Abstract
Cancer stem cells (CSCs) are a small subset of cells in cancers that are thought to initiate tumorous transformation and promote metastasis, recurrence, and resistance to treatment. Growing evidence has revealed the existence of CSCs in various types of cancers and suggested that CSCs differentiate into diverse lineage cells that contribute to tumor progression. We may be able to overcome the limitations of cancer treatment with a comprehensive understanding of the biological features and mechanisms underlying therapeutic resistance in CSCs. This review provides an overview of the properties, biomarkers, and mechanisms of resistance shown by CSCs. Recent findings on metabolic features, especially fatty acid metabolism and ferroptosis in CSCs, are highlighted, along with promising targeting strategies. Targeting CSCs is a potential treatment plan to conquer cancer and prevent resistance and relapse in cancer treatment.
Keywords: Cancer stem cells, therapeutic resistance, metabolism, immunology, biomarkers
Introduction
Cancer stem cells (CSCs) are a specific subpopulation of tumor cells with stem cell-like capacities of self-renewal and differentiation that were originally proposed to exist by Mackillop in 19831. The CSC theory hypothesizes that tumor initiation, metastasis, and recurrence are favored by a small number of CSCs present in tumors2. Over the years studies have identified CSCs in various types of cancers, including leukemia3, breast cancer4, colorectal cancer (CRC)5, and lung cancer6. CSCs are mostly, but not necessarily found in a mitotically dormant or quiescent state. CSCs can potentially differentiate into different lineage cells, such as cancer cells, vascular endothelial cells, pericytes, and erythroblasts7–10. Recent studies have also revealed the phenotypic and functional heterogeneity of CSCs during tumor progression11. Therefore, targeting CSCs could be an effective therapeutic approach to eradicate the source of cancer cells and combat therapeutic resistance, ultimately transforming the therapeutic paradigm for cancers and improving patient prognosis.
This review discusses recent advances pertaining to CSCs, including biological features, biomarkers, and mechanisms underlying resistance to different therapies. We have focused on the metabolic and immunologic aspects of CSCs and the potential therapeutic implications of targeting CSCs to overcome resistance to cancer treatment.
Biological properties of CSCs
Several studies have investigated the characteristics of CSCs in distinct types of cancers, as thoroughly reviewed elsewhere12–14. In this section we will focus on recent discoveries on the immunologic and metabolic properties of CSCs.
Immunologic properties of CSCs
A previous study showed that epigenetic immunoediting may drive an acquired immune evasion program in the most aggressive mesenchymal glioblastoma multiforme (GBM) subtype by modifying the tumor immune microenvironment15,16. Recent research indicates heterogeneous immunomodulatory molecules in CSCs and crosstalk between CSCs and stromal cells in the tumor microenvironment (TME)17. This section summarizes the immunomodulatory molecules found in CSCs, with a particular focus on major histocompatibility complex (MHC) molecules, natural killer (NK) ligands, and immune checkpoints (Figure 1). The interplay between CSCs and immune cells, as well as other stromal cells, which have been extensively reviewed in other sources, are also briefly discussed18–21.
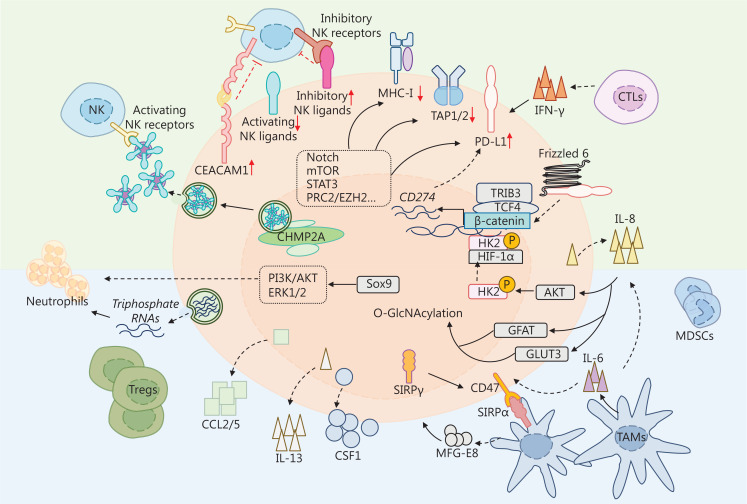
Figure 1.
Immunologic properties of cancer stem cells (CSCs). CSCs escape immune surveillance by altering immunomodulatory molecules. CSCs avoid recognition by immune cells by downregulating MHC class I (MHC-I) molecules and antigen processing machinery (APM) molecule [antigen processing 1/2 (TAP1/2)]. CSCs evade NK-mediated killing by decreasing the expression of and shedding activating NK ligands (ULBP1-6, MICA/B, Nkp30L, and Nkp44L) and increasing inhibitory NK ligands (KIR2DL1-5 and KIR3D1-3), and carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1). High levels of MICA/B are secreted through membrane vesicles facilitated by increased levels of charged multivesicular body protein (CHMP2A) in CSCs. CSCs also express high levels of immune checkpoints, such as programmed death-ligand 1 (PD-L1), which interact with corresponding receptors on immune cells and thereby impede the activation and proliferation of immune cells. The expression and stability of PD-L1 is regulated by multiple signaling pathways, including Notch, mTOR, phosphoinositide 3-kinase (PI3K)/AKT, signal transducer, and activator of transcription-3 (STAT3), as well as interleukin and interferon-γ (IFN-γ). IFN-γ is released by CSCs or other immune cells. Interleukin-8 (IL-8) can be secreted by CSCs and tumor-associated macrophages (TAMs). IL-8 induces phosphorylation and nuclear translocation of hexokinase 2 (HK2) by AKT. Phosphorylated HK2 and hypoxia-inducible factor-1 alpha (HIF-1α) promote the transcription of CD274 by binding the promoter. PD-L1 also binds to Frizzled 6 to activate β-catenin signaling and further upregulates PD-L1. IL-8 enhances O-GlcNAcylation by upregulating glucose uptake transporter 3 (GLUT3) and glutamine fructose-6-phosphate aminotransferase (GFAT), which contribute to stemness in CSCs. Moreover, CSCs secrete cytokines, chemokines, and triphosphate RNAs to recruit immunosuppressive immune cells. Sox9/C-C motif chemokine ligand 1 (CCL1) axis recruits neutrophils through PI3K/AT and ERK1/2 signaling. CSCs release triphosphate RNAs to recruit neutrophils. Additionally, CSCs secrete CCL2/5, IL-13, and CSF1 to recruit inhibitory Tregs and TAMs. TAMs release milk-fat globule-epidermal growth factor-VIII (MFG-E) and IL-6 to maintain the stemness of CSCs. Moreover, the increasing CD47 in CSCs interacts with signal regulatory protein alpha (SIRPα) to exert a “don’t eat me” signal. SIRγ is highly expressed in CSCs to provoke a “don’t eat me” signal.
Surface immunoregulatory molecules in CSCs
CD8+ cytotoxic T cells have a vital role in eliminating tumors by recognizing and killing tumor cells that display foreign antigens presented by MHC-I molecules. Several studies have shown that dysregulation of antigen presentation-related molecules in CSCs contributes to immune evasion. In various types of cancers, such as melanoma, lung cancer, GBM, and head and neck squamous cell carcinoma (HNSCC), CSCs exhibit a reduction or deficiency of MHC-I/II molecules via an in vitro tumorsphere formation assay22–25. The CSC-enriched tumorspheres from murine TC-1 lung cancer cells have lower expression of surface MHC-I molecules than other tumorspheres, which makes the tumorspheres resistant to human papillomavirus (HPV) 16 E6/E7 peptide vaccine-mediated killing. In a tumorsphere-bearing mouse model, less CD8+ CTL infiltration is found in CSC-enriched tumorspheres23,25. Similarly, in HNSCC cell lines, CD44+ CSCs exhibit low levels of HLA-A2, HLA-II, and antigen processing 2 (TAP2) expression, making it difficult for cytotoxic T cells (CTLs) or NK cells to identify the CD44+ CSCs25,26. An analysis of tumor-associated antigens (TAAs) and the antigen processing and presentation molecule (APM) in tumorspheres from 12 human solid tumor cell lines indicated that weak or deficient expression of HLA-I/II molecules was detected in 9 cell lines, whereas the increasing expression of APMs, such as low molecular mass protein-2/7 (LMP2/7), multi-catalytic endopeptidase complex subunit 1 (MECL-1), and TAP1/2, was observed in 12 cell lines26. Further studies in immunocompetent and immunocompromised mice have demonstrated that aldehyde dehydrogenase (ALDH)+, but not CD44+CD24− breast CSCs, have TAP1 genes and the co-stimulatory molecule, CD80, that are downregulated by DNA hypermethylation, which causes impairment in T cell-mediated killing27. Lastly, a genome-wide CRISPR/Cas9 screen has revealed that polycomb repressive complex 2 (PRC2) epigenetically downregulates the MHC-I in an EZH2-dependent manner, prompting resistance to T cell-mediated killing28.
Generally, cells with low expression or absence of MHC-I molecules are susceptible to attack by NK cells, which suggests the potential for NK cell-mediated CSC killing29–31. However, CSCs in GBM exhibit resistance to lysis mediated by resting NK cells due to MHC class I molecule expression, as reported by Avril et al.32 Activation of NK cells by lectins restores GSC sensitivity to NK lysis. Additionally, upregulated expression of NKG2DL augments NK-mediated killing in glioma CSCs and drug-resistant ovarian cancer cells33,34. CSCs isolated from CRC patients express lower levels of MHC-I molecules and higher levels of the activating NK ligands, Nkp30L and Nkp44L, making the CSCs isolated from CRC patients more susceptible to NK cell-mediated killing35. According to a report by Luna et al.36, the proteasome inhibitor, bortezomib, induces the expression of CSC-related genes and the activation of NKG2DL MHC class I chain-related molecules A/B (MICA/B) in ALDH+ and ALDH− cells from GBM, synovial sarcoma, and pancreatic adenocarcinoma cell lines. Bortezomib sensitizes ALDH+ cells to NK cell-mediated killing in in vitro and in vivo models36. A mechanistic study involving reprogrammed CSCs from liver cancer revealed that CD44 mRNA functions as a competing endogenous RNA (ceRNA) that specifically binds microRNA (miR)-34a, thereby preventing the activating NKG2DL, UL16 binding protein 2 (ULBP2), from degradation. Stable expression of ULBP2 facilitates NK cells to kill liver CSCs37; however, several studies have indicated that CSCs evade the innate immune response by increasing inhibitory NK ligands, which decreases the expression of activating NK ligands or shedding activating NK ligands38–41. Tumors remove natural killer group 2D (NKG2D) ligands, which is one of the primary mechanisms responsible for subverting NKG2D-mediated immunosurveillance in leukemia stem cells (LSCs)39. Similarly, MICA/B downregulation is modulated by oncogenic miR-20a and leads to the resistance of CSCs to NK cell cytotoxicity, as well as lung cancer metastasis, in breast CSCs42. Furthermore, CD24−/low/CD44+ CSCs isolated from radiotherapy-resistant triple-negative breast cancer (TNBC) cells exhibit reduced MICA/B expression and profound expression of the inhibitory NKG2A ligand, HLA-E. In an in vivo mouse model, CD24−/low/CD44+ CSCs recruit NK cells to the peritumoral area but deprive CD24−/low/CD44+ CSCs of cytotoxicity43. Additionally, EpCAMhigh hepatocellular carcinoma (HCC) cells that express high levels of carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) display CSC properties and are not susceptible to NK killing44. Upregulated CEACAM1 is also detected in tumorspheres with CSC properties developed from liver cancer cells. The tumorspheres express low levels of ULBP1 and MICA/B on the cell surface, whereas elevated levels of soluble MICA are detected in conditioned medium from tumorspheres, which impedes NK cell-mediated killing40. The whole genome CRISPR–Cas9 screening system has identified the vital role of chromatin-modifying protein/charged multivesicular body protein (CHMP2A) in desensitizing CSCs to NK-mediated killing in HNSCC and GBM cells. Specifically, CHMP2A induces CSCs to secrete extracellular vesicles (EVs) expressing MICA/B and tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL), thereby inducing apoptosis in NK cells41.
High levels of immune checkpoint molecules have been detected in CSCs, which hamper the immune response45. PD-L1 expression is higher in CSCs from various types of cancer, including breast cancer, squamous cell carcinoma, endometrial cancer, CRC, and non-small cell lung cancer (NSCLC)45–49. Numerous studies have identified different mechanisms underlying PD-L1 regulation in cancer, which have been thoroughly reviewed50. Hsu et al.51 reported that inhibition of PD-L1 expression with etoposide leads to an increase in tumor-infiltrating T cells. PD-L1 is modulated by diverse signaling pathways in CSCs, such as Wnt, phosphoinositide 3-kinase (PI3K)/AKT, Notch, mTOR, signal transducer and activator of transcription-3 (STAT3), and epigenetic signaling46,51–56. Notch 3 induces PD-L1 expression through mTOR and maintains stemness in PD-L1high CSCs from breast cancer46. Sun et al.52 reported that interleukin-8 (IL-8) derived from gastric mesenchymal stem cells promotes PD-L1 expression via the STAT3/mTOR/c-Myc axis in CSCs from gastric cancer. IL-8 derived from gastric mesenchymal stem cells, along with AKT, promotes the phosphorylation and nuclear localization of HK2, which binds to HIF-1α and facilitates PD-L1 transcription56. Studies in CSCs from CRC organoids have suggested that tribble pseudokinase (TRIB3) recruits transcription factor 4 (TCF4) and β-catenin to the promoters of Wnt target genes, which in turn induces TRIB3 expression and maintains their stemness57. Administration of selective Wnt inhibitors or activators leads to a reduction and increase in PD-L1 expression in ALDH+/CD44+ CSCs from TNBCs, respectively, indicating the positive regulation of PD-L1 by Wnt53. The Wnt/β-catenin and PI3K/AKT pathways cooperate to promote tumorigenesis and resistance to therapy in CSCs from leukemia. β-catenin binds to loci on multiple immune checkpoint genes, including PD-L1, T-cell immunoglobulin domain mucin domain 3 (TIM3), and CD24. Targeting AKT inhibits this process, leading to a decrease in PD-L1, TIM3, and CD24 expression55. Moreover, PD-L1 promotes the activation of β-catenin and β-catenin CSC-associated target genes via an interaction with the receptor, Frizzled 648. The expression and stability of PD-L1 is regulated in an epigenetic and posttranslational manner, respectively51,54,58,59; however, the expression landscape of immunomodulatory molecules varies between cancer types and individuals. The underlying regulatory mechanisms are also sophisticated and heterogeneous. Further understanding of molecular mechanisms can contribute to an improvement in CSC-targeted immunotherapy efficacy.
Crosstalk between CSCs and the TME
The interplay between CSCs and immune cells in the TME has a crucial role in the evasion of immune surveillance, which enables the survival and growth of CSCs. CD44+ CD90+ CSCs have a higher tendency for lymph node metastasis in small-cell lung cancer (SCLC), which promotes the response of cytotoxic T lymphocytes (CTLs). The expression of PD-L1, which is promoted by the secretion of IFN-γ by activated CTLs, leads to adaptive resistance capacity, resulting in prolonged inflammation and upregulation of regulatory ligands that ultimately impair the proliferative capacity and cytotoxicity of CTLs60. Research has shown that regulatory T cells (Tregs) promote cancer stemness in gliomas through the TGF-β/NF-κB/IL-6/STAT3 signaling axis. The anti-IL-6 receptor antibody, tocilizumab, shows efficacy in inhibiting tumor growth and stemness induced by Tregs in glioma xenograft models60,61 Tregs are subpopulations of CD4+ T cells that suppress the immune response by inhibiting the activation of NK cells and the cytotoxic function of CD8+ T cells62. Other subpopulations of CD4+ T cells include Th1, Th2, and Th17 cells. Th1 cells secrete IL-2 and interferons, which activate the proliferation of CD8+ T cells and NK cells. Th2 cells promote the maturation and clonal proliferation of B cells by secreting cytokines, such as IL-4 and IL-662. CSCs secrete the chemokines, C-C motif chemokine ligand 1 (CCL1), CCL2, and CCL5, which recruit Tregs19. The balance of Th17/Treg cells is closely related to tumor immunity and has a critical role in tumor progression63. Cytokines secreted by CSCs, including CCL5, MKN-45, IL-6, and IL-8, have been shown to affect the Th17/Treg balance, which has been reviewed elsewhere64.
In the context of cancer, M2 macrophages are commonly regarded as tumor-associated macrophages (TAMs)65,66. Activation of Yes-associated protein (YAP) in hepatocellular CSCs leads to tumorigenesis and TAM recruitment67. CSCs secrete chemokines, such as CCL2, CCL5, colony stimulation factor 1 (CSF1), growth differentiation factor 15 (GDF15), IL-13, and transforming growth factor-β (TGFβ), as well as periostin and Wnt-induced signaling protein 1 [WISP1 (also known as CCN4)], which may impact the polarization state of TAMs and promote tumorigenesis19. TAMs secrete IL-6, which promotes HCC carcinogenesis by stimulating CSC-like characteristics. Tocilizumab disrupts TAM-enhanced CSC expansion in HCC68. The cytokine, IL-8, which is produced by various cells, including macrophages and monocytes, acts as a chemotactic cytokine to bring neutrophils to inflammatory or injured sites69. IL-8 has been shown to enhance O-GlcNAcylation, but not glycolysis mediated by the upregulation of glucose uptake transporter 3 (GLUT3) and glutamine fructose-6-phosphate aminotransferase (GFAT), which promotes the generation and maintenance of CSCs in colon and lung cancer cells. The effect of O-GlcNAcylation on CSCs has not been fully elucidated70. TAMs have been shown to secrete CCL5, which mediates the self-renewal of prostatic CSCs and metastases71. Jinushi and colleagues72 reported that CD44+/ALDH+ colon CSCs and CD133+/ALDH1+ lung CSCs induce secretion of the milk-fat globule-epidermal growth factor-VIII (MFG-E8) by TAMs, which with IL-6 maintains the activity and promotes the therapeutic resistance of CSCs. Additionally, the “don’t eat me” signal, CD47, is upregulated in CSCs from several cancer types. CD47 binds to myeloid-specific signal regulatory protein alpha (SIRPα) on macrophages, impeding phagocytosis and allowing immune evasion73–75. Our previous study identified a population of cancer cells expressing SIRPγ. SIRPγhi cancer cells display stemness-related properties and contribute to immune escape signals by sustaining CD47 expression, which halts macrophage-mediated phagocytosis in SIRPγhi and SIRPγlo/− tumor cells6.
Several studies have explored the immunosuppressive role of neutrophils in cancer progression. One study by the Szczerba group76 identified an interaction between neutrophils and circulating cancer cells that contributes to metastasis in patients and mouse models of breast cancer. Circulating cancer cells have stem cell characteristics and are precursors of metastasis77. Sox9 has been identified as a CSC marker in HCC. The Sox9/CXCL5 axis activates PI3K/AKT and ERK1/2 signaling, promoting the proliferation and invasion of HCC cells, as well as infiltration of intratumoral Ly6G+ neutrophils in the F4/80+ macrophage orthotopic xenograft model78. Hwang et al.79 discovered that exosomes released by colorectal CSCs prolong the lifespan of neutrophils by activating PRR/NF-κB signaling through exosomal triphosphate RNAs, leading to the expression of IL-1β and accelerating tumorigenesis.
Myeloid-derived suppressor cells (MDSCs) are a heterogeneous lineage of immature myeloid cells that can be divided into two major subsets [polymorphonuclear (PMN)-MDSCs and monocytic (M)-MDSCs]. MDSCs enhance ovarian cancer stemness by upregulating miR-101 and downregulating C-terminal binding proteins (CtBP2)80. Shidal et al.81 showed that miR-92a enhances integrin and TGF-β expression in CD133+ melanoma CSCs and leads to increased immunosuppressive cell phenotypes, including granulocytic MDSCs (gMDSCs) and Tregs. Another study found that ALDH1A1 promotes MDSC expansion by stimulating the secretion of GM-CSF, which is activated by the TGF-β-activated kinase 1 (TAK1)/NF-κB signaling pathway82.
Dendritic cells (DCs) also have a role in CSC maintenance. The interaction between C-X-C motif chemokine receptor 4 (CXCR4) expressed by follicular lymphoma (FL) cells with CSC-like activities and CXCL12, which is secreted by follicular DCs, facilitates chemotherapy resistance and tumorigenicity83. Natural killer T (NKT) cells are a specialized subtype of T cells that can be categorized into two types of cells [invariant NKT cells (iNKT cells) and type II NKT cells]. Although the role of NKT cells in CSCs has not been reported, iNKT cells release a variety of proinflammatory and anti-inflammatory cytokines that affect DCs, macrophages, neutrophils, NK cells, and T cells, which exert effects on CSCs84. B lymphocytes have a critical role in promoting and inhibiting tumor development. For example, B lymphocytes secrete cytokines, such as IL-10, TGF-β, and IL-35, and exhibit inhibitory effects by interacting with tumor tissues and lymphocytes, such as T cells, APCs, Tregs, and MDSCs85.
Cancer-associated fibroblasts (CAFs) and adipocytes also affect the stemness of CSCs. Studies conducted in vitro have shown that CAFs promote the expression of stem cell markers [CD44, SRY-box 2 (Sox2), and Bmi-1], as well as the self-renewal and expansion of CSCs by secreting cytokines, growth factors, androgen receptor-regulated factors, and exosomes86–90. CAFs also induce the tumorsphere-forming phenotype in breast cancer cells by producing CCL2, which activates the Notch signaling pathway91. CAFs boost breast CSC proliferation by secreting stromal-derived-factor-1 (SDF-1), which activates the Wnt/β-catenin and PI3K/AKT signaling pathways92. Adipocytes maintain the stemness of CSCs by secreting more resistin, which upregulates stemness-related transcription factors [Octamer-binding transcription factor 4 (Oct4), Sox2, Nanog homeobox (Nanog), and ALDH1] and activates stemness-related pathways (Notch and Wnt/β-catenin)93–96. Adipocytes also shield breast CSCs treated with doxorubicin by secreting more resistin, which mediates the activation of the AMPK/mTOR and JNK pathways97.
The TME is an intricate and dynamic system comprising tumor cells, immune cells, fibroblasts, extracellular matrix, and other interconnected components. The interaction between CSCs and infiltrated immune cells is particularly important. However, the specific mechanism of immune evasion in CSCs and the interplay with the TME, especially NKT cells, B lymphomas, and neutrophils, have not been extensively explored and warrant further investigation.
Metabolic properties of CSCs
Tumors are highly adaptive to metabolic perturbations98. Under hypoxic conditions, mitochondrial oxidative phosphorylation (OXPHO) is replaced by glycolysis to compensate for deficient mitochondrial machinery99. During nutrient deficiency, autophagy is one of the essential strategies for preserving cell viability100–102. Mounting evidence has revealed the unique metabolic features of CSCs, such as aberrant glucose consumption, excessive lactate production, and inefficient ATP production103,104. Depending on the availability of oxygen and nutrients, as well as other stromal cells in the TME, CSCs show heterogeneity in different tissues105. Glioma CSCs exhibit high metabolic plasticity because glioma CSCs switch metabolism to glycolytic metabolism when OXPHOS is blocked106. Studies involving different tumors, including osteosarcoma, GBM, breast cancer, lung cancer, ovarian cancer, nasopharyngeal carcinoma (NPC), HCC, and colon cancer, suggest that CSCs have higher glycolytic potential and less mitochondrial oxidative metabolism than other differentiated tumor cells103,107–113. The mitochondrial circRNA for translocating phosphoglycerate kinase 1 (mcPGK1) is in high levels in liver CSCs. mcPGK1 has a crucial role in regulating cell metabolism by inhibiting OXPHOS and promoting glycolysis, which changes the levels of specific chemicals, such as α-ketoglutaric acid and lactic acid. These changes, in turn, activate the Wnt/β-catenin pathway and promote self-renewal of liver CSCs. Additionally, mcPGK1 helps introduce PGK1 to the mitochondria by interacting with TOM40 and reprograms cell metabolism from oxidative phosphorylation to glycolysis via the PGK1-PDK1-PDH axis114. Gu et al.115 reported that the absence of miR-192-5p increases glycolysis by upregulating glucose transporter type 1 (GLUT1), 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase3 (PFKFB3), and c-Myc, which inhibit the transcription of miR-192-5p and maintain high glycolytic activity in HCC cells. High glycolytic activity in HCC cells produces excess lactic acid, which activates ERK phosphorylation in co-cultured LX2 and THP1 via the N-Myc downstream regulatory gene 3 (NDRG3) and monocarboxylate transporters 1 (MCT1), and promotes tumor stemness115. Another study showed that HectH9 is an activator of glucose metabolism. HectH9 does this by mediating the K63-linked ubiquitin of hexokinase 2 (HK2), which then regulates the location of HK2 in mitochondria. Regulation of HK2 in mitochondria is essential in inducing glycolysis and preventing apoptosis. Conversely, blocking the HectH9/HK2 pathway leads to an increase in reactive oxygen species (ROS), which inhibits CSC expansion and the development of tumors116. Although controversial, some investigations have shown that CSCs rely more on mitochondrial oxidative metabolism117–123. Pancreatic CSCs derived from patient-derived xenograft (PDX) models have a preference for mitochondrial metabolism to survive124. Similarly, ovarian CSCs highly express genes related to mitochondrial OXPHOS and fatty acid oxidation121. In breast cancer, increasing mitochondrial bulk in CSCs maintains stem-like characteristics, metastatic potential, and resistance to DNA damage125. Moreover, some subpopulations of cells from various tumors, such as CD133+ CSCs in GBM and pancreatic ductal adenocarcinoma, ROSlow quiescent cells in leukemia, and side population cells in lung and breast cancer, have been profiled to express the OXPHOS phenotype118,119,122,126. Notably, both OXPHOS and glycolysis are active in ovarian CSCs121,127.
