Abstract
Reverse transcription of human immunodeficiency virus type-1 is primed by cellular tRNALys3, which is selectively packaged into viral particles where it is bound at its 3′ terminus to a complementary sequence of viral RNA termed the primer binding site (PBS). Since cellular tRNALys3 is highly conserved, it might conceivably serve as a good target for novel antagonists to block reverse transcriptase (RT) activity. In this study, we have examined a number of antisense oligodeoxyribonucleotides (ODNs) that are complementary to different parts of the tRNA primer and, therefore, may interfere with the initiation of RT-mediated DNA synthesis. We found that the stability of complexes between synthetic tRNALys3 and ODNs was significantly increased when binding occurred via sequences involved in tertiary interactions of the tRNA. In particular, ODNs with complementarity to both the variable and TΨC stem–loop of tRNALys3 bound with high affinity to both free tRNALys3 as well as to the binary tRNALys3/RNA complex. As a result, the initiation of DNA synthesis was severely compromised under these conditions. Moreover, RT-associated RNase H activity recognized the tRNA within this ternary tRNALys3/RNA/ODN complex as an RNA template and initiated its degradation. Both this RNase H degradation of tRNALys3 as well as the altered structure of the tRNA/RNA complex, due to the binding of the ODN, contributed to the inhibition of synthesis of viral DNA. The initiation of RT activity was almost completely blocked when using ODNs that interfered with intermolecular tRNA/RNA interactions that involved both the PBS and sequences outside the PBS. Similar findings were obtained with natural preparation of tRNALys3.
INTRODUCTION
A key step in the life cycle of all retroviruses is the conversion of single-stranded genomic RNA into double-stranded DNA. This process, termed reverse transcription, requires the virus-encoded reverse transcriptase (RT), a multifunctional enzyme that possesses DNA- and RNA-dependent polymerase activities, as well as a ribonuclease H (RNase H) activity that specifically degrades the viral RNA template (1). As with other retroviruses, human immunodeficiency virus type-1 (HIV-1) recruits a cellular tRNA primer in order to initiate DNA synthesis (2–4).
Due to its central role in HIV replication, HIV-1 RT is an important target for anti-retroviral drugs. Nucleoside analog RT-inhibitors such as 3′-azido-3′-dideoxythymidine (AZT) and 2′,3′-dideoxy-3′-thiacytidine (3TC), as well as non-nucleoside RT-inhibitors, such as nevirapine, are used in the treatment of HIV disease (5). Although combinations of currently available drugs can often reduce viral burden to below the limit of detection (6), these compounds cannot prevent the emergence of mutations in the RT or protease genes that confer drug resistance (7). Therefore, an ongoing search exists for alternative strategies, that include gene therapy approaches based on antisense nucleic acids, decoy RNAs and ribozymes (8–18). To avoid the problem of viral mutability and resistance, the targets of such approaches should include highly conserved sequences. The HIV-1 specific initiation complex fulfills this criterion, since it involves interactions among viral genomic RNA and a cellular component, i.e. primer tRNA.
The tRNALys3 primer of reverse transcription is selectively packaged into HIV-1 virions, where it is found hybridized through 18 terminal nucleotides at its 3′ end to a complementary primer binding site (PBS) near the 5′ end of viral RNA (19,20). The RT enzyme recognizes this binary tRNA/RNA complex and initiation of reverse transcription ensues. In vitro studies have shown that the mature viral nucleocapsid protein as well as the Pr55gag precursor can promote annealing between tRNALys3 and the PBS (21–23). However, PBS complementarity is not the only factor that determines specific tRNALys3 primer usage. Enzymatic and chemical probing of the binary tRNALys3/RNA complex have revealed the existence of viral RNA sequences outside the PBS that can make contact with the tRNA primer (24,25). For example, a 5′-USUU-3′ sequence in the anticodon loop of tRNALys3 may be able to interact with an A-rich loop in the U5 region of viral RNA, located ~15 nt upstream of the PBS. The importance of this latter intermolecular loop–loop interaction is supported by results showing that mutated viruses with deletions in the A-rich loop are impaired in both synthesis of viral DNA and viral replication (26). Other cellular tRNAs may also serve as replication primers, as long as the HIV genome is mutated to provide complementarity to the 3′ ends as well as to the anticodon loops of these tRNA species (27–33). Thus, it seems likely that the PBS and adjacent sequences can form a structural environment that facilitates initiation of reverse transcription by tRNALys3.
In this study, we have employed oligodeoxyribonucleotides (ODNs) that are complementary to tRNALys3 to aggravate formation of the specific binary tRNA/RNA complex as a strategy to inhibit synthesis of the first discrete DNA product of reverse transcription. This DNA fragment, termed minus strand strong-stop DNA [(–) strand ssDNA], remains attached to the tRNA primer, and is later transferred to the 3′ end of viral RNA to permit the completion of synthesis of (–) strand DNA (1,4). We have demonstrated in cell-free assays that efficient inhibition of initiation of reverse transcription requires ODNs that destabilize interactions between tRNALys3 and the PBS as well as the extended contact between primer tRNA and viral RNA.
