Abstract
Wide-scale DNA testing requires the development of small, fast and easy-to-use devices. This article describes the preparation, operation and applications of biosensors and gene chips, which provide fast, sensitive and selective detection of DNA hybridization. Various new strategies for DNA biosensors and gene chips are examined, along with recent trends and future directions. The integration of hybridization detection schemes with the sample preparation process in a ‘Lab-on-a-Chip’ format is also covered. While the use of DNA biosensors and gene chips is at an early stage, such devices are expected to have an enormous effect on future DNA diagnostics.
INTRODUCTION
Few scientific areas have witnessed dramatic changes of the magnitude observed recently in DNA diagnostics. With the completion of the human genome, we are just at the beginning of a revolution in genetic analysis. The information obtained from the project opens the door to tremendous analytical opportunities ranging from diagnostic tests for mutations to the assessment of medical treatment. To continue these advances, to exploit these opportunities, and to address the growing market needs in the 21st Century, future devices must link high performance, with speed, simplicity and low cost.
The aim of the present article is to review recent efforts in the areas of DNA biosensors, gene chips and miniaturized (‘Lab-on-a-Chip’) DNA analyzers. The goal is to connect recent activities in these closely-related areas, and to introduce the concepts of DNA biosensors and ‘Lab-on-a-Chip’ to the readers of this journal. DNA biosensors and gene chips are of considerable recent interest due to their tremendous promise for obtaining sequence-specific information in a faster, simpler and cheaper manner compared to traditional hybridization assays. Recent advances in developing such devices thus opens up new opportunities for DNA diagnostics. Although an attempt has been made to cover in this review the range of latest activities and trends, this is not a comprehensive review.
DNA BIOSENSORS
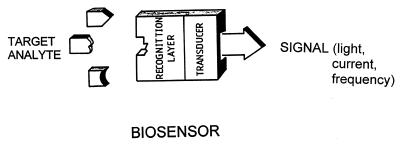
Biosensors are small devices which utilize biological reactions for detecting target analytes (1). Such devices intimately couple a biological recognition element (interacting with the target analyte) with a physical transducer that translates the biorecognition event into a useful electrical signal (Fig. 1). Common transducing elements, including optical, electrochemical or mass-sensitive devices, generate light, current or frequency signals, respectively. There are two types of biosensors, depending on the nature of the recognition event. Bioaffinity devices rely on the selective binding of the target analyte to a surface-confined ligand partner (e.g. antibody, oligonucleotide). In contrazst, in biocatalytic devices, an immobilized enzyme is used for recognizing the target substrate. For example, sensor strips with immobilized glucose oxidase have been widely used for personal monitoring of diabetes.
Figure 1.
Biosensors: the intimate coupling of biorecognition and signal transduction.
DNA biosensors, based on nucleic acid recognition processes, are rapidly being developed towards the goal of rapid, simple and inexpensive testing of genetic and infectious diseases (2,3), and for the detection of DNA damage and interactions (4). Unlike enzyme or antibodies, nucleic acid recognition layers can be readily synthesized and regenerated for multiple use.
Sequence-specific hybridization biosensors
Hybridization biosensors rely on the immobilization of a single-stranded (ss) DNA probe onto the transducer surface. The duplex formation can be detected following the association of an appropriate hybridization indicator or through other changes accrued from the binding event.
Surface chemistry and biochemistry. The immobilization of the nucleic acid probe onto the transducer surface plays an important role in the overall performance of DNA biosensors and gene chips. The immobilization step should lead to a well-defined probe orientation, readily accessible to the target (5,6). The environment of the immobilized probes at the solid surface depends upon the mode of immobilization and can differ from that experienced in the bulk solution. Depending upon the nature of the physical transducer, various schemes can be used for attaching the DNA probe to the surface. These include the use of thiolated DNA for self assembly onto gold transducers (gold electrodes or gold-coated piezoelectric crystals), covalent linkage to the gold surface via functional alkanethiol-based monolayers, the use of biotylated DNA for complex formation with a surface-confined avidin or strepavidin, covalent (carbodiimide) coupling to functional groups on carbon electrodes, or a simple adsorption onto carbon surfaces. As in solution-based hybridization assays, conditions for interfacial hybridization events (e.g. ionic strength, temperature, presence of accelerators) have to be optimized. Chemical and thermally-induced dehybridization of the resulting duplex is often used for regenerating the interface.
