Abstract
The ecto-5′-nucleotidase (CD73)/adenosine signaling pathway has been reported to regulate tumor epithelial-mesenchymal transition (EMT), migration and proliferation. However, little is known about the metabolic mechanisms underlying its role in trophoblast proliferation and migration. In this study, we aimed to investigate the metabolic role of the CD73/adenosine signaling pathway on the proliferation and migration of trophoblast. We found that CD73 levels were upregulated in preeclamptic placentas compared with the placentas of normotensive pregnant women. EMT and migration of HTR-8/SVneo cells were enhanced when treated with a CD73 inhibitor (100 μM) in vitro. Conversely, excessive adenosine (25 or 50 μM) suppressed trophoblast cell EMT, migration and proliferation. RNA-seq, metabolomics and seahorse findings showed that adenosine treatment resulted in increased expression of PDK1, suppression of aerobic respiration, glycolysis and amino acids synthesis, as well as increased utilization of short-chain fatty acids (SCFAs). Furthermore, the 13C-adenosine isotope tracking experiment demonstrated that adenosine served as a carbon source for the tricarboxylic acid (TCA) cycle. Our results reveal the role of adenosine in regulating trophoblast energy metabolism is like a double-edged sword - either inhibiting aerobic respiration or supplementing carbon sources into metabolic flux. CD73/adenosine signaling regulated trophoblast EMT, migration, and proliferation by modulating energy metabolism. This study indicates that CD73/adenosine signaling potentially plays a role in the occurrence of placenta-derived diseases, including preeclampsia.
Keywords: CD73, Adenosine, Epithelial-mesenchymal transition, Energy metabolism, Extravillous trophoblast
1. Introduction
Successful placental formation is essential for a healthy pregnancy and depends on well-functioning trophoblasts. In early gestation, villous cytotrophoblasts experience an epithelial-mesenchymal transition (EMT), differentiating into extravillous trophoblasts (EVTs). During this differentiation, they acquire migratory and invasive phenotypes [1]. Subsequently, EVTs migrate into the decidua and remodel the spiral arteries to supply adequate nutrients and oxygen to the fetus [2]. EVTs are one of the most important functional cells in the placenta. Impaired proliferation, migration and EMT progression of EVTs are pivotal contributors to the pathogenesis of placenta-derived diseases, such as intrauterine growth restriction, miscarriage, and preeclampsia [3].
Ecto-5′-nucleotidase (CD73) is a cell surface enzyme-producing extracellular adenosine from adenosine monophosphate (AMP) [4]. CD73/adenosine signaling has been shown to regulate EMT in various cancer cell types, including gastric, endometrial, and ovarian cancer cells. However, the cellular specificity appears to determine the different roles of CD73 in mediating EMT. For instance, elevated CD73/adenosine signaling promotes gastric cancer cell migration by positively modulating EMT [5]. On the contrary, loss of CD73 enhances endometrial tumor progression [6,7]. CD73-derived adenosine suppresses the cellular migration of ovarian carcinoma SKOV-3 cells by inducing the relocation of E-cadherin at the cellular contact zone to form adherens junctions, suggesting an epithelial-like phenotype [8]. Although contradictory results have been observed in different cancer types, CD73/adenosine signaling is increasingly recognized as one of the major pathways involved in the regulation of EMT. Owing to first-trimester trophoblast cells sharing similarities with tumor cells, it has been proposed that they use similar mechanisms for regulating migration [9]. However, whether CD73/adenosine signaling is associated with the EMT in EVT remains unclear.
In addition, it has been found that CD73-mediated adenosine production regulates energy metabolism by restricting glucose uptake in T lymphocytes [10]. Energy metabolism is a potent regulator of cellular function [11,12]. Apart from providing essential energy substrates, cellular energy metabolism provides the materials required to support biosynthesis for cell proliferation [13]. Indeed, elevated levels of adenosine have been observed in the maternal and fetal circulation and placentas of preeclampsia patients [[14], [15], [16]]. Previous studies have shown that extracellular adenosine suppressed trophoblast migration by deactivating the MAPK signaling pathway and reducing proMMP-2 levels [17,18]. However, the association between CD73/adenosine signaling and energy metabolism of EVTs remains unclear. Therefore, this study focused on the metabolic role of the CD73-adenosine axis on the cellular function of the EVTs and how this axis may be involved in the pathogenesis of placenta-derived diseases, including preeclampsia.
2. Material and methods
2.1. Collection of human placenta samples
The collection of human placenta tissue was approved by the Ethics Committee of Chongqing Medical University (Ethic No. 2014034). Informed consent was obtained from all participants. In this study, placenta tissues were collected from women with normotensive pregnancies (39.00 ± 1.24 weeks' gestation, n = 12) and preeclampsia patients (37.14 ± 1.52 weeks' gestation, n = 12) diagnosed in accordance with the American College of Obstetrician and Gynecologists (ACOG) guidelines [19]. The exclusion criteria included diagnosed gestational diabetes, pregestational diabetes mellitus, cardiovascular disease, collagen disorder, chronic renal disease, chronic hypertension, and multiple pregnancies. Participants in the preeclampsia and control groups were matched for age, body mass index (BMI), and gravidity. Table 1 shows the clinical features of the individuals. Placental tissue was obtained during cesarean section, placed in a sterile container, and transported to our laboratory for further preparation. Placenta tissue specimens, approximately 1 cm × 1 cm, were dissected from 5 randomly maternal sites of the placenta. The samples were partitioned into small pieces and washed three times with phosphate buffer saline (PBS). Some specimens were frozen in liquid nitrogen and stored at −80 °C for western blotting, while the rest were fixed in paraffin for immunohistochemical examination. All samples were collected within 40 min after delivery.
Table 1.
Clinical characteristics of the human participants.
| Category | Normal (n = 12) | Preeclampsia (n = 12) | P-value |
|---|---|---|---|
| Age (years) | 29.25 ± 1.82 | 29.50 ± 3.85 | 0.841a |
| Body mass index (BMI; kg/m2) | 27.87 ± 1.59 | 28.04 ± 2.44 | 0.839a |
| Gravidity | 2.50 ± 1.00 | 2.58 ± 0.90 | 0.832a |
| Gestational age at delivery (weeks) | 39.00 ± 1.24 | 37.14 ± 1.52 | 0.125 b |
| Placental weight (g) | 534.17 ± 56.16 | 420.00 ± 120.91 | 0.009a |
| Neonatal birth weight (g) | 3352.93 ± 210.37 | 2652.50 ± 745.18 | 0.003 b |
| Proteinuria (g/24 h) | 0.00 ± 0.00 | 2.75 ± 1.22 | <0.001 b |
| Systolic blood pressure (mmHg) | 112.33 ± 8.98 | 155.08 ± 19.68 | <0.001a |
| Diastolic blood pressure (mmHg) | 72.83 ± 9.20 | 98.25 ± 12.79 | <0.001a |
Values are shown as the mean ± SD; aStudent T-test; and bMann-Whitney test.
2.2. Immunohistochemistry
The placenta tissues were washed with PBS and fixed overnight with 4 % paraformaldehyde (P0099, Beyotime, China) at room temperature. The tissues were then dehydrated and embedded in paraffin before sectioning into 4-mm sections. To perform the immunohistochemical analysis, the sections were deparaffinized in xylene, rehydrated in a serial ethanol gradient, blocked with 3 % H2O2 for 10 min, and then microwaved in 10 mM citric sodium (pH 6.0) for 15 min to retrieve antigens. After that, the sections were incubated with primary rabbit antibody against CD73 (1:200; #ab175396, Abcam, U.K.) at 4 °C overnight. Following incubation, a goat-anti-rabbit IgG secondary antibody (1:500; #ab97051, Abcam, U.K.) was applied for 30 min at room temperature. The signals were displayed with a diaminobenzidine solution. Sections stained with rabbit IgG served as negative controls.
2.3. Cell culture and chemical treatments
The HTR-8/SVneo immortalized human trophoblast cell line used in this study was obtained from the American Type Culture Collection (ATCC, USA). The HTR-8/SVneo cell line was cultured in RPMI 1640 medium (C11875500BT; Gibco, USA) supplemented with 10 % fetal bovine serum (FBS, ST30-2602; Pansera, Germany) and 1 % penicillin-streptomycin (C0222, Beyotime, China) at 37 °C under 5 % CO2. The specific CD73 inhibitor α,β-methylene adenosine diphosphate (APCP, M3763; Sigma, USA, 100 μM) or AB-680 (HY-1125286; MCE, China, 100 nM), adenosine (ADO, HY-B0228; MCE, China, 25 or 50 μM), 5′-N-ethylcarboxamidoadenosine (NECA; HY-103173; MCE, China, 30 nM), 8-cyclopentyl-1,3-dipropylxanthine (DPCPX, HY-100937; MCE, China, 100 nM), ZM 241385 (HY-19532; MCE, China, 10 nM), and MRS 1706 (HY-103186; MCE, China, 10 nM) were used in this study.