MATERIALS AND METHODS
Nucleic acids and enzymes
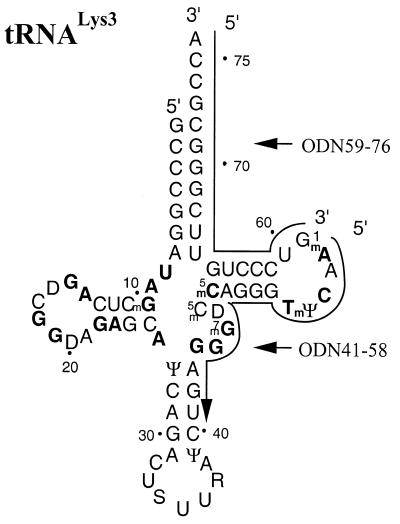
All ODNs used in this study were chemically synthesized and purchased from Gibco BRL (Canada). These ODNs are complementary to different segments of human tRNALys3, including the 3′ terminus, the TΨC stem–loop and the variable loop. Sequences of antisense ODNs used are listed below; numbering refers to base positions of the tRNA to which the ODN shows complementarity and position 76 represents the 3′ terminal residue of tRNALys3 (Fig. 1): ODN59–76, TGG CGC CCG AAC AGG GAC; ODN41–58, TTG AAC CCT GGA CCC TCA; ODN57–76, TGG CGC CCG AAC AGG GAC TT; ODN55–76, TGG CGC CCG AAC AGG GAC TTG A; ODN53-76, TGG CGC CCG AAC AGG GAC TTG AAC; ODN47–76, TGG CGC CCG AAC AGG GAC TTG AAC CCT GGA; ODN41–76, TGG CGC CCG AAC AGG GAC TTG AAC CCT GGA CCC TCA.
Figure 1.
Secondary structure of human tRNALys3. Bases that involve tertiary interactions are highlighted in bold. The antisense ODNs 59–76 and 41–58 are complementary to the acceptor arm and TΨC-loop of tRNALys3, respectively, as indicated.
Natural tRNALys3 was prepared and purified from human placenta as previously described (34). In addition, synthetic tRNALys3 was synthesized in vitro using the plasmid pT7hLys3 as template. This construct contains the 76-bp fragment that represents mature human tRNALys3 under control of the T7 RNA polymerase promotor. A FokI restriction site was introduced downstream of the tRNA gene to generate a DNA template that ensures run-off transcription of the tRNA with the correct 3′ CCA terminus (35). In vitro transcription with T7 RNA polymerase was performed overnight at 37°C using 5 µg of the digested plasmid which was incubated in a total volume of 100 µl containing 40 mM Tris–HCl (pH 8.0), 25 mM MgCl2, 50 mM NaCl, 1 mM spermidine, 50 mM DTT, 4 mM NTPs (each), 200 U of T7 polymerase (MBI, Fermentas, Canada) and 20 U of the placental RNase inhibitor RNasin (Pharmacia, Canada). The tRNA reaction product was purified using 8% polyacrylamide gels that contained 7 M urea and 50 mM Tris–borate–EDTA (TBE), and was visualized under UV light, prior to elution from isolated gel slices using a solution of 0.5 M sodium acetate and 0.1% SDS. Radiolabeled tRNA was prepared using the same procedure, except that 4 mM UTP was replaced with a mixture of 50 µM UTP and 2.5 µCi/µl [α-32P]UTP. An RNA template, containing the 5′ terminal sequence of genomic RNA, including the PBS, was prepared in similar fashion, after digestion of the plasmid pHIV-PBS with BssHII (38). Heterodimeric HIV-1 RT (p66/p51) was prepared and purified as previously described (37).
Band shift assays
The ability of different antisense ODNs to bind to tRNALys3 was analyzed under native conditions using 5% polyacrylamide gels. A constant amount of tRNALys3 (80 nM) was incubated with various concentrations of the ODN tested in a buffer containing 50 mM Tris–HCl (pH 7.8) and 50 mM NaCl. Reactions were allowed to proceed for 30 min at 37°C. To test the influence of divalent cations on affinity between tRNA and ODNs, this assay was also performed in the presence of 6 mM MgCl2. Products were analyzed on 5% polyacrylamide gels containing 50 mM TBE or, alternatively, 50 mM TB and 500 µM MgCl2 to assay ODN–tRNA interactions in the presence of the divalent cations (Figs 2 and 3). Results were analyzed by molecular image analysis. The ability of ODNs to bind to tRNALys3 in the presence of the RNA template was analyzed using the same procedure (Fig. 4).
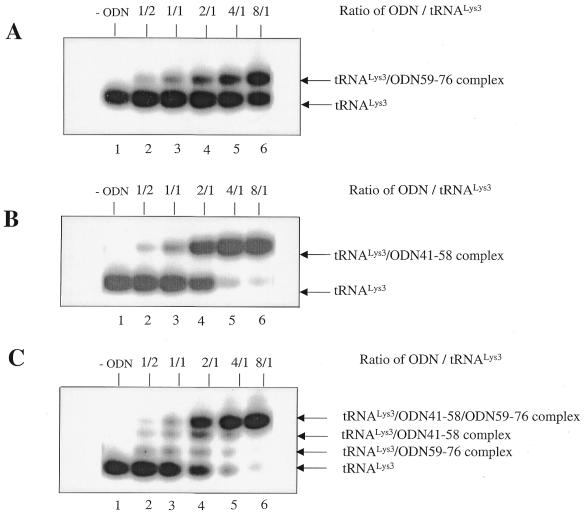
Figure 2.
Binding of ODNs to stRNALys3 in the absence of Mg2+. Lanes 1–6 represent experiments performed in the absence of Mg2+, with a constant concentration of tRNALys3 and increasing amounts of ODN, i.e. molar ODN/tRNA ratios 0, 1/2, 1/1, 2/1, 4/1 and 8/1. (A) Binding of ODN59–76 to tRNALys3. (B) Binding of ODN41–58 to tRNALys3. (C) Binding of both ODN59–76 and ODN41–58 to tRNALys3. (D) The graph summarizes the data shown in (A–C).
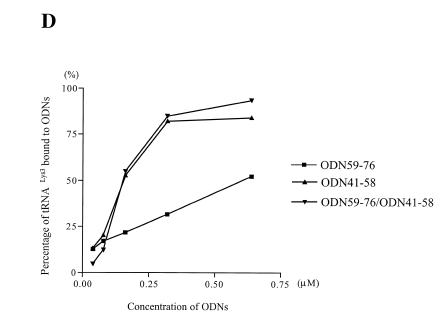
Figure 3.