Recent advances in nucleic acid recognition can enhance the power of DNA biosensors. For example, the introduction of peptide nucleic acid (PNA) has opened up exciting opportunities for DNA biosensors. PNA is a DNA mimic in which the sugar–phosphate backbone is replaced with a pseudopeptide one. The unique structural, hybridization and recognition features of solution-phase PNA (7) can be readily extrapolated onto transducer surfaces in connection with the design of highly-selective DNA biosensors. Such use of surface-confined PNA recognition layers imparts remarkable sequence specificity onto DNA biosensors (including detection of single-base mismatches) and offers other attractive advantages (including greater latitude in the selection of experimental conditions) (8).
DNA dendrimers can be used for imparting higher sensitivity onto DNA biosensors. These tree-like superstructures possess numerous single-stranded arms that can hybridize to their complementary DNA sequence. A greatly increased hybridization capacity and hence a substantially amplified response is achieved by immobilizing these dendritic nucleic acids onto the physical transducer (9).
Optical biosensors. Optical detection of DNA hybridization has attracted considerable attention. DNA optical biosensors commonly rely on a fiber optic to transduce the emission signal of a fluorescent label. Fiber optics are devices that carry light from one place to another by a series of internal inflections. The operation of fiber-optic DNA biosensors typically involves placement of a ssDNA probe at the end of the fiber and monitoring the fluorescent changes resulting from the association of a fluorescent compound (indicator) with the double-stranded (ds) DNA hybrid. The first DNA optical biosensor, developed by Krull and coworkers (10), relied on the use of an ethidium bromide indicator. Extremely low (femtomolar) detection limits have been achieved in connection with other fluorescent indicators (e.g. PicoGreen). Walt’s group (11) developed a fiber-optic DNA sensor array for the simultaneous detection of multiple DNA sequences. Hybridization of fluorescently-labeled complementary olgonucleotides was monitored by observing the increase in fluorescence that accompanied binding. A different optical transduction, based on evanescent wave devices, can offer real-time label-free optical detection of DNA hybridization (12,13). These biosensors rely on monitoring changes in surface optical properties (shift in resonance angle due to change in the interfacial refractive index) resulting from the surface binding reaction. Such devices thus combine the simplicity of surface plasmon resonance with the sensitivity of wave guiding devices. The coupling of chemiluminescence with sandwich hybridization, magnetic bead capture and flow injection operation has been used for the rapid detection of hepatitis B virus DNA (14).
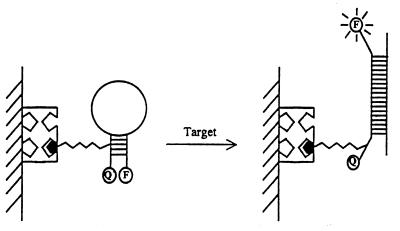
In situ, label-free, optical detection can be achieved through changes in other optical properties. For example, a novel nanoparticle-based colorimetric detection offers great promise for direct detection of DNA hybridization (15). In this case, a distance change, accrued from the hybridization event, results in changes of the optical properties of the aggregated functional gold nanoparticles. Another new innovative approach for the direct fluorescent detection of DNA hybridization relies on the use of molecular beacons (MBs) (16). MBs are oligonucleotides with a stem-and-loop structure, labeled with a fluorophore and a quencher on the two ends of the stem, that become fluorescent upon hybridization (Fig. 2). In addition to their direct monitoring capability, MB probes offer high sensitivity and specificity.
Figure 2.
The operation of MB based optical DNA biosensors. The hybridization event induces conformational reorganization that separates the quencher from the fluorophore. [Reprinted with permission from (16). Copyright 1999 American Chemical Society.]