2.4. Western blotting analysis
The total protein from placenta tissues or HTR-8/SVneo cells was extracted using a radio-immunoprecipitation assay (RIPA) lysis buffer (P0013B, Beyotime Institute of Biotechnology, China) that contained a protease and phosphatase inhibitor cocktail (P1045, Beyotime Institute of Biotechnology, China). Total protein concentration was determined using a Bicinchoninic Acid kit (P0012, Beyotime Institute of Biotechnology, China). Protein from each sample (30 μg) was loaded and separated by 10 % SDS-PAGE gel electrophoresis and transferred to the polyvinylidene fluoride membrane (03010040001, Roche, Switzerland). After blocking in 5 % skim milk in TBST (20 mM Tris-HCL, 1500 mM NaCl, and 0.1 % Tween 20) at room temperature for 1.5 h, the membrane was washed three times with TBST for 30 min and subsequently incubated at 4 °C overnight with the primary antibody including E-cadherin (1:1000, #3195, Cell Signaling Technology, USA), N-cadherin (1:1000, #13116, Cell Signaling Technology, USA), Vimentin (1:1000, #5741, Cell Signaling Technology, USA), CD73 (1:1000, #13160S, Cell Signaling Technology, USA), GAPDH (1:1000, #5174, Cell Signaling Technology, USA) and β-actin (1:5000, Catalog#: GB11001, Servicebio, China). Subsequently, the membrane was washed three times with TBST for 30 min and incubated with the horseradish peroxidase (HRP)-conjugated secondary antibody (1:5000, Proteintech, China) in a blocking solution for an hour at room temperature. Immunoreactive bands were illuminated by enhanced chemiluminescence reagents and were analyzed using Fusion analysis software (FX5; Vilber Lourmat, France). All experiments were repeated independently three times.
2.5. Measurement of CD73 activity
We assessed the CD73 enzyme activity of HTR-8/SVneo cells by measuring the conversion rate from AMP to adenosine. In brief, HTR-8/SVneo cells were incubated with Hank's balanced salt solution (HBSS) in the presence or absence of APCP for 30 min. After washing 3 times with HBSS, cells were stimulated with 100 μM AMP for 10 min. Then, the supernatants (100 μl) were subjected to liquid chromatograph-mass spectrometry (LC-MS) analysis (Agilent 1290–6495C, California, USA). An Ultimate® HILIC Amphion column (2.1 × 100 mm, 3 μm, Welch Materials, China) was used for separation. The mobile phase consisted of (A) 0.1 % formic acid solution and (B) acetonitrile, 0–2.0 min, 2 % A; 2.0–6.0 min, 70 % A; 6.0–7.0 min, 70 % A; 7.0–8.0 min, 2 % A. The flow rate was 0.3 ml/min, and the injection volume was 2 μl. Adenosine was monitored with an MRM transition of 268.11 to 136.1 m/z, fragmentor of 166 V and collision energy of 17 V. AMP was monitored with an MRM transition of 348.07 to 136.1 m/z, fragmentor of 166 V and collision energy of 21 V. Data were processed with the Agilent MassHunter Quantitative Analysis software (version 10.1, Agilent, USA). The enzyme activity of CD73 was expressed as adenosine production per mg protein for 10 min.
2.6. RNA-sequencing and data analysis
RNA sequencing was completed by Novogene Inc. (Beijing, China). Total intracellular RNA was extracted from HTR-8/SVneo cells using TRIzol reagent (15596026, Invitrogen, USA) according to the manufacturer's instructions. The integrity of total RNA was measured using an Agilent 2100 Bioanalyzer. The cDNA library was constructed via the NEBNext Ultra Directional RNA Library Prep Kit for Illumina. Qubit2.0 Fluorometer and quantitative RT-PCR (qRT-PCR) were performed to determine the effective concentration of the cDNA library. After library pooling, sequencing was performed using Illumina technology. High-quality reads were mapped to the reference genome utilizing the HISAT2. Differential genes were selected following the criteria: |log2(FoldChange)| ≥ 0 and padj≤0.05. GO Enrichment Analysis was applied to the differential genes. RNA sequencing was performed with three biological replicates from each group.
2.7. qRT-PCR
Total RNA was isolated from HTR-8/SVneo cell lines with the TRIzol reagent (Invitrogen), according to the manufacturer's instructions. A NanoDrop 2000 (Thermo Fisher Scientific) was used to determine extracted RNA concentration and quality. Extracted RNA was reverse transcribed to cDNA using an Evo M-MLV RT Kit (#AG11728, ACCURATE BIOTECHNOLOGY(HUNAN)CO.,LTD, ChangSha, China). qPCR was performed using a SYBR Green Premix Pro Taq HS qPCR Kit (#AG11701, ACCURATE BIOTECHNOLOGY(HUNAN)CO.,LTD, ChangSha, China) and a CFX96 Real-Time System (Bio-Rad, USA). Relative gene expression was calculated using the 2−ΔΔCt method, and the GAPDH was used as a housekeeping gene. Primer pairs to perform qRT-PCR were as follows: N-cadherin: forward: 5′ GGACAGCCTCTTCTCAATGT 3′, reverse: 5′ GCCACTTTTCCTGGGTCTC 3′; vimentin: forward: 5′ GCACACAGCAAGGCGAT 3′, reverse: 5′ GATTCAAGTCTCAGCGGGC 3′; MMP9: forward: 5′ CAGTACCGAGAGAAAGCCTATT 3′, reverse: 5′ CAGGATGTCATAGGTCACGTAG 3′; PDK1: forward: 5′ AACCGACACAATGATGTCATTC 3′, reverse: 5′ ATGCGACTCATGTAGAATCGAT 3′; HK-II: forward: 5′ CGACAGCATCATTGTTAAGGAG 3′, reverse: 5′ GCAGGAAAGACACATCACATTT 3′; LDHA: forward: 5′ AGGTGATCAAACTCAAAGGCTA 3′, reverse: 5′ CCCAAAATGCAAGGAACACTAA 3′; PFK: forward: 5′ GCGAGACCAAGATAAATATCGC 3′, reverse: 5′ CCTGTCGTTCTAGCTCCATTAT 3′; PDK4: forward: 5′ CAAGATGCCTTTGAGTGTTCAA 3′, reverse: 5′ GGTCTTCTTTTCCCAAGACAAC 3′; GAPDH: forward: 5′ GCACCGTCAAGGCTGAGAAC 3′, reverse: 5′ TGGTGAAGACGCCAGTGGA 3′.
2.8. Enzyme-linked immunosorbent assay (ELISA)
The concentration of intracellular acetyl-CoA (# JL32777; Jianglai Biology, China), extracellular ATP (#14432H1; Jiangsu Meimian Industrial Co., Ltd, China), AMP (#2039H1; Jiangsu Meimian Industrial Co., Ltd, China), and adenosine (#1913H1; Jiangsu Meimian Industrial Co., Ltd, China) were determined with ELISA Kits, analyzed according to manufacturer's instructions.
2.9. Cell proliferation assay
HTR-8/SVneo cells were seeded into 96/well plates at 5 × 10 3 cells per well, precultured for 24 h, and then combined with different concentrations of adenosine (0, 10, 25, 50, 75, 100 and 125 μM) for another 24 h. A 10 μl aliquot of CCK-8 solution (HY–K0301; MCE, China) was added to each well, and the cells were incubated at 37 °C for 4 h. A Multiskan Go microplate reader (Thermo Fisher Scientific) was used to measure the absorbance at 450 nm. All experiments were repeated independently three times.
2.10. Cell cycle assay
HTR-8/SVneo cells treated with or without adenosine were detached using trypsin, fixed with 500 μL iced 70 % ethanol, and stained using a Coulter DNA-Prep Reagents Kit (Beckman Coulter, USA). The stained HTR-8/SVneo cells were then analyzed using a CytoFlex Flow Cytometer (Beckman Coulter, USA). All experiments were repeated independently three times.
2.11. Cell wound-healing assay
HTR-8/SVneo cells were incubated in a 6-well plate at a density of 5 × 105 cells per well. When the cell confluence reached 80 %, the cells were scratched with a 200 μl pipette tip, washed three times with PBS, and then RPMI 1640 medium containing 2 % FBS was added. The scratch sizes of cells at 0 h, 12 h, and 24 h were recorded with a Z1 inverted microscope (Carl Zeiss, USA). The percentage of wound healing was analyzed using ImageJ software (version 1.53; National Institutes of Health, USA). All experiments were repeated independently three times.
