The Nf1-PS rats have the earliest onset tumors (8 weeks), followed by Nf1-IFPS (10 weeks), followed by Nf1-IF (1 year). Before performing energetic and metabolic analyses, we aimed to characterize our novel MCF7, fibroblast, and tumor cell lines. To evaluate the impact of NF1-deficiency on cell proliferation, cells were grown for 48hr with 10 nM estradiol (E2) in a Sartorius IncuCyte, and cell doublings were measured via confluence over time. Of note, unless otherwise stated, all assays were conducted in the presence of estradiol to better replicate an in vivo environment. Compared to NF1-WT controls (NF1-EV and Nf1-WT), NF1-deficient cells proliferate faster in 2D (Figure 1C). When compared to the NF1-EV control, NF1-45 developed larger 3D spherical colonies, indicating greater tumor potential (Figure 1D). Since neurofibromin is the primary RAS-GAP, alteration of neurofibromin leads to dysregulated RAS-pathway signaling, and RAS pathway signaling can activate the parallel PI3K-AKT pathway (Figure 1E). Neurofibromin can also indirectly activate and directly inhibit ER signaling and alter cellular localization (Figure 1E) [53]. Given the relationship between, neurofibromin, ER, RAS, and PI3K-AKT signaling, we performed western blot analysis to characterize neurofibromin expression and ER, RAS, and PI3K-AKT signaling (Figure 1F and G). Neurofibromin expression was evaluated using several distinct antibodies that recognize the 125 kDa and 250 kDa isoforms in human and rat cells (Figure 1F, Supplementary Fig. 1). In the MCF7 cells, NF1 deficiency resulted in decreased expression of the neurofibromin 250 kDa isoform (Figure 1F). Next neurofibromin expression was measured in mammary tumor cells and mammary fibroblast cells isolated from the Nf1-deficient rat model. Similar to what we previously observed, the 125 kDa isoform was strongly expressed in the Nf1-deficient mammary tumor cells, whereas in the Nf1-deficient mammary fibroblasts we observed a decrease in the 250 kDa isomer [1]. When we measured ER signaling via phosphorylation at the ERK activation-associated S118 site, we saw that NF1-45 cells had decreased active and total ERα activation at baseline and following estradiol (E2) stimulation (Figure 1G). Conversely, when we evaluated ERK signaling, we found that NF1 deficiency dramatically increased active T202/Y204-phosphorylated ERK, which is indicative of increased RAS-pathway signaling (Figure 1G). We then probed against the activation-associated AKT S473 phosphorylation sites and saw a modest increase in PI3K-AKT pathway signaling (Figure 1G). To summarize our phenotyping results, NF1 deficiency increases 2D and 3D cell proliferation, alters neurofibromin isoform expression, decreases ER signaling, and increases RAS and AKT signaling.
3.2. NF1 deficiency alters global transcription and metabolism-related transcriptomic networks
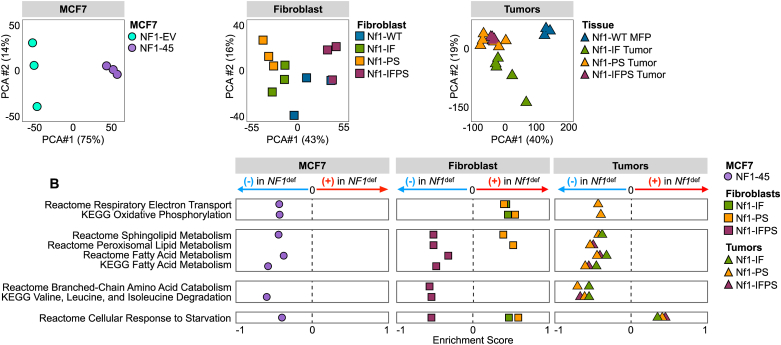
To better understand the impact of NF1 deficiency on global and metabolism-related transcription, we performed RNAseq and utilized principal component analysis (PCA) to visualize the variability across NF1-WT and NF1-deficient cells (Figure 2A). PCA displayed the global shift in gene expression in the NF1-deficient cells compared to the NF1-WT controls (Figure 2A). To evaluate the effect of NF1 deficiency on metabolism-related transcription, we performed gene set enrichment analysis (GSEA). We queried the data for metabolism-related KEGG and REACTOME pathways that significantly increased or decreased in enrichment across our NF1-deficient models. We found that NF1 deficiency altered oxidative phosphorylation-related (OXPHOS) transcription (Figure 2B), a finding that is phenotypically paralleled in our energetic analyses (Figure 3). We also found that NF1 deficiency decreased fatty acid (FA) metabolism, membrane lipid production-related transcription, and branched-chain amino acid degradation-related transcription (Figure 2B). Our RNAseq analysis demonstrates that NF1 indels alter metabolism in breast cancer. Next, we sought to define the impact of NF1 mutations on cellular energetics (glycolysis and OXPHOS) and amino acid and lipid metabolism.
Figure 2.
NF1 deficiency alters global transcription and metabolism-related transcriptomic networks. A) NF1-deficient cells are transcriptionally distinct across cell types and models (n = 3). PCA coordinates were generated using iDEP 9.1. B) NF1 deficiency alters the metabolic transcriptome. GSEA enrichment analysis for metabolism-related KEGG and Reactome pathways. Each bubble represents the pathway enrichment score for each NF1-deficient vs. NF1-WT comparison. The list of metabolism-related KEGG and REACTOME pathways was manually curated from a list of all KEGG and REACTOME pathways in all models.