Several studies have revealed that fatty acid metabolism, especially the mevalonate pathway, is fundamental to the maintenance of stemness in CSCs121,128–130. Fatty acid oxidation (FAO) helps overcome glucose starvation and contributes to chemotherapeutic resistance in epithelial ovarian CSCs131. Consumption of the dietary fat palmitic acid has also been linked to increased metastatic potential in CSCs of oral squamous cell carcinoma (OSCC)132. Sterol regulatory-element binding protein 1 (SREBP-1) regulates genes involved in lipid metabolism, such as acetyl-CoA carboxylase (ACC) and fatty acid synthase (FASN). According to a study in cisplatin-resistant NSCLC cells, SREBP-1/SCAP/FASN signaling lowers CSC sensitivity to cisplatin. Treatment with fatostatin, an SREBP inhibitor, reverses cisplatin resistance and hampers stemness133. Dysregulation of lipid metabolism is associated with the maintenance of CSC stemness and poor survival. Further research should seek to clarify the function of SREBP-1/SCAP/FASN signaling in cisplatin resistance. Polyunsaturated fatty acids (PUFAs) and monounsaturated fatty acids (MUFAs) are the primary lipid metabolites and key regulators of ferroptosis, a form of cell death triggered by disturbances in metabolic networks, such as iron metabolism, mitochondrial metabolism, and lipid metabolism, and characterized by the accumulation of markers of lipid peroxidation134. Ferroptosis can be triggered by drugs, ionizing radiation, and cytokines, thereby suppressing tumor growth. However, ferroptosis can also encourage tumor growth by promoting inflammation-associated immunosuppression and other signaling pathways135. Recent research has highlighted the critical role and targeting potential of ferroptosis in CSCs (Figure 2). For example, a study reported that ferroptosis inducers selectively kill a mesenchymal breast cancer subpopulation with CSC properties through a non-apoptotic mechanism of action mediated by ROS in an iron-dependent manner136. Furthermore, Turcu et al.137 reported that the natural compound, salinomycin, selectively kills CD44high/CD24low CSCs from breast cancer by interacting with lysosomal iron, which promotes ROS production and causes lysosomal membrane permeabilization. Intracellular ferric iron is primarily bound to transferrin (TF) and is imported via transferrin receptor 1 (TFR1). CSCs from breast and ovarian carcinomas express higher levels of TFRs, which induce iron uptake and sensitize CSCs to agents inducing ferroptosis138,139. The six-transmembrane epithelial antigen of prostate 3 (STEAP3) is a ferrireductase located at the plasma membrane that catalyzes the reduction of Fe3+ to Fe2+ after lysosome-endosome fusion. Fe2+ is then released from the endosome into a labile iron pool (LIP) in the cytoplasm, which is regulated by divalent metal transporter protein 1 (DMT1) and contributes to iron homeostasis. Elevated STEAP3 promotes the proliferation and stemness of CSCs in gliomas140. Inhibition of DMT1 also causes accumulation of lysosomal iron and ROS, leading to ferroptosis137; however, a study involving non-CSC glioblastoma cells demonstrated that temozolomide induces ferroptosis by upregulating DMT1 expression and increasing iron content141. Ferritin is composed of a heavy chain (FTH) and a light chain (FTL), and can store > 4,000 iron atoms and convert Fe2+ to Fe3+. Ferritinophagy is the process by which nuclear receptor coactivator 4 (NCOA4) releases the iron stored in ferritin to LIP and triggers ferroptosis142. A study conducted on osteosarcoma CSCs showed that exposure to a static magnetic field stimulates NCOA4-mediated ferritinophagy, promoting CSC proliferation and self-renewal143. Higher levels of glutathione peroxidase 4 (GPX4) and cystine/glutamate antiporter solute carrier family 7 member 11 (SLC7A11) protect esophageal CSCs from ferroptosis, which is induced by increased intracellular iron content. In vitro experiments have shown that heat shock protein 27 (Hsp27) positively regulates SLC7A11/GPX4 by downregulating p53144. Epithelial-to-mesenchymal transition (EMT) leads to the acquisition of CSC properties. FTH-mediated ROS dysregulation promotes C-X-C motif ligand 12 (CXCL12)/C-X-C motif chemokine receptor 4 (CXCR4) axis activation and EMT in erythroleukemia K562 cells145. Studies have also reported that FTH expression modulates EMT in in vitro models of breast and lung cancer146,147. The results of another study suggested that FTH silencing leads to an imbalance in the metabolism of unsaturated fatty acids and overexpression of stem cell markers in ovarian cancer148. Furthermore, gastric cancer (GC)-secreted exosomal lnc-ENDOG-1:1 (lncFERO) promotes stearoyl-CoA-desaturase (SCD1) translation by recruiting heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1), then inhibits ferroptosis and enhances stemness in gastric CSCs in vitro and in vivo149. The roles of fatty acid metabolism and ferroptosis in CSCs from various types of cancer are clearly controversial.
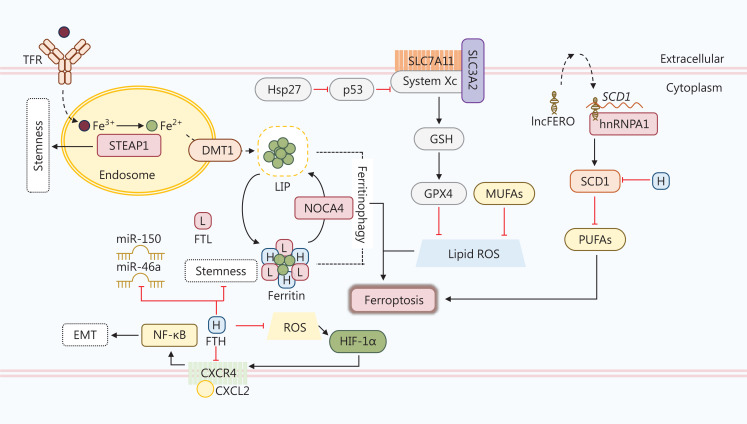
Figure 2.
Ferroptosis in CSCs. The role of ferroptosis in CSCs is controversial. First, enzymes and transporters in the ferroptosis pathway are altered in some CSCs, including sterol regulatory-element binding protein 1 (STEAP1), divalent metal transporter protein 1 (DMT1), and nuclear receptor coactivator (NOCA4). The ferritin heavy chain (FTH) is downregulated in CSCs. The absence of FTH induces the expression of stemness-related genes, microRNA-150 (miR-150), and miR-46a, which maintain the stemness of CSCs. FTH also inhibits the activation of CXCR4/CXCL2 signaling and subsequent NF-κB-mediated EMT directly or by blocking ROS-regulated HIF-1α. A high level of SLC7A11, which is promoted by heat shock protein 27 (Hsp27)-mediated inhibition of p53, protects CSCs from ferroptosis via the GSH/GPX4 axis. Moreover, lnc-ENDOG-1:1 (lncFERO) secreted by CSCs interacts with stearoyl-CoA-desaturase (SCD1) mRNA and recruits hnRNPA1 to facilitate the translation of SCD1. SCD1 inhibits PUFA synthesis and thereby curbs ferroptosis. SCD1 is also inhibited by FTH in CSCs.
In addition to glucose and fatty acid metabolism, other metabolic signaling pathways, such as glutamine metabolism and lysine catabolism, are enhanced in CSCs. CSCs rely heavily on glutamine in pancreatic cancer, so limiting the availability of CSCs can reduce the limited self-renewal of CSCs and enhance the sensitivity of CSCs to radiation therapy, followed by an increased level of ROS in cell lines and mouse models150. Elevated lysine catabolism has been detected in colon adenocarcinoma circulating tumor cells, which exhibit a high capacity for colonization in the liver and an active Wnt signaling pathway151. Despite the findings mentioned above, the full extent of metabolic networks in CSCs is not fully understood. In recent decades, altered metabolism in cancer and non-cancer cells in the TME has been considered a potential target for cancer treatment and relapse prevention. In this regard, a thorough understanding of CSC metabolism is essential for developing effective treatments and preventing cancer relapse.
Biomarkers of CSCs
Because CSCs have a vital role in tumorigenesis and therapeutic resistance, it is essential to identify this specific population in cancer tissues. Numerous studies have investigated molecular biomarkers for CSCs in cancer cells, mouse models, and patient tissues. In addition to the classic biomarkers, such as CD133, CD44, epithelial cell adhesion molecule (EpCAM), and CD90, other biomarkers have also been studied in specific cancers. In this section, we will summarize the current knowledge about CSC biomarkers in different types of cancers, including cell-surface molecules, transcription factors, and other CSC markers (Table 1).
Table 1.
CSC biomarkers in different cancer types
| Biomarkers | Cancer types |
|---|---|
| CD133 | Lung cancer152–154, colon cancer155,156, prostate cancer157, ovarian cancer158, melanoma159, osteosarcoma160,161, leukemia162, hepatocellular carcinoma (HCC)163, pancreatic cancer164, and oral squamous cell carcinoma (OSCC)165 |
| CD44 | Colorectal cancer (CRC)166–168, pancreatic cancer169, ovarian cancer170, gastric cancer171,172, prostate cancer157,173, non-small cell lung cancer (NSCLC)174, OSCC175, nasopharyngeal carcinoma (NPC)176, HCC168,177,178 |
| CD90 | Murine breast cancer179, HCC180, gastric cancer181, esophageal squamous cell carcinoma (ESCC)182, lung cancer183, pancreatic cancer184, glioma185,186, insulinoma187, HCC188 |
| EpCAM | Breast cancer189,190, CRC166, HCC188,191, NPC176, pancreatic cancer192,193 |
| Lgr5 | Gastric cancer194,195, pancreatic cancer196,197, HCC198,199, CRC200,201, ovarian cancer202, cervical cancer203, breast cancer204 |
| Oct4 | Glioma205, pancreatic cancer206, HCC207, breast cancer208, prostate cancer209,210, bladder cancer211, ovarian cancer212, lung cancer213 |
| Sox2 | Head and neck squamous cell carcinoma (HNSCC)214, medulloblastoma215, glioma216, breast cancer217, gastric cancer218, CRC219, lung cancer220, cervical cancer221, melanoma222, osteosarcoma223, ovarian cancer224, pancreatic cancer225, bladder cancer226, skin cancer227 |
| Klf4 | Leukemia228, anaplastic meningioma229, CRC230, gastric cancer231, NSCLC232, HCC233, bladder cancer234, ESCC235 |
| Nanog | Glioblastoma (GBM)236, leukemia237, lung cancer238, breast cancer239, ESCC240, gastric cancer241, CRC242, ovarian cancer243, prostate cancer244, HCC245, HNSCC246, renal cancer247 |
| c-Myc | Neuroblastoma248, lung cancer249, CRC250, breast cancer251 |
| ALDH | Ovarian cancer252,253, lung cancer254–256, breast cancer257,258, cervical cancer259, HCC260, colon cancer261,262 |
| Bmi-1 | Gastric cancer263, HNSCC5,264, CRC265, ESCC266–268, hematopoietic neoplasm269, glioma cancer270,271 |
| Nestin | Neurogenic tumors272–276, rhabdomyosarcoma277, osteosarcoma278, chondrosarcoma160, fibrosarcoma160, ovarian279–281, OSCC282, prostate cancer283,284, gallbladder cancer285, lung cancer286, colon cancer287, breast cancer288,289, gastric cancer290, pancreatic cancer291 |
| Msi1 | CRC292, OSCC293,294, neuronal cancer295 |
| Tim-3 | Small-cell lung cancer (SCLC)60, acute myelogenous leukemia (AML)296 |
| CXCR4 | Glioma297, prostate cancer298 |
Cell surface molecules
Cell surface molecules are practical for isolating CSCs by flow cytometry or magnetic sorting, as well as specific targeting. Many surface markers have been identified in CSCs, such as CD133, CD44, CD90, EpCAM, leucine-rich repeat-containing G protein-coupled receptor 5 (Lgr5), CD13, CD19, CD20, CD24, CD26, CD27, and CD34. Among the surface markers, CD133, CD44, CD90, EpCAM, and Lgr5 are the most investigated markers in CSCs.
CD133
CD133 is a transmembrane glycoprotein initially characterized as a hematopoietic stem cell and neuroepithelial stem cell marker299,300. In 2004 Singh et al.273 isolated a subpopulation of cells expressing CD133, possessing self-renewal potential, and the ability to recapitulate the original tumor from brain tumors. In situ injection of 100 CD133+ tumor cells in non-obese, diabetic, severe combined immunodeficient (NOD-SCID) mouse brains successfully produced a tumor. CD133 is a CSC biomarker of various cancer types, including lung cancer152–154, colon cancer155,156,177, prostate cancer157, ovarian cancer158, melanoma159, osteosarcoma160,161,278, leukemia162, HCC163, pancreatic cancer164, and OSCC165. CD133 alone might not be sufficient to identify CSCs, so other biomarkers are required. A few studies have also reported that CD133+ cells fail to recapitulate the original tumor and that CD133− cells have the potential to produce tumors in mouse models167,301–303.
CD44
CD44 is a transmembrane glycoprotein that was first used as a CSC marker in breast cancer189. In 9 of 10 breast cancer patients there is a subpopulation of cancer cells expressing high CD44 and low or no CD24, that enables formation of a tumor in vivo limiting dilution assay189. CD44 has also been identified as a CSC marker in CRC166–168, pancreatic cancer169, ovarian cancer170, gastric cancer171,172, prostate cancer157,173, NSCLC174, OSCC175, and NPC176. The combination of CD133 and CD44 more specifically defines CSCs in CRC and HCC168,177,178. In fact, this combination has been used to define a subpopulation of HCC with high intrahepatic or lung metastatic capacity304. Several splicing variants of CD44 have been generated through alternative splicing in the membrane-proximal stem region. Exons 1-5 and 6-20 are spliced together and translated into the standard isoform, CD44s. Alternatively, exons 6-15 can be spliced to yield variant isoforms (labeled CD44v) along with the standard isoform305. CD44v8-10 have been identified as human gastric CSC markers that contribute to tumor initiation306. CD44v6+ has also been reported as a CSC marker for colon cancer and HCC90,307. CD44s, CD44v4, and CD44v9 at the invasive tumor front are associated with poor prognosis in gastric cancer patients and CD44s-expressing CSCs exhibit mesenchymal properties308. CD44s has also been suggested to contribute to mesenchymal properties and metastasis in breast cancer and CRC309,310.
EpCAM
EpCAM is a cell–cell adhesion molecule expressed in healthy epithelial cells. Increasing evidence has shown EpCAM to be a CSC marker for numerous cancers, such as breast cancer189,190, CRC166, HCC188,191, NPC176, and pancreatic cancer192,193.
CD90
CD90, also known as thymocyte differentiation antigen-1 (Thy-1), is a glycosylphosphatidylinositol (GPI)-anchored glycoprotein belonging to the immunoglobulin superfamily. CD90 and other markers, such as Oct4, Sox2, and ALDH1, are upregulated in enriched CSCs when tumor cells are cultured in tumorsphere-forming conditions. This finding suggests a vital role for CD90 as a marker for CSCs181. CD90 has been identified in CSCs of several cancers, such as murine breast cancer179, HCC180, gastric cancer181, esophageal squamous cell carcinoma (ESCC)182, lung cancer183, pancreatic cancer184, gliomas185,186, and insulinomas187. According to the study by Yamashita et al.188, EpCAM+ and CD90+ CSCs in HCC have distinct phenotypes and metastatic potential. Other studies have revealed that CD90+/CD44+ HCC cells are more aggressive and likely to metastasize to the lung. The combination of CD90+/CXCR4+ is more specific for defining circulating CSCs in HCC180,311.
Lgr5
Lgr5 is a transmembrane receptor belonging to the rhodopsin family of G protein-coupled receptors. Lgr5 has a pivotal role in normal embryonic development that was first known as a marker of intestinal stem cells312. Lgr5 is also highly expressed in various cancer tissues and enhances tumorigenesis, cancer cell mobility, and EMT in breast cancer cells by activating multiple pathways, such as Wnt/β-catenin and Notch signaling. Recent evidence also suggests that Lgr5 has a significant role in maintaining CSCs, making Lgr5 a CSC biomarker in numerous types of cancers, such as gastric cancer194,195, pancreatic cancer196,197, HCC198,199, CRC200,201, ovarian cancer202, cervical cancer203, and breast cancer204. de Sousa e Melo and colleagues313 reported that proliferative Lgr5- CRC cells replenish Lgr5+ CSCs, resulting in rapid tumor recurrence upon treatment cessation. By analyzing the stemness properties of Lgr5+/CD44+/EpCAM+, Lgr5+/CD44+/EpCAM−, Lgr5+/CD44-/EpCAM+, Lgr5−/CD44+/EpCAM+, and Lgr5-/CD44-/EpCAM− cells, Leng and colleagues314 concluded that Lgr5+ cells have greater potential for colony formation, self-renewal, differentiation, and tumorigenicity than Lgr5− cells. The combination of Lgr5+/CD44+/EpCAM+ is a more specific marker of human CRC CSCs.
Transcription factors
Numerous studies have revealed that multiple stemness-related transcription factors are abnormally expressed in cancers and associated with both CSCs and poor prognosis. Stem cells highly express approximately 25 transcription factors not found in healthy somatic cells.
Oct4
Oct4 is a transcription factor encoded by the Pou5f1 gene. Oct4 belongs to the POU-homeodomain family and binds to an octamer motif, ATGCAAAT. Oct4 has a crucial role in maintaining pluripotency and self-renewal of both embryonic stem cells (ESCs) and CSCs315–318. Additionally, Oct4 induces tumorsphere formation, EMT, tumorigenesis, and resistance to chemo- or radio-therapy238,319,320. Oct4 is highly expressed in CSCs of various human cancers, such as glioma205, pancreatic cancer206, HCC207, breast cancer208, prostate cancer209,210, bladder cancer211, ovarian cancer212, and lung cancer213.
Sox2
Sox2 is one of the core transcription factors associated with pluripotency. Sox2 has a vital role in maintaining self-repair and proliferation of CSCs in various human cancers321. Sox2 also has oncogenic roles322,323. In lung cancer, Sox2 is highly linked with the ‘lineage-specific survival mechanism’ in lung cancer. Sox2, with or without mutated Lkb1, promotes mouse lung adenocarcinoma progression into squamous cell carcinoma (SCC) through pathologically mixed intermediates324. SOX2 expression characterizes CSCs in various cancers, including HNSCC214, medulloblastoma215, glioma216, breast cancer217, gastric cancer218, CRC219, lung cancer220, cervical cancer221, melanoma222, osteosarcoma223, ovarian cancer224, pancreatic cancer225, bladder cancer226, and skin cancer227.
Krüppel-like factor 4 (Klf4)
Klf4 is one of four crucial transcription factors involved in maintaining pluripotency in embryonic cells325. Klf4 is a bifunctional transcription factor that belongs to the Krüppel-like factor family. In the intestinal and gastric epithelium, Klf4 acts as a tumor suppressor326. Klf4 expression declines in leukemia228, anaplastic meningioma229, CRC230, gastric cancer231, NSCLC232, HCC233, bladder cancer234, and ESCC235. In melanoma and canine mammary tumors, Klf4 promotes tumorigenesis. In melanoma xenografts, Klf4 knockdown inhibits tumor growth in vivo327. In canine mammary tumors, highly overexpressed Klf4 is related to a more aggressive phenotype328. These findings indicate that Klf4 has a complex role in CSCs.
Nanog
Nanog has a crucial role in maintaining pluripotency329. Nanog functions in tandem with other regulators in CSCs, such as Sox2, Oct4, kinases, and miRNAs, to mediate the stemness phenotype through several signaling pathways, such as TGF-β, Wnt/β-catenin, JAK/STAT, Notch, and Hedgehog330–332. Overexpression of Nanog combined with Wnt1 leads to the initiation of breast tumors, but Nanog alone does not lead to tumorigenesis333. Nanog is often used as a stemness-associated reporter and is ubiquitously found in tumors, including GBM236, leukemia237, lung cancer238, breast cancer239, ESCC240, gastric cancer241, CRC242, ovarian cancer243, prostate cancer244, HCC245, HNSCC246, and renal cancer247.
c-Myc
The Myc gene family comprises three members (C-Myc, N-Myc, and L-Myc). These members exert an oncogenic role by regulating various cellular processes, such as the cell cycle, cellular survival, proliferation, and metabolic reprogramming334–338. c-MYC is expressed in CSCs of multiple cancers, including neuroblastoma248, lung cancer249, CRC250, and breast cancer251.
Other markers in CSCs
In addition to cell surface molecules and transcription factors, studies have identified other CSC markers, including ALDH, Bmi-1, Nestin, Musashi-1, T-cell immunoglobulin mucin-3 (TIM-3), and CXCR4.
ALDH
ALDH is an enzyme that is involved in intracellular aldehyde detoxification and retinoic acid synthesis. ALDH is a single marker of CSCs in HNSCC and lung cancer339,340. ALDH+ cells exhibit signatures of both leukemia stem cells and hematopoietic stem cells in acute myeloid leukemia (AML), whereas ALDH− cells mainly show progenitor cell signatures, indicating that ALDH+ AML originates from stem cells341. The ALDH family is composed of 19 members with ambiguous functions in cancer. Increasing evidence has shown that ALDH1A1 can be used as a CSC marker in a panel of cancers, including ovarian cancer252,253, lung cancer254,255, breast cancer257, cervical cancer259, and HCC260. ALDH1A3 is another CSC marker found in cancers of the breast258, lung256, and colon261. Moreover, ALDH1B1 has been referred to as a CSC marker in colon cancer262.
Bmi-1
Bmi-1 is a member of polycomb repressor complex I and is considered a proto-oncogene predominantly expressed in CSCs and essential for self-renewal and clonal expansion342,343. Bmi-1 overexpression leads to EMT and enhances cancer stemness in the NSCLC cell line, A549344. Activation of Bmi-1 has also been found in breast CSCs characterized by CD44+/CD24−/low/Lin−345. Targeting Bmi-1+ CSCs inhibits the growth and eliminates chemotherapy resistance in HNSCC342. The cancers characterized by Bmi-1 include gastric cancer263, HNSCC5,264, CRC265, ESCC266–268, hematopoietic neoplasms269, and glioma270,271.
Nestin
Nestin, an intermediate filament protein, was initially described as a neuronal stem cell or progenitor cell marker in 1990346. Nestin is often co-expressed with other stem cell markers, such as CD133, Oct3/4, and Sox-2 in various human solid tumors, including neurogenic tumors272–276, rhabdomyosarcoma277, osteosarcoma278, chondrosarcoma160, fibrosarcoma160, ovarian cancer279–281, OSCC282, prostate cancer283,284, gallbladder cancer285, lung cancer286, colon cancer287, breast cancer288,289, gastric cancer290 and pancreatic cancer291.
Musashi-1 (Msi1)
The RNA-binding protein, Msi1, is involved in post-transcriptional gene regulation by competing with eukaryotic translation initiation factor 4G (eIF4G). Kanemura et al.347 and Toda et al.348 reported the prognostic significance of Msi1 and MIB1 in human gliomas. Msi1 is considered a stem cell marker regulating homeostasis between self-renewal and differentiation349. Msi1 is expressed in CRC292, OSCC293,294, and neuronal cancer CSCs295.
Tim-3
Tim-3 signaling regulates immune responses by downregulating interferon production, thereby inducing T-cell exhaustion350,351. CD44+CD90+ CSC-like cells and T cells exhibit increased TIM-3 in SCLC60. TIM-3 is highly expressed on LSCs from most AML patients, but not those with acute promyelocytic leukemia, and is generally not expressed on normal hematopoietic stem cells296.
CXCR4
CXCR4 belongs to an important subfamily of chemokine receptors that consist of seven mutually parallel, tightly arranged transmembrane-spanning segments that are closely related to cancer progression and prognosis352,353. CXCR4 is a GPCR chemokine receptor regulating leukocyte trafficking, stem cell mobilization, and homing of stem cells354,355. CXCR4 keeps stem cells in niches by interacting with SDF-1356. The expression of CD44 and CD133 is associated with a high level of CXCR4 in prostate CSCs357. CD133+/CXCR4+ CSCs promote metastasis in CRC358. The same subpopulation has also been identified in pancreatic cancer, where CD133+/CXCR4+ CSCs have a high tendency to metastasize164. Moreover, CXCR4 is a CSC marker in glioma297 and prostate cancer298.
Novel biomarkers of CSCs include SIRYγ and OSMR in lung cancer and glioblastoma, respectively6,359. These new biomarkers help us identify specific CSC subpopulations in distinct cancers that express different phenotypic markers for CSCs with higher accuracy. However, current biomarkers are not specific enough, so comprehensive studies are needed to identify more accurate biomarkers, either individually or in combination, for identifying CSCs.
Mechanisms of therapeutic resistance in CSCs
Multiple stemness-related signaling pathways initially found in normal stem cells have been validated in CSCs, including the Hedgehog, Wnt, Notch, and NF-κB pathways, which also have crucial roles in therapeutic resistance and have been extensively reviewed elsewhere360,361. Mechanisms of therapeutic resistance are complex and involve activation of survival signaling, evasion of apoptosis, high activities of acetaldehyde dehydrogenase, cell dormancy, disrupted cell differentiation, abnormal DNA damage/repair, altered epigenetic modification, immune suppression, inhibition of ROS, and hypoxia. For example, ROS scavengers in CSCs partially block the increase in ROS and protect CSCs from permanent damage to DNA, RNA, and other biomacromolecules362. The earliest finding in this regard was that ATP-binding cassette efflux transporters (ABC transporters) are the membrane proteins in bacteria. ABC transporters were subsequently shown to have an essential role in resistance to chemotherapy by efflux of drugs in humans363,364. ABC transporters (ABCB1, ABCC2, and ABCG2) overexpressed in CSCs is regarded as a CSC biomarker and the predictor of chemotherapeutic resistance365,366. Vasculogenic mimicry (VM), referring to the replacement of endothelial cells by tumor cells and the creation of a vessel with a lumen, can be induced by VM-related molecules secreted by CSCs, such as VE-cadherin protein, which ultimately confer resistance to antiangiogenic therapies and other anti-cancer therapies367–370. In this section, we mainly review the mechanisms underlying resistance to radio- and chemo-therapy.