2.12. Oxygen consumption rate and extracellular acidification rate measurements
The Cell Mito Stress Test Kit (#103707-100; Agilent, USA) was used to measure oxygen consumption rates (OCRs). The HTR-8/SVneo cells were seeded in Seahorse micro-well plates in 80 μL of standard growth media. The cells were pretreated with different treatments and incubated at 37 °C and 5 % CO2 for 24 h. One μM oligomycin, 0.5 μM FCCP, and one μM Rot/AA, were added according to the manufacturer's instructions. The cells were analyzed using the XFp Extracellular Flux Analyzer (Agilent, USA).
The pH Xtra Glycolysis Assay Kit (# PH-100; Luxcel, Ireland) was used to measure the extracellular acidification rates (ECARs). The HTR-8/SVneo cells pretreated with different treatments were incubated in a CO2-free incubator at 37 °C for 2 h, washed with respiration buffer, and then had an addition of 150 μl of respiration buffer containing 1 μM pH probe. A further 0.02 μM of DMSO, 1 μM oligomycin, and 50 mM 2-DG were added to the corresponding wells. The cells were analyzed using a CLARIOstar Plus plate reader (BMG Labtech, Germany). All experiments were repeated independently three times.
2.13. Metabolomic analysis
2.13.1. The extraction of intracellular and extracellular metabolites
The cellular metabolism of HTR-8/SVneo cells was terminated using liquid nitrogen. Before extraction, 2,3,3,3-d4-alanine (0.3 μmol/sample) was added to each sample as an internal standard. Intracellular metabolites were extracted with 1.5 ml of a fresh extraction solution (methanol/chloroform, 9:1 v/v). The intracellular metabolite extracts and 0.5 ml of culture medium containing extracellular metabolites were centrifuged at 2000×g for 10 min. The isolated supernatants were then dried for 4–5 h with a Labconco CentriVap® SpeedVac concentrator (Labconco Corporation) and stored at −80 °C until chemical derivatization.
2.13.2. Methyl chloroformate (MCF) derivatization and gas chromatography-mass spectrometry (GC-MS) analysis
Dried intracellular and extracellular metabolite extracts were resuspended in 200 μl of sodium hydroxide (1 M). The MCF derivatization step was performed according to the method published by Han et al. [20]. All MCF-derivatized samples were analyzed using an Agilent GC7890B chromatography system coupled to a MSD5975 mass spectrometer (Agilent, California, USA) operating at 70 eV. The GC column was a ZB-1701 GC capillary column (30 m × 250 μm id × 0.15 μm with a 5 m guard column, Phenomenex, USA). GC-MS parameters and temperature programs were set according to the protocol reported in Han et al. [20].
2.13.3. Metabolite identification, data mining, and statistical analysis
For metabolite identification and deconvolution, the Automated Mass Spectral Deconvolution & Identification System (AMDIS) software was used. The metabolites were identified by comparing MS fragmentation patterns and GC retention time to an in-house MS library built from chemical standards. A commercial NIST mass spectrum library was used to identify the remaining chemicals. Relative abundance of the metabolites was determined using the MassOmics XCMS R-based script by measuring the peak height of the most substantial fragmented ion mass. These values were normalized by the internal standard (2,3,3,3-d4-alanine) and subsequently by the total concentration in each sample. The Metaboanalyst 3.0 package for R was used to perform the principal component analysis (PCA). Tukey's honest significant difference test was used in R to test for significant differences between the treatment and control groups. P values < 0.05 were considered statistically significant. The Pathway Activity Profiling (PAPi) algorithm [21] was used to estimate and compare various metabolic pathways in HTR-8/SVneo cells with different treatments. Graphical illustrations of the findings were generated using ggplot2 R packages. The metabolomic analysis was repeated with three biological replicates from each group.
2.13.4. 13C adenosine isotope tracking experiment
In 13C labeling experiments, HTR-8/SVneo cells were cultivated with or without 50 μM adenosine or 50 μM 13C-labelled adenosine (#CLM-3678-0.05, Cambridge Isotope Laboratories, USA) and harvested after 24 h. Extraction of intracellular and extracellular metabolites followed the identical procedure outlined in the non-labelling metabolomic experiment, as described in section 2.13.1. The distribution of 13C-labelling from adenosine in the identified metabolite pool was calculated using the 13C/12C ratio in the major mass fragments. Stable isotope labelled metabolites were identified by their metabolite-specific fragmentation and retention time using AMDIS. The labelling degree of metabolites was assessed by the difference in the fragmentation patterns between 13C-labelled metabolites and their counterpart 12C mass spectrum. All observed 13C-labelled metabolites from samples without additionally labelled substrates were corrected for natural isotope abundances. Finally, the percentage of isotope labelling enrichment in metabolites derived from the catabolism of 13C labelled adenosine was calculated by subtracting natural abundance from total 13C enrichment. The calculation was based on a formula published in Han et al. [20].
2.14. Statistical analysis
The data are presented as mean ± standard deviation (SD) or standard error of the mean (SEM). The data were analyzed using GraphPad Prism software (version 8, GraphPad Software, USA) and SPSS software (version 22.0, SPSS Inc., USA). Data were examined for normal distribution using Kolmogorov–Smirnov test. The Mann-Whitney test was used for non-normally distributed data, while the unpaired two-tailed t-test was applied for the normally distributed data to compare the two groups. Alternatively, the one-way analysis of variance (ANOVA) with Tukey post-hoc tests was used to compare multiple groups. A p-value <0.05 was considered to be statistically significant.
3. Results
3.1. Clinical characteristics
The clinical characteristics of the study subjects are shown in Table 1. The age, BMI, and gravidity were similar between the preeclampsia and the normotensive pregnancy groups. Women suffering from preeclampsia had significantly higher proteinuria, systolic blood pressure, and diastolic blood pressure, but placental weight and neonatal birth weight were lower, as compared to women with normotensive pregnancies.
3.2. CD73 expression was upregulated in preeclampsia human placentas
To determine the involvement of CD73 in the development of preeclampsia, we evaluated the expression of CD73 in human placental tissue using immunohistochemistry and western blotting. As illustrated in Fig. 1A, CD73 was expressed in various types of placental trophoblast cells, including cytotrophoblasts (CTB), syncytiotrophoblasts (STB), and EVTs. Western blotting demonstrated that CD73 protein levels were increased in preeclampsia placentas (Fig. 1B).
Fig. 1.
CD73 was upregulated in preeclampsia placental tissues (A) Immunohistochemical staining of CD73 in human term-placentas in normal pregnancies (n = 3 biological replicates) and preeclampsia patients (n = 3 biological replicates). The distribution of CD73 in women with normal or preeclamptic pregnancies was localized in trophoblast cells (400 × ; Scale bars: 100 μm). Serial sections of the same samples immunostained with nonspecific IgG were shown as a negative control. EVT, extravillous trophoblast; CTB, cytotrophoblast; STB, syncytiotrophoblast; PE, preeclampsia. (B) Representative western blotting analysis of CD73 in human term-placentas from normal pregnancies (n = 12 biological replicates) and preeclampsia patients (n = 12 biological replicates). PE: preeclampsia. The original blots were provided in supplementary data as Supplementary Fig. S5A. Results are shown as mean ± SEM, **p < 0.01, Student's t-test.
3.3. The effects of APCP treatment on the extracellular levels of ATP, AMP and adenosine in HTR-8/SVneo cells
We first assessed the effect of APCP treatment on the enzymatic activity of CD73 by measuring the amount of 5′-AMP hydrolyzed to adenosine using LC-MS. A representative LC-MS chromatogram was presented in Fig. S1A. The results showed that the production of adenosine with the treatment of 50 μM and 100 μM APCP was significantly decreased by 55.44 % and 70.37 % (Fig. S1A), respectively. We then investigated the changes in the extracellular ATP, AMP, and adenosine concentration. Results showed that the APCP treatment increased the extracellular concentration of ATP and AMP and decreased that of adenosine (Fig. S1B).