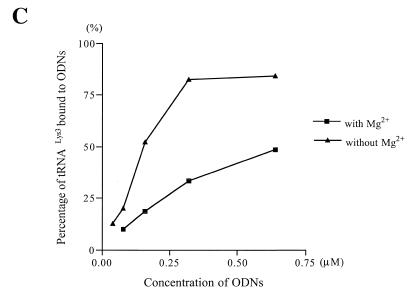
Effect of MgCl2 on the affinity of ODNs for stRNALys3. This experiment was performed as described in the legend to Figure 2. Binding of ODN41–58 to tRNALys3 in the absence (A) or presence (B) of Mg2+. (C) The graph shows differences in the affinity of ODN41–58 for tRNALys3 in the presence and absence of MgCl2.
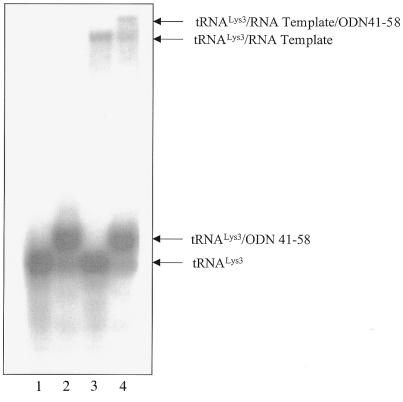
Figure 4.
Gel retardation with stRNALys3, RNA template and ODN41–58. Lane 1, tRNALys3 alone; lane 2, formation of complex of tRNALys3/ODN41–58 with molar ratios of 1/5; lane 3, formation of binary complex of tRNALys3 and viral RNA template with molar ratio of 1/2; lane 4, formation of ternary tRNALys3/viral RNA/ODN41–58 complex with molar ratio of 1/2/5. This experiment was performed in the presence of Mg2+ (0.5 mM) and the components of this reaction were not heat-annealed.
Inhibition of synthesis of (–) strand ssDNA
We used a cell-free assay to study the effects of different ODNs on the efficiency of synthesis of (–) ssDNA. Reverse transcription when initiated from the PBS of the RNA template normally yields a DNA product of 178 nt that is joined to the 76 nt tRNALys3 primer (38). The yield of this (–) ssDNA product is significantly increased when the tRNA primer is heat-annealed to the RNA template prior to initiation of DNA synthesis. This procedure ensures complete hybridization and was performed in a 10 µl reaction mixture containing 50 mM Tris–HCl (pH 7.8), 50 mM NaCl, 80 nM tRNALys3 and 160 nM template RNA. This mixture was incubated for 2 min at 95°C followed by 20 min at 70°C. Synthesis of (–) ssDNA was initiated by addition of 240 nM RT in the presence or absence of 400 nM ODN, also in the presence of 6 mM MgCl2 and 200 µM dNTPs (each). Aliquots of 2 µl were removed at different time points, and reactions were terminated in 8 µl of a solution containing 80% formamide and 40 mM EDTA. As indicated in the figure legends, reactions were monitored using radiolabeled tRNALys3 or, alternatively, by including [α-32P]dGTP in the reaction mixture. The latter method allowed us to study the inhibition of DNA synthesis in the presence of the different ODNs, and, as well, the usage of ODNs as primers for the reverse transcription of the tRNA. In contrast, usage of internally labeled tRNALys3 yielded information regarding the efficiency of RNase H-mediated degradation of the tRNA primer. Products were separated on 8% polyacrylamide–7 M urea gels and analyzed as described above.
RESULTS
The aim of this study was to identify ODNs that could bind efficiently to the tRNALys3 primer in order to antagonize reverse transcription. Two ODNs were specifically designed for this purpose. One of these, ODN59–76, is complementary to the 3′ end of tRNALys3 and a second, ODN41–58, is complementary to the TΨC- and the variable loop of tRNA (Fig. 1). While the first ODN may aggravate interactions with the PBS, the latter may be required to destabilize intermolecular contacts outside the PBS. These ODNs were tested, both individually and in combination, for their ability to bind to tRNALys3 and to inhibit synthesis of (–) strand ssDNA.
Efficient binding to tRNALys3 requires ODN sequences that are complementary to tRNALys3 nucleotide sequences involved in tertiary interactions
To evaluate the affinities of ODN59–76 and ODN41–58 for tRNALys3, we first performed band-shift experiments in which a constant amount of free, synthetic tRNALys3 (stRNALys3) was incubated at 37°C with increasing concentrations of ODN. The isolated tRNA and complexes consisting of both tRNALys3 and ODN were then separated on native gels. The results in Figure 2 show that ODN41–58 possessed a higher affinity for tRNALys3 than did ODN59–76 (Fig. 2A and B). A 2-fold molar excess of ODN41–58 over tRNA was required to obtain 50% complex formation, while the same level of band-shifting with ODN59–76 required an 8-fold excess. The simultaneous presence of both ODNs did not further improve the efficiency of complex formation between tRNA and the ODN complexes (Fig. 2C and D).
