Abstract
Fear-, anxiety- and stress-related disorders are among the most frequent mental disorders. Given substantial rates of insufficient treatment response and often a chronic course, a better understanding of the pathomechanisms of fear-, anxiety- and stress-related disorders is urgently warranted. Epigenetic mechanisms such as histone modifications - positioned at the interface between the biological and the environmental level in the complex pathogenesis of mental disorders - might be highly informative in this context. The current state of knowledge on histone modifications, chromatin-related pharmacology and animal models modified for genes involved in the histone-related epigenetic machinery will be reviewed with respect to fear-, anxiety- and stress-related states. Relevant studies, published until 30th June 2022, were identified using a multi-step systematic literature search of the PubMed and Web of Science databases. Animal studies point towards histone modifications (e.g., H3K4me3, H3K9me1/2/3, H3K27me2/3, H3K9ac, H3K14ac and H4K5ac) to be dynamically and mostly brain region-, task- and time-dependently altered on a genome-wide level or gene-specifically (e.g., Bdnf) in models of fear conditioning, retrieval and extinction, acute and (sub-)chronic stress. Singular and underpowered studies on histone modifications in human fear-, anxiety- or stress-related phenotypes are currently restricted to the phenotype of PTSD. Provided consistent validation in human phenotypes, epigenetic biomarkers might ultimately inform indicated preventive interventions as well as personalized treatment approaches, and could inspire future innovative pharmacological treatment options targeting the epigenetic machinery improving treatment response in fear-, anxiety- and stress-related disorders.
Keywords: Epigenetics, chromatin, histone, HDAC, anxiety, fear, stress, Bdnf
1. INTRODUCTION
Fear-, anxiety- and stress-related disorders such as specific phobias, agoraphobia, social anxiety disorder, generalized anxiety disorder, panic disorder or post-traumatic stress disorder (PTSD) constitute the most frequent mental disorders with a cumulative 12-month prevalence of ~16% and confer a substantial individual as well as socioeconomic burden [1-6]. The first-line treatment of fear-, anxiety- and stress-related disorders comprises psychopharmacological options as well as psychotherapy. However, non-response rates are considerable, and in about 30% of the cases, the disorders take an intermittently relapsing or even chronic course [7].Thus, a better understanding of the pathomechanisms of fear-, anxiety- and stress-related symptoms is urgently warranted and can be expected to inform innovative and personally tailored treatment approaches.
The etiology of fear-, anxiety- and stress-related disorders is complex with an interaction of biological factors and environmental influences [3, 5]. Epigenetic mechanisms comprising histone modifications, deoxyribonucleic acid (DNA) methylation as well as non-coding RNAs (ncRNAs) have been suggested to be positioned at the interface between the biological and the environmental level and to dynamically broker adaptation or maladaptation, respectively, to environmental conditions or challenges [8-10]. Thus, epigenetic mechanisms constitute prime candidates in the search for fear-, anxiety- and stress-related disorder biomarkers informing indicated preventive interventions as well as personalized treatment approaches, and possibly also inspiring future innovative pharmacological treatment options targeting the epigenetic machinery.
As an introduction, the present paper provides a propaedeutic overview of the terminology and basic mechanisms of epigenetics spotlighting histone modifications. The subsequent main focus will be on reviewing the current state of knowledge on histone modifications, chromatin-related pharmacology and animal models mutant for genes involved in the histone-related epigenetic machinery with respect to fear-, anxiety- and stress-related states in an effort to complement and extend existing reviews [11-13]. For a comprehensive overview of studies on the role of DNA methylation and ncRNAs in this context several previous reviews may be referred e.g., [8, 9, 11, 14-23].
1.1. Propaedeutics on Epigenetics
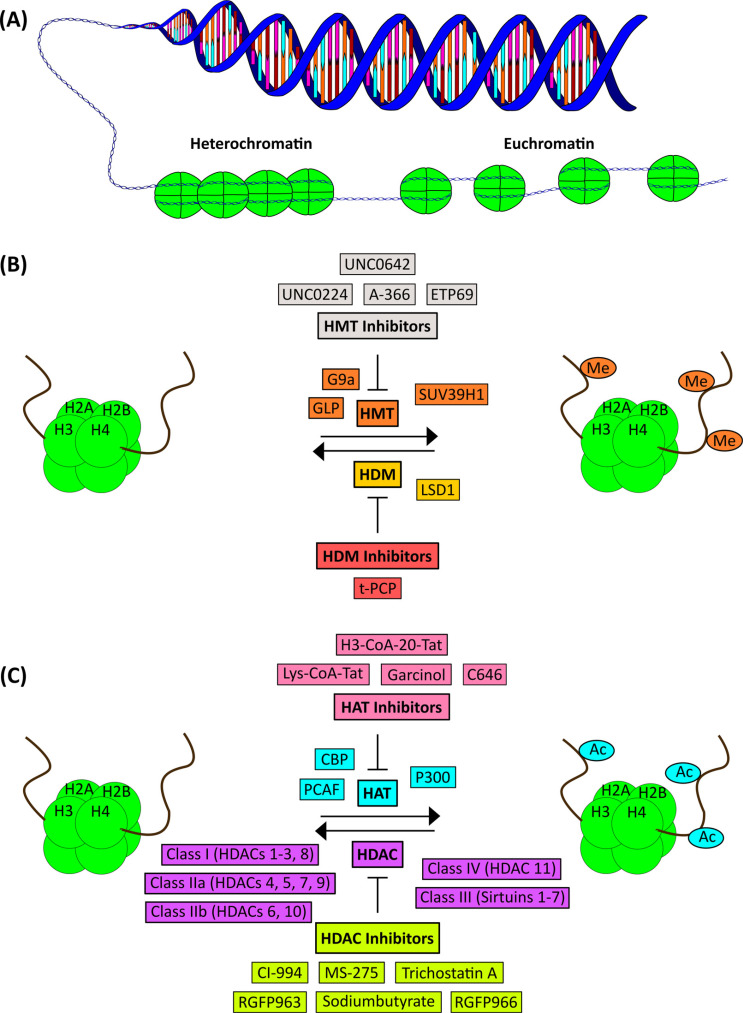
The genetic structure rests on DNA which is composed of nucleotides consisting of deoxyribose, a phosphate group and the four nitrogenous bases (nucleobases) guanine (G), cytosine (C), adenine (A) and thymine (T) that form specific pairs of A/T and G/C on opposite strands of the DNA double helix. Sequences of these nucleic acids directly encode genetic information. The DNA strands are wrapped around histone (H2A, H2B, H3, H4) octamers composed of two H2A-H2B dimers and a H3-H4 tetramer forming a packaging unit called nucleosome. Nucleosomes build threads or fibers with so-called linker DNA organized by the linker histone H1 in between them to form the primary structure of chromatin. Depending on the packaging density, chromatin can exist in a compact, i.e. closed state (‘heterochromatin’), which limits the accessibility of the genomic DNA for proteins like transcription factors (TFs) and thus lead to a transcriptionally ‘silent’ state. The looser, i.e. open state (‘euchromatin’) facilitates binding of the transcriptional machinery and subsequent gene transcription (Fig. 1A) [24]. Chromatin is organized in a hierarchical fashion inside the nucleus, and the highest hierarchical level results in the chromosomes [25].
Fig. (1).
Chromatin modifications and regulators targetable by currently available chemical compounds. Schematic illustration of (A) the DNA double-strand wrapped around histone (H2A, H2B, H3, H4) octamers composed of two H2A-H2B dimers and two H3-H4 dimers to form the primary structure of chromatin, which can exist in a compact, i.e. closed state (‘heterochromatin’), or in an open state (‘euchromatin’); (B) mechanisms of histone methylation (HMT: histone methyltransferase) and demethylation (HDM: histone demethylase) as well as exemplary pharmacological HMT and HDM inhibitors; (C) mechanisms of histone acetylation (HAT: histone acetyltransferase) and deacetylation (HDAC: histone deacetylase) as well as exemplary pharmacological HAT and HDAC inhibitors; for further details and abbreviations see main manuscript (1.1. and 3.3.).
The term epigenetics literally translated means ‘above genetics’. This term was first introduced in 1942 by the embryologist Conrad Waddington who defined it as “the branch of biology that studies the causal interactions between genes and their products which bring the phenotype into being” [26]. Nowadays, the study of epigenetics is concerned with heritable changes that influence gene expression without affecting the DNA sequence itself. Epigenetic factors act on different cellular levels and can have either activating or repressing properties on gene expression profiles [27]. Such alterations can be transferred through mitosis to the next cell generation or through meiosis between organismal generations [28, 29]. Epigenetic change is a regular and natural occurrence mostly observed during early embryonic development and cell differentiation but remains partly malleable throughout the lifespan. Epigenetic processes have been suggested to be related to a vast amount of biological processes as well as environmental stressors, chemicals, drugs, aging, diet, mental state, social interaction or physical workouts [29, 30]. The three major players initiating and sustaining epigenetic change are histone modifications, DNA methylation as well as ncRNAs along with histone variants such as H3.3 and H2A.Z (for review see [31].
Post-translational histone modifications crucially govern DNA packaging density around histone proteins and mainly comprise methylation or acetylation (Figs. 1B and 1C), but also for instance phosphorylation, GlcNAcylation, biotinylation, butyrilation, citrullination, crotonylation, carbonylation, ubiquitination, SUMOylation, ADP-ribosylation or isomerization targeting terminal tails of histone proteins [32]. The nomenclature of histone modifications starts with a capital ‘H’ for ‘histone’ followed by a number indicating the amino acid position of the modification, and ends with the type (e.g., acetylation) of the modification itself. For example, acetylation at lysine 27 of histone 3 would read ‘H3K27ac’. Histone modifications are dynamically deposited or removed at the histone tails by enzymatic ‘writers’ or ‘erasers’, after being recognized by ‘reader’ proteins [33]. Histone modification ‘writers’ comprise histone methyltransferases (HMTs) or histone acetyltransferases (HATs) that confer methylation or acetylation to tails of histones, respectively (Figs. 1B and 1C). HMTs exemplarily comprise the euchromatin histone-lysine N-methyltransferase 2 (G9a), the G9a/G9a-like protein (GLP) and the Suppressor of Variegation 3-9 Homolog 1 (SUV39H1). Prominent representatives of HATs are the E1A binding protein P300, the CREB binding protein (CBP) and the p300/CREB-binding protein-associated factor (PCAF). The counterparts of ‘writers’ are termed ‘erasers’ and remove methyl or acetyl groups from histone tails. Erasers comprise histone demethylases (HDMs) such as the lysine-specific histone demethylase 1A (LSD1) and various classes of histone deacetylases (HDACs; classes I [HDACs 1-3 and 8], IIa [HDACs 4, 5, 7 and 9], IIb [HDACs 6 and 10], III [Sirtuins 1-7], IV [HDAC 11]) (Figs. 1B and 1C). Methylation mainly occurs at lysines of histone H3. Lysine methylation can have either activating or repressing effects on gene transcription depending on the methylation site [34]. Additionally, lysines can be mono-, di- or tri-methylated conferring either activating or repressing transcriptional effects. For example, H3K4me3 is involved in active gene transcription, while H3K9me3 or H3K27me3 constitute repressive marks [35, 36]. Arginine residues on H3 and H4 can also be methylated, and this modification is generally linked to transcriptional activation. Acetylation of histone tails occurs at lysine residues, which thereby lose their positive charge resulting in the neutralization of the charge on the respective histone and consequently in the weakening of the binding to the negatively charged DNA. This weaker histone-DNA interaction contributes to an ‘open’ euchromatin state significantly increasing gene expression [37]. For a summary of the most prominent histone modifications and their studied function as well as preferred locations (Table 1).
Table 1.
Overview of the most prominent histone modifications, their associated function and preferred location.
| Histone Modification | Function | Location |
|---|---|---|
| H3K4me1 | Activation | Enhancers |
| H3K4me3 | Activation | Promoters, bivalent domains |
| H3K9me1 | Activation | Promoters |
| H3K9me2, H3K9me3 | Repression | Repetitive sequences |
| H3K27me2, H3K27me3 | Repression | Promoters, developmental regulators, bivalent domains |
| H3K36me3, H3K79me2 | Activation | Gene bodies |
| H3K9ac, H3K27ac | Activation | Enhancers, promoters |
| H3K14ac | Activation | Gene bodies |
| H4K5ac | Activation | TSS, Gene bodies |
| H4K16ac | Activation | Housekeeping genes |
Abbreviations: ac: acetylation, H: histone (H3 or H4), K: lysine, Kx: amino acid position of lysine counting from N-terminus, me1: monomethylation, me2: dimethylation, me3: trimethylation, TSS: Transcription start site; also see 1.1.
2. METHODS
A multi-step systematic literature search of the PubMed and Web of Science databases was performed by independent researchers ME and KD applying the following broad search terms: (“histone” OR “histone modification” OR “acetylation”) AND (“anxiety” OR “fear” OR “extinction” OR “startle” OR “stress” OR “anxiety disorder*” OR “stress-related disorder” OR “stress-related” OR “post-traumatic stress disorder” OR “PTSD” OR “panic disorder” OR “panic” OR “social phobia” OR “social anxiety disorder” OR “agoraphobia” OR “specific phobia*”). Identified articles published until 30th June 2022 were screened based on title and abstract reading. Only peer-reviewed studies published in English reporting on original research or review articles were considered eligible to be included in the present review. After the exclusion of articles irrelevant to the topic investigated, full texts of the remaining articles were further assessed for inclusion. Searching reference lists of selected articles and pertinent review articles revealed additional eligible articles. Disagreements in search and selection were resolved through discussion and consensus.
3. RESULTS
3.1. Histone Modifications in Animal Models of Fear and Anxiety
Paradigms such as contextual fear conditioning, partly combined with single-prolonged stress, auditory fear conditioning or fear extinction following conditioning in animals serve as mechanistic models for mostly fear- and anxiety-related disorders and their treatment [38], but have also been shown to constitute valid models for stress-related disorders such as PTSD in humans [39]. Epigenetic studies analyzing dynamic chromatin remodeling processes based on histone modifications in animal models have mostly been conducted on tissue harvested from brain regions centrally engaged in the brain fear circuit including the hippocampus, entorhinal cortex, anterior cingulate cortex (ACC), prefrontal cortex (PFC), amygdala or bed nucleus of the stria terminalis (BNST) [40, 41].
Eight studies investigated the role of histone modifications related to contextual fear conditioning. Following a standard contextual fear conditioning paradigm, H3K4me3 was observed to be significantly increased in the hippocampus, entorhinal cortex and ACC one hour after conditioning [42-45]. There are, however, conflicting results on whether increased H3K4me3 is persistent over a time period of 24 hours [44], returns to baseline [43], or even decreases below naïve levels after 24 hours [45]. Interestingly, in the CA1 hippocampal area as well as the ACC, H3K4me3 – accompanied by increased H3K9ac and decreased H3K27me3 – was only increased in genic regions, while in intergenic regions H3K4me3 was decreased along with increased H3K27me3 [42]. Contextual fear conditioning has furthermore been shown to lead to temporary increases of global H3K14ac levels [46] and transiently increased global levels of H4K5ac predominantly at promoter regions [47] one hour after conditioning. At random time points within an eight-minute training period, contextual fear conditioning was accompanied by globally increased H2BK5ac, H3K9K14ac and H4K12ac levels in the hippocampus, with H2BK5ac and H3K12ac being most specifically linked to the context-shock association, i.e. to learned fear [48]. On a candidate gene level, most studies have focused on the gene coding for the brain-derived neurotrophic factor (BDNF), which is crucially involved in brain development, neural plasticity, memory formation and particularly the acquisition, consolidation and extinction of conditioned fear [49-51]. Two hours after contextual fear conditioning, partly in interaction with single prolonged stress, levels of H3ac and H4ac at Bdnf promoters I and IV were found to be increased in the hippocampus along with elevated total Bdnf mRNA levels [52]. Accordingly, another study observed Bdnf promoter IV, histone H3 acetylation as well as phosphoacetylation to be increased two hours after contextual fear conditioning along with increased Bdnf exon IV mRNA and decreased DNA methylation at Bdnf exons I and IV. At Bdnf promoter II, however, H4 acetylation was discerned to be decreased [53].
