Abstract
The purpose of this study was to determine if polymerase chain reaction (PCR) could be used to detect FeLV proviral DNA in bone marrow samples of cats with varying suspicion of latent infection. Blood and bone marrow samples from 50 cats and bone marrow from one fetus were collected, including 16 cats with diseases suspected to be FeLV-associated. Serum enzyme-linked immunosorbent assay (ELISA), blood and bone marrow immunofluorescent antibody test (IFA), and blood and bone marrow PCR were performed on each cat, and IFA and PCR on bone marrow of the fetus. Forty-one cats were FeLV negative. Five cats and one fetus were persistently infected with FeLV. Four cats had discordant test results. No cats were positive on bone marrow PCR only. It appears persistent or latent FeLV infection is not always present in conditions classically associated with FeLV.
Latent FeLV infections, in which provirus is present in a non-replicating form in myelomonocytic progenitor cells, are suspected to be associated with diseases such as lymphoma, leukaemia, and cytopenias (Rojko & Hardy 1994, Rojko et al 1982, Jackson et al 1996a). Although such conditions are often called FeLV-associated disease, many affected cats are negative on traditional FeLV tests (Rojko & Hardy 1994). Although FeLV latency may account for this discrepancy, clinical tests capable of diagnosing latent infection are not readily available.
A number of methods are available for detection of FeLV infection. Enzyme-linked immunosorbent assay (ELISA) is the most common testing method used for detecting both transient and persistent FeLV infections. Immunofluorescent antibody test (IFA) detects the presence of FeLV structural antigens in the cytoplasm of FeLV infected leukocytes and platelets. A positive IFA is diagnostic of persistent marrow-origin viremia. Performed correctly, 98% of IFA positive cats are also positive on virus isolation (VI) (Hardy & Zuckerman 1991a, Hardy et al 1973, Hardy & Zuckerman 1991b, Hardy 1991). Rare discordancies between IFA and VI are likely due to early infection, prior to full infection of the marrow.
Polymerase chain reaction (PCR) is a technique for amplifying specific sequences of DNA (Sninsky 1990). Major dissimilarities between endogenous and exogenous FeLV-related sequences are found within the U3 region of the long terminal repeat (LTR) sequence (Rojko & Hardy 1994, Casey et al 1981, Fulton et al 1990). The U3 region is highly conserved among exogenous FeLV isolates, with an overall sequence homology of at least 95% (Fulton et al 1990). These properties make the U3 region an appropriate target for the PCR reaction. Oligonucleotide primers targeting sequences of variable base pair size within the U3 region have successfully identified FeLV proviral DNA in peripheral blood, corneal tissue, and formalin-fixed samples (Herring et al 1998, Jackson et al 1996a, Jackson et al 1993, Jackson et al 1996b, Miyazawa & Jarrett 1997).
The only reported method for detection of latent FeLV infection involves bone marrow culture (Pederson & Meric 1984). Use of polymerase chain reaction (PCR) to detect FeLV proviral DNA in bone marrow, the suspected site of the majority of latent infections, has not been reported. However, PCR is used in both human and veterinary medicine to detect latency with other viral infections (Sninsky 1990, Borchers et al 1999, Egyed & Barth 1998, Miyoshi et al 1999). Specific viral infections in which PCR is used to detect latency include human immunodeficiency virus-1, bovine immunodeficiencylike virus, equine herpes virus-4 (EHV-4), and canine herpes virus (CHV) (Sninsky 1990, Shimazage et al 1994, Borchers et al 1999, Egyed & Barth 1998, Miyoshi et al 1999). If PCR can detect untranscribed FeLV proviral DNA in bone marrow samples, it may provide a sensitive method of detecting latent FeLV infections.
The purpose of this study was to determine if PCR of feline bone marrow samples could be used to detect latent infections in cats with varying suspicion of FeLV infection. Additionally, agreement between test results of PCR and FeLV ELISA and IFA was to be determined.
Materials and methods
Sample collection
Blood and bone marrow samples from 50 cats and bone marrow from one fetus were collected. Eleven of the cats were born and raised in a laboratory facility, tested FeLV negative as kittens, and housed together without contact with other cats for 8 years. Thirteen cats were from a humane society. Ten cats and one fetus were brought to the Veterinary Teaching Hospital (VTH) (n=9 cats, n=1 fetus) or seen at another practice (n=1) as strays. Sixteen cats were evaluated at the VTH (n=15) or other practice (n=1). Age ranged from 2–11 years, with a mean of 5.6 years. These 16 cats had FeLV, or diseases classically associated with FeLV-infection, that were diagnosed by use of haematology, cytological, and histopathological evaluation, and laboratory information using VTH references values. These diseases included granulocytic leukaemia (n=3), lymphocytic leukaemia (n=2), non-regenerative anaemia (n=2), lymphoma (n=3), pancytopenia (n=1), leukopenia (n=1), neutropenia (n=1), haemolytic anaemia with thrombocytopenia (n=1), glomerulonephritis with renal failure (n=1), and thrombocytopenia with tetraparesis (n=1). Experimental design and procedures of this project were approved by the University Animal Care and Use Committee of Virginia Tech.