3.4. RNA-seq data analysis of the HTR-8/SVneo cells in the control and adenosine treatments
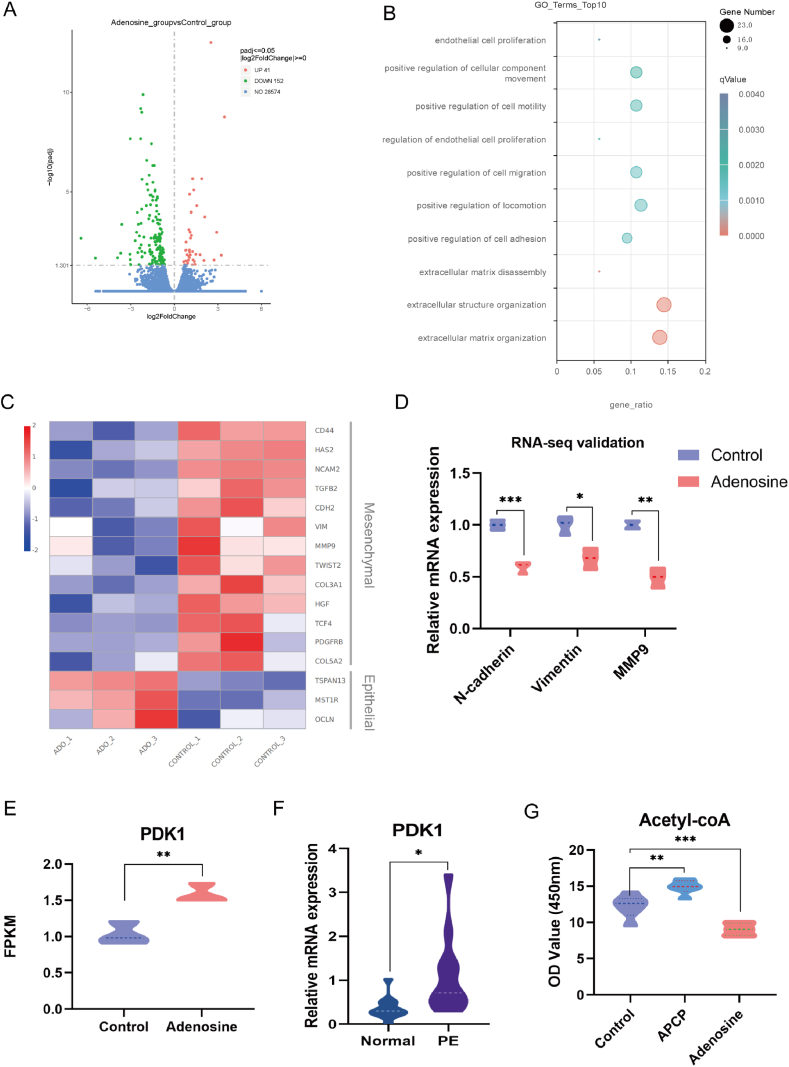
To characterize the impact of CD73/adenosine signaling on the transcriptome, we performed an RNA-seq analysis and compared HTR-8/SVneo cells with and without adenosine treatment. A total of 193 significant differential genes were discovered in the HTR-8/SVneo cells treated with adenosine, including 41 upregulated genes and 152 down-regulated genes (Fig. 2A). Analysis of these differential genes revealed that biological processes associated with extracellular adenosine signaling were enriched onto gene ontology terms such as extracellular matrix organization, positive regulation of cell adhesion, positive regulation of cell migration, and regulation of endothelial cell proliferation (Fig. 2B). Consistent with these enriched Gene Ontology terms, the expression of many mesenchymal marker genes, including vimentin, N-cadherin, and MMP9 decreased in a panel of EMT-related genes (Fig. 2C). We confirmed the expression levels of these genes using a qRT-PCR assay. The levels of vimentin, N-cadherin and MMP9 mRNA were significantly decreased in the adenosine-treated cells (Fig. 2D), displaying a similar expression trend to the RNA-seq data. Notably, we found that the transcription of pyruvate dehydrogenase kinase 1 (PDK1) was significantly increased (P < 0.01) through RNA-seq data mining (Fig. 2E). We then examined the expression level of PDK1 mRNA in human samples by qRT-PCR, and the results showed that preeclamptic placentas expressed increased levels of PDK1 mRNA as compared to normal placentas (Fig. 2F). Given that PDK1 is a key factor of energy metabolism, curbing the formation of acetyl-CoA from pyruvate [22], an ELISA assay was performed to determine whether adenosine alters acetyl-CoA levels in HTR-8/SVneo cells. As illustrated in Fig. 2G, the concentration of intracellular acetyl-CoA was significantly decreased in the cells treated with adenosine and increased in the cells treated with APCP. These findings suggest that CD73/adenosine may be involved in regulating proliferation, EMT, migration and energy metabolism of trophoblasts.
Fig. 2.
Adenosine signaling regulates gene expression of HTR-8/SVneo cells and the transcription of important EMT markers (A) HTR-8/SVneo cells were treated with or without adenosine (50 μM) for 24 h and RNA-Seq (n = 3 biological replicates per group) was performed. The volcano plot illustrates the 152 downregulated genes (green) and 41 upregulated genes (red) using RNA-seq data. (B)The Gene Ontology (GO) enrichment analysis of differentially expressed genes (DEGs) from RNA-seq data. The ordinate indicates the enriched pathways, and the abscissa gene ratio indicates the ratio of the DEGs number to the total number of genes in a certain pathway. The size of the dot indicates the number of genes with biological process terms, while the color represents the adjusted p value. (C) Heat map with some EMT markers in control and adenosine-treated samples from RNA-Seq. (D) Validation of RNA-seq data for some EMT markers by real-time PCR. (E) FPKM expression by RNA-seq between the control and adenosine groups. (F) PDK1 mRNA level in human term-placentas of pregnant women with or without preeclampsia by using qRT-PCR. PE: preeclampsia. Results are shown as mean ± SD, n = 12 biological replicates, *p < 0.05, Student's t-test. (G) HTR-8/SVneo cells were treated with or without adenosine (50 μM) or APCP (100 μM) for 24 h, and the concentrations of Acetyl-coA in HTR-8/SVneo cells were measured using ELISA. n = 3 biological replicates for each group. *p < 0.05, **p < 0.01, ***p < 0.001; One-way ANOVA. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. High concentrations of adenosine (ADO) inhibited the proliferation of HTR-8/SVneo cells
We performed a CCK-8 assay and flow cytometry analysis to investigate the effect of adenosine on the proliferation of HTR-8/SVneo cells. The CCK8 results showed that the relative number of HTR-8/SVneo cells gradually decreased with the increase of adenosine concentration, in a dose-dependent manner (Fig. 3A). At 50 μM adenosine, the viability of HTR-8/SVneo cells decreased by nearly 50 % relative to the control. Consistent with this finding, flow cytometry analysis revealed that 50 μM adenosine treatment of HTR-8/SVneo cells for 24 h led to S phase arrest. Compared with the control, adenosine treatment caused an increase in the percentage of cells in the S phase, which was accompanied by a decrease in the percentage of cells in the G2/M-phase (Fig. 3B).
Fig. 3.
CD73/adenosine signaling affected the viability, cell cycle, migration, and induced an epithelial phenotype in HTR-8/SVneo cells. (A) Cell viability was tested using Cell Counting Kit 8 (HY–K0301; MCE, China) at 24 h after treatment with 0, 10, 25, 50, 75, 100, 125 μM adenosine in HTR-8/SVneo Cells. n = 3 biological replicates for each group. (B) The cell cycle of HTR-8/SVneo cells treated with or without 50 μM adenosine were detected using flow cytometry. n = 3 biological replicates for each group. (C) Wound-healing assays for HTR-8/SVneo cells in the presence of 100 μM APCP or 25 μM adenosine. Images were taken at 0 h, 12 h and 24 h after treatment. n = 3 biological replicates for each group. (D) Western blotting of N-cadherin, E-cadherin and Vimentin in the aforementioned groups of cells. n = 3 biological replicates for each group. The original blots were provided in supplementary data as Supplementary Fig. S5B. (E) HTR-8/SVneo cells were treated with or without 50 μM adenosine, 30 nM 5′-N-ethylcarboxamido adenosine (NECA; non-selective adenosine receptor agonist), 100 nM DPCPX (A1-specific antagonist),10 nM ZM 241385 (A2A-specific antagonist), and 10 nM MRS 1706 (A2B-specific antagonist). Western blotting of N-cadherin, E-cadherin and vimentin in the aforementioned groups of cells. n = 3 biological replicates for each group. The original blots were provided in supplementary data as Supplementary Fig. S5C. Results are shown as mean ± SD, One-way ANOVA with post-hoc Tukey HSD, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
3.6. CD73/adenosine signaling suppressed migration and epithelial-mesenchymal transition in HTR-8/SVneo cells
To investigate the migration of trophoblast cells under the conditions of activation or inhibition of the CD73-adenosine pathway, we conducted the evaluation of the wound-healing assay on HTR-8/SVneo cells. It was found that the migratory capability of HTR-8/SVneo cells was suppressed in the presence of adenosine treatment but could be enhanced by APCP treatment (Fig. 3C). Previous studies found that CD73 was related to the EMT process and migration in cancer cell lines [5,8]. Therefore, it is important to understand whether CD73/adenosine signaling has an effect on the EMT process in HTR8-S/Vneo cells. Western blotting showed that the inhibition of CD73 (APCP or AB-680) significantly decreased the expression of E-cadherin (epithelial marker) and increased protein expression of N-cadherin (mesenchymal marker). By contrast, adenosine treatment resulted in the increased protein expression of E-cadherin (Fig. 3D and Fig. S3).