Retrieval of fear memory after contextual fear conditioning evoked differential epigenetic changes depending on the time frame of memory formation and the respective brain area. After retrieval of a recent (one hour) contextual fear memory, H3K4me3 was found to be increased in the hippocampal CA1 region both on a global level and in the coding region of the Npas4 but not the C-fos gene. Yet, following the retrieval of a remote fear memory 30 days after memory acquisition, increased H3K4me3 levels were observed in CpG-enriched coding regions of the C-fos gene in the ACC [44] suggesting differential and brain region-specific gene targets of increased H3K4 trimethylation following retrieval of recent or remote fear memory, respectively.
Two studies report on histone modifications in the context of auditory fear conditioning paradigms. On a genome-wide level, H3K9me2 was increased in the lateral amygdala both one hour and 24 hours after conditioning [54]. On a candidate gene level, auditory fear conditioning was furthermore shown to lead to increased H3ac levels at Bdnf promoters I and IV in the PFC, while fear extinction training reversed increased H3ac at Bdnf promoter I to below naïve levels and was furthermore associated with increased H4ac at Bdnf promoter IV along with increased Bdnf exon I and IV mRNA expression in the PFC [55]. In a similar vein, on a genome-wide level, following fear learning H3ac and H4ac increased in the amygdala and the prelimbic PFC, but not in the infralimbic PFC, while extinction learning led to increased H3ac in both the prelimbic and infralimbic PFC and increased H4ac in the infralimbic PFC only [56]. In addition, another study on histone modifications in BNST sub-regions observed fear conditioning to be accompanied by increased H3ac and H4ac in the anteromedial and ventrolateral BNST, while following extinction H4ac increased in the anterolateral BNST specifically [57].
Finally, one study focused on the quality of fear extinction, measured two hours after extinction, dependent on the time of extinction after memory acquisition. Immediate extinction ten minutes after fear memory acquisition did not lead to retention of fear extinction whereas delayed extinction 24 hours after fear memory acquisition led to the retention of fear extinction, measured by the relative freezing response to the previously conditioned fear. In both extinction paradigms, global levels of H3K9ac were increased in the prelimbic and infralimbic PFC along with elevated levels of c-Fos and CBP in both regions. However, delayed extinction resulted in a stronger increase in H3K9ac and CBP levels in the infralimbic PFC as compared to immediate extinction (Table 2) [58].
Table 2.
Dynamic histone modifications in animal models of fear/anxiety.
| Species | Paradigm | Measurement Post-training | Brain Region | Genomic Region | Histone Modification | Additional Findings | References |
|---|---|---|---|---|---|---|---|
| Rat | CFC and exposure alone | 1 h | Hip (CA1) | Genome-wide | H3K9me2↑ H3K4me3↑ | - | Gupta-Agarwal et al., 2012 [45] |
| Rat | CFC and exposure alone | 1 h | EC | Genome-wide | H3K9me2↑ H3K4me3↑ | - | Gupta-Agarwal et al., 2012 [45] |
| Rat | CFC and exposure alone | 24 h | EC | Genome-wide | H3K4me3↓ | - | Gupta-Agarwal et al., 2012 [45] |
| Rat | CFC and exposure alone | 1 h 24 h |
Hip (CA1) | Genome-wide | H3K9me2↑ H3K9me2↓ |
- | Gupta et al., 2010 [43] |
| Rat | CFC | 1 h | Hip (CA1) | Genome-wide | H3K4me3↑ | Returned to baseline after 24h |
Gupta et al., 2010 [43] |
| Mouse | CFC | 1 h | Hip (CA1), ACC | Genome-wide | H3K9ac↑ H3K4me3↑ H3K27me3↓ |
DNAme↓ (ACC) | Halder et al., 2016 [42] |
| Rat | CFC | 1 h | Hip (CA1) | Genome-wide | H3K14ac↑ | Returned to baseline after 24 h | Levenson et al., 2004 [46] |
| Mouse | CFC | 1 h | Hip | Genome-wide | H4K5ac↑ | Lasting for 2-4 h depending on the amount of training |
Park et al., 2013 [47] |
| Rat | CFC | Immediately | Hip | Genome-wide | H2BK5ac↑ H3K9K14ac↑ H4K12ac↑ | - | Bousiges et al., 2013 [48] |
| Rat | CFC (SPS) | 2 h | Hip | Bdnf promoter I and IV | H3ac↑ H4ac↑ |
Bdnf mRNA↑ | Takei et al., 2011 [52] |
| Rat | CFC | 2 h | Hip (CA1) |
Bdnf promoter IV Bdnf promoter II |
H3ac↑, H3phospho-ac↑ H4ac↓ |
Bdnf exon IV mRNA↑ DNAme at Bdnf exons I and IV↓ DNAme at Bdnf exon VI↑ |
Lubin et al., 2008 [53] |
| Rat | Memory retrieval after CFC | 1 h | Hip (CA1), BLA |
Genome-wide | H3K4me3↑ (CA1) H3K9me2↑ (BLA) |
H3K4me3 returned to baseline after 24 h 5hmC↑ (CA1) |
Webb et al., 2017 [44] |
| Rat | Memory retrieval after CFC | 1 h | Hip (CA1) | Npas4 | H3K4me3↑ | 5hmC↑ | Webb et al., 2017 [44] |
| Rat | Memory retrieval after CFC | 30 days | ACC | C-fos | H3K4me3↑ | 5hmC↑ | Webb et al., 2017 [44] |
| Rat | AFC | 1 h | Lateral amygdala | Genome-wide | H3K9me2↑ | G9a mRNA↓ | Gupta-Agarwal et al., 2014 [54] |
| Rat | AFC | 24 h | Lateral amygdala |
Genome-wide | H3K9me2↓ | - | Gupta-Agarwal et al., 2014 [54] |
| Mouse | AFC | 26 h | PFC | Bdnf promoter I and IV | H3ac↑ | - | Bredy et al., 2007 [55] |
| Mouse | AFC + extinction | 2 h | PFC | Bdnf promoter I | H3ac↓ | Bdnf I and IV mRNA↑ | Bredy et al., 2007 [55] |
| Mouse | AFC + extinction | 2 h | PFC | Bdnf promoter IV | H4ac↑ | Bdnf I and IV mRNA↑ | Bredy et al., 2007 [55] |
| Rat | Fear memory (5x CS+US) |
2 h | PFC, amygdala | Genome-wide | H3ac↑ and H4ac↑ in BA, LA, CeM, CeL, PL-PFC | C-fos + CBP expression↑ | Siddiqui et al., 2017 [56] |
| Rat | Extinction | Immediately | PFC, amygdala | Genome-wide | H3ac↑ in IL-PFC and PL-PFC H4ac↑ in IL-PFC |
C-fos + CBP expression↑ | Siddiqui et al., 2017 [56] |
| Rat | Fear memory (5x CS+US) |
2 h | STMA, STLV (neurons) |
Genome-wide | H3ac↑, H4ac↑ | CBP↑ | Ranjan et al., 2017 [57] |
| Rat | Extinction | Immediately | STLP (neurons) |
Genome-wide | H3ac↑, H4ac↑ | CBP↑ | Ranjan et al., 2017 [57] |
| Rat | Immediate fear extinction (10 min) |
2 h | IL-PFC, PLC |
Genome-wide | PLC: H3K9ac↑ IL-PFC: H3K9ac↑ |
Retention of fear extinction↓ PLC: c-Fos↑ IL-PFC: c-Fos↑ PLC: CBP↑ IL-PFC: CBP↑ |
Singh et al., 2018 [58] |
| Rat | Delayed fear extinction (24 h) |
2 h | IL-PFC, PLC |
Genome-wide | PLC: H3K9ac↑ IL-PFC: H3K9ac↑↑ |
Retention of fear extinction↑ PLC: c-Fos↑ IL-PFC: c-Fos↑ PLC: CBP↑ IL-PFC: CBP↑↑ |
Singh et al., 2018 [58] |
Abbreviations: ACC: Anterior cingulate cortex, AFC: Auditory fear conditioning, Bdnf: Brain Derived Neurotrophic Factor, BLA: Basolateral amygdala, CA1: First region in the hippocampal circuit, CBP: CREB-binding protein, CFC: Contextual fear conditioning, C-fos: Proto-oncogene, CS: Conditioned stimulus, EC: Entorhinal cortex, G9a: Euchromatin histone-lysine N-methyltransferase 2 (EHMT2), Hip: Hippocampus, IL-PFC: Infralimbic prefrontal cortex, Npas4: Neuronal PAS Domain Protein 4, PFC: Prefrontal cortex, PLC: Prelimbic cortex, SPS: Single prolonged stress, STLP: Antero-lateral bed nucleus of the stria terminalis, STLV: Lateral-ventral bed nucleus of the stria terminalis, STMA: Antero-medial bed nucleus of the stria terminalis, UCS: Unconditioned stimulus.
3.2. Histone Modifications in Animal Models of Acute and (Sub-)Chronic Stress
The role of epigenetic mechanisms in moderating the response to acute or (sub-)chronic stressful environmental influences and thus ultimately in possibly conferring risk of fear-, anxiety- and stress-related disorders has been investigated in animal models during different time windows of development (pre-conception, pre-/post-natal, juvenile, adult age) in fear- or anxiety-relevant brain regions (e.g., hippocampus, limbic system, cortex) using paradigms such as restraint stress, social defeat, forced swimming, predator stress, acute glucocorticoid treatment, chronic unpredictable stress or social isolation [59].
3.2.1. Acute Stress
Six studies were identified that explicitly investigated the effect of acute stress on histone modifications in rodent models (Table 3). Following acute restraint stress for 30 minutes, increased H3K9me3 as well as decreased H3K27me3 and H3K9me1 were observed on a genome-wide level in hippocampal tissue [60]. Against the background of a history of chronic restraint stress, two and 24 hours after acute restraint the histone acetyltransferase P300 was found to be involved in dynamic up-regulation (two hours) and down-regulation (24 hours), respectively, of metabotropic glutamate receptor 2 expressions in the hippocampus via differential acetylation of H3K27 bound to the metabotropic glutamate receptor 2 gene (Grm2) promoter [61]. Another study investigating the effects of a two-hours-restraint stress paradigm on histone modifications at the Bdnf gene discerned a significant decrease in acetylated H3 at Bdnf gene promoters I, IV and VI two hours post-stress along with a decrease in total Bdnf mRNA levels as well as Bdnf exon I-, and exon IV-specific mRNA, which, however, could both not be observed 24 hours post-stress [62]. Following the repeated social defeat, global levels of H3 acetylation in the hippocampus, amygdala, and PFC increased 30 minutes after the last defeat [63]. In a study applying forced swimming, predator, ether exposure as well as cold stress, only forced swimming and predator stress evoked increased genome-wide H3-phosphorylation [64]. Finally, acute glucocorticoid treatment was shown to decrease H3K9me3 on a genome-wide level at B2 short interspersed nuclear elements in the hippocampus [65].
Table 3.
Dynamic histone modifications in animal models of acute stress.
| Species | Paradigm | Measurement Post-stress | Brain Region | Genomic Region | Histone Modification | Further Results | References |
|---|---|---|---|---|---|---|---|
| Rat | Restraint stress (30 min) | Immediately | Hip (DG, CA1) | Genome-wide | H3K9me3↑ | - | Hunter et al., 2009 [60] |
| Rat | Restraint stress (30 min) | Immediately | Hip (DG, CA1) | Genome-wide | H3K27me3↓ | - | Hunter et al., 2009 [60] |
| Rat | Restraint stress (30 min) | Immediately | Hip (DG, CA1) | Genome-wide | H3K9me1↓ | - | Hunter et al., 2009 [60] |
| Mouse | Acute restraint in chronic restraint stress model (21 days) | 2 h | Hip (DG) | Grm2 promoter | P300↑ --> H3K27ac↑ | mGlu2 receptors↑ | Nasca et al., 2015 [61] |
| Mouse | Acute restraint in chronic restraint stress model (21 days) | 24 h | Hip (DG) | Grm2 promoter | P300↓ --> H3K27ac↓ | mGlu2 receptors↓ | Nasca et al., 2015 [61] |
| Rat | Restraint stress (2 h) | 2 h | Hip | Bdnf promoter I, IV, VI | H3ac↓ | Bdnf mRNA↓ | Fuchikami et al., 2009 [62] |
| Rat | Restraint stress (2 h) | 24 h | Hip | Bdnf promoter I, IV and IV | No sign. changes in H3ac |
- | Fuchikami et al., 2009 [62] |
| Rat | Social defeat (4 x 15 min) | 30 min | Hip | Genome-wide | H3ac↑ | - | Hollis et al., 2010 [63] |
| Rat | Forced swim (30 min) | 1 day | Hip (DG) | Genome-wide | H3ph↑ | - | Bilang-Bleuel et al., 2005 [64] |
| Rat | Predator stress (15 min) | 1 day | Hip (DG) | Genome-wide | H3ph↑ | - | Bilang-Bleuel et al., 2005 [64] |
| Rat | Ether exposure (3 min)/ cold stress (4 h) | 1 day | Hip (DG) | Genome-wide | No change in H3ph | - | Bilang-Bleuel et al., 2005 [64] |
| Rat | Acute glucocorticoid treatment | 1 h | Hip | Genome-wide | H3K9me3↓ | Especially at B2 short interspersed nuclear elements | Bartlett et al., 2021 [65] |
Abbreviations: Bdnf: Brain Derived Neurotrophic Factor, CA1: First region in the hippocampal circuit, DG: Dentate gyrus, Grm2: Glutamate metabotropic receptor 2, Hip: Hippocampus, mGlu2: Metabotropic glutamate receptor 2, P300: E1A binding protein p300.
3.2.2. (Sub-)Chronic Stress
(Sub-)chronic stress can be modelled in animals by the application of restraint stress over longer time periods (Table 4). After subchronic restraint stress for seven days, hippocampal genome-wide H3K4me3 and H3K27me3 levels were observed to be decreased and H3K9me3 to be increased [60]. Subchronic restraint stress was furthermore shown to decrease levels of H3K9me2 bound to the promoter region of the gene coding for the E3 ubiquitin-protein ligase, also known as neural precursor cell expressed developmentally down-regulated protein 4 (Nedd4-1) and to increase general HDAC2 levels in the PFC [66].
Table 4.