Blood was collected via jugular venipuncture, and was equally divided between an EDTA tube and a serum separator tube. Laboratory cats and clinical case cats were either sedated with ketamine (Vetamine; Schering Plough, Kenilworth, NJ, USA) (10 mg/kg bodyweight, intravenously —IV) and diazepam (Valium; Elkins-Sinn, Philadelphia, PA, USA) (0.2 mg/kg bodyweight, IV) or were placed under general anaesthesia with thiopental (Pentothal; Abbot Laboratories, Abbot Park, IL, USA) (10 mg/kg bodyweight, IV) for induction and maintained with isoflurane (Isovet; Mallinckrodt Veterinary, Mundelein, IL, USA) in oxygen for bone marrow collection. Bone marrow samples were obtained from these cats via humerus using a 14-gauge bone marrow needle. Bone marrow was collected from the random source cats within 3 min of euthanasia, using rongeurs to fracture the humerus, and the marrow carefully dissected. Bone marrow was obtained in this manner from the fetus after euthanasia of the queen; blood could not be obtained from this fetus. Marrow samples were placed in EDTA tubes. Air-dried smears were made both from blood and marrow samples. EDTA tubes with samples were placed on ice within 5 min of being obtained. Thirty min after collection, serum separator tubes were centrifuged at 10 000 rpm for 5 min. Serum was separated and placed on ice. Blood, serum, and marrow samples were then stored at −70°C until analysis. Samples from two cats from referring veterinarians were obtained and stored in EDTA tubes using unknown methods prior to sending them to the VTH. Stray cats were euthanised via an overdose of intravenous barbiturates.
A commercial ELISA test (Feline Leukemia Virus Antigen/Feline Immunodeficiency Virus Antibody Test Kit; IDEXX, Westbrook, ME, USA) was performed on serum samples to detect the presence of free FeLV p27 antigen on 49 cats. The two cats from private practices were tested with unknown ELISA test kits. FeLV immunofluorescent antibody test for intracellular p27 was performed on the air-dried blood and bone marrow smears (National Veterinary Laboratory).
Genomic deoxyribonucleic acid (DNA) was extracted from 200 μl aliquots of blood and bone marrow. DNA was extracted using commercial extraction kits (QIAgen tissue kit; QIAGEN, Inc, Valencia, CA, USA). Blood extraction protocols were used to extract DNA from blood samples while a tissue protocol was used to extract DNA from bone marrow and stored FL74 cell culture. FL74, a persistently FeLV-infected cell line, was used as a positive control. Confirmation of DNA extraction was performed. Three microlitres of the genomic DNA extraction was run on a low-melt 1.5% agarose gel with added ethidium bromide. A 100 base pair (bp) DNA ladder was included on each gel as a size standard. Electrophoresis in 1X Tris-Borate-EDTA (TBE) buffer was performed at 5V/cm for approximately 45 min. Following electrophoresis, the gel was visualised and photographed under UV illumination.
Oligonucleotide primers targeting a 495 bp sequence in the FeLV U3 LTR region were used. The sequences were:
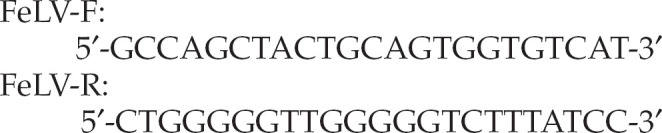 |
Oligonucleotide primers for a mitochondrial DNA copy of the 16 s RNA gene were utilised as positive controls to test DNA integrity. The primer sequences were:
 |
Ready-to-go PCR beads (0.2 ml) (Amersham-Pharmacia Biotech, Piscataway, NJ, USA) were used following the manufacturer's recommended protocol. For each reaction, a 1 μl sample of DNA template, 2 μl of each primer, and 20 μl of HPLC grade water were added to the PCR bead. Polymerase chain reaction was performed in a DNA thermal cycler (PCR Express; Hybaid, Franklin, MA, USA). The PCR protocol included an initial denaturation step at 94°C, which was performed for 2 min. This was followed by 35 cycles of 20 s denaturation at 94°C, 20 s primer annealing at 60°C, and 45 s extension at 72°C. A two-minute final extension at 72°C was performed, and then samples were held at 4°C until further analysis. Samples that failed to amplify FeLV DNA at 60°C annealing temperature were repeated at an annealing temperature of 56°C, with other cycling parameters remaining constant to allow more non-specific binding, and therefore decrease the chance of a false negative result. Two cats (cat 50 and 51) had bands detected under UV illumination on both blood and bone marrow samples that were not of the expected base pair size. These samples were repeated using Q-Solution in PCR, which changes the melting behaviour of DNA for systems not working well under standard conditions.