Mechanisms of radiotherapeutic resistance in CSCs
A multicenter study has shown that CD44, a CSC marker in laryngeal cancer, predicts an increase in the CSC population and acquires radio-resistance and recurrence after radiotherapy in patients at an early stage371. In HER2-expressing breast CSCs (HER2+/CD44+/CD24−/low), a significant reduction in cell sensitivity to irradiation (IR)-induced apoptosis and increased clonogenic survival were observed when compared to wild-type MCF7 cells372. Similarly, in lung cancer cell lines, exposure to a single 4 Gy of radiation enriched CD133+ CSCs with increased DNA repair. Homologous recombination (HR)-associated proteins [exonuclease 1 (Exo1) and RAD51] have been identified as key regulators of radio-resistance in CSCs (Figure 3)373. Ataxia–telangiectasia mutated (ATM) and ATM-RAD3-related (ATR) are well known as upstream proteins of checkpoints that respond to different types of DNA damage. ATM, ATR, and the downstream checkpoint kinases (Chk1 and Chk2) mediate radio-resistance. Rapid induction of Chk1 is associated with enhanced G2/M cell cycle checkpoint activation in gastric CSCs. Chk1 inhibition sensitizes CSCs to IR374–376. Chk1 and Chk2 facilitate the DNA damage response by initiating cell cycle checkpoint control and activating the corresponding DNA repair pathways. Enhanced ATM signaling that contributes to the specific DNA repair phenotype is also observed in glioma CSCs with radio-resistance377. Chk1 knockdown in CD133+/CD44+ prostatic CSCs abrogates radiation-induced G2/M arrest, inhibits DNA damage repair, and promotes premature mitosis, leading to increased apoptosis378. Furthermore, inhibition of ATM overcomes radio-resistance in breast CSCs379. A mechanistic study reported that breast CSCs have a 7-fold higher ATM phosphorylation in response to 2 Gy radiation over non-CSCs379. Cyclin D2 (CCND2), a member of the cyclin protein family, has an important role in promoting colorectal CSCs to survive after radiation by activating cell cycle progression, DNA replication, and DNA repair. Targeting the JAK2/STAT3/CCND2 pathway overcomes radio-resistance by resisting radiation-induced apoptosis380. Inhibition of autophagy by silencing beclin1 and autophagy-related 5 (ATG5) or bafilomycin A1 increases the sensitivity of CD133+ gastric CSCs to radiation381. HIFs mainly regulate CSCs from gastric cancer and CRC in the hypoxic microenvironment, which causes chemo- and radio-resistance382,383. Additionally, miR-99a modulates breast CSC self-renewal by suppressing HIF-1α and mTOR signaling384, while miR-18a-5p overexpression increases the radiosensitivity of CD133+ lung CSCs by downregulating HIF-1α and ATM in vitro and in vivo385. Furthermore, the ΔNp63α/ribosome S6 protein kinase 4 (RSK4)/glycogen synthase kinase 3β (GSK-3β) axis contributes to CSC properties and radio-resistance in ESCC, suggesting that RSK4 is a promising therapeutic target386. The roles of different signaling pathways associated with CSCs (STAT3, PI3K/Akt/mTOR, ERK, VEGF, Notch, and Wnt/β-catenin pathways) in radio-resistance have been summarized by Chang et al.387.
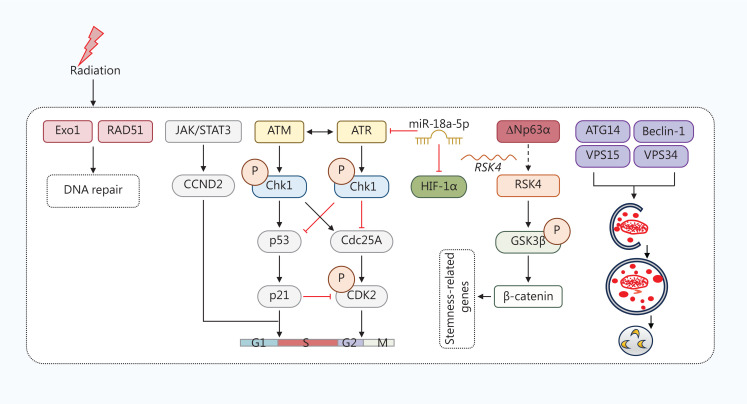
Figure 3.
The mechanisms underlying radio-resistance in CSCs. Upon radiation, multiple signaling pathways are triggered to induce resistance and maintain stemness in CSCs. First, radiation induces the expression of Exo1 and RAD51, which are involved in DNA repair. The JAK/STAT3 pathway is triggered by radiation, then activates the following CCND2 signaling and subsequent G1-to-S transition. Radiation also alters the cell cycle through ATM/ATR-associated signaling. In CSCs, miR-18a-5p overexpression degrades ATR and HIF-1α, thereby overcoming resistance to radiation. Additionally, ΔNp63α promotes the transcription of ribosome S6 protein kinase 4 (RSK4). Then, RSK4 phosphorylates GSK-3β at Ser9, which leads to the transcription of β-catenin-mediated stemness-related genes. Moreover, autophagy-related proteins are upregulated in some CSCs, where CSCs are involved in radio-resistance.
Mechanisms of chemoresistance in CSCs
During the process of treatment, chemotherapeutic agents often induce CSCs enrichment366. For example, doxorubicin treatment elevates the proportion of EpCAM+/CD133+ cells in HCC Huh7388. CSCs expressing Sox2 are resistant to tamoxifen, an antagonist of the estrogen receptor, in breast cancer through activation of the Wnt signaling pathway389. The intrinsic and induced enrichment of CSCs contributes to chemotherapeutic resistance. The mechanisms of chemoresistance in CSCs vary by cancer type. Here, we mainly focus on chemoresistant mechanisms in CSCs from lung cancer (Figure 4).
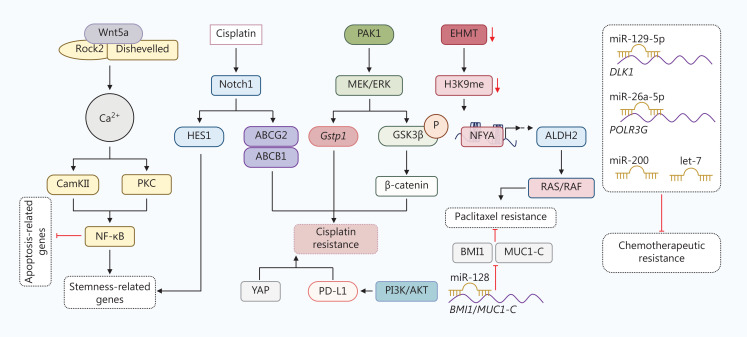
Figure 4.
The mechanisms underlying chemoresistance in lung CSCs. Extracellular Wnt5a initializes the Ca2+ pathway by interacting with Rho-kinase 2 (Rock2) and dishevelled on the membrane. Ca2+ then activates CamKII and protein kinase C (PKC), which induce nuclear factor kappa B (NF-κB)-mediated transcription of stemness-related genes and inhibit the transcription of apoptosis-related genes. Upon cisplatin treatment, Notch1 is upregulated in CSCs. Notch1 promotes stemness in a hairy and enhancer of split (HES1)-dependent manner and triggers cisplatin resistance by increasing ATP-binding cassette transporter G2 (ABCG2) and ABCB1. p21-activated kinase 1 (PAK1) activates the MEK/ERK signaling pathway in CSCs, followed by the elevated transcription of Gstp1 or activation of β-catenin signaling regulated by phosphorylation of GSK3β at Ser9, which leads to cisplatin resistance. Moreover, Yes1-associated transcriptional regulator (YAP1) and PI3K/AKT-induced PD-L1 are reported to cause cisplatin resistance. Euchromatic histone lysine methyltransferase (EHMT), which catalyzes the methylation of H3K9, is reduced in CSCs. Nuclear transcription factor Y subunit A (NFYA) is then recruited to DNA and initializes transcription of ALDH2, which results in paclitaxel resistance through RAS/RAF signaling. Several microRNAs (miRNAs), including miR-128, miR-129-5p, miR-26a-5p, miR-200, and let-7, impede chemoresistance in CSCs.
Mesenchymal NSCLC cells are resistant to epithelial growth factor receptor (EGFR), tyrosine kinase inhibitors (TKIs), erlotinib, and cisplatin compared to parental cells390. Platinum-based chemotherapy, paclitaxel, and etoposide are commonly used as first-line chemotherapy for lung cancer. A study showed that the development of cisplatin and etoposide resistance is associated with increased expression of the stem marker, CD133, in vitro391. ALDH1+ cells display greater resistance to the chemotherapeutic drugs, including cisplatin, gemcitabine, doxorubicin, daunorubicin, vinorelbine, and docetaxel, than ALDH1− cells392. The CSC biomarker, ALDH2, has been shown to contribute to paclitaxel resistance through the RAS/RAF pathway in lung cancer. The ALDH2 inhibitors, daidzin (DZN) and disulfiram (DSF), reverse paclitaxel resistance in xenograft models by promoting cell apoptosis and blocking the RAS/RAF pathway393. Cisplatin elevates the proportion of CD133+ cells by activating Notch1 signaling and upregulating ABCG2 and ABCB1 expression, which lead to cross-resistance to doxorubicin and paclitaxel394. Hashida et al.395 reported that established afatinib-resistant cells exhibit characteristics of EMT and stemness. Afatinib-resistant cells also exhibit amplification of MET genes, express high levels of ALDH1A1 and ABCB1, and are resistant to chemotherapeutic agents. Upregulated ABCG2 also facilitates resistance by effluxing gefitinib in NSCLC cells396. Several signaling pathways modulate chemoresistance in CSCs, such as YAP, Erk, and Notch.
Knockdown of YAP sensitizes A549 tumorspheres to cisplatin in NSCLC cells397. p21-activated kinase 1 (PAK1) confers cisplatin resistance in NSCLC cells. PAK1 activates MEK/ERK signaling, which promotes phosphorylation of GSK3β at Ser9 and subsequent β-catenin-mediated stemness398. Zhang and colleagues399 have detected a CD166+CD49fhighCD104−Lin− subpopulation with CSC properties from patient-derived sphere-forming assays. This subpopulation exhibits a high level of Notch1 and its ligand, delta-like canonical Notch ligand 4 (DLL4), which maintains self-renewal and platinum resistance through the Notch1 intracellular domain (NICD1)/hairy and enhancer of split (HES1)/STAT3 axis and survival regulators, respectively399. MEK/ERK signaling confers cisplatin resistance by transcriptionally inducing glutathione S-transferase Pi (Gstp1) in murine lung cancer cell LLC-derived CSCs400. Furthermore, activation of Wnt/β-catenin signaling is a mechanism resulting in cisplatin resistance. Silencing β-catenin sensitizes A549 cells to cisplatin by blocking Bcl-xl401. The non-canonical Wnt5a/PKC signaling pathway promotes stemness and cisplatin resistance in cisplatin-resistant A549 cells402. Recent studies have shown that upregulation of PD-L1 by the PI3K/AKT pathway is the main cause of cisplatin resistance in lung cancer cells403. The Wnt/β-catenin and Shh signaling pathways are commonly hyperactivated in CSCs. Furthermore, PD-L1 overexpression in CSCs contributes to immune evasion. Therefore, we speculate that Wnt/β-catenin signaling, the Shh signaling pathway, and PD-L1 might also have a role in cisplatin resistance in CSCs. Moreover, downregulated miR-129-5p, miR-26a-5p, miR-128, miR-200, and let-7 family miRNAs contribute to stemness and chemoresistance in stem-like NSCLC cells390,404–406. In silico prediction and in vitro experiments have identified the Notch signaling receptor, delta-like 1 homolog (DLK1), and RNA polymerase III subunit G (POLR3G) as targets of miR-129-5p and miR-26a-5p, respectively405,406. miR-128 suppresses BMI1 and MUC1-C expression, thereby weakening CSC-related traits in paclitaxel-resistant lung CSCs404. BRM270 extracted from herbal plants has been reported to induce miR-128 expression in chemoresistant CSCs, which overcomes paclitaxel resistance407.
Prospects of targeting CSCs
Because CSCs are the major cell population giving rise to therapeutic resistance, many studies have explored effective therapeutic strategies for targeting CSCs. Inhibitors targeting stem-associated signaling pathways (Shh, Wnt/β-catenin, and Hippo) have been evaluated preclinically and clinically, and summarized elsewhere408. Surface molecules are not only critical biomarkers for CSC isolation but also potential targets for treatment409. Additionally, the interplay between CSCs, the TME, and metabolism has given us promising therapeutic targets. In this section we mainly summarize the current treatments targeting the TME and CSC metabolism.
Targeting the TME of CSCs
Effective treatment for immunosuppressive tumor phenotypes can be challenging due to poor T-cell priming or immunologic ignorance. Single agents blocking PD-1 or PD-L1 performed poorly at converting “cold” tumors to “hot” tumors410. A PD1-based CSC vaccine has been shown to inhibit tumor growth in an animal colon cancer model411. Combining immune checkpoint blockade and CSC targeting therapies could be a promising treatment for immunosuppressive tumor phenotypes.
Various immune-based therapeutic strategies have been investigated to target CSCs412 (Table 2). T cell-based therapies, such as adoptive cell transfer therapy (ACT), have proven effective at fighting cancer. This form of personalized cancer treatment involves the administration of ex vivo expanded autologous tumor-infiltrating lymphocytes to cancer patients. CAR-T cells are patient-derived T cells engineered to express antibodies against desired cell surfaces. Human CAR-T cells targeting EpCAM have the potential to eradicate established tumor xenografts without causing toxicity in mouse models413,414. CAR-T cells expressing EpCAM accumulate in prostate tumors and eradicate CSCs in PC3M and PC3413. The adoptive transfer of γδ and CD8+ T cells also upregulates MHC class I and CD54/ICAM-1 on CSCs and activates antigen-specific T-cell killing415.
Table 2.
Ongoing clinical trials of investigational CSC-directed immunotherapeutic approaches
| Agent | Disease | Sample size | Phase | NCT number | Current status |
|---|---|---|---|---|---|
| CD19 CAR-T | Relapsed/refractory non-Hodgkin lymphoma (NHL) | 82 | II | NCT04089215 | Recruiting |
| CD123 CAR-T | Relapsed/refractory acute myelogenous leukemia (AML) | 40 | I/II | NCT04272125 | Recruiting |
| CD133 CAR-T | Relapsed/refractory advanced malignancies | 20 | I/II | NCT02541370 | Completed |
| CD22 CAR-T | B cell malignancies | 20 | I/II | NCT03262298 | Recruiting |
| Childhood leukemia | 100 | II | NCT04340167 | Recruiting | |
| CD33 CAR-T | Relapsed/refractory AML | 25 | I/II | NCT04835519 | Recruiting |
| CD38 CAR-T | Relapsed B-cell acute lymphocytic leukemia (ALL) after CD19 CAR-T adoptive cellular immunotherapy | 80 | I/II | NCT03754764 | Unknown |
| BCMA-CART | Relapsed/refractory multiple myeloma (MM) | 150 | I | NCT04394650 | Active, not recruiting |
| Mesothelin CAR-T | Advanced refractory solid tumors | 12 | I | NCT04981691 | Recruiting |
| Mesothelioma | 30 | I | NCT04577326 | Recruiting | |
| LeY CAR-T | Advanced solid tumors | 20 | I | NCT03851146 | Completed |
| CD30 CAR-T | Relapsed/refractory Hodgkin lymphoma | 97 | II | NCT04268706 | Active, not recruiting |
| CD30-expressing lymphomas | 26 | I | NCT03049449 | Completed | |
| Relapsed/refractory Hodgkin and T-cell lymphoma | 30 | I/II | NCT04653649 | Recruiting | |
| CD70 CAR-T | CD70-positive malignant hematologic diseases | 108 | I | NCT04662294 | Recruiting |
| CD71 CAR-T | CD70-positive advanced/metastatic solid tumors | 48 | I | NCT05468190 | Recruiting |
| CD171 CAR-T | Neuroblastoma/ganglioneuroblastoma | 65 | I | NCT02311621 | Active, not recruiting |
| CXCR4 CAR-T | Refractory/relapsed MM | I | NCT04727008 | not recruiting | |
| AMG 119 (anti-DLL3) CAR-T |
Small-cell lung cancer (SCLC) | 6 | I | NCT03392064 | Suspended |
| c-Met/PD-L1 CAR-T | Hepatocellular carcinoma (HCC) | 50 | I | NCT03672305 | Unknown |
| GD2 CAR-T | Relapsed/refractory neuroblastoma or other GD2-positive solid tumors | 42 | I/II | NCT03373097 | Recruiting |
| Diffuse intrinsic pontine gliomas (DIPG) and spinal diffuse midline glioma (DMG) | 54 | I | NCT04196413 | Recruiting | |
| Lung cancer | 24 | I | NCT05620342 | Not yet recruiting | |
| GPC3 CAR-T | Advanced HCC | 38 | I | NCT05003895 | Recruiting |
| GPC4 CAR-T | Pediatric HCC | 10 | I | NCT02932956 | Active, not recruiting |
| MOv19-BBz CAR-T | aFR expressing recurrent high grade serous ovarian, fallopian tube, or primary peritoneal cancer | 18 | I | NCT03585764 | Recruiting |
| P-MUC1C-ALLO1 CAR-T | Advanced or metastatic solid tumors | 100 | I | NCT05239143 | Recruiting |
| IL13Ralpha2 CAR T | Leptomeningeal glioblastoma, ependymoma, or medulloblastoma | 30 | I | NCT04661384 | Recruiting |
| EpCAM CAR-T | Advanced gastric cancer with peritoneal metastasis | 40 | I | NCT03563326 | Recruiting |
| EpCAM-positive cancer | 60 | I/II | NCT03013712 | Completed | |
| BCMA CAR-T | BCMA-positive relapsed/refractory MM | 28 | I | NCT03338972 | Completed |
| NK cells | Advanced lung adenocarcinoma (LUAD) with an EGFR mutation | 10 | I | NCT03662477 | Unknown |
| Recurrent glioblastoma multiform patients | 5 | I | NCT05108012 | Recruiting | |
| HCC | 18 | I | NCT02399735 | Unknown | |
| Cytokine-induced killer cell (CIK) | After resection of liver cancer | 200 | III | NCT00769106 | Completed |
| SCLC | 60 | II | NCT01592422 | Completed | |
| Urinary bladder neoplasms | 1500 | II | NCT02489890 | Active, not recruiting | |
| DC vaccine ALDH | Colorectal cancer (CRC) | 40 | I/II | NCT02176746 | Completed |
| Ovarian cancer | 40 | I/II | NCT02178670 | Completed | |
| Breast cancer | 40 | I/II | NCT02063893 | Completed | |
| Lung cancer | 40 | I/II | NCT02084823 | Completed | |
| Nasopharyngeal carcinoma (NPC) | 40 | I/II | NCT02115958 | Completed | |
| Pancreatic cancer | 40 | I/II | NCT02074046 | Completed | |
| Liver cancer | 40 | I/II | NCT02089919 | Completed | |
| Multiantigen DNA plasmid-based vaccine (CD105, Yb-1, SOX2, CDH3, and MDM2) | HER2-negative breast cancer | 42 | I | NCT02157051 | Active, not recruiting |
| DCs pulsed with lysate from allogeneic glioblastoma stem-like cell line | Newly diagnosed or recurrent glioblastoma | 39 | I | NCT02010606 | Completed |
| CSC vaccine | Pancreatic cancer | 40 | I/II | NCT02074046 | Completed |
| NPC | 40 | I/II | NCT02115958 | Completed | |
| Lung cancer | 40 | I/II | NCT02084823 | Completed | |
| HCC | 40 | I/II | NCT02089919 | Completed | |
| CRC | 40 | I/II | NCT02176746 | Completed | |
| Ovarian cancer | 40 | I/II | NCT02178670 | Completed | |
| Breast cancer | 40 | I/II | NCT02063893 | Completed | |
| OH2 oncolytic viral therapy | Solid tumors | 300 | I/II | NCT03866525 | Recruiting |
| Pancreatic cancer | 25 | I/II | NCT04637698 | Recruiting | |
| Advanced bladder carcinoma | 45 | II | NCT05248789 | Recruiting | |
| Central nervous system tumors | 28 | I/II | NCT05235074 | Recruiting | |
| Wild-type reovirus | Metastatic melanoma | 23 | II | NCT00651157 | Completed |
| MV-NIS | Recurrent medulloblastoma or recurrent atypical teratoid rhabdoid tumor (ATRT) | 46 | I | NCT02962167 | Recruiting |
| Bladder cancer who are undergoing radical cystectomy | 16 | I | NCT03171493 | Recruiting | |
| Oncolytic adenovirus Ad5-DNX-2401 | Recurrent high-grade glioma | 36 | I | NCT03896568 | Recruiting |
| Oncolytic adenovirus ONCOS-102 and pembrolizumab | Advanced or unresectable melanoma progressing after PD1 blockade | 21 | I | NCT03003676 | Completed |
| Oncolytic adenovirus VCN-01 | Refractory retinoblastoma | 13 | I | NCT03284268 | Recruiting |
| Oncolytic adenovirus ColoAd1 |
CRC/non-small cell lung cancer (NSCLC)/urothelial cell cancer (UCC)/renal cell carcinoma (RCC) | 17 | I | NCT02053220 | Completed |
| Oncolytic adenovirus OBP-301 |
Advanced esophageal cancer and are not candidates for surgery | 12 | I | NCT04391049 | Recruiting |
| HSV-1716 | Malignant pleural mesothelioma | 12 | I/II | NCT01721018 | Completed |
| Non-central nervous system (non-CNS) solid tumors | 18 | I | NCT00931931 | Completed | |
| HSV-M032 | Recurrent malignant glioma | 24 | I | NCT02062827 | Active, not recruiting |
| Hu5F9-G4 | Solid tumor | 88 | I | NCT02216409 | Completed |
| Hematologic malignancies | 20 | I | NCT02678338 | Completed | |
| SRF231 | Advanced solid and hematologic cancers | 148 | I | NCT03512340 | Completed |
| TTI-621 | Hematologic malignancies and selected solid tumors | 250 | I | NCT02663518 | Active, not recruiting |
| TTI-621 with or without rituximab | Advanced hematologic malignancies, including lymphoma, leukemia, and MM | 476 | I | NCT03530683 | Recruiting |
| ALX148 | Advanced solid and hematologic cancers | 60 | I | NCT02367196 | Completed |
| ALX149 | Advanced solid tumors and lymphoma | 174 | I | NCT03013218 | Active, not recruiting |
| Reparixin | Metastatic breast cancer | 33 | I | NCT02001974 | Completed |
| Metastatic triple-negative breast cancer (TNBC) | 194 | II | NCT02370238 | Completed | |
| KHK2823 | AML | 39 | I | NCT02181699 | Terminated |
| Talacotuzumab | AML | 30 | I | NCT01632852 | Completed |
| AML | 326 | II/III | NCT02472145 | Completed | |
| Flotetuzumab | AML | 246 | I/II | NCT02152956 | Terminated |
| XmAb14045 | CD123-expressing hematologic malignancies | 120 | I | NCT02730312 | Completed |
| JNJ-63709178 | Relapsed or refractory AML | 62 | I | NCT02715011 | Completed |
| MT110 | Solid tumors | 65 | I | NCT00635596 | Completed |
| AMG 757 | Neuroendocrine prostate cancer | 60 | I | NCT04702737 | Recruiting |
| Extensive stage SCLC | 340 | I | NCT05361395 | Recruiting | |
| CD133+ stem cell transplantation, busulfan, and melphalan | Children with solid tumors and lymphomas | 26 | Not applicable | NCT00152126 | Completed |
| CD133+ stem cell transplantation and portal vein embolization | Colorectal liver metastases | 4 | II | NCT03803241 | Completed |
| IFN-β therapy | Metastatic cutaneous melanoma or ocular melanoma | 21 | II | NCT00085306 | Completed |
| SBRT with anti-PD1 and anti-IL-8 | Multiple metastases in advanced solid tumors | 50 | I | NCT04572451 | Recruiting |
| Nivolumab with anti-IL-8 | NSCLC or HCC | 50 | II | NCT04123379 | Recruiting |
| MCLA-158 | Advanced solid tumors | 120 | I | NCT03526835 | Recruiting |
Oncolytic virus (OV) is another immunotherapy with low toxicity that targets and destroys tumor cells through cytopathic effects in a direct and indirect fashion416. The OV, GLV-1h68, can kill stem cell-like cancer cells (higher ALDH1 activity) in breast cancer. In cell culture, GLV-1h68 replicates in and kills breast CSCs417.
In addition to T cells, other immune cells, such as NK cells, B lymphomas, macrophages, and neutrophils, have roles in shaping the TME in cooperation with CSCs, as mentioned above. DCs were treated with CSC lysates and tumor-related antigens ex vivo to generate a DC-based vaccine, which was then injected back into cancer patients418. CSC lysate-pulsed DCs induced IFN-γ and IL-4 secretion in vaccinated mice with malignant melanoma, inhibiting tumor growth and prolonging survival in immunized mice419. Secretion of INF-γ and IL-2 induced by pancreatic CSC lysate-loaded DC vaccination promotes the function of lymphocytes in pancreatic cancer cells420. Similarly, DCs charged with Nanog peptides enhance the anti-tumor activity of T cells against CSCs in ovarian cancer421. DCs loaded with ALDHhigh SCC7 have also been reported to reduce recurrence and prolong survival in a murine HNSCC model422.
Moreover, the adoptive transfer of NK cells causes an improvement in MICA/B, Fas, and DR5 as NK cell-activating ligands on CSCs423. Anti-CD133 CAR-engineered NK-92 cells kill CD133+ ovarian CSCs in vitro and in vivo. Cisplatin treatment followed by anti-CD133 CAR-engineered NK-92 cells significantly augment the anti-tumor effect in a murine ovarian model424. Although previous evidence has suggested the essential role of crosstalk between CSCs and TAMs or neutrophils, specific strategies that target the interaction remain unknown due to the ambiguous mechanisms of the interplay in individuals.
Signaling pathways have been reported in immune cells and CSCs, and are also promising targets, such as STAT3 and PI3K. Multiple STAT3 inhibitors have been developed and processed for clinical trials425. Notably, the first-in-class antisense oligonucleotide (ASO) targeting STAT3 AZD9150 has chemical stability and anti-tumor activity in several cancers. The efficacy of AZD9150 in various cancers is still ongoing or pending426. The efficacy of PI3K inhibitors, including PX-866 (IND205), alpelisib (NCT02437318), PQR309 (PQR309), and pictilisib (GDC-0941), has been tested in several clinical trials (Table 2). These PI3K inhibitors show considerable efficacy in some settings, especially in combination with inhibitors of other pathways, such as MEK, and require further investigation. CAFs are also a potential target for tumor treatment. Chen et al.427 have reported a cancer cell-targeted nanoliposome system that specifically targets and delivers Navitoclax (Nav) to CAFs.
