Abstract
Minimally invasive liquid biopsies from the eye capture locally enriched fluids that contain thousands of proteins from highly specialized ocular cell types, presenting a promising alternative to solid tissue biopsies. The advantages of liquid biopsies include sampling the eye without causing irreversible functional damage, potentially better reflecting tissue heterogeneity, collecting samples in an outpatient setting, monitoring therapeutic response with sequential sampling, and even allowing examination of disease mechanisms at the cell level in living humans, an approach that we refer to as TEMPO (Tracing Expression of Multiple Protein Origins). Liquid biopsy proteomics has the potential to transform molecular diagnostics and prognostics and to assess disease mechanisms and personalized therapeutic strategies in individual patients. This review addresses opportunities, challenges, and future directions of high-resolution liquid biopsy proteomics in ophthalmology, with particular emphasis on the large-scale collection of high-quality samples, cutting edge proteomics technology, and artificial intelligence-supported data analysis.
Keywords: liquid biopsy proteomics, artificial intelligence, eye, aqueous humor, vitreous, precision medicine
Introduction
Examining diseases of the eye, particularly protein expression, is largely dependent on techniques such as immunohistochemistry of solid tissue biopsies. However, these biopsies are performed infrequently because much of the eye consists of nonregenerative cells that would be permanently damaged with a biopsy. As new molecular therapies continue to develop and significantly impact eye disease treatments,1 understanding protein expression has become more important. Liquid biopsies that measure proteins in locally enriched fluids from the eye allow for assessment of disease mechanisms and personalized therapeutic strategies in living humans and are emerging as an important alternative to solid tissue biopsies.2−4 These liquid biopsies offer significant advantages as they avoid direct disruption of nonregenerative tissues, may better reflect tissue heterogeneity, can be performed in a variety of clinical settings including outpatient clinics, and can be repeated allowing monitoring of therapeutic responses in individual patients. In addition, liquid biopsies from the eye can have higher sensitivity since they are molecularly enriched compared to the blood where protein biomarkers are diluted due to the high dynamic range.
Liquid biopsy proteomics has helped to substantially improve our understanding of human pathophysiology. In patients with neovascular age-related macular degeneration and diabetic macular edema, aqueous humor (AH) proteomics can help to predict the response to anti-VEGF (vascular endothelial growth factor) therapy,2,3 a significant medical need since about one-third of patients show persistent neovascular activity despite intensive anti-VEGF therapy.5,6 In patients with autoinflammatory eye disease, liquid biopsy proteomics can uncover personalized therapies that can be timed with specific disease activities and stages and can help prevent delivery of ineffective drugs.1 Although genomics has revolutionized cancer research,7 looking at proteins, the biological products of genes helps to better classify disease and predict therapeutic response.8−10
Protein detection is typically focused on a single protein like troponin or VEGF using an ELISA. Multiple proteins can be detected concurrently using a multiplex ELISA, but this requires knowing in advance which proteins are of interest. Advances in mass spectrometry, along with the development of highly accessible and user-friendly bioinformatic tools, have significantly advanced proteomic analysis, but resolution remains the main limitation of proteomics studies.11 The low volume and low protein concentration of liquid biopsies from the eye have been major technical barriers for proteomics analyses. Despite multiple advantages of proteomics over genomics,8−10 there is a significant need to analyze proteins at a scale comparable to genomics studies.
Analyzing high-resolution proteomics data covering thousands of different proteins requires sophisticated bioinformatic tools. We recently developed the tool TEMPO (Tracing Expression of Multiple Protein Origins) that integrates high-resolution liquid biopsy proteomics, artificial intelligence, and single-cell transcriptomics of all known cell types in the human eye, allowing us to trace thousands of AH proteins back to their cells of origin.4 Based on specific protein signatures from individual cell types, TEMPO enables the analysis of disease and aging mechanisms at the cell level in living humans, including in nonregenerative organs such as the retina where direct biopsies would cause irreversible functional damage.
This review focuses on the promises, limitations, and future directions of high-resolution liquid biopsy proteomics for personalized ophthalmology, covering the collection and biobanking of high-quality liquid specimens, state-of-the-art proteomics technology measuring thousands of proteins in microvolume samples, and cutting-edge artificial intelligence-supported bioinformatic tools.
Bioinformatic Analysis
With the rise of high-resolution proteomics technology, the ability to perform complex bioinformatics analyses is becoming increasingly relevant. Clinician scientists play a critical role in this process as they can combine clinical expertise with bioinformatic skills to formulate specific hypotheses and guide analysis.
Basic Data Analysis
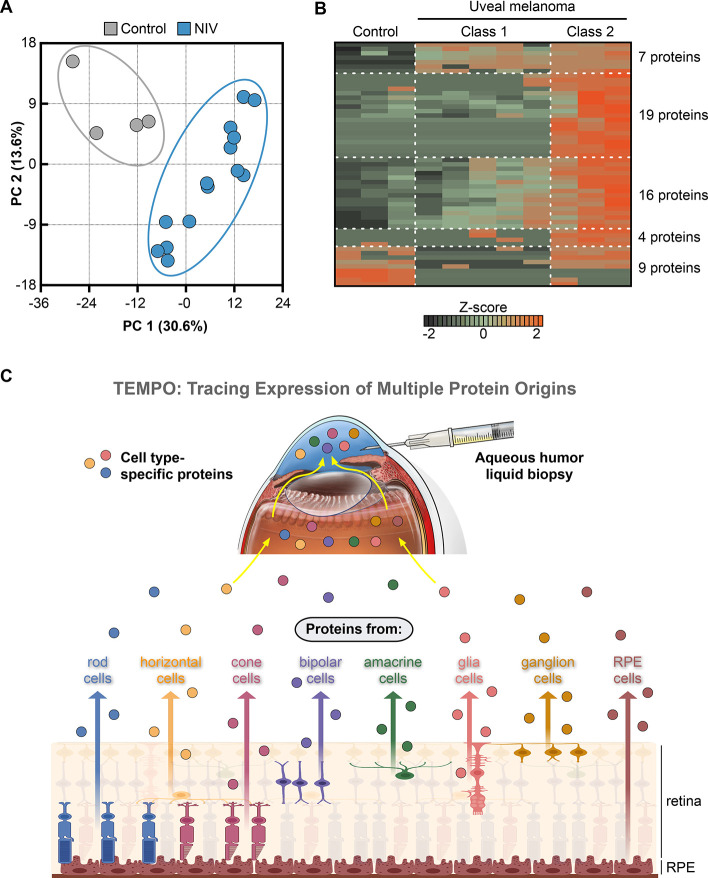
The steps in the early analysis aim to provide an unbiased overview of the samples based on all proteins identified. Principal component analysis (PCA) graphically clusters samples in a two-dimensional space based on two principal components, in which their protein profiles differ most. The distance between samples corresponds to the proteomic difference. The example in Figure 1A illustrates a clear difference in the vitreous proteomic profile between patients with neovascular inflammatory vitreoretinopathy (NIV) compared to controls (Figure adapted from Velez et al.12). A heatmap can be generated to visualize protein levels of multiple proteins in all samples. The example in Figure 1B shows more than 50 proteins that were significantly changed in the vitreous of patients with uveal melanoma (UM) compared to controls and even illustrates proteomic differences between UM with low (class 1) and high (class 2) metastatic risk (Figure adapted from Velez et al.13). Comparing the proteomic profiles between different groups (e.g., diseased vs healthy) can identify proteins that significantly differ between these populations. A pathway analysis based on these differentially expressed proteins can provide insights into the biological pathways affected by a disease. Several databases offer protein-to-pathway annotations, including Reactome pathways, WikiPathways, Gene Ontology, and the Kyoto Encyclopedia of Genes and Genomes (KEGG). More recently, open-access databases emerged, providing web-based, user-friendly, and intuitive insights into the molecular profiles of various ocular tissues and diseases without the need for advanced bioinformatics skills.14−16
Figure 1.
Bioinformatic analysis of ocular liquid biopsy proteomics data. (A) Principal component analysis (PCA) graphically clusters samples based on two principal components in which their protein profiles differ most. Each point is one sample. NIV: neovascular inflammatory vitreoretinopathy. Panel adapted from Velez et al., Sci. Rep., 2019. (B) A heatmap visualizes protein levels of 55 proteins in the vitreous of patients with uveal melanoma with high metastatic risk (Class 2), low metastatic risk (Class 1) and control samples. Each row is one protein, and each column is one sample. The z-score represents a protein’s abundance level in relation to its mean level in all samples by standard deviation units. Panel adapted from Velez et al., Mol. Cancer, 2021. (C) TEMPO (Tracing Expression of Multiple Protein Origins) allows one to trace the cellular origin of thousands of aqueous humor proteins, including specific proteins from individual retinal cell types, allowing cell level analyses in nonregenerative tissues such as the retina in living humans. TEMPO is based on the integration of high-resolution liquid biopsy proteomics with single-cell transcriptomics from all known cell types in the human eye. Panel adapted from Wolf et al., Cell, 2023.