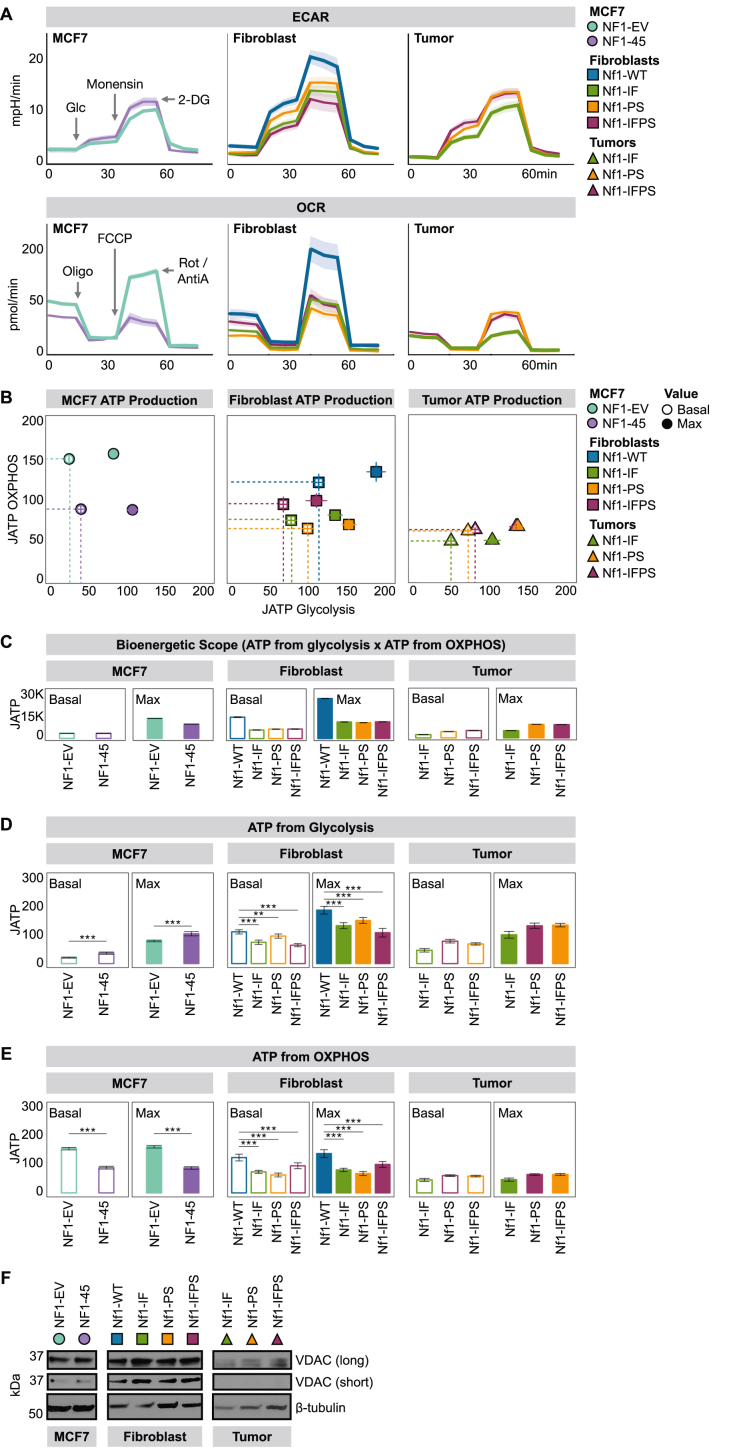
Figure 3.
NF1 deficiency constrains oxidative ATP production. A) NF1 deficiency decreases basal and maximal OCR across models. Monensin is an ionophore that pushes cells to the maximal glycolytic range. 2DG is a competitive hexokinase inhibitor. Oligomycin (oligo) is a complex V inhibitor, fluoro-carbonyl cyanide phenylhydrazone (FCCP) is an uncoupling agent, and rotenone and antimycin A (Rot/AntiA) are complex I and complex III inhibitors, respectively. B) NF1 deficiency restricts joules of ATP (JATP) production in MCF7 cells and rat mammary fibroblasts. JATP values were calculated using CEAS (R Package). Dotted lines outline the basal bioenergetic scope area. The symbols represent the geometric mean, and the lines represent the 95 % confidence interval. MCF7: n = 2, 12 technical replicates per biological replicate. Fibroblast: n = 2, 24 technical replicates per biological replicate. Tumor cells: n = 2, 28 technical replicates per biological replicate. C) Nf1 deficiency restricts max bioenergetic scope (JATP from glycolysis x JATP from OXPHOS) in MCF7 cells and rat mammary fibroblasts. The bar represents the geometric mean, and the black, horizontal lines represent the 95 % confidence interval boundaries. JATP values were calculated using CEAS (R Package). D) NF1 deficiency has a divergent effect on glycolytic JATP production based on cell type. JATP values were calculated using CEAS (R Package). The bar represents the geometric mean, and the black, horizontal lines represent the 95 % confidence interval boundaries. MCF7: n = 2, 12 technical replicates per biological replicate. Fibroblast: n = 2, 24 technical replicates per biological replicate. Tumor cells: n = 2, 28 technical replicates per biological replicate. E) NF1 deficiency restricts OXPHOS JATP production. JATP values were calculated using CEAS (R Package). The bar represents the geometric mean, and the black, horizontal lines represent the 95 % confidence interval boundaries. MCF7: n = 2, 12 technical replicates per biological replicate. Fibroblast: n = 2, 24 technical replicates per biological replicate. Tumor cells: n = 2, 28 technical replicates per biological replicate. F) NF1 deficiency impacts OCR without altering total mitochondrial load as measured by Western blot analysis of voltage-dependent anion channel (VDAC) expression. Immunoblotting was carried out using antibodies against VDAC and β-tubulin.
3.3. NF1 deficiency constrains oxidative ATP production
Cellular energetics describes how energy is captured and used by cells. Our analysis focused on core aspects of energy metabolism: glycolysis, the tricarboxylic acid cycle (TCA), and the electron transport chain (ETC). To measure cellular energetics in real-time, we used the Seahorse XF-96 Mito Stress Test and Glyco Stress Test assays. The Seahorse machine has two main outputs: extracellular acidification rate (ECAR) and oxygen consumption rate (OCR). ECAR measures media acidification and is a proxy of glycolysis. OCR measures media oxygen depletion and is a proxy of OXPHOS (O2 is the final electron transport chain [ETC] electron acceptor). Cellular energetics were modulated using the following compounds: glucose (glc) is a substrate for glycolysis, monensin is an ionophore, 2-deoxy-d-glucose (2DG) is a competitive hexokinase inhibitor, oligomycin (oligo) is a complex V inhibitor, fluoro-carbonyl cyanide phenylhydrazone (FCCP) is an uncoupling agent, and rotenone and antimycin A (Rot/AntiA) are complex I and complex III inhibitors, respectively. Across our human and rat models, NF1 deficiency decreased basal and maximal OXPHOS (Figure 3A). When we calculated the ATP produced from glycolysis and OXPHOS and visualized those values in two dimensions, we found that NF1 deficiency restricted energetic flexibility (Figure 3B). Despite oxidative constraints, only NF1-45 cells have glycolytic “compensation” at basal conditions, but all NF1-deficient lines have smaller max bioenergetic scopes, a metric calculated by multiplying the JATP glycolysis x JATP OXPHOS (Figure 3C). Bioenergetic scope describes the theoretical energetic space in which a cell can operate, so the larger the bioenergetic scope, the greater the energetic flexibility. When we compared NF1-WT and NF1-deficient ATP production from glycolysis and OXPHOS, we found that NF1-deficient cells have decreased OXPHOS ATP production (Figure 3D,E). Of note, not all NF1-deficient models had negative or significant Reactome respiratory electron transport and KEGG oxidative phosphorylation gene set enrichment scores despite their OXPHOS deficit (Figure 2B). These divergent RNAseq and OCR data harken to a fundamental question within the ETC field; how are the transcription, translation, assembly, and oxidative function of ETC proteins related? To understand whether this decrease in OXPHOS capacity was due to a decrease in total mitochondrial load, we performed Western Blot analysis and analyzed voltage-dependent anion channel (VDAC) expression. When we compared our NF1-deficient models to their WT controls, we saw no appreciable difference in VDAC expression (Figure 3F).