Electrochemical biosensors. Electrochemical devices have also proven very useful for sequence-specific biosensing of DNA (17,18). The inherent miniaturization of such devices and their compatibility with advanced microfabrication technology make them excellent candidates for DNA diagnostics. Electrochemical detection of DNA hybridization usually involves monitoring a current response under controlled potential conditions (in a manner analogous to most hand-held meters used by sufferers of diabetes for measuring their blood glucose level). The hybridization event is commonly detected via the increased current signal of a redox indicator (that recognizes the DNA duplex) or from other hybridization-induced changes in electrochemical parameters (e.g. conductivity or capacitance). Mikkelsen’s team, that pioneered the use of redox indicators, demonstrated its utility for detecting the cystic fibrosis ΔF508 deletion sequence associated with 70% of cystic fibrosis patients (19). A detection limit of 1.8 fmol was demonstrated for the 4000-base DNA fragment in connection to a Co(bpy)33+ indicator. High selectivity towards the disease sequence (but not to the normal DNA) was achieved by performing the hybridization at an elevated (43°C) temperature. Such use of the electrochemical transduction mode requires that proper attention be given to the choice of the indicator and its detection scheme. New redox indicators, offering greater discrimination between ss and dsDNA are being developed for attaining higher sensitivity. The use of a threading intercalator ferrocenyl naphthalene diimide (20) that binds to the DNA hybrid more tightly than usual intercalators and displays small affinity to the single-stranded probe has been very successful.
The use of enzyme labels also offers great promise for electrochemical detection of DNA hybridization. Heller’s group (21) demonstrated that a direct amperometric monitoring of the hybridization event can be achieved in connection to the use of horseradish-peroxidase labeled target. In this system, the hybridization event resulted in the ‘wiring’ of the enzyme to the transducer (via an electron-conducting redox hydrogel), hence leading in a continuous hydrogen-peroxide electroreduction current. Willner’s group (22) illustrated that multiple amplifications can be achieved by coupling of a peroxidase enzyme label with the surface accumulation of the phenol reaction product.
Increased attention has been given recently to new label-free electrochemical detection schemes which offer faster and simpler assays. For example, it is possible to exploit changes in the intrinsic electroactivity of DNA (e.g. the guanine oxidation peak) accrued from the hybridization event (23,24). To overcome the limitations of the probe sequences (absence of G), guanines in the probe sequence were substituted by inosine residues (pairing with Cs) and the hybridization was detected through the target DNA guanine signal (23). A greatly amplified guanine signal, and hence hybridization response, was obtained by using the Ru(bpy)3+2 redox mediator (24). Direct, label-free, electrical detection of DNA hybridization has also been accomplished by monitoring changes in the conductivity of conducting polymer molecular interfaces, e.g. DNA-modified polypyrrole films (25,26). Eventually, it would be possible to eliminate these polymeric interfaces and to exploit different rates of electron-transfer through ss and dsDNA for probing hybridization (including mutation detection via the perturbation in charge transfer through DNA). Recent activity in this direction is encouraging (27).
The electrochemical response of the G nucleobase is also very sensitive to the DNA structure and can thus be used for probing DNA damage or interactions. Changes in the guanine oxidation, and of other intrinsic DNA redox signals, have thus been used for detecting chemical and physical damage (4,28).
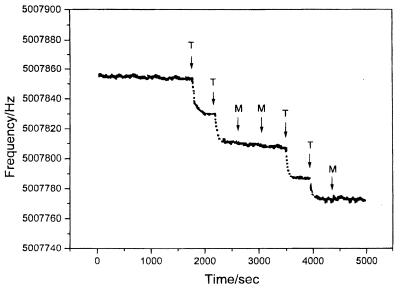
Mass-sensitive devices. Another useful indicator-free detection scheme relies on the use of quartz crystal microbalance (QCM) transducers. The QCM is an extremely sensitive mass-measuring device, that allows dynamic monitoring of hybridization events (29). QCM hybridization biosensors consist of an oscillating crystal with the DNA probe immobilized on its surface. The increased mass, associated with the hybridization reaction, results in a decrease of the oscillating frequency. This is illustrated in Figure 3 (30) where the high sensitivity of QCM transducers was coupled with the remarkable specificity of PNA probes towards the detection of a single-base alteration in the p53 gene. A highly-sensitive microgravimetric device was also developed for detecting the Tay–Sachs genetic disorder (31). QCM transducers have been used for investigating other DNA interactions, including real-time detection of protein–DNA binding (32) or monitoring of enzymatic cleavage reactions (33). Analogous acoustic wave devices were developed for monitoring the binding of anticancer drugs to DNA (34).