Tracing Expression of Multiple Protein Origins (TEMPO)
By integrating high-resolution liquid biopsy proteomics with single-cell transcriptomics from all known cell types in the human eye, we developed the TEMPO approach allowing us to trace the cellular origin of almost 6,000 proteins detected in AH.4 Based on hundreds of cell type-specific marker proteins, including for individual retinal cell types, TEMPO enables cell level analyses in nonregenerative tissues in living humans such as the retina (Figure 1C). In patients with diabetic retinopathy, the cells driving disease changed from blood vessel cells in the early stage to immune cells in the late stage. Diagnostic assessment of Parkinson’s disease (PD) is challenging, mainly because disease associated protein biomarkers can usually only be identified post-mortem. In the AH of patients with Parkinson’s disease, many brain proteins linked to the disease changed in the AH, indicating that AH protein biomarkers could help to diagnose PD or determine if a patient is responding to a therapy. TEMPO further revealed signs of molecular degeneration of the retina in PD patients, as previously indicated in imaging studies. The integration of two high-resolution methodologies, liquid biopsy proteomics and cell level transcriptomics, represents a major technical advance to examine disease mechanisms at the cell level in vivo. TEMPO could also be used to characterize other difficult-to-sample tissues. For example, liquid biopsies of cerebrospinal fluid could be used to study or diagnose the brain; synovial fluid could be used to study joints; and urine could be used to study the kidneys.
Role of Artificial Intelligence in Data Analysis
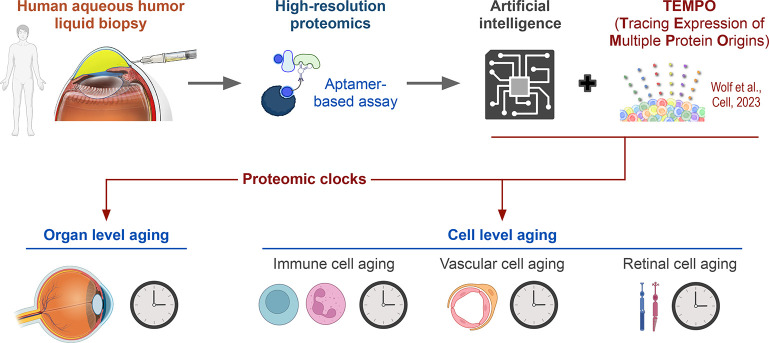
Artificial intelligence (AI) can learn rules and detect protein patterns including nonlinear features from large-scale proteomics data that may not be visible to human investigators. We recently developed an AI proteomic clock from the AH proteome that can predict a healthy person’s age based on a subset of 26 AH proteins (Figure 2).4 Because specific marker proteins of each cell were identified, AI clocks could predict the age of corresponding cell types such as blood vessels, retinal cells, or immune cells. Applying AI clocks to diseased eyes revealed that diseases such as diabetic retinopathy and uveitis cause accelerated aging within specific cell types. In patients with diabetic retinopathy, the degree of aging increased with disease progression and was accelerated by about 30 years in late-stage (proliferative) diabetic retinopathy. Even eyes that had been treated successfully, where the bleeding had stopped or the inflammation was controlled, showed accelerated aging, indicating that antiaging therapies may be needed to fully reverse damage from disease. In many patients with diabetes, there are no visible signs of eye disease. We found that in those eyes the age of the immune cells and retinal cells was about 10 years older than healthy eyes, suggesting that there is accelerated aging even before an ophthalmologist can see clinical signs of disease. Our findings indicate that targeted antiaging drugs could be the next step in preventative, personalized medicine.
Figure 2.
Artificial intelligence proteomic clocks assess age of the human eye. An artificial intelligence (AI) proteomic clock based on the aqueous humor proteome can predict a healthy person’s eye age. By integrating TEMPO, cell type-specific marker proteins were used to develop AI proteomic clocks to predict the age of a specific cell type, such as immune cells, vascular cells, and retinal cells. Adapted from Wolf et al., Cell, 2023.
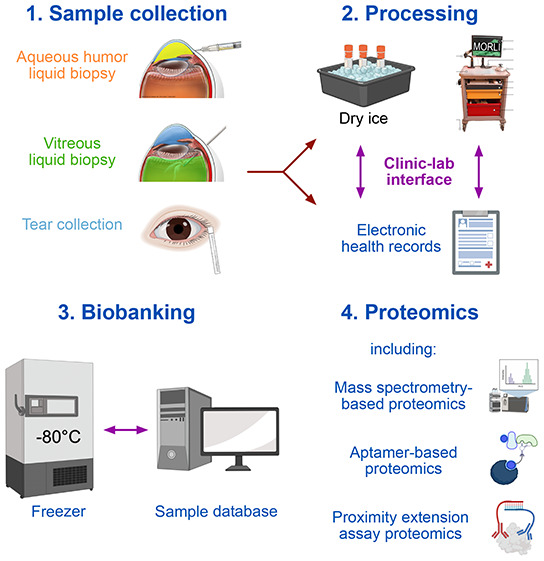
Sample Collection for Eye Proteomics
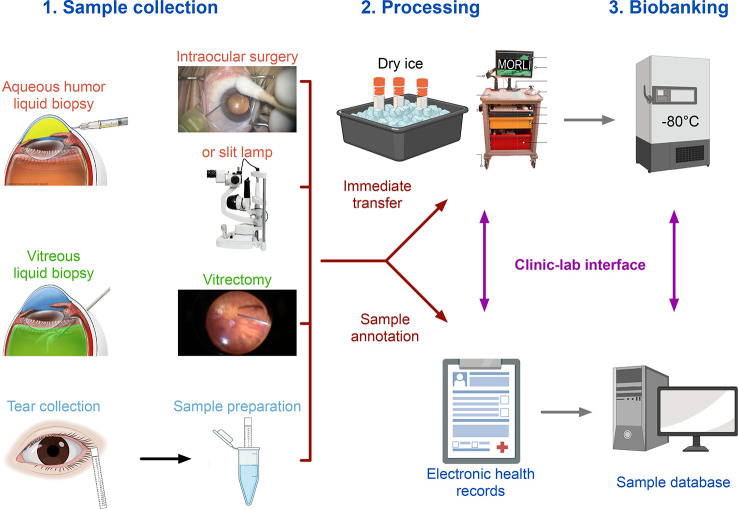
A variety of different fluids can be collected from the human eye and used for proteomic analyses, among them, samples from the vitreous in the posterior chamber of the eye, the aqueous humor (AH) from the anterior chamber of the eye, or tear samples (Figure 3). Using these specimens in translational research requires high sample quality and thorough sample annotation. It is critical that the specimens are snap-frozen on dry ice immediately after the collection and then stored at −80 °C until further analysis.17−19 Integration of proteomics data with clinical data is essential to obtaining clinically meaningful insights. This requires a viable and efficient interface between patient care in the operating room (OR) and the research laboratory that allows collection of clinical and demographic data at the site of sample collection.18 A mobile operating room lab interface (MORLI), consisting of a cart on wheels with a flat lab bench surface, a computer with a barcode scanner, lab supplies, and a box with dry ice, can help to achieve these goals.18,19
Figure 3.
Sample collection for eye proteomics. A variety of fluids can be collected from the human eye for proteomic analyses, including liquid biopsies from the aqueous humor (collected during intraocular surgery or using a slit lamp), vitrectomy (collected during vitrectomy), or tear samples. Immediately after the collection, the specimens are snap-frozen on dry ice and then stored at −80 °C until further analysis. A thorough sample annotation with links to the electronic health records of the patient is essential to obtain clinically meaningful insights. MORLI (mobile operating room lab interface), consisting of a cart on wheels with a flat lab bench surface, a computer with a barcode scanner, lab supplies, and a box with dry ice, helps with the processing of samples in the operating room.
Aqueous Humor Sample Collection
Aqueous humor (AH) samples can be safely collected in an outpatient clinic using a slit lamp or microscope;20−23 the fluid samples can then serve diagnostic purposes or in clinical trials to assess target engagement and biomarkers associated with clinical outcomes. Various studies have described a surgical technique performed at the slit lamp involving a 27- to 30-gauge needle attached to a syringe or an aqueous pipet that is inserted perpendicular to the limbus without preincision and without using a lid speculum.21,22 Other studies placed the patient in a supine position on the bed or a surgical chair in the outpatient clinic20,23 and used a lid speculum and a preincision with a 15° blade.20
With about 20 million cases each year, cataract surgery is one of the most frequently performed surgeries worldwide.24 At the beginning of this procedure and many other intraocular surgeries, including glaucoma, corneal, or vitreoretinal interventions, a small amount of AH fluid is collected but discarded. Collecting these samples would allow sample acquisition at a large scale.18 We recently demonstrated that AH fluid can be safely collected during intraocular surgery.25 Different surgical techniques can be applied to obtain samples during intraocular surgery. A 30-gauge needle connected to a 1 mL syringe can be inserted into the anterior chamber (AC) perpendicular to the limbus without prior incision to manually aspirate 50–100 μL of undiluted AH.18 Another option is to use the corneal incision, which is routinely performed at the beginning of the scheduled anterior segment surgery by using a 15° blade. Once the incision is established, an angled 30-gauge blunt cannula or a 30-gauge needle can be used to aspirate AH. Independent of the technique, direct visualization via the surgical microscope ensures that the tip of the needle or the cannula remains over the peripheral iris in the mid AC to avoid damage to intraocular structures, such as the corneal endothelium, iris, and lens.18 In vitrectomy cases, sclerotomies can be established before the AC biopsy to ensure the safe insertion of the trocars. The collected samples should immediately be snap-frozen and processed, as described previously.
Vitreous Sample Collection
Vitreous liquid biopsies can be safely collected at the beginning of a vitrectomy.26 After placing the 23-, 25-, or 27-G trocar cannulas and connecting the infusion cannula, the vitreous cutter is activated in the vitreous cavity without infusion, and 0.5 to 1 mL of undiluted vitreous is manually aspirated using a syringe that is connected to the vitreous extrusion cannula. Since the goal is to collect an undiluted vitreous sample, it is essential that the vitrectomy cutter is not primed with fluid. The collected specimens should immediately be snap-frozen and processed as described above.