The integrity of genomic DNA was tested using the 16 s primers. Samples which failed to amplify 16 s DNA were repeated with 0.001X, 0.1X, 1X, 2X, and 5X DNA template concentrations. PCR analyses were run concurrently with a water template as a negative control.
Following PCR, 4 μl of each product was electrophoresed on a low-melt 1.5% agarose gel with added ethidium bromide. A 100 bp DNA ladder was included on each gel as a size standard. Electrophoresis in 1X TBE buffer was performed at 5V/cm for approximately 45 min. Following electrophoresis, the gel was visualised and photographed under UV illumination. If a weak positive was noted in the area of the expected size of the amplified FeLV product, a punch sample of the agarose gel containing the weak positive band was obtained. The gel was liquified at 62°C with 150 ul of HPLC grade water for 10 min. A 1 μl sample of this was amplified at 60°C at 25 cycles, and fractionated on a 1.5% agarose gel as previously described. PCR test results were considered positive if a 495 bp band was visualised on ethidiumbromide-agarose gel electrophoresis viewed with UV transillumination.
Two randomly selected amplified bone marrow PCR products of the expected size (cats 12 and 36), and both blood and bone marrow PCR products of cat 51 were DNA sequenced (DNA Sequencing Facility; Virginia Tech). The PCR product was purified following the manufacturer's protocol for DNA purification (QIAuick PCR Purification Kit; QIAGEN, Inc).
Both strands of the purified DNA product from bone marrow of cat 36 were sequenced using the Thermosequenase Dye-Terminator Cycle Sequencing Kit (Amersham-Pharmacia Biotech), run on an automated fluorescent DNA sequencer (ALFExpress II; Amersham-Pharmacia Biotech), and aligned to sequences using Sequencher (GeneCodes). Both strands of the purified DNA product from bone marrow of cat 12 and blood and bone marrow of cat 51 were sequenced using standard methods on an ABI 377 automated DNA sequencer (PE Biosystems Big Dye Terminator chemistry; PE Biosystems, Foster City, CA, USA).
Kappa statistics were used to analyse agreement beyond that due to chance between the following tests: ELISA and IFA blood, ELISA and IFA bone marrow, ELISA and PCR blood, ELISA and PCR bone marrow, IFA blood and IFA bone marrow, IFA blood and PCR blood, IFA blood and PCR bone marrow, IFA marrow and PCR blood, IFA marrow and PCR marrow, and PCR blood and PCR marrow (Ott 1993). Additionally, kappa statistics were used for the above listed tests for the clinical case cats alone.
Results
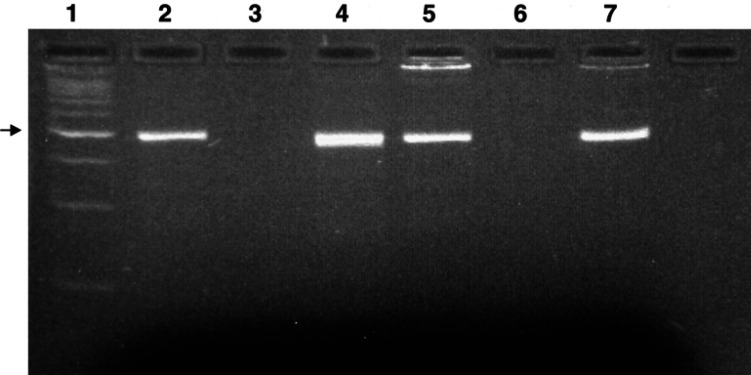
Blood and bone marrow samples from 11 laboratory cats, 23 random-source cats, one random-source fetus, and 16 clinical cases were analysed. DNA extracted from FL74 cells was consistently positive on PCR testing, and controls were consistently negative. Representative gels are shown in Fig. 1.
Fig 1.