Figure 3.
Frequency–time response of a PNA/QCM to additions of the target (T) and mismatch (M) oligonucleotides. The hybridization event results in decreased frequency, reflecting the increased mass of the crystal. [Reprinted with permission from (30). Copyright 1997 American Chemical Society.]
DNA MICROARRAYS
The analysis of complex DNA samples and acquisition of sequence and expression information would require the integration of multiple biosensors in connection with DNA microarrays (35,36). A number of terms, like DNA arrays, gene chips or biochips, are often being intermixed to describe these devices. The most attractive features of these devices are the miniaturization, speed and accuracy. Accordingly, this DNA microchip technology offers an enormous potential for rapid multiplex analysis of nucleic acid samples, including the diagnosis of genetic diseases, detection of infectious agents, measurements of differential gene expression, drug screening or forensic analysis (35,36). Such use of DNA microarrays is thus revolutionizing many aspects of genetic analysis.
Such hybridization chips are fabricated from glass, silicon or plastic supports, and comprise thousands of 10–100 µm reaction zones onto which individual oligonucleotides have been deposited. This results in high densities (up to 106 sites/cm2) in connection with typical 1–2 cm2-size chips. The exact number of probes varies in accordance with the application. The actual construction of gene chips involves the immobilization or synthesis of an array of DNA probes on a solid support. High-density DNA arrays often require the use of physical delivery (e.g. microjet deposition technology), involving the dispension of picoliter volumes onto discrete locations on the chip. It is essential to activate the surface for a covalent attachment of the oligonucleotide probes.
Successful implementation of DNA chip technology requires development of methods for fabricating the probe arrays, detecting the target hybridization, algorithms for analyzing the data, and reconstructing the target sequence. Such array technology thus integrates molecular biology, advanced microfabrication/micromachining technologies, surface chemistry, analytical chemistry, software, robotics and automation; i.e. a truly interdisciplinary field. The automation of gene chip systems greatly facilitates their production and accelerates their operation, while eliminating human errors (accrued from the sample handling). The detection of the DNA hybridization (at the individual spots) relies on the signal generated by the binding event. Unlike individual biosensors (described earlier), scanning or imaging the chip surface is essential for obtaining the complete hybridization pattern. Fluorescence imaging and mass spectroscopy are commonly used for such ‘reading’ of the chips. The former usually relies on confocal laser scanning. Bioinformatics tools are used to relate the complexity of the data into useful information. In the following sections, selected examples of DNA arrays developed by various major companies are described.
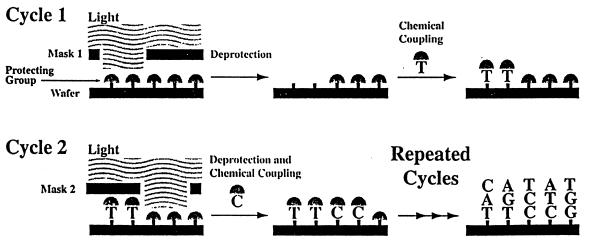
The GeneChip system of the array pioneer, Affymetrix (Santa Clara, CA), provides the sequence information by hybridization with a set of target DNA fragments prepared from one or more genes of interest (37,38). The system consists of four integrated parts: a disposable DNA probe array, a fluidic station for introducing the test sample, a scanner to read the data, and software to control the instrument and process the data. The probe array consists of an assembly of oligonucleotides of known sequence in a site-specific arrangement on a silicon surface. A photolithographic-combinatorial chemistry strategy is used to synthesize large numbers of DNA probes in specific locations on a chip (Fig. 4). The fluorescence intensity data, captured from the scanner, are used with computer files (containing the sequences and location of all the probes) to provide the DNA profiling of the test sample. Affymetrix’s DNA microarray chip technology is rapidly moving out from the feasibility phase into the application phase. The company has a number of GeneChips for screening for breast cancer, for detecting mutations in the HIV genome and in the p53 tumor-suppressor gene, or for identifying bacterial pathogens.