Tear Sample Collection
Collection of tear samples is a relatively simple and noninvasive procedure that does not require anesthesia.27 Tears can be collected using Schirmer’s strips, a glass microcapillary tube, or by a flush tear collection approach that instills ∼20–40 μL of 0.9% saline solution into the inferior palpebral fold of the eye.28−30 Following sample collection, samples can be stored in a −80 °C freezer for several months (or years) before sample preparation. However, analyzing fresh samples is preferred to increase the quality of the data and the number of identified peptides/proteins.30 Protein concentrations can be determined by the bicinchoninic acid (BCA) assay. Importantly, Schirmer’s strips and glass microcapillary tubes can vary between different manufacturers and batches and therefore, data reproducibility can be a challenge.30 Samples can later be processed using a lysis buffer containing 1% sodium dodecyl-sulfate (SDS), 200 mM HEPES (pH 7.6), and protease inhibitors,31 although several types of lysis solutions can be used depending on the type of analysis and instrumentation. Samples are then denatured, and the free cysteine is alkylated before trypsin digestion. After C18 chromatography, samples are analyzed by liquid chromatography and tandem mass spectrometry (LC-MS/MS).31
Analytical Instruments and Proteomics Methods
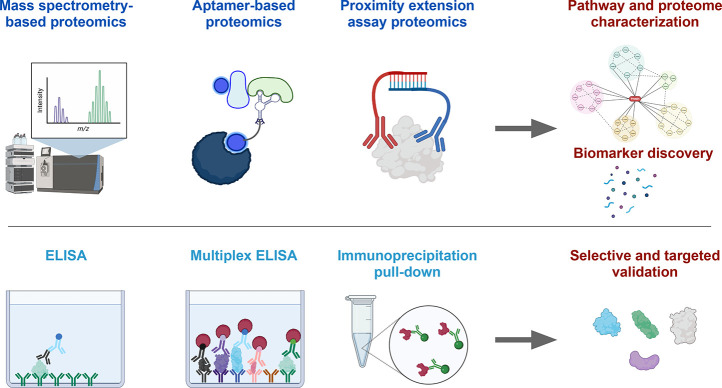
Quantitative proteomics allows the global evaluation, identification, and quantification of proteins in cells or tissues and helps to understand their composition, structure, interactions, and functions.32,33 Thus, it can be a great exploratory technique that identifies candidate proteins for disease diagnosis and drug development, leading to unbiased data-driven hypotheses.34−36 Most current proteomics techniques can be categorized in affinity-based or mass spectrometry-based identification37 (Figure 4). Affinity-based identification involves the specific binding between a target protein and an affinity agent such as antibodies, proteins, or small molecules. The affinity agent acts as the reporter itself or is followed by additional steps that ultimately lead to protein detection via colorimetric or fluorescence signals.38 The simplest example of affinity-based methods is an enzyme-linked immunosorbent assay (ELISA) that detects one protein with high sensitivity and specificity and is therefore widely used in research and clinical testing.39 More recently, multiplex ELISA and Proximity Extension Assay emerged as a high-throughput technique allowing the detection of hundreds of proteins simultaneously.1,2,13,40−42 Instead of antibodies, an aptamer-based assay uses synthetic DNA-based aptamers that specifically bind to their protein targets. This cutting-edge technology can measure over 7,000 different proteins in small volume liquid biopsies, such as AH and vitreous, representing a manifold higher proteomic resolution compared to previous mass spectrometry-based studies.4
Figure 4.
Analytical instruments for eye proteomics. Mass spectrometry-based, aptamer-based, and proximity extension assay-based proteomics allow the analysis of hundreds to thousands of different proteins in a single sample, allowing us to perform pathway and proteome characterization as well as biomarker discovery studies. An enzyme-linked immunosorbent assay (ELISA) and immunoprecipitation pull-down represent affinity-based methods that detect a single protein with high accuracy. A multiplex ELISA allows to detect tens to hundreds of different proteins in the same sample. These technologies are valuable for selective and targeted validation studies.
Mass spectrometry uses liquid chromatography coupled with a mass spectrometer to detect peptides based on their mass-to-charge ratio (m/z). One of the most significant advantages of the technique is that it allows for unsupervised analyses, meaning that researchers do not have to know which proteins to look for in advance. Mass spectrometry-based techniques detect thousands of proteins in a single experiment. However, the small volume of AH or vitreous samples represents a critical challenge for mass spectrometry-based studies. One solution may be to pool samples, but this then requires extensive labeling techniques to distinguish individual samples.43 Another challenge is that the high levels of albumin and immunoglobulins can mask lower abundant proteins, a problem that may be addressed by technologies that combine nanoparticle protein coronas with liquid chromatography-mass spectrometry.44 An aptamer-based assay recently demonstrated its potential for high-resolution proteomics in AH and vitreous specimens.4 Importantly, mass spectrometry allows the characterization of post-translational modifications (PTMs) of proteins, such as proteolysis,45 glycosylation,46 or phosphorylation,47 which requires specific and challenging enrichment steps that dictate the analytic avenue one might pursue.
Vitreous Liquid Biopsy Proteomics
Proteomic profiling of vitreous liquid biopsies from the human eye captures locally enriched fluid containing proteins secreted from highly specialized retinal cells. They allow the molecular characterization of eye diseases in living humans without the need for a direct tissue biopsy and offer the potential to identify novel diagnostic and therapeutic strategies. Vitreous liquid biopsies have been applied to a variety of retinal diseases including diabetic retinopathy and age-related macular degeneration.
Vitreous Proteomics in Diabetic Retinopathy
Studies using vitreous liquid biopsies in diabetic retinopathy (DR) mainly address proliferative DR (PDR),48 the late stage of the disease, because a vitrectomy is usually not performed in early disease stages. Apart from VEGF, several studies have confirmed the role of inflammatory mediators in PDR that may serve as alternative therapeutic targets, especially for nonresponders to anti-VEGF therapies.49−51 Other investigations demonstrated that the protein levels of VEGF and IL6 in the vitreous correlate with the severity of PDR52 and that the VEGF level after a vitrectomy may represent a predictor of late complications of PDR.53,54 Matrix metalloproteinases, found elevated in the vitreous of PDR patients, are involved in angiogenesis, photoreceptor loss, and breakdown of the blood-retinal barrier, suggesting them as disease biomarkers and potential therapeutic targets for PDR.55,56
Vitreous Proteomics in Age-Related Macular Degeneration
Vitreous proteomics has been widely applied in age-related macular degeneration (AMD) and has significantly advanced our understanding of its pathophysiology.57−59 Several biomarkers have been identified that can be useful to stratify disease stages, including dry and neovascular AMD.60 Multiple studies identified the complement system as a critical pathway in AMD,61,62 which led to the development and recent approval of complement targeting drugs for geographic atrophy.63,64
Vitreous Proteomics in Glaucoma
Thousands of different proteins in the vitreous and AH are released by a variety of specialized cell types in the eye, including retinal cells.4 A study in patients with glaucoma analyzed the proteome of paired retina and vitreous samples and found that about 80% of the more than 4,000 retina proteins were also detected in the vitreous.65 In addition, about one-third of the more than 350 proteins that significantly changed in the glaucomatous retina compared to healthy retina were also differentially expressed in the vitreous of the same patients, highlighting the value of liquid biopsies to capture disease mechanisms in adjacent nonregenerative tissues. Currently, the predominant treatment approach for glaucoma aims to reduce intraocular pressure, but modulating inflammation may also be important to prevent secondary glaucoma and mitigate glaucoma-associated damage.66 The vitreous proteome represents a promising tool to monitor inflammation during therapy.65
Vitreous Proteomics in Uveitis
In patients with uveitis, the vitreous proteome allows the differentiation between uveitis and primary vitreoretinal lymphoma and between disease stages and etiologies, such as sarcoid and tuberculosis-associated uveitis.12,67−70 Vitreous proteomics has further demonstrated its promise for personalized drug repurposing in autoinflammatory disease and has provided molecular evidence that can help to explain why prior conventional therapies have failed (e.g., drug target or drug-sensitive pathways not enriched).1
Vitreous Proteomics in Proliferative Vitreoretinopathy
Several studies have analyzed the vitreous proteome in patients with proliferative vitreoretinopathy (PVR) and provided new insights into the disease mechanisms.57 Among them, one study revealed stage-specific alterations of the vitreous proteome in patients with PVR.71 While cytokines involved in T-cell activation and proteins downstream of mTOR activation were increased in early PVR, cytokines driving monocyte response and stem-cell recruitment were enriched in late stage PVR, indicating that liquid biopsy proteomics can be beneficial to selecting appropriate targeted therapies for patients with PVR.
Vitreous Proteomics in Uveal Melanoma
In patients with uveal melanoma (UM), vitreous liquid biopsies have identified a variety of increased angiogenic and inflammatory proteins, some of them correlating with tumor size and with the presence of immune cell infiltrates.72−74 Despite aggressive therapeutic intervention, almost 50% of UM patients develop metastatic disease and survive less than one year.75 The metastatic risk can be assessed using a gene expression panel with high discriminatory power.76,77 However, this analysis requires a direct surgical biopsy of the tumor, which samples only a tiny fraction of a heterogeneous tumor,78,79 can damage sensitive tissues like the retina, and for clinical reasons, can only be performed once. Recently, it has been demonstrated that vitreous proteomics can assess the metastatic risk of UM patients.13 Compared to direct tumor biopsies, they offer several advantages, such as avoiding direct disruption of the tumor and possibly better reflecting tumor heterogeneity, indicating that liquid biopsies may complement prognostic assessment obtained by direct tumor biopsies.