Dynamic histone modifications in animal models of (sub-)chronic stress.
| Species | Paradigm | Measurement Post-stress | Brain Region | Genomic Region | Histone Modification | Further Results | References |
|---|---|---|---|---|---|---|---|
| Rat | Subchronic restraint stress (7 days) | 1 day | Hip (CA1) | Genome-wide | H3K4me3↓ | - | Hunter et al., 2009 [60] |
| Rat | Subchronic restraint stress (7 days) | 1 day | Hip (DG, CA1) | Genome-wide | H3K27me3↓ | - | Hunter et al., 2009 [60] |
| Rat | Subchronic restraint stress (7 days) | 1 day | Hip | Genome-wide | H3K9me3↑ | - | Hunter et al., 2009 [60] |
| Rat | Subchronic restraint stress (7 days) | 1 h | PFC | Nedd4-1 | H3K9me2↓ | HDAC↑ | Wei et al., 2016 [66] |
| Rat | CRS (21 days) | 1 day | Hip (DG) | Genome-wide | H3K4me3↑ | - | Hunter et al., 2009 [60] |
| Rat | CRS (21 days) | 1 day | Hip (DG) | Genome-wide | H3K9me3↓ | - | Hunter et al., 2009 [60] |
| Mouse | CRS (29 days) | - | Hip | Genome-wide | H3K9ac↓ H4K5ac↓ | HDAC2↑ in Hip | Wu et al., 2021 [67] |
| Mouse | Social defeat (10 days) | Immediately | PVN | Genome-wide | - | HDAC2↓ | Elliott et al., 2010 [68] |
| Mouse | Social defeat (10 days) | 1 day | NAc | Genome-wide | - | HDAC5↓ | Renthal et al., 2007 [69] |
| Mouse | Social defeat (10 days) | 10 days | NAc | Genome-wide | H3K9me2↓ | G9a↓, Glp↓, Suv39h1↓ | Covington et al., 2011 [70] |
| Mouse | Social defeat (10 days) | 24 h and 10 days | NAc | Genome-wide | H3K14ac↑ | HDAC2↓ | Covington et al., 2009 [71] |
| Mouse | Social defeat (10 days) | 28 days | Hip | Bdnf promoter III / IV | H3K27me2↑ | Bdnf III and IV mRNA↓ | Tsankova et al., 2006 [72] |
| Mouse | Social defeat (10 days) | 28 days | NAc | Genome-wide | H3K27me2↑ | - | Wilkinson et al., 2009 [73] |
| Mouse | Social isolation (10 days) | 28 days | NAc | Genome-wide | H3K27me2↑ H3K9me2↑ | - | Wilkinson et al., 2009 [73] |
| Mouse | Social defeat (21 days) | Immediately | Hip | Genome-wide | H3K9ac↓ | HAT↑ HDAC↓ in cortex | Montagud et al., 2016 [74] |
| Mouse | Social defeat (21 days) | 3 weeks | Hip | Genome-wide | H4K12ac↑ H3K4me3↑ | HAT↑ in Hip | Montagud et al., 2016 [74] |
| Rat | CVS(14 days) | 1 day | Hip (DG, CA3) | Genome-wide | H4K12ac↓ H3 phospho-acetylation (Lys9/Ser10)↓ | - | Ferland et al., 2011 [75] |
| Mouse | CVS (15 days) + acute stress (30 min) | Immediately | Hip (DG) | Genome-wide | Chronic stress: H4K12ac↓ Acute stress: H4K12ac↑ Chronic + acute stress: H4K12ac↑ | Chronic + acute stress: HPA activity↑ | Ferland et al., 2014 [76] |
| Rat | CVMS stress (14 days) | 1 day | Amygdala, BNST | - | - | HDAC5↓ in males (CeA) CBP↑ in females (BNST) | Sterrenburg et al., 2011 [77] |
| Rat | CVS (14 days) | 1 day | Hip (DG, CA3) | Genome-wide | H4K12ac↓ | - | Ferland et al., 2011 [75] |
| Rat | CUS (28 days) | 1 day | Hip | Genome-wide | H3K9ac↓ | HDAC5↑ | Liu et al., 2014 [78] |
| Rat | CUS (21 days) | 1 day | Hypothalamus | Crhr1 | H3K9me3↓ | - | Wan et al., 2014 [79] |
| Mouse | Prenatal CUS (4 weeks) | Immediately | Hip | Genome-wide | H3K14ac↓ in offspring (stronger in females) | Spatial memory↓ in offspring | Benoit et al., 2015 [80] |
| Mouse | Gestational stress (restraint and 24-h light disturbance) | - | Hip | Genome-wide | H3K14ac↓ in offspring | DNMT1↑, HDAC1↑, HDAC2↑ in offspring | Zheng et al., 2016 [81] |
| Mouse | Repeated cross-fostering | 72 days | Medulla oblongata | Genome-wide | H3ac↑ H3K4me3↑ H3K27me3↓ | - | Cittaro et al., 2016 [82] |
| Rat | PEPS (15 days) | 1 day, 2 weeks, 2 months | Hip (CA3) | Genome-wide | H4ac↑ (long lasting for 2 months) | MeCP2 activation | Baĭdo et al., 2009 [83] |
Abbreviations: Bdnf: Brain derived neurotrophic factor, BNST: Bed nucleus of the stria terminalis, CA1: First region in the hippocampal circuit, CA3: Third region in the hippocampal circuit, CBP: CREB-binding protein, CeA: Central nucleus of the amygdala, Crhr1: Corticotropin-releasing hormone receptor 1, CRS: Chronic restraint stress, CUS: Chronic unpredictable stress, CVMS: Chronic variable mild, CVS: Chronic variable stress, DG: Dentate gyrus, DNMT1: DNA Methyltransferase 1, G9a: Euchromatin histone-lysine N-methyltransferase 2 (EHMT2), Glp: Euchromatin histone-lysine N-methyltransferase 1 (EHMT1), HAT: Histone acetyltransferase, HDAC: Histone deacetylase, Hip: Hippocampus, HPA: Hypothalamic-pituitary-adrenal axis, MeCP2: Methyl-CpG Binding Protein 2, NAc: Nucleus accumbens, PEPS: Prolonged emotional and pain stress, PFC: Prefrontal cortex, PVN: Paraventricular nucleus, Suv39h1: Suppressor of Variegation 3-9 Homolog 1.
Conversely, genome-wide levels of hippocampal H3K4me3 increased and H3K9me3 decreased following the application of chronic restraint stress applied for 21 days [60]. Chronic restraint stress lasting for 29 days was furthermore accompanied by decreased genome-wide histone H3K9 and H4K5 acetylation levels in hippocampal neurons along with an upregulation of HDAC2 colocalized with glucocorticoid receptors [67].
Another model of chronic stress comprises social defeat over longer time periods (Table 4). Immediately and one day after a ten-day social defeat paradigm, HDAC2 and HDAC5 levels were found to be decreased in the paraventricular nucleus and the nucleus accumbens, respectively [68, 69]. Additionally, ten days after a ten-day social defeat stress paradigm, decreased global H3K9me2 levels along with decreased levels of the associated writer enzymes G9a and Glp and the repressive chromatin modifier Suv39h1 were observed in the nucleus accumbens [70]. In a related experiment, 24 hours and ten days after ten-day social defeat stress, global H3K14ac significantly increased in the nucleus accumbens along with a significant decrease in HDAC2 mRNA levels [71]. Twenty-eight days after ten-day social defeat stress, H3K27me2 levels were discerned to be increased both on a candidate gene level at the Bdnf P3 and P4 promoter in the hippocampus accompanied by downregulation in total Bdnf and particularly Bdnf III and IV mRNA levels [72] as well as on a genome-wide level in the nucleus accumbens [73]. In line with these findings, a similar model of chronic social stress (social isolation) investigating genome-wide histone modifications also 28 days after the stressor revealed H3K27me2 and H3K9me2 levels in the nucleus accumbens to be increased [73]. Finally, immediately following extended social defeat over 21 days, genome-wide H3K9ac was observed to be decreased in the hippocampus, accompanied by increased histone acetyltransferases and accordingly decreased histone deacetylases in the cortex. Three weeks after the last social defeat, H4K12ac, H3K4me3 and histone acetyltransferase activity were increased in the hippocampus [74].
Three studies were identified that applied chronic variable or mild stress for 14 to 15 days. Here, a significant genome-wide decrease in both H4K12ac and phospho-acetylation of H3 (Lys9/Ser10) in the CA3 region and dentate gyrus of the hippocampus was revealed [75]. Accordingly, a study combining chronic variable stress with subsequent acute stress for 30 minutes observed that chronic variable stress reduced genome-wide H4K12ac levels in the hippocampus, while acute stress both on its own or following chronic variable stress went along with increased H4K12ac [76]. Furthermore, chronic variable mild stress resulted in a sex-specific upregulation of histone acetyltransferase CBP mRNA levels in the BNST in female animals and a decrease in histone deacetylase 5 (HDAC5) mRNA levels in the central nucleus of the amygdala in males [77].
Chronic unpredictable stress for 21 to 28 days was shown to decrease H3K9ac levels along with increased HDAC5 expression in the hippocampus on a genome-wide level [78], and to decrease H3K9me3 on a candidate gene level at the promoter of the corticotropin-releasing hormone receptor 1 (Crh1) gene in the hypothalamus [79]. In the specific context of pregnancy, chronic unpredictable stress applied four weeks prior to, and then throughout pregnancy resulted in a global downregulation in H3K14ac in the hippocampus of the offspring with a stronger effect in female offspring, along with a decrease in spatial memory [80]. Accordingly, gestational stress induced by a combination of restraint and 24-hours light disturbance to pregnant mice also induced decreased genome-wide H3K14ac as well as increased expression of DNMT1, HDAC1 and HDAC2 in the offspring [81]. Repeated cross-fostering in mice, an early interference with the maternal environment, was on a descriptive level linked to mostly increases in H3K4me3 and H3ac as well as predominantly decreased H3K27me3 levels in the medulla oblongata in the offspring, possibly driving an oversensitivity to carbon dioxide [82].
Finally, in an animal model using prolonged emotional and pain stress for 15 days an increased activity of Methyl-CpG Binding Protein 2 (MeCP2), which acts as a transcriptional repressor in complex with histone deacetylases, as well as an increase of H4ac were discerned at two weeks post-stress and were still observable two months post-stress [83].
3.3. Pharmacological Compounds Targeting the Histone Acetylation/Methylation Machinery
Epigenetic mechanisms involving chromatin have been shown to be implicated in the mode of action of several established psychopharmacological agents such as valproic acid (VPA), which has been shown to decrease anxiety symptoms e.g. [84-87], or serotonin re-uptake inhibitors (SSRIs)/ serotonin and norepinephrine re-uptake inhibitors (SNRIs), which are approved for the treatment of anxiety- and stress-related disorders [88, 89]. In rodent models, innovative pharmacological compounds mainly targeting erasers (HDACs, HDMs) but also writers (HMTs, HATs) (Figs. 1B and 1C) are currently under research regarding anxiety, fear- and place memory formation or consolidation as well as the extinction of fear-related memories [90-92]. A selective review of these approaches is provided below and in Table 5.
Table 5.
Pharmacological compounds targeting the histone acetylation/methylation machinery in animal models of fear, anxiety and stress.
| Species | Paradigm | Treatment | Measurement Post-stress | Tissue | Genomic Region | Histone Modification | Further Results | References |
|---|---|---|---|---|---|---|---|---|
| Valproic Acid and SSRIs/SNRIs | ||||||||
| Mouse | Extinction | VPA | 2 h | PFC | Bdnf promoter I and IV | H4ac↑ | Long-term memory for extinction↑ | Bredy et al., 2007 [55] |
| Rat | Memory consolidation (2-day-FST) | VPA | Day 1 | VLO, Hip | Genome-wide | - | Memory consolidation↑ | Zhao et al., 2013 [93] |
| Rat | Memory consolidation (2-day-FST) | VPA | Day 2 | VLO, Hip | Genome-wide | - | Memory consolidation↑ Bdnf level in the VLO↓ phospho-ERK↓ | Zhao et al., 2013 [93] |
| Mouse | Memory reconsolidation (7 days) and extinction | VPA | 1 day | - | - | - | Consolidation / reconsolidation and extinction↑ | Bredy et al., 2008 [94] |
| Rat | Predator exposure/ psychosocial stress regimen (for 40 days) | VPA | Immediately | PFC, whole blood | Genome-wide | - | Superoxide↑ (whole blood) ROS↑ (whole blood) HDAC↑ (PFC) normalized to naive levels by VPA | Wilson et al., 2014 [95] |
| Rat | CUS (28 days) | VPA | 1 day | Hip | Genome-wide | Prevents H3K9ac↓ H4K12ac↓ | Prevents: HDAC5↑, anxiety- and depression-like behaviors↑ | Liu et al., 2014 [78] |
| Rat | CRS (21 days) | Fluoxetine (SSRI) | 1 day | Hip (DG) | Genome-wide | Rescues H3K9me3↓ | - | Hunter et al., 2009 [60] |
| Mouse | Chronic CORT stress (21 days) | Fluoxetine (SSRI) | Behavior: 7 days following stress Biochemical: 1 day after behavioral tests | Hip | Genome-wide | - | Anxiety- & depression-like behaviors↓ Prevents: serum CORT and serum ACTH↑, MDA↑ and Glutathion↓, TNF-α and IL-1ß levels↑, NF-κB and HDAC2 gene expression↑, NF-kB p65, COX-2, HDAC2 and p-JNK protein expression↑ | Kv et al., [96] |
| Rat | CUS (28 days) | Venlafaxine (SNRI) | Immediately | Hip | Genome-wide | Prevents: H3K9ac↓, H3K14ac↓ and H4K12ac↓ | Anxiety- & depression-like behavior↓ Prevents: HDAC5↑ | Qiao et al., 2019 [97] |
| Mouse | Fear extinction CUS (28 days) | Venlafaxine (SNRI) | 1 day | Whole brain | Genome-wide | H3K9me2↓ | Anxiety-like behavior↓ | Wang et al., 2018 [98] |
| Experimental Compounds Targeting Erasers (HDAC, HDM) | ||||||||
| Rat | Memory consolidation (CFC) | TSA/NaBu | 1 h | CA1 (Hip) | Genome-wide | H3ac↑ and H4ac↑ | Induction of LTP at Schaffer-collateral synapses↑ | Levenson et al., 2004 [46] |
| Mouse (nitric oxide synthase KO) | Fear memory consolidation | NaBu | Behavior: 1 h, 24 h and 7 days Biochemical analyses: 1 h | Hip, amygdala | Genome-wide | H3ac & H4ac↑ | Contextual fear conditioning↑ (24 h & 7 days) | Itzhak et al., 2012 [99] |
| Rat | Memory consolidation | TSA | 1.5, 3 or 6 h post training | BLA | - | - | Memory consolidation↑, hippocampal Bdnf level↑ | Valiati et al., 2017 [100] |
| Rat | Memory consolidation | TSA or MS-275 | Immediately after FC | Hip | - | - | Long-term object-location memory↑ | Hawk et al., 2011 [101] |
| Mouse | Anxiety (NIH; EZM) | NaBu acutely (3x on 1 day) or chronically (twice daily for 21 days) | 30 min | Hip | Genome-wide | Acute: H4ac↑ Chronic: H4ac↓ | Acute: anxiety↑ (only in NIH, not in EZM) Chronic: no anxiety-like behavior | Gundersen et al., 2009 [102] |
| Rat | Stress (CORT micropellets in CeA for 15 days) | TSA bilaterally infused in CeA (at day 8, for 7 days) | 1 day | CeA | Genome-wide | H3K9ac↑ | Anxiety-like behavior↓ mechanical somatic threshold↑ c-Fos, CRF and SIRT6 expression↓ GR expression↑ | Tran et al., 2015 [103] |
| Mouse | Fear extinction | NaBu | Behavior: 1 h, 24 h and 7 days Biochemical analyses: 1 h | Hip, amygdala | Genome-wide | H3ac↑ (amygdala) H4ac↑ | Speed of cued fear extinction↑ | Itzhak et al., 2012 [99] |
| Mouse | Fear extinction | NaBu (systemically), TSA (intrahippocampally) | Immediately | Hip | - | - | Fear extinction↑ | Lattal et al., 2007 [104] |
| Mouse | Fear extinction | NaBu (intrahippocampally) | 30 min | Infralimbic cortex | Genome-wide | H3K14ac↑ | Extinction levels↑ c-Fos expression↑ | Stafford et al., 2012 [105] |
| Rat | Spatial memory/ extinction (SPS) | NaBu | Immediately | - | - | Learning and memory↑ extinction of new memory↑ | Farani et al., 2020 [106] | |
| Mouse | RTT model (Mecp2 forebrain KO mice) | SAHA | Western Blot: Immediately Cue-dependent: 4 hrs after context | BLA | Genome-wide | H3ac↑ H4ac↑ | Anxiety-like behavior↑ cue-dependent memory ↓ | Adachi et al., 2009 [107] |
| Rat | CFC Fear extinction | SAHA | 1 day 2 h | -Hip | -Genome-wide | -H3ac↑ H4ac↑ | Fear extinction↑ | Fujita et al., 2012 [108] |
| Rat | SPS + CFC + extinction (2 days) | SAHA | 24 h after extinction (behavior) 2 h after injection (histones & proteins) | Hip | Genome-wide | H3ac↑ H4ac↑ | Fear extinction↑ Nr2B↑ CaMKII↑ | Matsumoto et al., 2013 [109] |
| Mouse | Chronic CORT stress (21 days) | SAHA treatment (14 days) | behavior: 7 days following stress biochemical: 1 day after behavioral tests | Hip | Genome-wide | - | Anxiety- & depression-like behaviors↓ Prevents: serum CORT and serum ACTH↑, MDA↑ and Glutathion↓, TNF-α and IL-1ß levels↑, NF-κB and HDAC2 gene expression↑, NF-kB p65, COX-2, HDAC2 and p-JNK protein expression↑ | Kv et al., 2018 [96] |
| Mouse | HAB vs. LAB | MS-275 | 2 h | Cingulate cortex neurons | - | Rescues H3ac↓ | - | Sah et al., 2019 [110] |
| Mouse | ZnR-induced fear extinction | MS-275 | 2 h | IL-PFC, BA neurons | Genome-wide | H4ac↑ | Fear extinction↑ | Whittle et al., 2016 [111] |
| Mouse | Fear extinction (1 to 30 days) + exposure | CI-994 1 h post recall | 1 day | - | - | - | Fear extinction↑ | Gräff et al., 2014 [112] |
| Mouse | Fear extinction | RGFP963 (5 min after extinction training) | 1 h | BLA, DG | Genome-wide | H3ac↓ and H4ac↓ (DG) | Consolidation of cued fear extinction↑ | Bowers et al., 2015 [113] |
| Mouse | Fear extinction | RGFP966 (5 min after extinction training) | 1 h | DG | Genome-wide | H3ac↑ (BLA) H3ac↓ and H4ac↓ | - | Bowers et al., 2015 [113] |
| Rat | Fear conditioning | t-PCP | 24 h | LA | - | H3K9me2↑ | Fear memory↑ | Gupta-Agarwal et al., 2014 [54] |
| Experimental Compounds Targeting Writers (HMTs, HATs) | ||||||||
| Rat | Fear conditioning | UNC0224 | 24 h | LA | - | H3K9me2↓ | Fear memory↓ | Gupta-Agarwal et al., 2014 [54] |
| Mouse | Fear extinction | UNC0642 A-366 | 1 day | Whole brain | Genome-wide | H3K9me2↓ | Anxiety-like behavior↓ | Wang et al., 2018 [98] |
| Rat | Memory consolidation | ETP69 | 24 h | Hip | Genome-wide | H3K9me2↓ | Anxiety-related behavior↓ objection location memory↑ | Snigdha et al., 2016 [114] |
| Mouse | Fear extinction | Lys-CoA-Tat | 24 h | IL-PFC | - | - | Fear extinction↑ | Marek et al., 2011 [117] |
| Mouse | Fear extinction | C646 | 24 h | IL-PFC | - | - | Fear extinction↑ | Marek et al., 2011 [117] |
| Rat | Memory reconsolidation after auditory fear conditioning | Garcinol | 90 min | LA | Genome-wide | H3ac↓ | Consolidation and reconsolidation↓ | Maddox et al., 2013 [115] |
| Rat | Memory reconsolidation | C646 | 90 min | LA | Genome-wide | H3ac↓ | Consolidation and reconsolidation↓ | Maddox et al., 2013 [115] |
| Mouse | CFC + extinction (24 h later) | - | 2h | IL-PFC | Genome-wide | H3K9me2↓ | MeCP2↑ HDAC2↓ PCAF↑ | Wei et al., 2012 [118] |
| Mouse | Fear extinction | SPV106 (PCAF activator) H3-CoA-20-Tat (PCAF inhibitor) | 2h | IL-PFC | - | - | SPV106: anxiety-behavior↓ fear extinction↑ H3-CoA-20-Tat: anxiety-behavior↑ | Wei et al., 2012 [118] |
Abbreviations: A-366: G9a/GLP Inhibitor, BA: Basal amygdala, Bdnf: Brain derived neurotrophic factor, BLA: Basolateral amygdala, C646: E1A binding protein p300 inhibitor, CA1: First region in the hippocampal circuit, CaMKII: Ca2+/calmodulin-dependent protein kinase II, CeA: central nucleus of the amygdala, CFC: contextual fear conditioning, C-fos: Proto-oncogene, CI-994: HDAC class I inhibitor tacedinaline, CORT: Corticosterone, CRF: Corticotropin Releasing Factor, CRS: Chronic restraint stress, CUS: Chronic unpredictable stress, DG: Dentate gyrus, ERK: extracellular-signal regulated kinases, EZM: Elevated zero maze, FC: Fear conditioning, FST: Forced swimming test, H3-CoA-20-Tat: PCAF inhibitor, HAB: High anxiety behavior, HAT: Histone acetyltransferase, HDAC: Histone deacetylase, HDM: Histone demethylase, Hip: Hippocampus, HMT: Histone methyltransferase, IL-PFC: Infralimbic prefrontal cortex, LA: Lateral amygdala, LAB: Low anxiety behavior, LTP: Long-term potentiation, Lys-CoA-Tat: E1A binding protein p300 inhibitor, MeCP2: Methyl-CpG Binding Protein 2, MS-275: HDAC class I inhibitor Entinostat, NaBu: Sodium butyrate, NIH: Novelty-induced hypophagia, Nr2B: N-methyl D-aspartate receptor subtype 2B, PCAF: P300/CBP-associated factor, PFC: Prefrontal cortex, RGFP963: HDAC class I inhibitor, RGFP966: HDAC3 inhibitor, ROS: Reactive oxygen species, RTT: Rett syndrome, SAHA: Suberoylanilide hydroxamic acid = vorinostat, SIRT6: Sirtuin 6, SNRIs: Serotonin and norepinephrine re-uptake inhibitors, SPS: Single prolonged stress, SPV106: PCAF activator, SSRIs: Selective serotonin re-uptake inhibitors, t-PCP: trans-2-phenylcyclopropylamine = histone lysine demethylase (HDM) 1 (LSD1) inhibitor, TSA: HDAC class I&2 inhibitor trichostatin A, UNC0224: selective G9a inhibitor, UNC0642: G9a/GLP Inhibitor, VLO: Ventrolateral orbitofrontal cortex, ZnR: Zinc restricted.