Results of polymerase chain reaction of feline leukaemia virus on ethidium-bromide stained gel. Expected size 495 bp. Lane 1, 100 bp molecular size marker, arrow indicates 500 bp; 2, PCR product from FL74 (persistently infected FeLV cell culture) used as positive control; 3, PCR product from negative control; 4, PCR product from blood of cat 12; 5, PCR product from bone marrow of cat 12; 6, PCR product from blood of cat 10; 7, PCR product from bone marrow of cat 35.
Mitochondrial 16 s DNA was detected from all DNA extractions from blood and bone marrow samples from the 11 laboratory cats. The 11 laboratory cats had negative FeLV ELISA, negative blood and bone marrow FeLV IFA, and negative blood and bone marrow FeLV PCR at 60°C and 56°C amplification (Table 1).
Table 1.
Summary of feline leukaemia virus test results for all cases
| Group of cats | All tests (–) | All tests (+) | ELISA (+)* | IFA BM (+)* | IFA BM (–)** |
|---|---|---|---|---|---|
| Laboratory cats (n=11) | 11 | 0 | 0 | 0 | 0 |
| Random source cats (n=24) | 18 | 5 | 1 | 0 | 0 |
| Clinical cases (n=16) | 12 | 1 | 1 | 1 | 1 |
All tests=serum enzyme linked immunosorbent assay (ELISA), immunofluorescent antibody test (IFA) on blood and bone marrow (BM), and polymerase chain reaction on blood and bone marrow. (–)=negative result; (+)=positive result;
=indicates that all other tests performed in these cats were negative.
= indicates that all other tests were positive.
Results from 23 random-source cats and one fetus are summarised in Table 1. Intact 16s mitochondrial DNA from blood and bone marrow extractions was detected from all random source cats and the fetus, except cat 13 which had degraded bone marrow DNA, seen as a smeared band when visualised under UV illumination. Eighteen of 23 random source cats (cats 17 through 34) had negative results on serum ELISA, blood and bone marrow IFA, and blood and bone marrow PCR (56°C and 60°C amplification). Three cats (cats 12, 14, and 15) were positive on ELISA, blood and bone marrow IFA, and blood and bone marrow PCR. Positive ELISA with negative blood and bone marrow IFA, and negative blood and bone marrow PCR (both 56°C and 60°C amplification) were noted in one cat (cat 16). One cat (cat 13) was positive on ELISA, blood and bone marrow IFA and blood PCR; DNA from the bone marrow was degraded and did not amplify. The fetus (cat 35) was positive on bone marrow IFA and bone marrow PCR.
Results of the 16 clinical cats are summarised in Tables 1 and 2. Mitochondrial 16 s DNA was detected from blood and bone marrow DNA extractions of all clinical case cats. Twelve of the 16 clinical case cats (cats 38, 40 through 47, 49 through 51) were negative on serum ELISA, blood and bone marrow IFA, and blood and bone marrow PCR (both 56°C and 60°C amplification). PCR on both blood and bone marrow of cats 50 and 51 revealed bands of approximately 700 bp, not the expected 495 bp, under UV illumination. Using Q-solution in PCR assays, cat 50 was negative on both blood and bone marrow PCR. Cat 51 had weak positive bands in the 600 bp region for both blood and bone marrow, however sequence analysis revealed that DNA was not FeLV. Therefore, cat 51 was considered negative on both blood and bone marrow PCR. One cat with lymphocytic leukaemia (cat 36) was positive on ELISA, weak positive on blood IFA, and negative on bone marrow IFA, with positive blood and bone marrow PCR. One cat with renal failure (cat 37) was positive on ELISA, and blood and bone marrow IFA, and both blood and bone marrow PCR. One cat (cat 39) was negative on serum ELISA, negative on IFA blood, positive on IFA bone marrow, and negative on both blood and bone marrow PCR (both 56°C and 60°C amplification). One cat (cat 48) was weakly positive on serum ELISA, negative on blood and bone marrow IFA, and negative on blood and bone marrow PCR (both 56°C and 60°C amplification).
Table 2.