Aqueous Humor Liquid Biopsy Proteomics
The AH is a highly accessible complex fluid that holds great promise for obtaining proteomic information from the human eye.4 Compared to the vitreous, AH biopsies are even more accessible, require less specialized instrumentation, and allow for serial sample collection in the outpatient setting. However, proteomic analysis of AH has been challenged by its low volume and low protein concentration, which is about 400 times lower than in plasma.80−82 We recently demonstrated that a DNA aptamer-based assay allows the detection of nearly 6,000 different proteins from all known ocular cell types, including retinal cells, in only 50 μL of AH. This proteomic resolution represents a significant improvement over prior lower resolution proteomics techniques, including LC-MS.4 Our analysis further revealed that almost 90% of vitreous proteins were also detected in the AH, indicating a substantial protein exchange between both compartments despite enzymatic and anatomic barriers. These findings indicate that AH proteomics represents a promising tool for diagnostic and prognostic assessment as well as for the monitoring of therapeutic response in living humans.
Aqueous Humor Proteomics in Diabetic Retinopathy
Several studies have investigated the proteome of AH specimens obtained from patients with diabetic retinopathy (DR), revealing new insights into DR’s pathophysiology and therapeutic response.48 In patients with proliferative DR (PDR), liquid chromatography-mass spectrometry (LC-MS) identified between almost 600 and 900 different proteins in the AH,83,84 including 30 proteins that significantly changed after intravitreal anti-VEGF therapy.83 An aptamer-based assay (SomaScan) recently demonstrated a much higher proteomic resolution in DR AH and TEMPO revealed that the cellular drivers of DR switched with disease stages from vascular cells in the early stage to immune cells in the late stage.4 Another critical advantage of analyzing AH is that it can be obtained from patients with early disease stages, e.g., during cataract surgery, where a vitrectomy is not indicated.4
Aqueous Humor Proteomics in Age-Related Macular Degeneration
In patients with AMD, various studies have used AH proteomics to investigate the molecular disease mechanisms in living patients.59 One study found that in 122 eyes with neovascular AMD, the AH proteome can predict therapeutic response to anti-VEGF therapy, whereas the VEGF levels alone failed to do so.2 One of the predictive proteins was ApoB100 that was found elevated in the AH from patients with a better therapeutic response. A mouse model with increased ApoB100 expression demonstrated smaller CNV lesions, confirming a protective role of ApoB100 in neovascular AMD. These findings highlight the value of liquid biopsy proteomics in identifying prognostic biomarkers and uncovering novel disease mechanisms with potential therapeutic translation.
Aqueous Humor Proteomics in Glaucoma
One study looking at patients with normal tension glaucoma found signs of neuroinflammation in the AH and further identified AH proteins that were correlated with functional and structural parameters, including visual field defects and OCT changes.85 Another study revealed that preoperative cytokine levels in the AH were associated with surgical failure and reduction of intraocular pressure one year after surgery,86 indicating that AH liquid biopsies may have prognostic value in glaucoma.
Aqueous Humor Proteomics in Uveitis
Diagnostic differentiation of infectious and noninfectious uveitis often represents a clinical challenge with important consequences for therapeutic decisions. A recent study applied an aptamer-based proteomic assay and identified thousands of AH proteins in patients with uveitis.87 Comparing the proteomic profiles of infectious and noninfectious uveitis revealed protein signatures that significantly differed between both etiologies. In contrast, the plasma proteome of the same patients did not reveal significant differences between both groups, further highlighting the value of liquid biopsies from locally enriched fluids, such as the AH compared to the blood.
Aqueous Humor Proteomics in Uveal Melanoma
Similar to vitreous liquid biopsies, recent studies demonstrated that the AH proteome is valuable to distinguish between low and high metastatic risk of patients with uveal melanoma.2,88,89 AH liquid biopsies are even more accessible, can be performed in a variety of clinical settings including outpatient clinics, and are repeatable allowing for individualized surveillance strategies after tumor treatment.74
Aqueous Humor Proteomics in Retinoblastoma
Direct tumor biopsies are considered contraindicated in patients with retinoblastoma due to the risk of metastatic spread. In contrast, AH liquid biopsies may represent a promising and safe alternative to capture molecular information from the tumor.90−92 A recent study analyzed the AH proteome from almost 50 retinoblastoma patients and revealed the molecular profile of different disease stages.93 Apart from molecular tumor analyses, AH liquid biopsies may provide the opportunity to monitor tumor dynamics in patients undergoing therapy and may guide personalized therapeutic management.
Tear Proteomics
Tear fluid represents a promising sample type for proteomics analyses. Several studies using tear fluid have provided new insights into common ocular surface and corneal diseases, such as dry eye disease,94,95 Sjogren’s syndrome,31 keratoconus,96 and inflammatory orbital disorders.97 Due to the high accessibility of tear fluid, tear proteomics represents a promising tool for molecular characterization of ocular surface disease that could lead to personalized therapies and the monitoring of therapeutic response. The low volume of only a few microliters represents an important challenge in tear proteomics studies, and new methods are continuously developing.
One emerging method is the isolation and analysis of exosomes from tear samples. Cells continuously release tiny vesicles known as exosomes into tears and contain a wealth of proteins, RNA, DNA, metabolic compounds, liposomes, and other biomolecules from the source cells. These exosomes hold remarkable promise for research and applications in diagnosing and treating diseases. In contrast to blood samples, tears offer numerous advantages, as they are less impure, convenient, and noninvasive, rendering them an ideal source of exosome samples. However, due to the limited volume, achieving high recovery rates of highly pure exosomes from small tear quantities remains a significant challenge. Recently, the method EXODUS was developed, which is an entirely automated system that swiftly isolates exosomes and extracellular vesicles (EVs) of various sizes with elevated yield and purity from just a few drops of tears (about 10 μL) within a mere 5 min.98 Remarkably, about 1,800 different proteins were identified in medium-sized EVs (ranging from 100 to 200 nm) in tears, including proteins related to maintaining retinal homeostasis and regulating inflammation.99 Through a comprehensive analysis, in conjunction with the Human Protein Atlas consensus data set, the sources of tear EV proteins across 37 tissues and 79 cell types were identified within the EV subtypes, with a marked selective enrichment of proteins associated with retinal neuronal cells, glial cells, blood cells, and immune cells. This opens the door for eye researchers to delve into the study of diseases such as uveitis, optic neuritis, diabetic retinopathy, macular disease, and ocular tumors. Moreover, tear exosomes can be leveraged to investigate changes associated with emotions, stress, and depression. This comprehensive exploration of the biological information and molecular functions of tear exosomes, while quantifying their molecular indicators, can unveil the pathogenesis and development of diseases, thereby refining precise molecular diagnostic strategies.
Conclusions
High-resolution proteomic profiling allows for the identification of thousands of different proteins in microvolume fluid samples from the human eye, holding considerable promise not only for translational research but also for diagnostics, prognostics, and personalized therapies. The integration of liquid biopsy proteomics with single cell transcriptomics of surrounding cells enables cell level analyses in nonregenerative tissues like the retina, representing a significant technical advancement in the study of cellular disease and aging mechanisms in living patients.
Challenges and Future Directions
Each year, millions of intraocular surgeries are performed worldwide, in which a small amount of fluid is removed from the eye and typically discarded. Collecting these samples holds great potential, but it remains a critical challenge to establish an efficient interface between the clinic and the laboratory to collect, annotate, and store high-quality specimens.18 Analyzing samples on a larger scale will allow for better differentiation of disease stages, better identification of prognostic protein patterns, and better understanding of the effect of specific mutations, sex, and ethnicity. Another critical challenge is proteomic resolution. Although the ability of the aptamer-based assay to measure more than 6,000 different proteins in AH is a significant improvement over prior lower resolution proteomics techniques, it still only represents a fraction of the total proteome, and many eye-specific proteins are not represented on the array. Increasing the number of proteins, including an improved representation for specific organs, will further enhance the approach. The integration with cell level transcriptomics from mainly healthy eyes proved its potential. However, generating and integrating more gene expression data from (post-mortem) diseased eyes may further refine TEMPO and its ability to assess cellular mechanisms in diseased eyes in living patients. Another critical question that needs to be addressed in future studies is how well animal models reflect human pathophysiology. Animal models are essential in modeling the complexity of human pathology, which in vitro models still fail to accomplish. However, they may not reflect the entire complexity of a human disease. A fundamental question will be which specific molecular aspects of human disease are well reflected by each model. In addressing this question, comparing high-resolution human omics data with the molecular profile of the animal model can provide valuable insights when developing novel therapeutic strategies.100,200
Among the proteomic methods discussed, it is remarkable that liquid biopsies from the vitreous, AH, and even tears can measure proteins originally expressed in the retina. Unexpectedly, these proteins must traverse several anatomic and enzymatic barriers. Fluid from each compartment may have some advantages over another. We found, for example, that the AH did not detect some 700 proteins that were detectable in the vitreous on the Somalogic platform.4 So, in retinal diseases, the vitreous may be the optimal compartment to capture the most proteins in a discovery experiment. On the other hand, fluid collection from the AH and, especially, tears is less invasive and more likely to be performed sequentially on the same patient, say before and after a therapy. If the emphasis is on studying the time course of more specific molecules and addressing specific proteins, tear and AH proteomics may be preferable. Another advantage of analyzing AH or tears is that samples can be obtained from patients with early disease stages, such as nonproliferative DR, early AMD, or glaucoma, where a vitrectomy may not be indicated. Cross-platform comparisons can provide insights into how a single biopsy performs and what the differences are based on biopsy type.101,102 Altogether, the current technologies make it possible to address these questions, and investigators have a menu of strategies from which to choose.