3.3.1. VPA/SSRIs and SNRIs
In an animal model, VPA, known to exert inhibitory effects on HDACs, has been shown to increase the efficiency of extinction training and long-term extinction memory along with elevated acetylation levels of H4 at Bdnf promoters I and IV [55]. Furthermore, treatment with VPA before stress-related memory formation in a two-day forced swimming test resulted in increased memory consolidation along with a significant decrease in ERK-phosphorylation and Bdnf protein levels in the ventrolateral orbital cortex on the following days [93]. In line with these findings, another study testing the effects of pre-test treatment with VPA on memory consolidation, reconsolidation and extinction of cued-fear after seven days of memory acquisition, VPA applied before each paradigm was observed to enhance consolidation, reconsolidation and extinction of long-term memory for conditioned fear, respectively [94]. In a mouse model of PTSD applying a 40-day regimen of predator exposure/ psychosocial stress, elevated superoxide and reactive oxygen species levels were observed in whole blood along with a global increase in HDACs in the PFC. These effects returned to naïve levels when VPA was administered daily for 30 days following the stress regimen [95]. Administration of VPA furthermore prevented increased HDAC5 expression as well as anxiety- and depression-like behavior induced by chronic unpredictable stress (see 3.3.2. [78]) along with a rescue of decreased H3K9 and H4K12 acetylation [78].
Treatment with the SSRI fluoxetine immediately before each individual restraint during 21-day restraint stress blocked the decrease of hippocampal genome-wide H3K9me3 24 hours post-stress, that occurred without fluoxetine treatment [60]. In a study consecutively applying 21 days of corticosterone treatment accompanied by 14 days of fluoxetine treatment starting by day eight of corticosterone treatment, an increase in anxiety- and depression-like behavior invoked by corticosterone stress was prevented by the application of fluoxetine. Further, all the biochemical changes invoked by chronic corticosterone treatment, like increased serum corticosterone and adrenocorticotropic hormone levels and alterations in several immunological markers were not observed in fluoxetine-treated animals [96]. Further, continuous treatment with the SNRI venlafaxine for 28 consecutive days was observed to prevent an increase in hippocampal HDAC5 expression and subsequent decreases in H3K9ac, H3K14ac and H4K12ac observed after 30 days of chronic unpredicted stress, leading to a significant improvement in anxiety- and depression-like behaviors [97]. In another rodent model of fear extinction following chronic unpredicted stress, treatment with venlafaxine was accompanied by decreased H3K9me2 along with anxiolytic effects [98].
3.3.2. Experimental Compounds Targeting Erasers (HDAC, HDM)
Broad spectrum HDAC inhibitors such as sodium butyrate (NaBu) or trichostatin A have quite consistently been reported to enhance fear conditioning, memory formation and anxiety-like behavior. For instance, NaBu prompted a strong long-term potentiation at Schaffer-collateral synapses in vitro along with an increase in genome-wide acetylation of histones H3 and H4 in area CA1 of the hippocampus, and resulted in increased freezing behavior 24 hours, but not immediately after contextual fear conditioning when administered in vivo one hour prior to conditioning relative to saline treatment [46]. Likewise, systemic NaBu administration one hour prior to fear conditioning in nitric oxide synthase knockout mice led to genome-wide increases in acetylation of histones H3 and H4 in the hippocampus and amygdala as well as a significant increase in contextual fear conditioning 24 hours and seven days, respectively, but not immediately after training. However, no effect of NaBu treatment on fear conditioning and retention was observed in wildtype mice [99]. Trichostatin A was shown to enhance memory consolidation of inhibitory avoidance along with increased hippocampal Bdnf levels when infused into the rat basolateral amygdala 1.5, 3 or 6 hours post-training, but not when given pre-training [100], and to enhance long-term object-location memory, again when given post fear conditioning [101]. Furthermore, acute administration of NaBu resulted in increased anxiety-like behavior in the novelty-induced hypophagia paradigm accompanied by an increase in global acetylation levels of H4 in the hippocampus 30 minutes after NaBu treatment [102]. Chronic treatment with NaBu twice daily for 21 days, however, did not lead to any anxiogenic behavior in the novelty-induced hypophagia paradigm and was accompanied by a global decrease in hippocampal H4 acetylation levels [102]. Accordingly, chronic bilateral infusion of trichostatin A for seven days – starting at day 8 of 15-day corticosterone stress elicited by the implantation of corticosterone micropellets in the central nucleus of the amygdala - was able to attenuate a number of corticosterone-induced effects such as increased anxiety-like behavior, decreased mechanical somatic sensitivity, a genome-wide downregulation of H3K9ac, increased c-Fos, CRF and SIRT6 expression and decreased glucocorticoid receptor expression [103]. Regarding the role of broad-spectrum HDAC inhibitors in fear extinction, four independent studies reported mainly extinction-enhancing effects: Systemic NaBu administration was observed to increase genome-wide acetylation of histone H4 in the hippocampus and amygdala as well as H3 acetylation in the amygdala, and application prior to fear conditioning appeared to accelerate cued, but not contextual fear extinction - while not affecting fear acquisition (see above) - after five days of extinction training [99]. Accordingly, systemic administration of NaBu or intrahippocampal injection of trichostatin A prior to a short three-minute extinction training showed improved fear extinction usually only observed after a longer, 24-minute extinction session [104]. A similar result was obtained by intra-hippocampal NaBu injection following weak extinction - otherwise leading to an insufficient fear extinction - that induced improved levels of extinction along with increased infralimbic H3K14ac levels and enhanced c-Fos expression comparable to strong extinction effects [105]. Subcutaneous administration of NaBu for seven days was furthermore shown to alleviate impaired learning and memory in rats exposed to a single prolonged stress procedure and to enhance the extinction of newly formed fear memory [106].
Infusion of the HDAC1 and HDAC2 inhibitor suberoylanilide hydroxamic acid (SAHA), also known as vorinostat, into the basolateral amygdala over seven days resulted in a significant induction of global H3 and H4 acetylation along with increased anxiety-like behavior in the open-field test and impairments in cue-dependent memory in a fear-conditioning paradigm [107]. In another study testing vorinostat on fear consolidation and extinction when injected intraperitoneally after one day of fear conditioning or after the second day of extinction training following one day of fear conditioning, vorinostat was also shown to increase global acetylation levels of H3 and H4 after two hours while leading to facilitated fear extinction [108]. In a rat model of single prolonged stress followed by contextual fear conditioning, vorinostat injected after two days of fear extinction training led to enhanced fear extinction, again accompanied by increased global hippocampal levels of H3 and H4 acetylation [109]. Finally, a 14-day treatment with vorinostat was shown to exert the same effects following chronic corticosterone-induced stress as fluoxetine (see above, [96]), i.e. to prevent the upregulation of corticosterone-stress-induced biochemical changes including corticosterone-induced increased hippocampal HDAC2 gene and protein expression [96].
Systemic treatment with the specific HDAC class I inhibitor MS-275, targeting HDACs 1-3 and 8, effectively reduced the general decrease of acetylated histone H3 associated with inborn hyperanxiety in a high anxiety behavior (HAB) mouse model [110]. In the 129S1/SvlmJ (S1) mouse model characterized by deficient fear extinction, MS-275 furthermore promoted the dietary zinc restriction-induced rescue of deficient fear extinction and increased H4ac levels in the medial PFC and the amygdala [111]. Furthermore, long-term object-location memory - observed to be enhanced by administration of the broad-spectrum HDAC inhibitor trichostatin A after fear conditioning (see above) - could also be enhanced by administration of MS-275 [101]. Remote memory, not successfully extinguishable by reconsolidation paradigms alone, was successfully extinguished during extinction training by additional systemic, intraperitoneal administration of the class I HDAC inhibitor CI-994 one hour after memory recall [112].
The specific class I HDAC inhibitor RGFP963 was shown to enhance the consolidation of cued fear extinction when administered intraperitoneally five minutes after the last conditioned stimulus trial of extinction training and went along with decreased global H3ac and H4ac in the hippocampal dentate gyrus [113]. In the same study, the HDAC3-selective inhibitor RGFP966 increased H3ac in the basolateral amygdala and decreased global acetylation of histones H3 and H4 in the dentate gyrus when injected intraperitoneally one hour before extinction training, but – in contrast to the class I HDAC inhibitor RGFP963 (see above) - did not enhance consolidation of cued fear extinction [113].
Finally, blocking the histone lysine demethylase (HDM) 1 (LSD1) in the lateral amygdala by trans-2-phenylcyclo-propylamine (t-PCP) resulted in enhanced fear memory along with an increase in H3K9me2 levels [54].
3.3.3. Experimental Compounds Targeting Writers (HMTs, HATs)
Inhibition of the H3K9me2 histone lysine methyltransferase G9a by UNC0224 in the lateral amygdala was linked to significantly less freezing behavior 24 hours after auditory fear conditioning and to decreased H3K9me2 levels [54]. Likewise, inhibition of G9a and GLP (G9a-like protein) by UNC0642 and A-366 in adult mice during paradigms to assess anxiety-like behavior such as the elevated zero maze was linked to a reduction in anxiety-like behavior and a general decrease of H3K9me2 methylation in the brain [98]. Pharmacological inhibition of the H3K9me3-specific histone methyl transferase SUV39H1 by means of ETP69, however, resulted in decreased levels of H3K9me3 in the hippocampus, but, however, to increased freezing behavior in a fear conditioning task [114].
Inhibition of p300/CBP by means of C646 or inhibition of p300/CBP and PCAF with Garcinol in the lateral amygdala shortly (one hour) after conditioning or fear memory retrieval significantly reduced freezing behavior 21 hours after fear conditioning via prevention of H3 acetylation in the lateral amygdala [115, 116]. Infusion of the combined p300/CBP histone acetyltransferase inhibitors C646 or Lys-CoA-Tat directly into the PFC post-extinction training led to significantly enhanced fear extinction [117]. Quite conversely, but rather consistent with the effects of HDAC inhibition as detailed above (3.3.2.), systemic administration of the PCAF inhibitor H3-CoA-20-Tat increased fear-like behavior in a contextual fear conditioning paradigm, and systemic administration of the PCAF activator SPV106 resulted in increased fear extinction and decreased fear-like behavior [118].
3.4. Genetic Modifications Targeting the Histone Acetylation/Methylation Machinery
Another way of understanding the role of chromatin modifications in the pathogenesis of anxiety-, fear- and stress-related disorders and of discovering putative new epigenetic drugs in this field is by overexpressing or knocking down/knocking out specific genes of interest coding for the histone-related epigenetic machinery in rodent models.
Effects of specific HDAC classes were tested in the context of synaptic plasticity and memory formation with the help of conditional brain-specific Hdac4 (Hdac4bko) and constitutive Hdac5 knockout mice. In motor coordination, total locomotion and elevated plus maze tests, Hdac4b knockout mice displayed deficits in motor coordination as well as hyperactive and less anxiety-like behavior, which could not be observed in Hdac5 knockout mice. Additionally, Hdac4, but not Hdac5 knockout led to deficits in context-dependent learning and memory and displayed decreased LTP during theta burst stimulation on Schaffer collateral pathway, indicating a decrease in synaptic plasticity [119]. A study testing functional redundance of class I HDACs (HDAC1 and HDAC2) observed Hdac2 but not Hdac1 knockout mice – while not exhibiting a change in locomotor and anxiety-related behavior – to display enhanced performance in an attentional set-shifting task along with accelerated conditioned fear extinction [120], which is in line with enhanced fear extinction elicited by pharmacological HDAC inhibition as detailed above (3.3.2.). However, quite in contrast to the above-mentioned pharmacological studies on HDAC inhibition (3.3.2.), inhibition of HDAC1 via siRNA-mediated knockdown of Hdac1 in the hippocampus was observed to lead to impaired fear extinction, while viral HDAC1 overexpression resulted in increased extinction of contextual fear memories along with a decrease of H3K9ac and an increase of H3K9me3 at the c-Fos promoter region in the hippocampus [121].