3.4. NF1 deficiency increases intracellular lipid pools
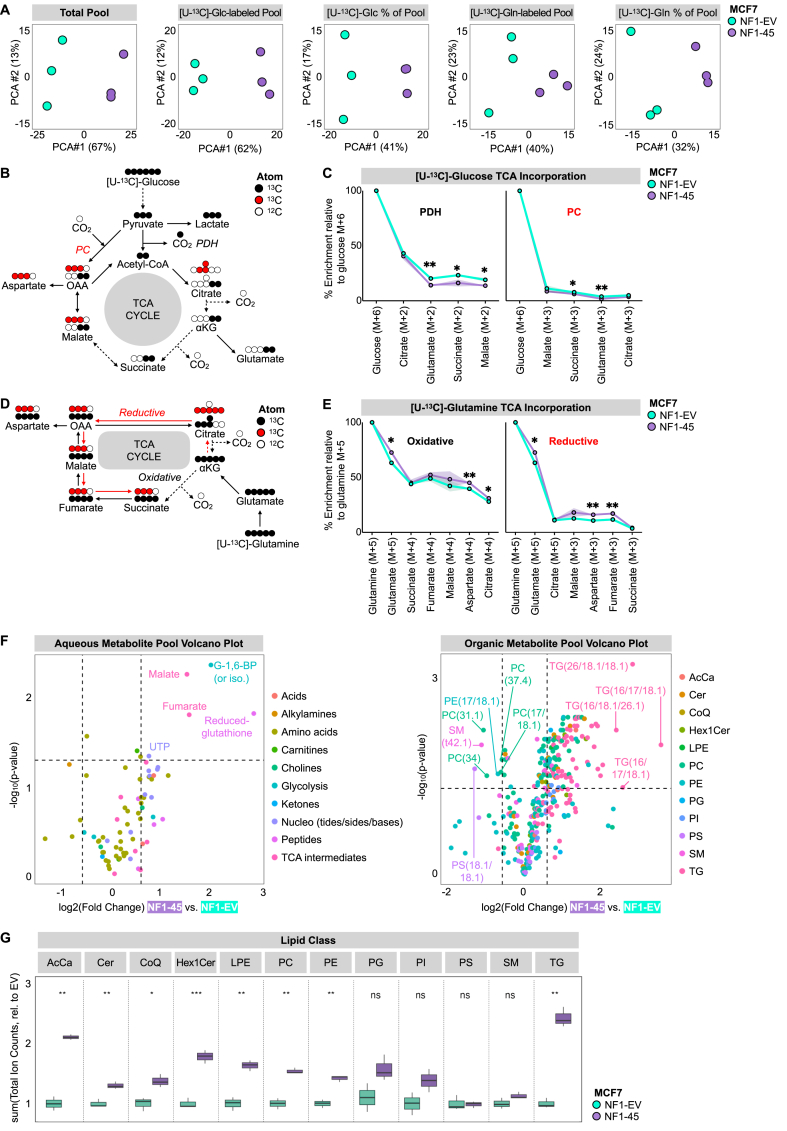
To define the NF1-deficient metabolome, we performed untargeted metabolomics and stable isotope labeling. We compared intracellular metabolite pools, labeled pool sizes (PS), and labeled pool fractional enrichment (FE) between NF1-WT and NF1-deficient MCF7 cells and visualized this difference with PCA analysis (Figure 4A). Across all queries, NF1-deficient cells are distinct, and this difference is most pronounced when comparing total metabolite pools (Figure 4A). Because we detected an NF1-driven OXPHOS loss, we wanted to understand how glucose and glutamine were incorporated into TCA intermediates via mass isotopologue distribution. [U–13C]-glucose TCA incorporation is facilitated by pyruvate dehydrogenase (PDH) and pyruvate carboxylase (PC), which respectively catalyze the conversion of pyruvate to acetyl-CoA and oxaloacetate. [U–13C]-glucose TCA entry via PDH leads to M+2 TCA intermediates, while entry via PC leads to M+3 TCA intermediates (Figure 4B). [U–13C]-glutamine is incorporated into glutamate and α-ketoglutarate (α-KG) as M+5, and α-KG oxidation leads to M+4 TCA labeling and reductive carboxylation results in M+3 TCA metabolites except for citrate, which is M+5 (Figure 4D). When we compared the ratio of M+3 glutamate to M+5 glutamate, we observed no difference between NF1-EV and NF1-45 cells, suggesting no difference in the rate of TCA cycling (Supplementary Fig. 2D). When we compared relative glucose incorporation into TCA intermediates via oxidative and reductive pathways, we found a small but significant decrease in glucose incorporation in NF1-45 cells (Figure 4C). Conversely, when we compared the relative glutamine incorporation into TCA intermediates via oxidative and reductive pathways, we saw a small but significant increase in glutamine incorporation in the NF1-45 cells (Figure 4E). This data suggests a small but statistically significant NF1-driven shift in TCA carbon source from glucose to glutamine. We next analyzed the total aqueous and organic metabolite pools and determined that NF1 deficiency increased select TCA and REDOX intermediates and increased levels of many lipid species, particularly triglycerides (TG) (Figure 4F). This NF1-dependent increase in intracellular TG is consistent with previous findings in NF1-deficient muscle cells [39]. Next, we wanted to understand the impact of NF1 deficiency on lipid class pools, and we found an increase in lipid levels across all classes except phosphatidylglycerol (PG), phosphatidylinositol (PI), phosphatidylserine (PS), and sphingomyelin (SM) (Figure 4G).