Acknowledgments
We thank Alton Szeto MFA for help with graphical illustrations and MaryAnn Mahajan BA for editorial assistance. Selected illustrations were created with BioRender.com. V.B.M. is supported by NIH grants (R01EY031952, R01EY031360, R01EY030151, and P30EY026877), the Stanford Center for Optic Disc Drusen, and Research to Prevent Blindness, New York, New York. P.M. and V.B.M. are supported by The Alan and Irene Adler Ocular Cancer Research Initiative. J.W. is supported by the Translational Research and Applied Medicine (TRAM) program at Stanford University and by the VitreoRetinal Surgery Foundation. For this manuscript, A.G.B. is supported by NIH grants R01EY030151 and R01EY031952. Role of the sponsor: the funding organizations had no role in design and conduct of the study; preparation, review, or approval of the paper; and decision to submit the paper for publication.
Author Contributions
V.B.M. takes responsibility for the integrity and the accuracy of the presented paper. J.W. conceived and wrote the paper with significant input from A.D., V.B.M., M.Z.D., J.F., and R.Y. F.L. contributed sections about tear exosomes. J.W., A.D., M.Z.D., R.Y., and J.F. designed and generated the figures. All authors critically revised the paper and agree with its final form.
The authors declare the following competing financial interest(s): V.B.M. has received speaker fees from Somalogic, Inc. The authors are not aware of any other affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Special Issue
Published as part of Journal of Proteome Researchvirtual special issue “Canadian Proteomics”.
References
- Velez G.; Bassuk A. G.; Colgan D.; Tsang S. H.; Mahajan V. B. Therapeutic drug repositioning using personalized proteomics of liquid biopsies. JCI Insight 2017, 2 (24), e97818 10.1172/jci.insight.97818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X.; Sanchez J. C.; Dinabandhu A.; Guo C.; Patel T. P.; Yang Z.; Hu M. W.; Chen L.; Wang Y.; Malik D. Aqueous proteins help predict the response of patients with neovascular age-related macular degeneration to anti-VEGF therapy. J. Clin Invest 2022, 132 (2), e144469 10.1172/JCI144469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udaondo P.; Hernandez C.; Brianso-Llort L.; Garcia-Delpech S.; Simo-Servat O.; Simo R. Usefulness of Liquid Biopsy Biomarkers from Aqueous Humor in Predicting Anti-VEGF Response in Diabetic Macular Edema: Results of a Pilot Study. J. Clin Med. 2019, 8 (11), 1841. 10.3390/jcm8111841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf J.; Rasmussen D. K.; Sun Y. J.; Vu J. T.; Wang E.; Espinosa C.; Bigini F.; Chang R. T.; Montague A. A.; Tang P. H.; et al. Liquid-biopsy proteomics combined with AI identifies cellular drivers of eye aging and disease in vivo. Cell 2023, 186 (22), 4868–4884. 10.1016/j.cell.2023.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wecker T.; Grundel B.; Reichl S.; Stech M.; Lange C.; Agostini H.; Bohringer D.; Stahl A. Anti-VEGF injection frequency correlates with visual acuity outcomes in pro re nata neovascular AMD treatment. Sci. Rep 2019, 9 (1), 3301. 10.1038/s41598-019-38934-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.; Zhao J.; Sun X. Resistance to anti-VEGF therapy in neovascular age-related macular degeneration: a comprehensive review. Drug Des Devel Ther 2016, 10, 1857–1867. 10.2147/DDDT.S97653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn W. C.; Bader J. S.; Braun T. P.; Califano A.; Clemons P. A.; Druker B. J.; Ewald A. J.; Fu H.; Jagu S.; Kemp C. J.; et al. An expanded universe of cancer targets. Cell 2021, 184 (5), 1142–1155. 10.1016/j.cell.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S.; Albrechtsen R.; Kronqvist P.; Cox J.; Mann M.; Geiger T. Proteomic maps of breast cancer subtypes. Nat. Commun. 2016, 7, 10259. 10.1038/ncomms10259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovich G.; Agmon H.; Harel M.; Sonnenblick A.; Peretz T.; Geiger T. Clinical Proteomics of Breast Cancer Reveals a Novel Layer of Breast Cancer Classification. Cancer Res. 2018, 78 (20), 6001–6010. 10.1158/0008-5472.CAN-18-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel M.; Ortenberg R.; Varanasi S. K.; Mangalhara K. C.; Mardamshina M.; Markovits E.; Baruch E. N.; Tripple V.; Arama-Chayoth M.; Greenberg E.; et al. Proteomics of Melanoma Response to Immunotherapy Reveals Mitochondrial Dependence. Cell 2019, 179 (1), 236–250. 10.1016/j.cell.2019.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omenn G. S.; Lane L.; Overall C. M.; Pineau C.; Packer N. H.; Cristea I. M.; Lindskog C.; Weintraub S. T.; Orchard S.; Roehrl M. H. A.; et al. The 2022 Report on the Human Proteome from the HUPO Human Proteome Project. J. Proteome Res. 2023, 22 (4), 1024–1042. 10.1021/acs.jproteome.2c00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velez G.; Yang J.; Li A. S.; Tsang S. H.; Bassuk A. G.; Mahajan V. B. Proteomic insight into the pathogenesis of CAPN5-vitreoretinopathy. Sci. Rep 2019, 9 (1), 7608. 10.1038/s41598-019-44031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velez G.; Nguyen H. V.; Chemudupati T.; Ludwig C. A.; Toral M.; Reddy S.; Mruthyunjaya P.; Mahajan V. B. Liquid biopsy proteomics of uveal melanoma reveals biomarkers associated with metastatic risk. Mol. Cancer 2021, 20 (1), 39. 10.1186/s12943-021-01336-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M.; Fagerberg L.; Hallstrom B. M.; Lindskog C.; Oksvold P.; Mardinoglu A.; Sivertsson A.; Kampf C.; Sjostedt E.; Asplund A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347 (6220), 1260419. 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Wolf J.; Boneva S.; Schlecht A.; Lapp T.; Auw-Haedrich C.; Lagreze W.; Agostini H.; Reinhard T.; Schlunck G.; Lange C. The Human Eye Transcriptome Atlas: A searchable comparative transcriptome database for healthy and diseased human eye tissue. Genomics 2022, 114 (2), 110286. 10.1016/j.ygeno.2022.110286. [DOI] [PubMed] [Google Scholar]
- Voigt A. P.; Whitmore S. S.; Lessing N. D.; DeLuca A. P.; Tucker B. A.; Stone E. M.; Mullins R. F.; Scheetz T. E. Spectacle: An interactive resource for ocular single-cell RNA sequencing data analysis. Exp. Eye Res. 2020, 200, 108204. 10.1016/j.exer.2020.108204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell B. L.; Yasui Y.; Li C. I.; Fitzpatrick A. L.; Lampe P. D. Impact of freeze-thaw cycles and storage time on plasma samples used in mass spectrometry based biomarker discovery projects. Cancer Inform 2005, 1 (1), 98–104. 10.1177/117693510500100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf J.; Chemudupati T.; Kumar A.; Rasmussen D. K.; Wai K. M.; Chang R. T.; Montague A. A.; Tang P. H.; Bassuk A. G.; Dufour A.; et al. Biobanking of Human Aqueous and Vitreous Liquid Biopsies for Molecular Analyses. J. Vis Exp 2023, ( (199), ). 10.3791/65804. [DOI] [PubMed] [Google Scholar]
- Skeie J. M.; Tsang S. H.; Zande R. V.; Fickbohm M. M.; Shah S. S.; Vallone J. G.; Mahajan V. B. A biorepository for ophthalmic surgical specimens. Proteomics Clin Appl. 2014, 8 (3–4), 209–217. 10.1002/prca.201300043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Lelij A.; Rothova A. Diagnostic anterior chamber paracentesis in uveitis: a safe procedure?. Br J. Ophthalmol 1997, 81 (11), 976–979. 10.1136/bjo.81.11.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung C. M.; Durrani O. M.; Murray P. I. The safety of anterior chamber paracentesis in patients with uveitis. Br J. Ophthalmol 2004, 88 (4), 582–583. 10.1136/bjo.2003.027219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi D.; Denniston A. K.; Murray P. I. Safety profile of anterior chamber paracentesis performed at the slit lamp. Clin Exp Ophthalmol 2011, 39 (8), 725–728. 10.1111/j.1442-9071.2011.02565.x. [DOI] [PubMed] [Google Scholar]
- Kitazawa K.; Sotozono C.; Koizumi N.; Nagata K.; Inatomi T.; Sasaki H.; Kinoshita S. Safety of anterior chamber paracentesis using a 30-gauge needle integrated with a specially designed disposable pipette. Br J. Ophthalmol 2017, 101 (5), 548–550. 10.1136/bjophthalmol-2016-309650. [DOI] [PubMed] [Google Scholar]