These data suggest that destabilization of crucial tertiary structures within tRNALys3 is necessary to obtain a stable ODN/tRNA complex under conditions that do not involve heat-annealing of the various reaction components. As a consequence, we reasoned that factors that stabilize the L-shaped structure of tRNA, such as divalent cations, might inhibit binding of the ODN. Figure 3 shows that this was indeed the case and that formation of a stable tRNA/ODN complex was compromised by ~3-fold in the presence of 6 mM Mg2+.
tRNALys3 bound to ODN is cleaved by RT-associated RNase H activity
To test whether ODN41–58 might also target tRNALys3 as a component of the binary tRNA/PBS complex, band-shift assays were performed in which this ODN was added to a mixture of tRNALys3 and an RNA template, containing the PBS (Materials and Methods). Figure 4 demonstrates the existence of a ternary tRNA/PBS/ODN complex that was formed under native conditions at 37°C (compare lanes 3 and 4). Hence, under these circumstances, the initiation of DNA synthesis is expected to be severely compromised. Conceivably, binding between ODN41–58 and tRNALys3 might provoke structural changes within the tRNA/RNA complex that may aggravate this initiation process. In addition, the RT-associated RNase H domain may recognize the tRNA/ODN complex as an RNA/DNA substrate and initiate degradation of the tRNA. This activity can be used in a functional assay to further elucidate the efficiency of binding of ODNs to tRNALys3.
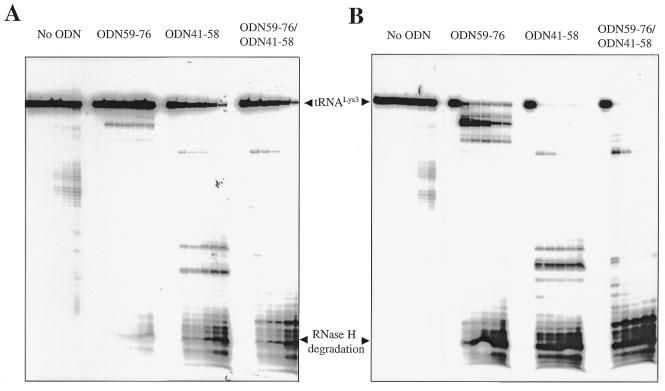
Therefore, RNase H degradation was analyzed in the presence of either ODN59–76 or ODN41–58 in time-course experiments. The ODN was added under native conditions to the pre-formed binary complex in which the tRNA was quantitatively hybridized with the RNA template (Materials and Methods). Heat-annealing of tRNALys3 and the RNA template was performed in the absence as well as in the presence of MgCl2 to study the effects of divalent metal ions on the formation of the ternary tRNA/RNA/ODN complex. Figure 5 shows that ODN59–76 was unable to initiate RNase H degradation of tRNALys3, under conditions in which the tRNA/RNA complex was pre-formed in the presence of Mg2+ (Fig. 5A). Cleavages with considerable efficiency were seen only when MgCl2 was omitted from the pre-incubation mixture and when these divalent metal ions were added to start the reaction (Fig. 5B). Thus, ODN59–76 can compete with the PBS of genomic RNA in the absence but not presence of Mg2+, suggesting that divalent cations stabilize the binary tRNA/RNA complex, and, in turn, diminish binding of ODN59–76. In contrast, ODN41–58 was able to efficiently initiate RNase H degradation of tRNA, regardless of whether MgCl2 had been present during formation of the tRNA/RNA complex (Fig. 5A and B). Although the efficiency of initial RNase H cuts was lower as the tRNA/RNA complex was formed in the presence of Mg2+, it can be seen that tRNALys3 was almost completely degraded as reaction times were increased.
Figure 5.
Degradation of ODN-bound stRNALys3 by RT-associated RNase H activity. (A) A time-course experiment was performed by pre-forming the tRNALys3/RNA complex (Materials and Methods), and incubating this substrate with ODNs at 37°C in the presence of Mg2+. Reactions were initiated by the addition of RT. Samples were removed after 0, 1, 5, 10, 30 and 60 min. (B) In this experiment, the tRNA/RNA complex was preformed in the absence of Mg2+ and incubated with ODNs and RT at 37°C. Mg2+ (6 mM) was added to initiate the reaction. Each panel shows a time course of RNase H degradation, monitored with radiolabeled tRNALys3.
An ODN that is complementary to both the variable and TΨC stem–loops as well as the 3′ terminus of tRNALys3 can block initiation of DNA synthesis
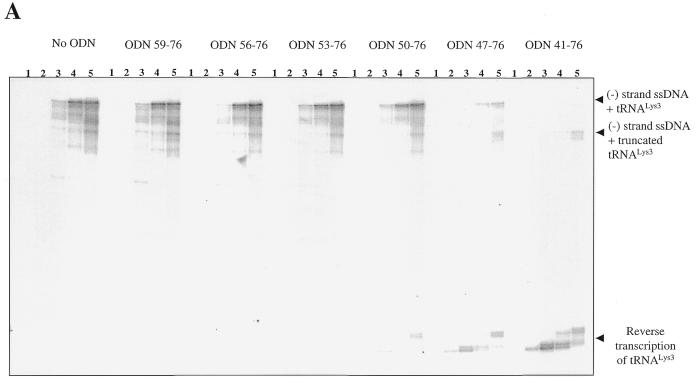
We next tested whether an ODN with complementarity to the PBS might be combined with the properties of ODN41–58 to inhibit the initiation of viral DNA synthesis. These studies employed a series of ODNs that contained the same 5′ end as that found in ODN59–76 but with an increased number of residues at the 3′ end. Considering the above results, this extension at the 3′ end could be expected to facilitate the binding of ODNs to the tRNA primer. The longest ODN used, i.e. ODN41–76, is a combination of both ODN59–76 and ODN41–58. Time-course experiments show that ODNs of 18, 21, 24 and 27 residues were unable to block synthesis of (–) strand ssDNA in comparison with controls from which ODNs were omitted (Fig. 6A). However, a set of bands of increased intensity, which corresponded to ~75 nt, were seen as longer ODNs were added into the system. This shows that the HIV-1 RT recognized the 3′ end of the ODN and copied the tRNA primer. Moreover, the longer the ODN employed, the better was the binding to the tRNA primer and the more efficient was its use as a template. These data are in good agreement with the efficient RNase H degradation of the tRNA observed in the presence of ODN41–58 (Fig. 5B).