Figure 4.
NF1 deficiency increases intracellular lipid pools. A) NF1 deficiency alters the ER + breast cancer metabolome. PCA analysis of NF1-EV and NF1-deficient NF1-45 MCF7 LC-MS data (n = 1, 3 technical replicates per biological replicate). B) Schematic of carbon flow from [U–13C]-glucose. C) NF1 deficiency decreases glucose incorporation into TCA intermediates. Relative metabolite enrichment compared to M+6 [U–13C]-glucose (n = 1, 3 technical replicates per biological replicate). D) Schematic of carbon flow from [U–13C]-glutamine. E) NF1 deficiency increases glutamine incorporation into TCA intermediates. Relative metabolite enrichment compared to M+5 [U–13C]-glutamine (n = 1, 3 technical replicates per biological replicate). F) NF1 deficiency increases TCA and REDOX intermediates and intracellular lipid species. Volcano plots describing aqueous and organic metabolite pool sizes between NF1-45 and NF1-EV control (n = 1, 3 technical replicates per biological replicate). The shape represents the geometric mean of the three technical replicates. G) NF1 deficiency increases total intracellular lipids. Boxplot of the sum of ion counts normalized to the mean ion count of the NF1-EV control within each lipid class. The line represents the geometric mean of the three technical replicates and the box boundaries at the 1st and 3rd quartile boundaries (n = 1, 3 technical replicates per biological replicate).
3.5. NF1 mutation increases synergy between RAS pathway and triglyceride synthesis inhibitors
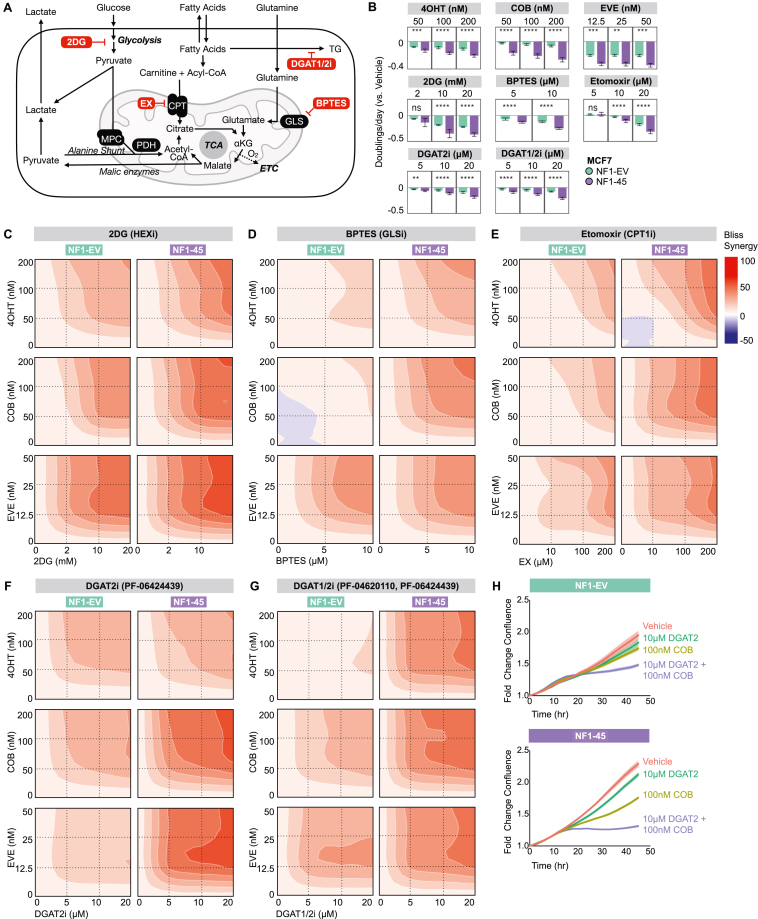
We next wanted to understand if metabolic inhibitors targeting NF1-driven metabolic changes synergize with traditional targeted inhibitors in NF1-deficient ER + breast cancer cells. For targeted RAS and PI3K/AKT inhibitors, we evaluated the antiproliferative effects of Tamoxifen (4OHT), Cobimetinib (COB), and Everolimus (EVE) (Figure 1E; Supplementary Fig. 3A). For metabolic inhibitors, we evaluated the antiproliferative effects of the following metabolic inhibitors: 2DG, bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl)ethyl sulfide (BPTES), Etomoxir (EX), PF-04620110 (DGAT1i), and PF-06424439 (DGAT2i) (Figure 5A). We tested the glycolysis inhibitor, 2DG, because we saw increased glycolysis in NF1-45 cells (Figure 3D). 2DG inhibits glycolysis via competitive hexokinase inhibition, so we anticipated increased 2DG sensitivity in NF1-45 cells. We tested the glutaminase (GLS) inhibitor, BPTES, because we saw increased glutamine incorporation into TCA intermediates in NF1-45 cells (Figure 4E). GLS catalyzes the conversion of glutamine to glutamate, and glutamate dehydrogenase (GDH) catalyzes the conversion of glutamate to α-ketoglutarate, which can then enter the TCA cycle, so we anticipated increased BPTES sensitivity in NF1-45 cells. We next tested the carnitine palmitoyl transferase 1 (CPT1) inhibitor, Etomoxir (EX), because we saw increased palmitoyl and oleoyl carnitine (AcCa) species, intermediates in mitochondrial FA transport, in NF1-45 cells (Figure 4G). This increase in AcCa species suggested increased or impeded mitochondrial FA transport, a process facilitated by CPT1, so we anticipated increased Etomoxir sensitivity in NF1-45 cells. Lastly, we chose to test the diacylglycerol o-acyltransferase 1 and 2 (DGAT1, DGAT2) inhibitors, PF-04620110 (DGAT1i), and PF-06424439 (DGAT2i), because we saw a ∼2.5X increase in total TG species in NF1-45 cells (Figure 4G). DGATs catalyze the final step in TG synthesis, so we anticipated increased DGAT1i and DGAT2i sensitivity in NF1-45 cells. Because DGAT2 had ∼2X higher expression than DGAT1 in MCF7 cells, we chose to test DGAT2i as a single agent and DGAT1/2i in combination (Supplementary Fig. 3B). When comparing single-agent antiproliferative effects, we found that NF1-45 cells were more sensitive to ERα inhibition by 4OHT, mitogen-activated protein kinase kinase (MEK) inhibition by COB, and mechanistic target of rapamycin (mTOR) inhibition by EVE (Figure 5B). NF1-45 cells were also more sensitive to each metabolic inhibitor tested (Figure 5B). Next, we combined each targeted and metabolic inhibitor to investigate genotype-driven synergy changes using a Bliss Synergy model and the SynergyFinder R package (Figure 5 and Supplementary Figs. 3C–G). A synergy score of less than −10 indicates that the drug-drug interaction is likely antagonistic, a synergy score between −10 and 10 indicates that the drug-drug interaction is likely additive, and a synergy score greater than 10 indicates that the drug-drug interaction is likely synergistic [54]. When we evaluated 2DG synergy with RAS, ER, and PI3K/AKT inhibitors, we saw a ∼1.5X NF1-dependent increase in synergy between 2DG and 4OHT, COB, and EVE (Figure 5C, Supplementary Fig. 3C). This NF1-dependent increase in 2DG-targeted inhibitor synergy could be due to increased glycolytic glucose use, but the loss of lipid, nucleotide, and TCA precursor species could also account for NF1-driven synergy. When we evaluated BTPES synergy with RAS, ER, and PI3K/AKT inhibitors, we saw ∼2X more synergy between BPTES and 4OHT and ∼3.5X more synergy between BPTES and COB in the NF1-deficient NF1-45 cells (Figure 5D, Supplementary Fig. 3D). When we evaluated Etomoxir synergy with RAS, ER, and PI3K/AKT inhibitors, we saw ∼1.5X more synergy Etomoxir and 4OHT, ∼2X more synergy between Etomoxir and COB, and a small increase in synergy between Etomoxir and EVE in NF1-45 cells (Figure 5E, Supplementary Fig. 3E). When we evaluated DGAT2i synergy with RAS, ER, and PI3K/AKT inhibitors, we saw a ∼1.5-2X increase in synergy between DGAT2i and COB and a ∼3X increase in synergy between DGAT2i and EVE in NF1-45 cells (Figure 5F, Supplementary Fig. 3F). When we evaluated DGAT1/2i synergy with RAS, ER, and PI3K/AKT inhibitors, we saw a ∼4X increase in synergy between DGAT1/2i and 4OHT, a ∼1.5X increase in synergy between DGAT1/2i and COB, and a 1.5X increase in synergy between DGAT1/2i and EVE (Figure 5G, Supplementary Fig. 3G). Taken together, these results underscore the NF1-driven energetic and metabolic changes seen in our Seahorse XF-96 and mass spectrometry experiments. We also provide evidence connecting NF1-driven metabolic reprogramming to inhibitor sensitivity and synergy. Indeed, NF1-deficient NF1-45 cells have increased glycolysis and increased glycolysis inhibitor sensitivity and synergy, increased TCA glutamine incorporation and increased GLS inhibitor sensitivity and synergy, increased AcCa pools and increased CPT1 inhibitor sensitivity and synergy, and increased TG pools and increased DGAT1/2 inhibitor sensitivity and synergy. When we compared the antiproliferative effects of all our combination strategies (Supplementary Figs. 4–5) and their respective Bliss synergies (Figure 5 and Supplementary Figs. 3C–G), we found that inhibition of RAS and triglyceride synthesis most effectively halted proliferation in NF1-deficient NF1-45 cells (Figure 5H). These findings pave the way for future in vivo experiments and highlight potential combination treatments for NF1-deficient, ER + breast cancer.
Figure 5.
NF1 mutation increases synergy between RAS pathway and triglyceride synthesis inhibitors. A) Schematic description of selected metabolic inhibitors and their respective enzymatic targets: 2-Deoxy-d-glucose (2DG), bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl)ethyl sulfide (BPTES), Etomoxir (EX), PF-04620110 (DGAT1i), PF-06424439 (DGAT2i). B) NF1 deficiency increases sensitivity to targeted and metabolic inhibitors. A crossbar plot describing the antiproliferative impact of 4OHT, COB, EVE, 2DG, BPTES, EX, DGAT2i, and DGAT1/2i on NF1-EV and NF1-45 (n = 2, 8 technical replicates per biological replicate). The bar represents the geometric mean, and the error bar boundaries represent the 95 % confidence interval boundaries. C) Bliss synergy plots describing the synergy between 2DG and 4OHT, COB, and EVE. Bliss synergy was calculated and visualized using the SynergyFinder R package (n = 2, 8 technical replicates per biological replicate). D) Bliss synergy plots describing the synergy between BPTES and 4OHT, COB, and EVE. Bliss synergy was calculated and visualized using the SynergyFinder R package (n = 2, 8 technical replicates per biological replicate). E) Bliss synergy plots describing the synergy between Etomoxir and 4OHT, COB, and EVE. Bliss synergy was calculated and visualized using the SynergyFinder R package (n = 2, 8 technical replicates per biological replicate). F) Bliss synergy plots describing the synergy between DGAT2i and 4OHT, COB, and EVE. Bliss synergy was calculated and visualized using the SynergyFinder R package (n = 2, 8 technical replicates per biological replicate). G) Bliss synergy plots describing the synergy between DGAT1i and DGAT2i combination treatment and 4OHT, COB, and EVE. Bliss synergy was calculated and visualized using the SynergyFinder R package (n = 2, 8 technical replicates per biological replicate). H) IncuCyte proliferation graphs of EV and MUT45 cells grown in 10 nM E2 and Vehicle, 10 μM DGAT2, 100 nM COB, or 10 μM DGAT2 + 100 nM COB. The geometric mean is visualized by the connecting line and the 95 % confidence interval by the shading (n = 2, 8 technical replicates per biological replicate).
4. Discussion
Historically, the tumor suppressor NF1 was studied in the context of NF-associated benign and malignant neuronal tumors, but recent genomic and epidemiological analyses have identified NF1 as a driver of inherited breast cancer, sporadic breast cancer, and endocrine resistance [[2], [3], [4], [5], [6], [7], [8], [9]]. Despite these recent analyses connecting NF1 and breast cancer, there is a fundamental need for a greater understanding of neurofibromin's role in breast cancer development, biology, and therapeutic response. One essential and previously unexplored area of study is the relationship between NF1 deficiency and breast cancer cell metabolism. Activated oncogenes and inactivated tumor suppressors are known to contribute to tumorigenesis by reprogramming cellular energetics and metabolism towards macromolecular synthesis – a phenomenon termed “metabolic reprogramming” [20]. Despite neurofibromin's known role as a breast cancer tumor suppressor, no study had evaluated NF1-driven metabolic reprogramming in ER+ breast cancer – a gap this study aimed to fill.