- Rossi T.; Romano M. R.; Iannetta D.; Romano V.; Gualdi L.; D’Agostino I.; Ripandelli G. Cataract surgery practice patterns worldwide: a survey. BMJ. Open Ophthalmol 2021, 6 (1), e000464 10.1136/bmjophth-2020-000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf J.; Chemudupati T.; Kumar A.; Franco J. A.; Montague A. A.; Lin C. C.; Lee W.-S.; Fisher A. C.; Goldberg J. L.; Mruthyunjaya P.; et al. Using electronic health record data to determine the safety of aqueous humor liquid biopsies for molecular analyses. medRxiv 2023, 2023.2011.2022.23298937. 10.1101/2023.11.22.23298937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra K.; Velez G.; Chemudupati T.; Tang P. H.; Mruthyunjaya P.; Sanislo S. R.; Mahajan V. B. Intraoperative Complications With Vitreous Biopsy for Molecular Proteomics. Ophthalmic Surg Lasers Imaging Retina 2023, 54 (1), 32–36. 10.3928/23258160-20221214-02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nattinen J.; Aapola U.; Nukareddy P.; Uusitalo H. Clinical Tear Fluid Proteomics-A Novel Tool in Glaucoma Research. Int. J. Mol. Sci. 2022, 23 (15), 8136. 10.3390/ijms23158136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieczyński J.; Szulc U.; Harazna J.; Szulc A.; Kiewisz J. Tear fluid collection methods: Review of current techniques. Eur. J. Ophthalmol 2021, 31 (5), 2245–2251. 10.1177/1120672121998922. [DOI] [PubMed] [Google Scholar]
- Bachhuber F.; Huss A.; Senel M.; Tumani H. Diagnostic biomarkers in tear fluid: from sampling to preanalytical processing. Sci. Rep 2021, 11 (1), 10064. 10.1038/s41598-021-89514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijs M.; Arumugam S.; van de Sande N.; Webers C. A. B.; Sethu S.; Ghosh A.; Shetty R.; Vehof J.; Nuijts R. M. M. A. Pre-analytical sample handling effects on tear fluid protein levels. Sci. Rep 2023, 13 (1), 1317. 10.1038/s41598-023-28363-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das N.; Menon N. G.; de Almeida L. G. N.; Woods P. S.; Heynen M. L.; Jay G. D.; Caffery B.; Jones L.; Krawetz R.; Schmidt T. A.; et al. Proteomics Analysis of Tears and Saliva From Sjogren’s Syndrome Patients. Front Pharmacol 2021, 12, 787193. 10.3389/fphar.2021.787193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam B.; Basit M.; Nisar M. A.; Khurshid M.; Rasool M. H. Proteomics: Technologies and Their Applications. J. Chromatogr Sci. 2017, 55 (2), 182–196. 10.1093/chromsci/bmw167. [DOI] [PubMed] [Google Scholar]
- Al-Amrani S.; Al-Jabri Z.; Al-Zaabi A.; Alshekaili J.; Al-Khabori M. Proteomics: Concepts and applications in human medicine. World J. Biol. Chem. 2021, 12 (5), 57–69. 10.4331/wjbc.v12.i5.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrecque A. A.; Roy S.; Young D.; Chen S. A.; Brechenmacher L.; Le D.; Cooper S.; Gibson P.; Clarke A. E.; Dufour A.; et al. Romiplostim drug presence in pregnancy and lactation. Blood 2023, 141 (20), 2537–2540. 10.1182/blood.2022019343. [DOI] [PubMed] [Google Scholar]
- Mainoli B.; Hirota S.; Edgington-Mitchell L. E.; Lu C.; Dufour A. Proteomics and Imaging in Crohn’s Disease: TAILS of Unlikely Allies. Trends Pharmacol. Sci. 2020, 41 (2), 74–84. 10.1016/j.tips.2019.11.008. [DOI] [PubMed] [Google Scholar]
- Pettersen V. K.; Antunes L. C. M.; Dufour A.; Arrieta M. C. Inferring early-life host and microbiome functions by mass spectrometry-based metaproteomics and metabolomics. Comput. Struct Biotechnol J. 2022, 20, 274–286. 10.1016/j.csbj.2021.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor Z.; Ahn S. B.; Baker M. S.; Ranganathan S.; Mohamedali A. Mass spectrometry-based protein identification in proteomics-a review. Brief Bioinform 2021, 22 (2), 1620–1638. 10.1093/bib/bbz163. [DOI] [PubMed] [Google Scholar]
- Brody E. N.; Gold L.; Lawn R. M.; Walker J. J.; Zichi D. High-content affinity-based proteomics: unlocking protein biomarker discovery. Expert Rev. Mol. Diagn 2010, 10 (8), 1013–1022. 10.1586/erm.10.89. [DOI] [PubMed] [Google Scholar]
- Ahsan H. Monoplex and multiplex immunoassays: approval, advancements, and alternatives. Comp Clin Path 2022, 31 (2), 333–345. 10.1007/s00580-021-03302-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Moujahed A.; Velez G.; Vu J. T.; Lima de Carvalho J. R.; Levi S. R.; Bassuk A. G.; Sepah Y. J.; Tsang S. H.; Mahajan V. B. Proteomic analysis of autoimmune retinopathy implicates NrCAM as a potential biomarker. Ophthalmol Sci. 2022, 2 (2), 100131. 10.1016/j.xops.2022.100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tighe P. J.; Ryder R. R.; Todd I.; Fairclough L. C. ELISA in the multiplex era: potentials and pitfalls. Proteomics Clin Appl. 2015, 9 (3–4), 406–422. 10.1002/prca.201400130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmet C.; Blum L. J.; Marquette C. A. Multiplex microarray ELISA versus classical ELISA, a comparison study of pollutant sensing for environmental analysis. Environ. Sci. Process Impacts 2013, 15 (10), 1876–1882. 10.1039/c3em00296a. [DOI] [PubMed] [Google Scholar]
- Trzeciecka A.; Pattabiraman P.; Piqueras M. C.; Toris C.; Bhattacharya S. K. Quantitative Proteomic Analysis of Human Aqueous Humor Using iTRAQ 4plex Labeling. Methods Mol. Biol. 2018, 1695, 89–95. 10.1007/978-1-4939-7407-8_9. [DOI] [PubMed] [Google Scholar]
- Blume J. E.; Manning W. C.; Troiano G.; Hornburg D.; Figa M.; Hesterberg L.; Platt T. L.; Zhao X.; Cuaresma R. A.; Everley P. A.; et al. Rapid, deep and precise profiling of the plasma proteome with multi-nanoparticle protein corona. Nat. Commun. 2020, 11 (1), 3662. 10.1038/s41467-020-17033-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Main K.; Wang H.; Julien O.; Dufour A. Biochemical Tools for Tracking Proteolysis. J. Proteome Res. 2021, 20 (12), 5264–5279. 10.1021/acs.jproteome.1c00289. [DOI] [PubMed] [Google Scholar]
- De Leoz M. L. A.; Duewer D. L.; Fung A.; Liu L.; Yau H. K.; Potter O.; Staples G. O.; Furuki K.; Frenkel R.; Hu Y.; et al. NIST Interlaboratory Study on Glycosylation Analysis of Monoclonal Antibodies: Comparison of Results from Diverse Analytical Methods. Mol. Cell Proteomics 2020, 19 (1), 11–30. 10.1074/mcp.RA119.001677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G.; Chen L.; Zhang B.; Song Q.; Yu Y.; Moore C.; Wang T. L.; Shih I. M.; Zhang H.; Chan D. W.; et al. Proteome-wide Tyrosine Phosphorylation Analysis Reveals Dysregulated Signaling Pathways in Ovarian Tumors. Mol. Cell Proteomics 2019, 18 (3), 448–460. 10.1074/mcp.RA118.000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midena E.; Frizziero L.; Midena G.; Pilotto E. Intraocular fluid biomarkers (liquid biopsy) in human diabetic retinopathy. Graefes Arch Clin Exp Ophthalmol 2021, 259 (12), 3549–3560. 10.1007/s00417-021-05285-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs K.; Marra K. V.; Yu G.; Wagley S.; Ma J.; Teague G. C.; Nandakumar N.; Lashkari K.; Arroyo J. G. Angiogenic and Inflammatory Vitreous Biomarkers Associated With Increasing Levels of Retinal Ischemia. Invest Ophthalmol Vis Sci. 2015, 56 (11), 6523–6530. 10.1167/iovs.15-16793. [DOI] [PubMed] [Google Scholar]
- Boss J. D.; Singh P. K.; Pandya H. K.; Tosi J.; Kim C.; Tewari A.; Juzych M. S.; Abrams G. W.; Kumar A. Assessment of Neurotrophins and Inflammatory Mediators in Vitreous of Patients With Diabetic Retinopathy. Invest Ophthalmol Vis Sci. 2017, 58 (12), 5594–5603. 10.1167/iovs.17-21973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Lu Q.; Lu P. Quantitative proteomics analysis of vitreous body from type 2 diabetic patients with proliferative diabetic retinopathy. BMC Ophthalmol 2018, 18 (1), 151. 10.1186/s12886-018-0821-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funatsu H.; Yamashita H.; Noma H.; Mimura T.; Nakamura S.; Sakata K.; Hori S. Aqueous humor levels of cytokines are related to vitreous levels and progression of diabetic retinopathy in diabetic patients. Graefes Arch Clin Exp Ophthalmol 2005, 243 (1), 3–8. 10.1007/s00417-004-0950-7. [DOI] [PubMed] [Google Scholar]