Feline leukaemia virus test results and clinical diagnosis of 16 clinical case cats (36 through 51)
| Cat No. | ELISA | IFA BLD | IFA BM | PCR BLD | PCR BM | Diagnosis |
|---|---|---|---|---|---|---|
| 36 | + | +(w) | – | + | + | Lymphocytic leukaemia |
| 37 | + | + | + | + | + | Glomerulonephrits with renal failure |
| 38 | – | – | – | – | – | Pancytopenia |
| 39 | – | – | + | – | – | Granulocytic leukaemia |
| 40 | – | – | – | – | – | Leukopenia |
| 41 | – | – | – | – | – | Lymphoblastic leukaemia |
| 42 | – | – | – | – | – | Non-regenerative anaemia |
| 43 | – | – | – | – | – | Alimentary lymphoma |
| 44 | – | – | – | – | – | Non-regenerative anaemia |
| 45 | – | – | – | – | – | Hemolytic anaemia, TCP |
| 46 | – | – | – | – | – | Hepatic/splenic lymphoma |
| 47 | – | – | – | – | – | TCP, tetraparesis |
| 48 | +(w) | – | – | – | – | Multicentric lymphoma |
| 49 | – | – | – | – | – | Neutropenia |
| 50 | – | – | – | – | – | Granulocytic leukaemia |
| 51 | – | – | – | – | – | Granulocytic leukaemia |
ELISA=peripheral blood enzyme-linked immunosorbent assay; IFA BLD=peripheral blood immunofluorescent antibody; IFA BM=bone marrow immunofluorescent antibody; PCR BLD=blood polymerase chain reaction; PCR BM=bone marrow polymerase chain reaction; (–)=negative result; (+)=positive result; +(w)=weak positive result; TCP=thrombocytopenia.
Polymerase chain reaction products of bone marrow from two of six randomly selected FeLV positive animals (cats 12 and 36) were sequenced. Comparison of each sequence with a previously published FeLV provirus 5′ LTR-gag gene (Altschul et al 1997) sequence revealed 97.5% homology.
Kappa values (Table 3) for all cats revealed perfect agreement between PCR blood and PCR bone marrow, IFA blood and PCR blood, and IFA blood and PCR bone marrow (k=1.0). Almost perfect agreement was noted between ELISA and IFA blood (k=0.84), ELISA and PCR blood (k=0.83), ELISA and PCR marrow (k=0.83), IFA bone marrow and PCR blood (k=0.81), IFA bone marrow and PCR bone marrow (k=0.81), and IFA blood and IFA bone marrow (k=0.81). Substantial agreement between ELISA and IFA bone marrow (k=0.69) was noted.
Table 3.
Kappa statistics values for test comparisons and interpretation for all cats and for clinical case cats
| FeLV test comparison | Kappa value for all cats (n=51) | Kappa value for clinical cases (n=16) |
|---|---|---|
| IFA blood and PCR blood | 1 | 1 |
| IFA blood and PCR bone marrow | 1 | 1 |
| PCR blood and PCR bone marrow | 1 | 1 |
| ELISA and IFA blood | 0.84 | 0.77 |
| ELISA and PCR blood | 0.83 | 0.76 |
| ELISA and PCR bone marrow | 0.83 | 0.76 |
| IFA blood and IFA bone marrow | 0.81 | 0.47 |
| IFA bone marrow and PCR blood | 0.81 | 0.42 |
| IFA bone marrow and PCR bone marrow | 0.81 | 0.42 |
| ELISA and IFA bone marrow | 0.69 | 0.29 |
| Kappa statistic interpretation a Kappa value | Strength of agreement | |
| 0 | No better than chance | |
| 0.01–0.20 | Slight | |
| 0.21–0.40 | Fair | |
| 0.41–0.60 | Moderate | |
| 0.61–0.80 | Substantial | |
| 0.81–0.99 | Almost perfect | |
| 1 | Perfect | |
ELISA=enzyme-linked immunosorbent assay; IFA=immunofluorescent antibody; PCR=polymerase chain reaction.
, see Holton et al (1998).
Kappa values performed on results from the clinical case cats revealed perfect agreement with PCR blood and PCR bone marrow, IFA blood and PCR blood, and IFA blood and PCR bone marrow (k=1). Substantial agreement was present between ELISA and IFA blood (k=0.77), ELISA and PCR blood (k=0.76), and ELISA and PCR bone marrow (k=0.76). Moderate agreement was noted between, IFA blood and IFA bone marrow (k=0.47), IFA bone marrow and PCR blood (k=0.42), and IFA bone marrow and PCR bone marrow (k=0.42). Fair agreement was noted between ELISAand IFAbone marrow (k=0.29). A scale for interpretation of kappa values can be found in Table 3 (Holton et al 1998).
Discussion
Results indicate that PCR detects FeLV proviral DNA in both blood and bone marrow samples of infected cats. However, no latent infections (positive bone marrow sample PCR only) were detected. Although PCR techniques have not been previously evaluated for diagnosis of FeLV latency, PCR currently is used for this purpose in diagnosing latent infections in both human and veterinary medicine (Sninsky 1990, Shimazage et al 1994, Borchers et al 1999, Egyed & Barth 1998, Miyoshi et al 1999). Therefore, PCR would be expected to be useful for FeLV latency diagnosis.