Localized knockdown of MeCP2, which acts as a transcriptional repressor in complex with histone deacetylases, in the basolateral amygdala led to a genome-wide increase in H3ac accompanied by increased anxiety-like behavior and deficits in cue-dependent fear conditioning [107].
Transgenic mice expressing an inhibitory, truncated form of the histone acetyltransferase (HAT) P300, lacking the HAT and activation domains, displayed impaired long-term recognition memory and contextual fear memory as well as a genome-wide decrease of H3ac in the forebrain [122]. Results of a subsequent study of postnatal knockdown of P300 in mice, with restriction to subregions of the forebrain, support the pivotal role of P300 in the hippocampus and cortical areas for the formation of long-term recognition memory and long-term contextual memory [123]. Further, knockout of the histone acetyltransferase CBP in the same regions – critical for the acetylation of lysines on histones H3, H4 and H2B – led to impairments in LTP and long-term memory of contextual fear and object recognition [124], which in summary is consistent with the findings of pharmacological p300/CBP inhibition by means of C646 or Garcinol to impair consolidation and reconsolidation of fear memory (see 3.3.3.).
In the nucleus accumbens shell, overexpression of G9a, a histone methyltransferase specific for H3K9me2, mainly associated with repressive transcription resulted in an increase in H3K9me2 levels and subsequently increased addiction- and anxiety-like behaviors [125]. Vice versa, it was found that knocking down G9a expression in the nucleus accumbens shell led to a decrease in addiction- and anxiety-like behaviors [126], which is in line with pharmacological HMT inhibition by UNC0224 [54] or UNC0642 and A-366 [98] also leading to a reduction in anxiety-like behavior (see 3.3.3.). Selective ablation of lysine (K) methyltransferase 2a (Kmt2a), a HMT specific for H3K4, which is linked to active transcription, but not the ortholog Kmt2b, in adult ventral striatum/nucleus accumbens neurons, however, markedly increased anxiety-like behavior in multiple behavioral paradigms [127].
In another knockdown study in mice, where KAP1, an essential cofactor of the epigenetic repressor family of zinc finger proteins, was deleted in the forebrain, mice exhibited heightened levels of anxiety-like behavior and stress-induced alterations in memory and spatial learning [128]. These behavioral changes were accompanied by dysregulation of hippocampal genes caused by a decrease of H3K9me3 and increased general acetylation levels of H3 and H4 [128].
Finally, a conditional-inducible H2A.Z knock-out resulted in enhanced fear memory in males, reduced stress-induced sensitization of fear learning in females and suppressed non-stressful memory in both sexes [129].
3.5. Histone Modifications in Human Fear-, Anxiety- and Stress-Related Disorders
In a sample of 17 war veterans suffering from PTSD as compared to 16 healthy controls, the genome-wide signal distribution profile of H3K4me3, H3K27me3, H3K36me3 and H3K9me3 in peripheral blood mononuclear cells differed significantly between patients and controls pointing towards a shift in the genomic location of these marks [130]. In another study by the same group, ChIPseq on six peripheral blood mononuclear cell samples of patients with PTSD compared with six samples of healthy controls, however, failed to reveal genome-wide alterations in H3K4me3 in PTSD, whereas on a gene-specific level H3K4me3 was found to be increased in PTSD patients around the promoter regions of the WNT10B, WNT10A, WNT7A, DVL1 and TCF7 genes [131]. Authors speculate that the discrepant findings on a genome-wide level might be due to the fact that in the latter study only about 40,000 cells were used for ChIPseq analyses [131], while the previous analyses were based on >10 million cells [130].
4. DISCUSSION
This review article provides an overview of the relevant available literature on histone modifications in animal models of fear, anxiety and acute/chronic stress, on pharmacological compounds and genetic modifications targeting the histone acetylation/methylation machinery in animals as well as on histone modifications in human fear-, anxiety- and stress-related disorders. Animal studies point towards several histone modifications to be dynamically altered in models of fear conditioning, retrieval and extinction, acute and chronic stress. Furthermore, histone modification writers/erasers are suggested to be involved in these processes. Only very few and underpowered studies have been published on histone modifications in humans, restricted to the phenotype of PTSD and to the investigation of peripheral biomaterial given the non-accessibility of human brain tissue in vivo.
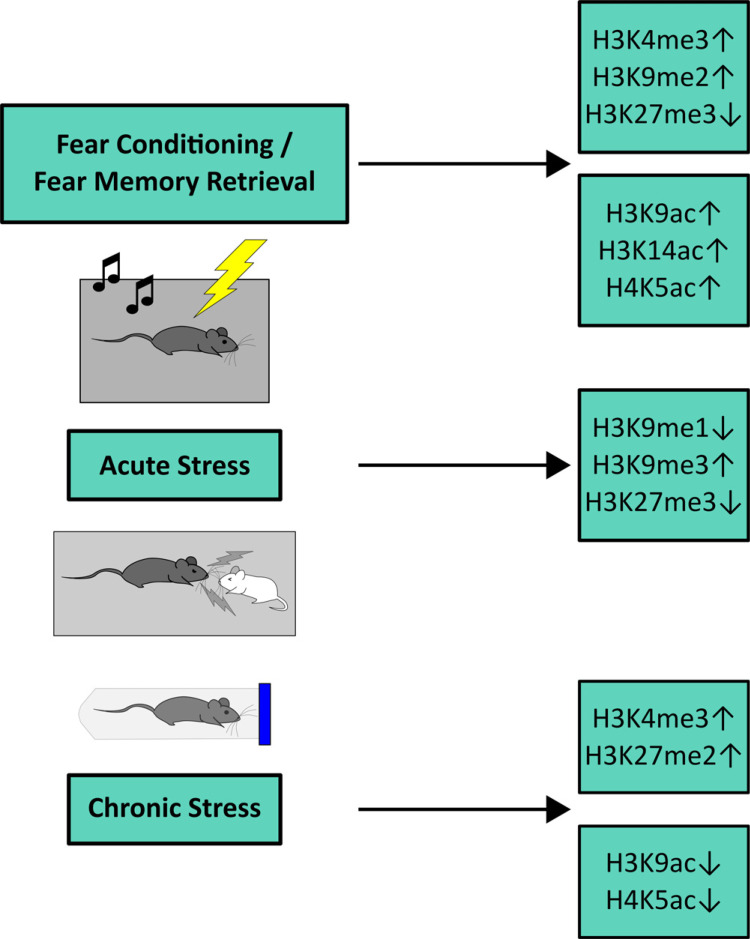
As illustrated in Fig. (2), the most consistent findings imply genome-wide increased H3K4me3 in fear conditioning and fear memory retrieval as well as chronic stress in animal models and gene-specifically in human PTSD. Furthermore, several findings point to globally increased H3K9me2 in animal models of fear conditioning and fear memory retrieval, increased H3K9me3 following acute stress, and increased H3K27me2 in animal models of chronic stress. Decreased H3K27me3, however, was observed quite consistently in animal models of fear conditioning and acute stress, and decreased H3K9me1 following acute stress. Finally, whereas fear conditioning in most studies resulted in increased H3K9ac, H3K14ac and H4K5ac, animal models of chronic stress or offspring of chronically stressed animals showed decreased H3K9ac and H4K5ac. Most of these studies have reported genome-wide data without further specifying the genic/intergenic regions particularly enriched for shifts in histone modification patterns. Very few studies, however, have additionally provided a more in-depth investigation for instance by ChIP-Seq based unbiased peak calling followed by gene ontology analysis as well as by using publicly available microarray data or conventional mRNA analyses e.g., [47, 82]. These data – though presently still scarce – are essential to uncover yet unknown key genes and assess the functional, i.e. transcriptional relevance of altered histone modification signatures at particular genes in anxiety, fear or stress-related phenotypes. This will also aid in elucidating whether epigenetic changes constitute a mechanistic component in its own right or rather catalyze downstream processes in conferring anxiety-, fear- or stress-related symptoms, which, however, is not necessarily mutually exclusive (cf. [92]).
Fig. (2).
Graphical illustration of the currently most consistently identified genome-wide histone modifications in fear-/anxiety- and stress-related animal models. Abbreviations: ac: acetylation; H: histone (H3 or H4); K: lysine; Kx: amino acid position of lysine counting from N-terminus; me1: monomethylation, me2: dimethylation, me3: trimethylation.
On a candidate gene level, BDNF is a promising candidate to be epigenetically studied in relation to fear, anxiety and stress as it plays a pivotal role in brain development, neural plasticity, memory formation and particularly the acquisition, consolidation and extinction of conditioned fear [49-51]. Accordingly, histone modifications have been found to be significantly altered in this context with for instance increased Bdnf promoter I H3K4me3 after fear conditioning, but decreased after extinction. Furthermore, increased Bdnf promoter IV H3 and H4 acetylation facilitating transcriptional activity was observed after fear conditioning but decreased H3ac was discerned at Bdnf promoters I, IV and VI after acute stress.
Furthermore, pharmacological inhibitors of CBP and PCAF, which act as histone acetyl transferases, have been reported to reduce freezing behavior after fear conditioning and enhance fear extinction [116-118]. These findings support previous evidence for a pivotal role of the transcription factor CREB (cAMP/Ca(2+) response element binding protein) in the formation of fear memories in general [132-134].
Despite these first promising findings on a genome-wide and candidate gene as well as pharmacological level it has to be noted that most studies are restricted to particular brain regions or even specific cell types and that the functional consequence remains to be elucidated. Also, some results could not be replicated or have even been contradicted by other studies, which, however, might be due to different paradigms applied across studies. A very important point to consider is the time course of histone modification dynamics in the context of fear, anxiety and stress. Several studies have shown that some effects observable one or two hours after a given event do not appear to persist after 24 hours, while other alterations seem to be detectable even after three weeks, which, however, might be specific to individual histone modifications and/or characteristic for particular paradigms. Finally, rodent models of fear, anxiety and stress most probably do not fully represent the complexity of human fear-, anxiety- and stress-related phenotypes [38] and thus might not accurately reflect their underlying epigenetic mechanisms.
The present review focused on the role of histone modifications and their regulators in fear-, anxiety- and stress-related phenotypes. However, other epigenetic mechanisms such as DNA methylation and non-coding RNAs have also been suggested to be involved in those states (e.g., [8, 9, 11, 14-23]. DNA methylation and histone modifications are highly interdependent with a significant bi-directional epigenetic crosstalk between histone-modifying and DNA methylation enzymes [135]. For instance, in a positive-feedback loop Methyl-CpG-Binding Domain Protein 1 (MBD1) bound to DNA methylation can recruit histone methylases like the SET Domain Bifurcated Histone Lysine Methyltransferase 1 (SETDB1) and thus provoke the methylation of H3K9. H3K9me3 in turn binds to DNMTs which leads to further DNA methylation and promotes heterochromatin formation [136]. It is hypothesized that DNA methylation might ’lock’ the repressive state previously gained by histone modifications [137]. Similarly, epigenetically relevant ncRNAs comprising micro-RNA (miRNA), small interfering RNA (siRNA), piwi-interacting RNA (piRNA) and long non-coding RNA (lncRNA) [138] mostly repressing gene transcription have been shown to influence heterochromatin formation in impacting histone modifications and DNA methylation [139, 140]. Thus, further research is warranted to understand the complex interplay of the whole array of epigenetic mechanisms in governing fear-, anxiety- and stress-related phenotypes. Furthermore, future studies should consider investigating mechanisms such as chromatin remodeling (e.g., changes in nucleosome positioning). First studies in that respect have suggested a role for aberrant regulation of CHD-type chromatin remodeling factors in learning and memory as well as contextual fear extinction in particular [141, 142], or of the ISWI-related remodeling enzyme SNF2H (also known as SMARCA5) in high anxiety behavior (HAB) mice [143].
A better understanding of the role of histone modifications and epigenetic mechanisms in general in conferring fear-, anxiety- and stress-related disorder risk might result in the definition of epigenetic biomarkers suited as both indicators and targets of preventive interventions [144]. As a prerequisite for the suitability of epigenetic marks as biosensors in humans, those markers would have to be stable enough to constitute a molecular signature of disease risk and to be measurable systemically as brain tissue is not accessible in vivo [145]. While many studies point towards a transient nature of chromatin modifications following learning or stressful experiences, there is some evidence for certain histone marks to persist following transcriptional events e.g., [146], and several studies suggest that certain peripheral DNA methylation marks might reflect central nervous system patterns (for review see 8; in silico databases such as BECon, IMAGE-CpG or Blood Brain DNA Methylation Comparison Tool), [147-149]. Given the general scarcity of studies on histone modifications in humans, to the best of our knowledge, no study so far has investigated the comparability of histone modification patterns in human peripheral material such as blood or saliva and human brain tissue. In this context, imaging tools such as positron emission tomography (PET) using tracers selective for histone deacetylases (e.g., [11C]Martinostat) or Lysine-Specific Demethylase 1 might prove to be highly useful in characterizing the histone modification machinery in the living human brain [150-155]. Apart from their potential as biomarkers of disorder risk, altered histone modifications might trigger downstream mechanisms such as mRNA, protein or structural changes eventually driving the pathogenesis of fear-, anxiety- or stress-related states. These downstream mechanisms remain to be investigated in future functional studies.
In analogy to the first studies demonstrating an intergenerational transmission of DNA methylation signatures associated with early life stress [156], the role of histone modifications in transporting the effects of stress in a transgenerational manner remains to be investigated. While many aspects of epigenetic intergenerational transmission in short-lived organisms, such as C. elegans and D. melanogaster, have been thoroughly described [29, 157, 158], the functions and mechanisms of epigenetic intergenerational transmission in mammals still remain poorly understood [29]. Recent studies in mammals suggest that histone post-translational modifications [159-161] could have important roles in the intergenerational transmission of information, but more work needs to be done for claiming causality [29, 162].
Pharmacological compounds targeting the histone acetylation/methylation machinery such as HDAC or HMT (G9a) inhibitors, which have been shown to possibly enhance fear-extinction processes or decrease anxiety-like behavior, respectively, in animal models, carry great promise also regarding the exposure-based and thus extinction-related treatment of human fear-, anxiety- and stress-related disorders. Such “epigenetic drugs” might constitute novel therapeutics in the treatment of anxiety and stress-related disorders by enhancing learning processes, fear extinction and neuronal plasticity [90-92, 163]. However, issues such as the lack of selectivity and specificity of currently available compounds, the partly antagonistic effects regarding fear memory consolidation and fear extinction in animal models and the risk of severe adverse effects when administered systemically remain to be solved before the field can proceed toward clinical trials in humans. As there is a burgeoning body of evidence for disease-associated DNA methylation patterns to “normalize” along with a response to psychotherapy [8], future studies might want to investigate whether histone modifications related to fear, anxiety or stress might also be malleable by psychotherapeutic interventions such as cognitive-behavioral therapy. On a related note, compounds targeting histone acetylation/methylation might be beneficial in augmenting extinction or extinction memory consolidation in a psychotherapeutic setting.
CONCLUSION
The translation of epigenetic findings into human research is hoped to expand the present state of knowledge not only on the pathomechanisms underlying anxiety and stress-related disorders but also on their prognosis, diagnosis and treatment. However, considerable gaps of knowledge remain concerning the intricate role of epigenetic mechanisms in anxiety and stress-related disorders given the numerous obstacles in translating findings from animal models to human research [38], rendering clinical application a long way off. Still, as evident by the promising first advances in the field thus far as outlined above, further detangling the role of the epigenetic machinery in fear, anxiety and stress opens up novel avenues towards gaining a better understanding of mental disorder pathogenesis, their prevention as well as optimized treatment.
ACKNOWLEDGEMENTS
MAS and KD are members of the Anxiety Disorders Research Network (ADRN) of the European College of Neuropsychopharmacology (ECNP).