Figure 4.
The light directed probe array synthesis process used for the preparation of Affymetrix’s GeneChip.
Hyseq Inc. (Sunnyvale, CA) is collaborating with Perkin-Elmer in designing a universal microarray DNA ‘Superchip’ system that facilitates sequencing by hybridization (SBH). In the Hychip system, capture probes on the chip surface hybridize to target DNA and are covalently bound to the labeled probes that also hybridize to the target DNA in an adjacent position. A fluorescence spot indicates the side-by-side binding of the unknown DNA with the probe and tagged probe. The process is repeated with another labeled probe and dedicated software figures out what the DNA sequence must be. The same chip can be used for detecting point mutations related to genetic diseases, cancer or infectious diseases.
Clinical Micro Sensors (Pasadena, CA) is developing a small, battery-operated instrument, based on electrochemical detection of DNA hybridization, for meeting the needs of point-of-care diagnosis. This electrochemical route is ideal for shrinking the hardware (compared to that used for generating fluorescence hybridization patterns). The company, that recently formed an alliance with Motorola, relies on DNA–label complexes that are connected to the electrode by phenylacetylene ‘molecular wires’ embedded in a self-assembled monolayer of alkane thiols. The layer also protects the surface, hence facilitating the analysis of complex biological samples.
DNA microarrays offer great promise for monitoring gene expression in humans. Technology developed by Brown and co-workers uses RNA expression in biochips to identify differential gene expression relevant to different biological states (39). The gene-expression arrays of Incyte Pharmaceuticals Inc. (Palo Alto, CA) are based on fluorescent-labeled probes and competitive hybridization; the differential gene expression is being measured through competitive hybridization of two mRNA sets isolated from normal and diseased samples, and labeled with different-colored fluorescent tags. Scanning for the two colors and using dedicated expression analysis software allow researchers to quantify expression changes in healthy versus diseased samples. An ink-jet printing technology is used for localizing the (500–5000 bp long) probes on a glass substrate. The AtlasTM microarrays of Clontech Inc. (Palo Alto, CA) also provide sensitive detection of gene expression in connection to fluorescent dyes and glass or Nylon substrates.
‘LAB-ON-A-CHIP’
Another active field is the integration of the sample preparation and DNA array detection in the so-called ‘Lab-on-a-Chip’ configuration (40,41). The goal of this technology is to fully integrate multiple processes, including sample collection and pretreatment with the DNA extraction, amplification, hybridization and detection, on a microfluidic platform. The ability to perform all the steps of the biological assay on a single self-contained microchip promises significant advantages in terms of speed, cost, sample/reagent consumption, contamination, efficiency and automation (including parallel sample processing). Such miniaturization of the analytical instrumentation will enable transportation of the laboratory to the sample source (as desired for point-of-care testing). The preparation of these credit-card sized microlaboratories commonly relies on advanced microfabrication and micromachining technologies, using processes common in the manufacture of electronic circuitry. Sophisticated devices have thus been fabricated with pumps, valves, heaters, filters, along with the corresponding fluidic network. On-chip fluid manipulations have been demonstrated for sequentially transporting nanoliter samples through a network of microchannels, mixing of sample and reagents, dispensing of samples, DNA restriction digestion and electrically-driven separations (42).