To further validate that the effects of extracellular adenosine occur through triggering adenosinergic signaling pathways, we treated cells with adenosine, NECA (non-selective adenosine receptor agonist), DPCPX (A1-specific antagonist), ZM 241385 (A2A-specific antagonist), and MRS 1706 (A2B-specific antagonist). Western blotting showed that NECA treatment increased the expression of E-cadherin and suppressed the expression of N-cadherin and vimentin, which was similar to adenosine treatment (Fig. 3E). The selective A2B antagonist, but not the A1 antagonist or A2A antagonist, prevented the NECA-mediated repression of EMT phenotype of HTR-8/SVneo cells. This demonstrated that the inhibitory effect of adenosine on EMT in HTR-8/SVneo cells is mainly mediated by the A2B receptor. Then, we determined the expression of E-cadherin, N-cadherin, and vimentin in human samples using western blotting. The results showed that preeclamptic placentas expressed an increased level of E-cadherin and decreased levels of N-cadherin and vimentin compared to normal placentas (Fig. S2). Together, these results indicate that CD73/adenosine signaling inhibited trophoblast migration through the EMT process.
3.7. Intracellular metabolite profiling
The PCA analysis plot of intracellular metabolite profiles of the three treatments revealed a clear separation among adenosine and APCP treatments and control cells (Fig. 4A). The mass spectral library identified a total of 131 metabolites, 37 of which were significantly different when comparing the two treatments with the control cells. The significant metabolites included eighteen amino acids or their derivatives, three tricarboxylic acid (TCA) cycle intermediates or their derivatives, one antioxidant and vitamin, four saturated fatty acids, four unsaturated fatty acids, one glyoxylate cycle intermediate, and six other metabolites (Fig. 4B). Fourteen amino acids (beta-alanine, glycine, threonine, tyrosine, asparagine, valine, leucine, 2-aminoadipic acid, isoleucine, tryptophan, alanine, phenylalanine, methionine, aspartic acid) were significantly decreased in the cells exposed to the adenosine treatment, whereas two saturated fatty acids and three unsaturated fatty acids were slightly increased. In contrast, the majority of intracellular metabolites (including nine amino acids, two TCA cycle intermediates, etc.) had a higher relative concentration in the APCP-treated cells compared with the control cells.
Fig. 4.
Effects of CD73/adenosine signaling on metabolite profiling of HTR-8/SVneo cells (A) HTR-8/SVneo cells were treated with PBS (control group) or 100 μM APCP (APCP group) or 50 μM adenosine (adenosine group) for 24 h, then intracellular and extracellular metabolites of each group (n = 3 biological replicates) were measured by gas chromatography mass spectrometry (GC-MS). PCA plot of the metabolite profiles from control, adenosine and APCP groups. (B) Heatmap of the intracellular metabolite profiles from adenosine and APCP treatment groups, when compared to the control group, respectively. Relative levels of intracellular metabolites are displayed as log2-fold changes. Red colors represent higher concentrations in the treatment than the control groups, while purple colors indicate lower metabolite levels, and the darker the color, the larger the difference. Tukey's honest significance test, only p < 0.05 are shown. (C) Relative levels of extracellular metabolites in three different groups of HTR-8/SVneo cells. The relative concentrations of extracellular metabolite were normalized using an internal standard (2,3,3,3-d4-alanine). Subsequently, each metabolite level was subtracted from the corresponding level in the blank culture medium. The normalized levels were further adjusted for cell numbers using biomass. The positive values represent the release of metabolites into the medium, and the negative values indicate the consumption of metabolites. Only metabolites found to be statistically significant between the control group and the treatment groups (Tukey's honest significance test, p < 0.05) were shown. Only the key differential extracellular metabolites are displayed; the remaining results can be found in Supplementary Fig. S4. (D) Activity of HTR-8/SVneo cell metabolic pathways based on comparisons of intracellular metabolomics data between the control group and the treatment groups. The metabolic pathway activities relative to the control group were visualized using a log2 scale. Positive values represent upregulated metabolic pathway activities, and negative values specify downregulated activities. The horizontal lines indicate the 95 % confidence intervals for a ratio of two means (Treatments/Control). Solid patterns represent no statistical significance, and hollow patterns indicate statistical significance (Tukey's honest significance test p < 0.05). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.8. Extracellular metabolite profiling
The PCA plot of extracellular metabolite profiles revealed a clear separation between cells treated with APCP and untreated control cells (Fig. 4A). Meanwhile, the adenosine-treated cells did not show a distinct disparity from the control cells. We detected a total of 141 extracellular metabolites in the HTR-8/SVneo cells culture medium, and 50 of the extracellular metabolites were significantly different (P < 0.05) when comparing the control and two treatments. The significant metabolites included two TCA cycle intermediates, three glycolytic intermediates, sixteen amino acids, two short-chain fatty acids, two medium-chain fatty acids, fifteen long-chain fatty acids, one antioxidant and nine other metabolites (Fig. 4C). In the adenosine-treated cells, the uptake of short-chain fatty acids and secretion of pyruvic acid was significantly increased (Fig. 4C). The adenosine-treated cells showed relatively smaller differences in metabolite levels when compared to the control cells, than the differences observed when comparing the APCP-treated cells to the control cells (Fig. 4C). A significant increase in the secretion of two TCA cycle intermediates (α-ketoglutaric acid and citric acid), two glycolytic intermediates (lactic acid and 2-phosphoenolpyruvic acid), six amino acids, two medium-chain fatty acids, three long-chain fatty acids and an antioxidant were observed when the HTR-8/SVneo cells were treated with APCP (Fig. 4C).
3.9. Metabolic pathways
To estimate the metabolic pathways that might be dysregulated in HTR-8/Svneo cells with different treatments, identified intracellular metabolites were mapped onto the KEGG metabolic network. We found that thirty-six metabolic pathways, related to the metabolism of energy, nucleotides, lipids, amino acids, cofactors and vitamins, and cell growth and death, appeared to have been dysregulated in HTR-8/SVneo cells as a result of the two different treatments (Fig. 4D). It is worth noting that in HTR-8/SVneo cells, APCP treatment significantly upregulated fourteen metabolic pathways including three pathways related to energy metabolism (Fig. 4D). In addition, there is a significant upregulation of lipid metabolism and a significant downregulation of amino acid metabolism in the adenosine-treated cells (Fig. 4D).
3.10. CD73/adenosine signaling suppressed the oxidative phosphorylation (OXPHOS) and glycolysis of HTR-8/SVneo cells
To further validate the effect of CD73/adenosine signaling on energy metabolism in HTR-8/SVneo cells, we measured the oxygen consumption rates (OCRs) and the extracellular acidification rates (ECARs). The results demonstrated that compared with the untreated control cells, the APCP treatment significantly increased basal respiration, maximal respiratory capacity, ATP production (Fig. 5A) and basal glycolysis (Fig. 5B) in HTR-8/Svneo cells, while adenosine treatment inhibited basal respiration, basal glycolysis and maximal glycolytic capacity of HTR-8/Svneo cells. We also evaluated the mRNA levels of key enzymes involved in glucose metabolism in HTR-8/SVneo cells: hexokinase II (HK-II), phosphofructokinase (PFK), lactate dehydrogenase (LDHA) and pyruvate dehydrogenase kinase 4 (PDK4). The results showed that HK-II and PFK mRNA were decreased, while LDHA and PDK4mRNA were increased in the adenosine-treated cells (Fig. 5C).
Fig. 5.
CD73 restricts mitochondrial respiration and glycolysis in HTR-8/SVneo cells (A) HTR-8/SVneo cells were pretreated with PBS (control group) or 100 μM APCP (APCP group) or 50 μM adenosine (adenosine group) for 24 h, and OCR value of each group was measured using a Seahorse analyzer. n = 3 biological replicates per group. (B) ECAR value of HTR-8/SVneo cells in the normal group, APCP group and adenosine group. n = 3 biological replicates per group. (C) Expression of hexokinase II (HK-II), phosphofructokinase (PFK), lactate dehydrogenase A (LDHA) and pyruvate dehydrogenase kinase 4 (PDK4) mRNAs were analyzed by real-time PCR in HTR-8/SVneo cells with different treatments (n = 3 biological replicates per group). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; One-way ANOVA. (D) Incorporation of 13C derived from 13C adenosine into several key metabolites from the HTR-8/SVneo cells with or without APCP treatment. The solid circle represents the 13C-labelled carbon atom, and the hollow circle represents the unlabeled carbon atom. The fractional abundance differences of 13C-labelled metabolites are shown between the unlabeled adenosine group (13C-Natural abundance) and 13C adenosine group (13C enrichment). M represents the main molecular ion of identified metabolites. M+1 is 1 m/z higher than the M. n = 3 biological replicates per group. Significance: *p < 0.05, **p < 0.01, ***p < 0.001; Student's t-test.