Figure 6.
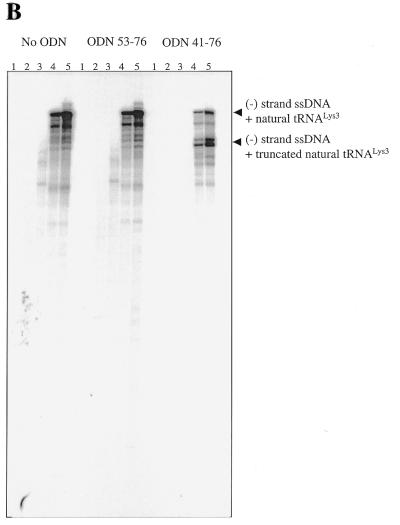
(A) Inhibition of synthesis of (–) ssDNA through use of a variety of ODNs of increasing length at the 3′ terminus. Each panel shows a time course of DNA synthesis, monitored by incorporation of [α-32P]dGTP into growing DNA chains. Reactions were initiated with stRNALys3 and stopped after 5, 15, 30 and 60 min (lanes 2–5); lane 1 is a control performed in the absence of dNTPs. (B) Inhibition of (–) ssDNA synthesis primed with natural tRNALys3. The reaction was monitored in the absence (left) or presence of ODN53–76 (middle) and ODN41–76 (right). (C) Time course of reverse transcription of the natural tRNALys3 primer and its synthetic counterpart, using 5′ end-labeled ODN41–76 as primer.
Concomitant with the reverse transcription of tRNALys3, the efficiency of synthesis of (–) strand ssDNA was drastically decreased, particularly when using ODNs that possessed complementarity to 30 or 36 residues at the 3′ end of the tRNA primer. Moreover, longer ODNs further modulated this diminished initiation reaction and yielded shorter DNA products than normally formed. This is a consequence of RNase H cuts in the tRNA that now resulted in a full-length (–) strand ssDNA covalently attached to a truncated version of tRNALys3.
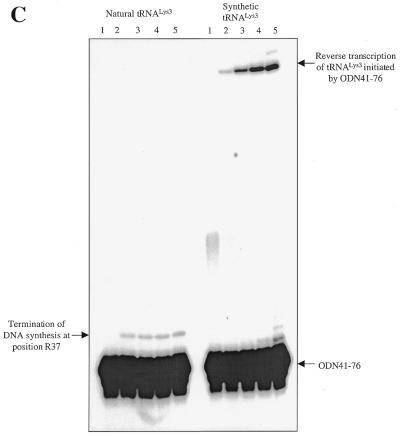
Finally, we have tested the effectiveness of the best inhibitor, i.e. ODN41–76, using natural tRNALys3 as a primer. Biochemical data have suggested that post-transcriptionally modified residues may contribute to the stability of the tRNA structure (39). Such increased stabilization could conceivably diminish complex formation between the natural tRNA and complementary ODNs, as compared to synthetic tRNA counterparts. Our results show that synthesis of (–) strand ssDNA, primed with natural human tRNALys3, was strongly inhibited in the presence of ODN41–76 (Fig. 6B). As described for reactions performed with the in vitro synthesized primer (Fig. 6A), the presence of a truncated version of the strong stop product points to the likelihood that RNase H had cleaved the natural tRNA. However, in contrast to reactions primed with synthetic primer, we did not observe bands attributable to transcription of tRNA. Hence, it is possible that hypermodified residues in the anticodon loop of tRNALys3 may have caused premature termination of DNA synthesis. To test this hypothesis directly, we labelled ODN41–76 at its 5′ end and looked for extension products in the presence of both RT and the preformed binary tRNA/RNA complex (Fig. 6C). Complete transcription of tRNALys3 using ODN41–76 was only seen with the synthetic molecule (Fig. 6C, right), consistent with the results of Figure 6A. In contrast, the natural tRNALys3 was specifically extended by only 3 nt (Fig. 6C, left, lanes 2–5), indicating that the hypermodified adenine, located at position 37 of the anticodon loop, caused premature termination of tRNA transcription. Collectively, these assays demonstrate that ODN41–76 inhibits synthesis of (–) strand ssDNA by interfering with both the natural initiation complex as well as those involving synthetic primers.
DISCUSSION
The emergence of HIV variants that display resistance to anti-retroviral drugs can limit the success of current therapeutic strategies (40). Thus, there is a need to identify novel targets that involve highly conserved mechanisms and structures, that may not easily undergo genetic alteration. The initiation of reverse transcription is a specific process that is sensitive to structural changes that involve participating molecules, i.e. RT, genomic RNA in the region of the PBS and primer tRNA (26,28–33,41,42). Both cell-free assays and tissue culture experiments have shown that the initiation of reverse transcription can be inhibited using antisense ODNs that bind to viral genomic RNA upstream of the PBS (14,36).
In this study, we have evaluated the possibility of directly targeting the primer tRNA, since this cellular component is unlikely to undergo genetic alteration. We have identified antisense ODNs that bind specifically to tRNALys3 and that are capable of inhibiting the initiation of (–) strand ssDNA synthesis in cell-free assays. Several factors must be considered to explain the observed diminution of viral DNA synthesis and to further evaluate the effectiveness of this approach in biological systems. First, our data show that the antisense ODN can bind to the free tRNALys3 primer as well as to tRNA that is complexed with genomic RNA. One cannot exclude that the first of these could result in toxic side effects (see below). However, the latter binding event is expected to induce changes in the structure of the binary tRNA/RNA complex and may, therefore, specifically inhibit the initiation of (–) strand ssDNA. Second, the three components, i.e. tRNALys3, the RNA template and certain ODNs, can indeed form a stable ternary complex, provided that the ODN is complementary to the TΨC- and variable loops of the tRNA. Under these circumstances, the RT enzyme can recognize both the 3′ end of the primer tRNA as well as the 3′ end of the ODN. Competition between these recognition events additionally contributes to the inhibitory effect of these ODNs. Finally, interactions between the polymerase active site of RT and the 3′ terminus of ODN position the RNase H active site over the tRNA primer; consequently, the latter serves as a template strand and can undergo RNase H degradation.