Targeting metabolism in CSCs
As mentioned above, heterogeneous metabolic patterns have been reported in CSCs of different types of tumors. Glycolysis is enhanced, which makes glycolysis a promising target in some CSCs. Targets of glycolysis include rate-limiting enzymes, transporters, and other complex regulators428–430. GLUT1, a glucose transporter, has an important role in the maintenance of pancreatic, ovarian, and GBM CSCs431. WZB117, a specific GLUT1 inhibitor, successfully inhibits the self-renewal and tumor-initiating capacity of the CSCs in vitro432. Silibinin, another GLUT1 inhibitor, causes the dual blockade of EMT and stemness of bladder CSCs via inactivation of β-catenin/ZEB1 signaling in vitro433. Phase I-II clinical trials have assessed the toxicity and efficacy of glucose transport inhibitors, such as WZB117, fisetin, phloretin, and silybin/silibinin, for advanced HCC and prostate cancer; however, these therapies have limitations due to side effects, such as hyperbilirubinemia and elevation of alanine aminotransferase (ALT)434,435. Blocking OXPHOS therapeutically suppresses CSC growth, including sphere and tumor formation potential122,436,437. Atovaquone, an U.S. FDA-approved anti-malarial drug and a selective OXPHOS inhibitor, has therapeutic efficacy against MCF7 breast cancer cells by targeting CoQ10-dependent mitochondrial complex III. Mitochondrial respiration is damaged, causing glycolysis to increase as compensation438. Mitochondrially-targeting antibiotics, including salinomycin, erythromycin, tetracyclines, and glycylcyclines, have been U.S. FDA-approved to reduce stemness characteristics in CSCs437,439–441. Metformin, an inhibitor of mitochondrial complex I, has been studied extensively for its potential to target CSCs. Metformin inhibits the self-renewal of CSCs in breast cancer by suppressing estrogen receptor-mediated Oct4 expression in vitro442. A recent study has suggested that targeting glutamine metabolism enhances the radiosensitization of prostate cancer cells by increasing DNA damage, shifting redox balance, and retarding CSC properties. Metformin also shows the capacity to inhibit both glutamine metabolism and autophagy in tumor cells443. Another approach to reversing the resistance of CSCs with an intermediate glycolytic/OXPHOS phenotype is the administration of menadione, an ROS inducer122.
Targeting CSCs through the inhibition of glutaminolysis, which is the process of converting glutamine-to-glutamate via the enzyme, glutaminase (GLS), is a promising metabolic interference strategy431. GLS1 is associated with different types of cancer, and the GLS1 inhibitor, BPTES, combined with the phosphodiesterase-5 inhibitor, zaprinast, increases the sensitivity of pancreatic CSCs to radiotherapy and promotes apoptosis by increasing the level of intracellular ROS431. The first glutaminase inhibitor, DON, was isolated from Peruvian soil and induces apoptosis of breast CSCs444. Additionally, nuclear factor-erythroid 2-related factor 2 (Nrf2), a redox-related transcription factor that regulates antioxidant enzymes for maintaining cellular redox status, has been linked to the regulation of CSCs445–448. In particular, the natural compound, honokiol, which is isolated from the wood of Cupressaceae trees, impedes the self-renewal, migration, and colony-forming ability of CSCs by inhibiting Nrf2 expression449. Similarly, the chestnut leaf inhibits sphere cell development and increases the chemosensitivity of breast CSCs to paclitaxel through inhibition of Nrf2 activity in vitro450.
FASN has a critical role in the production of endogenous fatty acids. Cerulenin is a natural antifungal antibiotic that has potent inhibitory properties against FASN451. Cerulenin curbs the self-renewal of gastric CSCs by suppressing adipogenesis452. Curcumin, in contrast, downregulates SCD1 and inhibits the self-renewal of breast CSCs453. Additionally, the SCD1 inhibitor, CAY10566, and the Δ6 desaturase inhibitor, SC-26196, inhibit the stemness of ovarian CSCs454. The vulnerability of cancer cells, particularly CSCs, to ferroptosis drives much effort to investigate the potential of ferroptosis as an anti-cancer strategy, although this vulnerability varies by cancer type. This finding has led to the investigation of many pathways, such as lipid metabolism, iron metabolism, and Nrf signaling, that regulate ferroptosis. Agents targeting these pathways have the potential to enhance the sensitivity of CSCs to ferroptosis. The combination of ferroptosis inducers with current treatments has been studied and reviewed by Lei et al.142 and Elgendy et al.455. The use of ferroptosis inducers requires careful investigation before clinical application because ferroptosis promotes tumor growth135.
Although there is still much to learn about metabolic patterns in CSCs, it is likely that targeting metabolism will be an effective strategy to treat cancer. Recent research suggests that drugs designed to target resistance regulatory pathways or abnormal proteins in CSCs could improve therapeutic outcomes. In addition to these therapies, the differentiation of tumor cells may be a promising approach. One such method is the use of all-trans retinoic acid (ATRA), which downregulates the stem cell markers, Oct4, Sox2, Nestin, and CD44, in HNSCC CSCs456. ATRA also enhances the chemosensitivity of HNSCC CSCs to cisplatin, suppresses the proliferation of CSCs in vitro and in vivo, and inhibits the Wnt/β-catenin pathway, resulting in a decrease in stemness456.
Conclusions
Numerous attempts have been made to develop biomaterial-based platforms to enrich and study CSCs with the goal of targeting CSCs457. The potential of translating CSC biological research into clinical practice is quite promising. First, some biomarkers can distinguish CSCs from normal adult stem cells and cancer cells, but there is no definitive way to differentiate one from the other. Therefore, CSCs should be identified not only by the molecular and cellular biological features of normal stem cells but also by tumor-specific biomarkers with higher accuracy. Additionally, growing evidence indicates that CSCs have distinctive metabolic features in individual cancers, which suggests altered metabolism as a potential target. Some old drugs used for metabolic disorders, such as diabetes and natural compounds, have proven to be effective at targeting CSCs in preclinical trials. However, our understanding of the metabolic landscape in different types of cancers remains unclear. The metabolic patterns of CSCs, differentiated cancer cells, and stromal cells in the TME are not fully understood, which necessitates further investigation before clinical targeting of metabolism can be translated into practice. Moreover, all targeting strategies should take every compartment, including quiescent and proliferating CSCs, as well as the stromal cells and the TME matrix, into account. A combination of immunotherapies and other therapies against CSCs may improve the prognosis of patients and be widely available for clinical use. Precise drug delivery to CSCs enhances therapeutic accuracy and efficacy while minimizing damage to normal cells, which might be achieved through findings in chemistry and materials science, such as nanotechnology458.
Funding Statement
This work was supported by the National Key Research and Development Program of China (Grant No. 2023YFC3402100) and the National Natural Science Foundation of China (Grant No. 92259102).
Conflict of interest statement
No potential conflicts of interest are disclosed.
Author contributions
Substantial contributions to conception: Shuo Zhang, Lanlin Hu, Chuan Xu.
Manuscript and tables: Shuo Zhang, Lanlin Hu, Rui Yang, Yujie Ouyang, Yang Shen.
Figures: Shuo Zhang, Lanlin Hu.
References
- 1.Mackillop WJ, Ciampi A, Till JE, Buick RN. A stem cell model of human tumor growth: implications for tumor cell clonogenic assays. J Natl Cancer Inst. 1983;70:9–16. [PubMed] [Google Scholar]
- 2.O’Brien CA, Kreso A, Dick JE. Cancer stem cells in solid tumors: an overview. Semin Radiat Oncol. 2009;19:71–7. doi: 10.1016/j.semradonc.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Vetrie D, Helgason GV, Copland M. The leukaemia stem cell: similarities, differences and clinical prospects in CML and AML. Nat Rev Cancer. 2020;20:158–73. doi: 10.1038/s41568-019-0230-9. [DOI] [PubMed] [Google Scholar]
- 4.Zhang R, Tu J, Liu S. Novel molecular regulators of breast cancer stem cell plasticity and heterogeneity. Semin Cancer Biol. 2022;82:11–25. doi: 10.1016/j.semcancer.2021.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Yu D, Liu Y, Yang J, Jin C, Zhao X, Cheng J, et al. Clinical implications of BMI-1 in cancer stem cells of laryngeal carcinoma. Cell Biochem Biophys. 2015;71:261–9. doi: 10.1007/s12013-014-0194-z. [DOI] [PubMed] [Google Scholar]
- 6.Xu C, Jin G, Wu H, Cui W, Wang YH, Manne RK, et al. SIRPγ-expressing cancer stem-like cells promote immune escape of lung cancer via Hippo signaling. J Clin Invest. 2022;132:e141797. doi: 10.1172/JCI141797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ricci-Vitiani L, Pallini R, Biffoni M, Todaro M, Invernici G, Cenci T, et al. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature. 2010;468:824–8. doi: 10.1038/nature09557. [DOI] [PubMed] [Google Scholar]
- 8.Soda Y, Marumoto T, Friedmann-Morvinski D, Soda M, Liu F, Michiue H, et al. Transdifferentiation of glioblastoma cells into vascular endothelial cells. Proc Natl Acad Sci U S A. 2011;108:4274–80. doi: 10.1073/pnas.1016030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng L, Huang Z, Zhou W, Wu Q, Donnola S, Liu JK, et al. Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell. 2013;153:139–52. doi: 10.1016/j.cell.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumon K, Afify SM, Hassan G, Ueno S, Monzur S, Nawara HM, et al. Differentiation of cancer stem cells into erythroblasts in the presence of CoCl(2) Sci Rep. 2021;11:23977. doi: 10.1038/s41598-021-03298-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang G, Tu J, Zhou L, Dong M, Fan J, Chang Z, et al. Single-cell transcriptomics reveal the heterogeneity and dynamic of cancer stem-like cells during breast tumor progression. Cell Death Dis. 2021;12:979. doi: 10.1038/s41419-021-04261-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou HM, Zhang JG, Zhang X, Li Q. Targeting cancer stem cells for reversing therapy resistance: mechanism, signaling, and prospective agents. Signal Transduct Target Ther. 2021;6:62. doi: 10.1038/s41392-020-00430-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang T, Song X, Xu D, Tiek D, Goenka A, Wu B, et al. Stem cell programs in cancer initiation, progression, and therapy resistance. Theranostics. 2020;10:8721–43. doi: 10.7150/thno.41648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ju F, Atyah MM, Horstmann N, Gul S, Vago R, Bruns CJ, et al. Characteristics of the cancer stem cell niche and therapeutic strategies. Stem Cell Res Ther. 2022;13:233. doi: 10.1186/s13287-022-02904-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beier CP, Kumar P, Meyer K, Leukel P, Bruttel V, Aschenbrenner I, et al. The cancer stem cell subtype determines immune infiltration of glioblastoma. Stem Cells Dev. 2012;21:2753–61. doi: 10.1089/scd.2011.0660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gangoso E, Southgate B, Bradley L, Rus S, Galvez-Cancino F, McGivern N, et al. Glioblastomas acquire myeloid-affiliated transcriptional programs via epigenetic immunoediting to elicit immune evasion. Cell. 2021;184:2454–70.e2426. doi: 10.1016/j.cell.2021.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson NM, Simon MC. The tumor microenvironment. Curr Biol. 2020;30:R921–5. doi: 10.1016/j.cub.2020.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen P, Hsu WH, Han J, Xia Y, DePinho RA. Cancer stemness meets immunity: from mechanism to therapy. Cell Rep. 2021;34:108597. doi: 10.1016/j.celrep.2020.108597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bayik D, Lathia JD. Cancer stem cell-immune cell crosstalk in tumour progression. Nat Rev Cancer. 2021;21:526–36. doi: 10.1038/s41568-021-00366-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou H, Tan L, Liu B, Guan XY. Cancer stem cells: recent insights and therapies. Biochem Pharmacol. 2023;209:115441. doi: 10.1016/j.bcp.2023.115441. [DOI] [PubMed] [Google Scholar]
- 21.Wu B, Shi X, Jiang M, Liu H. Cross-talk between cancer stem cells and immune cells: potential therapeutic targets in the tumor immune microenvironment. Mol Cancer. 2023;22:38. doi: 10.1186/s12943-023-01748-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramgolam K, Lauriol J, Lalou C, Lauden L, Michel L, de la Grange P, et al. Melanoma spheroids grown under neural crest cell conditions are highly plastic migratory/invasive tumor cells endowed with immunomodulator function. PLoS One. 2011;6:e18784. doi: 10.1371/journal.pone.0018784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrison BJ, Steel JC, Morris JC. Reduction of MHC-I expression limits T-lymphocyte-mediated killing of Cancer-initiating cells. BMC Cancer. 2018;18:469. doi: 10.1186/s12885-018-4389-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Tomaso T, Mazzoleni S, Wang E, Sovena G, Clavenna D, Franzin A, et al. Immunobiological characterization of cancer stem cells isolated from glioblastoma patients. Clin Cancer Res. 2010;16:800–13. doi: 10.1158/1078-0432.CCR-09-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chikamatsu K, Takahashi G, Sakakura K, Ferrone S, Masuyama K. Immunoregulatory properties of CD44+ cancer stem-like cells in squamous cell carcinoma of the head and neck. Head Neck. 2011;33:208–15. doi: 10.1002/hed.21420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao T, Kaufmann AM, Qian X, Sangvatanakul V, Chen C, Kube T, et al. Susceptibility to cytotoxic T cell lysis of cancer stem cells derived from cervical and head and neck tumor cell lines. J Cancer Res Clin Oncol. 2013;139:159–70. doi: 10.1007/s00432-012-1311-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sultan M, Vidovic D, Paine AS, Huynh TT, Coyle KM, Thomas ML, et al. Epigenetic silencing of TAP1 in Aldefluor(+) breast cancer stem cells contributes to their enhanced immune evasion. Stem Cells. 2018;36:641–54. doi: 10.1002/stem.2780. [DOI] [PubMed] [Google Scholar]
- 28.Burr ML, Sparbier CE, Chan KL, Chan YC, Kersbergen A, Lam EYN, et al. An evolutionarily conserved function of polycomb silences the MHC class I antigen presentation pathway and enables immune evasion in cancer. Cancer Cell. 2019;36:385–401.e388. doi: 10.1016/j.ccell.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim GR, Ha GH, Bae JH, Oh SO, Kim SH, Kang CD. Metastatic colon cancer cell populations contain more cancer stem-like cells with a higher susceptibility to natural killer cell-mediated lysis compared with primary colon cancer cells. Oncol Lett. 2015;9:1641–6. doi: 10.3892/ol.2015.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ames E, Canter RJ, Grossenbacher SK, Mac S, Chen M, Smith RC, et al. NK cells preferentially target tumor cells with a cancer stem cell phenotype. J Immunol. 2015;195:4010–9. doi: 10.4049/jimmunol.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dianat-Moghadam H, Mahari A, Heidarifard M, Parnianfard N, Pourmousavi-Kh L, Rahbarghazi R, et al. NK cells-directed therapies target circulating tumor cells and metastasis. Cancer Lett. 2021;497:41–53. doi: 10.1016/j.canlet.2020.09.021. [DOI] [PubMed] [Google Scholar]
- 32.Avril T, Vauleon E, Hamlat A, Saikali S, Etcheverry A, Delmas C, et al. Human glioblastoma stem-like cells are more sensitive to allogeneic NK and T cell-mediated killing compared with serum-cultured glioblastoma cells. Brain Pathol. 2012;22:159–74. doi: 10.1111/j.1750-3639.2011.00515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh SJ, Yang JI, Kim O, Ahn EJ, Kang WD, Lee JH, et al. Human U87 glioblastoma cells with stemness features display enhanced sensitivity to natural killer cell cytotoxicity through altered expression of NKG2D ligand. Cancer Cell Int. 2017;17:22. doi: 10.1186/s12935-017-0397-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li G, Nikkhoi SK, Hatefi A. Stem cell-assisted enzyme/prodrug therapy makes drug-resistant ovarian cancer cells vulnerable to natural killer cells through upregulation of NKG2D ligands. Med Oncol. 2023;40:110. doi: 10.1007/s12032-023-01975-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tallerico R, Todaro M, Di Franco S, Maccalli C, Garofalo C, Sottile R, et al. Human NK cells selective targeting of colon cancer-initiating cells: a role for natural cytotoxicity receptors and MHC class I molecules. J Immunol. 2013;190:2381–90. doi: 10.4049/jimmunol.1201542. [DOI] [PubMed] [Google Scholar]
- 36.Luna JI, Grossenbacher SK, Sturgill IR, Ames E, Judge SJ, Bouzid LA, et al. Bortezomib augments natural killer cell targeting of stem-like tumor cells. Cancers (Basel) 2019;11:85. doi: 10.3390/cancers11010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weng J, Han X, Liu K, Yang J, Wei S, Zhang Y, et al. CD44 3′-Untranslated region functions as a competing endogenous RNA to enhance NK sensitivity of liver cancer stem cell by regulating ULBP2 expression. Int J Biol Sci. 2019;15:1664–75. doi: 10.7150/ijbs.35216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohme M, Riethdorf S, Pantel K. Circulating and disseminated tumour cells - mechanisms of immune surveillance and escape. Nat Rev Clin Oncol. 2017;14:155–67. doi: 10.1038/nrclinonc.2016.144. [DOI] [PubMed] [Google Scholar]
- 39.Paczulla AM, Rothfelder K, Raffel S, Konantz M, Steinbacher J, Wang H, et al. Absence of NKG2D ligands defines leukaemia stem cells and mediates their immune evasion. Nature. 2019;572:254–9. doi: 10.1038/s41586-019-1410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimura Y, Tsunedomi R, Yoshimura K, Matsukuma S, Shindo Y, Matsui H, et al. Immune evasion of hepatoma cancer stem-like cells from natural killer cells. Ann Surg Oncol. 2022;29:7423–33. doi: 10.1245/s10434-022-12220-w. [DOI] [PubMed] [Google Scholar]
- 41.Bernareggi D, Xie Q, Prager BC, Yun J, Cruz LS, Pham TV, et al. CHMP2A regulates tumor sensitivity to natural killer cell-mediated cytotoxicity. Nat Commun. 2022;13:1899. doi: 10.1038/s41467-022-29469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang B, Wang Q, Wang Z, Jiang J, Yu SC, Ping YF, et al. Metastatic consequences of immune escape from NK cell cytotoxicity by human breast cancer stem cells. Cancer Res. 2014;74:5746–57. doi: 10.1158/0008-5472.CAN-13-2563. [DOI] [PubMed] [Google Scholar]
- 43.Jin H, Kim HJ. NK cells lose their cytotoxicity function against cancer stem cell-rich radiotherapy-resistant breast cancer cell populations. Int J Mol Sci. 2021;22:9639. doi: 10.3390/ijms22179639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park DJ, Sung PS, Kim JH, Lee GW, Jang JW, Jung ES, et al. EpCAM-high liver cancer stem cells resist natural killer cell-mediated cytotoxicity by upregulating CEACAM1. J Immunother Cancer. 2020;8:e000301. doi: 10.1136/jitc-2019-000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang C, Li Y, Jia L, Kim JK, Li J, Deng P, et al. CD276 expression enables squamous cell carcinoma stem cells to evade immune surveillance. Cell Stem Cell. 2021;28:1597–613.e1597. doi: 10.1016/j.stem.2021.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mansour FA, Al-Mazrou A, Al-Mohanna F, Al-Alwan M, Ghebeh H. PD-L1 is overexpressed on breast cancer stem cells through notch3/mTOR axis. Oncoimmunology. 2020;9:1729299. doi: 10.1080/2162402X.2020.1729299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin S, Guo Y, Wen X, Zeng H, Chen G. Increased expression of PD-L1 in endometrial cancer stem-like cells is regulated by hypoxia. Front Biosci (Landmark Ed) 2022;27:23. doi: 10.31083/j.fbl2701023. [DOI] [PubMed] [Google Scholar]
- 48.Fu L, Fan J, Maity S, McFadden G, Shi Y, Kong W. PD-L1 interacts with Frizzled 6 to activate beta-catenin and form a positive feedback loop to promote cancer stem cell expansion. Oncogene. 2022;41:1100–13. doi: 10.1038/s41388-021-02144-2. [DOI] [PubMed] [Google Scholar]
- 49.Koh YW, Han JH, Haam S. Expression of PD-L1, cancer stem cell and epithelial-mesenchymal transition phenotype in non-small cell lung cancer. Pathology. 2021;53:239–46. doi: 10.1016/j.pathol.2020.07.009. [DOI] [PubMed] [Google Scholar]
- 50.Yamaguchi H, Hsu JM, Yang WH, Hung MC. Mechanisms regulating PD-L1 expression in cancers and associated opportunities for novel small-molecule therapeutics. Nat Rev Clin Oncol. 2022;19:287–305. doi: 10.1038/s41571-022-00601-9. [DOI] [PubMed] [Google Scholar]
- 51.Hsu JM, Xia W, Hsu YH, Chan LC, Yu WH, Cha JH, et al. STT3-dependent PD-L1 accumulation on cancer stem cells promotes immune evasion. Nat Commun. 2018;9:1908. doi: 10.1038/s41467-018-04313-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun L, Wang Q, Chen B, Zhao Y, Shen B, Wang H, et al. Gastric cancer mesenchymal stem cells derived IL-8 induces PD-L1 expression in gastric cancer cells via STAT3/mTOR-c-Myc signal axis. Cell Death Dis. 2018;9:928. doi: 10.1038/s41419-018-0988-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Castagnoli L, Cancila V, Cordoba-Romero SL, Faraci S, Talarico G, Belmonte B, et al. WNT signaling modulates PD-L1 expression in the stem cell compartment of triple-negative breast cancer. Oncogene. 2019;38:4047–60. doi: 10.1038/s41388-019-0700-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darvin P, Sasidharan Nair V, Elkord E. PD-L1 expression in human breast cancer stem cells is epigenetically regulated through posttranslational histone modifications. J Oncol. 2019;2019:3958908. doi: 10.1155/2019/3958908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perry JM, Tao F, Roy A, Lin T, He XC, Chen S, et al. Overcoming Wnt-β-catenin dependent anticancer therapy resistance in leukaemia stem cells. Nat Cell Biol. 2020;22:689–700. doi: 10.1038/s41556-020-0507-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang C, Chen B, Wang X, Xu J, Sun L, Wang D, et al. Gastric cancer mesenchymal stem cells via the CXCR2/HK2/PD-L1 pathway mediate immunosuppression. Gastric Cancer. 2023;26:691–707. doi: 10.1007/s10120-023-01405-1. [DOI] [PubMed] [Google Scholar]
- 57.Hua F, Shang S, Yang YW, Zhang HZ, Xu TL, Yu JJ, et al. TRIB3 interacts with beta-catenin and TCF4 to increase stem cell features of colorectal cancer stem cells and tumorigenesis. Gastroenterology. 2019;156:708–21.e715. doi: 10.1053/j.gastro.2018.10.031. [DOI] [PubMed] [Google Scholar]
- 58.Zhou S, Zhu J, Xu J, Gu B, Zhao Q, Luo C, et al. Anti-tumour potential of PD-L1/PD-1 post-translational modifications. Immunology. 2022;167:471–81. doi: 10.1111/imm.13573. [DOI] [PubMed] [Google Scholar]
- 59.Ma X, Wu J, Wang B, Liu C, Liu L, Sun C. Epigenetic modifications: critical participants of the PD-L1 regulatory mechanism in solid tumors (Review) Int J Oncol. 2022;61:134. doi: 10.3892/ijo.2022.5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kursunel MA, Taskiran EZ, Tavukcuoglu E, Yanik H, Demirag F, Karaosmanoglu B, et al. Small cell lung cancer stem cells display mesenchymal properties and exploit immune checkpoint pathways in activated cytotoxic T lymphocytes. Cancer Immunol Immunother. 2022;71:445–59. doi: 10.1007/s00262-021-02998-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu S, Zhang C, Wang B, Zhang H, Qin G, Li C, et al. Regulatory T cells promote glioma cell stemness through TGF-beta-NF-kappaB-IL6-STAT3 signaling. Cancer Immunol Immunother. 2021;70:2601–16. doi: 10.1007/s00262-021-02872-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu X, Zhang Z, Zhao G. Recent advances in the study of regulatory T cells in gastric cancer. Int Immunopharmacol. 2019;73:560–7. doi: 10.1016/j.intimp.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 63.Qianmei Y, Zehong S, Guang W, Hui L, Lian G. Recent advances in the role of Th17/Treg cells in tumor immunity and tumor therapy. Immunol Res. 2021;69:398–414. doi: 10.1007/s12026-021-09211-6. [DOI] [PubMed] [Google Scholar]
- 64.Rezalotfi A, Ahmadian E, Aazami H, Solgi G, Ebrahimi M. Gastric cancer stem cells effect on Th17/Treg balance; a bench to beside perspective. Front Oncol. 2019;9:226. doi: 10.3389/fonc.2019.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Madden EC, Gorman AM, Logue SE, Samali A. Tumour cell secretome in chemoresistance and tumour recurrence. Trends Cancer. 2020;6:489–505. doi: 10.1016/j.trecan.2020.02.020. [DOI] [PubMed] [Google Scholar]