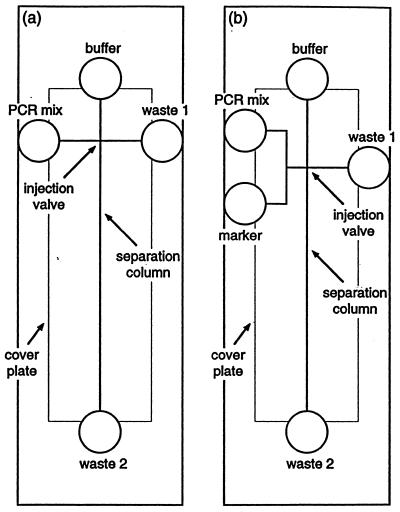
A variety of microstructures have been proposed for on-chip PCR amplification (43,44). For example, PCR amplification has been performed by continuously flowing the sample through three well-defined temperature controlled zones on a glass microchip (43). The pattern of the chip layout determined the relative time a fluid element is exposed to each temperature zone. An array of PCR microchips, based on multiple reaction chambers with resistive heaters was developed at the Lawrence Livermore National Laboratory (44). A microchip device for cell lysis, multiplex PCR and electrophoretic sizing has also been described (Fig. 5; 42). Voltages, applied through electrodes placed within the individual reservoirs (circles in Fig. 5), are used to drive the electroosmotic flow. Such electroosmotic ‘pumping’ obviates the need for mechanical pumps or valves, with the channel intersections serving as ‘virtual injection valves’.
Figure 5.
Schematic of microchips used for cell lysis, PCR amplification and electrophoretic analysis (a), and for sizing of PCR products with a marker (b). [Reprinted with permission from (42). Copyright 1998 American Chemical Society.]
Nanogen Inc. (San Diego, CA) has developed an electronic sample preparation process. The company is addressing each step in the sample-to-result process on microfabricated chip-based devices (45). This includes the integration of electronic cell separation, electronic sample transport, electronically-accelerated hybridization and electronic denaturation. Controlled electric fields have been used for discriminating among oligonucleotide hybrids with varying binding strengths (including between complete match and single-base mismatch) and to expel the unhybridized DNA. Such fine-tuned electronic stringency selection obviates the need for extensive washing. The electronically-regulated sample preparation process was demonstrated for the dielectrophoretic separation of Eschericia coli from blood cells (46). After the isolation, the bacteria were lysed by a series of high-voltage pulses. A variety of microelectronic chips including 25, 100, 400, 1600 and 10 000 addressable test sites have been fabricated by Nanogen Inc. The 10 000 test site chip is being developed for drug discovery applications.
Microfabricated devices are also attractive for high-throughput electrically-driven DNA separations. For example, Woolley et al. (47) described capillary electrophoresis microchips that allow DNA sizing of 12 samples in parallel with a resolution of 10 bp and higher throughputs than conventional techniques. The first commercially available product based on microfluidic technology is the HP 2100 Bioanalyzer (Hewlett Packard). This instrument integrates the sample handling and electrophoretic separation. The disposable LabChipTM has multiple interconnected reservoirs for samples, sizing ladder, sieving matrix and buffers. Assays are available for analyzing restriction digestions with DNA fragments 100–12 000 bases long, and for determining size and concentration information for DNA fragments 100–7500 bases long.
CONCLUSIONS
Over the past decade we have witnessed intense activity aimed at developing DNA biosensors and gene chips. Such devices offer considerable promise for obtaining the sequence-specific information in a faster, simpler and cheaper manner compared to traditional hybridization assays. These DNA microarray and biosensor technologies are rapidly advancing and applications ranging from genetic testing to gene expression and drug discovery have been demonstrated. Further scaling down, particularly of the support instrumentation, should lead to hand-held DNA analyzers. Innovative efforts, coupling fundamental biological and chemical sciences with technological advances in the fields of micromachining and microfabrication should lead to even more powerful devices that will accelerate the realization of large-scale genetic testing. A wide range of new gene chips and DNA biosensors are thus expected to reach the market in the coming years. While offering remarkable tools for genetic analysis, proper applications of these new devices would still require a solid intellectual input.
Acknowledgments
ACKNOWLEDGEMENTS
DNA work in the author’s laboratory is funded by NIH (grant RR 14549-01) and by the US Army Medical Research.
REFERENCES
- 1.Taylor R.F. and Schultz,J.S. (1996) Handbook of Chemical and Biological Sensors. Institute of Physics Publishing, Bristol, UK.