The ODN that showed the strongest inhibitory effect in regard to DNA synthesis, i.e. ODN41–76, possesses complementarity to the 3′ end of tRNA as well as to its TΨC- and variable loops. Shorter ODNs that were truncated at their 3′ ends did not effectively block the initiation of DNA synthesis. These data suggest that effective ODNs must interfere with interactions between tRNALys3 and the PBS, as well as those occurring outside the PBS. In particular, ODNs that were too short to bind to the variable loop, which is implicated in extended intermolecular interactions (25), did not inhibit (–) ssDNA synthesis (Fig. 7). Moreover, ODNs that bound to the variable loop effectively initiated RNase H degradation of the tRNA primer. Another consequence of this RNase H activity is that the low amount of (–) ssDNA that was generated was no longer attached to full-length tRNA. This shortened DNA product, that contains a truncated version of tRNALys3, could conceivably have an inhibitory effect on the first strand transfer event that follows synthesis of (–) ssDNA. Indeed, previous studies have shown that tRNALys3 facilitates this first strand transfer far more efficiently than a primer that contains only the first 18 nt of tRNALys3 (38,43). The effects seen with the various ODNs used in this study are summarized in the model shown in Figure 8.
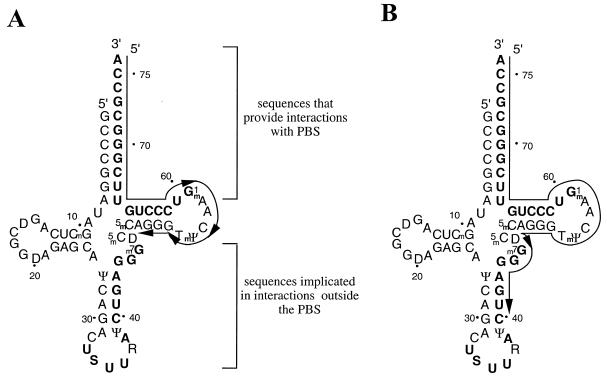
Figure 7.
Interactions of ODNs with residues of the tRNA primer that are implicated in binding to viral RNA. These residues are highlighted in bold. (A) ODNs ≤ ODN50–76 that are complementary to the 3′ terminus and TΨC stem–loop fail to inhibit (–) ssDNA synthesis. (B) ODNs ≥ ODN47–76 may interfere with extended interactions between tRNALys3 and viral genomic RNA. The latter ODNs severely compromised synthesis of (–) ssDNA.
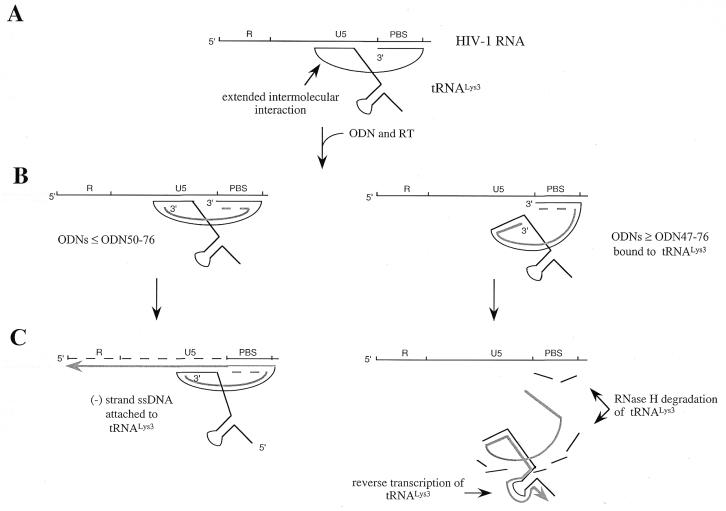
Figure 8.
Possible mechanisms involved in inhibition of (–) ssDNA synthesis by antisense ODNs ≥ ODN47–76. (A) Schematic representation of intermolecular interactions seen in the binary tRNA/viral RNA complex. (B) ODNs ≥ ODN47–76 can interfere with interactions outside the PBS (right), while shorter ODNs do not contact residues of the variable-loop or the anticodon stem–loop. The nature of the interaction between the 3′ terminus of tRNALys3 and ODNs is uncertain, since the antisense molecules compete with the PBS. This is shown by the dashed line. (C) The formation of a stable ODN/tRNALys3 complex, with ODNs ≥ ODN47–76 (right), can initiate both reverse transcription as well as RNase H degradation of tRNALys3. Thus, the tRNA primer serves as a template under these circumstances. Reverse transcription is indicated by the arrowheads. ODNs ≤ ODN50–76 are not sufficiently long to inhibit (–) ssDNA synthesis (left).
In summary, we have identified an ODN that effectively blocks the initiation of synthesis of HIV-1 (–) ssDNA, by targeting the primer tRNA in cell-free assays. Of course, targeting of tRNALys3 can interfere with cellular functions normally performed by this species and lead to cellular toxicity. However, it is possible that other tRNALys isoacceptors, e.g. tRNALys1,2,4,5 (44,45), may be able to replace tRNALys3, since the regions that are targeted by the ODNs used in this study differ among these tRNA species.