- 66.Larionova I, Cherdyntseva N, Liu T, Patysheva M, Rakina M, Kzhyshkowska J. Interaction of tumor-associated macrophages and cancer chemotherapy. Oncoimmunology. 2019;8:1596004. doi: 10.1080/2162402X.2019.1596004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo X, Zhao Y, Yan H, Yang Y, Shen S, Dai X, et al. Single tumor-initiating cells evade immune clearance by recruiting type II macrophages. Genes Dev. 2017;31:247–59. doi: 10.1101/gad.294348.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wan S, Zhao E, Kryczek I, Vatan L, Sadovskaya A, Ludema G, et al. Tumor-associated macrophages produce interleukin 6 and signal via STAT3 to promote expansion of human hepatocellular carcinoma stem cells. Gastroenterology. 2014;147:1393–404. doi: 10.1053/j.gastro.2014.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Standiford TJ, Kunkel SL, Basha MA, Chensue SW, Lynch JP, 3rd, Toews GB, et al. Interleukin-8 gene expression by a pulmonary epithelial cell line. A model for cytokine networks in the lung. J Clin Invest. 1990;86:1945–53. doi: 10.1172/JCI114928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shimizu M, Tanaka N. IL-8-induced O-GlcNAc modification via GLUT3 and GFAT regulates cancer stem cell-like properties in colon and lung cancer cells. Oncogene. 2019;38:1520–33. doi: 10.1038/s41388-018-0533-4. [DOI] [PubMed] [Google Scholar]
- 71.Huang R, Wang S, Wang N, Zheng Y, Zhou J, Yang B, et al. CCL5 derived from tumor-associated macrophages promotes prostate cancer stem cells and metastasis via activating β-catenin/STAT3 signaling. Cell Death Dis. 2020;11:234. doi: 10.1038/s41419-020-2435-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jinushi M, Chiba S, Yoshiyama H, Masutomi K, Kinoshita I, Dosaka-Akita H, et al. Tumor-associated macrophages regulate tumorigenicity and anticancer drug responses of cancer stem/initiating cells. Proc Natl Acad Sci U S A. 2011;108:12425–30. doi: 10.1073/pnas.1106645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cioffi M, Trabulo S, Hidalgo M, Costello E, Greenhalf W, Erkan M, et al. Inhibition of CD47 effectively targets pancreatic cancer stem cells via dual mechanisms. Clin Cancer Res. 2015;21:2325–37. doi: 10.1158/1078-0432.CCR-14-1399. [DOI] [PubMed] [Google Scholar]
- 74.Xi Q, Zhang J, Yang G, Zhang L, Chen Y, Wang C, et al. Restoration of miR-340 controls pancreatic cancer cell CD47 expression to promote macrophage phagocytosis and enhance antitumor immunity. J Immunother Cancer. 2020;8:e000253. doi: 10.1136/jitc-2019-000253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tahk S, Vick B, Hiller B, Schmitt S, Marcinek A, Perini ED, et al. SIRPalpha-alphaCD123 fusion antibodies targeting CD123 in conjunction with CD47 blockade enhance the clearance of AML-initiating cells. J Hematol Oncol. 2021;14:155. doi: 10.1186/s13045-021-01163-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Szczerba BM, Castro-Giner F, Vetter M, Krol I, Gkountela S, Landin J, et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature. 2019;566:553–7. doi: 10.1038/s41586-019-0915-y. [DOI] [PubMed] [Google Scholar]
- 77.Aramini B, Masciale V, Arienti C, Dominici M, Stella F, Martinelli G, et al. Cancer stem cells (CSCs), circulating tumor cells (CTCs) and their interplay with cancer associated fibroblasts (CAFs): a new world of targets and treatments. Cancers (Basel) 2022;14:2408. doi: 10.3390/cancers14102408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ren Z, Chen Y, Shi L, Shao F, Sun Y, Ge J, et al. Sox9/CXCL5 axis facilitates tumour cell growth and invasion in hepatocellular carcinoma. FEBS J. 2022;289:3535–49. doi: 10.1111/febs.16357. [DOI] [PubMed] [Google Scholar]
- 79.Hwang WL, Lan HY, Cheng WC, Huang SC, Yang MH. Tumor stem-like cell-derived exosomal RNAs prime neutrophils for facilitating tumorigenesis of colon cancer. J Hematol Oncol. 2019;12:10. doi: 10.1186/s13045-019-0699-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cui TX, Kryczek I, Zhao L, Zhao E, Kuick R, Roh MH, et al. Myeloid-derived suppressor cells enhance stemness of cancer cells by inducing microRNA101 and suppressing the corepressor CtBP2. Immunity. 2013;39:611–21. doi: 10.1016/j.immuni.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shidal C, Singh NP, Nagarkatti P, Nagarkatti M. MicroRNA-92 expression in CD133(+) melanoma stem cells regulates immunosuppression in the tumor microenvironment via integrin-dependent activation of TGFbeta. Cancer Res. 2019;79:3622–35. doi: 10.1158/0008-5472.CAN-18-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu C, Qiang J, Deng Q, Xia J, Deng L, Zhou L, et al. ALDH1A1 activity in tumor-initiating cells remodels myeloid-derived suppressor cells to promote breast cancer progression. Cancer Res. 2021;81:5919–34. doi: 10.1158/0008-5472.CAN-21-1337. [DOI] [PubMed] [Google Scholar]
- 83.Lee CG, Das B, Lin TL, Grimes C, Zhang X, Lavezzi T, et al. A rare fraction of drug-resistant follicular lymphoma cancer stem cells interacts with follicular dendritic cells to maintain tumourigenic potential. Br J Haematol. 2012;158:79–90. doi: 10.1111/j.1365-2141.2012.09123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nelson A, Lukacs JD, Johnston B. The current landscape of NKT cell immunotherapy and the hills ahead. Cancers (Basel) 2021;13:5174. doi: 10.3390/cancers13205174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shen M, Sun Q, Wang J, Pan W, Ren X. Positive and negative functions of B lymphocytes in tumors. Oncotarget. 2016;7:55828–39. doi: 10.18632/oncotarget.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liao CP, Chen LY, Luethy A, Kim Y, Kani K, MacLeod AR, et al. Androgen receptor in cancer-associated fibroblasts influences stemness in cancer cells. Endocr Relat Cancer. 2017;24:157–70. doi: 10.1530/ERC-16-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Burke CA. A statistical view of clinical trials in chronic hepatitis B. J Hepatol. 1986;(3 Suppl 2):S261–7. doi: 10.1016/s0168-8278(86)80130-1. [DOI] [PubMed] [Google Scholar]
- 88.Hasegawa T, Yashiro M, Nishii T, Matsuoka J, Fuyuhiro Y, Morisaki T, et al. Cancer-associated fibroblasts might sustain the stemness of scirrhous gastric cancer cells via transforming growth factor-β signaling. Int J Cancer. 2014;134:1785–95. doi: 10.1002/ijc.28520. [DOI] [PubMed] [Google Scholar]
- 89.Lau EY, Lo J, Cheng BY, Ma MK, Lee JM, Ng JK, et al. Cancer-associated fibroblasts regulate tumor-initiating cell plasticity in hepatocellular carcinoma through c-Met/FRA1/HEY1 signaling. Cell Rep. 2016;15:1175–89. doi: 10.1016/j.celrep.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 90.Todaro M, Gaggianesi M, Catalano V, Benfante A, Iovino F, Biffoni M, et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell. 2014;14:342–56. doi: 10.1016/j.stem.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 91.Tsuyada A, Chow A, Wu J, Somlo G, Chu P, Loera S, et al. CCL2 mediates cross-talk between cancer cells and stromal fibroblasts that regulates breast cancer stem cells. Cancer Res. 2012;72:2768–79. doi: 10.1158/0008-5472.CAN-11-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang M, Li Y, Zhang H, Nan F. Breast cancer stromal fibroblasts promote the generation of CD44+CD24- cells through SDF-1/CXCR4 interaction. J Exp Clin Cancer Res. 2010;29:80. doi: 10.1186/1756-9966-29-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wolfson B, Eades G, Zhou Q. Adipocyte activation of cancer stem cell signaling in breast cancer. World J Biol Chem. 2015;6:39–47. doi: 10.4331/wjbc.v6.i2.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li H, Feng Z, He ML. Lipid metabolism alteration contributes to and maintains the properties of cancer stem cells. Theranostics. 2020;10:7053–69. doi: 10.7150/thno.41388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mao J, Hu X, Xiao Y, Yang C, Ding Y, Hou N, et al. Overnutrition stimulates intestinal epithelium proliferation through β-catenin signaling in obese mice. Diabetes. 2013;62:3736–46. doi: 10.2337/db13-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen D, Liu S, Ma H, Liang X, Ma H, Yan X, et al. Paracrine factors from adipose-mesenchymal stem cells enhance metastatic capacity through Wnt signaling pathway in a colon cancer cell co-culture model. Cancer Cell Int. 2015;15:42. doi: 10.1186/s12935-015-0198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu Z, Shi A, Song D, Han B, Zhang Z, Ma L, et al. Resistin confers resistance to doxorubicin-induced apoptosis in human breast cancer cells through autophagy induction. Am J Cancer Res. 2017;7:574–83. [PMC free article] [PubMed] [Google Scholar]
- 98.Montrose DC, Galluzzi L. Drugging cancer metabolism: expectations vs. reality. Int Rev Cell Mol Biol. 2019;347:1–26. doi: 10.1016/bs.ircmb.2019.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cipolleschi MG, Marzi I, Santini R, Fredducci D, Vinci MC, D’Amico M, et al. Hypoxia-resistant profile implies vulnerability of cancer stem cells to physiological agents, which suggests new therapeutic targets. Cell Cycle. 2014;13:268–78. doi: 10.4161/cc.27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Aguilar E, Marin de Mas I, Zodda E, Marin S, Morrish F, Selivanov V, et al. metabolic reprogramming and dependencies associated with epithelial cancer stem cells independent of the epithelial-mesenchymal transition program. Stem Cells. 2016;34:1163–76. doi: 10.1002/stem.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen EI, Hewel J, Krueger JS, Tiraby C, Weber MR, Kralli A, et al. Adaptation of energy metabolism in breast cancer brain metastases. Cancer Res. 2007;67:1472–86. doi: 10.1158/0008-5472.CAN-06-3137. [DOI] [PubMed] [Google Scholar]
- 102.Sosa MS, Bragado P, Aguirre-Ghiso JA. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat Rev Cancer. 2014;14:611–22. doi: 10.1038/nrc3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Emmink BL, Verheem A, Van Houdt WJ, Steller EJ, Govaert KM, Pham TV, et al. The secretome of colon cancer stem cells contains drug-metabolizing enzymes. J Proteomics. 2013;91:84–96. doi: 10.1016/j.jprot.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 104.Hammoudi N, Ahmed KB, Garcia-Prieto C, Huang P. Metabolic alterations in cancer cells and therapeutic implications. Chin J Cancer. 2011;30:508–25. doi: 10.5732/cjc.011.10267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Peiris-Pagès M, Martinez-Outschoorn UE, Pestell RG, Sotgia F, Lisanti MP. Cancer stem cell metabolism. Breast Cancer Res. 2016;18:55. doi: 10.1186/s13058-016-0712-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Flavahan WA, Wu Q, Hitomi M, Rahim N, Kim Y, Sloan AE, et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat Neurosci. 2013;16:1373–82. doi: 10.1038/nn.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ciavardelli D, Rossi C, Barcaroli D, Volpe S, Consalvo A, Zucchelli M, et al. Breast cancer stem cells rely on fermentative glycolysis and are sensitive to 2-deoxyglucose treatment. Cell Death Dis. 2014;5:e1336. doi: 10.1038/cddis.2014.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liao J, Qian F, Tchabo N, Mhawech-Fauceglia P, Beck A, Qian Z, et al. Ovarian cancer spheroid cells with stem cell-like properties contribute to tumor generation, metastasis and chemotherapy resistance through hypoxia-resistant metabolism. PLoS One. 2014;9:e84941. doi: 10.1371/journal.pone.0084941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Palorini R, Votta G, Balestrieri C, Monestiroli A, Olivieri S, Vento R, et al. Energy metabolism characterization of a novel cancer stem cell-like line 3AB-OS. J Cell Biochem. 2014;115:368–79. doi: 10.1002/jcb.24671. [DOI] [PubMed] [Google Scholar]
- 110.Zhou Y, Zhou Y, Shingu T, Feng L, Chen Z, Ogasawara M, et al. Metabolic alterations in highly tumorigenic glioblastoma cells: preference for hypoxia and high dependency on glycolysis. J Biol Chem. 2011;286:32843–53. doi: 10.1074/jbc.M111.260935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shen YA, Wang CY, Hsieh YT, Chen YJ, Wei YH. Metabolic reprogramming orchestrates cancer stem cell properties in nasopharyngeal carcinoma. Cell Cycle. 2015;14:86–98. doi: 10.4161/15384101.2014.974419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen CL, Uthaya Kumar DB, Punj V, Xu J, Sher L, Tahara SM, et al. NANOG Metabolically reprograms tumor-initiating stem-like cells through tumorigenic changes in oxidative phosphorylation and fatty acid metabolism. Cell Metab. 2016;23:206–19. doi: 10.1016/j.cmet.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Feng W, Gentles A, Nair RV, Huang M, Lin Y, Lee CY, et al. Targeting unique metabolic properties of breast tumor initiating cells. Stem Cells. 2014;32:1734–45. doi: 10.1002/stem.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen Z, He Q, Lu T, Wu J, Shi G, He L, et al. mcPGK1-dependent mitochondrial import of PGK1 promotes metabolic reprogramming and self-renewal of liver TICs. Nat Commun. 2023;14:1121. doi: 10.1038/s41467-023-36651-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gu Y, Ji F, Liu N, Zhao Y, Wei X, Hu S, et al. Loss of miR-192-5p initiates a hyperglycolysis and stemness positive feedback in hepatocellular carcinoma. J Exp Clin Cancer Res. 2020;39:268. doi: 10.1186/s13046-020-01785-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee HJ, Li CF, Ruan D, He J, Montal ED, Lorenz S, et al. Non-proteolytic ubiquitination of Hexokinase 2 by HectH9 controls tumor metabolism and cancer stem cell expansion. Nat Commun. 2019;10:2625. doi: 10.1038/s41467-019-10374-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.De Luca A, Fiorillo M, Peiris-Pagès M, Ozsvari B, Smith DL, Sanchez-Alvarez R, et al. Mitochondrial biogenesis is required for the anchorage-independent survival and propagation of stem-like cancer cells. Oncotarget. 2015;6:14777–95. doi: 10.18632/oncotarget.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Janiszewska M, Suvà ML, Riggi N, Houtkooper RH, Auwerx J, Clément-Schatlo V, et al. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev. 2012;26:1926–44. doi: 10.1101/gad.188292.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013;12:329–41. doi: 10.1016/j.stem.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lamb R, Bonuccelli G, Ozsvári B, Peiris-Pagès M, Fiorillo M, Smith DL, et al. Mitochondrial mass, a new metabolic biomarker for stem-like cancer cells: understanding WNT/FGF-driven anabolic signaling. Oncotarget. 2015;6:30453–71. doi: 10.18632/oncotarget.5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pastò A, Bellio C, Pilotto G, Ciminale V, Silic-Benussi M, Guzzo G, et al. Cancer stem cells from epithelial ovarian cancer patients privilege oxidative phosphorylation, and resist glucose deprivation. Oncotarget. 2014;5:4305–19. doi: 10.18632/oncotarget.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sancho P, Burgos-Ramos E, Tavera A, Bou Kheir T, Jagust P, Schoenhals M, et al. MYC/PGC-1α balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell Metab. 2015;22:590–605. doi: 10.1016/j.cmet.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 123.Vlashi E, Lagadec C, Vergnes L, Reue K, Frohnen P, Chan M, et al. Metabolic differences in breast cancer stem cells and differentiated progeny. Breast Cancer Res Treat. 2014;146:525–34. doi: 10.1007/s10549-014-3051-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lonardo E, Cioffi M, Sancho P, Sanchez-Ripoll Y, Trabulo SM, Dorado J, et al. Metformin targets the metabolic achilles heel of human pancreatic cancer stem cells. PLoS One. 2013;8:e76518. doi: 10.1371/journal.pone.0076518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Farnie G, Sotgia F, Lisanti MP. High mitochondrial mass identifies a sub-population of stem-like cancer cells that are chemo-resistant. Oncotarget. 2015;6:30472–86. doi: 10.18632/oncotarget.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ye XQ, Li Q, Wang GH, Sun FF, Huang GJ, Bian XW, et al. Mitochondrial and energy metabolism-related properties as novel indicators of lung cancer stem cells. Int J Cancer. 2011;129:820–31. doi: 10.1002/ijc.25944. [DOI] [PubMed] [Google Scholar]
- 127.Anderson AS, Roberts PC, Frisard MI, Hulver MW, Schmelz EM. Ovarian tumor-initiating cells display a flexible metabolism. Exp Cell Res. 2014;328:44–57. doi: 10.1016/j.yexcr.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sánchez N, Marchesini M, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–32. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ginestier C, Monville F, Wicinski J, Cabaud O, Cervera N, Josselin E, et al. Mevalonate metabolism regulates Basal breast cancer stem cells and is a potential therapeutic target. Stem Cells. 2012;30:1327–37. doi: 10.1002/stem.1122. [DOI] [PubMed] [Google Scholar]
- 130.Kim WY. Therapeutic targeting of lipid synthesis metabolism for selective elimination of cancer stem cells. Arch Pharm Res. 2019;42:25–39. doi: 10.1007/s12272-018-1098-z. [DOI] [PubMed] [Google Scholar]
- 131.Dar S, Chhina J, Mert I, Chitale D, Buekers T, Kaur H, et al. Bioenergetic adaptations in chemoresistant ovarian cancer cells. Sci Rep. 2017;7:8760. doi: 10.1038/s41598-017-09206-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pascual G, Avgustinova A, Mejetta S, Martín M, Castellanos A, Attolini CS, et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature. 2017;541:41–5. doi: 10.1038/nature20791. [DOI] [PubMed] [Google Scholar]
- 133.Tiong TY, Weng PW, Wang CH, Setiawan SA, Yadav VK, Pikatan NW, et al. Targeting the SREBP-1/Hsa-Mir-497/SCAP/FASN oncometabolic axis inhibits the cancer stem-like and chemoresistant phenotype of non-small cell lung carcinoma cells. Int J Mol Sci. 2022;23:7283. doi: 10.3390/ijms23137283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zheng J, Conrad M. The metabolic underpinnings of ferroptosis. Cell Metab. 2020;32:920–37. doi: 10.1016/j.cmet.2020.10.011. [DOI] [PubMed] [Google Scholar]
- 135.Chen X, Kang R, Kroemer G, Tang D. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol. 2021;18:280–96. doi: 10.1038/s41571-020-00462-0. [DOI] [PubMed] [Google Scholar]
- 136.Taylor WR, Fedorka SR, Gad I, Shah R, Alqahtani HD, Koranne R, et al. Small-molecule ferroptotic agents with potential to selectively target cancer stem cells. Sci Rep. 2019;9:5926. doi: 10.1038/s41598-019-42251-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Turcu AL, Versini A, Khene N, Gaillet C, Caneque T, Muller S, et al. DMT1 inhibitors kill cancer stem cells by blocking lysosomal iron translocation. Chemistry. 2020;26:7369–73. doi: 10.1002/chem.202000159. [DOI] [PubMed] [Google Scholar]
- 138.Basuli D, Tesfay L, Deng Z, Paul B, Yamamoto Y, Ning G, et al. Iron addiction: a novel therapeutic target in ovarian cancer. Oncogene. 2017;36:4089–99. doi: 10.1038/onc.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mai TT, Hamai A, Hienzsch A, Caneque T, Muller S, Wicinski J, et al. Salinomycin kills cancer stem cells by sequestering iron in lysosomes. Nat Chem. 2017;9:1025–33. doi: 10.1038/nchem.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Han M, Xu R, Wang S, Yang N, Ni S, Zhang Q, et al. Six-transmembrane epithelial antigen of prostate 3 predicts poor prognosis and promotes glioblastoma growth and invasion. Neoplasia. 2018;20:543–54. doi: 10.1016/j.neo.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Song Q, Peng S, Sun Z, Heng X, Zhu X. Temozolomide drives ferroptosis via a DMT1-dependent pathway in glioblastoma cells. Yonsei Med J. 2021;62:843–9. doi: 10.3349/ymj.2021.62.9.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lei G, Zhuang L, Gan B. Targeting ferroptosis as a vulnerability in cancer. Nat Rev Cancer. 2022;22:381–96. doi: 10.1038/s41568-022-00459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhao B, Yu T, Wang S, Che J, Zhou L, Shang P. Static magnetic field (0.2-0.4 T) stimulates the self-renewal ability of osteosarcoma stem cells through autophagic degradation of ferritin. Bioelectromagnetics. 2021;42:371–83. doi: 10.1002/bem.22352. [DOI] [PubMed] [Google Scholar]
- 144.Liu CC, Li HH, Lin JH, Chiang MC, Hsu TW, Li AF, et al. Esophageal cancer stem-like cells resist ferroptosis-induced cell death by active Hsp27-GPX4 pathway. Biomolecules. 2021;12:48. doi: 10.3390/biom12010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chirillo R, Aversa I, Di Vito A, Salatino A, Battaglia AM, Sacco A, et al. FtH-mediated ROS dysregulation promotes CXCL12/CXCR4 axis activation and EMT-like trans-differentiation in erythroleukemia K562 cells. Front Oncol. 2020;10:698. doi: 10.3389/fonc.2020.00698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang KH, Tian HY, Gao X, Lei WW, Hu Y, Wang DM, et al. Ferritin heavy chain-mediated iron homeostasis and subsequent increased reactive oxygen species production are essential for epithelial-mesenchymal transition. Cancer Res. 2009;69:5340–8. doi: 10.1158/0008-5472.CAN-09-0112. [DOI] [PubMed] [Google Scholar]
- 147.Aversa I, Zolea F, Ierano C, Bulotta S, Trotta AM, Faniello MC, et al. Epithelial-to-mesenchymal transition in FHC-silenced cells: the role of CXCR4/CXCL12 axis. J Exp Clin Cancer Res. 2017;36:104. doi: 10.1186/s13046-017-0571-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lobello N, Biamonte F, Pisanu ME, Faniello MC, Jakopin Z, Chiarella E, et al. Ferritin heavy chain is a negative regulator of ovarian cancer stem cell expansion and epithelial to mesenchymal transition. Oncotarget. 2016;7:62019–33. doi: 10.18632/oncotarget.11495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang H, Wang M, He Y, Deng T, Liu R, Wang W, et al. Chemotoxicity-induced exosomal lncFERO regulates ferroptosis and stemness in gastric cancer stem cells. Cell Death Dis. 2021;12:1116. doi: 10.1038/s41419-021-04406-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Li D, Fu Z, Chen R, Zhao X, Zhou Y, Zeng B, et al. Inhibition of glutamine metabolism counteracts pancreatic cancer stem cell features and sensitizes cells to radiotherapy. Oncotarget. 2015;6:31151–63. doi: 10.18632/oncotarget.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wu Z, Wei D, Gao W, Xu Y, Hu Z, Ma Z, et al. TPO-induced metabolic reprogramming drives liver metastasis of colorectal cancer CD110+ tumor-initiating cells. Cell Stem Cell. 2015;17:47–59. doi: 10.1016/j.stem.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 152.Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15:504–14. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- 153.Tirino V, Camerlingo R, Franco R, Malanga D, La Rocca A, Viglietto G, et al. The role of CD133 in the identification and characterisation of tumour-initiating cells in non-small-cell lung cancer. Eur J Cardiothorac Surg. 2009;36:446–53. doi: 10.1016/j.ejcts.2009.03.063. [DOI] [PubMed] [Google Scholar]
- 154.Bertolini G, Roz L, Perego P, Tortoreto M, Fontanella E, Gatti L, et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci U S A. 2009;106:16281–6. doi: 10.1073/pnas.0905653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–10. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 156.Vermeulen L, Todaro M, de Sousa Mello F, Sprick MR, Kemper K, Perez Alea M, et al. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci U S A. 2008;105:13427–32. doi: 10.1073/pnas.0805706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–51. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 158.Kryczek I, Liu S, Roh M, Vatan L, Szeliga W, Wei S, et al. Expression of aldehyde dehydrogenase and CD133 defines ovarian cancer stem cells. Int J Cancer. 2012;130:29–39. doi: 10.1002/ijc.25967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Grasso C, Anaka M, Hofmann O, Sompallae R, Broadley K, Hide W, et al. Iterative sorting reveals CD133+ and CD133- melanoma cells as phenotypically distinct populations. BMC Cancer. 2016;16:726. doi: 10.1186/s12885-016-2759-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tirino V, Desiderio V, Paino F, De Rosa A, Papaccio F, Fazioli F, et al. Human primary bone sarcomas contain CD133+ cancer stem cells displaying high tumorigenicity in vivo. FASEB J. 2011;25:2022–30. doi: 10.1096/fj.10-179036. [DOI] [PubMed] [Google Scholar]
- 161.Fujiwara T, Katsuda T, Hagiwara K, Kosaka N, Yoshioka Y, Takahashi RU, et al. Clinical relevance and therapeutic significance of microRNA-133a expression profiles and functions in malignant osteosarcoma-initiating cells. Stem Cells. 2014;32:959–73. doi: 10.1002/stem.1618. [DOI] [PubMed] [Google Scholar]