We employed a novel NF1-deficient ER + breast cancer MCF7 model and previously published Nf1-deficient rat models to evaluate NF1-driven metabolic reprogramming in ER+ breast cancer.
We demonstrated that NF1 deficiency results in decreased NF1 mRNA and decreased neurofibromin 250 kDa isoform expression. This change in neurofibromin expression is accompanied by a decrease in estrogen pathway signaling and an increase in RAS and PI3K/AKT pathway signaling, which is expected given neurofibromin's RAS-GAP function. We also demonstrate that NF1 deficiency increases 2D and 3D cell proliferation while reducing basal and maximal oxidative ATP production twofold.
Using RNAseq, we found that NF1 deficiency globally altered transcription. When we investigated the impact of NF1 deficiency on metabolism-specific transcription, we noted a change in OXPHOS-related transcription and FA and lipid-related transcription. When we performed glucose and glutamine stable isotope labeling, we saw a decrease in glucose TCA incorporation and an increase in glutamine TCA incorporation, suggesting a shift in TCA intermediate carbon sourcing. Perhaps the most striking metabolic finding was a ∼1.5–2.5-fold increase in lipid species across classes. Of the enriched species, TGs were most dramatically and consistently increased.
Lastly, we demonstrated that NF1-driven metabolic changes introduce therapeutic vulnerabilities and drug synergies. We illustrated increased sensitivity to single-agent inhibitors against ER, RAS, and PI3K/AKT pathway members and increased sensitivity to glycolysis, glutamine, fatty acid, and lipid metabolism inhibitors that parallel NF1-driven pathway and metabolic changes. When we combined targeted and metabolic inhibitors, we saw broad NF1-driven synergy between glutamine, fatty acid mitochondrial transport, and triglyceride synthesis inhibitors and ER, RAS, and PI3K/AKT pathway inhibitors. When we evaluated our drug combinations for both antiproliferative and synergistic effects, we determined that RAS and triglyceride synthesis inhibition was the most effective strategy to inhibit proliferation in NF1-deficient breast cancer cells. This study is the first to combine RAS and triglyceride synthesis inhibitors in an RAS-active tumor model, let alone an NF1-deficient, ER+, RAS-active context.
We found a significant phenotypic overlap between NF1-deficiency and RAS-activation-driven metabolic reprogramming. In both of our NF1-deficient systems and published RAS-active systems, we see increased cellular proliferation, increased glutamine TCA incorporation, and increased neutral lipid accumulation (TG). We also saw increased glutaminase and CPT1 inhibitor sensitivity in our NF1-deficient models and published Nf1-deficient and RAS-active models [42,55]. We have demonstrated that NF1-deficient ER + breast cancer cells have impaired mitochondrial respiration without a change in total mitochondrial load, an observation reproduced in Nf1−/− mouse embryonic fibroblasts and Nf1-KO drosophila melanogaster [35,36]. Published data from RAS-active cells shows an overlapping energetic phenotype with impaired mitochondrial respiration but increased mitochondrial load. This discrepancy in mitochondrial load may speak to a difference in the degree of RAS pathway activation, specifically to the difference between an NF1-deficient RAS-dysregulated system and constitutively RAS-activated systems. The difference in mitochondrial load could also relate to the fundamental signaling distinction between an NF1-deficient and a RAS-active system: the impact of NF1 deficiency on ERα at the transcriptional activity. Neurofibromin is a known ERα transcriptional co-repressor, so NF1 deficiency can impact ER's transcriptional activity. In our NF1-deficient NF1-45 cells, we see decreased ERK-dependent S118 phosphorylation despite increased ERK T202/Y204 phosphorylation and decreased total ERα, highlighting the nuanced relationship between NF1 deficiency and estrogen signaling. Overall, want to underscore the importance of further metabolic inquiry in NF1-deficient contexts given the efficacy of metabolism-directed therapeutics in RAS-active cancers.
5. Conclusion
In conclusion, this study demonstrates that NF1 deficiency drives metabolic reprogramming in ER+ breast cancer. NF1-driven ER + breast cancer metabolic reprogramming is characterized by increased proliferation, constrained oxidative ATP production, increased glutamine TCA influx, and lipid pool expansion. These metabolic changes introduce novel single-agent sensitivity and metabolic-to-targeted inhibitor synergies. This study highlights neurofibromin's ability to alter cellular metabolism across cell types and model organisms.
Funding
This project was supported, in part, by generous donations from NF Michigan, the Bee Brave Foundation, and the Van Andel Foundation.
CRediT authorship contribution statement
Rachel (Rae) J. House – review & editing, Writing – original draft, Visualization, Validation, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Elizabeth A. Tovar: Writing – review & editing, Data curation. Curt J. Essenburg: Data curation. Patrick S. Dischinger: Resources, Data curation. Abigail E. Ellis: Data curation. Ian Beddows: Data curation. Ryan D. Sheldon: Validation, Supervision, Resources. Evan C. Lien: Writing – review & editing, Validation, Resources. Carrie R. Graveel: Writing – review & editing, Supervision, Funding acquisition, Conceptualization. Matthew R. Steensma: Writing – review & editing, Supervision, Funding acquisition, Conceptualization.
Declaration of competing interest
None.
Acknowledgements
We would like to thank the VAI Vivarium staff, Flow Cytometry Core, and Genomics Core for their valued expertise. We want to extend our deepest gratitude to all VAI Mass Spectrometry Core members for their expertise, camaraderie, and mentorship. We also want to thank Rae's thesis advisory committee members, Russel (Rusty) Jones, PhD, Ralph DeBerardinis, PhD, and Timothy Triche, PhD, for their direction and counsel throughout this project. Lastly, we would like to thank the VAI Department of Metabolism and Nutritional Programming for their metabolic support, especially the Lien Lab. Lastly, we would like to thank iNFiXion BioScience for providing the neurofibromin iNF-07E antibody and supporting supplementary data.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2024.101876.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
MCF7 RNAseq data is available at Gene Expression Omnibus (GEO) GSE251692, and rat fibroblast, mammary fat pad, and tumor RNAseq data is available at GEO GSE231603. Raw mass spectrometry files will be made available upon request.