In addition, one might take advantage of the fact that the structure of tRNALys3 in free form differs significantly from that which is bound to the RNA template (25,46). Despite earlier observations, that indicated a highly compact structure of the binary tRNA/RNA complex, our data make clear that antisense molecules can interfere with intermolecular interactions of the HIV-specific initiation complex. Inhibition of (–) strand DNA synthesis was seen regardless of whether reactions were initiated with in vitro synthesized tRNALys3 or with natural tRNALys3. At the same time it should be stressed that these data do not necessarily imply that the structures of complexes containing synthetic tRNALys3 or the natural primer are identical. This is important, as structural and functional studies have indicated that post-transcriptional modifications of human tRNALys3 are required for extended intermolecular tRNA/RNA interactions, which, in turn, may facilitate initiation and subsequent elongation of reverse transcription (47,48). Considering the proposed compact structure of the binary complex, it may seem surprising that ODN41–76 can bind to tRNALys3 and inhibit DNA synthesis of (–) strand DNA. However, since reverse transcription of this natural tRNA was specifically terminated at the first modified residue, that would need to be bypassed by RT, this shows that the binding of ODN41–76 was highly specific.
The mechanism of complex formation remains to be elucidated, and, it is conceivable that the presence of RT may help to facilitate binding. However, recent studies on the effects of nucleic acid structure on the formation of complexes between natural tRNAPhe and a library of ODNs point to the importance of a few single-stranded, and accessible, residues that help to nucleate the hybridization process (49). The structural model of the tRNA/RNA complex predicts that such a single-stranded region is located directly upstream from the 3′ terminus of tRNA that binds to the PBS (25).
Structural differences among free tRNALys3 and bound tRNA might be of considerable advantage, since putative toxic effects of antisense-based strategies may be limited when specifically targeting the tRNA that is bound to the RNA template. Such an approach may be superior to those that only target tRNALys3 in its free form (50). Conceivably, an antisense molecule that is tethered to specific HIV packaging signals may permit colocalization with tRNALys3, where it is bound to genomic RNA in the virion. Such an approach has been successful in the Moloney murine leukemia virus system, using a chimeric molecule that contained the Ψ-packaging signal as well as a ribozyme that specifically cleaved viral genomic RNA (51). Moreover, ribozymes fused to tRNALys3 had been identified within HIV particles (18).
Acknowledgments
ACKNOWLEDGEMENTS
This work was performed by X.W. in partial fulfillment of the requirements for a Ph.D. degree, Faculty of Graduate Studies and Research, McGill University, Montreal, Quebec, Canada. Support for this research was provided by the Medical Research Council of Canada.
REFERENCES
- 1.Telesnitsky A. and Goff,S.P. (1997) In Coffin,J.M., Hughes,S.H. and Varmus,H.E. (eds), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 121–161.
- 2.Marquet R., Isel,C., Eshresmann,C. and Ehresmann,B. (1995) Biochimie, 77, 113–124. [DOI] [PubMed] [Google Scholar]
- 3.Mak J. and Kleiman,L. (1997) J. Virol., 71, 8087–9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Götte M., Li X. and Wainberg,M.A. (1999) Arch. Biochem. Biophys., 365, 199–210. [DOI] [PubMed] [Google Scholar]
- 5.Emini E.A. and Fan,H.Y. (1997) In Coffin,J.M., Hughes,S.H. and Varmus,H.E. (eds), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 637–709.
- 6.Fischl M.A. (1999) AIDS, 13(Suppl. 1), 49–59. [PubMed] [Google Scholar]
- 7.Günthard H.F., Wong,J.K., Ignacio,C.C., Guatelli,J.C., Riggs,N.L., Havlir,D.V. and Richman,D.D. (1998) J. Virol., 72, 2422–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lisziewicz J., Sun,D., Smythe,J., Lusso,P., Lori,F., Louie,A., Markham,P., Rossi,J., Reitz,M. and Gallo,R.C. (1993) Proc. Natl Acad. Sci. USA, 90, 8000–8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lisziewicz J., Sun,D., Weichold,F.F., Thierry,A.R., Lusso,P., Tang,J., Gallo,R.C. and Agrawal,S. (1994) Proc. Natl Acad. Sci. USA, 91, 7942–7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee S.W., Gallardo,H.F., Gaspar,O., Smith,C. and Gilboa,E. (1995) Gene Ther., 2, 377–384. [PubMed] [Google Scholar]
- 11.Biasolo M.A., Radaelli,A., Del Pup,L., Franchin,E., De Giuli-Morghen,C. and Palu,G. (1996) J. Virol., 70, 2154–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jendis J., Strack,B., Volkmann,S., Boni,J. and Molling,K. (1996) AIDS Res. Hum. Retroviruses, 12, 1161–1168. [DOI] [PubMed] [Google Scholar]
- 13.Jendis J., Strack,B. and Moelling,K. (1998) AIDS Res. Hum. Retroviruses, 14, 999–1005. [DOI] [PubMed] [Google Scholar]
- 14.El Dirani-Diab R., Sarih-Cottin,L., Delord,B., Dumon,B., Moreau,S., Toulme,J.J., Fleury,H. and Litvak,S. (1997) Antimicrob. Agents Chemother., 41, 2141–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Y., Planelles,V., Li,X., Palaniappan,C., Day,B., Challita-Eid,P., Amado,R., Stephens,D., Kohn,D.B., Bakker,A., Fay,P., Bambara,R.A. and Rosenblatt,J.D. (1997) J. Biol. Chem., 272, 14523–14531. [DOI] [PubMed] [Google Scholar]