- Suzuki Y.; Suzuki K.; Kudo T.; Metoki T.; Nakazawa M. Level of Vascular Endothelial Growth Factor in the Vitreous Fluid of Proliferative Diabetic Retinopathy Patients and Prognosis after Vitrectomy. Ophthalmologica 2016, 236 (3), 133–138. 10.1159/000449261. [DOI] [PubMed] [Google Scholar]
- Petrovič M. G.; Korošec P.; Košnik M.; Hawlina M. Association of preoperative vitreous IL-8 and VEGF levels with visual acuity after vitrectomy in proliferative diabetic retinopathy. Acta Ophthalmol. 2010, 88 (8), e311–e316. 10.1111/j.1755-3768.2010.02030.x. [DOI] [PubMed] [Google Scholar]
- Opdenakker G.; Abu El-Asrar A. Metalloproteinases mediate diabetes-induced retinal neuropathy and vasculopathy. Cell. Mol. Life Sci. 2019, 76 (16), 3157–3166. 10.1007/s00018-019-03177-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu El-Asrar A. M.; Mohammad G.; Nawaz M. I.; Siddiquei M. M.; Van den Eynde K.; Mousa A.; De Hertogh G.; Opdenakker G. Relationship between vitreous levels of matrix metalloproteinases and vascular endothelial growth factor in proliferative diabetic retinopathy. PLoS One 2013, 8 (12), e85857 10.1371/journal.pone.0085857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos F. M.; Ciordia S.; Mesquita J.; de Sousa J. P. C.; Paradela A.; Tomaz C. T.; Passarinha L. A. P. Vitreous humor proteome: unraveling the molecular mechanisms underlying proliferative and neovascular vitreoretinal diseases. Cell. Mol. Life Sci. 2023, 80 (1), 22. 10.1007/s00018-022-04670-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F. M.; Mesquita J.; Castro-de-Sousa J. P.; Ciordia S.; Paradela A.; Tomaz C. T. Vitreous Humor Proteome: Targeting Oxidative Stress, Inflammation, and Neurodegeneration in Vitreoretinal Diseases. Antioxidants (Basel) 2022, 11 (3), 505. 10.3390/antiox11030505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten E.; Paun C. C.; Schellevis R. L.; Hoyng C. B.; Delcourt C.; Lengyel I.; Peto T.; Ueffing M.; Klaver C. C. W.; Dammeier S.; et al. Systemic and ocular fluid compounds as potential biomarkers in age-related macular degeneration. Surv Ophthalmol 2018, 63 (1), 9–39. 10.1016/j.survophthal.2017.05.003. [DOI] [PubMed] [Google Scholar]
- Schori C.; Trachsel C.; Grossmann J.; Zygoula I.; Barthelmes D.; Grimm C. The Proteomic Landscape in the Vitreous of Patients With Age-Related and Diabetic Retinal Disease. Invest Ophthalmol Vis Sci. 2018, 59 (4), AMD31–AMD40. 10.1167/iovs.18-24122. [DOI] [PubMed] [Google Scholar]
- Santos F. M.; Ciordia S.; Mesquita J.; Cruz C.; Sousa J.; Passarinha L. A.; Tomaz C. T.; Paradela A. Proteomics profiling of vitreous humor reveals complement and coagulation components, adhesion factors, and neurodegeneration markers as discriminatory biomarkers of vitreoretinal eye diseases. Front Immunol 2023, 14, 1107295. 10.3389/fimmu.2023.1107295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koss M. J.; Hoffmann J.; Nguyen N.; Pfister M.; Mischak H.; Mullen W.; Husi H.; Rejdak R.; Koch F.; Jankowski J.; et al. Proteomics of vitreous humor of patients with exudative age-related macular degeneration. PLoS One 2014, 9 (5), e96895 10.1371/journal.pone.0096895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe G. J.; Westby K.; Csaky K. G.; Mones J.; Pearlman J. A.; Patel S. S.; Joondeph B. C.; Randolph J.; Masonson H.; Rezaei K. A. C5 Inhibitor Avacincaptad Pegol for Geographic Atrophy Due to Age-Related Macular Degeneration: A Randomized Pivotal Phase 2/3 Trial. Ophthalmology 2021, 128 (4), 576–586. 10.1016/j.ophtha.2020.08.027. [DOI] [PubMed] [Google Scholar]
- Liao D. S.; Grossi F. V.; El Mehdi D.; Gerber M. R.; Brown D. M.; Heier J. S.; Wykoff C. C.; Singerman L. J.; Abraham P.; Grassmann F.; et al. Complement C3 Inhibitor Pegcetacoplan for Geographic Atrophy Secondary to Age-Related Macular Degeneration: A Randomized Phase 2 Trial. Ophthalmology 2020, 127 (2), 186–195. 10.1016/j.ophtha.2019.07.011. [DOI] [PubMed] [Google Scholar]
- Mirzaei M.; Gupta V. B.; Chick J. M.; Greco T. M.; Wu Y.; Chitranshi N.; Wall R. V.; Hone E.; Deng L.; Dheer Y.; et al. Age-related neurodegenerative disease associated pathways identified in retinal and vitreous proteome from human glaucoma eyes. Sci. Rep 2017, 7 (1), 12685. 10.1038/s41598-017-12858-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudouin C.; Kolko M.; Melik-Parsadaniantz S.; Messmer E. M. Inflammation in Glaucoma: From the back to the front of the eye, and beyond. Prog. Retin Eye Res. 2021, 83, 100916. 10.1016/j.preteyeres.2020.100916. [DOI] [PubMed] [Google Scholar]
- Schrijver B.; Kolijn P. M.; Ten Berge J. C. E. M.; Nagtzaam N. M. A.; van Rijswijk A. L. C. T.; Swagemakers S. M. A.; van der Spek P. J.; Missotten T. O. A. R.; van Velthoven M. E. J.; de Hoog J.; et al. Vitreous proteomics, a gateway to improved understanding and stratification of diverse uveitis aetiologies. Acta Ophthalmol 2022, 100 (4), 403–413. 10.1111/aos.14993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu H.; Usui Y.; Tsubota K.; Fujii R.; Yamaguchi T.; Maruyama K.; Wakita R.; Asakage M.; Shimizu H.; Yamakawa N.; et al. Comprehensive Proteomic Profiling of Vitreous Humor in Ocular Sarcoidosis Compared with Other Vitreoretinal Diseases. J. Clin Med. 2022, 11 (13), 3606. 10.3390/jcm11133606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velez G.; Roybal C. N.; Colgan D.; Tsang S. H.; Bassuk A. G.; Mahajan V. B. Precision Medicine: Personalized Proteomics for the Diagnosis and Treatment of Idiopathic Inflammatory Disease. JAMA Ophthalmol 2016, 134 (4), 444–448. 10.1001/jamaophthalmol.2015.5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepah Y. J.; Velez G.; Tang P. H.; Yang J.; Chemudupati T.; Li A. S.; Nguyen Q. D.; Bassuk A. G.; Mahajan V. B. Proteomic analysis of intermediate uveitis suggests myeloid cell recruitment and implicates IL-23 as a therapeutic target. Am. J. Ophthalmol Case Rep 2020, 18, 100646. 10.1016/j.ajoc.2020.100646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roybal C. N.; Velez G.; Toral M. A.; Tsang S. H.; Bassuk A. G.; Mahajan V. B. Personalized Proteomics in Proliferative Vitreoretinopathy Implicate Hematopoietic Cell Recruitment and mTOR as a Therapeutic Target. Am. J. Ophthalmol 2018, 186, 152–163. 10.1016/j.ajo.2017.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarkatti-Gude N.; Bronkhorst I. H.; van Duinen S. G.; Luyten G. P.; Jager M. J. Cytokines and chemokines in the vitreous fluid of eyes with uveal melanoma. Invest Ophthalmol Vis Sci. 2012, 53 (11), 6748–6755. 10.1167/iovs.12-10123. [DOI] [PubMed] [Google Scholar]
- Dunavoelgyi R.; Funk M.; Sacu S.; Georgopoulos M.; Zlabinger G.; Zehetmayer M.; Schmidt-Erfurth U. Intraocular activation of angiogenic and inflammatory pathways in uveal melanoma. Retina 2012, 32 (7), 1373–1384. 10.1097/IAE.0b013e318239e299. [DOI] [PubMed] [Google Scholar]
- Heiferman M. J.; Mahajan V. B.; Mruthyunjaya P. Proteomics in uveal melanoma. Curr. Opin Ophthalmol 2022, 33 (3), 202–210. 10.1097/ICU.0000000000000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliki S.; Shields C. L. Uveal melanoma: relatively rare but deadly cancer. Eye (Lond) 2017, 31 (2), 241–257. 10.1038/eye.2016.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onken M. D.; Worley L. A.; Char D. H.; Augsburger J. J.; Correa Z. M.; Nudleman E.; Aaberg T. M. Jr; Altaweel M. M.; Bardenstein D. S.; Finger P. T.; et al. Collaborative Ocular Oncology Group report number 1: prospective validation of a multi-gene prognostic assay in uveal melanoma. Ophthalmology 2012, 119 (8), 1596–1603. 10.1016/j.ophtha.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L.; Paez-Escamilla M.; Walter S. D.; Tarlan B.; Decatur C. L.; Perez B. M.; Harbour J. W. Gene Expression Profiling and PRAME Status Versus Tumor-Node-Metastasis Staging for Prognostication in Uveal Melanoma. Am. J. Ophthalmol 2018, 195, 154–160. 10.1016/j.ajo.2018.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez D. A.; Yang J.; Durante M. A.; Shoushtari A. N.; Moschos S. J.; Wrzeszczynski K. O.; Harbour J. W.; Carvajal R. D. Multiregional genetic evolution of metastatic uveal melanoma. NPJ. Genom Med. 2021, 6 (1), 70. 10.1038/s41525-021-00233-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maat W.; Jordanova E. S.; van Zelderen-Bhola S. L.; Barthen E. R.; Wessels H. W.; Schalij-Delfos N. E.; Jager M. J. The heterogeneous distribution of monosomy 3 in uveal melanomas: implications for prognostication based on fine-needle aspiration biopsies. Arch Pathol Lab Med. 2007, 131 (1), 91–96. 10.5858/2007-131-91-THDOMI. [DOI] [PubMed] [Google Scholar]