References
- 1.Dischinger P.S., Tovar E.A., Essenburg C.J., Madaj Z.B., Gardner E.E., Callaghan M.E., et al. NF1 deficiency correlates with estrogen receptor signaling and diminished survival in breast cancer. NPJ Breast Cancer. 2018;4:29. doi: 10.1038/s41523-018-0080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stephens P.J., Tarpey P.S., Davies H., Van Loo P., Greenman C., Wedge D.C., et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486(7403):400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Razavi P., Chang M.T., Xu G., Bandlamudi C., Ross D.S., Vasan N., et al. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell. 2018;34(3):427–438 e426. doi: 10.1016/j.ccell.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffith O.L., Spies N.C., Anurag M., Griffith M., Luo J., Tu D., et al. The prognostic effects of somatic mutations in ER-positive breast cancer. Nat Commun. 2018;9(1):3476. doi: 10.1038/s41467-018-05914-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sokol E.S., Feng Y.X., Jin D.X., Basudan A., Lee A.V., Atkinson J.M., et al. Loss of function of NF1 is a mechanism of acquired resistance to endocrine therapy in lobular breast cancer. Ann Oncol. 2019;30(1):115–123. doi: 10.1093/annonc/mdy497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearson A., Proszek P., Pascual J., Fribbens C., Shamsher M.K., Kingston B., et al. Inactivating NF1 mutations are enriched in advanced breast cancer and contribute to endocrine therapy resistance. Clin Cancer Res. 2020;26(3):608–622. doi: 10.1158/1078-0432.CCR-18-4044. [DOI] [PubMed] [Google Scholar]
- 8.Wallace M.D., Pfefferle A.D., Shen L., McNairn A.J., Cerami E.G., Fallon B.L., et al. Comparative oncogenomics implicates the neurofibromin 1 gene (NF1) as a breast cancer driver. Genetics. 2012;192(2):385–396. doi: 10.1534/genetics.112.142802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ratner N., Miller S.J. A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat Rev Cancer. 2015;15(5):290–301. doi: 10.1038/nrc3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uusitalo E., Kallionpaa R.A., Kurki S., Rantanen M., Pitkaniemi J., Kronqvist P., et al. Breast cancer in neurofibromatosis type 1: overrepresentation of unfavourable prognostic factors. Br J Cancer. 2017;116(2):211–217. doi: 10.1038/bjc.2016.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X., Teer J.K., Tousignant R.N., Levin A.M., Boulware D., Chitale D.A., et al. Breast cancer risk and germline genomic profiling of women with neurofibromatosis type 1 who developed breast cancer. Genes Chromosomes Cancer. 2018;57(1):19–27. doi: 10.1002/gcc.22503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seminog O.O., Goldacre M.J. Age-specific risk of breast cancer in women with neurofibromatosis type 1. Br J Cancer. 2015;112(9):1546–1548. doi: 10.1038/bjc.2015.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howell S.J., Hockenhull K., Salih Z., Evans D.G. Vol. 9. Dove Med Press; 2017. Increased risk of breast cancer in neurofibromatosis type 1: current insights; pp. 531–536. (Breast cancer). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seminog O.O., Goldacre M.J. Risk of benign tumours of nervous system, and of malignant neoplasms, in people with neurofibromatosis: population-based record-linkage study. Br J Cancer. 2013;108(1):193–198. doi: 10.1038/bjc.2012.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell P.M. Oncogenic Ras pushes (and pulls) cell cycle progression through ERK activation. Methods Mol Biol. 2014;1170:155–163. doi: 10.1007/978-1-4939-0888-2_9. [DOI] [PubMed] [Google Scholar]
- 17.von Lintig Friederike C., A.D.D, Varki Nissi M., Wallace Anne M., Casteel Darren E., a.G.R.B Ras activation in human breast cancer. Breast Cancer Res Treat. 2000;(62):51–62. doi: 10.1023/a:1006491619920. [DOI] [PubMed] [Google Scholar]
- 18.Sivaraman Vimala S., Wang H.Y., Nuovo Gerard J., Malbon Craig C. Hyperexpression of mitogen-activated protein kinase in human breast cancer. J Clin Invest. 1997;99(7):1478–1483. doi: 10.1172/JCI119309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bollag G., Clapp D.W., Shih S., Adler F., Zhang Y.Y., Thompson P., et al. Loss of NF1 results in activation of the Ras signaling pathway and leads to aberrant growth in haematopoietic cells. Nat Genet. 1996;12(2):144–148. doi: 10.1038/ng0296-144. [DOI] [PubMed] [Google Scholar]
- 20.Ward P.S., Thompson C.B. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phan L.M., Yeung S.C., Lee M.H. Cancer metabolic reprogramming: importance, main features, and potentials for precise targeted anti-cancer therapies. Cancer Biol Med. 2014;11(1):1–19. doi: 10.7497/j.issn.2095-3941.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boroughs L.K., DeBerardinis R.J. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol. 2015;17(4):351–359. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Dard L., Bellance N., Lacombe D., Rossignol R. RAS signalling in energy metabolism and rare human diseases. Biochim Biophys Acta Bioenerg. 2018;1859(9):845–867. doi: 10.1016/j.bbabio.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Moiseeva O., Bourdeau V., Roux A., Deschenes-Simard X., Ferbeyre G. Mitochondrial dysfunction contributes to oncogene-induced senescence. Mol Cell Biol. 2009;29(16):4495–4507. doi: 10.1128/MCB.01868-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laurent Dejean B.B., Bunoust O., Guérin B., Rigoulet M. Activation of Ras cascade increases the mitochondrial enzyme content of respiratory competent yeast. Biochem Biophys Res Commun. 2002;293(5):1383–1388. doi: 10.1016/S0006-291X(02)00391-1. [DOI] [PubMed] [Google Scholar]