- 16.Lee R., Kaushik,N., Modak,M.J., Vinayak,R. and Pandey,V.N. (1998) Biochemistry, 37, 900–910. [DOI] [PubMed] [Google Scholar]
- 17.Veal G.J., Agrawal,S. and Byrn,R.A. (1998) Nucleic Acids Res., 26, 5670–5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medina M.F. and Joshi,S. (1999) Nucleic Acids Res., 27, 1698–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang M., Mak,J., Ladha,A., Cohen,E., Klein,M., Rovinski,B. and Kleiman,L. (1993) J. Virol., 67, 3246–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mak J., Jiang,M., Wainberg,M.A., Hammarskjold,M.L., Rekosh,D. and Kleiman,L. (1994) J. Virol., 68, 2065–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barat C., Lullien,V., Schatz,O., Keith,G., Nugeyre,M.T., Gruninger-Leitch,F., Barre-Sinoussi,F., LeGrice,S.F. and Darlix,J.L. (1989) EMBO J., 8, 3279–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Rocquigny H., Gabus,C., Vincent,A., Fournie-Zaluski,M.C., Roques,B. and Darlix,J.L. (1992) Proc. Natl Acad. Sci. USA, 89, 6472–6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng Y.X., Campbell,S., Harvin,D., Ehresmann,B., Ehresmann,C. and Rein,A. (1999) J. Virol., 73, 4251–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isel C., Marquet,R., Keith,G., Ehresmann,C. and Ehresmann,B. (1993) J. Biol. Chem., 268, 25269–25272. [PubMed] [Google Scholar]
- 25.Isel C., Ehresmann,C., Keith,G., Ehresmann,B. and Marquet,R. (1995) J. Mol. Biol., 247, 236–250. [DOI] [PubMed] [Google Scholar]
- 26.Liang C., Li,X., Rong,L., Inouye,P., Quang,Y., Kleiman,L. and Wainberg,M.A. (1997) J. Virol., 71, 5750–5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wakefield J,K., Kang,S.M. and Morrow,C.D. (1996) J. Virol., 70, 966–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z., Kang,S.M., LeBlanc,A., Hajduk,S.L. and Morrow,C.D. (1996) Virology, 226, 306–317. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z., Kang,S.M., Li,Y. and Morrow,C.D. (1998) RNA, 4, 394–406. [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z., Kang,S.M. and Morrow,C.D. (1998) AIDS Res. Hum. Retroviruses, 14, 979–988. [DOI] [PubMed] [Google Scholar]
- 31.Kang S.M., Zhang,Z. and Morrow,C.D. (1997) J. Virol., 71, 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang S.M., Zhang,Z. and Morrow,C.D. (1999) Virology, 257, 95–105. [DOI] [PubMed] [Google Scholar]
- 33.Kang S.M. and Morrow,C.D. (1999) J. Virol., 73, 1818–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang M., Mak,J., Ladha,A., Cohen,E., Klein,M., Rovinski,B. and Kleiman,L. (1993) J. Virol., 67, 3246–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu M. and Horowitz,J. (1993) Biotechnology, 15, 264–266. [Google Scholar]
- 36.Bordier B., Perala-Heape,M., Degols,G., Lebleu,B., Litvak,S., Sarih-Cottin,L. and Helene,C. (1995) Proc. Natl Acad. Sci. USA, 92, 9383–9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le Grice S.F. and Gruninger-Leitch,F. (1990) Eur. J. Biochem., 187, 307–314. [DOI] [PubMed] [Google Scholar]
- 38.Arts E.J., Li,X., Gu,Z., Kleiman,L., Parniak,M.A. and Wainberg,M.A. (1994) J. Biol. Chem., 269, 14672–14680. [PubMed] [Google Scholar]
- 39.Perret V., Garcia,A., Puglisi,J., Grosjean,H., Ebel,J.P., Florentz,C. and Giege,R. (1990) Biochimie, 72, 735–743. [DOI] [PubMed] [Google Scholar]
- 40.Wainberg M.A. (1999) In Dalgleish,A.G. and Weiss,R.A. (eds), HIV and the New Viruses. Academic Press, London, UK, pp. 223–249.
- 41.Lanchy J.M., Ehresmann,C., Le Grice,S.F., Ehresmann,B. and Marquet,R. (1996) EMBO J., 15, 7178–7187. [PMC free article] [PubMed] [Google Scholar]
- 42.Arts E.J., Miller,J.T., Ehresmann,B. and Le Grice,S.F. (1998) J. Biol. Chem., 73, 14523–14532. [DOI] [PubMed] [Google Scholar]
- 43.Brulé F., Bec,G., Keith,G., Le Grice,S.F.J., Roques,B.P., Ehresmann,B., Ehresmann,C. and Marquet,R. (2000) Nucleic Acids Res., 28, 634–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raba M., Limburg,K., Burghagen,M., Katze,J.R., Simsek,M., Heckman,J.E., Rajbhandary,U.L. and Gross,H.J. (1979) Eur. J. Biochem., 97, 305–318. [DOI] [PubMed] [Google Scholar]
- 45.Das A.T., Klaver,B. and Berkhout,B. (1997) J. Gen. Virol., 78, 837–840. [DOI] [PubMed] [Google Scholar]
- 46.Skripkin E., Isel,C., Marquet,R., Ehresmann,B. and Ehresmann,C. (1996) Nucleic Acids Res., 24, 509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Isel C., Lanchy,J.M., Le Grice,S.F., Ehresmann,C., Ehresmann,B. and Marquet,R. (1996) EMBO J., 15, 917–924. [PMC free article] [PubMed] [Google Scholar]
- 48.Isel C., Westhof,E., Massire,C., Le Grice,S.F., Ehresmann,B., Ehresmann,C. and Marquet,R. (1999) EMBO J., 18, 1038–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mir K.M. and Southern,E. (1999) Nature Biotechnol., 17, 788–792. [DOI] [PubMed] [Google Scholar]
- 50.Shterman N., Elroy-Stein,O., Morad,I., Amitsur,M. and Kaufmann,G. (1995) Nucleic Acids Res., 23, 1744–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sullenger B.A. and Cech,T.R. (1993) Science, 262, 1566–1569. [DOI] [PubMed] [Google Scholar]