LIST OF ABBREVIATIONS
- ACC
Anterior Cingulate Cortex
- BDNF
Brain-derived Neurotrophic Factor
- DNA
Deoxyribonucleic Acid
- HAB
High Anxiety Behavior
- ncRNAs
Non-coding RNAs
- PFC
Prefrontal Cortex
- PTSD
Post-traumatic Stress Disorder
- SAHA
Suberoylanilide Hydroxamic Acid
- TFs
Transcription Factors
- VPA
Valproic Acid
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
This work was supported by the German Research Foundation (DFG; CRC-TRR58, projects C02 and Z02, to KD; DO 1241/8-1 to KD and SCHI 1607/2-1 to MAS) and the German Federal Ministry of Education and Research (BMBF) (Grant/Award Number: FKZ 01EE1402A, PROTECT-AD, project P5, to KD). MAS is supported by a CRC 992 - Young Investigator Fellowship (DFG, German Research Foundation, Project-ID 192904750). NI is supported by the Max Planck Society; Deutsche Forschungsgemeinschaft - Project ID 192904750 - CRC 992 Medical Epigenetics; Behrens-Weise Stiftung; EMBO YIP; CIBSS EXC-2189. This project has also received support from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 819941) ERC CoG, EpiRIME. The funding sources had no role in study design, in the collection, analysis and interpretation of data, in the writing of the report, or in the decision to submit the paper for publication.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Baxter A.J., Vos T., Scott K.M., Ferrari A.J., Whiteford H.A. The global burden of anxiety disorders in 2010. Psychol. Med. 2014;44(11):2363–2374. doi: 10.1017/S0033291713003243. [DOI] [PubMed] [Google Scholar]
- 2.Kessler R.C., Petukhova M., Sampson N.A., Zaslavsky A.M., Wittchen H.U. Twelve-month and lifetime prevalence and lifetime morbid risk of anxiety and mood disorders in the United States. Int. J. Methods Psychiatr. Res. 2012;21(3):169–184. doi: 10.1002/mpr.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maercker A., Cloitre M., Bachem R., Schlumpf Y.R., Khoury B., Hitchcock C., Bohus M. Complex post-traumatic stress disorder. Lancet. 2022;400(10345):60–72. doi: 10.1016/S0140-6736(22)00821-2. [DOI] [PubMed] [Google Scholar]
- 4.Olesen J., Gustavsson A., Svensson M., Wittchen H.U., Jönsson B. The economic cost of brain disorders in Europe. Eur. J. Neurol. 2012;19(1):155–162. doi: 10.1111/j.1468-1331.2011.03590.x. [DOI] [PubMed] [Google Scholar]
- 5.Penninx B.W.J.H., Pine D.S., Holmes E.A., Reif A. Anxiety disorders. Lancet. 2021;397(10277):914–927. doi: 10.1016/S0140-6736(21)00359-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wittchen H.U., Jacobi F., Rehm J., Gustavsson A., Svensson M., Jönsson B., Olesen J., Allgulander C., Alonso J., Faravelli C., Fratiglioni L., Jennum P., Lieb R., Maercker A., van Os J., Preisig M., Salvador-Carulla L., Simon R., Steinhausen H.C. The size and burden of mental disorders and other disorders of the brain in Europe 2010. Eur. Neuropsychopharmacol. 2011;21(9):655–679. doi: 10.1016/j.euroneuro.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 7.Solis E.C., van Hemert A.M., Carlier I.V.E., Wardenaar K.J., Schoevers R.A., Beekman A.T.F., Penninx B.W.J.H., Giltay E.J. The 9-year clinical course of depressive and anxiety disorders: New NESDA findings. J. Affect. Disord. 2021;295:1269–1279. doi: 10.1016/j.jad.2021.08.108. [DOI] [PubMed] [Google Scholar]
- 8.Schiele M.A., Gottschalk M.G., Domschke K. The applied implications of epigenetics in anxiety, affective and stress-related disorders - A review and synthesis on psychosocial stress, psychotherapy and prevention. Clin. Psychol. Rev. 2020;77:101830. doi: 10.1016/j.cpr.2020.101830. [DOI] [PubMed] [Google Scholar]
- 9.Schiele M.A., Domschke K. Epigenetics at the crossroads between genes, environment and resilience in anxiety disorders. Genes Brain Behav. 2018;17(3):e12423. doi: 10.1111/gbb.12423. [DOI] [PubMed] [Google Scholar]
- 10.Szyf M., Bick J. DNA methylation: A mechanism for embedding early life experiences in the genome. Child Dev. 2013;84(1):49–57. doi: 10.1111/j.1467-8624.2012.01793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dion A., Muñoz P.T., Franklin T.B. Epigenetic mechanisms impacted by chronic stress across the rodent lifespan. Neurobiol. Stress. 2022;17:100434. doi: 10.1016/j.ynstr.2022.100434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weaver I.C.G., Korgan A.C., Lee K., Wheeler R.V., Hundert A.S., Goguen D. Stress and the emerging roles of chromatin remodeling in signal integration and stable transmission of reversible phenotypes. Front. Behav. Neurosci. 2017;11:41. doi: 10.3389/fnbeh.2017.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marshall P.R., Bredy T.W. Neuroepigenetic mechanisms underlying fear extinction: Emerging concepts. Psychopharmacology (Berl.) 2019;236(1):133–142. doi: 10.1007/s00213-018-5084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hing B., Gardner C., Potash J.B. Effects of negative stressors on DNA methylation in the brain: Implications for mood and anxiety disorders. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2014;165(7):541–554. doi: 10.1002/ajmg.b.32265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malan-Müller S., Seedat S., Hemmings S.M.J. Understanding posttraumatic stress disorder: Insights from the methylome. Genes Brain Behav. 2014;13(1):52–68. doi: 10.1111/gbb.12102. [DOI] [PubMed] [Google Scholar]
- 16.Malan-Müller S., Hemmings S.M.J. The big role of small rnas in anxiety and stress-related disorders. Vitam. Horm. 2017;103:85–129. doi: 10.1016/bs.vh.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Gottschalk M.G., Domschke K., Schiele M.A. Epigenetics underlying susceptibility and resilience relating to daily life stress, work stress, and socioeconomic status. Front. Psychiatry. 2020;11:163. doi: 10.3389/fpsyt.2020.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zannas A.S., Provençal N., Binder E.B. Epigenetics of posttraumatic stress disorder: Current evidence, challenges, and future directions. Biol. Psychiatry. 2015;78(5):327–335. doi: 10.1016/j.biopsych.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Provençal N., Binder E.B. The effects of early life stress on the epigenome: From the womb to adulthood and even before. Exp. Neurol. 2015;268:10–20. doi: 10.1016/j.expneurol.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Klengel T., Pape J., Binder E.B., Mehta D. The role of DNA methylation in stress-related psychiatric disorders. Neuropharmacology. 2014;80:115–132. doi: 10.1016/j.neuropharm.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 21.Narayanan R., Schratt G. miRNA regulation of social and anxiety-related behaviour. Cell. Mol. Life Sci. 2020;77(21):4347–4364. doi: 10.1007/s00018-020-03542-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt U., Keck M.E., Buell D.R. miRNAs and other non-coding RNAs in posttraumatic stress disorder: A systematic review of clinical and animal studies. J. Psychiatr. Res. 2015;65:1–8. doi: 10.1016/j.jpsychires.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 23.Hommers L.G., Domschke K., Deckert J. Heterogeneity and individuality: MicroRNAs in mental disorders. J. Neural Transm. (Vienna) 2015;122(1):79–97. doi: 10.1007/s00702-014-1338-4. [DOI] [PubMed] [Google Scholar]
- 24.Svaren J., Klebanow E., Sealy L., Chalkley R. Analysis of the competition between nucleosome formation and transcription factor binding. J. Biol. Chem. 1994;269(12):9335–9344. doi: 10.1016/S0021-9258(17)37113-2. [DOI] [PubMed] [Google Scholar]
- 25.Hildebrand E.M., Dekker J. Mechanisms and functions of chromosome compartmentalization. Trends Biochem. Sci. 2020;45(5):385–396. doi: 10.1016/j.tibs.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waddington C.H. The epigenotype. Int. J. Epidemiol. 2012;41(1):10–13. doi: 10.1093/ije/dyr184. [DOI] [PubMed] [Google Scholar]
- 27.Feil R. Environmental and nutritional effects on the epigenetic regulation of genes. Mutat. Res. 2006;600(1-2):46–57. doi: 10.1016/j.mrfmmm.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 28.Iacobuzio-Donahue C.A. Epigenetic changes in cancer. Annu. Rev. Pathol. 2009;4(1):229–249. doi: 10.1146/annurev.pathol.3.121806.151442. [DOI] [PubMed] [Google Scholar]
- 29.Skvortsova K., Iovino N. Bogdanović O. Functions and mechanisms of epigenetic inheritance in animals. Nat. Rev. Mol. Cell Biol. 2018;19(12):774–790. doi: 10.1038/s41580-018-0074-2. [DOI] [PubMed] [Google Scholar]
- 30.Jirtle R.L., Skinner M.K. Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 2007;8(4):253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuebel K., Gitik M., Domschke K., Goldman D. Making sense of epigenetics. Int. J. Neuropsychopharmacol. 2016;19(11):pyw058. doi: 10.1093/ijnp/pyw058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y., Sun Z., Jia J., Du T., Zhang N., Tang Y., Fang Y., Fang D. Overview of histone modification. Adv. Exp. Med. Biol. 2021;1283:1–16. doi: 10.1007/978-981-15-8104-5_1. [DOI] [PubMed] [Google Scholar]
- 33.Yun M., Wu J., Workman J.L., Li B. Readers of histone modifications. Cell Res. 2011;21(4):564–578. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greer E.L., Shi Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012;13(5):343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A., Boyer L.A., Young R.A., Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S.A., Flynn R.A., Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470(7333):279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bannister A.J., Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bach D.R. Cross-species anxiety tests in psychiatry: Pitfalls and promises. Mol. Psychiatry. 2022;27(1):154–163. doi: 10.1038/s41380-021-01299-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bienvenu T.C.M., Dejean C., Jercog D., Aouizerate B., Lemoine M., Herry C. The advent of fear conditioning as an animal model of post-traumatic stress disorder: Learning from the past to shape the future of PTSD research. Neuron. 2021;109(15):2380–2397. doi: 10.1016/j.neuron.2021.05.017. [DOI] [PubMed] [Google Scholar]
- 40.Dresler T., Guhn A., Tupak S.V., Ehlis A.C., Herrmann M.J., Fallgatter A.J., Deckert J., Domschke K. Revise the revised? New dimensions of the neuroanatomical hypothesis of panic disorder. J. Neural Transm. (Vienna) 2013;120(1):3–29. doi: 10.1007/s00702-012-0811-1. [DOI] [PubMed] [Google Scholar]
- 41.Namkung H., Thomas K.L., Hall J., Sawa A. Parsing neural circuits of fear learning and extinction across basic and clinical neuroscience: Towards better translation. Neurosci. Biobehav. Rev. 2022;134:104502. doi: 10.1016/j.neubiorev.2021.12.025. [DOI] [PubMed] [Google Scholar]
- 42.Halder R., Hennion M., Vidal R.O., Shomroni O., Rahman R.U., Rajput A., Centeno T.P., van Bebber F., Capece V., Vizcaino J.C.G., Schuetz A.L., Burkhardt S., Benito E., Sala M.N., Javan S.B., Haass C., Schmid B., Fischer A., Bonn S. DNA methylation changes in plasticity genes accompany the formation and maintenance of memory. Nat. Neurosci. 2016;19(1):102–110. doi: 10.1038/nn.4194. [DOI] [PubMed] [Google Scholar]
- 43.Gupta S., Kim S.Y., Artis S., Molfese D.L., Schumacher A., Sweatt J.D., Paylor R.E., Lubin F.D. Histone methylation regulates memory formation. J. Neurosci. 2010;30(10):3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Webb W.M., Sanchez R.G., Perez G., Butler A.A., Hauser R.M., Rich M.C., O'Bierne A.L., Jarome T.J., Lubin F.D. Dynamic association of epigenetic H3K4me3 and DNA 5hmC marks in the dorsal hippocampus and anterior cingulate cortex following reactivation of a fear memory. Neurobiol Learn Mem. 2017;142(Pt A):66–78. doi: 10.1016/j.nlm.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 45.Gupta-Agarwal S., Franklin A.V., DeRamus T., Wheelock M., Davis R.L., McMahon L.L., Lubin F.D. G9a/GLP histone lysine dimethyltransferase complex activity in the hippocampus and the entorhinal cortex is required for gene activation and silencing during memory consolidation. J. Neurosci. 2012;32(16):5440–5453. doi: 10.1523/JNEUROSCI.0147-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levenson J.M., O’Riordan K.J., Brown K.D., Trinh M.A., Molfese D.L., Sweatt J.D. Regulation of histone acetylation during memory formation in the hippocampus. J. Biol. Chem. 2004;279(39):40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- 47.Park C., Rehrauer H., Mansuy I.M. Genome-wide analysis of H4K5 acetylation associated with fear memory in mice. BMC Genomics. 2013;14(1):539. doi: 10.1186/1471-2164-14-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bousiges O., Neidl R., Majchrzak M., Muller M.A., Barbelivien A., Pereira de Vasconcelos A., Schneider A., Loeffler J.P., Cassel J.C., Boutillier A.L. Detection of histone acetylation levels in the dorsal hippocampus reveals early tagging on specific residues of H2B and H4 histones in response to learning. PLoS One. 2013;8(3):e57816. doi: 10.1371/journal.pone.0057816. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Andero R., Ressler K.J. Fear extinction and BDNF: Translating animal models of PTSD to the clinic. Genes Brain Behav. 2012;11(5):503–512. doi: 10.1111/j.1601-183X.2012.00801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dincheva I., Lynch N.B., Lee F.S. The role of BDNF in the development of fear learning. Depress. Anxiety. 2016;33(10):907–916. doi: 10.1002/da.22497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Notaras M., van den Buuse M. Neurobiology of BDNF in fear memory, sensitivity to stress, and stress-related disorders. Mol. Psychiatry. 2020;25(10):2251–2274. doi: 10.1038/s41380-019-0639-2. [DOI] [PubMed] [Google Scholar]
- 52.Takei S., Morinobu S., Yamamoto S., Fuchikami M., Matsumoto T., Yamawaki S. Enhanced hippocampal BDNF/TrkB signaling in response to fear conditioning in an animal model of posttraumatic stress disorder. J. Psychiatr. Res. 2011;45(4):460–468. doi: 10.1016/j.jpsychires.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 53.Lubin F.D., Roth T.L., Sweatt J.D. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J. Neurosci. 2008;28(42):10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gupta-Agarwal S., Jarome T.J., Fernandez J., Lubin F.D. NMDA receptor- and ERK-dependent histone methylation changes in the lateral amygdala bidirectionally regulate fear memory formation. Learn. Mem. 2014;21(7):351–362. doi: 10.1101/lm.035105.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bredy T.W., Wu H., Crego C., Zellhoefer J., Sun Y.E., Barad M. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learn. Mem. 2007;14(4):268–276. doi: 10.1101/lm.500907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siddiqui S.A., Singh S., Ranjan V., Ugale R., Saha S., Prakash A. Enhanced histone acetylation in the infralimbic prefrontal cortex is associated with fear extinction. Cell. Mol. Neurobiol. 2017;37(7):1287–1301. doi: 10.1007/s10571-017-0464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ranjan V., Singh S., Siddiqui S.A., Tripathi S., Khan M.Y., Prakash A. Differential histone acetylation in sub-regions of bed nucleus of the stria terminalis underlies fear consolidation and extinction. Psychiatry Investig. 2017;14(3):350–359. doi: 10.4306/pi.2017.14.3.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh S., Siddiqui S.A., Tripathy S., Kumar S., Saha S., Ugale R., Modi D.R., Prakash A. Decreased level of histone acetylation in the infralimbic prefrontal cortex following immediate extinction may result in deficit of extinction memory. Brain Res. Bull. 2018;140:355–364. doi: 10.1016/j.brainresbull.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 59.Dunsmoor J.E., Cisler J.M., Fonzo G.A., Creech S.K., Nemeroff C.B. Laboratory models of post-traumatic stress disorder: The elusive bridge to translation. Neuron. 2022;110(11):1754–1776. doi: 10.1016/j.neuron.2022.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hunter R.G., McCarthy K.J., Milne T.A., Pfaff D.W., McEwen B.S. Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc. Natl. Acad. Sci. USA. 2009;106(49):20912–20917. doi: 10.1073/pnas.0911143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nasca C., Zelli D., Bigio B., Piccinin S., Scaccianoce S., Nisticò R., McEwen B.S. Stress dynamically regulates behavior and glutamatergic gene expression in hippocampus by opening a window of epigenetic plasticity. Proc. Natl. Acad. Sci. USA. 2015;112(48):14960–14965. doi: 10.1073/pnas.1516016112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fuchikami M., Morinobu S., Kurata A., Yamamoto S., Yamawaki S. Single immobilization stress differentially alters the expression profile of transcripts of the brain-derived neurotrophic factor (BDNF) gene and histone acetylation at its promoters in the rat hippocampus. Int. J. Neuropsychopharmacol. 2009;12(1):73–82. doi: 10.1017/S1461145708008997. [DOI] [PubMed] [Google Scholar]