- 27.Yao W., Yue P., Zhang G., Owonikoko T.K., Khuri F.R., Sun S.Y. Enhancing therapeutic efficacy of the MEK inhibitor, MEK162, by blocking autophagy or inhibiting PI3K/Akt signaling in human lung cancer cells. Cancer Lett. 2015;364(1):70–78. doi: 10.1016/j.canlet.2015.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaglio D., Soldati C., Vanoni M., Alberghina L., Chiaradonna F. Glutamine deprivation induces abortive s-phase rescued by deoxyribonucleotides in k-ras transformed fibroblasts. PLoS One. 2009;4(3) doi: 10.1371/journal.pone.0004715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saqcena M., Mukhopadhyay S., Hosny C., Alhamed A., Chatterjee A., Foster D.A. Blocking anaplerotic entry of glutamine into the TCA cycle sensitizes K-Ras mutant cancer cells to cytotoxic drugs. Oncogene. 2015;34(20):2672–2680. doi: 10.1038/onc.2014.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Son J., Lyssiotis C.A., Ying H., Wang X., Hua S., Ligorio M., et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496(7443):101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oba D., Inoue S.I., Miyagawa-Tomita S., Nakashima Y., Niihori T., Yamaguchi S., et al. Mice with an oncogenic HRAS mutation are resistant to high-fat diet-induced obesity and exhibit impaired hepatic energy homeostasis. EBioMedicine. 2018;27:138–150. doi: 10.1016/j.ebiom.2017.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Commisso C., Davidson S.M., Soydaner-Azeloglu R.G., Parker S.J., Kamphorst J.J., Hackett S., et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497(7451):633–637. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White E. Exploiting the bad eating habits of Ras-driven cancers. Genes Dev. 2013;27(19):2065–2071. doi: 10.1101/gad.228122.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schulte M.L., Fu A., Zhao P., Li J., Geng L., Smith S.T., et al. Pharmacological blockade of ASCT2-dependent glutamine transport leads to antitumor efficacy in preclinical models. Nat Med. 2018;24(2):194–202. doi: 10.1038/nm.4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masgras I., Ciscato F., Brunati A.M., Tibaldi E., Indraccolo S., Curtarello M., et al. Absence of neurofibromin induces an oncogenic metabolic switch via mitochondrial ERK-mediated phosphorylation of the chaperone TRAP1. Cell Rep. 2017;18(3):659–672. doi: 10.1016/j.celrep.2016.12.056. [DOI] [PubMed] [Google Scholar]
- 36.Tong J.J., Schriner S.E., McCleary D., Day B.J., Wallace D.C. Life extension through neurofibromin mitochondrial regulation and antioxidant therapy for neurofibromatosis-1 in Drosophila melanogaster. Nat Genet. 2007;39(4):476–485. doi: 10.1038/ng2004. [DOI] [PubMed] [Google Scholar]
- 37.Ghoshal P., Singla B., Lin H., Cherian-Shaw M., Tritz R., Padgett C.A., et al. Loss of GTPase activating protein neurofibromin stimulates paracrine cell communication via macropinocytosis. Redox Biol. 2019;27 doi: 10.1016/j.redox.2019.101224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bloomfield G., Traynor D., Sander S.P., Veltman D.M., Pachebat J.A., Kay R.R. Neurofibromin controls macropinocytosis and phagocytosis in Dictyostelium. Elife. 2015;4 doi: 10.7554/eLife.04940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sullivan K., El-Hoss J., Quinlan K.G., Deo N., Garton F., Seto J.T., et al. NF1 is a critical regulator of muscle development and metabolism. Hum Mol Genet. 2014;23(5):1250–1259. doi: 10.1093/hmg/ddt515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turner A.N., Johnson M.S., Brosius S.N., Yoder B.S., Yang K., Yang Q., et al. 2018. Neurofibromin is essential to maintain metabolic function and sustain life in the adult mouse. [Google Scholar]
- 41.Sheikh Tahir N., Cremers S., Schwartz Gary K. Targeted inhibition of glutaminase as a potential new approach for the treatment of NF1 associated soft tissue malignancies. Oncotarget. 2017;8(55):94054–94068. doi: 10.18632/oncotarget.21573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanchez-Macedo N., Feng J., Faubert B., Chang N., Elia A., Rushing E.J., et al. Depletion of the novel p53-target gene carnitine palmitoyltransferase 1C delays tumor growth in the neurofibromatosis type I tumor model. Cell Death Differ. 2013;20(4):659–668. doi: 10.1038/cdd.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Souza M.L., Jansen A.K., Martins A.S., Rodrigues L.O., de Rezende N.A. Nutrient intake in neurofibromatosis type 1: a cross-sectional study. Nutrition. 2015;31(6):858–862. doi: 10.1016/j.nut.2014.12.021. [DOI] [PubMed] [Google Scholar]
- 44.Koga M., Yoshida Y., Imafuku S. Nutritional, muscular and metabolic characteristics in patients with neurofibromatosis type 1. J Dermatol. 2016;43(7):799–803. doi: 10.1111/1346-8138.13218. [DOI] [PubMed] [Google Scholar]
- 45.Martins A.S., Jansen A.K., Rodrigues L.O.C., Matos C.M., Souza M.L.R., Miranda D.M., et al. Increased insulin sensitivity in individuals with neurofibromatosis type 1. Arch Endocrinol Metab. 2018;62(1):41–46. doi: 10.20945/2359-3997000000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tovar E.A., Sheridan R., Essenburg C.J., Dischinger P.S., Arumugam M., Callaghan M.E., et al. Dissecting the rat mammary gland: isolation, characterization, and culture of purified mammary epithelial cells and fibroblasts. Bio Protoc. 2020;10(22) doi: 10.21769/BioProtoc.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liberzon A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2(3) doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mookerjee S.A., Gerencser A.A., Nicholls D.G., Brand M.D. Quantifying intracellular rates of glycolytic and oxidative ATP production and consumption using extracellular flux measurements. J Biol Chem. 2017;292(17):7189–7207. doi: 10.1074/jbc.M116.774471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.House, R., Eapen, J., Shen, H., Graveel, C., Steensma, M., CEAS: cellular energetics analysis software [Computer software].
- 52.Bligh E.D., Wj A Rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8) doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 53.Zheng Z.Y., Anurag M., Lei J.T., Cao J., Singh P., Peng J., et al. Neurofibromin is an estrogen receptor-alpha transcriptional Co-repressor in breast cancer. Cancer Cell. 2020;37(3):387–402 e387. doi: 10.1016/j.ccell.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ianevski A., Giri A.K., Aittokallio T. SynergyFinder 3.0: an interactive analysis and consensus interpretation of multi-drug synergies across multiple samples. Nucleic Acids Res. 2022;50(W1):W739–W743. doi: 10.1093/nar/gkac382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quijano C., Cao L., Fergusson M.M., Romero H., Liu J., Gutkind S., et al. Oncogene-induced senescence results in marked metabolic and bioenergetic alterations. Cell Cycle. 2012;11(7):1383–1392. doi: 10.4161/cc.19800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MCF7 RNAseq data is available at Gene Expression Omnibus (GEO) GSE251692, and rat fibroblast, mammary fat pad, and tumor RNAseq data is available at GEO GSE231603. Raw mass spectrometry files will be made available upon request.