3.11. Isotope labelled adenosine tracing
To track the metabolism of adenosine in vitro, we supplied HTR-8/Svneo cells with 13C-labelled adenosine. In this study, 13C-labelling patterns of intracellular metabolites were similar to extracellular metabolites. We observed that many metabolites were significantly increased in two and/or three 13C-labelling in the HTR-8/Svneo cells incubated in a medium containing 13C-adenosine in relation to cells treated with 12C-adenosine. These included lactate (one 13C-labelled), glutamate (one to five 13C-labelled), proline (two to three 13C-labelled), and some TCA cycle intermediates including citrate (two to three 13C-labelled), succinate (one and three 13C-labelled), and fumarate (three 13C-labelled) (Fig. 5D).
4. Discussion
Cellular metabolic homeostasis is critical for maintaining normal cell function. Clarifying the regulatory mechanisms in trophoblast metabolism will further our understanding of the pathogenesis of gestational diseases related to trophoblast dysfunction. This study applied metabolomics and transcriptomics analysis to reveal the metabolic role of the CD73/adenosine signaling pathway on trophoblast function. Our results demonstrated that the expression of CD73 was significantly increased in the preeclamptic placenta. By treating HTR-8/SVneo cells with adenosine and CD73 inhibitors, we found that the CD73/adenosine signaling pathway restricted cellular energy metabolism and amino acid synthesis, and elevated the uptake of short-chain fatty acids in trophoblast cells. These metabolic alterations likely contribute to the inhibition of proliferation, migration, and EMT of trophoblast cells.
4.1. Adenosine serves as a double-edged sword in the regulation of the TCA cycle
Our previous publication reported that CD73 inhibitors reduced the production of extracellular adenosine in placental trophoblast cells [23]. Other studies also suggested that adenosine plays an important role in cellular energy metabolism and suppresses mitochondrial respiration [10,24]. Our RNA-seq and ELISA experiments demonstrated that adenosine promoted the expression of PDK1 (Fig. 2E) and decreased intracellular acetyl-CoA levels (Fig. 2G). It is known that PDK1 inactivates mitochondrial pyruvate dehydrogenase (PDH), which in turn decarboxylates pyruvate to acetyl-CoA and subsequently adds an acetyl group to TCA cycle intermediates to maintain mitochondrial function [25]. Although no study has reported a direct association between adenosine and PDK1, adenosine is an activator of Wnt signaling [26,27], which is an upstream singling pathway to promote the expression of PDK1 [28]. Based on these findings, we suggest that extracellular adenosine signaling may activate PDK1 and lead to downregulation of the TCA cycle.
However, adenosine may not only suppress respiratory metabolism but may also act as an alternative carbon source that feeds into the TCA cycle. Our 13C-labelled adenosine metabolic flux results revealed that extracellular adenosine could be incorporated into intracellular TCA cycle intermediates, lactic acid, and several amino acids (Fig. 5D). Although glucose is the primary source of carbon and energy for cells, adenosine may become an important energy supply when cells experience metabolic stress [29]. Indeed, extracellular adenosine is internalized via the equilibrative nucleoside transporters (ENTs), and then intracellular adenosine could be either phosphorylated to AMP by adenosine kinase (ADK) or deaminated to inosine through adenosine deaminase reaction [30]. Subsequently, inosine can be decomposed into ribose-1-phosphate by purine nucleoside phosphorylase (PNP) and enter glycolysis through the pentose phosphate pathway [29,31]. These data suggest that adenosine might be double-edged in regulating energy metabolism in trophoblast cells. On the one hand, adenosine inhibits the TCA cycle by upregulating PDK1; on the other hand, adenosine could be supplied into the metabolic flux as a carbon source.
4.2. CD73/adenosine pathway inhibits glycolytic metabolism but promotes the utilization of fatty acids in trophoblasts
Apart from influencing the TCA cycle, the CD73/adenosine pathway appears to regulate glycolytic metabolism and fatty acids utilization in the trophoblast. Our results showed that CD73 inhibition increased glycolytic capacity (Fig. 5B) and promoted the accumulation of extracellular lactic acid (Fig. 4C) in HTR-8/SVneo cells, while adenosine treatment decreased extracellular acidification rate (Fig. 5B) and promoted the secretion of pyruvate (Fig. 4C). Wang et al. showed the inhibitory effects of adenosine on glycolysis and they also found that excess adenosine suppressed the expression of glycolytic‐related enzymes (HK-II and PFK) by activation of adenosine receptor 2A (A2AR) [32]. Finegan et al. also demonstrated that adenosine treatment suppressed the rate of glycolysis and cellular acidosis [33]. It has been reported that pyruvate can be transported out of the cell by monocarboxylate transporters (MCTs) [[34], [35], [36], [37]], and these transporters were activated by elevated extracellular pH [38,39]. Thus, we speculate that the increased secretion of pyruvate might result from the downregulation of glycolysis by adenosine. Furthermore, Sivaprakasam et al. reported that MCTs are involved in the uptake of short-chain fatty acids (SCFAs) through the plasma membrane [40]. Li et al. found that several types of SCFAs were increased in patients with preeclampsia and are positively correlated with systolic, and diastolic blood pressure levels [41]. Our extracellular metabolite profiling pinpointed that the uptake of SCFA increased significantly when extracellular adenosine concentration increased (Fig. 4C). Compared to long-chain fatty acids, the entry of SCFAs into the mitochondrial membranes does not depend on the carnitine shuttle. Schönfeld and Wojtczak proposed that SCFAs can serve as a more immediate energy fuel than LCFAs [42]. Since fatty acid oxidation and pyruvate metabolism are the main routes for the production of acetyl-CoA [43], we propose that the uptake of SCFA may be a compensatory mechanism to facilitate the production of acetyl-CoA when pyruvic acid secretion is promoted by adenosine. Further studies would validate this hypothesis.
4.3. The adenosine-induced energy dysregulation reduces migration and proliferation of trophoblasts
Adenosine-related suppression of trophoblast migration seems to be associated with dysregulated energy metabolism. Our seahorse experiment showed that adenosine treatment decreased mitochondrial basal respiratory level and glycolysis level in HTR-8/SVneo cells (Fig. 5A and B). Additionally, we found that the migration of HTR-8/SVneo cells was compromised after adenosine treatment (Fig. 3C). Both RNA-seq analysis (Fig. 2C) and western blotting results (Fig. 3D) demonstrated that the expression of EMT-related genes was decreased after adenosine treatment. Evidence suggests that preeclamptic placentas presented with mitochondrial dysfunction [44] and glycolytic inhibition [45]. It has been reported that cell migration demands a large amount of energy generated from mitochondria [46], and that dysfunction in aerobic respiration attenuated trophoblast migration [47]. Liang et al. demonstrated that inhibition of aerobic glycolysis impaired EMT, migration and invasion of trophoblast [48]. These findings indicate that impaired energy metabolism may result in the suppression of EMT and migration of trophoblast. On the other hand, adenosine seems to suppress the proliferation of trophoblast through the downregulation of amino acid metabolism. Our metabolomics data showed that the concentration of fourteen amino acids was decreased after adenosine treatment, while the levels of nine amino acids were increased after CD73 inhibition (Fig. 4B). Amino acids are the essential building blocks for cell growth and proliferation, and the reduction of amino acid bioavailability restrains cellular proliferative capacity [49,50]. Taken together, these findings suggest that the upregulated CD73/adenosine signaling pathway can potentially disrupt the equilibrium of bioenergetic and biosynthetic processes, resulting in the inhibition of EMT phenotype, migration, and proliferation of trophoblast. Since trophoblast dysfunction leads to defective remodeling of spiral arteries, which is thought to be the leading pathomechanism in preeclampsia [51], the CD73/adenosine signaling pathway may participate in the pathogenesis of preeclampsia.
There are some limitations in this study that are worth noting. Firstly, for ethical reasons, we could not obtain clinical samples during early pregnancy and hence collected placenta during late pregnancy. Secondly, although the HTR-8/SVneo cell line is frequently used as an EVT replacement, it may not fully represent EVTs under normal physiological conditions.
In summary, this study revealed a novel role of the CD73/adenosine signaling pathway in the metabolic remodeling of trophoblast (Fig. 6). The CD73/adenosine signaling pathway suppressed energy metabolism and amino acids synthesis, and resulted in a compensatory increase in SCFA utilization, all of which affected the EMT, migration and proliferation of trophoblast cells. Upregulated CD73/adenosine signaling pathway may play an important role in the development and progression of preeclampsia, and is a possible therapeutic target for the disease. Future studies of primary trophoblast cells and live animal subjects are required to verify our findings.
Fig. 6.
CD73/adenosine signaling plays a role in the alteration of cellular metabolism.
Ethics statement
The collection of human placenta tissue was approved by the Ethics Committee of Chongqing Medical University (Ethic No. 2014034).