- 63.Hollis F., Wang H., Dietz D., Gunjan A., Kabbaj M. The effects of repeated social defeat on long-term depressive-like behavior and short-term histone modifications in the hippocampus in male Sprague–Dawley rats. Psychopharmacology (Berl.) 2010;211(1):69–77. doi: 10.1007/s00213-010-1869-9. [DOI] [PubMed] [Google Scholar]
- 64.Bilang-Bleuel A., Ulbricht S., Chandramohan Y., De Carli S., Droste S.K., Reul J.M.H.M. Psychological stress increases histone H3 phosphorylation in adult dentate gyrus granule neurons: Involvement in a glucocorticoid receptor-dependent behavioural response. Eur. J. Neurosci. 2005;22(7):1691–1700. doi: 10.1111/j.1460-9568.2005.04358.x. [DOI] [PubMed] [Google Scholar]
- 65.Bartlett A.A., DeRosa H., Clark M., Lapp H.E., Guffanti G., Hunter R.G. Corticosterone dynamically regulates retrotransposable element expression in the rat hippocampus and C6 cells. Neurobiol. Stress. 2021;15:100397. doi: 10.1016/j.ynstr.2021.100397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wei J., Xiong Z., Lee J.B., Cheng J., Duffney L.J., Matas E., Yan Z. Histone Modification of Nedd4 Ubiquitin Ligase Controls the Loss of AMPA Receptors and Cognitive Impairment Induced by Repeated Stress. J. Neurosci. 2016;36(7):2119–2130. doi: 10.1523/JNEUROSCI.3056-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu J., Liu C., Zhang L., He B., Shi W.P., Shi H.L., Qin C. Chronic restraint stress impairs cognition via modulating HDAC2 expression. Transl. Neurosci. 2021;12(1):154–163. doi: 10.1515/tnsci-2020-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Elliott E., Ezra-Nevo G., Regev L., Neufeld-Cohen A., Chen A. Resilience to social stress coincides with functional DNA methylation of the Crf gene in adult mice. Nat. Neurosci. 2010;13(11):1351–1353. doi: 10.1038/nn.2642. [DOI] [PubMed] [Google Scholar]
- 69.Renthal W., Maze I., Krishnan V., Covington H.E., III, Xiao G., Kumar A., Russo S.J., Graham A., Tsankova N., Kippin T.E., Kerstetter K.A., Neve R.L., Haggarty S.J., McKinsey T.A., Bassel-Duby R., Olson E.N., Nestler E.J. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56(3):517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 70.Covington H.E., III, Maze I., Sun H., Bomze H.M., DeMaio K.D., Wu E.Y., Dietz D.M., Lobo M.K., Ghose S., Mouzon E., Neve R.L., Tamminga C.A., Nestler E.J. A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron. 2011;71(4):656–670. doi: 10.1016/j.neuron.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Covington H.E., III, Maze I., LaPlant Q.C., Vialou V.F., Ohnishi Y.N., Berton O., Fass D.M., Renthal W., Rush A.J., III, Wu E.Y., Ghose S., Krishnan V., Russo S.J., Tamminga C., Haggarty S.J., Nestler E.J. Antidepressant actions of histone deacetylase inhibitors. J. Neurosci. 2009;29(37):11451–11460. doi: 10.1523/JNEUROSCI.1758-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tsankova N.M., Berton O., Renthal W., Kumar A., Neve R.L., Nestler E.J. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat. Neurosci. 2006;9(4):519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 73.Wilkinson M.B., Xiao G., Kumar A., LaPlant Q., Renthal W., Sikder D., Kodadek T.J., Nestler E.J. Imipramine treatment and resiliency exhibit similar chromatin regulation in the mouse nucleus accumbens in depression models. J. Neurosci. 2009;29(24):7820–7832. doi: 10.1523/JNEUROSCI.0932-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Montagud-Romero S., Montesinos J., Pascual M., Aguilar M.A., Roger-Sánchez C., Guerri C., Miñarro J., Rodríguez-Arias M. `Up-regulation of histone acetylation induced by social defeat mediates the conditioned rewarding effects of cocaine. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2016;70:39–48. doi: 10.1016/j.pnpbp.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 75.Ferland C.L., Schrader L.A. Regulation of histone acetylation in the hippocampus of chronically stressed rats: A potential role of sirtuins. Neuroscience. 2011;174:104–114. doi: 10.1016/j.neuroscience.2010.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ferland C.L., Harris E.P., Lam M., Schrader L.A. Facilitation of the HPA axis to a novel acute stress following chronic stress exposure modulates histone acetylation and the ERK/MAPK pathway in the dentate gyrus of male rats. Endocrinology. 2014;155(8):2942–2952. doi: 10.1210/en.2013-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sterrenburg L., Gaszner B., Boerrigter J., Santbergen L., Bramini M., Elliott E., Chen A., Peeters B.W.M.M., Roubos E.W., Kozicz T. Chronic stress induces sex-specific alterations in methylation and expression of corticotropin-releasing factor gene in the rat. PLoS One. 2011;6(11):e28128. doi: 10.1371/journal.pone.0028128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu D., Qiu H.M., Fei H.Z., Hu X.Y., Xia H.J., Wang L.J., Qin L.J., Jiang X.H., Zhou Q.X. Histone acetylation and expression of mono-aminergic transmitters synthetases involved in CUS-induced depressive rats. Exp. Biol. Med. (Maywood) 2014;239(3):330–336. doi: 10.1177/1535370213513987. [DOI] [PubMed] [Google Scholar]
- 79.Wan Q., Gao K., Rong H., Wu M., Wang H., Wang X., Wang G., Liu Z. Histone modifications of the Crhr1 gene in a rat model of depression following chronic stress. Behav. Brain Res. 2014;271:1–6. doi: 10.1016/j.bbr.2014.05.031. [DOI] [PubMed] [Google Scholar]
- 80.Benoit J.D., Rakic P., Frick K.M. Prenatal stress induces spatial memory deficits and epigenetic changes in the hippocampus indicative of heterochromatin formation and reduced gene expression. Behav. Brain Res. 2015;281:1–8. doi: 10.1016/j.bbr.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zheng Y., Fan W., Zhang X., Dong E. Gestational stress induces depressive-like and anxiety-like phenotypes through epigenetic regulation of BDNF expression in offspring hippocampus. Epigenetics. 2016;11(2):150–162. doi: 10.1080/15592294.2016.1146850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cittaro D., Lampis V., Luchetti A., Coccurello R., Guffanti A., Felsani A., Moles A., Stupka E., D’ Amato F.R., Battaglia M. Histone modifications in a mouse model of early adversities and panic disorder: Role for asic1 and neurodevelopmental genes. Sci. Rep. 2016;6(1):25131. doi: 10.1038/srep25131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baĭdo, A.I.; Diuzhikova, N.A.; Shiriaeva, N.V.; Sokolova, N.E.; Vshivtseva, V.V.; Savenko, IuN. Systemic control of the molecular, cell, and epigenetic mechanisms of long-lasting consequences of stress. Genetika. 2009;45(3):342–348. [PubMed] [Google Scholar]
- 84.Primeau F., Fontaine R., Beauclair L. Valproic acid and panic disorder. Can. J. Psychiatry. 1990;35(3):248–250. doi: 10.1177/070674379003500309. [DOI] [PubMed] [Google Scholar]
- 85.Keck P.E., Jr, Taylor V.E., Tugrul K.C., McElroy S.L., Bennett J.A. Valproate treatment of panic disorder and lactate-induced panic attacks. Biol. Psychiatry. 1993;33(7):542–546. doi: 10.1016/0006-3223(93)90010-B. [DOI] [PubMed] [Google Scholar]
- 86.Kinrys G., Pollack M.H., Simon N.M., Worthington J.J., Nardi A.E., Versiani M. Valproic acid for the treatment of social anxiety disorder. Int. Clin. Psychopharmacol. 2003;18(3):169–172. doi: 10.1097/01.yic.0000064261.66765.9f. [DOI] [PubMed] [Google Scholar]
- 87.Aliyev N.A., Aliyev Z.N. Valproate (depakine-chrono) in the acute treatment of outpatients with generalized anxiety disorder without psychiatric comorbidity: Randomized, double-blind placebo-controlled study. Eur. Psychiatry. 2008;23(2):109–114. doi: 10.1016/j.eurpsy.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 88.Lötsch J., Schneider G., Reker D., Parnham M.J., Schneider P., Geisslinger G., Doehring A. Common non-epigenetic drugs as epigenetic modulators. Trends Mol. Med. 2013;19(12):742–753. doi: 10.1016/j.molmed.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 89.Boks M.P., de Jong N.M., Kas M.J.H., Vinkers C.H., Fernandes C., Kahn R.S., Mill J., Ophoff R.A. Current status and future prospects for epigenetic psychopharmacology. Epigenetics. 2012;7(1):20–28. doi: 10.4161/epi.7.1.18688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Whittle N., Singewald N. HDAC inhibitors as cognitive enhancers in fear, anxiety and trauma therapy: Where do we stand? Biochem. Soc. Trans. 2014;42(2):569–581. doi: 10.1042/BST20130233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peedicayil J. The potential role of epigenetic drugs in the treatment of anxiety disorders. Neuropsychiatr. Dis. Treat. 2020;16:597–606. doi: 10.2147/NDT.S242040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stafford J.M., Lattal K.M. Is an epigenetic switch the key to persistent extinction? Neurobiol. Learn. Mem. 2011;96(1):35–40. doi: 10.1016/j.nlm.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhao Y., Xing B., Dang Y., Qu C., Zhu F., Yan C. Microinjection of valproic acid into the ventrolateral orbital cortex enhances stress-related memory formation. PLoS One. 2013;8(1):e52698. doi: 10.1371/journal.pone.0052698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bredy T.W., Barad M. The histone deacetylase inhibitor valproic acid enhances acquisition, extinction, and reconsolidation of conditioned fear. Learn. Mem. 2008;15(1):39–45. doi: 10.1101/lm.801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wilson C.B., McLaughlin L.D., Ebenezer P.J., Nair A.R., Francis J. Valproic acid effects in the hippocampus and prefrontal cortex in an animal model of post-traumatic stress disorder. Behav. Brain Res. 2014;268:72–80. doi: 10.1016/j.bbr.2014.03.029. [DOI] [PubMed] [Google Scholar]
- 96.Kv A., Madhana R.M., Js I.C., Lahkar M., Sinha S., Naidu V.G.M. Antidepressant activity of vorinostat is associated with amelioration of oxidative stress and inflammation in a corticosterone-induced chronic stress model in mice. Behav. Brain Res. 2018;344:73–84. doi: 10.1016/j.bbr.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 97.Qiao M., Jiang Q.S., Liu Y.J., Hu X.Y., Wang L.J., Zhou Q.X., Qiu H.M. Antidepressant mechanisms of venlafaxine involving increasing histone acetylation and modulating tyrosine hydroxylase and tryptophan hydroxylase expression in hippocampus of depressive rats. Neuroreport. 2019;30(4):255–261. doi: 10.1097/WNR.0000000000001191. [DOI] [PubMed] [Google Scholar]
- 98.Wang D., Kosowan J., Samsom J., Leung L., Zhang K., Li Y., Xiong Y., Jin J., Petronis A., Oh G., Wong A.H.C. Inhibition of the G9a/GLP histone methyltransferase complex modulates anxiety-related behavior in mice. Acta Pharmacol. Sin. 2018;39(5):866–874. doi: 10.1038/aps.2017.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Itzhak Y., Anderson K.L., Kelley J.B., Petkov M. Histone acetylation rescues contextual fear conditioning in nNOS KO mice and accelerates extinction of cued fear conditioning in wild type mice. Neurobiol. Learn. Mem. 2012;97(4):409–417. doi: 10.1016/j.nlm.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Valiati F.E., Vasconcelos M., Lichtenfels M., Petry F.S., de Almeida R.M.M., Schwartsmann G., Schröder N., de Farias C.B., Roesler R. Administration of a histone deacetylase inhibitor into the basolateral amygdala enhances memory consolidation, delays extinction, and increases hippocampal BDNF levels. Front. Pharmacol. 2017;8:415. doi: 10.3389/fphar.2017.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hawk J.D., Florian C., Abel T. Post-training intrahippocampal inhibition of class I histone deacetylases enhances long-term object-location memory. Learn. Mem. 2011;18(6):367–370. doi: 10.1101/lm.2097411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gundersen B.B., Blendy J.A. Effects of the histone deacetylase inhibitor sodium butyrate in models of depression and anxiety. Neuropharmacology. 2009;57(1):67–74. doi: 10.1016/j.neuropharm.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tran L., Schulkin J., Ligon C.O., Greenwood-Van Meerveld B. Epigenetic modulation of chronic anxiety and pain by histone deacetylation. Mol. Psychiatry. 2015;20(10):1219–1231. doi: 10.1038/mp.2014.122. [DOI] [PubMed] [Google Scholar]
- 104.Lattal K.M., Barrett R.M., Wood M.A. Systemic or intrahippocampal delivery of histone deacetylase inhibitors facilitates fear extinction. Behav. Neurosci. 2007;121(5):1125–1131. doi: 10.1037/0735-7044.121.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stafford J.M., Raybuck J.D., Ryabinin A.E., Lattal K.M. Increasing histone acetylation in the hippocampus-infralimbic network enhances fear extinction. Biol. Psychiatry. 2012;72(1):25–33. doi: 10.1016/j.biopsych.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mohammadi-Farani A., Pourmotabbed A., Ardeshirizadeh Y. Effects of HDAC inhibitors on spatial memory and memory extinction in SPS-induced PTSD rats. Res. Pharm. Sci. 2020;15(3):241–248. doi: 10.4103/1735-5362.288426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Adachi M., Autry A.E., Covington H.E., III, Monteggia L.M. MeCP2-mediated transcription repression in the basolateral amygdala may underlie heightened anxiety in a mouse model of Rett syndrome. J. Neurosci. 2009;29(13):4218–4227. doi: 10.1523/JNEUROSCI.4225-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fujita Y., Morinobu S., Takei S., Fuchikami M., Matsumoto T., Yamamoto S., Yamawaki S. Vorinostat, a histone deacetylase inhibitor, facilitates fear extinction and enhances expression of the hippocampal NR2B-containing NMDA receptor gene. J. Psychiatr. Res. 2012;46(5):635–643. doi: 10.1016/j.jpsychires.2012.01.026. [DOI] [PubMed] [Google Scholar]
- 109.Matsumoto Y., Morinobu S., Yamamoto S., Matsumoto T., Takei S., Fujita Y., Yamawaki S. Vorinostat ameliorates impaired fear extinction possibly via the hippocampal NMDA-CaMKII pathway in an animal model of posttraumatic stress disorder. Psychopharmacology (Berl.) 2013;229(1):51–62. doi: 10.1007/s00213-013-3078-9. [DOI] [PubMed] [Google Scholar]