- 2.Christopoulos T.K. (1999) Anal. Chem., 71, 425R–438R. [Google Scholar]
- 3.Zhai J., Hong,C. and Yang,R. (1997) Biotechnol. Adv., 15, 43–58. [DOI] [PubMed] [Google Scholar]
- 4.Palecek E., Fojta,M., Tomschick,M. and Wang,J. (1998) Biosens. Bioelectron., 13, 621–628. [DOI] [PubMed] [Google Scholar]
- 5.Herne T. and Tarlov,M.J. (1997) J. Am. Chem. Soc., 119, 8916–8920. [Google Scholar]
- 6.Piunno P.A., Watterson,J., Wust,C. and Krull,U. (1999) Anal. Chim. Acta, 400, 73–89. [Google Scholar]
- 7.Nielsen P. (1999) Acc. Chem. Res., 32, 624–630. [Google Scholar]
- 8.Wang J., Palecek,E., Nielsen,P., Rivas,G., Cai,X., Shiraishi,H., Dontha,N., Luo,D. and Farias,P. (1996) J. Am. Chem. Soc., 118, 7667–7671. [Google Scholar]
- 9.Wang J., Jiang,M., Nilsen,T. and Getts,R. (1998) J. Am. Chem. Soc., 120, 8281–8282. [Google Scholar]
- 10.Piunno P., Krull,U., Hudson,R., Damha,M. and Cohen,H. (1995) Anal. Chem., 67, 2635–2643. [DOI] [PubMed] [Google Scholar]
- 11.Ferguson J.A., Boles,T., Adams,C. and Walt,D. (1996) Nat. Biotechnol., 14, 1681–1684. [DOI] [PubMed] [Google Scholar]
- 12.Watts H., Yeung,D. and Parkes,H. (1995) Anal. Chem., 67, 4283–4289. [DOI] [PubMed] [Google Scholar]
- 13.Thiel A., Frutos,A., Jordan,C., Corn,R. and Smith,L. (1997) Anal. Chem., 69, 4984–4956. [Google Scholar]
- 14.Chen X., Zhang,X., Chai,Y., Hu,W., Zhang,Z., Zhang,X. and Cass,A.E. (1998) Biosens. Bioelectron., 13, 451–458. [DOI] [PubMed] [Google Scholar]
- 15.Storhoff J., Elghanian,R., Mucic,C., Mirkin,C. and Letsinger,R. (1998) J. Am. Chem. Soc., 120, 1959–1964. [Google Scholar]
- 16.Fang X., Liu,X., Schuster,S. and Tan,W. (1999) J. Am. Chem. Soc., 121, 2921–2922. [Google Scholar]
- 17.Mikkelsen S.R. (1996) Electroanalysis, 8, 15–19. [Google Scholar]
- 18.Wang J. (1999) Chem. Eur. J., 5, 1681–1685. [Google Scholar]
- 19.Millan K.M., Saraulo,A. and Mikkelsen,S.R. (1994) Anal. Chem., 66, 2943–2948. [DOI] [PubMed] [Google Scholar]
- 20.Takenaka S., Uto,Y., Saita,H., Yokoyama,M., Kondo,H. and Wilson,D. (1998) Chem. Commun., 1111–1112. [Google Scholar]
- 21.de Lumley T. Campbell,C. and Heller,A. (1996) J. Am. Chem. Soc., 118, 5504–5508. [Google Scholar]
- 22.Patolsky F., Katz,E., Bardea,A. and Willner,I. (1999) Langmuir, 15, 3703. [Google Scholar]
- 23.Wang J., Rivas,G., Fernandes,J., Paz,J.L., Jiang,M. and Waymire,R. (1998) Anal. Chim. Acta, 375, 197–203. [Google Scholar]
- 24.Johnston D.H., Glasgow,K. and Thorp,H.H. (1995) J. Am. Chem. Soc., 117, 8933–8938. [Google Scholar]
- 25.Korri-Youssoufi H., Garnier,F., Srivtava,P., Godillot,P. and Yassar,A. (1997) J. Am. Chem. Soc., 119, 7388–7391. [Google Scholar]
- 26.Wang J., Jiang,M., Fortes,A. and Mukherjee,B. (1999) Anal. Chim. Acta, 402, 7–12. [Google Scholar]