- 162.Cox CV, Diamanti P, Evely RS, Kearns PR, Blair A. Expression of CD133 on leukemia-initiating cells in childhood ALL. Blood. 2009;113:3287–96. doi: 10.1182/blood-2008-04-154187. [DOI] [PubMed] [Google Scholar]
- 163.Suetsugu A, Nagaki M, Aoki H, Motohashi T, Kunisada T, Moriwaki H. Characterization of CD133+ hepatocellular carcinoma cells as cancer stem/progenitor cells. Biochem Biophys Res Commun. 2006;351:820–4. doi: 10.1016/j.bbrc.2006.10.128. [DOI] [PubMed] [Google Scholar]
- 164.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–23. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 165.Ma Z, Zhang C, Liu X, Fang F, Liu S, Liao X, et al. Characterisation of a subpopulation of CD133(+) cancer stem cells from Chinese patients with oral squamous cell carcinoma. Sci Rep. 2020;10:8875. doi: 10.1038/s41598-020-64947-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104:10158–63. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Du L, Wang H, He L, Zhang J, Ni B, Wang X, et al. CD44 is of functional importance for colorectal cancer stem cells. Clin Cancer Res. 2008;14:6751–60. doi: 10.1158/1078-0432.CCR-08-1034. [DOI] [PubMed] [Google Scholar]
- 168.Abbasian M, Mousavi E, Arab-Bafrani Z, Sahebkar A. The most reliable surface marker for the identification of colorectal cancer stem-like cells: a systematic review and meta-analysis. J Cell Physiol. 2019;234:8192–202. doi: 10.1002/jcp.27619. [DOI] [PubMed] [Google Scholar]
- 169.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–7. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 170.Zhang S, Balch C, Chan MW, Lai HC, Matei D, Schilder JM, et al. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008;68:4311–20. doi: 10.1158/0008-5472.CAN-08-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Takaishi S, Okumura T, Tu S, Wang SS, Shibata W, Vigneshwaran R, et al. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells. 2009;27:1006–20. doi: 10.1002/stem.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chen W, Zhang X, Chu C, Cheung WL, Ng L, Lam S, et al. Identification of CD44+ cancer stem cells in human gastric cancer. Hepatogastroenterology. 2013;60:949–54. doi: 10.5754/hge12881. [DOI] [PubMed] [Google Scholar]
- 173.Patrawala L, Calhoun T, Schneider-Broussard R, Li H, Bhatia B, Tang S, et al. Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene. 2006;25:1696–708. doi: 10.1038/sj.onc.1209327. [DOI] [PubMed] [Google Scholar]
- 174.Leung EL, Fiscus RR, Tung JW, Tin VP, Cheng LC, Sihoe AD, et al. Non-small cell lung cancer cells expressing CD44 are enriched for stem cell-like properties. PLoS One. 2010;5:e14062. doi: 10.1371/journal.pone.0014062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Patil S, Al-Brakati A, Abidi NH, Almasri MA, Almeslet AS, Patil VR, et al. CD44-positive cancer stem cells from oral squamous cell carcinoma exhibit reduced proliferation and stemness gene expression upon adipogenic induction. Med Oncol. 2022;39:23. doi: 10.1007/s12032-021-01617-4. [DOI] [PubMed] [Google Scholar]
- 176.Hoe SLL, Tan LP, Abdul Aziz N, Liew K, Teow SY, Abdul Razak FR, et al. CD24, CD44 and EpCAM enrich for tumour-initiating cells in a newly established patient-derived xenograft of nasopharyngeal carcinoma. Sci Rep. 2017;7:12372. doi: 10.1038/s41598-017-12045-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Haraguchi N, Ohkuma M, Sakashita H, Matsuzaki S, Tanaka F, Mimori K, et al. CD133+CD44+ population efficiently enriches colon cancer initiating cells. Ann Surg Oncol. 2008;15:2927–33. doi: 10.1245/s10434-008-0074-0. [DOI] [PubMed] [Google Scholar]
- 178.Zhu Z, Hao X, Yan M, Yao M, Ge C, Gu J, et al. Cancer stem/progenitor cells are highly enriched in CD133+CD44+ population in hepatocellular carcinoma. Int J Cancer. 2010;126:2067–78. doi: 10.1002/ijc.24868. [DOI] [PubMed] [Google Scholar]
- 179.Cho RW, Wang X, Diehn M, Shedden K, Chen GY, Sherlock G, et al. Isolation and molecular characterization of cancer stem cells in MMTV-Wnt-1 murine breast tumors. Stem Cells. 2008;26:364–71. doi: 10.1634/stemcells.2007-0440. [DOI] [PubMed] [Google Scholar]
- 180.Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153–66. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 181.Jiang J, Zhang Y, Chuai S, Wang Z, Zheng D, Xu F, et al. Trastuzumab (herceptin) targets gastric cancer stem cells characterized by CD90 phenotype. Oncogene. 2012;31:671–82. doi: 10.1038/onc.2011.282. [DOI] [PubMed] [Google Scholar]
- 182.Tang KH, Dai YD, Tong M, Chan YP, Kwan PS, Fu L, et al. A CD90(+) tumor-initiating cell population with an aggressive signature and metastatic capacity in esophageal cancer. Cancer Res. 2013;73:2322–32. doi: 10.1158/0008-5472.CAN-12-2991. [DOI] [PubMed] [Google Scholar]
- 183.Yan X, Luo H, Zhou X, Zhu B, Wang Y, Bian X. Identification of CD90 as a marker for lung cancer stem cells in A549 and H446 cell lines. Oncol Rep. 2013;30:2733–40. doi: 10.3892/or.2013.2784. [DOI] [PubMed] [Google Scholar]
- 184.Shi J, Lu P, Shen W, He R, Yang MW, Fang Y, et al. CD90 highly expressed population harbors a stemness signature and creates an immunosuppressive niche in pancreatic cancer. Cancer Lett. 2019;453:158–69. doi: 10.1016/j.canlet.2019.03.051. [DOI] [PubMed] [Google Scholar]
- 185.He J, Liu Y, Zhu T, Zhu J, Dimeco F, Vescovi AL, et al. CD90 is identified as a candidate marker for cancer stem cells in primary high-grade gliomas using tissue microarrays. Mol Cell Proteomics. 2012;11:M111.010744. doi: 10.1074/mcp.M111.010744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Parry PV, Engh JA. CD90 is identified as a marker for cancer stem cells in high-grade gliomas using tissue microarrays. Neurosurgery. 2012;70:N23–4. doi: 10.1227/01.neu.0000413227.80467.92. [DOI] [PubMed] [Google Scholar]
- 187.Buishand FO, Arkesteijn GJ, Feenstra LR, Oorsprong CW, Mestemaker M, Starke A, et al. Identification of CD90 as putative cancer stem cell marker and therapeutic target in insulinomas. Stem Cells Dev. 2016;25:826–35. doi: 10.1089/scd.2016.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yamashita T, Honda M, Nakamoto Y, Baba M, Nio K, Hara Y, et al. Discrete nature of EpCAM+ and CD90+ cancer stem cells in human hepatocellular carcinoma. Hepatology. 2013;57:1484–97. doi: 10.1002/hep.26168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Mal A, Bukhari AB, Singh RK, Kapoor A, Barai A, Deshpande I, et al. EpCAM-mediated cellular plasticity promotes radiation resistance and metastasis in breast cancer. Front Cell Dev Biol. 2020;8:597673. doi: 10.3389/fcell.2020.597673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Tsui YM, Chan LK, Ng IO. Cancer stemness in hepatocellular carcinoma: mechanisms and translational potential. Br J Cancer. 2020;122:1428–40. doi: 10.1038/s41416-020-0823-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Fong D, Steurer M, Obrist P, Barbieri V, Margreiter R, Amberger A, et al. Ep-CAM expression in pancreatic and ampullary carcinomas: frequency and prognostic relevance. J Clin Pathol. 2008;61:31–5. doi: 10.1136/jcp.2006.037333. [DOI] [PubMed] [Google Scholar]
- 193.Hu F, Sun X, Li G, Wu Q, Chen Y, Yang X, et al. Inhibition of SIRT2 limits tumour angiogenesis via inactivation of the STAT3/VEGFA signalling pathway. Cell Death Dis. 2018;10:9. doi: 10.1038/s41419-018-1260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–82. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 195.Wang B, Chen Q, Cao Y, Ma X, Yin C, Jia Y, et al. LGR5 is a gastric cancer stem cell marker associated with stemness and the EMT signature genes NANOG, NANOGP8, PRRX1, TWIST1, and BMI1. PLoS One. 2016;11:e0168904. doi: 10.1371/journal.pone.0168904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Jiang X, Hao HX, Growney JD, Woolfenden S, Bottiglio C, Ng N, et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc Natl Acad Sci U S A. 2013;110:12649–54. doi: 10.1073/pnas.1307218110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Mizuno N, Yatabe Y, Hara K, Hijioka S, Imaoka H, Shimizu Y, et al. Cytoplasmic expression of LGR5 in pancreatic adenocarcinoma. Front Physiol. 2013;4:269. doi: 10.3389/fphys.2013.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Cao W, Li M, Liu J, Zhang S, Noordam L, Verstegen MMA, et al. LGR5 marks targetable tumor-initiating cells in mouse liver cancer. Nat Commun. 2020;11:1961. doi: 10.1038/s41467-020-15846-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Akbari S, Kunter I, Azbazdar Y, Ozhan G, Atabey N, Firtina Karagonlar Z, et al. LGR5/R-Spo1/Wnt3a axis promotes stemness and aggressive phenotype in hepatoblast-like hepatocellular carcinoma cell lines. Cell Signal. 2021;82:109972. doi: 10.1016/j.cellsig.2021.109972. [DOI] [PubMed] [Google Scholar]
- 200.Kemper K, Prasetyanti PR, De Lau W, Rodermond H, Clevers H, Medema JP. Monoclonal antibodies against Lgr5 identify human colorectal cancer stem cells. Stem Cells. 2012;30:2378–86. doi: 10.1002/stem.1233. [DOI] [PubMed] [Google Scholar]
- 201.Hirsch D, Barker N, McNeil N, Hu Y, Camps J, McKinnon K, et al. LGR5 positivity defines stem-like cells in colorectal cancer. Carcinogenesis. 2014;35:849–58. doi: 10.1093/carcin/bgt377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Schindler AJ, Watanabe A, Howell SB. LGR5 and LGR6 in stem cell biology and ovarian cancer. Oncotarget. 2018;9:1346–55. doi: 10.18632/oncotarget.20178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Cao HZ, Liu XF, Yang WT, Chen Q, Zheng PS. LGR5 promotes cancer stem cell traits and chemoresistance in cervical cancer. Cell Death Dis. 2017;8:e3039. doi: 10.1038/cddis.2017.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Yang L, Tang H, Kong Y, Xie X, Chen J, Song C, et al. LGR5 promotes breast cancer progression and maintains stem-like cells through activation of Wnt/β-catenin signaling. Stem Cells. 2015;33:2913–24. doi: 10.1002/stem.2083. [DOI] [PubMed] [Google Scholar]
- 205.Du Z, Jia D, Liu S, Wang F, Li G, Zhang Y, et al. Oct4 is expressed in human gliomas and promotes colony formation in glioma cells. Glia. 2009;57:724–33. doi: 10.1002/glia.20800. [DOI] [PubMed] [Google Scholar]
- 206.Gzil A, Zarębska I, Bursiewicz W, Antosik P, Grzanka D, Szylberg Ł. Markers of pancreatic cancer stem cells and their clinical and therapeutic implications. Mol Biol Rep. 2019;46:6629–45. doi: 10.1007/s11033-019-05058-1. [DOI] [PubMed] [Google Scholar]
- 207.Wang G, Zhou H, Gu Z, Gao Q, Shen G. Oct4 promotes cancer cell proliferation and migration and leads to poor prognosis associated with the survivin/STAT3 pathway in hepatocellular carcinoma. Oncol Rep. 2018;40:979–87. doi: 10.3892/or.2018.6491. [DOI] [PubMed] [Google Scholar]
- 208.Wu G, Wilson G, Zhou G, Hebbard L, George J, Qiao L. Oct4 is a reliable marker of liver tumor propagating cells in hepatocellular carcinoma. Discov Med. 2015;20:219–29. [PubMed] [Google Scholar]
- 209.Vaddi PK, Stamnes MA, Cao H, Chen S. Elimination of SOX2/OCT4-associated prostate cancer stem cells blocks tumor development and enhances therapeutic response. Cancers (Basel) 2019;11:1331. doi: 10.3390/cancers11091331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Miyazawa K, Tanaka T, Nakai D, Morita N, Suzuki K. Immunohistochemical expression of four different stem cell markers in prostate cancer: high expression of NANOG in conjunction with hypoxia-inducible factor-1α expression is involved in prostate epithelial malignancy. Oncol Lett. 2014;8:985–92. doi: 10.3892/ol.2014.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Hatefi N, Nouraee N, Parvin M, Ziaee SA, Mowla SJ. Evaluating the expression of oct4 as a prognostic tumor marker in bladder cancer. Iran J Basic Med Sci. 2012;15:1154–61. [PMC free article] [PubMed] [Google Scholar]
- 212.Ruan Z, Yang X, Cheng W. OCT4 accelerates tumorigenesis through activating JAK/STAT signaling in ovarian cancer side population cells. Cancer Manag Res. 2019;11:389–99. doi: 10.2147/CMAR.S180418. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 213.Lu CS, Shiau AL, Su BH, Hsu TS, Wang CT, Su YC, et al. Oct4 promotes M2 macrophage polarization through upregulation of macrophage colony-stimulating factor in lung cancer. J Hematol Oncol. 2020;13:62. doi: 10.1186/s13045-020-00887-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lee SH, Oh SY, Do SI, Lee HJ, Kang HJ, Rho YS, et al. SOX2 regulates self-renewal and tumorigenicity of stem-like cells of head and neck squamous cell carcinoma. Br J Cancer. 2014;111:2122–30. doi: 10.1038/bjc.2014.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Cox JL, Wilder PJ, Desler M, Rizzino A. Elevating SOX2 levels deleteriously affects the growth of medulloblastoma and glioblastoma cells. PLoS One. 2012;7:e44087. doi: 10.1371/journal.pone.0044087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Jeon HM, Sohn YW, Oh SY, Kim SH, Beck S, Kim S, et al. ID4 imparts chemoresistance and cancer stemness to glioma cells by derepressing miR-9*-mediated suppression of SOX2. Cancer Res. 2011;71:3410–21. doi: 10.1158/0008-5472.CAN-10-3340. [DOI] [PubMed] [Google Scholar]
- 217.Lengerke C, Fehm T, Kurth R, Neubauer H, Scheble V, Müller F, et al. Expression of the embryonic stem cell marker SOX2 in early-stage breast carcinoma. BMC Cancer. 2011;11:42. doi: 10.1186/1471-2407-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Tian T, Zhang Y, Wang S, Zhou J, Xu S. Sox2 enhances the tumorigenicity and chemoresistance of cancer stem-like cells derived from gastric cancer. J Biomed Res. 2012;26:336–45. doi: 10.7555/JBR.26.20120045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Fang X, Yu W, Li L, Shao J, Zhao N, Chen Q, et al. ChIP-seq and functional analysis of the SOX2 gene in colorectal cancers. Omics. 2010;14:369–84. doi: 10.1089/omi.2010.0053. [DOI] [PubMed] [Google Scholar]
- 220.Hussenet T, Dali S, Exinger J, Monga B, Jost B, Dembelé D, et al. SOX2 is an oncogene activated by recurrent 3q26.3 amplifications in human lung squamous cell carcinomas. PLoS One. 2010;5:e8960. doi: 10.1371/journal.pone.0008960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Liu XF, Yang WT, Xu R, Liu JT, Zheng PS. Cervical cancer cells with positive Sox2 expression exhibit the properties of cancer stem cells. PLoS One. 2014;9:e87092. doi: 10.1371/journal.pone.0087092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Santini R, Pietrobono S, Pandolfi S, Montagnani V, D’Amico M, Penachioni JY, et al. SOX2 regulates self-renewal and tumorigenicity of human melanoma-initiating cells. Oncogene. 2014;33:4697–708. doi: 10.1038/onc.2014.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Basu-Roy U, Seo E, Ramanathapuram L, Rapp TB, Perry JA, Orkin SH, et al. Sox2 maintains self renewal of tumor-initiating cells in osteosarcomas. Oncogene. 2012;31:2270–82. doi: 10.1038/onc.2011.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Bareiss PM, Paczulla A, Wang H, Schairer R, Wiehr S, Kohlhofer U, et al. SOX2 expression associates with stem cell state in human ovarian carcinoma. Cancer Res. 2013;73:5544–55. doi: 10.1158/0008-5472.CAN-12-4177. [DOI] [PubMed] [Google Scholar]
- 225.Herreros-Villanueva M, Zhang JS, Koenig A, Abel EV, Smyrk TC, Bamlet WR, et al. SOX2 promotes dedifferentiation and imparts stem cell-like features to pancreatic cancer cells. Oncogenesis. 2013;2:e61. doi: 10.1038/oncsis.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zhu F, Qian W, Zhang H, Liang Y, Wu M, Zhang Y, et al. SOX2 is a marker for stem-like tumor cells in bladder cancer. Stem Cell Rep. 2017;9:429–37. doi: 10.1016/j.stemcr.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Boumahdi S, Driessens G, Lapouge G, Rorive S, Nassar D, Le Mercier M, et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511:246–50. doi: 10.1038/nature13305. [DOI] [PubMed] [Google Scholar]
- 228.Yasunaga J, Taniguchi Y, Nosaka K, Yoshida M, Satou Y, Sakai T, et al. Identification of aberrantly methylated genes in association with adult T-cell leukemia. Cancer Res. 2004;64:6002–9. doi: 10.1158/0008-5472.CAN-04-1422. [DOI] [PubMed] [Google Scholar]
- 229.Tang H, Zhu H, Wang X, Hua L, Li J, Xie Q, et al. KLF4 is a tumor suppressor in anaplastic meningioma stem-like cells and human meningiomas. J Mol Cell Biol. 2017;9:315–24. doi: 10.1093/jmcb/mjx023. [DOI] [PubMed] [Google Scholar]
- 230.Leng Z, Li Y, Zhou G, Lv X, Ai W, Li J, et al. Krüppel-like factor 4 regulates stemness and mesenchymal properties of colorectal cancer stem cells through the TGF-β1/Smad/snail pathway. J Cell Mol Med. 2020;24:1866–77. doi: 10.1111/jcmm.14882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Cho YG, Song JH, Kim CJ, Nam SW, Yoo NJ, Lee JY, et al. Genetic and epigenetic analysis of the KLF4 gene in gastric cancer. APMIS. 2007;115:802–8. doi: 10.1111/j.1600-0463.2007.apm_643.x. [DOI] [PubMed] [Google Scholar]
- 232.Bianchi F, Hu J, Pelosi G, Cirincione R, Ferguson M, Ratcliffe C, et al. Lung cancers detected by screening with spiral computed tomography have a malignant phenotype when analyzed by cDNA microarray. Clin Cancer Res. 2004;10:6023–8. doi: 10.1158/1078-0432.CCR-04-0619. [DOI] [PubMed] [Google Scholar]
- 233.Li Q, Gao Y, Jia Z, Mishra L, Guo K, Li Z, et al. Dysregulated Krüppel-like factor 4 and vitamin D receptor signaling contribute to progression of hepatocellular carcinoma. Gastroenterology. 2012;143:799–810.e792. doi: 10.1053/j.gastro.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Ohnishi S, Ohnami S, Laub F, Aoki K, Suzuki K, Kanai Y, et al. Downregulation and growth inhibitory effect of epithelial-type Krüppel-like transcription factor KLF4, but not KLF5, in bladder cancer. Biochem Biophys Res Commun. 2003;308:251–56. doi: 10.1016/s0006-291x(03)01356-1. [DOI] [PubMed] [Google Scholar]
- 235.Luo A, Kong J, Hu G, Liew CC, Xiong M, Wang X, et al. Discovery of Ca2+-relevant and differentiation-associated genes downregulated in esophageal squamous cell carcinoma using cDNA microarray. Oncogene. 2004;23:1291–9. doi: 10.1038/sj.onc.1207218. [DOI] [PubMed] [Google Scholar]
- 236.Zbinden M, Duquet A, Lorente-Trigos A, Ngwabyt SN, Borges I, Ruiz i Altaba A. NANOG regulates glioma stem cells and is essential in vivo acting in a cross-functional network with GLI1 and p53. EMBO J. 2010;29:2659–74. doi: 10.1038/emboj.2010.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Cao J, Li L, Chen C, Lv C, Meng F, Zeng L, et al. RNA interference-mediated silencing of NANOG leads to reduced proliferation and self-renewal, cell cycle arrest and apoptosis in T-cell acute lymphoblastic leukemia cells via the p53 signaling pathway. Leuk Res. 2013;37:1170–7. doi: 10.1016/j.leukres.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 238.Chiou SH, Wang ML, Chou YT, Chen CJ, Hong CF, Hsieh WJ, et al. Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer Res. 2010;70:10433–44. doi: 10.1158/0008-5472.CAN-10-2638. [DOI] [PubMed] [Google Scholar]
- 239.Bourguignon LY, Spevak CC, Wong G, Xia W, Gilad E. Hyaluronan-CD44 interaction with protein kinase C(epsilon) promotes oncogenic signaling by the stem cell marker Nanog and the Production of microRNA-21, leading to down-regulation of the tumor suppressor protein PDCD4, anti-apoptosis, and chemotherapy resistance in breast tumor cells. J Biol Chem. 2009;284:26533–46. doi: 10.1074/jbc.M109.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Narusaka T, Ohara T, Noma K, Nishiwaki N, Katsura Y, Kato T, et al. Nanog is a promising chemoresistant stemness marker and therapeutic target by iron chelators for esophageal cancer. Int J Cancer. 2021;149:347–57. doi: 10.1002/ijc.33544. [DOI] [PubMed] [Google Scholar]
- 241.Ma X, Wang B, Wang X, Luo Y, Fan W. NANOGP8 is the key regulator of stemness, EMT, Wnt pathway, chemoresistance, and other malignant phenotypes in gastric cancer cells. PLoS One. 2018;13:e0192436. doi: 10.1371/journal.pone.0192436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Tamura S, Isobe T, Ariyama H, Nakano M, Kikushige Y, Takaishi S, et al. E-cadherin regulates proliferation of colorectal cancer stem cells through NANOG. Oncol Rep. 2018;40:693–703. doi: 10.3892/or.2018.6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Siu MK, Wong ES, Kong DS, Chan HY, Jiang L, Wong OG, et al. Stem cell transcription factor NANOG controls cell migration and invasion via dysregulation of E-cadherin and FoxJ1 and contributes to adverse clinical outcome in ovarian cancers. Oncogene. 2013;32:3500–9. doi: 10.1038/onc.2012.363. [DOI] [PubMed] [Google Scholar]
- 244.Jeter CR, Liu B, Lu Y, Chao HP, Zhang D, Liu X, et al. NANOG reprograms prostate cancer cells to castration resistance via dynamically repressing and engaging the AR/FOXA1 signaling axis. Cell Discov. 2016;2:16041. doi: 10.1038/celldisc.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Shan J, Shen J, Liu L, Xia F, Xu C, Duan G, et al. Nanog regulates self-renewal of cancer stem cells through the insulin-like growth factor pathway in human hepatocellular carcinoma. Hepatology. 2012;56:1004–14. doi: 10.1002/hep.25745. [DOI] [PubMed] [Google Scholar]
- 246.Bourguignon LY, Wong G, Earle C, Chen L. Hyaluronan-CD44v3 interaction with Oct4-Sox2-Nanog promotes miR-302 expression leading to self-renewal, clonal formation, and cisplatin resistance in cancer stem cells from head and neck squamous cell carcinoma. J Biol Chem. 2012;287:32800–24. doi: 10.1074/jbc.M111.308528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Zhang T, Sun H, Liu R, Cao W, Zhang T, Li E, et al. Nanog mediates tobacco smoke-induced enhancement of renal cancer stem cell properties. Environ Toxicol. 2020;35:1274–83. doi: 10.1002/tox.22992. [DOI] [PubMed] [Google Scholar]
- 248.Le Grand M, Mukha A, Püschel J, Valli E, Kamili A, Vittorio O, et al. Interplay between MycN and c-Myc regulates radioresistance and cancer stem cell phenotype in neuroblastoma upon glutamine deprivation. Theranostics. 2020;10:6411–29. doi: 10.7150/thno.42602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Hongwiangchan N, Sriratanasak N, Wichadakul D, Aksorn N, Chamni S, Chanvorachote P. Hydroquinone 5-O-cinnamoyl ester of renieramycin M suppresses lung cancer stem cells by targeting Akt and destabilizes c-Myc. Pharmaceuticals (Basel) 2021;14:1112. doi: 10.3390/ph14111112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Zhu Y, Wang C, Becker SA, Hurst K, Nogueira LM, Findlay VJ, et al. miR-145 antagonizes SNAI1-mediated stemness and radiation resistance in colorectal cancer. Mol Ther. 2018;26:744–54. doi: 10.1016/j.ymthe.2017.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Choi HS, Kim SL, Kim JH, Deng HY, Yun BS, Lee DS. Triterpene acid (3-O-p-coumaroyltormentic acid) isolated from aronia extracts inhibits breast cancer stem cell formation through downregulation of c-Myc protein. Int J Mol Sci. 2018;19:2528. doi: 10.3390/ijms19092528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Condello S, Morgan CA, Nagdas S, Cao L, Turek J, Hurley TD, et al. β-catenin-regulated ALDH1A1 is a target in ovarian cancer spheroids. Oncogene. 2015;34:2297–308. doi: 10.1038/onc.2014.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Meng E, Mitra A, Tripathi K, Finan MA, Scalici J, McClellan S, et al. ALDH1A1 maintains ovarian cancer stem cell-like properties by altered regulation of cell cycle checkpoint and DNA repair network signaling. PLoS One. 2014;9:e107142. doi: 10.1371/journal.pone.0107142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Gao F, Zhou B, Xu JC, Gao X, Li SX, Zhu GC, et al. The role of LGR5 and ALDH1A1 in non-small cell lung cancer: cancer progression and prognosis. Biochem Biophys Res Commun. 2015;462:91–8. doi: 10.1016/j.bbrc.2015.04.029. [DOI] [PubMed] [Google Scholar]
- 255.Ma Y, Li M, Si J, Xiong Y, Lu F, Zhang J, et al. Blockade of Notch3 inhibits the stem-like property and is associated with ALDH1A1 and CD44 via autophagy in non-small lung cancer. Int J Oncol. 2016;48:2349–58. doi: 10.3892/ijo.2016.3464. [DOI] [PubMed] [Google Scholar]
- 256.Shao C, Sullivan JP, Girard L, Augustyn A, Yenerall P, Rodriguez-Canales J, et al. Essential role of aldehyde dehydrogenase 1A3 for the maintenance of non-small cell lung cancer stem cells is associated with the STAT3 pathway. Clin Cancer Res. 2014;20:4154–66. doi: 10.1158/1078-0432.CCR-13-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Sun M, Zhao H, Xiao Q, Yu Z, Song Z, Yao W, et al. Combined expression of aldehyde dehydrogenase 1A1 and β-catenin is associated with lymph node metastasis and poor survival in breast cancer patients following cyclophosphamide treatment. Oncol Rep. 2015;34:3163–73. doi: 10.3892/or.2015.4273. [DOI] [PubMed] [Google Scholar]