Funding
This work was supported by the National Natural Science Foundation of China (No. 81971406, 82271715, 82071671, 81871185), The 111 Project (Yuwaizhuan (2016)32), Chongqing Science & Technology Commission (cstc2021jcyj-msxmX0213), Natural Science Foundation of Chongqing, China (CSTB2022NSCQ-MSX1679), Chongqing Municipal Education Commission (KJZD-K202100407), Chongqing Health Commission and Chongqing Science & Technology Commission (2021MSXM121, 2020MSXM101), Doctoral Innovation Project of the First Affiliated Hospital of Chongqing Medical University (CYYY-BSYJSCXXM-202316).
Ethics approval
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of Chongqing Medical University (Ethic No. 2014034).
Informed consent statement
Informed consent was obtained from all subjects involved in the study.
Data availability
The data that support the findings of this study are available from the corresponding author on reasonable request.
CRediT authorship contribution statement
Guangmin Song: Writing – review & editing, Writing – original draft, Visualization, Methodology, Investigation, Conceptualization. Dan Zhang: Writing – review & editing, Investigation. Jianan Zhu: Resources, Investigation, Conceptualization. Andi Wang: Methodology. Xiaobo Zhou: Methodology. Ting-Li Han: Writing – review & editing, Project administration, Formal analysis. Hua Zhang: Writing – review & editing, Supervision, Project administration, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e25252.
Contributor Information
Ting-Li Han, Email: tinglihan@cqmu.edu.cn.
Hua Zhang, Email: zh2844@gmail.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Davies J.E., Pollheimer J., Yong H.E., Kokkinos M.I., Kalionis B., Knöfler M., Murthi P. Epithelial-mesenchymal transition during extravillous trophoblast differentiation. Cell Adhes. Migrat. 2016;10(3):310–321. doi: 10.1080/19336918.2016.1170258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DaSilva-Arnold S., James J.L., Al-Khan A., Zamudio S., Illsley N.P. Differentiation of first trimester cytotrophoblast to extravillous trophoblast involves an epithelial-mesenchymal transition. Placenta. 2015;36(12):1412–1418. doi: 10.1016/j.placenta.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 3.Li Y., Yan J., Chang H.M., Chen Z.J., Leung PCK roles of TGF-β superfamily proteins in extravillous trophoblast invasion. Trends Endocrinol. Metabol.: TEM (Trends Endocrinol. Metab.) 2021;32(3):170–189. doi: 10.1016/j.tem.2020.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Dongre A., Rashidian M., Eaton E.N., Reinhardt F., Thiru P., Zagorulya M., Nepal S., Banaz T., Martner A., Spranger S., Weinberg R.A. Direct and indirect regulators of epithelial-mesenchymal transition-mediated immunosuppression in breast carcinomas. Cancer Discov. 2021;11(5):1286–1305. doi: 10.1158/2159-8290.CD-20-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Z., Gu C., Yao X., Guo W., Wang H., Lin T., Li F., Chen D., Wu J., Ye G., Zhao L., Hu Y., Yu J., Shi J., Li G., Liu H. CD73 promotes tumor metastasis by modulating RICS/RhoA signaling and EMT in gastric cancer. Cell Death Dis. 2020;11(3):202. doi: 10.1038/s41419-020-2403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowser J.L., Blackburn M.R., Shipley G.L., Molina J.G., Dunner K., Jr. & Broaddus RR Loss of CD73-mediated actin polymerization promotes endometrial tumor progression. J. Clin. Invest. 2016;126(1):220–238. doi: 10.1172/JCI79380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma X.L., Shen M.N., Hu B., Wang B.L., Yang W.J., Lv L.H., Wang H., Zhou Y., Jin A.L., Sun Y.F., Zhang C.Y., Qiu S.J., Pan B.S., Zhou J., Fan J., Yang X.R., Guo W. CD73 promotes hepatocellular carcinoma progression and metastasis via activating PI3K/AKT signaling by inducing Rap1-mediated membrane localization of P110β and predicts poor prognosis. J. Hematol. Oncol. 2019;12(1):37. doi: 10.1186/s13045-019-0724-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martínez-Ramírez A.S., Díaz-Muñoz M., Battastini A.M., Campos-Contreras A., Olvera A., Bergamin L., Glaser T., Jacintho Moritz C.E., Ulrich H., Vázquez-Cuevas F.G. Cellular migration ability is modulated by extracellular purines in ovarian carcinoma SKOV-3 cells. J. Cell. Biochem. 2017;118(12):4468–4478. doi: 10.1002/jcb.26104. [DOI] [PubMed] [Google Scholar]
- 9.Ferretti C., Bruni L., Dangles-Marie V., Pecking A.P., Bellet D. Molecular circuits shared by placental and cancer cells, and their implications in the proliferative, invasive and migratory capacities of trophoblasts. Hum. Reprod. Update. 2007;13(2):121–141. doi: 10.1093/humupd/dml048. [DOI] [PubMed] [Google Scholar]
- 10.Briceño P., Rivas-Yañez E., Rosemblatt M.V., Parra-Tello B., Farías P., Vargas L., Simon V., Cárdenas C., Lladser A., Salazar-Onfray F., Elorza A.A., Rosemblatt M., Bono M.R., Sauma D. CD73 ectonucleotidase restrains CD8+ T cell metabolic fitness and anti-tumoral activity. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.638037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arra M., Swarnkar G., Ke K., Otero J.E., Ying J., Duan X., Maruyama T., Rai M.F., O'Keefe R.J., Mbalaviele G., Shen J., Abu-Amer Y. LDHA-mediated ROS generation in chondrocytes is a potential therapeutic target for osteoarthritis. Nat. Commun. 2020;11(1):3427. doi: 10.1038/s41467-020-17242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rigaud V.O.C., Hoy R., Mohsin S., Khan M. Stem cell metabolism: powering cell-based therapeutics. Cells. 2020;9(11):2490. doi: 10.3390/cells9112490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaelin W.G., Jr., McKnight S.L. Influence of metabolism on epigenetics and disease. Cell. 2013;153(1):56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoneyama Y., Suzuki S., Sawa R., Yoneyama K., Power G.G., Araki T. Increased plasma adenosine concentrations and the severity of preeclampsia. Obstet. Gynecol. 2002;100(6):1266–1270. doi: 10.1016/s0029-7844(02)02247-0. [DOI] [PubMed] [Google Scholar]
- 15.Iriyama T., Sun K., Parchim N.F., Li J., Zhao C., Song A., Hart L.A., Blackwell S.C., Sibai B.M., Chan L.N., Chan T.S., Hicks M.J., Blackburn M.R., Kellems R.E., Xia Y. Elevated placental adenosine signaling contributes to the pathogenesis of preeclampsia. Circulation. 2015;131(8):730–741. doi: 10.1161/CIRCULATIONAHA.114.013740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espinoza J., Espinoza A.F., Power G.G. High fetal plasma adenosine concentration: a role for the fetus in preeclampsia? Am. J. Obstet. Gynecol. 2011;205(5):485.e24–485.e27. doi: 10.1016/j.ajog.2011.06.034. [DOI] [PubMed] [Google Scholar]
- 17.Escudero C., Roberts J.M., Myatt L., Feoktistov I. Impaired adenosine-mediated angiogenesis in preeclampsia: potential implications for fetal programming. Front. Pharmacol. 2014;5:134. doi: 10.3389/fphar.2014.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darashchonak N., Sarisin A., Kleppa M.J., Powers R.W., von Versen-Höynck F. Activation of adenosine A2B receptor impairs properties of trophoblast cells and involves mitogen-activated protein (MAP) kinase signaling. Placenta. 2014;35(9):763–771. doi: 10.1016/j.placenta.2014.06.369. [DOI] [PubMed] [Google Scholar]
- 19.ACOG practice bulletin Diagnosis and management of preeclampsia and eclampsia. Number 33, January 2002. American College of obstetricians and Gynecologists. Int. J. Gynaecol. Obstet.: the official organ of the International Federation of Gynaecology and Obstetrics. 2002;77(1):67–75. [PubMed] [Google Scholar]
- 20.Han T.L., Cannon R.D., Gallo S.M., Villas-Bôas S.G. A metabolomic study of the effect of Candida albicans glutamate dehydrogenase deletion on growth and morphogenesis. NPJ biofilms and microbiomes. 2019;5(1):13. doi: 10.1038/s41522-019-0086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aggio R.B., Ruggiero K., Villas-Bôas S.G. Pathway Activity Profiling (PAPi): from the metabolite profile to the metabolic pathway activity. Bioinformatics. 2010;26(23):2969–2976. doi: 10.1093/bioinformatics/btq567. [DOI] [PubMed] [Google Scholar]