- 27.Holmlim R.E., Dandliker,P.J. and Barton,J.K. (1997) Angnew Chem. Int. Ed. Engl., 36, 2714–2718. [Google Scholar]
- 28.Wang J., Rivas,G., Ozsoz,M., Grant,D., Cai,X. and Parrado,C. (1997) Anal. Chem., 69, 1457–1460. [DOI] [PubMed] [Google Scholar]
- 29.Okahata Y., Matsunobo,Y., Ijiro,K., Murakami,A. and Makino,M. (1992) J. Am. Chem. Soc., 114, 8299–8300. [Google Scholar]
- 30.Wang J., Nielsen,P.E., Jiang,M., Cai,X., Fernandes,J., Grant,D., Ozsoz,M., Beglieter,A. and Mowat,M. (1997) Anal. Chem., 69, 5200–5202. [DOI] [PubMed] [Google Scholar]
- 31.Bardea A., Dagan,A., Ben-Dov,I., Amit,B. and Willner,I. (1998) Chem. Commun., 839–841. [Google Scholar]
- 32.Nikura K., Nagata,Y. and Okahata,Y. (1996) Chem. Lett., 863–866. [Google Scholar]
- 33.Wang J., Jian,M. and Palecek,E. (1999) Bioelectrochem. Bioenerg., 48, 477–480. [DOI] [PubMed] [Google Scholar]
- 34.Su H., Williams,P. and Thompson,M. (1995) Anal. Chem., 67, 1010–1016. [DOI] [PubMed] [Google Scholar]
- 35.Service R. (1998) Science, 282, 396–399. [DOI] [PubMed] [Google Scholar]
- 36.Ramsey G. (1998) Nat. Biotechnol., 16, 40–45. [DOI] [PubMed] [Google Scholar]
- 37.Gette W. and Kreiner,T. (1997) Am. Lab., March issue, 15–17. [Google Scholar]
- 38.Lipshutz R., Morris,D., Chee,M., Hubbell,E., Kozal,M., Shah,N., Shen,N., Yang,R. and Fodor,S. (1995) Biotechnology, 19, 442–445. [PubMed] [Google Scholar]
- 39.Iyer V., DeRisi,J., Eisen,M., Ross,D., Spellman,P., Hudson,J., Schuler,G., Lashkari,D., Shalon,D., Botstein,D. and Brown,P. (1997) FASEB J., 11, 1571. [Google Scholar]
- 40.Burke D.T., Burns,M.A. and Mastrangelo,C. (1997) Genome Res., 7, 189–197. [DOI] [PubMed] [Google Scholar]
- 41.Harrison D.J., Fluri,K., Seiler,K., Fan,Z., Effenhauser,C. and Manz,A. (1993) Science, 261, 895–997. [DOI] [PubMed] [Google Scholar]
- 42.Waters L.C., Jacobson,S., Kroutchinina,N., Khandurina,J., Foote,R. and Ramsey,J.M. (1998) Anal. Chem., 70, 158–162. [DOI] [PubMed] [Google Scholar]
- 43.Kopp M.U., de Mello,A. and Manz,A. (1998) Science, 280, 1046–1050. [DOI] [PubMed] [Google Scholar]
- 44.Woolley A.T., Hadley,D., Landre,P., de Mello,A., Mathies,R.A. and Northrup,M.A. (1996) Anal. Chem., 68, 4081–4086. [DOI] [PubMed] [Google Scholar]
- 45.Sosnoweki R.G., Tu,E., Butler,W., O’Connell,J. and Heller,M.J. (1997) Proc. Natl Acad. Sci. USA, 94, 1119–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng J., Sheldon,E., Wu,L., Gerrue,L., Carrino,J., Heller,M. and O’Connell,J. (1998) Nat. Biotechnol., 16, 541–545. [DOI] [PubMed] [Google Scholar]
- 47.Woolley A.T., Senasbaugh,G. and Mathies,R. (1997) Anal. Chem., 69, 2181–2186. [DOI] [PubMed] [Google Scholar]