- 258.Thomas ML, de Antueno R, Coyle KM, Sultan M, Cruickshank BM, Giacomantonio MA, et al. Citral reduces breast tumor growth by inhibiting the cancer stem cell marker ALDH1A3. Mol Oncol. 2016;10:1485–96. doi: 10.1016/j.molonc.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Wang W, Li Y, Liu N, Gao Y, Li L. MiR-23b controls ALDH1A1 expression in cervical cancer stem cells. BMC Cancer. 2017;17:292. doi: 10.1186/s12885-017-3192-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Yan Z, Xu L, Zhang J, Lu Q, Luo S, Xu L. Aldehyde dehydrogenase 1A1 stabilizes transcription factor Gli2 and enhances the activity of Hedgehog signaling in hepatocellular cancer. Biochem Biophys Res Commun. 2016;471:466–73. doi: 10.1016/j.bbrc.2016.02.052. [DOI] [PubMed] [Google Scholar]
- 261.Feng H, Liu Y, Bian X, Zhou F, Liu Y. ALDH1A3 affects colon cancer in vitro proliferation and invasion depending on CXCR4 status. Br J Cancer. 2018;118:224–32. doi: 10.1038/bjc.2017.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Singh S, Arcaroli J, Chen Y, Thompson DC, Messersmith W, Jimeno A, et al. ALDH1B1 Is Crucial for Colon Tumorigenesis by Modulating Wnt/β-Catenin, Notch and PI3K/Akt Signaling Pathways. PLoS One. 2015;10:e0121648. doi: 10.1371/journal.pone.0121648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Wang X, Wang C, Zhang X, Hua R, Gan L, Huang M, et al. Bmi-1 regulates stem cell-like properties of gastric cancer cells via modulating miRNAs. J Hematol Oncol. 2016;9:90. doi: 10.1186/s13045-016-0323-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Jakob M, Sharaf K, Schirmer M, Leu M, Küffer S, Bertlich M, et al. Role of cancer stem cell markers ALDH1, BCL11B, BMI-1, and CD44 in the prognosis of advanced HNSCC. Strahlenther Onkol. 2021;197:231–45. doi: 10.1007/s00066-020-01653-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Wang J, Xing Y, Wang Y, He Y, Wang L, Peng S, et al. A novel BMI-1 inhibitor QW24 for the treatment of stem-like colorectal cancer. J Exp Clin Cancer Res. 2019;38:422. doi: 10.1186/s13046-019-1392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Islam F, Gopalan V, Wahab R, Smith RA, Lam AK. Cancer stem cells in oesophageal squamous cell carcinoma: identification, prognostic and treatment perspectives. Crit Rev Oncol Hematol. 2015;96:9–19. doi: 10.1016/j.critrevonc.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 267.Hwang CC, Nieh S, Lai CH, Tsai CS, Chang LC, Hua CC, et al. A retrospective review of the prognostic value of ALDH-1, Bmi-1 and Nanog stem cell markers in esophageal squamous cell carcinoma. PLoS One. 2014;9:e105676. doi: 10.1371/journal.pone.0105676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Liu W, Feng JQ, Shen XM, Wang HY, Liu Y, Zhou ZT. Two stem cell markers, ATP-binding cassette, G2 subfamily (ABCG2) and BMI-1, predict the transformation of oral leukoplakia to cancer: a long-term follow-up study. Cancer. 2012;118:1693–700. doi: 10.1002/cncr.26483. [DOI] [PubMed] [Google Scholar]
- 269.Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–60. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 270.Godlewski J, Nowicki MO, Bronisz A, Williams S, Otsuki A, Nuovo G, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–30. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 271.Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–7. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–8. [PubMed] [Google Scholar]
- 273.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 274.Hussein D, Punjaruk W, Storer LC, Shaw L, Othman R, Peet A, et al. Pediatric brain tumor cancer stem cells: cell cycle dynamics, DNA repair, and etoposide extrusion. Neuro Oncol. 2011;13:70–83. doi: 10.1093/neuonc/noq144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Zanini C, Ercole E, Mandili G, Salaroli R, Poli A, Renna C, et al. Medullospheres from DAOY, UW228 and ONS-76 cells: increased stem cell population and proteomic modifications. PLoS One. 2013;8:e63748. doi: 10.1371/journal.pone.0063748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Iacopino F, Angelucci C, Piacentini R, Biamonte F, Mangiola A, Maira G, et al. Isolation of cancer stem cells from three human glioblastoma cell lines: characterization of two selected clones. PLoS One. 2014;9:e105166. doi: 10.1371/journal.pone.0105166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Sana J, Zambo I, Skoda J, Neradil J, Chlapek P, Hermanova M, et al. CD133 expression and identification of CD133/nestin positive cells in rhabdomyosarcomas and rhabdomyosarcoma cell lines. Anal Cell Pathol (Amst) 2011;34:303–18. doi: 10.3233/ACP-2011-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Veselska R, Hermanova M, Loja T, Chlapek P, Zambo I, Vesely K, et al. Nestin expression in osteosarcomas and derivation of nestin/CD133 positive osteosarcoma cell lines. BMC Cancer. 2008;8:300. doi: 10.1186/1471-2407-8-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Bapat SA, Mali AM, Koppikar CB, Kurrey NK. Stem and progenitor-like cells contribute to the aggressive behavior of human epithelial ovarian cancer. Cancer Res. 2005;65:3025–9. doi: 10.1158/0008-5472.CAN-04-3931. [DOI] [PubMed] [Google Scholar]
- 280.Ma L, Lai D, Liu T, Cheng W, Guo L. Cancer stem-like cells can be isolated with drug selection in human ovarian cancer cell line SKOV3. Acta Biochim Biophys Sin (Shanghai) 2010;42:593–602. doi: 10.1093/abbs/gmq067. [DOI] [PubMed] [Google Scholar]
- 281.He QZ, Luo XZ, Wang K, Zhou Q, Ao H, Yang Y, et al. Isolation and characterization of cancer stem cells from high-grade serous ovarian carcinomas. Cell Physiol Biochem. 2014;33:173–184. doi: 10.1159/000356660. [DOI] [PubMed] [Google Scholar]
- 282.Chiou SH, Yu CC, Huang CY, Lin SC, Liu CJ, Tsai TH, et al. Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin Cancer Res. 2008;14:4085–95. doi: 10.1158/1078-0432.CCR-07-4404. [DOI] [PubMed] [Google Scholar]
- 283.Guzmán-Ramírez N, Völler M, Wetterwald A, Germann M, Cross NA, Rentsch CA, et al. In vitro propagation and characterization of neoplastic stem/progenitor-like cells from human prostate cancer tissue. Prostate. 2009;69:1683–93. doi: 10.1002/pros.21018. [DOI] [PubMed] [Google Scholar]
- 284.Liu T, Xu F, Du X, Lai D, Liu T, Zhao Y, et al. Establishment and characterization of multi-drug resistant, prostate carcinoma-initiating stem-like cells from human prostate cancer cell lines 22RV1. Mol Cell Biochem. 2010;340:265–73. doi: 10.1007/s11010-010-0426-5. [DOI] [PubMed] [Google Scholar]
- 285.Shi CJ, Gao J, Wang M, Wang X, Tian R, Zhu F, et al. CD133(+) gallbladder carcinoma cells exhibit self-renewal ability and tumorigenicity. World J Gastroenterol. 2011;17:2965–71. doi: 10.3748/wjg.v17.i24.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Janikova M, Skarda J, Dziechciarkova M, Radova L, Chmelova J, Krejci V, et al. Identification of CD133+/nestin+ putative cancer stem cells in non-small cell lung cancer. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2010;154:321–6. doi: 10.5507/bp.2010.048. [DOI] [PubMed] [Google Scholar]
- 287.Li Y, Wang L, Pappan L, Galliher-Beckley A, Shi J. IL-1β promotes stemness and invasiveness of colon cancer cells through Zeb1 activation. Mol Cancer. 2012;11:87. doi: 10.1186/1476-4598-11-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Zhao Z, Lu P, Zhang H, Xu H, Gao N, Li M, et al. Nestin positively regulates the Wnt/β-catenin pathway and the proliferation, survival and invasiveness of breast cancer stem cells. Breast Cancer Res. 2014;16:408. doi: 10.1186/s13058-014-0408-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Apostolou P, Toloudi M, Chatziioannou M, Ioannou E, Papasotiriou I. Cancer stem cells stemness transcription factors expression correlates with breast cancer disease stage. Curr Stem Cell Res Ther. 2012;7:415–9. doi: 10.2174/157488812804484639. [DOI] [PubMed] [Google Scholar]
- 290.Dhingra S, Feng W, Brown RE, Zhou Z, Khoury T, Zhang R, et al. Clinicopathologic significance of putative stem cell markers, CD44 and nestin, in gastric adenocarcinoma. Int J Clin Exp Pathol. 2011;4:733–41. [PMC free article] [PubMed] [Google Scholar]
- 291.Matsuda Y, Yoshimura H, Ueda J, Naito Z, Korc M, Ishiwata T. Nestin delineates pancreatic cancer stem cells in metastatic foci of NOD/Shi-scid IL2Rγ(null) (NOG) mice. Am J Pathol. 2014;184:674–85. doi: 10.1016/j.ajpath.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Sureban SM, May R, George RJ, Dieckgraefe BK, McLeod HL, Ramalingam S, et al. Knockdown of RNA binding protein musashi-1 leads to tumor regression in vivo. Gastroenterology. 2008;134:1448–58. doi: 10.1053/j.gastro.2008.02.057. [DOI] [PubMed] [Google Scholar]
- 293.Ravindran G, Devaraj H. Aberrant expression of CD133 and musashi-1 in preneoplastic and neoplastic human oral squamous epithelium and their correlation with clinicopathological factors. Head Neck. 2012;34:1129–35. doi: 10.1002/hed.21896. [DOI] [PubMed] [Google Scholar]
- 294.Varun BR, Jayanthi P, Ramani P. Cancer stem cells: a comprehensive review on identification and therapeutic implications. J Oral Maxillofac Pathol. 2020;24:190. doi: 10.4103/jomfp.JOMFP_336_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Kaneko Y, Sakakibara S, Imai T, Suzuki A, Nakamura Y, Sawamoto K, et al. Musashi1: an evolutionally conserved marker for CNS progenitor cells including neural stem cells. Dev Neurosci. 2000;22:139–53. doi: 10.1159/000017435. [DOI] [PubMed] [Google Scholar]
- 296.Kikushige Y, Akashi K. TIM-3 as a therapeutic target for malignant stem cells in acute myelogenous leukemia. Ann N Y Acad Sci. 2012;1266:118–23. doi: 10.1111/j.1749-6632.2012.06550.x. [DOI] [PubMed] [Google Scholar]
- 297.Flüh C, Hattermann K, Mehdorn HM, Synowitz M, Held-Feindt J. Differential expression of CXCR4 and CXCR7 with various stem cell markers in paired human primary and recurrent glioblastomas. Int J Oncol. 2016;48:1408–16. doi: 10.3892/ijo.2016.3354. [DOI] [PubMed] [Google Scholar]
- 298.Miki J, Furusato B, Li H, Gu Y, Takahashi H, Egawa S, et al. Identification of putative stem cell markers, CD133 and CXCR4, in hTERT-immortalized primary nonmalignant and malignant tumor-derived human prostate epithelial cell lines and in prostate cancer specimens. Cancer Res. 2007;67:3153–61. doi: 10.1158/0008-5472.CAN-06-4429. [DOI] [PubMed] [Google Scholar]
- 299.Yin AH, Miraglia S, Zanjani ED, Almeida-Porada G, Ogawa M, Leary AG, et al. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90:5002–12. [PubMed] [Google Scholar]
- 300.Lee A, Kessler JD, Read TA, Kaiser C, Corbeil D, Huttner WB, et al. Isolation of neural stem cells from the postnatal cerebellum. Nat Neurosci. 2005;8:723–9. doi: 10.1038/nn1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Beier D, Hau P, Proescholdt M, Lohmeier A, Wischhusen J, Oefner PJ, et al. CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–5. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- 302.Shmelkov SV, Butler JM, Hooper AT, Hormigo A, Kushner J, Milde T, et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133- metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118:2111–20. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Nair RM, Balla MM, Khan I, Kalathur RKR, Kondaiah P, Vemuganti GK. In vitro characterization of CD133(lo) cancer stem cells in Retinoblastoma Y79 cell line. BMC Cancer. 2017;17:779. doi: 10.1186/s12885-017-3750-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Hou Y, Zou Q, Ge R, Shen F, Wang Y. The critical role of CD133(+)CD44(+/high) tumor cells in hematogenous metastasis of liver cancers. Cell Res. 2012;22:259–72. doi: 10.1038/cr.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Thapa R, Wilson GD. The importance of CD44 as a stem cell biomarker and therapeutic target in cancer. Stem Cells Int. 2016;2016:2087204. doi: 10.1155/2016/2087204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Lau WM, Teng E, Chong HS, Lopez KA, Tay AY, Salto-Tellez M, et al. CD44v8-10 is a cancer-specific marker for gastric cancer stem cells. Cancer Res. 2014;74:2630–41. doi: 10.1158/0008-5472.CAN-13-2309. [DOI] [PubMed] [Google Scholar]
- 307.Chen W, Wang R, Zhao Y, Li Y, Wang X, Peng W, et al. CD44v6+ hepatocellular carcinoma cells maintain stemness properties through Met/cJun/Nanog signaling. Stem Cells Int. 2022;2022:5853707. doi: 10.1155/2022/5853707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Kodama H, Murata S, Ishida M, Yamamoto H, Yamaguchi T, Kaida S, et al. Prognostic impact of CD44-positive cancer stem-like cells at the invasive front of gastric cancer. Br J Cancer. 2017;116:186–94. doi: 10.1038/bjc.2016.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, et al. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest. 2011;121:1064–74. doi: 10.1172/JCI44540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Mashita N, Yamada S, Nakayama G, Tanaka C, Iwata N, Kanda M, et al. Epithelial to mesenchymal transition might be induced via CD44 isoform switching in colorectal cancer. J Surg Oncol. 2014;110:745–51. doi: 10.1002/jso.23705. [DOI] [PubMed] [Google Scholar]
- 311.Zhu L, Zhang W, Wang J, Liu R. Evidence of CD90+CXCR4+ cells as circulating tumor stem cells in hepatocellular carcinoma. Tumour Biol. 2015;36:5353–60. doi: 10.1007/s13277-015-3196-6. [DOI] [PubMed] [Google Scholar]
- 312.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 313.de Sousa e Melo F, Kurtova AV, Harnoss JM, Kljavin N, Hoeck JD, Hung J, et al. A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer. Nature. 2017;543:676–80. doi: 10.1038/nature21713. [DOI] [PubMed] [Google Scholar]
- 314.Leng Z, Xia Q, Chen J, Li Y, Xu J, Zhao E, et al. Lgr5+CD44+EpCAM+ strictly defines cancer stem cells in human colorectal cancer. Cell Physiol Biochem. 2018;46:860–72. doi: 10.1159/000488743. [DOI] [PubMed] [Google Scholar]
- 315.Pesce M, Schöler HR. Oct-4: gatekeeper in the beginnings of mammalian development. Stem Cells. 2001;19:271–8. doi: 10.1634/stemcells.19-4-271. [DOI] [PubMed] [Google Scholar]
- 316.Bigdelou Z, Mortazavi Y, Saltanatpour Z, Asadi Z, Kadivar M, Johari B. Role of Oct4-Sox2 complex decoy oligodeoxynucleotides strategy on reverse epithelial to mesenchymal transition (EMT) induction in HT29-ShE encompassing enriched cancer stem-like cells. Mol Biol Rep. 2020;47:1859–69. doi: 10.1007/s11033-020-05280-2. [DOI] [PubMed] [Google Scholar]
- 317.Zhao Y, Li C, Huang L, Niu S, Lu Q, Gong D, et al. Prognostic value of association of OCT4 with LEF1 expression in esophageal squamous cell carcinoma and their impact on epithelial-mesenchymal transition, invasion, and migration. Cancer Med. 2018;7:3977–87. doi: 10.1002/cam4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Zeineddine D, Hammoud AA, Mortada M, Boeuf H. The Oct4 protein: more than a magic stemness marker. Am J Stem Cells. 2014;3:74–82. [PMC free article] [PubMed] [Google Scholar]
- 319.Wang YD, Cai N, Wu XL, Cao HZ, Xie LL, Zheng PS. OCT4 promotes tumorigenesis and inhibits apoptosis of cervical cancer cells by miR-125b/BAK1 pathway. Cell Death Dis. 2013;4:e760. doi: 10.1038/cddis.2013.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Kristensen DM, Nielsen JE, Kalisz M, Dalgaard MD, Audouze K, Larsen ME, et al. OCT4 and downstream factors are expressed in human somatic urogenital epithelia and in culture of epididymal spheres. Mol Hum Reprod. 2010;16:835–45. doi: 10.1093/molehr/gaq008. [DOI] [PubMed] [Google Scholar]
- 321.Mamun MA, Mannoor K, Cao J, Qadri F, Song X. SOX2 in cancer stemness: tumor malignancy and therapeutic potentials. J Mol Cell Biol. 2020;12:85–98. doi: 10.1093/jmcb/mjy080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Wuebben EL, Rizzino A. The dark side of SOX2: cancer - a comprehensive overview. Oncotarget. 2017;8:44917–43. doi: 10.18632/oncotarget.16570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Weina K, Utikal J. SOX2 and cancer: current research and its implications in the clinic. Clin Transl Med. 2014;3:19. doi: 10.1186/2001-1326-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Mukhopadhyay A, Berrett KC, Kc U, Clair PM, Pop SM, Carr SR, et al. Sox2 cooperates with Lkb1 loss in a mouse model of squamous cell lung cancer. Cell Rep. 2014;8:40–9. doi: 10.1016/j.celrep.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 326.Flandez M, Guilmeau S, Blache P, Augenlicht LH. KLF4 regulation in intestinal epithelial cell maturation. Exp Cell Res. 2008;314:3712–23. doi: 10.1016/j.yexcr.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Riverso M, Montagnani V, Stecca B. KLF4 is regulated by RAS/RAF/MEK/ERK signaling through E2F1 and promotes melanoma cell growth. Oncogene. 2017;36:3322–33. doi: 10.1038/onc.2016.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Tien YT, Chang MH, Chu PY, Lin CS, Liu CH, Liao AT. Downregulation of the KLF4 transcription factor inhibits the proliferation and migration of canine mammary tumor cells. Vet J. 2015;205:244–53. doi: 10.1016/j.tvjl.2014.12.031. [DOI] [PubMed] [Google Scholar]
- 329.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–55. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 330.Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, et al. Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem. 2005;280:24731–7. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 331.Alemohammad H, Asadzadeh Z, Motafakker Azad R, Hemmat N, Najafzadeh B, Vasefifar P, et al. Signaling pathways and microRNAs, the orchestrators of NANOG activity during cancer induction. Life Sci. 2020;260:118337. doi: 10.1016/j.lfs.2020.118337. [DOI] [PubMed] [Google Scholar]
- 332.Najafzadeh B, Asadzadeh Z, Motafakker Azad R, Mokhtarzadeh A, Baghbanzadeh A, Alemohammad H, et al. The oncogenic potential of NANOG: an important cancer induction mediator. J Cell Physiol. 2021;236:2443–58. doi: 10.1002/jcp.30063. [DOI] [PubMed] [Google Scholar]
- 333.Lu X, Mazur SJ, Lin T, Appella E, Xu Y. The pluripotency factor nanog promotes breast cancer tumorigenesis and metastasis. Oncogene. 2014;33:2655–64. doi: 10.1038/onc.2013.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Mukherjee B, Morgenbesser SD, DePinho RA. Myc family oncoproteins function through a common pathway to transform normal cells in culture: cross-interference by Max and trans-acting dominant mutants. Genes Dev. 1992;6:1480–92. doi: 10.1101/gad.6.8.1480. [DOI] [PubMed] [Google Scholar]
- 335.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–90. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 336.Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med. 2013;3:a014217. doi: 10.1101/cshperspect.a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–64. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 338.Chen H, Liu H, Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct Target Ther. 2018;3:5. doi: 10.1038/s41392-018-0008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Clay MR, Tabor M, Owen JH, Carey TE, Bradford CR, Wolf GT, et al. Single-marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head Neck. 2010;32:1195–201. doi: 10.1002/hed.21315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Sullivan JP, Spinola M, Dodge M, Raso MG, Behrens C, Gao B, et al. Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling. Cancer Res. 2010;70:9937–48. doi: 10.1158/0008-5472.CAN-10-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Blume R, Rempel E, Manta L, Saeed BR, Wang W, Raffel S, et al. The molecular signature of AML with increased ALDH activity suggests a stem cell origin. Leuk Lymphoma. 2018;59:2201–10. doi: 10.1080/10428194.2017.1422862. [DOI] [PubMed] [Google Scholar]
- 342.Chen D, Wu M, Li Y, Chang I, Yuan Q, Ekimyan-Salvo M, et al. Targeting BMI1(+) cancer stem cells overcomes chemoresistance and inhibits metastases in squamous cell carcinoma. Cell Stem Cell. 2017;20:621–34.e626. doi: 10.1016/j.stem.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Siddique HR, Saleem M. Role of BMI1, a stem cell factor, in cancer recurrence and chemoresistance: preclinical and clinical evidences. Stem Cells. 2012;30:372–8. doi: 10.1002/stem.1035. [DOI] [PubMed] [Google Scholar]
- 344.Shen HT, Chien PJ, Chen SH, Sheu GT, Jan MS, Wang BY, et al. BMI1-mediated pemetrexed resistance in non-small cell lung cancer cells is associated with increased SP1 activation and cancer stemness. Cancers (Basel) 2020;12:2069. doi: 10.3390/cancers12082069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, Jackson KW, et al. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66:6063–71. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Lendahl U, Zimmerman LB, McKay RD. CNS stem cells express a new class of intermediate filament protein. Cell. 1990;60:585–95. doi: 10.1016/0092-8674(90)90662-x. [DOI] [PubMed] [Google Scholar]
- 347.Kanemura Y, Mori K, Sakakibara S, Fujikawa H, Hayashi H, Nakano A, et al. Musashi1, an evolutionarily conserved neural RNA-binding protein, is a versatile marker of human glioma cells in determining their cellular origin, malignancy, and proliferative activity. Differentiation. 2001;68:141–52. doi: 10.1046/j.1432-0436.2001.680208.x. [DOI] [PubMed] [Google Scholar]
- 348.Toda M, Iizuka Y, Yu W, Imai T, Ikeda E, Yoshida K, et al. Expression of the neural RNA-binding protein Musashi1 in human gliomas. Glia. 2001;34:1–7. doi: 10.1002/glia.1034. [DOI] [PubMed] [Google Scholar]
- 349.Glazer RI, Vo DT, Penalva LO. Musashi1: an RBP with versatile functions in normal and cancer stem cells. Front Biosci (Landmark Ed) 2012;17:54–64. doi: 10.2741/3915. [DOI] [PubMed] [Google Scholar]
- 350.Tang D, Lotze MT. Tumor immunity times out: TIM-3 and HMGB1. Nat Immunol. 2012;13:808–10. doi: 10.1038/ni.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Cheng L, Ruan Z. Tim-3 and Tim-4 as the potential targets for antitumor therapy. Hum Vaccin Immunother. 2015;11:2458–62. doi: 10.1080/21645515.2015.1056953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Mollica Poeta V, Massara M, Capucetti A, Bonecchi R. Chemokines and chemokine receptors: new targets for cancer immunotherapy. Front Immunol. 2019;10:379. doi: 10.3389/fimmu.2019.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Do HTT, Lee CH, Cho J. Chemokines and their receptors: multifaceted roles in cancer progression and potential value as cancer prognostic markers. Cancers (Basel) 2020;12:287. doi: 10.3390/cancers12020287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Bachelerie F, Ben-Baruch A, Burkhardt AM, Combadiere C, Farber JM, Graham GJ, et al. International Union of Basic and Clinical Pharmacology. [corrected]. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacol Rev. 2014;66:1–79. doi: 10.1124/pr.113.007724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Hopman RK, DiPersio JF. Advances in stem cell mobilization. Blood Rev. 2014;28:31–40. doi: 10.1016/j.blre.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–88. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 357.Dubrovska A, Elliott J, Salamone RJ, Telegeev GD, Stakhovsky AE, Schepotin IB, et al. CXCR4 expression in prostate cancer progenitor cells. PLoS One. 2012;7:e31226. doi: 10.1371/journal.pone.0031226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Zhang SS, Han ZP, Jing YY, Tao SF, Li TJ, Wang H, et al. CD133(+)CXCR4(+) colon cancer cells exhibit metastatic potential and predict poor prognosis of patients. BMC Med. 2012;10:85. doi: 10.1186/1741-7015-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Sharanek A, Burban A, Laaper M, Heckel E, Joyal JS, Soleimani VD, et al. OSMR controls glioma stem cell respiration and confers resistance of glioblastoma to ionizing radiation. Nat Commun. 2020;11:4116. doi: 10.1038/s41467-020-17885-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Clara JA, Monge C, Yang Y, Takebe N. Targeting signalling pathways and the immune microenvironment of cancer stem cells - a clinical update. Nat Rev Clin Oncol. 2020;17:204–32. doi: 10.1038/s41571-019-0293-2. [DOI] [PubMed] [Google Scholar]
- 361.Manni W, Min W. Signaling pathways in the regulation of cancer stem cells and associated targeted therapy. MedComm (2020) 2022;3:e176. doi: 10.1002/mco2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Wang K, Zhang T, Dong Q, Nice EC, Huang C, Wei Y. Redox homeostasis: the linchpin in stem cell self-renewal and differentiation. Cell Death Dis. 2013;4:e537. doi: 10.1038/cddis.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Chen Z, Shi T, Zhang L, Zhu P, Deng M, Huang C, et al. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family in multidrug resistance: a review of the past decade. Cancer Lett. 2016;370:153–64. doi: 10.1016/j.canlet.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 364.Kazi JU. Mechanisms of anticancer therapy resistance: the role of cancer stem cells. Int J Mol Sci. 2020;21:9006. doi: 10.3390/ijms21239006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Hou H, Sun H, Lu P, Ge C, Zhang L, Li H, et al. Tunicamycin potentiates cisplatin anticancer efficacy through the DPAGT1/Akt/ABCG2 pathway in mouse Xenograft models of human hepatocellular carcinoma. Mol Cancer Ther. 2013;12:2874–84. doi: 10.1158/1535-7163.MCT-13-0201. [DOI] [PubMed] [Google Scholar]