The laboratory cats, which had tested FeLV negative as kittens, and had been housed together without contact with other cats for 8 years, were negative on all FeLV tests performed. These cats were not expected to have latent infections, as there had been no exposure to infected cats.
Of the random-source cats, animals 12, 14, and 15 were positive on all FeLV tests (ELISA, IFA blood and bone marrow, and PCR blood and bone marrow). This is the expected finding in persistently infected cats. Cat 13 had a degraded bone marrow sample, and DNA could not be amplified with the PCR method used in this study. This bone marrow sample was not placed on ice or refrigerated for 72 h prior to long-term storage, accounting for degradation. All FeLV tests performed on cat 13 (ELISA, IFA blood and bone marrow, and PCR blood) were positive, consistent with persistent infection. Cat 35, the fetus of a persistently infected queen (cat 14), was positive on bone marrow IFA and PCR. The fetus had likely become persistently infected transplacentally. Cat 16 had discordant test results, with a positive ELISA and negative IFA and PCR on both blood and bone marrow. It is possible that this was a false positive ELISA result due to technician or test error. It is also possible, but unlikely, that both IFA and PCR tests were falsely negative. Other possible causes of such discordancy include intermittent viremia and sequestered infections. False negative IFA test results can be associated with technician error, leukopenia, or latency. PCR false negatives can occur with technician error or test error. Random source cats 17 through 34 were negative on all FeLV tests performed. Due to the unknown history of these cats, recent infection and latent infection were considered possible. However, PCR did not detect proviral FeLV DNA, and therefore no latent infection existed in this group of cats. The detection of anti-FeLV antibodies would have been helpful to ascertain if viral exposure had occurred, increasing the chance of latency. However, this test was not performed in this study.
The 16 clinical case cats represented a wide age range, had diseases traditionally associated with FeLV infection, and therefore persistent and latent FeLV infections were considered possible. Cats 38, 40 through 47, and 49 through 51 tested FeLV negative on ELISA, IFA blood and bone marrow, and PCR blood and bone marrow. Therefore, these cats were neither persistently nor latently infected. In these cases, FeLV infection does not appear to be related to their clinical disease. Cat 36, diagnosed with lymphocytic leukaemia, which had a positive ELISA, weak positive blood IFA, negative bone marrow IFA, and was positive on both blood and bone marrow PCR, was likely persistently infected. It is unusual for IFA to be positive on blood and negative on bone marrow, as the marrow has a greater concentration of white blood cells. The discordancy in the IFA test results may have resulted from technician or test error. Cat 37, an animal with renal failure, was positive on all FeLV tests, and was, therefore, persistently infected with FeLV. Cat 39, an animal with granulocytic leukaemia, also had discordant results. FeLV IFA bone marrow was positive, and all other tests were negative. The discordancy is likely due to false positive bone marrow IFA, as it is unlikely that all four other tests would be falsely negative. Cat 48, an animal with multicentric lymphoma, had a weak positive ELISA, and negative IFA and PCR results. Weak positive ELISA results generally represent a lower antigen burden. The discordancy noted in this case likely resulted from a transient viremia with low viral burden, or a false positive ELISA from technician or test error. However, the latter was considered more likely as PCR blood should be positive with a true positive ELISA.
Using standard PCR methods, cats 50 and 51 had PCR products amplified from the blood and bone marrow that were larger than expected. Although these were technically negative results, these samples were further evaluated to confirm these cats were not infected with FeLV. One explanation for obtaining a larger PCR product than expected include non-specific primer annealing at different sites of DNA if the target sequence is not present. Though a PCR product in the 600 bp range was obtained from DNA from blood and bone marrow from cat 51, after additional techniques were performed, this cat was considered negative.
Cats 12 and 36, which had PCR products of the expected size, were chosen at random to be sequenced. Sequence analysis of these products revealed 97.5% homology to a previously published FeLV sequence. This high degree of homology revealed that these were true FeLV amplicons. Though not all positive PCR results were DNA sequenced, the remainder of positive PCR samples all had distinct bands of the expected size on gel electrophoresis when run under standard conditions, and are assumed to have similar degrees of homology.