- 22.Kim J.W., Tchernyshyov I., Semenza G.L., Dang C.V. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metabol. 2006;3(3):177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Zhu J., Song G., Zhou X., Han T.L., Yu X., Chen H., Mansell T., Novakovic B., Baker P.N., Cannon R.D., Saffery R., Chen C., Zhang H. CD39/CD73 dysregulation of adenosine metabolism increases decidual natural killer cell cytotoxicity: implications in unexplained recurrent spontaneous abortion. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.813218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chambers A.M., Wang J., Lupo K.B., Yu H., Atallah Lanman N.M., Matosevic S. Adenosinergic signaling alters natural killer cell functional responses. Front. Immunol. 2018;9:2533. doi: 10.3389/fimmu.2018.02533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owen O.E., Kalhan S.C., Hanson R.W. The key role of anaplerosis and cataplerosis for citric acid cycle function. J. Biol. Chem. 2002;277(34):30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 26.Kim J., Shin J.Y., Choi Y.H., Lee S.Y., Jin M.H., Kim C.D., Kang N.G., Lee S. Adenosine and cordycepin accelerate tissue remodeling process through adenosine receptor mediated Wnt/β-Catenin pathway stimulation by regulating GSK3b activity. Int. J. Mol. Sci. 2021;22(11):5571. doi: 10.3390/ijms22115571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Procopio M.C., Lauro R., Nasso C., Carerj S., Squadrito F., Bitto A., Di Bella G., Micari A., Irrera N., Costa F. Role of adenosine and purinergic receptors in myocardial infarction: focus on different signal transduction pathways. Biomedicines. 2021;9(2):204. doi: 10.3390/biomedicines9020204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pate K.T., Stringari C., Sprowl-Tanio S., Wang K., TeSlaa T., Hoverter N.P., McQuade M.M., Garner C., Digman M.A., Teitell M.A., Edwards R.A., Gratton E., Waterman M.L. Wnt signaling directs a metabolic program of glycolysis and angiogenesis in colon cancer. EMBO J. 2014;33(13):1454–1473. doi: 10.15252/embj.201488598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giannecchini M., Matteucci M., Pesi R., Sgarrella F., Tozzi M.G., Camici M. Uptake and utilization of nucleosides for energy repletion. Int. J. Biochem. Cell Biol. 2005;37(4):797–808. doi: 10.1016/j.biocel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Yegutkin G.G., Boison D. ATP and adenosine metabolism in cancer: exploitation for therapeutic gain. Pharmacol. Rev. 2022;74(3):797–822. doi: 10.1124/pharmrev.121.000528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bzowska A., Kulikowska E., Shugar D. Purine nucleoside phosphorylases: properties, functions, and clinical aspects. Pharmacol. Therapeut. 2000;88(3):349–425. doi: 10.1016/s0163-7258(00)00097-8. [DOI] [PubMed] [Google Scholar]
- 32.Wang M., Jia J., Cui Y., Peng Y., Jiang Y. CD73-positive extracellular vesicles promote glioblastoma immunosuppression by inhibiting T-cell clonal expansion. Cell Death Dis. 2021;12(11):1065. doi: 10.1038/s41419-021-04359-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finegan B.A., Lopaschuk G.D., Coulson C.S., Clanachan A.S. Adenosine alters glucose use during ischemia and reperfusion in isolated rat hearts. Circulation. 1993;87(3):900–908. doi: 10.1161/01.cir.87.3.900. [DOI] [PubMed] [Google Scholar]
- 34.Vanderheyden N., Wong J., Docampo R. A pyruvate-proton symport and an H+-ATPase regulate the intracellular pH of Trypanosoma brucei at different stages of its life cycle. Biochem. J. 2000;346(Pt 1):53–62. Pt 1. [PMC free article] [PubMed] [Google Scholar]
- 35.Nicolae A., Wahrheit J., Bahnemann J., Zeng A.P., Heinzle E. Non-stationary 13C metabolic flux analysis of Chinese hamster ovary cells in batch culture using extracellular labeling highlights metabolic reversibility and compartmentation. BMC Syst. Biol. 2014;8:50. doi: 10.1186/1752-0509-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Contreras-Baeza Y., Sandoval P.Y., Alarcón R., Galaz A., Cortés-Molina F., Alegría K., Baeza-Lehnert F., Arce-Molina R., Guequén A., Flores C.A. San Martín A & Barros LF Monocarboxylate transporter 4 (MCT4) is a high affinity transporter capable of exporting lactate in high-lactate microenvironments. J. Biol. Chem. 2019;294(52):20135–20147. doi: 10.1074/jbc.RA119.009093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang B., Jin Q., Xu L., Li N., Meng Y., Chang S., Zheng X., Wang J., Chen Y., Neculai D., Gao N., Zhang X., Yang F., Guo J., Ye S. Cooperative transport mechanism of human monocarboxylate transporter 2. Nat. Commun. 2020;11(1):2429. doi: 10.1038/s41467-020-16334-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Futagi Y., Narumi K., Furugen A., Kobayashi M., Iseki K. Molecular characterization of the orphan transporter SLC16A9, an extracellular pH- and Na(+)-sensitive creatine transporter. Biochem. Biophys. Res. Commun. 2020;522(2):539–544. doi: 10.1016/j.bbrc.2019.11.137. [DOI] [PubMed] [Google Scholar]
- 39.Benson P.J., Purcell-Meyerink D., Hocart C.H., Truong T.T., James G.O., Rourke L., Djordjevic M.A., Blackburn S.I., Price G.D. Factors altering pyruvate excretion in a glycogen storage mutant of the cyanobacterium, synechococcus PCC7942. Front. Microbiol. 2016;7:475. doi: 10.3389/fmicb.2016.00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sivaprakasam S., Bhutia Y.D., Yang S., Ganapathy V. Short-chain fatty acid transporters: role in colonic homeostasis. Compr. Physiol. 2017;8(1):299–314. doi: 10.1002/cphy.c170014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J., Wang L., Chen H., Yang Z., Chen S., Wang J., Zhou Y., Xuan R. The diagnostic potential of gut microbiota-derived short-chain fatty acids in preeclampsia. Frontiers in pediatrics. 2022;10 doi: 10.3389/fped.2022.878924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schönfeld P., Wojtczak L. Short- and medium-chain fatty acids in energy metabolism: the cellular perspective. J. Lipid Res. 2016;57(6):943–954. doi: 10.1194/jlr.R067629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuramoto K., Yamamoto M., Suzuki S., Togashi K., Sanomachi T., Kitanaka C., Okada M. Inhibition of the lipid droplet-peroxisome proliferator-activated receptor α Axis suppresses cancer stem cell properties. Genes. 2021;12(1):99. doi: 10.3390/genes12010099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marín R., Chiarello D.I., Abad C., Rojas D., Toledo F., Sobrevia L. Oxidative stress and mitochondrial dysfunction in early-onset and late-onset preeclampsia. Biochim. Biophys. Acta, Mol. Basis Dis. 2020;1866(12) doi: 10.1016/j.bbadis.2020.165961. [DOI] [PubMed] [Google Scholar]
- 45.Bloxam D.L., Bullen B.E., Walters B.N., Lao T.T. Placental glycolysis and energy metabolism in preeclampsia. Am. J. Obstet. Gynecol. 1987;157(1):97–101. doi: 10.1016/s0002-9378(87)80354-x. [DOI] [PubMed] [Google Scholar]
- 46.LeBleu V.S., O'Connell J.T., Gonzalez Herrera K.N., Wikman H., Pantel K., Haigis M.C., de Carvalho F.M., Damascena A., Domingos Chinen L.T., Rocha R.M., Asara J.M., Kalluri R. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat. Cell Biol. 2014;16(10) doi: 10.1038/ncb3039. 992-1003, 1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y., Han T.L., Luo X., Bai Y., Chen X., Peng W., Xiong X., Baker P.N., Tong C., Qi H. The metabolic role of LncZBTB39-1:2 in the trophoblast mobility of preeclampsia. Genes & diseases. 2018;5(3):235–244. doi: 10.1016/j.gendis.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang X., Tang S., Song Y., Li D., Zhang L., Wang S., Duan Y., Du H. Effect of 2-deoxyglucose-mediated inhibition of glycolysis on migration and invasion of HTR-8/SVneo trophoblast cells. J. Reprod. Immunol. 2023;159 doi: 10.1016/j.jri.2023.104123. [DOI] [PubMed] [Google Scholar]
- 49.Schulze A., Harris A.L. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491(7424):364–373. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- 50.Maddocks O.D., Berkers C.R., Mason S.M., Zheng L., Blyth K., Gottlieb E., Vousden K.H. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493(7433):542–546. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rana S., Lemoine E., Granger J.P., Karumanchi S.A. Preeclampsia: pathophysiology, challenges, and perspectives. Circ. Res. 2019;124(7):1094–1112. doi: 10.1161/CIRCRESAHA.118.313276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on reasonable request.