- 366.Hu L, Liang Y, Wu K, Wang C, Zhang T, Peng R, et al. Repressing PDCD4 activates JNK/ABCG2 pathway to induce chemoresistance to fluorouracil in colorectal cancer cells. Ann Transl Med. 2021;9:114. doi: 10.21037/atm-20-4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Yao XH, Ping YF, Bian XW. Contribution of cancer stem cells to tumor vasculogenic mimicry. Protein Cell. 2011;2:266–72. doi: 10.1007/s13238-011-1041-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Liu TJ, Sun BC, Zhao XL, Zhao XM, Sun T, Gu Q, et al. CD133+ cells with cancer stem cell characteristics associates with vasculogenic mimicry in triple-negative breast cancer. Oncogene. 2013;32:544–53. doi: 10.1038/onc.2012.85. [DOI] [PubMed] [Google Scholar]
- 369.Zheng N, Zhang S, Wu W, Zhang N, Wang J. Regulatory mechanisms and therapeutic targeting of vasculogenic mimicry in hepatocellular carcinoma. Pharmacol Res. 2021;166:105507. doi: 10.1016/j.phrs.2021.105507. [DOI] [PubMed] [Google Scholar]
- 370.Sun H, Yao N, Cheng S, Li L, Liu S, Yang Z, et al. Cancer stem-like cells directly participate in vasculogenic mimicry channels in triple-negative breast cancer. Cancer Biol Med. 2019;16:299–311. doi: 10.20892/j.issn.2095-3941.2018.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.de Jong MC, Pramana J, van der Wal JE, Lacko M, Peutz-Kootstra CJ, de Jong JM, et al. CD44 expression predicts local recurrence after radiotherapy in larynx cancer. Clin Cancer Res. 2010;16:5329–38. doi: 10.1158/1078-0432.CCR-10-0799. [DOI] [PubMed] [Google Scholar]
- 372.Duru N, Fan M, Candas D, Menaa C, Liu HC, Nantajit D, et al. HER2-associated radioresistance of breast cancer stem cells isolated from HER2-negative breast cancer cells. Clin Cancer Res. 2012;18:6634–47. doi: 10.1158/1078-0432.CCR-12-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Desai A, Webb B, Gerson SL. CD133+ cells contribute to radioresistance via altered regulation of DNA repair genes in human lung cancer cells. Radiother Oncol. 2014;110:538–45. doi: 10.1016/j.radonc.2013.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 375.Ahmed SU, Carruthers R, Gilmour L, Yildirim S, Watts C, Chalmers AJ. Selective inhibition of parallel DNA damage response pathways optimizes radiosensitization of glioblastoma stem-like cells. Cancer Res. 2015;75:4416–28. doi: 10.1158/0008-5472.CAN-14-3790. [DOI] [PubMed] [Google Scholar]
- 376.Wu J, Lai G, Wan F, Xiao Z, Zeng L, Wang X, et al. Knockdown of checkpoint kinase 1 is associated with the increased radiosensitivity of glioblastoma stem-like cells. Tohoku J Exp Med. 2012;226:267–74. doi: 10.1620/tjem.226.267. [DOI] [PubMed] [Google Scholar]
- 377.Carruthers R, Ahmed SU, Strathdee K, Gomez-Roman N, Amoah-Buahin E, Watts C, et al. Abrogation of radioresistance in glioblastoma stem-like cells by inhibition of ATM kinase. Mol Oncol. 2015;9:192–203. doi: 10.1016/j.molonc.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Wang X, Ma Z, Xiao Z, Liu H, Dou Z, Feng X, et al. Chk1 knockdown confers radiosensitization in prostate cancer stem cells. Oncol Rep. 2012;28:2247–54. doi: 10.3892/or.2012.2068. [DOI] [PubMed] [Google Scholar]
- 379.Yin H, Glass J. The phenotypic radiation resistance of CD44+/CD24(− or low) breast cancer cells is mediated through the enhanced activation of ATM signaling. PLoS One. 2011;6:e24080. doi: 10.1371/journal.pone.0024080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Park SY, Lee CJ, Choi JH, Kim JH, Kim JW, Kim JY, et al. The JAK2/STAT3/CCND2 axis promotes colorectal Cancer stem cell persistence and radioresistance. J Exp Clin Cancer Res. 2019;38:399. doi: 10.1186/s13046-019-1405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Lomonaco SL, Finniss S, Xiang C, Decarvalho A, Umansky F, Kalkanis SN, et al. The induction of autophagy by gamma-radiation contributes to the radioresistance of glioma stem cells. Int J Cancer. 2009;125:717–22. doi: 10.1002/ijc.24402. [DOI] [PubMed] [Google Scholar]
- 382.Seidel S, Garvalov BK, Wirta V, von Stechow L, Schanzer A, Meletis K, et al. A hypoxic niche regulates glioblastoma stem cells through hypoxia inducible factor 2 alpha. Brain. 2010;133:983–95. doi: 10.1093/brain/awq042. [DOI] [PubMed] [Google Scholar]
- 383.Nagaraju GP, Bramhachari PV, Raghu G, El-Rayes BF. Hypoxia inducible factor-1alpha: its role in colorectal carcinogenesis and metastasis. Cancer Lett. 2015;366:11–8. doi: 10.1016/j.canlet.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 384.Yang Z, Han Y, Cheng K, Zhang G, Wang X. miR-99a directly targets the mTOR signalling pathway in breast cancer side population cells. Cell Prolif. 2014;47:587–95. doi: 10.1111/cpr.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Chen X, Wu L, Li D, Xu Y, Zhang L, Niu K, et al. Radiosensitizing effects of miR-18a-5p on lung cancer stem-like cells via downregulating both ATM and HIF-1alpha. Cancer Med. 2018;7:3834–47. doi: 10.1002/cam4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Li MY, Fan LN, Han DH, Yu Z, Ma J, Liu YX, et al. Ribosomal S6 protein kinase 4 promotes radioresistance in esophageal squamous cell carcinoma. J Clin Invest. 2020;130:4301–19. doi: 10.1172/JCI134930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Chang L, Graham P, Hao J, Ni J, Deng J, Bucci J, et al. Cancer stem cells and signaling pathways in radioresistance. Oncotarget. 2016;7:11002–17. doi: 10.18632/oncotarget.6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Karabicici M, Alptekin S, Fırtına Karagonlar Z, Erdal E. Doxorubicin-induced senescence promotes stemness and tumorigenicity in EpCAM-/CD133- nonstem cell population in hepatocellular carcinoma cell line, HuH-7. Mol Oncol. 2021;15:2185–202. doi: 10.1002/1878-0261.12916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Piva M, Domenici G, Iriondo O, Rábano M, Simões BM, Comaills V, et al. Sox2 promotes tamoxifen resistance in breast cancer cells. EMBO Mol Med. 2014;6:66–79. doi: 10.1002/emmm.201303411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Ahmad A, Maitah MY, Ginnebaugh KR, Li Y, Bao B, Gadgeel SM, et al. Inhibition of Hedgehog signaling sensitizes NSCLC cells to standard therapies through modulation of EMT-regulating miRNAs. J Hematol Oncol. 2013;6:77. doi: 10.1186/1756-8722-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Sarvi S, Mackinnon AC, Avlonitis N, Bradley M, Rintoul RC, Rassl DM, et al. CD133+ cancer stem-like cells in small cell lung cancer are highly tumorigenic and chemoresistant but sensitive to a novel neuropeptide antagonist. Cancer Res. 2014;74:1554–65. doi: 10.1158/0008-5472.CAN-13-1541. [DOI] [PubMed] [Google Scholar]
- 392.Jiang F, Qiu Q, Khanna A, Todd NW, Deepak J, Xing L, et al. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res. 2009;7:330–8. doi: 10.1158/1541-7786.MCR-08-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Wang W, Wang J, Liu S, Ren Y, Wang J, Liu S, et al. An EHMT2/NFYA-ALDH2 signaling axis modulates the RAF pathway to regulate paclitaxel resistance in lung cancer. Mol Cancer. 2022;21:106. doi: 10.1186/s12943-022-01579-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Liu YP, Yang CJ, Huang MS, Yeh CT, Wu AT, Lee YC, et al. Cisplatin selects for multidrug-resistant CD133+ cells in lung adenocarcinoma by activating Notch signaling. Cancer Res. 2013;73:406–16. doi: 10.1158/0008-5472.CAN-12-1733. [DOI] [PubMed] [Google Scholar]
- 395.Hashida S, Yamamoto H, Shien K, Miyoshi Y, Ohtsuka T, Suzawa K, et al. Acquisition of cancer stem cell-like properties in non-small cell lung cancer with acquired resistance to afatinib. Cancer Sci. 2015;106:1377–84. doi: 10.1111/cas.12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Chen YJ, Huang WC, Wei YL, Hsu SC, Yuan P, Lin HY, et al. Elevated BCRP/ABCG2 expression confers acquired resistance to gefitinib in wild-type EGFR-expressing cells. PLoS One. 2011;6:e21428. doi: 10.1371/journal.pone.0021428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Song J, Xie LX, Zhang XY, Hu P, Long MF, Xiong F, et al. Role of YAP in lung cancer resistance to cisplatin. Oncol Lett. 2018;16:3949–54. doi: 10.3892/ol.2018.9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Chen MJ, Wu DW, Wang YC, Chen CY, Lee H. PAK1 confers chemoresistance and poor outcome in non-small cell lung cancer via β-catenin-mediated stemness. Sci Rep. 2016;6:34933. doi: 10.1038/srep34933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Zhang Y, Xu W, Guo H, Zhang Y, He Y, Lee SH, et al. NOTCH1 signaling regulates self-renewal and platinum chemoresistance of cancer stem-like cells in human non-small cell lung cancer. Cancer Res. 2017;77:3082–91. doi: 10.1158/0008-5472.CAN-16-1633. [DOI] [PubMed] [Google Scholar]
- 400.Li J, Ye T, Liu Y, Kong L, Sun Z, Liu D, et al. Transcriptional activation of Gstp1 by MEK/ERK signaling confers chemo-resistance to cisplatin in lung cancer stem cells. Front Oncol. 2019;9:476. doi: 10.3389/fonc.2019.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Zhang J, Liu J, Li H, Wang J. β-catenin signaling pathway regulates cisplatin resistance in lung adenocarcinoma cells by upregulating Bcl-xl. Mol Med Rep. 2016;13:2543–51. doi: 10.3892/mmr.2016.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Yang J, Zhang K, Wu J, Shi J, Xue J, Li J, et al. Wnt5a increases properties of lung cancer stem cells and resistance to cisplatin through activation of Wnt5a/PKC signaling pathway. Stem Cells Int. 2016;2016:1690896. doi: 10.1155/2016/1690896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Zhang P, Ma Y, Lv C, Huang M, Li M, Dong B, et al. Upregulation of programmed cell death ligand 1 promotes resistance response in non-small-cell lung cancer patients treated with neo-adjuvant chemotherapy. Cancer Sci. 2016;107:1563–71. doi: 10.1111/cas.13072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Koh H, Park H, Chandimali N, Huynh DL, Zhang JJ, Ghosh M, et al. MicroRNA-128 suppresses paclitaxel-resistant lung cancer by inhibiting MUC1-C and BMI-1 in cancer stem cells. Oncotarget. 2017;8:110540–51. doi: 10.18632/oncotarget.22818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Ma Z, Cai H, Zhang Y, Chang L, Cui Y. MiR-129-5p inhibits non-small cell lung cancer cell stemness and chemoresistance through targeting DLK1. Biochem Biophys Res Commun. 2017;490:309–16. doi: 10.1016/j.bbrc.2017.06.041. [DOI] [PubMed] [Google Scholar]
- 406.Park CR, Lee M, Lee SY, Kang D, Park SJ, Lee DC, et al. Regulating POLR3G by MicroRNA-26a-5p as a promising therapeutic target of lung cancer stemness and chemosensitivity. Noncoding RNA Res. 2023;8:273–81. doi: 10.1016/j.ncrna.2023.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Chandimali N, Koh H, Kim J, Lee J, Park YH, Sun HN, et al. BRM270 targets cancer stem cells and augments chemo-sensitivity in cancer. Oncol Lett. 2020;20:103. doi: 10.3892/ol.2020.11964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang J, et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct Target Ther. 2020;5:8. doi: 10.1038/s41392-020-0110-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Lee JW, Lee HY. Targeting cancer stem cell markers or pathways: a potential therapeutic strategy for oral cancer treatment. Int J Stem Cells. 2021;14:386–99. doi: 10.15283/ijsc21084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 411.Kaumaya PTP, Guo L, Overholser J, Penichet ML, Bekaii-Saab T. Immunogenicity and antitumor efficacy of a novel human PD-1 B-cell vaccine (PD1-Vaxx) and combination immunotherapy with dual trastuzumab/pertuzumab-like HER-2 B-cell epitope vaccines (B-Vaxx) in a syngeneic mouse model. Oncoimmunology. 2020;9:1818437. doi: 10.1080/2162402X.2020.1818437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Rekoske BT, McNeel DG. Immunotherapy for prostate cancer: false promises or true hope? Cancer. 2016;122:3598–607. doi: 10.1002/cncr.30250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Deng Z, Wu Y, Ma W, Zhang S, Zhang YQ. Adoptive T-cell therapy of prostate cancer targeting the cancer stem cell antigen EpCAM. BMC Immunol. 2015;16:1. doi: 10.1186/s12865-014-0064-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Zhang BL, Li D, Gong YL, Huang Y, Qin DY, Jiang L, et al. Preclinical evaluation of chimeric antigen receptor-modified T cells specific to epithelial cell adhesion molecule for treating colorectal cancer. Hum Gene Ther. 2019;30:402–12. doi: 10.1089/hum.2018.229. [DOI] [PubMed] [Google Scholar]
- 415.Chen HC, Joalland N, Bridgeman JS, Alchami FS, Jarry U, Khan MWA, et al. Synergistic targeting of breast cancer stem-like cells by human γδ T cells and CD8(+) T cells. Immunol Cell Biol. 2017;95:620–9. doi: 10.1038/icb.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Cook M, Chauhan A. Clinical application of oncolytic viruses: a systematic review. Int J Mol Sci. 2020;21:7505. doi: 10.3390/ijms21207505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Wang H, Chen NG, Minev BR, Szalay AA. Oncolytic vaccinia virus GLV-1h68 strain shows enhanced replication in human breast cancer stem-like cells in comparison to breast cancer cells. J Transl Med. 2012;10:167. doi: 10.1186/1479-5876-10-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Tomasicchio M, Semple L, Esmail A, Meldau R, Randall P, Pooran A, et al. An autologous dendritic cell vaccine polarizes a Th-1 response which is tumoricidal to patient-derived breast cancer cells. Cancer Immunol Immunother. 2019;68:71–83. doi: 10.1007/s00262-018-2238-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Dashti A, Ebrahimi M, Hadjati J, Memarnejadian A, Moazzeni SM. Dendritic cell based immunotherapy using tumor stem cells mediates potent antitumor immune responses. Cancer Lett. 2016;374:175–85. doi: 10.1016/j.canlet.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 420.Yin T, Shi P, Gou S, Shen Q, Wang C. Dendritic cells loaded with pancreatic Cancer Stem Cells (CSCs) lysates induce antitumor immune killing effect in vitro. PLoS One. 2014;9:e114581. doi: 10.1371/journal.pone.0114581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Wefers C, Schreibelt G, Massuger L, de Vries IJM, Torensma R. Immune curbing of cancer stem cells by CTLs directed to NANOG. Front Immunol. 2018;9:1412. doi: 10.3389/fimmu.2018.01412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Hu Y, Lu L, Xia Y, Chen X, Chang AE, Hollingsworth RE, et al. Therapeutic efficacy of cancer stem cell vaccines in the adjuvant setting. Cancer Res. 2016;76:4661–72. doi: 10.1158/0008-5472.CAN-15-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Guo F, Zhang Y, Bai L, Cui J. Natural killer cell therapy targeting cancer stem cells: old wine in a new bottle. Cancer Lett. 2023;570:216328. doi: 10.1016/j.canlet.2023.216328. [DOI] [PubMed] [Google Scholar]
- 424.Klapdor R, Wang S, Hacker U, Büning H, Morgan M, Dörk T, et al. Improved killing of ovarian cancer stem cells by combining a novel chimeric antigen receptor-based immunotherapy and chemotherapy. Hum Gene Ther. 2017;28:886–96. doi: 10.1089/hum.2017.168. [DOI] [PubMed] [Google Scholar]
- 425.Hu X, Li J, Fu M, Zhao X, Wang W. The JAK/STAT signaling pathway: from bench to clinic. Signal Transduct Target Ther. 2021;6:402. doi: 10.1038/s41392-021-00791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Shiah JV, Grandis JR, Johnson DE. Targeting STAT3 with proteolysis targeting chimeras and next-generation antisense oligonucleotides. Mol Cancer Ther. 2021;20:219–28. doi: 10.1158/1535-7163.MCT-20-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Chen B, Dai W, Mei D, Liu T, Li S, He B, et al. Comprehensively priming the tumor microenvironment by cancer-associated fibroblast-targeted liposomes for combined therapy with cancer cell-targeted chemotherapeutic drug delivery system. J Control Release. 2016;241:68–80. doi: 10.1016/j.jconrel.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 428.Annibaldi A, Widmann C. Glucose metabolism in cancer cells. Curr Opin Clin Nutr Metab Care. 2010;13:466–70. doi: 10.1097/MCO.0b013e32833a5577. [DOI] [PubMed] [Google Scholar]
- 429.Krasnov GS, Dmitriev AA, Snezhkina AV, Kudryavtseva AV. Deregulation of glycolysis in cancer: glyceraldehyde-3-phosphate dehydrogenase as a therapeutic target. Expert Opin Ther Targets. 2013;17:681–93. doi: 10.1517/14728222.2013.775253. [DOI] [PubMed] [Google Scholar]
- 430.Ceradini DJ, Kulkarni AR, Callaghan MJ, Tepper OM, Bastidas N, Kleinman ME, et al. Progenitor cell trafficking is regulated by hypoxic gradients through HIF-1 induction of SDF-1. Nat Med. 2004;10:858–64. doi: 10.1038/nm1075. [DOI] [PubMed] [Google Scholar]
- 431.Kao TW, Chuang YC, Lee HL, Kuo CC, Shen YA. Therapeutic targeting of glutaminolysis as a novel strategy to combat cancer stem cells. Int J Mol Sci. 2022;23:15296. doi: 10.3390/ijms232315296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Shibuya K, Okada M, Suzuki S, Seino M, Seino S, Takeda H, et al. Targeting the facilitative glucose transporter GLUT1 inhibits the self-renewal and tumor-initiating capacity of cancer stem cells. Oncotarget. 2015;6:651–61. doi: 10.18632/oncotarget.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Wu K, Ning Z, Zeng J, Fan J, Zhou J, Zhang T, et al. Silibinin inhibits β-catenin/ZEB1 signaling and suppresses bladder cancer metastasis via dual-blocking epithelial-mesenchymal transition and stemness. Cell Signal. 2013;25:2625–33. doi: 10.1016/j.cellsig.2013.08.028. [DOI] [PubMed] [Google Scholar]
- 434.Flaig TW, Gustafson DL, Su LJ, Zirrolli JA, Crighton F, Harrison GS, et al. A phase I and pharmacokinetic study of silybin-phytosome in prostate cancer patients. Invest New Drugs. 2007;25:139–46. doi: 10.1007/s10637-006-9019-2. [DOI] [PubMed] [Google Scholar]
- 435.Shen YA, Chen CC, Chen BJ, Wu YT, Juan JR, Chen LY, et al. Potential therapies targeting metabolic pathways in cancer stem cells. Cells. 2021;10:1772. doi: 10.3390/cells10071772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Roesch A, Vultur A, Bogeski I, Wang H, Zimmermann KM, Speicher D, et al. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell. 2013;23:811–25. doi: 10.1016/j.ccr.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Lamb R, Ozsvari B, Lisanti CL, Tanowitz HB, Howell A, Martinez-Outschoorn UE, et al. Antibiotics that target mitochondria effectively eradicate cancer stem cells, across multiple tumor types: treating cancer like an infectious disease. Oncotarget. 2015;6:4569–84. doi: 10.18632/oncotarget.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Fiorillo M, Lamb R, Tanowitz HB, Mutti L, Krstic-Demonacos M, Cappello AR, et al. Repurposing atovaquone: targeting mitochondrial complex III and OXPHOS to eradicate cancer stem cells. Oncotarget. 2016;7:34084–99. doi: 10.18632/oncotarget.9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Lamb R, Harrison H, Hulit J, Smith DL, Lisanti MP, Sotgia F. Mitochondria as new therapeutic targets for eradicating cancer stem cells: quantitative proteomics and functional validation via MCT1/2 inhibition. Oncotarget. 2014;5:11029–37. doi: 10.18632/oncotarget.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.An H, Kim JY, Oh E, Lee N, Cho Y, Seo JH. Salinomycin promotes anoikis and decreases the CD44+/CD24- stem-like population via inhibition of STAT3 activation in MDA-MB-231 Cells. PLoS One. 2015;10:e0141919. doi: 10.1371/journal.pone.0141919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Chu DJ, Yao DE, Zhuang YF, Hong Y, Zhu XC, Fang ZR, et al. Azithromycin enhances the favorable results of paclitaxel and cisplatin in patients with advanced non-small cell lung cancer. Genet Mol Res. 2014;13:2796–805. doi: 10.4238/2014.April.14.8. [DOI] [PubMed] [Google Scholar]
- 442.Jung JW, Park SB, Lee SJ, Seo MS, Trosko JE, Kang KS. Metformin represses self-renewal of the human breast carcinoma stem cells via inhibition of estrogen receptor-mediated OCT4 expression. PLoS One. 2011;6:e28068. doi: 10.1371/journal.pone.0028068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Mukha A, Kahya U, Dubrovska A. Targeting glutamine metabolism and autophagy: the combination for prostate cancer radiosensitization. Autophagy. 2021;17:3879–81. doi: 10.1080/15548627.2021.1962682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Jariyal H, Gupta C, Andhale S, Gadge S, Srivastava A. Comparative stemness and differentiation of luminal and basal breast cancer stem cell type under glutamine-deprivation. J Cell Commun Signal. 2021;15:207–22. doi: 10.1007/s12079-020-00603-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Zhu J, Wang H, Sun Q, Ji X, Zhu L, Cong Z, et al. Nrf2 is required to maintain the self-renewal of glioma stem cells. BMC Cancer. 2013;13:380. doi: 10.1186/1471-2407-13-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Zhu J, Wang H, Fan Y, Hu Y, Ji X, Sun Q, et al. Knockdown of nuclear factor erythroid 2-related factor 2 by lentivirus induces differentiation of glioma stem-like cells. Oncol Rep. 2014;32:1170–8. doi: 10.3892/or.2014.3320. [DOI] [PubMed] [Google Scholar]
- 447.Ryoo IG, Choi BH, Kwak MK. Activation of NRF2 by p62 and proteasome reduction in sphere-forming breast carcinoma cells. Oncotarget. 2015;6:8167–84. doi: 10.18632/oncotarget.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Ryoo IG, Lee SH, Kwak MK. Redox modulating NRF2: a potential mediator of cancer stem cell resistance. Oxid Med Cell Longev. 2016;2016:2428153. doi: 10.1155/2016/2428153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Ouyang WC, Liao YW, Chen PN, Lu KH, Yu CC, Hsieh PL. Hinokitiol suppresses cancer stemness and oncogenicity in glioma stem cells by Nrf2 regulation. Cancer Chemother Pharmacol. 2017;80:411–9. doi: 10.1007/s00280-017-3381-y. [DOI] [PubMed] [Google Scholar]
- 450.Woo Y, Oh J, Kim JS. Suppression of Nrf2 activity by chestnut leaf extract increases chemosensitivity of breast cancer stem cells to paclitaxel. Nutrients. 2017;9:760. doi: 10.3390/nu9070760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Cheng G, Palanisamy AP, Evans ZP, Sutter AG, Jin L, Singh I, et al. Cerulenin blockade of fatty acid synthase reverses hepatic steatosis in ob/ob mice. PLoS One. 2013;8:e75980. doi: 10.1371/journal.pone.0075980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Van de Sande T, De Schrijver E, Heyns W, Verhoeven G, Swinnen JV. Role of the phosphatidylinositol 3’-kinase/PTEN/Akt kinase pathway in the overexpression of fatty acid synthase in LNCaP prostate cancer cells. Cancer Res. 2002;62:642–6. [PubMed] [Google Scholar]
- 453.Colacino JA, McDermott SP, Sartor MA, Wicha MS, Rozek LS. Transcriptomic profiling of curcumin-treated human breast stem cells identifies a role for stearoyl-coa desaturase in breast cancer prevention. Breast Cancer Res Treat. 2016;158:29–41. doi: 10.1007/s10549-016-3854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Li J, Condello S, Thomes-Pepin J, Ma X, Xia Y, Hurley TD, et al. Lipid desaturation is a metabolic marker and therapeutic target of ovarian cancer stem cells. Cell Stem Cell. 2017;20:303–14.e305. doi: 10.1016/j.stem.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Elgendy SM, Alyammahi SK, Alhamad DW, Abdin SM, Omar HA. Ferroptosis: an emerging approach for targeting cancer stem cells and drug resistance. Crit Rev Oncol Hematol. 2020;155:103095. doi: 10.1016/j.critrevonc.2020.103095. [DOI] [PubMed] [Google Scholar]
- 456.Lim YC, Kang HJ, Kim YS, Choi EC. All-trans-retinoic acid inhibits growth of head and neck cancer stem cells by suppression of Wnt/β-catenin pathway. Eur J Cancer. 2012;48:3310–18. doi: 10.1016/j.ejca.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 457.Luo C, Ding Z, Tu Y, Tan J, Luo Q, Song G. Biomaterial-based platforms for cancer stem cell enrichment and study. Cancer Biol Med. 2021;18:458–69. doi: 10.20892/j.issn.2095-3941.2020.0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Li R, Chen Z, Dai Z, Yu Y. Nanotechnology assisted photo- and sonodynamic therapy for overcoming drug resistance. Cancer Biol Med. 2021;18:388–400. doi: 10.20892/j.issn.2095-3941.2020.0328. [DOI] [PMC free article] [PubMed] [Google Scholar]