Kappa statistics revealed perfect agreement between test results for PCR blood and PCR bone marrow, IFA blood and PCR blood, and IFA blood and PCR bone marrow. All remaining kappa statistics for all cases revealed almost perfect or at least substantial agreement. Some degree of test result variation is expected between cases as the various tests may detect FeLV infection at different stages, but overall a high agreement between tests was present. In the cases of clinically affected cats, kappa statistics did not reveal as strong agreement for some test comparisons. Therefore, in cats presenting with a high suspicion of FeLV-related disease, it appears that test result agreements may be lower, particularly results of IFA bone marrow. A bone marrow IFA is generally only performed in cats that are leukopenic, as it may be difficult to detect leukocytes in a blood smear, and test results may be inconclusive. In these cases, bone marrow smears, which have a higher leukocyte concentration, can be used. In both cases in which bone marrow IFA results did not agree with other tests, the cat was leukaemic, and therefore a FeLV bone marrow IFA would not likely have been performed in a normal clinical situation. In this study, PCR results were similar to IFA blood results, and therefore PCR testing did not provide any additional information. To determine if the results of IFA and PCR are in fact similar, a larger number of clinical cases with FeLV-associated diseases should be evaluated.
In the present study, no evidence of latency was detected in any group of cats, which is lower than expected compared to these previously reported experimental studies (Madewell & Jarret 1983, Pacitti & Jarret 1985). This is also unusual as the diseased cats were highly suspected to have FeLV-related conditions. A decrease in prevalence of FeLV infection has been noted from the Tufts Veterinary Diagnostic Laboratory. At this laboratory, a gradual decline has been noted in the number of positive ELISA test results, from 8% in 1989 to 4% in 1995 (Cotter 1998). Previous studies have anecdotally noted that fewer FeLV positive cats are affected by lymphoma than was noted 10–15 years ago. In a recent study of feline lymphoma, only 25% of lymphoma cases were associated with FeLV antigenemia, compared with the earlier published 60–70% rates (Vail et al 1998). Possible causes for this decrease in prevalence include routine testing of kittens with subsequent eradication, and the development of FeLV vaccinations.
The lack of evidence of FeLV latency in any group in this study indicates these cats were either not latently infected, the PCR technique used in this study was not sensitive enough, or FeLV infection may be sequestered in areas other than the bone marrow. Further studies on a larger number of cats with high suspicion for naturally-occurring FeLV-related disease are needed to determine if these results can be verified. Additionally, future investigations to perform quantitative PCR would be needed to determine the detection threshold of FeLV infected cells from bone marrow samples. It is possible that culture of bone marrow cells prior to PCR may increase the detection of latency; further studies would be needed to determine the utility of cell culture prior to PCR. Although FeLV latency is reportedly responsible for a variety of diseases, and that bone marrow is the suspected primary site of latent infection, it appears this is not true in all cases.
Acknowledgements
The authors would like to acknowledge the Virginia Veterinary Medical Association Pet Memorial Fund and the Office of Research and Graduate Studies, Virginia Maryland Regional College of Veterinary Medicine for their funding of this project.
References
- Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acid Research 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchers K, Wolfinger U, Ludwig H. (1999) Latency-associated transcripts of equine herpesvirus type 4 in trigeminal ganglia of naturally infected horses. Journal of General Virology 80, 2165–2171. [DOI] [PubMed] [Google Scholar]
- Casey JW, Roach A, Mullins JI, Burck KB, Nicolson MO, Gardner MB, Davidson N. (1981) The U3 portion of feline leukaemia virus DNA identifies horizontally acquired proviruses in leukemic cats. Proceedings of National Academy of Science USA 7778–7782. [DOI] [PMC free article] [PubMed]
- Cotter S. (1998) Feline viral neoplasia. In: Infectious Diseases of the Dog and Cat. Greene C. (ed). Philadelphia, WB Saunders, pp. 71–84. [Google Scholar]
- Egyed L, Bartha A. (1998) PCR studies on the potential sites for latency of BHV-4 in calves. Veterinary Research Communications 22, 209–216. [DOI] [PubMed] [Google Scholar]
- Fulton R, Plumb M, Shield L, Neil JC. (1990) Structural diversity and nuclear protein binding sites in the long terminal repeats of feline leukemia virus. Journal of Virology 64, 1675–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy WDJ. (1991) General principles of retrovirus immunodetection tests. Journal of the American Veterinary Medical Association 199, 1282–1287. [PubMed] [Google Scholar]
- Hardy WDJ, Hirshaut Y, Hess P. (1973) Detection of the feline leukaemia virus and other mammalian oncornaviruses by immunofluorescence. In: Unifying Concepts of Leukemia. Dutcher RM, Chieco-Bianchi L. (eds). New york, Karger and Basel, pp. 778–799. [DOI] [PubMed] [Google Scholar]
- Hardy WD, Zuckerman EE. (1991) Ten year study comparing enzyme-linked immunosorbent assays with the immunofluorescent antibody test for the detection of feline leukemia virus infection in cats. Journal of the American Veterinary Medical Association 199, 1365–1373. [PubMed] [Google Scholar]
- Hardy WDJ, Zuckerman EE. (1991) Development of the immunofluorescent antibody test for detection of feline leukemia virus infection in cats. Journal of the American Veterinary Medical Association 199, 1327–1334. [PubMed] [Google Scholar]
- Herring IP, Troy GC, Pickett JP, Toth T, Champagne ES. (1998) Detection of feline leukemia virus in corneal tissues of infected cats by polymerase chain reaction and immuno-histochemistry. 29th Annual Proceedings of the American College of Veterinary Ophthalmologists, Seattle, WA, p. 65.