- 110.Sah A., Sotnikov S., Kharitonova M., Schmuckermair C., Diepold R.P., Landgraf R., Whittle N., Singewald N. Epigenetic mechanisms within the cingulate cortex regulate innate anxiety-like behavior. Int. J. Neuropsychopharmacol. 2019;22(4):317–328. doi: 10.1093/ijnp/pyz004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Whittle N., Maurer V., Murphy C., Rainer J., Bindreither D., Hauschild M., Scharinger A., Oberhauser M., Keil T., Brehm C., Valovka T., Striessnig J., Singewald N. Enhancing dopaminergic signaling and histone acetylation promotes long-term rescue of deficient fear extinction. Transl. Psychiatry. 2016;6(12):e974. doi: 10.1038/tp.2016.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gräff J., Joseph N.F., Horn M.E., Samiei A., Meng J., Seo J., Rei D., Bero A.W., Phan T.X., Wagner F., Holson E., Xu J., Sun J., Neve R.L., Mach R.H., Haggarty S.J., Tsai L.H. Epigenetic priming of memory updating during reconsolidation to attenuate remote fear memories. Cell. 2014;156(1-2):261–276. doi: 10.1016/j.cell.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bowers M.E., Xia B., Carreiro S., Ressler K.J. The Class I HDAC inhibitor RGFP963 enhances consolidation of cued fear extinction. Learn. Mem. 2015;22(4):225–231. doi: 10.1101/lm.036699.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Snigdha S., Prieto G.A., Petrosyan A., Loertscher B.M., Dieskau A.P., Overman L.E., Cotman C.W. H3K9me3 inhibition improves memory, promotes spine formation, and increases BDNF levels in the aged hippocampus. J. Neurosci. 2016;36(12):3611–3622. doi: 10.1523/JNEUROSCI.2693-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Maddox S.A., Watts C.S., Doyère V., Schafe G.E. A naturally-occurring histone acetyltransferase inhibitor derived from Garcinia indica impairs newly acquired and reactivated fear memories. PLoS One. 2013;8(1):e54463. doi: 10.1371/journal.pone.0054463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Maddox S.A., Watts C.S., Schafe G.E. p300/CBP histone acetyltransferase activity is required for newly acquired and reactivated fear memories in the lateral amygdala. Learn. Mem. 2013;20(2):109–119. doi: 10.1101/lm.029157.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Marek R., Coelho C.M., Sullivan R.K.P., Baker-Andresen D., Li X., Ratnu V., Dudley K.J., Meyers D., Mukherjee C., Cole P.A., Sah P., Bredy T.W. Paradoxical enhancement of fear extinction memory and synaptic plasticity by inhibition of the histone acetyltransferase p300. J. Neurosci. 2011;31(20):7486–7491. doi: 10.1523/JNEUROSCI.0133-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wei W., Coelho C.M., Li X., Marek R., Yan S., Anderson S., Meyers D., Mukherjee C., Sbardella G., Castellano S., Milite C., Rotili D., Mai A., Cole P.A., Sah P., Kobor M.S., Bredy T.W. p300/CBP-associated factor selectively regulates the extinction of conditioned fear. J. Neurosci. 2012;32(35):11930–11941. doi: 10.1523/JNEUROSCI.0178-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kim M.S., Akhtar M.W., Adachi M., Mahgoub M., Bassel-Duby R., Kavalali E.T., Olson E.N., Monteggia L.M. An essential role for histone deacetylase 4 in synaptic plasticity and memory formation. J. Neurosci. 2012;32(32):10879–10886. doi: 10.1523/JNEUROSCI.2089-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Morris M.J., Mahgoub M., Na E.S., Pranav H., Monteggia L.M. Loss of histone deacetylase 2 improves working memory and accelerates extinction learning. J. Neurosci. 2013;33(15):6401–6411. doi: 10.1523/JNEUROSCI.1001-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bahari-Javan S., Maddalena A., Kerimoglu C., Wittnam J., Held T., Bähr M., Burkhardt S., Delalle I., Kügler S., Fischer A., Sananbenesi F. HDAC1 regulates fear extinction in mice. J. Neurosci. 2012;32(15):5062–5073. doi: 10.1523/JNEUROSCI.0079-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Oliveira A.M.M., Wood M.A., McDonough C.B., Abel T. Transgenic mice expressing an inhibitory truncated form of p300 exhibit long-term memory deficits. Learn. Mem. 2007;14(9):564–572. doi: 10.1101/lm.656907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Oliveira A.M.M., Estévez M.A., Hawk J.D., Grimes S., Brindle P.K., Abel T. Subregion-specific p300 conditional knock-out mice exhibit long-term memory impairments. Learn. Mem. 2011;18(3):161–169. doi: 10.1101/lm.1939811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Barrett R.M., Malvaez M., Kramar E., Matheos D.P., Arrizon A., Cabrera S.M., Lynch G., Greene R.W., Wood M.A. Hippocampal focal knockout of CBP affects specific histone modifications, long-term potentiation, and long-term memory. Neuropsychopharmacology. 2011;36(8):1545–1556. doi: 10.1038/npp.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Anderson E.M., Larson E.B., Guzman D., Wissman A.M., Neve R.L., Nestler E.J., Self D.W. Overexpression of the histone dimethyltransferase G9a in nucleus accumbens shell increases cocaine self-administration, stress-induced reinstatement, and anxiety. j. neurosci. 2018;38(4):803–813. doi: 10.1523/JNEUROSCI.1657-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Anderson E.M., Sun H., Guzman D., Taniguchi M., Cowan C.W., Maze I., Nestler E.J., Self D.W. Knockdown of the histone di-methyltransferase G9a in nucleus accumbens shell decreases cocaine self-administration, stress-induced reinstatement, and anxiety. Neuropsychopharmacology. 2019;44(8):1370–1376. doi: 10.1038/s41386-018-0305-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shen E.Y., Jiang Y., Javidfar B., Kassim B., Loh Y.H.E., Ma Q., Mitchell A.C., Pothula V., Stewart A.F., Ernst P., Yao W.D., Martin G., Shen L., Jakovcevski M., Akbarian S. Neuronal deletion of Kmt2a/Mll1 histone methyltransferase in ventral striatum is associated with defective spike-timing-dependent striatal synaptic plasticity, altered response to dopaminergic drugs, and increased anxiety. Neuropsychopharmacology. 2016;41(13):3103–3113. doi: 10.1038/npp.2016.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jakobsson J., Cordero M.I., Bisaz R., Groner A.C., Busskamp V., Bensadoun J.C., Cammas F., Losson R., Mansuy I.M., Sandi C., Trono D. KAP1-mediated epigenetic repression in the forebrain modulates behavioral vulnerability to stress. Neuron. 2008;60(5):818–831. doi: 10.1016/j.neuron.2008.09.036. [DOI] [PubMed] [Google Scholar]
- 129.Ramzan F., Baumbach J., Monks A.D., Zovkic I.B. Histone H2A.Z is required for androgen receptor-mediated effects on fear memory. Neurobiol. Learn. Mem. 2020;175:107311. doi: 10.1016/j.nlm.2020.107311. [DOI] [PubMed] [Google Scholar]
- 130.Bam M., Yang X., Zhou J., Ginsberg J.P., Leyden Q., Nagarkatti P.S., Nagarkatti M. Evidence for epigenetic regulation of pro-inflammatory cytokines, interleukin-12 and interferon gamma, in peripheral blood mononuclear cells from PTSD patients. J. Neuroimmune Pharmacol. 2016;11(1):168–181. doi: 10.1007/s11481-015-9643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bam M., Yang X., Busbee B.P., Aiello A.E., Uddin M., Ginsberg J.P., Galea S., Nagarkatti P.S., Nagarkatti M. Increased H3K4me3 methylation and decreased miR-7113-5p expression lead to enhanced Wnt/β-catenin signaling in immune cells from PTSD patients leading to inflammatory phenotype. Mol. Med. 2020;26(1):110. doi: 10.1186/s10020-020-00238-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Josselyn S.A. Continuing the search for the engram: Examining the mechanism of fear memories. J. Psychiatry Neurosci. 2010;35(4):221–228. doi: 10.1503/jpn.100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Carlezon W., Jr, Duman R., Nestler E. The many faces of CREB. Trends Neurosci. 2005;28(8):436–445. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 134.McCullough K.M., Chatzinakos C., Hartmann J., Missig G., Neve R.L., Fenster R.J., Carlezon W.A., Jr, Daskalakis N.P., Ressler K.J. Genome-wide translational profiling of amygdala Crh-expressing neurons reveals role for CREB in fear extinction learning. Nat. Commun. 2020;11(1):5180. doi: 10.1038/s41467-020-18985-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.D’Alessio A.C.D.A.C., Szyf M. Epigenetic tête-à-tête: The bilateral relationship between chromatin modifications and DNA methylation. Biochem. Cell Biol. 2006;84(4):463–476. doi: 10.1139/o06-090. [DOI] [PubMed] [Google Scholar]
- 136.Stewart M.D., Li J., Wong J. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol. Cell. Biol. 2005;25(7):2525–2538. doi: 10.1128/MCB.25.7.2525-2538.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.O’Neill C. The epigenetics of embryo development. Anim. Front. 2015;5(1):42–49. doi: 10.2527/af.2015-0007. [DOI] [Google Scholar]
- 138.Cao J. The functional role of long non-coding RNAs and epigenetics. Biol. Proced. Online. 2014;16(1):42. doi: 10.1186/1480-9222-16-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Frías-Lasserre D., Villagra C.A. The importance of ncRNAs as epigenetic mechanisms in phenotypic variation and organic evolution. Front. Microbiol. 2017;8:2483. doi: 10.3389/fmicb.2017.02483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Peschansky V.J., Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics. 2014;9(1):3–12. doi: 10.4161/epi.27473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schoberleitner I., Mutti A., Sah A., Wille A., Gimeno-Valiente F., Piatti P., Kharitonova M., Torres L., López-Rodas G., Liu J.J., Singewald N., Schwarzer C., Lusser A. Role for chromatin remodeling factor Chd1 in learning and memory. Front. Mol. Neurosci. 2019;12:3. doi: 10.3389/fnmol.2019.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wille A., Maurer V., Piatti P., Whittle N., Rieder D., Singewald N., Lusser A. Impaired contextual fear extinction learning is associated with aberrant regulation of CHD-type chromatin remodeling factors. Front. Behav. Neurosci. 2015;9:313. doi: 10.3389/fnbeh.2015.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wille A., Amort T., Singewald N., Sartori S.B., Lusser A. Dysregulation of select ATP-dependent chromatin remodeling factors in high trait anxiety. Behav. Brain Res. 2016;311:141–146. doi: 10.1016/j.bbr.2016.05.036. [DOI] [PubMed] [Google Scholar]
- 144.Domschke K. Prevention in psychiatry: A role for epigenetics? World Psychiatry. 2021;20(2):227–228. doi: 10.1002/wps.20854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Abi-Dargham M.S., Farzana A., DeLorenzo C., Domschke K., Horga G., Jutla A., Paulus M.P., Rubio J.M., Veenstra-VanderWeele J., Krystal J.H. The search for biomarkers in neuropsychiatric disorders. World Psychiatry. under review. [Google Scholar]
- 146.McGarvey K.M., Fahrner J.A., Greene E., Martens J., Jenuwein T., Baylin S.B. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 2006;66(7):3541–3549. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- 147.Edgar R.D., Jones M.J., Meaney M.J., Turecki G., Kobor M.S. BECon: A tool for interpreting DNA methylation findings from blood in the context of brain. Transl. Psychiatry. 2017;7(8):e1187. doi: 10.1038/tp.2017.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hannon E., Lunnon K., Schalkwyk L., Mill J. Interindividual methylomic variation across blood, cortex, and cerebellum: Implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics. 2015;10(11):1024–1032. doi: 10.1080/15592294.2015.1100786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Braun P.R., Han S., Hing B., Nagahama Y., Gaul L.N., Heinzman J.T., Grossbach A.J., Close L., Dlouhy B.J., Howard M.A., III, Kawasaki H., Potash J.B., Shinozaki G. Genome-wide DNA methylation comparison between live human brain and peripheral tissues within individuals. Transl. Psychiatry. 2019;9(1):47. doi: 10.1038/s41398-019-0376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gilbert T.M., Zürcher N.R., Catanese M.C., Tseng C.E.J., Di Biase M.A., Lyall A.E., Hightower B.G., Parmar A.J., Bhanot A., Wu C.J., Hibert M.L., Kim M., Mahmood U., Stufflebeam S.M., Schroeder F.A., Wang C., Roffman J.L., Holt D.J., Greve D.N., Pasternak O., Kubicki M., Wey H.Y., Hooker J.M. Neuroepigenetic signatures of age and sex in the living human brain. Nat. Commun. 2019;10(1):2945. doi: 10.1038/s41467-019-11031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Koole M., Van Weehaeghe D., Serdons K., Herbots M., Cawthorne C., Celen S., Schroeder F.A., Hooker J.M., Bormans G., de Hoon J., Kranz J.E., Van Laere K., Gilbert T.M. Clinical validation of the novel HDAC6 radiotracer [18F]EKZ-001 in the human brain. Eur. J. Nucl. Med. Mol. Imaging. 2021;48(2):596–611. doi: 10.1007/s00259-020-04891-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Matsuda S., Hattori Y., Matsumiya K., McQuade P., Yamashita T., Aida J., Sandiego C.M., Gouasmat A., Carroll V.M., Barret O., Tamagnan G., Koike T., Kimura H. Design, synthesis, and evaluation of [18F]T-914 as a novel positron-emission tomography tracer for lysine-specific demethylase 1. J. Med. Chem. 2021;64(17):12680–12690. doi: 10.1021/acs.jmedchem.1c00653. [DOI] [PubMed] [Google Scholar]
- 153.Pascoal T.A., Chamoun M., Lax E., Wey H.Y., Shin M., Ng K.P., Kang M.S., Mathotaarachchi S., Benedet A.L., Therriault J., Lussier F.Z., Schroeder F.A., DuBois J.M., Hightower B.G., Gilbert T.M., Zürcher N.R., Wang C., Hopewell R., Chakravarty M., Savard M., Thomas E., Mohaddes S., Farzin S., Salaciak A., Tullo S., Cuello A.C., Soucy J.P., Massarweh G., Hwang H., Kobayashi E., Hyman B.T., Dickerson B.C., Guiot M.C., Szyf M., Gauthier S., Hooker J.M., Rosa-Neto P. [11C]Martinostat PET analysis reveals reduced HDAC I availability in Alzheimer’s disease. Nat. Commun. 2022;13(1):4171. doi: 10.1038/s41467-022-30653-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tseng C.E.J., Gilbert T.M., Catanese M.C., Hightower B.G., Peters A.T., Parmar A.J., Kim M., Wang C., Roffman J.L., Brown H.E., Perlis R.H., Zürcher N.R., Hooker J.M. In vivo human brain expression of histone deacetylases in bipolar disorder. Transl. Psychiatry. 2020;10(1):224. doi: 10.1038/s41398-020-00911-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Turkman N., Liu D., Pirola I. Design, synthesis, biochemical evaluation, radiolabeling and in vivo imaging with high affinity class-IIa histone deacetylase inhibitor for molecular imaging and targeted therapy. Eur. J. Med. Chem. 2022;228:114011. doi: 10.1016/j.ejmech.2021.114011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Stenz L., Schechter D.S., Serpa S.R., Paoloni-Giacobino A. Intergenerational transmission of DNA tethylation signatures associated with early life stress. Curr. Genomics. 2018;19(8):665–675. doi: 10.2174/1389202919666171229145656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zenk F., Loeser E., Schiavo R., Kilpert F. Bogdanović O.; Iovino, N. Germ line-inherited H3K27me3 restricts enhancer function during maternal-to-zygotic transition. Science. 2017;357(6347):212–216. doi: 10.1126/science.aam5339. [DOI] [PubMed] [Google Scholar]
- 158.Gaydos L.J., Wang W., Strome S. H3K27me and PRC2 transmit a memory of repression across generations and during development. Science. 2014;345(6203):1515–1518. doi: 10.1126/science.1255023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Inoue A., Jiang L., Lu F., Suzuki T., Zhang Y. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature. 2017;547(7664):419–424. doi: 10.1038/nature23262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Dahl J.A., Jung I., Aanes H., Greggains G.D., Manaf A., Lerdrup M., Li G., Kuan S., Li B., Lee A.Y., Preissl S., Jermstad I., Haugen M.H., Suganthan R., Bjørås M., Hansen K., Dalen K.T., Fedorcsak P., Ren B., Klungland A. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature. 2016;537(7621):548–552. doi: 10.1038/nature19360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang B., Zheng H., Huang B., Li W., Xiang Y., Peng X., Ming J., Wu X., Zhang Y., Xu Q., Liu W., Kou X., Zhao Y., He W., Li C., Chen B., Li Y., Wang Q., Ma J., Yin Q., Kee K., Meng A., Gao S., Xu F., Na J., Xie W. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature. 2016;537(7621):553–557. doi: 10.1038/nature19361. [DOI] [PubMed] [Google Scholar]
- 162.Bohacek J., Mansuy I.M. A guide to designing germline-dependent epigenetic inheritance experiments in mammals. Nat. Methods. 2017;14(3):243–249. doi: 10.1038/nmeth.4181. [DOI] [PubMed] [Google Scholar]
- 163.Fuchikami M., Yamamoto S., Morinobu S., Okada S., Yamawaki Y., Yamawaki S. The potential use of histone deacetylase inhibitors in the treatment of depression. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2016;64:320–324. doi: 10.1016/j.pnpbp.2015.03.010. [DOI] [PubMed] [Google Scholar]