- Chowdhury U. R.; Madden B. J.; Charlesworth M. C.; Fautsch M. P. Proteome analysis of human aqueous humor. Invest Ophthalmol Vis Sci. 2010, 51 (10), 4921–4931. 10.1167/iovs.10-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen E. T.; Lumi X.; Hansen A. K.; Enghild J. J.; Petrovski G. Protein Composition of the Subretinal Fluid Suggests Selective Diffusion of Vitreous Proteins in Retinal Detachment. Transl Vis Sci. Technol. 2020, 9 (11), 16. 10.1167/tvst.9.11.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeman M.; Choi J.; Hansson S.; Storm M. U.; Nilsson L. Proteins and antibodies in serum, plasma, and whole blood-size characterization using asymmetrical flow field-flow fractionation (AF4). Anal Bioanal Chem. 2018, 410 (20), 4867–4873. 10.1007/s00216-018-1127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T.; Chen H.; Du X.; Li N.; Chen Y.; Min H. Differences in aqueous humor protein profiles in patients with proliferative diabetic retinopathy before and after conbercept treatment. J. Proteomics 2023, 276, 104838. 10.1016/j.jprot.2023.104838. [DOI] [PubMed] [Google Scholar]
- Xiao H.; Xin W.; Sun L. M.; Li S. S.; Zhang T.; Ding X. Y. Comprehensive Proteomic Profiling of Aqueous Humor Proteins in Proliferative Diabetic Retinopathy. Transl Vis Sci. Technol. 2021, 10 (6), 3. 10.1167/tvst.10.6.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H.; Jung J. H.; Park T. K.; Moon C. E.; Han K.; Lee J.; Lee H. K.; Ji Y. W.; Kim C. Y. Proteome alterations in the aqueous humor reflect structural and functional phenotypes in patients with advanced normal-tension glaucoma. Sci. Rep 2022, 12 (1), 1221. 10.1038/s41598-022-05273-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos-Blasco B.; Vidal-Villegas B.; Saenz-Frances F.; Fernandez-Vigo J. I.; Andres-Guerrero V.; Espino L.; Garcia-Feijoo J.; Martinez-de-la-Casa J. M. Cytokine profile in tear and aqueous humor of primary open-angle patients as a prognostic factor for trabeculectomy outcome. Eur. J. Ophthalmol 2022, 32 (5), 2994–3004. 10.1177/11206721211055965. [DOI] [PubMed] [Google Scholar]
- Pessuti C. L.; Medley Q. G.; Li N.; Huang C. L.; Loureiro J.; Banks A.; Zhang Q.; Costa D. F.; Ribeiro K. S.; Nascimento H.; et al. Differential Proteins Expression Distinguished Between Patients With Infectious and Noninfectious Uveitis. Ocul Immunol Inflamm 2023, 1–8. 10.1080/09273948.2022.2150224. [DOI] [PubMed] [Google Scholar]
- Wierenga A. P. A.; Cao J.; Mouthaan H.; van Weeghel C.; Verdijk R. M.; van Duinen S. G.; Kroes W. G. M.; Dogrusöz M.; Marinkovic M.; van der Burg S. S. H.; et al. Aqueous Humor Biomarkers Identify Three Prognostic Groups in Uveal Melanoma. Invest Ophthalmol Vis Sci. 2019, 60 (14), 4740–4747. 10.1167/iovs.19-28309. [DOI] [PubMed] [Google Scholar]
- Peng C. C.; Sirivolu S.; Pike S.; Kim M. E.; Reiser B.; Li H. T.; Liang G.; Xu L.; Berry J. L. Diagnostic Aqueous Humor Proteome Predicts Metastatic Potential in Uveal Melanoma. Int. J. Mol. Sci. 2023, 24 (7), 6825. 10.3390/ijms24076825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. E.; Xu L.; Prabakar R. K.; Shen L.; Peng C. C.; Kuhn P.; Gai X.; Hicks J.; Berry J. L.. Aqueous Humor as a Liquid Biopsy for Retinoblastoma: Clear Corneal Paracentesis and Genomic Analysis. J. Vis Exp 2021, ( (175), ). 10.3791/62939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. T.; Xu L.; Weisenberger D. J.; Li M.; Zhou W.; Peng C. C.; Stachelek K.; Cobrinik D.; Liang G.; Berry J. L. Characterizing DNA methylation signatures of retinoblastoma using aqueous humor liquid biopsy. Nat. Commun. 2022, 13 (1), 5523. 10.1038/s41467-022-33248-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J. L.; Xu L.; Polski A.; Jubran R.; Kuhn P.; Kim J. W.; Hicks J. Aqueous Humor Is Superior to Blood as a Liquid Biopsy for Retinoblastoma. Ophthalmology 2020, 127 (4), 552–554. 10.1016/j.ophtha.2019.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galardi A.; Stathopoulos C.; Colletti M.; Lavarello C.; Russo I.; Cozza R.; Romanzo A.; Carcaboso A. M.; Locatelli F.; Petretto A.; et al. Proteomics of Aqueous Humor as a Source of Disease Biomarkers in Retinoblastoma. Int. J. Mol. Sci. 2022, 23 (21), 13458. 10.3390/ijms232113458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L.; Zhang T.; Ma H.; Pan Y.; Wang S.; Liu X.; Dai X.; Zheng Y.; Lee L. P.; Liu F. Discovering the Secret of Diseases by Incorporated Tear Exosomes Analysis via Rapid-Isolation System: iTEARS. ACS Nano 2022, 16 (8), 11720–11732. 10.1021/acsnano.2c02531. [DOI] [PubMed] [Google Scholar]
- Kannan R.; Das S.; Shetty R.; Zhou L.; Ghosh A.; Deshpande V. Tear proteomics in dry eye disease. Indian J. Ophthalmol 2023, 71 (4), 1203–1214. 10.4103/IJO.IJO_2851_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Lopez M.; Regueiro U.; Bravo S. B.; Chantada-Vazquez M. D. P.; Pena C.; Diez-Feijoo E.; Hervella P.; Lema I. Shotgun Proteomics for the Identification and Profiling of the Tear Proteome of Keratoconus Patients. Invest Ophthalmol Vis Sci. 2022, 63 (5), 12. 10.1167/iovs.63.5.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazaei H.; Khazaei D.; Verma R.; Ng J.; Wilmarth P. A.; David L. L.; Rosenbaum J. T. The potential of tear proteomics for diagnosis and management of orbital inflammatory disorders including Graves’ ophthalmopathy. Exp. Eye Res. 2021, 213, 108813. 10.1016/j.exer.2021.108813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Zhu Q.; Cheng L.; Wang Y.; Li M.; Yang Q.; Hu L.; Lou D.; Li J.; Dong X.; et al. Exosome detection via the ultrafast-isolation system: EXODUS. Nat. Methods 2021, 18 (2), 212–218. 10.1038/s41592-020-01034-x. [DOI] [PubMed] [Google Scholar]
- Hu L.; Liu X.; Zheng Q.; Chen W.; Xu H.; Li H.; Luo J.; Yang R.; Mao X.; Wang S.; et al. Interaction network of extracellular vesicles building universal analysis via eye tears: iNEBULA. Sci. Adv. 2023, 9 (11), eadg1137 10.1126/sciadv.adg1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf J.; Schlecht A.; Rosmus D. D.; Boneva S.; Agostini H.; Schlunck G.; Wieghofer P.; Lange C. Comparative transcriptome analysis of human and murine choroidal neovascularization identifies fibroblast growth factor inducible-14 as phylogenetically conserved mediator of neovascular age-related macular degeneration. Biochim Biophys Acta Mol. Basis Dis 2022, 1868 (4), 166340. 10.1016/j.bbadis.2022.166340. [DOI] [PubMed] [Google Scholar]
- Pauleikhoff L.; Boneva S.; Boeck M.; Schlecht A.; Schlunck G.; Agostini H.; Lange C.; Wolf J. Transcriptional Comparison of Human and Murine Retinal Neovascularization. Invest. Ophthalmol. Vis. Sci. 2023, 64 (15), 46. 10.1167/iovs.64.15.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeie J. M.; Brown E. N.; Martinez H. D.; Russell S. R.; Birkholz E. S.; Folk J. C.; Boldt H. C.; Gehrs K. M.; Stone E. M.; Wright M. E.; et al. Proteomic analysis of vitreous biopsy techniques. Retina 2012, 32 (10), 2141–2149. 10.1097/IAE.0b013e3182562017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velez G.; Wolf J.; Dufour A.; Mruthyunjaya P.; Mahajan V. B. Cross-Platform Identification and Validation of Uveal Melanoma Vitreous Protein Biomarkers. Invest Ophthalmol Vis Sci. 2023, 64 (14), 14. 10.1167/iovs.64.14.14. [DOI] [PMC free article] [PubMed] [Google Scholar]