- Holton LL, Scott EM, Nolan AM, Reid J, Welsh E, Flaherty D. (1998) Comparison of three methods used for assessment of pain in dogs. Journal of the American Veterinary Medical Association 212, 61–66. [PubMed] [Google Scholar]
- Jackson ML, Haines DM, Meric SM, Misra V. (1993) Feline leukemia virus detection by immunohistochemistry and polymerase chain reaction in formalin-fixed, paraffinembedded tumor tissue from cats with lymphosarcoma. Canadian Journal of Veterinary Research 57, 269–276. [PMC free article] [PubMed] [Google Scholar]
- Jackson ML, Haines DM, Taylor SM, Misra V. (1996a) Feline leukemia virus detection by ELISA and PCR in peripheral blood from 68 cats with high, moderate, or low suspicion of having FeLV-related disease. Journal of Veterinary Diagnostic Investigation 8, 25–30. [DOI] [PubMed] [Google Scholar]
- Jackson ML, Wood SL, Misra V, Haines DM. (1996b) Immunohistochemical identification of B and T lymphocytes in formalin fixed, paraffin-embedded feline lymphosarcomas: Relation to feline leukemia virus status, tumor site, and patient age. Canadian Journal of Veterinary Research 60, 199–204. [PMC free article] [PubMed] [Google Scholar]
- Madewell BR, Jarret O. (1983) Recovery of feline leukaemia virus from non-viraemic cats. Veterinary Record 112, 339–342. [DOI] [PubMed] [Google Scholar]
- Miyazawa T, Jarrett O. (1997) Feline leukaemia virus proviral DNA detected by polymerase chain reaction in antigenemic but non-viraemic (‘discordant') cats. Archives of Virology 142, 323–332. [DOI] [PubMed] [Google Scholar]
- Miyoshi M, Ishii Y, Takiguchi M, Takada A, Yasuda J, Hashimoto A, Okazaki A, Kida K. (1999) Detection of canine herpesvirus DNA in the ganglionic neurons and the lymph node lymphocytes of latently infected dogs. Journal of Veterinary Medical Science 61, 375–9. [DOI] [PubMed] [Google Scholar]
- Ott LR. (1993) An Introduction to Statistical Methods and Data Analysis. 4th ed. Belmont, Wadsworth Publishing Company. [Google Scholar]
- Pacitti AM, Jarret O. (1985) Duration of the latent state in feline leukaemia virus infections. Veteterinary Record 117, 472–474. [DOI] [PubMed] [Google Scholar]
- Pederson N, Meric S. (1984) The clinical significance of latent feline leukaemia virus infection in cats. Feline Practice 14, 32–48. [Google Scholar]
- Rojko JL, Hardy WD. (1994) Feline leukemia and other retroviruses. In: The Cat: Diseases and clinical management (2nd ed), Sherding RG. (ed). Philadelphia, WB Saunders, pp. 263–432. [Google Scholar]
- Rojko JL, Hoover EA, Quackenbush SL, Olsen RG. (1982) Reactivation of latent feline leukemia virus infection. Nature 298, 385–388. [DOI] [PubMed] [Google Scholar]
- Shimazage J, Tsubota K, Sawa M, Kinoshita S, Ohkura T, Honda M. (1994) Detection of human immunodeficiency virus, hepatitis B virus, and hepatitis C virus in donor eyes using polymerase chain reaction. British Journal of Ophthalmology 78, 859–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sninsky JJ. (1990) The polymerase chain reaction (PCR): A valuable method for retroviral detection. Lymphology 23, 92–97. [PubMed] [Google Scholar]
- Vail D, Moore A, Ogilvie G, Volk LM. (1998) Feline lymphoma (145 cases): Proliferation indices, cluster of differentiation 3 immunoreactivity, and their association with prognosis in 90 cats. Journal of Veterinary Internal Medicine 12, 349–354. [DOI] [PubMed] [Google Scholar]